Abstract
With increasing complexity of expression studies and the repertoire of characterized sequences, combinatorial cloning has become a common necessity. Techniques like BioBricks and Golden Gate aim to standardize and speed up the process of cloning large constructs while enabling sharing of resources. The BioBricks format provides a simplified and flexible approach to endless assembly with a compact library and useful intermediates but is a slow process, joining only two parts in a cycle. Golden Gate improves upon the speed with use of Type IIS enzymes and joins several parts in a cycle but requires a larger library of parts and logistical inefficiencies scale up significantly in the multigene format. We present here a method that provides improvement over these techniques by combining their features. By using Type IIS enzymes in a format like BioBricks, we have enabled a faster and efficient assembly with reduced scarring, which performs at a similarly fast pace as Golden Gate, but significantly reduces library size and user input. Additionally, this method enables faster assembly of operon-style constructs, a feature requiring extensive workaround in Golden Gate. Our format allows such inclusions resulting in faster and more efficient assembly.
Keywords: BioBricks, Golden Gate, Type IIS enzymes, operon cloning, bacteria, chloroplast
1. Introduction
Synthetic biology, often starting with coding complex designs in the form of DNA constructs, aspires to make or modulate aspects of biology. With increasingly complex schemes in synthetic biology and the growing repertoire of characterized expression elements (commonly referred to as parts), advanced cloning methods frequently become essential for generating combinatorial or multigene constructs.
There are a variety of cloning approaches to choose from that enable speedy and specific assembly of multiple segments of DNA in a short time (1). Some of these strategies also aim to improve efficiency by enabling reutilization and sharing of the parts across the community, a vision that is supported by iGEM, BioBricks Foundation and several Biofoundries across the globe (2–4). A vetted library of parts is useful not only for quick turnaround times and low cost but also enables community science and labs in developing countries.
Amongst the approaches that enable parts to be reused, BioBricks and Golden Gate are two popular strategies for assembling short DNA fragments into a large construct, with many iterations published (1). The BioBricks cloning strategy was one of the initial attempts to bring standardization to traditional cloning methods (3). It defines a BioBrick part as a DNA sequence flanked by two specific restriction enzymes on each side, one of which produces a compatible overhang with the restriction site on the other end (5). This strategy enables the combining of two parts in the desired order and the product itself becomes a BioBrick part, hence enabling endless assembly with the addition of one part to the front or rear in each cycle. This makes it a fairly simple and flexible approach with a compact library where a sequence of interest (SOI) needs to be represented only once. Although this method provides the advantage of a standard design for endless and specific assembly, it has a few caveats. Notably, each joining event also produces an eight-base pair scar sequence from the disabled mixed restriction sites, which can be problematic for fusion protein assembly (6). Also, it is a relatively slower assembly method, restricted to the addition of one part in each cycle. Furthermore, the four restriction enzymes present on the flank cannot be present in the SOI itself, necessitating domestication of such sites if present.
The Golden Gate cloning strategy overcomes the speed limitations of BioBricks by using BsaI, a Type IIS restriction enzyme (7). Type IIS restriction enzymes cut outside the recognition sequence and provide the opportunity to choose the overhang sequence, with the advantage of making it non-palindromic. A non-palindromic sequence prevents self-ligation of the part and can greatly improve the efficiency of end-joining reaction, especially when combining many parts in one cycle. With these two features, Golden Gate strategy enables multi-part assembly, including fusion proteins, while significantly reducing scar sequences (Figure 1). An optimized choice of overhang sequences and ligation protocol has demonstrated the assembly of up to 24 parts in one cycle with greater than 90% accuracy enabling rapid assembly of large constructs (8). However, this strategy does not allow addition of more parts to the final construct, i.e. endless assembly.
Figure 1.

Advantage of Type IIS ligation: sticky ends of Type IIS enzymes can be designed to be non-palindromic, which prevents the issue of self-ligation responsible for several unintended byproducts.
