SUMMARY
Highly specific expression patterns can be caused by the overlapping activities of activator and repressor sequences in enhancers. However, few studies illuminate how these sequences evolve in the origin of new enhancers. Here, we show that expression of the bond gene in the semicircular wall epithelium (swe) of the Drosophila melanogaster male ejaculatory bulb (EB) is controlled by an enhancer consisting of an activator region that requires Abdominal-B driving expression in the entire EB and a repressor region that restricts this expression to the EB swe. Although this expression pattern is independently gained in the distantly related Scaptodrosophila lebanonensis and does not require Abdominal-B, we show that functionally similar repressor sequences are present in Scaptodrosophila and also in species that do not express bond in the EB. We suggest that during enhancer evolution, repressor sequences can precede the evolution of activator sequences and may lead to similar but independently evolved expression patterns.
Graphical Abstract
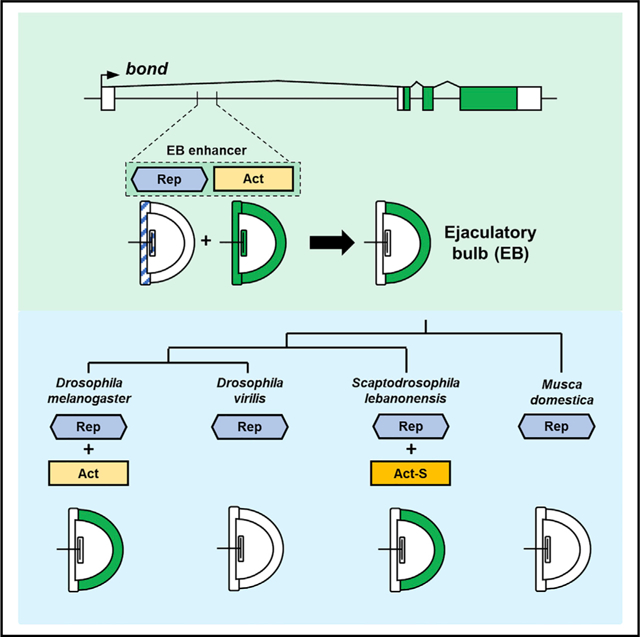
In brief
Pu et al. show that the independent gain of a highly specific expression pattern across distantly related species may be because of the preexistence of repressor sequences that precedes the diversification of these species. This may reflect a general mechanism underlying the evolution of highly specific enhancers.
INTRODUCTION
Highly specific gene expression patterns are central to the development and evolution of multicellular organisms. These expression patterns are regulated by the action of modular cis-regulatory elements called enhancers (Small and Arnosti, 2020), which are controlled at different levels by transcription factors (TFs), such as pioneer factors that open up chromatin and initiate DNA binding by other factors (Zaret and Mango, 2016), transcriptional activators that drive gene expression, and transcriptional repressors that inhibit gene expression (Small and Arnosti, 2020), as well as other transcriptional factors controlled by specific signaling pathways (Barolo and Posakony, 2002). The overlapping and combinatorial actions of these different transcriptional factors within an enhancer control highly specific gene expression in time and space (Spitz and Furlong, 2012). A well-studied example of this logic is the even-skipped (eve) stripe 2 enhancer, where the combinatorial actions of two activators and three repressors define a single stripe of eve expression in the developing insect embryo (Andrioli et al., 2002; Arnosti et al., 1996; Ludwig et al., 2005; Small and Arnosti, 2020).
In the past few decades, numerous studies have demonstrated that changes in enhancer activities can lead to novel expression patterns and evolutionary innovations in morphology and physiology (Carroll, 2008; Rebeiz and Tsiantis, 2017; Wittkopp and Kalay, 2011). These evolutionary changes in the enhancers include the gain (Jeong et al., 2006) and loss (Jeong et al., 2008; Kvon et al., 2016) of activator-binding sequences, as well as the gain (Preger-Ben Noon et al., 2016) and loss (Sumiyama and Saitou, 2011) of repressor-binding sequences. Although these studies clearly show that evolutionary changes in either activator or repressor sequences can lead to phenotypic changes, fundamental questions remain regarding how these activator and repressor sequences interact during the birth of enhancers that control highly specific expression patterns.
In one of the first studies of cis-regulatory changes and evolutionary novelty, the gain of a novel enhancer that underlies a highly specific wing spot in males of Drosophila biarmipes is hypothesized to be due to recruitment of at least one activator and one repressor (Arnoult et al., 2013; Gompel et al., 2005; Xin et al., 2020). How do enhancers that rely on the combinatorial effects of both activator and repressor sequences evolve? A reasonable hypothesis would be that activator sequences evolve first, driving a broad expression pattern prior to the appearance of repressor sequences that restricts expression of the gene. However, one caveat of this prediction is that for many pleiotropic genes, gains in broad expression patterns may lead to negative fitness effects in the organism, as a result of possible misexpression of these genes in key tissues. Many studies in model organisms, such as Drosophila, mice, and zebrafish, have shown that misexpression of key genes in the wrong tissues can lead to negative phenotypic effects on the organism (Furuchi et al., 1996; Morgan et al., 1992; Ungar et al., 1995; Zhang and Odenwald, 1995). In humans, the mis-regulation of genes underlies many disease conditions (Lee and Young, 2013). Therefore, these negative fitness effects may lead to new broad expression patterns being selected against before the gain of repressor sequences. An alternate hypothesis is that repressor sequences precede the evolution of activator sequences, leading to the gain of a specific expression pattern without having evolved broad expression first, thus mitigating the negative fitness effects of broad gene expression patterns. However, the hypothesis also poses a potential quandary: can repressor sequences evolve before activator sequences if no gene expression is driven by repressors alone? How would repressor sequences be maintained during evolution if there is no gene expression output? Determining the evolutionary dynamics between activator and repressor sequences can lead to a better understanding of how enhancers originate and how highly specific expression patterns in nature are generated during evolution.
In this study, we investigated the cis-regulatory evolution of the fatty acyl-CoA elongase gene bond, a pleiotropic gene involved in Drosophila spermatogenesis (Szafer-Glusman et al., 2008), as well as the biosynthesis of the male anti-aphrodisiac, CH503, a male-specific pheromone produced in the ejaculatory bulb (EB) by several Drosophila species, which is transferred to the female during mating to prevent remating by other males (Ng et al., 2014; Yew et al., 2009). The specific expression of bond in the EB is necessary for the production of CH503. However, species such as D. erecta and D. ananassae do not produce CH503 but express bond in the EB, suggesting that bond may also have other unknown roles in the EB of these species (Ng et al., 2015). We investigated the evolution of EB-specific expression of bond across a broad phylogeny of 21 Drosophila and other related species. We show that there are two independent evolutionary gains of bond expression across these species. In D. melanogaster, bond expression is controlled by an enhancer consisting of an activator region (Act) that drives bond expression in the entire EB and a repressor region (Rep) that restricts expression to a specific part of the EB. Interestingly, this Rep region is present even in D. willistoni and D. virilis, as well as the distantly related housefly, Musca domestica, where EB expression of bond is not detected. We further show that although bond EB expression in Drosophila requires the TF Abdominal-B (Abd-B), bond EB expression in Scaptodrosophila does not require Abd-B. Taken together, our experiments suggest that the evolution of the repressor sequences can precede the evolution of highly specific expression patterns. Repeated evolution of similar specific patterns across lineages may be because of the preexistence of similar repressor sequences, even if the activator sequences may be different.
RESULTS
Two independent evolutionary gains of bond EB expression across the Drosophila genus and related species
The EB in D. melanogaster is made of distinct parts, including a bulbar cavity devoid of cells that holds the ejaculate (Cohen and Wolfner, 2018). Expression of bond in the D. melanogaster EB is restricted to a specific epithelial region, which we named the semicircular wall epithelium (swe) (Figure 1A). To determine the evolutionary history of bond expression in the EB, we used in situ hybridization of specific bond RNA probes to examine a broad phylogeny of 21 different species, including 16 Drosophila species, 3 Scaptodrosophila species, 1 Chymomyza species, and the housefly, Musca domestica. EB expression of bond is detected in seven Drosophila species, D. melanogaster, D. simulans, D. erecta, D. yakuba, D. ananassae, D. pseudoobscura, and D. subobscura, as well as three Scaptodrosophila species, S. latifasciaeformis, S. lebanonensis, and S. rufifrons (Figure 1B; Figure S1). Because the species expressing bond in the EB are divided across two lineages in the phylogenetic tree, we reconstructed the ancestral status of bond EB expression using a maximum likelihood model to determine whether EB expression is gained or lost multiple times during the evolutionary history of these species. Our analysis showed there are likely two independent gains of bond EB expression across our phylogeny: one in the lineage before the split of the melanogaster group and the obscura group in the Drosophila genus, and one in the lineage before the diversification of the Scaptodrosophila genus (Figure 1B; Figure S1).
Figure 1. Two independent evolutionary gains of bond ejaculatory bulb (EB) expression across 21 species.
