Abstract
Background
Xanthomonas citri pv. fuscans (Xcf) and Xanthomonas phaseoli pv. phaseoli (Xpp) are the causal agents of common bacterial blight of bean (CBB), an important disease worldwide that remains difficult to control. These pathogens belong to distinct species within the Xanthomonas genus and have undergone a dynamic evolutionary history including the horizontal transfer of genes encoding factors probably involved in adaptation to and pathogenicity on common bean. Seed transmission is a key point of the CBB disease cycle, favouring both vertical transmission of the pathogen and worldwide distribution of the disease through global seed trade.
Taxonomy
Kingdom: Bacteria; phylum: Proteobacteria; class: Gammaproteobacteria; order: Lysobacterales (also known as Xanthomonadales); family: Lysobacteraceae (also known as Xanthomonadaceae); genus: Xanthomonas; species: X. citri pv. fuscans and X. phaseoli pv. phaseoli (Xcf‐Xpp).
Host range
The main host of Xcf‐Xpp is the common bean (Phaseolus vulgaris). Lima bean (Phaseolus lunatus) and members of the Vigna genus (Vigna aconitifolia, Vigna angularis, Vigna mungo, Vigna radiata, and Vigna umbellata) are also natural hosts of Xcf‐Xpp. Natural occurrence of Xcf‐Xpp has been reported for a handful of other legumes such as Calopogonium sp., Pueraria sp., pea (Pisum sativum), Lablab purpureus, Macroptilium lathyroides, and Strophostyles helvola. There are conflicting reports concerning the natural occurrence of CBB agents on tepary bean (Phaseolus acutifolius) and cowpea (Vigna unguiculata subsp. unguiculata).
Symptoms
CBB symptoms occur on all aerial parts of beans, that is, seedlings, leaves, stems, pods, and seeds. Symptoms initially appear as water‐soaked spots evolving into necrosis on leaves, pustules on pods, and cankers on twigs. In severe infections, defoliation and wilting may occur.
Distribution
CBB is distributed worldwide, meaning that it is frequently encountered in most places where bean is cultivated in the Americas, Asia, Africa, and Oceania, except for arid tropical areas. Xcf‐Xpp are regulated nonquarantine pathogens in Europe and are listed in the A2 list by the European and Mediterranean Plant Protection Organization (EPPO).
Genome
The genome consists of a single circular chromosome plus one to four extrachromosomal plasmids of various sizes, for a total mean size of 5.27 Mb with 64.7% GC content and an average predicted number of 4,181 coding sequences.
Disease control
Management of CBB is based on integrated approaches that comprise measures aimed at avoiding Xcf‐Xpp introduction through infected seeds, cultural practices to limit Xcf‐Xpp survival between host crops, whenever possible the use of tolerant or resistant bean genotypes, and chemical treatments, mainly restricted to copper compounds. The use of pathogen‐free seeds is essential in an effective management strategy and requires appropriate sampling, detection, and identification methods.
Useful websites
https://gd.eppo.int/taxon/XANTPH, https://gd.eppo.int/taxon/XANTFF, and http://www.cost.eu/COST_Actions/ca/CA16107.
Keywords: common bacterial blight of bean, Phaseolus vulgaris, Xanthomonas
This pathogen profile summarizes the current knowledge on Xanthomonas phaseoli pv. phaseoli and Xanthomonas citri pv. fuscans, two phylogenetically distant groups of strains that cause common bacterial blight of bean.

1. HISTORY AND TAXONOMY
Common bacterial blight of bean (CBB) was first described in 1893, and the causal agent was isolated, identified, and named Bacillus phaseoli by Smith in 1897 (Zaumeyer, 1930). Variant strains isolated by Burkholder from beans grown in Switzerland in 1924 produced a brown pigment on tyrosine‐containing medium and were thus described as fuscous strains. This brown pigment results from the secretion and subsequent oxidation of homogentisic acid (Goodwin & Sopher, 1994), possibly due to a single‐nucleotide deletion that led to pseudogenization of the hmgA gene encoding a homogentisate oxygenase in the fuscous variants (Aritua et al., 2015; Darrasse et al., 2013). Following the description of the genus Xanthomonas by Dowson in 1939 to gather gram‐negative rods that are motile by the means of a single polar flagellum and that form yellow mucoid colonies, the bacteria causing CBB were renamed Xanthomonas phaseoli (Corey & Starr, 1957). Following revisions of the taxonomy, the fuscous and nonfuscous strains were grouped under the name Xanthomonas campestris pv. phaseoli (Dye et al., 1980). Then, an overhaul of the genus transfered this pathovar into the species Xanthomonas axonopodis as X. axonopodis pv. phaseoli (Vauterin et al., 1995). Molecular divergence between fuscous and nonfuscous strains has long been reported (Birch et al., 1997; Chan & Goodwin, 1999; El‐Sharkawy & Huisingh, 1971; Lazo et al., 1987), but it was not until 2005 that a taxonomic distinction between the two was proposed. First, repetitive element polymerase chain reaction (rep‐PCR) analyses led to splitting X. axonopodis into six rep‐PCR groups (9.1 to 9.6), with fuscous strains belonging to group 9.6 while nonfuscous strains belonged to group 9.4 (Rademaker et al., 2005). Then, using a combination of DNA/DNA reassociation, intergenic spacer sequences, and phenotypic traits, Schaad et al. (2005) proposed to reallocate the fuscous strains into a new species–subspecies combination, namely Xanthomonas fuscans subsp. fuscans, while the nonfuscous strains conserved the name X. axonopodis pv. phaseoli. Amplified fragment length polymorphism (AFLP) analysis, rep‐PCR, and multilocus sequence analysis (MLSA) led to the description of three genetic lineages (GL1, GL2, and GL3) for X. axonopodis pv. phaseoli (Alavi et al., 2008; Mhedbi‐Hajri et al., 2013; Mkandawire et al., 2004), which was problematic because lineages GL2 and GL3 grouped with X. fuscans subsp. fuscans within rep‐PCR group 9.6, while GL1 was phylogenetically distant from the others. On the basis of a polyphasic analysis, Constantin et al. (2016) proposed a phylogenetically coherent revision of the species X. axonopodis that led to reallocation of existing pathovars into four species (X. citri, X. phaseoli, X. euvesicatoria, and X. axonopodis). Consequently, the four lineages of bacterial pathogens responsible for CBB are currently distributed across two species within the Xanthomonas genus (Figure 1a). Lineage GL1 corresponds to X. phaseoli pv. phaseoli (Xpp), while the other three lineages (GL2, GL3, and fuscans) form X. citri pv. fuscans (Xcf). Additional strains previously described as “X. axonopodis pv. phaseoli” isolated from lablab bean (Lablab purpureus) in Sudan or Zimbabwe form a distinct genetic lineage (“lablab”) within the X. citri species (Aritua et al., 2015). This lineage should probably be considered as a new pathovar, distinct from Xcf, because these isolates were less pathogenic on Phaseolus vulgaris than Xcf and were not pathogenic on Vigna unguiculata, while Xcf was virulent (Sabet, 1959); however, no formal taxonomic proposal has been published so far.
FIGURE 1.
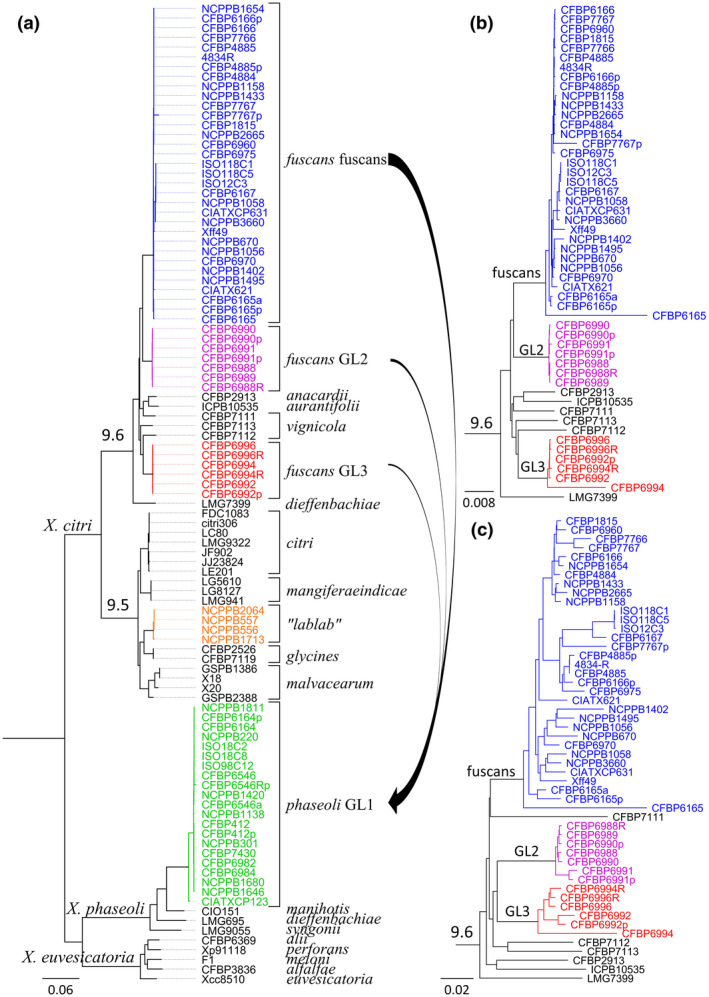
Phylogeny of common bacterial blight of bean (CBB) agents. (a) Phylogenetic tree based on the core genome of Xanthomonas, constructed with the PanX pipeline (Ding et al., 2018). Briefly, 1,335 single‐copy core genes were aligned and variable positions were extracted to construct a core‐genome SNP matrix. The SNP matrix was then used to build a phylogenetic tree using FastTree2 (Price et al., 2010), which was further refined by RaxML v. 8 (Stamatakis, 2014). Arrows represent the main horizontal gene transfer events detected between Xanthomonas citri pv. fuscans (Xcf) and Xanthomonas phaseoli pv. phaseoli (Xpp) ancestors. Rooting was done using Xanthomonas vasicola strain NCPPB702, Xanthomonas campestris strain NCPPB4381, and Xanthomonas oryzae strains KACC10331, MAFF 311018, PXO99A, BAI15, BAI20, BAI21, and BLS256 (not shown) as an outgroup. (b, c) Subtrees of rep‐PCR group 9.6, from trees constructed using CVtree (Qi et al., 2004) on the same strain dataset as for the tree presented in (a). These trees are based on the frequency of appearance of overlapping oligopeptides of length K = 6. The tree in (b) was constructed based on the same 1,335 core genes as in (a), while the tree in (c) was constructed based on the pan‐genome consisting of all predicted coding sequences. Topologies in (a) and (b) are congruent with each other
2. AVAILABLE GENOMIC DATA
Since the first whole‐genome sequence published in 2013 (Darrasse et al., 2013), 70 Xcf‐Xpp genomes have been released to date at the National Center for Biotechnology Information (Table 1). These genomes represent a diversity of 51 Xcf‐Xpp strains, 13 of which were sequenced more than once. Most sequenced strains were isolated from common bean except for Xpp CIAT XCP123, which was isolated from lima bean, which lacks a XopJ5 homolog encoded in other Xpp and is phylogenetically an outlier within GL1 (Figure 1a; Aritua et al., 2015). According to PacBio assemblies, the genome consists of a single chromosome plus one to four extrachromosomal plasmids of various sizes. The mean total size is 5.27 Mb with 64.7% GC content and an average of 4,181 predicted coding sequences. Genome sequence analyses have identified a large range of putative virulence factors, including type II secreted enzymes, type III effectors (T3E), and various fibrillar and nonfibrillar adhesins (Darrasse et al., 2013).
TABLE 1.
