Abstract
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. N-terminal asparagine and glutamine are tertiary destabilizing residues, in that they are enzymatically deamidated to yield secondary destabilizing residues aspartate and glutamate, which are conjugated to arginine, a primary destabilizing residue. N-terminal arginine of a substrate protein is bound by the Ubr1-encoded E3α, the E3 component of the ubiquitin-proteasome-dependent N-end rule pathway. We describe the construction and analysis of mouse strains lacking the asparagine-specific N-terminal amidase (NtN-amidase), encoded by the Ntan1 gene. In wild-type embryos, Ntan1 was strongly expressed in the branchial arches and in the tail and limb buds. The Ntan1−/− mouse strains lacked the NtN-amidase activity but retained glutamine-specific NtQ-amidase, indicating that the two enzymes are encoded by different genes. Among the normally short-lived N-end rule substrates, only those bearing N-terminal asparagine became long-lived in Ntan1−/− fibroblasts. The Ntan1−/− mice were fertile and outwardly normal but differed from their congenic wild-type counterparts in spontaneous activity, spatial memory, and a socially conditioned exploratory phenotype that has not been previously described with other mouse strains.
A multitude of regulatory circuits involve conditionally or constitutively short-lived proteins (26, 27, 44, 48, 49, 64). Features of proteins that confer metabolic instability are called degradation signals, or degrons (37, 63). The essential component of one degradation signal, termed the N-degron, is a destabilizing N-terminal residue of a protein (3). A set of N-degrons containing different N-terminal residues which are destabilizing in a given cell yields a rule, termed the N-end rule, which relates the in vivo half-life of a protein to the identity of its N-terminal residue. An N-end rule pathway is present in all organisms examined, from mammals and plants to fungi and prokaryotes (63).
In eukaryotes, an N-degron comprises two determinants: a destabilizing N-terminal residue and an internal lysine of a substrate protein (4, 32, 60). The Lys residue is the site of formation of a substrate-linked multiubiquitin chain (15, 49). The N-end rule pathway is thus one pathway of the ubiquitin (Ub) system (25–27). Ub is a 76-residue eukaryotic protein that exists in cells either free or covalently conjugated to many other proteins. The Ub system plays a role in a vast range of processes, including cell growth, division, differentiation, and responses to stress. In most of these processes, Ub acts through routes that involve the degradation of Ub-protein conjugates by the 26S proteasome, an ATP-dependent multisubunit protease (10, 17, 20, 51).
(Throughout the text, the names of mouse genes are in italics, with the first letter uppercase. The names of human and Saccharomyces cerevisiae genes are also in italics, all uppercase. If human and mouse genes are named in the same sentence, the mouse gene notation is used. The names of S. cerevisiae proteins are roman, with the first letter uppercase and an extra lowercase “p” at the end. The names of mouse and human proteins are the same, except that all letters but the last “p” are uppercase. The latter usage is a modification of the existing convention [58], to facilitate simultaneous discussions of yeast, mouse, and human proteins. In some citations, the abbreviated name of a species precedes the gene's name.)
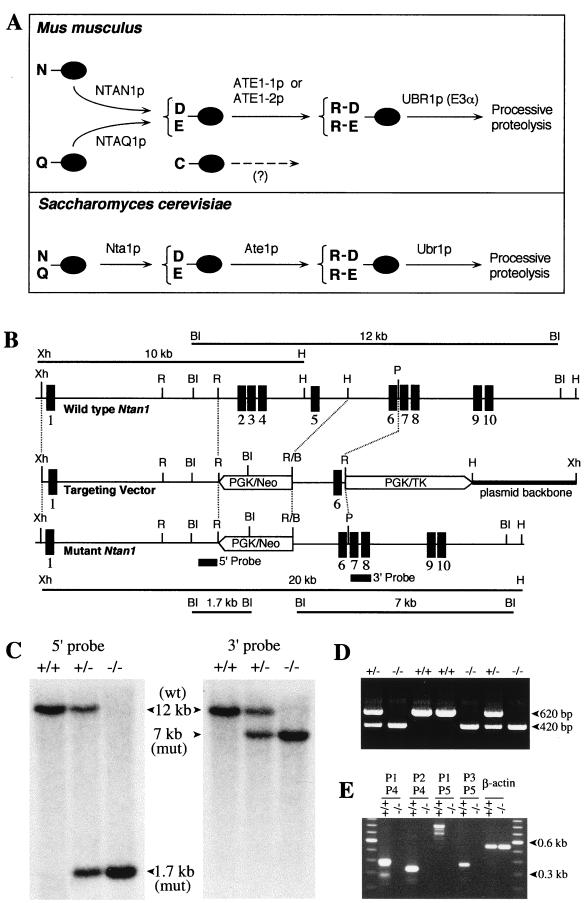
The N-end rule has a hierarchic structure. In the yeast S. cerevisiae, Asn and Gln are tertiary destabilizing N-terminal residues in that they function through their conversion, by the NTA1-encoded N-terminal amidohydrolase (Nt-amidase), into the secondary destabilizing N-terminal residues Asp and Glu (6). Destabilizing activity of N-terminal Asp and Glu requires their conjugation, by the S. cerevisiae ATE1-encoded Arg-tRNA protein transferase (R-transferase) (8, 41), to Arg, one of the primary destabilizing residues (Fig. 1A). In mammals, the deamidation step is mediated by two Nt-amidases, NtN-amidase and NtQ-amidase, which are specific, respectively, for N-terminal Asn and Gln (Fig. 1A) (24, 59). The mammalian counterpart of the yeast R-transferase Ate1p exists as two distinct species, ATE1-1p and ATE1p-2, which are produced through alternative splicing of Ate1 pre-mRNA (34). In vertebrates, the set of secondary destabilizing residues contains not only Asp and Glu but also Cys, which is a stabilizing residue in yeast (Fig. 1A) (18, 23). The primary destabilizing N-terminal residues are bound directly by the UBR1-encoded N-recognin, the targeting (E3) component of the N-end rule pathway. In S. cerevisiae, Ubr1p is a 225-kDa protein which recognizes potential N-end rule substrates through its type 1 and type 2 substrate-binding sites. The type 1 site binds the basic N-terminal residues Arg, Lys, and His. The type 2 site binds the bulky hydrophobic N-terminal terminal residues Phe, Leu, Trp, Tyr, and Ile (35, 63). Ubr1p contains yet another substrate-binding site that targets proteins such as Cup9p and Gpa1p, which bear internal (non-N-terminal) degrons (12, 54). The Ubr1 genes encoding mouse and human N-recognins, also called E3α, have been cloned (36), and mouse strains lacking Ubr1 have recently been constructed (Y. T. Kwon and A. Varshavsky, unpublished data).
FIG. 1.
Deletion-disruption of the mouse Ntan1 gene. (A) Comparison of enzymatic reactions that underlie the activity of tertiary and secondary destabilizing residues in the yeast S. cerevisiae and the mouse. N-terminal residues are indicated by single-letter abbreviations for amino acids. The ovals denote the rest of a protein substrate. The Ntan1-encoded mammalian NtN-amidase converts N-terminal Asn to Asp. N-terminal Gln is deamidated by NtQ-amidase, which remains to be isolated (see text). In contrast, the yeast Nt-amidase Nta1p can deamidate either N-terminal Asn or Gln (6). The secondary destabilizing residues Asp and Glu are arginylated by the mammalian ATE1-1p or ATE1-2p R-transferase (34). A Cys-specific mammalian R-transferase (23) remains to be identified. N-terminal Arg, one of the primary destabilizing residues, is recognized by N-recognin, the E3 component of the N-end rule pathway (63). (B) Targeting strategy. Top, partial restriction map of the mouse Ntan1 gene; middle, structure of the targeting vector; bottom, structure of the deletion-disruption Ntan1− allele. Exons are denoted by solid vertical bars. The directions of transcription of the neo and tk genes are indicated. Homologous recombination resulted in the replacement of the Ntan1 exons 2 to 5 with the neo cassette. Probes for Southern hybridization are indicated by solid rectangles. Restriction sites: Xh, XhoI; R, EcoRI; BI, BamHI; H, HindIII; P, PstI. (C) Southern analysis of BamHI-digested tail DNA from wild-type (+/+), heterozygous (Ntan1+/−), and Ntan1−/− mice. The 5′ probe yielded the 12- and 1.7-kb Ntan1 fragments for the wild-type (wt) and mutant (mut) Ntan1 alleles, respectively; the 3′ probe detected 12- and 7-kb fragments. The organization of the deletion-disruption allele was independently verified by Southern analysis of the XhoI-HindIII-digested tail DNA (data not shown). (D) PCR analysis of tail DNA. The primers were 5′-GCCACTTGTGTAGCGCCAAGTGCCAGC (for neo, forward), 5′-CTTCCCACCAAGCCTGACTGTTGATC (for Ntan1, forward) and 5′-CTTCAATTTCTGTGCTCAGCTAAGCTC (for Ntan1, reverse). (E) RT-PCR analysis of the total RNA isolated from +/+ and Ntan1−/− EF cells, using primers P1 (for exon 1), P2 (exon 2), P3 (exon 6), P4 (exon 5), and P5 (exon 10). β-Actin mRNA was used as a control, at the 20-fold-lower primer concentration in comparison to other lanes.
The known functions of the N-end rule pathway include the control of peptide import in S. cerevisiae, through the degradation of Cup9p, a transcriptional repressor of PTR2, which encodes the peptide transporter (1, 12); a mechanistically undefined role in regulating the Sln1p-dependent phosphorylation cascade that mediates osmoregulation in S. cerevisiae (47); the degradation of alphaviral RNA polymerases and other viral proteins in infected metazoan cells (19, 38); and the degradation of Gpa1p, a Gα protein of S. cerevisiae (43, 54). Physiological N-end rule substrates were also identified among the proteins secreted into the mammalian cell's cytosol by intracellular parasites such as the bacterium Listeria monocytogenes. Short half-lives of these bacterial proteins are required for the efficient presentation of their peptides to the immune system (56). Inhibition of the N-end rule pathway was reported to interfere with mammalian cell differentiation (28) and to delay limb regeneration in amphibians (61). Studies of the Ub-dependent proteolysis of endogenous proteins in muscle extracts suggested that the N-end rule pathway plays a role in catabolic states that result in muscle atrophy (39, 57). A crush injury to the rat sciatic nerve was reported to result in a ∼10-fold increase in the rate of arginine conjugation to the N termini of proteins in the nerve's region upstream of the crush site, suggesting an injury-induced increase in the concentration of R-transferase substrates and/or an enhanced activity of the N-end rule pathway (65).