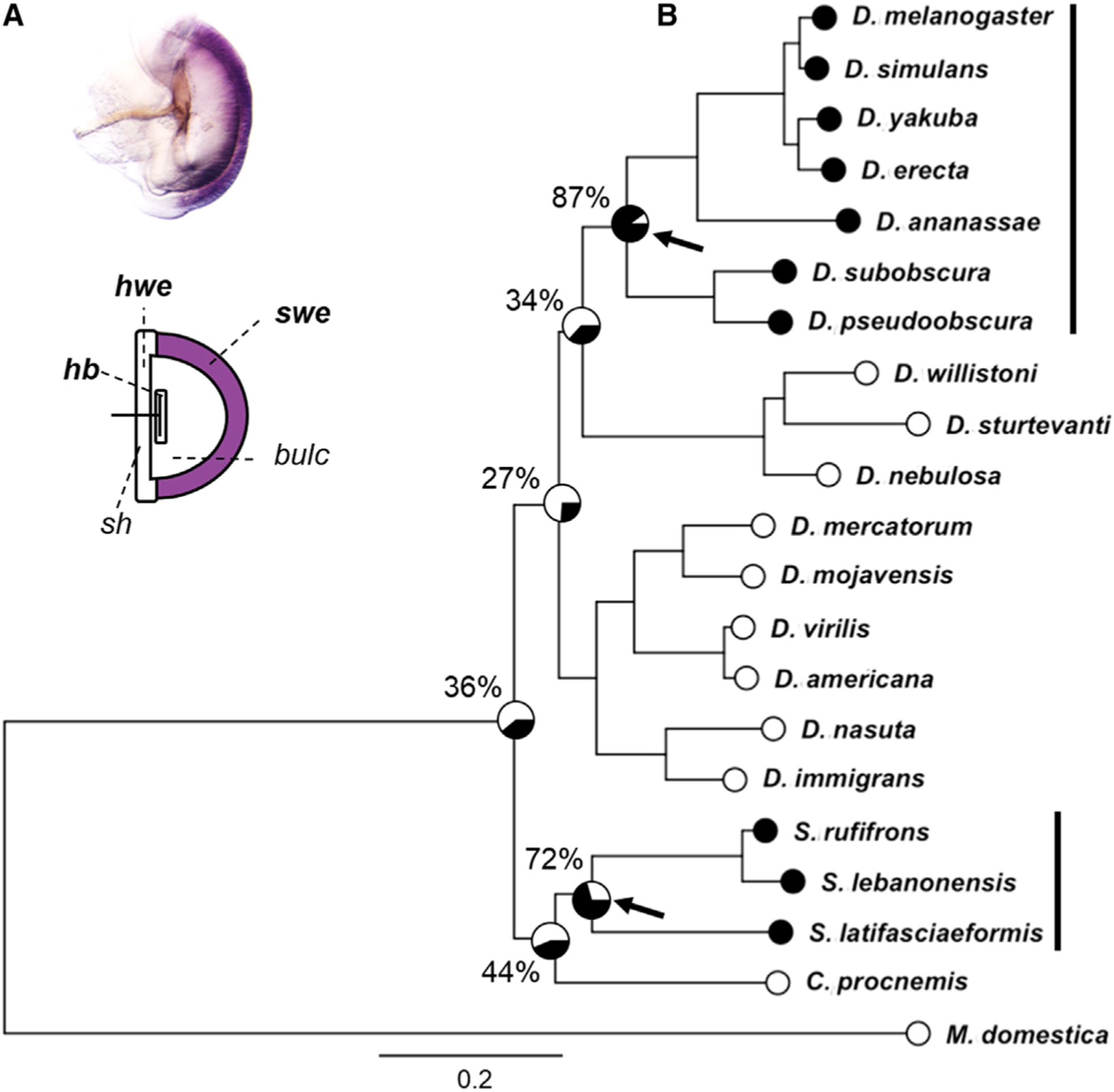
(A) Schematic of the EB in D. melanogaster. The bulbar cavity is devoid of cells and holds the ejaculate. bond is specifically expressed in the swe of the EB in Drosophila melanogaster.
(B) Ancestral trait reconstruction using a maximum likelihood model suggests two independent evolutionary gains of bond EB expression across the phylogeny. Arrows indicate likely gains in bond EB expression: one in the lineage leading to the melanogaster group and the obscura group in the Drosophila genus, and the other in the lineage leading to the Scaptodrosophila genus. Black and white circles for each species indicate the presence or absence of bond EB expression, respectively. Percentages on key nodes indicate probability of bond EB expression in the ancestor. Phylogeny was constructed using data from Finet et al. (2021). Scale bar indicates the number of nucleotide changes per site.
bc, bulbar cavity; hb, handle base; hwe, horn wall epithelium; sh, sclerite handle; swe, semicircular wall epithelium.
cis-regulatory evolution underlies the differential expression of bond in different species
To investigate how the expression of bond in EB arose, we first sought to locate the enhancer driving this expression pattern in D. melanogaster. GFP reporter constructs using non-coding DNA sequences around the bond gene (3R: 22547847..22558341, Flybase version: FB2021_01) in D. melanogaster showed that the enhancer responsible for driving gene expression in the EB swe lies in the first intron of bond (Figure 2A; Figure S2). Homologous regions from D. simulans, D. yakuba, D. erecta, D. ananassae, and S. lebanonensis drove similar GFP expression in the EB swe, but homologous regions from D. willistoni, D. virilis, and M. domestica did not (Figure 2B). This observation is consistent with our in situ hybridization results for these species (Figure S1), suggesting that differences in bond EB expression across these species are due to the evolution of cis-regulatory sequences present in the first intron of bond.
Figure 2. Evolution of cis-regulatory sequences in the first intron of bond underlies differences in EB expression across species.
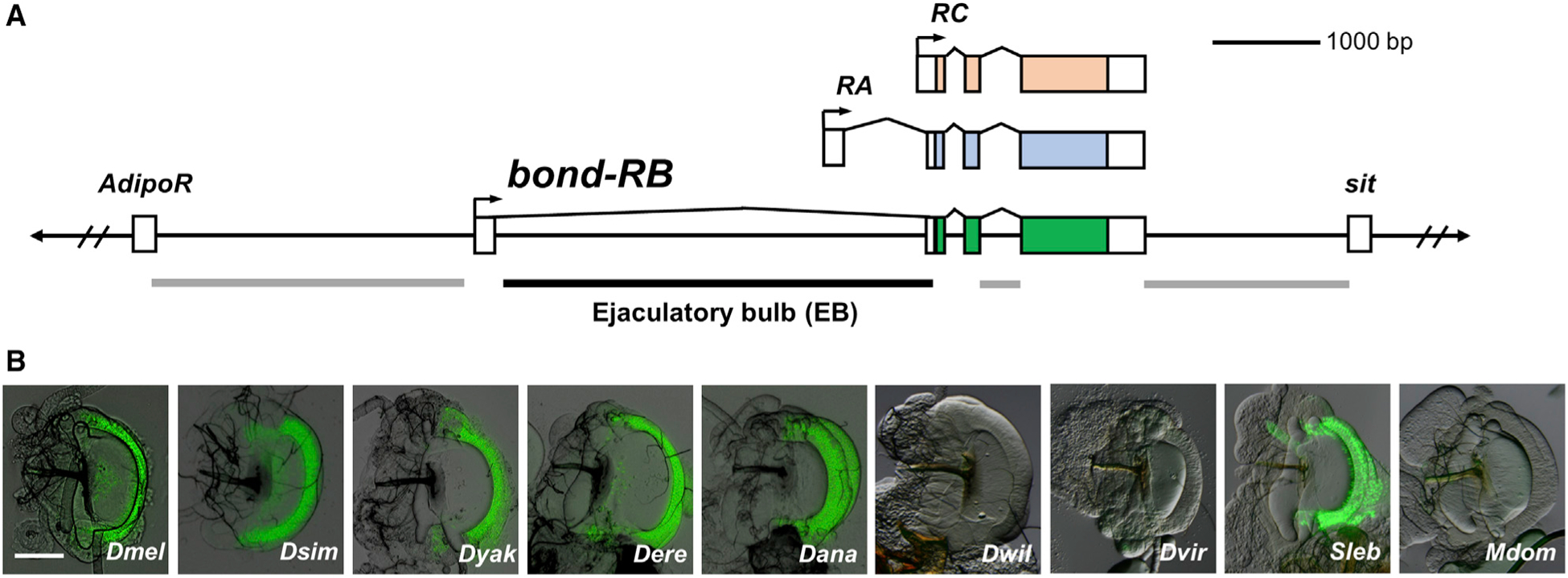
(A) The non-coding regions around the D. melanogaster bond locus were screened for enhancer activity that is able to drive GFP reporter protein expression in the EB. One fragment (black line), the first intron, was able to drive GFP expression in the EB. The other fragments (gray lines) did not drive EB expression.
(B) Intron 1 of bond from D. melanogaster, D. simulans, D. yakuba, D. erecta, D. ananassae, and S. lebanonensis drove GFP expression in the EB swe. Homologous fragments from D. willistoni, D. virilis, and M. domestica did not. Scale bar, 100 μm.
A 285-bp enhancer, composed of both activator and repressor sequences, recapitulates specific expression of bond in the swe of the D. melanogaster EB
Having confirmed that cis-regulatory differences underlie bond expression differences in the swe of the EB, we sought to identify the evolutionary changes in the cis-regulatory sequences that led to the differential expression patterns. Our first step was to determine the minimum enhancer region specific for bond expression in the EB swe. We performed a systematic dissection of the first intron of D. melanogaster to delimit smaller regions for the enhancer by creating smaller overlapping GFP constructs (Figure S2). We first narrowed down a 404-bp region (bc fragment) that recapitulates the expression pattern driven by the full intron (Figure 3A; Figure S2). Further dissection of this region allowed us to narrow down an even smaller 285-bp fragment (construct bc23) that expresses GFP in the EB swe (Figures 3A and 3B). We named this the EB swe enhancer. We next set out to identify sequences in this minimal enhancer that potentially underlie bond expression differences between species. We initially divided the 285-bp enhancer fragment into overlapping constructs, bc2 and bc3. bc2 did not drive any GFP expression, but notably, bc3 drove expression in the entire EB, not just the swe of the EB (Figures 3A and 3B). This result suggests that repressor sequences that are present in the bc2 fragment repress ectopic expression in the horn wall epithelium (hwe) and handle base (hb) of the EB driven by the bc3 fragment (Figure 3B). Dissection of bc3 into two smaller overlapping constructs, bc3i and bc3ii, shows that although bc3i drove expression in the hb, bc3ii did not drive any GFP expression. This result suggests that bc3ii contains activator sequences necessary to drive expression in the whole EB in conjunction with bc3i but cannot independently drive expression. Together, our data show that the bond EB swe enhancer comprises a Rep region and an Act region, which has at least two different transcriptional inputs (Ac1 and Ac2) (Figure 3C).
Figure 3. The combination of repressor and activator sequences in the D. melanogaster bond EB enhancer drives specific expression in the swe.
