Abstract
Objective:
In AIDS Clinical Trials Group study A5338, concomitant rifampicin, isoniazid, and efavirenz was associated with more rapid plasma medroxyprogesterone acetate (MPA) clearance compared to historical controls without tuberculosis or HIV therapy. We characterized the pharmacogenetics of this interaction.
Methods:
In A5338, women receiving efavirenz-based HIV therapy and rifampicin plus isoniazid for tuberculosis underwent pharmacokinetic evaluations over 12 weeks following a 150-mg intramuscular injection of depot MPA. Data was interpreted with nonlinear mixed-effects modelling. Associations between individual pharmacokinetic parameters and polymorphisms relevant to rifampicin, isoniazid, efavirenz, and MPA were assessed.
Results:
Of 62 A5338 participants in four African countries, 44 were evaluable for pharmacokinetic associations, with 17 CYP2B6 normal, 21 intermediate, and 6 poor metabolizers, and 5 NAT2 rapid, 20 intermediate, and 19 slow acetylators. There were no associations between either CYP2B6 or NAT2 genotype and MPA Cmin at week 12, apparent clearance, Cmax, AUC0–12wk or half-life, or unexplained interindividual variability in clearance, and uptake rate constant or mean transit time of the slow-release fraction (p>0.05 for each). In exploratory analyses, none of 28 polymorphisms in 14 genes were consistently associated with MPA pharmacokinetic parameters, and none withstood correction for multiple testing.
Conclusions:
Study A5338 suggested that more frequent depot MPA dosing may be appropriate for women receiving rifampicin, isoniazid, and efavirenz. The present results suggest that knowledge of CYP2B6 metabolizer or NAT2 acetylator status does not inform individualized DMPA dosing in this setting.
Introduction
Tuberculosis and human immunodeficiency virus (HIV) are leading causes of infection-related deaths worldwide [1]. Women comprise more than half of the estimated 37 million persons living with HIV and are disproportionately affected by new infections among individuals over 15 years of age in sub-Saharan Africa [2]. Coinfection with M. tuberculosis and HIV in pregnancy markedly increases risk of maternal and child morbidity and mortality [3–5], thus access to effective contraception is critical. Depot medroxyprogesterone acetate (DMPA) is an intermediate-acting progesterone-only injectable contraceptive commonly used globally, including in sub-Saharan Africa because of its efficacy and convenience of administration [6]. Following a 150-mg intramuscular dose of DMPA, medroxyprogesterone acetate (MPA) concentrations exceed the therapeutic target (0.1 ng/mL) for approximately 12 weeks and inhibit ovulation for up to 14 weeks [7]. The probability of ovulation increases when MPA concentrations fall below 0.1 ng/mL [8, 9]. MPA undergoes metabolism by hepatic CYP isoforms, primarily CYP3A4 [10].
Rifampicin and isoniazid are cornerstone drugs for treating drug-sensitive tuberculosis, while efavirenz is recommended for women of childbearing potential as an alternative to dolutegravir as component of first-line antiretroviral therapy in sub-Saharan Africa [11]. Rifampicin, isoniazid, and efavirenz are often used concomitantly in sub-Saharan Africa in patients living with HIV and tuberculosis. Rifampicin and efavirenz are potent inducers of cytochrome (CYP) P450 enzymes that metabolize MPA [10, 12], and have been shown to reduce concentrations and compromise efficacy of some hormonal contraceptives [13, 14]. This raised concern that concomitant rifampicin and efavirenz would increase risk for contraceptive failure with DMPA, as compared to DMPA without such drug-drug interactions.
Study A5338 (NCT02412436) of the AIDS Clinical Trials Group (ACTG) was a 12-week, phase II, open-label, single-arm study of steady-state pharmacokinetic interactions among HIV and tuberculosis coinfected women receiving efavirenz-containing ART, and continuation-phase tuberculosis treatment that included rifampicin and isoniazid [15]. Study A5338 tested the hypothesis that clearance of MPA would increase when given with rifampicin and efavirenz, increasing risk of ovulation. Among 42 pharmacokinetic-evaluable women from four African countries, the study showed that of the exposure of MPA was substantially decreased, with the AUC0–12wks being 33% lower than in historical controls [16, 17], and with MPA concentrations below the therapeutic target of 0.1 ng/mL at week 12 in 12% of women, suggesting that more frequent DMPA dosing may be appropriate.
Plasma exposure of isoniazid and efavirenz vary significantly between individuals due to human genetic polymorphisms. Isoniazid acetylation by hepatic N-acetyltransferase 2 generates hydrazine metabolites [18], and there are well-described loss-of-function NAT2 alleles [18–21]. Individuals who carry one or two copies of such alleles have intermediate or slow acetylator phenotypes, respectively, and progressively greater plasma isoniazid exposure [18–21]. Plasma efavirenz exposure is predicted by CYP2B6 polymorphisms [22], especially CYP2B6 516G→T (rs3745274) [23–25], 983T→C (rs28399499) [25–27], and 15582C→T (rs4803419) [25].
