Key Points
Maternal–fetal immune repertoire analyses provide insights to preterm labor.
Clonal convergence suggests a response to shared stimuli.
Abstract
Preterm labor (PTL) is the leading cause of neonatal morbidity and mortality worldwide. Whereas many studies have investigated the maternal immune responses that cause PTL, fetal immune cell activation has recently been raised as an important contributor to the pathogenesis of PTL. In this study, we analyzed lymphocyte receptor repertoires in maternal and cord blood from 14 term and 10 preterm deliveries, hypothesizing that the high prevalence of infection in patients with PTL may result in specific changes in the T cell and B cell repertoires. We analyzed TCR β-chain (TCR-β) and IgH diversity, CDR3 lengths, clonal sharing, and preferential usage of variable and joining gene segments. Both TCR-β and IgH repertoires had shorter CDR3s compared with those in maternal blood. In cord blood samples, we found that CDR3 lengths correlated with gestational age, with shorter CDR3s in preterm neonates suggesting a less developed repertoire. Preterm cord blood displayed preferential usage of a number of genes. In preterm pregnancies, we observed significantly higher prevalence of convergent clones between mother/baby pairs than in term pregnancies. Together, our results suggest the repertoire of preterm infants displays a combination of immature features and convergence with maternal TCR-β clones compared with that of term infants. The higher clonal convergence in PTL could represent mother and fetus both responding to a shared stimulus like an infection. These data provide a detailed analysis of the maternal–fetal immune repertoire in term and preterm patients and contribute to a better understanding of neonate immune repertoire development and potential changes associated with PTL.
Introduction
Preterm birth (delivery before 37 weeks’ gestational age) is a worldwide clinical concern with ∼15 million cases annually and is the leading cause of neonatal deaths worldwide (1). Preterm premature rupture of membranes (PPROM) is a common cause of spontaneous preterm labor (PTL) and is associated with intraamniotic infection in many cases (2, 3). Despite remarkable improvements in prenatal care over the past three decades, rates of PPROM and subsequent preterm delivery have worsened (2). Several epidemiological and clinical factors are considered precursors to PPROM, including reproductive tract infections, behavioral factors, and obstetric complications (4–6). Environmental factors (e.g., stress and toxin exposure) and genetic predisposition have also been proposed (4–7). Approximately 70% of PPROM cases are associated with intraamniotic infection, as documented by positive amniotic fluid cultures or by clinical evidence of infection (2, 3). Whereas many studies have focused on investigating changes associated with maternal immune responses during PTL, recent studies have indicated an important role of the fetal immune system in the pathogenesis of PPROM (8, 9).
To further investigate neonatal immunity in the context of PTL, we have focused on the adaptive immune responses in term and preterm neonates and its association with PTL pathogenesis. We recently demonstrated that fetal T cells have a robust, proliferative, and proinflammatory response to maternal Ags, leading to myometrium contractility in cases of PTL, including PPROM, and revealing the fetal immune system as an important contributor to the pathogenesis of PTL (8). In addition, we reported a transcriptomic meta-analysis of term and preterm mothers and neonates, identifying genetic signatures in preterm neonates associated with spontaneous preterm birth (10), and leveraged that signature for drug discovery (11). In another study, we further demonstrated via DNA sequencing the presence of relevant microbial cell–free DNA in maternal plasma and umbilical cord plasma from patients with clinical PPROM, suggesting in utero fetal exposure to pathogens associated with PPROM (12). These studies highlight the significance of sequencing-based methodologies in the field and how they may contribute to the knowledge advancement of fetal immune activation and PTL.
We have previously studied the adaptive immunity of infants to environmental factors and infectious diseases (13). Adaptive immunity relies on the expression of a largely diversified set of Ag-specific TCRs and Igs or BCRs, defining the immune repertoire (13–15). The BCR contains a pair of H chains (IgH) and L chains (Igκ or Igλ), whereas the TCR consists of a paired set of α- and β-chains (TCR β-chains [TCR-β]) or γ- and δ-chains. The constant domains of these receptors are encoded by germline DNA and are thus invariable. Production of TCR and BCR molecules requires V(D)J recombination, which involves genomic rearrangement of variable (V), diversity (D; present in IgH and TCR-β), and joining (J) gene elements at the TCR and Ig loci (16). These receptors are heterodimeric proteins that can be divided into constant domains, which encode effector function, and variable domains, which endow Ag specificity.
There are five different Ab isotypes: the first Abs to be produced in a humoral immune response are always IgM and quickly progress to the production of all the different isotypes, IgD, IgA, IgG, and IgE. IgD can be coexpressed with IgM. The primary sequence of the variable domains of BCR and TCR can be divided into four framework regions (FRs) of relatively conserved sequence and three CDRs (CDR1, CDR2, and CDR3) of hypervariable sequence. Together, the six CDRs of the two-paired chains form the classic Ag-binding site. The CDRs rest on top of a scaffold that is created by the folding of the FRs. CDR1 and CDR2 are entirely encoded by the V gene segment and are located on the perimeter of the Ag-binding site, whereas CDR3 (the region containing part of the V gene, all of the D gene, and part of the J gene) lies at the center of the Ag-binding site and thus plays an essential role in defining the specificity of the receptor. Therefore, features of TCR/BCR gene arrangements, such as sequence, length, and gene segment usage, can be studied during immune response to pathogens or self-antigens.
Previous analysis has suggested that term neonate TCR and BCR repertoires have shorter CDR3 lengths and skewed distribution of gene segment usage at varying developmental stages when compared with adults (17). Furthermore, a recent immune BCR repertoire study in extremely preterm neonates (24–28 weeks) demonstrated slower postnatal maturation of the IgG IgH repertoires than what was observed in term neonates (18). Preterm neonate repertoire studies have focused mainly on extremely preterm cases not associated with PPROM. However, we hypothesized that the high prevalence of infection in patients with PPROM may result in specific changes in the TCR and BCR repertoires.
In this study, we performed a pilot study of 24 pregnancies to analyze TCR-β and IgH repertoires in maternal and cord blood from 14 term and 10 preterm deliveries due to PPROM, focusing on TCR-β and IgH diversity, somatic hypermutation (SHM) rates, average CDR3 lengths, clonal sharing, and preferential usage of V, D, and J gene segments. Our findings provide evidence of unique immune repertoire features in infants born preterm secondary to PPROM when compared with term infants, which we hope to probe further in follow-up studies.
Materials and Methods
Specimen collection
Maternal blood samples and cord blood samples from 14 term pregnancies and 10 PPROM pregnancies were collected at the University of California, San Francisco, at the time of delivery after Institutional Review Board approval (10-00350). PBMCs were isolated by Ficoll-Paque PLUS (GE Healthcare) density gradient centrifugation and cryopreserved in 90% FBS and 10% DMSO in liquid nitrogen.
