Abstract
Conversion of the germ line micronuclear genome into the genome of a somatic macronucleus in Tetrahymena thermophila requires several DNA rearrangement processes. These include (i) excision and subsequent elimination of several thousand internal eliminated sequences (IESs) scattered throughout the micronuclear genome and (ii) breakage of the micronuclear chromosomes into hundreds of DNA fragments, followed by de novo telomere addition to their ends. Chromosome breakage sequences (Cbs) that determine the sites of breakage and short regions of DNA adjacent to them are also eliminated. Both processes occur concomitantly in the developing macronucleus. Two stage-specific protein factors involved in germ line DNA elimination have been described previously. Pdd1p and Pdd2p (for programmed DNA degradation) physically associate with internal eliminated sequences in transient electron-dense structures in the developing macronucleus. Here, we report the purification, sequence analysis, and characterization of Pdd3p, a novel developmentally regulated, chromodomain-containing polypeptide. Pdd3p colocalizes with Pdd1p in the peripheral regions of DNA elimination structures, but is also found more internally. DNA cross-linked and immunoprecipitated with Pdd1p- or Pdd3p-specific antibodies is enriched in IESs, but not Cbs, suggesting that different protein factors are involved in elimination of these two groups of sequences.
Developmentally programmed excision and subsequent degradation of specific germ line DNA sequences have been reported to occur in a variety of species, including humans (4). In some organisms, programmed DNA rearrangements are essential steps in somatic development and differentiation of certain cell types. Examples include rearrangements in immunoglobulin and T-cell receptor genes (reviewed in reference 11), surface antigen variation in trypanosomes (27), and switching of mating type in yeast (1).
Partial elimination of the germ line genome is an important process in somatic nuclear differentiation in ciliated protozoa (6, 25). Like most ciliates, Tetrahymena thermophila contains two types of nuclei: a diploid, transcriptionally inert germ line micronucleus, responsible for storage and transmission of the genetic information, and a polyploid, transcriptionally active somatic macronucleus whose function is to express the genetic information (reviewed in reference 13). The sequence complexity of macronuclear DNA is 15 to 20% lower than that of micronuclear DNA, owing to the loss of micronucleus-specific germ line DNA sequences that occurs in the developing macronuclei during conjugation (31). Conjugation is a sexual pathway during which two cells mate and exchange gametic micronuclei, which then fuse to form a zygotic nucleus that divides twice (22). Products of this division differentiate into two micronuclei and two developing macronuclei, often referred to as anlagen (24). Two major DNA rearrangement events occur in developing anlagen, resulting in loss of germ line sequences: (i) excision and elimination of internal eliminated sequences (IESs) (31) and (ii) processing of the micronuclear chromosomes (n = 5) into 200 to 300 macronuclear chromosomes (33). There are approximately 6,000 IESs dispersed throughout the micronuclear genome, consisting of both single-copy and repetitive sequences ranging in size from hundreds to several thousands of base pairs (reviewed in reference 6). Although IES excision occurs with high precision, no consensus excision signals have been identified at or near IES boundaries (35). In contrast, breakage, the first step of chromosome processing, requires chromosome breakage sequences (Cbs), a highly conserved motif of 15 bp (34). Telomeres are added to the DNA fragments created by chromosome breakage (36), while Cbs and about 40 bp of DNA adjacent to them are eliminated (33). Elimination of both IESs and Cbs occurs during a very short period of time, and it remains unclear whether the same trans-acting factors are involved. At approximately the same time, the old parental macronucleus is degraded through mechanisms resembling apoptotic DNA degradation (8).
To date, two polypeptides have been associated with DNA elimination events in Tetrahymena: Pdd1p and Pdd2p (for programmed DNA degradation). Both proteins are found in the degrading old macronucleus as well as in the new macronucleus, where they colocalize in specialized electron-dense structures that also contain IESs (20, 29). Both Pddps have been shown to associate with IESs in chromatin immunoprecipitation assays (29). Pdd1p possesses three chromodomains, suggestive of its association with the germ line DNA packaged in a specialized heterochromatin form (20). No conserved domains or other homology was detected between Pdd2p and other known proteins (29). Interestingly, expression of Pdd1p and Pdd2p begins early in development, well ahead of anlagen formation and DNA elimination, raising the possibility that these polypeptides have other, yet unidentified, function(s). Consistent with this possibility, inhibition of pre-anlagen expression of Pdd1p and Pdd2p by disrupting their genes in the somatic macronucleus leads to impairment of the DNA rearrangements in anlagen, resulting in lethality (7, 23).
In this study, we describe the properties of a new developmentally regulated protein, Pdd3p. The derived sequence of the gene encoding this polypeptide predicts that it is a chromodomain-containing protein. Immunoblotting experiments demonstrated that expression of Pdd3p differs from that of Pdd1p and Pdd2p inasmuch as it is restricted to the anlagen stage of Tetrahymena development, peaking at the time when DNA rearrangements are known to occur (3). Immunofluorescence analysis showed that Pdd3p initially colocalizes with Pdd1p in the old macronucleus and in anlagen. At later stages, in addition to colocalization with Pdd1p at the periphery of the specialized DNA elimination structures, Pdd3p is detected in the central area of these structures. These data suggest that Pdd3p has a unique function in the DNA degradation process. Analysis of anlagen DNA coimmunoprecipitated with either Pdd3p- or Pdd1p-specific antibodies demonstrated that it was enriched in IESs but not Cbs, suggesting that different trans-factors are involved in these major DNA elimination processes.