Summary of whole‐genome assemblies available at the NCBI (as of 2021‐03‐19)
| Release date | Reference | Sequencing strategy | Number of genomes 1 | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Xcf‐fuscans | Xpp‐GL1 | Xcf‐GL2 | Xcf‐GL3 | Xcf‐lablab | ||||
| 2013 | Darrasse et al. (2013) | Illumina, 454, Sanger | 1 | ‐ | ‐ | ‐ | ‐ | 1 |
| 2014 | Indiana et al. (2014) | Illumina | 1 | ‐ | ‐ | ‐ | ‐ | 1 |
| 2015 | Aritua et al. (2015) | Illumina | 13 | 9 | ‐ | ‐ | 4 | 26 |
| 2015 | Constantin et al. (2016) | Illumina | 1 | ‐ | ‐ | ‐ | ‐ | 1 |
| 2017 | Ruh et al. (2017b) | PacBio | 6 | 4 | 5 | 2 | ‐ | 17 |
| 2017 | Chen, Serres‐Giardi, et al. (2018) | Illumina | 6 | 5 | 3 | 3 | ‐ | 17 |
| 2017 | Kremer et al. (2019) | Illumina | 1 | ‐ | ‐ | ‐ | ‐ | 1 |
| 2019 | Xie et al. (2016) | PacBio, Illumina | 3 | 3 | ‐ | ‐ | ‐ | 6 |
| Total | 32 | 21 | 8 | 5 | 4 | 70 | ||
Note. Xcf, Xanthomonas citri pv. fuscans; Xpp, Xanthomonas phaseoli pv. phaseoli.
3. ELUCIDATING THE PATHOLOGICAL CONVERGENCE OF CBB AGENTS
The polyphyletic nature of CBB agents raises the question of the molecular determinants underlying the pathological convergence observed between Xpp (i.e., GL1) and Xcf (i.e., fuscans, GL2, and GL3). In 2008, suppression subtractive hybridization (SSH) analyses identified possible horizontal gene transfers between Xpp and Xcf (Alavi et al., 2008). Subsequently, a PCR‐based analysis showed that Xpp and Xcf clustered together in a dendrogram based on the presence/absence of T3Es (Hajri et al., 2009), suggesting that horizontal transfers of T3E occurred between the ancestors of Xpp and Xcf. More recently, genome‐wide analyses unveiled dozens of alleles, or portions of genes, sharing 100% identity between Xpp and Xcf (Aritua et al., 2015; Chen, Serres‐Giardi, et al., 2018). Most of these alleles, disseminated over the chromosome and plasmids, were transferred from the fuscans lineage of Xcf to the ancestor of Xpp. This suggests that Xpp acquired the ability to cause CBB through extensive horizontal gene transfers from Xcf. A large part of these genes encodes different proteins putatively involved in pathogenicity, including TonB‐dependent transporters (TBDTs) and proteins related to two‐component regulatory systems or the type III, type IV, and type VI secretion systems (Chen, Serres‐Giardi, et al., 2018). These include T3Es, among which are two transcription activator‐like effectors (TALEs), Xfutal1 and Xfutal2 (Ruh et al., 2017a). Significantly, a c.44 kb plasmidic region was entirely transferred from Xcf to Xpp. This region comprises 28 genes, including Xfutal1, and seven genes encoding proteins from the type IV conjugal transfer system. In all, whole‐genome data revealed that the pathological convergence observed between Xcf and Xpp is associated with horizontal gene transfers and highlighted the role of plasmids in these transfers. Functional validation is still needed to confirm the role of these genes in pathogenicity. No evidence of horizontal gene transfer was observed between Xcf‐Xpp and lablab‐associated strains, indicating that they have undergone an evolutionary history distinct from the other CBB strains (Aritua et al., 2015).
Although the dichotomy between Xpp and Xcf has clearly been established, the phylogenetic positions of the fuscans, GL2, and GL3 lineages have never been clearly elucidated with respect to other strains from rep‐PCR group 9.6 (including members of pathovars vignicola, anacardii, aurantifolii, and dieffenbachia). Phylogenetic trees obtained using partial sequences of seven housekeeping genes (atpD, dnaK, efp, fyuA, glnA, gyrB, and rpoD) (Mhedbi, 2010) showed different topologies for each gene (explained by intrastrain recombination). However, vignicola, anacardii, aurantifolii, and dieffenbachia strains were interleaved between Xcf lineages in every tree, ruling out the hypothesis of Xcf being monophyletic. Using whole‐genome data, we hereby show that GL2 and fuscans lineages group together in two trees based on the core genome of Xanthomonas (Figure 1a,b), suggesting that both lineages form a monophyletic group separated from the GL3 lineage. Moreover, GL2 and GL3 group together and split from the fuscans lineage when using the pan‐genome (Figure 1c). The incongruence between core and pan‐genomes could be explained by important recombination events having occurred within the accessory genome of GL2 and GL3 ancestors. Interestingly, in contrast to the Xcf fuscans lineage and Xpp, which have a worldwide distribution, the GL2 and GL3 lineages were isolated only from bean plants grown in La Réunion and Tanzania. Thus, recombinations could have been facilitated by geographical proximity between both lineages. Whether GL2 and GL3 strains exist elsewhere or are endemic to this region of the world remains uncertain. In all, these observations indicate that Xcf itself is a polyphyletic pathovar within a complex of recombinant pathovars formed by Xcf and X. citri pathovars vignicola, anacardii, aurantifolii, and dieffenbachia.
4. IMPACT AND DISTRIBUTION
CBB is one of the five major diseases of common bean (Broughton et al., 2003). Yield losses up to 45% were reported in susceptible genotypes (Saettler, 1989; Wallen & Jackson, 1975; Yoshii et al., 1975). In the field, CBB directly affects yield either by degrading edible fresh materials or by reducing the area of photosynthetic tissues. Materials with symptoms become unsaleable, and additional losses are due to the time and costs involved in controlling the disease. A major threat posed by CBB concerns seed quality, impacting both the seed industry and edible seed production. In particular, infected seed lots cannot be sold in disease‐free areas, even in the absence of symptoms. According to the EPPO global database, CBB is widely distributed over 104 countries across the five continents (Figure 2). CBB was reported in most regions where common bean is cultivated except in dry tropical areas. To our knowledge, the most recent disease outbreak was reported in 2019 in Belgium (Bultreys & Gheysen, 2020).
FIGURE 2.
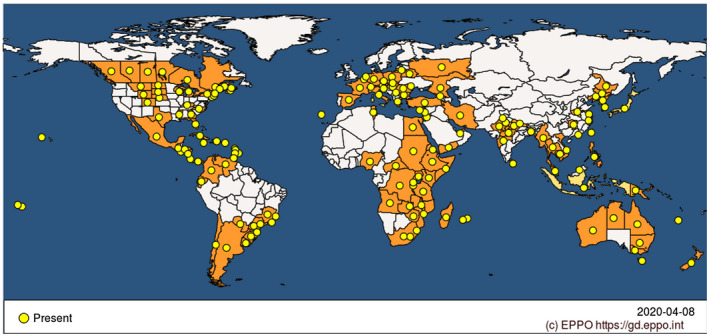
Worldwide distribution of common bacterial blight of bean (CBB) agents. The distribution includes both Xanthomonas citri pv. fuscans and Xanthomonas phaseoli pv. phaseoli. Modified from https://gd.eppo.int/taxon/XANTPH/distribution (EPPO, 2021)
5. HOST RANGE
Phaseolus vulgaris is the main host of Xpp and Xcf. Natural infections have been reported for both Xpp and Xcf on diverse other legume species such as Calopogonium sp., L. purpureus, Macroptilium lathyroides, Phaseolus acutifolius, Phaseolus coccineus, Phaseolus lunatus, Pisum sativum, Pueraria sp., and Strophostyles helvola, as well as various Vigna species including V. aconitifolia, V. angularis, V. mungo, V. radiata, V. umbellata, and V. unguiculata, (Bradbury, 1986; Gilbertson & Maxwell, 1992). Additional hosts of CBB agents were reported after artificial inoculation. This was the case for Amaranthus retroflexus, Ambrosia artemisiifolia, Beta vulgaris, Chenopodium album, Echinochloa crus‐galli, Glycine max, Helianthus annuus, Lupinus polyphyllus, Solanum nigrum, and Zea mays (Bradbury, 1986). Asymptomatic carriage of CBB agents was reported for Digitaria scalarum and Senna hirsuta (Opio et al., 1996).
There are conflicting reports concerning the natural occurrence of CBB agents on tepary bean (P. acutifolius) and cowpea (V. unguiculata). Indeed, Gilbertson and Maxwell (1992) considered them as hosts, while Bradbury (1986) considered them as susceptible hosts only after artificial inoculation. In 1987, Zapata and Vidaver (1987) described P. acutifolius as “a natural host of X. campestris pv. phaseoli”, which is supported by the existence of races of Xpp‐Xcf displaying differential phenotypic reactions on susceptible or resistant varieties of tepary beans (Opio et al., 1996; Zapata & Vidaver, 1987). These observations led to the hypothesis of a gene‐for‐gene relationship between CBB agents and tepary bean (Opio et al., 1996), which is consistent with P. acutifolius being a natural host having coevolved with Xcf‐Xpp. Gilbertson and Maxwell (1992) reported unpublished data of fuscous strains isolated from cowpea in Puerto Rico that were pathogenic on common bean. Although this finding still requires molecular identification of the strains, it supports the idea that cowpea may be a natural host of Xcf. Interestingly, the observation that Xpp strains were nonpathogenic on cowpea suggests that Xpp and Xcf may be differentiated by their host range (Gilbertson & Maxwell, 1992; Vakili et al., 1974). Furthermore, phylogenetic proximity between Xcf and X. citri pv. vignicola (a pathogen of cowpea), together with the fact that both are pathogenic on common bean and cowpea, challenges the boundaries between these two pathovars. A finer description of the symptoms naturally induced by each pathovar on each host could bring essential elements to enlighten this question. Moreover, recent whole‐genome sequencing data indicate that the TALE‐encoding genes from X. citri pv. vignicola are very different from those found in Xcf and Xpp (Ruh et al., 2017b; unpublished data). In particular, Xfutal1 is absent in vignicola strains while it is shared by all Xcf‐Xpp strains sequenced so far (Ruh et al., 2017a), indicating differential adaptation of these pathovars to either common bean or cowpea. More in‐depth analyses combining whole‐genome sequencing, phylogeny, and crosspathogenicity assays are required to ascertain the differences observed between pathovars phaseoli, fuscans, and vignicola.
6. SYMPTOMS
CBB symptoms produced by Xcf and Xpp are indistinguishable from each other. All aerial parts of bean plants, that is, seedling, leaf, stem, pod, and seed, can present symptoms of CBB (Gilbertson & Maxwell, 1992; Zaumeyer, 1930). Examples of symptoms on leaves, seeds, and pod are shown in Figure 3. Leaf symptoms occur as water‐soaked spots on the limb usually beginning at hydathodes, subsequently evolving into dry and brown necrotic lesions surrounded by a narrow yellow halo (Chupp & Sherf, 1960). These spots may merge, giving the leaf a burnt appearance, resulting in the most severe cases in plant defoliation and death. A reddish‐brown discolouration of the veins with a water‐soaking of adjoining interveinal areas may be observed in systemic infection. Infected stems present reddish longitudinal streaks. Symptoms on pods appear as water‐soaked spots, later evolving in dark red‐brown lesions, slightly depressed circular spots, with possible bacterial ooze. Shrinking and death of pods may occur in case of severe infection. Seed symptoms appear as butter yellow spots that turn into brown spots and localize according to the infection pathway: on the hilum area in case of vascular transmission, at the micropyle in case of floral infection, and on the entire surface of the seed coat in case of infection by contact (Darrasse et al., 2010; Maude, 1997). Seed may be shrivelled when strongly infected, severely affecting their germination rate and vigour (Darrasse et al., 2018). Under warm temperatures and when the infection is serious, the whole plant may die. Infected seeds and plants may also be symptomless (Darrasse et al., 2007; Jacques et al., 2005; Weller & Saettler, 1980a, 1980b).
FIGURE 3.