Extraction, library preparation, and high-throughput sequencing of IGH and TCR-β samples
Total RNA was extracted from frozen PBMC pellets using the AllPrep DNA/RNA kit (QIAGEN) according to manufacturer’s guidelines. For each sample, total RNA was reverse-transcribed to cDNA using Superscript III RT (Invitrogen) with random hexamer primers (Promega). cDNA corresponding to 100 ng of total RNA was used for each of the isotype PCRs using IGHV FR1 primers based on the BIOMED-2 design (19) and isotype specific primers located in the first exon of the C region for each isotype category (IgM, IgD, IgE, IgA, and IgG). Primers contain additional sequence representing the first part of the Illumina linkers. The different isotypes were amplified in separate reaction tubes. Eight-nucleotide barcode sequences were included in the primers to indicate sample identity. Four randomized bases were included upstream of the barcodes on the C region primer for Illumina clustering. PCR was carried out with AmpliTaq Gold (Applied Biosystems) following the manufacturer’s instructions and used a program of 95°C for 7 min; 35 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 120 s; and final extension at 72°C for 10 min. A second round of PCR using Qiagen Multiplex PCR Kit was performed to complete the Illumina sequencing adapters at the 5′ and 3′ ends of amplicons. Cycling conditions were 95°C for 15 min; 12 cycles of 95°C for 30 s, 60°C for 45 s, and 72°C for 90 s; and final extension at 72°C for 10 min.
cDNA corresponding to 100 ng of total RNA was used for each TCR-β library, and libraries were prepared using TCR-β primers from the ImMunoGeneTics database. As previously described, PCR amplification was performed by using a multiplex PCR kit (QIAGEN) and used a program of 95°C for 15 min; 30 cycles of 94°C for 30 s, 59°C for 90 s, and 72°C for 120 s; and final extension at 72°C for 10 min (20). For both IgH and TCR-β libraries, PCR products were subsequently pooled, gel purified (QIAGEN), and quantified with the Qubit fluorometer (Invitrogen). Samples were sequenced on the Illumina MiSeq: PE300 for IgH and PE150 for TCR-β, using 600 and 300 cycle kits, respectively.
Sequence quality assessment, filtering, and analysis
Paired-end reads were merged using FLASH (21), demultiplexed (100% barcode match), and primer trimmed. The V, D, and J gene segments and V–D (N1) and D–J (N2) junctions were identified using the IgBLAST alignment program (22). For cDNA-templated IgH reads, isotypes and subclasses were called by exact matching to the C region gene sequence upstream from the primer. Sequences accepted for further analysis were productive reads (sequences with no internal frame shift and no in-frame stop codons) with identified V and J segments and a CDR3 region, with a minimum V gene alignment score of 200. Alignment scores were calculated by counting alignment matches and mismatches (+5 for match, −4 for mismatch) along the V region (22), with higher scores denoting better alignment between input sequences and reference sequences. For IgH, SHM events were determined by counting mismatches in the IGHV sequence. Clonal identities within each subject were inferred using single-linkage clustering and the following definitions: for same V and J genes (disregarding allele call), equal CDR3 length, and minimum 90 or 95% CDR3 nucleotide identity for IgH or TCR-β, respectively. In the context of IgH, the threshold is set to 90% to account for SHM; for TCR-β, 95% is to account for potential mismatches due to sequencing error.
Splitting IgH into isotypes and unmutated/mutated
The IgH data also recorded isotype information, with reads present for each of IgA, IgD, IgE, IgG, and IgM. To separately analyze sequences likely to be derived from naive and memory B cells, we used a cutoff in the rate of SHM based on prior work (23). In this study, we used the rate of SHM of the nucleotides of the IGHV region to split IgD and IgM into two groups—unmutated (SHM rate < 0.01) and mutated (SHM rate ≥ 0.01).
Downsampling read count
The number of total reads and clones per sample varied (Supplemental Tables I, II). To adjust for read depth, we applied a downsampling procedure. For TCR-β, we used the minimum number of reads (205,164) as a baseline and randomly sampled that number of reads from each sample. For BCR, we repeated the same procedure for each isotype split: IgA (1,268 reads), IgD unmutated (20,491), IgD mutated (4,760), IgG (1,649), IgM unmutated (10,514), and IgM mutated (6,185). Because of variation in the number of reads, one preterm mother/baby pair was excluded from the IgM data. For IgA, six term pairs were excluded because of low read counts, whereas for IgG, one term pair and one preterm pair were excluded. We repeated the downsampling procedure 10 additional times (for a total of 11) and aggregated the results by taking the mean value from each repetition.
Diversity analysis—entropy, rate of SHM, and CDR3 length
As a measure of the diversity of the repertoires, we calculated Shannon entropy (H) for the clonal landscape of each sample, given by H = − log pi, where pi was the proportion of reads belonging to the ith clone. We also calculated the number of SHMs for each of the IGH reads. This measure was calculated by comparing mismatches in nucleotides. The CDR3 length was measured as the number of nucleotides in the CDR3 region. We calculated the average CDR3 length for each sample by taking the mean CDR3 length across all reads for that sample. We used the Wilcoxon rank-sum test to test for significant differences between distributions (e.g., term maternal versus term fetal).
Gene usage
We calculated gene usages for each sample, with usage defined as the number of unique clones expressing a particular gene divided by the total number of unique clones. To calculate this, we used the full dataset without downsampling and reduced the data down to only unique clones (regardless of read count). We calculated the V gene usage, D gene usage, and J gene usage of each sample per isotype. For comparisons between groups of samples, we calculated p values using the Mann–Whitney U test and adjusted using the Benjamini–Hochberg procedure. Clustering of gene usage was performed by hierarchical clustering (with Euclidean distance as the measure).
Public clone analysis
We examined the presence of public clones in samples within the two groups (preterm or term) as well as clones present in paired mother-infant samples. To identify clones across individuals, we defined similar clones as having the same V and J genes, equal CDR3 length, and an 85% (IgH) or 100% (TCR) matching amino acid identity. We used amino acids in this study as clones of different individuals that can be similar because they recognize the same Ags. The lower threshold for IgH was used to account for SHMs. To test for significance, we performed permutation analysis by randomly shuffling the tags of the samples. After shuffling, the public clones’ measures were once again calculated among the new, synthetic groups. We repeated this procedure 10,000 times to assemble a distribution of scores expected by chance, with p values calculated as the likelihood of a random score being greater than or equal to the actual score.
Results
Maternal blood samples and cord blood samples were collected at delivery for 14 term and 10 PPROM pregnancies (Fig. 1, Table I). Both TCR-β and IgH (Fig. 2A) were sequenced and aligned, with initial quality control steps used to filter the data resulting in a dataset of over 35 million reads (Supplemental Table I). IgH libraries for each isotype (IgA, IgD, IgE, IgG, and IgM) were amplified, sequenced, and analyzed separately. IgE read counts were close to zero for every sample and were excluded from further analysis. IgD and IgM were further split into “unmutated” (<1% V gene SHM) and “mutated” groups to distinguish between sequences likely derived from naive and memory B cells, respectively. Data per sample were downsampled to adjust for read depth. For each of the read transcripts, we extracted the associated V, D, and J gene and nucleotide length of the CDR3 (Fig. 2A). The rate of SHM in IGHV gene segments was also calculated for IgH data. Reads were assigned to clones, with two reads belonging to the same clone if they had the same V gene, same J gene, identical CDR3 length, and a minimum threshold of matching CDR3 nucleotides (90% for IgH and 95% for TCR). The results of the analyses are summarized in Tables II and III.
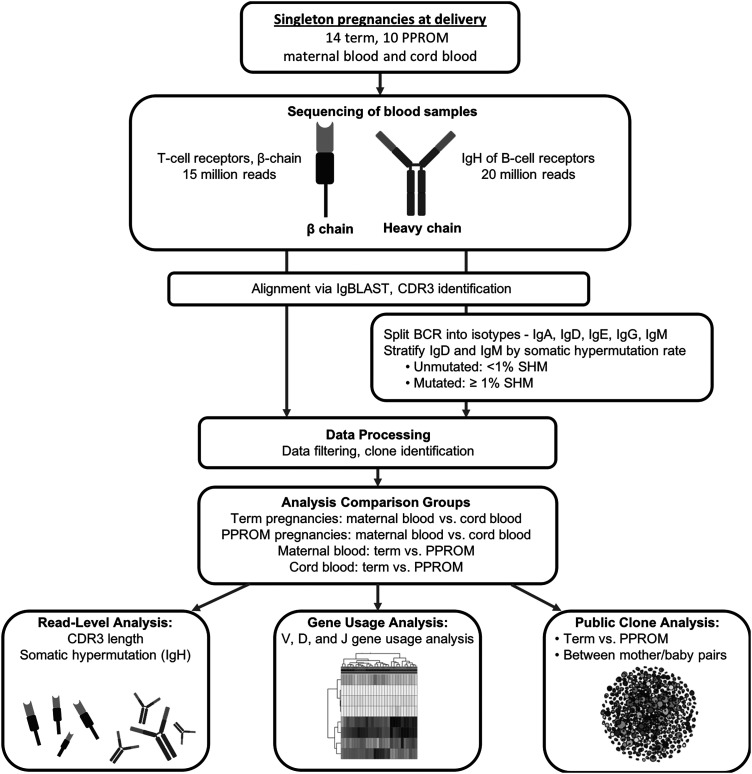
FIGURE 1.
