Abstract
The functions of ubiquitin‐conjugating enzymes (E2) in plant immunity are not well understood. In this study, OsUBC26, a rice ubiquitin‐conjugating enzyme, was characterized in the defence against Magnaporthe oryzae. The expression of OsUBC26 was induced by M. oryzae inoculation and methyl jasmonate treatment. Both RNA interference lines and CRISPR/Cas9 null mutants of OsUBC26 reduced rice resistance to M. oryzae. WRKY45 was down‐regulated in OsUBC26 null mutants. In vitro E2 activity assay indicated that OsUBC26 is an active ubiquitin‐conjugating enzyme. Yeast two‐hybrid assays using OsUBC26 as bait identified the RING‐type E3 ligase UCIP2 as an interacting protein. Coimmunoprecipitation assays confirmed the interaction between OsUBC26 and UCIP2. The CRISPR/Cas9 mutants of UCIP2 also showed compromised resistance to M. oryzae. Yeast two‐hybrid screening using UCIP2 as bait revealed that APIP6 is a binding partner of UCIP2. Moreover, OsUBC26 working with APIP6 ubiquitinateds AvrPiz‐t, an avirulence effector of M. oryzae, and OsUBC26 null mutation impaired the proteasome degradation of AvrPiz‐t in rice cells. In summary, OsUBC26 plays important roles in rice disease resistance by regulating WRKY45 expression and working with E3 ligases such as APIP6 to counteract the effector protein AvrPiz‐t from M. oryzae.
Keywords: AvrPiz‐t, Magnaporthe oryzae, OsUBC26, plant immunity, rice, ubiquitin‐conjugating enzyme, WRKY45
Rice ubiquitin‐conjugating enzyme OsUBC26 is essential for immunity to blast fungus by regulating WRKY45 expression and working with APIP6 to promote the ubiquitination and degradation of AvrPiz‐t.
1. INTRODUCTION
The ubiquitin‐26S proteasome system (UPS) involves the sequential activities of three enzymes, namely ubiquitin activating enzyme (E1), ubiquitin‐conjugating enzyme (E2), and ubiquitin protein ligase enzyme (E3). First, ubiquitin is activated by E1 in an ATP‐dependent manner. Then, the ubiquitin (Ub)‐tagged E1 delivers Ub to E2 through transesterification. Ultimately, the ubiquitinated E2 targets E3, which recruits substrates for ubiquitination (Hershko, 2005; Smalle & Vierstra, 2004).
In plants, ubiquitination plays critical roles in responses to biotic and abiotic stimuli (Dielen et al., 2010; Marino et al., 2012; Stone, 2014; Zeng et al., 2006). During rice–pathogen interactions, E3 ubiquitin ligase plays an important role to counteract pathogens (Ning et al., 2016). For example, rice Spl11 is an E3 ligase and functions as a negative regulator of programmed cell death and disease resistance (Zeng et al., 2004). Rice APIP6 and APIP10 are RING‐type E3 ligases that interact with AvrPiz‐t effector from Magnaporthe oryzae and promote the ubiquitination and degradation of AvrPiz‐t, therefore positively regulating rice immunity to blast fungus (Park et al., 2012, 2016). Rice Xa21‐binding protein 3 (XB3) is another RING‐type E3 ligase that is required in Xa21‐mediated resistance to Xanthomonas oryzae pv. oryzae (Wang et al., 2006).
In addition to ubiquitin E3 ligases, a number of plant E2 genes have been identified and functionally characterized in diverse biological processes. The overexpression of VrUBC1, AhUBC2, and GmUBC2 enhanced the drought tolerance of transgenic Arabidopsis thaliana (Chung et al., 2013; Wan et al., 2011; Zhou et al., 2010). Ectopic expression of wild rice, Oryza grandiglumis, ubiquitin‐conjugating enzyme 1 (OgUBC1) in Arabidopsis increases resistance to Botrytis cinerea infection (Jeon et al., 2012). Triticum aestivum ubiquitin‐conjugating enzyme 4 (TaU4) is a negative regulator of wheat defence against Septoria, and virus‐induced gene silencing of TaU4 results in delayed disease symptom development (Millyard et al., 2016). A group of E2 enzymes in tomato is essential for plant immunity, and gene silencing of the homologs in tobacco reduced pattern‐triggered immunity (Zhou et al., 2017). Although these reports indicate that E2 enzymes are involved in plant defence, their specific mechanisms still need to be revealed.
There are 48 members of E2s in rice plants that have been classified into 15 groups according to Arabidopsis and human E2s (Bae & Kim, 2014). However, none of them have been characterized in rice immunity. In this study, we characterize a group VII E2 enzyme, OsUBC26, in rice immunity by using reverse genetic and biochemical methods. CRISPR/Cas9 mutants of OsUBC26 significantly compromised rice resistance to M. oryzae. We also screened a rice cDNA library and found that RING‐type E3 ligase UCIP2 is a working partner of OsUBC26. In addition, OsUBC26 could work with APIP6 and ubiquitinate AvrPiz‐t in vitro. Our work revealed a ubiquitin‐conjugating enzyme in rice that is essential for rice immunity.
2. RESULTS
2.1. OsUBC26 expression was induced by M. oryzae infection and methyl jasmonate treatment
Our previous study indicated that the expression of OsUBC26 can be induced by M. oryzae, X. oryzae pv. oryzae, and Rhizoctonia solani infection (Zhao et al., 2008). To further investigate the expression profile of OsUBC26, quantitative reverse transcription PCR (RT‐qPCR) was used to quantify the expression level of OsUBC26 at different time points during rice blast infection. As illustrated in Figure 1a, OsUBC26 responds to rice blast fungus from a very early stage, as the expression level of OsUBC26 increased quickly after the challenge by M. oryzae with a peak at 6 hr postinoculation (hpi) and then gradually decreased. As salicylic acid (SA) and jasmonic acid (JA) are two major phytohormones involved in disease resistance, SA and methyl jasmonate (Me‐JA) were used to treat rice seedlings. We found that SA had no obvious effect on the expression of OsUBC26 (data not shown). On the other hand, the expression of OsUBC26 increased to the highest level at 12 hours post‐treatment (hpt) and was approximately 13 times higher than the expression level observed at 0 hpt after the Me‐JA treatment (Figure 1b). This result suggests that OsUBC26 selectively participates in the Me‐JA‐mediated defence response of rice.
FIGURE 1.