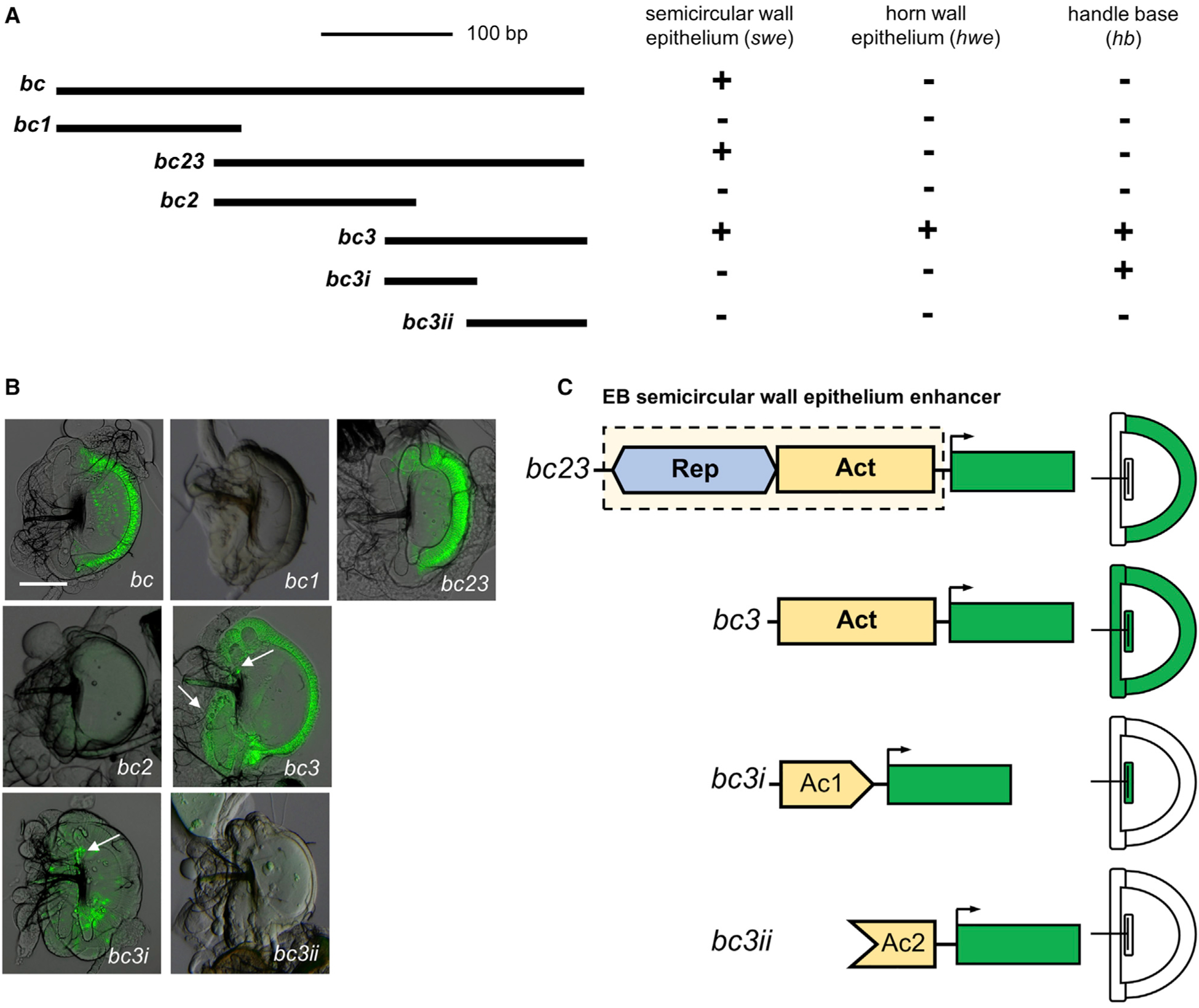
(A) Schematic of the overlapping GFP constructs of the EB enhancer. The bc23 fragment is the minimum region that can recapitulate the EB expression of bond in the swe. The bc3 fragment shows the ectopic GFP expression in horn wall epithelium (hwe) and handle base (hb). Plus sign (+) indicates the presence of expression; minus sign (−) indicates the absence of detectable expression.
(B) GFP reporter protein expression in the EB corresponding to the different overlapping constructs. Arrows indicate the expression in hwe and hb. Scale bar, 100 μm.
(C) The D. melanogaster bond EB swe enhancer contains two modular regions. The activator region (Act) contains activator sequences that drive expression in the whole EB (in hwe, hb, and swe). The repressor region (Rep) represses GFP activity in the hb and hwe of EB and restricts activity to the swe of the EB. The Act region can be divided into two different inputs, Ac1 and Ac2, both of which are needed to drive expression in the whole EB. Ac1 on its own can drive GFP expression in the hb, but Ac2 alone does not drive any expression.
Activator sequences for EB expression are present in D. willistoni, even though bond is not expressed in the EB of this species
To trace the evolutionary origins of this enhancer in Drosophila, we examined the activity of the homologous sequences from three other species, D. ananassae, D. willistoni, and D. virilis, based on their bond expression in the EB and their phylogenetic relationships (Figure 1B). Our results indicate that the bc fragments from these three species are able to recapitulate the EB expression of bond (in the case of D. willistoni and D. virilis, no expression) (Figure 4A). Our a priori expectation is that creating smaller fragments of the 285-bp enhancer in D. melanogaster and D. ananassae would allow us to narrow the region involved in enhancer evolution, and we expected that there would be no GFP expression driven by the smaller fragments in D. willistoni and D. virilis. Although we did not detect GFP expression in any of the D. virilis constructs, to our surprise, the bc3i fragment of D. willistoni was able to drive GFP expression in the hb of the EB, similar to homologous fragments in D. melanogaster and D. ananassae (Figure 4A). This result suggests that there are activator sequences in D. willistoni that can drive partial GFP expression in the EB. Our observation that the D. willistoni bc3i fragment can drive GFP expression in the hb of the EB, but not the full D. willistoni bc fragment, suggests that repressor sequences similar to the Rep region present in D. melanogaster may also be present in D. willistoni (Figure 4A).
Figure 4. Repressor sequences that can repress bond expression in the EB hwe and hb are present in all species tested, including species that do not express bond in the EB.
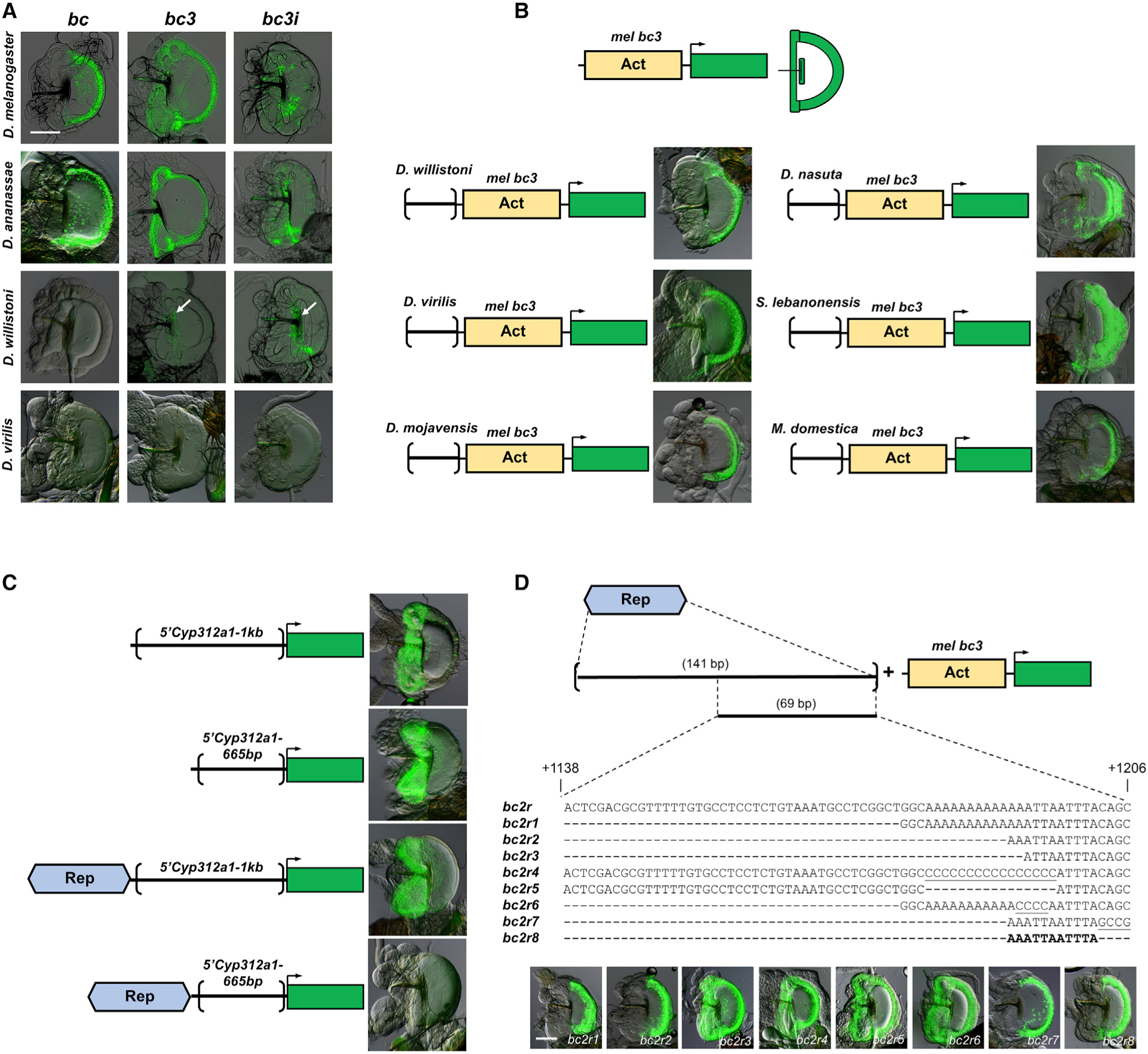
(A) The larger bc fragment recapitulates the native expression of bond in the EB in D. melanogaster and D. ananassae and showed no EB GFP expression with D. willistoni and D. virilis homologous constructs, similar to their native expression. However, the smaller D. willistoni bc3 and bc3i constructs could drive GFP expression in the handle base (white arrows), similar to the bc3i construct of D. melanogaster, suggesting the presence of repressor sequences in the larger bc construct of this species. Scale bar, 100 μm.
(B) Homologous sequences to the D. melanogaster Rep region from six other species, D. willistoni, D. virilis, D. mojavensis, D. nasuta, S. lebanonensis, and M. domestica, can repress GFP expression in the hwe and hb driven by the mel bc3 fragment, suggesting that these regions contain repressor sequences similar to the D. melanogaster Rep region.
(C) The D. melanogaster Rep region can repress EB GFP expression in the hwe and hb driven by another activator sequences from another gene, Cyp312a1, in a distance-dependent manner.
(D) A series of deletion and mutation constructs are made for the 141-bp Rep region of D. melanogaster. The mutated nucleotides are underscored. A 11-bp sequence (5′-AAATTAATTTA-3′) is able to repress GFP expression in the hwe and hb driven by the mel bc3 fragment. Scale bar, 100 μm.