Overview of analytical pipeline. Maternal blood and cord blood from singleton pregnancies were sequenced for TCRs and BCRs. After applying initial quality control steps (such as requiring a minimum V gene alignment score), read transcripts were assigned to clones based on V gene and J gene identities and CDR3 similarities. The samples were split into different groupings for comparative analysis: term maternal blood versus term cord blood, term maternal blood versus PPROM maternal blood, and term cord blood versus PPROM cord blood. We analyzed CDR3 length and rate of SHM of the read transcripts, gene usage of the unique clones, and the presence of highly similar, public clones across samples.
Table I.
Demographics of control (n = 14) and PPROM patients (n = 10)
| Control | PPROM | p Value | |
|---|---|---|---|
| Maternal age (y) | 35.3 (31–39.6) | 30 (24.1–35.9) | 0.1097 (Mann–Whitney) |
| Race/ethnicity (%) | 0.35 (χ2) | ||
| African American | 0 | 10 | |
| White | 64 | 50 | |
| Hispanic | 0 | 30 | |
| Asian | 14 | 10 | |
| Other/mixed | 0 | 0 | |
| Unknown | 21 | 0 | |
| Primigravid | 7 (50%) | 2 (20%) | <0.0001 (Fisher) |
| Gestational age at delivery (wk) | 39.65 (38.69–40.61) | 34 (32.4–35.2) | <0.0001 (Fisher) |
| Cesarean delivery | 1 (7%) | 3 (30%) | 0.2721 (Fisher) |
| Birth weight (g) | 3396 (2934–3823) | 2190 (1926–2640) | <0.0001 (Mann–Whitney) |
| Gender (male) | 7 (50%) | 6 (60%) | 0.6968 (Fisher) |
Demographics table from clinical cases in which maternal blood and cord blood samples were collected for repertoire analysis. Data presented as N (%) or median (interquartile range). The p values are calculated using Fisher exact or Mann–Whitney U test.
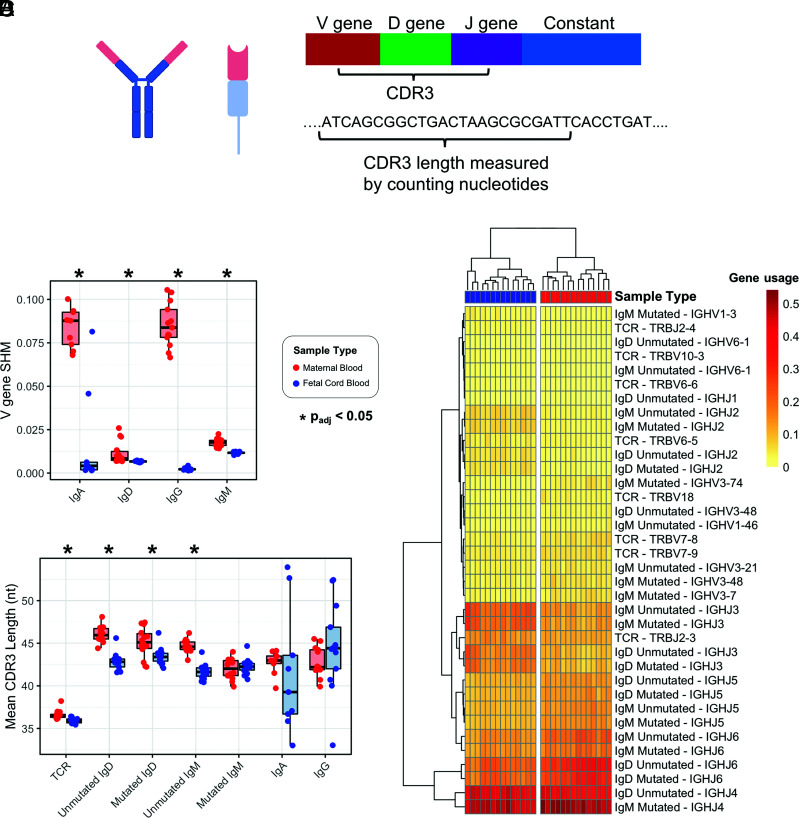
FIGURE 2.
Term maternal versus cord blood TCR-β and IgH comparison for SHM rates, CDR3 length, and gene usage. (A) Zoom-in of V region for TCR-β and IgH. (B) V gene SHM rate in maternal versus cord blood. (C) Mean CDR3 length in maternal versus cord blood. *Adjusted p value < 0.05 by Wilcoxon rank-sum test and adjusted for multiple comparisons using the Benjamini–Hochberg procedure. (D) Applying hierarchical clustering to the V and J genes with significantly different usage results in perfect separation of maternal blood and cord blood samples into two clusters.
Table II.
