Abstract
Xanthomonas phaseoli pv. manihotis (Xpm) and X. cassavae (Xc) are two bacterial pathogens attacking cassava. Cassava bacterial blight (CBB) is a systemic disease caused by Xpm, which might have dramatic effects on plant growth and crop production. Cassava bacterial necrosis is a nonvascular disease caused by Xc with foliar symptoms similar to CBB, but its impacts on the plant vigour and the crop are limited. In this review, we describe the epidemiology and ecology of the two pathogens, the impacts and management of the diseases, and the main research achievements for each pathosystem. Because Xc data are sparse, our main focus is on Xpm and CBB.
Keywords: cassava, cassava bacterial blight, cassava bacterial necrosis, quantitative resistance, Xanthomonas
This pathogen profile describes the hallmarks of the Xanthomonas phaseoli pv. manihotis–cassava pathosystem and compiles the available data for the nonvascular cassava pathogen Xanthomonas cassavae.
1. INTRODUCTION
Cassava is an important staple crop as a source of food and income for hundreds of millions of people in tropical countries. This major crop is threatened by several pests and pathogens that significantly affect productivity. Among bacterial pathogens, the vascular and systemic Xanthomonas phaseoli pv. manihotis (Xpm) has received considerable attention due to its devastating potential in the tropics and scientific importance worldwide (Mansfield et al., 2012). This pathogen profile describes the main hallmarks of this pathosystem and updates several reviews and original articles on the subject. Although more bacterial pathogens infect cassava, information in the literature is scarce. In this regard, we also present a compilation of information for a second xanthomonad infecting cassava, the nonvascular pathogen X. cassavae.
2. DISEASE DESCRIPTION AND EPIDEMIOLOGY
2.1. Cassava bacterial blight symptoms
Cassava bacterial blight (CBB), which is caused by the bacterial pathogen Xpm, is characterized by a range of symptoms that mainly affect leaves, petioles, and stems, frequently leading to plant death (Figure 1). Early symptoms appear as brown to dark‐brown water‐soaked translucent angular spots on the leaf tissue browning at later stages, occasionally surrounded by a chlorotic halo. Veins around these spots discolour and affected tissues frequently produce creamy white and later yellow‐to‐orange exudates on the lower side of the leaf. Blight results from spot coalescence, which creates necrotic areas that become dry and curl the leaflets, giving them the aspect of a superficial burn. As disease progresses, bacteria access the xylem vessels from the mesophyll and move towards the stem through the petioles, which become brown and collapse, causing the leaf to wilt. Vessel colonization in the stem allows Xpm to systemically move upwards and downwards. When infection reaches the plant upper part where stem tissues are greener and less lignified, stem rotting leads to dieback characterized by shoot apex wilting. New sprouts can grow from buds located in more basal zones, giving the plant the appearance of a candle stick. However, if these buds are also contaminated by Xpm they will eventually wilt. Infected fruits also show water‐soaked spots and the resulting seeds suffer cotyledon and endosperm necrosis, and seed deformation. Roots from highly susceptible cultivars can show delayed symptoms restricted to the vascular tissues, with discoloured vascular strands surrounded by dry and rotten spots (Boher et al., 1995; Lozano, 1986; Lozano & Sequeira, 1974b; Maraite & Meyer, 1975).
FIGURE 1.
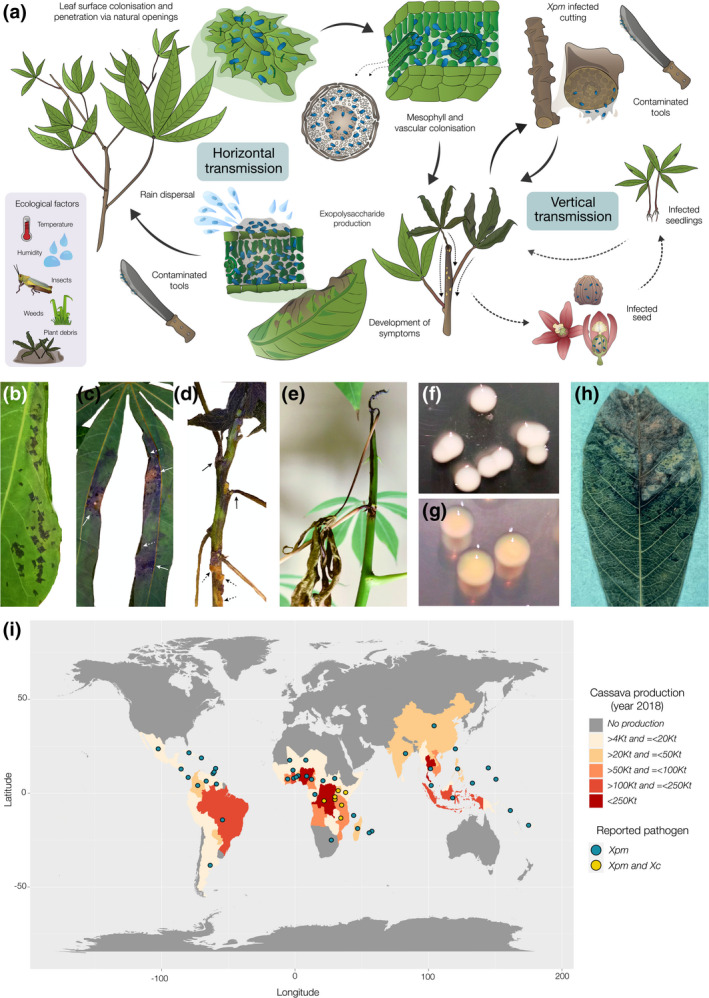
Aetiology, ecology, and distribution of Xanthomonas phaseoli pv. manihotis (Xpm) and Xanthomonas cassavae (Xc). (a) Cassava bacterial blight disease cycle. Dashed arrows indicate processes that are only relevant for traditional farming systems, that is, where unmanaged sexual reproduction may lead to the incorporation of seedlings into local germplasm. The inset indicates environmental and ecological factors that affect the spread or development of disease. (b) Angular leaf spots caused by Xpm. (c) Blight (solid white arrows) and leaflet curling (dashed white arrows) caused by Xpm. (d) Collapsed petioles from wilted leaves (solid black arrows) and gum exudation (dashed black arrows) from stems caused by Xpm infection. (e) Shoot apex wilting and dieback caused by Xpm. (f) Typical colonies of Xpm on LPGA medium. (g) Typical colonies of Xc on LPGA medium. (h) Leaf spots caused by Xc. (i) Worldwide cassava production (FAO, 2020) by country (missing data for South Africa, Guam, and Palau) for 2018 and distribution of Xpm and Xc (CABI, 2020)
Symptoms of cassava bacterial necrosis (CBN) caused by X. cassavae (Xc) might be similar at a first sight (Figure 1h), but the outcome of the infection is not as devastating (Mostade & Butare, 1979). Infections caused by Xc initially produce rounded water‐soaked spots surrounded by a yellow chlorotic halo and radial necrosis of the veins (Maraite & Perreaux, 1979). As the disease progresses, bacteria colonize the adjacent mesophyll tissues, lesions expand, and yellow exudates can be observed in them (Maraite, 1993). Leaves wilt, collapse, and dry, but the pathogen is not able to colonize vascular tissues and there is no formation of secondary spots that turn into extended blight areas (Lozano & Sequeira, 1974b; Maraite, 1993; Maraite & Perreaux, 1979; Van Den Mooter et al., 1987; Verdier et al., 1994).
2.2. Disease cycle
Xpm has an initial epiphytic phase where it is able to colonize the surface of cassava aerial tissues. When conditions are favourable, especially high humidity, multiplication of the pathogen increases around trichomes and penetration of the outer layer takes place (Daniel & Boher, 1985a). Xpm accesses the plant through wounds and natural openings of the leaf, potentially primarily via adaxial stomates (Kemp et al., 2004). Although it is common for other vascular Xanthomonas to enter through hydathodes, their role in the initial stages of infection for Xpm is not known. Internal tissue colonization is accompanied by the production of highly hygroscopic bacterial exopolysaccharides (EPS), which are exuded from lesions and then hydrated and carried by raindrops. Wind and rain‐mediated splashing of these bacterial suspensions is considered the main natural way of horizontal pathogen transmission from one plant to another (Lozano & Sequeira, 1974a). Once Xpm reaches new cassava leaves, the disease cycle restarts. Because this crop is propagated from cuttings, presence of the pathogen on the propagative material and working tools is the major factor for disease spread (Lozano & Sequeira, 1974a). Plants sprouted from contaminated cuttings quickly develop the disease and are a major source for secondary infections in the field. Bacteria can also be transmitted inside the sexual seed (Daniel & Boher, 1985b; Elango & Lozano, 1980). Stems can get infected by the dispersal of wind‐driven sand or hail (Maraite, 1993).
2.3. Ecology of the pathogen
Xpm shows two different lifestyles in the field: an epiphytic phase (Daniel & Boher, 1985b; Elango & Lozano, 1981) and a biotrophic parasitic phase that starts when environmental conditions facilitate pathogen growth and entrance into the mesophyll apoplast of cassava leaves (Verdier et al., 1990). The epiphytic stage plays an important role ensuring the natural persistence of the pathogen between cropping cycles. Epiphytic populations of Xpm have been found on symptomless leaves in fields where CBB has been reported. Bacterial titres vary according to environmental conditions, humidity and temperature being two of the most important factors. The optimal temperature for CBB development is 30 °C, whereas that for CBN is 25 °C (Maraite & Perreaux, 1979). There is a marked increase of epiphytic populations and a shift to the parasitic phase during the rainy season. Conversely, symptoms of CBB are less frequent and epiphytic populations decrease during the dry season (Daniel & Boher, 1985a, 1985b). Xpm has also been detected in several weeds that occur naturally in cassava fields in South America, suggesting that the pathogen can survive epiphytically on them between cropping cycles (Elango & Lozano, 1981). Artificial inoculation of several cassava crop‐associated weeds (nonhost interactions in all cases) highlighted different degrees of bacterial survival and pathogen viability for up to 54 days (Fanou et al., 2017; Marcano & Trujillo, 1984).
Infected plant debris and soil are also suggested to play a role in Xpm field persistence. However, survival of Xpm in a free‐living state in the soil has been experimentally proven to be limited to up to 3 weeks (Fanou et al., 2017). In contrast, Xpm was shown to survive in slow‐decaying dry debris for more than 2 months and 1 year under field and controlled conditions, respectively (Daniel & Boher, 1985b; Fanou et al., 2017). Insect‐mediated dissemination of Xpm has been reported. The African grasshopper pest Zonocerus variegatus recovered from diseased cassava fields harboured a significant number of infective bacterial cells (Daniel & Boher, 1985b; Zandjanakou‐Tachin et al., 2007). The coreid bug Pseudotheraptus devastans has also been reported as a facilitator of infection through the generation of punctures through which the bacterium gains access to internal tissue (Maraite & Meyer, 1975).
2.4. Distribution of the pathogen
The botanical, geographical, and domestication origin of cassava is a controversial topic. A widely accepted theory suggests that the cultivated cassava species Manihot esculenta subsp. esculenta arose from the wild M. esculenta subsp. flabellifolia in the Amazon basin, and its domestication began 7,000 to 9,000 years ago in the southern Amazon border region (Allem, 2002; Olsen & Schaal, 1999). Cassava was introduced to Africa by Portuguese traders during the 16th century and was adopted as a staple crop across several countries (Jones, 1959). In Asia it is believed to have been introduced from Mexico to the Philippines during the 17th century. In agreement with the geographical origin of the crop, genetic diversity of Xpm populations in South America has been shown to be high (Bart et al., 2012; Verdier et al., 2004).
CBB was first reported in Brazil in 1912 by Bondar and then identified in different South American countries during and after the 1970s. The disease and the pathogen were detected in Africa for the first time in Nigeria in 1972, and then systematically detected in several sub‐Saharan countries (Hillocks & Wydra, 2002; Persley, 1976). To date, Xpm has been reported in 49 countries located all over in the tropics (CABI, 2020; Taylor et al., 2017). In contrast, Xc has only been reported in the East African countries of Burundi, the Democratic Republic of the Congo, Kenya, Malawi, Rwanda, Tanzania, and Uganda (Maraite, 1993; Mostade & Butare, 1979; Verdier et al., 1994). Figure 1 shows cassava production areas and the distribution of both pathogens.
3. DISEASE IMPACTS AND MANAGEMENT
3.1. Prevalence, incidence, and losses associated to CBB
Historically CBB is reminiscent of a famine period from 1970 to 1975 in the Democratic Republic of the Congo, where cassava crop diversity was low and food and economic dependence on cassava was considerable (Hillocks & Wydra, 2002; Lozano, 1986; Maraite & Meyer, 1975). Several studies have addressed the distribution and effects of the pathogen on the crop. Figure 2 (and Tables S1, S2, and S3) summarizes data from several reports where incidence, severity, and/or yield losses were assessed, mainly in farms or field trials. Incidence varies according to environmental factors, but most of the reports show ranges between 30% and 90%, with an incidence peak between 60% and 70%. Disease severity reports show systemic symptoms in all the surveys (values greater than 2 in 1–5 scales), even for measurements performed within the first 3 months after planting (MAP). Severity seems higher between the third and the sixth MAP, and there is a slight decrease in the following months, but complete dieback and plant stunting have been recorded in all the surveys performed after the third MAP.
FIGURE 2.
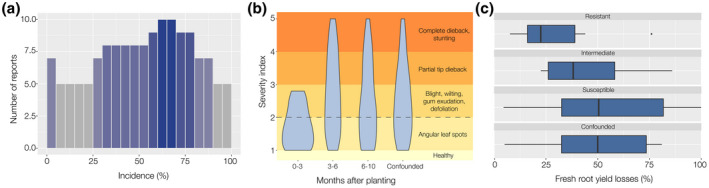
Graphical meta‐analysis of incidence, severity, and losses reported for cassava bacterial blight (CBB). (a) Histogram of the incidence ranges reported by 11 studies (Table S1). The colour scale correlates with the frequency of the reported range. (b) Violin plots showing severity ranges recorded by 15 studies (Table S2) according to measurement timepoints after planting. The confounded variable groups data from reports that did not include timepoint information. A harmonized severity grading scale is presented at the right side of the plot. The dashed line indicates that values above 2 reflect systemic disease. (c) Boxplots showing fresh root yield losses ranges described by seven reports (some studies reported several ranges, Table S3) according to the designated resistance status of the plants. The confounded variable groups data from reports that did not include the designated resistance status of the evaluated plants
Yield losses are more difficult to measure accurately mainly due to two reasons: (a) cassava fields are not only affected by CBB in most of the cases, and (b) as for other diseases, outcomes are dependent on environmental conditions, that is, plants can counterbalance the negative effects of CBB when favourable growth conditions are given (Harris et al., 2015; Zinsou et al., 2005). However, available metadata suggest that fresh root yield losses (Figure 2) can reach up to 100%, with a median of about 50% in susceptible varieties, while it reaches up to 76% in resistant varieties, but the median is below 25%. This is an indication of the high potential of Xpm to cause important losses.
