Abstract
Chili plants are affected by the hemibiotrophic ascomycota fungus Colletotrichum capsici causing Anthracnose. Infection results in yield and marketability loss due to a decrease in the quality of fruits. The study of morphological symptom development in two cultivars, Bullet, and Beldanga, showed very different disease expression pattern. To understand the reasons behind such differential response, we investigated, in a time-dependent manner, biochemical activities of important defense enzymes, PR proteins, like peroxidase, polyphenol-oxidase, phenylalanine ammonia lyase, β-glucanase, chitinase, catalase, as well as phenols, flavonoids, chlorophyll and the key signaling molecule nitric oxide in their leaves. We further performed real-time nitric oxide (NO) detection studies. The results showed striking differences in the activity profile of these defense molecules through the course of the study. We monitored the gene expression levels of 12 important defense-related genes under in vivo condition. The transcription levels were mostly increased in the tolerant cultivar till 7 days post-infection (DPI), while downregulation of some of the genes were observed in the susceptible one. These data indicated that disease manifestation is a simulated response of these defense molecules which can nullify the effect of the pathogen and its products, when resistance occurs. Alternatively, the pathogen suppresses the host defense when the disease develops.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11738-021-03334-x.
Keywords: Pathogenesis, Disease development, Anthracnose, Chili, Colletotrichum, Tolerant, Susceptible, Enzyme assay
Introduction
Chili, Capsicum annum, is an important cash crop of the world and through centuries, it has made its place, as an important condiment, in the cuisines of many cultures of the world, especially because of its tangy-pungent spicy taste of capsaicinoids (Mishra et al., 2017). Nevertheless, it has been found to have rich nutritional status, particularly in its content of vitamin A and C, phenolic compounds, carotenoids, and flavonoids (Mateos et al., 2013; Mishra et al., 2017). However huge economic loss is incurred globally with respect to its production and yield, due to profound damage caused by anthracnose disease (Sharma et al., 2005). The pathogen has been widely identified as the ascomycota fungus, Colletotrichum species (Than et al., 2008), Colletotrichum capsici (Sydow) Butler, and Bisby, being the most devastating species (Taylor et al., 2007). The disease appears as small sunken necrotic lesions (Isaac 1992), in leaves, stems, fruits during pre and post-harvest stage, and also within seeds during storage (Than et al. 2008).
The hemibiotrophic pathogen infects nearly all plant parts- leaves, stem, fruits, nodes, flowers in all stages of crop development (Isaac 1992) and can even propagate in affected plant debris lying in the field and gets propagated with fresh plantations, thus amplifying the damage (Saxena et al., 2019). It can also remain quiescent in the seeds of shelved fruits during storage (Than et al., 2008). A more threatening reason is anthracnose infected chili fruits become an easy target for secondary infection due to Aspergillus sp. which produces aflatoxin (Roy et al. 2013). Thus studies regarding its pathogenicity become necessary not only to control yield and marketability loss but also in the broader picture of global food security.
The rapidity and promptness with which a plant fights back the pathogen depends on a number of factors and takes place along multiple lines of action. Presence of pathogen or pathogen sensing on plants is aided by pathogen-associated molecular pattern (PAMP) (Monaghan and Zipfel, 2012) which are recognized by host cell surface receptors called pathogen recognition receptors or PRRs, (Du et al., 2015), resulting in PAMP triggered immunity (PTI) (Monaghan and Zipfel, 2012). This triggers signal transduction cascades via several downstream effecter molecules. PTI is followed by effecter triggered immunity (ETI) that occurs in the cytoplasm (Singla et al., 2019). The immediate host response involves the generation of reactive oxygen species or ROS (Heller et al., 2011), and initiates a localized hypersensitive response (HR), wherein the plant chooses to kill its own cells, i.e., necrosis to contain the pathogen within the infection spot by a series of controlled cellular events resulting in PCD or programmed cell death (Mbengue et al., 2016; Prasad et al. 2020). A more potent response is achieved through increased expression and activation of antioxidative enzymes and defense enzymes, such as catalase, peroxidase, polyphenol oxidase, phenylalanine ammonia lyase; pathogenesis-related proteins such as β,1–3, glucanase, and chitinase; and phenolics and other antimicrobial compounds such as defensing and phytoalexins. (Yadav et al., 2020; Malik et al. 2020). Nitric oxide (NO) (Mur et al., 2013; Nasir et al., 2020) has also been reported as a key regulator in these signaling pathways. Eventually, the signal may be transmitted to distal parts of the plant via phloem resulting in systemic acquired resistance or SAR (Saxena et al., 2019).