To enable extension of assembly, maintain universal compatibility of parts and sharing across labs, a standard for overhang sequence was formulated for Golden Gate, named MoClo, with the addition of one more Type IIS enzyme BpiI (9, 10). In the MoClo standard, a level 0 part is a basic unit or plasmid that hosts a SOI like a promoter, UTR, CDS, etc. Such level 0 parts are combined in level 1 assembly, which can take one part of any number of compatible types (11, 12). This assembly frequently represents one transcriptional unit (TU). The level 1 assembly host also determines the position of the TU in the next round of super assembly of multiple level 1 TUs into a level 2 multigene construct. This approach has a few limitations. Level 1 assemblies cannot have multiple parts of the same type and hence disallow operons out of the box. Second, level 1 parts are preset for their order in level 2 assembly and this order cannot be changed, limiting their reuse or sharing. Finally, it necessitates multiple level 0 parts of the same element for each type of fusion partner. This problem and the previous issue combined usually result in a much larger library of parts and intermediate plasmids that are not very useful. The GoldenBraid 2.0 standard was introduced for Golden Gate assembly to reduce the problem of fixed intermediates in MoClo; however, it requires additional cycles of cloning and three restriction enzymes, which leads to a higher domestication load (13).
There are also several polymerase-based methods, most prominently Gibson assembly and its variations (1, 14). These techniques, however, do not generate parts that are easily reusable and have significant one-time use investment of resources. They often require resource investment in designing and ordering specific primers and subsequently validating constructs for the absence of PCR-based errors. Additionally, most PCR-based methods have issues with repetitive sequences, very small sequences, sequences with strong secondary structures and often non-specific amplification. Some of these issues are frequently encountered in regulatory sequences.
With advances in synthetic biology, many regulatory elements are characterized, and the repertoire of SOIs is constantly increasing. Standardized protocols and several efforts to contribute such validated parts hold promise for improving speed while cutting down significantly on the costs making such experiments accessible for a larger community (15–18). Our interest in improving multi-gene cloning is derived from our effort to engineer chloroplasts, which are lucrative sites for synthetic biology, both for transgene expression and metabolic tweaking, and are now targets for expression of large pathways (19–23). Current techniques like BioBricks fall short because of their slow speed when it comes to assembling a large number of genes. Golden Gate’s speed is best utilized when making one transcription unit, but not an operon, from several fragments. In a multigene and combinatorial scenario, it quickly becomes logistically complex. Considering the limited variety of well-defined chloroplast regulatory elements across plant species, researchers will frequently have to resort to optimizing their constructs in operon formats and combinatorial trials for such endeavors. Next-generation efforts to improve or add novel metabolite pathways linked to food security or biotech applications require increasingly complex expression systems.
Here, we are presenting a strategy, GoldBricks, that strikes a balance between simplicity and flexibility of BioBricks and the speed of Golden Gate approaches by including a Type IIS enzyme in the Biobrick. This strategy greatly improves upon the speed of BioBricks and at the same time has advantages over Golden Gate by enabling operon-based constructs with reduced library size and user input. While this cloning method is an improved approach to do multigene and combinatorial cloning in most common practical scenarios, we believe it would be of exceptional interest in the engineering not only of chloroplast or bacterial operons, but would also be useful for constructing multi-gene assemblies.
2. Materials and methods
2.1. Bacterial strains, growth conditions and reagents
E. coli DH5α cells were used for cloning and transformation. Cells were grown in Luria-Bertani medium at 37°C with ampicillin 100 µg mL−1 or kanamycin 50 µg mL−1 for selection. All the fast digest restriction enzymes and Fast alkaline phosphatase (Fast AP cat. #EF0652) used in this study were purchased from ThermoFisher. The molecular weight marker used in gels is Generuler 1 kb plus from ThermoFisher. PCR amplifications were performed using NEB Q5 High fidelity 2× Master mix with the primers listed in the supplementary Genbank files. Plasmid preparations were performed using the Macherey-Nagel Nucleospin plasmid kit. For DNA or PCR product purification, Macherey-Nagel Nucleospin Gel and PCR Clean-up kit were used. For ligation cloning, T4 ligase from Invitrogen was used. All reagents were used according to the manufacturer’s protocol. For three-part and five-part cloning, parts were added in approximately equimolar ratios including the destination vector. We have found that an equal amount of plasmids digested provides a good approximation for part sizes smaller than 1 kb.