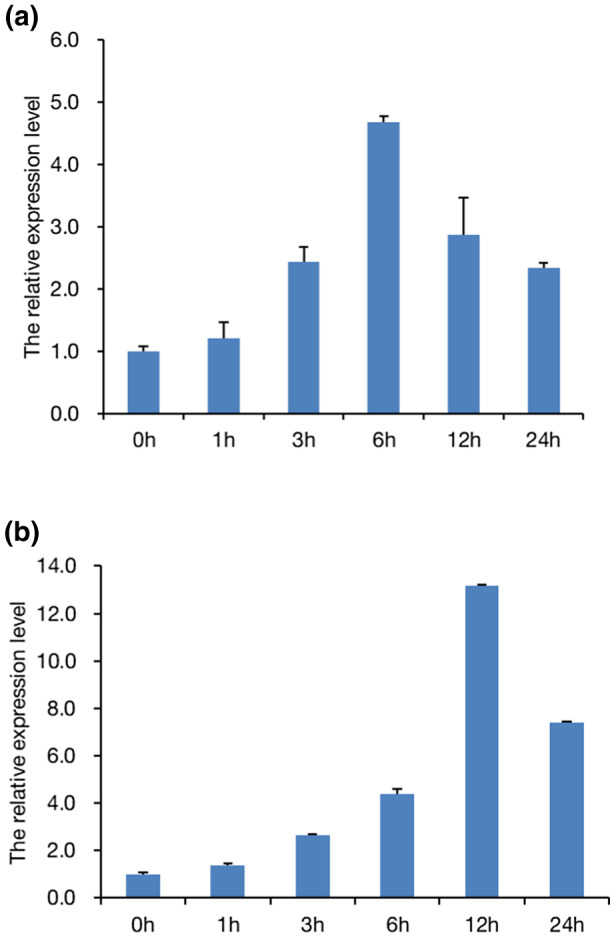
The expression of OsUBC26 was induced by Magnaporthe oryzae inoculation and methyl jasmonate (Me‐JA) treatment. (a) Expression pattern of OsUBC26 after inoculation with Guy11. (b) Expression pattern of OsUBC26 after treatment with 100 μM Me‐JA. Time is hours post‐treatment (hpt). Each experiment was done at least three times. Values are the means of three replicates and the error bars represent SEM
2.2. OsUBC26 is important for rice immunity against M. oryzae infection
To characterize the role of OsUBC26 in rice resistance to M. oryzae OsUBC26 RNAi lines were generated in the background of Kitaake. Transcription analysis showed that the expression of OsUBC26 in the RNAi lines was approximately half that of wild‐type plants (Figure 2a). M. oryzae inoculation assay demonstrated that the disease symptoms became more severe in OsUBC26 RNAi lines with a larger lesion area than wild‐type plants (Figure 2b,c). Hence, RNA interference of OsUBC26 diminished rice resistance to M. oryzae. To further characterize the function of OsUBC26, gene null mutants were generated by CRISPR/Cas9 technology. A 2 bp deletion and a 1 bp deletion in target 1 cause a premature stop codon (Figure S1). Homozygous mutants were selected in the T3 generation. The punch inoculation method was used for disease resistance evaluation (Ono et al., 2001; Park et al., 2012). The result showed that the lesion size and relative fungal biomass were significantly larger and greater in OsUBC26 null mutants compared to wild‐type Nipponbare (Figure 2d,e). Thus, OsUBC26 is an important factor for rice immunity against M. oryzae infection.
FIGURE 2.
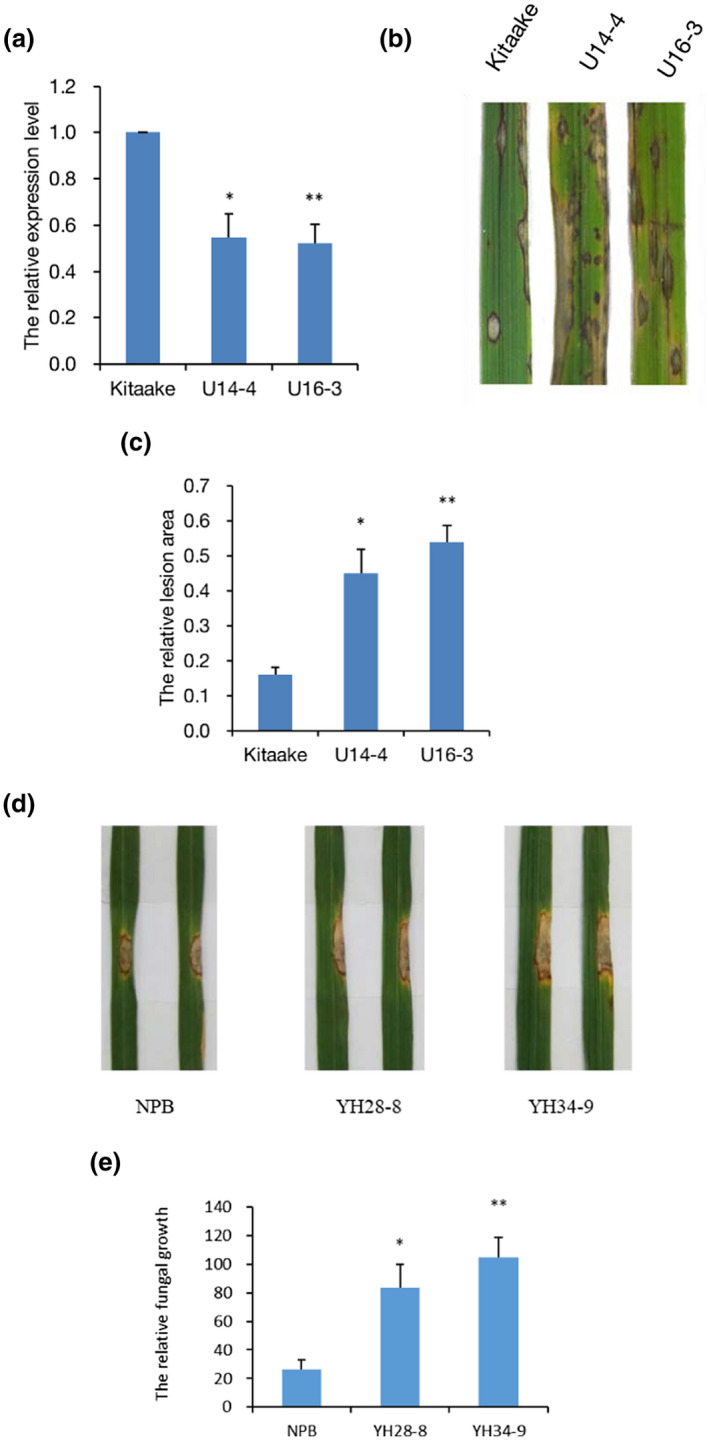
OsUBC26 RNAi and null mutation reduce rice resistance to Magnaporthe oryzae. (a) The relative expression level of OsUBC26 in its RNAi lines. (b) Inoculation results of OsUBC26 RNAi lines and corresponding wild‐type plants. Three‐week‐old rice seedlings were inoculated with a conidial spore suspension of M. oryzae isolate Guy11. Pictures were taken 7 days postinoculation (dpi). (c) Relative lesion area of RNAi lines and corresponding wild‐type plants. The relative lesion area was measured by ImageJ software. (d) Punch inoculation of OsUBC26 null mutants and its background cultivar Nipponbare (NPB). Leaves of 6‐week‐old rice plants were inoculated with virulent isolate FJ86‐CT. The leaves were photographed at 7 dpi. (e) Relative fungal growth (2[ C t( OsUBQ )− C t( MoPOT 2)] × 100) was measured at 7 dpi. Each experiment was done at least three times. Values are the means of three replicates and error bars represent SEM. Significant difference, *p < 0.05, **p < 0.01
2.3. Null mutation of OsUBC26 down‐regulated the expression of WRKY45
To reveal why CRISPR/Cas9 null mutants of OsUBC26 compromised rice resistance to M. oryzae, RNA‐Seq of the OsUBC26 null mutant and its wild‐type Nipponbare was conducted. A total of 237 genes were up‐regulated and 215 genes were down‐regulated in the OsUBC26 null mutant (Tables S1 and S2). The lower expression of WRKY45 in OsUBC26 null mutant attracted our attention. WRKY45 is a WRKY transcription factor that positively regulates rice resistance to the blast fungus and its function is affected by nuclear ubiquitin proteasome degradation (Matsushita et al., 2013; Shimono et al., 2007, 2012). The down‐regulation of WRKY45 in the OsUBC26 null mutants was further confirmed by RT‐qPCR (Figure 3).