Physiological substrates of either yeast or metazoan Nt-amidases and R-transferases are unknown. Engineered N-end rule substrates, including substrates of Nt-amidases and R-transferases, can be produced in vivo through the Ub fusion technique, in which a Ub-X reporter fusion is cleaved by deubiquitylating enzymes (DUBs) (66) after the last residue of Ub, yielding a reporter bearing the desired N-terminal residue X (3, 63).
The mouse Asn-specific NtN-amidase is encoded by the 17-kb Ntan1 gene. The 1.6-kb Ntan1 mRNA specifies the 310-residue NtN-amidase (24). In the present work, we characterized the expression and intracellular localization of NtN-amidase. We also constructed mouse strains bearing a homozygous deletion-disruption of Ntan1 and showed that these mice lacked both NtN-amidase and the Asn-specific branch of the N-end rule pathway. The Ntan1−/− mice were fertile and outwardly normal but were found to differ from their congenic wild-type counterparts in spontaneous activity and spatial memory. Among these differences was a socially conditioned exploratory phenotype of Ntan1−/− mice that has not been previously described, to our knowledge, with other mouse strains.
MATERIALS AND METHODS
Construction of mouse strains lacking NtN-amidase.
Genomic Ntan1 DNA fragments were subcloned from the P1 phage DNA containing the strain C129-derived mouse Ntan1 gene (24). The 1,462-bp HindIII-PstI fragment containing exon 6 and its flanking introns was used as the vector's short homology arm. The 7.2-kb XhoI-NotI fragment containing exon 1 and its flanking intron was used as the long homology arm. To construct the targeting vector, a 1,462-bp HindIII-PstI fragment containing exon 6 and its flanking introns (the vector's short arm) and the phosphoglycerate kinase gene (PGK)-thymidine kinase (tk) cassette of pPNT (a gift from R. C. Mulligan, Harvard Medical School, Boston, Mass.) were inserted into pPGKRN containing the wild-type PGK-neo (neomycin resistance gene) cassette (a gift from R. Jaenisch, Whitehead Institute, Cambridge, Mass.), yielding pPGK-SA. The vector's 7.2-kb long arm was produced by joining, within pPGK-SA, a 4.8-kb XhoI-EcoRI fragment to its flanking 2.6-kb EcoRI-EcoRI fragment containing exon 1 and flanking intron pPGK-SA, yielding the Ntan1 targeting vector pNTAN1-KO (Fig. 1B). The XhoI-linearized targeting vector (Fig. 1B) was electroporated into CJ7 embryonic stem (ES) cells. Selection with G418 (at 0.4 mg/ml) and 1-(2′-deoxy, 2′ fluoro-β-d-arabinofuranosyl)-5-iodouracil (FIAU; at 0.4 μM) was started 24 h after electroporation. Correctly targeted ES cells were identified by PCR and Southern hybridization with the 5′ and 3′ probes (Fig. 1C and D). Cells of the 12 independent ES cell clones were injected into 3.5-days-postcoitum C57BL/6J blastocysts. The resulting male chimeras were bred with C57BL/6J females to test for germ line transmission of the mutated Ntan1 gene. The Ntan1+/− mice resulting from this cross (6 out of 12 independent ES clones were found to populate germ line in these tests) were intercrossed to produce Ntan1−/− mice. Alternatively, the initial male chimeras were mated with 129/SvEv females, yielding, through the analogous series of steps, Ntan1−/− mice in the strain 129 background. All of the behavioral tests were carried out with Ntan1−/− mice in the strain 129 background. For genotyping, the tail-derived DNA was analyzed by PCR, or digested with either BamHI or HindIII/XhoI, and analyzed by Southern hybridization. A 0.75-kb PstI fragment and a 0.9-kb PstI fragment (indicated in Fig. 1B) were used as the 5′ and 3′ hybridization probes, respectively.
Northern and RT-PCR analyses of RNA.
Total RNA was isolated from brain, testis, and embryonic fibroblasts (EF cells) of wild-type and Ntan1−/− mice as described elsewhere (34). RNA was fractionated by electrophoresis in 1% formaldehyde-agarose gels, blotted onto Hybond N+ (Amersham), and hybridized with 32P-labeled probes specific for different regions of the Ntan1 cDNA (GenBank accession no. U57692): probe a, nucleotide (nt) 34 to 900; probe b, nt 118 to 450; probe c, nt 118 to 900; probe d, nt 34 to 450; probe e, nt 470 to 670; and probe f, nt 680 to 900. Alternatively, reverse transcription-PCR (RT-PCR) was carried out (2), with first-strand cDNA synthesized using Superscript II polymerase (GIBCO, Frederick, Md.). PCR was done using primers specific for different regions of the Ntan1 cDNA: primer P1, 5′-ATGCCACTGCTGGTGGATGG-GCAG (forward); P2, 5′-GAGCCAGACTTCTCAGAGGTCAG (forward); P3, 5′-GACA-TTCACTTAGTGACATTATG (forward); P4, 5′-TTCTGTGACAGCTGCCTGTCATC (reverse); and P5, 5′-CATCAAGGTAGATCTAATATGTTC (reverse). The ratio of mouse Ate1-1 and Ate1-2 mRNAs was determined as described elsewhere (34).
Whole-mount in situ hybridization.
Wild-type and Ntan1−/− embryos were staged, fixed, and processed for in situ hybridization as described elsewhere (16). A 0.3-kb fragment of the Ntan1 cDNA (nt 108 to 448) that was encompassed by the deleted region in the Ntan1−/− allele was subcloned into XbaI-XhoI sites of pBluescript II SK+ (Stratagene), and the resulting plasmid, pMR27, was used as a template for synthesizing antisense RNA probe labeled with digoxigenin (Roche Molecular Biochemicals, Indianapolis, Ind.).
Localization of NTAN1p-GFP.
The mouse Ntan1 open reading frame (ORF) was subcloned into the XhoI and AgeI sites of pEGFP-N1 (Clontech), yielding pNTAN1-GFP, which expressed the NTAN1p green fluorescent protein (GFP) fusion from the cytomegalovirus promoter. NIH 3T3 cells (ATCC 1658-CRL) were grown as monolayers in Dulbecco's modified Eagle medium (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum. Cells were grown to ∼15% confluence on glass coverslips for 24 h prior to transfection with either the GFP-expressing control vector pEGFP-N1 or pNTAN1-GFP, using Lipofectamine (GIBCO) and the manufacturer-supplied protocol. Cells were incubated for 5 h at 37°C in serum-free DMEM containing DNA and Lipofectamine. An equal volume of medium containing 20% serum was then added, and the cells were grown for another 12 to 20 h at 37°C. Cells were fixed with 2% formaldehyde in phosphate-buffered saline, and GFP fluorescence was examined in a Zeiss Axiophot microscope.
Immortalization of EF cells, transfection, and pulse-chase analysis.
EF cells were isolated from either Ntan1−/− or +/+ 13.5-day-old (e13.5) embryos as described elsewhere (52) and grown in DMEM–F-12 (GIBCO) supplemented with 15% fetal bovine serum, antibiotics, and 2 mM l-glutamine. They were immortalized using simian virus 40 (14) and then transiently transfected, using Lipofectamine PLUS (GIBCO), with plasmid pRC/dhaUbXnsP4βgal, which expressed DHFR (dihydrofolate reductase)-HA (hemagglutinin)-UbR48-X-nsP4βgal test proteins (X = Met, Asn, Gln, or Arg) (40) (see Results). Cells were labeled with Tran35S-label (New England Nuclear, Boston, Mass.), followed by a chase for 0, 1, and 2 h in the presence of cycloheximide, preparation of extracts, immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), autoradiography, and quantitation with PhosphorImager, essentially as described elsewhere (40).
X-DHFR test proteins.
35S-labeled X-DHFR proteins (X = Asn, Gln, or Asp) were prepared as described elsewhere (24). Briefly, plasmids pSG4, pSG41, and pSG44, expressing, respectively, Ub-Asn-DHFR, Ub-Asp-DHFR, and Ub-Gln-DHFR from the Ptrc promoter (24), were transformed into Escherichia coli JM101 carrying pJT184, which expressed Ubp1p, a DUB of S. cerevisiae (62). Cells of a 50-ml culture grown at 37°C to an A600 of ∼0.9 in Luria broth plus ampicillin (40 μg/ml) and chloramphenicol (20 μg/ml) were pelleted and resuspended in 50 ml of M9 supplemented with glucose (0.2%), thiamine (2 μg/ml), ampicillin (40 μg/ml), 1 mM isopropylthio-β-d-galactoside (IPTG), and methionine assay medium (Difco). The suspension was shaken for 1 h at 37°C, followed by the addition of 1 mCi of Tran35S-label (ICN) and further incubation for 1 h at 37°C. Unlabeled l-methionine was then added to 1 mM, and shaking was continued for another 10 min. The cells were harvested, and lysates were prepared by the addition of lysozyme and Triton X-100, followed by centrifugation (24). The supernatant, containing [35S]X-DHFR (X = Asn, Asp, or Glu) was purified by affinity chromatography on a methotrexate column (Pierce) (0.5-ml bed volume). [35S]X-DHFRs were examined by SDS-PAGE and found to be greater than 95% pure.
IEF assay for amidase activity.
Extracts from wild-type or Ntan1−/− EF cells were prepared by homogenization in a mixture containing 0.01% Triton X-100, 10% glycerol, 125 mM KCl, 7.5 mM MgCl2, 1 mM dithiothreitol, 0.25 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 130 mM Tris-HCl (pH 7.5), and the protease inhibitors antipain, chymostatin, leupeptin, pepstatin, and aprotinin (Sigma), each at 25 μg/ml. For the deamidation assay, 5 μl of 35S-labeled X-DHFR (0.5 mg/ml in storage buffer; X = Asn, Asp, or Glu) was mixed with 20 μl of the extract, incubated for 2 h at 37°C, and placed on ice. Samples (5 μl) were applied onto isoelectrofocusing (IEF)-PAGE plates (pH 3.5 to 9.5; Pharmacia, Piscataway, N.J.) precooled to 10°C. IEF was carried out for 80 min at 30 W in a cooled IEF apparatus (Hoefer, San Francisco, Calif.). The plates were soaked in 100 ml of 10% CCl3COOH–5% 5-sulfosalicylic acid, stained with Coomassie blue to detect IEF markers (Pharmacia), and autoradiographed. 35S in the bands of more acidic (deamidated) and more basic (initial) X-DHFR species was quantitated with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Behavioral tests.