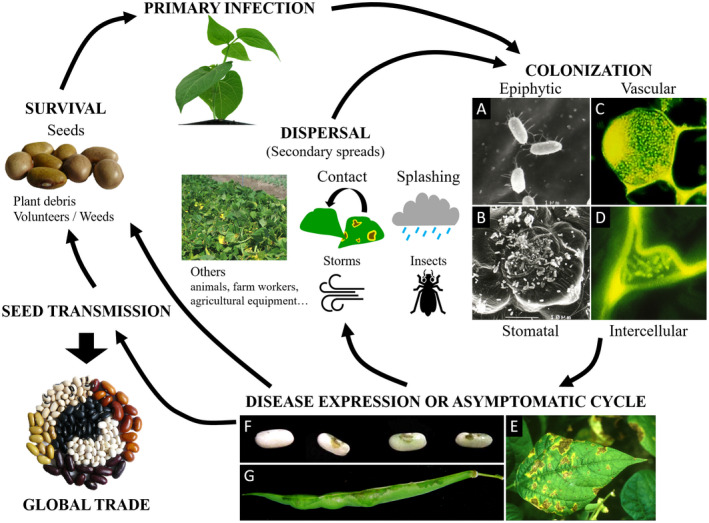
Common bacterial blight of bean (CBB) disease cycle and symptoms. Primary infection occurs mainly by vertical transmission through contamined seeds or from infected plant debris, volunteers, or weeds. Colonization starts with an epiphytic stage where bacteria multiply and aggregate into biofilms, followed by entry through openings such as stomata, hydathodes, or wounds. Bacteria colonize the host tissues and multiply in both the intercellular space and the vessels. Symptoms can occur on all aerial parts of the plant. Secondary spreads occur by direct contact and splashing or transport by storms and insects. Long‐distance transmission usually results from global seed trade. (a–d) Microscopy images of bacterial colonization of common bean. (a) Scanning electron microscopy (SEM) micrograph of solitary cells of epiphytic Xanthomonas citri pv. fuscans (Xcf) on bean leaves. (b) SEM micrograph of Xcf cells aggregated in biofilm in open stomata, the guard and neighbouring cells of which are largely colonized by epiphytic cells. (c) Confocal laser‐scanning microscopy (CLSM) image showing green fluorescent protein (GFP)‐tagged cells of Xcf aggregated in a biofilm adhering to the inner surface of a xylem vessel in the stem of a bean plantlet and plugging the vessel. (d) CLSM image showing GFP‐tagged cells of Xcf in the intercellular space of bean leaf parenchyma. (e–g). Images of disease expression on different plant organs. (e) Typical symptoms on leaf, showing brown necrotic lesions surrounded by a yellow halo. (f) Healthy seed (left) and three symptomatic seeds (right) showing brownish symptoms at the hilum area. (g) Pod displaying greasy symptoms spreading from the pod suture
Symptoms of CBB on pods and/or leaves are very similar to those caused by Pseudomonas savastanoi pv. phaseolicola, the causative agent of halo blight. To date, the only means to ascertain that the observed symptoms are caused by Xpp or Xcf are bacterial isolation or molecular detection, for example, using specific PCR primers (Audy et al., 1994; Boureau et al., 2013; Grimault et al., 2014). Phenotyping of the disease has been assessed using different methods of inoculation and symptom quantification, each method having its own strengths and weaknesses (Aggour et al., 1989; Cafati & Saettler, 1980a; Opio et al., 1993; Pastor‐Corrales et al., 1981; Zapata, 2006). In the last decade, image‐based methods were developed, allowing better standardization and objectivization of phenotyping. One method is based on conventional imaging (Xie et al., 2012), while another is based on chlorophyll fluorescence imaging (Rousseau et al., 2013).
7. DISEASE CYCLE
7.1. Sources of inoculum
The main source of CBB inoculum corresponds to infected seeds. Indeed, CBB agents can reside on both sides of the seed coat as well as on the surface of the embryo (Darrasse et al., 2018; Zaumeyer, 1930), allowing overwintering and long‐term survival (up to 30 years) in infected seeds (Saettler, 1989; Schuster & Sayre, 1967). The variation of survival time in seeds depends on storage conditions (temperature and humidity) (Marques et al., 2005). Disease outbreak is possible from as little as one seed in a lot of 10,000 to 30,000 (Darrasse et al., 2007; Opio et al., 1993; Sutton & Wallen, 1970; Zaumeyer & Thomas, 1957). In susceptible genotypes, a minimum of 100 bacteria per seed is required for successful plantlet colonization (Darrasse et al., 2007; Opio et al., 1993; Weller & Saettler, 1980b). Colonization of seeds from resistant genotypes is also possible (Cafati, 1980; Mabagala, 1997).
In tropical and subtropical areas, survival on weeds and crop residues represents a particularly effective strategy for CBB dissemination (Arnaud‐Santana, 1991; Fininsa & Tefera, 2001; Fininsa & Yuen, 2002). Bacterial survival for more than 7 months has been observed in debris on or near the soil surface (Arnaud‐Santana, 1991; Chávez & Granada, 1988). Under a temperate climate, the survival capacity of CBB agents in crop residues is not clear. In some cases, CBB agents did not survive after 3 months in plant debris (Saettler et al., 1986; Wimalajeewa & Nancarrow, 1980). However, overwintering (up to 7 months) was recorded in Nebraska and Wisconsin (Gilbertson et al., 1990; Schuster & Coyne, 1974). In both tropical and temperate areas, burial of residues was shown to be effective in reducing the survival of pathogens to less than 5 weeks (Chávez & Granada, 1988; Wimalajeewa & Nancarrow, 1980).
CBB agents have been detected as epiphytes on various alternative hosts and weeds (Angeles‐Ramos et al., 1991; Cafati & Saettler, 1980b; Gent et al., 2005; Karavina et al., 2011), and their role as inoculum sources for CBB has been assessed under greenhouse conditions and in the field. Reciprocal secondary spread has been observed in the field between common bean and lambsquarters (Chenopodium album) or pigweed (Amaranthus retroflexus) 12 days after inoculation (Cafati & Saettler, 1980b). These observations are particularly important in the context of crop rotation. For example, epiphytic CBB agents were recovered from onion plants in fields cultivated for dry beans in the previous year, but not after the cultivation of other nonhost plants such as maize, sugar beet, or winter wheat (Gent et al., 2005).
7.2. Infection process
Primary infection usually starts with an epiphytic phase where CBB agents grow on the plant surface without invading the internal tissues and await favourable conditions for infection (Weller & Saettler, 1980a). Epiphytic survival is facilitated by aggregation in biofilms where bacterial populations grow and stabilize (Jacques et al., 2005). After growing on the bean leaf surface, bacteria penetrate into the host tissues through openings such as stomata, hydathodes, or woundings (Rudolph, 1993; Zaumeyer, 1930). Inside the host tissues, the bacteria multiply exponentially and may express their pathogenicity when the population achieves a threshold of about 106 bacterial cells per cm2 (Weller & Saettler, 1980a). The time of appearance and severity of symptoms are aggravated at optimal temperatures of 28 to 32 °C and a relative humidity above 80% (Darrasse et al., 2007; Weller & Saettler, 1980a). The progression of bacteria within the host leads to colonization of the vascular tissues, which can lead to wilting of the plant in the most severe cases (Vidaver, 1993). In nonoptimal conditions for bacterial multiplication, the bacteria colonize the plant symptomlessly (Darrasse et al., 2007; Weller & Saettler, 1980a).
7.3. Spread capacity
The dissemination of CBB agents is multifactorial. The spread of disease as well as its incidence and severity are favoured under warm temperature and high‐humidity conditions (Saettler, 1991). Bacteria present at the surface of infected plants or plant debris can be carried over short distances by wind‐blown rain or splashing. Thus, overhead irrigation is not recommended as it mimics rainfall and favours secondary spread of the bacteria (Akhavan et al., 2013). Dissemination of bacteria can also be caused by transportation via animals, farm workers, or agricultural equipment (Belete & Bastas, 2017; Saettler, 1991).
The role of insects as CBB vectors is still understudied, but has been reported for a long time (Sackett, 1905; Zaumeyer & Thomas, 1957). CBB agents were detected on the bodies of different bean‐feeding insect species such as Chalcodermus ebeninus, Empoasca sp., Nezara viridula, Cerotoma ruficornis, and Diaprepes abbreviata (Kaiser & Vakili, 1978). The bacteria were retrieved in faeces of C. ruficornis and D. abbreviata after being fed on infected leaves, and both species were able to transmit the disease at feeding sites in controlled trials. Further investigations are needed to assess the role of insects in CBB epidemics.
Transport of infected seed by human exchanges is a major way for Xcf and Xpp to travel long distances and spread around the world (Zaumeyer & Thomas, 1957). Thus, one can imagine that the worldwide distribution of CBB is related both to its ability to be transmitted by seeds and to the expansion of international trade, which relies on global exports and imports of potentially contaminated seed lots.
8. SEED TRANSMISSION OF CBB AGENTS: EVIDENCE AND PROCESSES
CBB agents were demonstrated to be seed‐transmitted as early as 1930 (Zaumeyer, 1930): Contaminated seeds harvested from diseased plants were sown among healthy seeds and developed disease symptoms that spread in the field.
8.1. The different routes of seed contamination
Contamination of the seed can be achieved through different pathways: (a) by direct contact of the seed either with tissues from the fruit with symptoms or with contaminated plant debris, (b) by the vascular pathway that passes through the funicle to the hilum, and (c) by the floral pathway through the stigma and style to the micropyle (Maude, 1997).
It has long been known that CBB seed contamination can occur by direct contact with tissues with symptoms in the field (Zaumeyer, 1932) or during threshing and postharvest treatments (Zaumeyer, 1930). Weller and Seattler (1980b) artificially mimicked this pathway of contamination by mixing healthy seeds with contaminated debris and showed that superficially contaminated seed can lead to disease outbreaks in the field.
CBB contamination via the vascular route has been known for a long time as well (Burkholder, 1921). This was illustrated by drawings of microscopic examinations showing bacterial disintegration of the funicle tissue and bacterial masses within the seed coat (Zaumeyer, 1930). Almost 100 years later, the vascular pathway was detailed using confocal laser scanning microscopy (CLSM) to visualize a green fluorescent protein (GFP)‐labelled Xcf strain within the seed tissues (Figure 4; Darrasse et al., 2018). Localization of the bacteria was assessed along the entire vascular route, from the funicle tissues to the hilar region, the vessels of the tracheid bar, and the intercellular spaces of seed coat parenchyma. Vascular seed infection by CBB agents was demonstrated by infiltrations in the pod pedicel or in the cotyledon node of 20‐day‐old seedlings (Weller & Saettler, 1980b) or by spraying plants whose reproductive organs were encapsulated in cellophane bags until harvest (Darsonval et al., 2008). Using directed mutagenesis, the vascular route was shown to require a functional type III secretion system (T3SS) and to be dependent on adhesins involved in the formation of bacterial aggregates (Figure 4; Darsonval et al., 2008, 2009).
FIGURE 4.
