SUMMARY
Sex chromosomes are generally derived from a pair of classical type-A chromosomes, and relatively few alternative models have been proposed up to now.1,2 B chromosomes (Bs) are supernumerary and dispensable chromosomes with non-Mendelian inheritance found in many plant and animal species3,4 that have often been considered as selfish genetic elements that behave as genome parasites.5,6 The observation that in some species Bs can be either restricted or predominant in one sex7–14 raised the interesting hypothesis that Bs could play a role in sex determination.15 The characterization of putative B master sex-determining (MSD) genes, however, has not yet been provided to support this hypothesis. Here, in Astyanax mexicanus cavefish originating from Pachón cave, we show that Bs are strongly male predominant. Based on a high-quality genome assembly of a B-carrying male, we characterized the Pachón cavefish B sequence and found that it contains two duplicated loci of the putative MSD gene growth differentiation factor 6b (gdf6b). Supporting its role as an MSD gene, we found that the Pachón cavefish gdf6b gene is expressed specifically in differentiating male gonads, and that its knockout induces male-to-female sex reversal in B-carrying males. This demonstrates that gdf6b is necessary for triggering male sex determination in Pachón cavefish. Altogether these results bring multiple and independent lines of evidence supporting the conclusion that the Pachón cavefish B is a “B-sex” chromosome that contains duplicated copies of the gdf6b gene, which can promote male sex determination in this species.
Graphical abstract

In brief
B chromosomes (Bs) are supernumerary chromosomes. Their implication in sex determination has been often suggested, but expressional and functional proofs are scarce. Imarazene et al. show that Pachón cavefish, Astyanax mexicanus, males have a predominant B-sex chromosome that contains a gene that can promote male sex determination in this species.
RESULTS AND DISCUSSION
Pachón cavefish B chromosomes are male-predominant B chromosomes
Supernumerary B chromosomes (Bs) are generally thought to arise from the duplication and assembly of A chromosome sequences,16–19 and their relationship to sex chromosomes has often been suspected and discussed.15 Some hypotheses state that Bs are derived from sex chromosomes or, alternatively, evolved to become sex chromosomes.15,20–23 Because Bs have been described in A. mexicanus,19,24,25 we performed cytogenetic analyses in 17 males and 11 females of a laboratory population of Pachón cavefish (Pachón) to investigate whether Pachón Bs could be sex restricted. We found that Pachón Bs are euchromatic mitotically unstable microchromosomes (Figures 1A, 1B, and S1) that are present in one to three copies in most male metaphases (mean number ± SD of Bs per male metaphase = 1.08 ± 0.41), contrasting with a barely detectable B occurrence in female metaphases (mean number ± SD of Bs per female metaphase = 0.05 ± 0.08; Figure 1C; Data S1A). Chromosomal mapping by fluorescence in situ hybridization (FISH) using probes generated from microdissected male Bs painted Bs in males and even the rare B in females, supporting that Pachón male-predominant Bs and the low-occurrence female Bs share similar genomic DNA (gDNA) content (Figure 1D). In addition, weaker FISH signals were also detected on different terminal parts of some A chromosomes (see small white arrows in Figure 1D), suggesting that Pachón Bs are made up of many duplicated fragments of A chromosomes,16–19 and/or that they share repetitive DNAs with the A chromosomes.21,24,26 Female or male sex-restricted or sex-predominant Bs have been described in many fishes–for instance, in some cichlids12–14 and characiforms.8,9,27,28 In A. mexicanus, Bs were recently described as being restricted only in some, but not all, males,19 suggesting population differences in the frequency and sex linkage of Bs that have also been reported in many species.4–6,29 In addition to metaphase spreads (Figures 1E and 1F), we also detected Bs in pachytene chromosome spreads from testes of two Pachón males (n = 52 and n = 25). Single Bs were found in most cells (44/52 and 23/25) and always as unpaired chromosomes (Figures 1G and 1H). Hence, Pachón Bs are present in meiotic germ cells and they do not pair with A chromosomes, in line with what has been found in other species with Bs.30 The question of whether Pachón Bs can pair to each other remains open because we detected no case of multiple Bs in these pachytene chromosome spreads.
Figure 1. Karyological characterization of male-predominant supernumerary B chromosomes (Bs) in Pachón cave Astyanax mexicanus.

(A and B) Representative C-banding patterns of single B+ male (A) and B− female (B) from Pachón cave. The Bs (black arrow) lack C-bands, suggesting that Pachón cavefish Bs are largely euchromatic. See also Figure S1.
(C) Boxplots of the average number of Bs per metaphase in Pachón cavefish males and females. Horizontal lines indicate the median, the box indicates the interquartile range (IQR), and the whiskers the range of values that are within 1.5× IQR. Statistical significance between males and females was tested with the Wilcoxon rank test (***p < 0.001). See also Data S1A.
(D) Fluorescence in situ hybridization (FISH) of male Pachón cave mitotic metaphase labeled with combined microdissected B probes. Yellow arrows point to the strong labeling of Bs and the small white arrows to the lighter labeling of some different parts of A chromosomes.
(E and F) FISH co-labeling of a 1B male (E) and a 1B female (F) metaphase with microdissected B (green) and gdf6b-specific (red) probes. Bs are indicated by yellow arrows and the white arrowheads point to pairs of A chromosome sister chromatids labeled by the gdf6b probe. Only one A-chromosome gdf6b signal was detected in (E). The two gdf6b signals (see inset in E) that were often visible on male metaphases cannot be interpreted as the two different B-gdf6b loci due to their genomic proximity (see Data S1C) and the FISH resolution.
(G and H) Synaptonemal complex (SC) analysis showing that Pachón cave Bs (yellow arrow) do not pair with the 25 fully synapsed standard bivalents of A chromosomes. SCs were visualized by anti-SYCP3 antibody (green), the recombination sites were identified by anti-MLH1 antibody (red), and chromosomes were counterstained by DAPI (blue). (G) Merged image. (H) SYCP3 visualization only.
Scale bars, 5 μm.
Characterization of the Pachón cavefish B chromosome sequence
Because the publicly available Pachón genome assembly was obtained from a female,31 we sequenced the genome of a B+ Pachón male to assemble its B sequence. Bs are notoriously difficult to assemble,32,33 due to their complex mosaic composition of A chromosome fragments16–19 and their high-repeat content,17,19 and most of the B sequence information is from short-read sequencing of purified Bs17,18 or B+ versus B-devoid (B−) individuals.17–19,34 To accurately assemble a high-quality Pachón B sequence, we used a combination of HiFi PacBio and Oxford nanopore long-reads, 10X genomics Illumina short-linked reads, and a Hi-C chromosome contact map. The resulting whole-genome assembly (see assembly metrics in Data S1B) contains 25 large scaffolds corresponding to the 25 Pachón A-chromosome pairs19,25 and 170 remaining unplaced scaffolds (2.12% of the total assembly size).
To identify the Pachón B sequence, we used its male-predominant feature and a pool sequencing (pool-seq) approach to contrast whole-genome sequences of a gDNA pool of 91 phenotypic males versus a gDNA pool of 81 phenotypic females. By remapping these male and female pool-seq reads on our male Pachón genome assembly, we identified a single 2.97 Mb unplaced scaffold (HiC_Scaffold_28) that displays a clear male-biased read coverage profile (Figures 2A and 2A’). The sequence analysis of HiC_Scaffold_28 revealed that it is made up from a complex mosaic of numerous duplicated fragments of A chromosomes (Figure 2B) including complete but also truncated duplicates of A chromosome genes (Data S1C). The B also displays a repeat content that is markedly different from A chromosomes (Figures 2C, 2D, S2A, and S2B; Data S1D). Both its sex-biased profile and its sequence characteristics indicate that HiC_Scaffold_28 is the Pachón B. The contribution of many A chromosome regions to the structure of Pachón Bs is in line with the recent findings that A. mexicanus Bs contain a large number of transposable elements.19 The high B proportion of satellite DNA (Figure S2A; Data S1D) was also reported in other species.35 Our manually curated B gene annotation (Data S1C) identified 63 genes on the Pachón B. Of these, 20 show high-quality annotation over their full length, 11 are truncated compared to their conserved homologs in other fishes, and one is a chimeric gene. Five genes show multiple copies and constitute almost one-third of the B gene content (Data S1C). These results contrast with earlier studies that reported a much higher number of B genes in A. mexicanus.19 These annotation differences are likely due to indirect assessment of the B gene content based on short-read sequencing of very few B+ versus B− individuals.19 This comparison clearly illustrates the need for better, complete, and high-quality B assemblies like we provided here for Pachón cavefish to better understand the structure and gene content of Bs generally.
Figure 2. Genomic characterization of Pachón cavefish B chromosome (B).
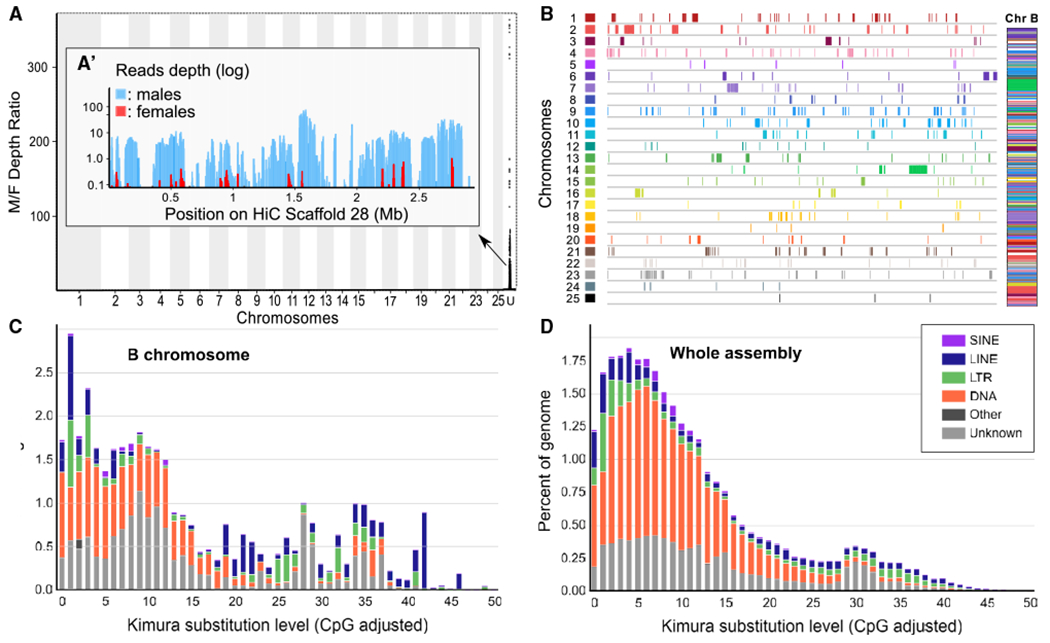
(A) Read depth ratio of male and female Pachón genomic pools showing a strong coverage bias in a single scaffold, Hi_scaffold_28 (enlarged in A’ inset showing male and female read coverage).
(B) Karyoplots of the A chromosome regions duplicated on the Pachón cavefish B (Chr B) showing that the Pachón B is made from a complex mosaic of duplicated A chromosomal fragments.
(C and D) Comparison of the repeat landscapes of the Pachón B (C) and whole genome including the B (D), showing that the Pachón B has a very different repeat element (color code provided in inset of D) content compared to A chromosomes. Short interspersed repeated sequences, SINEs; long interspersed nuclear elements, LINEs; long terminal repeats, LTRs; DNA repeat elements, DNAs; terminal inverted repeat sequences, TIRs.
See also Figures S2A and S2B and Data S1D for additional details.
The Pachón cavefish B contains two copies of a putative master sex-determining gene
Among the B genes with well-supported annotation evidence (Data S1C), we identified two duplicated loci of the A-chromosome-3 growth differentiation factor 6b (gdf6b) gene (located at Chr03:863,919-866,170), which is the teleost ohnolog (teleost whole-genome duplication36,37 paralogous copy) of the gdf6a gene (Figure 3A). Gdf6 genes belong to the TGF-β superfamily within which many master sex-determining (MSD) genes have been found, including TGF-β receptors38–40 and ligands.41–44 Of note, gdf6a on the Y chromosome (gdf6aY) has been characterized as the MSD gene in the turquoise killifish, Nothobranchius furzeri.44 The Pachón B gdf6b genes (B-gdf6b = B-gdf6b-1 and B-gdf6b-2) were thus retained as potential candidate B MSD genes. Chromosome FISH with a gdf6b locus probe revealed a gdf6b hybridization signal on the Pachón Bs, along with a single gdf6b labeling on a single A chromosome pair, both in the male-predominant Bs and the low-occurrence female Bs (Figures 1E and 1F and inset in Figure 1E).
Figure 3. Gdf6b protein evolution and structure.
