Abstract
The yeast Saccharomyces cerevisiae transcription factor Ste12p is responsible for activating genes in response to MAP kinase cascades controlling mating and filamentous growth. Ste12p is negatively regulated by two inhibitor proteins, Dig1p (also called Rst1p) and Dig2p (also called Rst2p). The expression of a C-terminal Ste12p fragment (residues 216 to 688) [Ste12p(216–688)] from a GAL promoter causes FUS1 induction in a strain expressing wild-type STE12, suggesting that this region can cause the activation of endogenous Ste12p. Residues 262 to 594 are sufficient to cause STE12-dependent FUS1 induction when overexpressed, and this region of Ste12p was found to bind Dig1p but not Dig2p in yeast extracts. In contrast, recombinant glutathione S-transferase–Dig2p binds to the Ste12p DNA-binding domain (DBD). Expression of DIG2, but not DIG1, from a GAL promoter inhibits transcriptional activation by an Ste12p DBD-VP16 fusion. Furthermore, disruption of dig1, but not dig2, causes elevated transcriptional activation by a LexA–Ste12p(216–688) fusion. Ste12p has multiple regions within the C terminus (flanking residue 474) that can promote multimerization in vitro, and we demonstrate that these interactions can contribute to the activation of endogenous Ste12p by overproduced C-terminal fragments. These results demonstrate that Dig1p and Dig2p do not function by redundant mechanisms but rather inhibit pheromone-responsive transcription through interactions with separate regions of Ste12p.
Ste12p activates signal-responsive transcription in Saccharomyces cerevisiae. In haploid yeasts, Ste12p is required for the response to mating pheromone produced by the opposite mating type and for invasive growth, possibly in response to limiting nutrients (13). In diploids, Ste12p regulates pseudohyphal development in response to nitrogen starvation (13). In each case, Ste12p induces the transcription of genes necessary to produce the appropriate cell cycle progression and morphological alterations. Since Ste12p is necessary for responses to separate signals that cause substantial changes in the organism, its activity must be tightly regulated. Part of the differential function of Ste12p in regulating separate classes of genes is mediated through interactions with different DNA-binding partners. Ste12p binds cooperatively with itself, Mcm1p, and α1p to regulate pheromone-responsive genes (36, 37, 42, 43) and with Tec1p to activate genes required for filamentous growth (22).
Regulation of Ste12p's function in pheromone and filamentous responses appears to involve overlapping signaling mechanisms that control the MAP kinases Fus3p and Kss1p, respectively (23). In response to mating pheromone, Fus3p is thought to phosphorylate substrates that mediate the activation of pheromone-responsive transcription and cause the transient G1 cell cycle arrest required for mating. Downstream targets of Fus3p may include Ste12p (9, 16, 38), the two inhibitors of Ste12p encoded by DIG1 (also called RST1) and DIG2 (also called RST2) (3, 40), and Far1p, which inhibits Cdc28-G1 cyclin complexes and promotes pheromone-responsive cell cycle arrest (9, 26, 41). The unactivated form of Fus3p has also been shown to inhibit inappropriate activation of Kss1p by the pheromone response pathway (23). Similarly, the unactivated form of Kss1p inhibits filamentous response element-dependent transcription, while active Kss1p is required for the expression of filamentous response genes (2, 23). Like Fus3p, Kss1p is known to phosphorylate Dig1p and Dig2p (5), but the functional significance of these phosphorylations has not been determined.
Two inhibitors of Ste12p encoded by DIG1 and DIG2 were identified in two-hybrid screens with Kss1p (5) and Cln1p and Cln2p (40) and have been shown to be present in complexes that also contain Ste12p and Kss1p and/or Fus3p (5, 40). Dig1p and Dig2p appear to negatively regulate the function of Ste12p in both filamentous growth and pheromone response (3, 5, 40). Pheromone treatment causes phosphorylation of Dig1p and Dig2p (40), and it has been suggested that the activation of Ste12p may be mediated through inhibition of the function of these negative regulators (40). Consistent with this model, a minimal pheromone-responsive segment of Ste12p was shown to interact with Dig1p and Dig2p in a two-hybrid analysis (27). Dig1p and Dig2p are 22% identical over their entire sequences and share a 60-amino-acid segment with 64% similarity. Disruption of DIG1 or DIG2 individually has no apparent effect on cell morphology or pheromone response, but yeasts bearing disruptions of both dig1 and dig2 form extensive filaments and show elevated expression of pheromone-responsive genes (5, 30, 40). Because of their sequence similarity and apparent phenotypic redundancy, these two inhibitors have generally been considered to have similar, if not identical, functions (3, 5, 40). However, DIG1 is expressed constitutively, whereas DIG2 has a cluster of upstream pheromone response elements and is induced approximately twofold in response to pheromone (5, 30).
Because an understanding of Ste12p regulation is complicated by its interaction with multiple regulatory proteins and DNA-binding partners, we have sought to simplify analysis of the functions of Dig1p and Dig2p by examining their effects on the pheromone-responsive gene FUS1, which Ste12p can activate on its own (14). We have found that Dig1p and Dig2p bind to separate regions of Ste12p; Dig1p interacts directly with the Ste12p central region (residues 309 to 547), while Dig2p interacts with the DNA-binding domain (DBD) (residues 21 to 195). These interactions are necessary to inhibit pheromone-responsive transcription by Ste12p in vivo. These results demonstrate that Dig1p and Dig2p are not mechanistically redundant but rather must inhibit Ste12p function through independent mechanisms.
MATERIALS AND METHODS
Plasmids, yeast strains, and yeast techniques.
The yeast strains used for these experiments are listed in Table 1. WHY4 is an SY2585 derivative in which ste12 was disrupted by use of plasmid pSUL16 (11). The dig1Δ173 disruption was produced with pIS173, which is a URA3 two-step disruption plasmid that removes DIG1 nucleotides −125 to +1050. The ste12Δ12 disruption was generated similarly with plasmid pAO012, which deletes nucleotides −493 to +640. Disruptions were confirmed by PCR and Southern blotting. Plasmids pJL1 and pYe/STE12ΔXba, which express GAL-inducible wild-type (WT) Ste12p and Ste12p without the DBD (Ste12pΔDBD) (residues 216 to 688), respectively, have been described previously (16). Ste12p deletion mutants (see Fig. 2A) were produced by amplification in vitro using the oligonucleotides listed in Table 2 and cloned into pYeDP8-1/2 (7) as KpnI/EcoRI fragments. Plasmids p10, p11, and p12 were constructed by subcloning BamHI deletion fragments from p2 into p5, p1 into p5, and p3 into p6, respectively. The LEU2 GAL-STE12 deletion plasmids pIS222 (residues 216 to 688), pIS224 (356 to 688), and pIS225 (216 to 500) contain a LEU2 BglII fragment (made blunt) inserted into the EcoRV site of URA3 in their respective parents described above. His6-Ste12p DBD expression plasmids were produced by cloning EcoRI/BamHI fragments produced by amplification with combinations of the oligonucleotides indicated in Table 2 into pRSET-A. pAO003, which expresses Ste12p DBD from a GAL promoter, contains an EcoRI/BamHI fragment from pRSET-A (residues 1 to 215) subcloned into pYeDP8-1/2. The Ste12p DBD-VP16 fusion was created by cloning a DBD-encoding fragment from pSTE12-7 (16) into the EcoRI site of pM3VP16 (31). An Ste12p DBD-VP16 XhoI/HindIII fragment was then made blunt with PolIK and cloned into the BamHI (made blunt) site of pYeDP8-1/2 to produce pSTVP/235. pG4-DBD, for expression of His6-Gal4p DBD (residues 1 to 147) in Escherichia coli, contains a HindIII/EcoRI fragment from pMA241 (21) in pRSET-B. Plasmids for the expression of Dig1p and Dig2p from GAL promoters in yeasts (pG1T and pG2T, respectively) and as glutathione S-transferase (GST) fusions in E. coli (pT580 and pT581, respectively) were as described previously (40). GST-Ste12p E. coli expression plasmids pGT11 (residues 216 to 594), pGT12 (216 to 500), pGT16 (262 to 688), pGT14 (356 to 688), and pGT15 (450 to 688) contain KpnI/EcoRI fragments as described above cloned into pGT10, which is pGEX4-T3 with a KpnI linker inserted into the BamHI (made blunt) site. pMHLex and pIS181 are TRP1 ARS-CEN plasmids expressing LexA from the ADH1 promoter, followed by multiple cloning sites (39). LexA-Ste12p yeast expression plasmids pIS196 (residues 1 to 688) and pIS182 (215 to 688) contain an EcoRI fragment and pIS183 (215 to 473) contains an EcoRI/BamHI fragment from corresponding Gal4 fusion plasmids (38) cloned into pMHLex. LexA-Ste12p expression plasmids pIS184 (residues 216 to 500), pIS187 (262 to 688), pIS188 (356 to 688), pIS189 (403 to 688), and pIS194 (450 to 688) contain KpnI/EcoRI fragments as described above in pIS181. His6-Gal4p (residues 1 to 93) fusions were expressed in E. coli using pRJR1 (29).