Mice were kept on a 0600- to 1800-h light cycle and tested during the light period. For the rotarod, weight retention, coat hanger, and platform-leaving tests, strain 129 Ntan1−/− mice and their congenic +/+ littermates (produced through matings of Ntan1−/+ mice) were used. Nonlittermates were also used for the platform-leaving test (see below). For the shuttlebox and passive avoidance tests, the elevated plus-maze test, the open-field test, the Morris maze, the radial arm maze, and the Lashley maze, strain 129 +/+ and congenic Ntan1−/− mice (produced, respectively, through +/+ × +/+ and Ntan1−/− × Ntan1−/− matings) were used. The same series of tests were repeated 6 weeks or more after the initial experiment, using both male and female mice. To determine whether the Ntan1 genotype affected long-term memory, mice were retested in the Morris maze, Lashley maze, and shuttlebox avoidance test at least 7 weeks after their original learning. The data were evaluated by analysis of variance. Significance was set at the 0.05 level; all behavioral differences described in Results were at this or a higher level of statistical significance.
(i) Rotarod test.
The apparatus (UGO, Basile, Italy) consisted of a motor-driven rod 3 cm in diameter that carried five compartments, separated by walls 25 cm in height. One pair of littermates was placed on the rotating rod in each compartment. Mice were placed on the rod at either 10 or 20 rpm; the cutoff time was 2 min. Each mouse was tested on the rod six times, at 1-h intervals.
(ii) Weight retention.
Mice were suspended by the tails and made to grab a wire loop (wire diameter, 2 mm; weight, 40 g) 70 cm above the floor. Time elapsed before the animal released the loop was measured. Each mouse was tested five times a day, at 1-h intervals, for a total of 3 days.
(iii) Coat hanger test.
A wire coat hanger (wire diameter, 2 mm; length of side bars, 29 cm; length of the horizontal bar, 45 cm) suspended 75 cm above the table was used to examine motor coordination (11). A trial began by allowing the mouse to grab the middle of the horizontal bar with two front paws. The time elapsed before the animal grabbed the horizontal bar with all four paws and the time before the two front paws reached one of the side bars were determined. Each mouse in the trial was tested twice a day, at 6-h intervals, for 9 days.
(iv) Shuttlebox avoidance, passive avoidance, and elevated plus-maze tests.
For the shuttlebox and passive avoidance tests, the Gemini avoidance system (San Diego Instruments, San Diego, Calif.) was used. The procedures are described in Results.
(v) Open-field test.
For the open-field activity measurements, an Omnitech Digiscan animal activity monitor was used. Each mouse was placed into a square chamber (20 by 20 cm) for one 9-min session, and the movements were tabulated along the x and y axes, using infrared sensors.
(vi) Morris maze.
A mouse was placed into a circular black water tub (diameter, 123 cm; temperature, 19 to 21°C) from one of the four quadrants and had to find a platform (diameter, 23 cm) submerged 1 cm below the surface of water and set 23 cm from the wall. There were no cues within the maze, and so the animal had to use extramaze spatial cues in guiding itself to the platform in repeated trials (46). An animal was given four trials per day (maximum of 45 s per trial), one from each of the four locations, their order determined quasi-randomly. Each mouse was tested for 5 days, with the platform in a fixed position. On day 6, the platform was moved to the quadrant diagonally opposite its previous location, and each mouse was given four trials, as before. This regimen is referred to as reversal learning. The mice were retested (with the platform in the original position) 7 or 8 weeks later. The animal's path was traced on an electronic digitizing tablet containing a template of the maze (22). The following parameters were recorded: total time, distance traveled, average speed, and percentage of time spent in each quadrant of the maze.
(vii) Spatial radial arm maze.
The apparatus and testing procedure were described previously (30). Hidden escape platforms were placed at the ends of four of the eight identical arms which radiated from the central area. During training the animal was released from a start arm and had to find one of the escape platforms. Once a platform was found, the arm containing that platform was blocked. The training session consisted of four trials in which a mouse had to locate all four platforms. This was followed by 11 testing sessions which differed from the training session only by removing the platform while the mouse was in its home cage between trials. Thus, on successive trials, the animal had to remember which alleys it had previously entered and not enter those alleys again. The score was total errors obtained by summing together Working Memory Correct errors, Working Memory Incorrect errors, and Reference Memory errors (30).
(ix) Nonspatial radial arm maze learning.
The nonspatial swimming radial arm maze apparatus and testing procedure were similar to the above spatial version, except for the following. Each of the arms was painted in a different black/white pattern: solid black, solid white, vertical stripes, horizontal stripes, black with white polka dots, white with black polka dots, zigzags, and the galvanized steel gray color (the start arm). The patterned arms which contained platforms were different for each mouse but remained constant throughout the testing of a given mouse. For the training session (session 1), the entrance to the arm with the just chosen platform was blocked; in addition, the maze was quasi-randomly rotated, so that each patterned arm now pointed toward a different place in space. As in the spatial version, the test sessions (sessions 2 to 12) were the same as the training session, except that while the mouse was in its home cage, each platform it found was removed from the maze, and the arm's entrance remained open for the remainder of the session. Over eight trials, or two sessions, each patterned arm pointed toward each of the eight spatial locations in the testing room. The errors were scored as above.
(x) Lashley maze learning.
This maze contained cul-de-sacs that an animal had to learn to avoid, along with choices that required the animal to learn making correct left or right turns (T-choices). A water version of the maze was used, with the temperature at 19 to 20°C (21). All mice were given one trial per day for 5 days. An animal can learn this maze using two mutually nonexclusive strategies: by memorizing extramaze spatial cues, or by memorizing the sequence of correct left and right turns. The measures of learning included the learning index, defined as the number of correct entries divided by total number of entries, the number of cul entries (entries into dead ends) when swimming forward (i.e., toward the goal), number of forward T-choice errors, and the total number of backward errors. Retention testing was done 7 weeks later for experiment 1 and 8 weeks later for experiment 2.
(xi) Platform-leaving test.
To compare the exploratory activities of two previously untested mice on the same platform, we devised the platform-leaving test. One Ntan1−/− mouse of strain 129 and one congenic +/+ mouse of the same gender (either a littermate or a nonlittermate) were placed, at the same time, on the platform (16 by 22 cm) 2.7 cm above the surface of a laboratory bench. For each test, the platform was covered with fresh white paper. Mice were allowed to either explore or step down from the platform up until the cutoff time of 3 min. (Step-down was recorded when all four paws of a mouse were no longer in contact with platform.) The number and genotypes of mice leaving the platform first, and the number of mice not leaving by 3 min, were recorded. A total of three trials, at 1-h intervals, were carried out for every pair of mice tested on a given day. Each set of tests employed ∼10 pairs of mice 2 to 4 months old. Four independent sets of tests, with previously untested mice, were carried out over a period of 6 months.
RESULTS
Construction of Ntan1−/− mice.
A deletion-disruption allele of the ∼17-kb mouse Ntan1 gene was constructed by replacing Ntan1 exons 2 to 5 with a neo cassette (Fig. 1B). Exons 2 to 5 encoded a 118-residue region of the 310-residue NTAN1p (NtN-amidase) that was significantly conserved among the NTAN1p proteins of different species, including the plant Arabidopsis thaliana, the fly Drosophila melanogaster, and mammals (data not shown). Of the ∼1,000 ES cell clones (strain 129/CJ7) resistant to both G418 and FIAU, 33 clones contained the expected mutation, as verified by PCR and Southern analyses (data not shown). Twelve of these correctly targeted ES cell clones were used to generate male chimeras, and in six of them the Ntan1− allele was transmitted through the germ line. Male chimeras were mated with either 129/SvEv or C57BL/6 females, yielding Ntan1+/− heterozygotes. Intercrosses of heterozygous mice produced Ntan1−/− progeny at the expected frequency of approximately one in four (Fig. 1C and D), indicating that the absence of NTAN1p did not increase embryonic lethality. The behavioral tests described below used exclusively Ntan1−/− mice produced through matings in the strain 129 background.
Expression of Ntan1 and intracellular localization of NTAN1p.
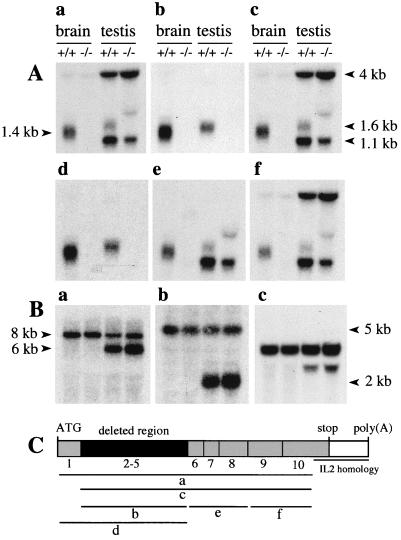
Several regions of Ntan1 transcripts were analyzed using RT-PCR with RNA from wild-type and Ntan1−/− EF cells. The level of Ntan1-derived RNA in the Ntan1−/− EF cells was below the RT-PCR detection threshold (Fig. 1E and data not shown). Northern hybridization analyses, using probes encompassing either exons 2 to 5 (the deleted region) or exons 1 to 5 detected the 1.4- and 1.6-kb Ntan1 mRNAs, respectively, in the brains and testes of +/+ mice; the +/+ testes also contained the Ntan1-derived RNAs of 1.1 and 4 kb (Fig. 2A). No Ntan1 transcripts were detected in the brains and testes of Ntan1−/− mice with these probes (Fig. 2Ab and d).
FIG. 2.
Northern hybridization analysis of the total RNA isolated from +/+ and Ntan1−/− brains and testes. (A) Hybridization using probes a to f that encompassed different regions of the Ntan1 cDNA (indicated in panel C). (B) The same Northern blots were hybridized with probes specific, respectively, for the mouse Ubr1, Ate1, and β-actin cDNAs. (C) Exons of Ntan1 and the hybridization probes used. The sequence of a 208-bp segment of the Ntan1 cDNA (nt 896 to 930), termed the IL-2 homology region, is 98.6% identical to the sequence of a 206-bp segment in the 3′-flanking untranslated region of the mouse Il2 gene, which encodes IL-2 (24).