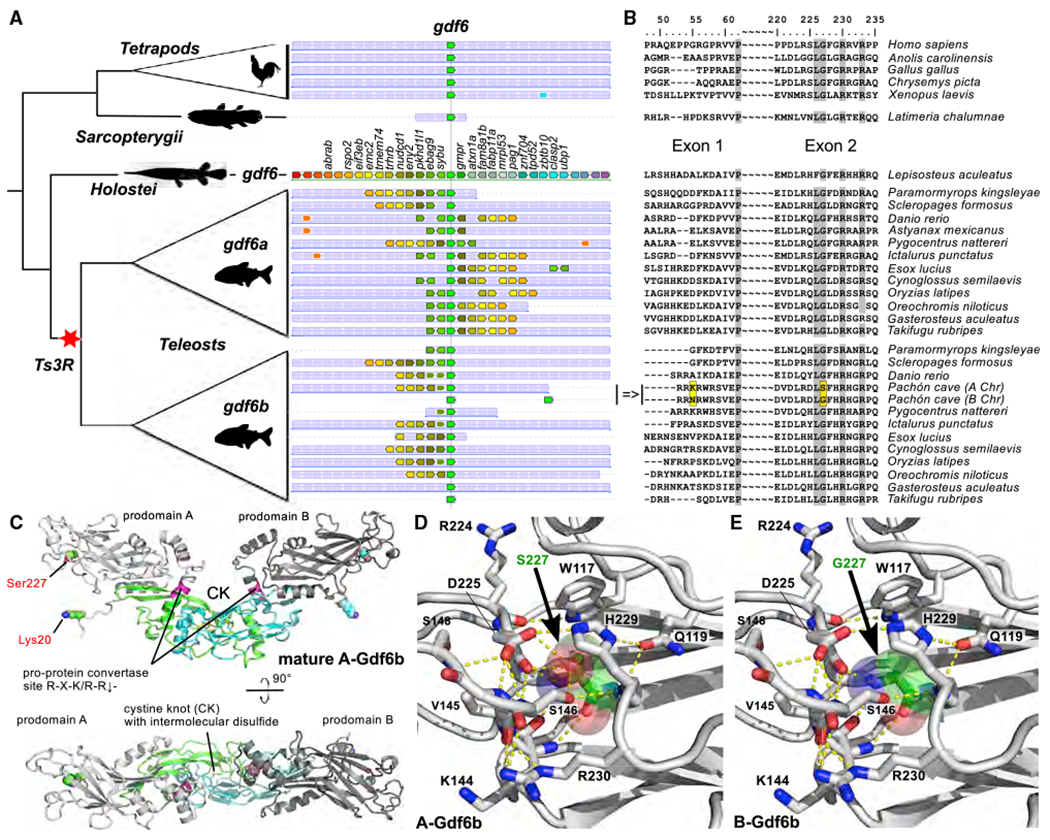
(A) Simplified phylogeny (left panel) and synteny (middle panel) relationships of the gdf6a and gdf6b genes, with the additional B-gdf6b Pachón cavefish duplication, showing that they are duplicated paralogs stemming from the teleost whole-genome duplication (Ts3R). Species names are given on the right side of (B).
(B) Corresponding multiple alignments of Gdf6 protein-coding sequences around the A-Gdf6b lysine to B-Gdf6b asparagine switch in Exon 1 (p.Lys60Asn) and the A-Gdf6b serine to B-Gdf6b glycine switch in Exon 2 (p.Ser227Gly).
(C) Ribbon plot homology model of A-Gdf6b proprotein dimer (bottom panel view is rotated by 90° around the x axis). The prodomains are shown in light and dark gray, the furin processing site (R274-R-K-R-R278) is indicated in magenta, and the activity-containing mature C-terminal domain is shown in green and cyan. The two residues differing between A-Gdf6b and B-Gdf6b are presented as spheres with their carbon atoms colored in green and cyan.
(D and E) Comparative magnifications of the structure of A-Gdf6b (D) and B-Gdf6b (E) around the p.Ser227Gly switch. Amino acid residues interacting with Ser227 or Gly227 are shown as sticks, and hydrogen bonds as yellow stippled lines. As shown, the side chain hydroxyl group of Ser227 (shown with carbon atoms colored in green and transparent spheres highlighting the van der Waals spheres of the atoms) engages in several hydrogen bonds with surrounding residues, e.g., Ser146 and Asp225, thereby stabilizing the tertiary and secondary structure in this region. Upon exchange of Ser227 with a glycine as in B-Gdf6b, these hydrogen bonds are lost, thereby potentially destabilizing this region in the prodomain.
The two B-gdf6b loci are 99.6% identical in the 21.6 kb region shared by the two genes, and 100% identical in their coding sequences (CDS), with B-gdf6b-2 being derived from an internal B duplication of the B-gdf6b-1 locus (Figure S2C). Such internal B duplications and insertions indicate that the origin of the B structure can be more complex than initially thought. Comparison of these B-gdf6b loci with the overlapping sequence of their A chromosome counterpart (A-gdf6b) revealed numerous differences in their proximal promoters and also their intron that contains two B-gdf6b specific insertions (Figure S2D). However, differences within the gdf6b CDS were limited to a T-to-C (A-gdf6b-to-B-gdf6b) synonymous substitution (c.591T>C) and two nonsynonymous substitutions, i.e., a T-to-G (A-gdf6b-to-B-gdf6b) transversion (c.180T>G) in exon 1 that switches a A-Gdf6b lysine into a B-Gdf6b asparagine (p.Lys60Asn) and a G-to-A (A-gdf6b-to-B-gdf6b) transition (c.679G>A) in exon 2 that switches the A-Gdf6b serine into a B-Gdf6b glycine (p.Ser227Gly) (Figure S2E). The Lys60Asn does not impact a conserved amino acid position of Gdf6b proteins, in contrast to the Ser227Gly that impacts a glycine of the TGF-β/BMP propeptide domain that is conserved in Pachón B-Gdf6b and in all vertebrate Gdf6 proteins, but not in Pachón A-Gdf6b (Figure 3B). It is interesting to note that this Ser227Gly modification suggests that the A-gdf6b acquired this mutation after the B-gdf6b copy was duplicated on the B. This non-conserved Ser227 engages in a central hydrogen bond network at a looptip region in the TGF-β/BMP propeptide domain (Figure 3C) that could affect the stability of this pro-domain. The Gly227Ser exchange in Pachón A-Gdf6b leads to the gain of several hydrogen bonds that are due to the side chain hydroxyl group of the Ser227 residue (Figures 3D and 3E). The gain of hydrogen bonds can confer a higher folding stability of the proprotein complex and thereby might affect activation of Gdf6b as this requires release of the mature C-terminal domain from the proprotein complex. The mature C-terminal growth factor domain, however, is likely to be unaffected by this mutation. Whether these conformation differences between the A-Gdf6b and B-Gdf6b proproteins could provide a potential functional explanation for a sex-determining role of the male-predominant Pachón B-Gdf6b remains to be explored, but point mutations in other MSD genes like amhr2Y in Takifugu rubripes and amhY in Oreochromis niloticus have been described to be directly responsible for male sex determination.39,41
Based on these B-gdf6b and A-gdf6b loci differences, we developed several B-specific PCR genotyping tests on fin clips (Figure S3A). In our Pachón laboratory population, we found a complete (100%) association between B-specific amplifications and the male phenotype in 723 males, with all the 787 tested females being negative (p value of association with sex <2.2e–16). We also found the same complete association in wild-caught Pachón individuals (20 males and 20 females, recognized by external sex-specific traits without sacrifice; p value of association with sex = 1.87e–09) (Data S1F), showing that this male-predominant B is not the result of a domestication effect. These results strengthen our cytological observations of a male-predominant B. The absence of B-specific amplifications in females despite the cytogenetic detection of rare Bs in females is probably the result of a PCR sensitivity issue as increasing the number of PCR cycles allows the detection of a faint PCR fragment in Pachón females (Figure S3B).
Evidence supporting gdf6b as a potential MSD gene in Pachón cavefish
Sex-specific expression patterns during the sex differentiation period and alteration of gonadal development upon knockout are key arguments for the evaluation of a candidate MSD gene. Due to the high sequence identity of the B-gdf6b and A-gdf6b cDNAs, we were not able to specifically quantify the B-gdf6b expression. Quantification of the expression of gdf6b (B-gdf6b and/or A-gdf6b) showed that it has both a predominant expression in developing and adult male gonads (as well as in male brain, intestine, kidney, and swim-bladder; Figure S4A) and a strong sexually dimorphic expression during early differentiation of B+ individuals (Figure 4A). Using in situ hybridization, the expression of gdf6b during the early differentiating period was restricted to gonads of B+ individuals (at 15, 21, 30, and 60 days post-fertilization), with no strict colocalization with the gonadal soma-derived factor gene (gsdf) (Figures 4B, 4C, and S4B). Gsdf is a well-known gonad-restricted, somatic-supporting cell lineage marker45–47 that has also been described as one of the earliest Pachón gonadal sex differentiation marker genes.48 This result demonstrates that gdf6b mRNA (B-gdf6b and/or A-gdf6b) has an expression profile compatible with a male (B+) MSD function, being expressed in the right place, i.e., only in the differentiating testis, and at the right time, i.e., during early testicular differentiation.
Figure 4. Gene expression and functional evidence supporting a role of gdf6b as a potential master sex-determining gene in Pachón cavefish.
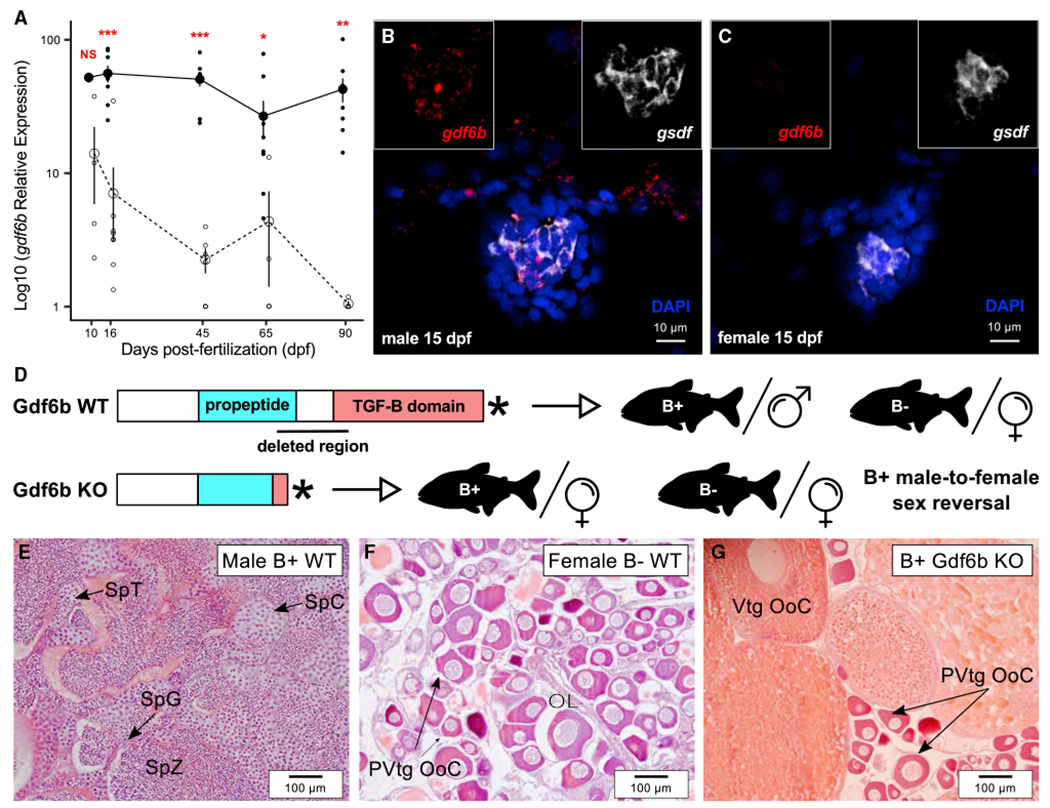
(A) Expression profiles of gdf6b in male and female trunks during early development from 10 to 90 days post-fertilization (dpf; males, solid line; females, dashed line) showing a significant overexpression in males compared to females starting from 16 dpf. Results are presented as log10 mean ± standard errors; black and white dots represent the individual values of relative expression in males and females, respectively. Statistical significance between males and females was tested with the Wilcoxon rank-sum test (Wilcoxon-Mann-Whitney test) and only significant differences are shown (***p < 0.01; **p < 0.01; *p < 0.05). See also Figure S4A.
(B and C) Gonadal expression of gdf6b (in red) and the Sertoli and granulosa supporting cell marker gsdf (in white) in male (B) and female (C) differentiating gonads at 15 dpf showing that gdf6b is specifically expressed in male gonads with no strict colocalization with gsdf in male. See Figure S4B for additional stages of development. Nuclei were stained with DAPI (in blue). Scale bar, 10 μm.
(D) Schematic representation of the wild-type (WT) and knockout (KO) Gdf6b proteins and the resulting phenotypes of B+ males and B− females.
(E–G) Representative gonadal histology of WT males (E), WT females (F), and Gdf6b KO B+ males showing that Gdf6b KO induces male-to-female sex reversal (G) with ovaries containing vitellogenic (Vtg Ooc) and previtellogenic oocytes (PVtg Ooc), like WT ovaries (F), contrasting with the testis in WT males (E).