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| SY2585 | MATa leu2 trp1 ura3 ade2 mfa2Δ::FUS1-LacZ his3::FUS1-HIS3 | C. Boone |
| WHY4 | MATa leu2 trp1 ura3 ade2 mfa2Δ::FUS1-LacZ his3::FUS1-HIS3 ste12::LEU2 | This study |
| ISY37 | MATa ade2 his3 leu2 trp1 ura3 can1 dig1Δ173 | This study |
| W303-1A | MATa leu2 trp1 ura3 ade2 his3 can1 | H. Ronne |
| YCN7 | MATa leu2 trp1 ura3 ade2 his3 can1 ste12Δ12 dig1Δ173 dig2::HIS3 | This study |
| MT1147 | MATa leu2 trp1 ura3 ade2 his3 can1 dig2::HIS3 | 40 |
| MT1154 | MATa leu2 trp1 ura3 ade2 his3 can1 dig1::TRP1 | 40 |
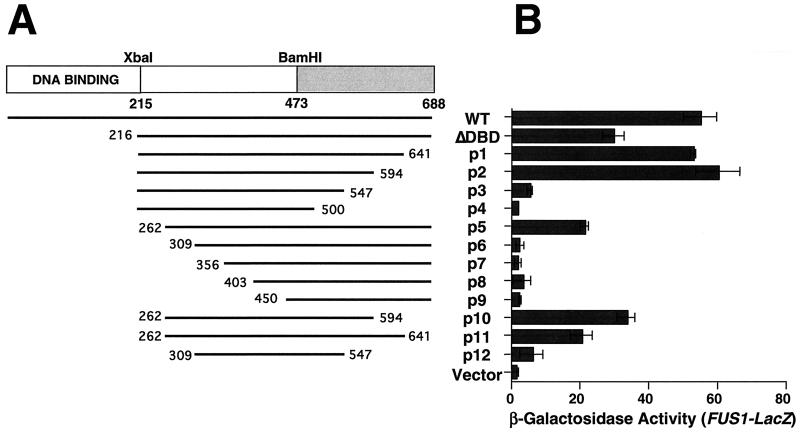
FIG. 2.
Ste12p residues 262 to 594 cause elevated FUS1 transcription when overexpressed in STE12 yeast. (A) Strain SY2585 (STE12) bearing plasmids expressing Ste12p fragments from a GAL promoter or pYEDP8-1/2 (vector) were grown to mid-log phase and induced with galactose for 2 h. (B) Relative FUS1-LacZ transcription was measured by assaying β-galactosidase activity.
TABLE 2.
Oligonucleotides
| Name | Ste12p amino acida | Sequence (5′ to 3′) |
|---|---|---|
| AO1 | 215F | GGTACCATGTCTAGAAGACCATCTAGTACAACA |
| AO7 | 262F | GGTACCATGCCCTCTCAAATTAATGATTTTATT |
| AO8 | 309F | GGTACCATGGACTATTTTCTTGTATCTGTTGAA |
| AO9 | 351F | GGTACCATGTCTCTTCTTAATAGATACCCCTAT |
| AO10 | 403F | GGTACCATGGACCCTACCAGCTACATGAAGTAT |
| AO11 | 450F | GGTACCATGCAATCTTACCCAAACGGAATGGTT |
| AO12 | 500R | GAATTCTCATTGTGGATACAGCATATTGTTATC |
| AO13 | 547R | GAATTCTCACTGCATGGAATTTGAACTTTGCAT |
| AO14 | 594R | GAATTCTCAATTTCCTTGTGAAGACTTCATTCC |
| AO15 | 641R | GAATTCTCAAGAATCTTCGTCACCAGCACTTGG |
| AO4 | 688R | GAATTCTCAGGTTGCATCTGGAAGGTTTTTATC |
| VT1 | 1F | CTGGATCCATGAAAGTCCAAATAACCAATAGT |
| SBD1 | 21F | CTGGATCCATGGAAAACGATGAAGTCAGTAAAGCT |
| AO29 | 45F | CTGGATCCATGTTCTTTTTAGCCACAGCG |
| VT2 | 215R | TCGAATTCTCATCTAGAATCTAAATGTTGAAGTAA |
| SDB2 | 195R | TCGAATTCTCATGAAAAAGATAAGGCGGGCTCATT |
| AO27 | 170R | TCGAATTCTCATTCCAACGCATCCG |
Terminal Ste12p amino acid residue encoded by amplified DNA fragments produced with the indicated oligonucleotide. Priming direction: F, forward; R, reverse.
Unless indicated otherwise, cells were grown in minimal selective medium to an optical density at 600 nm of 0.8 and induced with 2% galactose or 2 μg of α-factor (Sigma) per ml. β-Galactosidase activity in permeabilized cells was determined as described previously (1). RNA was extracted by the phenol-freeze technique (35), and specific transcripts were measured by Northern blotting (1).
Recombinant proteins and antibodies.
GST and His6 fusion proteins were expressed in E. coli and batch purified with glutathione-agarose (Sigma) and Ni-agarose, respectively (1). Extracts from E. coli RR1 expressing TRPE-Ste12p from plasmid pTES216 (16) were prepared as described previously (34). A recombinant baculovirus for expressing native WT Ste12p was produced by cotransfection of Autographa californica nuclear polyhedrosis virus (AcMNPV) DNA into SF9 cells with pBVS12, which contains the STE12 open reading frame cloned into the EcoRV/BamHI sites of pACYM1 (25). Extracts from infected cells were produced by Dounce homogenization in SF9 lysis buffer (20 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 3 mM dithiothreitol [DTT], 0.7 mM leupeptin, 2 μM pepstatin, 2 mM benzamidine, 2 μg of chymostatin per ml, 100 μg of tolylsulfonyl phenylalanyl chloromethyl ketone [TPCK] per ml) and clarified by centrifugation at 12,000 × g for 20 min. For measuring interactions between recombinant proteins, 5 μg of GST-Gal4 or His6-Gal4 fusion protein was mixed in GST lysis buffer (1 mM DTT, 0.1% Nonidet P-40, 250 mM NaCl, 50 mM NaF, 5 mM EDTA, 50 mM Tris [pH 7.5], 1 mM PMSF, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml, 10 μg of soybean trypsin inhibitor per ml, 10 μg of TPCK per ml, 0.6 mM dimethylaminopurine) with His6-Ste12p DBD, His6-Gal4p DBD, or 100 μg of E. coli lysates containing TRPE-Ste12p or was mixed in GST lysis buffer supplemented with 1 mg of bovine serum albumin per ml and 100 μg of total protein from infected SF9 extracts expressing WT Ste12p. Recombinant GST fusions and associated proteins were recovered with glutathione-agarose as described below and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting. Rabbit anti-histidine tag antibodies were purchased from Santa Cruz. Rabbit anti-Gal4p and anti-Ste12p (residues 216 to 688) [Ste12p(216–688)] antibodies have been described previously (33). Rabbit anti-Ste12p DBD antibodies were produced against His6-Ste12p(1–215) using standard techniques (15).
