Abstract
Purpose:
To develop and validate a natural language processing (NLP) algorithm to extract qualitative descriptors of microbial keratitis (MK) from electronic health records (EHR).
Methods:
In this retrospective cohort study, patients with MK diagnoses from two academic centers were identified using EHR records. An NLP algorithm was created to extract MK centrality, depth, and thinning. A random sample of MK patient encounters were used to train the algorithm (400 encounters of 100 patients) and compared to expert chart review. The algorithm was evaluated in internal (n=100) and external validation datasets (n=59) in comparison to masked chart review. Outcomes were sensitivity and specificity of the NLP algorithm to extract qualitative MK features as compared to masked chart review performed by an ophthalmologist.
Results:
Across datasets, gold-standard chart review found centrality was documented in 64.0–79.3% of charts, depth in 15.0–20.3%, and thinning in 25.4–31.3%. Compared to chart review, the NLP algorithm had a sensitivity of 80.3%, 50.0%, and 66.7% for identifying central MK, 85.4%, 66.7%, and 100% for deep MK, and 100.0%, 95.2%, and 100% for thin MK, in the training, internal and external validation samples, respectively. Specificity was 41.1%, 38.6%, and 46.2% for centrality, 100%, 83.3%, and 71.4% for depth, and 93.3%, 100%, and was not applicable (n=0) to the external data for thinning, in the samples, respectively.
Conclusions:
MK features are not documented consistently showing a lack of standardization in recording MK exam elements. NLP shows promise but will be limited if available clinical data is missing from the chart.
Keywords: cornea, ulcer, measurement, slit lamp imaging
Microbial keratitis (MK) is an acute corneal infection that causes severe pain, decreases quality of life, and has potential for vision loss. Patients require immediate, intense treatment to minimize visual insults and risk of complications. Management of MK involves empiric topical antibiotic therapy, culture and gram stain, and occasionally, corneal transplant.1 Appropriate treatment is based on the severity of the infection. Studies have shown that physicians treat MK in several different ways.2–4 Providers evaluate features of MK, including the infiltrate’s location, size, depth, thinning, and multifocal nature and document this information in the exam portion of the clinical note.5 Certain features of MK morphology are associated with more severe disease and worse prognosis. Vital et al. described a system that classified keratitis as potentially sight threatening when infiltrates were more central, larger, or caused more inflammation.6 Other studies have reported that centrality, depth, and corneal thinning all play a role in MK severity.7–9
As clinicians use electronic health record (EHR) systems to document findings, it provides the opportunity to study documentation and build automated systems to extract data from the EHR. Manual extraction of clinical data by chart review is a time-consuming task; natural language processing (NLP) can process the same volume of information automatically and more efficiently.10 Algorithms that extract key clinical features have been used to guide medical decision making and improve quality of care.11–14 Such NLP algorithms have been used in several fields to identify diseases and their potential risk factors, adverse effects of drugs, and resistance to treatments.15–19 In ophthalmology, NLP of the EHR has been used to identify diseases, extract intraoperative complications, and quantitative data related to MK.20–22 However, the use of these algorithms in ophthalmology is currently limited and prone to challenges, due to domain-specific vocabulary, structure of the data, and abbreviations used in documentation.10
The purpose of this study was to use NLP to automate the extraction of clinical morphological features of MK documented by ophthalmologists in the EHR and evaluate the extent of the documentation across patients. A prior study by the authors illustrated an NLP algorithm that successfully extracts quantitative measurement information related to MK from the EHR.22 In the current study, an NLP algorithm was created to extract specific key features that are important in assessing the severity of MK. Using an automated process to extract key data has potential to be invaluable in identifying patterns and triaging patients with severe MK, recognize gaps in EHR documentation, and improve quality of care.
