Abstract
The expression of the replication-dependent histone mRNAs is tightly regulated during the cell cycle. As cells progress from G1 to S phase, histone mRNA levels increase 35-fold, and they decrease again during G2 phase. Replication-dependent histone mRNAs are the only metazoan mRNAs that lack polyadenylated tails, ending instead in a conserved stem-loop. Much of the cell cycle regulation is posttranscriptional and is mediated by the 3′ stem-loop. A 31-kDa stem-loop binding protein (SLBP) binds the 3′ end of histone mRNA. The SLBP is necessary for pre-mRNA processing and accompanies the histone mRNA to the cytoplasm, where it is a component of the histone messenger RNP. We used synchronous CHO cells selected by mitotic shakeoff and HeLa cells synchronized at the G1/S or the M/G1 boundary to study the regulation of SLBP during the cell cycle. In each system the amount of SLBP is regulated during the cell cycle, increasing 10- to 20-fold in the late G1 and then decreasing in the S/G2 border. SLBP mRNA levels are constant during the cell cycle. SLBP is regulated at the level of translation as cells progress from G1 to S phase, and the protein is rapidly degraded as they progress into G2. Regulation of SLBP may account for the posttranscriptional component of the cell cycle regulation of histone mRNA.
The replication-dependent histone mRNAs are tightly regulated during the cell cycle, increasing 35-fold as cells progress from G1 to S phase (18). There is only a three- to five-fold increase in the rate of transcription of the histone genes (7, 19), indicating that much of the regulation is posttranscriptional. The posttranscriptional component of cell cycle regulation is mediated by the 3′ end of the histone mRNA (32, 35), which is a highly conserved stem-loop (34). The only processing step necessary for formation of the mature histone mRNA is an endonucleolytic cleavage to form the 3′ end (14). Cleavage is directed by two cis-acting elements, the stem-loop which binds to the hairpin binding factor (37, 38), and a purine-rich sequence about 10 nucleotides (nt) 3′ of the cleavage site that binds the 5′ end of U7 snRNA (3, 39, 55). Hairpin binding factor is composed of a single 31-kDa protein, the stem-loop binding protein (SLBP) (10, 60) or hairpin binding protein (33), which is required for processing in vivo (44) and which remains with the mature mRNA as a component of the cytoplasmic messenger RNP (mRNP) (9, 17). At least one additional factor, a heat-labile factor, is required for processing, but this factor has not been well defined biochemically (15).
Two posttranscriptional regulatory steps contribute to the cell cycle regulation of histone mRNA concentrations. Processing is regulated as cells progress from G1 to S phase, and the half-life of histone mRNA is reduced to about 10 min at the end of S phase (18), resulting in the destruction of histone mRNA prior to mitosis. We (60) and others (33) recently cloned the cDNA for the SLBP from several species using the yeast three-hybrid system (50). Here we show that SLBP, a major trans-acting factor necessary for histone pre-mRNA processing, is regulated during the cell cycle by both posttranscriptional and posttranslational processes. Regulation of SLBP is likely a critical component of the cell cycle regulation of histone mRNA.
MATERIALS AND METHODS
Cell culture and synchronization.
CHO cells were grown in McCoy's 5A medium with 10% calf serum and Pen-Strep as previously described (18). To select synchronous cells, an automated shakeoff device capable of simultaneously shaking 12 T75 flasks was constructed (11). Mitotic cells were collected by shaking every 10 min and stored at 4°C. Cells collected over a 4-h period were pooled and plated (0.5 × 106 to 1 × 106 cells per 10-mm plate). The number of cells in S phase was assessed by labeling cells with 5′-bromo-2′-deoxyuridine (BrdU) using the In Situ Cell Proliferation Kit (Boehringer Mannheim, Indianapolis, Ind.). Nuclei that had incorporated BrdU into DNA during the labeling period were visualized by staining with FastRed (Boehringer Mannheim).
To obtain populations of cells in G2 phase, HeLa cells were arrested by double thymidine block. Cells were blocked for 18 h with 2 mM thymidine, released for 9 h by washing out the thymidine, and then blocked again with 2 mM thymidine for 17 h to arrest all the cells at the beginning of S phase. The cells were released by washing out the thymidine, and lysates were prepared each hour after release from thymidine. The cells progressed through G2 and mitosis synchronously. HeLa cells were also synchronized by arresting them at the G1-S border with 2 mM thymidine for 18 h, followed by a 3-h release, and then arrested at mitosis with 100 ng of nocodazole per ml for 12 h. The cells were released from the nocodazole block with two washes of fresh medium and allowed to progress through G1 and into S phase. Any cells that had not attached 6 h after release from nocodazole were discarded. The cell cycle position of the cells was determined by propidium iodide staining and flow cytometry at the University of North Carolina Flow Cytometry facility.
Preparation of cell lysates and detection of SLBP and cyclin proteins.
CHO cells were collected, washed once in phosphate-buffered saline (PBS), and lysed by incubation for 10 min in NP-40 lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% NP-40, 1 mM dithiothreitol). The insoluble material was pelleted by centrifugation at 16,000 × g in a microcentrifuge. The HeLa cells were lysed the same way or in some experiments were lysed in 1% sodium dodecyl sulfate (SDS) and boiled for 5 min. Western blots were performed by standard protocols (60). Typically, 25 μg of total cell protein was resolved on an SDS–10% polyacrylamide gel and transferred to nitrocellulose. SLBP was detected with antibodies raised to the C terminus of the mouse SLBP (60). Mobility shift assays were performed with whole-cell lysates as previously described (36, 43). Antibodies to human cyclin A and cyclin B raised against glutathione-S-transferase fusion proteins were a gift from Yue Xiong (61).
Preparation of RNA and analysis of histone mRNA and SLBP mRNA.
Total cell RNA was prepared with the Ultraspec RNA isolation system (Biotecx Laboratories, Inc., Houston, Tex.). Concentrations of several mRNAs were determined by Northern blotting. Briefly, 10 μg of total RNA was denatured in 50% formamide plus 2.2 M formaldehyde, resolved on a 1% agarose–formaldehyde gel, transferred to a nylon membrane (Hybond; Amersham Pharmacia Biotech, Piscataway, N.J.) via capillary action, and cross-linked using a Stratalinker (Stratagene, La Jolla, Calif.). The blots were probed with either the mouse SLBP cDNA (NcoI-SpeI fragment), the rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA, or the mouse histone H3.2-614 gene. The appropriate restriction fragments were purified by agarose gel electrophoresis and labeled by random priming in the presence of [α-32P]dCTP. Blots were hybridized in Quikhyb (Stratagene) for 4 h at 55°C. U7 snRNA Northern blots were performed on total RNA resolved by electrophoresis on a 10% polyacrylamide–7 M urea gel using a riboprobe complementary to the mouse U7 snRNA (10). Where indicated, the hamster histone H3 mRNA was mapped in an S1 nuclease assay with the mouse H3.2-614 gene, labeled at the SalI site (16, 18).
Labeling of cells and analysis of SLBP synthesis rate.
CHO cells were labeled in Dulbecco's modified Eagle's medium (DMEM) without methionine and cysteine (ICN Pharmaceuticals, Costa Mesa, Calif.) supplemented with 10% dialyzed fetal bovine serum (Gibco-BRL, Gaithersburg, Md.) (24). For each point, a separate dish (25 mm) containing 106 cells was used. Cells were preincubated in DMEM without methionine and cysteine for 1 h prior to labeling to deplete the intracellular stores of methionine and cysteine. The cells were labeled with 1 mCi of [35S]methionine-cysteine (NEN Life Science Products, Boston, Mass.) in 2 ml of medium for 15 min. The label was removed, and the cells were washed in 1× PBS and lysed in NP-40 lysis buffer. Equal numbers of trichloroacetic acid-A precipitable counts (5 × 106 cpm) were used for each immunoprecipitation. Lysates were precleared with protein A-agarose beads and then incubated with affinity-purified anti-SLBP for 1 h. The SLBP-antibody complex was recovered by binding to protein A-agarose beads, and the beads were washed extensively with NP-40 lysis buffer. SLBP was eluted by heating the samples in SDS loading buffer and resolved by gel electrophoresis. The amount of labeled SLBP was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Analysis of distribution of mRNAs in polyribosomes.