3.2. Economic and social importance of the pathogen
In a recent report of the Evans School Policy Analysis and Research (Harris et al., 2015) for the Bill and Melinda Gates Foundation, economic impacts of CBB and postharvest physiological deterioration were weighed in the context of main constraints for cassava production. Despite its well‐known potential to cause important losses, the current impacts of CBB on the crop are masked by the lack of recent surveys due to limited research on the disease, underestimations due to the lack of farmers’ training for CBB identification, and the influence of environmental factors like drought and coinfections on the onset and severity of the disease. A 2‐year follow‐up of farmers’ practices and crop outcomes in Uganda and Kenya revealed that despite the high concern of farmers about the impacts of diseases like CBB and cassava anthracnose disease (CAD), the factors impacting yield losses the most were soil fertility and weed management (Fermont et al., 2009). This latter could result from the cumulative experience of agricultural control measures applied to cassava farming since the 1980s, which may have buffered the impact of CBB (Lozano, 1986).
Conversely, CBB dynamics seem to be altered by social behaviours linked mainly to trading cuttings, which favours the distribution of the pathogen over short and long distances (Restrepo et al., 2004; Restrepo & Verdier, 1997; Trujillo, Arias‐Rojas, et al., 2014, Trujillo, Ochoa, et al., 2014). In a case study in Colombia, disease incidence was positively correlated with use of agrochemicals, land ownership, and propagative material sharing. Land ownership limitations due to internal conflict and inequity in cassava farmer communities hamper the long‐term establishment of crops and force intercycle renewal of propagative material, which leads to the use of any available sources of stakes that frequently lack phytosanitary controls (Pérez, personal communication).
3.3. Control strategies
Efficient CBB control is based on three main pillars: sanitary controls, cultural practices, and deployment of tolerant or resistant varieties. Control practices have been deployed based on our knowledge about Xpm and its ecology. Sanitary controls of propagative materials and seeds are large‐scale measures that aim to stop pathogen dispersal. In general, these measures comprise the deployment of disease‐free materials and treatments to eliminate the pathogen, establish quarantine, and Xpm detection tools (Chavarriaga Aguirre et al., 2016; Frison & Feliu, 1991).
Cultural practices for CBB management include crop rotation, intercropping, fallowing, removal or burying of crop debris, weed management, delayed planting (at the end of the rainy season), and use of clean propagative materials. Crop rotation and fallowing aim to deplete the pathogen inoculum sources through time; they are deemed to be effective at buffering the impact of CBB in successive crop cycles (Lozano, 1986; Persley, 1978). Because weeds are reservoirs of the pathogen, their management during the entire crop cycle, as well as debris treatments before planting, are strongly recommended. Fanou et al. (2017) showed that Xpm survival is markedly reduced when debris is covered with soil or buried (less than 30 days versus more than 120 days for unburied debris under dry conditions). Moreover, Wydra and Fanou (2015) showed that removal of symptomatic leaves can reduce CBB severity and improve the quality of the crop as a source of propagative material; however, this practice did not show effects on yields in their study. Usually, planting during the second half of the rainy season decreases disease incidence and severity, while it maintains or improves yields (Ambe, 1993; Umemura & Kawano, 1983; Zinsou et al., 2004).
Considering that cassava is devoted to sustaining mainly low‐income farmers, deployment of tolerant or resistant varieties is considered the main solution for CBB control. Clonal selection and breeding for cassava disease resistance started in the 1940s and were mainly focused on CMD. Resistance against CBB is essentially due to genes introgressed from wild Manihot species, like the ceara rubber tree M. glaziovii (Nassar, 2007). Several studies addressed the performance of diverse cassava varieties against CBB (Dixon et al., 2002; Fokunang et al., 2000; Lamptey et al., 1998; Lozano & Laberry, 1982; Restrepo et al., 2000b; Umemura & Kawano, 1983; Wydra et al., 2004, 2007; Zinsou et al., 2005) (Table S2). In summary, three main aspects condition the successful deployment of CBB resistant varieties: (a) interactions of cassava with environmental factors profoundly affect the fitness and resistance of some cultivars; (b) the need for co‐selection of agronomical traits of value for farmers and the local market; and (c) the resistance against CBB is polygenic, additively inherited (Umemura & Kawano, 1983), pathotype‐specific (Wydra et al., 2004), and its molecular basis needs to be elucidated.
Regarding the first aspect, several studies (Dixon et al., 2002; Restrepo et al., 2000b; Wydra et al., 2007; Zinsou et al., 2005) demonstrated that the environment has a significantly greater influence on CBB disease incidence than the genotypic component, and that the interaction of both factors can mask differences between genotypes. For instance, the highly resistant variety TMS30572 performed as resistant in two edaphoclimatic zones of Benin, while it was moderately resistant in four other climatic zones (Zinsou et al., 2005). Therefore, researchers draw attention to test resistance in relevant conditions, with high disease pressure, and in parallel with greenhouse settings where pathogenicity assays are optimal (Restrepo et al., 2000b; Zinsou et al., 2005). Regarding the second aspect, surveys have shown that smallholders prioritize higher yields, taste, and good cooking qualities over diseases tolerance when selecting materials, which is highly relevant when engineering disease resistance (Harris et al., 2015). The latter aspect—cellular, genetic, and molecular bases of the resistance—is addressed later in this paper.
Despite the above‐mentioned caveats and to summarize the attempts to find high‐performing cassava varieties (Table S5), the outstanding resistance against CBB of genotype TMS30572 has been widely demonstrated in several trials in African countries. This genotype comes from the CBB‐resistant parent 58308, a low‐productive interspecific hybrid with M. glaziovii, and the susceptible, but high‐yielding Brazilian variety Branca Caterina de Santa (https://seedtracker.org/cassava/; Nassar, 2007; Umanah, 1977). Although performance of cassava varieties tested in South American fields has been highly variable, genotypes CMC40, MECU82, MCOL1916, MPAN19, MPAN12B, MBRA685, MBRA886, MBRA902, MNGA2, CM523‐7, and CM6438‐14 were considered resistant against different Xpm strains in a set of studies (Table S5).
4. PATHOGEN DESCRIPTION
4.1. Pathogen identification
Xpm and Xc are gammaproteobacteria that belong to the Xanthomonadaceae family. Xpm is a gram‐negative rod with a polar flagellum that forms shiny, slimy, convex, and circular colonies with entire margins when glucose or sucrose are present in the medium, without any pigmentation (Figure 1f) (Lozano & Sequeira, 1974b; Maraite & Meyer, 1975; Van Den Mooter et al., 1987). The characteristic white colour is due to the absence of xanthomonadin pigment, a rare trait also observed in Xanthomonas citri pv. mangiferaeindicae and Xanthomonas campestris pv. viticola (Midha & Patil, 2014). Colonies of the gram‐negative rod Xc are slimy, convex, and circular with entire margins and a deep yellow colour (Figure 1g). According to Van Den Mooter et al. (1987), these two pathogens can be differentiated by colony colour, growth on d‐saccharic acid (positive for Xc, but not for Xpm), hydrolysis of Tween 60, and growth on dl‐glyceric acid (positive for Xpm, but not for Xc).
4.2. Pathogen classification
First described by Bondar in 1912 the causal agent of CBB was initially named Bacillus manihotis (Arthaud‐Berthet). After several taxonomical reclassifications (Burkholder, 1942; Dedal et al., 1980; Lozano & Booth, 1974; Maraite & Meyer, 1975; Vauterin et al., 1995), the most recent classification based on a polyphasic taxonomic approach (a seven‐gene multilocus sequence analysis, average nucleotide identity, and biochemical analyses) assigned this pathogen to the species X. phaseoli, with the definite nomenclature X. phaseoli pv. manihotis (Constantin et al., 2016). As to the causative pathogen of CBN, it was first described by Wiehe and Dowson in 1953 in Malawi and named Xanthomonas cassavae (Wiehe & Dowson, 1953), which was further confirmed upon DNA–DNA hybridization later on (Vauterin et al., 1995). Figure 3 shows the taxonomic position of the two cassava pathogens among most known Xanthomonas species.
FIGURE 3.
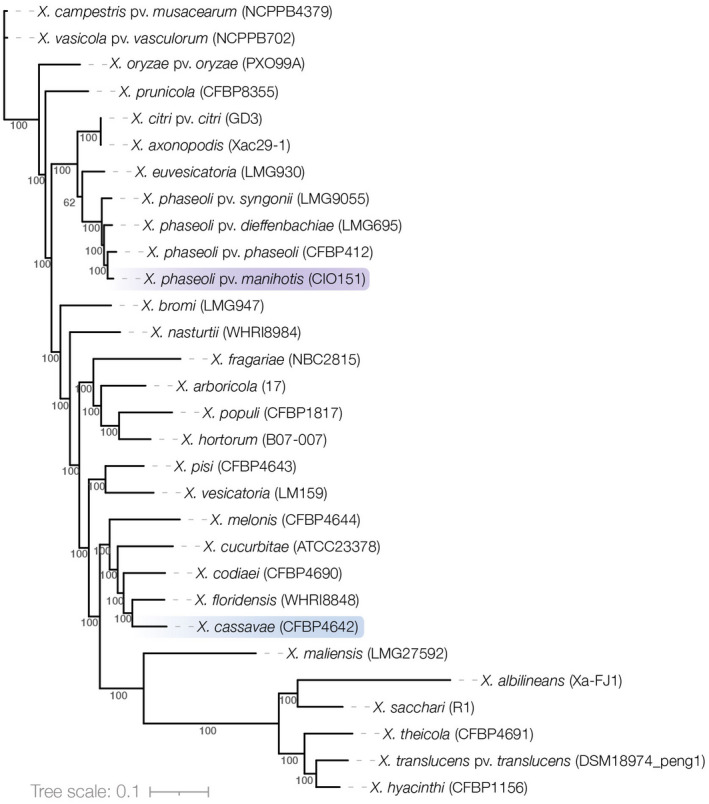
Taxonomic position of Xanthomonas phaseoli pv. manihotis (Xpm) and Xanthomonas cassavae (Xc). Phylogeny of 30 representative Xanthomonas, including 27 species and four pathovars of X. phaseoli (Table S4). Strain code is indicated in parentheses. Xpm and Xc are highlighted in purple and blue, respectively. Phylogeny was constructed from RefSeq complete genomes using the bioinformatic workflow PhaME (Shakya et al., 2020). Core genome alignments resulted in 155,507 aligned single nucleotide polymorphism (SNP) positions that covered coding and noncoding regions. Trees were reconstructed with the GTRGAMMAI model of RAxML and consensus tree was calculated from 100 bootstraps; results higher than 80 are shown above branches
4.3. Host range
Alternative hosts can play a key role in pathogen persistence in the environment, acting as reservoirs during long periods. To date, five Euphorbiaceous plants have been reported to be symptomatically affected by Xpm, supporting its multiplication and release to the environment: the three cassava wild relative species M. glaziovii, Manihot palmata, and Manihot aipi (Lozano & Sequeira, 1974b), Euphorbia pulcherrima and Pedilanthus tithymaloides (Dedal et al., 1980). Moreover, the limited distribution of Xc in Africa and its apparent absence in South America (origin of cassava) suggests that coevolution with cassava is rather short and that this plant may not be the main host of this bacterium (Hayward, 1993). No alternative host has been reported for this pathogen so far.
4.4. Diversity of the pathogen
Xpm diversity has been analysed by different methods during the last three decades, including restriction fragment length polymorphisms (RFLPs) (Berthier et al., 1993; Restrepo et al., 2004; Restrepo & Verdier, 1997; Verdier et al., 1993, 1994; Verdier, Restrepo et al., 2001), repetitive element sequence‐based PCR (rep‐PCR) (Chege et al., 2017; Restrepo et al., 2000c), random amplified polymorphic DNA (RAPD) (Ogunjobi et al., 2006, 2007), and amplified fragment length polymorphisms (AFLPs) (Gonzalez et al., 2002; Ogunjobi et al., 2010; Restrepo et al., 2000c; Trujillo, Ochoa, et al., 2014). Recent studies have shown that a slightly better discriminatory power is achieved using multiple locus variable‐number tandem repeat analysis (MLVA) (Rache et al., 2019; Trujillo, Arias‐Rojas, et al., 2014). An MLVA scheme with 15 variable number tandem repeats (VNTRs) amplified by multiplex PCR was developed to assess the pathogen diversity at a local scale and this scheme was tested in the northern coast of Colombia. This method has several advantages: it does not require DNA extraction, it can be easily formatted for high‐throughput settings, the amplification of up to four loci is performed in parallel in a unique PCR, and outputs show high reproducibility and portability (Rache et al., 2019).
As reviewed previously (López Carrascal & Bernal, 2012), the first studies on African isolates highlighted a rather clonal population context. Diversity seemed to be increasing from the 1980s to the 2010s, probably as a result of the introduction of new cassava varieties to African countries. More recently, high genetic relatedness of isolates from different cassava‐growing regions in Kenya using rep‐PCR has been reported, suggesting that despite the high prevalence of the disease, the genetic diversity of Xpm is still low in some African countries (Chege et al., 2017).
In contrast, Xpm diversity is higher in South America, in agreement with the hypothesis of the origin of the pathogen. Studies on Xpm populations isolated in several regions in Colombia showed no differentiated geographic structure, traces of pathogen migration, yearly changes of pathogen population structure in the same field, and indications of plant host pressure on the pathogen (reviewed by López Carrascal & Bernal, 2012). Recent studies highlighted two contrasting scenarios. Most haplotypes of Xpm were not structured geographically in northern regions of Colombia when using AFLPs (Trujillo, Ochoa, et al., 2014) or MLVA (Rache et al., 2019), suggesting an important role of pathogen migration through contaminated propagative material. However, a centre of origin of several Xpm haplotypes was discovered in one of the studied locations, which acts as a potential source of new founder pathogens (Trujillo, Ochoa, et al., 2014). In contrast, AFLP and MLVA‐based analysis showed that Xpm populations of the eastern plains were structured geographically, but still show some degree of genetic flow between distant regions. The authors considered that these contrasting scenarios reflect different agricultural practices, crop intensiveness, and distribution of lands dedicated to the crop (Trujillo, Arias‐Rojas, et al., 2014).
