Abstract
Viral double-stranded RNA (dsRNA) generated during the course of infection leads to the activation of a latent transcription factor, dsRNA-activated factor 1 (DRAF1). DRAF1 binds to a DNA target containing the type I interferon-stimulated response element and induces transcription of responsive genes. DRAF1 is a multimeric transcription factor containing the interferon regulatory factor 3 (IRF-3) protein and one of the histone acetyl transferases, CREB binding protein (CBP) or p300 (CBP/p300). In uninfected cells, the IRF-3 component of DRAF1 resides in the cytoplasm. The cytoplasmic localization of IRF-3 is dependent on a nuclear export signal, and we demonstrate IRF-3 recognition by the chromosome region maintenance 1 (CRM1) (also known as exportin 1) shuttling receptor. Following infection and specific phosphorylation, IRF-3 accumulates in the nucleus where it associates with CBP and p300. We identify a nuclear localization signal (NLS) in IRF-3 that is critical for nuclear accumulation. Mutation of the NLS abrogates nuclear localization even following infection. The NLS appears to be active constitutively, but it is recognized by only a subset of importin-α shuttling receptors. Evidence is presented to support a model in which IRF-3 normally shuttles between the nucleus and the cytoplasm but cytoplasmic localization is dominant prior to infection. Following infection, phosphorylated IRF-3 can bind to the CBP/p300 proteins resident in the nucleus. We provide the evidence of a role for CBP/p300 binding in the nuclear sequestration of a transcription factor that normally resides in the cytoplasm.
Cells respond to viral infection with the activation of latent transcription factors that function in host survival. During the course of viral infection with many DNA or RNA viruses, viral double-stranded RNA (dsRNA) is generated during transcription and/or replication. The dsRNA is a potent intracellular signal that stimulates the defense responses of the cell. One of the signal transduction pathways activated by dsRNA leads to transcriptional induction of type I interferon (IFN) genes (18, 24, 43, 48). IFNs are cytokines that have the unique ability to confer resistance to viral infections (15). Most of the biological effects of IFNs have been analyzed as paracrine hormones. Our investigations have led to the discovery of another defense response of the primary infected cell that is independent of autocrine IFN. The presence of dsRNA activates a latent cellular transcription factor designated the dsRNA-activated factor 1 (DRAF1) that directly induces a subset of genes stimulated by type I IFN (11, 12, 52).
Analyses of the composition of DRAF1 have identified the interferon regulatory factor 3 (IRF-3) protein and one of the histone acetylases, CREB binding protein (CBP) or p300 (henceforth CBP/p300) to be present in the complex (3, 6, 19, 32, 44, 51, 52, 57). IRF-3 normally resides in the cytoplasm of the cell, but accumulates in the nucleus following infection in association with CBP/p300 to form the DRAF1 transcription factor. CBP and p300 are nuclear acetyl transferases that can modify histones, resulting in chromatin remodeling and increased access of transcription factors to DNA (4, 38). They have also been reported to acetylate transcription factors and can interact with other acetyl transferases, general transcription factors, and the RNA polymerase II holoenzyme via RNA helicase A (5, 26, 35, 36, 49). DRAF1 binds to DNA target sites containing the IFN-stimulated response element (ISRE), but the DNA binding specificity of DRAF1 is such that only a subset of IFN-stimulated genes are induced (11, 12).
The function of most proteins is dependent on appropriate cellular localization. Latent transcription factors resident in the cytoplasm of the cell can respond to external signals and subsequently transmit them to the nucleus. A growing number of transcription factors function by such a regulated nuclear-cytoplasmic translocation mechanism to induce specific gene expression rapidly and transiently. Members of diverse transcription factor families, including STAT (signal transducer and activator of transcription), NF-κB (nuclear factor of immunoglobulin kappa B cells), and NFAT (nuclear factor of activated T cells), and steroid receptors receive an activating signal in the cytoplasm and are rapidly shuttled into the nucleus (9, 13, 14, 23, 45). Continuous presence in the nucleus may be undesirable because the factors serve the purpose of signal detection in the cytoplasm and/or because persistent activation of target genes may be detrimental to the cell.
Transport of proteins in and out of the nucleus relies on their recognition by soluble receptors that mediate movement across nuclear pore complexes (25, 29, 33, 37, 41, 42, 50, 53). Shuttling receptors recognize an amino acid sequence corresponding to a nuclear localization signal (NLS) or a nuclear export sequence (NES) in proteins destined for nuclear or cytoplasmic localization (16, 20, 54). The IRF-3 subunit of DRAF1 displays regulated nuclear-cytoplasmic localization in response to viral infection and the presence of viral dsRNA (52). In this report, we demonstrate that the localization of IRF-3 is mediated by the function of both an NLS and an NES that are recognized by distinct shuttling receptors. The results suggest that the NLS and NES in IRF-3 are constitutively active, but nuclear export is normally dominant. Following infection, IRF-3 accumulates in the nucleus, and this accumulation relies both on the function of its NLS and on its acquired ability to bind CBP/p300. The ability of IRF-3 to associate with CBP/p300 when it enters the nucleus depends on its modification by serine phosphorylation during infection (31, 52, 57). Binding to CBP/p300 appears to sequester IRF-3 in the nucleus so that the NES is no longer dominant. CBP/p300 binding thereby regulates the nuclear localization of IRF-3 and the formation of DRAF1 to induce specific gene expression.
MATERIALS AND METHODS
Cell culture, transfections, and infections.
Human endometrial carcinoma cells, HEC-1B (ATCC), were maintained in Dulbecco modified Eagle medium with 8% fetal bovine serum. DNA transfections were performed with calcium phosphate-DNA coprecipitates (52). Infections with Newcastle disease virus (NJ-LaSota-1946) (NDV) were performed typically at a multiplicity of 100 HA units/ml for 6 h (11).
Plasmid constructs.