FIGURE 3.
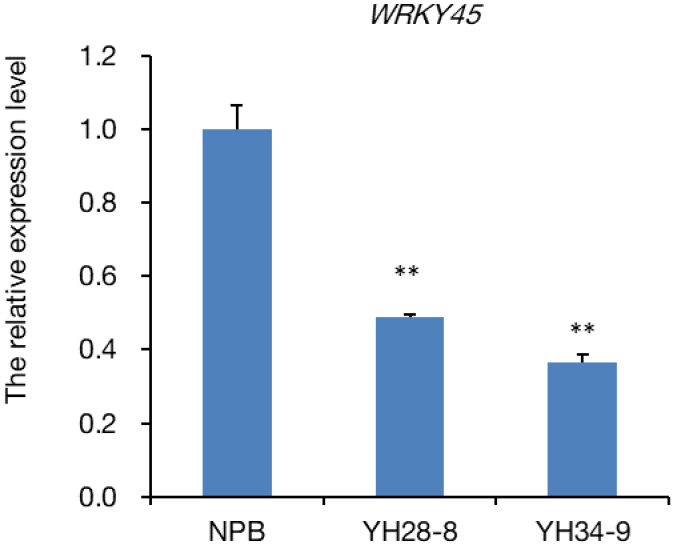
WRKY45 is down‐regulated in OsUBC26 null mutants. Values are the means of three replicates and error bars represent SEM. Significant difference, **p < 0.01
2.4. OsUBC26 is an active ubiquitin‐conjugating enzyme
In the presence of ATP, ubiquitin is activated by ubiquitin activating enzyme (E1), then transferred to the active site cysteine in E2 to form a thioester bond between ubiquitin and E2 (Smalle & Vierstra, 2004; Varshavsky, 1997; Wenzel et al., 2011; Ye & Rape, 2009). This thioester bond is sensitive to the thiol‐reducing agent dithiothreitol (DTT) and this characteristic can be used for E2 activity assay (Kraft et al., 2005). In an effort to prove that OsUBC26 is an active ubiquitin‐conjugating enzyme, histidine‐tagged Arabidopsis E1 component His:AtE1 and ubiquitin component His:ubiquitin, a histidine‐tagged rice E2 component His:OsUBC26, were expressed in a prokaryotic expression system and purified separately. Under nonreduced (without DTT) and reduced (with DTT) conditions, western blot analysis demonstrated that ubiquitin bound to His:OsUBC26 under nonreduced conditions, but not reduced conditions (Figure 4). This experiment demonstrates that OsUBC26 is an active ubiquitin‐conjugating enzyme.
FIGURE 4.
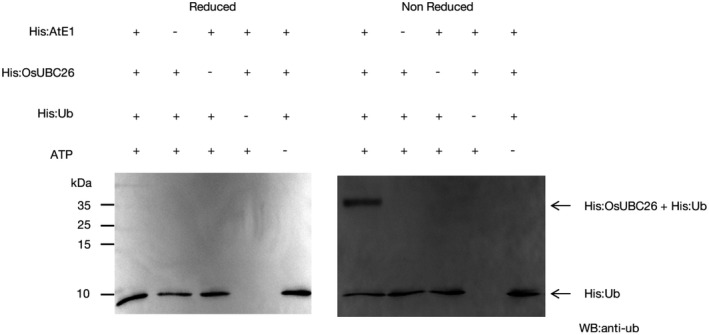
In vitro E2 activity assay of OsUBC26. E2 charging assays of His:OsUBC26 under nonreduced and reduced conditions. Ubiquitin bound to OsUBC26 under nonreduced conditions but not reduced conditions
2.5. OsUBC26 interacts with UCIP2
To further reveal the working partner of OsUBC26, a yeast two‐hybrid screening was conducted using OsUBC26 as bait. Several E3 ligases that interact with OsUBC26 were identified from a rice cDNA library in the yeast two‐hybrid assay (Table 1). Among them, RING‐type E3 ligase UCIP2 (OsUBC26 Interact Protein 2) represented 17 of 44 of the identified interacting clones. We cloned the full‐length coding sequence of UCIP2 and confirmed the interaction between OsUBC26 and UCIP2 in yeast two‐hybrid assay (Figure 5a). In addition, glutathione S‐transferase (GST)‐tagged OsUBC26 and maltose‐binding protein (MBP)‐tagged UCIP2 were prepared in a prokaryotic expression system. A GST pull‐down assay showed that GST:OsUBC26 interacted with MBP:UCIP2 (Figure 5b). Thus, OsUBC26 may have preference for UCIP2 and mediated the ubiquitination degradation of UCIP2‐recruited substrates. Interestingly, the protein ID of UCIP2 (LOC_Os01g47740) is the same as OsZFP1, which has been reported as a working partner of OsDjA6 (Zhong et al., 2018). OsDjA6, a chaperone DnaJ protein, negatively regulates WRKY45 expression and rice immunity to M. oryzae (Zhong et al., 2018). Thus, OsUBC26 as a conjugating enzyme works with E3 ligase UCIP2/OsZFP1 and chaperone DnaJ protein OsDjA6 to modulate rice innate immunity through regulation of WRKY45 expression.
TABLE 1.