At a larger geographical scale, 65 Xpm strains, mainly from Africa and South America, were sequenced by Illumina (Bart et al., 2012). Phylogenies based on more than 12,000 chromosomal single‐nucleotide polymorphisms (SNPs) show that African, Colombian, and some Brazilian strains share a common ancestor, in line with the idea of the introduction of the pathogen to Africa as a consequence of the slave trade and or Portuguese missions. The authors highlight that Xpm populations are evolving independently, but also show genetic flow between geographically distant places, which should be taken into consideration for surveillance and CBB control purposes.
4.5. Diagnostic tools
Early detection and diagnostic tools relied on in vitro isolation of the pathogen coupled with phage typing, cassava reinoculation, and/or serological assays on cassava extracts. With the popularization of more straightforward molecular methods, standard (Verdier et al., 1998) and nested PCR (Ojeda & Verdier, 2000), dot blots (Verdier & Mosquera, 1999), and an ELISA were developed (Verdier, Ojada et al., 2001). Three PCR‐based methods were reported for Xpm detection recently. The first method used an optimized version of a previous assay (Verdier et al., 1998) resulting in improved Xpm detection (Cerqueira‐Melo et al., 2019). The second approach was based on a multiplexed nested PCR including a broadly conserved fragment of Xpm transcription activator‐like (TAL) effector genes and a semispecific region of rpoB, resulting in a wider detection potential for Xpm strains (Bernal‐Galeano et al., 2018). The third strategy, consisting of a duplex PCR amplifying highly conserved chromosomal regions in Xpm and Xc to detect and distinguish both cassava pathogens, proved to be highly selective and sensitive (Flores et al., 2019).
5. DISEASE PHENOTYPING TOOLS UNDER CONTROLLED CONDITIONS
Generally, Xpm must first overcome plant defences in the leaf apoplast before accessing the vessels and migrating towards the stem. Defence mechanisms in the mesophyll are different from those exerted by the plant in the leaf and stem vessels, adding barriers of a different nature that are surpassed only by successful pathogens (Kpémoua et al., 1996; Restrepo et al., 2000b; Wydra et al., 2007). This multilayer tissue‐specific resistance might be the explanation for the quantitative and additive nature of resistance in cassava, which increases the complexity of disease phenotyping. Moreover, the propagation of cassava through vegetative cuttings results in developmentally unsynchronized plants with variations in their physiology that could affect the outcomes of phenotyping methods (Mutka et al., 2016). Hence, disease phenotyping tools in cassava and their relationship to real‐world plant performance are not trivial subjects.
Bacterial virulence is usually assessed on infiltration of leaves of adult 2‐ to 4‐month‐old plants through bacterial growth analysis, bacterial movement through the leaf, and symptom development (compilation of protocols in Cohn et al., 2015). Bacterial growth and movement within the host quantitatively reflect the ability to grow locally and to migrate through the xylem, while symptom development is a qualitative estimation of the Xpm capacity to induce water‐soaking lesions, which might improve bacterial fitness. Symptom development can also be quantified by measuring lesion areas around a perforated hole or on leaf clipping or leaf spraying (Pacumbaba, 1987; Verdier et al., 1994; Zandjanakou‐Tachin et al., 2007). More recently, image‐based phenotyping of Xpm with luminescent reporters efficiently allowed the disease to be quantitatively tracked in time and space (Mutka et al., 2016).
Leaf inoculation may be questionable for resistance phenotyping because discrepancies were observed between leaf lesion measurements and scoring on stem inoculation (Muñoz‐Bodnar et al., 2015; Restrepo et al., 2000a). However, others have successfully detected symptom differences when infiltrating leaves of susceptible and resistant cultivars at low densities (from 102 to 105 cfu/ml; Flood et al., 1995; Wydra et al., 2004, 2007).
In the stem, the most discriminant methodology consists of inoculating one internode of the apical region by puncturing with a sharp tool with bacteria then evaluating disease development over a 30‐day period using a standardized severity scale. Measurements from several replicates are mathematically integrated to obtain a dimensionless quantity (area under the disease progression curve, AUDPC) that can be compared even among experimental sets with similar conditions. Resistance and susceptibility is established from scores (on a 0–5 scale, >4 is deemed as susceptible and <3 is deemed resistant) or by setting thresholds when performing AUDPC analyses, this latter being the preferred method to assess resistance (Jorge & Verdier, 2002; Restrepo, Duque, et al., 2000b).
To avoid the recurrent problem of space limitation when phenotyping cassava (plants are large), two studies recently reported the use of cassava plants grown in vitro. A comparative resistance screening between plants grown in pots and in vitro showed that the latter did not wilt although some symptoms developed, indicating that the AUDPC methodology can be applied to assign resistance/susceptibility categories (Mbaringong et al., 2017). In another study Mora et al. (2019) developed methods to quantify AUDPC values and perform bacterial growth analysis, highlighting contrasted phenotypes between susceptible and resistant cassava varieties in terms of disease progression and bacterial growth. These technical advances, in combination with traditional and more recent tools, open a new era in cassava phenotyping research.
6. VIRULENCE MECHANISMS OF THE PATHOGEN
6.1. Genomics of the pathogen
The advent of next‐generation sequencing technologies allowed the sequencing of the genome of 66 Xpm strains mainly originating from Africa and South America (Arrieta‐Ortiz et al., 2013; Bart et al., 2012). Assemblies resulted in fragmented draft genomes that were used for genomic and phylogenetic analyses, highlighting two main clusters. Grouping of South American and African strains in the same cluster agrees with the hypothesis formulated years ago on the possible introduction of Xpm from South America to Africa. The authors also identified the invariable occurrence of the well‐known pathogen‐associated molecular patterns (PAMPs) fliC and ax21, the presence of 14 to 22 Xanthomonas outer proteins (Xop) per strain, with a core set of nine of them, and reported that all strains harbour at least one TAL effector (Bart et al., 2012). The first manually annotated and high‐quality draft genome for Xpm was obtained through 454 pyrosequencing technology of the Colombian strain CIO151 (Arrieta‐Ortiz et al., 2013). Analysis of the genomic sequence confirmed the presence of fully functional pathogenicity mechanisms such as type II, III, IV, and VI secretion systems, an exopolysaccharide production cluster, core and accessory type III effectors, and chemotaxis, type IV pili, flagella, siderophore biosynthesis, and putative polyketide synthesis genes. This study also explored for the first time the possibility of using an MLVA scheme for Xpm diversity studies and reported on 16 potential VNTR loci. Third‐generation long‐read sequencing has resulted in several high‐quality nonfragmented xanthomonad genomes (Booher et al., 2015; Cox et al., 2017; Denancé et al., 2018; Gochez et al., 2018; Peng et al., 2016; Ruh et al., 2017; Showmaker et al., 2017; Tran et al., 2018; Wilkins et al., 2015). A first attempt to sequence Xpm with Pacific Biosciences technology highlighted a high error rate on reads preventing the assembly of a high‐quality draft genome (Bart et al., 2012). Regarding Xc, a draft genome of strain CFBP4642 (= NCPPB101, ICMP204, LMG673), isolated in Malawi in 1951, highlighted the presence of a canonical type III secretion system (T3SS) and some type III effectors including TAL effectors (Bolot et al., 2013). Presently our team is producing high‐quality reference genomes for both Xpm and Xc using long‐read sequencing technologies.
6.2. Secretion systems
The type I secretion system (T1SS) allows the active transport of hydrolases (e.g., proteases, phosphatases, glucanases, nucleases, and lipases) and toxins from the cytoplasm to the extracellular matrix (reviewed by Delepelaire, 2004). The canonical type II secretion system (T2SS) allows the secretion of periplasmic enzymes (e.g., cellulases, pectin methylesterases, cellobiosidases, and polygalacturonases) to the extracellular matrix (reviewed by Cianciotto & White, 2017). The presence of canonical versions of these two systems (T1SS and T2SS) in Xpm was reported by Arrieta‐Ortiz and coworkers, highlighting two slightly different clusters (xps and x c s) for the T2SS in the CIO151 genome (Arrieta‐Ortiz et al., 2013). Type IV secretion systems (T4SS) translocate proteins and DNA–protein complexes to surrounding eukaryotic host or other prokaryotic cells, playing an important role for competition against other gram‐negative bacteria (reviewed by Sgro et al., 2019). Likewise, the type VI secretion system (T6SS) is a contractile apparatus that injects toxic effectors to accompanying eukaryotic and prokaryotic cells (reviewed by Bayer‐Santos et al., 2019). Xpm carries at least one class of T4SS (Arrieta‐Ortiz et al., 2013; Sgro et al., 2019) and one T6SS cluster encoded by 15 genes in strain CIO151, but their functionality remains to be shown (Arrieta‐Ortiz et al., 2013).
The type III secretion system (T3SS) is a translocation machinery that allows the injection of effector proteins into the host. Delivered effectors play a major role in counteracting the host defences and hijacking the cellular mechanisms of the invaded cell (reviewed by Timilsina et al., 2020). In Xpm, the T3SS is encoded by 26 genes of the hrp2 family cluster (Arrieta‐Ortiz et al., 2013) and its role in pathogenesis has been demonstrated through the mutagenesis of hrpX, a key regulator of the T3SS (Medina et al., 2018).
6.3. Type 3 effectors
Xanthomonas T3Es can be classified in two major groups considering their molecular structure, function, and interactors/targets. The first group forms a heterogeneous group of so‐called Xanthomonas outer proteins (Xops), that is, effectors with a wide range of enzymatic activities whose targets and associated physiological effects mainly take place in the host cytoplasm and their mechanism of action relies on protein–protein interactions. The second group is only composed of TAL effectors, which are modular proteins that share an unusual architecture combined with eukaryotic motifs that allow them to act as bona fide transcription factors inside the host nucleus. Most Xops are involved in disturbing plant defence through alteration of PAMP‐/damage‐associated molecular pattern (DAMP)‐triggered immunity (PTI/DTI) or effector‐triggered immunity (ETI) pathways, while TAL effectors act as transcription factors (TFs) that reshape the host cellular metabolism through the activation of susceptibility (S) genes to create better niches for bacteria (Timilsina et al., 2020).
Xpm strains harbour from 13 to 23 Xop effectors, of which nine are conserved among all the reported Xpm genomes and termed as Xpm core effectors: HpaA, HrpF, XopE1, XopV, Hpa2, XopAK, XopL, XopN, and XopAE (a.k.a HpaF), the latter five being almost completely monomorphic among the surveyed strains (Bart et al., 2012). AvrBs2, XopAO1, XopZ, and XopX are important for virulence; strains mutated in these T3Es do not multiply properly in planta, and vascular colonization and/or symptom formation is impacted for some of them. XopK plays a dual role by increasing the developmental rate of symptoms but limiting the spread of the pathogen, while XopN and XopQ seem to have a redundant pathogenicity function. Expression in heterologous systems showed that XopR, AvrBs2, and XopAO1 interfere with PTI, while XopAO1 and XopE4 interfere with ETI (Medina et al., 2018; Mutka et al., 2016).
The crucial role of TAL effectors in Xpm pathogenicity had been known for a long time and was finally reported recently (Castiblanco et al., 2013; Cohn et al., 2014; Cohn et al., 2016). Several diagnostics and diversity tools were unintentionally based on TAL effector detection, the results highlighting their high conservation within Xpm (reviewed by Verdier et al., 2004). The screening of 65 strains through RFLP showed that all contained between one and five TAL effectors with 12.5, 13.5, 14.5, 19.5, 20.5, or 21.5 repeats of the repeat variable diresidue (RVD), and most of them were plasmid‐borne (Bart et al., 2012). However, our understanding of Xpm TAL effector diversity is rather poor because it is limited to only seven genes from three strains (Castiblanco et al., 2013; Cohn et al., 2014, 2016).
Castiblanco et al. (2013) described the genomic context, gene structure, and function of TALE1Xam (a.k.a pthB), showing that XpmΔTALE1Xam is affected in bacterial growth and movement in planta. A combination of transcriptomics and in silico prediction for TAL effector binding sites led to the identification of a heat shock transcription factor as a potential target of TALE1Xam in cassava (Muñoz‐Bodnar et al., 2014). Cohn et al. (2014) analysed the respective role of the five TAL effectors present in the strain Xam668. This study showed that mutation of TAL14Xam668 (which differs by only one RVD from TALE1Xam ) and TAL20Xam668 both significantly affect pathogen growth in planta. Moreover, disruption of TAL20Xam668 leads to the abolishment of water‐soaked symptoms. TAL20 Xam668 transcriptionally activates MeSWEET10a, which encodes a clade‐III sugar transporter from the SWEET family that acts as a susceptibility (S) gene, as reported in other pathosystems (reviewed by Perez‐Quintero & Szurek, 2019). Despite being a major virulence factor in Xpm, the identification and validation of the targeted S gene(s) have been hindered because TALE14 induces transcription of an elevated number of genes (Cohn et al., 2016). Pérez‐Quintero et al. (2017) developed a web‐based platform called daTALbase, where available information on Xpm TAL effectors and genomics and transcriptomics resources of the host allows the in silico research of TALE‐targeted genes. Figure 4 summarizes our current understanding about Xpm T3Es roles in planta.
FIGURE 4.
