Abstract
Assessing immune responses and cell death in Nicotiana benthamiana leaf agro‐infiltration assays is a powerful and widely used experimental approach in molecular plant pathology. Here, we describe a reliable high‐throughput protocol to quantify strong, macroscopically visible cell death responses in N. benthamiana agro‐infiltration assays. The method relies on measuring the reduction of leaf autofluorescence in the red spectrum upon cell death induction and provides quantitative data suitable for straightforward statistical analysis. Two different well‐established model nucleotide‐binding and leucine‐rich repeat domain proteins (NLRs) were used to ensure the genericity of the approach. Its accuracy and versatility were compared to visual scoring of the cell death response and standard methods commonly used to characterize NLR activities in N. benthamiana. A discussion of the advantages and limitations of our method compared to other protocols demonstrates its robustness and versatility and provides an effective means to select the best‐suited protocol for a defined experiment.
Keywords: Agrobacterium, cell death, fluorescence imaging, immune response, Nicotiana benthamiana, quantitative analysis
We provide a high‐throughput method to quantify cell death in Nicotiana benthamiana agro‐infiltration assays relying on the measurement of leaf red fluorescence decline upon induction of cell death.
1. INTRODUCTION
Plant cell death is a universal phenomenon in plant–pathogen interactions. It can be a consequence of the infection strategy of the pathogen and relies on the action of cell death‐inducing effector proteins or secondary metabolites. It is also involved in the multicomponent immune response that plants elicit against biotrophic or hemibiotrophic pathogens. This programmed cell death, also known as the hypersensitive response, has long been recognized as a hallmark of plant immunity (Coll et al., 2011). Due to the fundamental role of plant cell death, its precise documentation and description are critical in research on plant–pathogen interactions and plant immunity.
Nicotiana benthamiana is the leading model system for the investigation of pathogen effector proteins and plant immune components. It allows efficient expression of single genes or their combinations by infiltrating leaves with Agrobacterium tumefaciens strains carrying appropriate constructs and monitoring the localization or activity of the corresponding gene products (Bally et al., 2018; Clemente, 2006). Measuring cell death in these infiltrated leaves is particularly valuable in research on nucleotide‐binding and leucine‐rich repeat domain proteins (NLRs). NLRs represent the major class of disease resistance proteins in crops and act as immune receptors detecting specific pathogen effector proteins (Cesari, 2018; Kourelis et al., 2018). Cell death assays in N. benthamiana are essential for the investigation of NLR‐mediated effector recognition and for the analysis of the molecular mechanisms induced during NLR‐triggered immune activation. Studying these phenomena in homologous systems is generally challenging and time consuming because it largely relies on the stable transformation of the host plant and/or the pathogen. For instance, testing the specific recognition of an effector by an NLR often requires the complementation of a pathogen isolate lacking this effector and subsequent investigation of its phenotype in interaction with the host plant expressing the NLR. In addition to the time‐consuming nature of stable transformation, many pathogens, especially biotrophs, are not transformable. On the contrary, agro‐infiltration assays allow simple coexpression of effectors and NLRs and observation of cell death associated with the immune response induced in the event of specific recognition.
Different methods are used to assess cell death in N. benthamiana agro‐infiltration assays. The simplest relies on qualitative observation of cell death symptoms visible under normal light, such as leaf chlorosis and partial or confluent cell death (Oh et al., 2010). By assigning scores based on the severity of the phenotypes, statistical analysis can be performed on these qualitative data allowing comparisons of responses to contrasting treatments. A similar approach is the qualitative estimation of leaf autofluorescence under ultraviolet (UV) light (De la Concepcion et al., 2018). The accumulation of phenolic compounds, which are fluorescent under UV light, is a well‐established immune response that is used as a proxy for immune activation and cell death. Staining of infiltrated leaves with trypan blue is another technique for the qualitative detection of immune responses as this colourimetric dye marks cells with defects in membrane integrity (Mulaosmanovic et al., 2020). Trypan blue staining can detect weak immune responses that do not necessarily trigger macroscopically visible cell death (Bai et al., 2012). However, this method requires harmful chemicals (e.g., crystal phenol) and is particularly polluting.
These qualitative methods allow an accurate evaluation of cell death and immune responses, but they rely on human evaluation and may be subjective. Therefore, it is good practice to supplement or replace them by methods measuring chemical or physical properties altered by immune activation. The most popular among them is measurement of ion leakage (Hatsugai & Katagiri, 2018). This sensitive method enables the detection of early immune responses, before the development of visible cell death, and of weak responses without macroscopic phenotypes. Another method consists in the spectrophotometric quantification of chlorophyll that monitors chlorosis caused by immune responses or necrotrophic effectors. Quantitative methods are often time consuming and not easily applicable at high throughput.
Here we describe a novel method to assess immune activation by quantifying cell death in the N. benthamiana model system. The method relies on measuring the reduction of leaf autofluorescence in the red spectrum upon cell death induction and provides quantitative data suitable for straightforward statistical analysis.
2. RESULTS AND DISCUSSION
The objective of this study was to establish a robust, high‐throughput, and nonsubjective method for the quantification of cell death in N. benthamiana agro‐infiltration assays and to compare it with other approaches widely used for this in plant immune research. To test the versatility of the method, we used two different types of cell death activators. The rice NLR RGA4 served as an autoactive immune component, while specific effector recognition was assessed using the rice NLR pair Pikp‐1/Pikp‐2, which triggers immune responses in the presence of the Magnaporthe oryzae effector AVR‐PikD (Césari et al., 2014; Maqbool et al., 2015).
2.1. Visual scoring, an effective method for the qualitative assessment of immune responses
To test the immune responses induced by RGA4 or by Pikp‐1/Pikp‐2 following the recognition of AVR‐PikD, we first carried out a qualitative analysis based on visual scoring of the phenotypes. Indeed, expression of cell death inducers in leaves of N. benthamiana generally causes phenotypes ranging from chlorosis to confluent cell death. To measure these phenotypes, we inferred a score ranging from 0 (no observable response) to 4 (confluent cell death) (Figure 1a).
FIGURE 1.