Summary of OsUBC26‐interacting proteins identified by yeast two‐hybrid screening
| Gene Code | Gene_locus | Gene_description | No. of independent clones |
|---|---|---|---|
| UCIP1 | LOC_Os08g42640 | Zinc finger, RING‐type domain containing protein | 3 |
| UCIP2 | LOC_Os01g47740 | Zinc finger, RING‐type domain containing protein | 17 |
| UCIP3 | LOC_Os04g10680 | Similar to protein containing C‐terminal RING‐finger | 2 |
| UCIP4 | LOC_Os09g33670 | Zinc finger, RING‐type domain containing protein | 5 |
| UCIP5 | LOC_Os12g39640 | Similar to transfactor‐like protein (MYB) | 1 |
| UCIP6 | LOC_Os02g44599 | Protein kinase‐like domain containing protein | 1 |
| UCIP7 | LOC_Os04g47770 | Similar to cis‐zeatin O‐glucosyltransferase 1 | 1 |
| UCIP8 | LOC_Os03g44900 | Ccr4‐Not complex, subunits 3 and 5 family protein | 1 |
| UCIP9 | LOC_Os04g33590 | Esterase/lipase/thioesterase domain containing protein | 2 |
| UCIP10 | LOC_Os01g56580 | Similar to casein kinase‐like protein | 1 |
| UCIP11 | LOC_Os03g63950 | Similar to plastid‐specific 30S ribosomal protein1 | 1 |
| UCIP12 | LOC_Os12g31850 | Similar to ureide permease 1 (AtUPS1) | 1 |
| UCIP13 | LOC_Os10g27340 | Prolyl 4‐hydroxylase, alpha subunit domain containing protein | 1 |
| UCIP14 | LOC_Os06g07350 | Similar to RNA‐binding protein‐like | 1 |
| UCIP15 | LOC_Os05g48360 | Membrane attack complex component/perforin/ complement C9 family protein | 1 |
| UCIP16 | LOC_Os08g26870 | Protein of unknown function DUF151 domain containing protein | 1 |
| UCIP17 | LOC_Os08g44820 | Similar to CUC2 | 1 |
| UCIP18 | LOC_Os01g52110 | RING‐type domain containing protein | 1 |
| UCIP19 | LOC_Os06g06760 | U‐box domain containing protein | 2 |
FIGURE 5.
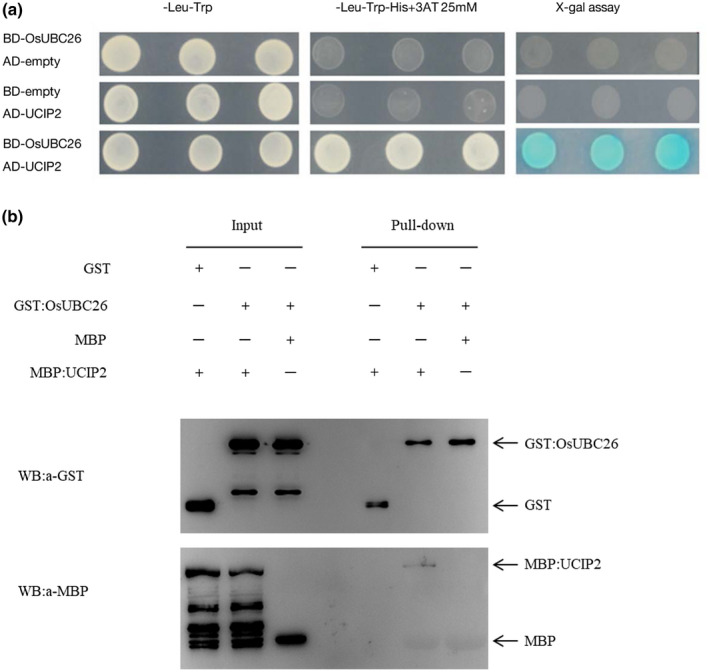
OsUBC26 interacts with UCIP2. (a) OsUBC26 interacts with UCIP2 in yeast two‐hybrid assay. The full‐length coding sequence of UCIP2 was cloned and fused to the GAL4‐activation domain. Cotransformation with BD‐UBC26 further confirmed the interaction between OsUBC26 and UCIP2. (b) OsUBC26 interacts with UCIP2 in glutathione S‐transferase (GST) pull‐down assay. GST‐tagged OsUBC26 was expressed in Escherichia coli and conjugated to glutathione magarose beads, which were used for maltose‐binding protein (MBP):UCIP2 pull‐down assay
2.6. UCIP2 plays a positive role in resistance to M. oryzae
To reveal the role of UCIP2 in rice immunity to M. oryzae, the CRISPR/Cas9 null mutants of UCIP2 were generated. A homozygous mutant was selected in the T2 generation with an 11 bp deletion of the target 1 sequence (Figure S2). The 11 bp deletion causes a premature stop codon in UCIP2. Punch inoculation of the UCIP2 null mutant showed compromised resistance to M. oryzae with larger lesion size and greater relative fungal biomass compared to Nipponbare (Figure 6).
FIGURE 6.
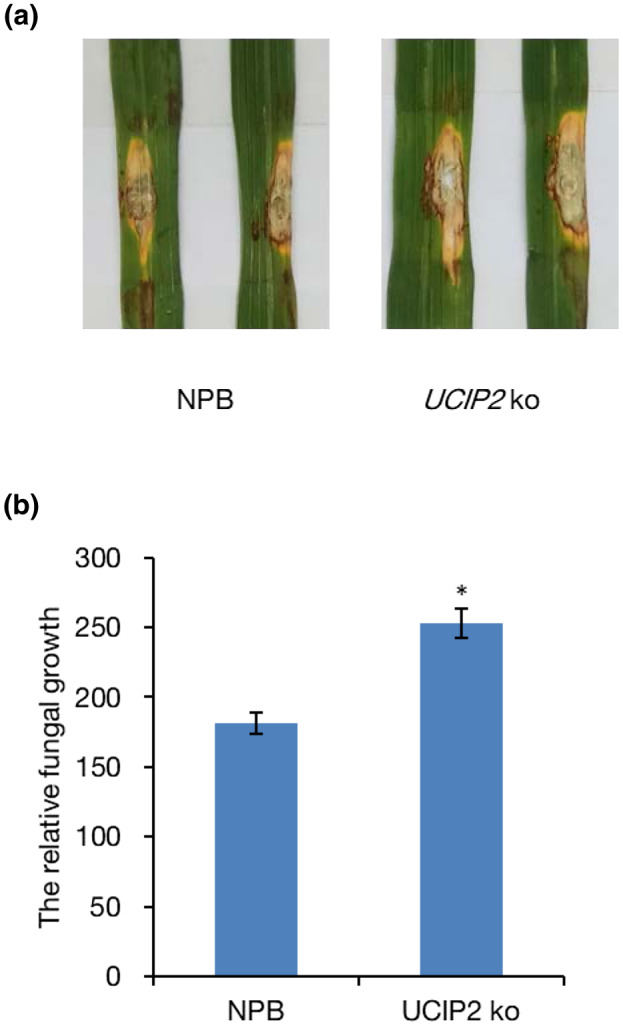
Null mutation of UCIP2 compromises resistance to Magnaporthe oryzae. (a) Punch inoculation of UCIP2 null mutants and its background cultivar Nipponbare (NPB). Leaves of 6‐week‐old rice plants were inoculated with virulent isolate FJ86‐CT. The leaves were photographed at 7 days postinoculation (dpi). (b) Relative fungal growth (2[ C t( OsUBQ )− C t( MoPOT 2)] × 100] was measured at 7 dpi. Values are the means of three replicates and error bars represent SEM. Significant difference, *p < 0.05
2.7. OsUBC26 could work with APIP6 and ubiquitinate AvrPiz‐t
During rice–pathogen interaction, E3 ligase activity is widely used by the host to reprogramme the cell to counteract pathogens (Ning et al., 2016). AvrPiz‐t is an avirulence effector from M. oryzae that targets a rice E3 ligase APIP6 to suppress PTI. In contrast, APIP6 can ubiquitinate AvrPiz‐t and promote the proteasome degradation of AvrPiz‐t (Park et al., 2012). In an effort to find substrate proteins of UCIP2, we screened the rice cDNA library using UCIP2 as bait and found that APIP6 is a binding partner of UCIP2. An in vitro ubiquitination assay showed that in the presence of Ub, AtE1, OsUBC26, and both APIP6 and UCIP2 were ubiquitinated (Figure S3), which suggests that OsUBC26 may also work with APIP6. We therefore checked whether OsUBC26 could work with APIP6 and ubiquitinate AvrPiz‐t by using a bacterial‐based synthetic approach (Han et al., 2017). This assay demonstrated that ubiquitination of AvrPiz‐t required the presence of AtE1, OsUBC26, APIP6, and ubiquitin (Figure 7). This indicates that OsUBC26 could work with APIP6 and ubiquitinate AvrPiz‐t in vitro.