HeLa cells were synchronized as described above, and polyribosomes were analyzed by a modification of published methods (5, 25). Just prior to harvesting, the cells were treated with cycloheximide (0.1 mM) to freeze the mRNAs on polyribosomes. The cells were washed in PBS and then lysed by gentle shaking in 0.5% NP-40–0.1 M NaCl–10 mM MgCl2–2 mM dithiothreitol–50 mM Tris-HCl (pH 7.5) containing 200 U of RNasin, 100 μg of cycloheximide, 200 μg of heparan, and 10 μl of protease inhibitor cocktail (catalog no. P-8340; Sigma Chemical, St. Louis, Mo.) per ml. The nuclei and membranes were removed by centrifugation at 10,000 × g for 10 min. The lysates were layered on 9-ml 15 to 40% (wt/vol) sucrose gradients in 0.15 M NaCl–5 mM MgCl2–25 mM Tris-HCl (pH 7.5) and centrifuged for 140 min at 35,000 rpm in an SW41 rotor. The gradients were fractionated with a Densiflow apparatus (Buchler Instruments, Lawrence, Kans.), and 0.6-ml fractions were collected. RNA was prepared from each fraction using TRIzol (Gibco-BRL). The RNA was resolved by agarose gel electrophoresis and detected by staining with ethidium bromide. A second aliquot of RNA from each fraction was treated with formaldehyde, resolved by agarose gel electrophoresis, transferred to nitrocellulose, and analyzed for SLBP and GAPDH mRNA as described above.
Chemicals.
Hydroxyurea and N-acetyl-leucyl-leucyl-methioninal (LLM) were obtained from Sigma. Aphidicolin and carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (Mg132) were obtained from Calbiochem.
RESULTS
Histone mRNAs are tightly regulated during the cell cycle at multiple steps. Much of the regulation is due to the 3′ end of histone mRNA (18, 56). The protein which binds the 3′ end of histone mRNA, SLBP, participates in many steps of histone mRNA metabolism in both the nucleus (9) and the cytoplasm (17). Since SLBP is the only factor that interacts with the 3′ end of histone mRNA, it is a possible regulator of histone mRNA during the cell cycle. In the nucleus, SLBP is present as a free protein, which can be detected both by mobility shift assays and by Western blotting (43). SLBP is also present in the cytoplasm on the polyribosomes associated with histone mRNA (9, 17). In contrast to the nuclear SLBP, the cytoplasmic SLBP that is stably bound to histone mRNA cannot be detected by mobility shift assay (17) but can be detected by Western blotting.
To understand the relationship between cell cycle progression and histone mRNA regulation, it is important to study continuously cycling cells. Most studies of cell cycle regulation have utilized cells arrested at different points of the cell cycle, either using “natural” conditions, such as serum starvation, or drugs which arrest cells at various points in the cell cycle. For example, studying the release of serum-starved cells from G0 by stimulation with serum results in analysis of a resting-to-growing transition as well as cell cycle progression.
SLBP is a cell cycle-regulated protein.
Few studies have analyzed cell cycle regulation in a system where one can obtain continuously cycling cells. Analysis of CHO cells selected by mitotic shakeoff allows the study of a highly synchronous population of cells whose growth has been minimally perturbed. To obtain enough cells for biochemical measurements, we constructed an automated shakeoff apparatus that allows us to collect 107 mitotic cells in 4 h (11). Mitotic cells are collected every 10 min and can be stored for up to 4 h at 4°C (18, 47). Cells are in metaphase when selected and progress into G1 phase within 30 min after plating. Between 80 and 95% of the cells plated entered S phase, as determined by labeling with BrdU. The cells started to enter S phase after 3 to 5 h, depending on the experiment.
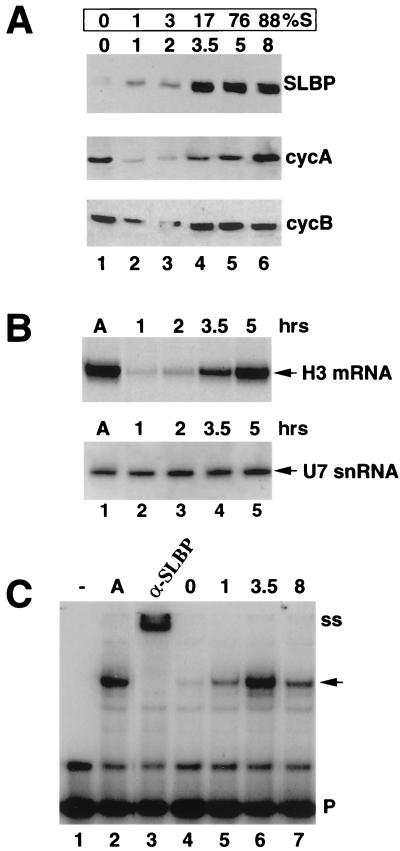
We analyzed the levels of SLBP during G1 and S phase by Western blot with an antibody that we prepared against a synthetic 13-amino-acid peptide corresponding to the C terminus of SLBP (60). No SLBP was detected in mitotic cells by Western blotting, and small amounts were detected in G1 cells 1 or 2 h after plating (Fig. 1A, lanes 1 to 3). As cells entered S phase (3.5 to 5 h), SLBP levels increased approximately 15- to 20-fold and remained constant throughout S phase (Fig. 1A, lanes 4 to 6). As a control, we measured the levels of the mitotic cyclins A and B by Western blotting of the same extracts (Fig. 1A). Cyclins A and B are present in mitotic extracts (which are from metaphase cells) but are rapidly degraded as cells complete mitosis. The levels of histone H3 mRNA in the same experiment were measured by S1 nuclease mapping. As reported previously (18), the increase in histone mRNA levels closely parallels the entry of cells into S phase (Fig. 1B). In contrast, the levels of U7 snRNA, another component of the histone pre-mRNA processing machinery (39, 55), were constant as cells progressed from G1 to S phase (Fig. 1B). Note that SLBP levels were maximal by 3.5 h, when less than 20% of the cells had entered S phase, and that SLBP levels peaked before histone mRNA levels peaked (Fig. 1A and B, lanes 4 and 5).
FIG. 1.
SLBP is cell cycle regulated. CHO cells were selected by mitotic shakeoff, and whole-cell extracts or total RNA was prepared at the times (hours) indicated above each lane. All panels used extracts and RNA from the same experiment. (A) Cells were harvested in mitosis (0 h), G1 phase (1 and 2 h), or S phase (3.5 to 8 h). SLBP (top), cyclin A (middle), and cyclin B (lower) levels were determined by Western blotting with extracts prepared at the indicated times. The percentage of cells in S phase at each time is shown above each lane. (B) Five micrograms of total RNA, prepared either from asynchronous cells (A) or from cells plated for the number of hours indicated above each lane, was analyzed for histone mRNA and U7 snRNA. Hamster histone H3 mRNA was measured with a 3′-end S1 nuclease assay with the mouse H3.2-614 gene, labeled at the SalI site, which maps all the hamster H3 histone mRNAs from codon 58 to the stop codon (18). The levels of U7 snRNA were determined by Northern blot analysis with a riboprobe complementary to the mouse U7 snRNA (10). In other experiments, the levels of U7 and histone mRNA in the mitotic cells (0 h) were similar to those at the 1-h time point. (C) The activity of SLBP in total-cell extracts was determined by mobility shift assay with the histone stem-loop as a probe (P). SLBP present in asynchronous cells (A) forms a specific complex which can be supershifted (ss) with antibodies to the C terminus of the SLBP (lanes 2 and 3). The activity of the SLBP was determined in cells plated for the times (hours) indicated above each lane. The SLBP:RNA complex is indicated by an arrow. The band migrating slightly above the probe is probably a dimer of the probe. There was not enough 5-h extract to analyze in this experiment, but in other experiments the 5-h and 8-h extracts showed similar activity.