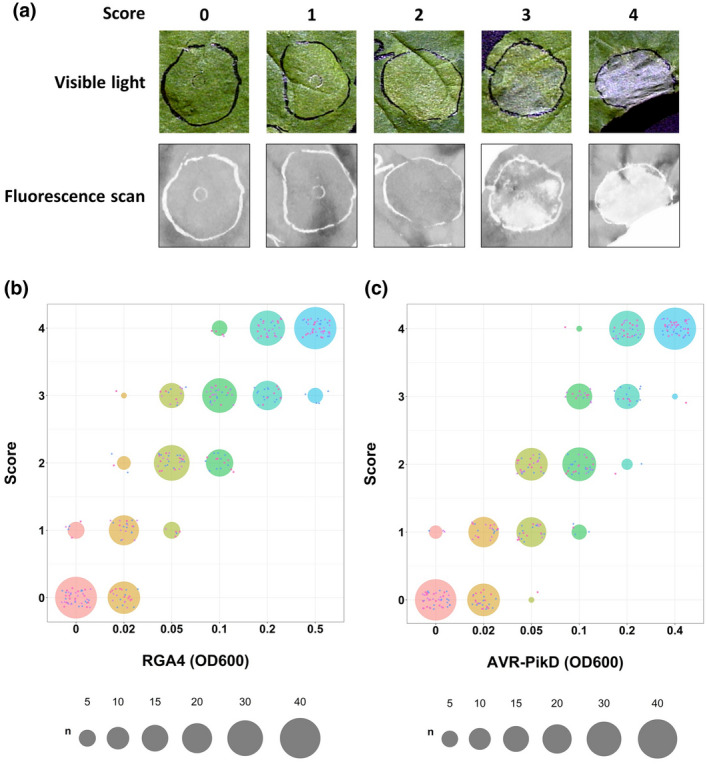
Scoring of NLR‐mediated immune responses in Nicotiana benthamiana. (a) Top panel: Leaf images representative of the five different classes of phenotypes observed following agro‐infiltration with the constructs used in our study and their related visual scores: 0 = no response, 1 = weak chlorosis, 2 = strong chlorosis, 3 = patchy cell death, and 4 = strong confluent cell death. Bottom panel: The same leaves were imaged using the Typhoon FLA 9000 laser scanner set up with a 635‐nm laser diode and with a long‐pass red filter (665 nm) to collect the red fluorescence. All images were taken 5 days after agro‐infiltration. (b) Quantification of macroscopic responses induced by different concentrations of agrobacteria carrying the RGA4:HA construct shown as dot plots. Different bacterial concentrations are displayed with distinct colours (from pink for an OD600 of 0 to blue for an OD600 of 0.5). The three biological replicates are represented by small dots of distinct colour and shape (i.e., purple triangle, pink square, and dark blue circle). The size of each circle is proportional to the number of replicates matching the corresponding score for each condition. The total number of replicates for each condition is 45. (c) Same as in panel (b) for the responses triggered by different concentrations of agrobacteria carrying the AVR‐PikD:HA construct coinfiltrated with steady amounts of agrobacteria expressing the Pikp‐1:Flag/Pikp‐2:HA construct (OD600 of 0.15)
To analyse a broad spectrum of immune responses, we infiltrated increasing concentrations of agrobacteria carrying the constructs responsible for inducing immune responses. Thus, infiltration of N. benthamiana leaves with increasing amounts of agrobacteria carrying an RGA4:HA construct resulted in increased cellular levels of RGA4 (Figure S1a). This activated macroscopically visible immune responses ranging from weak chlorosis, in the presence of low levels of RGA4 (OD600 of 0.02), to confluent cell death at high concentrations (OD600 ≥ 0.2) (Figure 1b).
Likewise, infiltration of the leaves with increasing amounts of bacteria carrying the AVR‐PikD:HA construct in the presence of steady amounts of Pikp‐1:Flag/Pikp‐2:HA agrobacteria (OD600 of 0.15) led to increasing levels of AVR‐PikD protein (Figure S1b). While the expression of Pikp‐1 and Pikp‐2 alone did not elicit any response, the presence of AVR‐PikD activated immune responses that ranged, depending on the expression level of the effector, from chlorosis (OD600 of 0.02 and 0.05) to confluent cell death (OD600 ≥ 0.2) (Figure 1c).
Taken together, these results show that visual scoring can be a powerful method for measuring qualitative phenotypes. This method is very efficient for the analysis of immune responses induced in N. benthamiana because it allows scoring subtle phenotypes such as weak chlorosis. Resulting data can be further analysed by statistical tests based on graphic estimation methods (Ho et al., 2019), which have been adapted to qualitative data sets (De la Concepcion et al., 2019) (Figure S2). However, this method relies on human decisions because the scoring is directly influenced by the judgement of the experimenter. Therefore, quantitative measurements of chemical or physical properties associated with immune responses are needed to corroborate visual scoring results.
2.2. Leaf fluorescence scanning allows quantification of strong cell death responses
Previous studies have shown that cell death induced by NLR receptors is associated with a reduction in red fluorescence emitted by N. benthamiana leaves upon excitation with red light (Césari et al., 2014; Ortiz et al., 2017). Based on this observation, we developed a simple method to quantify red fluorescence in agro‐infiltrated N. benthamiana leaves using a laser fluorescence scanner.
Five days after infiltration, when the immune responses are fully established (Figure 2a), red fluorescence was recorded by scanning the detached leaves using a 635‐nm laser and by quantifying the light emitted after passing through a long‐pass red filter (665 nm) (Figure 2b). Up to 20 leaves can be scanned simultaneously, allowing image acquisition at high throughput. The resulting images were processed with ImageJ software (Schneider et al., 2012) in order to quantify the fluorescence in the infiltrated areas (Suppl. Methods). For this, the infiltrated areas were manually delimited using the oval selection tool (Figure 2c) and shadows caused by the leaf veins and leaf shape were removed by adjusting the threshold (Figure 2d). This image processing is high throughput because the treatment of one scan with 20 leaves, corresponding to 80 data points, takes less than 10 mins.
FIGURE 2.