FIGURE 7.
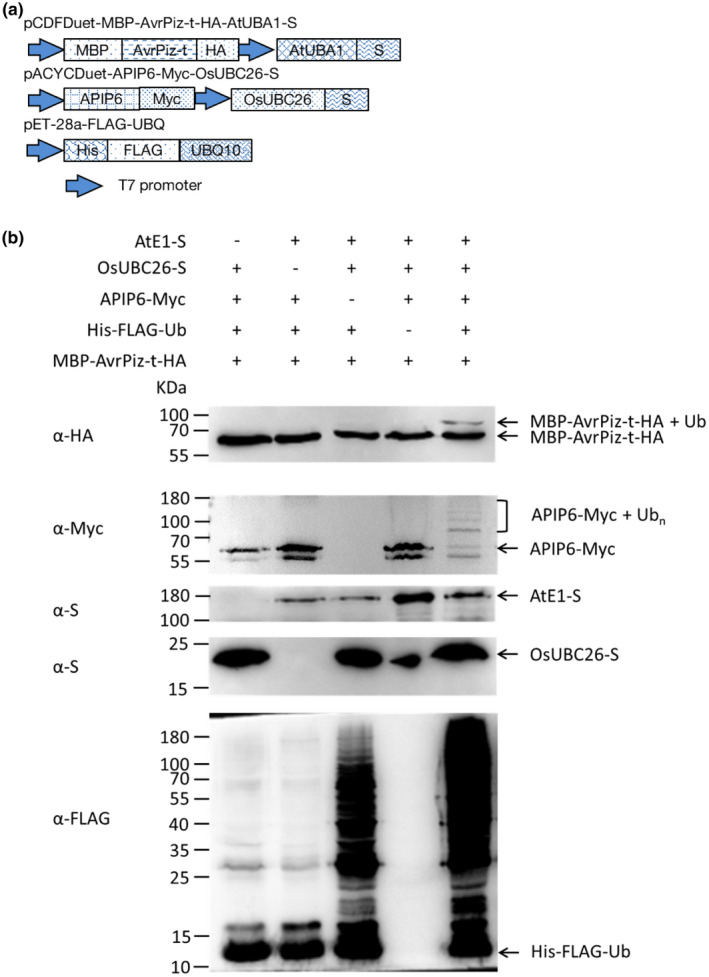
OsUBC26 could work with APIP6 and ubiquitinate AvrPiz‐t in vitro. (a) Schematic representation of the plasmids pCDFDuet‐MBP‐AvrPiz‐t‐HA‐AtUBA1‐S, pACYCDuet‐APIP6‐Myc‐OsUBC26‐S, and pET‐28a‐FLAG‐UBQ. (b) Detection of ubiquitination of AvrPiz‐t. The crude total proteins were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis and visualized by western blot with corresponding antibodies
2.8. OsUBC26 null mutation impairs the proteasome degradation of AvrPiz‐t
As OsUBC26 could work with APIP6 and mediate the ubiquitination of AvrPiz‐t, we wanted to know whether OsUBC26‐APIP6 mediates the degradation of AvrPiz‐t. Cell‐free degradation was conducted and the results showed that the proteasome degradation of AvrPiz‐t was significantly impaired in the OsUBC26 null mutant group compared to the wild‐type Nipponbare group (Figure 8). When MG132, a specific proteasome inhibitor, was added to the Nipponbare group the degradation of AvrPiz‐t was blocked. This experiment indicated that the degradation of AvrPiz‐t in rice cells was via the 26S proteasome system and OsUBC26 was involved in the ubiquitination and degradation of AvrPiz‐t.
FIGURE 8.
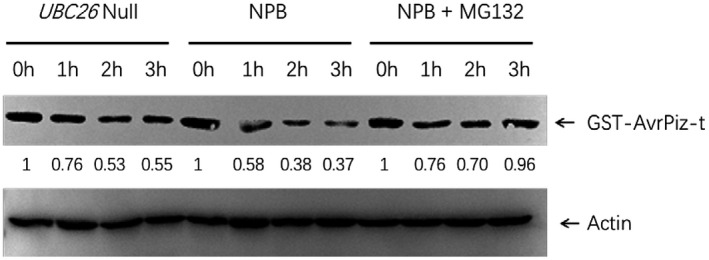
OsUB26 null mutation impairs proteasome degradation of AvrPiz‐t in rice cells. Total protein was extracted from OsUBC26 null mutant and Nipponbare with or without MG132. Purified proteins GST‐AvrPiz‐t from Escherichia coli BL21 (DE3) were incubated in rice total protein supernatant with the addition of 1 mM ATP and cultured at 28 °C for different times. Actin was used as a loading control. Band intensity was calculated by ImageJ software
3. DISCUSSION
The host ubiquitin proteasome system plays an important role in defence against pathogens. Compared to the deep understanding of the role of ubiquitin E3 ligases in plant immunity, relatively little is known about the role of E2 conjugating enzyme in immunity. In this study, we characterized a ubiquitin‐conjugating enzyme in rice defence. Knockdown and null mutation of OsUBC26 compromised rice resistance to M. oryzae, which indicates that OsUBC26 is essential for rice immunity to this fungus.