Ol, ovarian lamellae; SpC, spermatocytes; SpG, spermatogonia; SpT, spermatids; SpZ, spermatozoa. Scale bar, 100 μm.
To bring additional and functional evidence that gdf6b could act as an MSD gene, we generated gdf6b knockouts in Pachón cavefish using the genome-editing CRISPR-Cas9 system with two guide RNAs in order to remove a large part of the gdf6b CDS (Figure 4D). This large deletion (~470 bp) includes most of the TGF-β propeptide region and the beginning of the TGF-β-like domain, resulting in a truncated, likely nonfunctional, Gdf6b protein (Figure S2F). Among 200 first-generation microinjected individuals (which are mosaic for the genome-edited loci), eighteen B+ individuals (i.e., genotypic males) had an ~470 bp deletion in their A- and/or B-gdf6b exon 2 (Figure S3C), and they were all sex-reversed into phenotypic females (Figure 4G). In contrast, all B+ males and B− females without the gdf6b deletion developed normal testes (B+) or ovaries (B−) (Figures 4E and 4F). This result shows that gdf6b is necessary to trigger Pachón testicular development in B+ individuals and brings further functional evidence that gdf6b could be used as a male MSD gene in Pachón cavefish. However, because of the high similarity of the B-gdf6b loci with the A-gdf6b locus, we have not been able to specifically knock out the Pachón B-gdf6b. Further studies will bring more functional proof supporting the role of the B-gdf6b genes in sex determination, including the specific B-gdf6b knockout in B+ individuals and the overexpression of gdf6b by transgenesis in B− fish.
Conclusions
Altogether our results bring new pieces of evidence to support a role of some B chromosomes in sex determination. The potential implication of Bs in sex determination had been suspected in many species including some fishes.14,15 Up to now, however, the characterization of potential B MSD genes along with strong functional evidence has only been provided for the bacterial-derived haploidizer gene in the jewel wasp, Nasonia vitripennis B chromosome (named PSR for paternal sex ratio chromosome),49 although maleness in this haplo-diploid organism is determined through elimination of the paternal A chromosome set.50 Here, combining a variety of approaches we discovered that Pachón cavefish of the species A. mexicanus carry male-predominant Bs that contain two copies of the gdf6b gene, which itself behaves as an excellent MSD candidate gene. This indeed brings the interesting hypothesis that these Pachón Bs could be considered as “B-sex” chromosomes. However, the question remains open whether Pachón Bs are predominant in males because they are eliminated from female tissues or whether they are by themselves necessary and sufficient to trigger maleness. Despite being male predominant, Pachón Bs are also found in some females, albeit only in very few metaphases (21.6 times less abundant than in males on average) and most often as a single B copy. B frequencies have been described as being highly variable between species, sexes, individuals of the same population, and even in different cells of a single individual.4–6,29 This variation is assumed to result from meiotic and/or mitotic instability of Bs that can be present only in some organs and absent from others.4,29,51–53 In the plant Aegilops speltoides, a mechanism of programmed elimination of Bs occurs specifically in the roots.54 It results from a B chromatid nondisjunction during mitosis, leading to the micronucleation of Bs and their subsequent degradation at early stages of the proto-root embryonic tissue differentiation.54 Such a mechanism would potentially explain a specific B elimination in Pachón female organs, as it has been hypothesized in another Astyanax species with male-restricted Bs.11 Further studies are now needed to better understand this sex-specific B drive mechanism, and if it reflects a cause or a consequence of sex determination in Pachón cavefish. Our results also lay a high-quality genome-based foundation in an important emerging fish model for studying the genomic evolution of Bs, including the micro- and macro-evolution of this B in line with the evolution of sex chromosomes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yann Guiguen (yann.guiguen@inrae.fr).
Materials availability
To request Pachón fish lines or constructs created in this study, please contact the lead contact.
Data and code availability
Raw sequences and the whole genome assembly of Pachón cavefish have been deposited in the National Center for Biotechnology Information DDBJ/ENA/GenBank databases under the BioProject PRJNA734455. This accession number is listed in the key resources table. This study did not generate new unique code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Primary: rabbit anti-SYCP3 | Abcam | Cat# ab15093; RRID: AB_301639 |
| Primary: mouse anti-MLH1 | Abcam | Cat# ab14206: RRID: AB_300987 |
| Secondary: goat anti-rabbit IgG H&L Alexa488 | Abcam | Cat# ab150077; RRID: AB_2630356 |
| Secondary: goat anti-mouse IgG H&L Alexa555 | Abcam | Cat# ab150114; RRID: AB_2687594 |
| Chemicals, peptides, and recombinant proteins | ||
| Tricaine methanesulfonate (MS 22) | Sigma-Aldrich | Cat# E10521 |
| 2-phenoxyethanol | Sigma-Aldrich | Cat# 77699 |
| Ribonuclease A from bovine pancreas | Sigma-Aldrich | Cat# R6513 |
| Formamide | Sigma-Aldrich | Cat# 47671 |
| Cas9 protein | Tacgene | No reference |
| Hematoxylin | Microm Microtech | Cat# F/HAQ999 |
| Mountant and DAPI (4′,6-diamidino-2-phenolindole) | Cambio | Cat# 1124-MD-1250 |
| Spectrum-Orange-dUTP | Vysis | Cat# 02N33-050 |
| Spectrum-Green-dUTP | Vysis | Cat# 02N32-050 |
| Bovine Serum Albumin (BSA) | Vector | Cat# SP-5050 |
| Pepsin | Chemos CZ | Cat# A4289.0100 |
| Giemsa stain | Giemsa stain | Giemsa stain |
| Critical commercial assays | ||
| NucleoSpin plasmid DNA purification kit | Machery-Nagel | Cat# 740588.50 |
| NucleoSpin Kits for Tissue | Machery-Nagel | Cat# 740952.50 |
| HiDi Taq DNA polymerase | myPOLS Biotec | No reference |
| AccuPrime Taq DNA polymerase | Thermofisher | Cat# 12339016 |
| AccuPrime Taq DNA Polymerase, High Fidelity | Thermofisher | Cat# 12346086 |
| TOPO TA cloning Kit XL | Thermofisher | Cat# K8050-20 |
| ProLong Gold Antifade Mountant | Thermofisher | Cat# P10144 |
| MAXIscript T7 Transcription Kit | Thermofisher | Cat# AM1312 |
| Jumpstart Taq DNA polymerase | Sigma-Aldrich | Cat# D9307 |
| Chromium Genome Reagent Kits v2 | 10X genomics | Cat# PN-120258 |
| KAPA library Amplification kit | Roche Diagnostics | Cat# KK2620 |
| Kapa Library Quantification Kit | Roche Diagnostics | Cat# KK4824 |
| Illumina HiSeq3000 sequencing kits | Illumina | Cat# FC-410-101 |
| Illumina NovaSeq Reagent Kits | Illumina | Cat# 20012860 |
| Truseq DNA PCR-Free kit | Illumina | Cat# 20015962 |
| TruSeq Nano DNA HT Library Prep Kit | Illumina | Cat# 20015965 |
| SP flow cell | Illumina | Cat# 20027464 |
| S4 Novaseq flow cell | Illumina | Cat #20012866 |
| AMPure XP beads | Beckman Coulter | Cat# A63881 |
| Nanopore Ligation Sequencing Kit | Oxford Nanopore | Cat# SQK-LSK109 |
| SMRTbell Express Template Prep Kit 2.0 | PacBio | Cat# 101-685-400 |
| Binding kit 2.0 kit | PacBio | Cat# 102-089-000 |
| Sequencing kit 2.0 | PacBio | Cat# 101-829-200 |
| Arima-HiC kit | Arima | Cat# 510008 |
| Cy3 NT Labeling Kit | Jena Bioscience | Cat# PP-305L-CY3 |
| Multiplex Fluorescent Reagent Kit v2 | ACD Biotechne | Cat# 323100 |
| MagAttract HMW DNA kit (48) | QIAGEN | Cat# 67563 |
| Deposited data | ||
| Structure of proprotein complex of human GDF2 | 55 | PDB: 4YCG |
| Structure of the proprotein complex of human activin A | 56 | PDB: 5HLY |
| Structure of the growth factor domain of human GDF5 in complex with repulsive guidance molecule B | 57 | PDB: 6Z3J |
| Structure of the growth factor domain of human GDF5 in complex with repulsive guidance molecule B and neogenin | 57 | PDB: 6Z3M |
| Structure of the growth factor domain of human GDF5 in complex with BMP receptor 1A | 58 | PDB: 3QB4 |
| Pachón cavefish, Astyanax mexicanus, genome assembly and annotation | This study | GenBank: PRJNA734455 |
| Experimental models: Organisms/strains | ||
| A. mexicanus Pachón cavefish wild-type | Jeffery laboratory, University of Maryland, College Park, MD | N/A |
| A. mexicanus Pachón cavefish gdf6b KO | This study | N/A |
| Oligonucleotides | ||
| Primers for the amplification of the gdf6b FISH Probe, see Data S1E | Eurofins | N/A |
| Primers for B genotyping, see Data S1E | Eurofins | N/A |
| Primers for screening gdf6b-KO, see Data S1E | Eurofins | N/A |
| Primers for RT-QPCR for gdf6b, see Data S1E | Eurofins | N/A |
| Forward primer containing sgRNAs target sequences for #site 1 GAAATTAATACGACTCACTATAGGAGTCTGAAACCGGTTCTGGTTTTAGAGCTAGAAATAGCAAG | Eurofins | N/A |
| Forward primer containing sgRNAs target sequences for #site 2 GAAATTAATACGACTCACTATAGGGAGCTGGGCTGGGACGACGTTTTAGAGCTAGAAATAGCAAG | Eurofins | N/A |
| Universal Reverse primer: AAAAGCACCGACTCGGTGCCACT | Eurofins | N/A |
| Truseq DNA UD Indexes | Illumina | Cat# 20020590 |
| DOP primer (10 μM) GAGGATGAGGTTGAGNNNNNNGTGG | KRD, Prague, Czech Republic | N/A |
| RNAScope probe for cavefish Gdf6b | ACD Biotechne | N/A |
| RNAScope probe for cavefish gsdf | ACD Biotechne | N/A |
| Recombinant DNA | ||
| DR274 vector | 59 | Addgene plasmid #42250 |
| Gdf6b genomic FISH probe | This study | N/A |
| Software and algorithms | ||
| Hifiasm version 0.9 | 60 | https://github.com/chhylp123/hifiasm |
| Long Ranger v2.1.1 | 10X Genomics | https://support.10xgenomics.com/genome-exome/software/pipelines/latest/advanced/other-pipelines |
| Juicer | 61 | https://github.com/aidenlab/juicer |
| 3D-DNA pipeline | 62 | https://github.com/aidenlab/3d-dna |
| Juicebox | 62 | https://github.com/aidenlab/Juicebox/wiki/Juicebox-Assembly-Tools |
| Minimap2 v2.11 | 63 | https://github.com/lh3/minimap2 |
| RepeatModeler | Smit, AFA, Hubley, R. Unpublished | http://www.repeatmasker.org/ |
| RepeatMasker | Smit, AFA, Hubley, R & Green, P. Unpublished | http://www.repeatmasker.org/ |
| Genewise | 64 | https://www.ebi.ac.uk/seqdb/confluence/display/THD/GeneWise |
| Exonerate | European Bioinformatics Institute. Unpublished | https://www.ebi.ac.uk/about/vertebrate-genomics/software/exonerate |
| Hisat | 65 | http://www.ccb.jhu.edu/software/hisat/index.shtml |
| StringTie | 66 | https://ccb.jhu.edu/software/stringtie/ |
| Trinity | 67 | https://github.com/trinityrnaseq/trinityrnaseq/wiki |
| Pasa | 68 | https://github.com/PASApipeline/PASApipeline/wiki |
| Augustus | 69 | https://github.com/Gaius-Augustus/Augustus |
| karyoploteR | 70 | https://bernatgel.github.io/karyoploter_tutorial/ |
| Primer3web software version 4.1.0 | 71 | https://primer3.ut.ee/ |
| NovaSeq Control Software and Real-Time Analysis component (v3.4.4) | PacBio | N/A |
| Illumina’s conversion software (bcl2fastq v2.20) | Illumina | N/A |
| FastQC (v0.11.8) | 72 | https://github.com/s-andrews/FastQC |
| BWA mem version 0.7.17 | 73 | https://github.com/lh3/bwa |
| Picard tools version 2.18.2 | Broad Institute | http://broadinstitute.github.io/picard |
| samtools mpileup version 1.8 | 74 | https://sourceforge.net/projects/samtools/files/samtools/ |
| popoolation2 | 75 | https://sourceforge.net/projects/popoolation2/ |
| PSASS version 2.0.0 | Feron R. unpublished | https://doi.org/10.5281/zenodo.2615936 |
| Yasara | 76 | http://www.yasara.org |
| Yasara hm_build.mcr macro | 76 | http://www.yasara.org/hm_build.mcr |
| RStudio (Open Source version) | RStudio team | http://www.rstudio.com/ |
| Ikaros software | MetaSystems | N/A |
| DP Manager imaging software | Olympus | N/A |
| Olympus Acquisition Software | Olympus | N/A |
| Adobe Photoshop CS6 | Adobe | N/A |
| ZiFiT software | 77 | http://zifit.partners.org/ZiFiT/Disclaimer.aspx |
| Other | ||
| Nalgene Freezing Container (Mr. Frosty) | Merck | Cat# C1562-1EA |
| Megaruptor1 system | Diagenode | Cat# B06010001 |
| Qubit3 fluorometer Invitrogen | ThermoFisher | Cat# Q33216 |
| poly-l-lysine slides | ThermoFisher | Cat# J2800AMNZ |
| BluePippin Size Selection system | Sage Science | No reference |
| M220 Focused-ultrasonicator | Covaris | Cat# 500295 |
| Sequence data a whole genome assembly of Pachón | This study | PRJNA734455 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cavefish breeding and sampling
Laboratory stocks of A. mexicanus Pachón cavefish were obtained in 2004 from the Jeffery laboratory at the University of Maryland, College Park, MD. Fish were raised as previously described. Fertilized eggs were provided by CNRS cavefish experimental facilities (Gif sur Yvette, France) and maintained at 24°C until the hatching stage occurring around 24 ± 2 h post-fertilization (hpf).78 Subsequently, larvae were transferred and raised in the Fish Physiology and Genomics laboratory experimental facilities (LPGP, INRAE, Rennes, France) under standard photoperiod (12 h light / 12 h dark) and at two different temperatures: 21 ± 1°C and 28 ± 1°C. Animals were fed twice a day, first with live artemia (Ocean Nutrition) until 15 days post-fertilization (dpf), then with a commercial diet (BioMar) until adult stage. For animal dissections and organ sampling, fish were euthanized with a lethal dose of tricaine methanesulfonate (MS 222, 400 mg/l), supplemented by 150 mg/l of sodium bicarbonate. Phenotypic sex of individuals was determined at 4 months and more, either by macro-scopical examination of the gonads when they were enough differentiated, or by histology when gonads were not totally differentiated.48 Caudal fin clips were collected from all individuals and stored in ethanol 90% at 4°C before genomic DNA (gDNA) extraction. For the chromosome contact map (Hi-C), 80 μL of blood was sampled from three males using a syringe rinsed with EDTA 2%. The fresh blood was slowly frozen in a Freezing Container (Mr. Frosty, Nalgene) after addition of 15% of dimethyl sulfoxide (DMSO). Karyotypic analyses were carried out in 17 males and 11 females of Pachón cave Astyanax mexicanus. Fin samples (a narrow strip of the tail fin) were taken from the live specimens anesthetized by MS-222 (Merck KGaA, Darmstadt, Germany), while for direct preparations (chromosomes from kidneys and gonads), fishes were euthanized first using 2-phenoxyethanol (Sigma-Aldrich, St. Louis, MO, USA).