Rifampicin is a potent inducer of hepatic CYP isoforms, and in a study of 11 healthy, HIV-negative volunteers modestly and variably reduced efavirenz plasma exposure [28]. However, some patients receiving tuberculosis therapy that includes isoniazid with rifampicin experience increased plasma efavirenz exposure, particularly in the presence of CYP2B6 and/or NAT2 loss-of-function polymorphisms [29–31]. This effect is likely mediated by isoniazid, as isoniazid alone has also been shown to reduce plasma efavirenz clearance among CYP2B6 poor metabolizers [30]. The mechanism has been suggested to involve isoniazid inhibition of CYP2A6, a minor pathway for efavirenz elimination that assumes greater importance in CYP2B6 poor metabolizers [30–34].
The present study seeks to determine whether selected human genetic polymorphisms were associated with plasma pharmacokinetics of MPA among women living with HIV, and who were receiving concomitant isoniazid, rifampicin and efavirenz during participation in A5338.
METHODS
Study Population
Study A5338 was a 12-week, phase II, open-label, single-arm study of steady-state pharmacokinetic interactions among HIV and tuberculosis coinfected women receiving efavirenz-based ART and rifampicin plus isoniazid for treatment of tuberculosis [15]. Eligible participants were 18 to 46 years of age, non-pregnant, had been on efavirenz plus two nucleoside reverse transcriptase inhibitors (NRTIs) for at least 28 days prior to study entry, and were receiving rifampicin (600 mg) and isoniazid (300 mg) at least 5 days per week during the continuation phase of tuberculosis treatment. Participants were excluded if they had received DMPA or other injectable contraceptives within 180 days, any other hormonal therapies within 30 days of study entry, were taking CYP3A4 inducers or inhibitors within 30 or 7 days, respectively, before study entry, were pregnant or breastfeeding, or had a contraindication to DMPA administration. Participants provided written informed consent. Institutional review boards of the participating institutions approved the study, and participants gave written informed consent.
Procedures
A detailed description of A5338 procedures and primary results is provided elsewhere [15]. Briefly, at study entry DMPA was administered as a single 150-mg intramuscular injection. Plasma samples for MPA assays were obtained predose and 2, 4, 6, 8, 10, and 12 weeks post-dose. Adherence to HIV and tuberculosis medications was assessed at all study visits using an ACTG self-report questionnaire [15]. Assays for MPA were performed at the University of Cape Town Pharmacology Specialty Laboratory, using a validated liquid-liquid extraction method and liquid chromatography tandem mass spectrometry analysis (LC-MS/MS) with an AB Sciex API 5500Q mass spectrometer.
Genetic Polymorphisms
Human DNA extracted from whole blood was used to genotype 48 polymorphisms of interest, including 1 in ANO2 (anoctamin 2), 1 in CSK (C-terminal Src kinase), 2 in CYP19A1 (aromatase), 2 in CYP1A1, 7 in CYP1A2,1 in CYP2A6, 3 in CYP2B6, 3 in CYP3A4, 1 in CYP3A43, 2 in CYP3A5, 9 in NAT2, 12 in SLCO1B1 (which encodes organic anion transporting polypeptide 1B1), 1 in TRIM4, 1 in UGT1A1, 1 in LOC101927066, and 1 intergenic. These included CYP2B6 polymorphisms (rs3745274, rs28399499 and rs4803419) that predict plasma efavirenz exposure, and NAT2 polymorphisms (rs1801279, rs1801280, rs1799930 and rs1799931) that predict plasma isoniazid exposure. The other polymorphisms were selected based on showing genome-wide association (P < 5.0 × 10−8) with any estradiol trait in the GWAS Catalog, and CYP3A4, CYP3A5 and CYP1A1 polymorphisms genome-wide associated with any trait in the GWAS Catalog [35]. We also included functional polymorphisms in genes involved in estrogen metabolism [36], including CYP1A1, CYP1A2, CYP1B1, CYP3A4 and CYP3A5. Genotyping was done in VANTAGE (Vanderbilt Technology for Advanced Genomics) using MassARRAY® iPLEX Gold (Agena Bioscience™, California, USA) and Taqman (ThermoFisher Scientific, Massachusetts, USA). Final assay design is available upon request. Ample blank assays were included to assure validity, and all samples were assayed in duplicate.
We excluded 13 monomorphic loci, and 7 with minor allele frequencies less than 5%. Genotyping efficiency was 100% for all polymorphisms in all participants. All polymorphisms were in Hardy-Weinberg equilibrium (p>0.05). Association analyses ultimately included the 28 remaining polymorphisms (Supplemental Material).
Statistical analysis
A population pharmacokinetic model was developed to describe the concentrations on MPA using nonlinear mixed effects modelling in the software NONMEM applying first-order conditional estimation algorithm with eta-epsilon interaction (FOCE-I) [37]. The detailed description of the model is provided elsewhere [15, 38]. Briefly, MPA pharmacokinetics was modelled using a one-compartment disposition model with first-order elimination. To model the release of MPA from the depot in the injection site, a bi-phasic absorption model was used, with a fraction of the dose immediately available for uptake into the bloodstream with a first-order rate, while the remaining fraction is slowly released from the formulation crystals into the injection site, and only then is becomes available for uptake. The slower release process was modelled using a transit compartment model [39]. The model included the effect of body weight on all disposition parameters using allometric scaling [40], and random effects were included on the pharmacokinetic parameters to account for variability between subjects or visits. The model also included drug-drug interaction effects on MPA clearance for anti-tuberculosis treatment plus efavirenz, efavirenz alone, nelfinavir, and lopinavir/ritonavir. These effects were included as a categorical fixed effect for each arm, so all the values of clearance included in this analysis were centered around the typical value for the anti-tuberculosis treatment plus efavirenz arm.
