Abstract
Colletotrichum gloeosporioides is a hemibiotrophic ascomycete fungus that causes anthracnose on numerous plants worldwide and forms a specialized infection structure known as an appressorium in response to various plant surface signals. However, the associated mechanism of host surface signal recognition remains unclear. In the present study, three putative sensors, namely the mucin Msb2, the membrane sensor protein Sho1, and the G‐protein‐coupled receptor Pth11, were identified and characterized. The results showed that CgMsb2 plays a major role in the recognition of various host surface signals; deletion of CgMsb2 resulted in significant defects in appressorium formation, appressorium penetration, cellophane membrane penetration, and pathogenicity. CgSho1 plays a minor role and together with CgMsb2 cooperatively regulates host signal recognition, cellophane membrane penetration, and pathogenicity; deletion of CgSho1 resulted in an expansion defect of infection hyphae. Deletion of CgPth11 in wildtype, ΔCgMsb2, and ΔCgSho1 strains only resulted in a slight defect in appressorium formation at the early stage, and CgPth11 was dispensable for penetration and pathogenicity. However, exogenous cAMP failed to restore the defect of appressorium formation in ΔCgPth11 at the early stage. CgMsb2 contributed to the phosphorylation of the mitogen‐activated protein kinase CgMk1, which is essential for infection‐associated functions, while CgSho1 was unable to activate CgMk1 alone but rather cooperated with CgMsb2 to activate CgMk1. These data suggest that CgMsb2 contributes to the activation of CgMk1 and has overlapping functions with CgSho1 in plant surface sensing, appressorium formation, and pathogenicity.
Keywords: appressorium, Colletotrichum gloeosporioides, host surface signals, mitogen‐activated protein kinase, pathogenicity, sensors
The sensor protein CgMsb2 contributes to the activation of CgMk1 and has overlapping functions with CgSho1 in plant surface sensing, appressorium formation, and pathogenicity.
![]()
1. INTRODUCTION
Colletotrichum is an economically important plant‐pathogenic genus worldwide that currently comprises over 190 accepted species and is one of the top 10 fungal plant pathogens listed by Molecular Plant Pathology (Dean et al., 2012; Jayawardena et al., 2016). The hemibiotrophic ascomycete fungus Colletotrichum gloeosporioides is the causal agent of poplar anthracnose, which results in enormous economic losses in north‐east China (Li et al., 2012). C. gloeosporioides forms a specialized structure known as an appressorium that is required for infection (Gomes et al., 2009; Weir et al., 2012). Foliar infection by C. gloeosporioides starts with the adhesion of conidia to the cuticle of leaves. Subsequently, after a conidium germinates, a germ tube is generated from one side of the spore, and the dome‐shaped appressorium becomes differentiated from the tip of the germ tube. A mature appressorium contains a melanized layer that enables the accumulation of enormous turgor pressure and forms a penetration peg to mechanically rupture the cuticle of leaves. The penetration peg then differentiates into the primary and secondary hyphae that kill mesophyll cells, resulting in necrotic lesions on poplar leaves that are a typical symptom of anthracnose (Pereira et al., 2009; Zhang et al., 2018b). On the plant surface, the cues responsible for appressorium differentiation range from chemical signals, such as epicuticular waxes and cutin monomers, to the physical nature of the surface, such as its hydrophobicity, hardness, and topography (Kumamoto, 2008; Mendoza‐Mendoza et al., 2009). It is thought that these stimuli are perceived by fungal cell surface sensors that subsequently engage in signalling through downstream pathways.
The surfaces of plants represent multifunctional interfaces between the host and pathogens. For foliar pathogens, the penetration of the plant cuticle and cell wall is a crucial step to initiate infectious growth. The epidermis is the primary surface of nearly all terrestrial plants, with the outermost epidermal layer referred to as the cuticle (Holloway, 1994; Jeffree, 2007). Among all cuticular components, waxes are responsible for the primary function of the cuticle, with epicuticular waxes performing stabilization and barrier functions, while the intracuticular waxes are impregnated with a biopolymer called cutin (Koch et al., 2009, 2010). Cutin is composed of hydroxyl and hydroxyepoxy fatty acids, the major wax components being primary and secondary alcohols, ketones, fatty acids, and aldehydes, with alkanes being elementary substances in plant waxes (Baker, 1982; Jetter et al., 2007; Kunst & Samuels, 2003). After recognizing and responding to various host surface cues, phytopathogens form specialized infection structures such as appressoria or hyphopodia, initiating the infection process (Carver & Gurr, 2007; Wilson & Talbot, 2009). To elucidate the mechanisms of plant surface signal recognition in phytopathogens, a few transcriptomic studies on ascomycete fungi have been performed. In classic appressorium‐forming fungi, transcriptome analysis at the appressorium formation stage showed that the sensing of plant surface cues induces the expression of various genes. In Botrytis cinerea, on a hard and wax‐coated surface, early conidial development is accompanied by rapid shifts in gene expression that prepare the fungus for germ tube outgrowth, appressorium formation, and host cell invasion (Leroch et al., 2013). On a hydrophobic surface, large‐scale gene expression changes are observed during appressorium development in Magnaporthe oryzae (Soanes et al., 2012). In Ustilago maydis, transcriptional profiling revealed transcriptional changes in up to 20% of chromosomally encoded genes during the sensing of plant surface cues in vitro, responses that are dependent on the putative sensors UmSho1 and UmMsb2 (Lanver et al., 2014; Mendoza‐Mendoza et al., 2009). The results of these studies indicate that the sensing of plant surface cues is a crucial process for infection by phytopathogens, and putative sensors play an important role during this process.
The first examined receptor of a plant‐pathogenic fungus was the noncanonical G‐protein‐coupled receptor (GPCR) Pth11 in M. oryzae, which has been suggested to be a surface sensor functioning upstream of the cyclic adenosine monophosphate (cAMP) pathway. Moreover, MoPth11 mutants are impaired in appressorium formation in response to both cutin monomers and hydrophobic surface cues (DeZwaan et al., 1999). MoPth11 has a common in fungal extracellular membrane (CFEM) domain that has been shown to be required for proper development of appressoria, appressorium‐like structures, and pathogenicity (Kou et al., 2017). In Saccharomyces cerevisiae, Msb2 and Sho1 were functionally characterized in detail, and Msb2 was shown to have overlapping functions with Sho1 in regulating filamentous growth (FG)/invasive growth and hyperosmotic stress responses (Tatebayashi et al., 2007; Yang et al., 2009). In many plant pathogens, homologues of the yeast proteins Msb2 and Sho1 have been identified as putative sensors involved in plant surface signal recognition, pathogenicity, and activation of the mitogen‐activated protein kinase (MAPK) Pmk1 (Jiang et al., 2018). Pmk1 is orthologous to Fus3 and Kss1, which are key MAPKs involved in the pheromone response and the FG pathway in S. cerevisiae (Chen & Thorner, 2007). Activation of the MAPK Pmk1 occurs through the MAPK kinase Ste7 and the MAPK kinase kinase Ste11, which are homologues of the S. cerevisiae Ste7 MAPK/eRK kinase (MeK) and Ste11 MeK kinase, respectively (Zhao et al., 2005). The Pmk1 homologue is required for infection in all plant pathogens studied (Turrà et al., 2014). Intriguingly, Msb2 and Sho1 have been identified as upstream components of the Pmk1 pathway. In Fusarium oxysporum, Msb2 regulates a subset of Fmk1‐dependent functions, and both Msb2 and Sho1 are required for Fmk1 activation (Perez‐Nadales & Di Pietro, 2011, 2015). In M. oryzae and U. maydis, Msb2 and Sho1 also regulate the phosphorylation of Pmk1 (Kpp2 Kpp6), while the expression of a dominant active MST7/MEK7 allele partially suppressed the defects exhibited by the Msb2 deletion mutant (Lanver et al., 2010; Liu et al., 2011). In addition, the orthologous MAPK in C. gloeosporioides, CgMk1, is essential for appressorium formation and pathogenicity on Populus (He et al., 2017). However, the putative upstream sensors of CgMk1 remain unidentified.
In the present study, three putative sensors responsible for host surface signal recognition in C. gloeosporioides, namely CgMsb2, CgSho1, and CgPth11, were identified and functionally characterized. We demonstrated the significant role of CgMsb2 in signal perception and pathogenicity, and that of CgSho1 in invasive growth. Novel insight into the function of CgPth11 was obtained.
2. RESULTS
2.1. Characterization and deletion of Msb2, Sho1, and Pth11 orthologues in C. gloeosporioides
Msb2, Sho1, and Pth11 orthologues were identified by BLASTp searches of the genome database of C. gloeosporioides (http://genome.jgi.doe.gov/Gloci1/Gloci1.home.html) using the sequences of S. cerevisiae Msb2 (NP_011528.3), Sho1 (NP_011043.1), and M. oryzae Pth11 (XP_003711700.1) as queries. CgMsb2 encodes a hypothetical protein of 988 amino acids, while an alignment showed the overall sequence identity of CgMsb2 with other homologues was rather low (Figure S1a; Table 1). In particular, the alignment of Msb2 homologues showed diversity of amino acid sequences at the N‐terminus (Figure S1a), indicating that the noncytoplasmic domain sequence is not conserved among different fungi. Phylogenetic analysis showed a high degree of similarity among Msb2 homologues (Figure S1b). CgSho1 encodes a hypothetical protein of 287 amino acids, and alignment of the CgSho1 amino acid sequence revealed a high level of identity with Sho1 homologues (Figure S2a; Table 1), and phylogenetic analysis showed a high level of similarity among Sho1 homologues (Figure S2b). CgPth11 encodes a hypothetical protein of 446 amino acids, and also shared identity with Pth11 homologues, but the overall sequence identity was rather low (Figure S3a; Table 1), and phylogenetic analysis showed a high level of similarity among Pth11 homologues (Figure S3b). CgMsb2 and CgPth11 possess a signal peptide at the N‐terminus, and CgMsb2, CgSho1, and CgPth11 possess cytoplasmic domains at the C‐terminus (Figure S4). The cytoplasmic domain of CgMsb2 shares high sequence identity with homologues in M. oryzae (67.5%), Fusarium graminearum (66.4%), and F. oxysporum (67.5%) (Table 1). CgMsb2 possesses one transmembrane (TM) domain between the noncytoplasmic and the cytoplasmic domain (Figure S4). CgSho1 is well conserved in filamentous fungi and encodes a protein with four TM domains near the N‐terminus and one SH3 domain at the C‐terminus (Figure S4). The sequence of the SH3 domain in CgSho1 also shares high identity with the homologues in M. oryzae (62.5%), F. graminearum (58.2%), and F. oxysporum (56.2%) (Table 1). In addition, CgPth11 also possesses seven TM domains, which is typical of GPCR proteins, and a CFEM domain at the N‐terminus (Figure S4). However, the CFEM domain of CgPth11 shows low sequence identity with other homologues (Table 1). To determine the roles of CgMsb2, CgSho1, and CgPth11, the encoding genes were fully knocked out using the split marker method (Figure S5a,b). Subsequently, the full sequences of CgMsb2, CgSho1, and CgPth11 were reintroduced into the ΔCgMsb2, ΔCgSho1, and ΔCgPth11 strains for the complementation assays, respectively (Figure S5c). The double deletion mutants ΔCgMsb2Sho1, ΔCgMsb2Pth11, and ΔCgSho1Pth11 were also generated (Figure S5d). Additional strain construction details are provided in the Experimental Procedures section.
TABLE 1.
Sequence identity to homologues of CgMsb2, CgSho1, and CgPth11
| Protein name | Plant pathogen | Sequence identity (%) | Domain and sequence identity (%) | GenBank accession numbers | |
|---|---|---|---|---|---|
| Msb2 | Magnaporthe oryzae | 41.1 | Cytoplasmic domain | 67.5 | XP_003711882.1 |
| Botrytis cinerea | 46.5 | 52.6 | XP_024552048.1 | ||
| Fusarium graminearum | 46.3 | 66.4 | EYB29490.1 | ||
| Fusarium oxysporum | 26.8 | 67.5 | XP_018246369.1 | ||
| Saccharomyces cerevisiae | 33.8 | 24.0 | NP_011528.3 | ||
| Ustilago maydis | 29.2 | 14.3 | XP_756627.1 | ||
| Sho1 | M. oryzae | 71.9 | SH3 domain | 62.5 | XP_016845977.1 |
| B. cinerea | 64.5 | 52.2 | XP_024550493.1 | ||
| F. graminearum | 75.0 | 58.2 | XP_011328302.1 | ||
| F. oxysporum | 73.8 | 56.0 | TVY72941.1 | ||
| S. cerevisiae | 36.9 | 31.7 | NP_011043.1 | ||
| U. maydis | 41.6 | 34.7 | XP_011389617.1 | ||
| Pth11 | M.oryzae | 42.4 | CFEM domain | 25.8 | XP_003711700.1 |
| Colletotrichum higginsianum | 48.2 | 24.1 | XP_018159285.1 | ||
| Colletotrichum fructicola | 95.8 | 75.8 | KAF4934592.1 | ||
| Verticillium dahliae | 56.0 | 32.2 | PNH28039.1 | ||
| F. oxysporum | 49.8 | 32.9 | RKK60341.1 | ||
| Btp1 | B. cinerea | 31.7 | 7 transmembrane region | 26.1 | CAE55153.1 |
2.2. Msb2 plays a critical role in the recognition of artificial hydrophobic surfaces
Hydrophobic surfaces are crucial signals for the stimulation of appressorium formation in various fungi (Kumamoto, 2008). The artificial hydrophobic surface of a GelBond membrane was used to test the functions of CgMsb2, CgSho1, and CgPth11 in hydrophobic surface recognition. At 5 days postinoculation (dpi) on potato dextrose agar (PDA) plates, conidiation was similar among all strains (Table 2). An equal volume of conidial suspension (105 conidia/ml) from each strain was inoculated onto the hydrophobic and hydrophilic side of the GelBond membrane. On the hydrophobic surface, the wildtype (WT) strain formed numerous appressoria at 5 hr postinoculation (hpi) (>60%; Figure 1a,b). However, appressorium formation in ΔCgSho1 and ΔCgPth11 was significantly decreased compared with that observed for the WT and complementation strains, and was fully blocked in ΔCgMsb2 at 5 hpi (Figure 1a,b). At 12 hpi, approximately 90% of germ tubes in WT, ΔCgSho1, and ΔCgPth11 formed appressoria, but fewer than 10% of ΔCgMsb2 germ tubes formed appressoria (Figure 1a,b). Consistent with the results in the single mutants at 5 hpi, appressorium formation was abolished in ΔCgMsb2Sho1 and ΔCgMsb2Pth11, and appressorium formation in ΔCgSho1Pth11 was similar to that of ΔCgPth11 (Figure 1a,b). At 12 hpi, ΔCgMsb2Sho1 barely formed appressoria, and appressorium formation was significantly decreased in ΔCgMsb2Pth11 and slightly reduced in ΔCgSho1Pth11 compared to that of the WT (Figure 1a,b). On the hydrophilic surface at 12 hpi, appressorium formation in ΔCgMsb2, ΔCgMsb2Sho1, and ΔCgMsb2Pth11 was fully blocked (Figure 1c,d). However, ΔCgSho1, ΔCgPth11, and ΔCgSho1Pth11 showed similar appressorium formation to that of the WT (Figure 1c,d). A previous report showed that exogenous cAMP treatment can fully rescue the appressorium defect of a ΔMoPth11 mutant, suggesting that MoPth11 functions upstream of the cAMP pathway in M. oryzae (Kou et al., 2017). To test whether cAMP has a similar effect on C. gloeosporioides, a nontoxic concentration of cAMP (1.5 mM) was added to the conidial suspensions of WT and ΔCgPth11; however, exogenous cAMP treatment failed to restore the appressorium formation defects of the ΔCgPth11 mutant at the early stage (5 hpi; Figure 1e), indicating that CgPth11 is involved in the formation of appressoria in a cAMP‐independent manner. Collectively, these results indicated that CgMsb2 plays a critical role in the recognition of hydrophobic surfaces, CgSho1 and CgPth11 only play a minor role at the early stage, and CgMsb2 and CgSho1 have complementary roles in this process.