Similar analyses but using probes encompassing exons 1 to 10 or 2 to 10 (except for the interleukin-2 [IL-2] similarity region of exon 10 [24]) detected at most trace amounts of Ntan1-derived transcripts in the brains of Ntan1−/− mice (Fig. 2Aa and c). However, in the testes of Ntan1−/− mice, the same probes detected 1.1- and 4-kb Ntan1-derived, testis-specific transcripts but not the 1.6-kb transcript (Fig. 2Aa and c). Further mapping, using the probes shown in Fig. 2C, indicated that the 1.1- and 4-kb transcripts hybridized to Ntan1 exons 6 to 10 and exons 9 and 10, respectively (Fig. 2Ae and f). These transcripts were detected with antisense but not sense probes (data not shown), indicating a direction of transcription identical to that for the full-length Ntan1 mRNA. Thus, the testes but not the brains of Ntan1−/− mice contained a set of Ntan1-derived transcripts that lacked the 5′ half of the Ntan1 ORF (including the region deleted in Ntan1−/− mice) and encompassed its 3′ half. Apparently the same transcripts were present in the +/+ testes (Fig. 2Ae and f). Whether these testis-specific transcripts are physiologically relevant remains to be determined.
We also examined by Northern analysis whether the expression of other components of the N-end rule pathway was altered in the Ntan1−/− mice. No changes in the expression of either Ubr1 (encoding the E3 of the N-end rule pathway) or Ate1 (encoding R-transferase [34]; see the introduction) were detected in the brains and testes of Ntan1−/− mice (Fig. 2B). The expression of Ubr2 and Ubr3 (homologs of Ubr1; Kwon and Varshavsky, unpublished data) and of mHR6B, encoding the E214K Ub-conjugating (E2) enzyme, was also unchanged in the Ntan1−/− mice (data not shown). In addition, no alterations in the ratio of Ate1-1 mRNA to Ate1-2 mRNA, which encode two splicing-derived forms of the mouse R-transferase (34), was detected in the brains, testes, and EF cells of Ntan1−/− mice (data not shown).
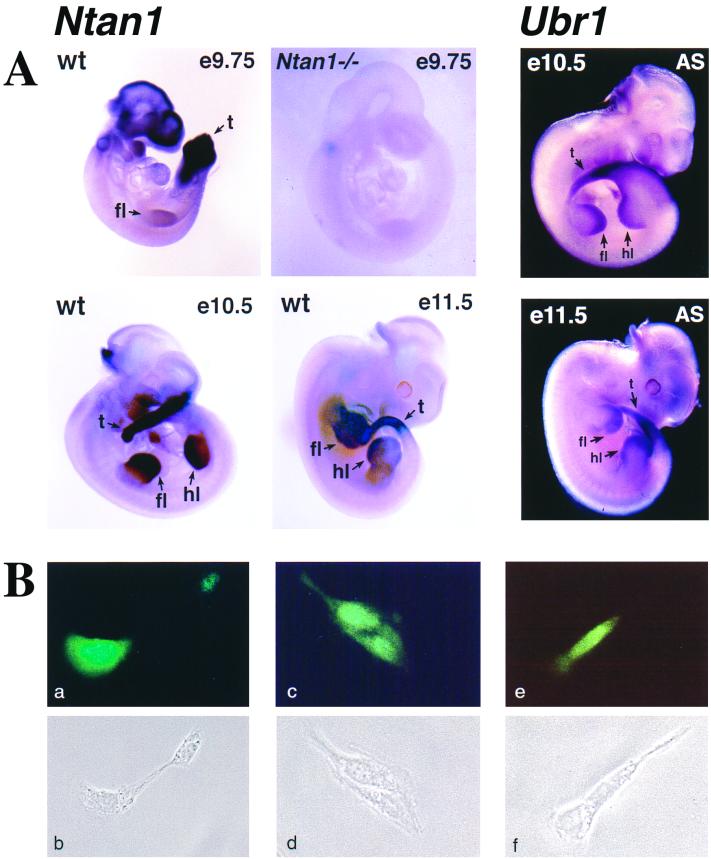
Ntan1 was expressed in most if not all tissues of adult mice (24). Whole-mount in situ hybridization was used to examine the expression of Ntan1 in +/+ mouse embryos. In e9.75 embryos, Ntan1 was most strongly expressed in the branchial arches, the tail bud, and the forelimb buds; the same probe detected no signal in the Ntan1−/− embryos (Fig. 3A). In e10.5 and e11.5 embryos, the expression of Ntan1 became high in the hindlimb buds as well; this expression pattern was indistinguishable from that of Ubr1 (Fig. 3A), consistent with the assignment of NTAN1p and UBR1p to the same pathway.
FIG. 3.
Expression of Ntan1 mRNA and localization of NTAN1p. (A) Whole-mount in situ hybridization of wild-type (wt) and Ntan1−/− embryos (left four panels, light background) with an antisense RNA probe derived from a 0.3-kb fragment of the Ntan1 cDNA (nt 108 to 448) that was absent from the Ntan1−/− allele. The regions of high Ntan1 expression in the tail buds (t), forelimb buds (fl), and hindlimb buds (hl) are indicated. The right two panels (dark background) show the results of in situ hybridization with an antisense (AS) Ubr1 cDNA probe. (B) Intracellular localization of mouse NTAN1p. NIH 3T3 cells were transiently transfected with a plasmid expressing the NTAN1p-GFP fusion (see Materials and Methods). Typical fluorescence patterns (a, c, and e) and the matching phase-contrast images (b, d, and f) are shown (see text).
To determine the intracellular location of NTAN1p, it was expressed in mouse 3T3 cells as a fusion to the N terminus of GFP. Whereas the free 26-kDa GFP was distributed uniformly throughout the cell, the 65-kDa NTAN1p-GFP was enriched in the nucleus, was also present in the nucleus-proximal cytoplasm, but was apparently much less abundant at the cells' periphery (Fig. 3B and data not shown).
Ntan1−/− mice lack NtN-amidase and the asparagine-specific branch of the N-end rule pathway.
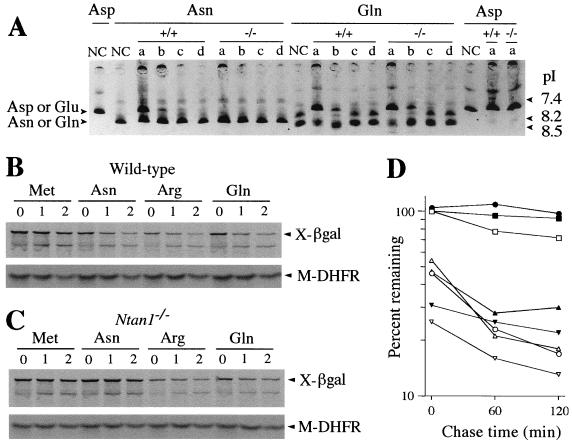
To determine whether Ntan1−/− EF cells lacked NtN-amidase activity, we used a previously described assay (24). The 35S-labeled X-DHFR test proteins (X = Asn, Gln, or Asp) were incubated with a whole-cell extract and thereafter fractionated by IEF, which separated Asn-DHFR and Gln-DHFR from Asp-DHFR and Glu-DHFR, respectively. Incubation of Asn-DHFR with the extract from +/+ EF cells shifted the pI of this protein to that of Asp-DHFR. In contrast, the pI of Asn-DHFR remained unchanged after an otherwise identical incubation with the same amount of extract from Ntan1−/− EF cells (Fig. 4A). The pI of Gln-DHFR shifted to that of Glu-DHFR after incubations with either +/+ or Ntan1−/− extracts (Fig. 4A). Thus, Ntan1−/− EF cells lacked NtN-amidase activity but contained wild-type amounts of the Gln-specific NtQ-amidase. The latter result indicated that the putative mammalian NtQ-amidase (which remains to be isolated and cloned) is encoded by a gene distinct from Ntan1.
FIG. 4.
Mouse Ntan1−/− cells lack NtN-amidase and are unable to degrade the normally short-lived N-end rule substrates bearing N-terminal Asn. (A) 35S-labeled, purified X-DHFR test proteins (X = Asn, Gln, or Asp) were incubated for 2 h at 37°C with buffer alone (negative controls [NC]) or with extracts from either wild-type or Ntan1−/− EF cells, followed by IEF and autoradiography. The assays were carried using either the initial extracts (lanes a) or the same extracts diluted with buffer 10-, 100-, and 1,000-fold (lanes b, c, and d, respectively). The IEF positions of X-DHFRs bearing N-terminal Asp or Glu versus Asn or Gln are shown on the left. The corresponding pH values are indicated on the right. (B) Immortalized +/+ and Ntan1−/− EF cells (see Materials and Methods) were transiently transfected with plasmid pRC/dhaUbXnsP4βgal, which expressed DHFR-HA-UbR48-X-nsP4βgal test proteins (X = Met, Asn, Gln, or Arg) (40). These proteins were cotranslationally cleaved in vivo by DUBs, yielding the long-lived reference protein DHFR-HA-UbR48 and the test protein X-nsP4βgal (X = Met, Asn, Gln, or Arg) (see text). Cells were labeled with [35S]methionine-cysteine, followed by a chase for 0, 1, and 2 h (as indicated at the top) in the presence of cycloheximide, preparation of extracts, immunoprecipitation, SDS-PAGE, autoradiography, and quantitation, essentially as described elsewhere (40). The bands of X-nsP4βgal proteins and the DHFR-HA-UbR48 reference protein are indicated on the right as X-βgal and DHFR. (C) Same as panel B but with immortalized Ntan1−/− EF cells. (D) Quantitation of the in vivo degradation of X-nsP4βgal test proteins using the reference-based pulse-chase patterns in panels B and C (see Materials and Methods). The amounts of 35S in an X-nsP4βgal protein, relative to 35S in the DHFR-HA-UbR48 reference protein at the same time points, were plotted as percentages of this ratio for Met-nsP4βgal at time zero. (Met-nsP4βgal bore a stabilizing N-terminal residue.) Open and closed symbols, wild-type and Ntan1−/− EF cells, respectively; □ and ■, Met-nsP4βgal; ○ and ●, Asn-nsP4βgal; ▿ and ▾, Arg-nsP4βgal; ▵ and ▴, Gln-nsP4βgal.