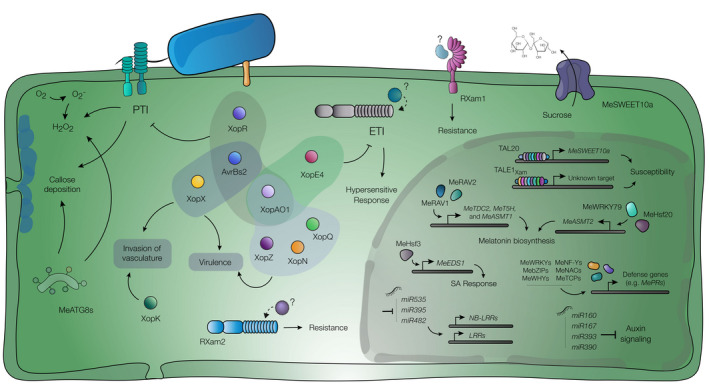
Roles of Xops, host determinants of susceptibility, and defence responses in the cassava–Xanthomonas phaseoli pv. manihotis (Xpm) interaction. Xop effectors (coloured circles) are injected by Xpm into the plant cytoplasm, where they interfere with host cellular processes. Type III effectors (T3Es) are grouped (shading) according to their predicted involvement in PAMP‐triggered immunity (PTI), effector‐triggered immunity (ETI), and/or other virulence‐related phenotypes (see section 6.3). The upper section of the depicted nucleus outlines the contribution of TAL effectors to cassava susceptibility, in which transcriptional activation of the S gene MeSWEET10a leads to an over‐accumulation of sugar transporters (purple, anchored to the membrane) and sugar export (see section 6.3). The RXam1 RLK (lilac, in the membrane) and the RXam2 NBS‐LRR (blue, in the cytoplasm) are associated with the defence response triggered against Xpm, but their matching elicitors are unknown. Calcium signalling and autophagy elements are represented in the cytoplasm as part of the anti‐Xpm defence mechanisms. The roles of miRNA and transcription factors are schematized in the nucleus (see section 7.5). H2O2 and O2 − represent reactive oxygen species (ROS) accumulation. SA, salicylic acid
Isolation and sequencing of TAL effector genes remain challenging due to their repetitive architecture and the existence of multiple copies and/or variants in the genome. Third‐generation sequencing technologies like Pacific Biosciences Single Molecule Real Time (PacBio SMRT) and Nanopore have been widely used to sequence genomes in other Xanthomonas species and eventually recovered high‐quality TALome sequences (Booher et al., 2015; Cox et al., 2017; Denancé et al., 2018; Gochez et al., 2018; Peng et al., 2016; Ruh et al., 2017; Showmaker et al., 2017; Tran et al., 2018). Targeted cloning techniques have also been optimized to isolate and study TAL effector function, diversity, and evolution systematically (Li et al., 2019; Yu et al., 2015). No Xpm genomes generated by these techniques have been reported to date, and the only two TALomes available so far were isolated from cosmid genomic libraries (Bart et al., 2012). Our team has sequenced several Xpm genomes by Pacific Biosciences SMRT technology and cloned more than 50 TAL effectors to assess their diversity and function among Xpm strains (Zárate‐Chaves et al., 2021). This information will allow TAL effector evolutionary analyses in Xpm and searches for other S genes in cassava.
6.4. Toxins
On bacterial entry into the leaves, the first symptoms to appear are water‐soaked angular spots. Generally, discrete spots begin to coalesce, and eventually surrounding and distal areas start to blight. This characteristic blight phenotype may be due to the diffusion into the laminar tissues around the infection foci of a small molecule that acts as a toxin. Perreaux and collaborators isolated from Xpm cultures a small organic acid, 3‐methyl thiopropionic acid, which induces the blight symptom when infiltrated alone into cassava leaves (Perreaux et al., 1982). Bacterial metabolism leads to a transamination coupled to a decarboxylation of methionine, resulting in the formation of 3‐methyl thiopropionic acid (Ewbank & Maraite, 1990). Concentrations of this toxin rise in leaves along with bacterial multiplication and reach a maximum just before the onset of the blight symptom (Perreaux et al., 1986). The exact mechanism of action of this small acid is not known. However, evidence for the role of this compound as a toxin is debated because concentrations of free methionine and the potential toxin in leaves are very low, and in vitro assays could have been biased by the acidic nature of the compound (Cooper et al., 2001).
6.5. Signalling and metabolic routes potentially involved in regulation of pathogenicity
Recently, an RNA‐seq study of a quorum‐sensing (QS)‐insensitive mutant of Xpm versus wild type grown in vitro was reported (Botero et al., 2020). The authors concluded that the QS system controls several subsequent signalling routes, including a number of phosphorylation sensor and transduction pathways, some of which share a c‐di‐GMP phosphodiesterase activity (HD‐GYP domain). Metabolic routes that were affected by QS included the already reported xanthan biosynthesis route and the newly reported NAD(P)+ balance, and fatty acid elongation, which should be further studied.
7. HOST GENETICS AND INTERACTION
7.1. Tools for Xpm–cassava interaction research
Development of adapted durable resistance must consider the local pathogen population structure and the host response variability. Pathotyping schemes can condense this complex interaction data to cluster pathogens according to virulence (Restrepo, Duque, et al., 2000b; Trujillo, Ochoa, et al., 2014; Verdier et al., 2004) and classify cassava genotypes based on susceptibility (Wydra et al., 2004). Inoculation of in vitro plants (Mora et al., 2019) and image‐based phenotyping methods (Mutka et al., 2016; Veley et al., 2021) are two important advances that will greatly facilitate the study of this pathosystem by leveraging high‐throughput applications and measuring the plant response in a more comprehensive way.
Cassava genomic resources include the read collection from 58 Illumina‐sequenced cassava or cassava relatives accessions (Bredeson et al., 2016), four high‐quality sequenced genomes from cultivars AM560‐2 (an MCOL‐1505‐derived partially inbred cultivar) (Bredeson et al., 2016; Prochnik et al., 2012), KU50 (Wang et al., 2014), TME3, and the highly CBB‐susceptible cultivar TMS60444, which is amenable to genetic transformation (Kuon et al., 2019). It also includes at least four high‐density genetic maps (International Cassava Genetic Map Consortium (ICGMC), 2015; Rabbi, Hamblin, Gedil, et al., 2014; Rabbi, Hamblin, Kumar, et al., 2014; Soto et al., 2015), one of them with an integrated physical map that includes immunity‐related genes (Soto et al., 2015), and bacterial artificial chromosome (BAC) libraries from cultivars TMS30001 and MECU72 (Tomkins et al., 2004). Transcriptomic resources for cassava challenged by Xpm include expressed sequenced tags (ESTs), simple sequence repeats (SSRs) (López Carrascal et al., 2004, 2007), a microarray (López Carrascal et al., 2005), transcript‐derived fragments (TDFs) (Santaella et al., 2004), and differentially expressed genes (DEGs) catalogs (Cohn et al., 2014; Gómez‐Cano et al., 2019; Muñoz‐Bodnar et al., 2014). The website www.cassavagenome.org provides tools incorporating the data from some of the above‐described resources such as transcriptomic and SSR data displayed on the AM560‐2 genome. As mentioned earlier, the daTALbase allows the research of potential virulence targets of TAL effectors through prediction of effector‐binding element (EBE) sequences in the host promoterome, and cross‐referencing to host transcriptomics and polymorphism data (Pérez‐Quintero et al., 2017).
7.2. Host susceptibility to Xpm
The susceptibility (S) gene MeSWEET10a is a clade III member of the well‐known SWEET family of phloem‐loading efflux carriers (reviewed by Chen, 2014), MeSWEET10a is transcriptionally activated by the TAL effector TAL20 Xam668 , and the resulting protein was found to export glucose and sucrose using Xenopus oocytes (Cohn et al., 2014). Increased susceptibility is probably related to increase of glucose accumulation in the apoplast, where it supports bacterial growth (Cohn et al., 2014), and/or by increasing osmotic water influx into the intercellular spaces that facilitates bacterial movement (El Kasmi et al., 2018; Schwartz et al., 2017). Cassava has 27 putative SWEET genes that are scattered in the genome (except for three clusters with five, three, and two SWEET genes), and account for at least five clade III members that could potentially also act as S genes, as shown in rice (Streubel et al., 2013).
Mora and coworkers compared the expression of MeSWEET10a during a compatible and an incompatible interaction, highlighting its upregulation in the susceptible cultivar TMS60444, but not in the resistant cultivar CM6438‐14, 50 hr after inoculation of a strain carrying TAL20Xam668 . Inspection of the MeSWEET10a promoter in both varieties showed conservation of the EBE, suggesting the immune response to stop susceptibility pathways at early stages of the infection (Mora et al., 2019).
7.3. Structural and cellular features of cassava resistance against Xpm
A mechanism associated with anti‐Xpm defence in cassava is the occlusion of adaxial stomata with wax, as a way to reduce pathogen entry into the leaf. Interestingly, the number of occluded stomata seems higher in resistant varieties (Cooper et al., 2001; Zinsou et al., 2006). Once in the substomatal spaces, Xpm reaches the mesophyll; colonization is accompanied by the formation of a fibrillar matrix made of bacterial exopolysaccharides and loosening of plant cell walls through enzymatic lysis. These modifications allow the pathogen to become vascular by accessing the xylem vessels and migrating to other parts of the plant (Boher et al., 1995, 1997; Kemp et al., 2004). Little is known about the resistant reactions taking place in the leaf mesophyll, but it has been suggested that these responses may be overcome by Xpm (Kpémoua et al., 1996; Verdier et al., 1994). Cassava resistance against Xpm is mediated by several cellular and molecular mechanisms, and the promptness of this response is the key difference between a susceptible and a resistant variety (Kpémoua et al., 1996). Resistance in the stem is characterized by the initial secretion of bactericidal phenolic compounds into intercellular spaces and vessel lumen, followed by a deposition of lignin, suberin, and callose in the paramural spaces. Later, pectic‐rich tyloses are formed to occlude vessels and to secrete more phenolic compounds, while bacteria are trapped in lysis pockets through lignification and suberization (Boher et al., 1995). Moreover, resistant genotypes are able to metabolically cope better with infection, with the maintenance of average stomatal resistance, water potential, and proline levels, factors that are markedly altered in compatible interactions (Restrepo Rubio et al., 2017).
7.4. Mapping resistance to Xpm
Due to the quantitative nature of CBB resistance, and the complex interaction between the host genetic background and the environment, the search for resistance sources against Xpm in cassava is mainly restricted to the identification of quantitative trait loci (QTLs) (Jorge et al., 2000, 2001; López Carrascal et al., 2007; Soto‐Sedano et al., 2017; Tappiban et al., 2018; Wydra et al., 2004). Identified QTLs were strain‐specific in all cases, explaining up to 61% of the resistance variance (reviewed by López & Bernal, 2012). Recently, thanks to a high‐density cassava genetic map (Soto et al., 2015), Soto‐Sedano and coworkers associated five novel resistance QTLs, accounting for 16%–22% of the variance phenotype and which colocalized with 29 genes potentially involved in defence (Soto‐Sedano et al., 2017). Furthermore, three of these QTLs were shown to be significantly influenced by environmental conditions, especially humidity. More recently, using an SSR‐based genetic map derived from the F1 offspring of an Asian cassava cultivar cross, 10 QTLs were associated to CBB resistance, explaining up to 26.5% of the genotypic variance, and five colocalized genes were shown to be differentially upregulated in resistant genotypes (Tappiban et al., 2018). Table S6 summarizes all the cassava QTLs (more than 100) reported to be involved in CBB resistance.
7.5. Genetic and molecular aspects of cassava defence against Xpm and Xc
Most of our knowledge about the molecular aspects of the cassava–Xpm interaction comes from studies of resistant varieties (Figure 4). However, challenging susceptible genotypes with Xpm also induces defence‐related genes, but expression is delayed when compared to resistant varieties (López Carrascal et al., 2005; Santaella et al., 2004). Several studies have identified pathogenesis‐related (PR) proteins (Li et al., 2017; Román et al., 2014; Yoodee et al., 2018) and various microRNA families potentially involved in plant defence (Pérez‐Quintero et al., 2012). The role of TF families in response to Xpm has been addressed by coexpression network analyses and functional studies by TF families. Specific cassava heat stress TFs (MeHSfs) (Wei et al., 2017; Wei, Liu, et al., 2018), related to ABI3 AND VP1 (MeRAVs) (Wei, Change, et al., 2018), Nuclear Factor Y (MeNF‐Ys) (He et al., 2019), Whirly (MeWHYs) (Liu et al., 2018), NAM/ATAF/CUC2 (NACs) (Gómez‐Cano et al., 2019), basic leucine zipper (bZIPs) (Gómez‐Cano et al., 2019; Li et al., 2017), WRKYs (Gómez‐Cano et al., 2019; Liu et al., 2018; Wei et al., 2017; Yan et al., 2017; Yoodee et al., 2018), and TB1/CIN)/PCF (TCPs) TFs (Gómez‐Cano et al., 2019) were shown to play important roles in cassava immunity against Xpm through activation of defences. For example, MeWHY1/2/3 (through interaction with MeWRKY75), MeNF‐YA1/3, MeNF‐YB11/16, and MeNF‐YC11/12 are crucial for upregulation of defences against Xpm (He et al., 2019; Liu et al., 2018).
Salicylic acid (SA), calcium signalling, and melatonin synthesis also play important roles in anti‐Xpm defence. The upregulation of MeHsf3 in response to Xpm infection triggers the activation of the SA pathway through transcriptional activation of MeEDS1, also triggering defence by activating the non‐SA‐related MePR4 gene (coding for a defence‐related protein containing an SCP domain) (Wei, Liu, et al., 2018). Several calcineurin B‐like proteins (CBLs) and CBL‐interacting protein kinases (MeCIPKs), which are, respectively transducers of CIPKs signalling and sensors of calcium signalling, have been found upregulated during the interaction. Among them, MeCIPK23 physically interacts with MeCBL1 and MeCBL9 to modulate defence against the pathogen, potentially as a response to calcium signalling (Yan et al., 2018). During infection Xpm also upregulates the melatonin biosynthesis pathway, leading to melatonin accumulation, activation of defence‐ and reactive oxygen species (ROS)‐related genes, and increased callose deposition (Wei et al., 2016). MeRAV1 and MeRAV2 TFs directly activate three genes of the melatonin biosynthesis pathway (MeTDC2, MeT5H, and MeASMT1), while MeWRKY79 and MeHsf20 increase the transcription rate of MeASMT2 (a second N‐acetylserotonin O‐methyltransferase gene involved in the melatonin biosynthesis pathway), all resulting in increased defence responses against Xpm (Wei et al., 2018, 2017, Wei, Change, et al., 2018).
Autophagy regulation is altered during Xpm infection, which significantly impacts immunity. Four members of the cassava autophagy‐related protein 8 (MeATG8) family, including MeATG8f which colocalizes with a resistance QTL (Tappiban et al., 2018) and whose product was reported to interact with MeWRKY20 to induce defence against Xpm (Yan et al., 2017). Furthermore, MeATG8b and Me8ATG8e have antagonistic roles against the two cassava glyceraldehyde‐3‐phosphate dehydrogenases MeGAPC4 and MeGAPC6, whose downregulation results in higher autophagic activity, higher H2O2 levels, and increased callose deposition (Zeng et al., 2018). Little attention has been paid to the cassava–Xc interaction, but analysis of an incompatible interaction showed that, unlike for Xpm, phenylalanine ammonia‐lyase (PAL) transcription and activity, and cell wall‐bound peroxidase activity are markedly increased in response to the pathogen (Pereira et al., 1999, 2000).