All animal protocols were carried out in strict accordance with the French and European legislations (French decree 2013-118 and directive 2010-63-UE) applied for ethical use and care of laboratory animals used for scientific purposes. Sylvie Retaux and CNRS institutional authorizations for maintaining and handling A. mexicanus in experimental procedures were 91-116 and 91272105, respectively. For karyotypic analysis, all handling of fish individuals followed European standards in agreement with §17 of the Act No. 246/1992 to prevent fish suffering. The procedures involving fish were supervised by the Institutional Animal Care and Use Committee of the Institute of Animal Physiology and Genetics CAS, v.v.i., the supervisors permit number CZ 02361 certified and issued by the Ministry of Agriculture of the Czech Republic. Several sampling campaigns were carried out in the field in Mexico between 2013 and 2019, resulting in a collection of tail fin clips from wild-caught individuals from the Pachón cave. In the field, the phenotypic sex of animals was determined by checking the presence or absence of denticles on the anal fins as described previously,79 a small fin clip was gently taken and fish were rapidly returned to their natural pond. In addition, pictures of each individual sampled were taken to confirm the phenotypic sex, back in the laboratory, based on the morphological criteria described previously.79 The permits for field sampling (02241/13, 02438/16, 05389/17 and 1893/19) were delivered by the Mexican authorities (Mexican Secretaría del Medio 16 Ambiente y Recursos Naturales) to Sylvie Rétaux and Patricia Ornelas-Garcia (UNAM, Mexico).
METHOD DETAILS
DNA extraction
For fish genotyping, gDNA was extracted from fin clips stored in 90% ethanol, after lysis in 5% chelex80 and 10 mg Proteinase K at 55°C for 2 h, followed by 10 min at 99°C. Following extraction, samples were centrifuged and the supernatant containing the gDNA was transferred in clean tubes and stored at −20°C. For pool-sequencing and TaqMan assay, gDNA was extracted using NucleoSpin Kits for Tissue (Macherey-Nagel, Düren, Germany) according to the supplier’s recommendations. For long-read male genome sequencing, high molecular weight (HMW) gDNA was extracted from a mature testis grounded in liquid nitrogen and lysed in TNES-Urea buffer (TNES-Urea: 4M urea; 10 mM Tris-HCl, pH 7.5; 120 mM NaCl; 10 mM EDTA; 5% SDS) for two weeks at room temperature. For HMW gDNA extraction the TNES-Urea solution was supplemented with Proteinase K at a final concentration of 150 μg/mL and incubated at 37°C overnight. HMW gDNA was extracted with a modified phenol-chloroform protocol as previously described.43 The gDNA concentrations for both pool-seq and genome sequencing were quantified with Qubit3 fluorometer (Invitrogen, Carlsbad, CA) and HMW gDNA quality and purity were assessed using spectrophotometry, fluorometry, and capillary electrophoresis.
Primers and probe design
All primers used in this study including PCR genotyping, qPCR gene expression and cDNA cloning were designed using Primer3web software71 version 4.1.0 and are listed in Data S1E.
B polymerase chain reaction (PCR) genotyping
Genetic sex of Pachón cave individuals was determined by PCR tests using the fact that males have a B-predominant chromosome with three different sets of primers (see Data S1E for primer sequences) based on differences between the A-gdf6b and B-gdf6b loci. Three sets of primers (P) were designed (see Figure S3A) with two primer sets designed to amplify specifically the two B-gdf6b copies based either on a single base variation between the A/B gdf6b CDS at position 679 bp (P1-P2), or based on primers located on both sides of the A/B breakpoints downstream of the B-gdf6b gene (P3-P4). The third set of primers (P5-P6) was designed on gaps/indels variations between A-gdf6b and the two B-gdf6b genes in the proximal promoter of gdf6b genes. Another set of primers (P9-P10) was designed as a PCR positive control with primers located on both sides of the A/B breakpoints downstream of the A-gdf6b gene. Primer sets P1-P2 and P3-P4 produce a single PCR fragment only in males (B+), primer set P5-P6 amplifies two bands in males (all B+) and only a single band in females (all B−), and primer set P9-P10 amplifies a single band in all individuals. For PCR reactions with the P1-P2 primer set, HiDi Taq DNA polymerase (myPOLS Biotec) was used for detecting a single nucleotide variation. PCRs were performed in a total volume of 12.5 μL containing 0.2 μM of each primer, a final concentration of 20 ng/μL gDNA, 200 μM of dNTPs mix, 1X of HiDi buffer (10X), and 2.5 U per reaction of HiDi DNA polymerase. Cycling conditions were as follows: 95°C for 2 min, then 35 cycles of (95°C for 15 s (secs) + 60°C for 10 s + 72°C for 30 s), and 72°C for 5 min. For PCR reactions with the P3-P4 and P9-P10 primer sets, PCR reactions were performed in a final volume of 50 μL containing 0.5 μM of each primer, a final concentration of 20 ng/μL gDNA, 10 μM dNTPs mix, 1X of 10X AccuPrime buffer, and 0.5 μL per reaction of AccuPrime Taq DNA polymerase. Cycling conditions were as follows: 94°C for 2 min, then 35 cycles of (94°C for 30 s + 58°C for 30 s + 68°C for 1 min and 30 s). For PCR reactions with the P5-P6 primer set, PCR reactions were performed in a final volume of 25 μL containing 0.5 μM of each primer, a final concentration of 20 ng/μL gDNA, 10 μM dNTPs mixture, 1X of Jumpstart buffer (10X), and 0.5 μL per reaction of Jumpstart Taq DNA polymerase. Cycling conditions were as follows: 95°C for 2 min, then 35 cycles of (95°C for 1 min + 60°C for 30 s + 72°C for 1 min), and 72°C for 5 min.
10 × Genomics sequencing
10X Chromium Library was prepared according to 10X Genomics protocols using the Genome Reagent Kits v2. Optimal performance has been characterized on input gDNA with a mean length greater than 50 kb (~144Kb). GEM reactions were performed on 0.625 ng of genomic DNA, and DNA molecules were partitioned and amplified into droplets to introduce 16-bp partition barcodes. GEM reactions were thermally cycled (30°C for 3 h and 65°C for 10 min; held at 4°C) and after amplification, the droplets were fractured. P5 and P7 primers, read 2, and sample index were added during library construction. The library was amplified using 10 cycles of PCR and the DNA was subsequently size selected to 450 bp by performing a double purification on AMPure Xp beads. Library quality was assessed using a Fragment Analyzer and quantified by qPCR using the Kapa Library Quantification Kit. Sequencing has been performed on an Illumina HiSeq3000 using a paired-end read length of 2x150 bp with the Illumina HiSeq3000 sequencing kits.
Oxford nanopore genome sequencing
High molecular weight gDNA purification steps were performed using AMPure XP beads (Beckman Coulter). Library preparation and sequencing were performed using Oxford Nanopore (Oxford Nanopore Technologies) Ligation Sequencing Kit SQK-LSK109 according to manufacturer’s instructions “1D gDNA selecting for long reads (SQK-LSK109).” Five μg of DNA was purified then sheared at 20 kb using the megaruptor1 system (diagenode). A one-step DNA damage repair + END-repair + dA tail of double-stranded DNA fragments was performed on 2 μg of sample. Then adapters were ligated to the library. Library was loaded onto 1 R9.4.1 flowcell and sequenced on a PromethlON instrument at 0.02 pM within 72 h.
PacBio HiFi genome sequencing
Library preparation and sequencing were performed according to the manufacturer’s instructions “Procedure & Checklist Preparing HiFi SMRTbell Libraries using SMRTbell Express Template Prep Kit 2.0.” Fifteen μg of DNA were purified and then sheared at 15 kb using the Megaruptor3 system (Diagenode). Using SMRTbell Express Template prep kit 2.0, a Single strand overhangs removal and then a DNA and END damage repair steps were performed on 10 μg of the sample. Then blunt hairpin adapters were ligated to the library. The library was treated with an exonuclease cocktail to digest unligated DNA fragments. A size selection step using a 12 kb cutoff was performed on the BluePippin Size Selection system (Sage Science) with “0.75% DF Marker S1 3-10 kb Improved Recovery” protocol. Using Binding kit 2.0 kit and sequencing kit 2.0, the primer V2 annealed and polymerase 2.0 bounded library was sequenced by diffusion loading onto 2 SMRT cells on Sequel2 instrument at 50 pM with a 2 h pre-extension and a 30 h movie.
Hi-C sequencing
Hi-C data was generated using the Arima-HiC kit (Ref. 510008), according to the manufacturer’s protocols using 10 μL of blood as starting material, the Truseq DNA PCR-Free kit and Truseq DNA UD Indexes (Illumina, ref. 20015962, ref. 20020590), and the KAPA library Amplification kit (Roche, ref. KK2620). Hi-C library was sequenced in paired-end 2x150 bp mode on Novaseq6000 (Illumina), using half a lane of a SP flow cell (ref. 20027464). Image analyses and base calling were performed using the Illumina NovaSeq Control Software and Real-Time Analysis component (v3.4.4). Demultiplexing was performed using Illumina’s conversion software (bcl2fastq v2.20). The quality of the raw data and potential contaminants was assessed using FastQC (v0.14.0)72 from the Babraham Institute and the Illumina software SAV (Sequencing Analysis Viewer).
Genome assembly
Pacbio HiFi reads were assembled with hifiasm60 version 0.9 using standard parameters. The genome assembly fasta file was extracted from the principal gfa assembly graph file using an awk command line. This assembly was then scaffolded using Hi-C and 10X as a source of linking information. 10X reads were aligned using Long Ranger v2.1.1 (10x Genomics). Hi-C reads were aligned to the draft genome using Juicer61 with default parameters. A candidate assembly was then generated with 3D de novo assembly (3D-DNA) pipeline62 with the -r 0 and–polisher-input-size 100000 parameters. Finally, the candidate assembly was manually reviewed using the Juicebox assembly tools.81 Due to the specific structure of the Pachón cave B chromosome, both Hi-C and 10X signals show some uncertainties in the order and orientation of the contigs. To improve the quality of the B chromosome assembly, ONT reads were then aligned to the final version of the genome using minimap263 v2.11 with -x map-ont parameter. Both reads spanning contig junctions and reads showing supplementary alignments linking contigs belonging to the B chromosome were analyzed to resolve these ambiguities.