2.2. Construction of parts and assembly
Parts were picked from our GoldBricks library that demonstrated a good range of sizes and had useful restriction sites to visually demonstrate the order of assembly by restriction digestion. These parts belong to our catalog for carboxysome expression in chloroplasts and consist of chloroplast regulatory elements and bacterial coding sequences (CDSs). The full sequence and primers used are provided as Genbank files in supplementary data. Tailed PCR primers were used to generate part amplicons with specific restrictions sites. These amplicons were purified and digested with NotI and MauBI and cloned into pStart-III to make the holding vector of the part. For the three-parts demonstration of assembly, parts were excised from the holding vectors using restriction enzymes marked in a dashed box in Figure 2 and ligated to pStart-IV linearized with NotI and MauBI without part purification. Destination vector was column purified after digestion to remove short DNA sequences. For the five-part assembly, parts were excised using the restriction enzymes marked in Figure 4A. For parts with one end phosphorylated, reactions were performed sequentially. First, the end to be dephosphorylated was treated with a corresponding restriction enzyme and Fast AP. After incubation, the enzyme mix was heat-inactivated. In the same mix, the second restriction enzyme was added and incubated. For parts with both ends dephosphorylated, both enzymes and Fast AP were added simultaneously. Digested parts were used without purification and used in a ligation-based assembly in equimolar concentration. The ligation mix was transformed into high-efficiency chemically competent DH5α cells (NEB catalog #C2987H) and Luria-Bertani agar plates with kanamycin (50 µg mL−1) were used for selection.
Figure 2.

A basic scheme for defining a GoldBrick part and a three-part assembly: (A) Here each part has a four-enzyme flank and part is typified by the overhang sequence of the Type IIS site. Each part has specific compatibility for the suffix and the prefix part. For an example of the three-part assembly, the parts can be excised using sites marked in the box and subsequently ligated to give a strictly ordered product. If the destination vector has different antibiotic marker, parts do not need to be purified for assembly. (B) Sequence format to show directionality of Type IIS enzymes.
Figure 4.

Demonstration of the five-part assembly: (A) Schematic for two assemblies 5P-A and 5P-B is shown. Each part was cut in its holding vector using the enzyme shown in the figure (N = NotI, Ec = Eco31I, Es = Esp3I and M = MauBI). Part ends that were dephosphorylated are marked by *. These five parts were then ligated into pStart-IV and selected on kanamycin. (B) Six colonies that were PCR positive for inserts were randomly picked and DNA prepared. These were validated for correct assembly using restriction digestion according to the schematic. All colonies showed the correct insertion. Specific fragment sizes are marked in blue.
2.3. Validation of assembly
For three-part assembly, eight colonies from the selection plate were randomly picked, cultured and plasmids were extracted. Plasmids obtained were digested using PagI, ScaI and MauBI and run on agarose gel electrophoresis for analysis. For five-part assembly, colonies from the selection plates were checked by colony PCR using NEB OneTaq quick load 2× master mix and manufacturer’s recommendation. The primer pair that was used flanked the NotI and MauBI sites. PCR positive colonies with appropriate size were further verified by restriction digestion to ensure components and their orientation.
3. Results and discussion
3.1. GoldBricks merges BioBricks and Golden Gate
The GoldBrick strategy uses a four-restriction enzyme scheme for defining a part. Like BioBricks there are two restriction sites on each side, where the inner flank is made up of Type IIS sites and the outer flank is made of Type II sites. Unlike BioBricks, where there are no part types, GoldBricks typifies parts based on overhangs and only compatible parts can be ligated. The use of Type IIS restriction enzymes provides an opportunity to use a non-palindromic overhang sequence, which prevents self-ligation of parts and improves efficiency and fidelity of end-joining reaction (Figure 1). This allows more parts to be included in the assembly. Our approach aims to reduce library size by avoiding redundant cloning of a SOI into different part types by using common overhangs whenever possible (Supplementary Figure S1A).
The required specificity in such a scenario is provided by Type II sites and occasionally by end-specific phosphorylation, which is possible by the use of two Type IIS enzymes. Furthermore, specificity of the order of parts and endless extensions are provided by the outer flank of Type II enzymes. BioBricks assembly also produces 8 bp scars at junctions, which are unsuitable for certain purposes such as fusion proteins or when sequence sanctity is mandatory. Our strategy reduces scar sequences and is on par with Golden Gate. Further, our method avoids the use of linkers, and associated scars, for inserting parts in most scenarios due to greater cross-compatibility of parts.