Park et al. (2012) found that APIP6 interacts with AvrPiz‐t and promotes the degradation of AvrPiz‐t in Nicotiana benthamiana leaves. When they tested the stability of AvrPiz‐t in rice cells, no signal was detected, which suggests that AvrPiz‐t was targeted for degradation in rice cells. However, the degradation of AvrPiz‐t in rice cells via the ubiquitin–26S proteasome system remains uncharacterized. Moreover, which ubiquitin‐conjugating enzyme works with APIP6 and mediates the ubiquitination of AvrPiz‐t in rice cells remains unknown. Here we found that OsUBC26 could work with APIP6 and ubiquitinate AvrPiz‐t in vitro. In addition, OsUBC26 null mutation impaired the proteasome degradation of AvrPiz‐t in rice cells, which suggests that OsUBC26 may work with APIP6 and promote the ubiquitination and degradation of AvrPiz‐t via the 26S proteasome in rice cells, and help the rice plant to counteract AvrPiz‐t secreted from M. oryzae.
In the rice genome there are 48 ubiquitin‐conjugating enzymes and over 1,000 E3 ligases (Bae & Kim, 2014; Zhou et al., 2009). Generally, one ubiquitin‐conjugating enzyme interacts with more than one E3 ligase. Yeast two‐hybrid screening discovered several E3 ligases that interact with OsUBC26 (Table 1). At least four RING‐type E3 ligases, UCIP1, UCIP2, UCIP3, and UCIP4, interact with OsUBC26. We further confirmed the interaction between OsUBC26 and UCIP2 in yeast using the full‐length coding sequence of UCIP2 and then GST pull‐down assay. Our work also revealed that OsUBC26 could work with APIP6 to ubiquitinate AvrPiz‐t in vitro. Bae and Kim (2014) characterized the interaction modes of rice E2s with ARM‐U‐box E3 ligases by yeast two‐hybrid assays and showed that OsUBC26 also interacts with SPL11, OsPUB33, OsPUB36, OsPUB38, and OsPUB41. Hence, OsUBC26 may interact with other E3 ligases in addition to UCIP2.
In addition to E3 ligases, there were some interesting additional candidate binding partners of OsUBC26 identified in the library screening. UCIP7, a cis‐zeatin O‐glucosyltransferase, converts cis‐zeatin to its inactive form. Cis‐zeatin functions as a kind of cytokinin, which indicates that OsUBC26 may have interplay with cytokinin signalling and regulate cytokinin activity by O‐glucosylation. UCIP9, a thioesterase domain‐containing protein, may play a role in the ubiquitin binding status of OsUBC26 as ubiquitin is bound to E2 through a thioester bond. UCIP6 and UCIP10 are kinase domain‐containing proteins, which indicates that the activity of OsUBC26 may be regulated by phosphorylation. UCIP5 and UCIP17 are transcription factors, which suggests that OsUBC26 may regulate UCIP5 and UCIP17 transcriptional activity by directly interacting with them.
Our RT‐qPCR results showed that the expression of OsUBC26 was induced by M. oryzae inoculation and Me‐JA treatment but not SA. This may imply that the JA pathway is involved in OsUBC26. However, null mutation of OsUBC26 lowered the expression of WRKY45, and WRKY45 is a transcription factor involved in rice resistance via the SA signalling pathway that is regulated by the ubiquitin proteasome system (Matsushita et al., 2013). With regard to the high innate level of SA in rice plants, both JA and SA signalling pathways may be involved in OsUBC26‐mediated rice immunity.
The results of Zhong et al. (2018) indicated that OsDjA6 is a negative regulator of WRKY45 expression, and OsZFP1 (UCIP2) was discovered as a working partner of OsDjA6. However, the expression of WRKY45 in UCIP2 null mutant shows no significant difference compared to wild‐type plants. OsUBC26‐UCIP2 did not ubiquitinate OsDjA6 in vitro (data not shown) so the regulation of WRKY45 expression by OsUBC26 may be a separate way. Further study is required to uncover how OsUBC26 and OsDjA6 regulate the expression of WRKY45.
4. EXPERIMENTAL PROCEDURES
4.1. Plant materials and growth conditions
Rice seeds were soaked into water for 48 hr at room temperature and the water was changed every 12 hr. The seeds were pregerminated in an incubator at 37 °C for 1 day. After germination, the seeds were planted in small pots and kept in a greenhouse at 28 °C light/23 °C dark and 70% relative humidity under a photoperiod of 14 hr light/ 10 hr dark for growth.
4.2. Spraying inoculation and Me‐JA treatment
M. oryzae isolate Guy11 was pregrown on rice bran medium for 10 days in the dark. The aerial hyphae were flattened using a sterilized glass slide and the growth plates were incubated under light to encourage a uniform crop of conidia. The concentration of conidia was adjusted to 1–2 × 105 spores/ml with 0.02% Tween‐20. Three‐week‐old rice seedlings were used for spray inoculation. After spraying, rice seedlings were kept in dark and high humidity conditions for 24 hr. The seedlings were then moved to a solar greenhouse with high humidity and grown for 7 days. ImageJ software was used to quantify the lesion area. For Me‐JA treatment, 3‐week‐old rice seedlings were sprayed with 100 µM Me‐JA, then kept in an incubator at 28 °C with high humidity. Samples were taken at time points of 0, 1, 3, 6, 12, and 24 hpt for RT‐qPCR analysis.
4.3. Punch inoculation and disease resistance evaluations
For punch inoculation, M. oryzae isolate FJ86‐CT was used. The concentration of the fungal spores was adjusted to 5 × 105 spores/ml with 0.02% Tween‐20 and 7 µl of spore suspension was used for punch inoculation as described by Zhong et al. (2018). After punch inoculation, the seedlings were kept in a growth chamber at 28 °C light/25 °C dark and 90% relative humidity under a photoperiod of 12 hr light/12 hr dark. After 7–9 days’ growth, the diseased leaves were sampled for relative fungal biomass quantitation using the DNA‐based quantitative PCR method as described by Park et al. (2012).
4.4. RNA isolation and RT‐qPCR
RNA was extracted by using a total RNA extraction kit (Promega) following the manufacturer's instructions. The RNA concentration was determined by NanoDrop 2000 machine (Eppendorf). A PrimeScript RT‐PCR Kit (Takara) was used for reverse transcription and TB Green Premix Ex Taq (Takara) was used for qPCR analysis following the manufacturer's instructions. Primers are shown in Table S3.
4.5. RNAi
For building of the OsUBC26 RNAi line, the pTCK‐303 plasmid and primer pair UBC26‐RNAi‐F/UBC26‐RNAi‐R was used. Sense sequence ligation using SpeI/SacI enzyme digestion sites and subsequent KpnI/BamHI enzyme digestion sites was used to obtain an insert for antisense sequence ligation. Agrobacterium tumefaciens LBA4404 was used for the transformation of rice cultivar Kitaake callus. All the transgenic seedlings were confirmed by PCR and β‐glucuronidase (GUS) staining analysis. The T2 generation of RNAi lines was used for the RT‐qPCR and inoculation experiments.