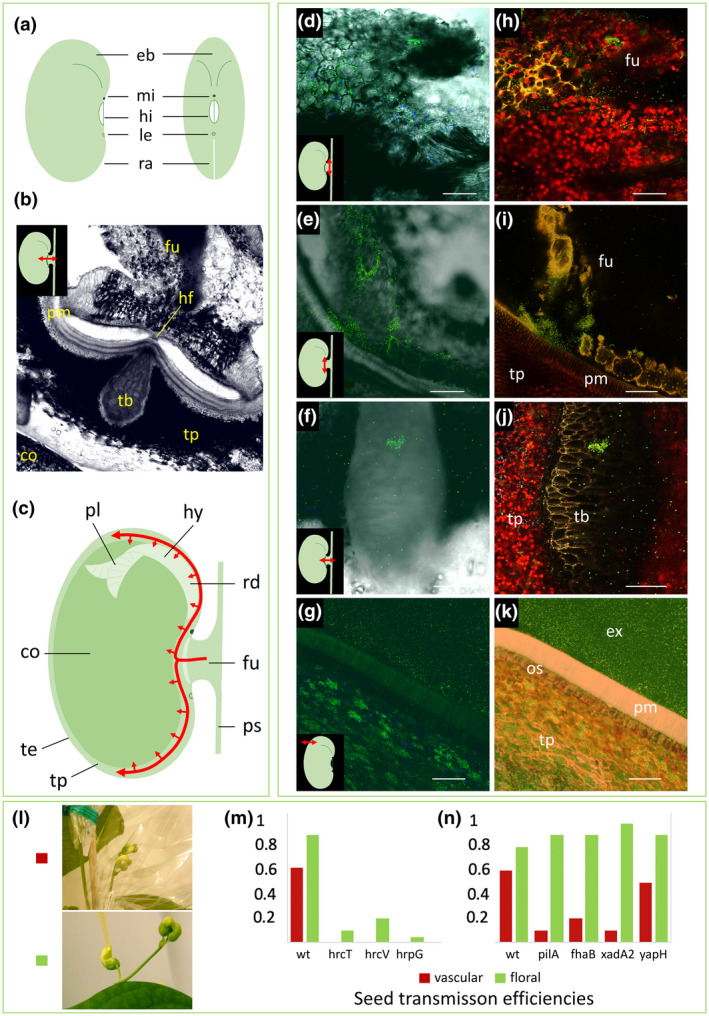
Evidence of seed transmission through the vascular and floral pathways. Tissues or structures are indicated by letters as follows: cotyledon (co), embryo bulge (eb), external side of seed (ex), funicle (fu), hilar fissure (hf), hilum (hi), hypocotyl (hy), lens (le), micropyle (mi), osteosclerides (os), plumule (pl), palisade of macrosclereids (pm), pod suture (ps), raphe (ra), radicule (rd), tracheid bar (tb), testa (te), testa parenchyma (tp). For microscopy images, we have enhanced luminosity by 40%. (a) Schematic views of the seed. (b) Transversal section of seed and funicle imaged using confocal laser‐scanning microscopy (CLSM). (c) Sagittal plane view of a seed attached by a funicle to a pod suture. Red curved arrows represent the most probable route of vascular infection for CBB agents according to the observations made (d–k), where bacteria are mainly retrieved in the testa parenchyma. (d–k) CLSM images of a green fluorescent protein (GFP)‐tagged Xanthomonas citri pv. fuscans (Xcf) strain in bean seeds (d–g). Images were generated by merging channels 488 nm and transmitted light to observe fluorescent objects within plant tissues, or under hyperspectral detector mode (excitation at 488 and 405 nm and signal reception with all channels). In some cases, autofluorescence of noncontaminated tissues can be observed. (h–k) The same images as in (d–g) under hyperspectral mode, respectively. Plant compounds fluoresce in yellow, red, or blue (probably corresponding to cell walls, chloroplasts, and amyloplasts, respectively), confirming that the green objects observed are indeed GFP‐tagged cells. Bacteria invade the funicle parenchyma, as shown (d, h) in a transversal section of the funicle or (e, i) in a frontal section of a seed at the hilum level. (f, j) Transversal section under the hilum, showing a bacterial aggregate in a tracheid‐like cell of the tracheid bar and scattered bacteria in intercellular spaces of the testa parenchyma. (g, k) Transversal section of a mature seed showing bacterial masses of GFP‐tagged cells within the testa parenchyma. (l) Frequencies of seed transmission of Xcf wild‐type strain (wt) and mutants in the type III secretion system (hrpG: Xcf::hrpG; hrcT: Xcf::hrcT; hrcTV: Xcf::hrcV) or adhesins (pilA: Xcf∆pilA; fhaB: Xcf::fhaB; xadA2: Xcf::xadA2; yapH: Xcf::yapH). Bacterial transmission to seeds through the vascular or floral pathways was tested, respectively, by spray inoculation of leaves, keeping the reproductive organs protected, or by depositing inoculum directly in the flower buds. Thirty plants per strain and treatment were harvested individually and their seeds were analysed for the presence of Xcf (data modified from histograms presented by Darsonval et al., 2008, 2009)
The floral route was evidenced by depositing the bacterial inoculum directly into the flower buds and taking care to extract the mature seeds without any contact with potentially contaminated external tissues. By this pathway, T3SS mutants, unable to contaminate seeds through the vascular pathway, were weakly transmissible to seeds, while mutants in genes encoding adhesins were transmitted as efficiently as the wild‐type strain (Figure 4; Darsonval et al., 2008, 2009). This indicates that floral organs can be an efficient and permissive entry point for pathogens, at least in growth chambers after inoculation. Yet, further analyses are needed to evaluate the importance of this pathway for the epidemiology of CBB agents in fields.
8.2. Transmission to the seedling
The localization and number of bacteria in seed can be decisive for successful transmission from the seed to the seedling. Evidence for this is often indirect because seed testing and microscopy are destructive methods. Contamination is generally restricted to the seed coat parenchyma. However, disruption of the testa and contamination of the embryo can be observed in some seeds with symptoms, together with bacterial aggregates and tissue disintegration. After seed imbibition, Xcf cells can be observed on the surface of the radicle and cotyledons and in the plumule (Darrasse et al., 2018). Zaumeyer (1930) reported that infections surrounded the embryo, which remains intact until germination. At this point, bacteria invade the tissues of the embryo, in particular the intercellular spaces of the cotyledons, and colonize the epicotyl and hypocotyl. By analysing several subsamples of seed lots, Weller and Saettler (1980a) showed that superficial contamination is more frequent than internal contamination and that contaminated seeds are not homogeneously distributed in the lots. They reported the existence of contaminated seed lots with and without symptoms. Nonintuitively, seeds with severe symptoms do not constitute a good primary inoculum because they rot after imbibition, fail to germinate, or give abnormal seedlings that do not develop further (Darrasse et al., 2018; Weller & Saettler, 1980b), and are generally eliminated during seed sorting operations. In contrast, symptomless seeds or seeds with less severe symptoms (hilum‐spotted seeds) lead to contaminated seedlings with normal development, allowing further transmission of the bacteria. Upon germination, bacteria multiply abundantly and their T3SS is not necessary to colonize the plantlet until 14 days. This may be favoured by the release of nutrients occurring during germination (Darrasse et al., 2010). This hypothesis is supported by gene expression analyses where Xcf do not repress plant defence pathways (e.g., the salicylic acid pathway) during seedling emergence, while this is the case in infiltrated seedlings or leaves.
9. DISEASE MANAGEMENT
9.1. Regulation
As mentioned above, CBB is endemic nearly everywhere where beans are cultivated, resulting in limited regulation. Nevertheless, quarantine was implemented in some pest‐free states of the USA such as Washington State (Washington State Bean Seed Quarantine Rules, 2019). In the European Union (EU), CBB agents have been regulated as nonquarantine pests (RNQPs) since December 2019. Before this date and since the beginning of the 21st century, CBB agents were listed as quarantine organisms in Annex II.A.2. of the EU council directive 2000/29/EC. This status was regarded as having contributed to limiting the introduction of contaminated seeds from third countries to CBB‐free EU member states (EFSA, 2014). However, CBB is now established in some EU countries (Figure 2). CBB agents are recognized as having a clear taxonomic identity, being present in the EU territory, being transmitted mainly through seeds, having an unacceptable direct economic impact at the place of production, and for which feasible and effective control measures are available. They hence comply with the RNQP definition (ISPM#2002). In consequence, in the EU, CBB agents are nowadays only regulated on bean seeds and there is no requirement for eradication in fields.
9.2. Sanitary practices
Only very limited chemical and biological options are available to efficiently control bacterial plant pathogens; also, control methods in use are limited to cultural practices and whenever possible the use of tolerant or resistant genotypes. Concerning CBB, the use of pathogen‐free seeds is certainly the most efficient management strategy. To ensure the availability of pathogen‐free seeds, bean seeds are produced in specified areas that are either free of CBB agents (de Boisgrollier, 1993) or whose climatic conditions are nonconducive for the disease (Gilbertson & Maxwell, 1992). In quarantine areas, imported seed lots are routinely tested using a method from the International Seed Testing Association involving isolation of bacterial strains, pathogenicity tests, and specific PCR assays (Audy et al., 1994; Boureau et al., 2013; Grimault et al., 2014). Seed lots are then rejected and/or eliminated if contaminated (at a rate of one seed out of 5,000), which is an efficient way to control the introduction of the disease in these countries.
9.3. Chemical and biological control
Chemical control is restricted to the application of copper‐based solutions in fields (Schwartz et al., 2005; Weller, 1975; Weller & Saettler, 1976). However, with CBB agents being vascular pathogens, the efficacy of foliar copper treatments is limited. Antibiotics such as streptomycin or kasugamycin are used in a few countries including the USA (Liang et al., 1992; Webster, 1983). Several alternative solutions for CBB control have been reported, including the application of a manganese‐based foliar fertilizer reducing the disease by more than 47% (Viecelli & Moerschbächer, 2013). Biological control using antagonistic bacteria such as Pseudomonas sp., Rahnella aquatilis, Rhodococcus fascians, or Bacillus spp. can also reduce CBB disease (Giorgio et al., 2016; Goncalves da Silva et al., 2008; Sallam, 2011; Zanatta et al., 2007) through induction of host defences, as shown by increased production of phenolic compounds and peroxidase activity that was observed in inoculated plants (Sallam, 2011). In vitro experiments showed that essential oils from multiple plants, including marigold (Tagetes minuta), blue pea (Clitoria ternatea), gingko (Ginkgo biloba), wintergreen (Gaultheria procumbens), oregano (Origanum vulgare), and lemongrass (Cymbopogon flexuosus), showed antimicrobial properties against CBB agents (Gakuubi et al., 2016; Kelemu et al., 2004; Sati & Joshi, 2011; Todorović et al., 2016). For example, eugenol is an essential oil component showing potential usefulness for disinfection of common bean seeds infected by Xcf (Lo Cantore et al., 2009). Likewise, a lipopeptide isolated from Paenibacillus polymyxa had a significant impact against Xpp (Mageshwaran et al., 2012).
9.4. Cultural practices
Given the paucity of efficient treatments, cultural practices are essential to control CBB. It is of particular importance to reduce the initial inoculum because CBB agents can survive within bean plant debris for several months (Arnaud‐Santana, 1991) as well as in dust on contaminated equipment (Belete & Bastas, 2017). Thus, regular cleaning of harvesting equipment and seed containers is a means to limit primary infection. Likewise, it is recommended to eliminate weeds, infected beans, and other potential hosts (Gilbertson et al., 1990; Saettler et al., 1986). The disposal of contaminated debris can significantly reduce the survival of bacteria responsible for the primary inoculum. Also, CBB epidemics can effectively be reduced through employing long crop‐rotations of 3 years or more to limit the risk of contamination of common bean crops by pathogens surviving on alternate hosts (Schwartz et al., 2005). However, the choice of the other crop is very important. For example, onion should be avoided, because it can serve as inoculum by symptomless epiphytic colonization (Gent et al., 2005). Intercropping can also help in reducing CBB incidence. For example, maize can serve as a physical barrier to the spreading of Xcf and Xpp (Fininsa, 1996).
9.5. Genetic resistance
The use of resistant varieties is an effective and environmentally sound approach to control CBB. Breeding resistance to CBB is a complex process that has been very well documented and reviewed (Miklas et al., 2006; Singh & Miklas, 2015; Tugume et al., 2018; Viteri, Terán et al., 2014; Yu et al., 2012). Despite two examples of resistance segregations corresponding to single genes (Adams et al., 1988; Zapata et al., 2010), CBB resistance is mainly quantitative and polygenic (Singh & Miklas, 2015; Singh & Schwartz, 2010; Yu et al., 2012). Moderate resistances to CBB exist within the Mesoamerican gene pool, while so far no resistances have been reported in the Andean gene pool (Singh & Miklas, 2015). Interestingly, studies focused on resistance (R) genes from the NOD‐like receptor (NLR) family highlighted large R gene clusters at chromosome ends, where elevated recombination led to distinct R gene repertoires in Andean and Mesoamerican common beans (Chen, Thareau, et al., 2018; David et al., 2009; Meziadi et al., 2016). It is, therefore, tempting to assume that the different resistance patterns between Andean and Mesoamerican gene pools could result from large divergences in R gene content within each gene pool. High resistances were found in other Phaseolus relatives such as P. coccineus, P. lunatus, and P. acutifolius. Thus, interspecific breeding has been a major way to introduce novel resistances in common beans.
To date, the identification of at least 27 quantitative trait loci (QTLs) of resistance to CBB spread across the 11 common bean chromosomes offers a solid source of resistances (Table 2). These QTLs do not appear to differentiate Xcf and Xpp and no significant crossover interactions between strains and common bean genotypes bearing different QTLs have been confirmed so far (Duncan et al., 2011; Mutlu et al., 2008; Opio et al., 1996; Viteri, Cregan, et al., 2014). Since the early 2000s, SAP6, a major QTL deriving from the Great Northern landrace cultivar Montana No. 5, has been effectively used for generating CBB‐resistant varieties grown in North and South America (Miklas et al., 2003). This QTL comprises 25 genes, 10 of which are associated with defence mechanisms (Zhu et al., 2016). In lines derived from crosses between P. vulgaris and P. acutifolius, two major resistance QTLs have been identified (BC420 and SU91) (Jung et al., 1997; Pedraza García et al., 1997; Shi, Navabi et al., 2011; Yu et al., 2000a), and analysis of their gene content highlighted several R gene candidates that still require functional validation (Perry et al., 2013; Shi, Chaudhary, et al., 2011).