Metabolic labeling, immunoprecipitation, and protein affinity precipitation.
Cells bearing GAL1-STE12 expression plasmids were starved for 20 min in Met-negative medium prior to labeling at 30°C with 1.2 mCi of [35S]methionine per ml in the presence of 2% galactose for 2 h. Immunoprecipitation with anti-Ste12p polyclonal antibody was done as described previously (16). Labeled lysates used to assay interactions with recombinant fusion proteins were prepared in GST lysis buffer as for immunoprecipitation. The lysates were precleared by incubation with 20 μg of GST and 50 μl of glutathione-agarose per ml for 1 h at 4°C, followed by microcentrifugation at 2,000 × g for 2 min. Clarified lysates were incubated with 5 μg of recombinant GST, GST-Dig1p, or GST-Dig2p for 1 h on ice. Following the addition of 25 μl of glutathione-agarose, the samples were incubated for an additional 1 h at 4°C with gentle agitation. The beads were washed three times with GST lysis buffer, and bound proteins were eluted by incubation for 30 min in GST lysis buffer plus 5 mM glutathione. Labeled proteins were resolved by SDS–10% PAGE and detected by autofluorography.
RESULTS
Overexpression of Ste12p C-terminal fragments causes the induction of a pheromone-responsive gene in the absence of pheromone.
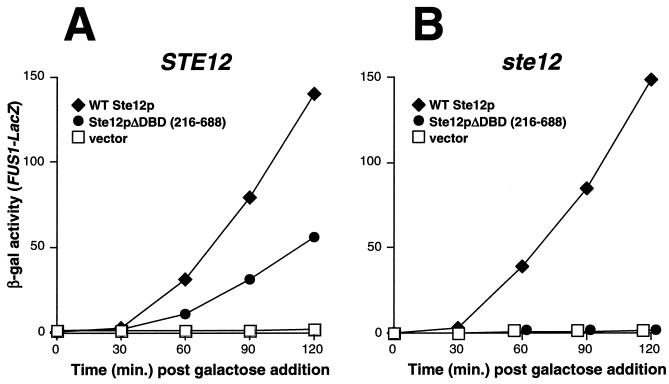
The ability of Ste12p to activate transcription is inhibited in the absence of pheromone by at least two negative regulators, Dig1p and Dig2p (5, 40). To identify the region(s) of Ste12p that is necessary for inhibition by these proteins, we initiated a strategy similar to that used to examine the regulation of Gal4p by the inhibitor Gal80p. The overproduction of Gal4p C-terminal fragments containing the major region of interaction with Gal80p causes the induction of GAL transcription in the absence of galactose by competing for the binding of Gal80p with endogenous WT Gal4p (17, 20). Consistent with previous results (38), we found that the overexpression of WT Ste12p from a galactose-inducible promoter in either WT or ste12 yeast caused elevated transcription from a pheromone-responsive promoter (FUS1-LacZ) in the absence of pheromone (Fig. 1). Also, similar to the results obtained with Gal4p, we found that FUS1-LacZ transcription could be induced in the absence of pheromone by overexpression of a truncated Ste12p derivative [Ste12p(216 to 688)] lacking the DBD in yeast cells expressing endogenous WT STE12 (Fig. 1A) but not in cells bearing an ste12 disruption (Fig. 1B). This result demonstrates that endogenous Ste12p can be activated in the absence of pheromone by overproduction of the Ste12p C terminus (residues 216 to 688). In view of our original rationale, one interpretation of this result is that the Ste12p C terminus might bind one or more inhibitors and that its overproduction causes induction by competing for the binding of inhibitors with endogenous Ste12p. However, we demonstrate below that the Ste12p C terminus has several segments that can promote multimerization. Therefore, overexpression of residues 216 to 688 likely causes activation by a mechanism involving a direct interaction with endogenous Ste12p, in addition to competition for the binding of negative regulatory proteins.
FIG. 1.
Overexpression of Ste12p residues 216 to 688 causes STE12-dependent activation of FUS1-LacZ transcription. Yeast strains SY2585 (STE12) (A) and WHY4 (ste12) (B) bearing plasmids pYeDP8-1/2 (vector), pJL1 (WT Ste12p), and pYe/STE12ΔXba (Ste12pΔDBD) were induced with galactose, and FUS1-LacZ expression was determined by measurement of β-galactosidase (β-gal) activity at the indicated times.
To identify the region of Ste12p necessary for causing the induction of FUS1-LacZ expression when overproduced, we expressed a set of C-terminal deletions from the GAL1 promoter (Fig. 2A). We found that sequences C terminal to residue 594 or N terminal to residue 262 could be deleted from overexpressed Ste12p without preventing the elevation of FUS1-LacZ reporter gene expression (Fig. 2B). In contrast, overexpression of Ste12p fragments bearing C-terminal truncations to residues 547 and N-terminal truncations to 309 did not cause a significant elevation of FUS1-LacZ transcription (Fig. 2B). Consistent with these results, expression of a fragment spanning residues 262 to 594 (Fig. 2B, p10) caused the activation of FUS1-LacZ expression, whereas a smaller fragment spanning residues 309 to 547 had no effect (Fig. 2B, p12). These observations indicate that endogenous Ste12p can be activated in the absence of pheromone by overproduction of a C-terminal Ste12p fragment spanning residues 262 to 594.
Dig1p and Dig2p interact with separate regions of Ste12p.
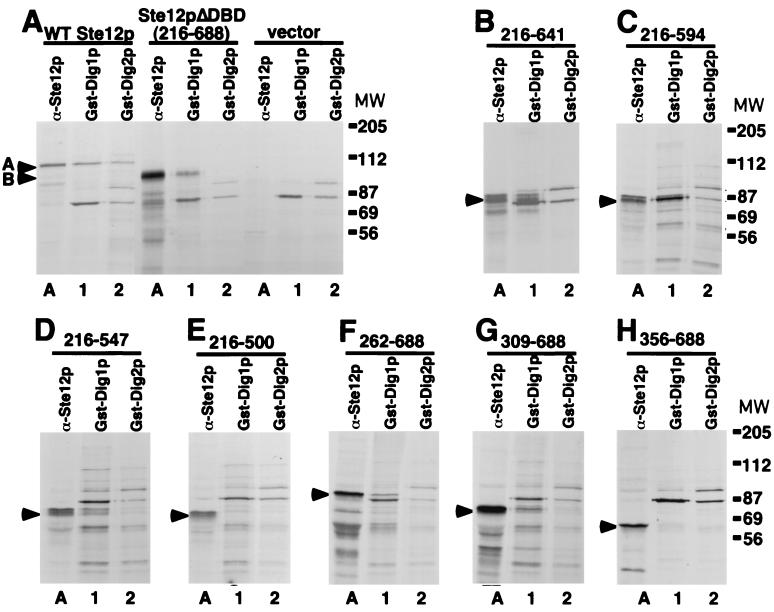
Dig1p and Dig2p are present in complexes with Ste12p and Kss1p or Fus3p in vivo (5, 40). Since overexpression of Ste12p residues 262 to 594 causes activation of endogenous Ste12p, we determined whether Dig1p or Dig2p interacts directly with this region. We examined whether Ste12p deletion fragments (Fig. 2A) could be recovered from labeled total yeast extracts by protein affinity precipitation with recombinant GST-Dig1p and GST-Dig2p. As shown previously (16), WT Ste12p and Ste12pΔDBD are readily detected by immunoprecipitation with polyclonal anti-Ste12p antibodies when expressed from a galactose-inducible promoter in [35S]methionine-labeled cells (Fig. 3A, lanes A). Recombinant GST-Dig1p mixed with [35S]methionine-labeled extracts prepared from ste12 cells and recovered with glutathione-agarose bound a single labeled protein of approximately 80 kDa (Fig. 3A, vector, lane 1), while similarly treated GST-Dig2p bound two labeled proteins of approximately 80 and 92 kDa (Fig. 3A, vector, lane 2). The identities of these 80- and 92-kDa proteins are unknown. Using this technique, we found that WT Ste12p could be recovered from labeled extracts by affinity precipitation with both GST-Dig1p and GST-Dig2p (Fig. 3A, WT Ste12p, lanes 1 and 2, respectively). In contrast, we found that the Ste12pΔDBD derivative could be recovered from extracts with GST-Dig1p but not GST-Dig2p (Fig. 3A, Ste12pΔDBD, compare lanes 1 and 2). These results demonstrate that recombinant Dig1p and Dig2p specifically interact with Ste12p plus several additional proteins in labeled yeast extracts. Furthermore, we found that both Dig1p and Dig2p bound WT Ste12p but that only Dig1p was capable of interacting with the Ste12pΔDBD derivative. This result indicates that these inhibitors must bind to different regions of Ste12p. Dig2p requires the DBD (residues 1 to 215) for interaction with Ste12p, whereas Dig1p can interact with residues 216 to 688.