The disease-causing virulent pathogen releases plant cell wall-degrading enzymes and toxins, and can silence or even evade off the defense response (Anand et al., 2009; Mbengue et al., 2016). Each successful disease establishment involves a plethora of molecular and biochemical events between the host plant and its pathogen, which involves defense genes and enzymes, viz., PO, PPO, PAL, β-glucanase, chitinase, catalase, etc. (Ramamoorthy and Samiyappan, 2001). This particular arena is well documented in the literature and has been found in a wide range of crops such as tomato and rice (Chandrasekhar and Umesha, 2012; Suthin et al. 2019). Higher activity of these enzymes has also been reported in resistant green chili fruits against Alternaria alternata as well as C. capsici (Anand et al., 2009) as well as in conjunction with induced systemic resistance (Banu et al., 2019).
For a clearer understanding of chili defense mechanisms, we monitored the activity levels of the defense enzymes and PR proteins in two cultivars of chili, Bullet and Beldanga, in response to Colletotrichum capsici infection from 0 to 3 days post-infection (DPI). We report changes in the activity levels of the defense molecules with time and striking differences in the two cultivars which are commensurate with the differences in their morphological disease symptom expression. The comparative study of disease resistance and susceptibility in the two cultivars against the pathogen was carried out by wound inoculation with the pathogen. This approach might surpass some of the initial resistance mechanisms of the plant, yet it results in more discrete disease development and thus helps in studies. Our studies focused on morphological symptom development after pathogen inoculation, assayed the defense enzymes and PR proteins biochemically, and concomitantly studied the gene expression levels in the two cultivars. Based on the morphological differences of symptom development on leaves of these two varieties, we chose a specific time frame, 0, 1, 2, 3 DPI, to assess the differences in host response. We evaluated the activity levels of important molecules of plant defense—PO, PPO, PAL, β-glucanase, chitinase, catalase, chlorophylls a and b, total phenol and flavonoids, and NO levels, as well as studied the gene expression profile. This is the first time course report on the comparative response of disease resistance and disease susceptibility in chili leaves against anthracnose pathogen to the best of our knowledge.
Materials and method
Plant material
Seeds of two cultivars of chilli were obtained from Balagar in West Bengal, India, and seedlings raised in natural conditions in the experimental garden in a completely randomized pattern. Young leaves from 45 days old healthy chili plants were surface sterilized with 0.1% sodium hypochlorite followed by several washes with sterile distilled water (SDW). The detached leaves were subjected to pathogen challenge by standard blotter method or SBM (ISTA, 2005). The leaves were kept on three layered sterile moist blotting paper bed supplemented with 0.1% 6-benzylaminopurine (BAP) in 20 cm Petri plates, thus maintaining moist chambers. In these conditions, biochemical experiments were carried out from 0 to 3 DPI.
Fungal Isolate and Inoculation
Pure cultures of virulent Colletotrichum capsici purchased from the culture collection unit of Agharkar Research Institute, Pune (NFCCI No. 1660), were maintained in the laboratory in potato dextrose agar or PDA slants by subculturing. From 8-day-old PDA slant cultures, spore suspension was made with SDW (Sharma et al., 2005) and diluted with SDW to a concentration of 2 × 107cells/ml using a hemocytometer. From this spore suspension, 10 μl was used for each infection spot made in between two veins by wound pricking. 10 μl of SDW was added at the wound pricked spot on the control sets.
Morphological disease progression studies
Seeds were isolated from ripe mature red chili fruits and seedlings raised therefrom. Fully expanded leaves from 45 days old seedlings were excised and surface sterilized with 0.1% NaOCl, followed by repeated SDW wash. Leaves in humid chambers (prepared as mentioned above) were observed for disease symptom development. The percent disease area was calculated and disease scoring was done following Inglis et al. (1988) with little modifications in scale for chili leaves in ex vivo condition. A disease scale from 0 to7 was assigned (Table 1).
Table 1.