From the final model, we extracted the individual values of the pharmacokinetic parameters (i.e., the empirical Bayesian estimates), and the associated “unexplained” variability random effect, which describe the differences between subjects after adjusting for the effect of body weight (which was included in the model as a fixed effect). Additionally, we obtained the individual values of the exposure of MPA in terms of AUC0–12wk, terminal half-life, peak concentration (Cmax), and concentration at 12 weeks (Cmin), i.e., when the next injection of DMPA was administered.
These individual pharmacokinetic parameter values, random effects, and exposure metrics were then tested against the genotypes using linear regression models. Associations of CYP2B6 metabolizer group and NAT2 acetylator group with each individual pharmacokinetic parameter were assessed by Spearman’s rank correlation test using STATA version 15.1 (StataCorp, College Station, Texas, USA). Associations with the 28 individual polymorphisms were assessed by linear regression using PLINK version 1.07 [41].
Composite CYP2B6 genotype was defined based on combinations of three polymorphisms as follows: normal metabolizer (1: 15582CC-516GG-983TT or 2: 15582CT-516GG-983TT); intermediate metabolizer (3: 15582TT-516GG-983TT; 4: 15582CC-516GT-983TT; 5: 15582CC-516GG-983CT; 6: 15582CT-516GT-983TT; or 7: 15582CT-516GG-983CT); and poor metabolizer (8: 15582CC-516TT-983TT; 9: 15582CC-516GT-983CT; 10: 15582CC-516GG-983CC [25]. For NAT2, genotypes were categorized based on combinations of rs1801280 (NAT2*5), rs1799930 (NAT2*6), rs1799931 (NAT2*7), and rs1801279 (NAT2*14), as slow if homozygous for the variant allele at any locus (i.e., AA, CC, AA, AA, respectively), or heterozygous at 2 or more loci; intermediate if heterozygous at a single locus; or extensive if no variant allele at any locus [42]. Associations of pharmacokinetic measures with CYP2B6 and NAT2 genotype groups were assessed using the Jonckheere-Terpstra test for ordered alternatives. Associations with the 28 individual polymorphisms were assessed by linear regression. For associations with CYP2B6 and NAT2 genotype groups, we did not correct for multiple comparisons, as these were our primary focus. We used Bonferroni correction for multiple comparisons for the 28 polymorphisms, giving a significance threshold of p = 1.8×10−3. We did not correct for comparing the multiple pharmacokinetic parameters, as these tended to correlate with each other. Two-sided tests were used.
RESULTS
Participant characteristics
Study A5338 enrolled 62 participants in Botswana (n=7), Zimbabwe (n=8), Kenya (n=12), Durban, South Africa (n=17) and Johannesburg, South Africa (n=18), from whom genotype data were available for all 62, and pharmacokinetic-evaluable data were available for 44. Of the 44 participants, 17 (39%) were CYP2B6 normal metabolizers, 21 (48%) were CYP2B6 intermediate metabolizers, and 6 (14%) were CYP2B6 poor metabolizers. In addition, 5 (11%) were NAT2 rapid acetylators, 20 (45%) were NAT2 intermediate acetylators, and 19 (43%) were NAT2 slow acetylators. At weeks 10 and 12, all women reported 100% adherence to their HIV and tuberculosis medications. Two participants excluded from the primary A5338 publication because they lacked pharmacokinetic results at either week 10 or week 12 [15] were included in the present analyses, as we allowed for participants with results for at least one of the two weeks.
Genetic associations with MPA pharmacokinetics
Among the 44 evaluable participants, there were no significant associations between CYP2B6 metabolizer status and any primary pharmacokinetic parameter for MPA, including Cmin at week 12 (p=0.48), apparent clearance (p=0.60), Cmax (p=0.59), AUC0–12wk (p=0.99) and half-life (p=0.19). Similarly, considering only the interindividual variability in pharmacokinetic parameters after adjusting for covariate effects, there were no associations with CYP2B6 metabolizer status, including for clearance (p=0.56), uptake rate (p=0.19) and mean transit time of the slow release (p=0.89). The relationship between CYP2B6 metabolizer status, MPA Cmin and MPA clearance is shown in the Figure. Individuals with CYP2B6 poor metabolizer genotypes were no more likely to have subtherapeutic MPA concentrations
Figure 1. Relationships between CYP2B6 metabolizer status, NAT2 acetylator status, and selected medroxyprogesterone acetate pharmacokinetic parameters among 44 participants.