MATERIALS AND METHODS
Strains and cell culture.
T. thermophila CU428 [mpr1-1/mpr1-1 (MPR1, mp-s, VII)] and CU427 [chx-1/chx-1 (CHX1, cy-s, VI)] were kindly provided by P. J. Bruns (Cornell University, Ithaca, N.Y.). Cells were grown in 1× SPP medium containing 1% Proteose Peptone (12). For conjugation, cells of different mating types were washed, starved (16 to 24 h, 30°C), and mated in 10 mM Tris-HCl (pH 7.5), as described in reference 2. Pairing efficiency was estimated by counting pairs at 2 h postmixing and was at least 85%. Labeling of conjugating cells with [3H]lysine at 1 μCi/ml of cell culture was performed between 9 and 10 h postmixing.
Preparation of nuclei.
Macronuclei, micronuclei, and anlagen were isolated from Tetrahymena at 10 h postmixing as described in reference 12, except that the nucleus isolation buffer contained 1 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10 mM sodium butyrate, but not spermidine. Purification of nuclei by sedimentation at unit gravity was performed according to the method of Allis and Dennison (2).
Purification of p32 and peptide sequencing.
Purified anlagen were resuspended in 2 M sodium perchlorate at a concentration of 2 × 106 nuclei/ml, sonicated (Sonifier Cell Disruptor 200; Branson) on ice with five 10-s bursts at an output of 3 and a duty cycle of 30%, and extracted for 1 h at 4°C. After centrifugation at 12,000 × g for 20 min, the supernatant was loaded onto a butylsilane-bonded silica gel solid-phase extraction cartridge (J. T. Baker), and proteins were eluted with 60% acetonitrile containing 0.1% trifluoroacetic acid (TFA) and fractionated by high-performance liquid chromatography (HPLC) on an octylsilane-bonded silica column with a linear gradient of 25 to 55% acetonitrile in 0.1% TFA over a period of 60 min. HPLC fractions were resolved on 12% sodium dodecyl sulfate (SDS)–polyacrylamide gel, and bands corresponding to p32, previously identified as a major newly synthesized anlagen protein (19), were localized by fluorography and excised from the gel. Approximately 100 pmol of p32 was digested by Lys-C protease at 35°C for 24 h. Peptides were extracted by sonication, fractionated on a Pharmacia SMART HPLC system, and sequenced with an Applied Biosystems model 477 A sequencer.
Cloning of the PDD3 gene.
First-strand cDNA was synthesized by a standard reverse transcription procedure with oligonucleotide LT [5′-GGTCAGAGCTAGGCCTCAAAGCTTCTCGAGGGATCCGAGTC(T)18-3′] and total RNA (1 μg) extracted from mating cells 10 h postmixing. This cDNA was used in the first round of PCR with primers Pep1 (5′-AGAGGWGGWATYCCYTTYGGYATG-3′) and ST (5′-GGTCAGAGCTAGGCCTCA-3′). PCR products were diluted 1:100 and used in the second round of PCR with primers Pep2 (5′-TGGGAACCYGAATGGAAT-3′) and IT (5′-AAGCTTCTCGAGGGATCCGAGTC-3′). The sequence of the Pep1 and Pep2 primers was derived from the sequence of p32-specific peptides. In both cases, PCR consisted of 30 cycles. Each cycle was composed of a melting step at 94°C for 45 s, an annealing step at 52°C for 1 min, and an elongation step at 72°C for 1 min. To obtain information about the 5′ end of the p32 gene cDNA, we used inverse PCR (16). Briefly, first-strand cDNA obtained as indicated above was incubated with T4 RNA ligase (New England Biolabs) according to the manufacturer's instructions to obtain single-stranded circular DNA molecules. After phenol-chloroform extraction and ethanol precipitation, ligation products were used in PCR with the following oligonucleotides: ECAR (5′-TGGGAACCTGAATGGAATC-3′) and RACE (5′-GTATTTAAATGATCTGCATAGCCTTTCC-3′). PCR products were diluted 1: 100 and used in the second round of PCR with primers SEQ1 (5′-TAGTGAAGTATCTGATG-3′) and RACE1 (5′-GAGTTAATCATCATACTTAGTC-3′). In both cases, PCR consisted of 35 cycles. Each cycle was composed of a melting step at 94°C for 45 s, an annealing step at 52°C for 1 min, and an elongation step at 72°C for 1 min. Products obtained in the secondary PCR were sequenced with a sequencing kit from U.S. Biochemicals.
Codon alteration and expression of recombinant p32.