METHODS:
All patients in the University of Michigan (UM) EPIC EHR from August 1, 2012 to March 30, 2018 were explored to identify the subset of subjects who interacted with an eye care provider. This study was approved by the Institutional Review Board at University of Michigan. The data from Henry Ford was de-identified and was approved as exempt. Of these patients, those with International Classification of Diseases codes related to MK (ICD-9 370.0; ICD-10 H16.0) were included for training or validating the NLP algorithm. MK patients with all their clinical encounters were identified and data pertaining to patient demographics, current procedural terminology (CPT) codes, diagnoses based on ICD billing codes, and the free text in the corneal examination from the physician note were extracted into a data set for study. To train the algorithm, patients with four encounters in the first 14 days of active MK were identified. This specific group was selected because such patients were likely to have active, changing features between encounters, hence a high yield of documentation. A random sample of 100 of these patients (each with 4 encounters) was selected to form the training dataset for chart review. A separate set of 100 MK patients with 1 random encounter each, from April to December 2018, were identified and chart review was performed to serve as an internal validation set for the NLP algorithm. Finally, an external validation set of 59 patients from the Henry Ford Health System (HFHS) diagnosed with MK from April 1, 2016 to May 1, 2018, identified in the EPIC EHR by study team members (AH, SA) in the HFHS ophthalmology department, was chart reviewed for MK features. Thorough chart review, by a study team member trained in research related to MK (NM), with oversight by a cornea specialist (MW), was performed on these samples to extract key features of MK. Chart review included review of the free text cornea part of the exam, review of any drawings, other text within the exam portion of the record, and the assessment and plan of patients’ charts to identify clinical features of MK. The key clinical features of MK included centrality, depth, and thinning. Each patient encounter was categorized for each feature as central or not central (where paracentral, peripheral, and “out of visual axis” were considered not central), deep or not deep (where depth ≥50% was considered deep), and thin or not thin (where thinning ≥50% was considered thin). Additionally, any references in the chart to perforation, glue, or seidel positive were categorized as being thin and deep. The study team member was masked to the algorithm results at the time of chart review.
Development of the natural language processing algorithm
The NLP algorithm was created using the Python programming language, with use of the re, nltk, spaCy, and pandas (release 0.23.1) libraries. The development of the NLP algorithm was an iterative process using the free text in the cornea examination field of the EHR for all MK encounters in the training set. First, the cornea specialist reviewed notes in the training set to identify key phrases or word patterns that revealed the qualitative aspects of MK. Next, these were encoded using formal pattern descriptions known as “regular expressions.” These regular expressions were then applied to the training set and compared against the ophthalmologist-determined gold standard. Any errors in the algorithm’s identification of qualitative aspects were shown to the ophthalmologist for correction. This process was repeated until the ophthalmologist determined that any remaining errors were related to unusual phrasings that would not be expected to be generalizable. Thus, capturing such phrases using regular expressions may improve algorithm performance in the training set, but would not be expected to improve performance more generally and may introduce additional errors when applied beyond the training set.
Once the iterative development of the algorithm was complete, the algorithm was finalized and used to extract MK features from each samples using the following steps: 1) pre-processing to handle abbreviations and synonyms (e.g., replacing “endothelium pigment” with “endopigment”), 2) splitting of sentences and paragraphs into sentence fragments, 3) part-of-speech tagging and syntactic dependency parsing to identify modifying words (e.g., identifying adjectives that describe the word “scar”), 4) application of regular expressions to identify qualitative aspects of the lesion, and 5) extraction of numerical percentages describing the degree of thinning and lesion depth, and 6) aggregation of findings within sentence fragments at the note level. The code for the final algorithm is available at https://github.com/ML4LHS/cornea-nlp-mk-qualitative.
Validation of the natural language processing algorithm
After the algorithm was completely trained, its performance was evaluated on both an internal and external validation set. Accuracy of the algorithm was determined by agreement with gold-standard, manual chart review extracted details. Agreement occurred when both methods extracted the same centrality, depth, or thinning details, or also when neither method found any information regarding the features individually. Disagreement in MK information occurred when chart review recorded a feature that NLP did not, NLP recorded a feature that chart review did not, or chart review and NLP recorded details that were different. When the NLP algorithm was inaccurate, the study team evaluated the case to identify a possible reason underlying the error.
Statistical analysis
Descriptive statistics were used to summarize patient demographics within each training or validation sample. Categorical measures were summarized with frequencies and percentages and continuous measures were summarized with means and standard deviations. Samples were compared for differences with one-way analysis of variance (ANOVA), Chi-squared, and Fisher’s exact test. Ulcer features were compared between NLP extraction and chart review for agreement. Accuracy, defined as the percent of features that agreed between the NLP algorithm and chart review extractions, was calculated. To account for chance agreement between chart review and the NLP algorithm, Kappa statistics are reported with 95% confidence intervals (CI). Sensitivity, specificity, and positive predictive value were also calculated and reported with 95% CIs, for each of the ulcer features. All analyses were stratified by the UM training sample, the UM validation sample, and the external HFHS validation sample. SAS version 9.4 (SAS Institute, Cary, NC) and R 3.6.1 (Vienna, Austria) were used for all statistical analyses.