To create green fluorescent protein (GFP)-IRF-3, the enhanced GFP gene (Clontech) was positioned in frame upstream of human IRF-3 cDNA by using a fragment generated by PCR with Pfu polymerase (Stratagene) (52). All mammalian expression systems used the cytomegalovirus immediate-early promoter. Repositioning of the IRF-3 NES in the GFP–IRF-3 plasmid (NES-wt and NES-IL/MM [IL/MM designating the replacement of amino acids (a.a.) IL with MM]) was performed by inserting an oligonucleotide corresponding to the IRF-3 amino acid sequence DILDELLGNMVL between GFP and IRF-3. The GFP–IRF-3–CBP plasmid was constructed by inserting a DNA sequence encoding GFP and IRF-3 (1 to 327 a.a.) upstream and in frame with the CBP gene. Site-directed mutagenesis was performed by using the Stratagene Quickchange site-directed mutagenesis kit. Deletion constructs were created at specific restriction enzyme sites. IRF-3 was subcloned into the bacterial expression plasmid pGEX2T (Pharmacia) to generate the glutathione S-transferase (GST) fusion GST–IRF-3. The GST-NES construct that encodes the NES of the protein kinase A inhibitor (PKI) was kindly provided by Susan Taylor (University of California, San Diego). The IRF-3/5D construct was a gift from John Hiscott (Lady Davis Institute). CBP and p300 cDNAs were gifts from Richard Goodman (Oregon Health Sciences University), and p300 constructs for in vitro translation were a gift from David Livingston (The Dana-Farber Cancer Institute). The human chromosome region maintenance 1 (CRM1) (also known as exportin 1) cDNA was a gift from Gerard Grosveld (St. Jude Children's Research Hospital) and was subcloned by PCR into pCDNA3 (Invitrogen). The importin-α constructs were subcloned into pCDNA3. Qip-1 was a gift from Takemi Enomoto (Tohoku University), hSRPα/Rch1 was a gift from Karsten Weis (University of California at Berkeley), hSRP1/NPI1 was a gift from Patricia Cortes (The Rockefeller University), and KPNA3 was a gift from Y. Hirai (Otsuka GEN Research Institute).
Protein analysis.
Immunoprecipitations were performed with cell extracts prepared following lysis in 50 mM Tris (pH 7.5), 400 mM NaCl, 10% glycerol, 50 mM sodium fluoride, 5 mM EDTA, 10 mM sodium phosphate, 1 mM β-mercaptoethanol, and 0.5% Nonidet P-40. Electrophoretic mobility shift assays were performed with the ISG15 dsDNA target site (5′-GGGAAAGGGAAACCGAAACTGAA-3′) (12). Antibodies to IRF-3 were described (52); antibodies to CBP (A-22) and p300 (N-15) were purchased from Santa Cruz Biotech, Inc.; and antibodies to GFP were purchased from Clontech. Proteins were synthesized in vitro with a coupled transcription-translation system (Promega) in the presence of [35S]methionine. Cells were fixed for GFP fluorescence with 4% paraformaldehyde, and microscopic images were captured with the use of Adobe Photoshop 4.0. Leptomycin B was a kind gift from Barbara Wolff-Winiski (Novartis). Images of autoradiographs are presented with use of Adobe Photoshop 4.0.
CRM1 binding.
Fifteen micrograms of bacterially expressed GST, GST–IRF-3, or GST-NES were bound to glutathione agarose beads (Sigma) that were preblocked with 1% bovine serum albumin at 4°C. The beads were washed in binding buffer and were incubated with human CRM1 translated in vitro in the presence of [35S]methionine and 5 μg of purified Ran Q69L in a 50-μl reaction of binding buffer (50 mM HEPES [pH 7.9], 200 mM KCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, 0.4% Tween-20, 0.4% milk, 2 mM GTP) for 2 h at 4°C. Ran Q69L was expressed in bacteria and was purified as described (17). The peptide corresponding to the NES of the human immunodeficiency virus type 1 Rev protein (CLPPLERLTL) was synthesized by Research Genetics, Inc. (Huntsville, Ala.), and was used at 1 mM in competitions (20). Following incubation, the beads were washed in binding buffer, and proteins were eluted and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and fluorography.
Importin-α binding.
Importin-α proteins were synthesized in vitro in the presence of [35S]methionine and were incubated with either bacterially expressed GST or GST–IRF-3 or with NLS peptides cross-linked to agarose beads. The proteins were incubated in a buffer containing 20 mM HEPES (pH 7.3), 110 mM K-acetate, 5 mM Na-acetate, 1.5 mM Mg-acetate, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.2% Tween 20, and 1% nonfat dry milk. Peptides containing the NLS from the simian virus 40 (SV40) T antigen (CGGGPKKKRKVED), the NLS of IRF-3 (CDLPTWKRNFRSALNRKEG), and a control peptide (RLQLGRLDYLPTCFMHSFH) were synthesized by Research Genetics, Inc. Peptides were chemically cross-linked to agarose beads (2). Following incubation, the complexes were washed, and proteins were eluted and analyzed by SDS-PAGE and fluorography.
IRF-3 binding to CBP or p300.
Fragments of p300 synthesized in vitro in the presence of [35S]methionine were incubated with immunocomplexes formed with control antibodies or antibodies to GFP or IRF-3 and whole-cell extracts (1 mg of protein). Alternatively, fragments of IRF-3 synthesized in vitro were incubated with immunocomplexes formed with control antibodies or antibodies to CBP. Binding was performed in 20 mM HEPES (pH 7.9), 10% glycerol, 200 mM KCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.5% NP-40, and 1% nonfat dry milk for 1 h at room temperature. The complexes were washed, and proteins were eluted and analyzed by SDS-PAGE and fluorography.
RESULTS
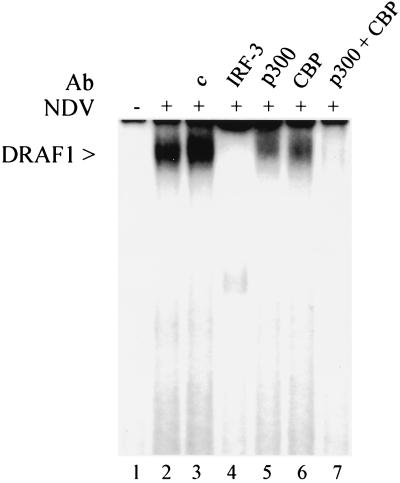
Following viral infection, DRAF1 can be detected in the nucleus by its ability to bind to a DNA sequence containing the ISRE. Activation of DRAF1 is independent of the IFN signal transduction pathway since it occurs in cells that do not respond to IFN, such as HEC-1B or cells derived from mice lacking the type I IFN receptor (11, 12, 52). This can be demonstrated by an electrophoretic mobility shift assay with nuclear extracts isolated from HEC-1B cells infected with NDV (Fig. 1). IRF-3 and either of the two histone acetyl transferases (CBP and p300) are components of the DRAF1 DNA binding complex (52). Addition of antibodies specific for IRF-3 to the DNA binding reaction inhibits the appearance of the DRAF1-DNA complex (Fig. 1, lane 4). Antibodies specific for either CBP or p300 significantly reduce the amount of complex detected (lanes 5 and 6), while the addition of both antibodies completely prevents the appearance of the DRAF1 complex (lane 7). This suggests that heterogeneous complexes of IRF-3 with CBP or p300 may exist on the DNA in the form of DRAF1.
FIG. 1.
Appearance of DRAF1 in the nucleus following viral infection. Electrophoretic mobility shift assay was performed with nuclear extracts from HEC-1B cells either uninfected (lane 1) or infected with NDV for 6 h (lanes 2 to 7). The following specific antibodies were included in the DNA binding reactions: lane 3, 1 μg of control rabbit serum; lane 4, 1 μg of anti-IRF-3 antibody; lane 5, 1 μg of anti-p300 antibody; lane 6, 1 μg of anti-CBP antibody; and lane 7, 0.5 μg of anti-CBP antibody and 0.5 μg of anti-p300 antibody.