To examine the in vivo degradation of N-end rule substrates in the absence of NtN-amidase, the immortalized +/+ and Ntan1−/− EF cell lines were transiently transfected with plasmids that expressed X-nsP4βgal test proteins (X = Met, Asn, Gln, or Arg). An X-nsP4βgal was expressed as part of a fusion containing the HA-tagged reference protein DHFR. In this previously developed UPR (Ub protein reference) technique (40, 60), a DHFR-HA-UbR48-X-nsP4βgal fusion is cotranslationally cleaved by DUBs at the Ub-X-nsP4βgal junction, yielding the long-lived DHFR-HA-UbR48 reference protein and the test protein X-nsP4βgal. In the UPR technique, the reference protein serves as an internal control for the levels of expression, immunoprecipitation yields, sample volumes, and other sources of sample-to-sample variation, thereby increasing the accuracy of pulse-chase assays (40, 60).
The previously introduced term IDx (initial decay, i.e., the extent of degradation of a protein during the pulse of x minutes [40]) was used to describe the in vivo decay curves of test proteins. The term IDx is superfluous in the case of a strictly first-order decay, which is defined by a single half-life. However, the in vivo degradation of most proteins deviates from first-order kinetics. Specifically, the rate of degradation of short-lived proteins can be much higher during the pulse, in part because a newly labeled (either nascent or just completed) polypeptide is conformationally immature and consequently may be targeted for degradation more efficiently than its mature counterpart. This enhanced early degradation, previously termed the zero-point effect (5), is described by the parameter IDx (40). It was found that a large fraction of the zero-point effect results from the cotranslational degradation of nascent (being synthesized) polypeptides, which never reach their mature size before their destruction by processive proteolysis (G. Turner and A. Varshavsky, unpublished data).
Asn-nsP4βgal was metabolically unstable in +/+ EF cells: its ID10 (percent degradation during the 10-min pulse) was ∼45%, in comparison to ID10 of the long-lived Met-nsP4βgal, taken as 100% (Fig. 4B and D). However, the same Asn-nsP4βgal was completely (and reproducibly) stabilized in the Ntan1−/− EF cells, its pulse-chase pattern becoming indistinguishable from that of the normally long-lived Met-nsP4βgal. In contrast, both Gln-nsP4βgal and Arg-nsP4βgal were comparably short-lived in both +/+ and Ntan1−/− cells (Fig. 4B to D). Taken together with the measurements of NtN-amidase enzymatic activity (Fig. 4A), these data indicated that the Ntan1−/− EF cells, in contrast to congenic wild-type cells, lacked the Asn-specific branch of the N-end rule pathway but retained the rest of this pathway.
Motor coordination and spontaneous activity of Ntan1−/− mice.
The Ntan1−/− mice were fertile, apparently healthy, and of similar size and weight as +/+ littermates, and they cared for their offspring. These mice oriented to sound; their limb movements and behavior appeared to be indistinguishable from those of congenic +/+ mice. Histological examination of Ntan1−/− tissues (small intestine, liver, pancreas, adrenal gland, thyroid gland, kidney, ovary, testis, heart, spleen, thymus, skeletal muscle, brain, and sciatic nerve) did not detect abnormalities. No significant differences were observed between +/+ and Ntan1−/− thymocytes in the ability to undergo apoptosis in response to either radiation or dexamethasone, suggesting that Ntan1−/− mice were not impaired in apoptotic responses. Thus far, the only nonbehavioral differences between +/+ and congenic Ntan1−/− mice were a weaker mitogenic response of Ntan1−/− splenocytes and thymocytes to phytohemagglutinin and hypersensitivity of Ntan1−/− mice to bacterial lipopolysaccharide (data not shown). In addition, over the last 1.5 years the frequency of natural death among Ntan1−/− mice was ∼50% higher than among congenic +/+ mice.
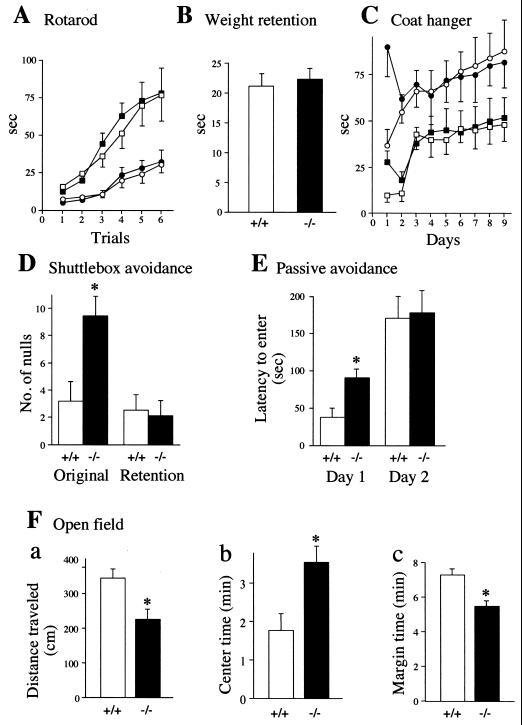
Motor coordination of Ntan1−/− and congenic +/+ mice was compared using the rotarod apparatus. No difference in the ability of mice to stay on a rotating horizontal rod was observed between the Ntan1−/− and +/+ strains (Fig. 5A). Comparisons of physical strength (weight retention test), of both coordination and strength (coat hanger test), and of walking patterns (hindpaw footprint test) also did not distinguish between Ntan1−/− and +/+ mice (Fig. 5B and C and data not shown). These and related observations indicated that Ntan1−/− mice were not impaired in either motor coordination, physical strength, or the learning ability required for improved performance in repeated trials (Fig. 5A and C).
FIG. 5.
Normal motor coordination and reduced spontaneous activity of Ntan1−/− mice. Open symbols: +/+ mice; solid symbols: congenic Ntan1−/− mice (see Materials and Methods). Statistically significant differences (P < 0.05) are indicated by ∗. Experiments used 24 Ntan1−/− mice and their congenic +/+ littermates (A to C) or nonlittermates (D to G). (A) Rotarod test. Data show time elapsed before the animals fell from a horizontal rod rotating at 10 rpm (squares) or 20 rpm (circles). No significant differences were found between +/+ and Ntan1−/− mice. (B) Weight retention test. Data show time elapsed before the animals released a hook. The height of each bar represents the time averaged from 360 trials. There were no significant differences between +/+ and Ntan1−/− mice. (C) Coat hanger test. Squares, time elapsed before the animal grabbed the horizontal bar with both rear paws; circles, time elapsed before the front two paws reached one of the side bars. Each data point is an average of 48 trials. There were no significant differences between +/+ and Ntan1−/− mice. (D) Shuttlebox avoidance test. Shown are the number of null responses per 50 daily trials in which the mouse remained in the original compartment of the shuttlebox and received 20 s of electric shock during the original learning and retention testing 7 to 10 weeks later. (E) Passive avoidance test, measuring time before entering a dark chamber after an automated guillotine door opened, exposing the dark chamber. On day 1 the mice were shocked after entering the dark chamber; 13 Ntan1−/− mice and 12 congenic +/+ mice were used. (F) Open-field test. Shown are the total distance traveled by the animal during the observation time of 9 min (a), time (out of 9 min in total) that the animal spent in the center area (b), and time (out of 9 min in total) that the animal spent within 1 cm of the walls. This set of tests used 28 Ntan1−/− mice and 27 congenic +/+ mice.
The locomotor activity of Ntan1−/− mice was assayed in the open-field test. By several criteria, Ntan1−/− mice exhibited significantly lower spontaneous activity than the congenic wild-type mice (Fig. 5F). Specifically, Ntan1−/− mice traveled significantly less distance (Fig. 5Fa), spent significantly more time near the field's center (Fig. 5Fb), and consequently spent less time within 1 cm from the walls (Fig. 5Fc). These findings will be considered in conjunction with data from the platform-leaving test (see Fig. 6 and Discussion).
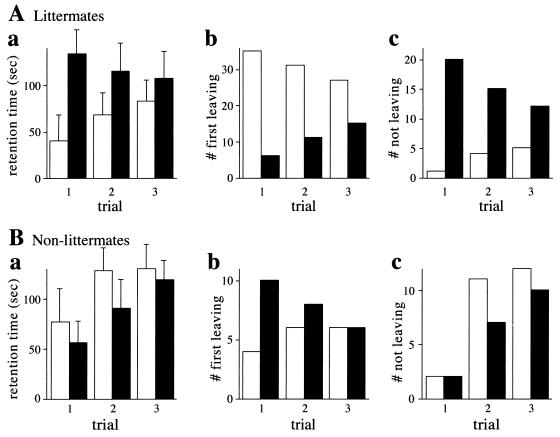
FIG. 6.
Socially conditioned differences in exploratory behavior between +/+ and congenic Ntan1−/− mice. Open bars, +/+ mice; solid bars, congenic Ntan1−/− mice. In the platform-leaving test, one Ntan1−/− mouse and one +/+ mouse (either a littermate or a nonlittermate) were placed, at the same time, on a 16- by 22- by 2.7-cm platform. Mice were allowed to either explore or step down from the platform until the cutoff time of 180 s. Bars show the time by which each animal left the platform (a), the number of animals, of each genotype, leaving first (b), and the number of animals, of each genotype, not leaving by the cutoff time (c). Four independent sets of tests (at least 10 pairs of mice per test) were carried out over 6 months, using 44 pairs of +/+ and Ntan1−/− littermates, which were produced through heterozygous matings and identified by genotyping ∼700 mice. The results differed by less than 15% among the independent tests. (B) Same as panel A except with pairs of nonlittermates (24 +/+ mice and 24 Ntan1−/− mice).
Active and passive avoidance learning.
The shuttlebox avoidance test compared shock-motivated learning in +/+ and congenic Ntan1−/− mice. In this active avoidance learning procedure, a light-conditioned stimulus precedes, by 5 s, the unconditioned stimulus of electric shock, which remains on for 20 s. The animal can (i) cross to the safe compartment within the 5-s conditioned-stimulus interval, thus avoiding shock; (ii) cross over during the 20-s unconditioned-stimulus interval, to escape the ongoing shock; or (iii) not cross over and receive the full 20 s of shock (the latter outcome is scored as the null response). Neither +/+ nor Ntan1−/− mice showed any evidence of learning over the 5 days of testing. However, there were performance differences in the distribution of their responses. The two groups averaged only four avoidance responses per day, but the +/+ mice exhibited more escape responses and therefore fewer null responses than the Ntan1−/− mice (Fig. 5D). Neither group showed evidence of learning during the 5 days of retention testing, but the number of null responses by Ntan1−/− mice decreased significantly in comparison to their performance during learning (data not shown), indicating that memory consolidation had occurred in this group between the original testing and retesting 7 to 10 weeks later. Strain 129 mouse substrains are known to perform poorly in active avoidance tests (7, 53).