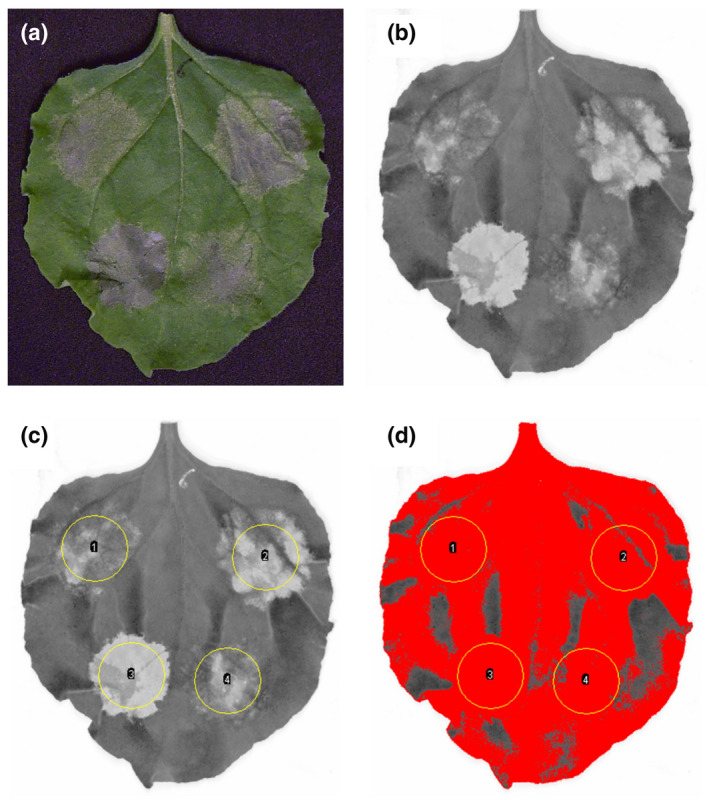
Quantification of cell death in Nicotiana benthamiana by red fluorescence scanning. (a) Picture of a leaf taken 5 days after agro‐infiltration with constructs inducing cell death. (b) The leaf was scanned using the Typhoon FLA 9000 laser scanner. A 635‐nm laser was used for excitation and the light emitted by the leaf was quantified after passing through a 665‐nm long‐pass red filter. (c) Infiltrated areas of the leaf were delimited using the oval selection tool of ImageJ and the ROI (region of interest) manager tool. Parameters were set to quantify the mean grey value of each ROI after threshold adjustment. (d) A threshold was applied to the image to exclude dark areas caused by leaf veins and leaf shape. Within each ROI, the mean grey value is calculated using exclusively the pixels that appear in red. Equivalent parameters were used for all leaf images within each replicate of an experiment
Statistical analysis of results from several independent experiments showed a significant reduction in red fluorescence specific to the infiltrated areas showing strong cell death (Figures 1a and 3a,b). Indeed, infiltration of agrobacteria strains carrying RGA4:HA or AVR‐PikD:HA at OD600 ≥ 0.2 showed a significant reduction in red fluorescence (Figure 3a,b; Figure S3). Infiltrated areas without symptoms, or exhibiting weak or strong chlorosis levels, showed a level of fluorescence similar to reference areas infiltrated with an Agrobacterium isolate carrying CFP, which does not fluoresce in the red‐light spectrum analysed.
FIGURE 3.
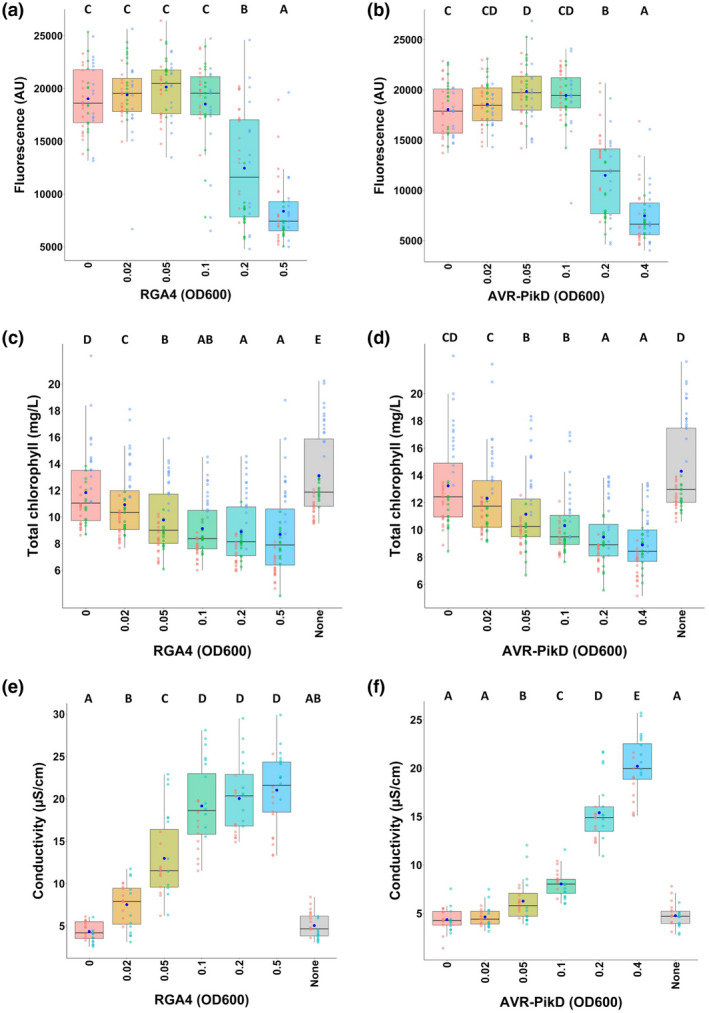
Comparison of three methods for the quantification of immune responses. (a) Quantification of immune responses induced by different concentrations of agrobacteria carrying the RGA4:HA construct using red fluorescence scanning. Leaves were harvested 5 days after infiltration and imaged using a fluorescence scanner with a 635‐nm laser used for excitation and a long‐pass red filter (wavelength range ≥ 665 nm) to detect the red fluorescence of the leaves. Images were processed with ImageJ to measure the red fluorescence by quantifying the mean grey value for each infiltrated area (expressed as arbitrary units [AU]). Collected data were plotted and dots with different colours (i.e., red, green, and light blue) correspond to three biological replicates. The total number of replicates for each condition is 45. The boxes represent the first quartile, median, and third quartile and the dark blue dot indicates the mean value. Differences of red fluorescence levels were assessed by one‐way analysis of variance followed by a Tukey honestly significant difference test. Groups with the same letter (A to C) are not statistically different (p > .05). (b) Same as in panel (a) for the responses triggered by different concentrations of agrobacteria carrying the AVR‐PikD:HA construct coinfiltrated with steady amounts of agrobacteria expressing the Pikp‐1:Flag/Pikp‐2:HA construct (OD600 of 0.15). (c,d) Quantification of immune responses induced by distinct concentrations of agrobacteria carrying RGA4:HA (c) or AVR‐PikD:HA (with constant levels of Pikp‐1:Flag/Pikp‐2:HA) (d) by chlorophyll measurements. Chlorophyll was extracted from infiltrated areas 5 days after infiltration and chlorophyll quantification was performed by spectrophotometry. The absorbance was measured at λ = 649 nm and λ = 665 nm and total chlorophyll content was calculated as the sum of chlorophyll a (Chl a = 12.19 × A665 − 3.45 × A649) and chlorophyll b (Chl b = 21.99 × A649 − 5.32 × A665) content (Slootweg et al., 2017; Tait & Hik, 2003). The total number of replicates for each condition is 45. The experiment was carried out three times and data were plotted and analysed as in panel (a). (e,f) Quantification of immune responses induced by various concentrations of agrobacteria carrying RGA4:HA (e) or AVR‐PikD:HA (with constant levels of Pikp‐1:Flag/Pikp‐2:HA) (f) by ion leakage measurements. Samples were harvested from infiltrated areas 48 hr after infiltration, and the conductivity was measured after 20 hr incubation in deionized water. The experiment was carried out twice and the total number of replicates for each condition in each experiment is 24. Data were plotted and analysed as in panel (a)