IRF-3 interaction with CBP/p300.
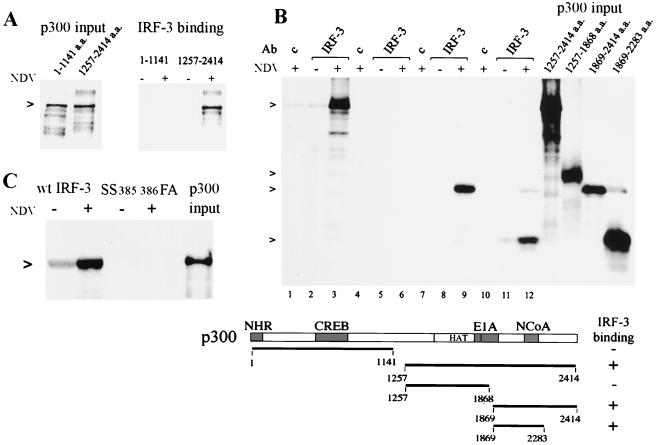
Since association of IRF-3 with CBP/p300 is critical for DRAF1 formation, we analyzed the domain of p300 that interacts with IRF-3. CBP/p300 can exist in multimeric complexes with distinct proteins in vivo, and for this reason we used an assay with in vitro-synthesized p300 molecules (19). Initially, two fragments of the p300 protein were tested for binding, the amino-terminal portion and the carboxyl-terminal portion (Fig. 2A). p300 fragments were synthesized in the presence of [35S]methionine and were incubated with immunocomplexes containing endogenous IRF-3 from uninfected or NDV-infected cells. The amino-terminal p300 fragment did not bind to IRF-3. The carboxyl-terminal p300 fragment did associate with IRF-3, but only when the source of IRF-3 was infected cells. To further delineate the region of p300 interaction, a series of carboxyl-terminal fragments of p300 was tested (Fig. 2B). Immunocomplexes were prepared with control serum or antibody to IRF-3 from uninfected or infected cells and were reacted with the p300 carboxyl fragments. IRF-3 from infected cells demonstrated specific binding to a region of p300 that includes a domain previously described to bind nuclear receptor coactivator NCoA-1 (also known as SRC-1) (40). This region is downstream of the histone acetyl transferase domain and the adenoviral E1A binding domain.
FIG. 2.
Specific binding of IRF-3 to the carboxyl region of p300 in vitro. (A) Amino-terminal (a.a. 1 to 1141) or carboxyl-terminal (a.a. 1257 to 2414) p300 fragments were synthesized in vitro in the presence of [35S]methionine. Lanes on left display relative input of p300. IRF-3 was isolated by immunoprecipitation from HEC-1B cells uninfected or infected with NDV, and the immunocomplexes were incubated with p300. (B) Various carboxyl-terminal fragments of p300 were generated (relative input is shown on the right) and were tested for binding to IRF-3 as in panel A: lanes 1 to 3, a.a. 1257 to 2414; lanes 4 to 6, a.a. 1257 to 1868; lanes 7 to 9, a.a. 1869 to 2414; lanes 10 to 12, a.a. 1869 to 2283. A diagrammatic representation of the results is shown in the lower panel. Domains of p300 known to bind to nuclear hormone receptors (NHR), cyclic AMP response element binding protein (CREB), adenoviral E1A oncoprotein (E1A), and NcoA are shown. The histone acetyl transferase domain is also noted (HAT). (C) In vitro binding to p300 (a.a. 1257 to 2414) was tested for the IRF-3 mutation (SS385/386FA). The right lane displays relative input of p300.
The ability of IRF-3 from infected cells to bind with p300 appears to be due to IRF-3 serine phosphorylation (31, 52, 57). Substitution of carboxyl-terminal serine residues at a.a. positions 385 and 386 in IRF-3 produces a protein that is not activated in response to viral infection in vivo (57). For this reason, we tested the effect of substitution of the serine residues at a.a. positions 385 and 386 in IRF-3 with phenylalanine and alanine, respectively, in binding to the in vitro-translated p300 (Fig. 2C). The IRF-3 wild type (wt) or serine mutant (SS385-386FA) was cloned into a mammalian expression vector as a carboxyl fusion with the GFP and was transfected into HEC-1B cells. The wt and mutant GFP–IRF-3 proteins were immunoprecipitated with antibody to GFP and were used in the p300 binding assay. The GFP–wt IRF-3 efficiently bound to p300 following viral infection, whereas the serine mutant of GFP–IRF-3 was not capable of binding to p300 in vitro. There was a low amount of detectable association of p300 with GFP–wt IRF-3 from uninfected cells. This appears to be due to some activation of GFP–wt IRF-3 during the transient transfection (data not shown). The expression of both GFP–IRF-3 proteins was confirmed by fluorescence and Western blotting (data not shown).
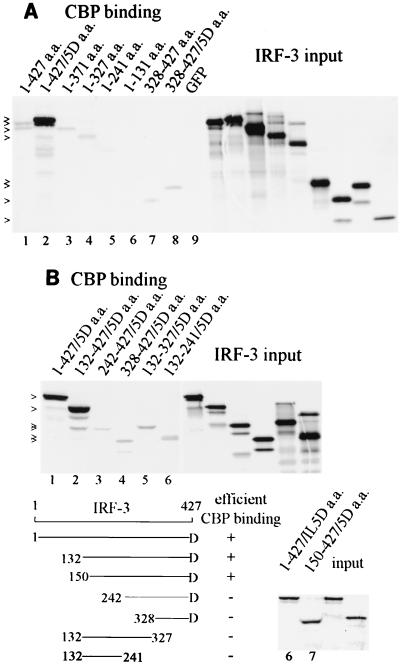
We next examined the region of the IRF-3 protein responsible for binding to CBP and tested various fragments of IRF-3 in vitro. This approach eliminated the effect of IRF-3 mutations on in vivo phosphorylation, cellular translocation, or possible dimerization with endogenous IRF-3. CBP was collected on immunocomplexes from uninfected cells with specific antibody. IRF-3 protein fragments were generated by in vitro translation in the presence of [35S]methionine and were tested for binding to CBP (Fig. 3). GFP–IRF-3 constructs were used to produce in vitro-translated proteins, since the GFP sequence contributed a translation initiation site for the carboxyl-terminal fragments and also provided additional methionines for radiolabeling. The wt IRF-3 (1 to 427 a.a.) demonstrated a weak but detectable binding to CBP in comparison to the GFP control (Fig. 3A, lane 1). Since it was reported previously that a constitutively active IRF-3 could be produced by replacing five carboxyl serine-threonine residues with the phosphomimetic aspartic acid (a.a. positions 396, 398, 402, 404, and 405), we tested this form of IRF-3 (IRF-3/5D) (31). The IRF-3/5D mutated protein bound with high affinity to CBP in this assay system (lane 2). The slower migration of IRF-3/5D versus the wt is apparently due to the amino acid substitutions. The difference in binding of wt IRF-3 and IRF-3/5D (lanes 1 and 2) appeared to indicate that the binding domain was resident in the modified carboxyl terminus. To examine this possibility, we generated carboxyl-terminal fragments (328 to 427 a.a.) of either wt IRF-3 or IRF-3/5D and tested their ability to bind to CBP. These carboxyl fragments demonstrated weak and nearly equivalent binding to CBP irrespective of the 5D substitutions, indicating the carboxyl terminus of activated IRF-3 is not sufficient for high-affinity binding to CBP (lanes 7 and 8). Various carboxyl deletions of wt GFP–IRF-3 were also tested (lanes 3 to 6). The amino-terminal fragment of IRF-3 containing a.a. 1 to 131 completely lacked the ability to bind CBP (lane 6), and the other protein fragments showed weak but detectable binding (lanes 3 to 5).