Pathogen perception relies on receptor‐like kinases (RLKs), receptor‐like proteins (RLPs), and nucleotide binding site‐leucine‐rich repeat (NBS‐LRRs) proteins. Cassava contains at least 253 RLK‐ (Soto et al., 2015) and 327 NBS‐LRR‐encoding genes. Most NBS‐LRRs are grouped in 39 clusters, with two superclusters with 43 and 19 of the genes located on chromosomes 16 and 17, respectively (Lozano et al., 2015). Mapping for resistance QTLs allowed the identification of the two resistance gene candidates RXam1 and RXam2, respectively, coding for an RLK and an NBS‐LRR (López et al., 2003). RXam1 shows similarity to the anti‐Xanthomonas oryzae pv. oryzae rice resistance gene Xa21, and was recently shown to be involved in defence against Xpm strain CIO136 (Díaz‐Tatis et al., 2018). RXam2 is a typical non‐TIR NB‐LRR located in a QTL explaining 61% of the resistance variance to Xpm strain CIO151. Recently, it was demonstrated that this gene confers partial and broad‐spectrum resistance to different Xpm strains (P. A. Díaz‐Tatis, personal communication).
7.6. Resistance engineering in cassava
Stable transformation of cassava is mainly achieved through bacterial‐mediated transformation of embryogenic calli and many transgenic cassava lines are reported in the literature (reviewed by Chavarriaga Aguirre et al., 2016). However, the incipient efforts to engineer CBB resistance are a reflection of the limited information on genetic determinants for resistance to Xpm. Stable transformants of the cassava cultivar 60444 overexpressing and silencing RXam1 were generated to study the function of this resistance gene candidate (Chavarriaga Aguirre et al., 2016; Díaz‐Tatis et al., 2018), as well as overexpressing the pepper R gene Bs2, which did not provide resistance against Xpm (Díaz‐Tatis et al., 2019).
Because TAL effectors recognize S genes through interaction with DNA, genome editing is a nice tool to disturb or favour the TAL–DNA interaction and induce resistance or prevent susceptibility (reviewed by Schornack et al., 2013). This approach was applied recently to help rice against X. oryzae pv. oryzae by editing five EBEs in the promoter of three SWEET genes targeted by X. oryzae pv. oryzae in elite varieties, resulting in a robust and broad‐spectrum resistance in the field (Oliva et al., 2019). This strategy could also be applied in cassava to engineer resistance against Xpm, given the requirement of MeSWEET10a for successful disease development.
8. CONCLUSION AND FUTURE DIRECTIONS
Whilst the application of measures based on the ecology of the pathogen has helped to mitigate the impact of CBB, Xpm remains a devastating pathogen causing important yield losses, but genetic resistance remains the best option to control the disease. To be effective and durable, breeding efforts must consider among other things the local diversity of the pathogen populations, the impact of other biotic and abiotic stresses, farming practices, and growers’ preferences. Although cassava–Xpm is considered an orphan pathosystem, research efforts lead by several institutions and researchers have resulted in robust knowledge and applied tools for either diagnosis or pathogen diversity assessment, which constitute important contributions to better understand some epidemiological aspects of the disease. Moreover, new image‐based techniques coupled to fluorescent or luminescent markers, the use of plants grown in vitro allowing for high‐throughput phenotyping, and third‐generation sequencing data will certainly help to progress genome‐wide association studies. Quantitative resistance studies have revealed that several genetic factors involved in resistance are still waiting to be more deeply characterized. On the other hand, functional analysis of Xpm TAL effectors has allowed a SWEET sugar transporter to be unmasked as a major susceptibility determinant. This opens a new perspective for CBB control by loss of susceptibility, as was successfully implemented in rice against bacterial leaf blight, resulting in plants with reduced susceptibility to X. oryzae pv. oryzae. More research is needed, however, to pinpoint the function of other major virulence TAL effectors and their targets. By exploiting this marked dependence, genome editing is a powerful tool to decrease susceptibility and improve resistance against this pathogen, but parallel efforts need to be made to understand the durability and farmers' acceptability of these strategies, taking into account the high diversity of the pathogen and its genomic repertoires. Further research should be deployed to better understand the lifestyle of Xc and also its phylogenetic relationship to Xpm. This could be an excellent opportunity and contribution to unveil the mechanisms that differentiate vascular from nonvascular diseases in plants.
Supporting information
TABLE S1 Incidence data extracted from several studies on cassava bacterial blight (CBB). Incidence was defined in these studies as the percentage of symptomatic plants (number of symptomatic plants × 100/number of evaluated plants) recorded in cassava fields that were naturally infected by Xanthomonas phaseoli pv. manihotis (Xpm) (no artificial inoculations). Incidence ranges (minimum and maximum), timepoint of the incidence evaluation, and country where the study was performed were extracted from each report. Min., minimum; Max., maximum; MAP, months after planting; nd, missing data
TABLE S2 Severity data extracted from several studies on cassava bacterial blight (CBB). Severity was measured in these studies using similar scales based on symptom recording in cassava fields that were naturally infected by Xanthomonas phaseoli pv. manihotis (Xpm) (no artificial inoculations). Severity ranges (minimum and maximum), timepoint of the severity evaluation, country where the study was performed, and grading scales were extracted from each report. The transformation column indicates if the values presented as minimum and maximum were mathematically transformed (see text after table) to obtain values of the same type and scale. Min., minimum; Max., maximum; MAP, months after planting; nd, missing data. Grading scales showed slight modifications, but all of them reflected the same damage extension with the corresponding grading numbers. Most of the scales were grouped within Scale A: 1, no disease symptoms; 2, only angular leaf spots on leaves; 3, leaf blight, leaf wilt, defoliation, gum exudation in petioles and stems; 4, extensive leaf blight, wilt, defoliation, and partial stem die‐back; 5, complete defoliation, stem die‐back, and stunting in most of the plants. Only one study used a similar grading system with different scale numbers, Scale B: 0, no symptoms; 1, symptoms on leaves only – blight; 2, presence of necrotic lesions on the stem or petiole; 3, most severe symptoms on leaves and/or the presence of necrotic lesions with gum exudation; 4, complete loss of leaves with apical death or death of the plant
TABLE S3 Losses data extracted from several studies on cassava bacterial blight (CBB). Only losses reported as the percentage of fresh root weight lost were considered. All the studies were carried out in cassava fields naturally or artificially infected by Xanthomonas phaseoli pv. manihotis (Xpm). Loss ranges (minimum and maximum), resistance status of the tested varieties, and country or region where the study was performed were extracted from each report. Min., minimum; Max., maximum
TABLE S4 Reference genomes used for phylogeny reconstruction. Phylogeny was constructed using the nucleotide concatenated sequence of five housekeeping genes (atpD, dnaK, efp, glnA, gyrB, and rpoD)
TABLE S5 Result compilation of cassava resistance assessments reported in literature
TABLE S6 Quantitative trait loci (QTLs) associated with cassava bacterial blight (CBB) resistance. QTL name, name of the QTL associated with CBB resistance; ExpVar, percentage of phenotypic variance explained by a given QTL; linkage group, linkage group where a given QTL can be found, linkage groups depend on the map used for associations; strain, strain used to inoculate the assessed plants (in most cases, QTLs are strain‐specific); natural inoculum, the study was performed in the field and the quantified disease resulted from infections with naturally occurring strains; undefined strain, the study states that only one strain was used to inoculate the assessed plants, but authors do not provide the strain identifier; technique, disease quantitation technique used to associate phenotypes with markers; AUDPC, area under the disease progression curve; 100‐DI, percentage of diseased plants (100‐disease incidence); MDR, mean disease rating; marker, peak marker associated with a given QTL; map reference, cassava map used to associate QTLs. The maps developed by Fregene et al. (1997) and Soto et al. (2015) derived from the female parental TMS 30572 and the male parental CM 2177–2; the map developed by Wydra et al. (2004) derived from five F1 male individuals (CM7857‐4, CM7857‐10, CM7857‐51, CM7857‐77, and CM7857‐115) and the female recurrent parent TMS30572; the map developed by Tappiban et al. (2018) derived from the parentals Huay Bong 60 (HB60) and Hanatee (HN). Study reference, the reference to the study where QTLs were found
ACKNOWLEDGEMENTS
The authors thank the Research Program on Roots, Tubers and Bananas, and the Agropolis Foundation (project #1403‐073) for funding support, and the Institut de Recherche pour le Développement for the doctoral fellowship awarded to C.A.Z.C. The authors acknowledge the IRD i‐Trop HPC (South Green Platform) for providing HPC resources that have contributed to the results reported within this paper (https://bioinfo.ird.fr/‐ http://www.southgreen.fr). The authors want to thank Ndomassi Tando and Julie Orjuela for their support in bioinformatics. Likewise, the authors are thankful to C. Bragard and H. Maraite (UCL, Belgium) for providing pictures of CBN.
Zárate‐Chaves CA, Gómez de la Cruz D, Verdier V, López CE, Bernal A, Szurek B. Cassava diseases caused by Xanthomonas phaseoli pv. manihotis and Xanthomonas cassavae . Mol Plant Pathol. 2021;22:1520–1537. 10.1111/mpp.13094
Contributor Information
Adriana Bernal, Email: abernal@uniandes.edu.co.
Boris Szurek, Email: boris.szurek@ird.fr.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Allem, A.C. (2002) The origins and taxonomy of cassava. In: Hillocks, R.J. , Thresh, J.M. & Bellotti, A. (Eds.) Cassava: Biology, Production and Utilization. Wallingford, UK: CABI Publishing, pp. 1–16. [Google Scholar]
- Ambe, J.T. (1993) The effect of planting dates on three cassava diseases in Cameroon. International Journal of Pest Management, 39, 309–311. [Google Scholar]
- Arrieta‐Ortiz, M.L. , Rodríguez‐R, L.M. , Pérez‐Quintero, Á.L. , Poulin, L. , Díaz, A.C. , Rojas, N.A. et al. (2013) Genomic survey of pathogenicity determinants and VNTR markers in the cassava bacterial pathogen Xanthomonas axonopodis pv. manihotis strain CIO151. PLoS One, 8, e79704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart, R. , Cohn, M. , Kassen, A. , McCallum, E.J. , Shybut, M. , Petriello, A. et al. (2012) High‐throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proceedings of the National Academy of Sciences of the United States of America, 109, E1972–E1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer‐Santos, E. , Ceseti, L. de M. , Farah, C.S. & Alvarez‐Martinez, C.E. (2019) Distribution, function and regulation of Type 6 secretion systems of Xanthomonadales . Frontiers in Microbiology, 10, 1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal‐Galeano, V. , Ochoa, J.C. , Trujillo, C. , Rache, L. , Bernal, A. & López Carrascal, C.E. (2018) Development of a multiplex nested PCR method for detection of Xanthomonas axonopodis pv. manihotis in cassava. Tropical Plant Pathology, 43, 341–350. [Google Scholar]
- Berthier, Y. , Verdier, V. , Guesdon, J.L. , Chevrier, D. , Denis, J.B. , Decoux, G. et al. (1993) Characterization of Xanthomonas campestris pathovars by rRNA gene restriction patterns. Applied and Environmental Microbiology, 59, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boher, B. , Kpemoua, K. , Nicole, M. , Luisetti, J. & Geiger, J.P. (1995) Ultrastructure of interactions between cassava and Xanthomonas campestris pv. manihotis: Cytochemistry of cellulose and pectin degradation in a susceptible cultivar. Phytopathology, 85, 777–788. [Google Scholar]
- Boher, B. , Nicole, M. , Potin, M. & Geiger, J.P. (1997) Extracellular polysaccharides from Xanthomonas axonopodis pv. manihotis interact with cassava cell walls during pathogenesis. Molecular Plant‐Microbe Interactions, 10, 803–811. [DOI] [PubMed] [Google Scholar]
- Bolot, S. , Munoz Bodnar, A. , Cunnac, S. , Ortiz, E. , Szurek, B. , Noel, L.D. et al. (2013) Draft genome sequence of the Xanthomonas cassavae type strain CFBP 4642. Genome Announcements, 1, e00679‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher, N.J. , Carpenter, S.C.D. , Sebra, R.P. , Wang, L. , Salzberg, S.L. , Leach, J.E. et al. (2015) Single molecule real‐time sequencing of Xanthomonas oryzae genomes reveals a dynamic structure and complex TAL (transcription activator‐like) effector gene relationships. Microbial Genomics, 1, e000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero, D. , Monk, J. , Rodríguez Cubillos, M.J. , Rodríguez Cubillos, A. , Restrepo, M. , Bernal‐Galeano, V. et al. (2020) Genome‐scale metabolic model of Xanthomonas phaseoli pv. manihotis: an approach to elucidate pathogenicity at the metabolic level. Frontiers in Genetics, 11, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredeson, J.V. , Lyons, J.B. , Prochnik, S.E. , Wu, G.A. , Ha, C.M. , Edsinger‐Gonzales, E. et al. (2016) Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nature Biotechnology, 34, 562–570. [DOI] [PubMed] [Google Scholar]
- Burkholder, W.H. (1942) Three bacterial plant pathogens: Phytomonas earyophylli sp.n., Phytomonas alliicola sp. n., and Phytomonas manihotis (Arthaud‐Berthet et Sondar) Viégas. Phytopathology, 32, 141–149. [Google Scholar]
- CABI . (2020) Xanthomonas axonopodis pv. manihotis (cassava bacterial blight). In: Invasive species compendium. Available at: https://www.cabi.org/isc/datasheet/56952. [Accessed November 2020]. [Google Scholar]
- Castiblanco, L.F. , Gil, J. , Rojas, A. , Osorio, D. , Gutiérrez, S. , Muñoz‐Bodnar, A. et al. (2013) TALE1 from Xanthomonas axonopodis pv. manihotis acts as a transcriptional activator in plant cells and is important for pathogenicity in cassava plants. Molecular Plant Pathology, 14, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira‐Melo, R.C. , Dórea‐Bragança, C.A. , Nogueira‐Pestana, K. , Alves da Silva, H.S. , Fortes‐Ferreira, C. & Oliveira, S.A. (2019) Improvement of the specific detection of Xanthomonas phaseoli pv. manihotis based on the pthB gene. Acta Scientiarum. Agronomy, 41, 1–9. [Google Scholar]
- Chavarriaga Aguirre, P. , Brand, A. , Medina, A. , Prías, M. , Escobar, R. , Martinez, J. et al. (2016) The potential of using biotechnology to improve cassava: A review. In Vitro Cellular & Developmental Biology – Plant, 52, 461–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chege, M. , Wamunyokoli, F. , Kamau, J. & Nyaboga, E. (2017) Phenotypic and genotypic diversity of Xanthomonas axonopodis pv. manihotis causing bacterial blight disease of cassava in Kenya. Journal of Applied Biology and Biotechnology, 5, 38–44. [Google Scholar]
- Chen, L.‐Q. (2014) SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytologist, 201, 1150–1155. [DOI] [PubMed] [Google Scholar]
- Cianciotto, N.P. & White, R.C. (2017) Expanding role of Type II secretion in bacterial pathogenesis and beyond. Infection and Immunity, 85, e00014–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, M. , Bart, R.S. , Shybut, M. , Dahlbeck, D. , Gomez, M. , Morbitzer, R. et al. (2014) Xanthomonas axonopodis virulence is promoted by a transcription activator‐like effector–mediated induction of a SWEET sugar transporter in cassava. Molecular Plant‐Microbe Interactions, 27, 1186–1198. [DOI] [PubMed] [Google Scholar]
- Cohn, M. , Morbitzer, R. , Lahaye, T. & Staskawicz, B.J. (2016) Comparison of gene activation by two TAL effectors from Xanthomonas axonopodis pv. manihotis reveals candidate host susceptibility genes in cassava. Molecular Plant Pathology, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, M. , Shybut, M. , Dahlbeck, D. & Staskawicz, B. (2015) Assays to assess virulence of Xanthomonas axonopodis pv. manihotis on cassava. Bio‐Protocols, 5, e1522. [Google Scholar]
- Constantin, E.C. , Cleenwerck, I. , Maes, M. , Baeyen, S. , Malderghem, C.V. , Vos, P.D. et al. (2016) Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathology, 65, 792–806. [Google Scholar]
- Cooper, R.M. , Kemp, B. , Day, R. , Gomez‐Vasquez, R. & Beeching, J.R. (2001) Pathogenicity and resistance in xanthomonas blight of cassava. In: de Boer, S.H. (Ed.) Plant pathogenic bacteria. Dordrecht: Springer, pp. 319–323. [Google Scholar]
- Cox, K.L. , Meng, F. , Wilkins, K.E. , Li, F. , Wang, P. , Booher, N.J. et al. (2017) TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nature Communications, 8, 15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, J.‐F. & Boher, B. (1985a) Epiphytic phase of Xanthomonas campestris pathovar manihotis on aerial parts of cassava. Agronomie, 5, 111–116. [Google Scholar]
- Daniel, J.‐F. & Boher, B. (1985b) Etude des modes de survie de l’agent causal de la bacteriose vasculaire du manioc, Xanthomonas campestris pathovar manihotis . Agronomie, 5, 339–346. [Google Scholar]
- Dedal, O.I. , Palomar, M.K. & Napiere, C.M. (1980) Host range of Xanthomonas manihotis Starr. Annals of Tropical Research, 2, 149–155. [Google Scholar]
- Delepelaire, P. (2004) Type I secretion in gram‐negative bacteria. Biochimica et Biophysica Acta, 1694, 149–161. [DOI] [PubMed] [Google Scholar]
- Denancé, N. , Szurek, B. , Doyle, E.L. , Lauber, E. , Fontaine‐Bodin, L. , Carrère, S. et al. (2018) Two ancestral genes shaped the Xanthomonas campestris TAL effector gene repertoire. New Phytologist, 219, 391–407. [DOI] [PubMed] [Google Scholar]
- Díaz‐Tatis, P.A. , Herrera Corzo, M. , Ochoa Cabezas, J.C. , Medina Cipagauta, A. , Prías, M.A. , Verdier, V. et al. (2018) The overexpression of RXam1, a cassava gene coding for an RLK, confers disease resistance to Xanthomonas axonopodis pv. manihotis . Planta, 247, 1031–1042. [DOI] [PubMed] [Google Scholar]
- Díaz‐Tatis, P.A. , Ochoa, J.C. , García, L. , Chavarriaga, P. , Bernal, A.J. & López Carrascal, C.E. (2019) Interfamily transfer of Bs2 from pepper to cassava (Manihot esculenta Crantz). Tropical Plant Pathology, 44, 225–237. [Google Scholar]
- Dixon, A.G.O. , Ngeve, J.M. & Nukenine, E.N. (2002) Genotype × environment effects on severity of cassava bacterial blight disease caused by Xanthomonas axonopodis pv. manihotis . European Journal of Plant Pathology, 108, 763–770. [Google Scholar]
- Elango, F. & Lozano, J.C. (1980) Transmission of Xanthomonas manihotis in seed of cassava (Manihot esculenta). Plant Disease, 64, 784–786. [Google Scholar]
- Elango, F. & Lozano, J.C. (1981) Epiphytic survival of Xanthomonas manihotis on common weeds in Colombia. In: 5th Int. Conf. Plant Path. Bact. Cali, Colombia, pp. 203–209.