RESULTS:
A total of 100 patients each with 4 encounters (400 encounters total) were selected for the UM training set, 100 patients with 100 encounters for the UM validation, and 59 patients with 59 encounters for the external validation from HFHS. Across the samples, patients were on average 45–58 years old, 51–69% female, and 52–85% White (Table 1). Patients from the HFHS external validation set were significantly older (58.1 years, SD=19.7; p=0.0016) and a larger percentage were female (69.5%; p=0.0499) and Black (33.9%; p<0.0001) compared to patients from the UM training set (45.4 years, SD=20.9; 51% female; 11.0% Black) and those from the UM validation set (49.7 years, SD=22.5; 52.0% female; 8% Black).
Table 1.
Patient demographic characteristics by training and validation data sets
| UM Training (n=100 pts) | UM Validation (n=100 pts) | HFHS Validation (n=59 pts) | ||
|---|---|---|---|---|
| Continuous Variable | Mean (SD) | Mean (SD) | Mean (SD) | P-value* |
|
| ||||
| Age (years) | 45.4 (20.9) | 49.7 (22.5) | 58.1 (19.7) | 0.0016 |
| Categorical Variable | # (%) | # (%) | # (%) | P-value** |
|
| ||||
| Gender | ||||
| Male | 49 (49.0) | 48 (48.0) | 18 (30.5) | 0.0499 |
| Female | 51 (51.0) | 52 (52.0) | 41 (69.5) | |
| Race | ||||
| White | 85 (85.0) | 82 (82.0) | 31 (52.5) | <0.0001 |
| Black | 11 (11.0) | 8 (8.0) | 20 (33.9) | |
| Other | 4 (4.0) | 10 (10.0) | 8 (13.6) | |
UM, University of Michigan; HFHS, Henry Ford Health System; SD, Standard Deviation
one-way analysis of variance, ANOVA (post-hoc pairwise comparisons with Tukey adjustment show HFHS patients are significantly older than both UM samples, p<0.05)
Chi-Squared test (Gender), Fisher’s exact test (Race)
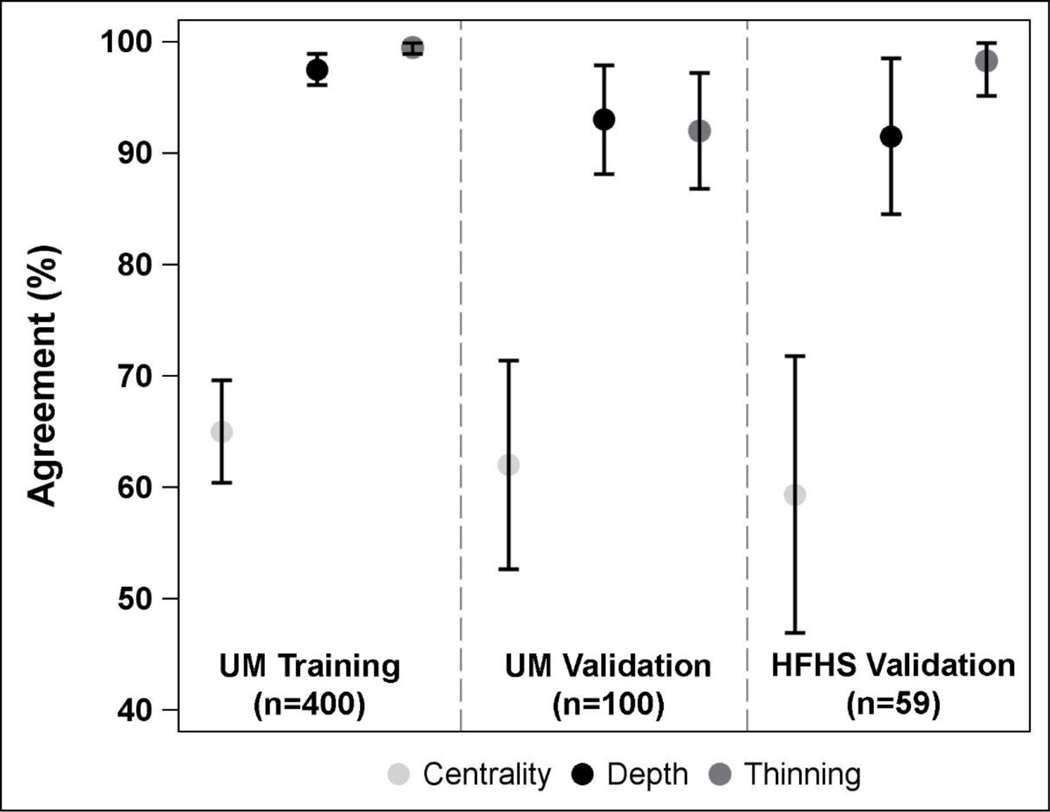
Centrality of MK was documented in the chart for 64.0%−79.3% of patients, depth for 15.0%−20.3%, and thinning for 25.4%−31.3% across the 3 samples (UM training, UM validation, HFHS validation). In comparison, the NLP algorithm found centrality of MK documented in the free text cornea part of the exam for 32.0%−50.9% of patients, depth for 16.0%−23.7%, and thinning for 27.1%−320% across the 3 samples. Agreement of information found between chart review and NLP ranged from 59.3% (95% CI, 46.8% to 71.9%) to 65.0% (95% CI, 60.3% to 69.7%) for ulcer centrality, 91.5% (95% CI, 84.4% to 98.6%) to 97.5% (95% CI, 96.0% to 99.0%) for ulcer depth, and 92.0% (95% CI, 86.7% to 97.3%) to 99.5% (95% CI, 98.8% to 100.0%) for ulcer thinning (Table 2, Figure 1). Similarly, kappa statistics ranged from 0.41 (95% CI, 0.23 to 0.58) to 0.50 (95% CI, 0.44 to 0.56) for ulcer centrality, 0.74 (95% CI, 0.57 to 0.91) to 0.92 (95% CI, 0.87 to 0.97) for ulcer depth, and 0.82 (95% CI, 0.70 to 0.94) to 0.99 (95% CI, 0.97 to 1.00) for ulcer thinning.