FIG. 3.
Domain of IRF-3 that binds to CBP in vitro. (A) CBP was immunoprecipitated from uninfected HEC-1B cells and was incubated with various fragments of GFP–wt IRF-3, GFP–IRF-3/5D, or GFP synthesized in vitro in the presence of [35S]methionine. Right panel displays relative input of IRF-3 protein. (B) Amino-terminal deletions of IRF-3/5D tested for binding to CBP as in panel A. Graphic illustration describes results.
Prior to activation in response to viral infection, intramolecular association of IRF-3 may conceal its CBP binding domain (32). Since the IRF-3/5D protein demonstrated strong binding in the in vitro assay, we tested amino-terminal deletion mutations of IRF-3/5D to map a binding domain (Fig. 3B). The amino-terminal deletion expressing 132-427/5Da.a. (lane 2) and 150-427/5Da.a. (lane 8) bound efficiently to CBP, but the fragment containing 242-427/5Da.a. had dramatically reduced binding (lane 3). If intramolecular associations of amino and carboxyl regions of latent IRF-3 conceal a CBP binding domain, an internal region may be able to bind CBP. However, evaluation of 132-327a.a. (lane 5) or 132-241a.a. (lane 6) did not reveal efficient binding. Therefore, it appears that binding to CBP may require a significant portion of IRF-3 or multiple sites of interaction within this region.
Cytoplasmic localization of IRF-3.
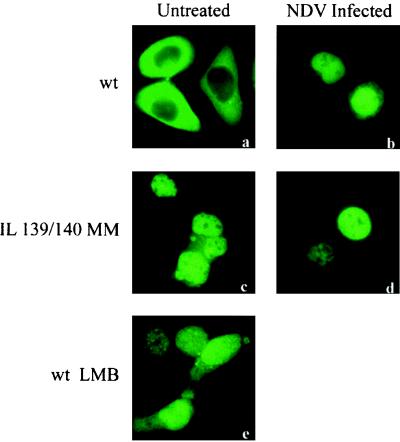
The DRAF1 transcription factor complex appears in the nucleus following viral infection. However, prior to infection, the DNA binding component, IRF-3, is located in the cytoplasm. Since IRF-3 phosphorylation during infection induces its ability to bind CBP/p300 in the nucleus, we investigated mechanisms that might regulate IRF-3 nuclear-cytoplasmic localization in response to viral infection. Proteins that are actively transported between the nucleus and cytoplasm possess targeting sequences that dictate their localization (16, 20, 21, 25, 33, 37, 50, 54). A subset of proteins that are actively exported from the nucleus have been characterized to possess an NES consisting of a leucine-rich stretch of amino acids (20, 54). In IRF-3, a leucine-rich sequence was identified to function as an NES (ILDELLGNMVL) spanning a.a. 139 to 149 (57). The localization effect of a targeted mutation in the NES can be visualized by fluorescent microscopy with a transfected GFP–IRF-3 fusion construct (Fig. 4). wt GFP–IRF-3 resides in the cytoplasm of uninfected cells and localizes to the nucleus only following modification during infection, whereas the NES mutated protein has a predominant nuclear presence constitutively. CBP/p300 does not appear to require the NES for IRF-3 binding, and, therefore, may not alter the function of the NES. A targeted mutation in the NES (IL139-140MM) of constitutively active IRF-3 (1-427/IL5Da.a.) or a complete deletion of the NES (150-427/5Da.a.) generates proteins that can still bind CBP efficiently in vitro (Fig. 3B, lanes 6 and 7).
FIG. 4.
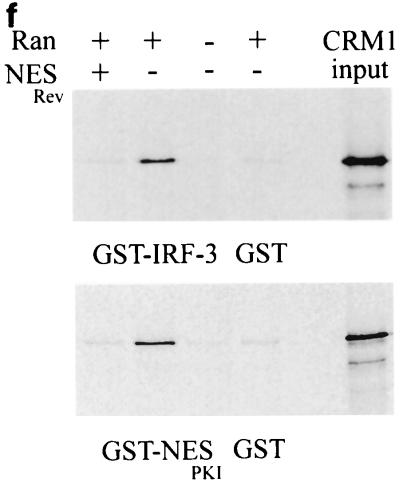
IRF-3 contains a nuclear export sequence that binds the exportin CRM1. HEC-1B cells were transfected with GFP–wt IRF-3 (wt) (a and b) or the NES mutation GFP–IRF-3 (IL 139/140 MM) (c and d) and were left untreated (a and c) or were infected with NDV as indicated (b and d). The effect of leptomycin (LMP) on localization of GFP–wt IRF-3 is shown (e). (f) CRM1 exportin binding to IRF-3 in vitro. CRM1 was synthesized in vitro in the presence of [35S]methionine (relative input shown on right) and was incubated with either GST, GST–IRF-3, or GST-NES from PKI. The binding was performed in the presence or absence of bacterially expressed Ran (Q69L) protein and a peptide corresponding to the NES of the Rev protein.
A shuttling receptor that appears to bind NESs and function in the export of proteins from the nucleus to the cytoplasm is CRM1 (21, 22). CRM1 can bind the small GTPase Ran and interact with nuclear pore complexes to effect translocation of NES-containing proteins. The antibiotic leptomycin B has been shown to bind CRM1 specifically and inhibit its export activity (30, 55). We tested the effect of leptomycin B on the cellular localization of IRF-3. Treatment of cells with leptomycin B resulted in the nuclear accumulation of IRF-3, indicating a direct role of CRM1 in export (57) (Fig. 4a).