To convert all TAG and TAA codons in the p32 gene to the conventional CAG and CAA codons, respectively, we performed single-stranded DNA mutagenesis (15) on the P32 cDNA cloned in pRSET-B expression vector (Invitrogen) in frame with a N-terminal polyhistidine (6xHis) tag. pRSET-B vector containing modified p32 cDNA (pRSET-B-P32-M) was introduced into the BL21 strain of Escherichia coli. Expression of the fusion protein from pRSET-B-P32-M was induced by adding 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) to the bacterial culture at an optical density at 600 nm of ≈ 0.5 for 4 h. Purification of the fusion 6xHis::p32 protein was carried out according to the manufacturer's instructions.
Generation of antibodies.
Two Pdd3p-specific peptides, peptide F (Y36LVKWKGYADHLNTWEPEWNL56-C) and peptide E (T106NKRKNDENSVSTRRSNK123-C), were synthesized containing an artificial cysteine on the C terminus. Peptides were conjugated to keyhole limpet hemocyanin via cysteine by standard protocols (Pierce). Rabbits were then immunized with injections of 1 mg of conjugated peptide of 1 mg of recombinant p32 (for details, see reference 18).
Electrophoresis and immunoblotting.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting analyses were performed as described previously (18). When whole-cell SDS lysates were used, aliquots of 106 cells were removed from a mating cell culture and processed as described previously (14). Rabbit polyclonal antibodies against Pdd1p were used as described previously (29).
Chromatin immunoprecipitation.
Conjugating Tetrahymena cells were collected by centrifugation at 14 h postmixing followed by isolation of nuclei. Chromatin cross-linking was carried out according to the method of Smothers et al. (29). Briefly, 5 × 106 to 10 × 106 anlagen were cross-linked by incubation for 1 h on ice in buffer containing 1% formaldehyde, 0.25 M sucrose, 10 mM HEPES (pH 7.5), 3 mM CaCl2, 1 mM MgCl2, 1 mM iodoacetamide, 10 mM butyric acid, and 1 mM PMSF. After fixation, nuclei were washed twice in the above buffer lacking formaldehyde, dispersed in buffer A (50 mM Tris-HCl [pH 8.6], 0.2% SDS, 5 mM EDTA), and sonicated on ice with five 10-s bursts with an output of 3 and a duty cycle of 30%. Sonicated material was cleared by centrifugation, and solubilized chromatin was diluted 10× in buffer B (1% Triton X-100, 0.15 M sodium chloride, 2 mM EDTA, 20 mM Tris-HCl [pH 8.6]) and incubated with antibodies overnight at 4°C. Antibodies were precipitated with protein A-Sepharose (Amersham Pharmacia Biotech AB) according to the manufacturer's instructions. Precipitated material was eluted from antibodies in a solution containing 100 mM sodium bicarbonate and 1% SDS, and cross-linking was reversed in the same buffer by incubation for 4 h at 65°C. DNA was then purified by phenol-chloroform extraction and precipitated with ethanol. DNA samples were quantitated by spectrophotometry and slot blotted to a Nylon membrane (Millipore). Fragments a and c obtained by HaeIII restriction digestion of the insert from plasmid pTt2512 (a gift from Meng-Chao Yao, Fred Hutchinson Cancer Center, Seattle, Wash.) were used as a probe for Tetrahymena IES DNA. For a loading control, we used a BTU-1 (β-tubulin) gene (10). All probes were labeled with [α-32P] dATP by random priming (26). Oligonucleotide CBS1 (5′TAAAGAAAGAGGTTGGTTTA-3′) (33) was used as a probe for Cbs. CBS1 was labeled with [γ-32P]ATP and hybridized as described previously (33). Autoradiographs were scanned and imported into Adobe Photoshop TM (version 4.0), and densitometric analysis was performed with the NIH Image program.
Indirect immunofluorescence analysis.
Conjugating cells were fixed and processed for indirect immunofluorescence as described previously (28). Cells were stained with the DNA-specific dye diamidinophenolindole (DAPI) at 0.3 μg/ml in Tris-buffered saline. Mouse polyclonal Pdd1p-specific antibodies were used as described previously (29).
Fluorescent and confocal microscopy.
Fluorescent microscopy was performed with an Olympus BH-2 microscope. Confocal images were obtained with a Leica TCS NT microscope, and digital images were processed with Adobe Photoshop (Adobe Systems, Inc., San Jose, Calif.).
Nucleotide sequence accession number.
The GenBank accession number for the PDD3 gene is AF226856.
RESULTS
p32 is a chromodomain-containing protein.