3.2. Format of GoldBricks
An entry vector with a part cloned in it forms the holding vector that is the basic unit of the library or collection of parts. The entry vector, pStart-III, is defined by NotI and MauBI sites and these two sites are the external flanking sites in all the part types (Figure 2A). Each part is flanked on the inner side by the two Type IIS enzymes, Esp3I and Eco31I, which define the compatibility of ligation of each part. Type IIS enzymes are oriented to cut towards the SOI (Figure 2B). A mix of universal overhangs and compatible overhangs define the order of ligation. For example, a promoter with a 5ʹUTR will have a Type IIS Eco31I site in its suffix that will generate a 5ʹTTAC overhang for complementarity to AATG start codon. To match, a CDS part will have a Esp3I site in the prefix generating a 5ʹAATG overhang. The CDS part’s Eco31I site in suffix produces 5ʹAAGC, which is complementary to the Terminator prefix Esp3I overhang, and so on. The full list of overhang compatibility of parts is listed in Supplementary Figure S1A. Some less common part types (e.g. N-tag or C-tag) have overhangs that can oligomerize; however, this potential problem can be easily remedied by reducing the number of parts in a ligation cycle or by end specific phosphorylation.
3.3. Three-part assembly
Parts are excised from the holding vectors using a combination of two enzymes. As shown in Figure 2, for an assembly of three parts, boxed sites are used to eject each part. Ligation has only one possible transformable outcome and hence a high-fidelity assembly is achieved. This scheme further reduces user effort by following optional recommendations. First, selection marker can be shuttled between holding vectors and destination vectors. When parts held in pStart-III with the ampicillin marker (blue backbone in Figure 2) are assembled in the destination vector of a different selection marker, pStart-IV with a kanamycin marker (red backbone in Figure 2), the user can avoid purification of parts for ligation. This is especially helpful with parts that are of very small size (<200 bp). Alternatively, when antibiotic shuttling is not possible due to mixed level 0 and level 1 parts, the backbone can be degraded by MbiI or other blunt cutters. Second, all restriction enzymes are heat-inactivable and heat-inactivated reaction mixtures are directly used in the ligation reaction, simplifying and reducing user effort.
For the demonstration of three-part assembly, two 5ʹUTR parts and a CDS were ligated according to the scheme (Figure 3A), a combination not possible in Golden Gate or BioBricks in one cycle. This reaction used antibiotic shuttling and hence purification of digested parts was not necessary. From the antibiotic selection plate, eight colonies were randomly inoculated and DNA isolated and digested to verify insertion. All the colonies showed the restriction pattern expected of correct assembly (Figure 3B). In our experience, this assembly has matched theoretical expectations and yields almost all colonies with valid constructs. As the resultant product is also a GoldBrick part, it can be utilized further for higher-level or multigene assembly by extending on either side, unlike MoClo. As MoClo level 1 assembly is defined for order and number of partners in the next assembly, it is not useful as a part but only as an intermediate. GoldBricks’ intermediate assembly is as flexible and hence useful as the parts that made them, a feature akin to BioBricks.
Figure 3.
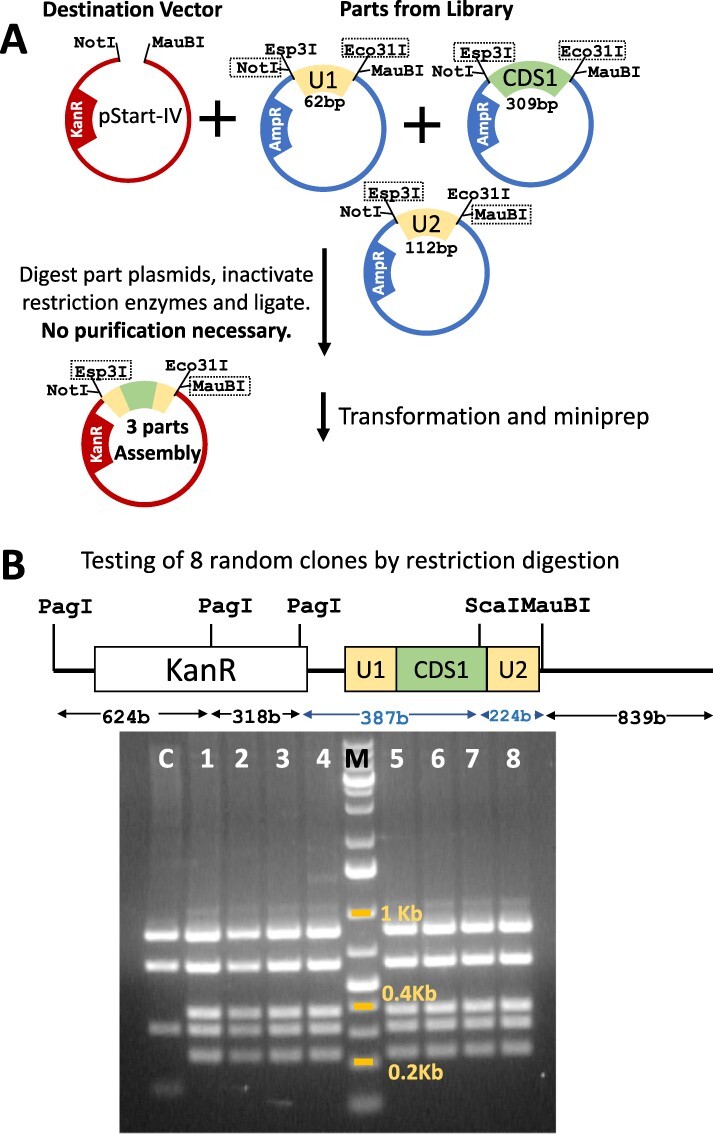