TABLE 2.
Conidiation of each strain at 5 days postinoculation
| Strain | Conidiation (×106 spores/plate) |
|---|---|
| Wild type | 7.9 ± 2.0 |
| ΔCgMsb2 | 7.4 ± 5.5 |
| ΔCgMsb2/MSB2 | 8.2 ± 3.5 |
| ΔCgSho1 | 8.4 ± 4.6 |
| ΔCgSho1/SHO1 | 7.9 ± 1.6 |
| ΔCgPth11 | 7.3 ± 0.5 |
| ΔCgPth11/PTH11 | 7.5 ± 1.5 |
| ΔCgMsb2Sho1 | 8.5 ± 0.8 |
| ΔCgMsb2Pth11 | 8.0 ± 2.7 |
| ΔCgSho1Pth11 | 7.7 ± 2.0 |
Conidiation of each strain was examined on potato dextrose agar plates; means and SD values were calculated from three independent experiments.
FIGURE 1.
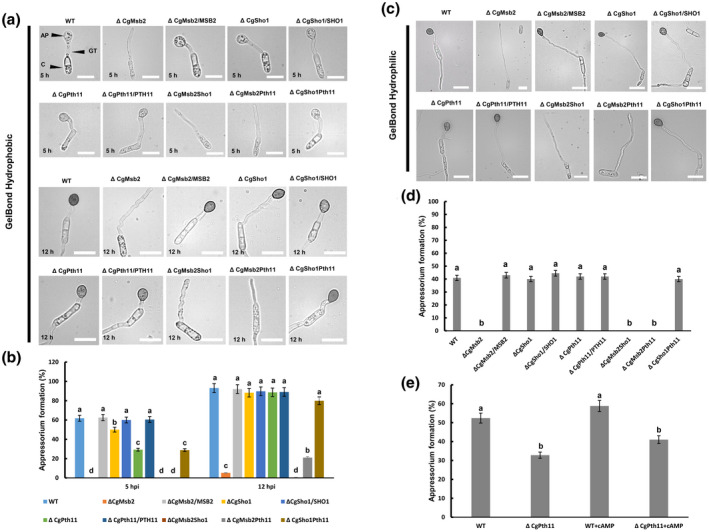
Appressorium formation assays on hydrophobic and hydrophilic membranes. (a) Equal volumes (30 μl) of conidial suspension (2 × 104/ml) of each strain were inoculated on the hydrophobic side of the GelBond membrane; the images were acquired at 5 and 12 hr postinoculation (hpi), respectively. AP, appressorium; GT, germ tube; C, conidia. Bars = 10 µm. (b) Bar chart showing the rate of appressorium formation on the hydrophobic side of the GelBond membrane at 5 and 12 hpi. At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test. (c) Equal volumes (30 μl) of conidial suspension (2 × 104/ml) of each strain were inoculated on the hydrophilic side of the GelBond membrane; the images were acquired at 12 hpi. Bars = 10 µm. (d) Bar chart showing the rate of appressorium formation on the hydrophilic side of the GelBond membrane at 12 hpi. At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test. (e) Exogenous cAMP was mixed with conidial suspensions of the wildtype (WT) and ΔCgPth11 at a final concentration of 1.5 mM. The bar chart shows the appressorium formation rate on the hydrophobic side of the GelBond membrane at 5 hpi. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test
2.3. Msb2 and Sho1 are required for appressorium penetration and invasive growth, respectively, and regulate cellophane membrane penetration cooperatively
In previous studies, penetration tests using onion epidermal cells have been extensively used in the analysis of phytopathogenic fungi (Park et al., 2004; Vanstreels et al., 2005). In the present study, onion epidermal cells were used to determine the penetration ability of the appressoria of each mutant. On the hydrophobic surfaces of onion epidermal cells at 12 hpi, similar defects in appressorium formation were observed for each mutant as those obtained for the GelBond membrane tests (Figure 2a,b). Moreover, the deletion of CgMsb2 resulted in both appressorium formation and penetration defects, as many appressoria formed by the ΔCgMsb2 mutant failed to develop infection hyphae (Figure 2a,b). CgSho1 or CgPth11 was dispensable for the formation of appressoria, and the penetration rate was not affected compared with that of the WT and complementation strains (Figure 2a,b). Deletion of CgPth11 in ΔCgMsb2 and ΔCgSho1 resulted in no further defects in appressorium formation or penetration compared to that of the respective single mutant strains (Figure 2a,b). However, the infection hyphae of the ΔCgSho1 mutant were shorter than those of the WT and complementation strains at 12 hpi (Figure 2a). At 24 hpi, the infection hyphae of the WT strain had spread from the initially infected cell into multiple cells, and infection hyphae successfully broke through the cell walls of adjacent cells (Figure 2c). In the ΔCgSho1 mutant, infection hyphae tended to be trapped in the initially infected cells, and the infection hyphae failed to break through the cell wall (Figure 2c,d). In the ΔCgMsb2Sho1 double mutant, very few appressoria formed, and those that did form showed defects in both appressorium penetration and invasive growth (Figure 2a–d). Reactive oxygen species (ROS) play an essential role in plant defence responses, and we previously revealed that the small G protein CgCdc42 and the endocytosis‐related protein CgEnd3 are also required for invasive growth, while exogenous treatment with the ROS inhibitor diphenyleneiodonium chloride (DPI) restored the defect efficiently (Wang et al., 2021a; Wang et al., 2018). Therefore, DPI was added during conidial germination at a final concentration of 5 µM. The trapped infection hyphae in ΔCgSho1 were successfully restored by exogenously added DPI (Figure 2c,d). This result indicated that CgSho1 is required for oxidant adaptation‐mediated invasive growth.
FIGURE 2.
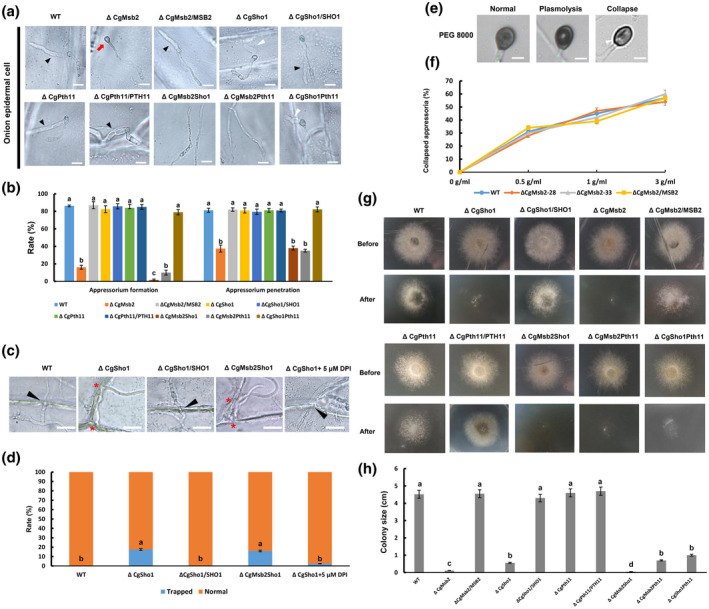
Appressorium formation, penetration, and cellophane membrane penetration assays. (a) Equal volumes (30 μl) of conidial suspension (2 × 104/ml) of each strain were inoculated on onion epidermal cells; the images were acquired at 12 hr postinoculation (hpi). A black triangle indicates an infection hypha. A red arrow indicates an appressorium without penetration. A white triangle indicates a short infection hypha. Bars = 10 µm. (b) Bar chart showing the rate of appressorium formation and penetration on onion epidermal cells at 12 hpi. At least 200 germinated conidia or appressoria from each strain were measured to calculate the appressorium formation or penetration rate, respectively. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test. (c) Infection hyphae of wildtype (WT), ΔCgSho1, ΔCgSho1/SHO1, ΔCgMsb2Sho1, and ΔCgSho1 supplied with 5 μM diphenyleneiodonium chloride in onion epidermal cells at 24 hpi. A black triangle indicates an infection hypha breaking through the cell wall of adjacent cells. A red asterisk indicates a trapped infection hypha. Bars = 25 µm. (d) The rate of normal or trapped infection hyphae of each strain at 24 hpi. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test. (e) Conidial suspensions (2 × 104/ml) from WT, ΔCgMsb2‐28, ΔCgMsb2‐33, and ΔCgMsb2/ MSB2 were inoculated on the hydrophobic side of a GelBond membrane. At 12 hpi, the water drop was removed by 30 µl of 0.5, 1, and 3 g/ml polyethylene glycol (PEG) 8000. After 5 min of treatment, the collapse rate of appressoria was recorded. There are three types of appressoria: normal, plasmolytic, and collapsed appressoria. A white triangle indicates a collapsed appressorium (longitudinal cavity). Bar = 2 μm. (f) Line chart showing the rate of collapsed appressoria under the treatment of different concentrations of PEG 8000. This experiment was repeated three times. At least 200 melanized appressoria from each strain were measured to calculate the proportion of collapsed appressoria. (g) The strains were grown on cellophane membranes overlaid on potato dextrose agar for 3 days at 25 °C (Before). The cellophane membrane was removed, and the resulting plates were incubated at 25 °C for an additional 2 days (After). (h) Bar chart showing the colony size of each strain at 2 days after removal of the cellophane membrane. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test
In many pathogenic fungi, the buildup of turgor pressure in the appressorium provides the driving force that enables fungi to mechanically break through the host tissue (Money & Howard, 1996). To determine whether the penetration defect in the ΔCgMsb2 mutant was caused by decreased turgor pressure, turgor pressure was tested using various concentrations of polyethylene glycol (PEG) 8000 (Howard et al., 1992). At 12 hpi, the collapse rate of appressoria was determined after a 5‐min treatment. No difference in collapse rate was observed between WT and ΔCgMsb2 mutants (Figure 2e,f), indicating that the penetration defect of the appressorium in ΔCgMsb2 was not caused by decreased turgor pressure. To further determine the roles of CgMsb2, CgSho1, and CgPth11 in penetration ability, WT, ΔCgMsb2, ΔCgSho1, ΔCgPth11, ΔCgMsb2Sho1, ΔCgMsb2Pth11, ΔCgSho1Pth11, and complementation strains were inoculated on cellophane membrane overlaid on PDA plates. Plates were cultured at 25 °C for 2 days (Before). At 2 dpi, the entire membrane with colony was removed, and the resulting plates were cultured at 25 °C for an additional 2 days (After). After the removal of the cellophane membrane, limited growth was observed for ΔCgSho1 and ΔCgMsb2, and a synergistic effect was observed in ΔCgMsb2Sho1 (Figure 2g,h). However, deletion of CgPth11 in WT, ΔCgMsb2, and ΔCgSho1 resulted in no further defects in cellophane membrane penetration (Figure 2g,h), indicating that CgPth11 is dispensable for cellophane membrane penetration. This result indicates that both CgMsb2 and CgSho1 are required for cellophane membrane penetration. Collectively, these results indicate that CgMsb2 and CgSho1 play roles in appressorium penetration and invasive growth, respectively, and have overlapping function in the penetration of cellophane membrane.