- Ewbank, E. & Maraite, H. (1990) Conversion of methionine to phytotoxic 3‐methylthiopropionic acid by Xanthomonas campestris pv. manihotis . Microbiology, 136, 1185–1189. [Google Scholar]
- Fanou, A. , Zinsou, V. & Wydra, K. (2017) Survival of Xanthomonas axonopodis pv. manihotis in weed species and in cassava debris: implication in the epidemiology of cassava bacterial blight. International Journal of Advanced Research, 5, 2098–2112. [Google Scholar]
- FAO . (2020) FAO.STAT crops. Available at: http://www.fao.org/faostat/en/#data/QC. [Accessed November 2020]. [Google Scholar]
- Fermont, A.M. , van Asten, P.J.A. , Tittonell, P. , van Wijk, M.T. & Giller, K.E. (2009) Closing the cassava yield gap: an analysis from smallholder farms in East Africa. Field Crops Research, 112, 24–36. [Google Scholar]
- Flood, J. , Cooper, R.M. , Deshappriya, N. & Day, R.C. (1995) Resistance of cassava (Manihot esculenta) to Xanthomonas blight in vitro and in planta. Aspects of Applied Biology, 42, 277–284. [Google Scholar]
- Flores, C. , Zarate, C. , Triplett, L. , Maillot‐Lebon, V. , Moufid, Y. , Kanté, M. et al. (2019) Development of a duplex‐PCR for differential diagnosis of Xanthomonas phaseoli pv. manihotis and Xanthomonas cassavae in cassava (Manihot esculenta). Physiological and Molecular Plant Pathology, 105, 34–46. [Google Scholar]
- Fokunang, C.N. , Akem, C.N. , Dixon, A.G.O. & Ikotun, T. (2000) Evaluation of a cassava germplasm collection for reaction to three major diseases and the effect on yield. Genetic Resources and Crop Evolution, 47, 63–71. [Google Scholar]
- Fregene, M. , Angel, F. , Gomez, R. , Rodriguez, F. , Chavarriaga, P. , Roca, W. et al. (1997) A molecular genetic map of cassava (Manihot esculenta Crantz). Theoretical and Applied Genetics, 95, 431–441. [Google Scholar]
- Frison, E. & Feliu, E. (1991) FAO/IBPGR technical guidelines for the safe movement of cassava germplasm. Rome, Italy: FAO/IBPGR. [Google Scholar]
- Gochez, A.M. , Huguet‐Tapia, J.C. , Minsavage, G.V. , Shantaraj, D. , Jalan, N. , Strauß, A. et al. (2018) Pacbio sequencing of copper‐tolerant Xanthomonas citri reveals presence of a chimeric plasmid structure and provides insights into reassortment and shuffling of transcription activator‐like effectors among X. citri strains. BMC Genomics, 19, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Cano, F. , Soto, J. , Restrepo, S. , Bernal, A. , López‐Kleine, L. & López Carrascal, C.E. (2019) Gene co‐expression network for Xanthomonas‐challenged cassava reveals key regulatory elements of immunity processes. European Journal of Plant Pathology, 153, 1083–1104. [Google Scholar]
- Gonzalez, C. , Restrepo, S. , Tohme, J. & Verdier, V. (2002) Characterization of pathogenic and nonpathogenic strains of Xanthomonas axonopodis pv. manihotis by PCR‐based DNA fingerprinting techniques. FEMS Microbiology Letters, 215, 23–31. [DOI] [PubMed] [Google Scholar]
- Harris, K.P. , Martin, A. , Novak, S. , Kim, S.‐H. , Reynolds, T. & Leigh, A. (2015) Cassava bacterial blight and postharvest physiological deterioration, production losses and control strategies. Available at: https://epar.evans.uw.edu/sites/default/files/EPAR_UW_Request_298_CBB%20%26%20PPD%20Production%20Losses%20and%20Control%20Strategies_3.23.15_0.pdf. [Accessed November 2020]. [Google Scholar]
- Hayward, A.C. (1993) The hosts of Xanthomonas . In: Swings, J.G. & Civerolo, E.L. (Eds.) Xanthomonas. Dordrecht: Springer, pp. 1–119. [Google Scholar]
- He, X. , Liu, G. , Li, B. , Xie, Y. , Wei, Y. , Shang, S. et al. (2019) Functional analysis of the heterotrimeric NF‐Y transcription factor complex in cassava disease resistance. Annals of Botany, 124, 1185–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillocks, R.J. & Wydra, K. (2002) Bacterial, fungal and nematode diseases. In: Hillocks, R.J. , Thresh, J.M. & Bellotti, A.C. (Eds.), Cassava: Biology, Production and Utilization. Wallingford, UK: CABI Publishing. pp. 261–280. [Google Scholar]
- International Cassava Genetic Map Consortium (ICGMC) (2015) High‐resolution linkage map and chromosome‐scale genome assembly for cassava (Manihot esculenta Crantz) from 10 populations. G3 Genes|Genomes|Genetics, 5, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, W. (1959) Manioc in Africa. Stanford, CA, USA: Stanford University Press. [Google Scholar]
- Jorge, V. , Fregene, M. , Duque, M.C. , Bonierbale, M.W. , Tohme, J. & Verdier, V. (2000) Genetic mapping of resistance to bacterial blight disease in cassava (Manihot esculenta Crantz). Theoretical and Applied Genetics, 101, 865–872. [Google Scholar]
- Jorge, V. , Fregene, M. , Vélez, C.M. , Duque, M.C. , Tohme, J. & Verdier, V. (2001) QTL analysis of field resistance to Xanthomonas axonopodis pv. manihotis in cassava. Theoretical and Applied Genetics, 102, 564–571. [Google Scholar]
- Jorge, V. & Verdier, V. (2002) Qualitative and quantitative evaluation of cassava bacterial blight resistance in F1 progeny of a cross between elite cassava clones. Euphytica, 123, 41–48. [Google Scholar]
- Kasmi, F.E. , Horvath, D. & Lahaye, T. (2018) Microbial effectors and the role of water and sugar in the infection battle ground. Current Opinion in Plant Biology, 44, 98–107. [DOI] [PubMed] [Google Scholar]
- Kemp, B.P. , Horne, J. , Bryant, A. & Cooper, R.M. (2004) Xanthomonas axonopodis pv. manihotis gumD gene is essential for EPS production and pathogenicity and enhances epiphytic survival on cassava (Manihot esculenta). Physiological and Molecular Plant Pathology, 64, 209–218. [Google Scholar]
- Kpémoua, K. , Boher, B. , Nicole, M. , Calatayud, P. & Geiger, J.P. (1996) Cytochemistry of defense responses in cassava infected by Xanthomonas campestris pv. manihotis . Canadian Journal of Microbiology, 42, 1131–1143. [Google Scholar]
- Kuon, J.‐E. , Qi, W. , Schläpfer, P. , Hirsch‐Hoffmann, M. , von Bieberstein, P.R. , Patrignani, A. et al. (2019) Haplotype‐resolved genomes of geminivirus‐resistant and geminivirus‐susceptible African cassava cultivars. BMC Biology, 17, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamptey, J.N.L. , Okoli, O.O. & Frimpong‐Manso, P.P. (1998) Incidence and severity of African cassava mosaic disease (ACMD) and cassava bacterial blight (CBB) on some local and exotic cassava varieties in different ecological zones of Ghana. Ghana Journal of Agricultural Science, 31, 35–43. [Google Scholar]
- Li, C. , Ji, C. , Huguet‐Tapia, J.C. , White, F.F. , Dong, H. & Yang, B. (2019) An efficient method to clone TAL effector genes from Xanthomonas oryzae using Gibson assembly. Molecular Plant Pathology, 20, 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Fan, S. , Hu, W. , Liu, G. , Wei, Y. , He, C. et al. (2017) Two cassava basic leucine zipper (bZIP) transcription factors (MebZIP3 and MebZIP5) confer disease resistance against cassava bacterial blight. Frontiers in Plant Science, 8, 2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Yan, Y. , Zeng, H. , Li, X. , Wei, Y. , Liu, G. et al. (2018) Functional characterization of WHY–WRKY75 transcriptional module in plant response to cassava bacterial blight. Tree Physiology, 38, 1502–1512. [DOI] [PubMed] [Google Scholar]
- López Carrascal, C.E. & Bernal, A.J. (2012) Cassava bacterial blight: Using genomics for the elucidation and management of an old problem. Tropical Plant Biology, 5, 117–126. [Google Scholar]
- López Carrascal, C.E. , Jorge, V. , Piégu, B. , Mba, C. , Cortes, D. & Restrepo, S. et al. (2004) A unigene catalogue of 5700 expressed genes in cassava. Plant Molecular Biology, 56, 541–554. [DOI] [PubMed] [Google Scholar]
- López Carrascal, C.E. , Quesada‐Ocampo, L.M. , Bohórquez, A. , Duque, M.C. , Vargas, J. , Tohme, J. et al. (2007) Mapping EST‐derived SSRs and ESTs involved in resistance to bacterial blight in Manihot esculenta . Genome, 50, 1078–1088. [DOI] [PubMed] [Google Scholar]
- López Carrascal, C.E. , Soto, M. , Restrepo, S. , Piégu, B. , Cooke, R. , Delseny, M. et al. (2005) Gene expression profile in response to Xanthomonas axonopodis pv. manihotis infection in cassava using a cDNA microarray. Plant Molecular Biology, 57, 393–410. [DOI] [PubMed] [Google Scholar]
- López Carrascal, C.E. , Zuluaga, A.P. , Cooke, R. , Delseny, M. , Tohme, J. & Verdier, V. (2003) Isolation of resistance gene candidates (RGCs) and characterization of an RGC cluster in cassava. Molecular Genetics and Genomics, 269, 658–671. [DOI] [PubMed] [Google Scholar]
- Lozano, J.C. (1986) Cassava bacterial blight: a manageable disease. Plant Disease, 70, 1089. [Google Scholar]
- Lozano, J.C. & Booth, R.H. (1974) Diseases of cassava (Manihot esculenta Crantz). PANS Pest Articles & News Summaries, 20, 30–54. [Google Scholar]
- Lozano, J.C. & Laberry, R. (1982) Screening for resistance to cassava bacterial blight. Plant Disease, 66, 316–318. [Google Scholar]
- Lozano, J.C. & Sequeira, L. (1974a) Bacterial blight of cassava in Colombia: Epidemiology and control. Phytopathology, 64, 83–88. [Google Scholar]
- Lozano, J.C. & Sequeira, L. (1974b) Bacterial blight of cassava in Colombia: Etiology. Phytopathology, 64, 74–82. [Google Scholar]
- Lozano, R. , Hamblin, M.T. , Prochnik, S. & Jannink, J.‐L. (2015) Identification and distribution of the NBS‐LRR gene family in the cassava genome. BMC Genomics, 16, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. et al. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology, 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraite, H. (1993) Xanthomonas campestris pathovars on cassava: cause of bacterial blight and bacterial necrosis. In: Swings, J.G. and Civerolo, E.L. (Eds.) Xanthomonas. London, UK: Springer Science + Business Media Dordrecht, pp. 18–25. [Google Scholar]
- Maraite, H. & Meyer, J. (1975) Xanthomonas manihotis (Arthaud‐Berthet) Starr, causal agent of bacterial wilt, blight and leaf spots of cassava in Zaire. International Journal of Pest Management, 21, 27–37. [Google Scholar]
- Maraite, H. & Perreaux, D. (1979) Comparative symptom development in cassava after infection by Xanthomonas manihotis or X. cassavae under controlled conditions. In: Terry, E.R. , Persley, G.J. and Cook, S.C.A. (Eds.) Cassava Bacterial Blight in Africa; Past, Present and Future. Report of an Interdisciplinary Workshop, IITA, Ibadan, Nigeria, 1978. London, UK: Centre for Overseas Pest Research, pp. 17–24. [Google Scholar]
- Marcano, M. & Trujillo, G. (1984) Papel de las malezas en relacion a la perpetuacion del anublo bacteria no de la yuca. Revista de la Facultad de Agronomia, 13, 167–181. [Google Scholar]
- Mbaringong, G. , Nyaboga, E. , Wang’ondu, V. & Kanduma, E. (2017) Evaluation of selected cassava (Manihot esculenta Crantz) cultivars grown in Kenya for resistance to bacterial blight disease. World Journal of Agricultural Research, 5, 94–101. [Google Scholar]
- Medina, C.A. , Reyes, P.A. , Trujillo, C.A. , Gonzalez, J.L. , Bejarano, D.A. , Montenegro, N.A. et al. (2018) The role of type III effectors from Xanthomonas axonopodis pv. manihotis in virulence and suppression of plant immunity. Molecular Plant Pathology, 19, 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midha, S. & Patil, P.B. (2014) Genomic insights into the evolutionary origin of Xanthomonas axonopodis pv. citri and its ecological relatives. Applied and Environmental Microbiology, 80, 6266–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora, R. , Rodriguez, M. , Gayosso, L. & López Carrascal, C.E. (2019) Using in vitro plants to study the cassava response to Xanthomonas phaseoli pv. manihotis infection. Tropical Plant Pathology, 44, 423–429. [Google Scholar]
- Mostade, J.M. & Butare, I. (1979) Symptomatologie et epidemiologie de la necrose bacterienne du manioc causée par Xanthomonas cassavae au Rwanda. In: Maraite, H. and Meyer, J.A. (Eds.) Disease of Tropical Food Crops. Proceedings of an International Symposium. Louvain‐la‐Neuve, Belgium: Universite Catholique de Louvain, pp. 95–117. [Google Scholar]
- Muñoz‐Bodnar, A. , Cruz‐Gómez, L.M. , Bernal, A. , Szurek, B. & López Carrascal, C.E. (2015) Comparing inoculation methods to evaluate the growth of Xanthomonas axonopodis pv. manihotis on cassava plants. Acta Biológica Colombiana, 20, 47–55. [Google Scholar]
- Muñoz‐Bodnar, A. , Perez‐Quintero, A.L. , Gomez‐Cano, F. , Gil, J. , Michelmore, R. , Bernal, A. et al. (2014) RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non‐pathogenic strains of Xanthomonas axonopodis pv. manihotis . Plant Cell Reports, 33, 1901–1912. [DOI] [PubMed] [Google Scholar]