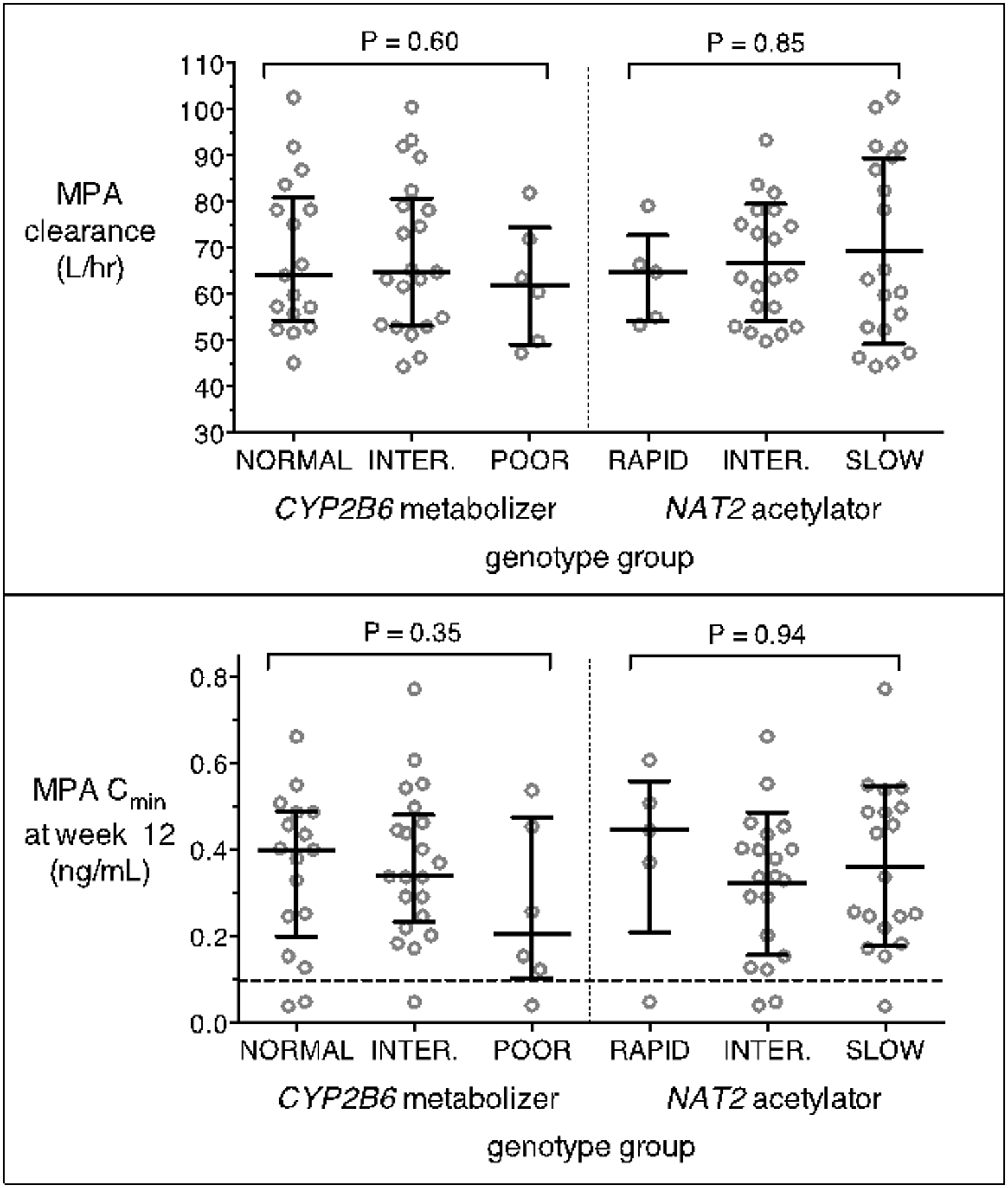
Top panel: associations of apparent clearance with CYP2B6 metabolizer status (left) and NAT2 acetylator status (right); Bottom panel: associations of trough concentration with CYP2B6 metabolizer status (left) and NAT2 acetylator status (right). The dashed line indicates the MPA therapeutic cut-off of 0.1 ng/mL. All participants received a single 150 mg intramuscular injection of depot medroxyprogesterone acetate. Error bars indicate median and interquartile range. Jonckheere-Terpstra test P-values are shown. MPA = medroxyprogesterone acetate; Cmin = trough concentration.
Among the 44 evaluable participants, there was no significant associations between NAT2 acetylator status and any primary pharmacokinetic parameter, including Cmin at week 12 (p=0.94), apparent clearance (p=0.85), Cmax (p=0.21), AUC0–12wk (p=0.62) and half-life (p=0.18). Considering only the unexplained interindividual variability in pharmacokinetic parameters, there were no associations with NAT2 acetylator status, including for clearance (p=0.65), uptake rate (p=0.18) and mean transit time of the slow release (p=0.14). The relationship between NAT2 acetylator status, MPA Cmin and MPA clearance is shown in the Figure.
Exploratory analyses beyond CYP2B6 metabolizer and NAT2 acetylator status included 28 polymorphisms. There were no consistent associations between any polymorphism and any MPA pharmacokinetic parameter, and none were statistically significant after correction for multiple comparisons. The lowest p-values for association were as follows: for Cmin at week 12, NAT2 rs1799929 (p=0.048); for apparent clearance, Cmax and AUC0–12wk, intergenic rs727428 (p=0.019, p=0.62 and p=0.0028, respectively); and for half-life, CYP2B6 rs28399499 (p=0.038). Considering unexplained interindividual variability in pharmacokinetic parameters, the lowest p-values for association were as follows: for clearance, CYP2B6 rs3745274 (p=0.036); for uptake rate, intergenic rs727428 (p=0.016); and for mean transit time of the slow release fraction, NAT2 rs1799929 (p=0.032). Complete association results for each MPA pharmacokinetic parameter with each polymorphism are in Supplemental Material.