Genome annotation
The cavefish whole genome assembly was annotated using a pipeline adapted from previous studies.82,83 In brief, RepeatModeler, RepeatProteinMask, and RepeatMasker (open-4.0.7, http://www.repeatmasker.org/) were first used to scan the genome and mask out repeats. Then protein-coding genes were annotated by collecting gene evidence from homology alignment, RNA-seq mapping, and ab initio prediction. For homology alignment, 464,144 protein sequences collected from NCBI were aligned to the assembly using Genewise64 and Exonerate respectively. RNA-seq data were mapped to the assembly in two independent parallel steps. First Hisat65 was used to align RNA-seq reads and then StringTie66 was used to predict the gene models; in the other step reads were first assembled into transcript sequences using Trinity67 and then PASA68 was used to map the transcripts to the assembly and model the gene structures. For ab initio prediction and final integrating, Augutus69 was first trained using the high-quality gene models and then ran in a hint-guide model.
B chromosome annotation
First, repeats were identified and masked from the B chromosome using RepeatModeler, RepeatProteinMask, and RepeatMasker (open-4.0.7, http://www.repeatmasker.org/). To annotate protein-coding genes, we collected all protein sequences of A. mexicanus annotated by NCBI (Genome ID: 13073) and Ensembl (release-104), and aligned them onto the repeat-masked B sequence using GeneWise64,84 and Exonerate respectively. For each query, the best hit was kept. To determine the best gene model when multiple ones compete for a splice site, we introduced RNA-seq data to evaluate the quality of these homology gene models. Hence RNA-seq data of A. mexicanus from the previous study85 were aligned to B using HISAT65 and parsed using StringTie.66 RNA-score of each homology gene model was then calculated as the match-extend of splice sites to that of StringTie prediction. When multiple homology gene models compete for a splice site, those with lower RNA-score were discarded. In cases when some genes failed to be identified using homology alignment, we also implemented an ab initio gene prediction using Augustus,69 where all the homology and transcriptome evidence were used as hints. The predicted results were included into the final gene set if 1) the splice sites are not occupied and 2) the splice sites match 100% to that of StringTie predictions (RNA-score = 100). To further evaluate the quality of the final gene set, we blasted their protein sequences to SWISSPROT (https://www.uniprot.org/) and NR (https://www.ncbi.nlm.nih.gov/), and took the alignment to the best hit to check how much of the query and subject was aligned, respectively (query coverage & subject coverage). Genes with query and subject coverage both > 90% were considered as being of good quality.
To characterize the A chromosome content of the B chromosome, sequences of the B chromosome assembly were aligned to the sequences of the 25 A chromosomes with minimap263 (v2.11) and the best match of each contig fragment was retained. Overlapping matches were manually filtered considering match lengths and similarities (cigarline and edit distance) in order to build the best nonoverlapping matching list. Karyoplots were then plotted using the R package karyoploteR70 (https://bernatgel.github.io/karyoploter_tutorial/). The median, minimum, and maximum sizes of the 628 B best matches on A chromosomes were respectively 1,087 bp, 44 bp and 41,908 bp.
Male and female Pool-sequencing
DNA was collected from 91 phenotypic Pachón cave males and 81 phenotypic Pachón cave females and was pooled as male and female pools separately. Before pooling, the DNA concentration was normalized in order to obtain an equal amount of each individual genome in the final pool. Pool-sequencing libraries were prepared using the Illumina TruSeq Nano DNA HT Library Prep Kit (Illumina, San Diego, CA) according to the manufacturer’s protocol. After the fragmentation of each gDNA pool (200 ng/pool) by sonication using an M220 Focused-ultrasonicator (COVARIS), the size selection was performed using SPB beads retaining fragments of 550 bp. Following the 3′ ends of blunt fragments mono-adenylation and the ligation to specific paired-end adaptors, the amplification of the construction was performed using Illumina-specific primers. Library quality was verified with a Fragment Analyzer (Advanced Analytical Technologies) and then quantified by qPCR using the Kapa Library Quantification Kit (Roche Diagnostics Corp, Indianapolis, IN). The enriched male and female pool libraries were then sequenced using a paired-end multiplexed sequencing mode on a NovaSeq S4 lane (Illumina, San Diego, CA), combining the two pools on the same lane and producing 2 × 150 bp with Illumina NovaSeq Reagent Kits according to the manufacturer’s instructions. Sequencing produced 288 million paired reads and 267 million paired reads for the male and female pool libraries, respectively.
Pool-sequencing analysis
Characterization of genomic regions enriched for sex-biased signals between Pachón cave males and females, consisting of coverage and Single Nucleotide Variations (SNVs) was performed as described previously.38,43 Pachón cave A. mexicanus paired-end reads from male and female pool-seq pools were mapped onto our own Pachón cave genome assembly using BWA mem version 0.7.17.73 The resulting BAM files were sorted and the duplicate reads due to PCR amplification during library preparation were removed using Picard tools version 2.18.2 (http://broadinstitute.github.io/picard) with default parameters. Then, for each pool and each genomic position, a file containing the nucleotide composition was generated using samtools mpileup74 version 1.8, and popoolation275 mpileup2sync version 1201. This file was then analyzed with custom software (PSASS version 2.0.0; https://doi.org/10.5281/zenodo.2615936) to compute: (a) the position and density of sex-specific SNVs, defined as SNVs heterozygous in one sex but homozygous in the other, and (b) the average read depths for male and female pools along the genome to look for regions present in one sex but absent in the other (i.e., sex-specific insertions). All PSASS analyses were run with default parameters except for the range of frequency for a sex-linked SNV in the homogametic sex,–range-hom, that was set to 0.01 instead of 0.05, and the size of the sliding window,–window-size, that was set at 50,000 instead of 100,000.
Chromosome conventional cytogenetics
Mitotic or meiotic chromosome spreads were obtained either from regenerating caudal fin tissue as previously described,86 with slight modifications87 and altered time of fin regeneration (one week), or by direct preparation from the cephalic kidney and gonads.88 In the latter, the quality of chromosomal spreading was enhanced by a previously described dropping method.89 Chromosomes were stained with 5% Giemsa solution (pH 6.8) (Merck, Darmstadt, Germany) for conventional cytogenetic analyses, or left unstained for other methods. For FISH, slides were dehydrated in an ethanol series (70%, 80% and 96%, 3 min each) and stored at −20°C before analysis. Constitutive heterochromatin was visualized by C-banding,90 with chromosomes being counterstained by 4′,6-diamidino-2-phenolindole (DAPI), 1.5 μg/mL in antifade (Cambio, Cambridge, United Kingdom).
gdf6b probe synthesis for FISH mapping
gDNA was extracted using NucleoSpin Kits for Tissue (Macherey-Nagel, Duren, Germany) as described above. A gdf6b fragment comprising the two exons, the intron and 2,260 bp of the proximal promoter (with a total size of 4,368 bp) was amplified by PCR in a total volume of 50 μL. The mixture contained 0.5 μM of each primer, a final concentration of 20 ng/μL gDNA, 1X of 10X AccuPrime PCR Buffer II, 1U/reaction of AccuPrime Taq DNA Polymerase, High Fidelity (Thermofisher), was adjusted to 50 μL with autoclaved and distilled water. Cycling conditions were as follows: 94°C for 45 s, then 35 cycles of (94° C for 15 s + 64°C for 30 s + 68° C for 5 min and 30 s), and 68°C for 5 min. The resulting PCR product was cloned into TOPO TA cloning Kit XL (Thermofisher) and after sequence verification it was purified using NucleoSpin plasmid DNA purification kit (Machery-Nagel, Düren, Germany) according to the supplier’s indications. This Pachón cave gdf6b cloned DNA fragment was labeled by nick translation with Cy3-dUTP using Cy3 NT Labeling Kit (Jena Bioscience, Jena, Germany). The optimal fragment size of the probe (approx. 200-500 bp) was achieved after 30 min of incubation at 15°C.
Chromosome microdissection and FISH mapping
Twelve copies of B chromosome from A. mexicanus male individual (male 10) and twelve copies encompassing two B chromosomes (per cell) from a male individual (male 9) were manually microdissected as previously described91 under an inverted microscope (Zeiss Axiovert 135) using a sterile glass needle attached to a mechanical micromanipulator (Zeiss). The chromosomes were subsequently amplified by degenerate oligonucleotide primed-PCR (DOP-PCR) following previously described protocols.92 One mL of the resulting amplification product was used as a template DNA for a labeling DOP-PCR reaction, with Spectrum Orange-dUTP and Spectrum Green-dUTP, for Male 9 (2B) and Male 10 (1B), respectively (both Vysis, Downers Grove, USA). The amplification was done in 30 cycles, following previously described protocols.93 Depending on the experimental scheme, the final probe mixture contained i) both painting probes (200 ng each) or ii) a single painting probe (200 ng) and a labeled 4,368 bp long fragment containing gdf6b gene and its promoter (300 ng; see below). To block the shared repetitive sequences, the probe also contained 4-5 mg of unlabelled competitive DNA prepared from female gDNA (on male preparations) or male gDNA (on female preparations). Male and female gDNAs were isolated from liver and spleen using MagAttract HMW DNA kit (QIAGEN) and C0t-1 DNA (i.e., fraction of gDNA enriched with highly and moderately repetitive sequences) was then generated from them according to previously described protocols.94 The complete B probe mixture was dissolved in the final volume 20 μL (in case of two painting probes) or 14 μL (in case of one painting probe and a gdf6b gene probe) of hybridization mixture (50% formamide and 10% dextran sulfate in 2 × SSC).
FISH and whole-chromosome painting
The FISH (Fluorescence in situ hybridization on chromosomes) experiments were done using a combination of two previously published protocols,87,95 with slight modifications. Briefly, the aging of slides took place overnight at 37°C and then 60 min at 60°C, followed by treatments with RNase A (200 μg/mL in 2 × SSC, 60–90 min, 37°C) (Sigma-Aldrich) and then pepsin (50 μg/mL in 10 mM HCl, 3 min, 37°C). Subsequently, the slides were incubated in 1% formaldehyde in PBS (10 min) to stabilize the chromatin structure. Denaturation of chromosomes was done in 75% formamide in 2 × SSC (pH 7.0) (Sigma-Aldrich) at 72°C, for 3 min. The hybridization mixture was denatured for 8 min (86°C) and then pre-hybridized at 37°C for 45 min to outcompete the repetitive fraction. After application of the probe cocktail on the slide, the hybridization took place in a moist chamber at 37°C for 72 h. Subsequently, non-specific hybridization was removed by post-hybridization washes: two times in 1 × SSC (pH 7.0) (65°C, 5 min each) and once in 4 × SSC in 0.01% Tween 20 (42°C, 5 min). Slides were then washed in PBS (1 min), passed through an ethanol series, and mounted in antifade containing 1.5 μg/mL DAPI (Cambio, Cambridge, United Kingdom).
Synaptonemal complex immunostaining
Pachytene chromosome spreads from two Pachón cave A. mexicanus males (male 2 and 4) were prepared from testes following the protocol for Danio rerio96,97 with some modifications. Briefly, dissected testes were suspended in 200-600 mL (based on the testes size and cell density) of cold PBS. Cell suspensions were applied onto poly-l-lysine slides (ThermoFisher), with 1:30 (v/v) dilution in hypotonic solution (PBS: H2O, 1:2 v/v). After 20 min at room temperature (RT), slides were fixed with freshly prepared cold 2% formaldehyde (pH 8.0 – 8.5) for 3 min at RT. Slides were then washed three times in 0.1% Tween-20 (pH 8.0-8.5), 1 min each, and left to dry (1 h). Afterward, immunofluorescence analysis of synaptonemal complexes took place, using antibodies against the proteins SYCP3 (lateral elements of synaptonemal complexes) and MLH1 (mismatch repair protein; marker for visualization of recombination sites). The primary antibodies – rabbit anti-SYCP3 (1:300; Abcam, Cambridge, UK) and mouse anti-MLH1 (1:50, Abcam) – were diluted (v/v) in 3% BSA (bovine serum albumin) in 0.05% Triton X-100/ PBS. After application onto the slides, the incubation was carried out overnight in a humid chamber at 37°C. Next day, slides were washed three times in 0.1% Tween-20 in PBS, 10 min each and secondary antibodies, diluted (v/v) in 3% BSA (bovine serum albumin) in 0.05% Triton X-100/ PBS, were applied. Specifically, we used goat anti-rabbit Alexa 488 (1:300; Abcam) and goat anti-mouse Alexa555 (1:100; Abcam), and the slides were incubated for 3 h at 37°C. Then, after washing in 0,1% Tween-20 in PBS (10 min) and brief washing in 0.01% Tween-20 in distilled H2O, slides were mounted in antifade containing DAPI, as described above.