2.4. Msb2 and Sho1 play complementary roles in virulence
In Colletotrichum species, appressorium formation is strongly correlated with pathogenicity. Considering the significant role of appressorium formation and penetration, leaf inoculation assays were performed to assess the roles of CgMsb2 and CgSho1 in C. gloeosporioides pathogenicity. Conidial suspensions (2 × 105/ml) from the WT, ΔCgMsb2 mutants, and ΔCgMsb2/ MSB2 complementation strains were inoculated onto detached poplar leaves. At 4–7 dpi, necrotic lesions were formed by the WT and ΔCgMsb2/ MSB2 complementation strains and spread at a steady rate, while the ΔCgMsb2 mutant did not induce lesion formation (Figure 3a,b). However, as the ΔCgMsb2 mutant was able to form a small number of appressoria on GelBond membranes and onion epidermal cells, we presumed that the ΔCgMsb2 mutant may retain weak pathogenicity, and prolonged observation (8 dpi) demonstrated that the pathogenicity of this strain was significantly decreased but not fully abrogated (Figure 3a,b). Unlike the ΔCgMsb2 mutant, the ΔCgSho1 mutant caused necrotic lesions similar to those formed by the WT strain at 4 dpi (Figure 3c,d). However, the expansion rate of lesions formed by the ΔCgSho1 mutant was slower than that observed for the WT and ΔCgSho1/ SHO1 complementation strains at 5–6 dpi (Figure 3c,d), indicating that the pathogenicity of the ΔCgSho1 mutant was also decreased, which was caused by the defect of invasive growth. Moreover, the double deletion of CgMsb2 and CgSho1 showed an exacerbated defect in pathogenicity, only causing a tiny lesion at 8 dpi (Figure 3e,f). However, the necrotic lesions caused by WT and ΔCgPth11 strains were comparable, and the deletion of CgPth11 also caused no exacerbated defect in the pathogenicity of the ΔCgMsb2 and ΔCgSho1 strains (Figure 3e,f). These results indicate that both CgMsb2 and CgSho1, but not CgPth11, are required for full virulence in C. gloeosporioides.
FIGURE 3.
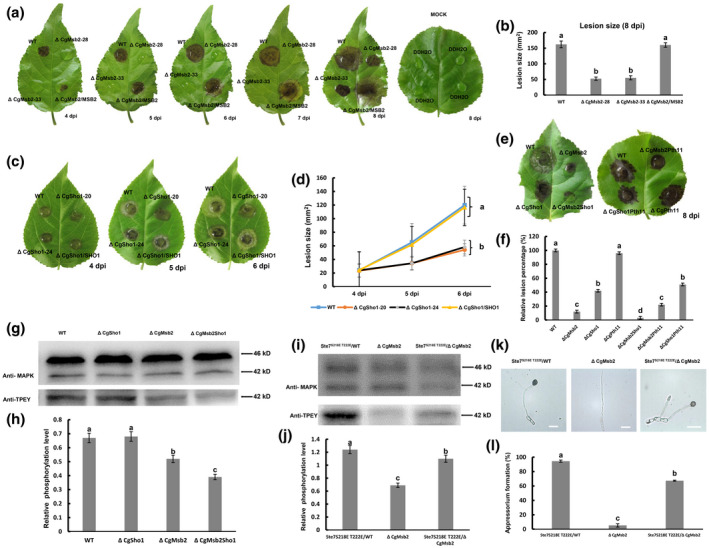
Plant infection and western blot assays. (a) Detached poplar leaves were inoculated with wildtype (WT), ΔCgMsb2, or ΔCgMsb2/MSB2 (complemented) strains or mock‐inoculated with water. Images were pictured at 4–8 days postinoculation (dpi). (b) Bar chart showing the lesion size after inoculation with WT, ΔCgMsb2, and ΔCgMsb2/MSB2 at 8 dpi. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test. (c) Detached poplar leaves were inoculated with WT, ΔCgSho1, and ΔCgSho1/SHO1. (d) Line chart showing the lesion size of WT, ΔCgSho1, and ΔCgSho1/SHO1 at 4–6 dpi. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test. (e) Detached poplar leaves were inoculated with WT, ΔCgMsb2, ΔCgSho1, ΔCgMsb2Sho1, ΔCgPth11, ΔCgMsb2Pth11, and ΔCgSho1Pth11. Images were taken at 8 dpi. (f) Bar chart showing the relative lesion size of each strain, normalized to the WT lesion size (100%). Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test. (g) Western blot analysis of the CgMk1 phosphorylation level in WT, ΔCgSho1, ΔCgMsb2, and ΔCgMsb2Sho1. Anti‐MAPK was used to detect the expression levels of CgMk1 (42 kDa) and putative CgSlt2 (46 kDa), and anti‐TPEY was used to detect the phosphorylation level of CgMk1 (42 kDa). (h) Bar chart showing the relative phosphorylation level of CgMk1 in WT, ΔCgSho1, ΔCgMsb2, and ΔCgMsb2Sho1 compared with that of the overall CgMk1 content in each respective strain. Data were taken from two biological replicates with similar results. Error bars represent the standard deviations based on two independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test. (i) Western blot analysis of the CgMk1 phosphorylation level in Ste7 S218E T222E/WT, ΔCgMsb2, and Ste7 S218E T222E/ΔCgMsb2. Anti‐MAPK was used to detect the expression levels of CgMk1 (42 kDa) and putative CgSlt2 (46 kD), and anti‐TPEY was used to detect the phosphorylation level of CgMk1 (42 kDa). (j) Bar chart showing the relative phosphorylation level of CgMk1 in Ste7 S218E T222E/WT, ΔCgMsb2, and Ste7 S218E T222E/ΔCgMsb2 compared with that of the overall CgMk1 content in each respective strain. Data were taken from two biological replicates with similar results. Error bars represent the standard deviations based on two independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test. (k) Appressorium formation of Ste7 S218E T222E/WT, ΔCgMsb2, and Ste7 S218E T222E/ΔCgMsb2 on the hydrophobic side of a GelBond membrane. Bars = 10 µm. (l) Bar chart showing the rate of appressorium formation in (k). Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test
2.5. Msb2 contributes to CgMk1 phosphorylation
In the present study, CgMsb2 was shown to regulate a subset of CgMk1‐related functions, including appressorium formation, appressorium penetration, and pathogenicity. To determine whether CgMsb2 functions upstream of the CgMk1 pathway, the phosphorylation and protein levels of CgMk1 were detected with anti‐TPEY and anti‐MAPK antibodies, respectively. The results showed no difference in CgMk1 (42 kDa) protein levels among the WT, ΔCgMsb2, ΔCgSho1, and ΔCgMsb2Sho1 strains (Figure 3g). However, reduced CgMk1 phosphorylation levels were detected in the ΔCgMsb2 mutant (Figure 3g,h). In the same western blot analysis, CgMk1 protein and phosphorylation levels were not affected in the ΔCgSho1 compared to the WT strain, whereas CgMk1 phosphorylation levels were further decreased in the ΔCgMsb2Sho1 mutant compared to those observed in the ΔCgMsb2 mutant (Figure 3g,h). These results indicate that CgMsb2 contributes to CgMk1 phosphorylation and that CgSho1 is unable to activate CgMk1 phosphorylation alone but rather cooperates with CgMsb2 to activate CgMk1.
To further demonstrate that CgMsb2 functions upstream from the CgMk1 pathway, the constitutively active allele of Ste7 S218E T222E was constructed and introduced into the WT and ΔCgMsb2 strains. A western blot assay showed that the phosphorylation of CgMk1 was significantly increased in the Ste7 S218E T222E/ΔCgMsb2 strain compared to that of the ΔCgMsb2 mutant (Figure 3i,j). Besides, appressorium formation by the Ste7 S218E T222E/ΔCgMsb2 strain was also partly restored compared to the ΔCgMsb2 mutant phenotype (Figure 3k,l), indicating that expression of the constitutively active Ste7 induces appressorium formation in the ΔCgMsb2 mutant. Collectively, these results indicate that CgMsb2 contributes to CgMk1 phosphorylation and functions upstream from CgMk1.
2.6. Msb2 and Sho1 regulate the recognition of multiple plant surface signals
In the present study, we show that CgMsb2 and CgSho1, but not CgPth11, are cooperatively involved in appressorium formation and pathogenicity. To determine whether CgMsb2 and CgSho1 are responsible for the recognition of plant surface signals, we first inoculated conidial suspensions of each strain on intact poplar leaves. To observe the initial stage of appressorium formation, each inoculated area was cut and stained with 1 µg/ml calcofluor white (CFW) at 24 hpi. During this stage, the WT and complementation strains produced abundant appressoria (>90%) (Figure 4a,c). However, only approximately 17.6% and 17.1% of the ΔCgMsb2 and ΔCgMsb2Sho1 mutant cells formed appressoria, respectively (Figure 4a,c), indicating that the loss of CgMsb2 and CgMsb2Sho1 resulted in defective appressorium formation on the host surface. In addition, the deletion of CgSho1 did not affect appressorium formation on plant leaves, as abundant appressoria (91.4%) were produced by the ΔCgSho1 strain, which was comparable with that observed for the WT and ΔCgSho1/ SHO1 strains (Figure 4a,c). These results indicated that the ΔCgSho1 mutant still fully responded to various cues present on poplar leaf surfaces for appressorium formation. In various fungi, the host surface wax has been shown to induce appressorium formation (Hwang & Kolattukudy, 1995; Lanver et al., 2010; Uchiyama & Okuyama, 1990). To test whether the surface wax of poplar leaves stimulates appressorium formation, epicuticular waxes were removed by hexane treatment. On dewaxed leaves, appressorium formation by the WT, ΔCgSho1, and complementation strains was essentially blocked, with these strains forming long germ tubes (Figure 4a,c). However, the lack of surface waxes did not affect appressorium formation by the ΔCgMsb2 mutant (Figure 4a,c), indicating that the recognition of surface waxes was blocked in the ΔCgMsb2 strain and that CgMsb2 is required for the recognition of leaf surface waxes. Notably, unlike the ΔCgMsb2 mutant, appressorium formation in the ΔCgMsb2Sho1 mutant was fully blocked on dewaxed leaves, and ΔCgMsb2Sho1 developed normal germ tubes that were similar to those formed by the ΔCgMsb2 mutant (Figure 4a,c). These results indicated that other host surface signals, such as hydrophobicity, may contribute to appressorium formation in the ΔCgMsb2 mutant on both intact and dewaxed host surfaces. The fully abrogated ability to recognize hydrophobicity and wax may have resulted in the inability of the ΔCgMsb2Sho1 mutant to form appressoria on dewaxed leaves.
FIGURE 4.
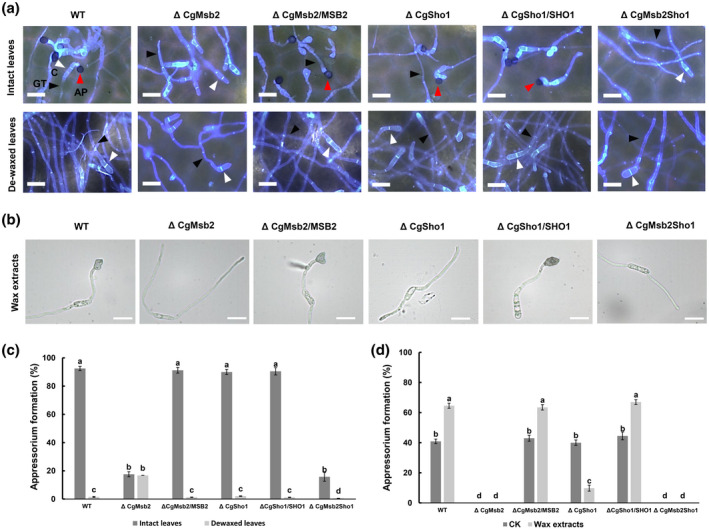
Appressorium formation on intact and dewaxed leaves and crude wax‐coated surfaces. (a) Equal concentrations of conidial suspension from wildtype (WT), ΔCgMsb2, ΔCgSho1, ΔCgMsb2Sho1, and complementation strains were inoculated on intact and dewaxed poplar leaves. The inoculation site of each strain was cut and stained using 1 μg/ml calcofluor white at 24 hr postinoculation. Images were acquired using fluorescence microscopy. A red triangle indicates a melanized appressorium. A black triangle indicates a germ tube. A white triangle indicates a conidium. AP, appressorium; GT, germ tube; C, conidium. Bars = 15 µm. (b) Equal concentrations of conidial suspension from WT, ΔCgMsb2, ΔCgSho1, ΔCgMsb2Sho1, and complementation strains were inoculated on glass slides coated with crude wax extracts. Bars = 10 µm. (c) Bar chart showing the appressorium formation rate in (a). At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate (including melanized and nonmelanized appressorium). Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different between different treatment groups (intact leaves and dewaxed leaves) at p < .05, as determined using Tukey's post hoc test. (d) Bar chart showing the appressorium formation rate on crude wax extract‐coated surfaces. CK indicates appressorium formation on glass slides. At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different between different treatment groups (CK and wax extracts) at p < .05, as determined using Tukey's post hoc test
To further assess the roles of CgMsb2 in the recognition of leaf surface waxes, crude wax was extracted using hexane. Crude extracted wax dissolved in chloroform was then coated on the surface of a glass slide (hydrophilic surface). For the WT and complementation strains, approximately 65% of germ tubes formed appressoria (Figure 4b,d). However, in the presence of wax extract, the ΔCgMsb2 and ΔCgMsb2Sho1 mutants completely failed to produce appressoria, and only 10% of ΔCgSho1 formed appressoria (Figure 4b,d). Collectively, these results indicate that CgMsb2 plays a crucial role in the recognition of surface wax.