DISCUSSION
Study A5338 was the first study to document the interaction between DMPA and rifampicin, albeit in combination with efavirenz [15]. It showed that, among HIV and tuberculosis coinfected women receiving efavirenz-containing ART and tuberculosis treatment that included rifampicin and isoniazid, and who were then administered as a single 150 mg intramuscular injection of DMPA, median clearance of MPA was substantially greater than in historical controls [16, 17], suggesting that more frequent DMPA dosing may be appropriate, most likely to every 8–10 weeks. The present study, based on 44 evaluable participants from A5338, found no substantial associations between either CYP2B6 metabolizer status or NAT2 acetylator status and any clinically-relevant MPA pharmacokinetic parameter. Exploratory analyses involving 28 individual polymorphisms found no consistent associations with MPA pharmacokinetic parameters, and no association withstood correction for multiple testing.
The lack of substantial associations of CYP2B6 metabolizer status with MPA pharmacokinetic parameters is unexpected. Efavirenz is a potent inducer of CYP450 enzymes that metabolize MPA [12, 13], and efavirenz has been shown to interact with some hormonal contraceptives [13, 14]. When efavirenz-based ART was combined with a levonorgestrel-releasing contraceptive implant, CYP2B6 poor metabolizer genotype was associated with lower levonorgestrel Cmax and AUC [43]. Similarly, in ACTG study A5316, efavirenz reduced plasma concentrations of etonogestrel and ethinyl estradiol, given as a vaginal ring [13], and among 24 women in the efavirenz group in that study, CYP2B6 poor metabolizer genotype was associated with greater reductions [44]. In contrast, ACTG study A5093, which compared pharmacokinetics of DMPA and selected ART regimens among women living with HIV, found no difference in MPA AUC, Cmax or clearance between study groups [16, 17], while a study by Nanda et al. demonstrated higher plasma MPA AUC values among women receiving ART with zidovudine, lamivudine and efavirenz than those not on ART [45]. It is possible that any modest induction of MPA clearance by efavirenz could not be discerned in the context of the greater induction of CYP3A4 activity by rifampicin [46].
The lack of substantial associations with NAT2 acetylator status is also somewhat unexpected. There is markedly greater plasma isoniazid exposure with NAT2 slow acetylator genotypes [18–21]. Although MPA is not a substrate of NAT2, isoniazid is a mechanism-based inhibitor of CYP3A4, CYP2A6, CYP1A2 and CYP2C19 [47], and MPA undergoes metabolism by hepatic CYP isoforms, primarily CYP3A4 [10]. Some patients receiving tuberculosis therapy that includes isoniazid with rifampicin experience increased plasma efavirenz exposure, particularly in the presence of CYP2B6 and/or NAT2 loss-of-function polymorphisms [29–31], which may involve isoniazid inhibition of CYP2A6 [30–32, 34]. In ACTG study A5279, which studied rifapentine plus isoniazid for preventing tuberculosis in patients living with HIV [48], NAT2 slow acetylators had higher plasma concentrations not only of efavirenz, but also of rifapentine, its 25-desacetyl rifapentine metabolite, and nevirapine [49], suggesting inhibitory effects of isoniazid beyond CYP2A6. In the present study, we observed no such increases in plasma MPA exposure among NAT2 slow acetylators. This study was not designed to distinguish inductive effects of rifampicin from possible inhibitory effects of isoniazid on MPA clearance.
The present study had limitations. Our sample size of 44 evaluable women limited our ability to more thoroughly define genetic associations. However, our small size should be sufficient for detecting pharmacokinetic associations with frequent polymorphisms with large effect sizes, as is true for CYP2B6 with efavirenz, and for NAT2 with isoniazid. For example, in ACTG study A5316, an analysis limited to 24 women in the efavirenz group showed a highly significant association (P = 6.7×10−4) between CYP2B6 poor metabolizer genotype and reduced plasma concentrations of etonogestrel and ethinyl estradiol, given as a vaginal ring [44]. The exploratory analyses only included 28 selected polymorphisms. Other variants may be associated with pharmacokinetics of MPA, including infrequent polymorphisms that were not included in this analysis. We did not assess whether particular NRTIs affect genetic associations or MPA pharmacokinetic parameters, which seems unlikely. We did not genotype several polymorphisms that were recently associated (although not significant after correcting for multiple testing) with plasma etonogestrel concentrations [50], which is structurally similar to MPA.
In summary, study A5338 suggested that more frequent DMPA dosing may be appropriate for HIV and tuberculosis coinfected women receiving efavirenz-containing ART, and whose tuberculosis treatment included rifampicin and isoniazid. The present analyses suggest that knowledge of CYP2B6 metabolizer status or NAT2 acetylator status will not be useful in individualizing DMPA dosing frequency in this setting.
Supplementary Material
Acknowledgments.