Table 2.
Agreement between chart review and natural language processing (NLP) for identifying features of microbial keratitis from the electronic health record.
| UM Training (n=400) | UM Validation (n=100) | HFHS Validation (n=59) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Chart Review | Chart Review | Chart Review | |||||||
| NLP | Yes | No | No Info | Yes | No | No Info | Yes | No | No Info |
|
| |||||||||
| # (% total) | # (% total) | # (% total) | # (% total) | # (% total) | # (% total) | # (% total) | # (% total) | # (% total) | |
| Central | |||||||||
| Yes | 102 (25.5) | 4 (1.0) | 3 (1.0) | 10 (10.0) | 3 (3.0) | 1 (1.0) | 12 (20.3) | 0 (0.0) | 3 (5.1) |
| No | 6 (1.5) | 78 (19.5) | 0 (0.0) | 1 (1.0) | 17 (17.0) | 0 (0.0) | 2 (3.4) | 12 (20.3) | 1 (1.7) |
| No Info | 19 (4.8) | 108 (27.0) | 80 (20.0) | 9 (9.0) | 24 (24.0) | 35 (35.0) | 4 (6.8) | 14 (23.7) | 11 (18.6) |
| Agreement: | 65.0% (60.3%, 69.7%) | 62.0% (52.5%, 71.5%) | 59.3% (46.8%, 71.9%) | ||||||
| Kappa: | 0.50 (0.44, 0.56) | 0.41 (0.29, 0.54) | 0.41 (0.23, 0.58) | ||||||
| Depth | |||||||||
| Yes | 35 (8.8) | 0 (0.0) | 2 (0.5) | 6 (6.0) | 1 (1.0) | 1 (1.0) | 5 (8.5) | 1 (1.7) | 2 (3.4) |
| No | 0 (0.0) | 35 (8.8) | 2 (0.5) | 1 (1.0) | 5 (5.0) | 2 (2.0) | 0 (0.0) | 5 (8.5) | 1 (1.7) |
| No Info | 6 (1.5) | 0 (0.0) | 320 (80.0) | 2 (2.0) | 0 (0.0) | 82 (82.0) | 0 (0.0) | 1 (1.7) | 44 (74.6) |
| Agreement: | 97.5% (96.0%, 99.0%) | 93.0% (88.0%, 98.0%) | 91.5% (84.4%, 98.6%) | ||||||
| Kappa: | 0.92 (0.87, 0.97) | 0.74 (0.57, 0.91) | 0.77 (0.59, 0.96) | ||||||
| Thinning | |||||||||
| Yes | 110 (27.5) | 1 (0.3) | 1 (0.3) | 20 (20.0) | 0 (0.0) | 7 (7.0) | 15 (25.4) | 0 (0.0) | 1 (1.7) |
| No | 0 (0.0) | 14 (3.5) | 0 (0.0) | 0 (0.0) | 5 (5.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| No Info | 0 (0.0) | 0 (0.0) | 274 (68.5) | 1 (1.0) | 0 (0.0) | 67 (67.0) | 0 (0.0) | 0 (0.0) | 43 (72.9) |
| Agreement: | 99.5% (98.8%, 100.0%) | 92.0% (86.7%, 97.3%) | 98.3% (95.0%, 100.0%) | ||||||
| Kappa: | 0.99 (0.97, 1.00) | 0.82 (0.70, 0.94) | 0.95 (0.87, 1.00) | ||||||
Note: Agreement is noted by highlighted cells, and overall agreement is reported as percent (95% confidence interval).
Figure 1.
Forest plot displaying percent agreement between chart review and natural language processing for extracting microbial keratitis features from the electronic health record. Agreement is stratified by sample and MK feature, and displayed with 95% confidence intervals. UM, University of Michigan; HFHS, Henry Ford Health System.
