Abstract
Environmental, maternal and early life microbial/immune networks program human developmental trajectories and health outcomes and strongly modify allergic disease risk. The effects of environmental microbiota are illustrated by the “farm effect” (the protection against asthma and allergy conferred by growing up on a traditional farm) and other natural experiments in populations exposed to microbe-rich environments. The role of gut microbiome maturation in the asthma/allergy trajectory is demonstrated by the most recent farm studies, which identified microbial metabolites specifically associated with asthma protection, and studies in other cohorts, which defined dynamic microbial community profiles associated with allergic disease phenotypes. Current and future studies in germ-free mice associated with gut microbiota from human disease states are providing novel mechanistic insights into the role of microbiota in shaping immune function and allergic disease susceptibility
Keywords: Allergic diseases, Asthma, Microbiota, Microbial metabolites, Microbiome maturation
Introduction
The prevalence of allergic diseases such as asthma, food allergy, rhinitis and eczema has increased dramatically over the past decades, especially in nations where westernized environmental exposures and lifestyles predominate [1]. Central to the effects of these exposures are maternal and early life microbial/immune networks that program developmental trajectories and health outcomes. A recent working model [2] suggests that environmental exposures during pregnancy shape the mother’s microbiome and immune function, which in turn affect immune and microbiome development in the fetus. These integrated influences train the neonatal innate immune system, calibrating its ability to respond to microbes that are vertically transmitted by the mother and colonize neonatal body habitats. Depending on their composition and functional properties, these “pioneer microbes” shape the composition and accumulation rate of exogenous microbes during the child’s first year of life, thereby regulating trajectories of microbiome and immune maturation, and ultimately allergic disease risk [3*]. Here we discuss the cross-talk between microbiota and allergic disease pathogenesis, focusing on data from human models.
Environmental microbiota and allergic diseases
Few epidemiologic findings demonstrate the effects of environmental microbial exposures on allergic disease risk as eloquently as the “farm effect” [4]. Children raised on traditional farms are strongly protected from asthma and allergies, and this protection is largely explained by the child’s contact with farm animals and their microbiota in early life [5,6]. The farm effect has been observed world-wide [4] but its multiple facets are especially clear in Amish and Hutterite children who share genetic ancestry and lifestyles but follow distinct farming practices (traditional among the Amish, industrialized among the Hutterites) and have sharply different asthma and allergy prevalence [4–5 times lower in the Amish]. This is accompanied by different levels of house dust endotoxin (7-fold higher among the Amish) and proportions, phenotypes and functions of innate and adaptive immune cells [7–9]. Importantly, inhalation of Amish, but not Hutterite, house dust was sufficient to protect mice against cardinal allergic asthma phenotypes through mechanisms that required innate immunity [8].
The asthma- and allergy-protective role of microbe-rich environments has been further dissected by modeling differences in house dust microbiota composition between farm and non-farm homes in Finnish birth cohorts. In children who grow up in non-farm homes, asthma risk decreases as the similarity of their home bacterial microbiota composition to that of farm homes increases. Protection was independent of microbial richness and total bacterial load and was associated with low abundance of Streptococcaceae and reduced production of proinflammatory cytokines in response to bacterial cell wall components. These findings were replicated in rural German children whose asthma risk was lower if they lived in non-farm homes with indoor microbiota more similar to Finnish farm homes. Thus, indoor dust microbiota composition appears to be a robust predictor of asthma risk [10].
Other “natural experiments” highlight the relation of environmental microbiota with microbiome maturation and composition in the host. For instance, Finnish and Russian Karelia represent socio-economically distinct but geo-climatically similar regions, and allergies are much more common in Finnish than Russian children. Microbial load in house dust and drinking water is associated with allergy protection in Russia [11,12], while genotype differences between Finns and Russians could not explain the allergy prevalence gap. In contrast, blood cell gene expression profiles and skin and nasal microbiota networks were richer and more diverse, while innate immunity pathways were suppressed, in Russian subjects. High abundance of Acinetobacter in these subjects correlated with suppression of innate immune responses [13].