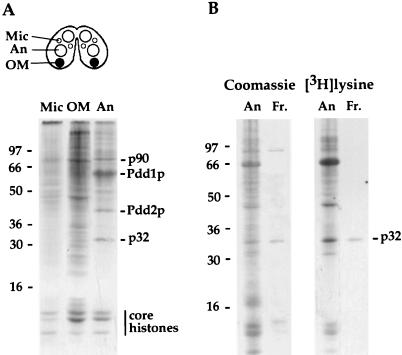
With a metabolic labeling protocol, several anlagen-specific polypeptides were detected (19) whose expression peaks during stages of conjugation when DNA rearrangements occur (macronuclear development II [MAC II] stage). Shown in Fig. 1A is a typical fluorograph of a 12% SDS gel of total nuclear protein extracted from purified micronuclei, old macronuclei, and new macronuclei or anlagen (An) isolated from mating cells pulse-labeled with [3H]lysine at the MAC II stage. Four polypeptides with apparent masses of 90, 65, 43, and 32 kDa are strongly labeled and selectively deposited into anlagen. Two of these proteins, Pdd1p and Pdd2p (Fig. 1A), were recently isolated and characterized (20, 29). Another polypeptide, with an apparent molecular mass of 32 kDa (p32), is also actively synthesized and deposited in anlagen at this stage. To isolate and characterize p32, total anlagen protein was extracted with 2 M sodium perchlorate and subjected to fractionation by reverse-phase HPLC, followed by separation from contaminating proteins on a 12% SDS gel (see Materials and Methods for details). p32 identified by incorporation of [3H]lysine (Fig. 1B) was excised from the gel and digested with Lys-C protease, and three HPLC-purified peptides were subjected to microsequence analysis. This analysis provided us with sufficient sequence to synthesize degenerate oligonucleotide primers. By using a combination of reverse transcription PCR and 5′- and 3′-rapid amplification of cDNA ends PCR, we isolated a P32 cDNA.
FIG. 1.
Polypeptides synthesized and targeted to different nuclei during late stages of Tetrahymena development. (A) Conjugating cells (schematically represented on the top of the figure) were pulse-labeled with [3H]lysine from 9 to 10 h postmixing. Nuclei were purified by sedimentation at 1 U of gravity (2). Total protein from different types of nuclei was resolved on a 12% SDS–polyacrylamide gel and analyzed by fluorography. Approximately 107 micronuclei (Mic), 106 old macronuclei (OM), and 3 × 106 anlagen (An) were used. Previously identified Tetrahymena polypeptides and protein molecular mass standards (kilodaltons) are shown. (B) p32 recovered from reverse-phase HPLC fractionation of proteins extracted from purified anlagen and resolved on 12% SDS–polyacrylamide gel. Proteins were visualized by staining with Coomassie blue, followed by fluorography to verify the position of p32 (shown on the right). An, total anlagen protein before the extraction; Fr, HPLC fraction containing p32. Protein molecular mass standards (kilodaltons) are designated on the left.
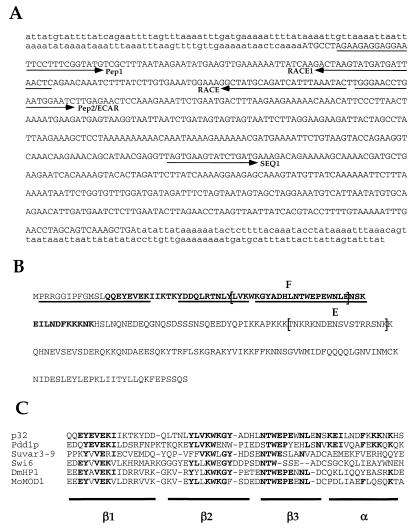
The predicted amino acid sequence corresponding to the longest open reading frame of the gene encoding p32 is shown in Fig. 2A. Several observations suggest that this sequence is correct. First, all three sequenced peptides are found in the predicted protein (Fig. 2A). Second, the calculated molecular mass of the deduced translation product open reading frame (≈26 kDa) is close to the apparent molecular mass of p32 on SDS gel (32 kDa). Finally, the predicted isoelectric point of p32 (9.1) is in a good agreement with that determined for p32 earlier (approximately 9.0) (19).
FIG. 2.
Analysis of p32 cDNA and amino acid sequence. (A) Nucleotide sequence of p32 cDNA. Nucleotides from the longest open reading frame are indicated by capital letters. The positions of oligonucleotides used in PCR are indicated by arrows (see Materials and Methods for details). (B) Derived amino acid sequence of the longest open reading frame of P32 cDNA. Underlined are peptides used for microsequencing. Shown in boldface is the chromodomain. Shown in brackets and designated by E and F are sequences of peptides used for antibody generation. (C) Alignment of the Pdd3p chromodomain with chromodomain from other proteins. Amino acid residues common with Pdd3p are shown in boldface. Horizontal bars define the type of secondary structure (β-sheet or α-helix) of the mouse MoMOD 1 protein (5). Note that Pdd1p and Pdd3p chromodomains share homology in the least-conserved α-helical region.
Comparison of the deduced amino acid sequence of p32 with those in the protein databases suggests that p32 contains a chromodomain (from chromatin organizing modifier) (Fig. 2C), a group of amino acids found predominantly in heterochromatin-associated proteins (17). Interestingly, the highest degree of homology to the p32 chromodomain is found in one of the three chromodomains of Pdd1p, a protein implicated in DNA elimination in Tetrahymena (described above). In both cases, the chromodomains are at the N-terminal end of the polypeptides.
p32 localizes to the DNA elimination structures in anlagen.