TABLE 2.
Quantitative trait loci of resistance to common bacterial blight of bean
| Chromosome | Genetic marker | Leaves | Pods | Seeds | Field | Reference |
|---|---|---|---|---|---|---|
| Resistance source from Phaseolus vulgaris | ||||||
| 2 | D0108 | x | Nodari et al. (1993) | |||
| 3 | PvSNP85p745405 | x | Xie et al. (2017) | |||
| 5 | D1081 | x | Nodari et al. (1993) | |||
| 7 | D1390 | x | Nodari et al. (1993) | |||
| 7 | AD4.1150 | x | Jung et al. (1996, 1999) | |||
| 7 | AA19.600‐L6.800 | x | x | Miklas et al. (2000) | ||
| 9 | D0157 | x | Nodari et al. (1993) | |||
| 10 | SAP6.820 | x | x | x | Miklas et al. (1996, 2003) | |
| 10 | BC409.1250 | x | x | Jung et al. (1996, 1999) | ||
| 10 | W10.550 | x | x | Ariyarathne et al. (1999) | ||
| 10 | Xap1 | x | Zapata et al. (2010) | |||
| 10 | BMp10s174/s244 | x | Zhu et al. (2016) | |||
| 11 | BNG25A | x | Yu et al. (1998) | |||
| 11 | BNG154 | x | Yu et al. (1998) | |||
| Resistance source from Phaseolus acutifolius | ||||||
| 1 | R20.1250 | x | x | Ariyarathne et al. (1999) | ||
| 2 | U8.1100 | x | Jung et al. (1996) | |||
| 2 | AM02.1500 | x | Jung et al. (1997) | |||
| 2 | A10.1750 | x | x | Ariyarathne et al. (1999) | ||
| 3 | BNG21 | x | x | Tar'an et al. (2001) | ||
| 3 | C1.1550 | x | Jung et al. (1996) | |||
| 4 | BNG71 | x | x | Tar'an et al. (2001) | ||
| 5 | PHVPVPK‐1 | x | x | Tar'an et al. (2001) | ||
| 6 | BC420.900 | x | Jung et al. (1997); Yu et al. (2000) | |||
| 6 | BC420‐CG14 | x | Shi et al. (2012) | |||
| 7 | AL7.1050 | x | x | x | Jung et al. (1997) | |
| 7 | G17.400 | x | x | Ariyarathne et al. (1999) | ||
| 7 | Pv‐tttc001 | x | Yu et al. (2004) | |||
| 8 | SU91 | x | Pedraza García et al. (1997) | |||
| 8 | R7313 | x | x | Bai et al. (1997) | ||
| 8 | SU91‐CG11 | x | Shi et al. (2012) | |||
| 8 | BC432.1000 | x | Jung et al. (1997) | |||
| 9 | Y4.1600 | x | Ariyarathne et al. (1999) | |||
| 11 | BC446.1200 | x | Jung et al. (1996) | |||
| 11 | SNP47467 | x | x | Viteri, Cregan, et al. (2014) | ||
| Resistance source from Phaseolus coccineus | ||||||
| 7 | BNG40 | x | Yu et al. (1998) | |||
| 8 | BNG139 | x | Yu et al. (1998) | |||
Numerous efforts were made to introduce CBB resistance into beans, but the resistance observed had variable heritability and level of expression (Singh & Schwartz, 2010). Indeed, resistance can be achieved to varying degrees depending on the environment, the genetic background, or the epidemic pressure (Miklas et al., 2006). Also, different genetic systems appear to control resistance in pods and leaves, and resistances may not be effective against the various strains responsible for CBB (Aggour et al., 1989; Duncan et al., 2011). For these reasons, a pyramiding strategy was used to ensure resistance under different conditions and/or in different tissues (Miklas et al., 2000; Mutlu et al., 2005; Viteri & Singh, 2014). For example, the bred line VAX6 exhibits high resistance to a large range of Xcf and Xpp isolates (Duncan et al., 2011; Mahuku et al., 2006; Singh & Muñoz, 1999). However, bearing several resistance QTLs does not necessarily lead to an addition of beneficial effects, as there may be epistasis between QTLs, as observed between BC420 and SU91 (O’Boyle et al., 2007; Vandemark et al., 2008). Moreover, resistance QTLs are not always compatible with other agronomic traits. For example, BC420 is associated with the V gene responsible for the darkening of flowers and seeds, which is not a favourable agronomic trait (Liu et al., 2008; Yu et al., 2012).
Marker‐assisted selection of CBB‐resistant lines has been facilitated by the development of genetic maps (Bai et al., 1997; Freyre et al., 1998; Meziadi et al., 2016; Shi et al., 2012; Yu et al., 2000b) and by the sequencing of Andean and Mesoamerican common bean genomes (Schmutz et al., 2014; Vlasova et al., 2016). However, expression studies related to CBB resistance are still limited (Cooper, 2015; Shi, Chaudhary, et al., 2011). Recently, a first description of whole‐transcriptome changes upon infection of resistant and susceptible plants showed that resistance was associated with up‐regulation of the salicylic acid pathway and down‐regulation of photosynthesis (Foucher et al., 2020). Interestingly, 26 genes had a large difference of expression between the resistant and susceptible genotypes and colocalized with resistance QTLs. Further functional analysis of these genes, together with additional expression studies, could bring new leads towards cloning CBB resistance genes.
10. FUTURE PROSPECTS
CBB is an important disease worldwide that remains difficult to control given the paucity of efficient treatments. Recent efforts in sequencing complete genomes have provided solid data supporting the diversity of CBB agents and revealing candidate genes potentially involved in bean colonization. However, functional validation is still restricted to few analyses on the T3SS or adhesins (Darsonval et al., 2008, 2009). Transcriptomics together with the development of single mutants or transposon insertion sequencing (Tn‐seq) strategies will provide key information on the genes involved in the pathogenicity and/or the pathological convergence of CBB agents. Besides, epidemiological survey of CBB agents is still restricted to their detection with few specific PCR primers (Audy et al., 1994; Boureau et al., 2013). The existence of numerous genomic regions sharing 100% identity among all Xcf‐Xpp lineages (Aritua et al., 2015; Chen, Serres‐Giardi, et al., 2018) suggests that the development of a generic multilocus variable copy number of tandem repeats analysis (MLVA) scheme would be a pivotal tool to study the epidemiology of CBB agents.
On the plant side, cloning of R genes has been hindered by the complexity of the genetic basis of resistance to CBB. Another way to develop resistance would be to use TALE‐based strategies (Boch et al., 2014; Schornack et al., 2013), either by finding TALE‐related resistances in common bean diversity or by genome editing, as has been done in rice and grapefruit (Blanvillain‐Baufumé et al., 2017; Hummel et al., 2012; Jia et al., 2016; Li et al., 2012; Wang et al., 2018; Zeng et al., 2015). However, genome editing still requires further improvements in common bean transformation and regeneration techniques (Hnatuszko‐Konka et al., 2014, 2019).
Joint efforts to better understand the epidemiology of the disease as well as the molecular basis of the pathogenicity of the bacteria and the resistance of the plant should lead to improvements in the prevention and management of CBB.
ACKNOWLEDGEMENTS
This article is based upon work from COST Action CA16107 EuroXanth, supported by COST (European Cooperation in Science and Technology). The authors declare that they have no competing interests.
Chen NWG, Ruh M, Darrasse A, et al. Common bacterial blight of bean: a model of seed transmission and pathological convergence. Mol Plant Pathol. 2021;22:1464–1480. 10.1111/mpp.13067
Contributor Information
Nicolas W. G. Chen, Email: nicolas.chen@agrocampus-ouest.fr.
Marie‐Agnès Jacques, Email: marie-agnes.jacques@inrae.fr.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Adams, M.W. , Kelly, J.D. & Saettler, A.W. (1988) A gene for resistance to common blight (Xanthomonas campestris pv. phaseoli). Annual Report of the Bean Improvement Cooperative, 31, 73–74. [Google Scholar]
- Aggour, A.R. , Coyne, D.P. & Vidaver, A.K. (1989) Comparison of leaf and pod disease reactions of beans (Phaseolus vulgaris L.) inoculated by different methods with strains of Xanthomonas campestris pv. phaseoli (Smith) dye. Euphytica, 43, 143–152. [Google Scholar]
- Akhavan, A. , Bahar, M. , Askarian, H. , Lak, M.R. , Nazemi, A. & Zamani, Z. (2013) Bean common bacterial blight: Pathogen epiphytic life and effect of irrigation practices. Springerplus, 2, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi, S.M. , Sanjari, S. , Durand, F. , Brin, C. , Manceau, C. & Poussier, S. (2008) Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI‐1 family by multiple suppression subtractive hybridizations. Applied and Environmental Microbiology, 74, 3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles‐Ramos, R. , Vidaver, A.K. & Flynn, P. (1991) Characterization of epiphytic Xanthomonas campestris pv. phaseoli and pectolytic xanthomonads recovered from symptomless weeds in the Dominican Republic. Phytopathology, 81, 677–681. [Google Scholar]
- Aritua, V. , Harrison, J. , Sapp, M. , Buruchara, R. , Smith, J. & Studholme, D.J. (2015) Genome sequencing reveals a new lineage associated with lablab bean and genetic exchange between Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans . Frontiers in Microbiology, 6, 1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyarathne, H.M. , Coyne, D.P. , Jung, G. , Skroch, P.W. , Vidaver, A.K. , Steadman, J.R. , Miklas, P.N. & Bassett, M.J. (1999) Molecular mapping of disease resistance genes for halo blight, common bacterial blight, and bean common mosaic virus in a segregating population of common bean. Journal of the American Society for Horticultural Science, 124, 654–662. [Google Scholar]
- Arnaud‐Santana, E. (1991) Longevity of Xanthomonas campestris pv. phaseoli in naturally infested dry bean (Phaseolus vulgaris) debris. Plant Disease, 75, 952. [Google Scholar]
- Audy, P. , Laroche, A. , Saindon, G. , Huang, H.C. & Gilbertson, R.L. (1994) Detection of the bean common blight bacteria, Xanthomonas campestris pv. phaseoli and X. c. phaseoli var. fuscans, using the polymerase chain reaction. Phytopathology, 84, 1185–1192. [Google Scholar]
- Bai, Y. , Michaels, T.E. & Pauls, K.P. (1997) Identification of RAPD markers linked to common bacterial blight resistance genes in Phaseolus vulgaris L. Genome, 40, 544–551. [DOI] [PubMed] [Google Scholar]
- Belete, T. & Bastas, K.K. (2017) Common bacterial blight (Xanthomonas axonopodis pv. phaseoli) of beans with special focus on Ethiopian condition. Journal of Plant Pathology & Microbiology, 8, 403 [Google Scholar]
- Birch, P.R.J. , Hyman, L.J. , Taylor, R. , Opio, A.F. , Bragard, C. & Toth, I.K. (1997) RAPD PCR‐based differentiation of Xanthomonas campestris pv. phaseoli and Xanthomonas campestris pv. phaseoli var. fuscans. European Journal of Plant Pathology, 103, 809–814. [Google Scholar]
- Blanvillain‐Baufumé, S. , Reschke, M. , Solé, M. , Auguy, F. , Doucoure, H. , Szurek, B. et al. (2017) Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14‐inducing TAL effectors. Plant Biotechnology Journal, 15, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. , Bonas, U. & Lahaye, T. (2014) TAL effectors—pathogen strategies and plant resistance engineering. New Phytologist, 204, 823–832. [DOI] [PubMed] [Google Scholar]
- de Boisgrollier, H. (1993) Bilan des zones indemnes de bactérioses. Bulletin de la Federation Nationale des Agriculteurs Multiplicateurs de Semences, 124, 41–44. [Google Scholar]
- Boureau, T. , Kerkoud, M. , Chhel, F. , Hunault, G. , Darrasse, A. , Brin, C. et al (2013) A multiplex‐PCR assay for identification of the quarantine plant pathogen Xanthomonas axonopodis pv. phaseoli . Journal of Microbiological Methods, 92, 42–50. [DOI] [PubMed] [Google Scholar]
- Bradbury, J.F. (1986) Guide to Plant Pathogenic Bacteria. Walllingford, UK: CAB International. [Google Scholar]
- Broughton, W.J. , Hernández, G. , Blair, M. , Beebe, S. , Gepts, P. & Vanderleyden, J. (2003) Beans (Phaseolus spp.)—Model food legumes. Plant and Soil, 252, 55–128. [Google Scholar]