If CRM1 functions as an export receptor for IRF-3, it should be able to bind to it directly. To test this possibility, we used an in vitro binding assay (Fig. 4). CRM1 was translated in vitro in the presence of [35S]methionine and was incubated with bacterially expressed GST–IRF-3 fusion protein bound to glutathione beads. The binding assay was performed in the presence or absence of RanQ69L produced and purified from bacteria (17). RanQ69L can bind but not hydrolyze GTP and thereby remains in an active GTP-bound state. In this assay, CRM1 was shown to bind IRF-3 in a Ran-dependent manner. Specific binding can be competed with the addition of an NES peptide corresponding to the human immunodeficiency virus Rev protein (20). GST protein was used as a negative control, and a GST-NES protein fragment containing the characterized NES of the PKI served as a positive control for Ran-dependent CRM1 binding (54). These results support the model that the constitutive function of the CRM1 exportin receptor is responsible for IRF-3 localization in the cytoplasm.
Identification of a functional NLS in IRF-3.
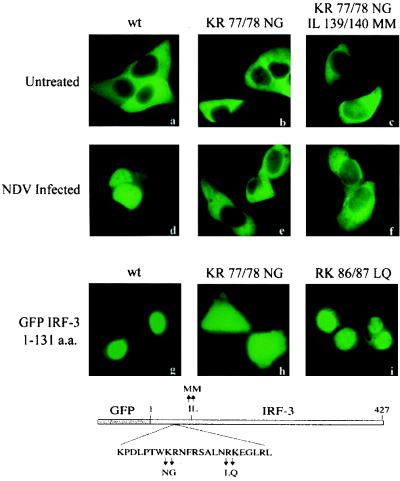
The result that IRF-3 accumulated in the nucleus if the NES was mutated (Fig. 4) suggested the existence of an NLS. A functional NLS may be intrinsic to IRF-3 or resident in an associated protein. The best-defined NLSs contain either a single stretch of basic amino acids or a bipartite sequence of basic amino acids spaced by nonconserved amino acids (16). Examination of the IRF-3 sequence revealed two pairs of basic residues situated amino terminal to the NES. Although the sequence and context did not fit a monopartite or bipartite consensus NLS, we performed site-directed mutagenesis to evaluate their contribution to nuclear accumulation. Mutation of the adjacent lysine and arginine residues at a.a. positions 77 and 78 in the GFP–IRF-3 construct to asparagine and glycine, respectively (designated KR77/78NG) produced a protein that remained in the cytoplasm both prior to and following viral infection (Fig. 5). When this mutation was evaluated in the context of the NES mutation (IL139/140MM), the predominant phenotype was the same, localization in the cytoplasm. The KR77/78NG mutation clearly inactivated an NLS function. To determine whether this pair of basic residues functioned as a bipartite NLS with a second pair of basic residues at a.a. positions 86 and 87, we mutated these downstream amino acids to leucine and glutamine, respectively (designated RK86/87LQ). The RK86/87LQ mutation did not disrupt the normal localization of IRF-3 and produced a protein with similar nuclear localization as the wt GFP–IRF-3 protein. These results suggest that the two basic residues at a.a. positions 77 and 78 (KR) serve within this amino acid context as an NLS in IRF-3.
FIG. 5.
Identification of a functional NLS in IRF-3. HEC-1B cells were transfected with GFP–wt IRF-3 (wt), GFP–IRF-3 (KR 77/78 NG), or GFP–IRF-3 (KR 77/78 NG plus IL 139/140 MM), and cells either were left untreated (a, b, and c) or were infected with NDV (d, e, and f). Cells were transfected with an IRF-3 deletion expressing GFP–wt IRF-3 a.a. 1 to 131 that lacks the NES (g) or with either of two mutations generated in this deletion construct, GFP–IRF-3 a.a. 1 to 131 (KR 77/78 NG) (h) and GFP–IRF-3 a.a. 1 to 131 (RK 86/87 LQ) (i). A diagrammatic representation of the mutations is shown below.
To determine whether this region of the IRF-3 protein is sufficient for nuclear localization, we evaluated the properties of an amino-terminal portion of IRF-3 lacking the NES. A GFP–IRF-3 construct containing a.a. 1 to 131 of IRF-3 produced a protein that accumulated predominantly in the nucleus (Fig. 5). If the KR77/78NG mutation is introduced into this fragment, fluorescence is seen in both nuclear and cytoplasmic compartments. The distribution of this mutated protein appears to reflect the absence of either an NES or NLS. Small molecules have been reported to transit the nuclear pore in an energy- and receptor-independent manner (41). Since the mass of the GFP–IRF-3 protein fragment is only approximately 42 kDa, it may be able to move in and out of the nuclear pore complex, but the presence of a functional NLS activity results in active transport and accumulation in the nucleus.
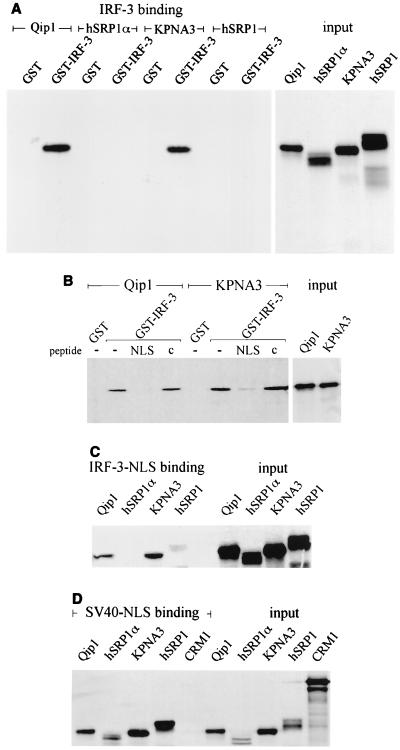
NLS sequences are specifically recognized by members of a family of cytoplasmic shuttling receptors (designated alpha importins) (25, 33, 37, 42). Importins-α in association with NLS cargo are recognized by a second shuttling protein, importin-β. Importin-β mediates translocation of the complex across the nuclear pore into the nucleus. Crystallographic analysis of the yeast importin-α revealed that the molecule contains tandem arrays of armadillo repeats that determine NLS association, and the amino terminus associates with importin-β (7). Since the NLS of IRF-3 does not conform to a consensus NLS, it is possible that IRF-3 nuclear transport is mediated by a specific subset of importin-α receptors. To address this possibility, we evaluated the ability of distinct importin-α receptors to recognize IRF-3 in an in vitro binding assay. The fact that the NES mutation results in nuclear accumulation suggests that the NLS within IRF-3 should be constitutively accessible. Four importin-α receptors, Qip1, hSRP1α/Rch1, KPNA3, and hSRP1/NPI1, were synthesized in vitro in the presence of [35S]methionine and were incubated with bacterially expressed GST–IRF-3 bound to glutathione beads (8, 10, 40, 46, 47) (Fig. 6A). Interaction of GST–IRF-3 with Qip1 and KPNA3 was readily detected, but interaction with hSRP1α/Rch1 or hSRP1/NPI1 was similar to that with the GST control. These results suggest that importin-α receptors may have unique functions in vivo and that the NLS of the IRF-3 protein is only recognized by a subset. The interaction of Qip1 and KPNA3 with IRF-3 is dependent on an NLS, as can be demonstrated by competition with a peptide containing the SV40 T antigen NLS, but not with a control peptide (Fig. 6B). The IRF-3 NLS can also specifically bind to Qip1 and KPNA3 as a single stretch of amino acids outside the context of IRF-3. The IRF-3 NLS peptide was covalently cross-linked to agarose beads and was used in the binding assay (Fig. 6C). The same specificity of importin-α binding was found with the NLS peptide as with the intact IRF-3 protein. To ensure that all the in vitro-synthesized importin-α receptors were functional, they were demonstrated to bind to the SV40 T antigen NLS peptide cross-linked to agarose beads (Fig. 6D).