FIG. 3.
Dig1p and Dig2p interact with different regions of Ste12p in yeast extracts. Strain WHY4 (ste12) bearing Ste12p expression plasmids pJL1 (A, WT Ste12p), pYe/STE12ΔXba (A, Ste12pΔDBD), and control pYeDP8-1/2 (A, vector) or plasmids expressing Ste12p deletions p1 (B), p2 (C), p3 (D), p4 (E), p5 (F), p6 (G), and p7 (H) was labeled with [35S]methionine in the presence of galactose. Labeled extracts were immunoprecipitated with Ste12p(216–688) polyclonal antibodies (lanes A) or analyzed by protein affinity precipitation with recombinant GST-Dig1p (lanes 1) or GST-Dig2 (lanes 2). Recovered proteins were resolved by SDS-PAGE and visualized by autofluorography. Migration of WT Ste12p and Ste12pΔDBD is indicated by arrowheads labeled A and B, respectively, in panel A. Migration of the Ste12p C-terminal fragments in panels B to H is indicated by an arrowhead. MW, molecular weight (in thousands).
To identify the region of Ste12p that binds GST-Dig1p, we examined interactions with the Ste12p C-terminal deletion fragments (Fig. 2A) by protein affinity precipitation from labeled yeast extracts. We found that Ste12p C-terminal truncations p1 (215 to 641) and p2 (215 to 594) were efficiently recovered from labeled yeast extracts with GST-Dig1p but not GST-Dig2p, while p3 (215 to 547) was recovered slightly less efficiently with GST-Dig1p (Fig. 2B, C, and D). In contrast, the C-terminal truncation p4 (215 to 500) did not interact with GST-Dig1p or Dig2p (Fig. 3E). Similarly, the Ste12p N-terminal truncations p5 (262 to 688) and p6 (309 to 688) were also recovered by GST-Dig1p (Fig. 3F and G), but the smaller N-terminal truncations p7 (356 to 688) (Fig. 3H) and p8 (403 to 688) and p9 (450 to 688) (data not shown) did not interact with either GST-Dig1p or GST-Dig2p. These results indicate that residues 309 to 547 of Ste12p are required for the most efficient interaction with recombinant Dig1p. Furthermore, the fact that none of the truncated Ste12p C-terminal fragments interacted with Dig2p supports the conclusion that the two inhibitors interact with separate regions of Ste12p.
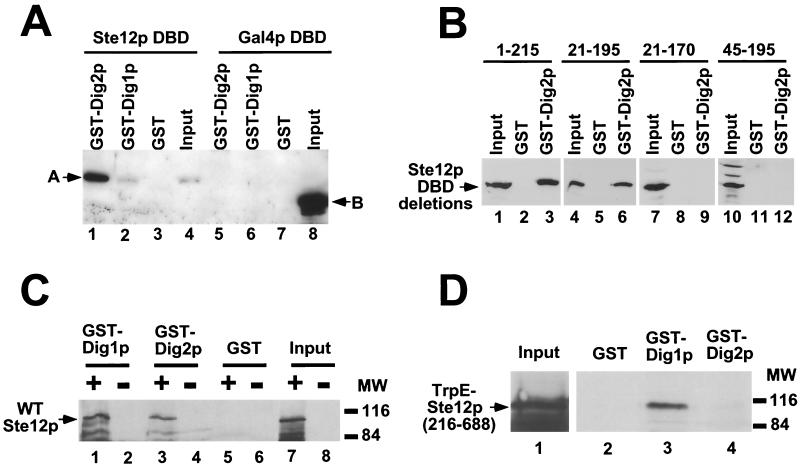
Ste12p is known to interact with other proteins in addition to Dig1p and Dig2p, including the transcription factors Mcm1p (10), α1p (43), and Tec1p (22) and the MAP kinases Kss1p and Fus3p (2, 3, 5, 40). We also observed at least two additional proteins, of 80 and 92 kDa, interacting with GST-Dig2p and one protein, of 80 kDa, interacting with GST-Dig1p in protein affinity precipitations of labeled yeast extracts (Fig. 3A). Therefore, it was necessary to determine whether Dig1p and Dig2p were capable of direct interactions with separate regions of recombinant Ste12p in the absence of additional yeast proteins. To examine whether GST-Dig2p interacts directly with the Ste12p DBD, we expressed this fragment (residues 1 to 215) in E. coli as a His6-tagged fusion (Fig. 4A, lane 4). We found that His6-Ste12p DBD is bound efficiently by GST-Dig2p (Fig. 4A, lane 1) but not significantly by GST-Dig1p (lane 2). Neither GST fusion protein interacted with His6-Gal4p DBD (Fig. 4A, lanes 5 to 8). Further deletion analysis demonstrated that residues 21 to 195 of Ste12p comprise the minimal Dig2p-binding region. GST-Dig2p interacted with His6-Ste12p(21–195) (Fig. 4B, lane 6) but not smaller fragments containing residues 21 to 170 or 45 to 195 of Ste12p (Fig. 4B, lanes 9 and 12). This result indicates that Dig2p binds to Ste12p at the same region required for DNA binding (42) (data not shown). GST-Dig1p did not interact with any of the Ste12p DBD deletion constructs (data not shown).
FIG. 4.
Dig1p and Dig2p interact with separate parts of recombinant Ste12p in vitro. (A) Recombinant His6-Ste12p DBD (lanes 1 to 4) and His6-Gal4p DBD (lanes 5 to 8) were mixed with GST-Dig2p (lanes 1 and 5), GST-Dig1p (lanes 2 and 6), or GST (lanes 3 and 7). Bound proteins were recovered with glutathione-agarose, resolved by SDS-PAGE, and detected by immunoblotting with anti-His6 tag antibodies. One-twelfth the input amount of His6-Ste12p DBD and His6-Gal4p DBD was analyzed directly by SDS-PAGE (lanes 4 and 8), and their migration is indicated as A and B, respectively. (B) Recombinant His6-Ste12p (DBD) fragments spanning residues 1 to 215 (lanes 1 to 3), 21 to 195 (lanes 4 to 6), 21 to 170 (lanes 7 to 9), or 45 to 195 (lanes 10 to 12) were assayed for interaction with GST (lanes 2, 5, 8, and 11) or GST-Dig2p (lanes 3, 6, 9, and 12). An equivalent amount of input Ste12p DBD was loaded in lanes 1, 4, 7, and 10. (C) Extracts from SF9 cells infected with WT Ste12p-expressing baculovirus (+, odd lanes), or control AcMNPV (−, even lanes) were analyzed directly by SDS-PAGE (lanes 7 and 8) or mixed with GST-Dig1p (lanes 1 and 2), GST-Dig2p (lanes 3 and 4), or GST (lanes 5 and 6). Bound proteins were recovered with glutathione-agarose and analyzed by SDS-PAGE and immunoblotting with Ste12p(216–688) antibodies. (D) E. coli lysates containing TRPE–Ste12p(216–688) (input, lane 1) were mixed with GST (lane 2), GST-Dig1p (lane 3), or GST-Dig2p (lane 4), and bound proteins were analyzed by SDS-PAGE and immunoblotting with Ste12p(216–688) antibodies. MW, molecular weight (in thousands).