Comparable observations were made in two populations of Mexican ancestry who reside only 70 miles apart on the two sides of the US/Mexico border but differ significantly in both their rates of childhood asthma (4-fold lower in Mexico) and their house dust and bacterial load (strikingly higher in Mexico) [14]. Moreover, a study of scholchildren from a Croatian county where drinking water is provided through public mains supply or individual wells found that cumulative bacterial load in drinking water was higher, and lifetime prevalence of allergic diseases was significantly lower, among children with individual supply. In a quantitative analysis, the risk of allergic diseases decreased significantly with increasing bacterial load in drinking water in the first year of life [15]. Another study tracked the transfer of environmental microbes to the human skin and respiratory tract following outdoor exposure to urban green space (air, soil, and leaves) from cities (Adelaide, Australia; Bournemouth, United Kingdom, and New Delhi, India) in different continents. Microbial richness and phylogenetic diversity in skin and nasal samples increased after urban green space exposure. Interestingly, the post-exposure microbial composition of skin samples became more similar to soil microbiota, while nasal samples became more similar to air samples [16].
The interactions between environmental microbes and allergic disease are not limited to children. In the first large-scale house dust microbiome study performed in an adult US farming population [17], the association between indoor bacterial community profiles and allergy outcomes also held in this age group, albeit with more subtle and variegated effects. Bacterial communities in house dust were associated with asthma, atopy, and hay fever, and were less diverse in homes of atopic individuals. Specific bacterial taxa were differentially abundant by current asthma or atopy outcomes: several taxa from Cyanobacteria, Bacteroidetes, and Fusobacteria were more abundant with asthma, atopy, or hay fever. In contrast, several taxa from Firmicutes were more abundant in homes of individuals with adequately controlled asthma, individuals without atopy, or individuals without hay fever. Thus, environmental microbial exposures are associated with risk of allergic diseases across a variety of environments and age groups.
Early-life microbiome and asthma development
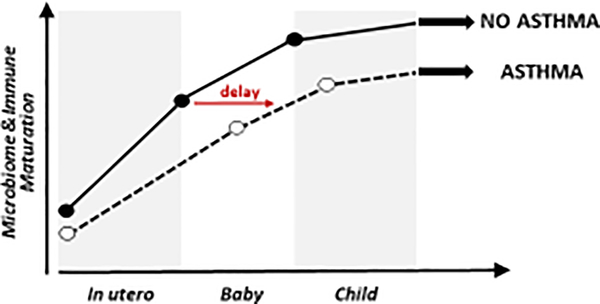
Immune and microbial development in early life are intimately linked, particularly in the gut, which houses the largest microbial diversity and burden [18]. The nascent gut microbiome integrates dietary [18] and antibiotic [19] encounters that decisively affect microbiome membership, productivity and development, and gut microbial products influence immune function both locally and at remote mucosal sites [2]; moreover, these early events are likely influenced by a combination of maternal, genetic [20], and epigenetic factors (DeVries at al., under review). Consistent with this hypothesis, data from several independent birth cohorts demonstrate that early life gut microbiota perturbations (often described as dysbiosis) are associated with increased risk of atopy and/or asthma later in childhood [21–23] (Fig. 1).
Fig. 1.
In utero and early life profiles of microbiome and immune maturation in children who do or do not develop asthma. Distinct developmental trajectories are shown as a dashed (asthma) or continuous (no asthma) line. The red horizontal arrow highlights the delay in immune and microbiome maturation that emerges at birth, persists through early life, and shifts the developmental trajectory to the right in children who later develop asthma.
These findings have been further refined by recent studies. Analyses in the Copenhagen Prospective Studies on Asthma in Childhood2010 have shown that 1-year-old children with an immature gut microbial composition characterized by lower relative abundance of Lachnospiraceae (including Roseburia) and Ruminococcaceae (including Ruminococcus and Faecalibacterium, a butyrate producer) have increased asthma risk at age 5 years. This association was only apparent among children born to asthmatic mothers, suggesting that inadequate microbial stimulation during the first year of life may license their inherited asthma risk, and conversely, adequate maturation of the gut microbiome in this time window may protect these predisposed children [24].