Leaf surface wax extracted with hexane is a mixture that may contain other chemical cues from the plant to stimulate appressorium formation. Therefore, in addition to the leaf crude wax, bee wax and paraffin wax were used to further assess whether CgMsb2 is involved in wax recognition. Conidial suspensions (105 spores/ml) of each strain were inoculated onto glass slides coated with bee or paraffin wax. Compared to bee wax, paraffin wax induced C. gloeosporioides appressorium formation more efficiently (Figure 5a,c). In the presence of paraffin wax, appressorium formation was significantly decreased in the ΔCgMsb2 mutant and slightly reduced in the ΔCgSho1 mutant compared with that of the WT and complementation strains, while the ΔCgMsb2Sho1 double mutant exhibited a stronger defect in appressorium formation (Figure 5a,c). Appressorium formation by each strain on bee wax was highly similar to that observed on intact poplar leaf surfaces (Figure 5a,c). Appressorium formation by the ΔCgMsb2 (8.82%) and ΔCgMsb2Sho1 (8.5%) mutants was significantly decreased, while appressorium formation by the ΔCgSho1 mutant was comparable to that observed for the WT and ΔCgSho1/SHO1 complementation strains (Figure 5a,c). Bee wax and paraffin wax render the glass slide surface hydrophobic. Therefore, there must be differences between bee and paraffin wax in the chemical components that stimulate appressorium formation. As described in a previous report, plant surface waxes primarily contain primary alcohols, secondary alcohols, alkanes, and long‐chain fatty acids (Kunst & Samuels, 2003). Bee wax consists of primary alcohol; paraffin wax mainly consists of long‐chain alkanes (Cwiertnia, 2015; Liu et al., 2011). To determine what compounds in wax are primarily responsible for the stimulation of appressorium formation, the primary alcohol 1‐octacosanol (C28) and hentricacontane alkanes (C31) were used as chemical cues. The WT and complementation strains more efficiently formed appressoria on hydrophilic surfaces coated with C28 than was observed for the C31 treatment (Figure 5b,d). However, C31 was unable to stimulate appressorium formation in ΔCgMsb2, and both C28 and C31 were unable to stimulate appressorium formation in the ΔCgMsb2Sho1 mutant (Figure 5b,d). Collectively, these results showed that CgMsb2 but not CgSho1 is required for the recognition of C28 and C31, and both primary alcohols and alkanes may act as stimulators of poplar leaves for appressorium formation by C. gloeosporioides.
FIGURE 5.
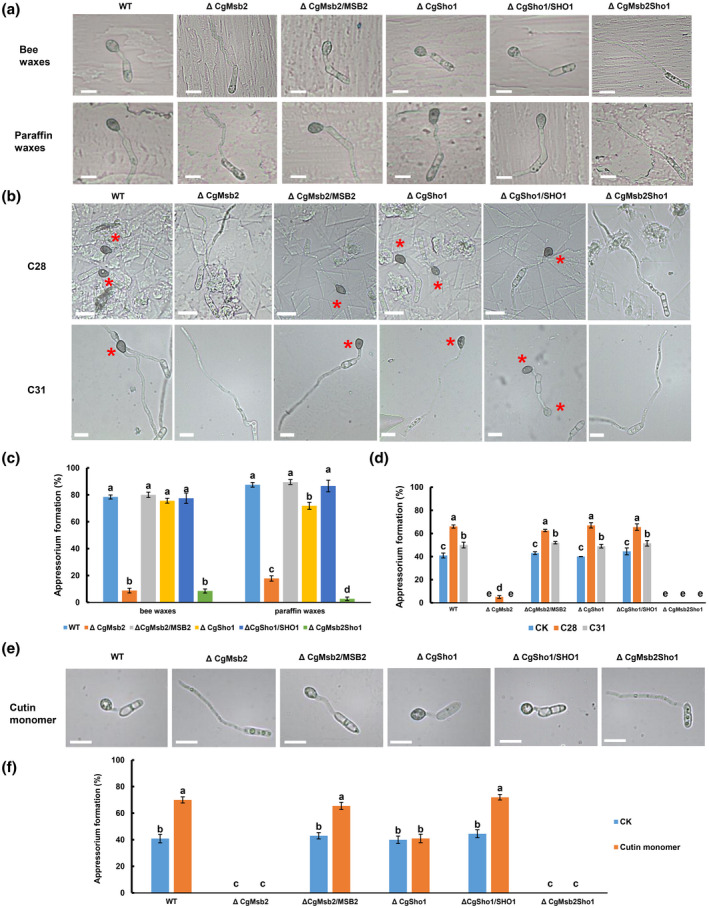
Appressorium formation on glass slides coated with waxes, 1‐octacosanol (C28), hentricacontane (C31), and cutin monomer cis‐9‐octadecen‐1‐ol. (a, b) Equal concentrations of conidial suspension from wildtype (WT), ΔCgMsb2, ΔCgSho1, ΔCgMsb2Sho1, and complementation strains were inoculated on glass sides coated with (a) bee waxes and paraffin waxes and (b) 4 mg/ml C28 and C31. Bars = 10 µm. The images were acquired at 12 hr postinoculation (hpi). A red asterisk indicates an appressorium. (c) Bar chart showing the appressorium formation rate in (a). At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate on bee wax and paraffin wax. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test. (d) Bar chart showing the appressorium formation rate in (b). At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate on C28‐ and C31‐coated surfaces. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different between different treatment groups (CK, C28, and C31) at p < .05, as determined using Tukey's post hoc test. CK indicates appressorium formation on glass slides. (e) Appressorium formation of each strain in the presence of 10 µM cutin monomer cis‐9‐octadecen‐1‐ol. Bars = 10 µm. The images were acquired at 12 hpi. (f) Bar chart showing the appressorium formation rate in (e). At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different between different treatment groups (CK and cutin monomer) at p < .05, as determined using Tukey's post hoc test. CK indicates appressorium formation on glass slides
In addition to hydrophobicity and waxes, cutin monomers are also known to serve as chemical cues for appressorium formation by various fungi (Gilbert et al., 1996; Lanver et al., 2010). In the presence of 10 µM cutin monomer cis‐9‐octadecen‐1‐ol, 70% of WT cells formed appressoria on glass slides, indicating that cutin monomers efficiently induced appressorium formation by C. gloeosporioides (Figure 5e,f). However, the appressorium formation of ΔCgMsb2, ΔCgSho1, and ΔCgMsb2Sho1 was not different from that on glass slides (CK) under the exogenous stimulation of cis‐9‐octadecen‐1‐ol (Figure 5e,f), indicating that cis‐9‐octadecen‐1‐ol had no stimulatory effect on ΔCgMsb2, ΔCgSho1, and ΔCgMsb2Sho1. Therefore, these results indicate that both CgMsb2 and CgSho1 are required for the recognition of cutin monomers.
2.7. Loss of Msb2 and loss of Sho1 cooperatively affect oxidant adaptation
In plants, ROS play a crucial role in the defence against infections by pathogens, and because oxidative bursts are among the earliest responses at the infection site (Apostol et al., 1989), oxidant adaptation is crucial for the infection of plant‐pathogenic fungi. In the present study, CgSho1 was determined to be required for oxidant adaptation‐mediated invasive growth (Figure 2c,d). First, to determine whether CgMsb2, CgSho1, and CgPth11 are involved in the oxidative stress response, each strain was inoculated onto PDA plates containing 5 or 10 mM H2O2. At 4 dpi, the colonies of different strains were comparable in size (Figure 6a,b). However, colonies of the ΔCgMsb2 and ΔCgSho1 strains treated with H2O2 were significantly smaller than WT and complementation strains, and an exacerbated defect was observed for the ΔCgMsb2Sho1 strain (Figure 6a,b). However, vegetative growth of ΔCgPth11 was comparable with that observed for the WT and ΔCgPth11/ PTH11 complementation strains in the presence of H2O2 (Figure 6a,b), and deletion of CgPth11 in the ΔCgSho1 and ΔCgMsb2 strains caused no further defects in the response to H2O2 oxidative stress compared to that of the respective single mutants (Figure 6a,b). These results indicate that both CgMsb2 and CgSho1 are required for the oxidative stress response and that they have overlapping functions. To determine whether deletion of CgMsb2 and CgSho1 affects the tolerance of C. gloeosporioides to ROS during infection, 3,3′‐diaminobenzidine (DAB) staining was performed to detect the accumulation of ROS on onion epidermal cells. At 12 hpi, ROS accumulation was observed around the appressoria, and the degree of staining for the appressoria of the ΔCgMsb2, ΔCgSho1, and ΔCgMsb2Sho1 strains was greater than that observed for the WT and complementation strains (Figure 6c,d). These results indicate that CgMsb2 and CgSho1 cooperatively affect oxidant adaptation.
FIGURE 6.
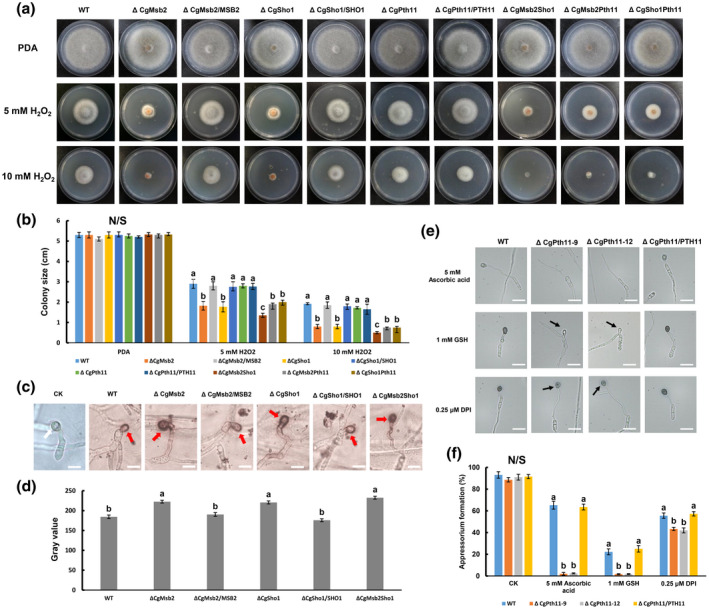
Responses to oxidative stress and antioxidants, and 3,3'‐diaminobenzidine (DAB) staining assays. (a) Colonies of wildtype (WT), ΔCgMsb2, ΔCgSho1, ΔCgPth11, ΔCgMsb2Sho1, ΔCgMsb2Pth11, ΔCgSho1Pth11, and complementation strains on potato dextrose agar (PDA) and PDA supplemented with 5 mM or 10 mM H2O2. Plates were incubated for 4 days at 25 °C. (b) Bar chart showing the colony size of strains in (a). Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test. N/S = difference not significant. (c) Accumulated reactive oxygen species (ROS) during infection of each strain were stained using 1 mg/ml DAB for 12 hr in darkness. DAB is converted to dark brown polymers in the presence of ROS. Images were captured by light microscopy. CK = image of appressorium on onion epidermal cells. A white arrow indicates an unstained appressorium. A red arrow indicates a site of ROS deposition. Bars = 10 µm. (d) The deposition of dark brown polymers was analysed using ImageJ. Data from at least 15 appressoria were collected from each strain in each replicate. Bar chart showing the grey value of each strain. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different at p < .05, as determined using Tukey's post hoc test. (e) Conidial suspensions of WT, ΔCgPth11, and ΔCgPth11/PTH11 complementation strain were mixed with antioxidants L‐glutathione (GSH), ascorbic acid, and diphenyleneiodonium chloride (DPI) at final concentrations of 1 mM, 5 mM, and 0.25 μM, respectively, and inoculated on the hydrophobic side of a GelBond membrane. Images were photographed at 12 hr postinoculation. A black arrow indicates an appressorium with reduced melanization. Bars = 10 µm. (f) Bar chart showing the appressorium formation rate of each strain in (e). At least 200 germinated conidia from each strain were measured to calculate the appressorium formation rate. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test. N/S = difference not significant
In M. oryzae, highly regulated ROS homeostasis is important for MoPth11‐mediated appressorium formation and host invasion. MagB and Pmk1 are also involved in appressorium differentiation in response to exogenous antioxidants. Intriguingly, antioxidants induce appressorium formation in MoPth11 deletion mutants (Kou et al., 2017). Although CgPth11 is dispensable for oxidant adaptation, we aimed to assess the effect of antioxidant treatment on appressorium formation of the ΔCgPth11 strain. To assess the effect of antioxidant treatment of the ΔCgPth11 strain, the antioxidants l‐glutathione (GSH, reduced), ascorbic acid, and DPI were individually tested (Figure 6e,f). Conidial suspensions of the WT, ΔCgPth11, and ΔCgPth11/PTH11 strains were mixed with GSH, ascorbic acid, and DPI at final concentrations of 1 mM, 5 mM, and 0.25 μM, respectively. However, different from the results obtained for the M. oryzae MoPth11 mutant, appressorium formation was significantly decreased in the ΔCgPth11 strain under GSH, ascorbic acid, and DPI treatment (Figure 6e,f), and appressorium with reduced melanization was observed in ΔCgPth11 under the treatment of GSH and DPI (Figure 6e). This result indicates that deletion of CgPth11 resulted in hypersensitivity to antioxidants.
2.8. Msb2 and Sho1 are required for vegetative growth under nitrogen‐limiting conditions
On PDA plates, the WT, ΔCgMsb2, ΔCgSho1, ΔCgMsb2Sho1, and ΔCgMk1 showed similar diameters at 4 dpi (Figure 7a), indicating that CgMsb2, CgSho1, and CgMk1 are not required for growth under nutrient‐rich conditions. However, a previous report showed that Msb2, Sho1, and Fmk1 are required for growth under nitrogen‐limiting conditions in F. oxysporum (Perez‐Nadales & Di Pietro, 2015). In addition, CgMk1 MAPK is also required for vegetative growth under nitrogen‐limited conditions (Wang et al., 2021b). In the present study, each strain was inoculated onto solid minimal medium (MM) supplemented with a single nitrogen source. The ΔCgMsb2, ΔCgSho1, ΔCgMsb2Sho1, and ΔCgMk1 strains showed reduced vegetative growth compared to that of the WT, ΔCgMsb2/ MSB2, and ΔCgSho1/ SHO1 strains on MM containing 10 mM or as nitrogen source, while this defect was not exacerbated in the ΔCgMsb2Sho1 strain (Figure 7a,b), indicating that both CgMsb2 and CgSho1 are involved, with no complementary roles, in growth under nitrogen‐limiting conditions.
FIGURE 7.