To determine the precision with which differences in cell death induction can be monitored by this method, responses to a dilution series of RGA4:HA agrobacteria of an OD600 ranging from 0.1 to 0.6 were analysed. Results from visual scoring and fluorescence scanning confirmed that strong cell death was associated with a decrease in red fluorescence and statistical analysis allowed to differentiate the cell death responses triggered by slightly different Agrobacterium concentrations (Figure S4). Thus, cell death triggered by RGA4 at an OD600 of 0.1 is significantly lower than that induced at an OD600 of 0.2. Likewise, the response to OD600 of 0.3 could be distinguished from the response to an OD600 of 0.5. Our leaf fluorescence scanning method therefore allows a nonsubjective quantification of the phenotypes associated with strong cell death.
Recently, another method relying on leaf fluorescence for the visualization of cell death has been described (Landeo Villanueva et al., 2021). While our method uses a 635‐nm laser for excitation and collects fluorescence signals ≥665 nm, this alternative protocol measures leaf fluorescence between 605 nm and 650 nm upon excitation with green light (495–570 nm). Therefore, the two protocols do not quantify the same fluorescence signals. Indeed, while our method quantifies a drastic drop of red fluorescence in dead tissues, the fluorescence signals measured by the method from Landeo Villanueva et al. increase upon cell death and may derive from increased chlorophyll fluorescence in plant tissue undergoing cell death (Landeo Villanueva et al., 2021).
2.3. Red fluorescence scanning does not reflect the chlorophyll content of the leaves
Most of the leaf red fluorescence measured in our method probably originates from chlorophyll. Chlorophyll loss is a well‐documented immune response that is triggered, among other stimuli, by activation of NLR receptors (Slootweg et al., 2017). To determine if the reduction in leaf red fluorescence during severe cell death is correlated with a loss of chlorophyll, we quantified chlorophyll spectrophotometrically in leaf areas expressing increasing levels of AVR‐PikD:HA (together with constant levels of Pikp‐1 and Pikp‐2) or RGA4:HA. We then compared these values with those measured for the reference areas expressing CFP.
Low levels of RGA4:HA or AVR‐PikD:HA (OD600 of 0.02) resulted in a significant reduction in the chlorophyll content that was further enforced as the amounts of RGA4:HA or AVR‐PikD:HA increased (Figure 3c,d). Interestingly, this chlorophyll loss did not correlate with the reduction in leaf red fluorescence or with the induction of strong cell death. It rather reflected the development of chlorotic symptoms determined by visual scoring (Figure 1). This is consistent with the definition of chlorosis as the loss of green leaf colouration due to chlorophyll and with our finding that chlorophyll loss reached a maximum as soon as all the infiltrated zones showed strong chlorosis or stronger phenotypes (for example RGA4:HA at an OD600 of 0.1) (Figure 3c; Figure S4d).
The lack of correlation between chlorophyll content and fluorescence may be due to the fact that the physiological and photochemical states of the leaf strongly influence the ratio between re‐emitted light and light consumed by photosystem II chemistry for photosynthesis (Baker, 2008). In the context of NLR‐triggered immune responses, the down‐regulation of photosynthesis genes and the damage of the electron transfer chain downstream of photosystem II strongly reduce photosynthesis (Mur et al., 2008). This raises the proportion of the absorbed light energy that is re‐emitted by fluorescence and thus compensates the reduced light absorption due to chlorophyll loss (Baker, 2008). The important reduction of leaf red fluorescence upon strong cell death may be a consequence of the complete destruction of chloroplasts during cell death, resulting in complex and drastic photochemical changes. For instance, the quantity of light absorbed by chlorophyll decreases due to destruction of the light‐harvesting complexes and chlorophyll fluorescence is reduced by changes in its chemical environment due to the disintegration of photosystem II, change of pH, and loss of water. In addition, large amounts of secondary metabolites accumulating during programmed cell death may further reduce residual chlorophyll fluorescence by quenching.