In the shuttlebox text, an electric shock is used to compel the animal to make an active response. By contrast, in passive learning, the shock is used to compel the animal to inhibit its behavior. The mouse was placed into a chamber. Ten seconds later, the chamber was illuminated and a guillotine door opened, exposing a dark chamber. After the animal moved into the dark (the natural behavior), the guillotine door closed and the animal received 0.4 mA of foot shock for 2 s. The time it took the mouse to enter the dark chamber (until the cutoff time of 400 s) was recorded. The mouse was put back in its home cage; 24 h later, the mouse was placed back into the original chamber, and all stimulus conditions were repeated except that no shock occurred when the mouse entered the dark chamber. The usual finding is that the mouse takes longer to enter the dark chamber on the second day because the prior shock experience inhibits its behavior. Figure 5E shows the time before entry into the dark chamber (latency time) on both days for the two genotypes. On the first day, the Ntan1−/− mice took significantly longer to enter the dark chamber than the +/+ animals. On the second day, the entry into the dark chamber by the Ntan1−/− mice was further delayed (indicating learning), while the entry by the +/+ mice was delayed even more, resulting in similar retention times for either genotype (Fig. 5E).
The longer retention time of the Ntan1−/− mice on the first day of passive learning, as well as their lower activity in the open-field test, suggested that +/+ and congenic Ntan1−/− mice might differ in their anxiety levels. To investigate this, they were tested in the elevated plus maze, which consists of two open arms and two closed arms radiating from a center platform. The open arms are more anxiety inducing, in that the animals with higher anxiety tend to decrease exploration of the open arms (42). Although the Ntan1−/− mice tended to explore the open arms less frequently than congenic +/+ mice, the observed difference was not statistically significant (data not shown).
A socially conditioned exploratory phenotype of Ntan1−/− mice.
To address the finding of lower activity in the Ntan1−/− mice (Fig. 5F) in a different way, we devised a simple test in which the exploratory activity was assessed in the context of a social interaction. In this platform-leaving test, two previously untested mice, Ntan1−/− and congenic +/+, are placed, close together and at the same time, onto the center of a 16- by 22-cm platform 2.7 cm in height. Thereafter one records the following variables: the time it takes each mouse to leave the platform, until the cutoff time of 3 min; the number and genotype of mice leaving the platform first (in independent trials with pairs of mice); and the number and genotypes of mice not leaving the platform by the cutoff time (see Materials and Methods). In contrast to the standard open-field test, a mouse in the platform-leaving test sees and smells the second mouse on the same platform and is otherwise influenced by the second mouse in the course of their peregrinations over the platform. Thus, the proclivity of a mouse to explore its environment is modulated, in the platform-leaving test, by interactions with another mouse.
A striking result of this test was the observation that an Ntan1−/− mouse paired with its congenic +/+ littermate tended to stay longer on the platform and almost never left it by the cutoff time, in contrast to the +/+ mouse (Fig. 6A). For example, in a trial with 44 pairs of previously untested Ntan1−/− and +/+ mice, only 1 Ntan1−/− mouse left the platform by the cutoff time, whereas 20 +/+ mice left it by that time (Fig. 6Ac). Repeated trials, at 1-h intervals, in the platform-leaving test progressively decreased this initial difference between the paired Ntan1−/− and +/+ mice (Fig. 6A). Note that this decrease resulted from both an increase in the exploratory activity of Ntan1−/− mice (in successive trials) and a decrease in the exploratory activity of their paired +/+ littermates (e.g., Fig. 6Ac). Thus, the strongly different initial responses of paired Ntan1−/− and +/+ mice to the novel environment (of which the other mouse was a part) became attenuated in subsequent trials (Fig. 6A) and nearly disappeared by the seventh trial (data not shown).
We then asked whether the observed difference between Ntan1−/− and +/+ mice was retained if the same test was performed with congenic nonlittermates. Strikingly, if the test of Fig. 6A was carried out with wild-type and mutant nonlittermates, the results were reproducibly different from those with littermates (Fig. 6B). Specifically, in this case Ntan1−/− mice did not stay longer on the platform: they left it, on average, more frequently than +/+ mice, a pattern opposite that seen with littermates (Fig. 6Bb; compare with Fig. 6Ab). In addition, the striking difference between Ntan1−/− and +/+ littermates in their frequency of not leaving the platform by the cutoff time was no longer seen in the otherwise identical tests with nonlittermates (Fig. 6Ac; compare with Fig. 6Bc).
In the control experiments, multiple pairs of littermates of the same genotype (either both +/+ or both Ntan1−/−) were subjected to this test. No statistically significant differences between the platform-leaving patterns of the two mice in a pair were observed (data not shown). The results presented in Fig. 6 were consistently reproduced in four independent sets of tests, carried out over 6 months with previously untested mice (at least 10 pairs of mice per test). This socially conditioned exploratory phenotype of Ntan1−/− mice (Fig. 6) has not been previously described, to our knowledge, with other mouse strains.
Spatial learning and memory.
To compare spatial learning of +/+ and congenic Ntan1−/− mice, we tested them on the hidden platform Morris maze (where a mouse had to find a platform submerged in water pool), using the reversal learning regimen (see Materials and Methods) and retesting 7 or 8 weeks later. Both +/+ and Ntan1−/− mice learned to find the platform and did not differ in either the time taken to do it or the distance traveled; they also did not differ on these measures in the subsequent reversal learning test, when the platform was moved to the diagonally opposite quadrant (data not shown). However, 7 to 8 weeks later, on the first day of retention testing, the Ntan1−/− mice were inferior to congenic +/+ mice, in that they took significantly longer time to find the platform and traveled a significantly longer distance before reaching it (data not shown), suggesting that the Ntan1−/− mice have a less effective spatial memory system.
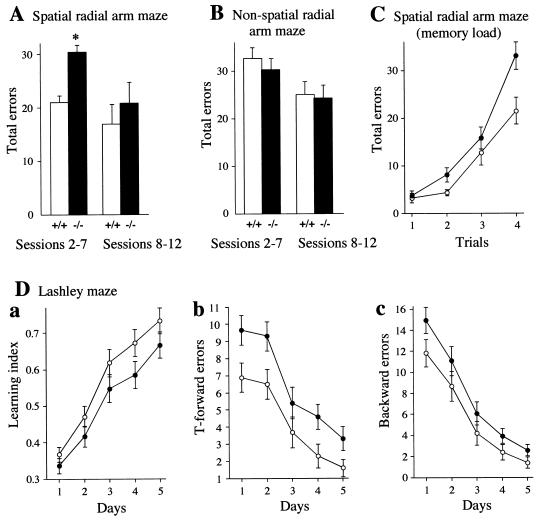
Another measure of spatial learning employs the water version of the radial arm maze, where four of the eight arms contain escape platforms (30). There are no intramaze cues available to the mouse, and it must use extramaze spatial information involving reference and working memory to remember which alleys to enter and which to avoid. The mice were given training for 1 day and tested for 11 consecutive days, receiving four trials each day. The measure used was the total number of errors. Ntan1−/− mice made significantly more errors in the acquisition phase (sessions 2 to 7) than the congenic +/+ mice (Fig. 7A). In the asymptotic phase (sessions 8 to 12), there were no significant differences between the two strains (Fig. 7A).
FIG. 7.
Spatial learning and memory in Ntan1−/− mice. Open symbols and bars represent +/+ mice; solid symbols and bars represent congenic Ntan1−/− mice. See the text for details. (A to C) Results for 10 +/+ mice and 10 Ntan1−/− mice. (A) Mean daily errors made by mice for each phase in the spatial radial arm maze. The data were separated into those for acquisition phase (sessions 2 to 7) and asymptotic phase (sessions 8 to 12). (B) Same as panel A except that the mice were tested in the nonspatial radial arm maze, in which the spatial cues were replaced by nonspatial cues. (C) Effects of memory load in the water version of the spatial radial arm maze. Shown are total errors per trial. (D) Performance of 29 +/+ mice and 30 congenic Ntan1−/− mice in the Lashley maze. Shown are the ratio of the number of correct path segments taken to the total number of segments (learning index) (a), errors in T-choices (b), and errors made by moving away from the goal (c).
The four trials within each daily session impose an increasing demand on memory. Thus, on the first trial, there are platforms in four of the eight alleys, and entering any of these terminates the trial. On the second trial, three of the alleys still contain platforms; the mouse has to remember which alley it had entered on the prior trial and not enter that alley again. Finally, on the fourth trial, the mouse has to remember which three alleys it had chosen earlier, so that it can find the one remaining alley containing a platform. Thus, by studying the error rate from trial to trial, summed over the sessions, one can compare the abilities of mice to cope with an increasing memory load. Figure 7C shows the error curves summed over the acquisition phase. As expected, errors increased as the memory load increased. However, the rate of increase was significantly greater for the Ntan1−/− mice, especially on trial 4 (Fig. 7C).
Unlike previously tested mouse strains (30), both +/+ and congenic Ntan1−/− mice exhibited poor learning over the 11 days of testing, consistent with inferior performance of strain 129-derived mice in several other learning regimens (7, 53). Thus, the significant effects described above (Fig. 7A and C) represent largely differences in performance but indicate little about the spatial learning ability of these mice. To determine whether the observed effects were related to spatial competence, the identical experiment was carried out using the same radial arm maze, except that a nonspatial version was devised, by having different visual patterns in each alley and rotating the maze between trials so that the extramaze cues were rendered meaningless. In four of the arms, escape platforms were placed, and the mice had to learn to associate a specific visual configuration (e.g., black dots on a white background, or horizontal black and white stripes) with the presence or absence of a platform (29). In contrast to the results with the spatial radial arm maze, no significant differences between the two genotypes were observed in the nonspatial radial arm maze, either in regard to the overall number of errors (Fig. 7B) or in regard to the error rate as a function of memory load (data not shown). Here, too, both +/+ and congenic Ntan1−/− mice did not exhibit significant learning over the 11 days of testing. Thus, no conclusions can be drawn about spatial or nonspatial learning. At the same time, it was possible to conclude that Ntan1−/− mice were less effective in dealing with spatial information than the congenic +/+ mice.