Repressor sequences are present in species that do not express bond in the EB
To confirm our observation that repressor sequences may be present in D. willistoni, we created GFP reporter constructs that fused the region from D. willistoni homologous to the Rep region in D. melanogaster with the D. melanogaster bc3 construct that drives expression in the entire EB. If the D. willistoni sequence functions as a repressor, it should spatially repress expression of the D. melanogaster bc3 fragment and restrict expression to the swe of the EB in D. melanogaster, similar to the repressor sequences in D. melanogaster (Figure 4A). Our results confirm this prediction: the D. willistoni fragment effectively repressed bc3-driven GFP expression in the hwe and the hb and restricted expression to the swe, thus confirming that repressor sequences are present in D. willistoni (Figure 4B). Because D. willistoni does not express bond in the EB and does not have sequences that can drive expression in the EB swe, we were intrigued that repressor sequences that can partially repress EB expression are present in D. willistoni. This observation motivated us to investigate whether these spatial EB repressor sequences are also present in other species where bond is not expressed in the EB. We tested three other Drosophila species, D. mojavensis, D. virilis, and D. nasuta, as well as a more distant species, M. domestica. We also tested the presence of repressor sequences in S. lebanonensis, a species that independently gains bond EB expression. Homologous regions from all five of these species can repress expression driven by the D. melanogaster bc3 construct, restricting GFP expression in the EB swe (Figure 4B). Taken together, these observations suggest that repressor sequences are present in these species and, based on the phylogeny, precede the evolution of the complete minimal EB swe enhancer.
The D. melanogaster bond EB Rep region can repress gene expression of another EB enhancer in a distance-dependent manner and is a short sequence in Drosophila species
The presence of similar spatial repression in different species led us to further characterize this repressor. First, we wanted to know if this repressor is modular, i.e., can it repress the expression of other genes expressed in the EB, or does it work only in the context of the bond gene? To determine this, we created GFP reporter constructs that fused the D. melanogaster Rep region to the 5′ regulatory region of another EB expressed gene, Cyp312a1 (Figure 4C). A 1-kb construct (5′Cyp312a1–1kb) and a 665-bp (5′Cyp312a1–665bp) construct of the 5′ regulatory region of Cyp312a1 drive GFP specifically in the hwe and hb of the EB (Figure 4C). When we fused the D. melanogaster Rep region to these two constructs, there was no change in spatial expression pattern in the 5′Cyp312a1–1kb construct. However, we did not detect GFP expression in the 5′Cyp312a1–665bp construct after being fused with the D. melanogaster Rep region (Figure 4C). Together, these results suggest that the Rep region is modular, because it is able to repress the EB expression driven by activator sequences of another gene. Moreover, the repressor activity is distance dependent because it can repress EB expression in the shorter 665-bp construct, but not the 1-kb construct. Next, we wanted to determine whether the Rep region is a large region or a short sequence capable of spatial repression in the hwe and hb of the EB. To narrow down the sequences involved in repression, we made smaller constructs of the D. melanogaster 141-bp Rep region and assayed for their ability to repress GFP expression driven by the bc3 construct (Act region) (Figure 4D). These constructs, coupled with additional site-directed mutagenesis experiments, narrowed the repressor sequence to an 11-bp sequence (5′-AAATTAATTTA-3′) that is able to recapitulate the repressor activity of the D. melanogaster Rep region (Figure 4D). A search on Flybase (Thurmond et al., 2019) showed that there are 634 hits to this sequence in the D. melanogaster genome, implying the non-uniqueness and pervasiveness of this sequence throughout the genome. An alignment of this region in some of the species tested in this study showed highly similar sequences in some species and dissimilar sequences in other species at the site of this 11-bp sequence (Figure S3). Experiments using the aligned 11-bp sequence from D. ananassae (5′-AAATTAAATTA-3′) and D. willistoni (5′-CTATTAATTTT-3′) showed that these sequences can repress expression in the hwe and hb of the EB, whereas the aligned 11-bp sequence from D. virilis (5′-ACAAAAAAAAA-3′) could not (Figure S4). Because the full D. virilis Rep region can repress expression in the hwe and hb of the EB (Figure 4B), the results suggest that the 11-bp sequences in distantly related species from D. melanogaster may be dissimilar in sequence and location but functionally similar in the ability to spatially repress expression in the EB.
Evolution of putative Abd-B activator binding sites is involved in the stepwise evolution of bond EB expression in Drosophila
We have established that repressor sequences are present in the several species that do not express bond in the EB (Figure 4B). One of these species, D. willistoni, has sequences homologous to the first transcriptional input (Ac1) in D. melanogaster that could drive partial expression in the EB when isolated from the repressor, similar to that of D. melanogaster and D. ananassae (Figure 4A). This result suggests that the gain of bond expression in the EB swe could be a stepwise evolutionary process. To investigate how the EB expression of bond evolved, we sought to identify potential transcriptional activators in this enhancer that can drive expression in the EB. We performed high-throughput transcriptomic sequencing of EBs from 8-day-old male D. melanogaster and identified putative TFs that are expressed in the EB (Table S1). Next, we carried out a reverse genetic screen by using an EB-specific GAL4 driver (EB-GAL4) to drive UAS::RNAi constructs of the most highly expressed TFs (Table S2) in order to ascertain which TF(s) can affect the mel bc3 construct-driven GFP expression (Figure 5A). Out of the 100 UAS::RNAi constructs tested, one line, driving RNAi knockdown of the homeobox gene, Abd-B, was able to disrupt bc3-driven GFP expression in the EB, suggesting that Abd-B may function as a transcriptional activator for the expression of bond in the EB (Figure 5B). The RNAi of Abd-B in the EB also led to morphological changes in the EB, suggesting that Abd-B may also be required for EB development. Bioinformatics analyses using JASPAR (Fornes et al., 2020) predicted three putative Abd-B binding sites (Figure 5C).
Figure 5. The evolution of two putative Abd-B binding sites is necessary for the expression of bond in the EB.
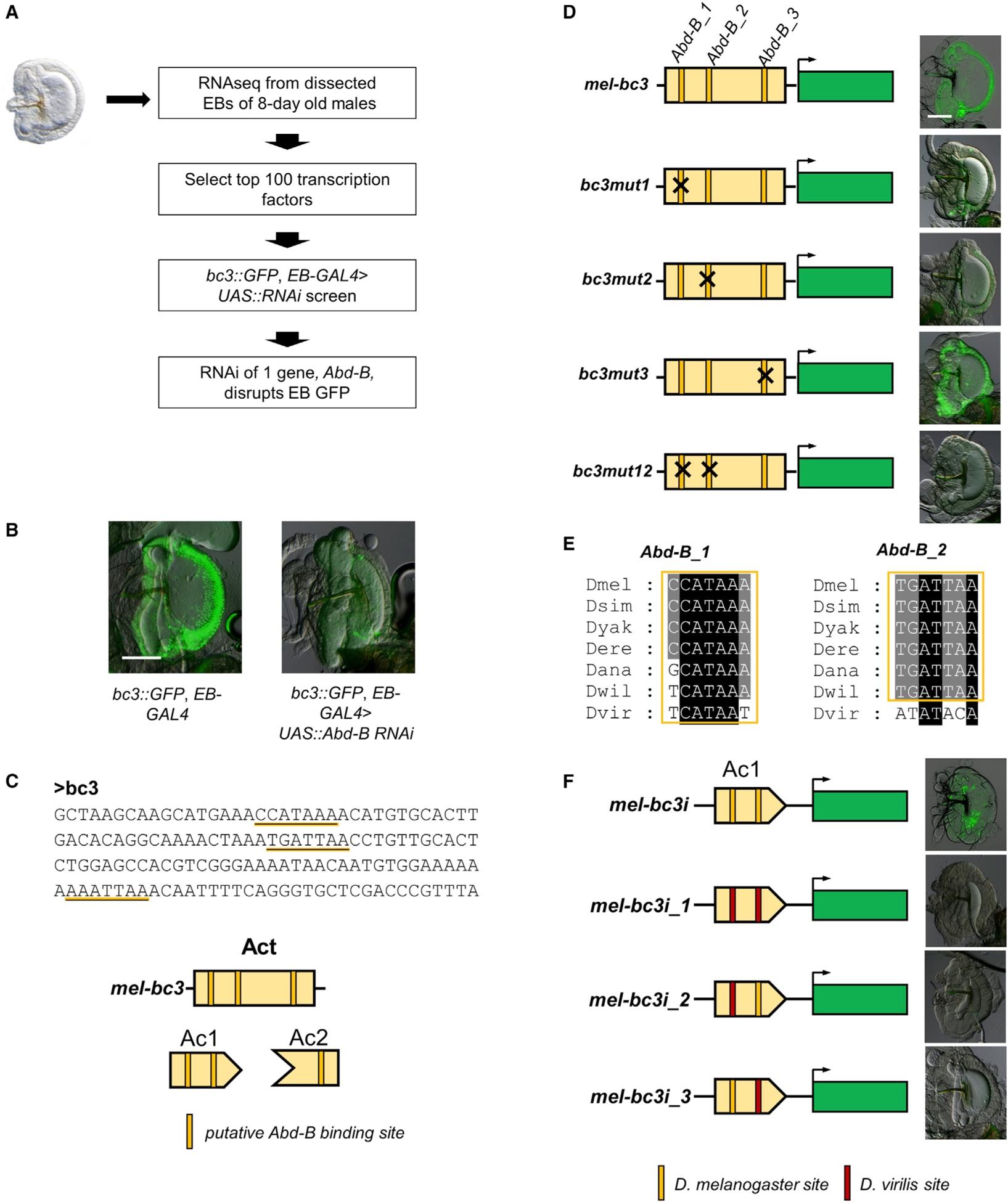
(A) An EB RNAi screen identifies Abd-B as a possible regulator of bond in the EB.
(B) RNAi knockdown of bond in the EB led to a big decrease in GFP expression driven by the D. melanogaster bc3 construct. Scale bar, 100 μm.
(C) JASPAR analysis identified three putative Abd-B binding sites in the D. melanogaster bc3 construct.
(D) Site-directed mutagenesis of these putative Abd-B binding sites individually shows that the Abd-B_1 and the Abd-B_2 sites are necessary for GFP expression driven by the bc3 construct, but not the Abd-B_3 site. Scale bar, 100 μm.
(E) Evolutionary analysis of the Abd-B_1 and the Abd-B_2 sites shows that all species have putative Abd-B binding sequences at Abd-B_1, and most species have putative Abd-B binding sequences at Abd-B_2 except D. virilis.
(F) Swapping in the D. virilis sequence at both sites either individually or in combination led to the loss of GFP expression in the hb of the EB driven by the D. melanogaster bc3i construct.
