Abstract
Enteropathogenic (EPEC) and enteroaggregative (EAEC) Escherichia coli are two of the major pathotypes of diarrheagenic E. coli causing disease worldwide. Here, we report a diarrheal outbreak caused by E. coli of serotype O3:H2, harboring virulence markers from EPEC (eae) and/or EAEC (aggR). This is likely the first E. coli diarrheal outbreak caused by a hybrid atypical-EPEC/EAEC clone reported in Brazil.
Keywords: Hybrid aEPEC/EAEC, Diarrhea and outbreak
Escherichia coli isolates associated with diarrheal disease are collectively termed diarrheagenic E. coli (DEC). On the basis of a set of molecular and phenotypic features, DEC can be divided into six main pathotypes: enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), and diffusely adherent E. coli (DAEC) [1–3].
EPEC produces a histopathological lesion in infected cells, termed attaching, and effacement (AE), which is characterized by intimate adherence of bacteria to the host cell, leading to microvillus effacement and accumulation of mainly F-actin but also other cytoskeleton elements, underneath adherent bacteria [4, 5]. All the proteins necessary for the AE lesion formation are encoded by genes located on a chromosomal pathogenicity island termed LEE (locus of enterocyte effacement) region [6]. Among these proteins, it is important to highlight the adhesin intimin encoded by the eae (E. coli attaching and effacing) gene [7].
One of the first virulence properties identified in EPEC isolates was the formation of very compact clusters of bacteria on the surface of infected epithelial cells (HeLa or Hep-2), which is referred as localized adherence (LA) pattern [8]. The adhesin responsible for the establishment of this phenotype is called bundle-forming pilus (BFP) and is encoded by a set of 14 genes (bfp operon) located in the EPEC adherence factor (EAF) plasmid [9, 10]. However, a study with adult volunteers showed that EPEC lacking the EAF plasmid can also cause diarrhea [11], thus leading to the subdivision of this pathotype into two subgroups: typical (tEPEC) and atypical (aEPEC), with the EAF (bfp+) plasmid present only in tEPEC [12–14]. Since aEPEC do not produce BFP, these isolates produce adherence patterns other than LA, such as the localized adherence-like (LAL) pattern, which differs from the LA pattern by the formation of less compact bacterial microcolonies on the surface of infected cells, detected only in assays performed with extended periods of bacteria-cell incubation [15]. Moreover, there are a large number of isolates that do not produce a characteristic adherence pattern or are non-adherent [16, 17].
The hallmark of EAEC is the production of a bacterial arrangement that resembles stacked bricks on epithelial cells, that is, known as aggregative adherence (AA) pattern [18], as well as biofilm formation on abiotic surface and intestinal mucosa [19, 20]. The formation of the AA pattern depends on the production of one of the five distinct chaperone-usher aggregative adherence fimbriae (AAF/I—AAF/V) described so far [21–25]. The genes encoding the proteins necessary for the AAFs biogenies are located in a plasmid (pAA), which also carries genes that encode other EAEC-associated virulence factors and the global virulence regulator AggR, which activates the expression of several plasmid and chromosomal virulence factor-encoding genes [26, 27].
In 2011, an outbreak of diarrhea in Germany, caused by a hybrid STEC/EAEC of serotype O104:H4, was responsible for the surprising number of diarrhea cases, namely 3816, of which 845 progressed to hemolytic-uremic syndrome, and a total of 54 deaths were registered [28, 29]. This outbreak drew attention to the emergence of hybrid DEC pathotypes and their high pathogenic potential, and since then, many other hybrid DEC pathotypes have been associated with diarrheal disease worldwide, such as: STEC/ETEC [30, 31] and aEPEC/ETEC [32].
A recently published epidemiological surveillance study carried out in Brazil between 2011 and 2016, involving a total of 5047 patients with acute diarrhea, demonstrated that aEPEC and EAEC appear as the most common DEC pathotypes in the Brazilian scenario [33].
Here, we report an investigation of a community-acquired diarrheal outbreak involving eight patients with acute diarrhea, occurring in São Paulo State, Brazil, during the year of 2010. Briefly, colony-forming bacteria, observed on MacConkey agar plates seeded with the stool samples collected from the diarrheal patients, were biochemically identified as E. coli. The presence of the molecular markers used for DEC pathotype identification (eae, bfpB, stx1, stx2, aggR, elt, est, and ipaH) was detected by PCR, using primers and conditions as previously described [33], and the somatic (O) and flagellar (H) antigens of the DEC isolates were serologically typed by tube agglutination assays employing absorbed somatic (O1-O188) and flagellar (H1-H56) antisera produced at Institute Adolfo Lutz (IAL). Furthermore, the adherence pattern produced in HeLa cells was determined, using 3 and 6 h of co-incubation of bacteria and epithelial cells, as previously described [15].