Another test of spatial learning and memory was the Lashley III maze. This maze contains (i) cul-de-sacs that an animal has to learn to avoid and (ii) T-choices. Both +/+ and Ntan1−/− mice were able to learn the maze, but Ntan1−/− mice were significantly less competent, as measured by the learning index, T-forward errors (making a wrong T-choice), and the total number of backward errors (Fig. 7D). When retested 7 or 8 weeks later, Ntan1−/− mice had lower scores on the first trial on all three measures, showing inferior long-term retention (data not shown). These results were similar to the findings with the Morris maze (see above), except that there was a significant difference in the original learning of the Lashley maze favoring the +/+ mice (Fig. 7D). This difference may stem from the fact that the Lashley maze can be learned using intramaze cues to memorize the pathway of left and right turns, in addition to the use of extramaze cues, which are present in both the Lashley and Morris maze tests. The results of the two radial arm studies (Fig. 7A to C) indicated that the Ntan1−/− mice used spatial information less effectively than the +/+ mice. In summary, the results of spatial memory tests (Fig. 7) strongly suggested that mouse NtN-amidase contributes to the processing of spatial information and to long-term retention of spatial learning.
DISCUSSION
Homologs of the 310-residue mammalian NtN-amidase, encoded by the Ntan1 gene (24, 59), are present in other vertebrates, in arthropods such as D. melanogaster, and in plants such as A. thaliana but are apparently absent from the nematode Caenorhabditis elegans (data not shown). The deamidation of N-terminal (and apparently only N-terminal) Asn residue in proteins or short peptides is the only known enzymatic activity of NtN-amidase. This enzyme was operationally defined as a component of the N-end rule pathway, because it converts, in vivo, the N-terminal Asn of engineered N-end rule substrates into N-terminal Asp, which is then conjugated by R-transferase to Arg, a primary destabilizing residue (34, 63). Physiological substrates of NtN-amidase remain to be identified. Physiological substrates are also unknown for the fungal Nt-amidase, which can deamidate either N-terminal Asn or N-terminal Gln and lacks significant sequence similarities to metazoan NtN-amidase (6). In the present work, mouse NtN-amidase was studied using several approaches, including targeted mutagenesis. We report the following results.
(i) Both copies of the mouse Ntan1 gene, which encodes NtN-amidase, were replaced by a deletion/disruption allele. The resulting Ntan1−/− mice were viable and fertile. Their behavioral phenotypes are described below.
(ii) EF cells from Ntan1−/− mice lacked NtN-amidase activity, in contrast to congenic +/+ EF cells. In addition, among the normally short-lived substrates of the N-end rule pathway, only those bearing N-terminal Asn, a tertiary destabilizing residue (see the introduction), became long-lived in the Ntan1−/− EF cells. Thus, at least these cells, and by inference the Ntan1−/− mice, lacked the asparagine branch of the N-end rule pathway.
(iii) The Ntan1−/− EF cells contained wild-type levels of NtQ-amidase, which mediates the activity of N-terminal Gln, another tertiary destabilizing residue in the N-end rule. Thus, NtN-amidase and NtQ-amidase (the latter enzyme remains to be isolated and characterized) are encoded by different genes.
(iv) The brains and testes of +/+ mice contained, respectively, the 1.4- and 1.6-kb Ntan1 mRNAs. In addition, the testes but not the brains of +/+ mice also contained the Ntan1-derived RNAs of 1.1 and 4 kb. The brains and testes of Ntan1−/− mice lacked, respectively, the 1.4- and 1.6-kb Ntan1 mRNAs. However, the testes of Ntan1−/− mice retained the testis-specific 1.1- and 4-kb Ntan1-derived RNAs, which were found to lack the 5′-half of the Ntan1 ORF (including the region deleted in Ntan1−/− mice) and to encompass the 3′ half of Ntan1 ORF. Whether these testis-specific transcripts are physiologically relevant in +/+ mice remains to be determined.
(v) Deletion-disruption of Ntan1 in the Ntan1−/− mice did not significantly change the levels of mRNAs encoding other components of the N-end rule pathway, such as ATE1p (R-transferase), UBR1p (E3α, the pathway's E3 component), UBR2p (a homolog of UBR1p), and mHR6Bp (E214K).
(vi) Ntan1, which is expressed, at different levels, in most if not all tissues of adult mice (24), was found through in situ hybridization to be particularly strongly expressed in the branchial arches, the tail bud, and the limb buds of E10 embryos. This expression pattern was indistinguishable from that of Ubr1 (36), consistent with the assignment of NTAN1p (NtN-amidase) and UBR1p (E3α) to the same pathway.
(vii) An NTAN1p-GFP fusion was enriched in the nucleus of transfected mouse 3T3 cells, in contrast to free GFP, which was uniformly distributed. NTAN1p-GFP was also present in the nucleus-proximal cytoplasm but was apparently much less abundant at the cells' periphery. This localization pattern of the mammalian NtN-amidase is quite different from that of the S. cerevisiae NTA1-encoded Nt-amidase, most of which is located in the yeast mitochondria (H. R. Wang and A. Varshavsky, unpublished data). In contrast to the mammalian NtN-amidase, the yeast Nt-amidase, which has no significant sequence similarities to the metazoan enzyme, can deamidate either N-terminal Asn or N-terminal Gln (6).
(viii) While the abnormal phenotypes of Ntan1−/− mice are apparently not confined to behavioral/cognitive alterations (see Results), it is the latter that have been analyzed in some detail. Briefly, the Ntan1−/− mice were indistinguishable from congenic +/+ mice in motor coordination and general physical performance but had reduced spontaneous activity and less effective spatial memory. Remarkably, the exploratory behavior of Ntan1−/− mice was found to be particularly different from that of congenic +/+ mice in the presence of social interactions. If a previously untested Ntan1−/− mouse was placed on a small, slightly elevated platform with its +/+ littermate (which it grew up with in the same cage), the Ntan1−/− mouse left the platform much more slowly than the +/+ mouse. In contrast, when an Ntan1−/− mouse was subjected to this test in the presence of a congenic nonlittermate +/+ mouse, it tended to leave the platform faster than the +/+ mouse, even though Ntan1−/− mice exhibited diminished exploratory activity when tested alone (Fig. 6).
Our working hypothesis is that Ntan1−/− mice are socially recessive, in relation to congenic +/+ mice, and in addition have a lower spontaneous exploratory activity. In a relatively unstressful setting, such as when it is placed on the platform side-by-side with a (familiar) littermate, the behavior of Ntan1−/− mice is governed largely by their inherently lower exploratory activity, yielding the observation that these mice leave the platform much more slowly than their +/+ littermates (Fig. 6A). By contrast, in an otherwise identical test with a +/+ nonlittermate, the postulated social recessiveness of Ntan1−/− mice becomes their behavior-governing trait. As a result, they leave the platform faster than the +/+ nonlittermates, to reduce the proximity-induced anxiety (Fig. 6B).
The socially conditioned exploratory phenotype of Ntan1−/− mice, identified through the platform-leaving test (Fig. 6), has not been previously described, to our knowledge, with other mouse strains. It is difficult to compare this finding with behavioral studies of other mouse mutants, because most of the earlier analyses of exploratory behavior used solitary mice. An example of differential effect of stress on a specific mutant is provided by the analysis of mice lacking the serotonin receptor 1A. These mice exhibited decreased exploratory activity and increased fear of aversive environments (in comparison to +/+ mice), as measured by the open-field test and the elevated plus-maze test, respectively. However, the mutant mice were indistinguishable from +/+ littermates in a less stressful setting such as their home cages (50).
A parsimonious molecular interpretation of our results (Fig. 5 to 7) is that a normally short-lived regulatory protein(s) that is targeted for degradation by the N-end rule pathway through the protein's N-terminal Asn becomes long-lived in Ntan1−/− mice. The resulting increase in the steady-state concentration of this protein(s) alters the functioning of relevant neural networks in the adult brain or, nonalternatively, changes these networks in the course of their establishment during development. That the inactivation of a Ub-dependent proteolytic pathway could affect cognitive functions is illustrated, for example, by the identification of UBE3A as a human gene encoding an E3 protein of the Ub system called E6AP. This gene is mutated in the Angelman's syndrome, the symptoms of which include motor dysfunction and mental retardation (33, 45). Mice lacking UBE3A exhibit deficits in contextual learning and long-term potentiation (31).
The absence of severe impairments in the Ntan1−/− mice should make them particularly suitable as vehicles for the approaches to conditional mutagenesis that use conditional destabilization of a protein of interest. One version of this approach could use a transgenic mouse strain that lacks endogenous Ntan1 and expresses this gene from one of the previously constructed promoters whose activity can be controlled by small molecules such as tetracycline or ecdysone (9). If a protein of interest is modified, through knock-in mutagenesis and the Ub fusion technique (63), to bear an Asn-containing N-degron, the resulting protein would be long-lived in the absence of the Ntan1-encoded NtN-amidase but short-lived, and therefore scarce, in the presence of NtN-amidase. In this method, the steady-state level, and hence the activity, of a protein of interest is controlled through Ntan1-dependent, regulated changes of the protein's metabolic stability. One advantage of this strategy is that it does not require alterations of a promoter that expresses a gene of interest, thereby avoiding potential perturbations of the promoter regulation during development and differentiation. This approach may be combined with the existing technologies for conditional mutagenesis of the mouse (13, 55), enhancing them through regulated degradation of a protein of interest.
Inasmuch as behavior is an emergent property of highly complex neural networks interacting with the musculoskeletal apparatus, a behavioral alteration that is caused by the absence of a specific protein from the brain is difficult to understand in terms of higher-order neural events even in the case of a protein (e.g., a neurotransmitter-gated ion channel) whose molecular function in the individual neurons is clear. The difficulty is further increased in the case of a protein such as NtN-amidase, whose physiological substrates remain to be identified. At the same time, the absence of a priori reasons to suspect a specific role for NtN-amidase in the brain's functions and the robustness of behavioral differences between the Ntan1−/− and congenic +/+ mice (Fig. 5 to 7) make these findings particularly intriguing. Further advances in this inquiry will require, at minimum, the identification of physiological substrates of NtN-amidase. The availability of Ntan1−/− mice and cell lines derived from them should facilitate the discovery of these substrates.