The same group then asked whether changes in gut microbiota mediate the association between cesarean section delivery and the risk of childhood asthma by age 6 years. Compared to vaginal delivery, delivery by cesarean section was indeed accompanied by marked changes in gut microbiota composition at 1 week and 1 month of age, but by 1 year of age only minor differences persisted. Interestingly, children born by cesarean section had increased asthma risk only if their gut microbiota composition at age 1 year still retained a cesarean section microbial signature, suggesting that appropriate maturation of the gut microbiota could mitigate the increased asthma risk associated with gut microbial changes linked to cesarean section delivery. It is also noteworthy that none of the taxa driving the cesarean section microbial signature at age 1 year were themselves associated with later asthma when tested individually. This finding suggests that microbial communities rather than single taxa drive the observed effects [25].
Finally, the farm studies again decisively illuminated the link between microbiome development and asthma protection by showing that gut microbiome maturation during the first year of life significantly contributes to the protective effect of farming on childhood asthma. Modeling gut microbiome maturation between 2 and 12 months of age in the Protection against Allergy: Study in Rural Environments (PASTURE) birth cohort showed that the estimated microbiome age at 12 months not only was strongly associated with previous farm exposure and reduced risk of asthma at school age, but also mediated a substantial proportion of the asthma protection associated with the farm effect [26].
Shifting the focus from the microbes to their metabolites
Metabolites produced by gut microbiota are both key determinants of host-microbe mutualism and critical mediators of a host-microbe crosstalk that influences inflammation in peripheral tissues, such as the lung - a concept referred to as the gut-lung axis (reviewed in [27]). For instance, elegant mouse studies demonstrated that dietary fermentable fiber content changed the composition of gut and lung microbiota and dampened lung allergic inflammation. Gut microbiota metabolized the fiber, thereby increasing the concentration of circulating short-chain fatty acids (SCFAs) that protected against allergic inflammation in the lung. Treatment of mice with the SCFA propionate altered bone marrow hematopoiesis and seeded the lungs with dendritic cells impaired in their ability to promote T helper type 2 cell responses [28].
Soon thereafter, studies in humans identified pro-inflammatory metabolites that were associated with compositionally distinct neonatal gut microbiota states and increased risk of childhood atopy and asthma. Ex vivo culture of adult human peripheral T cells with sterile fecal water from subjects carrying these microbiota expanded interleukin-4-producing CD4+ cells and reduced the relative abundance of Foxp3+CD25+CD4+ cells. 12,13 DiHOME, a metabolite that discriminated high from lower risk groups, recapitulated the immune effects of fecal water, pointing to a central role of neonatal gut microbial metabolites in driving atopy-associated immune dysfunction [22]. Moreover, introduction into the mouse gut microbiome of bacterial epoxide hydrolase genes responsible for 12,13 DiHOME production was sufficient to reduce airway regulatory T cell populations and exacerbate airway allergic inflammation [29], confirming the asthma-promoting role of this microbial lipid and linking early life gut microbiome metabolic productivity to airway immune alterations associated with allergy and asthma development.
At the opposite, protective end of the metabolite spectrum, the PASTURE study found inverse associations between asthma, levels of the fecal SCFA butyrate, the bacterial taxa that predict butyrate production (Roseburia and Coprococcus), and the relative abundance of the gene encoding butyryl-coenzyme A (CoA):acetate-CoA-transferase, a major enzyme in butyrate metabolism. However, changes in overall bacterial composition over time far outweighed the role of single taxa, none of which was protective in itself. These findings suggest that asthma-protective microbial communities act through the gut-lung metabolic axis [26]. In other studies, higher fecal levels of iso-butyric, iso-valeric and valeric acid at age 3 years (indicative of more rapid maturation of gut microbiota) were found in Swedish farm children compared to rural controls, and high levels of valeric acid were associated with low rate of eczema at 8 years of age [30]. Conversely, the microbiome of infants who developed allergic sensitization during childhood lacked genes encoding key enzymes for carbohydrate breakdown and butyrate production [31]. Studies in an Estonian/Finnish birth-cohort followed from 3 to 36 months of age also unveiled associations between profound changes in regulatory T cell frequency and activation and colonization with butyrate-producing bacteria, which occurred earlier in Estonian children at low risk of allergic disease [32].