- Mutka, A.M. , Fentress, S.J. , Sher, J.W. , Berry, J.C. , Pretz, C. , Nusinow, D.A. , et al. (2016) Quantitative, image‐based phenotyping methods provide insight into spatial and temporal dimensions of plant disease. Plant Physiology, 172, 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar, N.M.A. & Ortiz, R. (2007) Cassava improvement: challenges and impacts. Journal of Agricultural Science, 145, 163. [Google Scholar]
- Ogunjobi, A. , Dixon, A. & Fagade, O. (2007) Molecular genetic study of cassava bacterial blight casual agent in Nigeria using random amplified polymorphic DNA. Electronic Journal of Environmental, Agricultural and Food Chemistry, 6, 2364–2376. [Google Scholar]
- Ogunjobi, A. , Fagade, O. & Dixon, A. (2006) Molecular variation in population structure of Xanthomonas axonopodis pv. manihotis in the south eastern Nigeria. African Journal of Biotechnology, 5, 1868–1872. [Google Scholar]
- Ogunjobi, A.A. , Fagade, O.E. & Dixon, A.G.O. (2010) Comparative analysis of genetic variation among Xanthomonas axonopodis pv. manihotis isolated from the western states of Nigeria using RAPD and AFLP. Indian Journal of Microbiology, 50, 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda, S. & Verdier, V. (2000) Detecting Xanthomonas axonopodis pv. manihotis in cassava true seeds by nested polymerase chain reaction assay. Canadian Journal of Plant Pathology, 22, 241–247. [Google Scholar]
- Oliva, R. , Ji, C. , Atienza‐Grande, G. , Huguet‐Tapia, J.C. , Perez‐Quintero, A. , Li, T. et al. (2019) Broad‐spectrum resistance to bacterial blight in rice using genome editing. Nature Biotechnology, 37, 1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K.M. & Schaal, B.A. (1999) Evidence on the origin of cassava: Phylogeography of Manihot esculenta . Proceedings of the National Academy of Sciences of the United States of America, 96, 5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacumbaba, R.P. (1987) A screening method for detecting resistance against cassava bacterial blight disease. Journal of Phytopathology, 119, 1–6. [Google Scholar]
- Peng, Z. , Hu, Y. , Xie, J. , Potnis, N. , Akhunova, A. , Jones, J. et al. (2016) Long read and single molecule DNA sequencing simplifies genome assembly and TAL effector gene analysis of Xanthomonas translucens . BMC Genomics, 17, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, L.F. , Goodwin, P.H. & Erickson, L. (1999) The role of a phenylalanine ammonia lyase gene during cassava bacterial blight and cassava bacterial necrosis. Journal of Plant Research, 112, 51–60. [Google Scholar]
- Pereira, L.F. , Goodwin, P.H. & Erickson, L. (2000) Peroxidase activity during susceptible and resistant interactions between cassava (Manihot esculenta) and Xanthomonas axonopodis pv. manihotis and Xanthomonas cassavae . Journal of Phytopathology, 148, 575–578. [Google Scholar]
- Pérez‐Quintero, A.L. , Lamy, L. , Zarate, C.A. , Cunnac, S. , Doyle, E. , Bogdanove, A. et al. (2017) daTALbase: a database for genomic and transcriptomic data related to TAL effectors. Molecular Plant‐Microbe Interactions, 31, 471–480. [DOI] [PubMed] [Google Scholar]
- Pérez‐Quintero, Á.L. , Quintero, A. , Urrego, O. , Vanegas, P. & López Carrascal, C.E. (2012) Bioinformatic identification of cassava miRNAs differentially expressed in response to infection by Xanthomonas axonopodis pv. manihotis . BMC Plant Biology, 12, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Quintero, A.L. & Szurek, B. (2019) A decade decoded: spies and hackers in the history of TAL effectors research. Annual Review of Phytopathology, 57, 459–481. [DOI] [PubMed] [Google Scholar]
- Perreaux, D. , Maraite, H. & Meyer, J.A. (1982) Identification of 3(methylthio)‐propionic acid as a blight‐inducing toxin produced by Xanthomonas campestris pv. manihotis in vitro. Physiological Plant Pathology, 20, 313–319. [Google Scholar]
- Perreaux, D. , Maraite, H. & Meyer, J.A. (1986) Detection of 3(methylthio)‐propionic acid in cassava leaves infected by Xanthomonas campestris pv. manihotis . Physiological and Molecular Plant Pathology, 28, 323–328. [Google Scholar]
- Persley, G.J. (1976) Distribution and importance of cassava bacterial blight in Africa. In: Persley, J.G. , Terry, E.R. & MacIntyre, R. (Eds). Cassava bacterial blight, report on an interdisciplinary workshop. Ottawa, Canada: International Development Research Centre, pp. 9–14. [Google Scholar]
- Persley, G.J. (1978) Studies on the epidemiology and ecology of cassava bacterial bligth. In: Terry, E.R. , Persley, G.J. & Cook, S.C.A. (Eds.) Workshop on cassava bacterial blight in Africa, past, present and future. Ibadan, Nigeria: Centre for Overseas Pest Research, pp. 5–7. [Google Scholar]
- Prochnik, S. , Marri, P.R. , Desany, B. , Rabinowicz, P.D. , Kodira, C. , Mohiuddin, M. et al. (2012) The cassava genome: current progress, future directions. Tropical Plant Biology, 5, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbi, I. , Hamblin, M. , Gedil, M. , Kulakow, P. , Ferguson, M. , Ikpan, A.S. et al. (2014) Genetic mapping using genotyping‐by‐sequencing in the clonally propagated cassava. Crop Science, 54, 1384–1396. [Google Scholar]
- Rabbi, I.Y. , Hamblin, M.T. , Kumar, P.L. , Gedil, M.A. , Ikpan, A.S. , Jannink, J.‐L. et al. (2014) High‐resolution mapping of resistance to cassava mosaic geminiviruses in cassava using genotyping‐by‐sequencing and its implications for breeding. Virus Research, 186, 87–96. [DOI] [PubMed] [Google Scholar]
- Rache, L. , Blondin, L. , Flores, C. , Trujillo, C. , Szurek, B. , Restrepo, S. et al. (2019) An optimized microsatellite scheme for assessing populations of Xanthomonas phaseoli pv. manihotis . Phytopathology, 109, 859–869. [DOI] [PubMed] [Google Scholar]
- Restrepo, S. , Duque, M.C. & Verdier, V. (2000a) Characterization of pathotypes among isolates of Xanthomonas axonopodis pv. manihotis in Colombia. Plant Pathology, 49, 680–687. [Google Scholar]
- Restrepo, S. , Duque, M.C. & Verdier, V. (2000b) Resistance spectrum of selected Manihot esculenta genotypes under field conditions. Field Crops Research, 65, 69–77. [Google Scholar]
- Restrepo, S. , Vélez, C.M. & Verdier, V. (2000c) Measuring the genetic diversity of Xanthomonas axonopodis pv. manihotis within different fields in Colombia. Phytopathology, 90, 683–690. [DOI] [PubMed] [Google Scholar]
- Restrepo, S. , Velez, C.M. , Duque, M.C. & Verdier, V. (2004) Genetic structure and population dynamics of Xanthomonas axonopodis pv. manihotis in Colombia from 1995 to 1999. Applied and Environmental Microbiology, 70, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, S. & Verdier, V. (1997) Geographical differentiation of the population of Xanthomonas axonopodis pv. manihotis in Colombia. Applied and Environmental Microbiology, 63, 4427–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo Rubio, J.S. , López Carrascal, C.E. & Melgarejo, L.M. (2017) Physiological behavior of cassava plants (Manihot esculenta Crantz) in response to infection by Xanthomonas axonopodis pv. manihotis under greenhouse conditions. Physiological and Molecular Plant Pathology, 100, 136–141. [Google Scholar]
- Román, V. , Bossa‐Castro, A.M. , Vásquez, A.X. , Bernal‐Galeano, V. , Schuster, M. , Bernal, N. et al. (2014) Construction of a cassava PR protein‐interacting network during Xanthomonas axonopodis pv. manihotis infection. Plant Pathology, 63, 792–802. [Google Scholar]
- Ruh, M. , Briand, M. , Bonneau, S. , Jacques, M.‐A. & Chen, N.W.G. (2017) Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors. BMC Genomics, 18, 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaella, M. , Suárez, E. , López Carrascal, C.E. , González, C. , Mosquera, G. , Restrepo, S. et al. (2004) Identification of genes in cassava that are differentially expressed during infection with Xanthomonas axonopodis pv. manihotis . Molecular Plant Pathology, 5, 549–558. [DOI] [PubMed] [Google Scholar]
- Schornack, S. , Moscou, M.J. , Ward, E.R. & Horvath, D.M. (2013) Engineering plant disease resistance based on TAL effectors. Annual Review of Phytopathology, 51, 383–406. [DOI] [PubMed] [Google Scholar]
- Schwartz, A.R. , Morbitzer, R. , Lahaye, T. & Staskawicz, B.J. (2017) TALE‐induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proceedings of the National Academy of Sciences of the United States of America, 114, E897–E903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgro, G.G. , Oka, G.U. , Souza, D.P. , Cenens, W. , Bayer‐Santos, E. , Matsuyama, B.Y. et al. (2019) Bacteria‐killing Type IV secretion systems. Frontiers in Microbiology, 10, 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya, M. , Ahmed, S.A. , Davenport, K.W. , Flynn, M.C. , Lo, C.C. & Chain, P.S.G. (2020) Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life. Scientific Reports, 10, 1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showmaker, K.C. , Arick, M.A. , Hsu, C.‐Y. , Martin, B.E. , Wang, X. , Jia, J. et al. (2017) The genome of the cotton bacterial blight pathogen Xanthomonas citri pv. malvacearum strain MSCT1. Standards in Genomic Sciences, 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto, J.C. , Ortiz, J.F. , Perlaza‐Jiménez, L. , Vásquez, A.X. , Lopez‐Lavalle, L.A.B. , Mathew, B. et al. (2015) A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity‐related genes. BMC Genomics, 16, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto‐Sedano, J.C. , Mora‐Moreno, R.E. , Mathew, B. , Léon, J. , Gómez‐Cano, F.A. , Ballvora, A. et al. (2017) Major novel QTL for resistance to cassava bacterial blight identified through a multi‐environmental analysis. Frontiers in Plant Science, 8, 1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel, J. , Pesce, C. , Hutin, M. , Koebnik, R. , Boch, J. & Szurek, B. (2013) Five phylogenetically close rice SWEET genes confer TAL effector‐mediated susceptibility to Xanthomonas oryzae pv. oryzae . New Phytologist, 200, 808–819. [DOI] [PubMed] [Google Scholar]
- Tappiban, P. , Sraphet, S. , Srisawad, N. , Smith, D.R. & Triwitayakorn, K. (2018) Identification and expression of genes in response to cassava bacterial blight infection. Journal of Applied Genetics, 59, 391–403. [DOI] [PubMed] [Google Scholar]
- Taylor, R.K. , Griffin, R.L. , Jones, L.M. , Pease, B. , Tsatsia, F. , Fanai, C. et al. (2017) First record of Xanthomonas axonopodis pv. manihotis in Solomon Islands. Australasian Plant Disease Notes, 12, 49. [Google Scholar]
- Timilsina, S. , Potnis, N. , Newberry, E.A. , Liyanapathiranage, P. , Iruegas‐Bocardo, F. , White, F.F. et al. (2020) Xanthomonas diversity, virulence and plant–pathogen interactions. Nature Reviews Microbiology, 18, 415–427. [DOI] [PubMed] [Google Scholar]
- Tomkins, J. , Fregene, M. , Main, D. , Kim, H. , Wing, R. & Tohme, J. (2004) Bacterial artificial chromosome (BAC) library resource for positional cloning of pest and disease resistance genes in cassava (Manihot esculenta Crantz). Plant Molecular Biology, 56, 555–561. [DOI] [PubMed] [Google Scholar]
- Tran, T.T. , Pérez‐Quintero, A.L. , Wonni, I. , Carpenter, S.C. , Yu, Y. , Wang, L. et al. (2018) Functional analysis of African Xanthomonas oryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice. PLoS Pathogens, 14, e1007092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo, C. , Arias‐Rojas, N. , Poulin, L. , Medina, C. , Tapiero, A. , Restrepo, S. et al. (2014) Population typing of the causal agent of cassava bacterial blight in the Eastern Plains of Colombia using two types of molecular markers. BMC Microbiology, 14, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo, C. , Ochoa, J.C. , Mideros, M.F. , Restrepo, S. , López Carrascal, C.E. & Bernal, A. (2014) A complex population structure of the cassava pathogen Xanthomonas axonopodis pv. manihotis in recent years in the Caribbean region of Colombia. Microbial Ecology, 68, 155–167. [DOI] [PubMed] [Google Scholar]
- Umanah, E.E. (1977) Cassava research in Nigeria before 1972. In: Cock, J. , MacIntyre, R. & Graham, M. (Eds.) Fourth tropical root crops symposium, Ottawa, Canada: International Development Research Centre, pp. 137–141. [Google Scholar]
- Umemura, Y. & Kawano, K. (1983) Field assessment and inheritance of resistance to cassava bacterial blight 1. Crop Science, 23, 1127–1132. [Google Scholar]