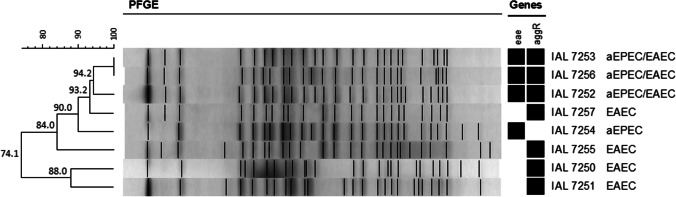
The presence of virulence factor-encoding genes revealed that one diarrheal patient was infected with aEPEC (eae+/bfpB−), four with EAEC (aggR+), and three with a hybrid aEPEC/EAEC (eae+/bfpB−/aggR+), with all DEC isolates obtained classified in the serotype O3:H2. Other enteropathogens, such as Salmonella spp., Shigella spp., Campylobacter spp., Aeromonas spp., Plesiomonas spp., Yersinia enterocolitica, rotavirus, and norovirus were not detected in the stool samples obtained from the 8 diarrheal patients evaluated in this study. One DEC isolate per patient was then selected and subjected to pulsed-field gel electrophoresis (PFGE) analysis, showing the DEC isolates of serotype O3:H2 with more than 74.1% similarity, regardless of the pathotype. Of importance, the three hybrid aEPEC/EAEC isolates showed more than 94.2% similarity (Fig. 1), thus confirming the first diarrheal outbreak caused by a hybrid aEPEC/EAEC of serotype O3:H2 in Brazil.
Fig. 1.
Dendrogram showing the relationships between EAEC, aEPEC, and hybrid aEPEC/EAEC isolates of serotype O3:H2 obtained during a diarrheal outbreak investigation. The XbaI DNA restriction profiles were analyzed in Bionumerics v7.5 (Applied Maths), and a dendrogram was generated by the unweighted pair group method with arithmetic mean (UPGMA) applying the Dice coefficient with optimization and tolerance set at 1.0%
EAEC of serotype O3:H2 has been identified in Brazil in diarrheal patients, healthy subjects [33–37], and bovine meat [38], thus pointing out the circulation of this pathogen in our setting, as well as in distinct case–control studies performed worldwide [39–42]. The occurrence of hybrid aEPEC/EAEC isolates appears to be rare in Brazil, but combination of virulence factor-encoding genes from these two DEC pathotypes has already been observed in E. coli isolates obtained from patients with acute diarrhea, from serotypes O142:H34 and O158:HNM [43, 44].
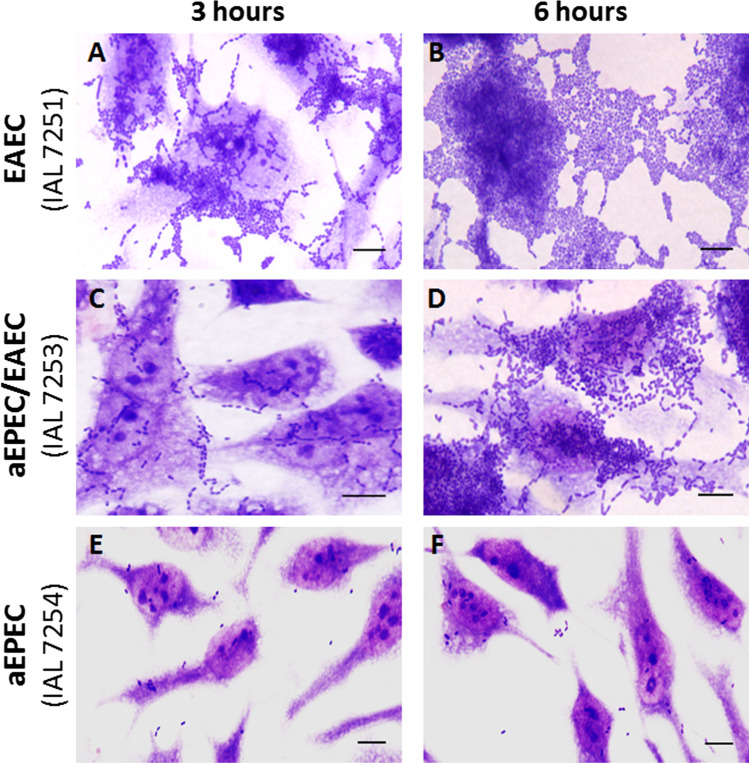
As expected, the four EAEC isolates displayed an AA pattern in HeLa cells in assays performed with 3 and 6 h of incubation. On the other hand, the three hybrid aEPEC/EAEC demonstrated only a trend to form the AA pattern in 3 h, with this phenotype being better observed in the 6-h assay, concomitantly with short chain-like adherence (CLA). In contrast, the aEPEC isolate did not produce a characteristic adherence pattern, adhering only sporadically to HeLa cells, even in assays performed with long incubation periods (Fig. 2).