Disease scoring scale of infected excised chili leaves
| Score | Affected area (% of whole leaf area) | Symptom details |
|---|---|---|
| 0 | 0 | No infection |
| 1 | Up to 5 | With a light colored water-soaked lesion |
| 2 | > 5–10 | Larger light brownish water-soaked lesion |
| 3 | > 10–20 | Necrotic lesions with limited chlorosis surrounding the lesions |
| 4 | > 20–30 | Necrotic lesions turn dark brown with chlorosis along the midrib |
| 5 | > 30–40 | Larger necrotic lesions with extensive chlorosis |
| 6 | > 40–50 | Raised pustule-like appearances, possibly acervuli, at wound-prick points within the necrotic lesions, extensive chlorosis and browning at the margins |
| 7 | > 50 | Fungal mycelium mats over the lesions, complete chlorosis and loss of tissue integrity |
Disease severity index was deduced according to Song et al., (2004) using the formula:
Biochemical assay of defense enzymes and PR proteins
Control water-inoculated and infected leaves were assayed for different defense enzymes and PR proteins, from 0 to 3 DPI. For each sample preparation, collected leaves were wiped with sterile blotting paper, weighed and leaf tissue was extracted in the chilling condition in different enzyme-specific extraction buffers each containing 0.1% polyvinylpyrrolidone (PVP) and 20 µl of 1 mM phenylmethylsulfonyl fluoride (PMSF). The resultant solutions were centrifuged at 4 °C, 1200 × g for 20 min. The supernatant was collected and used as the enzyme solution.
Peroxidase (PO) assay
PO was assayed following the protocol given by Hameda and Klein (1990). The control and infected leaves were weighed and each sample of 150 mg of leaf tissue was extracted in 3 ml of 0.1 M of sodium phosphate buffer (pH 7.0). Change in absorbance due to guiacol oxidation at 470 nm was monitored for 3 min in the reaction mixture.
Polyphenol oxidase (PPO) assay
PPO activity was assayed following the method outlined by Kumar and Khan (1982) with slight modifications. 150 mg leaf tissue was extracted in 3 ml of 0.1 M of sodium phosphate buffer (pH 7.0). The absorbance of purpurogallin formed was measured at 495 nm against the enzyme blank. PPO activity was measured as a change in absorbance (U) min−1 mg−1 of protein.
Phenylalanine ammonia lyase (PAL) assay
Quantitative estimation of PAL was carried out following the protocol given by Dickerson, (1984) with minor modifications. 150 mg of leaf tissue was extracted in 3 ml of 0.1 M sodium borate buffer (pH 8.7). Formation of trans-cinnamic from L-phenylalanine was determined from OD290.
β -1,3- glucanase assay
β-Glucanase activity was assayed as described by Pan et al., (1991). The absorbance of the red-colored solution formed due to reaction with laminarin was measured at 525 nm against a zero minute incubation blank set.
Chitinase
Chitinase activity was assayed following the protocol from Bansode and Bajekals, (2006) with little modifications. 100 mg of leaf tissue was crushed in 3 ml of 0.1 M of sodium acetate buffer (pH 5.0). The reaction mixture comprised 1 ml of 1% colloidal chitin, 2 ml of buffer, and 500 μL of enzyme solution and absorbance measured at 540 nm.
Catalase
Catalase activity was estimated following the protocol outlined by Anderson et al. 1995 with slight changes. 250 mg of leaf tissue was extracted in 5 ml of 0.067 M sodium phosphate buffer, pH 7. The reaction mixture comprised 3 ml of 0.1% of H2O2 solution, 3 ml of 0.067 M phosphate buffer (ph 7), and 60 μL of the enzyme extract. The decrease in absorbance at 240 nm was recorded for 1 min. The enzyme activity was calculated with € = 36 M−1 cm−1 and expressed as μmol min−1 g−1 of protein.
Estimation of total phenol
250 mg of leaf tissue from each sample was homogenized with 3 ml of 80% methanol, and the homogenate was kept at 65 ˚C in a dry incubator for 15 min. It was centrifuged and the supernatant collected for phenol estimation (Zeislin and Ben Zaken, 1993). To 5 ml of double-distilled water, 1 ml of the supernatant and 250 μL of Folin–Ciocalteu reagent were added, incubated for 30 min at room temperature, and OD value recorded at 725 nm. Using the standard curve of gallic acid, the phenol quantity was expressed as μg of gallic acid produced g−1 of tissue.