FIG. 6.
IRF-3 is recognized by specific importin-α proteins. (A) Importin-α proteins were synthesized in vitro in the presence of [35S]methionine (relative input shown on right). The importins were incubated with bacterially expressed GST or GST–IRF-3 protein bound to glutathionine beads. (B) Binding of IRF-3 to importin-α receptors is NLS specific. Qip-1 and KPNA-3 were translated in vitro (relative input on right) and were incubated with GST–IRF-3 in the absence of competitive peptide (−) or in the presence of the SV40 NLS peptide (NLS) or in the presence of a control peptide (c). (C) The NLS sequence of IRF-3 is responsible for binding to Qip-1 and KPNA-3. The importin-α receptors were translated in vitro (relative input on right) and were incubated with the IRF-3 NLS peptide cross-linked to agarose beads. (D) All the importin-α receptors tested can bind to the SV40 NLS. The importin-α receptors and the exportin CRM1 were translated in vitro (relative input on right) and were incubated with the SV40 NLS peptide cross-linked to agarose beads.
Regulated IRF-3 localization.
The mechanisms that regulate IRF-3 cellular localization may involve the gain of function of an NLS or NES, the masking of an NLS or NES, and/or the retention of IRF-3 in a cellular compartment by association with other proteins or by DNA binding. The hypothesis that both the NLS and the NES in IRF-3 are constitutively active in uninfected cells is supported by the fact that the NES mutation results in nuclear localization. This suggests that the IRF-3 protein normally shuttles between cytoplasmic and nuclear compartments and that export is the prevailing effect. Following infection, however, accumulation in the nucleus is the overall consequence. Nuclear accumulation does not appear to be due to DNA binding, since a protein deficient in DNA binding can localize to the nucleus following infection (57) (data not shown). The specific phosphorylation of IRF-3 appears to alter its conformation and allow it to bind to CBP/p300 in the nucleus (Fig. 2 and 3). These results suggest a model wherein CBP/p300 association with IRF-3 prevents its export to the cytoplasm.
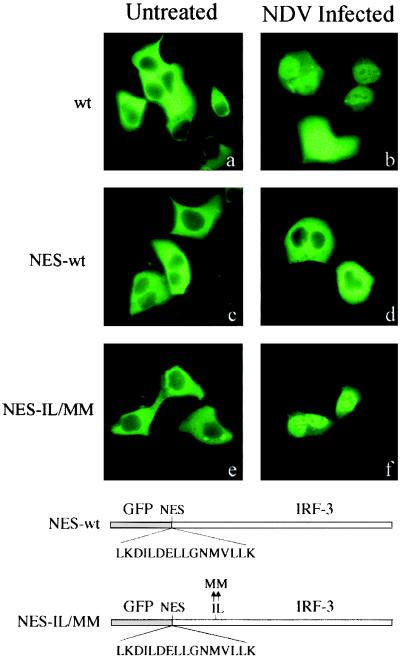
If the NES of IRF-3 is masked following activation and binding to CBP/p300, the DRAF1 complex will not shuttle, but remain nuclear. To distinguish between the possibility of NES masking by CBP/p300 binding and the possibility of nuclear retention due to sequestration by CBP/p300, we tested the effect of repositioning the IRF-3 NES at the very amino terminus of IRF-3, outside a region required for binding (Fig. 3). The IRF-3 NES was introduced into two different GFP–IRF-3 constructs, downstream of GFP and upstream of IRF-3. The sequence was inserted into wt IRF-3 (NES-wt) and also into the IRF-3 NES mutant IL139/140MM (designated NES-IL/MM). Cells were transfected, and localization of the proteins was monitored by fluorescent microscopy before and after infection (Fig. 7). Prior to infection, the wt IRF-3 is cytoplasmic, and following infection, it accumulates in the nucleus. Infected cells that express high levels of wt IRF-3 display some residual cytoplasmic fluorescence. This may be due to limiting amounts of CBP/p300 in the nucleus. The NES-wt displays a very different behavior, as it remains localized in the cytoplasm even following infection. Therefore, the new upstream NES is functional, and nuclear export is always dominant in the NES-wt. However, the NES-wt contains two NES, one at the new site and one at the native site, and this duplication may increase export efficiency. To evaluate IRF-3 containing a single relocated NES, a NES was inserted upstream, and the native NES site was eliminated in the NES-IL/MM. The NES-IL/MM behaved as wt IRF-3: it was cytoplasmic before infection and nuclear following infection. This result eliminates the possibility of NES masking by CBP/p300 binding as a mechanism of IRF-3 nuclear localization. In NES-IL/MM, the functional NES was repositioned upstream of the first amino acid of IRF-3 and outside the CBP/p300 binding region, but it was still rendered ineffective following infection. Together, these results provide evidence that IRF-3 localization in the nucleus following infection is due to its retention by CBP/p300.
FIG. 7.
Effect of relocation of the IRF-3 NES. The NES of IRF-3 was inserted in frame between GFP and the first amino acid of wt IRF-3 to create GFP–NES–IRF-3 (NES-wt) or it was inserted into the NES-deficient mutant (NES-IL/MM). The constructs were transfected into cells that were left untreated (a, c, e) or were infected with NDV (b, d, f), and fluorescent images are displayed. A diagrammatic representation of the constructs is shown.