We acknowledge and thank all the participants in ACTG A5338 and all ACTG personnel who made this study possible. We gratefully acknowledge the support of ACTG network and the scientific committees for their contribution during protocol development and study conduct. We especially want to acknowledge and thank Susan L. Rosenkranz, Yoninah Cramer and Kristine Coughlin for their contributions in study design, data monitoring, and interim analysis, and Xingye Wu for her work on the final analysis. We also wish to acknowledge the following study team members: Gary Maartens, Zephne Van Der Spuy, Akbar Shahkolahi, Jhoanna C. Roa, Laura Moran, Michelle Wildman, Ian S. Mugisa, Asuman Ssentamu, Vandana Kulkarni and Flavia Nakayima Miiro for their contributions. A sincere thank you to Jennifer Norman and her team at the UCT Speciality Laboratory for the analysis of MPA and progesterone samples. The research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers UM1 AI068634 (Statistical and Data Management Center of the AIDS Clinical Trials Group), UM1 AI068636, UM1 AI69471 and UM1 AI106701. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Disclaimer: The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the funding agencies. We acknowledge the following sites, site staff and investigators for performing all the work at their various sites:
Sr P Madlala and M Chikowore, Durban International Clinical Research Site (DICRS) CRS 11201, UM1 AI069432; Drs M Rassool and T Mwelase, University of the Witwatersrand Helen Joseph (WITS HJH) CRS 11101 UM1 AI069463 (CTU Grant Number) and AI068636 (ACTG Network Grant Number); Drs FA Mbata, E Ouma Kisumu and K Cain, Kisumu CRS 31460, UM1 AI068636. This work was supported in part by the Emory-CDC HIV/AIDS Clinical Trials Unit award number UM1 AI069418 from the NIH (NIAID) and the US Centers for Disease Control and Prevention (Division of HIV/AIDS Prevention); Dr U Chakalisa and MS Raesi Gaborone CRS 12701, UM1 AI069456 and Prof JG Hakim and Dr P Mukwekwerere, Parirenyatwa CRS 30313 UM1 AI069436. Additional support included AI069439, AI110527, AI077505, AI120790, and TR002243 (DWH). JAR was supported in part by grant funding by T32 GM066691. HM is funded by the Wellcome Trust (206379/Z/17/Z).
Footnotes
Declaration of Interests:
David W. Haas, Paxton Baker, Rosie Mngqibisa, Paxton Baker, Ayotunde Omoz-Oarhe, Susan E. Cohn, Jennifer A. Robinson, Helen McIlleron, Sharlaa Badal-Faesen, Paolo Denti, Susan L. Rosenkranz, Jose Francis, Sajeeda Mawlana, Wadzanai P. Samaneka: None
Contributor Information
David W. Haas, Department of Medicine, Vanderbilt University School of Medicine, Nashville, TN; Department of internal Medicine, Meharry Medical College, Nashville, TN
Rosie Mngqibisa, Enhancing Care Foundation, Durban International CRS, Durban, South Africa.
Jose Francis, Division of Clinical Pharmacology, Department of Medicine, University of Cape Town, South Africa.
Helen McIlleron, Division of Clinical Pharmacology, Department of Medicine, University of Cape Town, Mowbray, South Africa.
Jennifer A. Robinson, Department of Gynecology and Obstetrics, Johns Hopkins University School of Medicine, Baltimore, MD, USA
Michelle A. Kendall, Center for Biostatistics in AIDS Research, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Paxton Baker, Vanderbilt Technologies for Advanced Genomics, Vanderbilt University Medical Center, Nashville, TN, USA.
Sajeeda Mawlana, Enhancing Care Foundation, Durban International CRS, Durban, South Africa.
Sharlaa Badal-Faesen, Clinical HIV Research Unit, Department of Medicine, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa.
Francis Angira, Kenya Medical Research Institute, Center for Global Health (KEMRI/CGHR)/Emory-CDC CTU, Kisumu, Kenya.
Ayotunde Omoz-Oarhe, Botswana Harvard AIDS Institute Partnership, Gaborone, Botswana Clinical Trials Unit, Gaborone, Botswana.
Wadzanai P. Samaneka, Parirenyatwa CRS, Harare, Zimbabwe
Paolo Denti, Division of Clinical Pharmacology, Department of Medicine, University of Cape Town, South Africa.
Susan E. Cohn, Northwestern University Feinberg School of Medicine, Infectious Diseases Division, Chicago, IL, USA
REFERENCES
- 1.World Health Organization. Tuberculosis (TB). 2019. Available at: https://www.who.int/tb/areas-of-work/tb-hiv/en/. Accessed January 31, 2019.
- 2.UN Women. Facts and figures: HIV and AIDS. Secondary UN Women: Facts and figures: HIV and AIDS July 2018.; 2018. Available at: https://www.unwomen.org/en. Accessed 3/16/2020, 2020.
- 3.Sobhy S, Babiker Z, Zamora J, Khan KS, Kunst H. Maternal and perinatal mortality and morbidity associated with tuberculosis during pregnancy and the postpartum period: a systematic review and meta-analysis. BJOG. 2017;124(5):727–33. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. TB and Women Fact Sheet. Secondary TB and Women Fact Sheet 2015.; 2015. Available at: https://www.who.int/tb/publications/tb_women_factsheet.pdf. Accessed 3/16/2020, 2020.
- 5.Getahun H, Sculier D, Sismanidis C, Grzemska M, Raviglione M. Prevention, diagnosis, and treatment of tuberculosis in children and mothers: evidence for action for maternal, neonatal, and child health services. J Infect Dis. 2012;205 Suppl 2:S216–27. [DOI] [PubMed] [Google Scholar]
- 6.Darroch JE, Singh S. Trends in contraceptive need and use in developing countries in 2003, 2008, and 2012: an analysis of national surveys. Lancet. 2013;381(9879):1756–62. [DOI] [PubMed] [Google Scholar]
- 7.Pfizer. DepoProvera Contraceptive Injection (Depo-Provera CI) - Full Prescribing Information. 2010. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/020246s036lbl.pdf. Accessed 3/16/20, 2020.