To determine whether the putative Abd-B binding sites function in vivo, we systematically mutated each site in the bc3 GFP construct (Figure 5D). Our experiments show that site-directed mutagenesis of the first two putative Abd-B binding sites, Abd-B_1 and Abd-B_2, reduced GFP expression drastically when mutated individually and abolished GFP expression completely when mutated in combination. In contrast, mutation of the third putative Abd-B binding site, Abd-B_3, did not affect GFP expression (Figure 5D). Our results suggest that Abd-B_1 and Abd-B_2 sites are both necessary to drive bond expression in the EB of D. melanogaster. Because Abd-B_1 and Abd-B_2 are both in the Ac1 region (bc3i construct), we expect that these two sites will be conserved in all melanogaster group species tested and D. willistoni, but not D. virilis. To trace the evolution of Abd-B_1 and Abd-B_2 in these species, we performed an alignment of the bc23 region of these species (Figure S3). We used JASPAR to predict potential Abd-B binding sites in all these species and found that all Drosophila species have putative Abd-B binding sequences at Abd-B_1, and species in the Sophophora subgenus have putative Abd-B binding sequences at Abd-B_2, but not in D. virilis (Figure 5E). These predictions are consistent with the activity of homologous bc3i fragments (Ac1 region) from the four species, in which D. melanogaster, D. ananassae, and D. willistoni sequences drove GFP reporter expression in the hb of the EB, but D. virilis did not (Figure 4A). To determine whether the predicted difference at the Abd-B binding sites could underlie differences in expression driven by the D. melanogaster and D. virilis bc3i constructs, we swapped in the corresponding D. virilis sequences at the D. melanogaster Abd-B_1 and Abd-B_2 sites individually in the D. melanogaster bc3i construct. Our results show that the D. virilis sequence at both sites either individually or together led to the loss of GFP expression in the hb of the EB (Figure 5F). Taken together, the findings suggest that evolution in both Abd-B_1 and Abd-B_2 sites between these species could contribute toward the evolution of bond expression in the EB.
The S. lebanonensis enhancer does not require Abd-B to drive gene expression in the EB swe
Because the expression of bond in the EB swe is independently gained in two lineages, we wanted to determine whether the parallel evolution of this trait has a similar or different genetic basis. We had shown that the bond EB swe enhancer lies in intron 1 of bond in both lineages (Figure 2B). To investigate whether bond EB swe expression pattern in S. lebanonensis requires Abd-B, we use a S. lebanonensis bond intron 1 GFP construct that can drive EB swe GFP expression in transgenic D. melanogaster. The 3,112-bp S. lebanonensis B1 construct (Sle_B1) contains 64 putative Abd-B sites as predicted by JASPAR. However, when we drove RNAi of Abd-B in the EB of transgenic D. melanogaster carrying this S. lebanonensis B1 construct, we did not see obvious changes in GFP expression compared with the control (Figure 6). In contrast, RNAi of Abd-B drastically reduced GFP expression in a D. melanogaster bond EB swe enhancer-driven construct. In both cases, RNAi caused morphological changes to the EB, suggesting that EB RNAi of Abd-B is successful. Our results showed that bond EB expression in S. lebanonensis is likely to be Abd-B independent (Figure 6).
Figure 6. bond EB expression in S. lebanonensis is independent of Abd-B.
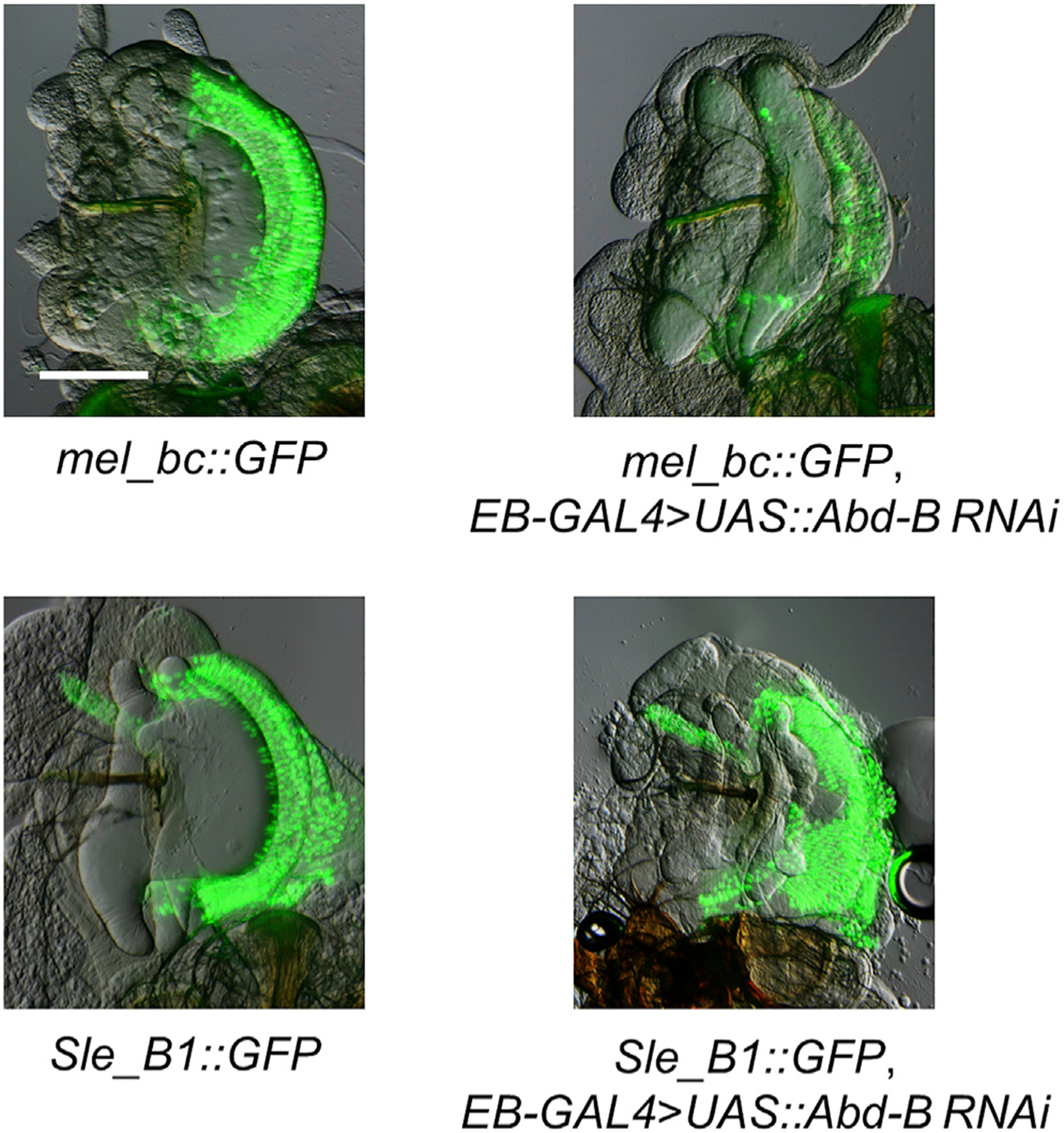
EB-specific RNAi of Abd-B greatly reduces expression of EB swe GFP driven by the D. melanogaster bc construct but has no observable effects on EB swe GFP driven by the S. lebanonensis B1 construct. Scale bar, 100 μm.
DISCUSSION
The generation of highly specific gene expression patterns is often due to the combinatorial actions of activators, repressors, and other transcriptional inputs that form a modular enhancer (Small and Arnosti, 2020; Spitz and Furlong, 2012). However, the evolutionary history that leads to the origin of these enhancers is not always clear. In this study, we have identified and characterized an enhancer that drives precise expression of the elongase gene bond in the male EB swe of D. melanogaster and several other species in the Drosophila genus. EB expression of bond has been shown to be necessary for the production of the male anti-aphrodisiac pheromone, CH503 (Ng et al., 2015). Our experiments show that the EB swe enhancer is made up of at least one Rep region and one Act region, which can be subdivided in two transcriptional inputs (Ac1 and Ac2). Together, the combined action of these transcriptional inputs restricts bond expression to the swe of the EB in D. melanogaster (Figure 3C). We further show that the Rep region containing sequences capable of repressing gene expression in parts of the EB is present in all Drosophila species tested in our study, as well as in distantly related species, Scaptodrosophila lebanonensis and Musca domestica, even though many of these species do not express bond in the EB swe (Figure 4B). These findings suggest that the spatial repressor precedes the evolution of the Act regions (Figure 7).
Figure 7. Repression precedes independent evolutionary gains of a highly specific gene expression pattern.
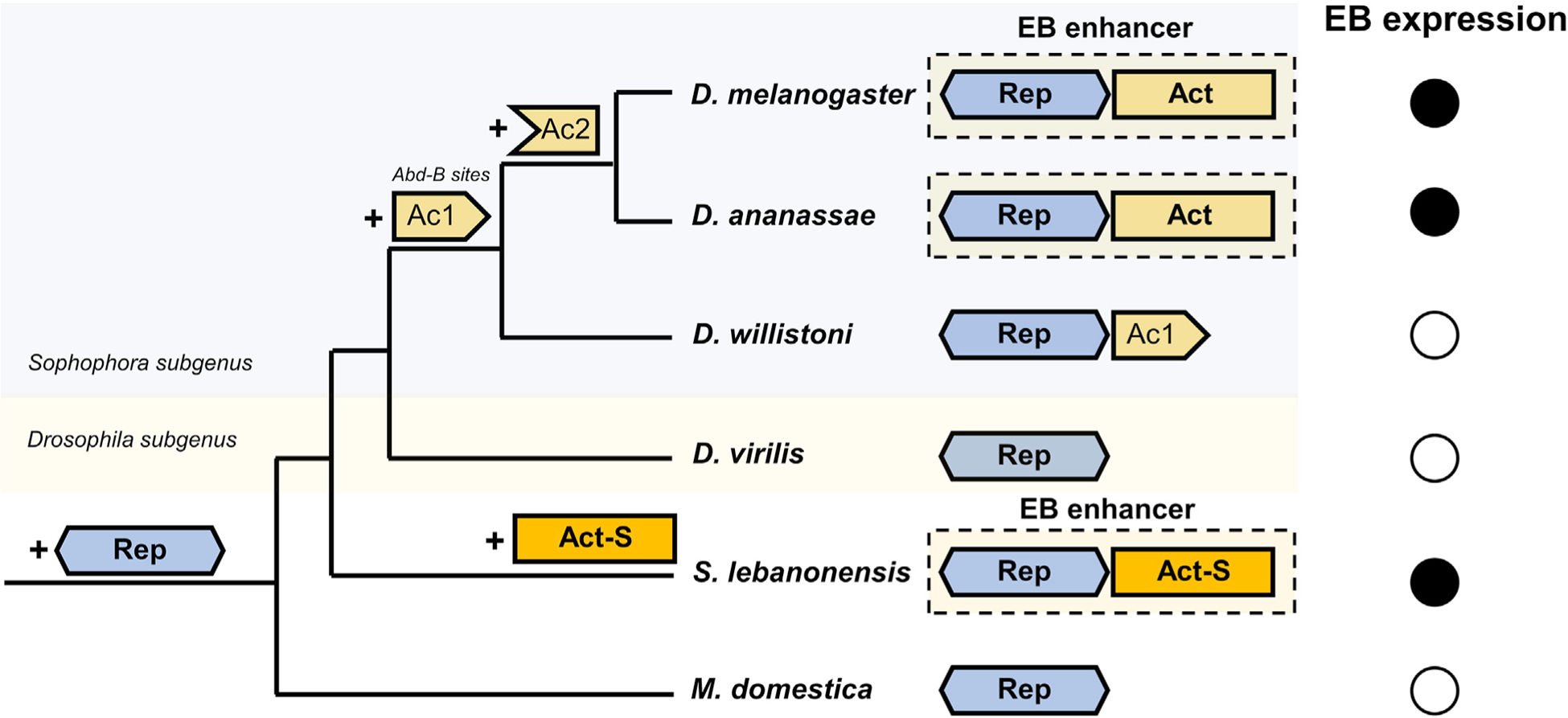
The EB swe enhancer present in several Drosophila species is made up of an Act region, which contains two distinct transcriptional inputs (Ac1 and Ac2) that drive expression in the whole EB (hb, hwe, and swe), and a Rep that represses expression in the hb and hwe, resulting in the expression of bond in the EB swe. The first transcriptional input (Ac1) evolved in the Sophophora subgenus, possibly because of the gain of Abd-B binding sites. The Ac1 region is present in D. willistoni, but there is no expression of bond in the EB of this species due to the presence of the Rep region. The second transcriptional input (Ac2) evolved before the split of the melanogaster group and the obscura group and can drive expression in the whole EB in conjunction with Ac1, but because of the presence of Rep, bond expression is restricted to the swe in these species. An independent gain of EB gene expression occurs in Scaptodrosophila species that do not require Abd-B as a transcriptional input. The Rep region is present in all species, including the housefly M. domestica, and precedes the evolution of the activator sequences.