Due to these differences between red fluorescence scanning and chlorophyll content measurement, it is possible to apply the two methods to two different types of immune responses induced by NLR receptors. The spectrophotometric chlorophyll assay quantifies chlorosis associated with a weak NLR‐mediated immune response, while red fluorescence scanning measures high‐intensity cell death responses caused for instance by strong activation of NLRs.
2.4. Ion leakage measurement enables the assessment of the temporal dynamics of the immune response
A widely used approach to quantify NLR‐triggered immune responses is the determination of electrolyte leakage (Hatsugai & Katagiri, 2018). It relies on the efflux of ions from cells establishing immune responses or undergoing cell death and consists in measuring the conductivity of a solution obtained by incubating infiltrated leaf discs in deionized water.
To compare this approach with leaf red fluorescence scanning, we determined ion leakage 48 hr after infiltration of the previously used dilution series of AVR‐PikD:HA‐carrying agrobacteria (in the presence of constant levels of Pikp‐1 and Pikp‐2) or RGA4:HA and compared it to reference spots expressing CFP (Figure 3e,f). In the case of RGA4:HA, electrolyte leakage was detected from the lowest Agrobacterium concentration (OD600 of 0.02) and increased until a concentration of 0.1, where it reached a plateau and did not significantly vary with increasing RGA4 levels (Figure 3e). With Pikp‐1/Pikp‐2/AVR‐PikD, ion leakage was first detected at an OD600 of 0.05 of AVR‐PikD:HA agrobacteria. It steadily increased up to the highest tested AVR‐PikD level (OD600 of 0.4) but remained much lower at intermediate OD600 levels than with RGA4 (Figure 3f).
Therefore, there is a clear difference in the ion leakage responses of RGA4 and Pikp‐1/Pikp‐2/AVR‐PikD that was not detected using the other methods tested. An explanation for this difference could be that the immune responses triggered by RGA4 appear faster than that of Pikp‐1/Pikp‐2/AVR‐PikD. Indeed, 48 hr after infiltration (time when ion leakage was measured), RGA4‐expressing areas already started to show visible signs of cell death, while no response was yet visible for Pikp‐1/Pikp‐2/AVR‐PikD.
Electrolyte leakage measurement therefore is appropriate to monitor the entire spectrum of NLR activation, from weak‐ to high‐intensity activation of immune signalling. However, to cover this full range of activities, recording of a time series with sampling of leaf discs before and after induction of macroscopically visible immune responses is required. Indeed, different intensities of strong immune activation can be differentiated at early time points, before symptoms appear, while weak responses are well differentiated at later time points.
However, this makes electrolyte leakage measurement a highly versatile method that requires numerous samples and plants and is, due to the associated workload, not as high‐throughput as leaf red fluorescence scanning.
3. CONCLUSION
Our study provides a detailed protocol for red fluorescence imaging of N. benthamiana leaves and demonstrates that it is a robust and versatile technique for the quantification of strong cell death in agro‐infiltration assays. This new method has the advantage of being high‐throughput because leaf scanning and image analysis require little hands‐on work (Table 1). Besides, it provides quantitative data suitable for statistical analysis. A limitation of the leaf red fluorescence quantification method is that it does not record weak immunity‐related phenotypes. Determination of chlorophyll loss monitors such weak immune responses and precisely quantifies chlorosis. However, it has a lower throughput because harvesting leaf discs is time consuming in large series. Ion leakage appears as an extremely versatile method that is low‐throughput as it is labour‐intensive and requires time series with multiple time points and large numbers of replicates to accurately document the entire range of immune response intensities (Table 1).
TABLE 1.
Comparison of the different methods to measure cell death and immune responses in Nicotiana benthamiana
| Method | Type | Workload | High‐throughput | Weak responses/chlorosis | Strong responses/cell death | Weakness |
|---|---|---|---|---|---|---|
| Visual scoring | Qualitative | + | Yes | + | + | Subjective |
| Red fluorescence | Quantitative | ++ | Yes | − | + | / |
| Chlorophyll | Quantitative | ++++ | No | + | − | / |
| Ion leakage | Quantitative | ++++ | No | + | + | / |
Taken together, our study provides a new, powerful, and robust method for the high‐throughput assessment of leaf cell death in N. benthamiana agro‐infiltration assays that relies on the scanning of leaf red fluorescence. This method fills a gap in the arsenal of already existing techniques for the quantification of immune responses in this model system.
4. EXPERIMENTAL PROCEDURES
4.1. Constructs
The RGA4 cDNA sequence was inserted by Gateway cloning in the pBIN19‐35S::GTW:3HA vector to generate the RGA4:HA construct (Cesari et al., 2013). The AVR‐PikD cDNA sequence without signal peptide was cloned in fusion with an HA tag into the pCambia1300 vector (Maqbool et al., 2015) and the Pikp‐1:Flag/Pikp‐2:HA construct (courtesy of Mark Banfield and Hannah Langlands) was generated in pICSL4723.
4.2. N. benthamiana growth and agro‐infiltration
N. benthamiana plants were grown in a growth chamber at 22 °C with a 16‐hr light period. All transformed A. tumefaciens GV3101‐pMP90 strains were grown in LB liquid medium containing 50 µg/ml kanamycin, 25 µg/ml gentamycin, and 25 µg/ml rifampicin. Agro‐infiltration experiments in N. benthamiana were performed as previously described (Cesari et al., 2013). The P19 suppressor of gene silencing was used in all infiltration assays at a final OD600 of 0.1 with only 50 µg/ml kanamycin for selection. In all relevant experiments the Agrobacterium strain carrying the Pikp‐1:Flag/Pikp‐2:HA construct was infiltrated at a final OD600 of 0.15. The final concentration of agrobacteria was equilibrated in each infiltrated mix using a strain carrying the CFP construct.