ACKNOWLEDGMENTS
We are grateful to members of the Caltech Transgenic Facility, especially S. Pease, B. Kennedy, and A. Granados, for the care of mice and expert technical help. We thank S. Grigoryev for helpful discussions at the beginning of this study, N. Barteneva for assistance with some of the early experiments, S. Offermanns for advice and help with preparation of embryonic fibroblasts, and R. C. Mulligan and R. Jaenisch for gifts of plasmids.
This work was supported by grants GM31530 and DK39520 from the National Institutes of Health to A.V.
REFERENCES
- 1.Alagramam K, Naider F, Becker J M. A recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:225–234. doi: 10.1111/j.1365-2958.1995.tb02237.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1998. [Google Scholar]
- 3.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 4.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 5.Baker R T, Varshavsky A. Inhibition of the N-end rule pathway in living cells. Proc Natl Acad Sci USA. 1991;87:2374–2378. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker R T, Varshavsky A. Yeast N-terminal amidase. A new enzyme and component of the N-end rule pathway. J Biol Chem. 1995;270:12065–12074. doi: 10.1074/jbc.270.20.12065. [DOI] [PubMed] [Google Scholar]
- 7.Balogh S A, McDowell C S, Stavnezer A, Denenberg V H. A behavioral and neuroanatomical assessment of an inbred substrain of 129 mice, with behavioral comparisons to C57BL/6J mice. Brain Res. 1999;836:38–48. doi: 10.1016/s0006-8993(99)01586-3. [DOI] [PubMed] [Google Scholar]
- 8.Balzi E, Choder M, Chen W, Varshavsky A, Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem. 1990;265:7464–7471. [PubMed] [Google Scholar]
- 9.Baron U, Schnappinger D, Helbl V, Gossen M, Hillen W, Bujard H. Generation of conditional mutants in higher eukaryotes by switching between the expression of two genes. Proc Natl Acad Sci USA. 1999;96:1013–1018. doi: 10.1073/pnas.96.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 11.Beaudin S, Lalonde R. The effects of pentobarbital on spatial learning, motor coordination, and exploration. Pharmacol Biochem Behav. 1997;57:111–114. doi: 10.1016/s0091-3057(96)00137-2. [DOI] [PubMed] [Google Scholar]
- 12.Byrd C, Turner G C, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capecchi M R. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 14.Chang S E, Keen J, Lane E B, Taylor-Papadimitriou J. Establishment and characterization of SV40-transformed human breast epithelial cell lines. Cancer Res. 1982;42:2040–2053. [PubMed] [Google Scholar]
- 15.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 16.Conlon R A, Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development. 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 17.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–817. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 18.Davydov I V, Patra D, Varshavsky A. The N-end rule pathway in Xenopus egg extracts. Arch Biochem Biophys. 1998;357:317–325. doi: 10.1006/abbi.1998.0829. [DOI] [PubMed] [Google Scholar]
- 19.deGroot R J, Rümenapf T, Kuhn R J, Strauss J H. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci USA. 1991;88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeMartino G N, Slaughter C A. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 21.Denenberg V H, Talgo N, Carroll D A, Freter S, Deni R. A computer-aided procedure for measuring Lashley III maze performance. Physiol Behav. 1991;50:857–861. doi: 10.1016/0031-9384(91)90031-i. [DOI] [PubMed] [Google Scholar]
- 22.Denenberg V H, Talgo N W, Waters N S, Kenner G H. A computer-aided procedure for measuring swim rotation. Physiol Behav. 1990;47:1023–1025. doi: 10.1016/0031-9384(90)90029-4. [DOI] [PubMed] [Google Scholar]
- 23.Gonda D K, Bachmair A, Wünning I, Tobias J W, Lane W S, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 24.Grigoryev S, Stewart A E, Kwon Y T, Arfin S M, Bradshaw R A, Jenkins N A, Copeland N G, Varshavsky A. A mouse amidase specific for N-terminal asparagine. The gene, the enzyme, and their function in the N-end rule pathway. J Biol Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- 25.Haas A J, Siepman T J. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 26.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;76:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 27.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 28.Hondermarck H, Sy J, Bradshaw R A, Arfin S M. Dipeptide inhibitors of ubiquitin-mediated protein turnover prevent growth factor-induced neurite outgrowth in rat pheochromocytoma PC12 cells. Biochem Biophys Res Commun. 1992;30:280–288. doi: 10.1016/0006-291x(92)91555-5. [DOI] [PubMed] [Google Scholar]
- 29.Hyde L A, Denenberg V H. BXSB mice can learn complex pattern discrimination. Physiol Behav. 1999;66:437–439. doi: 10.1016/s0031-9384(98)00310-2. [DOI] [PubMed] [Google Scholar]
- 30.Hyde L A, Hoplight B J, Denenberg V H. Water version of the radial-arm maze: learning in three inbred strains of mice. Brain Res. 1998;785:236–244. doi: 10.1016/s0006-8993(97)01417-0. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y H, Armstrong D, Albrecht U, Atkins C M, Noebels J L, Eichele G, Sweatt J D, Beaudet A L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 32.Johnson E S, Gonda D K, Varshavsky A. Cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990;346:287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- 33.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 34.Kwon Y T, Kashina A S, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon Y T, Lévy F, Varshavsky A. Bivalent inhibitor of the N-end rule pathway. J Biol Chem. 1999;274:18135–18139. doi: 10.1074/jbc.274.25.18135. [DOI] [PubMed] [Google Scholar]
- 36.Kwon Y T, Reiss Y, Fried V A, Hershko A, Yoon J K, Gonda D K, Sangan P, Copeland N G, Jenkins N A, Varshavsky A. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laney J D, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 38.Lawson T G, Gronros D L, Evans P E, Bastien M C, Michalewich K M, Clark J K, Edmonds J H, Graber K H, Werner J A, Lurvey B A, Cate J M. Identification and characterization of a protein destruction signal in the encephalomyocarditis virus 3C protease. J Biol Chem. 1999;274:9871–9880. [PubMed] [Google Scholar]
- 39.Lecker S H, Solomon V, Price S R, Kwon Y T, Mitch W E, Goldberg A L. Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J Clin Investig. 1999;104:1411–1420. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lévy F, Johnsson N, Rumenapf T, Varshavsky A. Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci USA. 1996;93:4907–4912. doi: 10.1073/pnas.93.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Pickart C. Inactivation of arginyl-tRNA protein transferase by a bifunctional arsenoxide: identification of residues proximal to arsenoxide site. Biochemistry. 1995;34:139–147. doi: 10.1021/bi00001a017. [DOI] [PubMed] [Google Scholar]
- 42.Lister R G. The use of a plus-maze to measure anxiety in the mouse. Psychopharmacology. 1987;92:180–185. doi: 10.1007/BF00177912. [DOI] [PubMed] [Google Scholar]
- 43.Madura K, Varshavsky A. Degradation of Gα by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 44.Maniatis T. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 45.Matsuura T, Sutcliffe J S, Fang P, Galjaard R J, Jiang Y H, Benton C S, Rommens J M, Beaudet A L. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 46.Morris R. Development of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 47.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 48.Peters J-M, King R W, Deshaies R J. Cell cycle control by ubiquitin-dependent proteolysis. In: Peters J M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 345–387. [Google Scholar]
- 49.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 50.Ramboz S, Oosting R, Amara D A, Kung H F, Blier P, Mendelsohn M, Mann J J, Brunner D, Hen R. Serotonin receptor 1A knockout: an animal model of anxiety-related disorder. Proc Natl Acad Sci USA. 1998;95:14476–14481. doi: 10.1073/pnas.95.24.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rechsteiner M. The 26S proteasome. In: Peters J M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 147–189. [Google Scholar]
- 52.Robertson E J. Embryo-derived stem cell lines. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford, United Kingdom: IRL Press; 1987. pp. 71–112. [Google Scholar]
- 53.Royce J R. Avoidance conditioning in nine strains of inbred mice using optimal stimulus parameters. Behav Genet. 1972;2:107–110. doi: 10.1007/BF01066739. [DOI] [PubMed] [Google Scholar]
- 54.Schauber C, Chen L, Tongaonkar P, Vega I, Madura K. Sequence elements that contribute to the degradation of yeast G-alpha. Genes Cells. 1998;3:307–319. doi: 10.1046/j.1365-2443.1998.00192.x. [DOI] [PubMed] [Google Scholar]
- 55.Schwenk F, Kuhn R, Angrand P O, Rajewsky K, Stewart A F. Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res. 1998;26:1427–1432. doi: 10.1093/nar/26.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sijts A J, Pilip I, Pamer E G. The Listeria monocytogenes-secreted p60 protein is an N-end rule substrate in the cytosol of infected cells. Implications for major histocompatibility complex class I antigen processing of bacterial proteins. J Biol Chem. 1997;272:19261–19268. doi: 10.1074/jbc.272.31.19261. [DOI] [PubMed] [Google Scholar]
- 57.Solomon V, Baracos V, Sarraf P, Goldberg A. Rates of ubiquitin conjugation increase when muscles atrophy, largely through activation of the N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:12602–12607. doi: 10.1073/pnas.95.21.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart A. Trends in genetics nomenclature guide. Cambridge, United Kingdom: Elsevier Science, Ltd.; 1995. [Google Scholar]
- 59.Stewart A E, Arfin S M, Bradshaw R A. The sequence of porcine protein NH2-terminal asparagine amidohydrolase. A new component of the N-end rule pathway. J Biol Chem. 1995;270:25–28. doi: 10.1074/jbc.270.1.25. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki T, Varshavsky A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 1999;18:6017–6026. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taban C H, Hondermarck H, Bradshaw R A, Boilly B. Effect of a dipeptide inhibiting ubiquitin-mediated protein degradation on nerve-dependent limb regeneration in the newt. Experientia. 1996;52:865–870. doi: 10.1007/BF01938871. [DOI] [PubMed] [Google Scholar]
- 62.Tobias J W, Varshavsky A. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12021–12028. [PubMed] [Google Scholar]
- 63.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y M, Ingoglia N A. N-terminal arginylation of sciatic nerve and brain proteins following injury. Neurochem Res. 1997;22:1453–1459. doi: 10.1023/a:1021998227237. [DOI] [PubMed] [Google Scholar]
- 66.Wilkinson K, Hochstrasser M. The deubiquitinating enzymes. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 99–126. [Google Scholar]