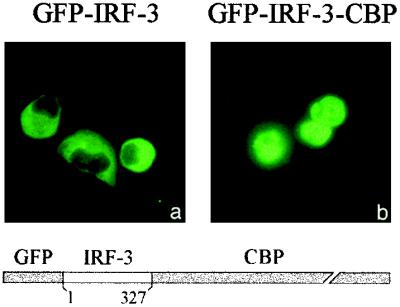
To definitively determine whether interaction with CBP/p300 is responsible for the nuclear accumulation of IRF-3, we tested the behavior of IRF-3 when recombinantly joined to CBP. The coding region of GFP–IRF-3 was joined in frame with full-length CBP. Transfection of the GFP–IRF-3–CBP expression plasmid into cells in the absence of infection demonstrated complete nuclear localization of the fusion protein (Fig. 8b). In contrast, the control GFP–IRF-3 protein is localized in the cytoplasm (Fig. 8a). To avoid any interference of the carboxyl terminus of IRF-3 with endogenous CBP/p300, we used a.a. 1 to 327 of IRF-3 to generate GFP–IRF-3–CBP. Production of the expected fusion protein was confirmed by Western blotting of nuclear extracts with appropriate antibodies (data not shown). This experiment provides direct evidence for the ability of CBP/p300 association to dictate IRF-3 cellular localization. Together, the results support the role of CBP/p300 interaction with phosphorylated IRF-3 in promoting nuclear accumulation of IRF-3 and formation of the DRAF1 complex following viral infection.
FIG. 8.
Localization of a recombinant IRF-3–CBP fusion protein to the nucleus. HEC-1B cells were transfected with expression plasmids encoding either (a) GFP–IRF-3 (a.a. 1 to 327) or (b) GFP–IRF-3 (a.a. 1 to 327)–CBP. Fluorescence microscopic images display protein localization. Diagrammatic representation of the GFP–IRF-3–CBP construct is shown.
DISCUSSION
We first identified DRAF1 as a cellular transcription factor that is activated in response to viral infection (11, 12). DRAF1 recognizes a DNA target that includes the ISRE and several adjacent adenine residues. The induction of a subset of the IFN-stimulated genes indicates that DRAF1 may provide a critical defense response to viral infection and contribute to host survival. This dsRNA-induced defense mechanism may have evolved prior to cytokine mediators of the immune system or may have evolved concurrently, as there is some evidence that IRF-3 may also participate in the induction of the IFN genes (28, 44, 51, 57).
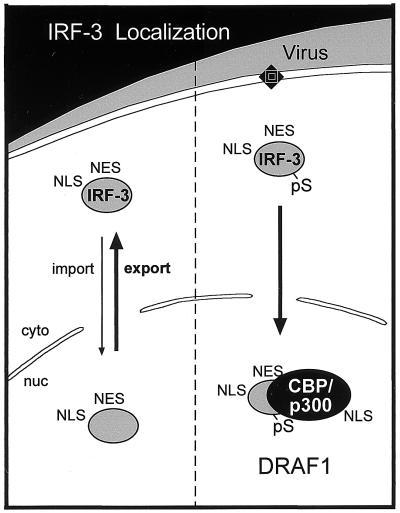
DRAF1 is now known to be a multimeric transcription factor containing the subunits IRF-3 and CBP or p300 (3, 31, 44, 51, 52, 57). IRF-3 is the key modulator of DRAF1 formation (Fig. 9). In this report, we present evidence that the IRF-3 protein normally shuttles between nuclear and cytoplasmic compartments. Resident in the cytoplasm, IRF-3 is phosphorylated in response to the presence of dsRNA. It subsequently accumulates in the nucleus in association with CBP or p300 to form DRAF1, which induces the transcription of responsive genes. The association of IRF-3 with p300 does not appear to require any virus-induced modification of p300. A carboxyl region of p300 protein synthesized in vitro demonstrates specific binding to IRF-3 (Fig. 2). This domain coincides with an interaction region of CBP that was shown to bind IRF-3 in vivo (31). This region contains four putative helical repeats that are differentially required for interaction with the NCoA-1/SRC-1 transcriptional coactivator of nuclear hormone receptors (34). Activated IRF-3 may successfully compete with NCoA-1 or other transcription factors for the limiting amounts of CBP and p300 that are present in cells (27, 56). For this reason, IRF-3 recruitment of CBP/p300 may be part of a cellular stress response to direct transcription specifically to genes involved in immune defense.
FIG. 9.
Conceptual model of IRF-3 cellular localization. Prior to infection, the dominant effect of the NES results in cytoplasmic localization of IRF-3 (left). Following infection, phosphorylation of IRF-3 results in nuclear association with CBP or p300. The CBP/p300 binding results in the sequestration of the IRF-3 in the nucleus to form the DRAF1 transcription factor (right).
IRF-3 isolated from the cytoplasm of uninfected cells cannot bind to CBP/p300. Modification of IRF-3 is necessary both for interaction with p300 and for binding to an ISRE-containing DNA target (Fig. 2 and 3) (52). Our group and others have shown that, following infection, an increase in serine phosphorylation of IRF-3 correlates with its activation (31, 52, 57). Although the precise sites of phosphorylation that exist prior to or following infection remain to be determined, mutational analyses indicate the importance of carboxyl-terminal serine residues (31, 57). We studied various domains of wt IRF-3 or the constitutively active IRF-3/5D synthesized in vitro for interaction with CBP/p300. This approach circumvents the in vivo requirement of proper cellular localization or activation for analysis. A relatively large domain of IRF-3 (150 to 427 a.a.) appears to be required for binding (Fig. 3). It has been suggested that prior to infection intramolecular interactions within IRF-3 prevent DNA binding or CBP/p300 binding, since an amino-terminal domain of IRF-3 (98 to 197 a.a.) can be coimmunoprecipitated with a carboxyl-terminal domain of IRF-3 (328 to 427 a.a.) when transiently expressed in cells (32). Our results are consistent with the hypothesis that the carboxyl-terminal modifications of IRF-3 do not serve primarily to bind CBP/p300, but serve to alter the conformation of IRF-3 to enable recognition of a previously concealed domain in wt IRF-3.
This biochemical characterization of the interaction between IRF-3 and CBP/p300 provided insight into the mechanisms regulating nuclear-cytoplasmic localization of IRF-3 in response to viral infection. The sequences that function in nuclear export and import of IRF-3 are located outside the CBP/p300 interaction region, and this interaction only occurs following viral infection and the resulting IRF-3 phosphorylation. Cellular localization is not a random event, but is precisely controlled by soluble shuttling receptors. These receptors bind to specific signal sequences and, by interaction with the small GTPase Ran and nuclear pore complexes, effect translocation across the nuclear pore (25, 33, 37, 41, 42, 50, 53). In this report, a site-directed mutational analysis was used to identify an NLS in IRF-3. The NLS appears to function constitutively and is responsible for trafficking IRF-3 to the nucleus. Nevertheless, the dominance of an NES results in IRF-3 accumulation in the cytoplasm in the absence of infection. The NLS sequences are recognized by a family of importin-α receptors. All of these receptors can bind to classical NLS sequences such as that found in the SV40 T antigen. However, in our analyses, only Qip1 and KPNA3 could bind to IRF-3 in an in vitro assay. Selectivity in importin-α recognition of NLSs has been reported in a yeast two-hybrid system with a bipartite NLS of DNA helicase Q1/RecQL (46). The NLS of IRF-3 appears to be a short basic sequence, and the importin-α family may have signal binding preferences for distinct NLSs. It is possible that this selectivity plays a physiological role in providing less-efficient nuclear import and the subsequent dominance of IRF-3 nuclear export to maintain high levels in the cytoplasm for signal reception. However, it is critical that the IRF-3 NLS constantly shuttle IRF-3 into the nucleus so that following infection and modification it will be in the correct cellular location to bind CBP/p300 and induce gene transcription.