4.3. Protein extraction and western blot
For each sample, three leaf discs were harvested 48 hr after agro‐infiltration to detect protein expression levels by western blot. Samples were ground in 100 μl NuPAGE LDS sample buffer with NuPAGE Reducing Agent as per the supplier's instructions (Life Technologies) and centrifuged at 15,700 × g for 10 min, and the supernatant was collected. After heating at 70 °C for 10 min, total protein extracts were separated on NuPAGE 4%–12% Bis‐Tris Mini Protein Gels (Thermo Fisher) and transferred to nitrocellulose membranes (Millipore) for immunoblotting. HA‐tagged proteins were detected using a horseradish peroxidase (HRP)‐conjugated anti‐HA antibody (clone 3F10; Roche), while the Pikp‐1:Flag protein was detected using an anti‐FLAG antibody (clone M2; Sigma‐Aldrich) and a secondary HRP‐conjugated anti‐mouse antibody (Sigma‐Aldrich). The Immobilon Western kit (Millipore) was used for detection.
4.4. Red fluorescence quantification
For red fluorescence quantification, each Agrobacterium mix was infiltrated on 15 different N. benthamiana leaves per experiment. For each assay, two to three completely independent experiments were carried out. Pictures of the detached leaves were taken 5 days after agro‐infiltration using a Samsung UF‐130 camera. Leaves were then scanned using the Typhoon FLA 9000 laser scanner (GE Healthcare) with their adaxial side facing the glass stage and their abaxial side facing the cover plate. The reading mode of the scanner was set to “fluorescence” and leaves were scanned using the 635‐nm laser diode for excitation and the long‐pass red (LPR) filter module (wavelength range, ≥665 nm) to collect the red fluorescence. To acquire the data, the photomultiplier tube was set to approximately 500 V and the pixel size to 200 μm. Images were saved as .tif and .gel format for further processing. Red fluorescence of the infiltrated leaf areas was measured using ImageJ software (Schneider et al., 2012) by quantifying the mean grey value within each area (see Suppl. Methods for a detailed protocol). Boxplots were generated using R v. 4.0.2 (R core Team, 2019) and the package tidyverse (Wickham, 2019). The script “QRedFluo.R” is available at https://www.github.com/vchochois/xi2021.
4.5. Chlorophyll measurements
For chlorophyll quantification, each Agrobacterium mix was infiltrated on 15 different N. benthamiana leaves in each experiment and two to three independent experiments were carried out. One disc (diameter, 8 mm) per infiltration spot was harvested 5 days after agro‐infiltration for chlorophyll extraction and quantification. Each leaf disc was individually incubated overnight at 28 °C in 500 μl of dimethyl sulphoxide under gentle shaking until the leaf discs became transparent. After a short centrifugation at 15,700 × g, 150 μl of the leaf extract was transferred to a 96‐well flat microplate (Greiner bio‐one) and the absorbance was measured at λ = 649 nm and λ = 665 nm using the SPARK multimode microplate reader (TECAN). Total chlorophyll content was calculated as the sum of chlorophyll a (Chl a = 12.19 × A665 − 3.45 × A649) and chlorophyll b (Chl b = 21.99 × A649 − 5.32 × A665) content (Slootweg et al., 2017; Tait & Hik, 2003). Boxplots were generated using R v. 4.0.2 (R core Team, 2019) and the package tidyverse (Wickham, 2019). The script “QChlorophyll.R” is available at https://www.github.com/vchochois/xi2021.
4.6. Ion leakage measurements
For ion leakage quantification, each Agrobacterium mix was infiltrated on 12 different N. benthamiana leaves per experiment and two completely independent experiments were carried out 48 hr after agro‐infiltration. One leaf disc (8 mm diameter) per infiltration spot was harvested and floated in 2 ml double distilled water for 30–45 min to remove the ion leakage caused by wounding. After a quick wipe, each leaf disc was incubated for 20 hr in 5 ml double distilled water in the growth chamber. The conductivity of this incubation solution was measured using a portable conductivity meter. Data were exported and plotted using R v. 4.0.2 (R core Team, 2019) and the package tidyverse (Wickham, 2019). The script “QIonLeakage.R” is available at https://www.github.com/vchochois/xi2021.
4.7. Data analysis
Qualitative cell death scores were analysed using estimation methods (De la Concepcion et al., 2019; Ho et al., 2019) and visualized with estimation graphics using in‐house scripts adapted from the besthr R library (MacLean, 2019). The script “EstimationGraphics.R” is available at https://www.github.com/vchochois/xi2021.
Differences of red fluorescence induced by the various concentrations of agrobacteria carrying the RGA4 or Pikp‐1/Pikp‐2/AVR‐PikD constructs were assessed by one‐way analysis of variance followed by a Tukey honestly significant difference (HSD) test. Similar analyses were applied to quantitative data from ion leakage and chlorophyll measurements. In the following cases, transformation of the data was applied prior to statistical analysis. Log transformation was used in the case of chlorophyll quantification following AVR‐PikD:HA expression and for the red fluorescence quantification after RGA4:HA infiltration (OD600 of 0 to 0.6). For the chlorophyll quantification following RGA4:HA expression (OD600 of 0 to 0.5), power transformation (x 0.25) was applied. Power transformation (x 0.4) was also used for ion leakage measurements after RGA4:HA expression (OD600 of 0 to 0.5).