Full-length recombinant WT Ste12p was also produced by expression in insect cells using a baculovirus vector (Fig. 4C, lane 7). Consistent with the results shown above (Fig. 3A), we found that recombinant WT Ste12p could be recovered from infected insect cell extracts with both GST-Dig1p (Fig. 4C, lane 1) and GST-Dig2p (lane 3). These results demonstrate that Dig1p and Dig2p are able to interact with Ste12p in the absence of other yeast proteins. However, since the unactivated form of Kss1p has been shown to bind and inhibit Ste12p (2), an effect that appears to require Dig1p or Dig2p (3), interpretation of the latter result may be complicated by the fact that insect cells express MAP kinases. To determine whether Dig1p interacts directly with the Ste12p central region, we used a recombinant TRPE–Ste12p(216–688) fusion produced in E. coli (16) (Fig. 4D, lane 1). Consistent with the results shown above, we found that TRPE–Ste12p(216–688) was bound by GST-Dig1p (Fig. 4D, lane 2) but not by GST-Dig2p (lane 4). These results demonstrate that Dig1p and Dig2p interact directly with separate regions of Ste12p. Dig2p binds to the Ste12p DBD (residues 1 to 215), while the strongest interaction of Dig1p with Ste12p requires a region spanning residues 309 to 547 (Fig. 3).
Dig1p and Dig2p inhibit Ste12p by separate interactions in vivo.
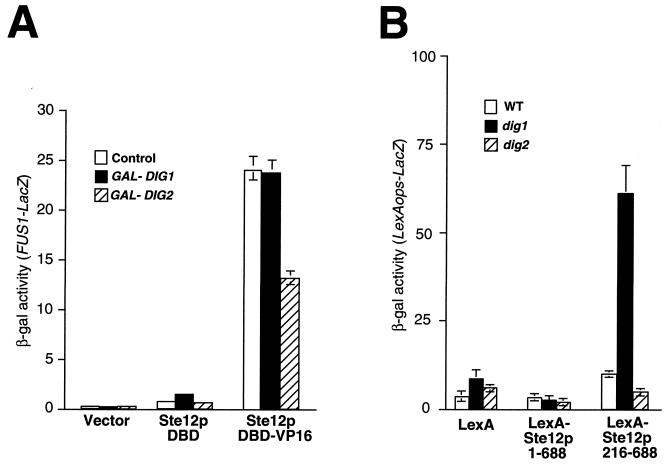
Since Dig1p and Dig2p interact with separate segments of Ste12p in vitro, we examined whether we could dissociate their inhibitory effects on Ste12p in vivo. We expressed the Ste12p DBD on its own (residues 1 to 215) and as a fusion to the strong transcriptional activation domain of herpes simplex virus type 1 VP16 (32) (Ste12p DBD-VP16) from GAL promoters. Ste12p DBD produced on its own caused a slight elevation of FUS1-LacZ transcription relative to the vector control (Fig. 5A), suggesting that the DBD might possess a weak transcriptional activation function. However, the Ste12p DBD-VP16 fusion activated FUS1-LacZ expression approximately 25-fold more than Ste12p DBD (Fig. 5A). We found that simultaneous expression of Dig2p but not Dig1p from a GAL promoter inhibited the activation of FUS1-LacZ expression by the Ste12p DBD-VP16 fusion (Fig. 5A). We also examined the effect of Ste12p DBD and Ste12p DBD-VP16 on the expression of the endogenous FUS1 gene by Northern blotting. Consistent with the results of Fig. 5A, we found that Ste12p DBD-VP16 strongly activated FUS1 transcription and that this effect could be inhibited by simultaneous overexpression of DIG2 but not DIG1 (data not shown). These results demonstrate that Dig2p inhibits Ste12p in vivo by its direct interaction with the DBD.
FIG. 5.
Dig1p and Dig2p inhibit Ste12p through interactions with different regions in vivo. (A) Strain WHY4 bearing plasmids pYeDP8-1/2 (vector), pAO003 (Ste12p DBD), and pSTVP/235 (Ste12p DBD-VP16) was cotransformed with YEplac112 (control), pG1T (GAL-DIG1), and pG2T (GAL-DIG2). Cells were grown to mid-log phase, and FUS1-LacZ expression was measured by assaying β-galactosidase (β-gal) activity 2 h after galactose addition. (B) Yeast strains W303-1A (WT), ISY37 (dig1), and MT1147 (dig2) bearing plasmid pSH18-34 (LexA ops-LacZ) were cotransformed with pMHLex (LexA), pIS196 [LexA-Ste12p(1–688)], or pIS182 [LexA-Ste12p(216–688)]. Transcriptional activation by LexA fusions was assayed by measuring β-galactosidase activity in cells grown to mid-log phase.
To examine whether Dig1p inhibits Ste12p by interaction with the central region, we examined activation by LexA-Ste12p fusions in dig1 and dig2 yeast strains (Fig. 5B). Consistent with previous observations (38), we found that LexA-Ste12p(1–688) and LexA-Ste12p(216–688) fusions were weak activators in the absence of pheromone (Fig. 5B), but activation could be stimulated by pheromone treatment in WT cells (data not shown). However, we found that activation by LexA-Ste12p(216–688) in the absence of pheromone was elevated in dig1 yeast cells relative to WT cells but not in dig2 yeast cells (Fig. 5B). In contrast, activation by the LexA–full-length Ste12p fusion [LexA-Ste12p(1–688)] was unaffected by disruption of either dig1 or dig2 in unstimulated cells (Fig. 5B). These results indicate that in the absence of pheromone, LexA-Ste12p(1–688) must be negatively regulated by both Dig1p and Dig2p, whereas LexA-Ste12p(216–688) is only inhibited by Dig1p. Together with the above results, these observations support the view that Dig1p and Dig2p inhibit Ste12p through interactions with separate regions.
Overexpression of Ste12p(216–688) does not cause the activation of endogenous Ste12p solely by competing for the binding of Dig1p.
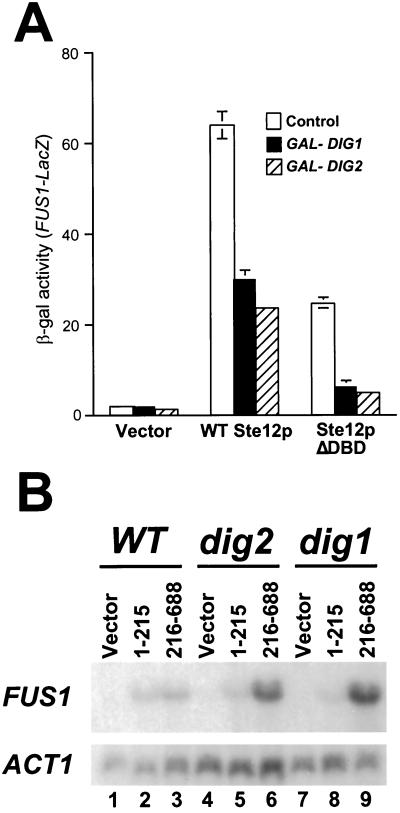
As indicated above, we initially examined the effect of overexpressed Ste12p C-terminal fragments with the rationale that they should cause the activation of endogenous Ste12p by competing for binding of the inhibitor proteins. However, several observations indicate that overexpressed Ste12p(216–688) cannot cause activation merely by competition for Dig1p. First, as shown above, overproduction of Ste12p(262–594) is required to cause the activation of FUS1 transcription by endogenous Ste12p (Fig. 2); in contrast, a smaller segment (residues 309 to 547) seems to be necessary for efficient interaction with Dig1p (Fig. 3). Second, Ste12p(216–688) does not interact directly with Dig2p (Fig. 3 and 4), yet overexpression of this region can activate endogenous Ste12p in a WT (DIG2) strain (Fig. 1). Furthermore, simultaneous overexpression of DIG2 inhibited transcriptional activation by both WT Ste12p and Ste12pΔDBD as efficiently as overexpression of DIG1 (Fig. 6A). We also directly examined whether Dig1p was required for the activation of FUS1 transcription by overexpressed Ste12p(216–688) (Fig. 6B). We found that overexpression of residues 216 to 688 caused much more extensive induction of FUS1 transcription in both dig1 and dig2 yeast cells than in WT cells (Fig. 6B, compare lanes 6 and 9 with lane 3). In contrast, overexpression of the Ste12p DBD (residues 1 to 215) on its own caused approximately equivalent levels of activation of FUS1 transcription in mutant cells and in WT cells (Fig. 6B, lanes 2, 5, and 8). This result demonstrates that overexpression of Ste12p(216–688) cannot cause FUS1 induction simply by competing with endogenous Ste12p for Dig1p.