During the divergence of the Sophophora and Drosophila subgenus in the genus Drosophila, the Ac1 activator regulatory input, containing putative binding sites for the TF Abd-B, evolved in the Sophophora subgenus. However, as seen in D. willistoni, the presence of Ac1 was not sufficient for bond expression in the EB due to the overlapping action of the repressor sequences. The Ac2 transcriptional input that evolved in the ancestor leading to the melanogaster and obscura groups allowed for expression in the entire EB with Ac1 but was not able to drive expression on its own (Figure 4A). Together, the combinatorial action of the Rep region and the full Act (Ac1 + Ac2) region form a modular enhancer driving specific expression in the swe of the EB in some Drosophila species (Figures 3C and 7). Based on our ancestral trait reconstruction analyses (Figure 1B), we show that there is likely to be an independent gain of bond EB expression in Scaptodrosophila species. We also showed that bond EB expression in S. lebanonensis is Abd-B independent, providing further evidence that it is likely to be an independent evolutionary gain.
The evolution of modular enhancers
The evolution of new enhancers activity can occur from at least four different evolutionary mechanisms: transposition, promoter switching, co-option, and de novo generation (Rebeiz and Tsiantis, 2017). Although the previous studies have provided evidence for the first three mechanisms (Chung et al., 2007; Guo et al., 2016; Koshikawa et al., 2015), de novo evolution of enhancers has been difficult to study because of their rarity (Rebeiz and Tsiantis, 2017). Identifying a de novo origin of an enhancer is challenging partly because these enhancers are usually composed of multiple binding sites or sequences that bind different transcriptional activators or repressors. It is therefore more parsimonious to evolve a new enhancer by co-opting existing regulatory sequences (Rebeiz et al., 2011). However, we suggest that the de novo generation of an enhancer is possible. Previous genome-wide studies in Drosophila and human suggest that a large fraction of these genomes is bound by TFs (ENCODE Project Consortium, 2012; Roy et al., 2010). Although there is a vigorous scientific debate on whether these TF-bound sequences are functional (Graur et al., 2013; Kellis et al., 2014), the fact that TF binding sites are short, degenerate, and dispersed widely and randomly across the genome suggests that a small number of mutations generating new TF binding sites around these existing TF-bound sequences could form a new enhancer based on the combinatorial effects of these sequences. Our study shows that the gain of the Ac2 transcriptional input led to the evolution of an EB swe enhancer in Drosophila species. However, Ac2 is unable to drive expression on its own, relying on the combinatorial activity with the Ac1 transcriptional input and the Rep region to drive expression specifically in the EB swe. Because we did not detect any other enhancer activities within 1 kb of this region, we suggest that this could be a case of de novo generation of a novel enhancer, although we cannot rule out that co-option could also be a possibility (see later discussion on the origin of the Rep region).
The presence of repressor sequences before the evolution of highly specific expression patterns
Numerous studies have demonstrated the significance of repressor binding sites or sequences in modular enhancers to constrain and shape gene expression (Gompel et al., 2005; Preger-Ben Noon et al., 2016; Small et al., 1992; Struffi et al., 2011). However, to our knowledge, there are no studies that investigate the order of whether activator sequences or repressor sequences evolved first in the origin of enhancers driving highly specific expression patterns. The presence of functionally similar repressor sequences (Rep) in multiple Drosophila species and Musca domestica that do not express bond in the EB suggest that these sequences may be present at least 150 million years ago, which is the estimated divergence time between Musca and Drosophila (Thomas et al., 2020). What could be the putative function(s) of these sequences before being part of the bond EB swe enhancer? We propose three different hypotheses.
The first hypothesis is that this repressor sequence could be part of another enhancer in the bond intron and is co-opted into the bond EB swe enhancer. However, our experiments in D. melanogaster showed that there are no other apparent enhancer activities within 1 kb of the Rep region aside from Ac1 and Ac2 (Figure S2). Our experiments show that the Rep region represses gene expression in a distance-dependent manner (Figure 4C) and is not able to repress gene expression that is more than 1 kb away. This observation could suggest that the repressor associated with the Rep region functions as distance-dependent “short-range repressors” like knirps and Krüppel, which can repress gene expression only around 100 bp away (Small and Arnosti, 2020), rather than “long-range repressors,” such as Hairy, which mediates repression of cis-regulatory sequences more than 1 kb away (Li and Arnosti, 2011). A recent paper showed that other cis-regulatory sequences, such as silencers, may have dual roles as transcriptional enhancers when the cellular context is different (Gisselbrecht et al., 2020). The Rep region identified in our study, which functions as a Rep region in the bond EB swe enhancer, may have other unidentified and undetermined roles in other cell types. We do not exclude the possibility that the Rep region of the bond EB swe enhancer was co-opted from another regulatory function that it may have, but determining other functions is beyond the scope of this paper.
A second hypothesis is the Rep region is evolutionarily conserved because of this region overlapping with the exon of an antisense non-coding RNA, CR44062, which resides on the opposite DNA strand to bond (Figure S5). Although the function of CR44062 is unknown, one possible scenario is that the Rep region is conserved because of potential functional constraint on the evolution of CR44062; i.e., evolutionary changes in the sequence of CR44062 may have negative fitness effects.
A third hypothesis is that binding sites for the transcriptional activators or repressors are usually very short (6–10 bp long) (Payne and Wagner, 2014; Stewart et al., 2012) and due to this short length are pervasive and randomly distributed throughout the genome. We narrowed down the putative repressor binding site to an 11-bp sequence in D. melanogaster (Figure 4D). Bioinformatics analyses showed that this exact 11-bp sequence has 634 complete matches in the D. melanogaster genome. In addition, homologous 11-bp sequences from D. ananassae and D. willistoni with inexact matches to the D. melanogaster 11-bp sequence also showed the ability to transgenically repress GFP expression in the hwe and hb of the D. melanogaster EB. Because the sequences of these binding sites are usually degenerate in nature, this may suggest that these sequences would be commonly distributed through the whole genome. Therefore, the likelihood of these short sequences randomly distributed across the genome without any apparent function is high. These sequences may not produce any pheno-types until activator sequences that produce an overlapping expression pattern with the repressor sequences evolve.
Regardless of whether each of the three hypotheses is correct, the phenomenon that repressor sequences precede the gain of activator sequences in enhancer evolution could be a common mechanism during the evolution of highly specific gene expression patterns.
The independent evolution of highly specific gene expression patterns
In this study, we showed a possible example of independent evolutionary gains of a highly specific expression pattern of bond in the EB swe of species in two distant lineages. Although the repressor sequences in these enhancers that drive this specific expression are likely to be ancestral, we found that the transcriptional activators are likely to be different; i.e., bond EB expression in Drosophila species requires the transcriptional factor Abd-B, but bond EB expression in Scaptodrosophila does not. Due to our limited RNAi screen, we did not identify all the TFs involved in driving expression in the Drosophila enhancer, so we could not determine whether some transcriptional inputs are similar between these two enhancers. However, we propose that preexisting spatial repressor sequences may preconfigure similar highly specific gene expression patterns in different lineages, even though other transcriptional inputs that led to the appearance of these patterns are different. This may reflect a general evolutionary mechanism for enhancer origins.
Limitations of the study
Our study is limited by the species we can obtain from stock centers, as well as the available genomes for the species. A more extensive sampling of species may shed more light into the origins of the repressor sequences and the independent gains of different activator sequences. In addition, our RNAi screen used only one RNAi resource (DRSC/Harvard TRiP) and focused on only the top 100 potential TFs that could affect the expression of bond in the EB. A more extensive screen, using both the DRSC/Harvard TRiP RNAi collection and the Vienna Drosophila Resource Center (VDRC) and focusing on more TFs identified in our EB transcriptome, may allow us to identify more transcriptional regulators that regulate the expression of bond in the EB. This can provide a more comprehensive picture of the evolutionary events described in this manuscript.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Henry Chung (hwchung@msu.edu)
Materials availability
All constructs and transgenic fly lines generated in this study are available from the Lead Contact upon request.