Entropy, CDR3 length, and SHM per analysis group
| Analysis Type | Term Maternal Blood Mean (SD) | PPROM Maternal Blood Mean (SD) | Term Cord Blood Mean (SD) | PPROM Cord Blood Mean (SD) | Term Maternal Blood versus Term Cord Blood p Value | PPROM Maternal Blood versus PPROM Cord Blood p Value | Term Maternal Blood versus PPROM Maternal Blood p Value | Term Cord Blood versus PPROM Cord Blood p Value | |
|---|---|---|---|---|---|---|---|---|---|
| TCR | Entropy (bit) | 12.87 (1.64) | 13.87 (0.93) | 13.53 (0.79) | 14.89 (0.99) | 3.52E−01 | 2.32E−02 | 1.54E−01 | 3.84E−03 |
| CDR3 length (nt) | 36.60 (0.53) | 36.44 (0.38) | 35.91 (0.30) | 35.37 (0.76) | 1.35E−05 | 1.30E−04 | 7.09E−01 | 2.20E−02 | |
| IgD | SHM (rate) | 1.16E−2 (6.38E−3) | 9.63E−3 (2.78E−3) | 6.69E−3 (3.9E−4) | 6.65E−3 (3.0E−4) | 2.53E−05 | 7.58E−05 | 7.52E−01 | 7.52E−01 |
| IgD unmutated | Entropy (bit) | 10.88 (1.88) | 12.20 (1.35) | 12.18 (1.02) | 12.90 (0.88) | 3.95E−02 | 1.65E−01 | 9.56E−02 | 5.59E−02 |
| CDR3 length (nt) | 46.06 (0.92) | 45.91 (0.74) | 42.81 (1.02) | 40.31 (0.90) | 9.47E−07 | 1.08E−05 | 7.96E−01 | 1.02E−06 | |
| IgD mutated | Entropy (bit) | 9.54 (2.19) | 10.66 (1.15) | 11.82 (0.36) | 11.98 (0.22) | 3.38E−04 | 1.05E−03 | 1.72E−01 | 1.92E−01 |
| CDR3 length (nt) | 45.07 (1.75) | 45.01 (1.44) | 43.49 (0.99) | 41.10 (1.09) | 1.22E−02 | 1.08E−05 | 7.96E−01 | 7.14E−06 | |
| IgM | SHM (rate) | 1.76E−2 (2.23E−3) | 1.77E−2 (2.36E−3) | 1.16E−2 (5.7E−4) | 0.0117 (6.4E−4) | 4.95E−08 | 4.11E−05 | 8.77E−01 | 8.77E−01 |
| IgM unmutated | Entropy (bit) | 11.20 (1.46) | 12.27 (0.98) | 12.41 (0.55) | 12.88 (0.4) | 2.41E−02 | 1.13E−01 | 4.56E−02 | 1.58E−02 |
| CDR3 length (nt) | 44.67 (0.75) | 44.82 (0.76) | 41.70 (0.90) | 39.70 (0.85) | 9.97E−08 | 4.11E−05 | 5.16E−01 | 7.34E−05 | |
| IgM mutated | Entropy (bit) | 10.26 (1.35) | 10.76 (0.99) | 12.33 (0.19) | 12.44 (0.16) | 3.34E−06 | 1.65E−04 | 5.57E−01 | 6.22E−02 |
| CDR3 length (nt) | 41.97 (1.19) | 42.43 (1.69) | 42.27 (0.92) | 40.43 (1.06) | 8.04E−01 | 5.63E−03 | 6.88E−01 | 4.70E−04 | |
| IgA | Entropy (bit) | 7.78 (1.12) | 8.00 (0.81) | 2.89 (2.54) | 3.59 (1.76) | 2.88E−04 | 4.11E−05 | 6.66E−01 | 6.66E−01 |
| SHM (rate) | 8.36E−2 (1.14E−2) | 8.07E−2 (7.9E−3) | 1.68E−2 (2.8E−2) | 5.21E−3 (9.08E−3) | 4.94E−04 | 4.11E−05 | 2.97E−01 | 2.97E−01 | |
| CDR3 length (nt) | 42.77 (1.39) | 41.00 (1.30) | 41.56 (7.38) | 39.41 (2.85) | 2.58E−01 | 1.13E−01 | 2.44E−02 | 9.31E−01 | |
| IgG | Entropy (bit) | 6.80 (1.26) | 7.69 (0.73) | 2.22 (1.74) | 3.75 (1.82) | 2.31E−06 | 8.23E−05 | 2.52E−02 | 7.08E−02 |
| SHM (rate) | 0.0852 (0.0126) | 0.0745 (0.0102) | 2.28E−3 (7.39E−4) | 2.96E−3 (1.34E−3) | 1.92E−07 | 4.11E−05 | 1.86E−01 | 1.86E−01 | |
| CDR3 length (nt) | 42.80 (1.73) | 42.36 (2.25) | 44.53 (5.22) | 42.32 (6.52) | 2.04E−01 | 6.05E−01 | 6.47E−01 | 1.26E−01 |
Summary of TCR-β and IgH repertoire analysis from term and preterm pregnancies grouped by entropy, CDR3 length, and SHM. Comparisons between groups were assessed using the Wilcoxon rank-sum test.
Table III.
Significant usage differences in term pregnancies (maternal blood versus cord blood)
| V Genes | D Genes | J Genes | |
|---|---|---|---|
| TCR | |||
| Higher cord blood usage | TRBV6-5 (1.42), TRBV6-6 (1.62), TRBV10-3 (1.31) | TRBD2 (1.07) | TRBJ2-3 (1.22), TRBJ2-4 (1.42) |
| Higher maternal blood usage | TRBV7-8 (1.56), TRBV7-9 (1.33), TRBV18 (1.64) | ||
| IgD unmutated | |||
| Higher cord blood usage | IGHV6-1 (2.71) | IGHD1-7 (1.74), IGHD1-26 (1.57), IGHD6-6 (1.93), IGHD7-27 (9.08) | IGHJ1 (1.47), IGHJ2 (2.49), IGHJ3 (1.88), IGHJ4 (1.09) |
| Higher maternal blood usage | IGHV3-48 (1.37) | IGHD3-16 (1.29), IGHD4-17 (1.59), IGHD6-19 (1.43) | IGHJ5 (1.35), IGHJ6 (1.55) |
| IgD mutated | |||
| Higher cord blood usage | IGHD1-26 (1.55), IGHD6-6 (2.03), IGHD7-27 (5.97) | IGHJ2 (2.53), IGHJ3 (1.83) | |
| Higher maternal blood usage | IGHD2-15 (1.45), IGHD5-24 (1.53), IGHD6-19 (1.42) | IGHJ5 (1.46), IGHJ6 (1.48) | |
| IgM unmutated | |||
| Higher cord blood usage | IGHV6-1 (2.81) | IGHD1-7 (1.59), IGHD1-26 (1.43), IGHD6-6 (1.93), IGHD6-13 (1.34), IGHD7-27 (7.52) | IGHJ2 (2.15), IGHJ3 (1.69) |
| Higher maternal blood usage | IGHV1-46 (1.43), IGHV3-21 (1.23) | IGHD2-15 (1.62), IGHD3-10 (1.29), IGHD4-17 (1.56), IGHD6-9 (1.42) | IGHJ5 (1.52), IGHJ6 (1.54) |
| IgM mutated | |||
| Higher cord blood usage | IGHV1-3 (1.83) | IGHD1-26 (1.40), IGHD6-6 (2.07), IGHD6-13 (1.51), IGHD7-27 (5.68) | IGHJ2 (2.29), IGHJ3 (1.75) |
| Higher maternal blood usage | IGHV3-7 (1.60), IGHV3-48 (1.56), IGHV3-74 (1.92) | IGHD2-15 (1.56), IGHD3-16 (1.33), IGHD4-17 (1.49), IGHD6-19 (1.53), IGHD6-25 (1.62) | IGHJ4 (1.06), IGHJ5 (1.58), IGHJ6 (1.37) |
Summary of term maternal versus neonate analysis for TCR-β and IgH V, D, and J gene segment usage. In pregnancies carried to term, a substantial number of genes had significantly higher usage in cord blood (compared with maternal blood) or in maternal blood (compared with cord blood), across TCR and various IgH isotypes. The usage difference in fold change is noted in parentheses next to each gene.
Maternal repertoires exhibit higher rates of SHM and longer CDR3s than matched cord blood samples in term pregnancies
From the reads for each sample, we calculated average SHM rates and CDR3 lengths. SHMs help shape the IgH repertoire, generating BCRs with the potential for high-affinity Ag binding (13, 24). Although this is a well-described process occurring in immune-competent individuals as part of successful adaptive immunity to a variety of pathogens, SHM during fetal development remains understudied. In term pregnancies, we measured and compared the rate of SHM between maternal blood and cord blood and found higher SHM rates in maternal blood for IgA, IgD, IgG, and IgM (Fig. 2B, Table II). We observed similar results in PPROM pregnancies (Supplemental Fig. 1).
The CDR3 regions of TCR-β and IgH receptors play a critical role in Ag binding and recognition. To characterize the CDR3 sequences between maternal blood and cord blood, we first performed a comparative analysis of the mean CDR3 nucleotide sequence lengths (Fig. 2C, Table II) and observed significantly shorter TCR-β CDR3 lengths in cord blood samples. For the IgH repertoire, the mean CDR3 lengths were significantly shorter for cord blood samples than for maternal blood samples in unmutated IgD, mutated IgD, and unmutated IgM.
V, D, and J gene usage differences are identified in maternal versus cord blood in term pregnancies
We next examined whether there was preferential usage of specific gene segments in TCR-β and IgH CDR3s and found skewed usage of specific gene segments. The analysis revealed a number of V, D, and J genes with either increased or decreased usage in term cord blood samples compared with that in term maternal blood samples. The significant (adjusted p value < 0.05) results are represented in Table III. Taken in combination, the V genes and J genes with significantly different usage result in two distinct sample clusters via hierarchical clustering, with maternal blood and cord blood samples perfectly separated (p = 1.55E−45) (Fig. 2D).