Estimation of Flavonoids content
Total flavonoids content was estimated following the protocol outlined by Chang et al., (2002), with little modifications. 250 mg of leaf tissue was homogenized in 3 ml of 80% aqueous ethanol, followed by dark incubation for half an hour at room temperature. To 1 ml of the supernatant collected after centrifugation, 4.3 ml of 80% aqueous ethanol was added along with 100 μL of 10% aluminum nitrate and 100 μL of 1 M sodium acetate solution, followed by 30 min of dark incubation, and absorbance was measured at 495 nm. Total flavonoids content was expressed as mg of quercitin g−1 of the tissue sample.
Estimation of chlorophyll content
For chlorophyll content estimation, 250 mg of leaf tissue was ground in 4 ml of 80% alkaline acetone solution (in 0.1 N NaOH solution). Following protocol outlined by Kamble et al., (2015), absorbance was recorded at 663 and 645 nm. Chlorophyll content was calculated according to the following formula:
Estimation of Total protein
The total protein content in each sample extract was estimated with Bradford assay (1976) using bovine serum albumin (BSA) as standard.
Nitric oxide (NO) estimation
Quantitative estimation of nitric oxide production was done by the hemoglobin assay method (Delledonne 2005). Each leaf sample was dipped in hemoglobin buffer comprising 10 mM l-arginine and 10 mM hemoglobin in a 0.1 M phosphate buffer solution. The OD of the reaction mixture was recorded every 2 h from 0 to 4 h. NO content in the reaction mixture was calculated using E = 38,600 M−1 cm−1 and expressed as moles g−1 of tissue h−1 (Salter and Knowles, 1998).
Real-time NO detection
The lower epidermal layer of leaves was peeled off and subjected to real-time NO detection with DAF-2DA (4,5-diaminofluorescein diacetate) (Bartha et al., 2005). Leaf peels from each sample were dark incubated for 20 min in a loading buffer comprising 10 mM KCl and 10 mM Tris– HCl (pH 7.2) with 10 μM final concentration of DAF-2DA. Real-time NO imaging in epidermal cells was observed owing to the cell-permeable fluorescent probe DAF-2DA, using Leica DMLS microscope at excitation and emission maxima of 490 nm and 515 nm, respectively.
Gene expression studies
Pathogen challenge to chili plants
45 days old plants of both the cultivars grown under natural greenhouse conditions were inoculated with 10 μl of spore suspension by wound pricking. Wounded spot mock inoculated with an equal volume of SDW served as control sets. Each infected leaf was covered with sterile plastic cups for 24 h to maintain aseptic isolation conditions. Leaf samples were collected at 0, 1, 3, and 7 DPI, immediately dipped in liquid nitrogen and stored at − 80 °C.
RNA isolation and semi-quantitative RT-PCR analysis
Total RNA was isolated from 100 mg leaf tissue (collected at different time points post-inoculation) using the RNeasy Plant Mini Kit (Qiagen, Germany) following the manufacturer’s protocol. First-strand cDNA synthesis was done from 1 µg of total RNA using RevertAid First-strand cDNA synthesizing kit [Thermo Scientific, (EU) Lithuania] according to the manufacturer’s instruction using an oligo-dT primer. The genes targeted for the semi-quantitative RT-PCR analysis and the primers used for the amplification of those genes are enlisted in the Supplementary Table S1. Actin was used as an internal control to normalize the sample amounts.
The semi-qRT-PCR was carried out in MJ Mini Personal Thermo Cycler (Bio-Rad, USA). The PCR cycling conditions were as follows: 95 °C for 2 min, followed by 35 cycles of 95 °C for 20 s, 55 °C for 20 s and 72 °C for 20 s; a final extension for 5 min at 72 °C was given at the end of the PCR reaction.
Statistical analysis
The plants were raised in a completely randomized design, with one plant per experimental pot. In each experiment, three replicates for each set were used. The experiments were repeated at least thrice. All values were means of three individual experiments each in triplicate. The individual bar represents the mean value of three replicates. All results obtained were subjected to one-way variance ANOVA using Dunnett’s multiple comparison test with GraphPad Prism software version 8.4.3. Comparison has been performed for the infected plants with respect to the corresponding control of their respective varieties. *p < 0.05, **p < 0.01 and ***p < 0.001 show statistically significant differences with control.