To determine the expression pattern and intranuclear localization of p32, we generated a set of p32-specific polyclonal antisera. First, two peptides (Fig. 2A) were synthesized based on the derived p32 sequence and were used to immunize rabbits. Second, the entire p32 fused with a histidine tag (30) was expressed in E. coli after changing TAG and TAA codons encoding glutamine in Tetrahymena to the conventional glutamine codons CAG and CAA, respectively (see Materials and Methods). Recombinant p32, when resolved on SDS gel, migrated more slowly than the original p32 (data not shown), probably because of the polyhistidine tag. Recombinant p32 was used to generate polyclonal antiserum in rabbits.
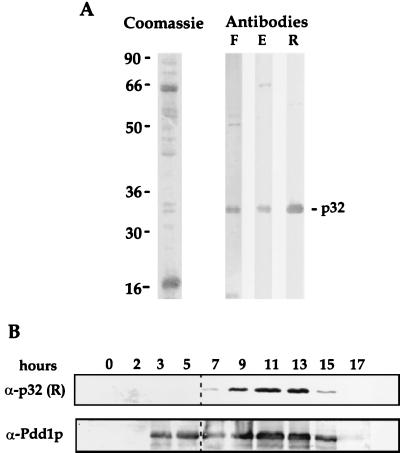
All three sera demonstrated high specificity toward p32 on Western blots containing total protein extracted from purified anlagen (Fig. 3A). Immunoblotting analysis of total protein from cells collected at different stages of conjugation and imunofluorescence data (not shown) revealed that, unlike Pdd1p (20) and Pdd2p (29), p32 is expressed only during later stages of conjugation, after anlagen are formed (roughly at 6 h [see dotted line in Fig. 3B]). The highest level of p32 is detected during stages of Tetrahymena development when DNA rearrangements were reported to occur (3), and the termination of its expression (as in case with Pdd1p and Pdd2p [19, 20, 29]) corresponds to the end of this process. These results were confirmed with several p32-specific antisera.
FIG. 3.
p32 is a developmentally regulated polypeptide. (A) Equal amounts of total anlagen protein from 14-h conjugating cells were loaded on 10% SDS–polyacrylamide gel. One part of the gel was stained with Coomassie blue, while the others were analyzed by Western blotting with antibodies generated against peptide F, peptide E, and recombinant p32 (designated by F, E, and R, respectively). (B) Total proteins from conjugating Tetrahymena cells at different times postmixing (indicated at the top in hours) were extracted and resolved on a 12% SDS–polyacrylamide gel, and distinct sections of the gel were probed in Western blots with serum against recombinant p32 and with previously obtained Pdd1p antibodies. Note the difference in the timing of expression of p32 and Pdd1p.
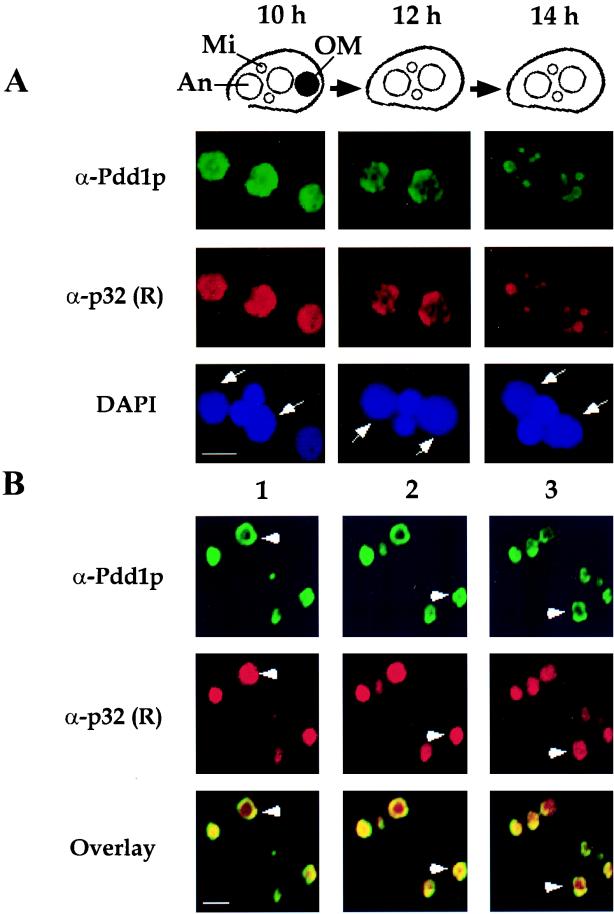
To determine the nuclear localization of p32 during macronuclear development, we performed indirect immunofluorescence analyses of Tetrahymena cells during conjugation by using antibodies against recombinant p32 (lane “R” in Fig. 3). As shown in Fig. 4A, p32 is uniformly distributed in new macronuclei as well as in the degenerating old macronucleus at 10 h postmixing. However, at about 12 h of conjugation the staining becomes uneven, culminating in clustering into characteristic circle-like intranuclear structures approximately 2 h later. Previous reports demonstrated that these structures contain micronucleus-specific DNA destined for elimination (20). Immunofluorescence experiments with Pdd1p and Pdd2p antibodies revealed that these proteins localize mostly at the periphery of these structures, resulting in characteristic “doughnut-like” images (19). It is noteworthy that the pattern of p32 distribution in anlagen coincides well with that of Pdd1p during most of macronuclear development, as evidenced by the double staining of anlagen with p32 rabbit and Pdd1p mouse polyclonal antibodies (Fig. 4A). However, as shown in Fig. 4B, the concentration of p32 inside DNA elimination structures is considerably higher than that of Pdd1p, suggesting that p32 plays a different role in the process of DNA degradation. Close association of p32 with DNA degradation structures motivated us to change its name to Pdd3p (for programmed DNA degradation).