Data and code availability
The RNA-seq datasets reported in this study has been deposited at NCBI GEO and is publicly available as of the date of publication. Accession number is listed in the Key resources table. The concatenated alignment and output tree files used to generate the phylogenetic tree are downloadable from Zenodo (Accession number is listed in the Key resources table).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| AscI | NEB | Cat# R0558L |
| SbfI | NEB | Cat# R0642L |
| TRIzol Reagent | Invitrogen | Cat# 15596026 |
| Critical commercial assays | ||
| TruSeq RNA Library Prep Kit v2 | Illumina | Cat# RS-122–2001 |
| Deposited data | ||
| Transcriptomic profiles obtained from 8-day old D. melanogaster male ejaculatory bulbs. | This paper | GEO: GSE185053 |
| The concatenated alignment and output tree files | This paper | 10.5281/zenodo.5238168 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: Canton-S | Laboratory of Sean B. Carroll Lab | N/A |
| D. melanogaster: Xout | Williams et al., 2008, Laboratory of Sean B. Carroll Lab | N/A |
| D. simulans: w 501 | National Drosophila Species Stock Center (Cornell University) | 14021–0251.195 |
| D. yakuba | National Drosophila Species Stock Center (Cornell University) | 14021–0261.00 |
| D. erecta | National Drosophila Species Stock Center (Cornell University) | 14021–0224.01 |
| D. ananassae | National Drosophila Species Stock Center (Cornell University) | 14024–0371.13 |
| D. pseudoobscura | National Drosophila Species Stock Center (Cornell University) | 14011–0121.94 |
| D. subobscura | National Drosophila Species Stock Center (Cornell University) | 14011–0131.05 |
| D. nebulosi | National Drosophila Species Stock Center (Cornell University) | 14030–0761.00 |
| D. sturtevanti | National Drosophila Species Stock Center (Cornell University) | 14043–0871.16 |
| D. willistoni | National Drosophila Species Stock Center (Cornell University) | 14030–0811.24 |
| D. immigrans | National Drosophila Species Stock Center (Cornell University) | 15111–1731.03 |
| D. nasuta | National Drosophila Species Stock Center (Cornell University) | 15112–1781.00 |
| D. Americana | National Drosophila Species Stock Center (Cornell University) | 15010–0951.00 |
| D. virilis | National Drosophila Species Stock Center (Cornell University) | 15010–1051.87 |
| D. mercatorum | National Drosophila Species Stock Center (Cornell University) | 15082–1521.38 |
| D. mojavensis | National Drosophila Species Stock Center (Cornell University) | 15081–1352.22 |
| S. latifasciaeformis | National Drosophila Species Stock Center (Cornell University) | 11030–0061.01 |
| S. lebanonensis | National Drosophila Species Stock Center (Cornell University) | 11010–0011.00 |
| S. rufifrons | National Drosophila Species Stock Center (Cornell University) | 11040–0071.00 |
| C. procnemis | National Drosophila Species Stock Center (Cornell University) | 20000–2631.01 |
| M. domestica | Josh’s Frogs (Owosso, MI) | B06XJ8DQ2X |
| D. melanogaster: Various GFP reporter constructs | This study | N/A |
| D. melanogaster: Various UAS-RNAi lines | Bloomington Drosophila Stock Center | See Table S2 |
| D. melanogaster: EB-GAL4 | Gift from Dr. Phillip Daborn (The University of Melbourne) | N/A |
| Oligonucleotides | ||
| Primers: Generation of GFP reporter constructs | This paper | See Table S3 |
| Primers: Generation of in situ hybridization probes | This paper | See Table S3 |
| Recombinant DNA | ||
| GFP reporter vector pS3aG | Williams et al., 2008, Laboratory of Sean B. Carroll Lab | N/A |
| Software and algorithms | ||
| MUSCLE | Edgar, 2004 | http://www.drive5.com/muscle |
| PhyML 3.0 | Guindon et al., 2010 | http://www.atgc-montpellier.fr/phyml/ |
| ‘Phytools’ package | Revell, 2013 | https://cran.r-project.org/web/packages/phytools/index.html |
| TopHat v2.0.9 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/index.shtml |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The Canton-S strain was used as the wild-type D. melanogaster strain. Fifteen other Drosophila species (D. simulans, D. yakuba, D erecta, D. ananassae, D. pseudoobscura, D. subobscura, D. nebulosa, D. sturtevanti, D. willistoni, D. immigrans, D. nasuta, D. americana, D. virilis, D. mercatorum, and D. mojavensis), three ScaptoDrosophila species (S. latifasciaeformis, S. lebanonensis, and S. rufifrons) and one Chymomyza species (C. procnemis) were obtained from the National Drosophila Species Stock Center at Cornell University. The housefly (Musca domestica) was obtained from Josh’s Frogs (Owosso, Michigan, USA). The EB-GAL4 D. melanogaster line drives GFP in the entire EB and is a gift from Dr. Phillip Daborn (University of Melbourne, Australia). UAS::RNAi transgenic flies from the Transgenic RNAi Project (TRiP) (Ni et al., 2009) were obtained from the Bloomington Drosophila Stock Center. The D. melanogaster Xout line was used in the production of transgenic lines carrying GFP reporter constructs (Williams et al., 2008). All flies were maintained at room temperature on standard Drosophila food (Bloomington formulation, Genesee Scientific). D. melanogaster GAL4/UAS-RNAi experiments were performed at 25°C.
METHOD DETAILS
Generation of GFP reporter constructs and transgenic flies
All GFP reporter constructs were generated by PCR amplification of the genomic fragments from different Drosophila species and cloned into the GFP reporter vector pS3aG via the AscI and SbfI site (All primers listed in Table S3). The initial screen to locate the D. melanogaster EB enhancer focused on four different regions based on bond-PB transcript: i) 5′ of the gene (3R: 22547847..22550679), intron 1 (3R: 22550873..22554636), intron 3 (3R: 22555014..22555385) and the 3′ non-coding region (3R: 22556495..22558341). All constructs were injected into the D. melanogaster Xout line and integrated into the genome using the PhiC31 integrase system.
In situ hybridization
Ejaculatory bulbs (EBs) from three-day old male adult flies were dissected in Phosphate-Buffered Saline (PBS). In situ hybridization was performed with RNA probes as described previously (Chung et al., 2009). Probes for bond in situ hybridization were synthesized from cDNA using species-specific primers (Table S3). The D. melanogaster probe was used for in situ hybridization to D. melanogaster, D. simulans, D. yakuba, and D. erecta. D. ananassae probe was used for D. ananassae. D. pseudoobscura probe was used for D. pseudoobscura and D. subobscura. D. willistoni probe was used for D. willistoni, D. sturtevanti, and D. nebulosa. S. lebanonensis probe was used to S. lebanonensis and S. rufifrons. C. procnemis probe was used for C. procnemis and S. latifasiaeformis. M. domestica probe was used for M. domestica.
Phylogenetic analyses
A multilocus dataset of 17 genes from 21 species was used for phylogenetic reconstruction. The genes included the 15 nuclear markers Amyrel (Amyrel), Distal-less (Dll), Dopa decarboxylase (Ddc), ebony (e), engrailed (en), even-skipped (eve), hedgehog (hh), Notum (Notum), patched (ptc), wingless (wg), 28S ribosomal RNA (28S), Alcohol dehydrogenase (Adh), Glycerol-3-phosphate dehydrogenase (Gpdh), Superoxide dismutase (Sod), Xanthine dehydrogenase (Xdh), and the two mitochondrial markers cytochrome oxidase subunit 1 (COI) and cytochrome oxidase subunit 2 (COII). Nucleotide sequences of the 20 drosophilid species were retrieved from the DrosoPhyla project (Finet et al., 2021), and nucleotide sequences of M. domestica were collected from the NCBI database. Alignments for each individual gene were generated using MUSCLE (Edgar, 2004) with default parameters. Unreliably aligned positions were excluded using trimAl with parameters −gt 0.5 and −st 0.001 (Capella-Gutiérrez et al., 2009). In-house Python scripts were used to concatenate the aligned sequences (Finet et al., 2021). Maximum-likelihood searches were performed using PhyML 3.0 (Guindon et al., 2010) under the GTR+Γ4+I model, and 100 bootstrap replicates were conducted for support estimation.
RNA sequencing and analysis
RNA from the EBs of approximately 200 eight-day old Canton-S D. melanogaster males was extracted using TRIzol Reagent according to manufacturer’s instructions. Indexed RNA-Seq libraries were prepared from ~1 μg of total RNA using the TruSeq RNA Library Prep Kit v2 (Illumina) according to manufacturer’s protocol. RNA quality and concentration were measured on an Agilent 2100 Bio-analyzer (Thermo Scientific). Paired end sequencing was performed on an NGS Illumina Hiseq 2000 with a 20 M read depth (75bp × 2; AITBiotech; Singapore). FastQ files were aligned to the Dmr6.05 Drosophila melanogaster reference genome (2012, r5.48) using TopHat v2.0.9 (Kim et al., 2013).
RNAi screen
Based on the expression level of the transcription factors (TFs) in EB and predicting TF binding sites of the bc3 fragment, 100 candidate TFs were used for the RNAi screen. Males from each UAS::RNAi line were crossed with virgin females of the bc3::GFP; EB-GAL4; + fly line. The EBs of three-day old males from the resulting crosses were dissected and imaged for GFP expression.
Imaging
All in situ hybridization and GFP images were captured using the Nikon SMZ18 dissecting stereo microscope system. For GFP images, EBs were dissected from three-day old males in 1 × PBS and mounted on slides with glycerol mountant [80% (vol/vol in water) glycerol, 0.1 M Tris (pH 8.0)].
QUANTIFICATION AND STATISTICAL ANALYSIS
To determine the evolution of bond expression in EB across the phylogeny, we reconstructed its ancestral state using the method with the ‘Phytools’ package in R (Revell, 2013). The maximum likelihood approach was used for discrete characters, based on the equal-rate model (Mooers and Schluter, 1999).
Supplementary Material
Highlights.