We also measured the amount of free SLBP, active in RNA binding, at different times during the cell cycle by a mobility shift assay with the histone stem-loop as a probe (43). We used whole-cell lysates in these experiments. All the RNA binding activity which formed a complex with the stem-loop was due to SLBP, since the complex was completely shifted by the antibody to the C-terminal peptide (Fig. 1C, lanes 2 and 3). Little SLBP binding activity was detected in mitotic cells or 1 h after mitosis (Fig. 1C, lanes 4 and 5), but the activity increased dramatically as cells entered S phase (Fig. 1C, lane 6). SLBP activity peaked at 3.5 h after mitosis, prior to the entry of most of the cells into S phase and before the maximum accumulation of histone mRNA (Fig. 1C, lane 6). The activity decreased threefold by 8 h, when 85 to 90% of the cells were in S phase (Fig. 1C, lane 7) and histone mRNA levels were maximal. The decrease in SLBP activity between 3.5 and 8 h probably reflects the accumulation of SLBP bound to histone mRNA, effectively reducing the amount of free SLBP available for binding the probe (17).
SLBP mRNA levels are constant during the cell cycle.
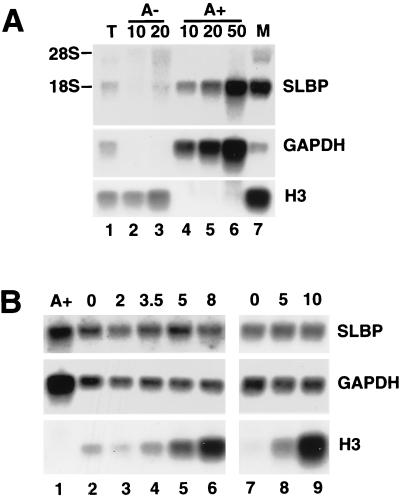
Mouse and human SLBP mRNAs are each about 1,730 nt, not including the poly(A) tail (60). We used the mouse SLBP cDNA as a probe to detect the hamster SLBP mRNA on a Northern blot. The mouse SLBP cDNA recognizes a 1.9-kb transcript in total hamster RNA by Northern blotting (Fig. 2), in good agreement with the expected size of SLBP mRNA. Since the transcript comigrates with 18S rRNA, total hamster RNA was fractionated into polyadenylated [poly(A)+] and poly(A)− fractions on oligo(dT)-cellulose (Fig. 2A). The great majority of the 1.9-kb transcript was detected in the poly(A)+ fraction (Fig. 2A, lanes 4 to 6) and comigrated with the mouse SLBP mRNA (Fig. 2A, lane 7), demonstrating that we were detecting the hamster SLBP mRNA with the mouse probe. Little cross-hybridization was observed to 10 and 20 μg of hamster poly(A)− RNA with the mouse SLBP probe (Fig. 2A, lanes 2 and 3). All of the histone H3 mRNA was in the poly(A)− fraction (Fig. 2A, lanes 2 and 3), and the GAPDH mRNA was in the poly(A)+ fraction.
FIG. 2.
SLBP mRNA is constant during the cell cycle. (A) Total RNA from exponentially growing CHO cells was fractionated into poly(A)− and poly(A)+ RNA by chromatography on oligo(dT)-cellulose. A 10-μg amount of total RNA (lane 1), 10 or 20 μg of poly(A)− RNA (lanes 2 and 3), and the equivalent amount of poly(A)+ RNA from 10, 20, or 50 μg of total cell RNA (lanes 4 to 6) was resolved by gel electrophoresis, transferred to a nylon membrane, and hybridized with the mouse SLBP cDNA (top), rat GAPDH cDNA (middle), or mouse histone H3-614 gene (bottom). Lane 7 is 10 μg of RNA from mouse myeloma cells. (B) Total-cell RNA was prepared from cells selected by mitotic shakeoff at the indicated times (hours) after plating (lanes 2 to 9). Two independent experiments are shown (lanes 2 to 6 and 7 to 9). Over 80% of these cells entered S phase by 8 h in the first experiment, and 95% of the cells entered S phase in the second experiment, as judged by BrdU labeling. A 10-μg amount of RNA was separated by agarose gel electrophoresis and analyzed as in panel A (lanes 2 to 9). Lane 1 is poly(A)+ RNA from the equivalent of 20 μg of total RNA from asynchronous cells.
We analyzed the levels of SLBP mRNA using total-cell RNA from CHO cells at different times after mitotic selection (Fig. 2B). Two independent experiments are shown. The levels of SLBP mRNA did not change significantly as cells progressed from mitosis to S phase (Fig. 2B, top panel, lanes 2 to 6 and lanes 7 to 9). SLBP mRNA was present in mitotic cells and remained constant into late S phase (10 h after mitosis) (Fig. 2B, lane 9). The blot was stripped and reprobed with the rat GAPDH gene as a loading control (Fig. 2B, middle panel). As a control for S-phase entry, the same blot was reprobed with the mouse histone H3 gene (Fig. 2B, lower panel). Since SLBP protein levels change 10- to 20-fold while SLBP mRNA levels are constant, the major regulation of SLBP must be at the translational or posttranslational level.
Regulation of SLBP during the G1-to-S-phase transition.
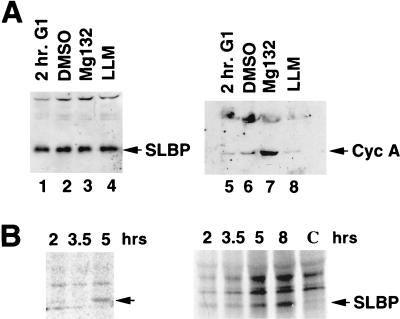
The low levels of SLBP in G1 cells could be due to synthesis and rapid degradation of the SLBP during G1 or to a failure to translate SLBP mRNA. The mitotic cyclins, for example, continue to be synthesized and rapidly degraded during G1 in both mammalian cells (4) and yeast cells (46). To determine whether the SLBP is rapidly degraded by the proteasome in G1, we treated G1 cells with the proteasome inhibitor Mg132 for 1 h. The levels of SLBP were unaffected by treatment of G1 cells with Mg132 (Fig. 3A, lanes 1 to 4), although cyclin A protein levels increased three- to fivefold (Fig. 3A, lane 7). An inactive analogue of the inhibitor, LLM, had no effect on either SLBP or cyclin A levels. Thus, rapid turnover of SLBP by the proteasome is not responsible for the low levels of SLBP in G1. We cannot rule out that a mechanism other than the proteasome could be responsible for rapidly degrading SLBP in G1.
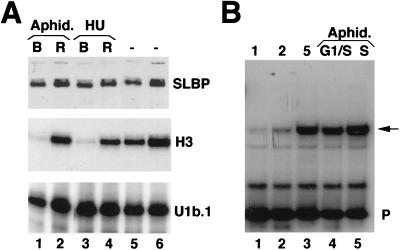
FIG. 3.