Fig. 2.
Adherence pattern displayed in HeLa cells, using 3 (A, C, and E) and 6 (B, D, and F) hours of incubation of cells and bacteria, by representatives EAEC (A and B), aEPEC (E and F), and hybrid aEPEC/EAEC (C and D) isolates of serotype O3:H2. (Bar scale = 10 μm)
Here, we report, to the best of our knowledge, the first diarrheal outbreak due to a hybrid aEPEC/EAEC of serotype O3:H2 in Brazil. Future whole genome sequencing studies will be necessary to better understand the genetic background of these hybrid isolates, the horizontal gene transfer events involved in their emergence, and how this particular combination of virulence factors can influence their pathogenic potential.
Funding
This study was supported in part by the São Paulo Research Foundation (FAPESP grant 2017/14821–7).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Elizabeth Andrade Marques
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper J, Nataro J, Mobley H. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26(4):822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41(3):1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knutton S, Baldwin T, Williams PH, McNeish AS. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57(4):1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92(5):1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerse AE, Yu J, Tall BD, Kaper JB. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A. 1990;87(20):7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scaletsky IC, Silva ML, Trabulsi LR. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45(2):534–536. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giron JA, Ho AS, Schoolnik GK. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254(5032):710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg MS, Girón JA, Nataro JP, Kaper JB. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 11.Levine MM, Nataro JP, Karch H, et al. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152(3):550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 12.Kaper JB. Defining EPEC. Rev Microbiol. 1996;27:130–133. [Google Scholar]
- 13.Trabulsi LR, Keller R, Gomes TAT. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8(5):508–513. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandes RT, Elias WP, Vieira MA, Gomes TA. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol Lett. 2009;297(2):137–149. doi: 10.1111/j.1574-6968.2009.01664.x. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues J, Scaletsky IC, Campos LC, Gomes TAT, Whittam TS, Trabulsi LR. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect Immun. 1996;64(7):2680–2686. doi: 10.1128/iai.64.7.2680-2686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe CM, Trabulsi LR, Blanco J, et al. Virulence features of atypical enteropathogenic Escherichia coli identified by the eae+ EAF-negative stx− genetic profile. Diagn Microbiol Infect Dis. 2009;64(4):357–365. doi: 10.1016/j.diagmicrobio.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 17.Vieira MA, Dias RC, dos Santos LF, Rall VL, Gomes TA, Hernandes RT. Diversity of strategies used by atypical enteropathogenic Escherichia coli to induce attaching and effacing lesion in epithelial cells. J Med Microbiol. 2019;68(6):940–951. doi: 10.1099/jmm.0.000998. [DOI] [PubMed] [Google Scholar]
- 18.Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, Levine MM. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J. 1987;6(9):829–831. doi: 10.1097/00006454-198709000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Hicks S, Candy DC, Phillips AD. Adhesion of enteroaggregative Escherichia coli to pediatric intestinal mucosa in vitro. Infect Immun. 1996;64(11):4751–4760. doi: 10.1128/iai.64.11.4751-4760.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheikh J, Hicks S, Dall’Agnol M, Phillips AD, Nataro JP. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol. 2001;41:983–997. doi: 10.1046/j.1365-2958.2001.02512.x. [DOI] [PubMed] [Google Scholar]
- 21.Nataro JP, Deng Y, Maneval DR, German AL, Martin WC, Levine MM. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect Immun. 1992;60(6):2297–2304. doi: 10.1128/iai.60.6.2297-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czeczulin JR, Balepur S, Hicks S, et al. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect Immun. 1997;65(10):4135–4145. doi: 10.1128/IAI.65.10.4135-4145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernier C, Gounon P, Le Bouguénec C. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun. 2002;70(8):4302–4311. doi: 10.1128/IAI.70.8.4302-4311.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun. 2008;76(7):3281–3292. doi: 10.1128/IAI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jønsson R, Struve C, Boisen N, et al. Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect Immun. 2015;83(4):1396–1405. doi: 10.1128/IAI.02820-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 27.Morin N, Santiago AE, Ernst RK, Guillot SJ, Nataro JP. Characterization of the AggR regulon in enteroaggregative Escherichia coli. Infect Immun. 2013;81(1):122–132. doi: 10.1128/IAI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bielaszewska M, Mellmann A, Zhang W, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis. 2011;11(9):671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 29.Kaper James B, O'Brien Alison D. Overview and historical perspectives. In: Sperandio V, Hovde CJ, editors. Enterohemorrhagic Escherichia coli and other Shiga toxin-producing E. coli. Washington, DC: ASM Press; 2015. pp. 1–13. [Google Scholar]