Compared to chart review, the NLP algorithm had a sensitivity of 80.3%, 50.0%, and 66.7% for identifying MK that was documented as central, 85.4%, 66.7%, and 100.0% for MK that was deep, and 100.0%, 95.2%, and 100% for MK that was thin, for the training, internal validation, and external validation samples, respectively (Figure 2, eTable 1 includes 95% CIs). Specificity was 41.1%, 38.6%, and 46.2% for the centrality feature, 100.0%, 83.3%, and 71.4% for the depth feature, and 93.3%, 100.0%, and not applicable (n=0) for the thin feature, for the training, internal validation, and external validation samples, respectively. Sensitivity and specificity of NLP algorithm for identifying MK features overall (i.e. combined central and non-central, deep and not deep, thin and not thin) are also reported in eTable 1.
Figure 2.
Forest plots to display sensitivity and specificity of natural language processing to identify microbial keratitis features from the electronic health record, compared to chart review. Sensitivity and specificity are stratified by sample and MK feature and are displayed with 95% confidence intervals. Confidence intervals are capped at 100%. UM, University of Michigan; HFHS, Henry Ford Health System.
Disagreement between chart review and the NLP for identifying MK features ranged from 0.5% to 40.7% depending on the sample and feature (Table 3). Disagreement was categorized into 4 main themes, including chart review finding information outside the corneal examination note, complex phrasing in the corneal examination note that NLP could not interpret, NLP failure, and chart review failure. For the centrality feature, over the 3 samples, 75.0%−89.3% of disagreement was due to information being found outside of the corneal exam note, 5.7%−16.7% was due to complex phrasing of the exam note, 4.3%−8.3% was due to NLP failures (e.g. segmentation issues or information returned for the unaffected eye), and 0.0–0.7% of disagreement was due to human error in chart review. For depth, most disagreement between chart review and NLP was due to either NLP failure (40.0%−42.9% across the 3 samples) or complex phrasing in the exam note (20.0%−42.9%), whereas less disagreement was found due to information found outside the cornea exam note (0.0%−14.3%) or human error in chart review (0.0%−40.0%). Lastly, for MK thinning, 100% of disagreement between chart review and NLP for the UM training sample (n=2) was found to be due to complex phrasing in the cornea exam note and 100% of disagreement for the HFHS validation sample (n=1) was found to be due to human error in chart review. For the UM validation sample, 25% disagreement in thinning was due to information outside of the cornea exam note, 37.5% was due to complex phrasing, and 37.5% was due to NLP failures. Examples of complex phrasing within the clinician note that the NLP algorithm was not able to accurately interpret are provided in eTable 2.