Translational control of SLBP during G1 phase. (A) Cells were selected by mitotic shakeoff. One hour after selection, cells were left untreated (2 hr. G1, lanes 1 and 5) or treated with either dimethyl sulfoxide (DMSO, lanes 2 and 6), 50 μM Mg132 in DMSO (lanes 3 and 7), or 50 μM LLM in DMSO (lanes 4 and 8). Lysates were prepared 1 h after treatment, and the amount of SLBP was determined by Western blotting (lanes 1 to 4). The same lysates were blotted for cyclin A (lanes 5 to 8). (B) Mitotic cells were selected and plated. At the indicated times after plating, cells were labeled with [35S]methionine for 15 min. The lysates, containing 5 × 106 cpm of labeled protein, were precipitated with affinity-purified anti-SLBP, and the precipitated proteins were analyzed by gel electrophoresis. The results of two experiments are shown. In lane C, the antigenic peptide was incubated with the antibody prior to precipitation of a sample labeled at 8 h after plating. The position of SLBP is indicated by the arrow.
A second possibility is that the translation rate of the SLBP is regulated. We analyzed the in vivo translation rate of SLBP mRNA by pulse labeling cells for 15 min with [35S]methionine and then immunoprecipitating the labeled proteins with anti-SLBP. Equal amounts of radioactive protein were assayed from each time point. Since translation of most mRNAs is inhibited during mitosis, the overall rate of methionine incorporation increased fourfold during the first 2 h after mitosis, but after 2 h the rate of overall protein synthesis had reached a constant level (not shown). In the labeling experiments, cells started to enter S phase only after 4 to 5 h, which is later than in other experiments, as a result of the depletion of methionine. G1 cells did not synthesize significant amounts of SLBP during the short pulse, while cells entering S phase or in S phase synthesized SLBP at a significantly higher rate (Fig. 3B). This result was observed in three independent experiments, and there was a three- to sixfold increase in labeling in each experiment. There is little change in SLBP mRNA levels during this time (Fig. 2B). We conclude that the SLBP is not rapidly degraded in G1 but that its translation rate is regulated between G1 and S phase. Since equal numbers of counts were loaded from each time point, the four- to fivefold increase in overall translation coupled with the three- to sixfold increase in selective translation of SLBP accounts for the 15- to 20-fold increase in SLBP protein levels observed as the CHO cells progress from mitosis through G1 into S phase. Further evidence for translation regulation is provided by analysis of polyribosomes in HeLa cells, shown below (see Fig. 9). We were not able to obtain enough cells by mitotic shakeoff to carry out the polyribosome analysis.
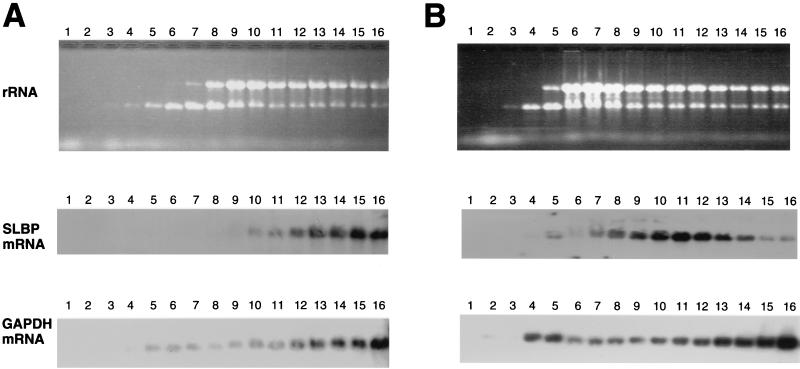
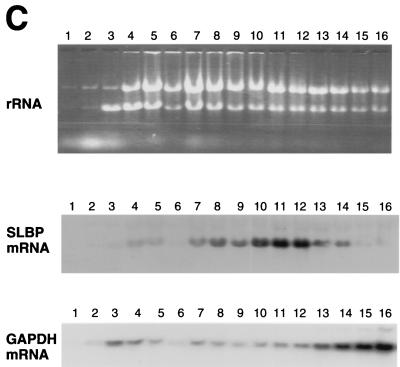
FIG. 9.
SLBP mRNA is translationally regulated during G1 phase. HeLa cells were synchronized either by double thymidine block (A and C) or by thymidine-nocodazole block (B), and cytoplasmic lysates were prepared 4 h after release (S phase, panel A), 6 h after release from a thymidine-nocodazole block (G1 phase, panel B), or 12 h after release from a double thymidine block (G1 phase, panel C). The cell cycle position was confirmed by flow cytometry. The lysates were fractionated on 15 to 40% sucrose gradients, and RNA was prepared from each fraction. The RNA was resolved by agarose gel electrophoresis and detected by staining with ethidium bromide (top panels in each group). The RNA was transferred to nylon and probed for either SLBP mRNA (middle panels) or GAPDH mRNA (bottom panels).
SLBP accumulates in late G1 phase.
To assess whether the increase in SLBP levels required ongoing DNA replication, we arrested the cells collected by mitotic shakeoff at the G1/S border by treatment with inhibitors of DNA synthesis. One hour after plating, when all the cells were still in G1, they were treated for 4 h with either aphidicolin, which blocks DNA polymerase α, or hydroxyurea, which inhibits ribonucleotide reductase. Most of the untreated control cells have entered S phase by this time (5 h after mitosis). In contrast, <1% of the cells treated with the inhibitors were synthesizing DNA 5 h after mitosis, as assayed by BrdU labeling.
The levels of SLBP protein and histone H3 mRNA were measured in these cells (Fig. 4A). SLBP has increased to control levels, although no histone mRNA has accumulated (Fig. 4A, lanes 1 and 3). When the inhibitor was removed, >80% of the cells entered S phase within 2 h. The levels of SLBP remained relatively constant, while histone mRNA accumulated rapidly (Fig. 4A, lanes 2 and 4).
FIG. 4.
SLBP accumulates independently of histone mRNA. (A) SLBP accumulates prior to the G1/S boundary. Mitotic cells were plated, and 1 h after plating they were treated with either aphidicolin (5 μg/ml) or hydroxyurea (5 mM) for 4 h to arrest cells at the G1/S boundary (lanes 1 and 3, B for blocked cells). The cells were then washed and incubated for 2 h in normal medium to allow them to enter S phase (lanes 2 and 4, R for released). As a control, cells were left untreated and harvested in parallel at 5 or 7 h after mitosis (lanes 5 and 6). Lysates and total-cell RNA were prepared from all cells. The levels of SLBP were measured by Western blotting (top), and the levels of histone H3 mRNA were measured by S1 nuclease protection analysis (middle). As a control for RNA recovery, the levels of U1b RNA were determined by an RNase protection assay (18) (bottom). (B) The SLBP RNA-binding activity in total-cell lysates was determined by mobility shift assay using the 26-nt histone stem-loop probe (P). Lysates were prepared from synchronous cell populations at the indicated times (hours) after mitosis (lanes 1 to 3), cells blocked at the G1/S boundary with aphidicolin (lane 4), or blocked cells 2 h after removal of the drug (lane 5). The SLBP-RNA complex is indicated by an arrow.
The amount of SLBP binding activity, measured by mobility shift assay, also increased between 1 and 5 h after plating, when cells have entered S phase (Fig. 4B, lanes 1 to 3). The binding activity was not affected by blocking cells at the G1/S border with aphidicolin (Fig. 4B, lane 4) or when cells were released from the block (Fig. 4B, lane 5). Identical results were obtained with lysates from cells treated with hydroxyurea (data not shown). Thus, accumulation of SLBP does not require DNA replication but is probably a result of a cell cycle signal that occurs prior to entry into S phase.
Half-life of SLBP in CHO cells.