- Bultreys, A. & Gheysen, I. (2020) First report of Xanthomonas phaseoli pv. phaseoli and Xanthomonas citri pv. fuscans causing common bacterial blight of bean in Belgium. New Disease Reports, 41, 6. [Google Scholar]
- Burkholder, W.H. (1921) The bacterial blight of the bean, a systemic disease. Phytopathology, 11, 61–69. [Google Scholar]
- Cafati, C.R. (1980) Transmission of Xanthomonas phaseoli in seed of resistant and susceptible Phaseolus genotypes. Phytopathology, 70, 638. [Google Scholar]
- Cafati, C.R. & Saettler, A.W. (1980a) Effect of host on multiplication and distribution of bean common blight bacteria. Phytopathology, 70, 675–679. [Google Scholar]
- Cafati, C.R. & Saettler, A.W. (1980b) Role of nonhost species as alternate inoculum sources of Xanthomonas phaseoli . Plant Disease, 64, 194–196. [Google Scholar]
- Chan, J.W.Y.F. & Goodwin, P.H. (1999) Differentiation of Xanthomonas campestris pv. phaseoli from Xanthomonas campestris pv. phaseoli var. fuscans by PFGE and RFLP. European Journal of Plant Pathology, 105, 867–878. [Google Scholar]
- Chávez, C.L. & Granada, G.A. (1988) Supervivencia de Xanthomonas campestris pv. phaseoli, agente causal de la bacteriosis del frijol, bajo condiciones del Valle del Cauca, Colombia. Fitopatologia Colombiana, 12, 9–14. [Google Scholar]
- Chen, N.W.G. , Serres‐Giardi, L. , Ruh, M. , Briand, M. , Bonneau, S. , Darrasse, A. et al. (2018) Horizontal gene transfer plays a major role in the pathological convergence of Xanthomonas lineages on common bean. BMC Genomics, 19, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N.W.G. , Thareau, V. , Ribeiro, T. , Magdelenat, G. , Ashfield, T. , Innes, R.W. et al. (2018) Common bean subtelomeres are hot spots of recombination and favor resistance gene evolution. Frontiers in Plant Science, 9, 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupp, C. & Sherf, A.F. (1960) Vegetable Diseases and Their Control. New York: Ronald Press Company. [Google Scholar]
- Constantin, E.C. , Cleenwerck, I. , Maes, M. , Baeyen, S. , Van Malderghem, C. , De Vos, P. et al. (2016) Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathology, 65, 792–806. [Google Scholar]
- Cooper, D.M. (2015) Identification and characterization of common bacterial blight resistance genes in the resistant common bean (Phaseolus vulgaris) variety OAC Rex. Ontario, Canada: University of Guelph. Available at: http://hdl.handle.net/10214/8952 [Accessed 23 March 2021]. [Google Scholar]
- Corey, R.R. & Starr, M.P. (1957) Colony types of Xanthomonas phaseoli . Journal of Bacteriology, 74, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrasse, A. , Barret, M. , Cesbron, S. , Compant, S. & Jacques, M. (2018) Niches and routes of transmission of Xanthomonas citri pv. fuscans to bean seeds. Plant and Soil, 422, 115–128. [Google Scholar]
- Darrasse, A. , Bureau, C. , Samson, R. , Morris, C.E. & Jacques, M.A. (2007) Contamination of bean seeds by Xanthomonas axonopodis pv. phaseoli associated with low bacterial densities in the phyllosphere under field and greenhouse conditions. European Journal of Plant Pathology, 119, 203–215. [Google Scholar]
- Darrasse, A. , Carrère, S. , Barbe, V. , Boureau, T. , Arrieta‐Ortiz, M.L. , Bonneau, S. et al. (2013) Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834‐R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics, 14, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrasse, A. , Darsonval, A. , Boureau, T. , Brisset, M.N. , Durand, K. & Jacques, M.A. (2010) Transmission of plant‐pathogenic bacteria by nonhost seeds without induction of an associated defense reaction at emergence. Applied and Environmental Microbiology, 76, 6787–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsonval, A. , Darrasse, A. , Durand, K. , Bureau, C. , Cesbron, S. & Jacques, M.‐A. (2009) Adhesion and fitness in the bean phyllosphere and transmission to seed of Xanthomonas fuscans subsp. fuscans . Molecular Plant‐Microbe Interactions, 22, 747–757. [DOI] [PubMed] [Google Scholar]
- Darsonval, A. , Darrasse, A. , Meyer, D. , Demarty, M. , Durand, K. , Bureau, C. et al. (2008) The type III secretion system of Xanthomonas fuscans subsp. fuscans is involved in the phyllosphere colonization process and in transmission to seeds of susceptible beans. Applied and Environmental Microbiology, 74, 2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, P. , Chen, N.W.G. , Pedrosa‐Harand, A. , Thareau, V. , Sévignac, M. , Cannon, S.B. et al. (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiology, 151, 1048–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, W. , Baumdicker, F. & Neher, R.A. (2018) panX: Pan‐genome analysis and exploration. Nucleic Acids Research, 46, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, R.W. , Singh, S.P. & Gilbertson, R.L. (2011) Interaction of common bacterial blight bacteria with disease resistance quantitative trait Loci in common bean. Phytopathology, 101, 425–435. [DOI] [PubMed] [Google Scholar]
- Dye, D.W. , Bradbury, J.F. , Goto, M. , Hayward, A.C. , Lelliott, R.A. & Schroth, M.N. (1980) International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Review of Plant Pathology, 59, 153–159. [Google Scholar]
- EFSA (2014) Scientific opinion on the pest categorisation of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans . EFSA Journal, 12, 3856. [Google Scholar]
- El‐Sharkawy, T.A. & Huisingh, D. (1971) Differentiation among Xanthomonas species by polyacrylamide gel electrophoresis of soluble proteins. Journal of General Microbiology, 68, 155–165. [Google Scholar]
- EPPO (2021) EPPO global database. Available at: https://gd.eppo.int [Accessed 29 March 2021]. [Google Scholar]
- Fininsa, C. (1996) Effect of intercropping bean with maize on bean common bacterial blight and rust diseases. International Journal of Pest Management, 42, 51–54. [Google Scholar]
- Fininsa, C. & Tefera, T. (2001) Effect of primary inoculum sources of bean common bacterial blight on early epidemics, seed yield and quality aspects. International Journal of Pest Management, 47, 221–225. [Google Scholar]
- Fininsa, C. & Yuen, J. (2002) Temporal progression of bean common bacterial blight (Xanthomonas campestris pv. phaseoli) in sole and intercropping systems. European Journal of Plant Pathology, 108, 485–495. [Google Scholar]
- Foucher, J. , Ruh, M. , Préveaux, A. , Carrère, S. , Pelletier, S. , Briand, M. et al. (2020) Common bean resistance to Xanthomonas is associated with upregulation of the salicylic acid pathway and downregulation of photosynthesis. BMC Genomics, 21, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyre, R. , Skroch, P.W. , Geffroy, V. , Adam‐Blondon, A.‐f. , Shirmohamadali, A. , Johnson, W.C. et al. (1998) Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theoretical and Applied Genetics, 97, 847–856. [Google Scholar]
- Gakuubi, M.M. , Wagacha, J.M. , Dossaji, S.F. & Wanzala, W. (2016) Chemical composition and antibacterial activity of essential oils of Tagetes minuta (Asteraceae) against selected plant pathogenic bacteria. International Journal of Microbiology, 2016, 10.1155/2016/7352509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent, D.H. , Lang, J.M. & Schwartz, H.F. (2005) Epiphytic survival of Xanthomonas axonopodis pv. allii and X. axonopodis pv. phaseoli on leguminous hosts and onion. Plant Disease, 89, 558–564. [DOI] [PubMed] [Google Scholar]
- Gilbertson, R.L. & Maxwell, D.P. (1992). Common bacterial blight of bean. In: Chaube, H.S. , Singh, U.S. & Mukhopadhay, A.N. (Eds.) Plant Diseases of International Importance. Volume II. Diseases of vegetables and oil seed crops. NJ, USA: Prentice Hall Inc., pp. 18–39. [Google Scholar]
- Gilbertson, R.L. , Rand, R.E. , Hagedorn, D.J. , and others. (1990) Survival of Xanthomonas campestris pv. phaseoli and pectolytic strains of X. campestris in bean debris. Plant Disease, 74, 322–327. [Google Scholar]
- Giorgio, A. , Cantore, P.L. , Shanmugaiah, V. , Lamorte, D. & Iacobellis, N.S. (2016) Rhizobacteria isolated from common bean in southern Italy as potential biocontrol agents against common bacterial blight. European Journal of Plant Pathology, 144, 297–309. [Google Scholar]
- Goncalves da Silva, E. , Bittencourt Moura, A. , Cardoso Deuner, C. & Farias, D.R. (2008) Estudo de mecanismos de biocontrole do crestamento bacteriano do feijoeiro por bactérias. Revista Ceres, 55, 377–383. [Google Scholar]
- Goodwin, P.H. & Sopher, C.R. (1994) Brown pigmentation of Xanthomonas campestris pv. phaseoli associated with homogentisic acid. Canadian Journal of Microbiology, 40, 28–34. [Google Scholar]
- Grimault, V. , Olivier, V. , Rolland, M. , Darrasse, A. & Jacques, M.‐A.A. (2014). Detection of Xanthomonas axonopodis pv. phaseoli on Phaseolus vulgaris . In: International rules for seed testing annexe to chapter 7: Seed health testing methods, 2014. Bassersdorf, Switzerland: International Seed Testing Association (ISTA). pp. 7‐021‐1‐20. [Google Scholar]
- Hajri, A. , Brin, C. , Hunault, G. , Lardeux, F. , Lemaire, C. , Manceau, C. et al. (2009) A “repertoire for repertoire” hypothesis: Repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . PLoS One, 4, e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnatuszko‐Konka, K. , Kowalczyk, T. , Gerszberg, A. , Glińska, S. & Grzegorczyk‐Karolak, I. (2019) Regeneration of Phaseolus vulgaris from epicotyls and hypocotyls via direct organogenesis. Scientific Reports, 9, 6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnatuszko‐Konka, K. , Kowalczyk, T. , Gerszberg, A. , Wiktorek‐Smagur, A. & Kononowicz, A.K. (2014) Phaseolus vulgaris—Recalcitrant potential. Biotechnology Advances, 32, 1205–1215. [DOI] [PubMed] [Google Scholar]
- Hummel, A.W. , Doyle, E.L. & Bogdanove, A.J. (2012) Addition of transcription activator‐like effector binding sites to a pathogen strain‐specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytologist, 195, 883–893. [DOI] [PubMed] [Google Scholar]
- Indiana, A. , Briand, M. , Arlat, M. , Gagnevin, L. , Koebnik, R. , Noel, L.D. , Portier, P. , Darrasse, A. & Jacques, M.A. (2014) Draft genome sequence of the flagellated Xanthomonas fuscans subsp. fuscans strain CFBP 4884. Genome Announcements, 2, 00966–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISPM#16 . (2002) Regulated non‐quarantine pests: concept and application. In: International Standards for Phytosanitary Measures. Rome, Italy: Secretariat of the International Plant Protection Convention, Food and Agriculture Organization of the United Nations. [Google Scholar]
- Jacques, M.‐A. , Josi, K. , Darrasse, A. & Samson, R. (2005) Xanthomonas axonopodis pv. phaseoli var. fuscans is aggregated in stable biofilm population sizes in the phyllosphere of field‐grown beans. Applied and Environmental Microbiology, 71, 2008–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, H. , Orbovic, V. , Jones, J.B. & Wang, N. (2016) Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4: DCsLOB1.3 infection. Plant Biotechnology Journal, 14, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G. , Coyne, D.P. , Skroch, P.W. , Nienhuis, J. , Arnaud‐Santana, E. , Bokosi, J. et al. (1996) Molecular markers associated with plant architecture and resistance to common blight, web blight, and rust in common beans. Journal of the American Society for Horticultural Science, 121, 794–803. [Google Scholar]