- 30.Bai X, Zhang J, Ambikan A, et al. Molecular characterization and comparative genomics of clinical hybrid Shiga toxin-producing and enterotoxigenic Escherichia coli (STEC/ETEC) strains in Sweden. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-42122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavalcanti A, Hernandes RT, Takagi EH, et al. virulence profiling and molecular typing of Shiga toxin-producing E. coli (STEC) from human sources in Brazil. Microorganisms. 2020;8(2):171. doi: 10.3390/microorganisms8020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazen TH, Michalski J, Luo Q, et al. Comparative genomics and transcriptomics of Escherichia coli isolates carrying virulence factors of both enteropathogenic and enterotoxigenic E. coli. Sci Rep. 2017;7(1):1–17. doi: 10.1038/s41598-017-03489-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ori E, Takagi E, Andrade T, et al. Diarrhoeagenic Escherichia coli and Escherichia albertii in Brazil: pathotypes and serotypes over a 6-year period of surveillance. Epidemiol Infect. 2019;147:E10. doi: 10.1017/S0950268818002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamboni A, Fabbricotti SH, Fagundes-Neto U, Scaletsky IC. Enteroaggregative Escherichia coli virulence factors are found to be associated with infantile diarrhea in Brazil. J Clin Microbiol. 2004;42(3):1058–1063. doi: 10.1128/JCM.42.3.1058-1063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno ACR, Fernandes Filho A, Gomes TAT. Etiology of childhood diarrhea in the northeast of Brazil: significant emergent diarrheal pathogens. Diagn Microbiol Infect Dis. 2010;66(1):50–57. doi: 10.1016/j.diagmicrobio.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 36.França FLS, Wells TJ, Browning DF, et al. Genotypic and phenotypic characterisation of enteroaggregative Escherichia coli from children in Rio de Janeiro Brazil. PLoS ONE. 2013;8(7):e69971. doi: 10.1371/journal.pone.0069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dias RC, Tanabe RH, Vieira MA, et al. Analysis of the virulence profile and phenotypic features of typical and atypical enteroaggregative Escherichia coli (EAEC) isolated from diarrheal patients in Brazil. Front Cell Infect Microbiol. 2020;10:144. doi: 10.3389/fcimb.2020.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelayo JS, Elias Junior AR, de Lima NR, Navarro A, da Rocha SPD. Detection of diarrheagenic Escherichia coli in bovine meat in the Northern Region of Paraná State Brazil. Braz Arch Biol Technol. 2019;62:e19180012. doi: 10.1590/1678-4324-2019180012. [DOI] [Google Scholar]
- 39.Vial PA, Robins-Browne R, Lior H, et al. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis. 1988;158(1):70–79. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]
- 40.Foster MA, Iqbal J, Zhang C, McHenry R, et al. Enteropathogenic and enteroaggregative E. coli in stools of children with acute gastroenteritis in Davidson County Tennessee. Diagn Microbiol Infect Dis. 2015;83(3):319–324. doi: 10.1016/j.diagmicrobio.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hebbelstrup Jensen B, Stensvold CR, Struve C, et al. Enteroaggregative Escherichia coli in daycare—a 1-year dynamic cohort study. Front Cell Infect Microbiol. 2016;6:75. doi: 10.3389/fcimb.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petro CD, Duncan JK, Seldina YI, et al. Genetic and Virulence Profiles of Enteroaggregative Escherichia coli (EAEC) isolated from deployed military personnel (DMP) with travelers’ diarrhea. Front Cell Infect Microbiol. 2020;10:200. doi: 10.3389/fcimb.2020.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghilardi ACR, Gomes TAT, Elias WP, Trabulsi LR. Virulence factors of Escherichia coli strains belonging to serogroups O127 and O142. Epidemiol Infect. 2003;131(2):815–821. doi: 10.1017/S095026880300877X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liebchen A, Benz I, Mellmann A, et al. Characterization of Escherichia coli strains isolated from patients with diarrhea in Sao Paulo, Brazil: identification of intermediate virulence factor profiles by multiplex PCR. J Clin Microbiol. 2011;49(6):2274–2278. doi: 10.1128/JCM.00386-11. [DOI] [PMC free article] [PubMed] [Google Scholar]