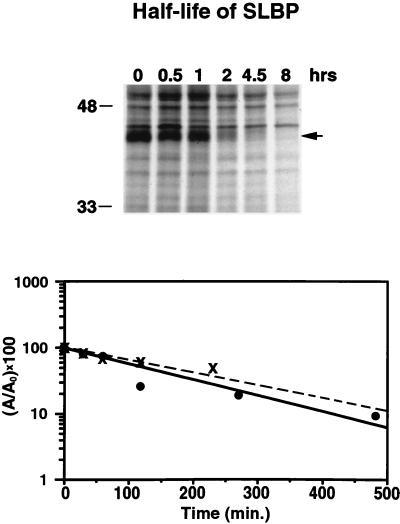
To assess the half-life of SLBP in CHO cells, we pulse-labeled both asynchronous cells and S-phase cells (5 h after plating) with [35S]methionine for 20 min, the label was washed out, and the cells were chased in medium containing excess unlabeled methionine for various amounts of time. The amount of labeled SLBP was determined by antibody precipitation followed by gel electrophoresis, as in Fig. 3. SLBP has a half-life of about 2 h in either asynchronous or S-phase cells (Fig. 5). Since there is at least a 10- to 20-fold decrease in SLBP levels in the 2 h between the end of S phase and entry into mitosis, the half-life of SLBP must be much shorter than 2 h between the end of S phase and mitosis. The data shown are for one of three experiments that gave similar results.
FIG. 5.
Half-life of SLBP in CHO cells. (A) Exponentially growing CHO cells were labeled with [35S]methionine for 30 min. The cells were washed and then chased with unlabeled medium, and cell extracts were prepared at the indicated times. The cell extracts were treated with anti-SLBP, and the immunoprecipitates were analyzed by SDS-polyacrylamide gel electrophoresis. The SLBP band is indicated with an arrow, and its identity was confirmed by competition with the antigenic peptide, as in Fig. 3B. (B) The data from exponentially growing cells (solid circles) and from S-phase cells (x's), labeled and chased as described for panel A, are plotted on a semilog plot to estimate the half-life of SLBP. A0 is the intensity of SLBP at zero time, and A is the intensity of SLBP at the indicated times.
The CHO cells collected by mitotic shakeoff are cells that become detached from the plate in a 15-min window (47), and these cells are in metaphase. SLBP is not present in these cells, while cyclin A and cyclin B are still present at high levels (Fig. 1A, lane 1). These cells complete mitosis within 30 min after plating, and the levels of the mitotic cyclins drop dramatically (Fig. 1A, lane 2). In synchronous CHO cells selected by mitotic shakeoff, SLBP levels are tightly regulated during the cell cycle, while SLBP mRNA levels are relatively constant. SLBP synthesis is activated at the level of translation in G1 phase, while SLBP is degraded sometime between S phase and mitosis.
Regulation of SLBP in HeLa cells.
It is not possible to directly collect G2-M cells by mitotic selection because the cells enter S phase over a 2-h period (3 to 5 h postmitosis) and hence do not synchronously exit S phase and progress through the short 2-h G2-M period asynchronously. To obtain a population of synchronous cells from late S phase through mitosis, we synchronized HeLa cells at the beginning of S phase by using a double thymidine block. HeLa cells were chosen because they arrest efficiently and, more important, essentially 100% of the cells reenter the cell cycle and progress synchronously through S phase and mitosis into G1. These arrested HeLa cells are not appropriate for studying the changes that occur as cells enter S phase because these cells have already progressed to the G1/S border, resulting in the accumulation of SLBP (Fig. 4A and Fig. 6B, lane 2). To obtain HeLa cells in G1, we arrested the cells with thymidine, released them, and then arrested them in mitosis with nocodazole. When these cells are released, they progress through G1 to S phase.
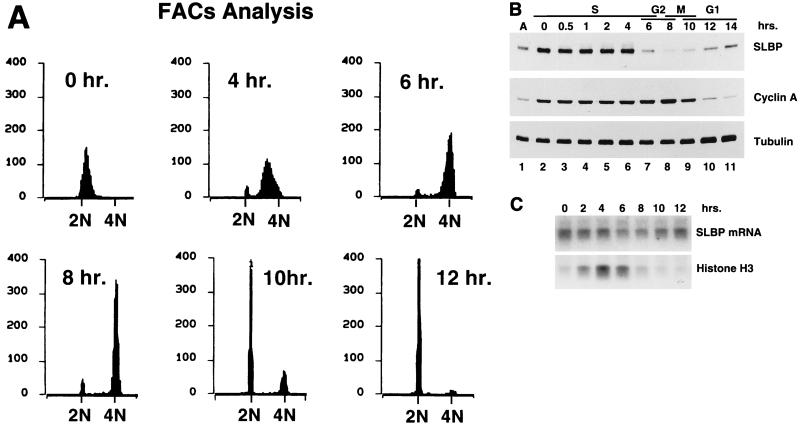
FIG. 6.
SLBP levels in synchronized HeLa cells. HeLa cells were arrested at the beginning of S phase by a double thymidine block. Samples were taken from arrested cells and from cells 30 min, 1 h, and 2 h after release and then at 2-h intervals. (A) Flow cytometry analysis of cells. The numbers of cells (arbitrary units) were plotted against DNA content. (B) Whole-cell extracts were prepared in SDS loading buffer, and the proteins were resolved by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose. The blot was incubated with anti-SLBP, stripped, and then incubated with anti-cyclin A and finally with anti tubulin. The cell cycle stage of each sample is shown above the lane. (C) Total-cell RNA prepared from cells every 2 h after release from the thymidine block (indicated above each lane) was resolved by agarose gel electrophoresis, and the amounts of SLBP mRNA and histone mRNA were determined by Northern blotting as in Fig. 2.
After releasing the HeLa cells from the thymidine block, the cells progress through S phase synchronously, as assessed by flow cytometry (Fig. 6A). More than 95% of the cells progress through S phase and enter G2 synchronously (Fig. 6A). The cells arrested at the G1/S border with the double thymidine block have accumulated high levels of SLBP (Fig. 6B, lane 2). SLBP levels stay high throughout S phase, and as cells exited S phase, SLBP protein levels dropped rapidly (Fig. 6B, lanes 6 and 7). The cells complete mitosis about 12 h after release, as evidenced by the degradation of cyclin A as the cells enter anaphase 10 to 12 h after release (Fig. 6B, lanes 9 and 10). SLBP is degraded about 4 to 6 h prior to the degradation of cyclin A. The 4N DNA content of the cells at 6 to 8 h after release and the timing of SLBP degradation relative to cyclin A degradation demonstrate that SLBP is degraded near the end of S phase or early in G2.
The levels of SLBP mRNA did not change significantly when cells were released from the thymidine block. In particular, there were high levels of SLBP mRNA in late G2/M cells (Fig. 6C, lane 9) and in G1 cells which had just completed mitosis (Fig. 6C, lane 10). By 12 h after release from the double thymidine block, most of the cells are in G1 phase. The level of SLBP protein is low in G1 cells (Fig. 6B, lanes 10 and 11), as is histone mRNA (Fig. 6C, lane 10). The levels of SLBP mRNA are relatively constant as the HeLa cells progress through S and G2 into G1 phase (Fig. 6C), varying no more than twofold. Thus, SLBP is regulated similarly in CHO cells and HeLa cells, and most of the regulation is at the posttranscriptional level.
SLBP is degraded by the proteasome at the end of S phase.
The levels of histone mRNA increased rapidly after release from the thymidine block and remained high as cells progressed through S phase and then declined dramatically (Fig. 6C), at about the same time as the levels of SLBP protein dropped precipitously.