Table 3.
Reasons for disagreement between chart review (CR) and natural language processing (NLP) for identifying features of microbial keratitis
| Ulcer Measurement | N disagree (%) | Information Outside Cornea Exam Note | Complex Phrasing | NLP Failure | CR Failure |
|---|---|---|---|---|---|
|
| |||||
| UM Training (n=400) | n (% of total sample) |
n (% of disagree) |
n (% of disagree) |
n (% of disagree) |
n (% of disagree) |
| Central | 140 (35.0%) | 125 (89.3) | 8 (5.7) | 6 (4.3) | 1 (0.7) |
| Deep | 10 (2.5%) | 0 (0.0) | 4 (40.0) | 4 (40.0) | 2 (20.0) |
| Thin | 2 (0.5%) | 0 (0.0) | 2 (100.0) | 0 (0.0) | 0 (0.0) |
| UM Validation (n=100) | |||||
| Central | 38 (38.0%) | 31 (81.6) | 5 (13.2) | 2 (5.3) | 0 (0.0) |
| Deep | 7 (7.0%) | 1 (14.3) | 3 (42.9) | 3 (42.9) | 0 (0.0) |
| Thin | 8 (8.0%) | 2 (25.0) | 3 (37.5) | 3 (37.5) | 0 (0.0) |
| HFHS Validation (n=59) | |||||
| Central | 24 (40.7%) | 18 (75.0) | 4 (16.7) | 2 (8.3) | 0 (0.0) |
| Deep | 5 (8.5%) | 0 (0.0) | 1 (20.0) | 2 (40.0) | 2 (40.0) |
| Thin | 1 (1.7%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) |
DISCUSSION:
Natural language processing has been introduced into many disciplines in medicine and has optimized data extraction for research studies.20,23 NLP algorithms allow more complete access to the EHR by accessing free text data. The rich detail in clinical notes, that houses the experience and knowledge of many physicians, can help in improving patient care.24 However, the complexity of data in the clinical record when written in text and without pre-specified fields, has limitations. Our NLP algorithm to identify key morphologic features of microbial keratitis in clinical notes was 50–80% sensitive at identifying centrality, 67–100% sensitive for identifying depth, and 95–100% sensitive for identifying thinning in an internal and external validation datasets. The lack of precise use of words in the chart required us to use quantitative numbers (e.g. >50% for depth or 90% thinning) to give even word descriptions clinical contextual meaning for the algorithm. Disagreement in feature identification between chart review and NLP was often due to information that was obtained from outside the cornea exam text note and from drawings within the chart. Clinicians often documented MK centrality in drawings. A study team member reviewing clinical charts could extract information from a drawing, but the NLP algorithm could only analyze text notes. Data on MK centrality was found in drawings or outside the cornea exam free text in approximately one third of encounters across samples. Data on MK depth and thinning was most frequently found in the free text corneal exam note, with few instances of these features found elsewhere in the EHR (only 1 case of depth and 2 of thinning were documented outside the corneal exam in the UM validation sample, and none noted outside the exam in the other samples).
NLP extraction was challenged by text and phrasing complexities, including abbreviations, misspellings, and semantic and syntax errors. In addition, clinicians used a wide vocabulary to describe similar features as has been shown in other domains,25 posing challenges for deciphering clinical notes. Abbreviations are not standardized and require interpretation based on the context. This poses a challenge not only for software-based interpretation, but also for clinicians when one patient is managed by multiple health care professionals over time.24 NLP algorithm failures, resulting from improper NLP segmentation or long phrasing separating key adjectives and nouns, occurred in a small portion of the sample (22 of 559 encounters across samples). Algorithm refinement to accommodate complex phrasing and specific failures, would likely overfit the training sample and potentially introduce additional errors. The NLP discordance with clinical data emphasizes a need for standardized terminology and language used to describe MK. Standardization would benefit clinicians, not just algorithms.