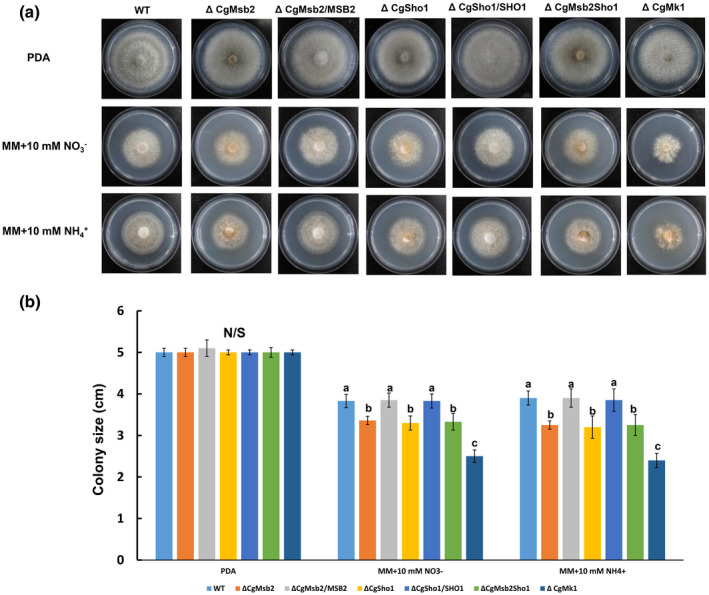
Vegetative growth under nitrogen‐limiting conditions. (a) Colonies of wildtype (WT), ΔCgMsb2, ΔCgSho1, ΔCgMsb2Sho1, ΔCgMk1, and complementation strains on potato dextrose agar (PDA) and minimal medium (MM) supplemented with 10 mM and , respectively. Plates were incubated for 4 days at 25 °C. (b) Bar chart showing the colony size of strains grown under different nitrogen sources. Error bars represent the standard deviations based on three independent replicates. The values indicated by different letters are significantly different within the same treatment at p < .05, as determined using Tukey's post hoc test. N/S = difference not significant
3. DISCUSSION
During the early interaction between pathogens and host plants, phytopathogens must sense and respond to the various signals associated with the host's surface, such as plant surface waxes, cutin monomers, surface hydrophobicity, and nutrient states. This process involves the use of cell surface sensors that subsequently activate downstream pathways. Herein, we found that two sensors, CgMsb2 and CgSho1, play a cooperative role in the recognition of various signals and contribute to the activation of CgMk1 MAPK. In summary, our results support a genetic network of host signal recognition–downstream pathway activation–appressorium formation in C. gloeosporioides (Wang et al., 2021b) (Figure 8a,b), and we determined the conserved and specific functions of CgMsb2 and CgSho1 (Figure 8c).
FIGURE 8.
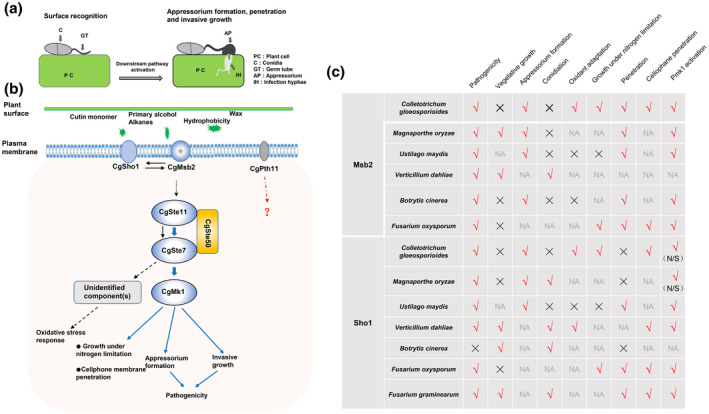
A proposed schematic model of the CgMsb2, CgSho1, and CgMk1 cascades in Colletotrichum gloeosporioides and the reported functions of Msb2 and Sho1 homologues. (a) Conidia of C. gloeosporioides germinate on a host cuticle and subsequently go through surface recognition, downstream pathway activation, appressorium formation, and penetration and invasive growth. PC, plant cell; C, conidium; GT, germ tube; AP, appressorium; IH, infection hypha. (b) The putative cell surface sensors CgMsb2 and CgSho1 recognize a variety of host signals and activate the downstream CgMk1 MAPK pathway; CgSte50, CgSte11, and CgSte7 subsequently activate CgMk1 through phosphorylation to regulate growth under nitrogen‐limiting conditions, cellophane membrane penetration, appressorium formation, invasive growth, and pathogenicity. Moreover, CgSte50–Ste11–Ste7 may activate unidentified component(s) of the oxidative stress response in a CgMk1‐independent manner (Wang et al., 2021b). Solid and dashed arrows indicate the verified and putative connections, respectively. (c) The conserved and species‐specific functions of reported Msb2 and Sho1 homologues in pathogenicity, vegetative growth, appressorium formation, conidiation, oxidant adaptation, growth under nitrogen‐limiting conditions, penetration, cellophane membrane penetration, and Pmk1 activation. √ and × indicate the homologues that are important or dispensable for specific functions, respectively. NA, not applicable; N/S, not significant
3.1. The activation relationship of CgMsb2 and downstream CgMk1
The MAPK Pmk1 plays a crucial role in infection‐related functions in various pathogenic fungi (Jiang et al., 2018; Wilson & Talbot, 2009). In the present study, CgMsb2 was shown to regulate a subset of CgMk1‐related functions and contribute to the phosphorylation of CgMk1. Thus, we suggest that CgMsb2 functions as a sensor upstream of CgMk1. Intriguingly, expression of Ste7 S218E T222E in ΔCgMsb2 did not fully rescue the defect in appressorium formation (Figure 3k,l). In M. oryzae, constitutively active Ste7 also failed to fully rescue the defect of Mst11 and Mst7 deletion mutants; a possible reason is that constitutively active Ste7 may perturb the pattern of Pmk1 activity during appressorium formation (Zhao et al., 2005). In M. oryzae and U. maydis, Msb2 and Sho1 act upstream of the MAPK Pmk1 (Kpp2 Kpp6), and a constitutively active version of the MEK Mst7/fuz7 was shown to induce appressorium formation in a Msb2 deletion mutant. The UmSho1 protein was also identified to interact with the MAPK Kpp6 in a yeast two‐hybrid assay, primarily through the Sho1 SH3 domain (Lanver et al., 2010; Liu et al., 2011). Most studies also claim that Msb2 and Sho1 are conserved sensors of the MAPK Pmk1 (Figure 8c). In F. graminearum, a mutant deleted for FgSho1 was defective in conidiation, pathogenicity, and deoxynivalenol biosynthesis, which is similar to the defect observed in the FgSte50, FgSte11, and FgSte7 mutants. In addition, FgSho1 also physically interacts with the MAPK module FgSte50–Ste11–Ste7 via its SH3 domain, and the level of FgGpmk1 phosphorylation exhibited a significant decrease in the ΔFgSho1 strain (Gu et al., 2014). In the soilborne vascular wilt fungus F. oxysporum, deletion of FoMsb2 and FoSho1 resulted in the characteristic defects of the Fmk1 mutant with respect to vegetative growth under nitrogen‐limiting conditions, penetration, and pathogenicity, and FoMsb2 and FoSho1 also regulate Fmk1 phosphorylation and the expression of Fmk1‐regulated effector genes (Gu et al., 2014; Perez‐Nadales & Di Pietro, 2011). In the present study, CgMsb2, CgSho1, and CgMk1 were also shown to contribute to vegetative growth under nitrogen‐limiting conditions (Figure 7). In S. cerevisiae, Verticillium dahliae, Aspergillus fumigatus, and Candida albicans, Sho1 homologues are involved in the oxidative stress response (Ma et al., 2008; Qi et al., 2016; Román et al., 2006; Singh, 2000). In the present study, we showed that both CgMsb2 and CgSho1 play significant roles in oxidant adaptation and that CgMsb2 cooperates with CgSho1 to promote this function (Figure 6). CgMk1 MAPKs are also involved in oxidant adaptation in C. gloeosporioides. CgSte50–CgSte11–CgSte7 deletion mutants were tested under H2O2 oxidative stress, and the results showed that the three MAPK mutants are hypersensitive to H2O2 oxidative stress (Wang et al., 2021b). In V. dahliae, VdSho1 was shown to be essential for both membrane penetration and melanin production, with the Vst50–Vst11–Vst7 module regulating VdSho1‐mediated plant penetration and melanin production, while VdSho1 interacts physically with the central conserved region of Vst50 through the SH3 domain (Li et al., 2019). In C. albicans, Msb2 cooperates with Sho1 to activate the MAPK Cek1 (a Pmk1 homologue) (Román et al., 2009). In B. cinerea, Msb2 regulates surface sensing and host penetration via BMP1–MAPK signalling (Leroch et al., 2015). In the present study, CgMsb2 alone or in cooperation with CgSho1 promoted the phosphorylation of CgMk1. However, determining whether CgSho1 acts as an upstream sensor of CgMk1 requires physical interaction assays between CgSho1 and the MAPK CgMk1, for example coimmunoprecipitation assays. Collectively, the results of these studies indicate a conserved function of Msb2 as an upstream sensor of the MAPK Pmk1 in various fungi.
3.2. Functional cooperation between CgMsb2 and CgSho1
In C. gloeosporioides, after adhesion of conidia to the cuticle of leaves, germ tubes germinate and subsequently form appressoria. During this process, various host surface signals are perceived by the cell membrane sensors of the pathogen and lead to the activation of downstream pathways, including MAPK pathways. It is important to better understand the molecular mechanisms associated with signal perception, the identification of sensors, their functions, and the clarification of the complementary relationship. In the present study, our results indicated that CgMsb2 and CgSho1 have multiple overlapping or cooperative functions in C. gloeosporioides. The evidence for a cooperative relationship between sensors is ubiquitous in fungi. In M. oryzae, MoMsb2 and MoSho1 have overlapping functions in the perception of various host surface signals, with MoMsb2 playing a major role in sensing surface hydrophobicity and cutin monomers, while MoSho1 has a more important role in wax recognition (Liu et al., 2011). In U. maydis, UmSho1 cooperates with the mucin UmMsb2 to regulate invasive growth and plant infection, and UmSho1 and UmMsb2 cooperatively regulate appressorium development (Lanver et al., 2010). In M. oryzae, F. graminearum, S. cerevisiae, and B. cinerea, there are also cooperative interactions between Sho1 and Sln1 homologues (Gu et al., 2014; O'Rourke et al., 2002; Ren et al., 2019; Zhang et al., 2010). Notably, in these studies, similar to the results observed for CgMsb2 and CgSho1, a mutant with a double deletion of Msb2 and Sho1 typically retained reduced abilities for surface signal recognition, appressorium formation, pathogenicity, and activation of CgMk1. Therefore, there must be other sensors required for host signal perception and an upstream sensor of CgMk1. For example, Wang et al. identified another mucin protein, MoCbp1, in M. oryzae, and a ΔMoMsb2Cbp1 mutant fully lost the ability to form appressoria on artificial hydrophobic surfaces and rarely formed appressoria on host surfaces. A MoMsb2Sho1Cbp1 triple mutant produced no appressoria on the host surface, and Pmk1 activation in the ΔMoMsb2Cbp1 mutant was fully blocked (Wang et al., 2015). These results indicated that host surface signal recognition involves several sensors on the fungal cell surface and that sensors typically cooperate with each other for specific functions or for the activation of downstream pathways. Thus, the identification and characterization of other sensors in C. gloeosporioides is needed.
In the present study, we showed that CgMsb2 and CgSho1 play a cooperative role in the recognition of various signals, and CgMsb2 plays a major role in the recognition of hydrophobicity, leaf surface waxes, bee waxes, and paraffin waxes. On the plant surface, surface hydrophobicity and waxes are the primary physical and chemical cues, respectively, that are recognized by pathogens. Cuticular waxes are mainly composed of primary and secondary alcohols, ketones, fatty acids, and aldehydes (Jetter et al., 2000; Kunst & Samuels, 2003). Intriguingly, CgMsb2 acts as the receptor of both hydrophobicity and leaf surface waxes in C. gloeosporioides. Our results showed that CgMsb2 is responsible for the recognition of both a primary C28 alcohol and a C31 alkane. In grass powdery mildew (Blumeria graminis), it has been reported that n‐hexacosanal (a C26 aldehyde) and n‐triacontanal (a C30 aldehyde), chemical constituents of the epicuticular wax layer of barley, are capable of effectively stimulating appressorium formation (Hansjakob et al., 2010). In M. oryzae, which shares a similar infection strategy with C. gloeosporioides, MoMsb2 is dispensable for the recognition of primary alcohols, which are the major components of surface waxes in grasses, and the formation of appressorium in MoMsb2 deletion mutants is fully rescued in the presence of leaf surface waxes (Liu et al., 2011). Hydrophobicity and waxes are two distinct signals of the host surface, and recognition of the major components of surface waxes is a prerequisite for the recognition of surface waxes in C. gloeosporioides and M. oryzae.