Results
Disease progression studies
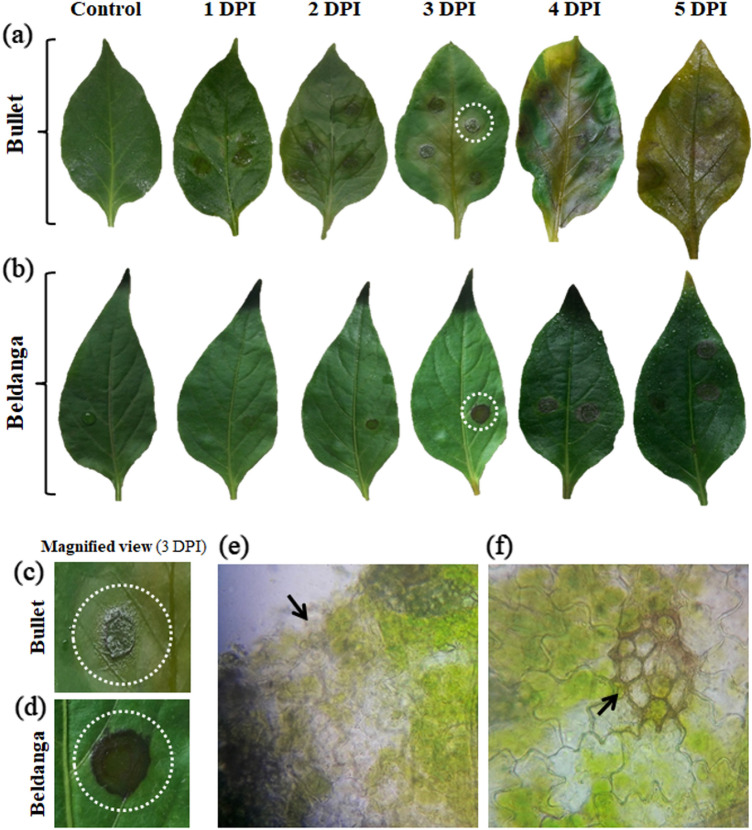
Chili leaves challenge inoculated with Colletotrichum capsici were studied for morphological expression of disease symptoms. Contrasting features were exhibited by leaves of two chili cultivars. The Bullet cultivar showed severe disease symptoms with sunken necrotic lesions with leaf discoloration surrounding each lesion and also along the midrib. Leaf integrity was lost somewhat near the margins after 3 DPI. However, Beldanga cultivar retained leaf color and integrity after 3 DPI. Localized deep brown-colored necrotic spots developed with raised margins on infection spots without much effect on the rest of the leaf (Fig. 1). The transverse section from near the lesions at 3 DPI revealed comparatively more localized fungal infestation in Beldanga. In Bullet, massive tissue damage and relatively more abundance of mycelium were observed at 3DPI.
Fig. 1.
Disease progression studies of the two cultivars. Images showing symptom development at different time intervals, i.e., 0 DPI, 1 DPI, 2 DPI, 3DPI, 4 DPI, 5 DPI in a Bullet cultivar (upper panel) and b Beldanga cultivar (lower panel),along with their corresponding controls; c and d magnified image of infection site at 3 DPI in Bullet and in Beldanga, respectively, where the infected site is highlighted with white dotted circle. e Microscopic view of a section through the infection lesion in Bullet and f in Beldanga. Identified infection sites are marked with black arrowheads
The disease scoring data revealed a 100% disease severity index (DSI) in Bullet at 5 DPI with a corresponding mean percent disease area of 66.29% (Supplementary Fig. 1). The DSI of Beldanga was calculated to be 28.58% with a corresponding mean percent disease area of 7.66%. The percent disease area in Bullet at 3 DPI was 33.06% with 71.42% DSI and that in Beldanga was 2.89% with 14.28% DSI.
Biochemical studies
Peroxidase (PO) assay
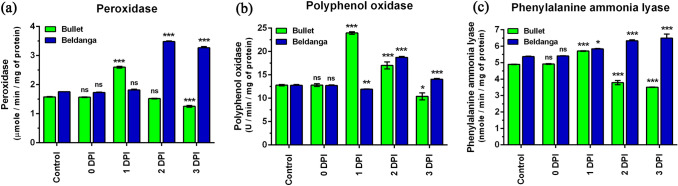
The PO activity levels in the pathogen challenged and unchallenged sets of Bullet and Beldanga through a course of 0 to 3 DPI differed considerably. PO activity in Bullet increased sharply at 1 DPI, thereafter it decreased and at 3 DPI the activity fell significantly. In the pathogen-infected Beldanga sets, a gradual increase in PO activity was observed. At 3 DPI, activity was maximum with an increase of about 1.86-fold compared to the control set of Beldanga. (Fig. 2a, b).