- 8.Mishell DR Jr., Pharmacokinetics of depot medroxyprogesterone acetate contraception. J Reprod Med. 1996;41(5 Suppl):381–90. [PubMed] [Google Scholar]
- 9.Smit J, Botha J, McFadyen L, Beksinska M. Serum medroxyprogesterone acetate levels in new and repeat users of depot medroxyprogesterone acetate at the end of the dosing interval. Contraception. 2004;69(1):3–7. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi K, Mimura N, Fujii H, Minami H, Sasaki Y, Shimada N, et al. Role of human cytochrome P450 3A4 in metabolism of medroxyprogesterone acetate. Clin Cancer Res. 2000;6(8):3297–303. [PubMed] [Google Scholar]
- 11.World Health Organization. Update of Recommendations on First- and Second-Line Antiretroviral Regimens. World Health Organization; 2019. Available at: https://apps.who.int/iris/bitstream/handle/10665/325892/WHO-CDS-HIV-19.15-eng.pdf. Accessed December 11, 2020, 2020.
- 12.Williamson B, Dooley KE, Zhang Y, Back DJ, Owen A. Induction of influx and efflux transporters and cytochrome P450 3A4 in primary human hepatocytes by rifampin, rifabutin, and rifapentine. Antimicrob Agents Chemother. 2013;57(12):6366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarsi KK, Cramer YS, Rosenkranz SL, Aweeka F, Berzins B, Coombs RW, et al. Antiretroviral therapy and vaginally administered contraceptive hormones: a three-arm, pharmacokinetic study. Lancet HIV. 2019;6(9):e601–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel RC, Onono M, Gandhi M, Blat C, Hagey J, Shade SB, et al. Pregnancy rates in HIV-positive women using contraceptives and efavirenz-based or nevirapine-based antiretroviral therapy in Kenya: a retrospective cohort study. Lancet HIV. 2015;2(11):e474–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mngqibisa R, Kendall MA, Dooley K, Wu XS, Firnhaber C, McIlleron H, et al. Pharmacokinetics and Pharmacodynamics of Depot Medroxyprogesterone Acetate in African Women Receiving Treatment for Human Immunodeficiency Virus and Tuberculosis: Potential Concern for Standard Dosing Frequency. Clin Infect Dis. 2020;71(3):517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watts DH, Park JG, Cohn SE, Yu S, Hitti J, Stek A, et al. Safety and tolerability of depot medroxyprogesterone acetate among HIV-infected women on antiretroviral therapy: ACTG A5093. Contraception. 2008;77(2):84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn SE, Park JG, Watts DH, Stek A, Hitti J, Clax PA, et al. Depo-medroxyprogesterone in women on antiretroviral therapy: effective contraception and lack of clinically significant interactions. Clin Pharmacol Ther. 2007;81(2):222–7. [DOI] [PubMed] [Google Scholar]
- 18.Huang YS, Chern HD, Su WJ, Wu JC, Lai SL, Yang SY, et al. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology. 2002;35(4):883–9. [DOI] [PubMed] [Google Scholar]
- 19.Huang YS, Chern HD, Su WJ, Wu JC, Chang SC, Chiang CH, et al. Cytochrome P450 2E1 genotype and the susceptibility to antituberculosis drug-induced hepatitis. Hepatology. 2003;37(4):924–30. [DOI] [PubMed] [Google Scholar]
- 20.Roy PD, Majumder M, Roy B. Pharmacogenomics of anti-TB drugs-related hepatotoxicity. Pharmacogenomics. 2008;9(3):311–21. [DOI] [PubMed] [Google Scholar]
- 21.Ramachandran G, Swaminathan S. Role of pharmacogenomics in the treatment of tuberculosis: a review. Pharmacogenomics and Personalized Medicine. 2012;5:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desta Z, Gammal RS, Gong L, Whirl-Carrillo M, Gaur AH, Sukasem C, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-Containing Antiretroviral Therapy. Clin Pharmacol Ther. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18(18):2391–400. [PubMed] [Google Scholar]
- 24.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genom. 2005;15(1):1–5. [DOI] [PubMed] [Google Scholar]
- 25.Holzinger ER, Grady B, Ritchie MD, Ribaudo HJ, Acosta EP, Morse GD, et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genom. 2012;22(12):858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyen C, Hendra H, Vogel M, Hoffmann C, Knechten H, Brockmeyer NH, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemo. 2008;61(4):914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Sonnerborg A, Rane A, Josephson F, Lundgren S, Stahle L, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genom. 2006;16(3):191–8. [DOI] [PubMed] [Google Scholar]