FIG. 4.
Intranuclear localization of p32. (A) Confocal sections of Tetrahymena cells fixed at the indicated time points and stained simultaneously with DAPI and with (i) mouse Pdd1p and fluorescein isothiocyanate-conjugated secondary antibodies and (ii) rabbit recombinant p32 (R) (lane R in Fig. 3) and rhodamine-conjugated secondary antibodies as shown. White arrows indicate anlagen, stained with DAPI. Shown above is a schematic representation of the late stages of conjugation: macronuclear development II (MAC II) and macronuclear development III (MAC III). Mi, An, and OM, micronucleus, anlagen, and old macronucleus (black circle), respectively. Only one of two paired cells is shown. Bar, 4 μm. Note that in approximately 30% of conjugations, we completely failed to detect p32 in nuclei at the time corresponding to the formation of DNA elimination structures, while Pdd1p in these cells was still readily detectable. Interestingly, p32 in these conjugations colocalizes with Pdd1p at earlier stages, and the amounts of p32 in anlagen purified from either type of conjugation as well as the duration of p32 expression in either case were similar, as evidenced by Western blotting (data not shown). Therefore, we attributed the absence of p32-specific staining to the inaccessibility of p32 to antibodies, which most likely is a result of its localization inside DNA degradation structures. (B) Consecutive confocal images (numbered 1 to 3; 0.4 μm apart) of a Tetrahymena cell at the MAC III stage, fixed, and stained as in panel A. White arrowheads indicate donut-like DNA degradation structures. Note the difference in the intranuclear distributions of Pdd1p and p32. Bar, 1 μm.
Pdd3p and Pdd1p associate with IES-specific, but not Cbs-specific, DNA.
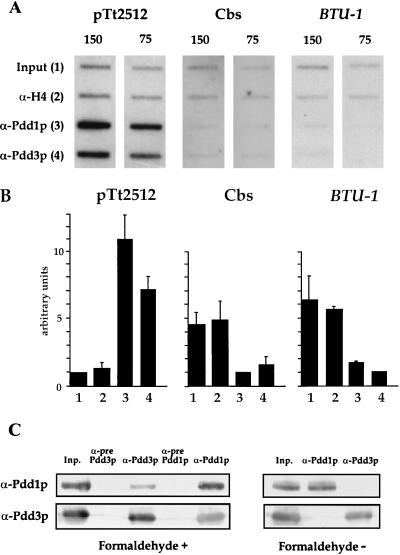
Localization of Pdd3p inside DNA degradation structures prompted us to analyze what DNA sequences might be physically associated with it. To this end, we obtained a nuclear fraction from 14-h conjugating Tetrahymena cells highly enriched in anlagen. At this time, anlagen contain multiple DNA elimination structures. Nuclei were then cross-linked with 1% paraformaldehyde and sonicated, and solubilized nuclear material was subjected to immunoprecipitation with various antibodies. Following cross-link reversal, immunoprecipitated DNA was purified, quantitated, slot blotted to the Nylon membrane, and probed sequentially with various sequences, stripping the membrane after each hybridization. To detect IES DNA, a fragment of pTt2512 was used that contains a sequence homologous to highly dispersed micronucleus-limited sequences that are repeated about 200 times in the micronuclear genome (32). To detect Cbs, a 20-bp oligonucleotide, CBS-1, that contains a consensus for Cbs sequences (33) was used. A PCR fragment of the Tetrahymena BTU-1 gene (10) was used as a control for loading.
The results of these hybridization analyses are shown in Fig. 5A and summarized in Fig. 5B. Chromatin immunoprecipitated with antibodies selective for Pdd3p or Pdd1p is clearly enriched with DNA hybridizing to the pTt2512 plasmid. In contrast, no enrichment over input DNA was observed in the same samples hybridized with the probe for Cbs or BTU. As expected, no enrichment for any probe was detected in DNA precipitated with histone H4 antibodies. Thus, we conclude that Pdd1p and Pdd3p associate with IES, but not Cbs, sequences in the anlagen genome at this stage of development.
FIG. 5.
Pdd3p associates with Pdd1p and with DNA enriched in IES, but not Cbs. (A) DNA samples obtained by chromatin immunoprecipitation with the antibodies shown on the left, as well as DNA isolated from solubilized, cross-linked chromatin before immunoprecipitation (input), were quantitated, slot blotted onto a Nylon membrane, and sequentially probed with the DNA sequences designated on the top of each section. The numbers above each column indicate the amount of loaded DNA in nanograms. (B) Relative densitometric signals for samples shown in panel A. Error bars indicate the standard deviation from average reading of signals in two groups: 150 and 75 ng. (C) Material immunoprecipitated with the antibodies designated on the top from either cross-linked (Formaldehyde +) or untreated (Formaldehyde −) solubilized chromatin from 14-h anlagen was resolved on the 12% SDS gel and probed by Western blotting with the antibodies shown on the left. α-prePdd1p and α-prePdd3p, preimmune Pdd1p and Pdd3p sera, respectively. Inp., input.