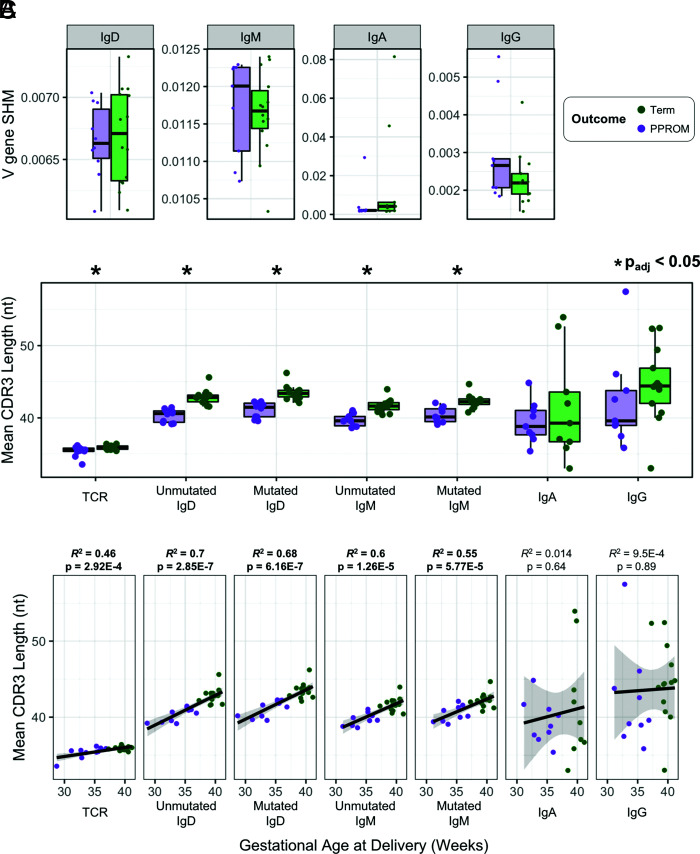
Term cord blood repertoires exhibit longer CDR3s in comparison with PPROM cord blood
Using the same procedures as when comparing term maternal blood with term cord blood, we characterized differences between term cord blood and PPROM cord blood to further investigate the role of the immune repertoire in PTL (Table II). We examined the IgH rates of SHM and observed no differences between term and PPROM cord blood samples for any of the IgH isotypes (Fig. 3A, Table II); however, shorter CDR3 lengths in PPROM samples were observed in all the naive-like B cell compartments (IgD and IgM) as well as in TCR-β (Fig. 3B). Furthermore, we found that these differences in length (IgD, IgM, and TCR-β) correlated significantly (p < 0.05) with gestational age recorded at the time of birth (Fig. 3C).
FIGURE 3.
Term versus PPROM cord blood TCR-β and IgH comparisons for rates of SHM and CDR3 length. (A) IgH V gene SHM rates for term and PPROM cord blood samples, separated by isotype. (B) For each cord blood sample, the mean CDR3 lengths for TCR-β and each IgH isotype were measured to investigate the differences between term and PPROM groups. (C) The mean CDR3 length of each sample was plotted against the gestational age at delivery.
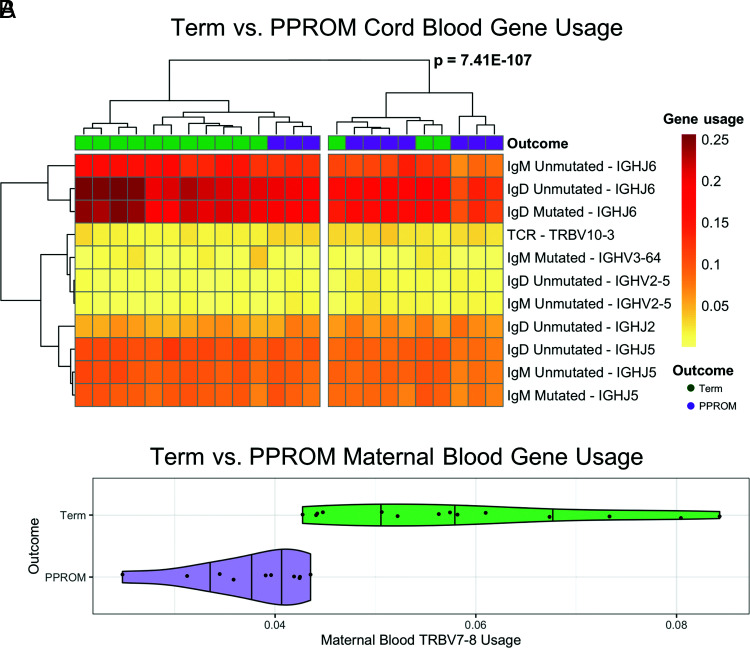
Significant differences in V, D, and J gene usage are identified in term versus PPROM
To further investigate the impact of PPROM deliveries in the cord blood immune receptor repertoire, we analyzed different V, D, and J gene segment usage for the TCR-β and IgH for term and PPROM cord blood. We found many V, D, and J genes with significantly (adjusted p value < 0.05) higher or lower usage in preterm samples relative to term samples (Table IV). Three V genes (TRBV10-3, IGHV2-5, and IGHV3-64) and three J genes (IGHJ2, IGHJ5, and IGHJ6) were used at significantly different rates between preterm and term. Applying hierarchical cluster analysis, we observed separation into two clusters (p = 7.41E−107), with significant (p = 0.035, Fisher exact test) separation between term and preterm outcomes for the significant V genes and the significant J genes (Fig. 4A).
Table IV.
Significant usage differences in cord bloom (term versus PPROM)
| V Genes | D Genes | J Genes | |
|---|---|---|---|
| TCR | |||
| Higher PPROM usage | TRBV10-3 (1.44) | ||
| Higher term usage | |||
| IgD unmutated | |||
| Higher PPROM usage | IGHV2-5 (2.03) | IGHD7-27 (1.55) | IGHJ2 (1.20) |
| Higher term usage | IGHJ5 (1.15), IGHJ6 (1.30) | ||
| IgD mutated | |||
| Higher PPROM usage | |||
| Higher term usage | IGHJ6 (1.28) | ||
| IgM unmutated | |||
| Higher PPROM usage | IGHV2-5 (2.07) | IGHD7-27 (1.48) | |
| Higher term usage | IGHJ5 (1.15), IGHJ6 (1.35) | ||
| IgM mutated | |||
| Higher PPROM usage | |||
| Higher term usage | IGHV3-64 (3.73) | IGHJ5 (1.14) |
Summary of term versus PPROM neonate analysis for TCR-β and IgH V, D, and J gene segment usage. A small number of genes were observed to have higher usage in PPROM cord blood (compared with term cord blood) or in term cord blood (compared with PPROM cord blood). The usage difference in fold change is noted in parentheses next to each gene.
FIGURE 4.
TCR-β and IgH gene usage heat map of term versus PPROM groups. (A) For cord blood samples, hierarchical clustering of V genes and J genes with significantly different usage resulted in two sample clusters. (B) Violin plot (with overlaid scatterplot) of the usage of TRBV7-8 in maternal blood samples. The shape of the plot reflects the probability distribution, and the vertical bars denote the quartile boundaries (25, 50, and 75%).
Next, we investigated the differences in the maternal immune repertoire between pregnancies carried to term and pregnancies ending with preterm birth. When comparing term maternal blood samples with PPROM maternal blood samples, we found no differences in the entropy, rate of SHM, and mean CDR3 length (Table II, Supplemental Fig. 2). In contrast, analysis of gene segment usage of TCR-β revealed significantly increased usage of TRBV7-8 in term pregnancies (Fig. 4B).