FIG. 6.
Activation of endogenous Ste12p by the overexpressed C terminus does not require DIG1. (A) Strain SY2585 (STE12) bearing Ste12p expression plasmid pJL1 (WT Ste12p), pYe/STE12ΔXba (Ste12pΔDBD), or control pYeDP8-1/2 (vector) was cotransformed with pG1T (GAL-DIG1), pG2T (GAL-DIG2), or Yeplac181 (control) (12). Cells were grown to mid-log phase, and FUS1-LacZ expression was measured by assaying β-galactosidase (β-gal) activity 2 h after galactose addition. (B) W303-1A (WT, lanes 1 to 3), MT1147 (dig2, lanes 4 to 6), and MT1154 (dig1, lanes 7 to 9) were transformed with pYeDP8-1/2 (vector, lanes 1, 4, and 7), pAO003 [GAL-Ste12p(1–215), lanes 2, 5, and 8], or pYe/STE12ΔXba [GAL-Ste12p(216–288), lanes 3, 6, and 9]. Cells were grown to mid-log phase and induced with galactose. RNA was extracted 2 h postinduction and analyzed by Northern blotting with FUS1 (top) and ACT1 (bottom) probes.
The overproduced Ste12p C terminus causes transcriptional activation through an interaction with endogenous Ste12p.
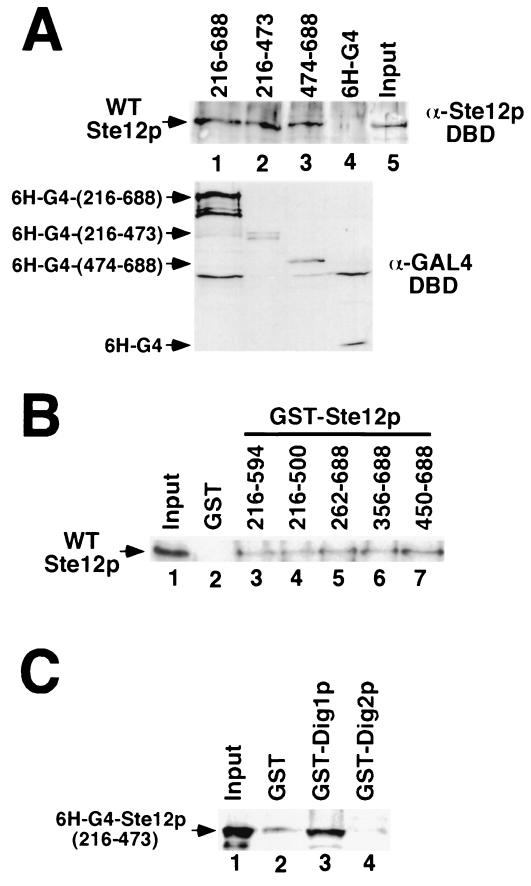
Since the activation of FUS1 transcription by overproduced residues 216 to 688 requires endogenous Ste12p (Fig. 1), we imagined that this effect might be mediated by direct interaction of this fragment with WT Ste12p. To examine this possibility, we determined whether a region(s) in the Ste12p C terminus could promote multimerization in vitro (Fig. 7). We found that recombinant WT Ste12p can interact in vitro with the Ste12p C terminus (residues 216 to 688) fused to the Gal4p DBD (Fig. 7A, lane 1) in coimmunoprecipitation experiments. Smaller Ste12p fragments, spanning residues 216 to 473 (Fig. 7A, lane 2) and 474 to 688 (lane 4), fused to Gal4p also interacted with WT Ste12p in vitro, indicating that multiple sites flanking residue 473 must be able to promote multimerization. We also examined the interaction of recombinant WT Ste12p with GST-Ste12p fusion proteins in vitro (Fig. 7B). Consistent with the results of Fig. 7A, we found that WT Ste12p interacted with GST fused to various Ste12p C-terminal fragments (Fig. 7B, lanes 3 to 7) but not with GST alone (lane 2). GST fused to Ste12p C-terminal fragments containing residues 216 to 500 (Fig. 7B, lane 4) or residues 450 to 688 (lane 7) interacted efficiently with WT Ste12p. Combined with the results of Fig. 7A, these results indicate that multiple segments within the Ste12p C terminus must promote multimerization.
FIG. 7.
Ste12p C-terminal regions cause multimerization in vitro. (A) WT Ste12p SF9 extracts (input, lane 5) were incubated with extracts from E. coli expressing His6-tagged Gal4p DBD (6H-G4, lane 4) or 6H-G4 fused to Ste12p residues 216 to 688 (lane 1), 216 to 473 (lane 2), or 474 to 688 (lane 3). Gal4p fusions were recovered by immunoprecipitation with GAL4 DBD monoclonal antibody, and the interacting Ste12 protein was detected by Western blotting with Ste12p(1–215) antibodies (top). Input 6H-G4 fusion protein was detected by Western blotting with Gal4p DBD antibodies (bottom). (B) WT Ste12p-containing extracts (input, lane 1) were incubated with recombinant GST (lane 2) or GST fused to residues 216 to 594 (lane 3), 216 to 500 (lane 4), 262 to 688 (lane 5), 356 to 688 (lane 6), or 450 to 688 (lane 7) of Ste12p. Bound WT Ste12p recovered with glutathione-agarose was analyzed by SDS-PAGE and immunoblotting with Ste12p DBD antibodies. (C) Extracts from E. coli expressing His6-Gal4p fused to Ste12p(216–473) (input, lane 1) were incubated with recombinant GST (lane 2), GST-Dig1p (lane 3), or GST-Dig2p (lane 4). Bound 6H-G4–Ste12p recovered with glutathione-agarose was detected by immunoblotting with Gal4p DBD antibodies.
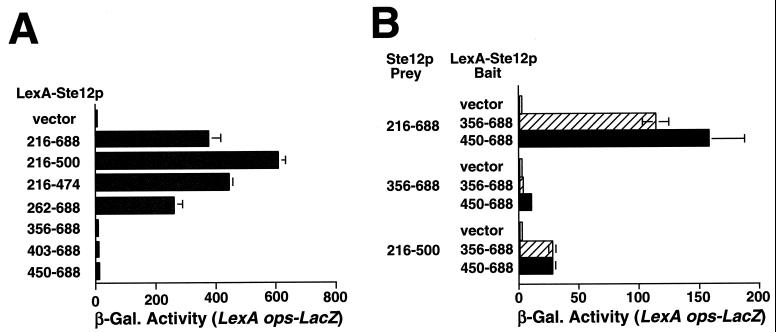
We examined whether overexpression of the Ste12p C terminus could cause activation by multimerization with WT Ste12p in vivo by using a modified two-hybrid system. For this purpose, we first needed to identify Ste12p C-terminal fragments that are incapable of activating transcription for use as bait fusions. We found that Ste12p(216–688) fused to LexA caused strong activation of transcription of a LexA-responsive reporter gene in untreated ste12 dig1 dig2 yeast cells (Fig. 8A, 216 to 688). Deletion of residues C terminal to Ste12p amino acid 474 did not prevent activation by LexA fusions (Fig. 8A, 216 to 474). However, deletion of residues N terminal to amino acid 356 prevented activation by LexA-Ste12p (Fig. 8A, 356 to 688). These results are consistent with previous observations (38) and indicate that the major activating segment of Ste12p resides between amino acids 216 and 356.
FIG. 8.