3.3. CgPth11 is dispensable for appressorium formation and pathogenicity
In various phytopathogens, including those that do or do not form appressoria, a great deal of research has focused on fungal sensor genes. The first identified sensor in a plant‐pathogenic fungus was MoPth11. In M. oryzae, Pth11 plays an important role in pathogenicity and appressorium formation in response to both cutin monomers and hydrophobic surface cues (DeZwaan et al., 1999), with further studies revealing that the CFEM domain of Pth11 is required for proper development of appressoria and appressoria‐like structures and pathogenicity (Kou et al., 2017). The subclass of Pth11 containing the CFEM domain is highly represented in M. oryzae (Kulkarni et al., 2005). Li et al. reported that MoEnd3 regulates the internalization of MoSho1 and MoPth11 by mediating endocytosis (Li et al., 2017). As the first identified sensor, and due to the significant role of MoPth11 in M. oryzae, few studies have characterized Pth11 in other pathogenic fungi, with the exception of BcBtp1 in B. cinerea. However, the deletion of CgPth11 in the WT, ΔCgSho1, and ΔCgMsb2 strains resulted in essentially no defects in appressorium formation and virulence. Furthermore, previous evidence showed that exogenous cAMP treatment can fully rescue the appressorium formation defect of a ΔMoPth11 mutant, suggesting that MoPth11 functions upstream of the cAMP pathway (Kou et al., 2017). In the present study, exogenous cAMP treatment also failed to restore the appressorium formation defects of the ΔCgPth11 mutant at the early stage (5 hpi) (Figure 1e), indicating that the function of CgPth11 is not conserved compared with that of MoPth11. In M. oryzae and C. gloeosporioides, a high level of endogenous ROS accumulates during appressorium differentiation (Egan et al., 2007; Wang et al., 2021b). The ROS intermediates have been suggested to be superoxides. Therefore, antioxidants such as the oxygen radical scavenger ascorbic acid and the superoxide dismutase MnTMPyP affect appressorium formation in M. oryzae efficiently. Appressoria formed in the presence of antioxidants are defective in their morphology (Egan et al., 2007). Intriguingly, constitutive expression of the CFEM domain of MoPth11 also results in defects in appressorial morphology, and antioxidants GSH, N‐acetyl‐l‐cysteine, and ascorbic acid induce appressorium formation in ΔMoPth11 efficiently, indicating that MoPth11 is required for appressorium formation under exogenous antioxidant treatment in M. oryzae (Kou et al., 2017). However, exogenous antioxidant treatment resulted in defects in appressorial morphology, but failed to induce appressorium formation in ΔCgPth11 (Figure 6e), indicating that the role of CgPth11 in ROS‐mediated appressorium formation is not conserved compared with that of MoPth11.
In B. cinerea, the first cloned putative receptor was the GPCR BcBtp1, which shows similarity with MoPth11. However, the deletion of BcBtp1 does not result in a significant defect in pathogenicity, and the sequence similarity to BcBtp1 and MoPth11 is restricted to the N‐terminal TM regions (Gronover et al., 2005). These results indicate that the functional difference between BcBtp1 and MoPth11 may result from sequence diversity. In M. oryzae, the CFEM domain of Pth11 is crucial to the surface‐sensing function of MoPth11, and the CFEM domain of C. albicans Csa1 fails to complement the loss of the CFEM function in MoPth11, indicating that the CFEM domain in MoPth11 serves a specific function (Kou et al., 2017). Interestingly, the CFEM domains from different proteins show remarkable sequence diversity (Kulkarni et al., 2003). Sequence alignment of the CFEM domains from CgPth11 and MoPth11 also showed low (25.8%) sequence identity (Table 1), leading us to presume that the differences in function between CgPth11 and MoPth11 may be caused by the sequence diversity of the CFEM domain. Jiang et al. reported a functional analysis of 105 GPCRs from F. graminearum. They identified and analysed 12 CFEM domain‐containing GPCRs from 105 GPCRs. However, deletion of each CFEM domain‐containing GPCR causes no obvious defect in growth, colony morphology, and pathogenicity (Jiang et al., 2019). In F. graminearum, there is an expanded subfamily of 22 pathogenicity‐related GPCRs. Among them, Giv1 is required for infection cushion formation, and Giv2 and Giv3 are required for infectious growth. Comparative analysis showed that most GPCRs in this subfamily are unique in F. graminearum (Jiang et al., 2019). Collectively, these results indicate that the functions of GPCRs or CFEM domain‐containing GPCRs are probably not conserved among different species.
Based upon the current and previously published data, this study revealed novel functions of CgMsb2 in the recognition of leaf surface waxes and illustrated the significant role of CgMsb2 in the recognition of both primary alcohol and alkanes, which are important components of leaf surface waxes. We also determined the novel function of CgSho1 in invasive growth. The CFEM domain‐containing GPCR CgPth11 was shown to play only a minor role in the early stage of appressorium formation, which is significantly different from the functions of Pth11 in the rice blast fungus. Further study of other putative sensors such as Cbp1 and Sln1 is needed to elucidate the cooperative relationships that occur during the early stage of signal perception in C. gloeosporioides.
4. EXPERIMENTAL PROCEDURES
4.1. Fungal strains and culture conditions
The C. gloeosporioides strain CFCC80308, which served as the WT strain throughout this work, was isolated from Populus × beijingensis in Beijing, China. Strains were grown on PDA for routine maintenance. TB3 liquid and solid media were used to prepare fresh mycelia for transformation and the selection of transformants, respectively. The liquid complete medium (CM) was used to culture mycelia for DNA extraction. Strains were also grown on Cove's glucose MM (GMM) with 1% (wt/vol) glucose as the sole carbon source, containing 0.52 g/L KCl, 0.52 g/L MgSO4.7H2O, 1.52 g/L KH2PO4 (Fernandez & Wilson, 2012). Various nitrogen sources were added to GMM as sole nitrogen sources at a final concentration of 10 mM for the test of nitrogen‐limiting conditions.
4.2. Isolation and phylogenetic analysis of CgMsb2, CgSho1, and CgPth11
The sequences of CgMsb2, CgSho1, and CgPth11 were obtained from the genome database of C. gloeosporioides (http://genome.jgi.doe.gov/Gloci1/Gloci1.home.html) after the use of the BlastP tool. CgMsb2, CgSho1, and CgPth11 amino acid sequences were aligned with other homologues using ClustalX v. 2.1. Phylogenetic analysis of Msb2, Sho1, and Pth11 homologues was conducted using MEGA v. 6.0 as previously described (Tamura et al., 2013). The domains in CgMsb2, CgSho1, and CgPth11 were predicted using the InterProScan tool (https://www.ebi.ac.uk/interpro).
4.3. Construction of single and double mutants and complementation strains
Gene functional analysis was achieved by the split marker method described in (Wilson et al., 2010). To generate the single deletion of CgMsb2, the upstream (c.1.5 kb) and downstream (c.1.5 kb) flanking sequences of CgMsb2 were amplified with primer pairs CgMsb2‐5Ffor/CgMsb2‐5Frev and CgMsb2‐3Ffor/CgMsb2‐3Frev, respectively. The hygromycin B resistance cassette was amplified with primer pair hygromycinfor and hygromycinrev, which include approximately 20 bp that overlap (M13F or M13R) with the 5′ and 3′ flanking sequences, respectively. The resulting upstream and downstream fragments were fused with two‐thirds of the hygromycin B resistance cassette amplified using primers CgMsb2‐5Ffor/HY‐R and YG‐F/CgMsb2‐3Frev, respectively, by using overlap PCR. The two fusion fragments were directly transformed into the protoplasts of the WT strain using the PEG‐mediated transformation, and the transformants were selected on TB3 medium (3 g/L yeast extract, 3 g/L casamino acids, 20% sucrose, and 0.7% agar) with 300 μg/ml hygromycin B. The putative CgMsb2 deletion mutants were screened by PCR assays with the primer pairs External‐CgMsb2for/External‐CgMsb2rev and Internal‐CgMsb2for/Internal‐CgMsb2rev, respectively. Targeted mutants of CgSho1 and CgPth11 were generated using a similar approach.
To generate the ΔCgMsb2Sho1 double mutant in the ΔCgMsb2 background, the sulphonylurea resistance‐conferring gene (Sur) carried by PCB1523 was applied (Sweigard et al., 1997; Wilson et al., 2010). Upstream and downstream fragments of CgSho1 were fused with two‐thirds of the Sur gene using the split marker method, the CgSho1 replacement construct was transformed into the CgMsb2 deletion mutants using PEG‐mediated protoplast transformation, and the putative CgMsb2Sho1 double mutants with both sulphonylurea and hygromycin B resistance were screened by PCR analysis. ΔCgMsb2Pth11 and ΔCgSho1Pth11 double mutants were generated using a similar approach (Figure S5). For the complementation assay, the phleomycin resistance cassette was used as a selective marker and the entire coding sequence of CgMsb2, CgSho1, or CgPth11 with c.1.5‐kb upstream region was transformed into each single deletion mutant. The complementation mutant was selected by primer pairs that bind to the coding sequence of target gene, and the complementation mutants were referred to as ΔCgMsb2/ MSB2, ΔCgSho1/ SHO1, and ΔCgPth11/ PTH11, respectively. All primers used in this study are listed in Table 3.
TABLE 3.
Primers used in this study
| Primer name | Sequence | Use in this study |
|---|---|---|
| CgMsb2‐5Ffor (1F) | TCAACTGAGGCGGTTATTCC | 5F flanking sequence |
| CgMsb2‐5Frev (2R) | GAGCGACTCGACGAGAAATC | |
| CgMsb2‐3Ffor (2F) | TACGCTTCGATACCCTTCGG | 3F flanking sequence |
| CgMsb2‐3Frev (3R) | CTTGGAGCGACAAGTTGGGA | |
| External‐CgMsb2for | TTCCTAGCGACCCTGTTGTT | External sequence used for validation of mutant |
| External‐CgMsb2rev | GCCCGGTGAGTAGAATGGTA | |
| Internal‐CgMsb2for | GCTATTGGAATCGGTGCTGT | Internal sequence used for validation of mutant |
| Internal‐CgMsb2rev | GGCGACTCCACCGTAGTTAG | |
| CgMsb2‐Compfor | TCAACTGAGGCGGTTATTCC | Complementary sequence |
| CgMsb2‐Comprev | GCCCGGTGAGTAGAATGGTA | |
| Hygromycinfor (HYG‐F) | CGCCAGGGTTTTCCCAGTCACGAC | Hygromycin cassette contains the region of M13F and M13R |
| Hygromycinrev (HYG‐R) | AGCGGATAACAATTTCACACAGGA | |
| YG‐F | GATGTAGGAGGGCGTGGATATGTCCT | The second/third portion of the hygromycin cassette |
| HY‐R | GATGTAGGAGGGCGTGGATATGTCCT | |
| SUR‐5′‐M13F | CGCCAGGGTTTTCCCAGTCACGACGTCGACGTGCCAACGCCACAG | Sur cassette contains the region of M13F and M13R |
| SUR‐3′‐M13R | AGCGGATAACAATTTCACACAGGAGTCGACGTGAGAGCATGCAAT | |
| SU‐SPLIT | CCAAGCATGTGCAGTGCCTTC | The second/third portion of the Sur cassette |
| UR‐SPLIT | GGAGGCCGACGTCATAGGCATC | |
| CgSho1‐5Ffor (1F) | AGTCTCCCACCTTCACTGTG | 5F flanking sequence |
| CgSho1‐5Frev (2R) | GTAGAATTGGCGAGGCAGTG | |
| CgSho1‐3Ffor (2F) | GATGGTTTTCGCGCTCAATG | 3F flanking sequence |
| CgSho1‐3Frev (3R) | GCGCAACATTCCTCACTCAA | |
| External‐CgSho1for | CACCACATTGCTGCTGTCTT | External sequence used for validation of mutant |
| External‐CgSho1rev | CTCTTGATGTTGCGAGGCTC | |
| Internal‐CgSho1for | TGGATCTTCTACTTCGGCTCA | Internal sequence used for validation of mutant |
| Internal‐CgSho1rev | GCGCCGAAGTGTACATTTGA | |
| CgSho1‐Compfor | AGTCTCCCACCTTCACTGTG | Complementary sequence |
| CgSho1‐Comprev | TAACAGGATCAGGTAGTTGC | |
| CgPth11‐5Ffor (1F) | TGGTCAAGTCTGTGGGTTGT | 5F flanking sequence |
| CgPth11‐5Frev (2R) | CAAGAAAAGCCGCTGACCAT | |
| CgPth11‐3Ffor (2F) | ATATCCCTGGCTTTGGTCGG | 3F flanking sequence |
| CgPth11‐3Frev (3R) | CAAGCCATGCCTTCCAGATC | |
| External‐CgPth11for | ATTCAACCGCCTCTTCTGGA | External sequence used for validation of mutant |
| External‐CgPth11rev | TATGTCACTCGATCTGGGGC | |
| Internal‐CgPth11 for | TTCTCAGCAACCTACGACGT | Internal sequence used for validation of mutant |
| Internal‐CgPth11rev | AAATCGATCGTCTCAGCGGA | |
| CgPth11‐Compfor | ATTCAACCGCCTCTTCTGGA | Complementary sequence |
| CgPth11‐Comprev | CCGACCAAAGCCAGGGATAT | |
| CgSte7S218E T222E for | AGAACTTATCAACGAGGTTGCAGACGAATTCGTTGGAACAT | Constitutively active allele of Ste7 |
| CgSte7S218E T222E rev | ATGTTCCAACGAATTCGTCTGCAACCTCGTTGATAAGTTCT |
4.4. Appressorium formation and penetration assays
Conidia were collected from colonies cultured on PDA at 5 dpi, and conidial suspensions of each strain were adjusted to 105 conidia/ml using distilled water. The hydrophobic side of a GelBond membrane (Lonza)/onion epidermal cell was coated on glass slides, 30 μl conidial suspension from each strain was inoculated on the GelBond membrane/onion epidermal cell, and glass slides were fixed onto filter paper and placed into 94‐mm Petri dishes containing 8 ml sterile water at 25 °C. The conidial germination and appressorium development at 12 hpi on GelBond membranes and penetration (formation and expansion of infection hyphae) of onion epidermal cells were monitored at 12 and 24 hpi using a light microscope. The ratio of appressorium formation was calculated based on the conidia that germinated. The penetration rate was calculated based on the conidia that formed an appressorium. This experiment was performed with three biological replicates and four technical replicates for each treatment. At least 200 conidia/appressoria from each strain were measured to calculate the appressorium formation and penetration rates, respectively.
4.5. Cellophane membrane penetration assays
To determine the penetration ability of each strain, cellophane membranes were cut into 3 × 3 cm squares and autoclaved at 120 °C for 20 min. PDA plates were used as culture medium; sterile cellophane membranes were placed in the centre of the medium, and hyphal blocks of each strain were inoculated on the cellophane membranes for 3 days at 25 °C. The colony on the membrane was then removed, and new colonies that developed on PDA were imaged and evaluated 2 days after removal. All penetration experiments were performed three times.