4.6. Gene editing
To generate the gene null transgenic plants, two targets were designed using the CRISPR‐GE toolkit (Xie et al., 2017). The two targets were ligated to promoters and inserted into the pYLCRISPR/Cas9‐MTmono vector (Ma et al., 2015). A. tumefaciens EHA105 was used for the transformation of rice cultivar Nipponbare callus. The gene editing result was further checked by sequencing of the amplified targeted sequence. Homozygotes were selected for phenotypic analysis.
4.7. E2 charging assay
For the E2 activity assay, pET28a‐ubiquitin, pET28a‐AtE1, and pET28a‐OsUBC26 were expressed in Escherichia coli BL21 (DE3) and purified using Ni‐NTA agarose (Qiagen) separately. The E2 charging assay was conducted according to protocols described elsewhere (Zhao et al., 2012). The final result was visualized using anti‐ubiquitin antibody.
4.8. Yeast two‐hybrid screening
Coding region sequences of OsUBC26 and UCIP2 were constructed in pDBLeu vector as bait. 3‐aminotriazole (3AT) sensitivity was determined by plate yeast transformants containing bait plasmid and empty prey plasmid onto SC−Leu−Trp−His plates containing 3AT at concentrations of 0, 10, 25, 50, 75, and 100 mM. Saccharomyces cerevisiae MaV203 (Invitrogen) containing bait plasmid was used for making competent cells. The cDNA library was transformed into competent cells and plated onto SC−Leu−Trp−His plus 25 mM 3AT plates to select positive clones.
4.9. GST pull‐down
For the GST pull‐down assay, the coding sequence of OsUBC26 was cloned into pGEX‐KG by using BamHI and EcoRI restriction sites. The open reading frame of UCIP2 was inserted into pMAL‐c5X by NotI and EcoRI restriction sites. Both constructions were expressed in E. coli BL21 (DE3). The ultrasonic lysate containing GST:OsUBC26 was bound to glutathione magarose beads overnight at 4 °C using GST binding buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). MBP binding buffer (20 mM Tris‐HCl, 200 mM NaCl, 1 mM EDTA, pH 7.4) was used for MBP:UCIP2 ultrasonic lysis. Beads were washed three times with GST binding buffer then MBP:UCIP2 ultrasonic lysate was added to the system and incubated 8–12 hr at 4 °C. Input and pull‐down proteins were visualized by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS‐PAGE) and western blot analysis with corresponding antibodies.
4.10. In vitro ubiquitination assays
Plasmids pCDFDuet‐MBP‐ABI3‐HA‐AtUBA1‐S, pACYCDuet‐AIP2‐Myc‐UBC8‐S, and pET‐28a‐FLAG‐UBQ were built by Han et al. (2017). We replaced AIP2 in the plasmid of pACYCDuet‐AIP2‐Myc‐UBC8‐S by APIP6 using BamHI and StuI restriction sites. Plasmids pCDFDuet‐MBP‐UCIP2‐HA‐AtUBA1‐S, pCDFDuet‐MBP‐AvrPiz‐t‐HA‐AtUBA1‐S, and pACYCDuet‐APIP6‐Myc‐OsUBC26‐S were constructed using the recombination‐type‐cloning method (ClonExpress II One Step Cloning Kit; Vazyme). For the deletion of AtUBA1 in the plasmid pCDFDuet‐MBP‐AvrPiz‐t‐HA‐AtUBA1‐S and deletion of OsUBC26 in the plasmid of pACYCDuet‐APIP6‐Myc‐OsUBC26‐S, PCR‐based amplification was used with specific restriction enzyme sites ClaI and SpeI, respectively. For the deletion of APIP6, pACYCDuet‐APIP6‐Myc‐OsUBC26‐S was digested by BamHI and StuI, and then blunt‐ended by T4 DNA polymerase (Takara).
4.11. Cell‐free degradation
Cell‐free degradation was performed as described by Kong et al. (2015). Briefly, total protein from 3‐week‐old rice seedlings was extracted with native extraction buffer (50 mM Tris‐MES, pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, pH 8.0, 5 mM dithiothreitol [DTT], and protease inhibitor cocktail) with or without 100 µM MG132. Insoluble debris was pelleted twice by centrifugation at 13,000 rpm for 10 min at 4 °C. For construction of GST‐AvrPiz‐t plasmid, pGEX‐KG was digested with SmaI and SalI, and AvrPiz‐t coding sequence was inserted in it using the recombination‐type‐cloning method (ClonExpress II One Step Cloning Kit; Vazyme). Purified proteins GST‐AvrPiz‐t from E. coli BL21 (DE3) were incubated in rice total protein supernatant with addition of 1 mM ATP and cultured at 28 °C for different times. Anti‐GST antibody was used to detect the GST‐AvrPiz‐t protein level by immunoblotting analysis.
Supporting information
FIGURE S1 Genome editing events occurring in OsUBC26 null mutants
FIGURE S2 Genome editing events occurring in UCIP2 null mutants
FIGURE S3 Both APIP6 and UCIP2 were ubiquitinated when coexpressed in Escherichia coli
TABLE S1 Up‐regulated genes in OsUBC26 null mutant
TABLE S2 Down‐regulated genes in OsUBC26 null mutant
TABLE S3 Primers used in this study
ACKNOWLEDGEMENTS
Plasmid pET28a‐AtE1 and pET28a‐UBQ were kindly provided by Dr Qi Xie of the Chinese Academy of Sciences. The rice cDNA library was kindly donated by Dr Guo‐Liang Wang at Ohio State University. Plasmids pCDFDuet‐MBP‐ABI3‐HA‐AtUBA1‐S, pACYCDuet‐AIP2‐Myc‐UBC8‐S, and pET‐28a‐FLAG‐UBQ were kindly donated by Dr Dongping Lu at the Chinese Academy of Sciences. This project was funded by the National Key Research and Development Program of China (2016YFD0100600) and the National Natural Science Foundation of China (NSFC) (31972251) to G.D.L.