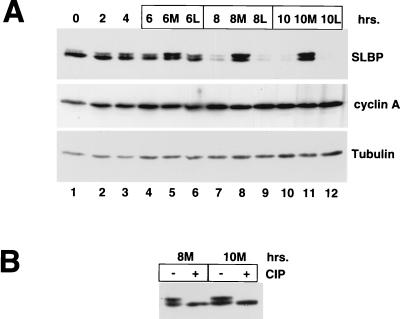
The degradation of SLBP at the end of S phase is prevented by treatment with proteasome inhibitors. Cells were synchronized at the G1/S boundary by double thymidine block, released, and allowed to progress into S phase. At 4 h after release, when cells are in mid- to late S phase, parallel plates of cells were treated with either the proteasome inhibitor Mg132 or the inactive analog LLM or left untreated. Cells were harvested at 2-h intervals after treatment with the proteasome inhibitor. The levels of SLBP increased in the cells treated with the proteasome inhibitor (Fig. 7A, lanes 5, 8, and 11), suggesting that SLBP is rapidly degraded by the proteasome as cells enter G2 phase. In addition, the major form of SLBP which accumulated in the presence of the proteasome inhibitor has a lower electrophoretic mobility than the bulk of the SLBP present at other phases of the cell cycle (Fig. 7A, lanes 5, 8, and 11). Treatment of these samples with calf intestinal phosphatase resulted in all the SLBP migrating as a single component (Fig. 7B, lanes 2 and 4). This result demonstrates that a phosphorylated form of SLBP accumulates in the presence of a proteasome inhibitor in late-S/G2 cells. It is possible that this phosphorylated SLBP is the form that is rapidly targeted for degradation by the proteasome. The rapid degradation of SLBP occurs only at the end of S phase, since treatment of asynchronous cells (not shown) or G1 cells (Fig. 3 and Fig. 8) did not result in a significant increase in SLBP concentrations or an increase in the amount of phosphorylated SLBP.
FIG. 7.
Effect of proteasome inhibitors on SLBP degradation in HeLa cells. (A) HeLa cells were synchronized at the G1/S border by double thymidine block and then released. Four hours after release of the cells, Mg132 was added to some samples and LLM was added to others. Samples were taken at 2-h intervals, and cell lysates were prepared and analyzed by Western blotting for SLBP. The blots were stripped and analyzed for cyclin A and for tubulin. Lanes 5, 8, and 11 are samples from cells treated with Mg132 (M), and lanes 6, 9, and 12 are samples from cells treated with LLM (L). (B) Extracts from the Mg132-treated cells shown in lanes 8 (8M) and 11 (10M) of panel A were treated with calf intestinal phosphatase (CIP) prior to analysis by Western blotting for SLBP.
FIG. 8.
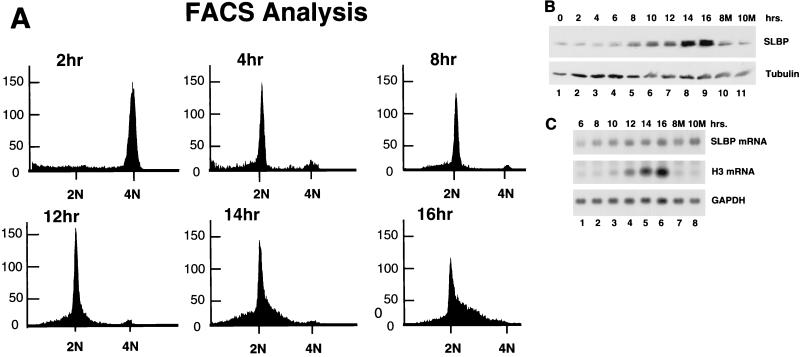
Regulation of SLBP during G1 in HeLa cells. HeLa cells were arrested in mitosis by blocking with thymidine followed by release and then blocking with nocodazole as described in the text. Samples were harvested every 2 h after release, the cell cycle stage was determined by flow cytometry, and total-cell extracts or total-cell RNA was prepared. Samples 8M and 10M (panel B, lanes 10 and 11; panel C, lanes 7 and 8) were prepared from cells which were treated with Mg132 starting 6 h after release from nocodazole and then harvested 2 and 4 h later, respectively. (A) Flow cytometric analysis of cells harvested at each time point. (B) Levels of SLBP and tubulin measured by Western blotting after separation of equal amounts of total cell protein by SDS-polyacrylamide gel electrophoresis. (C) Levels of SLBP mRNA, histone mRNA, and GAPDH mRNA measured by Northern blotting after separation of equal amounts of total-cell RNA by agarose gel electrophoresis.
In exponentially growing cells and in synchronous cells progressing through the cell cycle, a small amount of SLBP with a lower electrophoretic mobility is present. This form of SLBP is a result of phosphorylation of SLBP. There are many potential phosphorylation sites on SLBP (M. L. Whitfield, unpublished), and different sites may be phosphorylated when SLBP accumulates in S/G2 cells in the presence of proteasome inhibitors. There were no consistent changes in the phosphorylation of SLBP as detected by changes in electrophoretic mobility during the cell cycle.
Translation of SLBP mRNA is regulated in HeLa cells during G1.
The HeLa cells arrested by double thymidine block at the G1/S border have already accumulated SLBP. To obtain HeLa cells which progress through G1 into S phase, cells were synchronized by arrest at mitosis using nocodazole as described in Materials and Methods. HeLa cells were released from the mitotic block, and the SLBP levels were measured as the cells progressed through G1 into S phase. The cells complete mitosis 2 to 4 h after release from nocodazole and start to enter S phase about 12 to 14 h after release from nocodazole (Fig. 8A). As in the CHO cells, SLBP levels remained low during G1 and increased as cells entered S phase (Fig. 8B, lanes 1 to 7). We measured the levels of SLBP mRNA as cells progress through G1 (Fig. 8C, lanes 1 to 5). The level of SLBP mRNA was constant through late G1 and S phase. There was a decrease in SLBP mRNA in cells arrested with nocodazole (not shown). This is probably due to nocodazole's holding the cells in mitosis, which will greatly reduce the rate of transcription. In addition, a proportion of the cells never progress after being arrested with nocodazole, and these cells never adhere to the plate. These cells were discarded 6 h after release.
Note that HeLa cells allowed to progress continuously through mitosis did not show a decrease in SLBP mRNA (Fig. 6C, lanes 4 to 7), in agreement with the results obtained with CHO cells (Fig. 2B). Thus, as in CHO cells, there are large changes in SLBP protein levels as cells enter S phase, with very little (less than twofold) change in SLBP mRNA levels, indicating that SLBP is also regulated translationally or posttranslationally in HeLa cells.
To determine if SLBP is degraded by the proteasome in G1 of thymidine-nocodazole-synchronized HeLa cells, we treated G1 cells with Mg132 starting 6 h after release from nocodazole and measured the levels of SLBP at 2-h intervals. Mg132 had little effect on SLBP levels when G1 cells were treated (Fig. 8B, lanes 10 and 11), in contrast to the results when late-S-phase cells were treated (Fig. 7A). There was also no accumulation of a slower-migrating form of SLBP in the G1-phase cells treated with Mg132 (Fig. 8B, lanes 10 and 11). These results are essentially identical to those found with CHO cells (Fig. 3A).
To demonstrate that translation of SLBP mRNA is regulated during the progression from G1 to S phase, we analyzed the distribution of SLBP mRNA on polyribosomes at different stages of the cell cycle. Cytoplasmic lysates were prepared from S-phase cells (4 h after release from the thymidine block) and from G1 cells (either 12 h after release from the thymidine block or 6 h after release from the nocodazole block). The lysates were analyzed by sucrose gradient centrifugation, and the distribution of total RNA, SLBP mRNA, and GAPDH mRNA was determined in each sample. SLBP and GAPDH are similar in size, and thus if the two mRNAs are translated with similar efficiency, they should have a similar distribution on polyribosomes. In the S-phase cells (Fig. 9A), the SLBP mRNA was present near the bottom of the gradient with the heavy polyribosomes. The GAPDH mRNA showed a similar distribution, although there was some GAPDH mRNA higher up in the gradient, suggesting that the SLBP mRNA is translated even more efficiently than GAPDH mRNA in the S-phase cells (Fig. 9A). In contrast, in G1 cells, the SLBP mRNA sedimented with smaller polyribosomes (estimated to be in the di- to trisome range), while the GAPDH mRNA showed the same distribution as in S-phase cells (Fig. 9B and C). Similar results were found with G1 cells obtained 6 h after release from nocodazole (Fig. 9B) or 12 h after release from the double thymidine block (Fig. 9C). These results conclusively demonstrate that SLBP mRNA is translationally regulated as cells progress from G1 to S phase.