Fig. 2.
Specific activity of three enzymes in pathogen-challenged sets of Bullet and Beldanga cultivars at different time intervals, i.e., 0 DPI, 1 DPI, 2 DPI, and 3 DPI. a Peroxidase (PO), b polyphenol oxidase (PPO) and (c) phenylalanine ammonia lyase (PAL). Non-infected plants were used as a control for those experiments. Values represent mean ± SD of three independent experiments each in triplicate. Comparison was performed for the infected plants with respect to the corresponding control of their respective varieties. ns = not significant, *p < 0.05, **p < 0.01 and ***p < 0.001 representing statistically significant differences with control
Polyphenyl oxidase (PPO) assay
PPO activity differed in the two cultivars. In the pathogen-infected sets of Bullet, there was a sharp increase in PPO activity at 1 DPI (1.87 fold), followed by a gradual decrease and at 3 DPI, PPO activity was lesser than the control set. PPO level did not increase much at 1 DPI in the pathogen-infected sets of Beldanga, at 2 DPI it was highest (1.1 fold), and thereafter declined at 3 DPI (Fig. 2c, d).
Phenylalanine ammonia lyase (PAL) assay
The activity level of PAL in Bullet cultivar was highest at 1 DPI, when its increase was 1.16-fold, and then decreased. However, the decrease from 2 to 3 DPI was very little. In Beldanga cultivar, there was a gradual and steady increase in PAL activity from 0 to 3 DPI. It peaked at 3 DPI with a 1.2-fold increases (Fig. 2e, f).
β-Glucanase assay
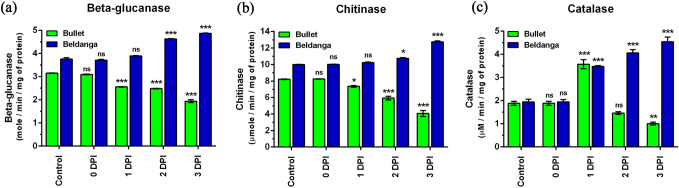
Bullet cultivar showed a decrease in the activity levels of β-glucanase post-inoculation. At 1 DPI the activity level fell and the decreased level was maintained at 2 DPI. At 3 DPI another significant drop in the activity level was noted. In contrast, the enzyme activity in Beldanga cultivar showed a progressive increase from 0 DPI. The initial level of β- glucanase activity in Beldanga was slightly higher than that in Bullet (1.19 times). At 2 DPI, it increased, with the highest level being at 3 DPI (1.3 fold) (Fig. 3a, b).
Fig. 3.
Specific activity of three enzymes in pathogen-challenged sets of Bullet and Beldanga cultivars at different time intervals, i.e., 0 DPI, 1 DPI, 2 DPI, and 3 DPI. a β-Glucanase, b chitinase, and c catalase. Non-infected plants were used as a control for those experiments. Values represent mean ± SD of three independent experiments each in triplicate. Comparison was performed for the infected plants with respect to the corresponding control of their respective varieties. ns = not significant, *p < 0.05, **p < 0.01 and ***p < 0.001 representing statistically significant differences with the control
Chitinase assay
The initial level of chitinase activity was found to be 1.21 times more in Beldanga than in Bullet. Its levels in Bullet decreased steadily post-infection, and the decrease was significant at 2 DPI and further decreased at 3 DPI. In the Beldanga cultivar, chitinase activity initially increased very slowly and a slight increase was observed at 2 DPI. However, there was a considerable increase at 3 DPI when it peaked (1.27 times) Fig. 3c and d).
Catalase assay
Catalase activity in Bullet cultivar rapidly increased at 1 DPI (2.2 fold) and then sharply decreased at 2 DPI compared to that of the corresponding control. Another sharp decline was observed at 3 DPI. Contrastingly, progressive enhancement in catalase activity was observed in Beldanga. From 0 to 1 DPI, the catalase activity showed a remarkable increase. At 3 DPI it was highest (2.6 fold) (Fig. 3e and f).