4.6. Pathogenicity tests
Conidial suspensions of the WT, mutant, and complementation strains were prepared using sterilized deionized water (2 × 105 conidia/ml). Thirty microlitres of conidial suspension of each strain was inoculated on the detached leaves of Populus × beijingensis. Leaves were all detached from 2‐week‐old water‐cultivated poplar branches. Inoculated leaves were fixed on filter paper and placed into a 94‐mm Petri dish containing 8 ml sterile water. Symptoms were photographed at 5–9 dpi. All pathogenicity experiments were performed three times.
4.7. Western blot analysis and the construction of the dominant active allele of Ste7
Protein isolation and CgMk1 MAPK activation assays were performed following the protocol in Zhang et al. (2018a). Vegetative hyphae were harvested from 2‐day‐old cultures in 5× YEG liquid medium (5 g yeast extract and 10 g glucose in 1 L double distilled water) grown at 150 rpm at 25 °C. About 200 mg of mycelia was ground into powder in liquid nitrogen and resuspended in 1 ml of extraction buffer (10 mM Tris‐HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Triton X‐100) with freshly added 1 mM phenylmethylsulfonyl fluoride (PMSF) and 10 μl protease inhibitor cocktail (Sigma). Total proteins were separated by 12% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to nitrocellulose membranes for western blot analysis. Phosphorylation and protein levels of CgMk1 MAPKs were detected with the phopho‐p44/42 and p44/42 MAPK antibodies, respectively (Cell Signaling Technology).
A putative constitutively active Ste7 allele was constructed by changing Ser‐218 to Glu and Thr‐222 to Glu using the fast site‐directed mutagenesis kit (TianGen) (Cowley et al., 1994; Zhao et al., 2005) following the manufacturer's instructions. The resulting vector was sequenced to confirm the exclusive mutagenesis of S218E and T222E. Primers used in this experiment are listed in Table 3.
4.8. CFW staining and microscopic observation
To monitor the germination process of C. gloeosporioides on poplar leaves, conidial suspensions of each strain were collected and adjusted to 2 × 105 conidia/ml using distilled water. Thirty microlitres of conidial suspension of each strain was inoculated on the detached leaves of Populus × beijingensis. Inoculated leaves were fixed on filter paper and placed into a 94‐mm Petri dish containing 8 ml sterile water. Each inoculated area was cut when the melanized appressorium formed at 24 hpi, and samples were stained with 1 μg/ml CFW for 1 min in the dark. Appressorium formation was observed using fluorescence microscopy (DM2500; Leica). This experiment was repeated three times, with four samples from each strain inoculated and analysed in each replicate.
4.9. Wax extraction and treatments
Nearly all the existing data on the chemical composition of plant waxes are based on solvent‐extracted waxes (Koch et al., 2010). In the present study, poplar leaves from a 2‐week‐old water‐cultivated poplar branch were dipped in hexane for 20 s, and epicuticular wax extracts dissolved in hexane were dried under a gentle flow of nitrogen (Chen et al., 2003; Hansjakob et al., 2010). Wax extraction experiments were performed with three biological replicates and four technical replicates for each treatment. For wax‐coated glass slides, 10 mg of the poplar leaf surface wax was dissolved in 3 ml chloroform. Fifty microlitres of crude wax extract was then dropped on microscope glass slides. Bee wax and paraffin wax were directly coated onto the glass surface. Thirty microlitres of conidial suspension (105 conidia/ml) was placed on the above wax‐coated surfaces and appressorium formation was assayed. Wax treatment was performed with three biological replicates and four technical replicates for each treatment. The C28 (1‐octacosanol, C28H58O) primary alcohol and C31 (hentricacontane, C31H64) alkane (Sigma) were dissolved to 4 mg/ml in chloroform.
4.10. H2O2 stress response and ROS staining assays
PDA plates were supplemented with H2O2 at a final concentration of 5 mM or 10 mM. A hyphal block of each strain was then inoculated on the centre of the plates, and colony size was measured at 4 dpi. This experiment was repeated three times, and four samples from each strain were inoculated and analysed in each replicate. Conidia of the WT, mutant, and complementation strains were resuspended in sterilized deionized water (105 conidia/ml). Conidial suspensions (30 µl) of each strain were inoculated on the hydrophobic onion epidermal cell surface. DAB was used to detect the accumulation of ROS. This experiment was repeated three times, and four samples from each strain were inoculated and analysed in each replicate. At least 15 appressoria from each strain were measured using ImageJ in each replicate. In the presence of ROS, DAB is converted to dark brown polymers. ROS were stained with 1 mg/ml DAB (30 µl) for 12 hr in darkness. The intensities of dark brown polymers were quantified with ImageJ.
4.11. Statistical analysis
Statistical analysis for phenotypic characterization was performed with data from at least three independent biological replicates. To determine the significance of differences, one‐way analysis of variance was conducted using SPSS v. 22.0 software followed by Tukey's post hoc test (p < .05).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
X.L.W. and C.M.T. designed the research. X.L.W. and L.D.X. performed the experiments. X.L.W. analysed the data and wrote the manuscript. All authors read and approved the manuscript.
Supporting information
FIGURE S1 Sequence analysis of CgMsb2 with homologues in other fungi. (a) Whole amino acid sequences of CgMsb2 (Protein ID: 1461327) and Msb2 homologues were aligned using ClustalX v. 2.1. Red frames indicate the signal peptide, noncytoplasmic domain, transmembrane (TM) domain, and cytoplasmic domains. (b) Phylogenetic tree of CgMsb2 and its homologues in other fungi. The phylogenetic tree was constructed using MEGA v. 6.0 with full‐length protein sequences and neighbour‐joining with 1,000 bootstrap replicates
FIGURE S2 Sequence analysis of CgSho1 with homologues in other fungi. (a) Whole amino acid sequences of CgSho1 (KAF 3805345.1) and Sho1 homologues were aligned using ClustalX v. 2.1. Red frames indicate the TM and SH3 domains. (b) Phylogenetic tree of CgSho1 and its homologues in other fungi. The phylogenetic tree was constructed using MEGA v. 6.0 with full‐length protein sequences and neighbour‐joining with 1,000 bootstrap replicates
FIGURE S3 Sequence analysis of CgPth11 with homologues in other fungi. (a) Whole amino acid sequences of CgPth11 (KAF 3797122.1) and Pth11 homologues were aligned using ClustalX v. 2.1. Red frames indicate the signal peptide, the CFEM domain, and seven transmembrane domains. (b) Phylogenetic tree of CgPth11 and its homologues in other fungi. The phylogenetic tree was constructed using MEGA v. 6.0 with full‐length protein sequences and neighbour‐joining with 1,000 bootstrap replicates
FIGURE S4 Schematic drawing of CgMsb2, CgSho1, and CgPth11 proteins. Full‐length amino acid sequences of CgMsb2, CgSho1, and CgPth11 were analysed using the InterproScan tool (https://www.ebi.ac.uk/interpro). Domains including the signal peptide, noncytoplasmic domain, SH3 domain, CFEM domain, transmembrane domain, and cytoplasmic domain were predicted and are marked in different colours. aa, amino acids
FIGURE S5 Generation of the deletion mutants in Colletotrichum gloeosporioides. The split marker method was used for single and double gene replacement. (a) Diagrammatic representation of the split marker targeted replacement strategy including location of primers. 5′: the upstream flank sequence of the target gene. 3′: the downstream flank sequence of the target gene. (b) In ΔCgMsb2 and ΔCgSho1, a hygromycin cassette (1.4 kb) replaced the fragment of CgMsb2 (3.0 kb) and CgSho1 (1.0 kb), respectively. In ΔCgPth11, a Sur cassette (2.2 kb) replaced the fragment of CgPth11 (1.5 kb). (c) Complementation of CgMsb2, CgSho1, and CgPth11 was achieved by reintroduction of the full coding sequence of each gene and confirmed using PCR. (d) A Sur cassette replaced the fragment of CgPth11 in ΔCgSho1 and ΔCgMsb2 to generate ΔCgSho1Pth11 and ΔCgMsb2Pth11, respectively. The Sur cassette replaced the fragment of CgSho1 in ΔCgMsb2 to generate the ΔCgMsb2Sho1 mutant
ACKNOWLEDGEMENTS
The research was supported by the National Key Research and Development Program (2017YFD0600100) and the National Natural Science Foundation of China (32071767).
Wang, X. , Lu, D. & Tian, C. (2021) Mucin Msb2 cooperates with the transmembrane protein Sho1 in various plant surface signal sensing and pathogenic processes in the poplar anthracnose fungus Colletotrichum gloeosporioides . Molecular Plant Pathology, 22, 1553–1573. 10.1111/mpp.13126
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Apostol, I. , Heinstein, P.F. & Low, P.S. (1989) Rapid stimulation of an oxidative burst during elicitation of cultured plant cells. Plant Physiology, 90, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, E.A. (1982) Chemistry and morphology of plant epicuticular waxes. The Plant Cuticle, 139–166. [Google Scholar]
- Carver, T. & Gurr, S. (2007) Filamentous fungi on plant surfaces. In: Riederer, M. & Müller, C. (Eds.) Biology of the plant cuticle. Annual Plant Reviews, Vol. 23. Oxford, UK: Blackwell Publishing Ltd, pp. 368–397. [Google Scholar]
- Chen, R.E. & Thorner, J. (2007) Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae . Biochimica et Biophysica Acta, 1773, 1311–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Goodwin, S. , Boroff, V. , Liu, X. & Jenks, M. (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. The Plant Cell, 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley, S. , Paterson, H. , Kemp, P. & Marshall, C. (1994) Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell, 77, 841–852. [DOI] [PubMed] [Google Scholar]
- Cwiertnia, E. (2015) Improved beeswax analysis to determine its origin: critical literature review and new methodological approach. Brussels, Belgium: Archives et Bibliothèques de Belgique. [Google Scholar]
- Dean, R. , Kan, J. , Pretorius, Z. , Hammond‐Kosack, K. , Pietro, A. , Spanu, P. et al. (2012) The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezwaan, T.M. , Carroll, A.M. , Valent, B. & Sweigard, J.A. (1999) Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. The Plant Cell, 11, 2013–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, M.J. , Wang, Z.Y. , Jones, M.A. , Smirnoff, N. & Talbot, N.J. (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proceedings of the National Academy of Sciences of the United States of America, 104, 11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J. & Wilson, R. (2012) Why no feeding frenzy? Mechanisms of nutrient acquisition and utilization during infection by the rice blast fungus Magnaporthe oryzae . Molecular Plant‐Microbe Interactions, 25, 1286–1293. [DOI] [PubMed] [Google Scholar]
- Gilbert, R. , Johnson, A. & Dean, R. (1996) Chemical signal responsible for appressorium formation in the rice blast fungus Magnaporthe grisea . Physiological and Molecular Plant Pathology, 48, 335–346. [Google Scholar]
- Gomes, S. , Prieto, P. , Martins‐Lopes, P. , Carvalho, T. , Martin, A. & Guedes‐Pinto, H. (2009) Development of Colletotrichum acutatum on tolerant and susceptible Olea europaea L. cultivars: a microscopic analysis. Mycopathologia, 168, 203–211. [DOI] [PubMed] [Google Scholar]
- Gronover, C.S. , Schumacher, J. , Hantsch, P. & Tudzynski, B. (2005) A novel seven‐helix transmembrane protein BTP1 of Botrytis cinerea controls the expression of GST‐encoding genes, but is not essential for pathogenicity. Molecular Plant Pathology, 6, 243–256. [DOI] [PubMed] [Google Scholar]
- Gu, Q. , Chen, Y. , Liu, Y. , Zhang, C. & Ma, Z. (2014) The transmembrane protein FgSho1 regulates fungal development and pathogenicity via the MAPK module Ste50‐Ste11‐Ste7 in Fusarium graminearum . New Phytologist, 206, 315–328. [DOI] [PubMed] [Google Scholar]
- Hansjakob, A. , Bischof, S. , Bringmann, G. , Riederer, M. & Hildebrandt, U. (2010) Very‐long‐chain aldehydes promote in vitro prepenetration processes of Blumeria graminis in a dose‐ and chain length‐dependent manner. New Phytologist, 188, 1039–1054. [DOI] [PubMed] [Google Scholar]
- He, P. , Wang, Y. , Wang, X. , Zhang, X. & Tian, C. (2017) The mitogen‐activated protein kinase CgMK1 governs appressorium formation, melanin synthesis, and plant infection of Colletotrichum gloeosporioides . Frontiers in Microbiology, 8, 2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway, P.J. (1994) Plant cuticles: physicochemical characteristics and biosynthesis. NATO ASI Series: Series G: Ecological Sciences, 36, 1–13. [Google Scholar]
- Howard, R.J. , Ferrari, M.A. , Roach, D.H. & Money, N.P. (1992) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proceedings of the National Academy of Sciences of the United States of America, 88, 11281–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, C. & Kolattukudy, P. (1995) Isolation and characterization of genes expressed uniquely during appressorium formation by Colletotrichum gloeosporioides conidia induced by the host surface wax. Molecular & General Genetics, 247, 282–294. [DOI] [PubMed] [Google Scholar]
- Jayawardena, R. , Hyde, K. , Damm, U. , Cai, L. , Liu, M. , Li, X.H. et al. (2016) Notes on currently accepted species of Colletotrichum . Mycosphere, 7, 1192–1260. [Google Scholar]
- Jeffree, C. (2007) The fine structure of the plant cuticle. In: Riederer, M. & Muller, C. (Eds.) Biology of the plant cuticle. Annual Plant Reviews, Vol. 23. Oxford, UK: Blackwell Publishing Ltd, pp. 11–125. [Google Scholar]
- Jetter, R. , Kunst, L. & Samuels, A. (2007) Composition of plant cuticular waxes. In: Riederer, M. & Muller, C. (Eds.) Biology of the plant cuticle. Annual Plant Reviews, Vol. 23. Oxford, UK: Blackwell Publishing Ltd, pp. 145–181. [Google Scholar]