Liu, X. , Song, L. , Zhang, H. , Lin, Y. , Shen, X. , Guo, J. , et al (2021) Rice ubiquitin‐conjugating enzyme OsUBC26 is essential for immunity to the blast fungus Magnaporthe oryzae . Molecular Plant Pathology, 22, 1613–1623. 10.1111/mpp.13132
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Bae, H. & Kim, W.T. (2014) Classification and interaction modes of 40 rice E2 ubiquitin‐conjugating enzymes with 17 rice ARM‐U‐box E3 ubiquitin ligases. Biochemical and Biophysical Research Communications, 444, 575–580. [DOI] [PubMed] [Google Scholar]
- Chung, E. , Cho, C.W. , So, H.A. , Kang, J.S. , Chung, Y.S. & Lee, J.H. (2013) Overexpression of VrUBC1, a mung bean E2 ubiquitin‐conjugating enzyme, enhances osmotic stress tolerance in Arabidopsis . PLoS One, 8, e66056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dielen, A.S. , Badaoui, S. , Candresse, T. & German‐Retana, S. (2010) The ubiquitin/26S proteasome system in plant–pathogen interactions: a never‐ending hide‐and‐seek game. Molecular Plant Pathology, 11, 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. , Sun, J. , Yang, J. , Tan, Z. , Luo, J. & Lu, D. (2017) Reconstitution of the plant ubiquitination cascade in bacteria using a synthetic biology approach. The Plant Journal, 91, 766–776. [DOI] [PubMed] [Google Scholar]
- Hershko, A. (2005) The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death and Differentiation, 12, 1191–1197. [DOI] [PubMed] [Google Scholar]
- Jeon, E.H. , Pak, J.H. , Kim, M.J. , Kim, H.J. , Shin, S.H. , Lee, J.H. et al. (2012) Ectopic expression of ubiquitin‐conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV‐B radiation and Botrytis infection in Arabidopsis thaliana . Biochemical and Biophysical Research Communications, 427, 309–314. [DOI] [PubMed] [Google Scholar]
- Kong, L. , Cheng, J. , Zhu, Y. , Ding, Y. , Meng, J. , Chen, Z. et al. (2015) Degradation of the ABA co‐receptor ABI1 by PUB12/13 U‐box E3 ligases. Nature Communications, 6, 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, E. , Stone, S.L. , Ma, L. , Su, N. , Gao, Y. , Lau, O.‐S. et al. (2005) Genome analysis and functional characterization of the E2 and RING‐type E3 ligase ubiquitination enzymes of Arabidopsis . Plant Physiology, 139, 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Marino, D. , Peeters, N. & Rivas, S. (2012) Ubiquitination during plant immune signaling. Plant Physiology, 160, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita, A. , Inoue, H. , Goto, S. , Nakayama, A. , Sugano, S. , Hayashi, N. et al. (2013) Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. The Plant Journal, 73, 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millyard, L. , Lee, J. , Zhang, C. , Yates, G. & Sadanandom, A. (2016) The ubiquitin conjugating enzyme, TaU4 regulates wheat defence against the phytopathogen Zymoseptoria tritici . Scientific Reports, 6, 35683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, Y. , Wang, R. , Shi, X. , Zhou, X. & Wang, G.L. (2016) A layered defense strategy mediated by rice E3 ubiquitin ligases against diverse pathogens. Molecular Plant, 9, 1096–1098. [DOI] [PubMed] [Google Scholar]
- Ono, E. , Wong, H.L. , Kawasaki, T. , Hasegawa, M. , Kodama, O. & Shimamoto, K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proceedings of the National Academy of Sciences USA, 98, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. et al. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 Ubiquitin Ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. The Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.H. , Shirsekar, G. , Bellizzi, M. , Chen, S. , Songkumarn, P. , Xie, X. et al. (2016) The E3 ligase APIP10 connects the effector AvrPiz‐t to the NLR receptor Piz‐t in rice. PLoS Pathogens, 12, e1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, M. , Koga, H. , Akagi, A. , Hayashi, N. , Goto, S. , Sawada, M. et al. (2012) Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Molecular Plant Pathology, 13, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, M. , Sugano, S. , Nakayama, A. , Jiang, C.‐J. , Ono, K. , Toki, S. et al. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole‐inducible blast resistance. The Plant Cell, 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J. & Vierstra, R.D. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annual Review of Plant Biology, 55, 555–590. [DOI] [PubMed] [Google Scholar]
- Stone, S.L. (2014) The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Frontiers in Plant Science, 5, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky, A. (1997) The ubiquitin system. Trends in Biochemical Sciences, 22, 383–387. [DOI] [PubMed] [Google Scholar]
- Wan, X. , Mo, A. , Liu, S. , Yang, L. & Li, L. (2011) Constitutive expression of a peanut ubiquitin‐conjugating enzyme gene in Arabidopsis confers improved water‐stress tolerance through regulation of stress‐responsive gene expression. Journal of Bioscience and Bioengineering, 111, 478–484. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Pi, L. , Chen, X. , Chakrabarty, P.K. , Jiang, J. , De Leon, A.L. et al. (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21‐mediated disease resistance. The Plant Cell, 18, 3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel, D. , Stoll, K. & Klevit, R. (2011) E2s: structurally economical and functionally replete. The Biochemical Journal, 433, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. , Ma, X. , Zhu, Q. , Zeng, D. , Li, G. & Liu, Y.G. (2017) CRISPR‐GE: A convenient software toolkit for CRISPR‐based genome editing. Molecular Plant, 10, 1246–1249. [DOI] [PubMed] [Google Scholar]
- Ye, Y. & Rape, M. (2009) Building ubiquitin chains: E2 enzymes at work. Nature Reviews Molecular Cell Biology, 10, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Qu, S. , Bordeos, A. , Yang, C. , Baraoidan, M. , Yan, H. et al. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U‐box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. The Plant Cell, 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L.R. , Vega‐Sanchez, M.E. , Zhu, T. & Wang, G.L. (2006) Ubiquitination‐mediated protein degradation and modification: an emerging theme in plant–microbe interactions. Cell Research, 16, 413–426. [DOI] [PubMed] [Google Scholar]
- Zhao, C.‐J. , Wang, A.‐R. , Shi, Y.‐J. , Wang, L.‐Q. , Liu, W.‐D. , Wang, Z.‐H. et al. (2008) Identification of defense‐related genes in rice responding to challenge by Rhizoctonia solani . Theoretical and Applied Genetics, 116, 501–516. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Liu, L. & Xie, Q. (2012) In vitro protein ubiquitination assay. Plant Signalling Networks, Methods in Molecular Biology. 876, Germany: Springer. pp.163–172. [DOI] [PubMed] [Google Scholar]
- Zhong, X. , Yang, J. , Shi, Y. , Wang, X. & Wang, G.L. (2018) The DnaJ protein OsDjA6 negatively regulates rice innate immunity to the blast fungus Magnaporthe oryzae . Molecular Plant Pathology, 19, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. , Mural, R.V. , Chen, X. , Oates, M.E. , Connor, R.A. , Martin, G.B. et al. (2017) A subset of ubiquitin‐conjugating enzymes is essential for plant immunity. Plant Physiology, 173, 1371–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D. , Zhou, X. , Li, L. & Su, Z. (2009) PlantsUPS: a database of plants' ubiquitin proteasome system. BMC Genomics, 10, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G.A. , Chang, R.Z. & Qiu, L.J. (2010) Overexpression of soybean ubiquitin‐conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress‐responsive gene expression in Arabidopsis . Plant Molecular Biology, 72, 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Genome editing events occurring in OsUBC26 null mutants
FIGURE S2 Genome editing events occurring in UCIP2 null mutants
FIGURE S3 Both APIP6 and UCIP2 were ubiquitinated when coexpressed in Escherichia coli
TABLE S1 Up‐regulated genes in OsUBC26 null mutant
TABLE S2 Down‐regulated genes in OsUBC26 null mutant
TABLE S3 Primers used in this study
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