Microscopy and image analysis
At least 50 metaphase spreads per individual were analyzed to confirm the diploid chromosome number (2n), karyotype structure, and FISH results. Giemsa-stained preparations were analyzed under Axio Imager Z2 microscope (Zeiss, Oberkochen, Germany), equipped with an automatic Metafer-MSearch scanning platform. Photographs of the chromosomes were captured under 100 × objective using CoolCube 1 b/w digital camera (MetaSystems, Altlussheim, Germany). The karyotypes were arranged using Ikaros software (MetaSystems, Altlussheim, Germany). Chromosomes were classified according to their centromere positions,98 modified as metacentric (m), submetacentric (sm), subtelocentric (st), or acrocentric (a). FISH preparations were inspected using an Olympus BX53 epifluorescence microscope (Olympus, Tokyo, Japan), equipped with an appropriate fluorescence filter set. Black-and-white images were captured under 100 × objective for each fluorescent dye with a cooled DP30BW CCD camera (Olympus) using Olympus Acquisition Software. The digital images were then pseudo-colored (blue for DAPI, red for Cy3, green for FITC) and merged in DP Manager (Olympus). Composed images were then optimized and arranged using Adobe Photoshop CS6.
Phylogeny and synteny of Gdf6 proteins
Phylogeny and synteny relationships of Gdf6 genes were inferred from a Genomicus instance99 in which synteny and phylogeny have been reconciled with the Scorpios pipeline100 on 4 tetrapod species, one sarcopterygii species, one holostei species and 13 teleosts (see Figure 3 for species names).
Three-dimensional protein modeling
Three-dimensional models for the proprotein of A. mexicanus A-Gdf6b and B-Gdf6b were obtained by homology modeling. The amino acid sequences of full-length A-Gdf6b and B-Gdf6b were submitted to automated homology modeling using the hm_build macro of the software package YASARA.76 Modeling includes alignment of the two target sequences against sequences from Uniprot via PSI-Blast to build a position-specific scoring function matrix/profile, which is then used to search the PDB structure data bank for suitable modeling targets. Five templates were identified with sufficiently high scores, i.e., PDB entries 4YCG (proprotein complex of GDF2 (alternative naming BMP9)),55 5HLY (proprotein complex of Activin A),56 6Z3J (the mature C-terminal growth factor domain of GDF5 in complex with repulsive guidance molecule B)57 as well as 6Z3M (which is like 6Z3J but includes neogenin in complex with GDF5 and the repulsive guidance molecule B),57 and 3QB4 (which is only the C-terminal growth factor domain of GDF5).58 Several initial models were built on the basis of these template structures, missing sequence elements were modeled in an automated procedure through YASARA on the basis of an indexed PDB structure database. By this scheme 13 models were built, three only covered the C-terminal growth factor domain comprising residues 293 to 398, while 10 models covered the proprotein complex consisting of residues 3 to 398 (models on the basis of the 5HLY entry) and of residues 66 to 398 (models on the basis of template 4YCG). These models were individually refined by a short molecular dynamics simulation in explicit water to optimize hydrogen bonding and protein packing. Due to the overall low Z-score of the individual models, YASARA used the various models to form a hybrid homology model combining the elements with the highest-scoring factors into a single 3D model. This hybrid model was then used for further analysis.
Expression analysis by Real-Time PCR
For gene expression studies, mRNA transcripts levels were quantified during gonadal development from 10 dpf to 90 dpf, male and female gametogenesis stages, and finally in 10 adult organs including gonads as described previously.48 All samples were frozen in liquid nitrogen and stored at −80°C until RNA extraction. Total RNA extraction from gonads, trunks, and adult organs, followed by cDNA synthesis, and expression analysis by RT-PCR were carried out as previously described.48 Specific primers were designed for gdf6b in the most divergent sequence regions between the two paralogous gdf6a and gdf6b genes.
RNAScope in situ hybridization of gdf6b
RNA in situ hybridization (ISH) assays have been carried out using the RNAscope technology (ACD Biotechne) on 7 μm cross-sections of 15, 21, 30 and 60 dpf Pachon cavefish fixed in paraformaldehyde 4% overnight at 4°C and embedded in paraffin after serial dehydration in increasing methanol solutions. Specific probes for Pachon cavefish gdf6b and gsdf were synthesized by ACD Biotechne. Sections were collected on Super frost+ slides, heated at 60°C for 1 h and dewaxed 2 × 5 min in xylene followed by 2× 2 min in ethanol 100% at RT. Fluorescent ISH was carried out with the Multiplex Fluorescent Reagent Kit v2 (ACD Biotechne, ref: 323100) according to manufacturer’s protocol. Following hybridization, nuclei were labeled with DAPI (4′,6-diamidino-2-phenylindole) staining and slides were mounted with ProLong Gold Antifade Mountant (Invitrogen) and observed with a Leica TCS SP8 laser scanning confocal microscope.
Knockout of Pachón A. mexicanus gdf6b
Pachón cave A. mexicanus inactivated for gdf6b were generated using the CRISPR/Cas9 method. Guide RNAs (sgRNAs) targeting two sites located in exon 2 of gdf6b were designed using ZiFiT software77 (http://zifit.partners.org/ZiFiT/Disclaimer.aspx). DR274 vector (Addgene #42250) containing the guide RNA universal sequence was first linearized with Bsa1, electrophoresed in a 2% agarose gel and purified. PCR amplifications were then performed using linearized DR274 as a template and two primers for each sgRNA. Forward primers containing sgRNAs target sequences (#site 1 and #site 2) (bolded and underlined) between the T7 promoter sequence in the 5′ end and the conserved tracrRNA domain sequence were as follows. Forward primer (#site 1): 5′-GAAATTAATACGACTCACTATAGGAGTCTGAAACCGGTTCTGGTTTTAGAGCTAGAAATAGCAAG-3′. Forward primer (#site 2): 5′-GAAATTAATACGACTCACTATAGGGAGCTGGGCTGGGACGACGTTTTAGAGCTAGAAATAGCAAG-3′. Universal Reverse primer: 5′-AAAAGCACCGACTCGGTGCCACT-3′. Subsequently, residual plasmid was digested with Dpn1 (renewed once) at 37°C for 3 h. The final product was purified and used as a DNA template for transcription. The sgRNAs were transcribed using the MAXIscript T7 Transcription Kit (Ambion) according to the manufacturer’s instructions. The sgRNAs were precipitated in 200 mL of isopropanol solution at −20°C, centrifuged and the supernatant was removed. The precipitated sgRNAs were resuspended in RNase-free water. The sgRNAs were co-injected with Cas9 protein. Synthesized RNAs were then injected into 1-cell stage A. mexicanus Pachón cave embryos at the following concentrations: 72 ng/μL for each sgRNA and 216 ng/μL for the Cas9 protein (kindly provided by Tacgene, MNHN, Paris). Genotyping was performed on gDNA from caudal fin-clips of adult fishes. CRISPR-positive fish were screened for mutations using a set of PCR primers (P7-P8) (Figure S3A; Data S1E) flanking the sgRNAs target sites leading to a ~470 bp deletion on the exon 2 of the gdf6b genes (Figure S3A). The genetic sex of the mutants was determined by specific primers (P5-P6) on the gdf6b promoter with gap/indel variation between A-gdf6b and B-gdf6b (see STAR Methods above and Figure S3A).
Histology
Gonads were fixed in Bouin’s fixative solution for 48 h and then dehydrated serially in aqueous 70% and 95% ethanol, ethanol/butanol (5:95), and butanol. Tissues were embedded in paraffin blocks that were cutted serially into 5 μm sections, and were stained with hematoxylin-eosin-safran (HES) (Microm Microtech, Brignais, France).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
For the sex genotyping marker based on the heterozygous and specific site of the B chromosome on the exon 2 (position 679 bp of the B-gdf6b CDS), the significance of the correlation between this polymorphism and the male phenotypic sex was tested with the Pearson’s Chi-square test with Yates’ continuity correction. For gene expression, normality of data residuals, homogeneity of variances and homoscedasticity were verified before performing parametric or non-parametric tests. Consequently, statistical analyses were carried out only with non-parametric tests using RStudio (Open Source version) considering the level of significance at p < 0.05. For comparisons between two groups, we used Wilcoxon signed rank test. All data are shown as Mean ± Standard Error of the Mean (SEM).
Supplementary Material
Highlights.
Pachón cavefish have supernumerary male-predominant B chromosomes (Bs)
This Pachón B contain two loci of the putative gdf6b master sex-determining gene
gdf6b is only expressed in Pachón male gonads and its knockout induces sex reversal
Pachón B is a “B-sex” chromosome containing a putative male sex determination gene
ACKNOWLEDGMENTS
We thank Victor Simon, Stéphane Père, Krystel Saroul, Pierre-Lo Sudan, and Amélie Patinote for taking care of and handling cavefish, and Manon Thomas and the LPGP TEFOR infrastructure platform for acquisition of the RNAScope confocal images. This project was supported by funds from the “Agence Nationale de la Recherche” (ANR/DFG, PhyloSex project, 2014-2016) to Y.G. and M.S. S.R. was supported by grants from an Equipe FRM (Fondation pour la Recherche Médicale, DEQ20150331745) and MITI CNRS (Mission pour les Initiatives Transverses et Interdisciplinaires). J.H.P. was supported by an NIH grant (R35 GM139635). Sequencing was supported by France Génomique as part of an “Investissement d’avenir” program managed by ANR (contract ANR-10-INBS-09) and by the GET-PACBIO program (Programme opérationnel FEDER-FSE MIDI-PYRENEES ET GARONNE 2014-2020). The CytoEvol platform at ISEM was supported by the Labex CeMEB. J.B., T.P., and A.S. were supported by RVO: 67985904 of IAPG CAS, Liběchov. T.P. was supported by the projects of the Czech Ministry of Education (SVV 260571/2021). S.A.S. was supported by the Russian Foundation for Basic Research (RFBR) (18-34-00638). B.I.’s PhD fellowship was supported by the Doctoral School of Ecology, Geosciences, Agronomy, Nutrition of the University of Rennes 1 and INRAE. We are grateful to the genotoul bioinformatics platform Toulouse Occitanie (Bioinfo Genotoul, https://doi.org/10.15454/1.5572369328961167E12) for providing help, computing, and storage resources. Funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.08.030.