Demonstration of the three-part assembly: (A) A three-part assembly was performed using parts U1, CDS1 and U2 that were 62 bp, 309 bp and 112 bp long, respectively. After transformation, eight colonies were selected randomly and DNA was prepared. (B) DNA was digested using PagI, ScaI and MauBI, which would give bands of the mentioned sizes from successful assemblies. The agarose gel of the digested DNA of empty pStart-IV (lane C) and eight randomly selected colonies (lanes 1–8) demonstrates that all clones harbored the correct assembly. Specific fragment sizes are marked in blue.
3.4. Five-part assembly
A modified approach allows the assembly of a greater number of parts while including parts of the same type in one assembly. As parts are excised by using two different restriction enzymes, it is possible to specifically remove the phosphate group from one end. All the restriction enzymes and alkaline phosphatase use a common reaction buffer and can be heat-inactivated. Such digestion and phosphatase reactions can be achieved within a tube without the need for additional purification. The scheme for specific assembly of five parts is shown in Figure 4A, which marks the restriction enzyme used to eject parts from holding vectors and phosphatase-treated ends. Two parts require end-specific phosphorylation. Such a scenario will have four legitimate Type IIS ligations and two illegitimate ligations. The illegitimate ligations, where #3 prefixes #2 or #4 prefixes #3, result in untransformable outcomes or require at least seven parts to circularize, making the likelihood extremely rare over five-part assembly. Resultant colonies from the ligation were screened by colony PCR for insert sizes. Colonies with inserts almost always had the correct size (Supplementary Figure S2). Plasmids were isolated from the PCR-positive colonies and were further checked by restriction digestion for order of the assembly, which is always found to be correct (Figure 4B).
3.5. Further manipulations
As these assemblies preserve the terminal restriction sites, it is possible to extend the construct on either side. We combined three transcription units in a cycle using the scheme shown in Figure 5A. This reaction imposes a strict ligation condition. The digested parts were used without purification in the ligation reaction to extend 5P-A assembly. Since the 5P-B backbone also had the kanamycin resistance, it was degraded using MbiI. MbiI is a blunt end cutter that cuts at two sites and hence there is a very low probability of recircularizing. Six colonies were randomly selected without screening, and DNA was prepared and tested by restriction digestion (Figure 5B). Similar to three-part assembly, all were found to be correct assemblies. A few common scenarios and extensions of intermediate assemblies are described in Supplementary Figure S3. Transcription units can be fused in any desired order. This also allows parallel assembly of parts, which can be fused in a second round for reduced time of assembly. Even though parts are typified, a GoldBrick part can bind to more part types than Golden Gate (Supplementary Figure S1); for example, the CDS can be prefixed by a 5ʹUTR or an N-tag and can be suffixed by a C-tag or a 3ʹUTR/Terminator. For certain non-standard scenarios, the cloning can still be achieved by using compatible restriction schemes and without linkers or recloning of SOI, for example, a fusion protein made of several pieces (Supplementary Figure S3F). Such scenarios are, however, presumed to be rare and can be easily accommodated most often without making new parts.
Figure 5.