- 28.Kwara A, Tashima KT, Dumond JB, Poethke P, Kurpewski J, Kashuba AD, et al. Modest but variable effect of rifampin on steady-state plasma pharmacokinetics of efavirenz in healthy African-American and Caucasian volunteers. Antimicrob Agents Chemother.55(7):3527–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwara A, Lartey M, Sagoe KW, Court MH. Paradoxically elevated efavirenz concentrations in HIV/tuberculosis-coinfected patients with CYP2B6 516TT genotype on rifampin-containing antituberculous therapy. AIDS. 2011;25(3):388–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dooley KE, Denti P, Martinson N, Cohn S, Mashabela F, Hoffmann J, et al. Pharmacokinetics of Efavirenz and Treatment of HIV-1 Among Pregnant Women With and Without Tuberculosis Coinfection. J Infect Dis. 2015;211(2):197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luetkemeyer AF, Rosenkranz SL, Lu D, Grinsztejn B, Sanchez J, Ssemmanda M, et al. Combined effect of CYP2B6 and NAT2 genotype on plasma efavirenz exposure during rifampin-based antituberculosis therapy in the STRIDE study. Clin Infect Dis. 2015;60(12):1860–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genom. 2009;19(4):300–9. [DOI] [PubMed] [Google Scholar]
- 33.Court MH, Almutairi FE, Greenblatt DJ, Hazarika S, Sheng H, Klein K, et al. Isoniazid mediates the CYP2B6*6 genotype-dependent interaction between efavirenz and antituberculosis drug therapy through mechanism-based inactivation of CYP2A6. Antimicrob Agents Chemother. 2014;58(7):4145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abel L, Fellay J, Haas DW, Schurr E, Srikrishna G, Urbanowski M, et al. Genetics of human susceptibility to active and latent tuberculosis: present knowledge and future perspectives. Lancet Infect Dis. 2018;18(3):e64–e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NHGRI-EBI. GWAS Catalog - The NHGRI-EBI Catalog of published genome-wide association studies. 2020. Available at: https://www.ebi.ac.uk/gwas/. Accessed August 20, 2020, 2020.
- 36.PharmGKB - The Pharmacogenomics Knowledgebase. 2021. Available at: https://www.pharmgkb.org/. 2021.
- 37.Beal S, Sheiner LBL, Boeckmann A, Bauer RRJ. NONMEM user’s guides (1989–2009). 2009. [Google Scholar]
- 38.Francis J, Mngqibisa R, McIlleron H, Kendall MA, Wu X, Dooley KE, et al. A semi-mechanistic pharmacokinetic model for depot medroxyprogesterone acetate and drug-drug interactions with antiretroviral and antituberculosis treatment. Clin Pharmacol Ther. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savic RM, Jonker DM, Kerbusch T, Karlsson MO. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J Pharmacokinet Pharmacodyn. 2007;34(5):711–26. [DOI] [PubMed] [Google Scholar]
- 40.Anderson BJ, Holford NH. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu Rev Pharmacol Toxicol. 2008;48:303–32. [DOI] [PubMed] [Google Scholar]
- 41.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boukouvala S. Database of arylamine N-acetyltransferases (NATs). Democritus University of Thrace; 2016. Available at: http://nat.mbg.duth.gr/. Accessed February 4, 2020, 2020.
- 43.Neary M, Lamorde M, Olagunju A, Darin KM, Merry C, Byakika-Kibwika P, et al. The Effect of Gene Variants on Levonorgestrel Pharmacokinetics When Combined With Antiretroviral Therapy Containing Efavirenz or Nevirapine. Clin Pharmacol Ther. 2017;102(3):529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas DW, Cramer YS, Godfrey C, Rosenkranz SL, Aweeka F, Berzins B, et al. Pharmacogenetic interactions between antiretroviral drugs and vaginally administered hormonal contraceptives. Pharmacogenet Genomics. 2020;30(3):45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nanda K, Amaral E, Hays M, Viscola MA, Mehta N, Bahamondes L. Pharmacokinetic interactions between depot medroxyprogesterone acetate and combination antiretroviral therapy. Fertil Steril. 2008;90(4):965–71. [DOI] [PubMed] [Google Scholar]
- 46.Hariparsad N, Nallani SC, Sane RS, Buckley DJ, Buckley AR, Desai PB. Induction of CYP3A4 by efavirenz in primary human hepatocytes: comparison with rifampin and phenobarbital. J Clin Pharmacol. 2004;44(11):1273–81. [DOI] [PubMed] [Google Scholar]
- 47.Wen X, Wang JS, Neuvonen PJ, Backman JT. Isoniazid is a mechanism-based inhibitor of cytochrome P450 1A2, 2A6, 2C19 and 3A4 isoforms in human liver microsomes. Eur J Clin Pharmacol. 2002;57(11):799–804. [DOI] [PubMed] [Google Scholar]
- 48.Swindells S, Ramchandani R, Gupta A, Benson CA, Leon-Cruz J, Mwelase N, et al. One Month of Rifapentine plus Isoniazid to Prevent HIV-Related Tuberculosis. N Engl J Med. 2019;380(11):1001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas DW, Podany AT, Bao Y, Swindells S, Chaisson RE, Mwelase N, et al. Pharmacogenetic interactions of rifapentine plus isoniazid with efavirenz or nevirapine. Pharmacogenet Genomics. 2021;31(1):17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazorwitz A, Aquilante CL, Oreschak K, Sheeder J, Guiahi M, Teal S. Influence of Genetic Variants on Steady-State Etonogestrel Concentrations Among Contraceptive Implant Users. Obstet Gynecol. 2019;133(4):783–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