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- 1.Wright AE, Dean R, Zimmer F, and Mank JE (2016). How to make a sex chromosome. Nat. Commun 7, 12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furman BLS, Metzger DCH, Darolti I, Wright AE, Sandkam BA, Almeida P, Shu JJ, and Mank JE (2020). Sex chromosome evolution: so many exceptions to the rules. Genome Biol. Evol 12, 750–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Ambrosio U, Alonso-Lifante MP, Barros K, Kovařík A, Mas de Xaxars G, and Garcia S (2017). B-chrom: a database on B-chromosomes of plants, animals and fungi. New Phytol. 216, 635–642. [DOI] [PubMed] [Google Scholar]
- 4.Jones N (2017). New species with B chromosomes discovered since 1980. Nucleus 60, 263–281. [Google Scholar]
- 5.Camacho JPM (2005). B chromosomes. In The Evolution of the Genome, Gregory TR, ed. (Academic Press; ), pp. 223–286. [Google Scholar]
- 6.Camacho JPM, Sharbel TF, and Beukeboom LW (2000). B-chromosome evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci 355, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beladjal L, Vandekerckhove TTM, Muyssen B, Heyrman J, de Caesemaeker J, and Mertens J (2002). B-chromosomes and male-biased sex ratio with paternal inheritance in the fairy shrimp Branchipus schaefferi (Crustacea, Anostraca). Heredity 88, 356–360. [DOI] [PubMed] [Google Scholar]
- 8.Favarato RM, Braga Ribeiro L, Ota RP, Nakayama CM, and Feldberg E (2019). Cytogenetic characterization of two metynnis species (Characiformes, Serrasalmidae) reveals B chromosomes restricted to the females. Cytogenet. Genome Res 158, 38–45. [DOI] [PubMed] [Google Scholar]
- 9.Néo DM, Filho OM, and Camacho JPM (2000). Altitudinal variation for B chromosome frequency in the characid fish Astyanax scabripinnis. Heredity 85, 136–141. [DOI] [PubMed] [Google Scholar]
- 10.Vicente VE, Moreira-Filho O, and Camacho JP (1996). Sex-ratio distortion associated with the presence of a B chromosome in Astyanax scabripinnis (Teleostei, Characidae). Cytogenet. Cell Genet 74, 70–75. [DOI] [PubMed] [Google Scholar]
- 11.Stange ER, and Almeida-toledo LF (1993). Supernumerary B chromosomes restricted to males in Astyanax scabripinnis (Pisces, Characidae). Rev. Bras. Genet 16, 601–615. [Google Scholar]
- 12.Clark FE, Conte MA, Ferreira-Bravo IA, Poletto AB, Martins C, and Kocher TD (2017). Dynamic sequence evolution of a sex-associated B chromosome in Lake Malawi cichlid fish. J. Hered 108, 53–62. [DOI] [PubMed] [Google Scholar]
- 13.Clark FE, and Kocher TD (2019). Changing sex for selfish gain: B chromosomes of Lake Malawi cichlid fish. Sci. Rep 9, 20213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida K, Terai Y, Mizoiri S, Aibara M, Nishihara H, Watanabe M, Kuroiwa A, Hirai H, Hirai Y, Matsuda Y, and Okada N (2011). B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 7, e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camacho JPM, Schmid M, and Cabrero J (2011). B chromosomes and sex in animals. Sex Dev. 5, 155–166. [DOI] [PubMed] [Google Scholar]
- 16.Hanlon SL, and Hawley RS (2018). B chromosomes in the Drosophila genus. Genes (Basel) 9, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martis MM, Klemme S, Banaei-Moghaddam AM, Blattner FR, Macas J, Schmutzer T, Scholz U, Gundlach H, Wicker T, Šimková H, et al. (2012). Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 109, 13343–13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valente GT, Conte MA, Fantinatti BEA, Cabral-de-Mello DC, Carvalho RF, Vicari MR, et al. (2014). Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol. Biol. Evol 31, 2061–2072. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad SF, Jehangir M, Cardoso AL, Wolf IR, Margarido VP, Cabral-de-Mello DC, O’Neill R, Valente GT, and Martins C (2020). B chromosomes of multiple species have intense evolutionary dynamics and accumulated genes related to important biological processes. BMC Genomics 21, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pansonato-Alves JC, Serrano É, Utsunomia R, Camacho JPM, da Costa Silva GJ, Vicari MR, et al. (2014). Single origin of sex chromosomes and multiple origins of B chromosomes in fish genus Characidium. PLoS ONE 9, e107169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano-Freitas ÉA, Silva DMZA, Ruiz-Ruano FJ, Utsunomia R, Araya-Jaime C, Oliveira C, et al. (2020). Satellite DNA content of B chromosomes in the characid fish Characidium gomesi supports their origin from sex chromosomes. Mol. Genet. Genomics 295, 195–207. [DOI] [PubMed] [Google Scholar]
- 22.Conte MA, Clark FE, Roberts RB, Xu L, Tao W, Zhou Q, Wang D, and Kocher TD (2021). Origin of a giant sex chromosome. Mol. Biol. Evol 38, 1554–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Q, Zhu HM, Huang QF, Zhao L, Zhang GJ, Roy SW, et al. (2012). Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics 13, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piscor D, and Parise-Maltempi PP (2016). Microsatellite organization in the B chromosome and A chromosome complement in Astyanax (Characiformes, Characidae) species. Cytogenet. Genome Res 148, 44–51. [DOI] [PubMed] [Google Scholar]
- 25.Kavalco KF, and De Almeida-Toledo LF (2007). Molecular cytogenetics of blind mexican tetra and comments on the karyotypic characteristics of genus Astyanax (Teleostei, Characidae). Zebrafish 4, 103–111. [DOI] [PubMed] [Google Scholar]
- 26.Ebrahimzadegan R, Houben A, and Mirzaghaderi G (2019). Repetitive DNA landscape in essential A and supernumerary B chromosomes of Festuca pratensis Huds. Sci. Rep 9, 19989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizoguchi SMHN, and Martins-Santos IC (1997). Macro- and microchromosomes B in females of Astyanax scabripinnis (Pisces, Characidae). Hereditas 127, 249–253. [Google Scholar]
- 28.de Brito Portela-Castro AL, Ferreira Júlio Júnior H, and Belini Nishiyama P (2000). New occurrence of microchromosomes B in Moenkhausia sanctaefilomenae (Pisces, Characidae) from the Parana River of Brazil: analysis of the synaptonemal complex. Genetica 110, 277–283. [DOI] [PubMed] [Google Scholar]
- 29.Houben A (2017). B chromosomes - a matter of chromosome drive. Front. Plant Sci 8, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones RN, González-Sánchez M, González-García M, Vega JM, and Puertas MJ (2008). Chromosomes with a life of their own. Cytogenet. Genome Res 120, 265–280. [DOI] [PubMed] [Google Scholar]
- 31.McGaugh SE, Gross JB, Aken B, Blin M, Borowsky R, Chalopin D, Hinaux H, Jeffery WR, Keene A, Ma L, et al. (2014). The cavefish genome reveals candidate genes for eye loss. Nat. Commun 5, 5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad SF, and Martins C (2019). The modern view of B chromosomes under the impact of high scale omics analyses. Cells 8, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blavet N, Yang H, Su H, Solanský P, Douglas RN, Karafiátová M, Šimková L, Zhang J, Liu Y, Hou J, et al. (2021). Sequence of the supernumerary B chromosome of maize provides insight into its drive mechanism and evolution. Proc. Natl. Acad. Sci. USA 118, e2104254118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro-Domínguez B, Ruiz-Ruano FJ, Cabrero J, Corral JM, López-León MD, Sharbel TF, et al. (2017). Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans. Sci. Rep 7, 45200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz-Ruano FJ, Cabrero J, López-León MD, Sánchez A, and Camacho JPM (2018). Quantitative sequence characterization for repetitive DNA content in the supernumerary chromosome of the migratory locust. Chromosoma 127, 45–57. [DOI] [PubMed] [Google Scholar]
- 36.Christoffels A, Koh EGL, Chia JM, Brenner S, Aparicio S, and Venkatesh B (2004). Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol 21, 1146–1151. [DOI] [PubMed] [Google Scholar]
- 37.Vandepoele K, De Vos W, Taylor JS, Meyer A, and Van de Peer Y (2004). Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. USA 101, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feron R, Zahm M, Cabau C, Klopp C, Roques C, Bouchez O, et al. (2020). Characterization of a Y-specific duplication/insertion of the anti-Mullerian hormone type II receptor gene based on a chromosome-scale genome assembly of yellow perch, Perca flavescens. Mol. Ecol. Resour 20, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, Mizuno N, et al. (2012). A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8, e1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rafati N, Chen J, Herpin A, Pettersson ME, Han F, Feng C, Wallerman O, Rubin C-J, Péron S, Cocco A, et al. (2020). Reconstruction of the birth of a male sex chromosome present in Atlantic herring. Proc. Natl. Acad. Sci. USA 117, 24359–24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Sun Y, Zhao J, Shi H, Zeng S, Ye K, et al. (2015).A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 11, e1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, Sakamoto T, Fernandino JI, Somoza GM, Yokota M, and Strüssmann CA (2012).A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 109, 2955–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan Q, Feron R, Yano A, Guyomard R, Jouanno E, Vigouroux E, et al. (2019). Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet. 15, e1008013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reichwald K, Petzold A, Koch P, Downie BR, Hartmann N, Pietsch S, Baumgart M, Chalopin D, Felder M, Bens M, et al. (2015). Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell 163, 1527–1538. [DOI] [PubMed] [Google Scholar]
- 45.Gautier A, Sohm F, Joly J-S, Le Gac F, and Lareyre J-J (2011). The proximal promoter region of the zebrafish gsdf gene is sufficient to mimic the spatio-temporal expression pattern of the endogenous gene in Sertoli and granulosa cells. Biol. Reprod 85, 1240–1251. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura S, Kobayashi D, Aoki Y, Yokoi H, Ebe Y, Wittbrodt J, and Tanaka M (2006). Identification and lineage tracing of two populations of somatic gonadal precursors in medaka embryos. Dev. Biol 295, 678–688. [DOI] [PubMed] [Google Scholar]
- 47.Sawatari E, Shikina S, Takeuchi T, and Yoshizaki G (2007). A novel transforming growth factor-β superfamily member expressed in gonadal somatic cells enhances primordial germ cell and spermatogonial proliferation in rainbow trout (Oncorhynchus mykiss). Dev. Biol 301, 266–275. [DOI] [PubMed] [Google Scholar]
- 48.Imarazene B, Beille S, Jouanno E, Branthonne A, Thermes V, Thomas M, et al. (2020). Primordial germ cell migration and histological and molecular characterization of gonadal differentiation in Pachón cavefish Astyanax mexicanus. Sex Dev. 14, 80–98. [DOI] [PubMed] [Google Scholar]
- 49.Dalla Benetta E, Antoshechkin I, Yang T, Nguyen HQM, Ferree PM, and Akbari OS (2020). Genome elimination mediated by gene expression from a selfish chromosome. Sci. Adv 6, z9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nur U, Werren JH, Eickbush DG, Burke WD, and Eickbush TH (1988). A “selfish” B chromosome that enhances its transmission by eliminating the paternal genome. Science 240, 512–514. [DOI] [PubMed] [Google Scholar]
- 51.Jones RN (2018).Transmission and drive involving parasitic B chromosomes. Genes (Basel) 9, E388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernardino ACS, Cabral-de-Mello DC, Machado CB, Palacios-Gimenez OM, Santos N, and Loreto V (2017). B chromosome variants of the grasshopper Xyleus discoideus angulatus are potentially derived from pericentromeric DNA. Cytogenet. Genome Res 152, 213–221. [DOI] [PubMed] [Google Scholar]
- 53.Stevens JP, and Bougourd SM (1994). Unstable B-chromosomes in a European population of Allium schoenoprasum L. (Liliaceae). Biol. J. Linn. Soc. Lond 52, 357–363. [Google Scholar]
- 54.Ruban A, Schmutzer T, Wu DD, Fuchs J, Boudichevskaia A, Rubtsova M, et al. (2020). Supernumerary B chromosomes of Aegilops speltoides undergo precise elimination in roots early in embryo development. Nat. Commun 11, 2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mi L-Z, Brown CT, Gao Y, Tian Y, Le VQ, Walz T, and Springer TA (2015). Structure of bone morphogenetic protein 9 procomplex. Proc. Natl. Acad. Sci. USA 112, 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Fischer G, and Hyvönen M (2016). Structure and activation of pro-activin A. Nat. Commun 7, 12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malinauskas T, Peer TV, Bishop B, Mueller TD, and Siebold C (2020). Repulsive guidance molecules lock growth differentiation factor 5 in an inhibitory complex. Proc. Natl. Acad. Sci. USA 117, 15620–15631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klammert U, Mueller TD, Hellmann TV, Wuerzler KK, Kotzsch A, Schliermann A, Schmitz W, Kuebler AC, Sebald W, and Nickel J (2015). GDF-5 can act as a context-dependent BMP-2 antagonist. BMC Biol. 13, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, and Joung JK (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol 31, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng H, Concepcion GT, Feng X, Zhang H, and Li H (2021). Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, and Aiden EL (2016). Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, and Aiden EL(2017).De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Birney E, Clamp M, and Durbin R (2004). GeneWise and Genomewise. Genome Res. 14, 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr., Hannick LI, et al. (2003). Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, and Morgenstern B (2006). AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gel B, and Serra E (2017). karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kõressaar T, Lepamets M, Kaplinski L, Raime K, Andreson R, and Remm M (2018). Primer3_masker: integrating masking of template sequence with primer design software. Bioinformatics 34, 1937–1938. [DOI] [PubMed] [Google Scholar]