- Jung, G. , Skroch, P.W. , Coyne, D.P. , Nienhuis, J. , Arnaud‐Santana, E. , Ariyarathne, H.M. et al. (1997) Molecular‐marker‐based genetic analysis of tepary bean‐derived common bacterial blight resistance in different developmental stages of common bean. Journal of the American Society for Horticultural Science, 122, 329–337. [Google Scholar]
- Jung, G. , Skroch, P.w. , Nienhuis, J. , Coyne, D.p. , Arnaud‐Santana, E. , Ariyarathne, H.M. et al. (1999) Confirmation of QTL associated with common bacterial blight resistance in four different genetic backgrounds in common bean. Crop Science, 39, 1448–1455. [Google Scholar]
- Kaiser, W.J. & Vakili, N.G. (1978) Insect transmission of pathogenic xanthomonads to bean and cowpea in Puerto Rico. Phytopathology, 68, 1057–1063. [Google Scholar]
- Karavina, C. , Mandumbu, R. , Parwada, C. & Tibugari, H. (2011) A review of the occurrence, biology and management of common bacterial blight. Journal of Agricultural Technology, 7, 1459–1474. [Google Scholar]
- Kelemu, S. , Cardona, C. & Segura, G. (2004) Antimicrobial and insecticidal protein isolated from seeds of Clitoria ternatea, a tropical forage legume. Plant Physiology and Biochemistry, 42, 867–873. [DOI] [PubMed] [Google Scholar]
- Kremer, F.S. , de Souza Junior, I.T. , Guimarães, A.M. , Woloski, R.S.D. , Moura, A.B. & da Silva, Pinto L. (2019) Genome sequence of Xanthomonas fuscans subsp. fuscans strain Xff49: a new isolate obtained from common beans in southern Brazil. Brazilian Journal of Microbiology, 50, 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo, G.R. , Roffey, R. & Gabriel, D.W. (1987) Pathovars of Xanthomonas campestris are distinguishable by restriction fragment‐length polymorphism. International Journal of Systematic Bacteriology, 37, 214–221. [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. & Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nature Biotechnology, 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Liang, L.Z. , Halloin, J.M. & Saettler, A.W. (1992) Use of polyethylene glycol and glycerol as carriers of antibiotics for reduction of Xanthomonas campestris pv. phaseoli in navy bean seeds. Plant Disease, 76, 857–879. [Google Scholar]
- Liu, S. , Yu, K. & Park, S.J. (2008) Development of STS markers and QTL validation for common bacterial blight resistance in common bean. Plant Breeding, 127, 62–68. [Google Scholar]
- Lo Cantore, P. , Shanmugaiah, V. & Iacobellis, N.S. (2009) Antibacterial activity of essential oil components and their potential use in seed disinfection. Journal of Agriculture and Food Chemistry, 57, 9454–9461. [DOI] [PubMed] [Google Scholar]
- Mabagala, R.B. (1997) The effect of populations of Xanthomonas campestris pv. phaseoli in bean reproductive tissues on seed infection of resistant and susceptible bean genotypes. European Journal of Plant Pathology, 103, 175–181. [Google Scholar]
- Mageshwaran, V. , Walia, S. & Annapurna, K. (2012) Isolation and partial characterization of antibacterial lipopeptide produced by Paenibacillus polymyxa HKA‐15 against phytopathogen Xanthomonas campestris pv. phaseoli M‐5. World Journal of Microbiology & Biotechnology, 28, 909–917. [DOI] [PubMed] [Google Scholar]
- Mahuku, G.S. , Jara, C. , Henriquez, M.A. , Castellanos, G. & Cuasquer, J. (2006) Genotypic characterization of the common bean bacterial blight pathogens, Xanthomonas axonopodis pv. phaseoli and Xanthomonas axonopodis pv. phaseoli var. fuscans by rep‐PCR and PCR‐RFLP of the ribosomal genes. Journal of Phytopathology, 154, 35–44. [Google Scholar]
- Marques, A.S. dos A. , Guimarães, P.M. , dos Santos, J.P. & Vieira, T.M. (2005) Survival and viability of Xanthomonas axonopodis pv. phaseoli associated with bean seeds stored under controlled conditions. Fitopatologia Brasileira, 30, 527–531. [Google Scholar]
- Maude, R.B. (1997) Seedborne diseases and their control: principles and practice. Wallingford, UK: CAB International. [Google Scholar]
- Meziadi, C. , Richard, M.M.S. , Derquennes, A. , Thareau, V. , Blanchet, S. , Gratias, A. et al. (2016) Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence. Plant Science, 242, 351–357. [DOI] [PubMed] [Google Scholar]
- Mhedbi, N. (2010) Approches cumulées de phylogénie et d’écologie pour déterminer les bases génétiques de la spécificité d’hôte des bactéries phytopathogènes, cas des Xanthomonas spp. Université d'Angers, PhD thesis. Available at: https://tel.archives‐ouvertes.fr/tel‐00648729 [Accessed 23 March 2021]. [Google Scholar]
- Mhedbi‐Hajri, N. , Hajri, A. , Boureau, T. , Darrasse, A. , Durand, K. , Brin, C. et al. (2013) Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis . PLoS One, 8, e58474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklas, P.N. , Coyne, D.P. , Grafton, K.F. , Mutlu, N. , Reiser, J. , Lindgren, D.T. et al. (2003) A major QTL for common bacterial blight resistance derives from the common bean great northern landrace cultivar Montana No. 5. Euphytica, 131, 137–146. [Google Scholar]
- Miklas, P.N. , Delorme, R. , Stone, V. , Daly, M.J. , Stavely, J.R. , Steadman, J.R. et al. (2000) Bacterial, fungal, and viral disease resistance loci mapped in a recombinant inbred common bean population ('Dorado’/XAN 176). Journal of the American Society for Horticultural Science, 125, 476–481. [Google Scholar]
- Miklas, P.N. , Johnson, E. , Stone, V. , Beaver, J.S. , Montoya, C. & Zapata, M. (1996) Selective mapping of QTL conditioning disease resistance in common bean. Crop Science, 36, 1344–1351. [Google Scholar]
- Miklas, P.N. , Kelly, J.D. , Beebe, S.E. & Blair, M.W. (2006) Common bean breeding for resistance against biotic and abiotic stresses: From classical to MAS breeding. Euphytica, 147, 105–131. [Google Scholar]
- Mkandawire, A.B.C. , Mabagala, R.B. , Guzmán, P. , Gepts, P. & Gilbertson, R.L. (2004) Genetic diversity and pathogenic variation of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) suggests pathogen coevolution with the common bean. Phytopathology, 94, 593–603. [DOI] [PubMed] [Google Scholar]
- Mutlu, N. , Miklas, P. , Reiser, J. & Coyne, D. (2005) Backcross breeding for improved resistance to common bacterial blight in pinto bean (Phaseolus vulgaris L.). Plant Breeding, 124, 282–287. [Google Scholar]
- Mutlu, N. , Vidaver, A.K. , Coyne, D.P. , Steadman, J.R. , Lambrecht, P.A. & Reiser, J. (2008) Differential pathogenicity of Xanthomonas campestris pv. phaseoli and X. fuscans subsp. fuscans strains on bean genotypes with common blight resistance. Plant Disease, 92, 546–554. [DOI] [PubMed] [Google Scholar]
- Nodari, R.O. , Tsai, S.M. , Guzmán, P. , Gilbertson, R.L. & Gepts, P. (1993) Toward an integrated linkage map of common bean. III. Mapping genetic factors controlling host–bacteria interactions. Genetics, 134, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle, P.D. , Kelly, J.D. & Kirk, W.W. (2007) Use of marker‐assisted selection to breed for resistance to common bacterial blight in common bean. Journal of the American Society for Horticultural Science, 132, 381–386. [Google Scholar]
- Opio, A.F. , Allen, D.J. & Teri, J.M. (1996) Pathogenic variation in Xanthonomas campestris pv. phaseoli, the causal agent of common bacterial blight in Phaseolus beans. Plant Pathology, 45, 1126–1133. [Google Scholar]
- Opio, A. , Teri, J. & Allen, D. (1993) Studies on seed transmission of Xanthomonas campestris pv. phaseoli in common beans in Uganda. African Crop Science Journal, 1, 59–67. [Google Scholar]
- Pastor‐Corrales, M.A. , Beebe, S.E. & Correa, F.J. (1981) Comparing 2 inoculation techniques for evaluating resistance in beans to Xanthomonas campestris pv. phaseoli . Fifth International Conference on Plant Pathogenic Bacteria, 493–503.
- Pedraza García, F. , Gallego, G.J. , Beebe, S.E. , & Tohme, M.J. (1997). Marcadores SCAR y RAPD para la resitencia a la bacteriosis comun (CBB). In: Singh, S.P. & Voysest, O. (Eds.) Taller de mejoramiento de frijol para el Siglo XXI: bases para una estrategia para America Latina. Cali, Colombia: International Center for Tropical Agriculture (CIAT), pp. 130–134. Available at: https://cgspace.cgiar.org/handle/10568/80268 [Accessed 23 March 2021]. [Google Scholar]
- Perry, G. , DiNatale, C. , Xie, W. , Navabi, A. , Reinprecht, Y. , Crosby, W. et al. (2013) A comparison of the molecular organization of genomic regions associated with resistance to common bacterial blight in two Phaseolus vulgaris genotypes. Frontiers in Plant Science, 4, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M.N. , Dehal, P.S. & Arkin, A.P. (2010) FastTree 2—Approximately maximum‐likelihood trees for large alignments. PLoS One, 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, J. , Luo, H. & Hao, B. (2004) CVTree: A phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Research, 32, 45–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker, J.L.W. , Louws, F.J. , Schultz, M.H. , Rossbach, U. , Vauterin, L. , Swings, J. et al. (2005) A comprehensive species to strain taxonomic framework for Xanthomonas . Phytopathology, 95, 1098–1111. [DOI] [PubMed] [Google Scholar]
- Rousseau, C. , Belin, E. , Bove, E. , Rousseau, D. , Fabre, F. , Berruyer, R. et al. (2013) High throughput quantitative phenotyping of plant resistance using chlorophyll fluorescence image analysis. Plant Methods, 9, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, K. (1993) Infection of the plant by Xanthomonas . In: Swings, J.G. & Civerolo, E.L. (Eds.) Xanthomonas. Dordrecht, Netherlands: Springer, pp. 193–264. [Google Scholar]
- Ruh, M. , Briand, M. , Bonneau, S. , Jacques, M.‐A. & Chen, N.W.G. (2017a) Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors. BMC Genomics, 18, 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruh, M. , Briand, M. , Bonneau, S. , Jacques, M. & Chen, N.W.G. (2017b) First complete genome sequences of Xanthomonas citri pv. vignicola strains CFBP7111, CFBP7112, and CFBP7113 obtained using long‐read technology. Genome Announcements, 5, e00813‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet, K.A. (1959) Studies in the bacterial diseases of Sudan crops: III. on the occurrence, host range and taxonomy of the bacteria causing leaf blight diseases of certain leguminous plants. Annals of Applied Biology, 47, 318–331. [Google Scholar]
- Sackett, W.G. (1905) Some bacterial diseases of plants prevalent in Michigan. Michigan State Agricultural College Experiment Station. [Google Scholar]
- Saettler, A.W. (1989) Common bacterial blight. Bean Production Problems in the Tropics, 2, 261–283. [Google Scholar]
- Saettler, A.W. (1991) Diseases caused by bacteria. In: Hall, R. (Ed.) Compendium of bean diseases. St Paul, MN, USA: APS Press, pp. 29–32. [Google Scholar]
- Saettler, A.W. , Cafati, C.R. & Weller, D.M. (1986) Nonoverwintering of Xanthomonas bean blight bacteria in Michigan. Plant Disease, 70, 285. [Google Scholar]
- Sallam, N.M. (2011) Biological control of common blight of bean (Phaseolus vulgaris) caused by Xanthomonas axonopodis pv. phaseoli by using the bacterium Rahnella aquatilis . Archives of Phytopathology and Plant Protection, 44, 1966–1975. [Google Scholar]