Supporting information
FIGURE S1 Protein expression in Nicotiana benthamiana. (a,c) The RGA4:HA construct was transiently expressed in N. benthamiana leaves using the indicated concentrations of agrobacteria. Total proteins were extracted 48 hr after infiltration and the RGA4:HA protein was detected by immunoblotting using an HRP‐conjugated anti‐HA antibody. Equal protein loading of each sample is shown by Ponceau staining of the RuBisCO large subunit. (b) The AVR‐PikD:HA construct was transiently coexpressed in N. benthamiana leaves with the Pikp‐1:Flag/Pikp‐2:HA construct. Indicated OD600 values correspond to the concentration of infiltrated agrobacteria carrying the AVR‐PikD:HA construct. The strain carrying the Pikp‐1:Flag/Pikp‐2:HA construct was infiltrated at a constant concentration (OD600 of 0.15 final). Proteins were extracted 48 hr after infiltration. HA‐tagged proteins were detected by western blotting using HRP‐conjugated anti‐HA while FLAG‐tagged Pikp‐1 was detected using anti‐FLAG and a secondary HRP‐conjugated anti‐mouse antibody
FIGURE S2 Estimation graphics for comparison of qualitative immune responses. Statistical analysis by estimation methods of the qualitative visual scoring assay for the different bacterial concentrations of RGA4:HA (a) and AVR‐PikD:HA (b). These analyses correspond to the data shown in Figure 1b and 1c. The mean scores for each replicate (n = 3) and each bacterial concentration (or treatment) were calculated and ranked. For each bacterial concentration, the left panel shows the ranked data (dots) and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations (count) with that specific value. The right panel shows the distribution of 1,000 bootstrap sample rank means for each OD600 > 0. The blue areas represent the 2.5% and 97.5% percentiles of the distribution. The responses to two different OD600 concentrations are considered significantly different if the rank mean of one (dotted line, left panel) does not fall between the 2.5% and 97.5% percentiles (grey area) of the mean distribution of the other (right panel)
FIGURE S3 Immune responses induced by RGA4 and AVR‐PikD recognition in Nicotiana benthamiana. Representative leaf images of immune responses induced by different concentrations of agrobacteria carrying the (a) RGA4:HA or (b) AVR‐PikD:HA constructs (with constant levels of Pikp‐1:Flag/Pikp‐2:HA) are shown. Pictures of the leaves taken under visible light are shown in the upper lane and their corresponding red fluorescence scanning images are shown in the lower lane. All images were taken 5 days after infiltration
FIGURE S4 Immune responses induced by high levels of RGA4 in Nicotiana benthamiana. (a) Quantification of macroscopic responses induced by different concentrations of agrobacteria carrying the RGA4:HA construct shown as dot plots. Different bacterial concentrations are displayed with distinct colours (from pink for an OD600 of 0 to light purple for an OD600 of 0.6). The two biological replicates are represented by little dots of distinct colour and shape (i.e., pink triangle and purple circle). The size of each circle is proportional to the number of replicates matching the corresponding score for each condition. The total number of replicates for each condition is 24. (b) Statistical analysis of strong cell death responses induced by RGA4 using estimation methods (see Figure S2). (c,d) Quantification of red fluorescence (c) and chlorophyll content (d) in leaf discs infiltrated with the indicated concentrations of RGA4:HA agrobacteria (data were plotted as in Figure 3a and c). Data from both panels (c) and (d) were statistically analysed with one‐way analysis of variance followed by a Tukey HSD test. Groups with the same letter are not statistically different (p > 0.05)
SUPPLEMENTARY MATERIAL Quantification of cell death responses in Nicotiana benthamiana by red fluorescence scanning and ImageJ
ACKNOWLEDGEMENTS
We thank Professor Mark Banfield and Hannah Langlands for providing the AVR‐PikD:HA and Pikp‐1:Flag/Pikp‐2:HA constructs. This study was supported by the Agence Nationale de la Recherche (ANR‐18‐CE20‐0016 MagMax), the European Research Council (ERC‐2019‐STG‐852482‐ii‐MAX), and the China Scholarship Council (CSC PhD grant 201806350131 to Y.X.). No competing interests are declared.
Xi, Y. , Chochois, V. , Kroj, T. & Cesari, S. (2021) A novel robust and high‐throughput method to measure cell death in Nicotiana benthamiana leaves by fluorescence imaging. Molecular Plant Pathology, 22, 1688–1696. 10.1111/mpp.13129
Thomas Kroj and Stella Cesari contributed equally to this work.
Contributor Information
Thomas Kroj, Email: thomas.kroj@inrae.fr.
Stella Cesari, Email: stella.cesari@inrae.fr.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- Bai, S. , Liu, J. , Chang, C. , Zhang, L. , Maekawa, T. , Wang, Q. et al. (2012) Structure‐function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathogens, 8, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N.R. (2008) Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annual Review of Plant Biology, 59, 89–113. [DOI] [PubMed] [Google Scholar]
- Bally, J. , Jung, H. , Mortimer, C. , Naim, F. , Philips, J.G. , Hellens, R. et al. (2018) The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annual Review of Phytopathology, 56, 405–426. [DOI] [PubMed] [Google Scholar]
- Cesari, S. (2018) Multiple strategies for pathogen perception by plant immune receptors. New Phytologist, 219, 17–24. [DOI] [PubMed] [Google Scholar]
- Césari, S. , Kanzaki, H. , Fujiwara, T. , Bernoux, M. , Chalvon, V. , Kawano, Y. et al. (2014) The NB‐LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO Journal, 33, 1941–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari, S. , Thilliez, G. , Ribot, C ., Chalvon, V. , Michel, C. , Jauneau, A. et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR‐Pia and AVR1‐CO39 by direct binding. The Plant Cell, 25, 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente, T. (2006) Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana) . In: Wang, K. (Ed.) Agrobacterium Protocols, Vol. 1. Methods in Molecular Biology, 2nd edition. Vol. 343, Totowa, NJ: Humana Press, pp. 143–154. [DOI] [PubMed] [Google Scholar]
- Coll, N.S. , Epple, P. & Dangl, J.L. (2011) Programmed cell death in the plant immune system. Cell Death and Differentiation, 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Concepcion, J.C. , Franceschetti, M. , Maclean, D. , Terauchi, R. , Kamoun, S. & Banfield, M.J. (2019) Protein engineering expands the effector recognition profile of a rice NLR immune receptor. eLife, 8, e47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Concepcion, J.C. , Franceschetti, M. , Maqbool, A. , Saitoh, H. , Terauchi, R. , Kamoun, S. et al. (2018) Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nature Plants, 4, 576–585. [DOI] [PubMed] [Google Scholar]