Assembly extension: Level 1 assemblies 5P-A, 5P-B and a tobramycin resistance cassette were combined using the restriction enzyme scheme shown in (A). Sites marked with ‘*’ were dephosphorylated. Six random clones of Super Assembly A (SA-A) were checked using restriction digestion according to schematic (B) and all were found to have correct assemblies. Specific fragment sizes are marked in blue.
3.6. Comparison of cloning methods
We have demonstrated that a relatively simple and BioBrick-like scheme can effectively ligate multiple parts. Our method aims to ligate up to five parts in a cycle, which we believe is good enough to form a TU or more under most common scenarios. We forfeit the ability to ligate a large number of parts in one cycle to allow compaction of the library, improve the flexibility of intermediates and reduce user effort. We would like to draw a comparison using hypothetical scenarios and show how our technique occupies an optimal position for most practical aspects. First, consider the library size requirement to construct multigene or combinatorial constructs. BioBricks does not typify parts and hence each SOI needs to be cloned only once. This keeps the cost and size of a library at a minimum. As shown in Supplementary Figure S1, GoldBricks combines the part definitions to reduce the number of part types compared to the MoClo standard (9). A MoClo scheme of 15 part types can be reduced to seven types in GoldBricks format, reducing redundant cloning of SOIs. Additionally, compared to the MoClo specification of nine overhang sequences, GoldBricks specifies only three overhangs, showing the increased cross-compatibility of parts. An additional layer of traditional enzymes and capping the maximum number of parts in a reaction to five provides markedly improved flexibility and reduced library size. For a hypothetical scenario of combinatorial cloning of four constructs using six SOIs, BioBrick will need six parts, GoldBricks will need eight parts and MoClo will need 12 parts (Supplementary Figure S4A). GoldBricks reduces the recloning of SOI in multiple types compared to MoClo, which in turn reduces the logistical requirements and complexity of the library of parts. GoldBricks enables operon-style constructs or concatemerization of parts (e.g. multimeric fusion proteins) without requiring linkers or unnecessary recloning of SOI.
Second, we need to consider the speed of the cloning in an operon format. We compare a hypothetical scenario of constructing a four-gene operon using GoldBricks and MoClo in Supplementary Figure S4B. As MoClo does not allow two parts of the same type in a cycle, such an assembly will need to be split into four level 1 assemblies, which are then combined in the next cycle to get the final construct (9, 11). On the other hand, GoldBricks is limited only by the number of parts in a cycle and similar constructs are achieved in the same amount of time but with less user input. Additionally, the GoldBricks intermediates have higher flexibility for reuse and do not have adapter/linker scars. BioBricks will take more time and more user input than either technique.
Third, we should evaluate the speed of cloning in the non-operon format. We compare a hypothetical scenario of combinatorial assembly of seven genes in form of seven TUs, where each TU is made of four parts (Supplementary Figure S4C). This represents a common cloning scenario for multigene nuclear expression constructs. In this scenario, MoClo is the quickest at accomplishing the given configuration in 4 days. GoldBricks takes 6 days and BioBricks takes 10 days. However, GoldBricks requires the least number of reactions to be set up, 10 vs. 50 for MoClo and vs. 27 in BioBricks. The intermediates generated here are capable of giving any combination of seven genes in any order required. The number of intermediates is only seven (1 per TU) for BioBricks and GoldBricks but is very high at 49 (7 per TU) for Golden Gate. Hence, GoldBricks presents a path of least effort and complexity at a slightly elevated requirement of time by generating more versatile intermediates. GoldenBraid 2.0, MOBIUS and LOOP methods have tried to address the inflexibility of level 1 assembly to organize into higher assembly by introducing variations in the collection of level 1 holding plasmids (13, 24, 25). GoldenBraid 2.0 increases the reusability of level 1 assembly at cost of speed and increased domestication load (Table 1). GoldenBraid drops down to combining two TUs in a cycle and with the requirement of occasional swapping of host-vector, the average is lower than 2 per cycle. MOBIUS ligates four TUs, partly relieves the domestication issue of GoldenBraid and adds visual indicators for tracking cloning, but it brings back rigid intermediates, linking sequence scars and speeds slower than MoClo. LOOP also uses a high number of intermediates and its advantages become apparent only when combining several TUs. While these methods provide tradeoffs suitable for certain applications, for these iterations to be backward compatible, they have not resolved the issues inherent to the Golden Gate scheme, primarily lack of out-of-the-box compatibility for operons and a larger library due to repetitive cloning of SOI. Our method does not need specific level 1 plasmids for organizing higher assemblies. Any vector with NotI/MauBI site can host at any stage.