DISCUSSION
In the present study, we report the purification and characterization of a new chromodomain-containing polypeptide, referred to as Pdd3p. Several lines of evidence suggest its involvement in the DNA elimination process in Tetrahymena. First, Pdd3p expression peaks during a short period corresponding to the major DNA rearrangement processes in the new macronucleus. Second, immunofluorescence data demonstrate that Pdd3p is uniquely localized throughout structures known to be associated with sequences undergoing elimination in anlagen. Finally, Pdd3p physically associates with at least one randomly selected family of micronucleus-specific DNA sequences destined for elimination.
There are several structural and biological similarities between Pdd3p and the founding member of this small family of proteins—Pdd1p. Pdd1p has three chromodomains (20), one of which shares especially high homology with the chromodomain of Pdd3p. Interestingly, the area of homology between these two proteins includes an α-helical region, which normally is one of the least conserved regions throughout chromodomains (Fig. 2C). Chromodomain proteins were shown to take part in transcriptional regulation (both activation and repression) and in maintenance of genomic stability (reviewed in references 5 and 17). The chromodomain is detected in proteins in conjunction with other domains, such as helicase, transposase, and Zn finger domains (17). Although the function of chromodomains has not yet been determined, it is generally believed that they are responsible for targeting proteins to specialized chromatin sites via protein-protein interactions (5). Since both Pdd1p and Pdd3p colocalize in the degenerating old macronucleus and in anlagen and are targeted to the same subnuclear structures in anlagen, it is likely that there is a shared mechanism of their recruitment to the eliminating DNA sequences. Our immunofluorescence and coimmunoprecipitation data suggest that both Pdd proteins exist in a complex with eliminated DNA, although the nature of our analysis does not allow us to conclude whether these proteins coimmunoprecipitate only when bound to IES elements.
On the other hand, several features of Pdd3p distinguish it from other members of the Pdd group. First, expression of Pdd3p, unlike that of Pdd1p and Pdd2p, has not been detected in the pre-anlagen stage of Tetrahymena development, suggesting that is not required for processes preceding anlagen differentiation. The precise link between these processes and DNA elimination in anlagen has not been established; however, pre-anlagen expression of Pdd1p and Pdd2p is essential for DNA elimination and nuclear differentiation (7, 23). Second, our immunofluorescence data suggest that the relative concentration of Pdd3p in the center of the DNA elimination structures is higher than that of other Pdd proteins. These results were obtained by using several previously characterized Pdd1p- and Pdd2p-specific antisera (19, 29) and polyclonal antisera against recombinant Pdd3p (R in Fig. 3) obtained from two rabbits.
Since the micronucleus-limited DNA sequences also localize to these regions, our data suggest that Pdd3p may be more directly in contact with eliminating DNA sequences than other Pdd proteins.
From the abundant nature of Pddps and the presence of chromodomains in two of them, we favor the view that chromatin structure and nucleostructural organization in addition to sequence specificity are responsible for directing excision of micronucleus-specific DNA—the first step in DNA elimination. This view is consistent with the observation that no consensual signals for IES excision have been identified yet. Also in accord with this hypothesis is the recent report that chromatin modifications are essential for another DNA rearrangement process: VDJ recombination in B and T cells (21).
IES excision and chromosome breakage, which includes excision and elimination of Cbs and adjacent regions, are two major DNA rearrangements occurring in anlagen during a short interval. This observation gave rise to the hypothesis that the same chromatin factors might be involved in both processes. Our experiments demonstrate that, unlike some IES, Cbs sequences are not enriched in DNA cross-linked to either Pdd1p or Pdd3p. These data are further supported by the finding that somatic disruption of the PDD1 gene impairs IES excision, but does not affect chromosome processing and telomere addition (7). The simplest interpretation of our data is that different sets of proteins are involved in two DNA rearrangement processes in the developing macronucleus.
ACKNOWLEDGMENTS
We are grateful to Jim Smothers and Craig Mizzen for their technical help, Richard Cook for protein sequencing, and Thomas Doak for his help in protein sequence analysis. We thank Nataliya Shulga for help with confocal microscopy, Jody Bowen for critical reading of the manuscript, and Jianxin Zhou for constant encouragement and suggestions throughout the work.
M.A.N. was a recipient of an Oncology Research Faculty Development Program Fellowship, NCI, NIH. This research was supported by grants from the National Institutes of Health to C.D.A. and M.A.G.