Higher abundance of public clones in PPROM pregnancies
Because of large recombinatorial biases of receptors, a small fraction of TCR α-chains, TCR-β, or IgH chains are shared among most individuals. These “public” clones have been found to be prevalent in some infectious, autoimmune, and other immune responses (25, 26) and among preterm neonates (27). The investigation of public clones in normal and pathological pregnancies remains relatively understudied, and it may provide new insights in immune dysregulation leading to disturbance of maternal–fetal tolerance and pathological delivery. We investigated the presence of public clones, defined as shared across at least two samples. Across individuals, clones can be similar by recognizing the same Ags and were stimulated to expand and appear at higher frequencies in the repertoires of the two individuals with common Ag exposure. For examining clones across samples, we use a similar clone identification procedure as before, but instead of using nucleotide identity matching, we instead use amino acid identity matching. We thus defined two sequences as belonging to the same public clone if they shared the same V gene, same J gene, equal CDR3 length, and a minimum 85% (IgH) or 100% (TCR-β) amino acid identity match, with a lower threshold for IgH to account for SHMs. To rule out sample contamination, we checked the clones for 100% identical nucleotide sequences and found no fully identical clones at the nucleotide level between matched pairs of maternal blood and cord blood.
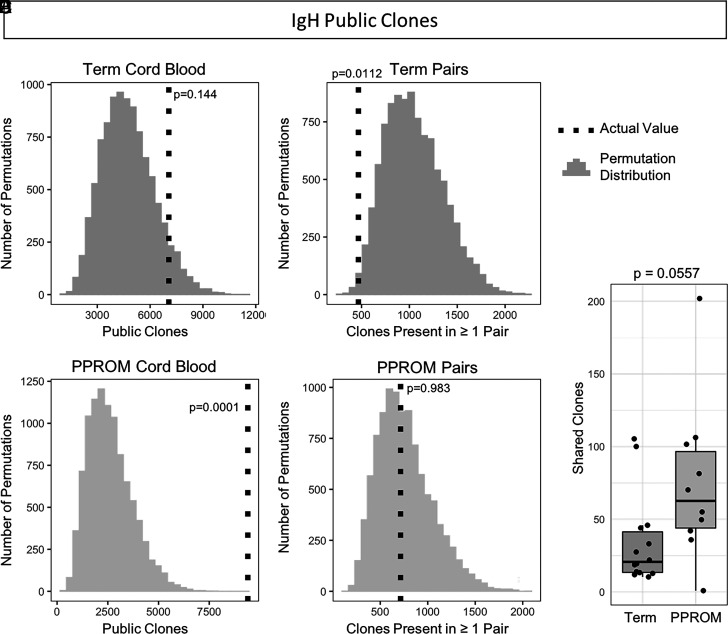
Across all term cord blood samples (n = 14) and aggregated over multiple downsampling repetitions, an average of 7062 IgH clones were present in at least two samples, whereas across all preterm cord blood samples (n = 10), there was an average of 9400 public clones. We compared these values with random distributions of expected public clone numbers generated by randomly shuffling (resampling) sample labels. For term cord blood samples, the number of public clones was on the upper end of the distribution, although not significant (p = 0.144) (Fig. 5A). However, for the PPROM cord blood samples, we observed significant enrichment in the number of public clones compared with the distribution obtained at random (p = 0.0001) (Fig. 5B). Next, we used a similar procedure to measure the number of public clones across matched mother/baby pairs, looking for public clones that were shared by both the maternal blood sample and the cord blood sample. In term pregnancies, we observed 468 unique clones present in both samples from at least one pair, significantly fewer compared with random (p = 0.0122) (Fig. 5C). In PPROM pregnancies, we observed an average of 714 unique clones from at least one pair (p = 0.983) (Fig. 5D). When comparing the distributions of shared clones between maternal blood and cord blood pairs that were term or PPROM, there was no significant difference between the two distributions (Fig. 5E).
FIGURE 5.
Enrichment of IgH public clones observed in preterm cord blood samples. (A) Observed value and expected distribution of IgH public clones in term cord blood. The expected distribution obtained from 10,000 permutations is shown as a histogram, and the actual observed value of public clones is overlaid as a vertical dotted line. The p value is determined by comparing the observed value to the permutation distribution. (B) Observed value and expected distribution of IgH public clones in PPROM cord blood. (C) IgH public clones observed in both the maternal and fetal samples of matched pairs in term pregnancies. (D) IgH public clones observed in match pairs for PPROM pregnancies. (E) Numbers of shared IgH clones between paired maternal blood and cord blood samples.
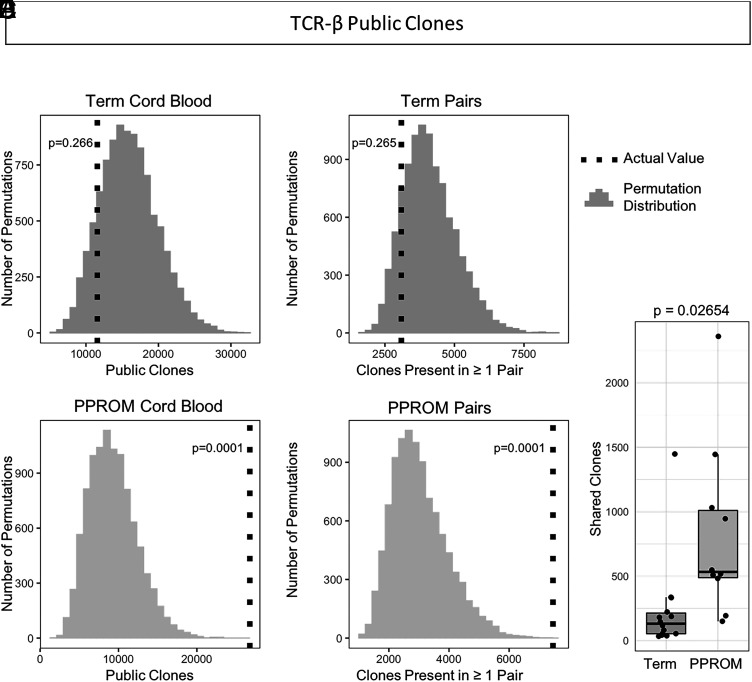
For public TCR clones in term cord blood, there was an average of 11,575 public clones (p = 0.266) (Fig. 6A). Across PPROM cord blood samples, there was an average of 26,751 public TCR clones, significantly more than random (p = 0.0016) (Fig. 6B). For public clones across paired samples, there were 3090 average clones across paired term samples (p = 0.265) (Fig. 6C) and 7459 average clones across paired PPROM samples (p = 0.0022) (Fig. 6D). Comparing the distributions of public clones in individual pairs, we observed significantly more public clones in the preterm samples over the term samples (p = 0.02654, Wilcoxon rank-sum test) (Fig. 6E).
FIGURE 6.
More TCR public clones observed in preterm cord blood samples and preterm paired samples. (A) Observed value and expected distribution of TCR public clones in term cord blood. The expected distribution obtained from 10,000 permutations is shown as a histogram, and the actual observed value of public clones is overlaid as a vertical dotted line. The p value is determined by comparing the observed value to the permutation distribution. (B) Observed value and expected distribution of TCR public clones in PPROM cord blood. (C) TCR public clones observed in both the maternal and fetal samples of matched pairs in term pregnancies. (D) TCR public clones observed in matched pairs for PPROM pregnancies. (E) Numbers of shared TCR clones between paired maternal blood and cord blood samples. Preterm pregnancies had more public clones shared by both the maternal and fetal samples than term pregnancies had.