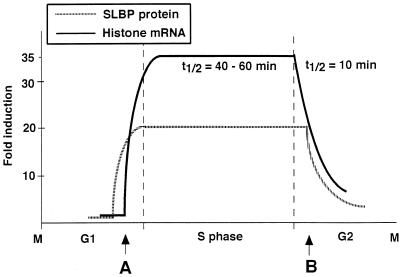
The results are summarized in Fig. 10, which shows the changes in histone mRNA and SLBP protein levels during the cell cycle. Like histone mRNA, SLBP is regulated by at least two different mechanisms during the cell cycle: by translation as cells progress from G1 to S, and by rapid degradation of the protein as cells progress from S phase into G2 (Fig. 10).
FIG. 10.
Model for SLBP and histone mRNA regulation during the cell cycle. The changes in histone mRNA and SLBP protein levels during the cell cycle are diagrammed. The primary step that is regulated at each transition (A, G1 to S phase; B, S to G2 phase) is indicated.
DISCUSSION
Histone mRNA levels increase and decrease rapidly about 35-fold during every cell cycle (18). To accomplish this dramatic regulation, histone gene expression is regulated at multiple steps (35, 49). Much of the regulation is posttranscriptional and is mediated by the 3′ end of histone mRNA. Since SLBP is a component of the histone mRNP (17) and hence must be present in stoichiometric amounts (1 molecule per molecule of histone mRNA), accumulation of histone mRNA without SLBP is not possible. It is likely that SLBP is a major trans-acting factor that contributes to the cell cycle regulation of histone mRNA. One major regulatory step is at the level of formation of the 3′ end of histone mRNA (18, 57). SLBP is essential for this reaction in vitro (9, 10, 44) and in vivo (44). Two possible mechanisms for regulating histone mRNA processing have been proposed: regulation of a heat-labile factor, which has not been well characterized, required for processing (15, 31), and the blocking of the 5′ end of U7 snRNA by a negative regulator (21). Other laboratories have not observed any alterations in U7 snRNP during the cell cycle (2). The low levels of SLBP in G1 cells would certainly prevent accumulation of cytoplasmic histone mRNA. The results reported here suggest that regulation of the SLBP could account for the posttranscriptional component of histone mRNA metabolism.
SLBP accumulates late in G1-phase and is present in both HeLa and CHO cells arrested at the G1/S border. These cells do not accumulate histone mRNA. It is possible that histone pre-mRNA processing has been activated in these cells by the accumulation of SLBP, but any histone mRNA formed would be very unstable because DNA synthesis has been inhibited (53). Alternatively, the cells arrested at the G1/S border might be lacking another factor required for histone pre-mRNA processing.
Although SLBP protein levels are regulated, the level of SLBP mRNA is not regulated in continuously growing cells (Fig. 2 and 6) or in serum-stimulated fibroblasts (M. L. Whitfield and W. F. Marzluff, unpublished results). Similar results were obtained for the SLBP mRNA in a study of global changes in steady-state mRNA levels in serum-stimulated cells using cDNA microarrays by Brown and coworkers (23). SLBP mRNA is readily detectable in all adult tissues (33), suggesting that the synthesis of SLBP protein is also regulated posttranscriptionally in normal somatic cells. Since regulation of SLBP is at the level of translation of preformed SLBP mRNA, SLBP concentrations increase rapidly once the signal to initiate SLBP synthesis is generated. The rapid synthesis of SLBP in late G1 results in a large amount of free, active SLBP which accumulates just prior to the maximal accumulation of histone mRNA (cf. Fig. 1A and C, lanes 4). Synthesis and export of histone mRNA from the nucleus is very rapid (48), suggesting that activation of SLBP mRNA translation would result in very rapid accumulation of cytoplasmic histone mRNA. Accumulation of SLBP occurs late in G1, after induction of cyclin E (unpublished results), and translation of SLBP mRNA may be activated by the same signals that ultimately result in entry into S phase.
Regulation of translation of selective mRNAs is probably critical for progression from G1 to S phase. Overexpression of eIF4E, one of the limiting factors for translation, results in transformation of cells (30), and one of the targets of this response is likely to be cyclin D mRNA (45). Activation of translation through activation of the Tor kinase, which results in phosphorylation of inhibitors of eIF4E, is an essential component of progression from G1 to S phase (29). Thus, translational regulation of key proteins required for cell cycle progression may be a common theme in cell cycle regulation.
At the end of S phase there is a rapid degradation of histone mRNA (13). The stem-loop at the 3′ end of the histone mRNA and presumably the bound SLBP are necessary for the change in histone mRNA half-life (42). Since SLBP levels remain constant when cells are treated with inhibitors of DNA replication (M. L. Whitfield and W. F. Marzluff, unpublished results) and histone mRNA is rapidly degraded (53), degradation of SLBP is not required for the degradation of histone mRNA. This result suggests that the degradation of SLBP is the result of a cell cycle signal generated at the end of S phase, while the degradation of histone mRNA occurs in response to cessation of DNA replication, either as a result of inhibition or at completion of S phase.
Other critical cell cycle transitions are driven by the rapid degradation of regulatory proteins (22, 27). During mitosis, the anaphase-promoting complex (APC) is activated, resulting in the degradation of cyclins A and B (20) as well as Pds1 (6) and probably other proteins necessary for mitosis. The APC recognizes a specific sequence, termed the destruction box, and directs ubiquitination and degradation of this set of proteins. The APC is activated in mitosis and remains active during the subsequent G1 phase (4). As cells progress from G1 to S phase, there is degradation of the Cdk inhibitor, p27, in mammalian cells (41) and degradation of Sic1 in yeast cells by a multiprotein complex termed the SCF (1, 12, 54, 58). The SCF is a multiprotein complex containing Skp-1, cullin/Cdc53, and a member of a diverse group of proteins containing an F-box motif. The F-box is a 30-amino-acid motif present in diverse proteins which interacts with Skp-1 and helps recruit phosphorylated substrates for ubiquitination (1, 12, 54, 58).
Subsequent purification and cloning of the subunits of the APC complex identified a subunit, APC-2, which has homology with the cullins (62, 64). Recently it was discovered that the APC and SCF contain an additional conserved subunit. APC11 (64) has considerable homology with ROC/Rbx1/HRT1 (26, 40, 51), a subunit of the SCF complex. The cullins represent a multigene family containing at least six members in mammalian cells (28), and there are at least two distinct roc genes (40), which may constitute a series of distinct ubiquitin ligase complexes (Y. Xiong, personal communication). These findings suggest that a conserved core is shared by different E3 ligases that are involved in regulation of diverse cellular processes (63). Many proteins are targeted for degradation during the G1-to-S transition by a regulatory phosphorylation (8, 52, 59). It is possible that other ubiquitin ligase complexes also regulate degradation of a set of proteins at the S-G2 transition and that SLBP might be one of the substrates targeted for degradation at this time.
There are distinct differences between the regulation of SLBP and the regulation of histone mRNA. SLBP accumulates in late G1, and there are high levels of SLBP in late G1 cells which contain little histone mRNA. Similarly, histone mRNA is rapidly degraded when DNA replication is inhibited, but SLBP is not degraded during this time. It is possible that processing of histone mRNA is activated in cells blocked at the G1/S border but the histone mRNA is very unstable (as it is in S-phase cells in which DNA synthesis is blocked). Clearly the signals regulating histone mRNA accumulation and SLBP accumulation are distinct. Histone mRNA levels are directly coupled to DNA replication, while SLBP is probably regulated by cell cycle signals which occur independently of DNA replication.