The last few years have also seen a rise in untargeted mass spectrometry-based metabolomic analyses that seek to correlate microbial metabolic profiles with allergic disease outcomes. Studies of serum samples from children with food allergy alone, asthma alone, or both food allergy and asthma unveiled disease-specific metabolomic signatures in patients with food allergy compared with both control subjects and asthmatic patients. Food allergy was uniquely associated with a marked decrease in sphingolipids and other lipid metabolites, while a comparison of food allergy and asthmatic patients revealed differences in aromatic amino acid and secondary bile acid metabolism. Among children with food allergy, a history of severe systemic reactions and multiple sensitizations were associated with changes in tryptophan metabolites, eicosanoids, plasmalogens, and fatty acids [33]. Another analysis, focused on the intestinal metabolome, identified inverse associations between asthma and polyunsaturated fatty acids and other lipids [34]. Finally, a study of fecal samples from a well-controlled cohort of twins concordant or discordant for food allergy revealed a bacterial signature of 64 operational taxonomic units, especially Clostridia, that distinguished healthy from allergic twins. Distinct metabolic pathways were enriched in each group, particularly diacylglycerol in healthy twins. Interestingly, twin pairs differed significantly in their fecal microbiomes and metabolomes through adulthood, suggesting that the gut microbiota may play a protective role in patients with food allergies beyond the infant stage [35].
The critical role of intestinal bacteria in regulating allergic responses to dietary antigens is also supported by the recent finding that colonization with dysbiotic fecal microbiota from infants with food allergy fails to protect germ-free mice in food allergy models [36*,37*]. These studies identified selected microbes from healthy subjects that protect against allergic responses to food, for instance Clostridiales such as Anaerostipes caccae [36*] or Subdoligranulum variabile [37], and bacterial consortia such as immunomodulatory Bacteroidales [37*]. Commensals were found to activate a MyD88/ROR-γt pathway in nascent Treg cells to protect against food allergy, while dysbiosis impairs this regulatory response to promote disease [37*]. As importantly, these results illustrate how studies in germ-free mice can help decipher the mechanisms underlying the interplay between human microbiota and immune responses in disease states.
What lies ahead
Our discussion of the relations between microbes and allergies is by no means exhaustive. Several emerging lines of investigation promise to further our mechanistic understanding of these relations: epigenetic remodeling as an effector of microbial exposures [38], the role of genetic background in constraining the spectrum of microbiome-triggered immune responses [39], the largely unexplored role of fungi and their products [40], and the growing list of immune events triggered by microbial metabolites through receptors considered “orphans” not too long ago [41]. The repertoire of allergy-relevant metabolites will also likely expand. On the analytical side, uniform data and reporting formats will be required to facilitate replication and meta-analysis efforts, and sample sizes will need to increase by orders of magnitude [42]. All in all, though, the evidence currently available already shows that the (microbial) company we keep is critical to determine trajectories of health and immune development. Truly, “we are (fortunately) not alone” [43].
Acknowledgements
DV was supported by the National Institutes of Health [P01AI148104, R21AI144722 and R25HL126140].
Footnotes
Conflicts of interest relevant to this work:
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.von Mutius E, Smits HH: Primary prevention of asthma: from risk and protective factors to targeted strategies for prevention. Lancet 2020, 396:854–866. [DOI] [PubMed] [Google Scholar]
- 2.Lynch SV, Vercelli D: Microbiota, epigenetics and trained Immunity: Convergent regulators of the asthma trajectory from pregnancy to childhood. Am J Respir Crit Care Med 2021, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilbert JA, Lynch SV: Community ecology as a framework for human microbiome research. Nat Med 2019, 25:884–889. * A powerful, elegant framework to understand human microbiome research.