The Ste12p C terminus can cause activation by multimerization in vivo. (A) Yeast strain YCN7 (ste12 dig1 dig2) bearing pSH18-34 (LexA ops-LacZ) was transformed with pMHLex expressing LexA (vector) or the LexA-Ste12p expression plasmids pIS182 (216 to 688), pIS184 (216 to 500), pIS183 (216 to 474), pIS187 (262 to 688), pIS188 (356 to 688), pIS189 (403 to 688), and pIS194 (450 to 688). Transcriptional activation by LexA fusions was assayed by measuring β-galactosidase (β-Gal) activity in cells grown to mid-log phase. (B) Yeast strain YCN7 bearing pSH18-34 and the LexA-Ste12p bait plasmids (as in panel A; LexA-Ste12p Bait) was cotransformed with vectors producing residues 216 to 288, 356 to 688, or 216 to 500 of Ste12p from a GAL promoter (Ste12p Prey). Cells were grown to mid-log phase, and expression from the LexA-responsive reporter was measured by assaying β-galactosidase activity 2 h after galactose addition.
We next determined whether Ste12p(216–688) could cause the activation of reporter gene expression in the presence of LexA-Ste12p fusions which are incapable of activating transcription on their own but which contain segments that can promote Ste12p multimerization. For this purpose, we used LexA fused to Ste12p(356–688) and Ste12p(450–688), two fragments that can interact with WT Ste12p in vitro as GST fusions (Fig. 7B, lanes 6 and 7). Consistent with the above results, coexpression of Ste12pΔDBD caused the activation of reporter gene expression in the presence of both LexA-Ste12p C-terminal fusions but not with LexA produced on its own (Fig. 8B, Ste12p Prey 216–688). In contrast, Ste12p(356–688) did not cause the activation of transcription when coexpressed with the LexA-Ste12p fusions (Fig. 8B, Ste12p Prey 356–688), indicating that the Ste12p activating region (Fig. 8A) is necessary for activation by overexpressed Ste12p C-terminal fragments. Additionally, coexpression of Ste12p(216–500) also caused much weaker activation in the presence of the LexA-Ste12p fusions (Fig. 8B, Ste12p Prey 216–500), a result which might reflect less efficient multimerization of this derivative in vivo. Note that Ste12p(216–500) can activate transcription efficiently when fused directly to LexA (Fig. 8A) but not when coexpressed with the LexA-Ste12p fusions in Fig. 8B or when produced in cells expressing WT Ste12p (Fig. 2, p4). These observations indicate that overexpression of Ste12p C-terminal fragments likely causes the activation of FUS1 transcription (Fig. 1 and 2) by forming complexes with endogenous Ste12p. In this view, the “activating” fragment must contain both the transcriptional activation region (residues 262 to 356) and sufficient C-terminal sequences to promote multimerization with endogenous WT Ste12p (residues 356 to 594).
Our observation that residues 309 to 547 of Ste12p are required for interaction with Dig1p (Fig. 3) are at odds with previous results indicating that a much smaller segment (residues 301 to 335) is sufficient for interaction with Dig1p in two-hybrid experiments (27). Because Ste12p appears to have C-terminal segments that can promote multimerization (Figs. 7 and 8), we wondered whether this discrepancy results from the fact that the Gal4p DBD forms a stable dimer (4, 24). If Dig1p interacts most efficiently with Ste12p multimers, then interactions in our experiments (Fig. 3) would require segments that can promote efficient multimerization in addition to the region of direct contact between these proteins. In contrast, since the Gal4p DBD itself forms a dimer, interactions of Ste12p fragments with Dig1p in a two-hybrid experiment should require only the region necessary for direct interaction. Consistent with this possibility, we found that GST-Dig1p could bind recombinant Gal4-Ste12p(216–473) in vitro (Fig. 8C, lane 3); in contrast, GST-Dig2p was unable to interact with recombinant Gal4-Ste12p(216–473) (lane 4). Considering that GST-Dig1p is unable to interact with Ste12p fragments lacking residues C terminal of residue 547 when produced as native fragments in yeast cells (Fig. 3), these observations suggest that Dig1p prefers to bind Ste12p multimers under the conditions of our experiments and previous two-hybrid analyses (27).
DISCUSSION
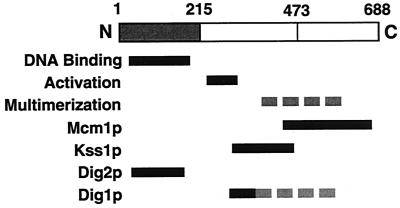
Ste12p is a transcriptional activator whose function involves interactions with multiple DNA-binding partners and that is negatively regulated by several inhibitory proteins (Fig. 9). In this work, we have examined the relationship between the regulators Dig1p and Dig2p and Ste12p's function in activating transcription of the pheromone-responsive gene FUS1. Although Dig1p and Dig2p have generally been considered to have overlapping, if not redundant, functions (2, 3, 5, 40), we demonstrate that they must regulate Ste12p by separate mechanisms. Dig1p binds directly to a central region of Ste12p (residues 309 to 547), while Dig2p binds to the Ste12p DBD (residues 1 to 215) (Fig. 9). These different interactions can account for their inhibitory effect on Ste12p in vivo. Overproduction of Dig2p but not Dig1p inhibits activation by an Ste12p DBD-VP16 fusion protein. In contrast, deletion of dig1 but not dig2 causes constitutive activation by a LexA-Ste12p(216–688) fusion. These observations demonstrate that Ste12p activity is regulated by two inhibitory proteins that function separately. Like Dig1p and Dig2p, the MAP kinases Kss1p and Fus3p were also initially thought to have redundant functions in the pheromone response (8) until it was recognized that these enzymes have inhibitory effects in their unactivated state (2, 23) and that Kss1p is preferentially required for regulation of the filamentous growth response (19).
FIG. 9.
Ste12p interacts with multiple transcription factors and regulatory proteins. Regions of Ste12p required for binding the pheromone response element (DNA binding) and for transcriptional activation are indicated by black bars. Several separate segments flanking residue 473 can promote multimerization (dashed grey bar). Regions required for interactions with Mcm1p, Kss1p, Dig2p, and Dig1p are indicated by black bars. An additional segment contributes to an interaction with Dig1p by causing Ste12p multimerization (dashed grey bar).
Dig1p and Dig2p inhibit Ste12p through interactions with separate regions.
A previous report indicated that both Dig1p and Dig2p interact with residues 301 to 335 of Ste12p, termed the pheromone induction domain (27). This region was shown to confer pheromone inducibility to Gal4p DBD fusions and to interact with Dig1p and Dig2p in two-hybrid assays. We found that a larger region of Ste12p, spanning residues 309 to 547, was required for an efficient interaction with GST-Dig1p. Furthermore, this region of Ste12p did not interact directly with Dig2p in our experiments (Fig. 3 and 8C). These discrepancies are likely related to the fact that Ste12p C-terminal segments can promote multimerization (Fig. 7 and 8). For example, the apparent interaction of Dig2p with the pheromone-responsive domain in previous two-hybrid analyses might be mediated through an interaction with endogenous Ste12p, since these experiments were performed with WT cells (27). Additionally, we found that GST-Dig1p could interact efficiently with Ste12p(216–474) fused to the Gal4p DBD in vitro (Fig. 8C) but not to Ste12p216–547 expressed on its own in yeast cells (Fig. 3). Considering that several regions in the Ste12p C terminus can cause multimerization (Fig. 7 and 8), this difference may be a consequence of the fact that the Gal4p DBD forms stable dimers (4, 24). One implication of this hypothesis is that Dig1p must preferentially interact with Ste12p multimers. However, since the stoichiometry of Ste12p and Dig1p-Ste12p complexes in uninduced and induced conditions has not been established, it is difficult to predict whether this apparent requirement for Dig1p interaction has any significance for the regulation of Ste12p. Nevertheless, in combination with the previous two-hybrid analyses (27), our results suggest that Dig1p directly interacts with residues 301 to 335 but that further C-terminal sequences to 547 contribute to the interaction, perhaps because they are necessary for multimerization (Fig. 9).