An enhancer controls a highly specific EB expression of bond in Drosophila
This enhancer contains activator and repressor sequences
A similar enhancer is independently gained in the distantly related S. lebanonensis
Similar repressor sequences are present in other species without bond EB expression
ACKNOWLEDGMENTS
We thank Caitlin Peffers, Mei Luo, Ian Paulsen, and Cole Richards for technical assistance and the Bloomington Drosophila Stock Center for fly stocks and reagents. We acknowledge critical comments to the manuscript by Dr. David Arnosti (Michigan State University) and Dr. Sean B. Carroll (University of Maryland, College Park, MD). J.Y.Y. and J.S.R.C. were supported by the Singapore National Research Foundation (NRF-RF2010-06). J.Y.Y. was also supported by the National Institutes of Health (Grant No. 1P20GM125508). H. Chung was supported by USDA NIFA via Michigan State University AgBioresearch (Umbrella project MICL02522).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109896.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Andrioli LPM, Vasisht V, Theodosopoulou E, Oberstein A, and Small S (2002). Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development 129, 4931–4940. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Barolo S, Levine M, and Small S (1996). The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122, 205–214. [DOI] [PubMed] [Google Scholar]
- Arnoult L, Su KF, Manoel D, Minervino C, Magriña J, Gompel N, and Prud’homme B (2013). Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339, 1423–1426. [DOI] [PubMed] [Google Scholar]
- Barolo S, and Posakony JW (2002). Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, and Gabaldón T (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36. [DOI] [PubMed] [Google Scholar]
- Chung H, Bogwitz MR, McCart C, Andrianopoulos A, Ffrench-Constant RH, Batterham P, and Daborn PJ (2007). Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Sztal T, Pasricha S, Sridhar M, Batterham P, and Daborn PJ (2009). Characterization of Drosophila melanogaster cytochrome P450 genes. Proc. Natl. Acad. Sci. USA 106, 5731–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AB, and Wolfner MF (2018). Dynamic changes in ejaculatory bulb size during Drosophila melanogaster aging and mating. J. Insect Physiol 107, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C, Kassner VA, Carvalho AB, Chung H, Day JP, Day S, Delaney EK, De Ré FC, Dufour HD, Dupim E, et al. (2021). DrosoPhyla: Resources for Drosophilid Phylogeny and Systematics. Genome Biol. Evol 13, evab179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, et al. (2020). JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuchi T, Masuko K, Nishimune Y, Obinata M, and Matsui Y (1996). Inhibition of testicular germ cell apoptosis and differentiation in mice misex-pressing Bcl-2 in spermatogonia. Development 122, 1703–1709. [DOI] [PubMed] [Google Scholar]
- Gisselbrecht SS, Palagi A, Kurland JV, Rogers JM, Ozadam H, Zhan Y, Dekker J, and Bulyk ML (2020). Transcriptional silencers in Drosophila serve a dual role as transcriptional enhancers in alternate cellular contexts. Mol. Cell 77, 324–337.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, and Carroll SB (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487. [DOI] [PubMed] [Google Scholar]
- Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, and Elhaik E (2013). On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol 5, 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, and Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gu X, Sheng Z, Wang Y, Luo C, Liu R, Qu H, Shu D, Wen J, Crooijmans RP, et al. (2016). A complex structural variation on chromosome 27 leads to the ectopic expression of HOXB8 and the muffs and beard pheno-type in chickens. PLoS Genet. 12, e1006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rokas A, and Carroll SB (2006). Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387–1399. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, and Carroll SB (2008). The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132, 783–793. [DOI] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, et al. (2014). Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA 111, 6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshikawa S, Giorgianni MW, Vaccaro K, Kassner VA, Yoder JH, Werner T, and Carroll SB (2015). Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 112, 7524–7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissières V, Pickle CS, Plajzer-Frick I, and Lee EA (2016). Progressive loss of function in a limb enhancer during snake evolution. Cell 167, 633–642.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, and Young RA (2013). Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LM, and Arnosti DN (2011). Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Curr. Biol 21, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, and Kreitman M (2005). Functional evolution of a cis-regulatory module. PLoS Biol. 3, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers AØ, and Schluter D (1999). Reconstructing ancestor states with maximum likelihood: support for one-and two-rate models. Syst. Biol 48, 623–633. [Google Scholar]
- Morgan BA, Izpisúa-Belmonte J-C, Duboule D, and Tabin CJ (1992). Targeted misexpression of Hox-4.6 in the avian limb bud causes apparent homeotic transformations. Nature 358, 236–239. [DOI] [PubMed] [Google Scholar]
- Ng SH, Shankar S, Shikichi Y, Akasaka K, Mori K, and Yew JY (2014). Pheromone evolution and sexual behavior in Drosophila are shaped by male sensory exploitation of other males. Proc. Natl. Acad. Sci. USA 111, 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WC, Chin JS, Tan KJ, and Yew JY (2015). The fatty acid elongase Bond is essential for Drosophila sex pheromone synthesis and male fertility. Nat. Commun 6, 8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. (2009). A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne JL, and Wagner A (2014). The robustness and evolvability of transcription factor binding sites. Science 343, 875–877. [DOI] [PubMed] [Google Scholar]
- Preger-Ben Noon E, Davis FP, and Stern DL (2016). Evolved repression overcomes enhancer robustness. Dev. Cell 39, 572–584. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, and Tsiantis M (2017). Enhancer evolution and the origins of morphological novelty. Curr. Opin. Genet. Dev 45, 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Jikomes N, Kassner VA, and Carroll SB (2011). Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc. Natl. Acad. Sci. USA 108, 10036–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ (2013). Two new graphical methods for mapping trait evolution on phylogenies. Methods Ecol. Evol 4, 754–759. [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. ; modENCODE Consortium (2010). Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, and Arnosti DN (2020). Transcriptional Enhancers in Drosophila. Genetics 216, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, and Levine M (1992). Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, and Furlong EE (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet 13, 613–626. [DOI] [PubMed] [Google Scholar]
- Stewart AJ, Hannenhalli S, and Plotkin JB (2012). Why transcription factor binding sites are ten nucleotides long. Genetics 192, 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struffi P, Corado M, Kaplan L, Yu D, Rushlow C, and Small S (2011). Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development 138, 4291–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama K, and Saitou N (2011). Loss-of-function mutation in a repressor module of human-specifically activated enhancer HACNS1. Mol. Biol. Evol 28, 3005–3007. [DOI] [PubMed] [Google Scholar]
- Szafer-Glusman E, Giansanti MG, Nishihama R, Bolival B, Pringle J, Gatti M, and Fuller MT (2008). A role for very-long-chain fatty acids in furrow ingression during cytokinesis in Drosophila spermatocytes. Curr. Biol 18, 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GWC, Dohmen E, Hughes DST, Murali SC, Poelchau M, Glastad K, Anstead CA, Ayoub NA, Batterham P, Bellair M, et al. (2020). Gene content evolution in the arthropods. Genome Biol. 21, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Mary-gold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. ; FlyBase Consortium (2019). FlyBase 2.0: the next generation. Nucleic Acids Res. 47, D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar AR, Kelly GM, and Moon RT (1995). Wnt4 affects morphogenesis when misexpressed in the zebrafish embryo. Mech. Dev 52, 153–164. [DOI] [PubMed] [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, and Carroll SB (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, and Kalay G (2011). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet 13, 59–69. [DOI] [PubMed] [Google Scholar]
- Xin Y, Le Poul Y, Ling L, Museridze M, Mühling B, Jaenichen R, Osipova E, and Gompel N (2020). Enhancer evolutionary co-option through shared chromatin accessibility input. Proc. Natl. Acad. Sci. USA 117, 20636–20644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew JY, Dreisewerd K, Luftmann H, Müthing J, Pohlentz G, and Kravitz EA (2009). A new male sex pheromone and novel cuticular cues for chemical communication in Drosophila. Curr. Biol 19, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Mango SE (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev 37, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-D, and Odenwald WF (1995). Misexpression of the white (w) gene triggers male-male courtship in Drosophila. Proc. Natl. Acad. Sci. USA 92, 5525–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets reported in this study has been deposited at NCBI GEO and is publicly available as of the date of publication. Accession number is listed in the Key resources table. The concatenated alignment and output tree files used to generate the phylogenetic tree are downloadable from Zenodo (Accession number is listed in the Key resources table).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| AscI | NEB | Cat# R0558L |
| SbfI | NEB | Cat# R0642L |
| TRIzol Reagent | Invitrogen | Cat# 15596026 |
| Critical commercial assays | ||
| TruSeq RNA Library Prep Kit v2 | Illumina | Cat# RS-122–2001 |
| Deposited data | ||
| Transcriptomic profiles obtained from 8-day old D. melanogaster male ejaculatory bulbs. | This paper | GEO: GSE185053 |
| The concatenated alignment and output tree files | This paper | 10.5281/zenodo.5238168 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: Canton-S | Laboratory of Sean B. Carroll Lab | N/A |
| D. melanogaster: Xout | Williams et al., 2008, Laboratory of Sean B. Carroll Lab | N/A |
| D. simulans: w 501 | National Drosophila Species Stock Center (Cornell University) | 14021–0251.195 |
| D. yakuba | National Drosophila Species Stock Center (Cornell University) | 14021–0261.00 |
| D. erecta | National Drosophila Species Stock Center (Cornell University) | 14021–0224.01 |
| D. ananassae | National Drosophila Species Stock Center (Cornell University) | 14024–0371.13 |
| D. pseudoobscura | National Drosophila Species Stock Center (Cornell University) | 14011–0121.94 |
| D. subobscura | National Drosophila Species Stock Center (Cornell University) | 14011–0131.05 |
| D. nebulosi | National Drosophila Species Stock Center (Cornell University) | 14030–0761.00 |
| D. sturtevanti | National Drosophila Species Stock Center (Cornell University) | 14043–0871.16 |
| D. willistoni | National Drosophila Species Stock Center (Cornell University) | 14030–0811.24 |
| D. immigrans | National Drosophila Species Stock Center (Cornell University) | 15111–1731.03 |
| D. nasuta | National Drosophila Species Stock Center (Cornell University) | 15112–1781.00 |
| D. Americana | National Drosophila Species Stock Center (Cornell University) | 15010–0951.00 |
| D. virilis | National Drosophila Species Stock Center (Cornell University) | 15010–1051.87 |
| D. mercatorum | National Drosophila Species Stock Center (Cornell University) | 15082–1521.38 |
| D. mojavensis | National Drosophila Species Stock Center (Cornell University) | 15081–1352.22 |
| S. latifasciaeformis | National Drosophila Species Stock Center (Cornell University) | 11030–0061.01 |
| S. lebanonensis | National Drosophila Species Stock Center (Cornell University) | 11010–0011.00 |
| S. rufifrons | National Drosophila Species Stock Center (Cornell University) | 11040–0071.00 |
| C. procnemis | National Drosophila Species Stock Center (Cornell University) | 20000–2631.01 |
| M. domestica | Josh’s Frogs (Owosso, MI) | B06XJ8DQ2X |
| D. melanogaster: Various GFP reporter constructs | This study | N/A |
| D. melanogaster: Various UAS-RNAi lines | Bloomington Drosophila Stock Center | See Table S2 |
| D. melanogaster: EB-GAL4 | Gift from Dr. Phillip Daborn (The University of Melbourne) | N/A |
| Oligonucleotides | ||
| Primers: Generation of GFP reporter constructs | This paper | See Table S3 |
| Primers: Generation of in situ hybridization probes | This paper | See Table S3 |
| Recombinant DNA | ||
| GFP reporter vector pS3aG | Williams et al., 2008, Laboratory of Sean B. Carroll Lab | N/A |
| Software and algorithms | ||
| MUSCLE | Edgar, 2004 | http://www.drive5.com/muscle |
| PhyML 3.0 | Guindon et al., 2010 | http://www.atgc-montpellier.fr/phyml/ |
| ‘Phytools’ package | Revell, 2013 | https://cran.r-project.org/web/packages/phytools/index.html |
| TopHat v2.0.9 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/index.shtml |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