- 72.Wingett SW, and Andrews S (2018). FastQ Screen: a tool for multigenome mapping and quality control. F1000Res. 7, 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, arXiv:1303.3997. https://arxiv.org/abs/1303.3997. [Google Scholar]
- 74.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kofler R, Pandey RV, and Schlötterer C (2011). PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27, 3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krieger E, and Vriend G (2014). YASARA View - molecular graphics for all devices-from smartphones to workstations. Bioinformatics 30, 2981–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, and Dobbs D (2010). ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 38, W462–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hinaux H, Pottin K, Chalhoub H, Père S, Elipot Y, Legendre L, et al. (2011). A developmental staging table for Astyanax mexicanus surface fish and Pachón cavefish. Zebrafish 8, 155–165. [DOI] [PubMed] [Google Scholar]
- 79.Elipot Y, Legendre L, Père S, Sohm F, and Rétaux S (2014). Astyanax transgenesis and husbandry: how cavefish enters the laboratory. Zebrafish 11, 291–299. [DOI] [PubMed] [Google Scholar]
- 80.Gharbi K, Gautier A, Danzmann RG, Gharbi S, Sakamoto T, Høyheim B, et al. (2006). A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172, 2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, and Aiden EL (2016). Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Powell DL, García-Olazábal M, Keegan M, Reilly P, Du K, Díaz-Loyo AP, Banerjee S, Blakkan D, Reich D, Andolfatto P, et al. (2020). Natural hybridization reveals incompatible alleles that cause melanoma in swordtail fish. Science 368, 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Du K, Stöck M, Kneitz S, Klopp C, Woltering JM, Adolfi MC, Feron R, Prokopov D, Makunin A, Kichigin I, et al. (2020). The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol 4, 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.She R, Chu JS-C, Wang K, Pei J, and Chen N (2009). GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 19, 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pasquier J, Cabau C, Nguyen T, Jouanno E, Severac D, Braasch I, Journot L, Pontarotti P, Klopp C, Postlethwait JH, et al. (2016). Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics 17, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Völker M, and Ràb P (2015). Direct chromosome preparation from regenerating fin tissue. In Fish Cytogenetic Techniques: Ray-Fin Fishes and Chondrichthyans, Ozouf-Costaz C, Pisano E, Foresti F, and de Almeida Toledo LF, eds. (CRC Press; ), pp. 37–41. [Google Scholar]
- 87.Sember A, Bohlen J, Šlechtová V, Altmanová M, Symonová R, and Ráb P (2015). Karyotype differentiation in 19 species of river loach fishes (Nemacheilidae, Teleostei): extensive variability associated with rDNA and heterochromatin distribution and its phylogenetic and ecological interpretation. BMC Evol. Biol 15, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ráb P, and Roth P (1988). Cold-blooded vertebrates. Methods of Chromosome Analysis (Czechoslovak Biological Society Publishers; ), pp. 115–124. [Google Scholar]
- 89.Bertollo L, Cioffi M, and Moreira-Filho O (2015). Direct chromosome preparation from freshwater teleost fishes. In Fish Cytogenetic Techniques: Ray-Fin Fishes and Chondrichthyans, Ozouf-Costaz C, Pisano E, Foresti F, and de Almeida Toledo LF, eds. (CRC Press; ), pp. 21–26. [Google Scholar]
- 90.Haaf T, and Schmid M (1984). An early stage of ZW/ZZ sex chromosome differentiation in Poecilia sphenops var. melanistica (Poeciliidae, Cyprinodontiformes). Chromosoma 89, 37–41. [Google Scholar]
- 91.Ahmed A-R, Leon BL, and Thomas L (2020). Glass needle-based chromosome microdissection–How to set up probes for molecular cytogenetics? Video J. Clin. Res 2, 100004VAM08AR2020. [Google Scholar]
- 92.Yang F, Trifonov V, Ng BL, Kosyakova N, and Carter NP (2009). Generation of paint probes by flow-sorted and microdissected chromosomes. In Fluorescence In Situ Hybridization (FISH) – Application Guide Springer Protocols Handbooks, Liehr T, ed. (Springer; ), pp. 35–52. [Google Scholar]
- 93.Yang F, and Graphodatsky AS (2009). Animal probes and ZOO-FISH. In Fluorescence In Situ Hybridization (FISH) – Application Guide Springer Protocols Handbooks, Liehr T, ed. (Springer; ), pp. 323–346. [Google Scholar]
- 94.Zwick MS, Hanson RE, Islam-Faridi MN, Stelly DM, Wing RA, Price HJ, and McKnight TD (1997). A rapid procedure for the isolation of C0t-1 DNA from plants. Genome 40, 138–142. [DOI] [PubMed] [Google Scholar]
- 95.Yano CF, Bertollo LA, Ezaz T, Trifonov V, Sember A, Liehr T, et al. (2017). Highly conserved Z and molecularly diverged W chromosomes in the fish genus Triportheus (Characiformes, Triportheidae). Heredity 118, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kochakpour N (2009). Immunofluorescent microscopic study of meiosis in zebrafish. Methods Mol. Biol 558, 251–260. [DOI] [PubMed] [Google Scholar]
- 97.Kochakpour N, and Moens PB (2008). Sex-specific crossover patterns in Zebrafish (Danio rerio). Heredity 100, 489–495. [DOI] [PubMed] [Google Scholar]
- 98.Levan A, Fredga K, and Sandberg AA (1964). Nomenclature for centromeric position on chromosomes. Hereditas 52, 201–220. [Google Scholar]
- 99.Louis A, Muffato M, and Roest Crollius H (2013). Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 41, D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parey E, Louis A, Cabau C, Guiguen Y, Roest Crollius H, and Berthelot C (2020). Synteny-guided resolution of gene trees clarifies the functional impact of whole-genome duplications. Mol. Biol. Evol 37, 3324–3337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences and the whole genome assembly of Pachón cavefish have been deposited in the National Center for Biotechnology Information DDBJ/ENA/GenBank databases under the BioProject PRJNA734455. This accession number is listed in the key resources table. This study did not generate new unique code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Primary: rabbit anti-SYCP3 | Abcam | Cat# ab15093; RRID: AB_301639 |
| Primary: mouse anti-MLH1 | Abcam | Cat# ab14206: RRID: AB_300987 |
| Secondary: goat anti-rabbit IgG H&L Alexa488 | Abcam | Cat# ab150077; RRID: AB_2630356 |
| Secondary: goat anti-mouse IgG H&L Alexa555 | Abcam | Cat# ab150114; RRID: AB_2687594 |
| Chemicals, peptides, and recombinant proteins | ||
| Tricaine methanesulfonate (MS 22) | Sigma-Aldrich | Cat# E10521 |
| 2-phenoxyethanol | Sigma-Aldrich | Cat# 77699 |
| Ribonuclease A from bovine pancreas | Sigma-Aldrich | Cat# R6513 |
| Formamide | Sigma-Aldrich | Cat# 47671 |
| Cas9 protein | Tacgene | No reference |
| Hematoxylin | Microm Microtech | Cat# F/HAQ999 |
| Mountant and DAPI (4′,6-diamidino-2-phenolindole) | Cambio | Cat# 1124-MD-1250 |
| Spectrum-Orange-dUTP | Vysis | Cat# 02N33-050 |
| Spectrum-Green-dUTP | Vysis | Cat# 02N32-050 |
| Bovine Serum Albumin (BSA) | Vector | Cat# SP-5050 |
| Pepsin | Chemos CZ | Cat# A4289.0100 |
| Giemsa stain | Giemsa stain | Giemsa stain |
| Critical commercial assays | ||
| NucleoSpin plasmid DNA purification kit | Machery-Nagel | Cat# 740588.50 |
| NucleoSpin Kits for Tissue | Machery-Nagel | Cat# 740952.50 |
| HiDi Taq DNA polymerase | myPOLS Biotec | No reference |
| AccuPrime Taq DNA polymerase | Thermofisher | Cat# 12339016 |
| AccuPrime Taq DNA Polymerase, High Fidelity | Thermofisher | Cat# 12346086 |
| TOPO TA cloning Kit XL | Thermofisher | Cat# K8050-20 |
| ProLong Gold Antifade Mountant | Thermofisher | Cat# P10144 |
| MAXIscript T7 Transcription Kit | Thermofisher | Cat# AM1312 |
| Jumpstart Taq DNA polymerase | Sigma-Aldrich | Cat# D9307 |
| Chromium Genome Reagent Kits v2 | 10X genomics | Cat# PN-120258 |
| KAPA library Amplification kit | Roche Diagnostics | Cat# KK2620 |
| Kapa Library Quantification Kit | Roche Diagnostics | Cat# KK4824 |
| Illumina HiSeq3000 sequencing kits | Illumina | Cat# FC-410-101 |
| Illumina NovaSeq Reagent Kits | Illumina | Cat# 20012860 |
| Truseq DNA PCR-Free kit | Illumina | Cat# 20015962 |
| TruSeq Nano DNA HT Library Prep Kit | Illumina | Cat# 20015965 |
| SP flow cell | Illumina | Cat# 20027464 |
| S4 Novaseq flow cell | Illumina | Cat #20012866 |
| AMPure XP beads | Beckman Coulter | Cat# A63881 |
| Nanopore Ligation Sequencing Kit | Oxford Nanopore | Cat# SQK-LSK109 |
| SMRTbell Express Template Prep Kit 2.0 | PacBio | Cat# 101-685-400 |
| Binding kit 2.0 kit | PacBio | Cat# 102-089-000 |
| Sequencing kit 2.0 | PacBio | Cat# 101-829-200 |
| Arima-HiC kit | Arima | Cat# 510008 |
| Cy3 NT Labeling Kit | Jena Bioscience | Cat# PP-305L-CY3 |
| Multiplex Fluorescent Reagent Kit v2 | ACD Biotechne | Cat# 323100 |
| MagAttract HMW DNA kit (48) | QIAGEN | Cat# 67563 |
| Deposited data | ||
| Structure of proprotein complex of human GDF2 | 55 | PDB: 4YCG |
| Structure of the proprotein complex of human activin A | 56 | PDB: 5HLY |
| Structure of the growth factor domain of human GDF5 in complex with repulsive guidance molecule B | 57 | PDB: 6Z3J |
| Structure of the growth factor domain of human GDF5 in complex with repulsive guidance molecule B and neogenin | 57 | PDB: 6Z3M |
| Structure of the growth factor domain of human GDF5 in complex with BMP receptor 1A | 58 | PDB: 3QB4 |
| Pachón cavefish, Astyanax mexicanus, genome assembly and annotation | This study | GenBank: PRJNA734455 |
| Experimental models: Organisms/strains | ||
| A. mexicanus Pachón cavefish wild-type | Jeffery laboratory, University of Maryland, College Park, MD | N/A |
| A. mexicanus Pachón cavefish gdf6b KO | This study | N/A |
| Oligonucleotides | ||
| Primers for the amplification of the gdf6b FISH Probe, see Data S1E | Eurofins | N/A |
| Primers for B genotyping, see Data S1E | Eurofins | N/A |
| Primers for screening gdf6b-KO, see Data S1E | Eurofins | N/A |
| Primers for RT-QPCR for gdf6b, see Data S1E | Eurofins | N/A |
| Forward primer containing sgRNAs target sequences for #site 1 GAAATTAATACGACTCACTATAGGAGTCTGAAACCGGTTCTGGTTTTAGAGCTAGAAATAGCAAG | Eurofins | N/A |
| Forward primer containing sgRNAs target sequences for #site 2 GAAATTAATACGACTCACTATAGGGAGCTGGGCTGGGACGACGTTTTAGAGCTAGAAATAGCAAG | Eurofins | N/A |
| Universal Reverse primer: AAAAGCACCGACTCGGTGCCACT | Eurofins | N/A |
| Truseq DNA UD Indexes | Illumina | Cat# 20020590 |
| DOP primer (10 μM) GAGGATGAGGTTGAGNNNNNNGTGG | KRD, Prague, Czech Republic | N/A |
| RNAScope probe for cavefish Gdf6b | ACD Biotechne | N/A |
| RNAScope probe for cavefish gsdf | ACD Biotechne | N/A |
| Recombinant DNA | ||
| DR274 vector | 59 | Addgene plasmid #42250 |
| Gdf6b genomic FISH probe | This study | N/A |
| Software and algorithms | ||
| Hifiasm version 0.9 | 60 | https://github.com/chhylp123/hifiasm |
| Long Ranger v2.1.1 | 10X Genomics | https://support.10xgenomics.com/genome-exome/software/pipelines/latest/advanced/other-pipelines |
| Juicer | 61 | https://github.com/aidenlab/juicer |
| 3D-DNA pipeline | 62 | https://github.com/aidenlab/3d-dna |
| Juicebox | 62 | https://github.com/aidenlab/Juicebox/wiki/Juicebox-Assembly-Tools |
| Minimap2 v2.11 | 63 | https://github.com/lh3/minimap2 |
| RepeatModeler | Smit, AFA, Hubley, R. Unpublished | http://www.repeatmasker.org/ |
| RepeatMasker | Smit, AFA, Hubley, R & Green, P. Unpublished | http://www.repeatmasker.org/ |
| Genewise | 64 | https://www.ebi.ac.uk/seqdb/confluence/display/THD/GeneWise |
| Exonerate | European Bioinformatics Institute. Unpublished | https://www.ebi.ac.uk/about/vertebrate-genomics/software/exonerate |
| Hisat | 65 | http://www.ccb.jhu.edu/software/hisat/index.shtml |
| StringTie | 66 | https://ccb.jhu.edu/software/stringtie/ |
| Trinity | 67 | https://github.com/trinityrnaseq/trinityrnaseq/wiki |
| Pasa | 68 | https://github.com/PASApipeline/PASApipeline/wiki |
| Augustus | 69 | https://github.com/Gaius-Augustus/Augustus |
| karyoploteR | 70 | https://bernatgel.github.io/karyoploter_tutorial/ |
| Primer3web software version 4.1.0 | 71 | https://primer3.ut.ee/ |
| NovaSeq Control Software and Real-Time Analysis component (v3.4.4) | PacBio | N/A |
| Illumina’s conversion software (bcl2fastq v2.20) | Illumina | N/A |
| FastQC (v0.11.8) | 72 | https://github.com/s-andrews/FastQC |
| BWA mem version 0.7.17 | 73 | https://github.com/lh3/bwa |
| Picard tools version 2.18.2 | Broad Institute | http://broadinstitute.github.io/picard |
| samtools mpileup version 1.8 | 74 | https://sourceforge.net/projects/samtools/files/samtools/ |
| popoolation2 | 75 | https://sourceforge.net/projects/popoolation2/ |
| PSASS version 2.0.0 | Feron R. unpublished | https://doi.org/10.5281/zenodo.2615936 |
| Yasara | 76 | http://www.yasara.org |
| Yasara hm_build.mcr macro | 76 | http://www.yasara.org/hm_build.mcr |
| RStudio (Open Source version) | RStudio team | http://www.rstudio.com/ |
| Ikaros software | MetaSystems | N/A |
| DP Manager imaging software | Olympus | N/A |
| Olympus Acquisition Software | Olympus | N/A |
| Adobe Photoshop CS6 | Adobe | N/A |
| ZiFiT software | 77 | http://zifit.partners.org/ZiFiT/Disclaimer.aspx |
| Other | ||
| Nalgene Freezing Container (Mr. Frosty) | Merck | Cat# C1562-1EA |
| Megaruptor1 system | Diagenode | Cat# B06010001 |
| Qubit3 fluorometer Invitrogen | ThermoFisher | Cat# Q33216 |
| poly-l-lysine slides | ThermoFisher | Cat# J2800AMNZ |
| BluePippin Size Selection system | Sage Science | No reference |
| M220 Focused-ultrasonicator | Covaris | Cat# 500295 |
| Sequence data a whole genome assembly of Pachón | This study | PRJNA734455 |