- Jetter, R. , Schffer, S. & Riederer, M. (2000) Leaf cuticular waxes are arranged in chemically and mechanically distinct layers: evidence from Prunus laurocerasus L. Plant, Cell & Environment, 23, 619–628. [Google Scholar]
- Jiang, C. , Shulin, C. , Wang, Z. , Xu, H. , Liang, J. , Liu, H. et al. (2019) An expanded subfamily of G‐protein‐coupled receptor genes in Fusarium graminearum required for wheat infection. Nature Microbiology, 4, 1582–1591. [DOI] [PubMed] [Google Scholar]
- Jiang, C. , Zhang, X. , Liu, H. & Xu, J.‐R. (2018) Mitogen‐activated protein kinase signaling in plant pathogenic fungi. PLoS Pathogens, 14, e1006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, K. , Bhushan, B. & Barthlott, W. (2009) Multifunctional surface structures of plants: an inspiration for biomimetics. Progress in Materials Science, 54, 137–178. [Google Scholar]
- Koch, K. , Bhushan, B. & Barthlott, W. (2010) Multifunctional plant surfaces and smart materials. Berlin, Germany: Springer. [Google Scholar]
- Kou, Y. , Tan, Y.H. , Ramanujam, R. & Naqvi, N.I. (2017) Structure‐function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytologist, 214, 330–342. [DOI] [PubMed] [Google Scholar]
- Kulkarni, R. , Kelkar, H. & Dean, R. (2003) An eight‐cysteine‐containing CFEM domain unique to a group of fungal membrane proteins. Trends in Biochemical Sciences, 28, 118–121. [DOI] [PubMed] [Google Scholar]
- Kulkarni, R.D. , Thon, M.R. , Pan, H. & Dean, R.A. (2005) Novel G‐protein‐coupled receptor‐like proteins in the plant pathogenic fungus Magnaporthe grisea . Genome Biology, 6, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto, C.A. (2008) Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nature Reviews Microbiology, 6, 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst, L. & Samuels, A. (2003) Biosynthesis and secretion of plant cuticular wax. Progress in Lipid Research, 42, 51–80. [DOI] [PubMed] [Google Scholar]
- Lanver, D. , Berndt, P. , Tollot, M. , Naik, V. , Vranes, M. , Warmann, T. et al. (2014) Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathogens, 10, e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver, D. , Mendoza‐Mendoza, A. , Brachmann, A. & Kahmann, R. (2010) Sho1 and Msb2‐related proteins regulate appressorium development in the smut fungus Ustilago maydis . The Plant Cell, 22, 2085–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroch, M. , Kleber, A. , Silva Moreno, E. , Coenen, T. , Koppenhfer, D. , Shmaryahu, A. et al. (2013) Transcriptome profiling of Botrytis cinerea conidial germination reveals upregulation of infection‐related genes during the prepenetration stage. Eukaryotic Cell, 12, 614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroch, M. , Mueller, N. , Hinsenkamp, I. & Hahn, M. (2015) The signalling mucin Msb2 regulates surface sensing and host penetration via BMP1 MAP kinase signalling in Botrytis cinerea: B. cinerea Msb2 involved in MAPK signalling. Molecular Plant Pathology, 16, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.J. , Zhou, L. , Yin, C.M. , Zhang, D.D. , Klosterman, S. , Wang, B.L. et al. (2019) The Verticillium dahliae Sho1‐MAPK pathway regulated melanin biosynthesis is required for cotton infection. Environmental Microbiology, 21, 4852–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Gao, C. , Li, L. , Liu, M. , Yin, Z. , Zhang, H. et al. (2017) MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae . PLoS Pathogens, 13, e1006449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Liang, Y.‐M. & Tian, C. (2012) Characterization of the causal agent of poplar anthracnose occurring in the Beijing region. Mycotaxon, 120, 277–286. [Google Scholar]
- Liu, W. , Zhou, X. , Li, G. , Li, L. , Kong, L. , Wang, C. et al. (2011) Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathogens, 7, e1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Qiao, J. , Liu, W. , Wan, Z. , Wang, X. , Calderone, R. et al. (2008) The Sho1 sensor regulates growth, morphology, and oxidant adaptation in Aspergillus fumigatus but is not essential for development of invasive pulmonary aspergillosis. Infection and Immunity, 76, 1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza‐Mendoza, A. , Berndt, P. , Djamei, A. , Weise, C. , Linne, U. , Marahiel, M. et al. (2009) Physical‐chemical plant‐derived signals induce differentiation in Ustilago maydis . Molecular Microbiology, 71, 895–911. [DOI] [PubMed] [Google Scholar]
- Money, N. & Howard, R. (1996) Confirmation of a link between fungal pigmentation, turgor pressure, and pathogenicity using a new method of turgor measurement. Fungal Genetics and Biology, 20, 217–227. [Google Scholar]
- O'Rourke, S. , Herskowitz, I. & O'Shea, E. (2002) Yeast go the whole HOG for the hyperosmotic response. Trends in Genetics, 18, 405–412. [DOI] [PubMed] [Google Scholar]
- Park, G. , Bruno, K. , Staiger, C. , Talbot, N. & Xu, J.‐R. (2004) Independent genetic mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus Magnaporthe grisea . Molecular Microbiology, 53, 1695–1707. [DOI] [PubMed] [Google Scholar]
- Pereira, I. , Abreu, M. , Alves, E. & Ferreira, J.B. (2009) Histopathological studies of the interaction Colletotrichum gloeosporioides–coffee trees. Bragantia, 68, 117–123. [Google Scholar]
- Perez‐Nadales, E. & di Pietro, A. (2011) The membrane mucin Msb2 regulates invasive growth and plant infection in Fusarium oxysporum . The Plant Cell, 23, 1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Nadales, E. & di Pietro, A. (2015) The transmembrane protein Sho1 cooperates with the mucin Msb2 to regulate invasive growth and plant infection in Fusarium oxysporum . Molecular Plant Pathology, 16, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X. , Zhou, S. , Shang, X. & Wang, X. (2016) VdSho1 regulates growth, oxidant adaptation and virulence in Verticillium dahliae . Journal of Phytopathology, 164.1064–1074. [Google Scholar]
- Ren, W. , Liu, N. , Yang, Y. , Yang, Q. , Chen, C. & Gao, Q. (2019) The sensor proteins BcSho1 and BcSln1 are involved in, though not essential to, vegetative differentiation, pathogenicity and osmotic stress tolerance in Botrytis cinerea . Frontiers in Microbiology, 10, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom, N.E. , Cottier, F. , Ernst, J. & Pla, J. (2009) Msb2 signaling mucin controls activation of Cek1 mitogen‐activated protein kinase in Candida albicans . Eukaryotic Cell, 8, 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom, N.E. , Nombela, C. & Pla, J. (2006) The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans . Molecular and Cellular Biology, 25, 10611–10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K. (2000) The Saccharomyces cerevisiae SLN1P‐SSK1P two‐component system mediates response to oxidative stress and in an oxidant‐specific fashion. Free Radical Biology & Medicine, 29, 1043–1050. [DOI] [PubMed] [Google Scholar]
- Soanes, D. , Chakrabarti, A. , Paszkiewicz, K. , Dawe, A. & Talbot, N. (2012) Genome‐wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae . PLoS Pathogens, 8, e1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard, J. , Carroll, A. , Farrall, L. & Valent, B. (1997) A series of vectors for fungal transformation. Fungal Genetics Reports, 44, 52–53. [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. & Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi, K. , Tanaka, K. , Yang, H.‐Y. , Yamamoto, K. , Matsushita, Y. , Tomida, T. et al. (2007) Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. The EMBO Journal, 26, 3521–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turr, D. , Segorbe, D. & Pietro, A. (2014) Protein kinases in plant‐pathogenic fungi: conserved regulators of infection. Annual Review of Phytopathology, 52, 267–288. [DOI] [PubMed] [Google Scholar]
- Uchiyama, T. & Okuyama, K. (1990) Participation of Oryza sativa leaf wax in appressoria formation by Pyricularia oryzae . Phytochemistry, 29, 91–92. [Google Scholar]
- Vanstreels, E. , Alamar, C. , Verlinden, B. , Enninghorst, A. , Loodts, J.K.A. , Tijskens, E. et al. (2005) Micromechanical behaviour of onion epidermal tissue. Postharvest Biology and Technology, 37, 163–173. [Google Scholar]
- Wang, G. , Li, G. , Zhang, S. , Jiang, C. , Qin, J. & Xu, J.R. (2015) Activation of the signalling mucin MoMsb2 and its functional relationship with Cbp1 in Magnaporthe oryzae . Environmental Microbiology, 17, 2969–2981. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Lu, D. & Tian, C. (2021a) CgEnd3 regulates endocytosis, appressorium formation, and virulence in the poplar anthracnose fungus Colletotrichum gloeosporioides . International Journal of Molecular Sciences, 22, 4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Lu, D. & Tian, C. (2021b) Mitogen‐activated protein kinase cascade CgSte50‐Ste11‐Ste7‐Mk1 regulates infection‐related morphogenesis in the poplar anthracnose fungus Colletotrichum gloeosporioides . Microbiological Research, 248, 126748. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Xu, X. , Liang, Y. , Wang, Y. & Tian, C. (2018) A Cdc42 homolog in Colletotrichum gloeosporioides regulates morphological development and is required for ROS‐mediated plant infection. Current Genetics, 64, 1153–1169. [DOI] [PubMed] [Google Scholar]
- Weir, B. , Johnston, P. & Damm, U. (2012) The Colletotrichum gloeosporioides species complex. Studies in Mycology, 73, 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.A. , Gibson, R.P. , Quispe, C.F. , Littlechild, J.A. & Talbot, N.J. (2010) An NADPH‐dependent genetic switch regulates plant infection by the rice blast fungus. Proceedings of the National Academy of Sciences of the United States of America, 107, 21902–21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.A. & Talbot, N.J. (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae . Nature Reviews Microbiology, 7, 185–195. [DOI] [PubMed] [Google Scholar]
- Yang, H.‐Y. , Tatebayashi, K. , Yamamoto, K. & Saito, H. (2009) Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. The EMBO journal, 28, 1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Liu, K. , Zhang, X. , Song, W. , Zhao, Q. , Dong, Y. et al. (2010) A two‐component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae . Current Genetics, 56, 517–528. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Bian, Z. & Xu, J.R. (2018a) Assays for MAP kinase activation in Magnaporthe oryzae and other plant pathogenic fungi. Methods in Molecular Biology, 1848, 93–101. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Zhang, J. , He‐Pu, H. , Wang, X. & Tian, C. (2018b) Histopathology study of poplar leaves infected by Colletotrichum gloeosporioides . Beijing Linye Daxue Xuebao [Journal of Beijing Forestry University], 40, 101–109. [Google Scholar]
- Zhao, X. , Kim, Y. , Park, G. & Xu, J.‐R. (2005) A mitogen‐activated protein kinase cascade regulating infection‐related morphogenesis in Magnaporthe grisea . The Plant Cell, 17, 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Sequence analysis of CgMsb2 with homologues in other fungi. (a) Whole amino acid sequences of CgMsb2 (Protein ID: 1461327) and Msb2 homologues were aligned using ClustalX v. 2.1. Red frames indicate the signal peptide, noncytoplasmic domain, transmembrane (TM) domain, and cytoplasmic domains. (b) Phylogenetic tree of CgMsb2 and its homologues in other fungi. The phylogenetic tree was constructed using MEGA v. 6.0 with full‐length protein sequences and neighbour‐joining with 1,000 bootstrap replicates
FIGURE S2 Sequence analysis of CgSho1 with homologues in other fungi. (a) Whole amino acid sequences of CgSho1 (KAF 3805345.1) and Sho1 homologues were aligned using ClustalX v. 2.1. Red frames indicate the TM and SH3 domains. (b) Phylogenetic tree of CgSho1 and its homologues in other fungi. The phylogenetic tree was constructed using MEGA v. 6.0 with full‐length protein sequences and neighbour‐joining with 1,000 bootstrap replicates
FIGURE S3 Sequence analysis of CgPth11 with homologues in other fungi. (a) Whole amino acid sequences of CgPth11 (KAF 3797122.1) and Pth11 homologues were aligned using ClustalX v. 2.1. Red frames indicate the signal peptide, the CFEM domain, and seven transmembrane domains. (b) Phylogenetic tree of CgPth11 and its homologues in other fungi. The phylogenetic tree was constructed using MEGA v. 6.0 with full‐length protein sequences and neighbour‐joining with 1,000 bootstrap replicates
FIGURE S4 Schematic drawing of CgMsb2, CgSho1, and CgPth11 proteins. Full‐length amino acid sequences of CgMsb2, CgSho1, and CgPth11 were analysed using the InterproScan tool (https://www.ebi.ac.uk/interpro). Domains including the signal peptide, noncytoplasmic domain, SH3 domain, CFEM domain, transmembrane domain, and cytoplasmic domain were predicted and are marked in different colours. aa, amino acids
FIGURE S5 Generation of the deletion mutants in Colletotrichum gloeosporioides. The split marker method was used for single and double gene replacement. (a) Diagrammatic representation of the split marker targeted replacement strategy including location of primers. 5′: the upstream flank sequence of the target gene. 3′: the downstream flank sequence of the target gene. (b) In ΔCgMsb2 and ΔCgSho1, a hygromycin cassette (1.4 kb) replaced the fragment of CgMsb2 (3.0 kb) and CgSho1 (1.0 kb), respectively. In ΔCgPth11, a Sur cassette (2.2 kb) replaced the fragment of CgPth11 (1.5 kb). (c) Complementation of CgMsb2, CgSho1, and CgPth11 was achieved by reintroduction of the full coding sequence of each gene and confirmed using PCR. (d) A Sur cassette replaced the fragment of CgPth11 in ΔCgSho1 and ΔCgMsb2 to generate ΔCgSho1Pth11 and ΔCgMsb2Pth11, respectively. The Sur cassette replaced the fragment of CgSho1 in ΔCgMsb2 to generate the ΔCgMsb2Sho1 mutant
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