Table 1.
Domestication requirement of GoldBricks and comparison with other techniques
| Domestication requirements of the restriction enzyme scheme | ||
|---|---|---|
| Goldbricks restriction enzymes and site length | ||
| Enzyme | Type | Restriction site length |
| NotI | Type II | 8 bp |
| MauBI | Type II | 8 bp |
| Esp3I | Type II-S | 6 bp |
| Eco31I | Type II-S | 6 bp |
| Comparison of domestication requirement in sample genomes | ||
| Strategy | Frequency of restriction sites in NtCp genome (150 kb) | Frequency of restriction sites in E.coli genome |
| BioBricks | 208 | 1448 |
| GoldBricks | 53 | 1581 |
| GoldenBraid 2.0 | 75 (100 for MoClo) | 6798 (3136 for MoClo) |
| Golden Gatea | 29 | 261 |
Of the four cloning methods listed here, Golden Gate cloning is not an endless cloning method.
NtCp, Nicotiana tabacum chloroplast.
GoldBricks uses a scheme of four enzymes, similar to BioBricks. However, the choice of enzymes does not cause a substantial change in the domestication requirement when compared with BioBricks and is significantly lower than Golden Gate versions that enable endless assembly (Table 1).
In summary, we present here a cloning strategy that is a hybrid approach of BioBricks and Golden Gate and brings in their best attributes. The GoldBricks strategy significantly improves the assembly speed compared to BioBricks. Like BioBricks, our strategy preserves the flexibility and idempotent nature of assemblies constructed. It also allows multiple parts of the same type in an assembly, hence significantly reducing the library size for constructing multigene operons compared to Golden Gate. By allowing a flexible partner choice, the parts and intermediates generated are more versatile leading to reduced library size and user input. These hybrid advantages are achieved by sacrificing the type-less structure of BioBricks and a very high number of unique part types that can be assembled in Golden Gate. Advantages gained by losing these features outweigh the disadvantages in most circumstances for general cloning and are especially useful for making multigene operons. Multigene operons are useful strategies for expressing a large number of genes in a compact format, especially for plastid gene expression studies, where the repertoire of well-characterized regulatory elements is small and reusing the same elements multiple times is prohibited by homologous recombination.
The major caveat of our strategy is that it is not backward compatible with the existing library of parts that are designed for BioBricks or Golden Gate, which is a common problem found amongst earlier iterations of those methods as well. Nevertheless, we believe that the approach we present strikes a balance between speed and flexibility in cloning, especially for multi-gene operon kind of constructs.
Supplementary Material
Acknowledgments
The authors are thankful to Dr. Myat Lin for his comments on the manuscript. V.C. would also like to acknowledge and thank his wife Vandana and newborn Avi as he borrowed time from them to revise this work. Their patience is deeply appreciated.
Contributor Information
Vishalsingh R Chaudhari, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY, USA.
Maureen R Hanson, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY, USA.
Supplementary data
Supplementary data are available at SYNBIO Online.
Data availability
DNA sequences for all the vectors, parts, primers and assemblies used in the paper are available at SYNBio online.
Material availability
Unique materials presented in the manuscript will be available from the authors upon reasonable request and through a materials transfer agreement.
Funding
National Science Foundation, USA [Systems and Synthetic Biology award MCB-1642386 to M.R.H.].
Conflict of interest statement.
None declared.
References
- 1. Chao R., Yuan Y. and Zhao H. (2015) Recent advances in DNA assembly technologies. FEMS Yeast Res., 15, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casini A., Storch M., Baldwin G.S. and Ellis T. (2015) Bricks and blueprints: methods and standards for DNA assembly. Nat. Rev. Mol. Cell Biol., 16, 568–576. [DOI] [PubMed] [Google Scholar]
- 3. Smolke C.D. (2009) Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol., 27, 1099–1102. [DOI] [PubMed] [Google Scholar]
- 4. Hillson N., Caddick M., Cai Y., Carrasco J.A., Chang M.W., Curach N.C., Bell D.J., Le Feuvre R., Friedman D.C., Fu X.. et al. (2019) Building a global alliance of biofoundries. Nat. Commun., 10, 2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knight T. (2007) The BioBricks Foundation: BBFRFC10. http://dspace.mit.edu/handle/1721.1/45138 (10 January 2021, date last accessed).