- 4.von Mutius E, Vercelli D: Farm living: effects on childhood asthma and allergy. Nat. Rev. Immunol. 2010, 10:861–868. [DOI] [PubMed] [Google Scholar]
- 5.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, Heederik D, Piarroux R, von Mutius E: Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 2011, 364:701–709. [DOI] [PubMed] [Google Scholar]
- 6.Pivniouk V, Gimenes Junior JA, Honeker LK, Vercelli D: The role of innate immunity in asthma development and protection: Lessons from the environment. Clin Exp Allergy 2020, 50:282–290. [DOI] [PubMed] [Google Scholar]
- 7.Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, Ledford JG, Marques Dos Santos M, Anderson RL, Metwali N, et al. : Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med 2016, 375:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gozdz J, Ober C, Vercelli D: Innate immunity and asthma risk. N. Engl. J. Med. 2016, 375:1897–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrusch CL, Stein MM, Gozdz J, Holbreich M, von Mutius E, Vercelli D, Ober C, Sperling AI: T cell phenotypes are associated with serum IgE levels in Amish and Hutterite children. J. Allergy Clin. Immunol. 2019, 144:1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirjavainen PV, Karvonen AM, Adams RI, Taubel M, Roponen M, Tuoresmaki P, Loss G, Jayaprakash B, Depner M, Ege MJ, et al. : Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat Med 2019, 25:1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, Karisola P, Auvinen P, Paulin L, Makela MJ, et al. : Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci U S A 2012, 109:8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haahtela T, Laatikainen T, Alenius H, Auvinen P, Fyhrquist N, Hanski I, von Hertzen L, Jousilahti P, Kosunen TU, Markelova O, et al. : Hunt for the origin of allergy - comparing the Finnish and Russian Karelia. Clin Exp Allergy 2015, 45:891–901. [DOI] [PubMed] [Google Scholar]
- 13.Ruokolainen L, Fyhrquist N, Laatikainen T, Auvinen P, Fortino V, Scala G, Jousilahti P, Karisola P, Vendelin J, Karkman A, et al. : Immune-microbiota interaction in Finnish and Russian Karelia young people with high and low allergy prevalence. Clin Exp Allergy 2020, 50:1148–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr TF, Beamer PI, Rothers J, Stern DA, Gerald LB, Rosales CB, Van Horne YO, Pivniouk ON, Vercelli D, Halonen M, et al. : Prevalence of asthma in school children on the Arizona-Sonora border. J Allergy Clin Immunol Pract 2017, 5:114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turkalj M, Drkulec V, Haider S, Plavec D, Banic I, Malev O, Erceg D, Woodcock A, Nogalo B, Custovic A: Association of bacterial load in drinking water and allergic diseases in childhood. Clin Exp Allergy 2020, 50:733–740. [DOI] [PubMed] [Google Scholar]
- 16.Selway CA, Mills JG, Weinstein P, Skelly C, Yadav S, Lowe A, Breed MF, Weyrich LS: Transfer of environmental microbes to the skin and respiratory tract of humans after urban green space exposure. Environ Int 2020, 145:106084. [DOI] [PubMed] [Google Scholar]
- 17.Lee MK, Wyss AB, Carnes MU, Richards M, Parks CG, Beane Freeman LE, Thorne PS, Umbach DM, Azcarate-Peril MA, Peddada SD, et al. : House dust microbiota in relation to adult asthma and atopy in a US farming population. J Allergy Clin Immunol 2021, 147:910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. : Human gut microbiome viewed across age and geography. Nature 2012, 486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patrick DM, Sbihi H, Dai DLY, Al Mamun A, Rasali D, Rose C, Marra F, Boutin RCT, Petersen C, Stiemsma LT, et al. : Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. Lancet Respir Med 2020, 8:1094–1105. [DOI] [PubMed] [Google Scholar]
- 20.Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, Igartua C, Morin A, Washington C 3rd, Nicolae D, et al. : A decade of research on the 17q12–21 asthma locus: Piecing together the puzzle. J Allergy Clin Immunol 2018, 142:749–764 e743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durack J, Kimes NE, Lin DL, Rauch M, McKean M, McCauley K, Panzer AR, Mar JS, Cabana MD, Lynch SV: Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat Commun 2018, 9:707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, et al. : Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016, 22:1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. : Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015, 7:307ra152. [DOI] [PubMed] [Google Scholar]
- 24.Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, Schoos AM, Kunoe A, Fink NR, Chawes BL, et al. : Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun 2018, 9:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokholm J, Thorsen J, Blaser MJ, Rasmussen MA, Hjelmso M, Shah S, Christensen ED, Chawes BL, Bonnelykke K, Brix S, et al. : Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Sci Transl Med 2020, 12. [DOI] [PubMed] [Google Scholar]
- 26. Depner M, Taft DH, Kirjavainen PV, Kalanetra KM, Karvonen AM, Peschel S, Schmausser-Hechfellner E, Roduit C, Frei R, Lauener R, et al. : Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat Med 2020, 26:1766–1775. ** The brilliant culmination of a research trajectory that began over 20 years ago with the epidemiology of the farm effect and has now linked asthma protection to the maturation and metabolic productivity of farm microbes.