- Sati, S.C. & Joshi, S. (2011) Antibacterial activities of Ginkgo biloba L. leaf extracts. Scientific World Journal, 11, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, N.W. , Postnikova, E. , Lacy, G.H. , Sechler, A. , Agarkova, I. , Stromberg, P.E. et al. (2005) Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) Dye 1978 as X. alfalfae subsp. alfalfae (ex Riker 1935) sp. nov. nom. rev.; and ‘‘var. fuscans’’ of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Systematic and Applied Microbiology, 28, 494–518. [DOI] [PubMed] [Google Scholar]
- Schmutz, J. , McClean, P.E. , Mamidi, S. , Wu, G.A. , Cannon, S.B. , Grimwood, J. et al. (2014) A reference genome for common bean and genome‐wide analysis of dual domestications. Nature Genetics, 46, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , Moscou, M.J. , Ward, E.R. & Horvath, D.M. (2013) Engineering plant disease resistance based on TAL effectors. Annual Review of Phytopathology, 51, 383–406. [DOI] [PubMed] [Google Scholar]
- Schuster, M.L. & Coyne, D.P. (1974) Survival mechanisms of phytopathogenic bacteria. Annual Review of Phytopathology, 12, 199–221. [Google Scholar]
- Schuster, M.L. , Sayre, R.M. , & others. (1967) A coryneform bacterium induces purple‐colored seed and leaf hypertrophy of Phaseolus vulgaris and other Leguminosae. Phytopathology, 57, 1064–1066. [Google Scholar]
- Schwartz, H.F. , Steadman, J.R. , Hall, R. & Forster, R.L. (2005) Compendium of bean diseases. St Paul, MN, USA: APS Press. [Google Scholar]
- Shi, C. , Chaudhary, S. , Yu, K. , Park, S.J. , Navabi, A. & McClean, P.E. (2011) Identification of candidate genes associated with CBB resistance in common bean HR45 (Phaseolus vulgaris L.) using cDNA‐AFLP. Molecular Biology Reports, 38, 75–81. [DOI] [PubMed] [Google Scholar]
- Shi, C. , Navabi, A. & Yu, K. (2011) Association mapping of common bacterial blight resistance QTL in Ontario bean breeding populations. BMC Plant Biology, 11, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, C. , Yu, K. , Xie, W. , Perry, G. , Navabi, A. , Peter Pauls, K. et al. (2012) Development of candidate gene markers associated to common bacterial blight resistance in common bean. Theoretical and Applied Genetics, 125, 1525–1537. [DOI] [PubMed] [Google Scholar]
- Singh, S.P. & Miklas, P.N. (2015) Breeding common bean for resistance to common blight : A review. Crop Science, 55, 971–984. [Google Scholar]
- Singh, S.P. & Muñoz, C.G. (1999) Resistance to common bacterial blight among Phaseolus species and common bean improvement. Crop Science, 39, 80–89. [Google Scholar]
- Singh, S.P. & Schwartz, H.F. (2010) Breeding common bean for resistance to diseases: A review. Crop Science, 50, 2199–2223. [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, M.D. & Wallen, V.R. (1970) Epidemiological and ecological relations of Xanthomonas phaseoli and X. phaseoli var. fuscans in southwestern Ontario, 1961–1968. Canadian Journal of Botany, 48, 1329–1334. [Google Scholar]
- Tar’an, B. , Michaels, T.E. & Pauls, K.P. (2001) Mapping genetic factors affecting the reaction to Xanthomonas axonopodis pv. phaseoli in Phaseolus vulgaris L. under field conditions. Genome, 44, 1046–1056. [PubMed] [Google Scholar]
- Todorović, B. , Potočnik, I. , Rekanović, E. , Stepanović, M. , Kostić, M. , Ristić, M. et al. (2016) Toxicity of twenty‐two plant essential oils against pathogenic bacteria of vegetables and mushrooms. Journal of Environmental Science and Health, Part B, 51, 832–839. [DOI] [PubMed] [Google Scholar]
- Tugume, J.K. , Tusiime, G. , Male, A. , Buruchara, R. & Mugisha, C. (2018) Diversity and interaction of common bacterial blight disease‐causing bacteria (Xanthomonas spp.) with Phaseolus L. Crop Journal, 7, 1–7. [Google Scholar]
- Vakili, N.G. , Kaiser, W.J. , Enrique Pérez, J. & Cortés‐Monllor, A. (1974) Bacterial blight of beans caused by two Xanthomonas pathogenic types from Puerto Rico. Phytopathology, 65, 401–403. [Google Scholar]
- Vandemark, G.J. , Fourie, D. & Miklas, P.N. (2008) Genotyping with real‐time PCR reveals recessive epistasis between independent QTL conferring resistance to common bacterial blight in dry bean. Theoretical and Applied Genetics, 117, 513–522. [DOI] [PubMed] [Google Scholar]
- Vauterin, L. , Hoste, B. , Kersters, K. & Swings, J. (1995) Reclassification of Xanthomonas . International Journal of Systematic Bacteriology, 45, 472–489. [Google Scholar]
- Vidaver, A.K. (1993) Xanthomonas campestris pv. phaseoli: cause of common bacterial blight of bean. In: Swings, J.G. & Civerolo, E.L. (Eds.) Xanthomonas. London, UK: Chapman & Hall, pp. 40–44. [Google Scholar]
- Viecelli, C.A. & Moerschbächer, T. (2013) Control of the common bacterial blight in the bean crop by using foliar fertilizers. Scientia Agraria Paranaensis, 12, 66–72. [Google Scholar]
- Viteri, D.M. , Cregan, P.B. , Trapp, J.J. , Miklas, P.N. & Singh, S.P. (2014) A new common bacterial blight resistance QTL in VAX 1 common bean and interaction of the new QTL, SAP6, and SU91 with bacterial strains. Crop Science, 54, 1598–1608. [Google Scholar]
- Viteri, D.M. & Singh, S.P. (2014) Response of 21 common beans of diverse origins to two strains of the common bacterial blight pathogen, Xanthomonas campestris pv. phaseoli . Euphytica, 200, 379–388. [Google Scholar]
- Viteri, D.M. , Terán, H. , Asensio‐s, M.C. , Asensio, C. , Porch, T.G. , Miklas, P.N. et al. (2014) Progress in breeding Andean common bean for resistance to common bacterial blight. Crop Science, 54, 2084–2092. [Google Scholar]
- Vlasova, A. , Capella‐Gutiérrez, S. , Rendón‐Anaya, M. , Hernández‐Oñate, M. , Minoche, A.E. , Erb, I. et al. (2016) Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biology, 17, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallen, V.R. & Jackson, H.R. (1975) Model for yield loss determination of bacterial blight of field beans utilizing aerial infrared photography combined with field plot studies. Phytopathology, 65, 942. [Google Scholar]
- Wang, J. , Zeng, X. , Tian, D. , Yang, X. , Wang, L. & Yin, Z. (2018) The pepper Bs4C proteins are localized to the endoplasmic reticulum (ER) membrane and confer disease resistance to bacterial blight in transgenic rice. Molecular Plant Pathology, 19, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington State bean seed quarantine rules . (2019). Available at: https://apps.leg.wa.gov/wac/default.aspx?dispo=true&cite=16‐301‐365 [Accessed 23 March 2021].
- Webster, D.M. (1983) Bacterial blights of snap beans and their control. Plant Disease, 67, 935. [Google Scholar]
- Weller, D.M. (1975) Chemical control of bean common and fuscous bacterial blights. Michigan State University. MS thesis. [Google Scholar]
- Weller, D.M. & Saettler, A.W. (1976) Chemical control of common and fuscous bacterial blights in Michigan navy (pea) beans. Plant Disease Reporter, 60, 793–797. [Google Scholar]
- Weller, D.M. & Saettler, A.W. (1980a) Colonization and distribution of Xanthomonas phaseoli and Xanthomonas phaseoli var. fuscans in field‐grown navy beans. Phytopathology, 70, 500–506. [Google Scholar]
- Weller, D.M. & Saettler, A.W. (1980b) Evaluation of seedborne Xanthomonas phaseoli and X. phaseoli var. fuscans as primary inocula in bean blights. Phytopathology, 70, 148–152. [Google Scholar]
- Wimalajeewa, D.L.S. & Nancarrow, R.J. (1980) Survival in soil of bacteria causing common and halo blights of French bean in Victoria. Australian Journal of Experimental Agriculture and Animal Husbandry, 20, 102–104. [Google Scholar]
- Xie, W. , Khanal, R. , McClymont, S. , Stonehouse, R. , Bett, K. , Yu, K. et al. (2017) Interaction of quantitative trait loci for resistance to common bacterial blight and pathogen isolates in Phaseolus vulgaris L. Molecular Breeding, 37, 55. [Google Scholar]
- Xie, W. , Smith, T. , Perry, G. & Pauls, P.K. (2016) Towards an understanding of interactions between bacterial virulence factors and genes in bean resistance QTL for breeding beans with durable common bacterial blight resistance. Annual Report of the Bean Improvement Cooperative (USA), 59, 13–14. [Google Scholar]
- Xie, W. , Yu, K. , Pauls, K.P. & Navabi, A. (2012) Application of image analysis in studies of quantitative disease resistance, exemplified using common bacterial blight–common bean pathosystem. Phytopathology, 102, 434–442. [DOI] [PubMed] [Google Scholar]
- Yoshii, K. , Galvez, G.E. & Alvarez, G. (1975) Estimation of yield losses in beans caused by common blight. Proceedings of the American Phytopathological Society, 3, 298. [Google Scholar]
- Yu, K. , Park, S.J. & Poysa, V. (2000a) Marker‐assisted selection of common beans for resistance to common bacterial blight: Efficacy and economics. Plant Breeding, 119, 411–415. [Google Scholar]
- Yu, K. , Park, S.J. , Poysa, V. & Gepts, P. (2000b) Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). Journal of Heredity, 91, 429–434. [DOI] [PubMed] [Google Scholar]
- Yu, K. , Park, S.J. , Zhang, B. , Haffner, M. & Poysa, V. (2004) An SSR marker in the nitrate reductase gene of common bean is tightly linked to a major gene conferring resistance to common bacterial blight. Euphytica, 138, 89–95. [Google Scholar]
- Yu, K. , Shi, C. & Zhang, B. (2012) Development and application of molecular markers to breed common bean (Phaseolus vulgaris L.) for resistance to common bacterial blight (CBB)—current status and future directions. Applied Photosynthesis, pp. 365–388. [Google Scholar]
- Yu, Z.H. , Stall, R.E. & Vallejos, C.E. (1998) Detection of genes for resistance to common bacterial blight of beans. Crop Science, 38, 1290–1296. [Google Scholar]
- Zanatta, Z.G.C.N. , Moura, A.B. , Maia, L.C. & Dos Santos, A.S. (2007) Bioassay for selection of biocontroller bacteria against bean common blight (Xanthomonas axonopodis pv. phaseoli). Brazilian Journal of Microbiology, 38, 511–515. [Google Scholar]
- Zapata, M. (2006) A proposal for a uniform screening procedure for the greenhouse evaluation of variability of Xanthomonas axonopodis pv. phaseoli and resistance on leaves of Phaseolus vulgaris . Annual Report of the Bean Improvement Cooperative, 49, 213–214. [Google Scholar]
- Zapata, M. , Beaver, J.S. & Porch, T.G. (2010) Dominant gene for common bean resistance to common bacterial blight caused by Xanthomonas axonopodis pv. phaseoli . Euphytica, 179, 373–382. [Google Scholar]
- Zapata, M. & Vidaver, A.K. (1987) Differenciation of Xanthomonas campestris pv. phaseoli into pathogenic races based on the tepary bean reactions. Phytopathology, 77, 1709. [Google Scholar]
- Zaumeyer, W.J. (1930) The bacterial blight of beans caused by Bacterium phaseoli . Technical Bulletin of the United States Department of Agriculture, 186, 1–34. [Google Scholar]
- Zaumeyer, W.J. (1932) Comparative pathological histology of three bacterial diseases of bean. Journal of Agricultural Research, 44, 1–35. [Google Scholar]
- Zaumeyer, W.J. & Thomas, H.R. (1957) A monographic study of bean diseases and methods for their control. United States Department of Agriculture, Economic Research Service. [Google Scholar]
- Zeng, X. , Tian, D. , Gu, K. , Zhou, Z. , Yang, X. , Luo, Y. et al. (2015) Genetic engineering of the Xa10 promoter for broad‐spectrum and durable resistance to Xanthomonas oryzae pv. oryzae . Plant Biotechnology Journal, 13, 993–1001. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Wu, J. , Wang, L. , Blair, M.W. , Zhu, Z. & Wang, S. (2016) QTL and candidate genes associated with common bacterial blight resistance in the common bean cultivar Longyundou 5 from China. The Crop Journal, 4, 344–352. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