Discussion
We studied the immune repertoires of mothers and babies delivered at term or prematurely by sequencing the TCR-β and IgH of maternal blood and fetal cord samples taken at delivery. As a pilot study, we collected a limited number of samples but were still able to find significant differences in immune repertoires between both maternal/cord blood groupings and term/PPROM groupings. In term pregnancies, we observed higher rates of SHM and longer CDR3s in maternal samples. For cord blood, fetal PPROM repertoires had shorter CDR3s than fetal term repertoires had. We also found a number of V, D, and J gene usage differences across groupings. Finally, in examining public clones expressed among different samples, we found enriched numbers of clones in PPROM cord blood samples, as well as higher numbers of public clones appearing in PPROM maternal blood/cord blood pairs.
We observed elevated rates of SHM in term maternal blood to term cord blood. These increases are consistent with previous results (28, 29). In past work studying the shaping of infant B cell repertoires, we observed that IgM and IgD reach adult average SHM frequencies by 1–2 y of age, whereas class-switched isotypes (IgA and IgG) only reach ∼60% of adult values by 3 y of age (13).
Our findings of shorter CDR3 lengths in term cord blood (versus term maternal blood) are similar to results previously described (17). These differences, although statistically significant, were small, especially in TCR-β, in which maternal blood and cord blood differed by ∼1 nt. For the CDR3 differences in IgH, the increased usage of IGHJ6 in term cord blood can explain in part the CDR3 length differences, as IGHJ6 is among the longest IGHJ gene segments. We observed large variation in both the IgA and the IgG CDR3 lengths in cord blood, which is most likely due to the low number of IgA and IgG clones observed (Supplemental Table I).
The gene segments with significantly different usage between term maternal blood samples and term cord blood samples include IGHV6-1 [a J-proximal gene reported to have higher usage in fetal and cord blood samples (30)] and IGHD7-27 [J-proximal and the shortest D gene with higher usage in cord blood samples (17) and fetal samples (31)]. These gene segments may represent combinatorial rearrangements specific to the fetal and neonate repertoire development. In comparing term cord blood and PPROM cord blood samples, we found three V genes with significantly different usage. Many of these V genes with significantly different usage in cord blood are novel in the context of preterm birth, including IGHV2-5 and TRBV10-3. We found few differences in maternal blood when comparing the lymphocyte receptor repertoires of term pregnancies with those of PPROM pregnancies. Only the V gene TRBV7-8 had significantly decreased usage in PPROM pregnancies (Fig. 4B). TRBV7-8 usage has been reported as gene segment involved in TCR autoreactivity toward rare self-phospholipids such as phosphatidylglycerol (32), which rises in concentration in amniotic fluid as the pregnancy approaches term, an indicator of fetal maturity. The increased usage near term may suggest specific TCR gene segment usage involved in the physiology of term birth. Further functional TCR repertoire studies investigating the maternal TCR near term may point to new insights in T cell response in term labor.
For paired mother/baby samples, we observed more clone sharing in the PPROM cases compared with term controls. Among PPROM cord blood samples, we observed a relative abundance of public clones. We speculate that the enriched abundance of public clones among PPROM infants might be due to a less developed repertoire and also as an increase in microchimerism, potentially leading to a breakdown of maternal–fetal tolerance in PPROM (8). The increased public sharing is consistent, with one report studying CD8 naive T cell repertoire in which preterm neonates showed higher clonal sharing when compared with term neonates (27). These results further support the hypothesis of increased maternal immune cell trafficking across the placenta and into the fetus in PTL cases, leading to increased microchimerism levels. This may disturb the immune balance between tolerance and rejection, leading to breakdown of maternal–fetal tolerance and PTL.
There were several limitations to our study that should be noted. This study was conceived as a pilot study, with the small number of samples collected used as a proof of concept for this type of analysis, limiting our power to detect subtle statistical and biological changes. Because of the small sample size, additional follow-up and verification is required especially for the preferential V, D, and J gene usages. Another limiting factor is the inability to reasonably study blood from unborn infants; the usage of cord blood as a surrogate may not be ideal in sequencing the immune repertoire of the baby at the time of delivery. We did not perform HLA typing, and there was a significant difference in the proportions of primigravid mother between the term and PPROM groups that could not be further resolved because of the small sample size. Although the downsampling approach ensured the same number of reads per sample, this method also resulted in small read counts, especially for certain BCR isotypes. Samples could also be studied with more-advanced recent technologies that allow BCR and TCR measurements at the single-cell level. The public clone analyses would greatly benefit from a restricted design to eliminate sample contamination. As the samples were collected at the time of delivery, this is difficult to achieve in practice. However, we have carried out extensive analysis on the nucleotide sequence level of the public clones to make sure that although the amino acid sequences are concordant, there are mutations on the nucleotide level that ensure that these observations are not due to contamination.
In studying maternal blood and cord blood samples from pregnancies delivered at term or delivered early because of PPROM, we observed numerous differences in the adult and neonatal immune repertoire. Overall, infant immune repertoires were much more diverse and less developed than maternal immune repertoires, as shown by increased SHM and longer CDR3s in the mothers. Moreover, the immune repertoires of term babies had evidence of more-developed repertoires compared with those of PPROM babies, as shown by longer TCR, IgD, and IgM CDR3s, which correlated with gestational age. We also found a number of V genes and J genes differentiated between term and PPROM pregnancies, primarily in cord blood. Future studies will examine a larger number of samples in addition to longitudinal maternal lymphocyte receptor profiling throughout pregnancy to gain a better understanding of the maternal and fetal immune repertoires during pregnancy and how they might impact early parturition. It will be of interest to explore the role that Ig genotypes of infants and mothers play in this analysis. Future studies should further examine whether repertoire changes associated with PTL may have an impact on adaptive immunity development.
Supplementary Material
Acknowledgments
We thank Daniel Bunis for useful discussions and Pooja Kathail for technical assistance.
This work was supported by the Burroughs Wellcome Fund (to T.C.M. and M.S.), the National Institutes of Health R01 AI116880 (to T.C.M.), and the Ulla og Mogens Folmer Andersens Fond (to S.C.A.N.).
B.L.L., R.S., S.D.B., T.C.M., and M.S. designed and coordinated the study. R.S., Q.-H.N., and T.C.M. collected the specimens. S.C.A.N., J.-Y.L., and S.D.B. sequenced the samples. B.L.L. and S.P. developed and implemented the analysis pipeline. All the authors contributed to making figures and writing and editing the manuscript.
The sequences presented in this article have been submitted to ImmPort (https://www.immport.org/shared/home) under accession number SDY1769.
The online version of this article contains supplemental material.
- FR
- framework region
- PPROM
- preterm premature rupture of membrane
- PTL
- preterm labor
- SHM
- somatic hypermutation
- TCR-β
- TCR β-chain
Disclosures
The authors have no financial conflicts of interest.