- 6. Cai Y., Wilson M.L. and Peccoud J. (2010) GenoCAD for iGEM: a grammatical approach to the design of standard-compliant constructs. Nucleic Acids Res., 38, 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engler C., Gruetzner R., Kandzia R., Marillonnet S. and Peccoud J. (2009) Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoSOne, 4, e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Potapov V., Ong J.L., Kucera R.B., Langhorst B.W., Bilotti K., Pryor J.M., Cantor E.J., Canton B., Knight T.F., Evans T.C.. et al. (2018) Comprehensive profiling of four base overhang ligation fidelity by T4 DNA ligase and application to DNA assembly. ACS Synth. Biol., 7, 2665–2674. [DOI] [PubMed] [Google Scholar]
- 9. Marillonnet S. and Werner S. (2020) Assembly of multigene constructs using the modular cloning system MoClo. Methods Mol. Biol., 2205, 125–141. [DOI] [PubMed] [Google Scholar]
- 10. Werner S., Engler C., Weber E., Gruetzner R. and Marillonnet S. (2012) Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs, 3, 38–43. [DOI] [PubMed] [Google Scholar]
- 11. Grutzner R. and Marillonnet S. (2020) Generation of MoClo standard parts using Golden Gate cloning. Methods Mol. Biol., 2205, 107–123. [DOI] [PubMed] [Google Scholar]
- 12. Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. and Peccoud J. (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS One, 6, e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarrion-Perdigones A., Vazquez-Vilar M., Palaci J., Castelijns B., Forment J., Ziarsolo P., Blanca J., Granell A. and Orzaez D. (2013) GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol., 162, 1618–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. and Smith H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- 15. Vafaee Y., Staniek A., Mancheno-Solano M. and Warzecha H. (2014) A modular cloning toolbox for the generation of chloroplast transformation vectors. PLoS One, 9, e110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lampropoulos A., Sutikovic Z., Wenzl C., Maegele I., Lohmann J.U. and Forner J. (2013) GreenGate—a novel,versatile,and efficient cloning system for plant transgenesis. PLoS One, 8, e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Engler C., Youles M., Gruetzner R., Ehnert T.-M., Werner S., Jones J.D.G., Patron N.J. and Marillonnet S. (2014) A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol., 3, 839–843. [DOI] [PubMed] [Google Scholar]
- 18. Occhialini A., Piatek A.A., Pfotenhauer A.C., Frazier T.P., Stewart C.N. and Lenaghan S.C. (2019) MoChlo: a versatile, modular cloning toolbox for chloroplast biotechnology. Plant Physiol., 179, 943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bock R. (2015) Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol., 66, 211–241. [DOI] [PubMed] [Google Scholar]
- 20. Lin M.T., Occhialini A., Andralojc P.J., Devonshire J., Hines K.M., Parry M.A. and Hanson M.R. (2014) Beta-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J., 79, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanson M.R., Lin M.T., Carmo‐Silva A.E. and Parry M.A.J. (2016) Towards engineering carboxysomes into C3 plants. Plant J., 87, 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenblueth M., Ormeño-Orrillo E., López-López A., Rogel M.A., Reyes-Hernández B.J., Martínez-Romero J.C., Reddy P.M. and Martínez-Romero E. (2018) Nitrogen fixation in cereals. Front. Microbiol., 9, 1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ivleva N.B., Groat J., Staub J.M., Stephens M. and Kumar S. (2016) Expression of active subunit of nitrogenase via integration into plant organelle genome. PLoSOne, 11, e0160951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andreou A.I., Nakayama N. and Isalan M. (2018) Mobius Assembly: a versatile Golden-Gate framework towards universal DNA assembly. PLoS One, 13, e0189892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pollak B., Cerda A., Delmans M., Álamos S., Moyano T., West A., Gutiérrez R.A., Patron N.J., Federici F. and Haseloff J. (2019) Loop assembly: a simple and open system for recursive fabrication of DNA circuits. New Phytol., 222, 628–640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences for all the vectors, parts, primers and assemblies used in the paper are available at SYNBio online.