REFERENCES
- 1.Abraham J, Nasmyth K A, Strathern J N, Klar A J S, Hicks J B. Regulation of mating type information in yeasts. Negative control requiring sequences both 5′ and 3′ to the regulated regions. J Mol Biol. 1984;176:307–311. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- 2.Allis C D, Dennison D K. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev Biol. 1982;93:519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- 3.Austerberry C F, Allis C D, Yao M C. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci USA. 1984;81:7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassnett S, Mataic D. Chromatin degradation in differentiating fiber cells of the eye lens. J Cell Biol. 1997;137:37–49. doi: 10.1083/jcb.137.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalli G, Paro R. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr Opin Cell Biol. 1998;10:354–360. doi: 10.1016/s0955-0674(98)80011-2. [DOI] [PubMed] [Google Scholar]
- 6.Coyne R S, Chalker D L, Yao M C. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- 7.Coyne R S, Nikiforov M A, Smothers J F, Allis C D, Yao M-C. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol Cell. 1999;4:865–872. doi: 10.1016/s1097-2765(00)80396-2. [DOI] [PubMed] [Google Scholar]
- 8.Davis M C, Ward J G, Herrick G, Allis C D. Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Dev Biol. 1992;154:419–432. doi: 10.1016/0012-1606(92)90080-z. [DOI] [PubMed] [Google Scholar]
- 9.Frohman M A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 10.Gaertig J, Gu L, Hai B, Gorovsky M A. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelert M. V(D)J recombination gets a break. Trends Genet. 1992;8:408–412. doi: 10.1016/0168-9525(92)90322-u. [DOI] [PubMed] [Google Scholar]
- 12.Gorovsky M A, Yao M C, Keevert J B, Pleger G L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 13.Gorovsky M A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- 14.Guttman S D, Glover C V, Allis C D, Gorovsky M A. Heat shock, deciliation and release from anoxia induce the synthesis of the same set of polypeptides in starved T. pyriformis. Cell. 1980;22:299–307. doi: 10.1016/0092-8674(80)90177-4. [DOI] [PubMed] [Google Scholar]
- 15.Hill D E. Mutagenesis of cloned DNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1988. pp. 8.0.1–8.5.9. [Google Scholar]
- 16.Huang S H. Inverse polymerase chain reaction. An efficient approach to cloning cDNA ends. Mol Biotechnol. 1994;2:15–22. doi: 10.1007/BF02789286. [DOI] [PubMed] [Google Scholar]
- 17.Koonin E V, Zhou S, Lucchesi J C. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin R, Leone J W, Cook R G, Allis C D. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madireddi M T, Davis M C, Allis C D. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev Biol. 1994;165:418–431. doi: 10.1006/dbio.1994.1264. [DOI] [PubMed] [Google Scholar]
- 20.Madireddi M T, Coyne R S, Smothers J F, Mickey K M, Yao M C, Allis C D. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- 21.McMurry M T, Krangel M S. A role for histone acetylation in the developmental regulation of V(D)J recombination. Science. 2000;287:495–498. [PubMed] [Google Scholar]
- 22.Nanney D L. Introduction. In: Gall J G, editor. Molecular biology of ciliated protozoa. Orlando, Fla: Academic Press; 1986. pp. 1–21. [Google Scholar]
- 23.Nikiforov M A, Smothers J F, Gorovsky M A, Allis C D. Excision of micronuclear-specific DNA requires parental expression of Pdd2p and occurs independently from DNA replication in Tetrahymena thermophila. Genes Dev. 1999;13:2852–2862. doi: 10.1101/gad.13.21.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orias E. Ciliate conjugation. In: Gall J G, editor. Molecular biology of ciliated protozoa. Orlando, Fla: Academic Press; 1986. pp. 45–84. [Google Scholar]
- 25.Prescott D M. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards E J. Preparation and analysis of DNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1988. pp. 2.0.1.–2.12.5.. [Google Scholar]
- 27.Robertson B D, Meyer T F. Genetic variation in pathogenic bacteria. Trends Genet. 1992;8:422–427. doi: 10.1016/0168-9525(92)90325-x. [DOI] [PubMed] [Google Scholar]
- 28.Smothers J F, Madireddi M T, Warner F D, Allis C D. Programmed DNA degradation and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. J Eukaryot Microbiol. 1997;44:79–88. doi: 10.1111/j.1550-7408.1997.tb05942.x. [DOI] [PubMed] [Google Scholar]
- 29.Smothers J F, Mizzen C A, Tubbert M M, Cook R G, Allis C D. Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development. 1997;124:4537–4545. doi: 10.1242/dev.124.22.4537. [DOI] [PubMed] [Google Scholar]
- 30.Tabor S. Protein expression. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1988. pp. 16.0.5–16.5.4. [Google Scholar]
- 31.Yao M C, Gorovsky M A. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48:1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- 32.Yao M C. Elimination of specific DNA sequences from the somatic nucleus of the ciliate Tetrahymena. J Cell Biol. 1982;92:783–789. doi: 10.1083/jcb.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao M C, Zheng K, Yao C H. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell. 1987;48:779–788. doi: 10.1016/0092-8674(87)90075-4. [DOI] [PubMed] [Google Scholar]
- 34.Yao M C, Yao C H, Monks B. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- 35.Yao M C. Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet. 1996;12:26–30. doi: 10.1016/0168-9525(96)81385-0. [DOI] [PubMed] [Google Scholar]
- 36.Yu G L, Blackburn E H. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]