- Hatsugai, N. & Katagiri, F. (2018) Quantification of plant cell death by electrolyte leakage assay. Bio‐Protocol, 8, e2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, J. , Tumkaya, T. , Aryal, S. , Choi, H. & Claridge‐Chang, A. (2019) Moving beyond P values: data analysis with estimation graphics. Nature Methods, 16, 565–566. [DOI] [PubMed] [Google Scholar]
- Kourelis, J. & van der Hoorn, R.A.L. (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. The Plant Cell, 30, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeo Villanueva, S. , Malvestiti, M.C. , van Ieperen, W. , Joosten, M.H.A.J. & van Kan, J.A.L. (2021) Red light imaging for programmed cell death visualization and quantification in plant–pathogen interactions. Molecular Plant Pathology, 22, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, D. (2019) TeamMacLean/besthr: Initial Release (0.1.0). Zenodo. 10.5281/zenodo.3374507 [DOI]
- Maqbool, A. , Saitoh, H. , Franceschetti, M. , Stevenson, C. , Uemura, A. , Kanzaki, H. et al. (2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife, 4, e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulaosmanovic, E. , Lindblom, T.U.T. , Bengtsson, M. , Windstam, S.T. , Mogren, L. , Marttila, S. et al. (2020) High‐throughput method for detection and quantification of lesions on leaf scale based on trypan blue staining and digital image analysis. Plant Methods, 16, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur, L.A.J. , Kenton, P. , Lloyd, A.J. , Ougham, H. & Prats, E. (2008) The hypersensitive response; The centenary is upon us but how much do we know? Journal of Experimental Botany, 59, 501–520. [DOI] [PubMed] [Google Scholar]
- Oh, C.S. , Pedley, K.F. & Martin, G.B. (2010) Tomato 14‐3‐3 protein 7 positively regulates immunity‐associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKKα. The Plant Cell, 22, 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, D. , de Guillen, K. , Cesari, S. , Chalvon, V. , Gracy, J. , Padilla, A. et al. (2017) Recognition of the Magnaporthe oryzae effector AVR‐Pia by the decoy domain of the rice NLR immune receptor RGA5. The Plant Cell, 29, 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2019) R: a language and environment for statistical computing. Vienna, Austria: R foundation for Statistical Computing. https://www.R‐project.org/ [Accessed 19 August 2021]. [Google Scholar]
- Schneider, C.A. , Rasband, W.S. & Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slootweg, E. , Koropacka, K. , Roosien, J. , Dees, R. , Overmars, H. , Lankhorst, R.K. et al. (2017) Sequence exchange between homologous NB‐LRR genes converts virus resistance into nematode resistance, and vice versa. Plant Physiology, 175, 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait, M.A. & Hik, D.S. (2003) Is dimethylsulfoxide a reliable solvent for extracting chlorophyll under field conditions? Photosynthesis Research, 78, 87–91. [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2019) Welcome to the tidyverse. Journal of Open Source Software, 4, 1686. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Protein expression in Nicotiana benthamiana. (a,c) The RGA4:HA construct was transiently expressed in N. benthamiana leaves using the indicated concentrations of agrobacteria. Total proteins were extracted 48 hr after infiltration and the RGA4:HA protein was detected by immunoblotting using an HRP‐conjugated anti‐HA antibody. Equal protein loading of each sample is shown by Ponceau staining of the RuBisCO large subunit. (b) The AVR‐PikD:HA construct was transiently coexpressed in N. benthamiana leaves with the Pikp‐1:Flag/Pikp‐2:HA construct. Indicated OD600 values correspond to the concentration of infiltrated agrobacteria carrying the AVR‐PikD:HA construct. The strain carrying the Pikp‐1:Flag/Pikp‐2:HA construct was infiltrated at a constant concentration (OD600 of 0.15 final). Proteins were extracted 48 hr after infiltration. HA‐tagged proteins were detected by western blotting using HRP‐conjugated anti‐HA while FLAG‐tagged Pikp‐1 was detected using anti‐FLAG and a secondary HRP‐conjugated anti‐mouse antibody
FIGURE S2 Estimation graphics for comparison of qualitative immune responses. Statistical analysis by estimation methods of the qualitative visual scoring assay for the different bacterial concentrations of RGA4:HA (a) and AVR‐PikD:HA (b). These analyses correspond to the data shown in Figure 1b and 1c. The mean scores for each replicate (n = 3) and each bacterial concentration (or treatment) were calculated and ranked. For each bacterial concentration, the left panel shows the ranked data (dots) and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations (count) with that specific value. The right panel shows the distribution of 1,000 bootstrap sample rank means for each OD600 > 0. The blue areas represent the 2.5% and 97.5% percentiles of the distribution. The responses to two different OD600 concentrations are considered significantly different if the rank mean of one (dotted line, left panel) does not fall between the 2.5% and 97.5% percentiles (grey area) of the mean distribution of the other (right panel)
FIGURE S3 Immune responses induced by RGA4 and AVR‐PikD recognition in Nicotiana benthamiana. Representative leaf images of immune responses induced by different concentrations of agrobacteria carrying the (a) RGA4:HA or (b) AVR‐PikD:HA constructs (with constant levels of Pikp‐1:Flag/Pikp‐2:HA) are shown. Pictures of the leaves taken under visible light are shown in the upper lane and their corresponding red fluorescence scanning images are shown in the lower lane. All images were taken 5 days after infiltration
FIGURE S4 Immune responses induced by high levels of RGA4 in Nicotiana benthamiana. (a) Quantification of macroscopic responses induced by different concentrations of agrobacteria carrying the RGA4:HA construct shown as dot plots. Different bacterial concentrations are displayed with distinct colours (from pink for an OD600 of 0 to light purple for an OD600 of 0.6). The two biological replicates are represented by little dots of distinct colour and shape (i.e., pink triangle and purple circle). The size of each circle is proportional to the number of replicates matching the corresponding score for each condition. The total number of replicates for each condition is 24. (b) Statistical analysis of strong cell death responses induced by RGA4 using estimation methods (see Figure S2). (c,d) Quantification of red fluorescence (c) and chlorophyll content (d) in leaf discs infiltrated with the indicated concentrations of RGA4:HA agrobacteria (data were plotted as in Figure 3a and c). Data from both panels (c) and (d) were statistically analysed with one‐way analysis of variance followed by a Tukey HSD test. Groups with the same letter are not statistically different (p > 0.05)
SUPPLEMENTARY MATERIAL Quantification of cell death responses in Nicotiana benthamiana by red fluorescence scanning and ImageJ
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.