- Van Den Mooter, M. , Maraite, H. , Meiresonne, L. , Swings, J. , Gillis, M. , Kersters, K. et al. (1987) Comparison between Xanthomonas campestris pv. manihotis (ISPP List 1980) and X. campestris pv. cassavae (ISPP List 1980) by means of phenotypic, protein electrophoretic, DNA hybridization and phytopathological techniques. Microbiology, 133, 57–71. [Google Scholar]
- Vauterin, G. , Hoste, B. , Kersters, K. & Swings, J. (1995) Reclassification of Xanthomonas. International Journal of Systematic Bacteriology, 45, 472–489. [Google Scholar]
- Veley, K.M. , Okwuonu, I. , Jensen, G. , Yoder, M. , Taylor, N.J. , Meyers, B.C. & Bart R.S. (2021) Gene tagging via CRISPR‐mediated homology‐directed repair in cassava. G3 Genes|Genomes|Genetics, 11, jkab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier, V. , Boher, B. , Maraite, H. & Geiger, J.‐P. (1994) Pathological and molecular characterization of Xanthomonas campestris strains causing diseases of cassava (Manihot esculenta). Applied and Environmental Microbiology, 60, 4478–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier, V. , Dongo, P. & Boher, B. (1993) Assessment of genetic diversity among strains of Xanthomonas campestris pv. manihotis . Journal of General Microbiology, 139, 2591–2601. [Google Scholar]
- Verdier, V. & Mosquera, G. (1999) Specific detection of Xanthomonas axonopodis pv. manihotis with a DNA hybridization probe. Journal of Phytopathology, 147, 417–423. [Google Scholar]
- Verdier, V. , Mosquera, G. & Assigbétsé, K. (1998) Detection of the cassava bacterial blight pathogen, Xanthomonas axonopodis pv. manihotis, by polymerase chain reaction. Plant Disease, 82, 79–83. [DOI] [PubMed] [Google Scholar]
- Verdier, V. , Ojeda, S. & Mosquera, G. (2001) Methods for detecting the cassava bacterial blight pathogen: A practical approach for managing the disease. Euphytica, 120, 103–107. [Google Scholar]
- Verdier, V. , Restrepo, S. , Mosquera, G. , Duque, M.C. , Gerstl, A. & Laberry, R. (2001) Genetic and pathogenic variation of Xanthomonas axonopodis pv. manihotis in Venezuela. Plant Pathology, 47, 601–608. [Google Scholar]
- Verdier, V. , Restrepo, S. , Mosquera, G. , Jorge, V. & López Carrascal, C.E. (2004) Recent progress in the characterization of molecular determinants in the Xanthomonas axonopodis pv. manihotis–cassava interaction. Plant Molecular Biology, 56, 573–584. [DOI] [PubMed] [Google Scholar]
- Verdier, V. , Schmit, J. & Lemattre, M. (1990) Étude en microscopie électronique à balayage de l’installation de deux souches de Xanthomonas campestris pv. manihotis sur feuilles de vitroplants de manioc. Agronomie, 10, 93–102. [Google Scholar]
- Wang, W. , Feng, B. , Xiao, J. , Xia, Z. , Zhou, X. , Li, P. et al. (2014) Cassava genome from a wild ancestor to cultivated varieties. Nature Communications, 5, 5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y. , Chang, Y. , Zeng, H. , Liu, G. , He, C. & Shi, H. (2018) RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. Journal of Pineal Research, 64, e12454. [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Liu, G. , Chang, Y. , He, C. & Shi, H. (2018) Heat shock transcription factor 3 regulates plant immune response through modulation of salicylic acid accumulation and signaling in cassava. Molecular Plant Pathology, 19, 2209–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y. , Hu, W. , Wang, Q. , Liu, W. , Wu, C. , Zeng, H. et al. (2016) Comprehensive transcriptional and functional analyses of melatonin synthesis genes in cassava reveal their novel role in hypersensitive‐like cell death. Scientific Reports, 6, 35029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y. , Liu, G. , Bai, Y. , Xia, F. , He, C. & Shi, H. (2017) Two transcriptional activators of N‐acetylserotonin O‐methyltransferase 2 and melatonin biosynthesis in cassava. Journal of Experimental Botany, 68, 4997–5006. [DOI] [PubMed] [Google Scholar]
- Wiehe, P.O. & Dowson, W.J. (1953) A bacterial disease of cassava (Manihot utilissima) in Nyasaland. Empire Journal of Experimental Agriculture, 21, 141–143. [Google Scholar]
- Wilkins, K.E. , Booher, N.J. , Wang, L. & Bogdanove, A.J. (2015) TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Frontiers in Plant Science, 6, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydra, K. , Banito, A. & Kpémoua, K.E. (2007) Characterization of resistance of cassava genotypes to bacterial blight by evaluation of leaf and systemic symptoms in relation to yield in different ecozones. Euphytica, 155, 337–348. [Google Scholar]
- Wydra, K. & Fanou, A. (2015) Removal of symptomatic cassava leaves as cultural practice to control cassava bacterial blight. International Journal of Plant Pathology, 3, 117–124. [Google Scholar]
- Wydra, K. , Zinsou, V. , Jorge, V. & Verdier, V. (2004) Identification of pathotypes of Xanthomonas axonopodis pv. manihotis in Africa and detection of quantitative trait loci and markers for resistance to bacterial blight of cassava. Phytopathology, 94, 1084–1093. [DOI] [PubMed] [Google Scholar]
- Yan, Y. , He, X. , Hu, W. , Liu, G. , Wang, P. , He, C. et al. (2018) Functional analysis of MeCIPK23 and MeCBL1/9 in cassava defense response against Xanthomonas axonopodis pv. manihotis . Plant Cell Reports, 37, 887–900. [DOI] [PubMed] [Google Scholar]
- Yan, Y. , Wang, P. , He, C. & Shi, H. (2017) MeWRKY20 and its interacting and activating autophagy‐related protein 8 (MeATG8) regulate plant disease resistance in cassava. Biochemical and Biophysical Research Communications, 494, 20–26. [DOI] [PubMed] [Google Scholar]
- Yoodee, S. , Kobayashi, Y. , Songnuan, W. , Boonchird, C. , Thitamadee, S. , Kobayashi, I. et al. (2018) Phytohormone priming elevates the accumulation of defense‐related gene transcripts and enhances bacterial blight disease resistance in cassava. Plant Physiology and Biochemistry, 122, 65–77. [DOI] [PubMed] [Google Scholar]
- Yu, Y.‐H. , Lu, Y. , He, Y.‐Q. , Huang, S. & Tang, J.‐L. (2015) Rapid and efficient genome‐wide characterization of Xanthomonas TAL effector genes. Scientific Reports, 5, 13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandjanakou‐Tachin, M. , Fanou, A. , Gall, P.L. & Wydra, K. (2007) Detection, survival and transmission of Xanthomonas axonopodis pv. manihotis and X. axonopodis pv. vignicola, causal agents of cassava and cowpea bacterial blight, respectively, in/by insect vectors. Journal of Phytopathology, 155, 159–169. [Google Scholar]
- Zárate‐Chaves, C.A. , Osorio‐Rodríguez, D. , Mora, R.E. , Pérez‐Quintero, Á.L. , Dereeper, A. , Restrepo, S. et al. (2021) TAL effector repertoires of strains of Xanthomonas phaseoli pv. manihotis in commercial cassava crops reveal high diversity at the country scale. Microorganisms, 9, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H. , Xie, Y. , Liu, G. , Lin, D. , He, C. & Shi, H. (2018) Molecular identification of GAPDHs in cassava highlights the antagonism of MeGAPCs and MeATG8s in plant disease resistance against cassava bacterial blight. Plant Molecular Biology, 97, 201–214. [DOI] [PubMed] [Google Scholar]
- Zinsou, V. , Wydra, K. , Ahohuendo, B. & Hau, B. (2004) Effect of soil amendments, intercropping and planting time in combination on the severity of cassava bacterial blight and yield in two ecozones of West Africa. Plant Pathology, 53, 585–595. [Google Scholar]
- Zinsou, V. , Wydra, K. , Ahohuendo, B. & Hau, B. (2005) Genotype × environment interactions in symptom development and yield of cassava genotypes with artificial and natural cassava bacterial blight infections. European Journal of Plant Pathology, 111, 217–233. [Google Scholar]
- Zinsou, V. , Wydra, K. , Ahohuendo, B. & Schreiber, L. (2006) Leaf waxes of cassava (Manihot esculenta Crantz) in relation to ecozone and resistance to xanthomonas blight. Euphytica, 149, 189–198. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 Incidence data extracted from several studies on cassava bacterial blight (CBB). Incidence was defined in these studies as the percentage of symptomatic plants (number of symptomatic plants × 100/number of evaluated plants) recorded in cassava fields that were naturally infected by Xanthomonas phaseoli pv. manihotis (Xpm) (no artificial inoculations). Incidence ranges (minimum and maximum), timepoint of the incidence evaluation, and country where the study was performed were extracted from each report. Min., minimum; Max., maximum; MAP, months after planting; nd, missing data
TABLE S2 Severity data extracted from several studies on cassava bacterial blight (CBB). Severity was measured in these studies using similar scales based on symptom recording in cassava fields that were naturally infected by Xanthomonas phaseoli pv. manihotis (Xpm) (no artificial inoculations). Severity ranges (minimum and maximum), timepoint of the severity evaluation, country where the study was performed, and grading scales were extracted from each report. The transformation column indicates if the values presented as minimum and maximum were mathematically transformed (see text after table) to obtain values of the same type and scale. Min., minimum; Max., maximum; MAP, months after planting; nd, missing data. Grading scales showed slight modifications, but all of them reflected the same damage extension with the corresponding grading numbers. Most of the scales were grouped within Scale A: 1, no disease symptoms; 2, only angular leaf spots on leaves; 3, leaf blight, leaf wilt, defoliation, gum exudation in petioles and stems; 4, extensive leaf blight, wilt, defoliation, and partial stem die‐back; 5, complete defoliation, stem die‐back, and stunting in most of the plants. Only one study used a similar grading system with different scale numbers, Scale B: 0, no symptoms; 1, symptoms on leaves only – blight; 2, presence of necrotic lesions on the stem or petiole; 3, most severe symptoms on leaves and/or the presence of necrotic lesions with gum exudation; 4, complete loss of leaves with apical death or death of the plant
TABLE S3 Losses data extracted from several studies on cassava bacterial blight (CBB). Only losses reported as the percentage of fresh root weight lost were considered. All the studies were carried out in cassava fields naturally or artificially infected by Xanthomonas phaseoli pv. manihotis (Xpm). Loss ranges (minimum and maximum), resistance status of the tested varieties, and country or region where the study was performed were extracted from each report. Min., minimum; Max., maximum
TABLE S4 Reference genomes used for phylogeny reconstruction. Phylogeny was constructed using the nucleotide concatenated sequence of five housekeeping genes (atpD, dnaK, efp, glnA, gyrB, and rpoD)
TABLE S5 Result compilation of cassava resistance assessments reported in literature
TABLE S6 Quantitative trait loci (QTLs) associated with cassava bacterial blight (CBB) resistance. QTL name, name of the QTL associated with CBB resistance; ExpVar, percentage of phenotypic variance explained by a given QTL; linkage group, linkage group where a given QTL can be found, linkage groups depend on the map used for associations; strain, strain used to inoculate the assessed plants (in most cases, QTLs are strain‐specific); natural inoculum, the study was performed in the field and the quantified disease resulted from infections with naturally occurring strains; undefined strain, the study states that only one strain was used to inoculate the assessed plants, but authors do not provide the strain identifier; technique, disease quantitation technique used to associate phenotypes with markers; AUDPC, area under the disease progression curve; 100‐DI, percentage of diseased plants (100‐disease incidence); MDR, mean disease rating; marker, peak marker associated with a given QTL; map reference, cassava map used to associate QTLs. The maps developed by Fregene et al. (1997) and Soto et al. (2015) derived from the female parental TMS 30572 and the male parental CM 2177–2; the map developed by Wydra et al. (2004) derived from five F1 male individuals (CM7857‐4, CM7857‐10, CM7857‐51, CM7857‐77, and CM7857‐115) and the female recurrent parent TMS30572; the map developed by Tappiban et al. (2018) derived from the parentals Huay Bong 60 (HB60) and Hanatee (HN). Study reference, the reference to the study where QTLs were found
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