- 27.Dang AT, Marsland BJ: Microbes, metabolites, and the gut-lung axis. Mucosal Immunol 2019, 12:843–850. [DOI] [PubMed] [Google Scholar]
- 28.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. : Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014, 20:159–166. [DOI] [PubMed] [Google Scholar]
- 29. Levan SR, Stamnes KA, Lin DL, Panzer AR, Fukui E, McCauley K, Fujimura KE, McKean M, Ownby DR, Zoratti EM, et al. : Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol 2019, 4:1851–1861. * A creative mechanistic study that definitively demonstrates the allergy-promoting role of a specific microbial lipid metabolite.
- 30.Gio-Batta M, Sjoberg F, Jonsson K, Barman M, Lundell AC, Adlerberth I, Hesselmar B, Sandberg AS, Wold AE: Fecal short chain fatty acids in children living on farms and a link between valeric acid and protection from eczema. Sci Rep 2020, 10:22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cait A, Cardenas E, Dimitriu PA, Amenyogbe N, Dai D, Cait J, Sbihi H, Stiemsma L, Subbarao P, Mandhane PJ, et al. : Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J Allergy Clin Immunol 2019, 144:1638–1647 e1633. [DOI] [PubMed] [Google Scholar]
- 32.Ruohtula T, de Goffau MC, Nieminen JK, Honkanen J, Siljander H, Hamalainen AM, Peet A, Tillmann V, Ilonen J, Niemela O, et al. : Maturation of Gut Microbiota and Circulating Regulatory T Cells and Development of IgE Sensitization in Early Life. Front Immunol 2019, 10:2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crestani E, Harb H, Charbonnier LM, Leirer J, Motsinger-Reif A, Rachid R, Phipatanakul W, Kaddurah-Daouk R, Chatila TA: Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J Allergy Clin Immunol 2020, 145:897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee-Sarwar KA, Kelly RS, Lasky-Su J, Zeiger RS, O'Connor GT, Sandel MT, Bacharier LB, Beigelman A, Laranjo N, Gold DR, et al. : Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol 2019, 144:442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao R, Hesser LA, He Z, Zhou X, Nadeau KC, Nagler CR: Fecal microbiome and metabolome differ in healthy and food-allergic twins. J Clin Invest 2021, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, Campbell E, Aitoro R, Nocerino R, Paparo L, et al. : Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med 2019, 25:448–453. *
- 37. Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, Wang L, Li N, Crestani E, Spielman S, et al. : Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat Med 2019, 25:1164–1174. * References 36 and 37 illustrate how reconstitution of germ-free mice with relevant human microbiota can elucidate the microbial and immune mechanisms underlying disease pathogenesis.
- 38.Morin A, McKennan CG, Pedersen CT, Stokholm J, Chawes BL, Malby Schoos AM, Naughton KA, Thorsen J, Mortensen MS, Vercelli D, et al. : Epigenetic landscape links upper airway microbiota in infancy with allergic rhinitis at 6 years of age. J Allergy Clin Immunol 2020, 146:1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. : Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555:210–215. [DOI] [PubMed] [Google Scholar]
- 40.Sharma A, Laxman B, Naureckas ET, Hogarth DK, Sperling AI, Solway J, Ober C, Gilbert JA, White SR: Associations between fungal and bacterial microbiota of airways and asthma endotypes. J Allergy Clin Immunol 2019, 144:1214–1227 e1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster SR, Hauser AS, Vedel L, Strachan RT, Huang XP, Gavin AC, Shah SD, Nayak AP, Haugaard-Kedstrom LM, Penn RB, et al. : Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors. Cell 2019, 179:895–908 e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y: Host variables confound gut microbiota studies of human disease. Nature 2020, 587:448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee YK, Mazmanian SK: Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 2010, 330:1768–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]