The accumulation of IRF-3 in the nucleus during infection appears to be due to the ability of CBP/p300 to retain it in the nucleus. The complex formed between IRF-3 and CBP/p300 following infection is resistant to high concentrations of salt and detergent lysis, indicating a relatively strong association (52). There is precedence in other systems for nuclear sequestration via interaction with proteins resident in the nucleus (1). CBP and p300 proteins are constitutively resident in the nucleus. p300 has been shown to possess a functional NLS within its amino terminus, a region distant from the IRF-3 binding site, and CBP possesses a similar sequence in a corresponding region (19). Our experiments shown in Fig. 7 address the possibility of physical masking of the IRF-3 NES by CBP/p300 binding or alternate mechanisms that lead to nuclear accumulation of IRF-3 following viral infection. The addition of a duplicate copy of the NES upstream of wt IRF-3 (NES-wt), outside the region of CBP/p300 binding, confers cytoplasmic localization even following infection. Since this effect is due to the concerted export function of both the new and native NESs, the result excludes an inhibitory modification of the native IRF-3 NES during infection. If a single NES is maintained in IRF-3 but is repositioned upstream of the IRF-3 NES mutant (NES-IL/MM), away from the CBP/p300 binding domain, it still accumulates in the nucleus following infection, as does wt IRF-3. Thus, when there is a single functional NES in IRF-3, even if it is distant from the CBP/p300 binding region, viral infection results in nuclear accumulation. These results exclude the possibility of physical masking of the NES by CBP/p300 binding as a mechanism for IRF-3 nuclear localization. Rather, it appears that IRF-3 nuclear accumulation following infection is due to sequestration by CBP/p300 resident in the nucleus. This is supported by the complete nuclear localization of a fusion protein encoding GFP–IRF-3–CBP in the absence of viral infection (Fig. 8). The CBP/p300 transcriptional coactivators cooperate with an ever-growing number of DNA binding factors. To our knowledge, this is the first report providing evidence for a function of CBP/p300 in nuclear sequestration, and future studies may reveal that it serves this function for other transcription factors that also receive activation signals in the cytoplasm.
ACKNOWLEDGMENTS
We thank the members of our laboratory who have provided helpful suggestions and Jennifer Gallub for her technical assistance. Our thanks extend to Richard Goodman for the gifts of CBP and p300 expression plasmids, Gerard Grosveld for the gift of CRM1 expression plasmid, David Livingston for the gift of p300 cDNA plasmids, John Hiscott for the gift of the IRF-3/5D plasmid, Barbara Wolff for the gift of leptomycin B, Connie Nguyen and Susan Taylor for the gift of the PKI clone, Karsten Weis for the gift of the hSRP1α clone, Takemi Enomoto for the gift of the Qip1 clone, Particia Cortes for the gift of the hSRP1 clone, and Y. Hirai for the gift of the KPNA3 clone.
This work was supported by grants from the National Institutes of Health (RO1CA50773 and PO1CA28146) to N.C.R. and by a scholarship from the Council for Tobacco Research to C.D.
REFERENCES
- 1.Abu-Shaar M, Ryoo H D, Mann R S. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam S A, Sterne-Marr R, Gerace L. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 1992;219:97–110. doi: 10.1016/0076-6879(92)19013-v. [DOI] [PubMed] [Google Scholar]
- 3.Au W C, Moore P A, Lowther W, Juang Y T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 6.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 7.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 8.Cortes P, Ye Z S, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crabtree G R. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 10.Cuomo C A, Kirch S A, Gyuris J, Brent R, Oettinger M A. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly C, Reich N C. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J Biol Chem. 1995;270:23739–23746. doi: 10.1074/jbc.270.40.23739. [DOI] [PubMed] [Google Scholar]
- 12.Daly C, Reich N C. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol Cell Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 14.DeFranco D B. Regulation of steroid receptor subcellular trafficking. Cell Biochem Biophys. 1999;30:1–24. doi: 10.1007/BF02737882. [DOI] [PubMed] [Google Scholar]
- 15.DeMaeyer E M, De Maeyer-Guignard J. Interferons and other regulatory cytokines. New York, N.Y: John Wiley & Sons, Inc.; 1988. [Google Scholar]
- 16.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 17.Dingwall C, Palacios I. In vitro systems for the reconstitution of snRNP and protein nuclear import. Methods Cell Biol. 1998;53:517–543. doi: 10.1016/s0091-679x(08)60893-6. [DOI] [PubMed] [Google Scholar]
- 18.Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 19.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 20.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 21.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 22.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh S, May M J, Kopp E B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 24.Goodbourn S, Zinn K, Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985;41:509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- 25.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 27.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juang Y, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koepp D M, Silver P A. A GTPase controlling nuclear trafficking: running the right way or walking RANdomly? Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- 30.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 31.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 34.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 36.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 38.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 39.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 40.O'Neil R E, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 41.Pante N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- 42.Pemberton L F, Blobel G, Rosenblum J S. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 43.Ryals J, Dierks P, Ragg H, Weissmann C. A 46-nucleotide promoter segment from an IFN-alpha gene renders an unrelated promoter inducible by virus. Cell. 1985;41:497–507. doi: 10.1016/s0092-8674(85)80023-4. [DOI] [PubMed] [Google Scholar]
- 44.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 45.Savory J G, Hsu B, Laquian I R, Giffin W, Reich T, Hache R J, Lefebvre Y A. Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol Cell Biol. 1999;19:1025–1037. doi: 10.1128/mcb.19.2.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seki T, Tada S, Katada T, Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- 47.Takeda S, Fujiwara T, Shimizu F, Kawai A, Shinomiya K, Okuno S, Ozaki K, Katagiri T, Shimada Y, Nagata M, Watanabe T, Takaichi A, Kuga Y, Suzuki M, Hishigaki H, Takahashi E, Shin S, Nakamura Y, Hirai Y. Isolation and mapping of karyopherin alpha 3 (KPNA3), a human gene that is highly homologous to genes encoding Xenopus importin, yeast SRP1 and human RCH1. Cytogenet Cell Genet. 1997;76:87–93. doi: 10.1159/000134521. [DOI] [PubMed] [Google Scholar]
- 48.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 49.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 50.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 51.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 52.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 54.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 55.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 56.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 57.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]