One of the striking aspects of EHR documentation highlighted in this study was the lack of documentation of several key features that describe MK. Notably, centrality of MK was documented in only 64%−79% of encounters, depth in 15%−20%, and thinning in 25%−31%, across the samples. Several studies have shown the burden on clinicians of EHR documentation.26,27 In this context, patients with MK are often complex and require urgent in-office needs, potentially hindering detailed documentation compared to other ophthalmic conditions.28 The fact that centrality is documented more than other key features likely highlights the prognostic importance of the ulcer’s location relative to the pupil center. The lack of documentation of depth and thinning may also reflect that morphologic features often are not documented in their absence; they are documented only when they are present. For example, if MK causes corneal thinning, it would likely be documented, but if there was no thinning, a clinician is less likely to document “no thinning.” These natural charting nuances result in less data for analysis of depth and thinning.
The strengths of this study include robust data and methods to train the algorithm, an external validation dataset to evaluate generalizability, masked analysis, and a detailed exploration of algorithm discordance with the clinician chart review. As we highlight in our partner paper in Ophthalmology,22 NLP has the potential to improve use of secondary data collected for other purposes, such as the Intelligent Research In Sight (IRIS) registry supported by the American Academy of Ophthalmology. The limitations of this study include sample size limitations due to missing chart data from both institutions, restriction to the cornea free text exam portion of the health record, and inability to parse long phrases of text effectively due to lack of standardization.
Natural language processing shows promise to identify prognostic features of MK from the EHR; however, missing data may somewhat limit these efforts. Standardization in physician documentation would aid in robust analyses, as well as help other clinicians in the co-management of the patient. Algorithms that can evaluate text and imaging data simultaneously would likely be able to overcome limitations of missing data. In particular, future efforts to detect qualitative features from ophthalmologic notes should focus on automated interpretation of clinician drawings, which are a core part of the documentation.
Supplementary Material
Sensitivity and specificity of natural language processing to identify information in the electronic health record related to microbial keratitis features, in comparison to chart review.
Examples of complex phrasing within the clinician note that the NLP algorithm was not able to accurately interpret
ACKNOWLEDGEMENTS:
We would like to thank Huan Tan, Autumn Valicevic, and Adharsh Murali for their contributions to this manuscript.
Financial support: This work was supported by a grant from the National Institutes of Health, Bethesda, MD (Woodward, NIH-1R01EY031033) and Blue Cross Blue Shield Foundation of Michigan (Woodward, Singh, AWD012094). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acronyms:
- ED
Epithelial defect
- SI
Stromal infiltrate
- NLP
Natural Language Processing
- EHR
Electronic health records
- MK,
Microbial keratitis
- UM
University of Michigan
- HFHS
Henry Ford Health System
- ICD
International Classification of Diseases
Footnotes
Conflict of interest: The authors have no proprietary or commercial interest in any of the materials discussed in this article.
REFERENCES:
- 1.McLeod SD, LaBree LD, Tayyanipour R, et al. The importance of initial management in the treatment of severe infectious corneal ulcers. Ophthalmol 1995;102:1943–1948. [DOI] [PubMed] [Google Scholar]
- 2.Daniell M, Mills R, Morlet N. Microbial keratitis: what’s the preferred initial therapy? Br J Ophthalmol 2003;87:1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelmus KR. Indecision about corticosteroids for bacterial keratitis: An evidence-based update. Ophthalmol 2002;109:835–842. [DOI] [PubMed] [Google Scholar]
- 4.McLeod SD, DeBacker CM, Viana MA. Differential care of corneal ulcers in the community based on apparent severity. Ophthalmol 1996;103:479–484. [DOI] [PubMed] [Google Scholar]
- 5.Vital MC, Belloso M, Prager TC, Lanier JD. Classifying the Severity of Corneal Ulcers by Using the “1, 2, 3” Rule. Cornea 2007;26:16–20. [DOI] [PubMed] [Google Scholar]
- 6.Al-Mujaini A, Al-Kharusi N, Thakral A, Wali UK. Bacterial keratitis: Perspective on epidemiology, Clinico-Pathogenesis, diagnosis and treatment. Sultan Qaboos Univ Med J 2009;9:184–195. [PMC free article] [PubMed] [Google Scholar]
- 7.Lin A, Rhee MK, Akpek EK, et al. Bacterial Keratitis Preferred Practice Pattern®. Ophthalmol 2019;126:P1–P55. [DOI] [PubMed] [Google Scholar]