However, SLBP must be synthesized each cell cycle to allow accumulation of histone mRNA. The regulation of SLBP accumulation is determined by a cell cycle signal that results in an increased rate of translation of SLBP mRNA as cells progress from G1 to S phase (Fig. 3). SLBP is then degraded near the S-G2 border, as a result of a second cell cycle signal, making new synthesis of SLBP necessary for the next round of histone mRNA synthesis (Fig. 1). Thus, as with histone mRNA, both the biosynthesis and stability of SLBP are regulated during the cell cycle. We are currently investigating the molecular mechanisms underlying these two regulatory events.
ACKNOWLEDGMENTS
L.-X.Z. and A.B. contributed equally to this work.
This work was supported by grants from the National Institutes of Health to W.F.M. (GM 29832) and M.M.H. (GM 467658).
We thank Beth Alexander for technical assistance, Zbig Dominski for the data on U7 snRNA, and Yue Xiong and Bob Duronio for critical review of the manuscript.
REFERENCES
- 1.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Bond U, Yario T A. The steady state levels and structure of the U7 snRNP are constant during the human cell cycle: lack of cell cycle regulation of histone mRNA 3′ end formation. Cell Mol Biol Res. 1994;40:27–34. [PubMed] [Google Scholar]
- 3.Bond U M, Yario T A, Steitz J A. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 4.Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C Y A, Xu N H, Shyu A B. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 7.DeLisle A J, Graves R A, Marzluff W F, Johnson L F. Regulation of histone mRNA production and stability in serum-stimulated mouse fibroblasts. Mol Cell Biol. 1983;3:1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshaies R J. Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 9.Dominski Z, Sumerel J, Hanson R J, Marzluff W F. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA. 1995;1:915–923. [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski Z, Zheng L-X, Sanchez R, Marzluff W F. The stem-loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol Cell Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliassen K A, Baldwin A, Sikorski E M, Hurt M M. Role for a YY1 binding site in replication-dependent mouse histone gene expression. Mol Cell Biol. 1998;18:7106–7118. doi: 10.1128/mcb.18.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 13.Gallwitz D. Kinetics of inactivation of histone mRNA in the cytoplasm after inhibition of DNA replication in synchronised HeLa cells. Nature. 1975;257:247–248. doi: 10.1038/257247a0. [DOI] [PubMed] [Google Scholar]
- 14.Gick O, Krämer A, Keller W, Birnstiel M L. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gick O, Krämer A, Vasserot A, Birnstiel M L. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc Natl Acad Sci USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graves R A, Wellman S E, Chiu I-M, Marzluff W F. Differential expression of two clusters of mouse histone genes. J Mol Biol. 1985;183:179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- 17.Hanson R J, Sun J-H, Willis D G, Marzluff W F. Efficient extraction and partial purification of the polyribosomal-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry. 1996;35:2146–2156. doi: 10.1021/bi9521856. [DOI] [PubMed] [Google Scholar]
- 18.Harris M E, Böhni R, Schneiderman M H, Ramamurthy L, Schümperli D, Marzluff W F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heintz N, Sive H L, Roeder R G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershko A. Mechanisms and regulation of ubiquitin-mediated cyclin degradation. Adv Exp Med Biol. 1996;389:221–227. doi: 10.1007/978-1-4613-0335-0_27. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann I, Birnstiel M L. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature. 1990;346:665–668. doi: 10.1038/346665a0. [DOI] [PubMed] [Google Scholar]
- 22.Hoyt M A. Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 23.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins C W, Xiong Y. Immunoprecipitations and immunoblotting in cell cycle studies. In: Pagano M, editor. Cell cycle: materials and methods. New York, N.Y: Springer-Verlag; 1995. pp. 250–263. [Google Scholar]
- 25.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, Harper J W, Conaway J W. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 27.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 28.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence J C, Jr, Abraham R T. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- 30.Lazaris-Karatzas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 31.Lüscher B, Schümperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lüscher B, Stauber C, Schindler R, Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3′-terminal part of the gene. Proc Natl Acad Sci USA. 1985;82:4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marzluff W F. Histone 3′ ends: essential and regulatory functions. Gene Expression. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- 35.Marzluff W F, Pandey N B. Multiple levels of regulation of histone mRNA concentrations. Trends Biochem Sci. 1988;13:49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- 36.Marzluff W F, Whitfield M L, Dominski Z, Wang Z-F. Identification of the protein that interacts with the 3′ end of histone mRNA. In: Richter J D, editor. mRNA formation and function. New York, N.Y: Academic Press; 1997. pp. 163–193. [Google Scholar]
- 37.Melin L, Soldati D, Mital R, Streit A, Schümperli D. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mowry K L, Oh R, Steitz J A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3′-end processing reaction. Mol Cell Biol. 1989;9:3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mowry K L, Steitz J A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 40.Ohta T, Michel J J, Schottelius A J, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 41.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 42.Pandey N B, Marzluff W F. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey N B, Sun J-H, Marzluff W F. Different complexes are formed on the 3′ end of histone mRNA in nuclear and polysomal extracts. Nucleic Acids Res. 1991;19:5653–5659. doi: 10.1093/nar/19.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandey N B, Williams A S, Sun J-H, Brown V D, Bond U, Marzluff W F. Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′-end formation. Mol Cell Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenwald I B, Kaspar R, Rousseau D, Gehrke L, Leboulch P, Chen J J, Schmidt E V, Sonenberg N, London I M. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem. 1995;270:21176–21180. doi: 10.1074/jbc.270.36.21176. [DOI] [PubMed] [Google Scholar]
- 46.Schneider B L, Patton E E, Lanker S, Mendenhall M D, Wittenberg C, Futcher B, Tyers M. Yeast G1 cyclins are unstable in G1 phase. Nature. 1998;395:86–89. doi: 10.1038/25774. [DOI] [PubMed] [Google Scholar]
- 47.Schneiderman M H, Dewey W C, Leeper D B, Nagasawa H. Use of the mitotic selection procedure for cell cycle analysis. Comparison between the X-ray and cycloheximide G2 markers. Exp Cell Res. 1972;74:430–438. doi: 10.1016/0014-4827(72)90398-9. [DOI] [PubMed] [Google Scholar]
- 48.Schodhetman G, Perry R P. Early appearance of histone messenger RNA in polyribosomes of cultured L cells. J Mol Biol. 1972;63:591–596. doi: 10.1016/0022-2836(72)90450-0. [DOI] [PubMed] [Google Scholar]
- 49.Schümperli D. Cell-cycle regulation of histone gene expression. Cell. 1986;45:571–572. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- 50.SenGupta D J, Zhang B L, Kraemer B, Prochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seol J H, Feldman R M R, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Shevchenko A, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 53.Sittman D B, Graves R A, Marzluff W F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci USA. 1983;80:1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 55.Soldati D, Schümperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol Cell Biol. 1988;8:1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stauber C, Lüscher B, Eckner R, Lotscher E, Schümperli D. A signal regulating mouse histone H4 mRNA levels in a mammalian cell cycle mutant and sequences controlling RNA 3′ processing are both contained within the same 80-bp fragment. EMBO J. 1986;5:3297–3303. doi: 10.1002/j.1460-2075.1986.tb04643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stauber C, Schümperli D. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 1988;16:9399–9413. doi: 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 59.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z-F, Whitfield M L, Ingledue T I, Dominski Z, Marzluff W F. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 61.Xiong Y, Zhang H, Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 62.Yu H T, Peters J M, King R W, Page A M, Hieter P, Kirschner M W. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 63.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 64.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]