Our finding that Dig1p and Dig2p interact with separate regions of Ste12p (Fig. 9) suggests that they inhibit transcription through independent mechanisms. Because Dig2p binds to the DBD and inhibits activation by an Ste12p DBD-VP16 fusion, the simplest model is that this inhibitor modulates the ability of Ste12p to bind to the pheromone response element. In support of this hypothesis, we found that binding of the Ste12p DBD (residues 1 to 215) to a pheromone response element is inhibited by equimolar amounts of recombinant GST-Dig2p but not GST-Dig1p in vitro (data not shown). Furthermore, in vivo footprinting analysis suggests that pheromone treatment causes filling of pheromone response elements on a multicopy FUS1 reporter template (not shown). These results suggest that some Ste12p may be sequestered in a complex with Dig2p prior to pheromone treatment, although we found that Dig2p was produced at considerably lower levels than Dig1p in unstimulated cells (not shown). It is also possible that Dig2p inhibits through a mechanism other than or in addition to prevention of Ste12p DNA binding. Dig1p, in contrast, interacts with a region spanning residues 309 to 547 of Ste12p (Fig. 8). This region also overlaps sequences that are necessary for transcriptional activation (38) (Fig. 8A). Therefore, one possibility is that Dig1p functions in a manner similar to that of Gal80p, which binds directly to the major activation domain of Gal4p and inhibits transcriptional activation in the absence of galactose (20). In this model, we would expect Dig1p to interact with DNA-bound Ste12p to prevent transcriptional activation in the absence of pheromone. However, the precise mechanism by which Dig1p inhibits Ste12p remains to be elucidated and, like that for Dig2p, may require an understanding of the involvement of Kss1p and Fus3p.
The MAP kinase gene KSS1 was initially identified in a multicopy suppressor screen for its ability to promote recovery from pheromone-induced growth arrest (6). It was discovered more recently that the unactivated form of Kss1p functions as a negative regulator of Ste12p's function in the filamentous response (5, 23). The inhibitory effect on filamentous response element-dependent transcription was shown to involve the direct binding of Kss1p to Ste12p (2). Unactivated Kss1p also inhibits the transcription of pheromone-responsive genes, in a manner which appears to be more dependent on Dig1p and Dig2p than on the inhibition of filamentous response element-dependent transcription (3). These observations suggest that the full inhibitory effect of Dig1p and Dig2p on pheromone-responsive transcription might require interactions with the unactivated MAP kinases Kss1p and Fus3p. Perhaps the interactions that we observed between Dig1p, Dig2p, and Ste12p are stabilized in vivo by the MAP kinases (3).
Regulation of Ste12p activity by pheromone-stimulated signaling.
Of the transcription factors that have been characterized to date, Ste12p may be unique in being negatively regulated by two proteins that inhibit through separate mechanisms. Our finding that Dig1p and Dig2p inhibit by nonredundant mechanisms is not surprising, considering the central role that Ste12p plays in coordinating cell fate in response to physiological signaling. Ste12p activity may be induced in response to pheromone through Fus3p-mediated phosphorylation of the inhibitors and/or Ste12p (5, 9, 16, 38, 40). However, since Dig1p and Dig2p inhibit Ste12p by interacting with different regions, it is likely that the full activation of Ste12p involves multiple mechanisms. It should also be noted that the induction of Ste12p activity may not necessarily require dissociation of these inhibitors. Most recent experiments investigating Gal4p indicate that GAL induction may occur without dissociation of the negative regulator Gal80p (18, 28). Therefore, elucidation of the mechanisms regulating pheromone-responsive transcription will require a better understanding of the interactions between Dig1p, Dig2p, and Ste12p as well as the MAP kinases.
ACKNOWLEDGMENTS
This work was supported by a grant to I.S. from the N.C.I.C., with funds from the Canadian Cancer Society. K.A.O. was supported by an N.S.E.R.C. postgraduate studentship.
We thank Mike Tyers, Stanley Fields, H. Ronne, and Malcolm Whiteway for plasmids and yeast strains. Karen Lund, Susan Goto, Martin Hirst, and several attentive anonymous reviewers provided helpful comments on the manuscript.
K. Amy Olson, Chris Nelson, and Ivan Sadowski contributed equally to the design and implementation of these experiments.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates/Wiley-Interscience; 1987. [Google Scholar]
- 2.Bardwell L, Cook J G, Voora D, Baggott D M, Martinez A R, Thorner J. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell L, Cook J G, Zhu-Shimoni J X, Voora D, Thorner J. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc Natl Acad Sci USA. 1998;95:15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 5.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 6.Courchesne W E, Kunisawa R, Thorner J. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell. 1989;58:1107–1119. doi: 10.1016/0092-8674(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 7.Cullin C, Pompon D. Synthesis of functional mouse cytochromes P-450 and chimeric P-450 P3-1 in the yeast Saccharomyces cerevisiae. Gene. 1988;65:203–217. doi: 10.1016/0378-1119(88)90457-x. [DOI] [PubMed] [Google Scholar]
- 8.Elion E A, Brill J A, Fink G R. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc Natl Acad Sci USA. 1991;88:9392–9396. doi: 10.1073/pnas.88.21.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elion E A, Satterberg B, Kranz J E. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errede B, Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989;3:1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- 11.Fields S, Herskowitz I. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell. 1985;42:923–930. doi: 10.1016/0092-8674(85)90288-0. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–535. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 13.Gustin M C, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagen D C, McCaffrey G, Sprague G F., Jr Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2952–2961. doi: 10.1128/mcb.11.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 16.Hung W, Olson K A, Breitklutz A, Sadowski I. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur J Biochem. 1997;245:241–251. doi: 10.1111/j.1432-1033.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- 17.Johnston S A, Hopper J E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leuther K, Johnston S A. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 21.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 22.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 23.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 24.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 25.Matsuura Y, Possee R D, Overton H A, Bishop D H. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J Gen Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 26.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 27.Pi H, Chien C T, Fields S. Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p. Mol Cell Biol. 1997;17:6410–6418. doi: 10.1128/mcb.17.11.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platt A, Reece R J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reece R J, Rickles R J, Ptashne M. Overproduction and single-step purification of GAL4 fusion proteins from Escherichia coli. Gene. 1993;126:105–107. doi: 10.1016/0378-1119(93)90596-u. [DOI] [PubMed] [Google Scholar]
- 30.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, Tyers M, Boone C, Friend S H. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 31.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 32.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 33.Sadowski I, Niedbala D, Wood K, Ptashne M. GAL4 is phosphorylated as a consequence of transcriptional activation. Proc Natl Acad Sci USA. 1991;88:10510–10514. doi: 10.1073/pnas.88.23.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadowski I, Pawson T. Catalytic and non-catalytic domains of the Fujinami sarcoma virus P130gag-fps protein-tyrosine kinase distinguished by the expression of v-fps polypeptides in Escherichia coli. Oncogene. 1987;1:181–191. [PubMed] [Google Scholar]
- 35.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengupta P, Cochran B H. MAT alpha 1 can mediate gene activation by a-mating factor. Genes Dev. 1991;5:1924–1934. doi: 10.1101/gad.5.10.1924. [DOI] [PubMed] [Google Scholar]
- 37.Sengupta P, Cochran B H. The PRE and PQ box are functionally distinct yeast pheromone response elements. Mol Cell Biol. 1990;10:6809–6812. doi: 10.1128/mcb.10.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song D, Dolan J W, Yuan Y L, Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- 39.Stone G, Sadowski I. GAL4 is regulated by a glucose-responsive functional domain. EMBO J. 1993;12:1375–1385. doi: 10.1002/j.1460-2075.1993.tb05782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 41.Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13:5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan Y L, Fields S. Properties of the DNA-binding domain of the Saccharomyces cerevisiae STE12 protein. Mol Cell Biol. 1991;11:5910–5918. doi: 10.1128/mcb.11.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Y L, Stroke I L, Fields S. Coupling of cell identity to signal response in yeast: interaction between the alpha 1 and STE12 proteins. Genes Dev. 1993;7:1584–1597. doi: 10.1101/gad.7.8.1584. [DOI] [PubMed] [Google Scholar]