- 8.Miedziak AI, Miller MR, Rapuano CJ, Laibson PR, Cohen EJ. Risk factors in microbial keratitis leading to penetrating keratoplasty Ophthalmol 1999; 106:1166–70. [DOI] [PubMed] [Google Scholar]
- 9.Bourcier T, Thomas F, Borderie V, Chaumeil C, Laroche L. Bacterial keratitis: predisposing factors, clinical and microbiological review of 300 cases. Br J Ophthalmol. 2003;87:834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. J Biomed Inform 2017;73:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wi C Il, Sohn S, Rolfes MC, et al. Application of a natural language processing algorithm to asthma ascertainment: An automated chart review. Am J Respir Crit Care Med 2017;196:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusum SM, Yue L, Heng Deconstructing intensive care unit triage: Identifying factors influencing admission decisions using natural language processing and semantic technology. Am J Respir Crit Care Med 2015;191:A5223. [Google Scholar]
- 13.Glaser AP, Jordan BJ, Cohen J, et al. Automated Extraction of Grade, Stage, and Quality Information From Transurethral Resection of Bladder Tumor Pathology Reports Using Natural Language Processing. JCO Clin Cancer Informatics 2018:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pai VM, Rodgers M, Conroy R, et al. Workshop on using natural language processing applications for enhancing clinical decision making: An executive summary. J Am Med Informatics Assoc 2014;21:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh K, Betensky RA, Wright A, et al. A Concept-Wide Association Study of Clinical Notes to Discover New Predictors of Kidney Failure. Clin J Am Soc Nephrol 2016;11:2150–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh K, Choudhry NK, Krumme AA, et al. A concept‐wide association study to identify potential risk factors for nonadherence among prevalent users of antihypertensives. Pharmacoepidemiol Drug Saf 2019;28:1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duke J, Chase M, Poznanski-Ring N, et al. Natural Language Processing to Improve Identification of Peripheral Arterial Disease in Electronic Health Data. J Am Coll Cardiol 2016;67:2280. [Google Scholar]
- 18.Patel R, Wilson R, Jackson R, et al. Cannabis use and treatment resistance in first episode psychosis: a natural language processing study. Lancet 2015;385:S79. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Jung K, Winnenburg R, Shah NH. A method for systematic discovery of adverse drug events from clinical notes. J Am Med Informatics Assoc 2015;22:1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein JD, Rahman M, Andrews C, et al. Evaluation of an Algorithm for Identifying Ocular Conditions in Electronic Health Record Data. JAMA Ophthalmol 2019;137:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Shorstein NH, Amsden LB, Herrinton LJ. Natural language processing to ascertain two key variables from operative reports in ophthalmology. Pharmacoepidemiol Drug Saf 2017;26:378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maganti N, Tan H, Niziol LM, et al. Natural Language Processing to Quantify Microbial Keratitis Measurements. Ophthalmol 2019;126:1722–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doan S, Maehara CK, Chaparro JD, et al. Building a Natural Language Processing Tool to Identify Patients with High Clinical Suspicion for Kawasaki Disease from Emergency Department Notes. Academic Emergency Medicine.; 2016;23:628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menasalvas E, Gonzalo-Martin C. Challenges of Medical Text and Image Processing: Machine Learning Approaches. In: Holzinger A, ed. Machine Learning for Health Informatics: State-of-the-Art and Future Challenges.Vol 9605 LNAI. Cham, Switzerland: Springer International Publishing; 2016:221–242. [Google Scholar]
- 25.Cawsey AJ, Webber BL, Jones RB. Natural Language Generation in Health Care. J Am Med Informatics Assoc 1997;4:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh RP, Bedi R, Li A, et al. The practice impact of electronic health record system implementation within a large multispecialty ophthalmic practice. JAMA Ophthalmol 2015;133:668–674. [DOI] [PubMed] [Google Scholar]
- 27.Chiang MF, Read-Brown S, Hribar MR, et al. Time requirements for pediatric ophthalmology documentation with electronic health records (EHRs): a time-motion and big data study. J Am Assoc Pediatr Ophthalmol Strabismus 2016;20:e2–e3. [Google Scholar]
- 28.Chiang MF, Boland M V., Brewer A, et al. Special requirements for electronic health record systems in ophthalmology. Ophthalmol 2011;118:1681–1687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity and specificity of natural language processing to identify information in the electronic health record related to microbial keratitis features, in comparison to chart review.
Examples of complex phrasing within the clinician note that the NLP algorithm was not able to accurately interpret