References
- 1. Blencowe H., Cousens S., Oestergaard M. Z., Chou D., Moller A.-B., Narwal R., Adler A., Vera Garcia C., Rohde S., Say L., Lawn J. E.. 2012. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 379: 2162–2172. [DOI] [PubMed] [Google Scholar]
- 2. Purisch S. E., Gyamfi-Bannerman C.. 2017. Epidemiology of preterm birth. Semin. Perinatol. 41: 387–391. [DOI] [PubMed] [Google Scholar]
- 3. Mercer B. M., Lewis R.. 1997. Preterm labor and preterm premature rupture of the membranes. Diagnosis and management. Infect. Dis. Clin. North Am. 11: 177–201. [DOI] [PubMed] [Google Scholar]
- 4. Macones G. A., Parry S., Elkousy M., Clothier B., Ural S. H., Strauss J. F. III. 2004. A polymorphism in the promoter region of TNF and bacterial vaginosis: preliminary evidence of gene-environment interaction in the etiology of spontaneous preterm birth. Am. J. Obstet. Gynecol. 190: 1504–1508, discussion 3A. [DOI] [PubMed] [Google Scholar]
- 5. Burns D. N., Landesman S., Muenz L. R., Nugent R. P., Goedert J. J., Minkoff H., Walsh J. H., Mendez H., Rubinstein A., Willoughby A.. 1994. Cigarette smoking, premature rupture of membranes, and vertical transmission of HIV-1 among women with low CD4+ levels. J Acquir Immune Defic Syndr (1988) 7: 718–726. [PubMed] [Google Scholar]
- 6. Silverman R. K., Wojtowycz M.. 1998. Risk factors in premature rupture of membranes. Prim. Care Update Ob. Gyns. 5: 181. [DOI] [PubMed] [Google Scholar]
- 7. Menon R. 2008. Spontaneous preterm birth, a clinical dilemma: etiologic, pathophysiologic and genetic heterogeneities and racial disparity. Acta Obstet. Gynecol. Scand. 87: 590–600. [DOI] [PubMed] [Google Scholar]
- 8. Frascoli M., Coniglio L., Witt R., Jeanty C., Fleck-Derderian S., Myers D. E., Lee T.-H., Keating S., Busch M. P., Norris P. J., et al. 2018. Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-γ and TNF-α. Sci. Transl. Med. 10: eaan2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Halkias J., Rackaityte E., Hillman S. L., Aran D., Mendoza V. F., Marshall L. R., MacKenzie T. C., Burt T. D.. 2019. CD161 contributes to prenatal immune suppression of IFNγ-producing PLZF+ T cells. J. Clin. Invest. 129: 3562–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vora B., Wang A., Kosti I., Huang H., Paranjpe I., Woodruff T. J., MacKenzie T., Sirota M.. 2018. Meta- analysis of maternal and fetal transcriptomic data elucidates the role of adaptive and innate immunity in preterm birth. Front. Immunol. 9: 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le B. L., Iwatani S., Wong R. J., Stevenson D. K., Sirota M.. 2020. Computational discovery of therapeutic candidates for preventing preterm birth. JCI Insight 5: e133761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Witt R. G., Blair L., Frascoli M., Rosen M. J., Nguyen Q.-H., Bercovici S., Zompi S., Romero R., Mackenzie T. C.. 2020. Detection of microbial cell-free DNA in maternal and umbilical cord plasma in patients with chorioamnionitis using next generation sequencing. PLoS One 15: e0231239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nielsen S. C. A., Roskin K. M., Jackson K. J. L., Joshi S. A., Nejad P., Lee J.-Y., Wagar L. E., Pham T. D., Hoh R. A., Nguyen K. D., et al. 2019. Shaping of infant B cell receptor repertoires by environmental factors and infectious disease. Sci. Transl. Med. 11: eaat2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rechavi E., Somech R.. 2017. Survival of the fetus: fetal B and T cell receptor repertoire development. Semin. Immunopathol. 39: 577–583. [DOI] [PubMed] [Google Scholar]
- 15. Nielsen S. C. A., Boyd S. D.. 2019. New technologies and applications in infant B cell immunology. Curr. Opin. Immunol. 57: 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schatz D. G., Ji Y.. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11: 251–263. [DOI] [PubMed] [Google Scholar]
- 17. Guo C., Wang Q., Cao X., Yang Y., Liu X., An L., Cai R., Du M., Wang G., Qiu Y., et al. 2016. High-throughput sequencing reveals immunological characteristics of the TRB-/IgH-CDR3 region of umbilical cord blood. J. Pediatr. 176: 69–78.e1. [DOI] [PubMed] [Google Scholar]
- 18. Zemlin M., Hoersch G., Zemlin C., Pohl-Schickinger A., Hummel M., Berek C., Maier R. F., Bauer K.. 2007. The postnatal maturation of the immunoglobulin heavy chain IgG repertoire in human preterm neonates is slower than in term neonates. J. Immunol. 178: 1180–1188. [DOI] [PubMed] [Google Scholar]
- 19. van Dongen J. J. M., Langerak A. W., Brüggemann M., Evans P. A., Hummel M., Lavender F. L., Delabesse E., Davi F., Schuuring E., García-Sanz R., et al. 2003. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17: 2257–2317. [DOI] [PubMed] [Google Scholar]
- 20. Qi Q., Liu Y., Cheng Y., Glanville J., Zhang D., Lee J.-Y., Olshen R. A., Weyand C. M., Boyd S. D., Goronzy J. J.. 2014. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl. Acad. Sci. USA 111: 13139–13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Magoč T., Salzberg S. L.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye J., Ma N., Madden T. L., Ostell J. M.. 2013. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41: W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Glanville J., Kuo T. C., von Büdingen H.-C., Guey L., Berka J., Sundar P. D., Huerta G., Mehta G. R., Oksenberg J. R., Hauser S. L., et al. 2011. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc. Natl. Acad. Sci. USA 108: 20066–20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z., Woo C. J., Iglesias-Ussel M. D., Ronai D., Scharff M. D.. 2004. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 18: 1–11. [DOI] [PubMed] [Google Scholar]
- 25. Fazilleau N., Delarasse C., Sweenie C. H., Anderton S. M., Fillatreau S., Lemonnier F. A., Pham-Dinh D., Kanellopoulos J. M.. 2006. Persistence of autoreactive myelin oligodendrocyte glycoprotein (MOG)-specific T cell repertoires in MOG-expressing mice. Eur. J. Immunol. 36: 533–543. [DOI] [PubMed] [Google Scholar]
- 26. Menezes J. S., van den Elzen P., Thornes J., Huffman D., Droin N. M., Maverakis E., Sercarz E. E.. 2007. A public T cell clonotype within a heterogeneous autoreactive repertoire is dominant in driving EAE. J. Clin. Invest. 117: 2176–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carey A. J., Hope J. L., Mueller Y. M., Fike A. J., Kumova O. K., van Zessen D. B. H., Steegers E. A. P., van der Burg M., Katsikis P. D.. 2017. Public clonotypes and convergent recombination characterize the naïve CD8+ T-cell receptor repertoire of extremely preterm neonates. Front. Immunol. 8: 1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hong B., Wu Y., Li W., Wang X., Wen Y., Jiang S., Dimitrov D. S., Ying T.. 2018. In-depth analysis of human neonatal and adult IgM antibody repertoires. Front. Immunol. 9: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prabakaran P., Chen W., Singarayan M. G., Stewart C. C., Streaker E., Feng Y., Dimitrov D. S.. 2012. Expressed antibody repertoires in human cord blood cells: 454 sequencing and IMGT/HighV-QUEST analysis of germline gene usage, junctional diversity, and somatic mutations. Immunogenetics 64: 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Es J. H., Raaphorst F. M., van Tol M. J., Meyling F. H., Logtenberg T.. 1993. Expression pattern of the most JH-proximal human VH gene segment (VH6) in the B cell and antibody repertoire suggests a role of VH6-encoded IgM antibodies in early ontogeny. J. Immunol. 150: 161–168. [PubMed] [Google Scholar]
- 31. Rechavi E., Lev A., Lee Y. N., Simon A. J., Yinon Y., Lipitz S., Amariglio N., Weisz B., Notarangelo L. D., Somech R.. 2015. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci. Transl. Med. 7: 276ra25. [DOI] [PubMed] [Google Scholar]
- 32. Shahine A., Van Rhijn I., Cheng T.-Y., Iwany S., Gras S., Moody D. B., Rossjohn J.. 2017. A molecular basis of human T cell receptor autoreactivity toward self-phospholipids. Sci. Immunol. 2: eaao1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.