Phenol estimation
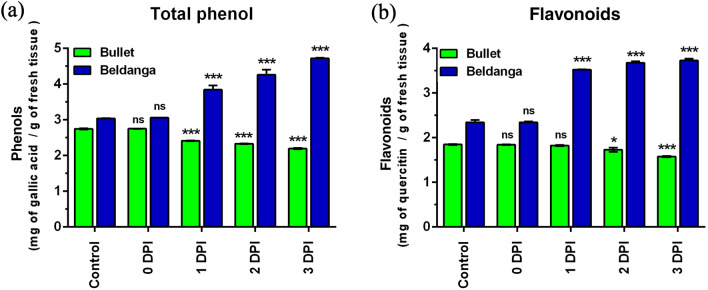
Phenol was estimated as mg of gallic acid/g of fresh tissue. The phenol profile of the two cultivars showed a complete contrasting pattern in the duration of the experiment. While the total phenol level in the Bullet gradually decreased from 0 to 1 DPI, the same in Beldanga showed progressive enhancement. After an initial significant dip in the phenol levels of Bullet from 0 to 1 DPI, the level continued to decrease slowly from 1 to 3 DPI. On the other hand, a sharp increase was observed from 0 to 1 DPI in the Beldanga cultivar. A further increase was observed from 1 to 3 DPI in this cultivar (Figs. 4a and b). The increase in Beldanga was 1.52 times the initial level when it peaked at 3 DPI.
Fig. 4.
Phenols and flavonoids estimation in pathogen-challenged sets of Bullet and Beldanga cultivars at different time intervals, i.e., 0 DPI, 1 DPI, 2 DPI, and 3 DPI. a Total phenol content and b flavonoids content. Non-infected plants were used as a control for those experiments. Values represent mean ± SD of three independent experiments each in triplicate. Comparison was performed for the infected plants with respect to the corresponding control of their respective varieties. ns = not significant, *p < 0.05, **p < 0.01 and ***p < 0.001 representing statistically significant differences with control
Flavonoids estimation
While Bullet showed a slow decrease in flavonoid levels from 1 to 3 DPI, the corresponding levels were increased in Beldanga, sharply at 1 DPI, and maintained with slight enhancement at each day (Fig. 4c, d). The decrease in Bullet was more pronounced at 3 DPI. In the pathogen-challenged sets of Beldanga, flavonoid level peaked at 3 DPI (1.58 times).
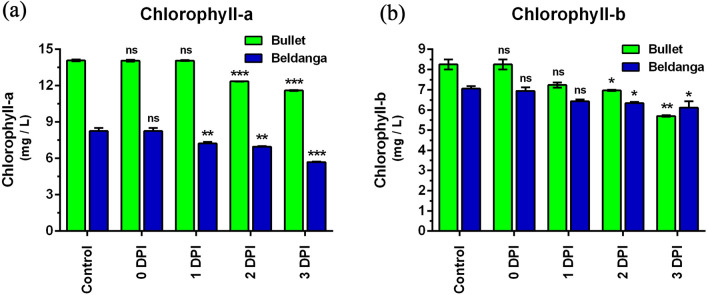
Chlorophyll
In the Bullet cultivar, chlorophyll a decreased post-inoculation, and the decrease was significant at 2 DPI. From there it again dropped at 3 DPI (Fig. 5a). In the Beldanga cultivar, the chlorophyll level decreased from 0 to 1 DPI, maintained the same levels at 2 DPI, and further dropped at 3 DPI (Fig. 5b). The initial chlorophyll a and b levels were observed to be higher in the Bullet cultivar. The decrease in infected sets of Bullet and Beldanga chlorophyll a levels at 3 DPI was 1.21 and 1.06 times of their initial levels, respectively.
Fig. 5.
Chlorophyll estimation in pathogen-challenged sets of Bullet and Beldanga cultivars at different time intervals, i.e., 0 DPI, 1 DPI, 2 DPI, and 3 DPI. a Chlorophyll a content and b chlorophyll b content. Non-infected plants were used as a control for those experiments. Values represent mean ± SD of three independent experiments each in triplicate. Comparison was performed for the infected plants with respect to the corresponding control of their respective varieties. ns = not significant, *p < 0.05, **p < 0.01 and ***p < 0.001 representing statistically significant differences with control
Chlorophyll b level in infected sets of Bullet cultivar was steady till 1 DPI with a slight decrease at 2 and 3 DPI (Fig. 5c). In Beldanga cultivar too there was a slight decrease in chlorophyll b levels from 0 to 3 DPI (Fig. 5d).
Overall, there were significant differences in the activity levels of the defense enzymes and PR proteins in the two cultivars throughout the experiment. In the Beldanga cultivar, in general, the levels were higher after pathogen challenge and never fell below the initial level in the course of the study. An increase in their activity levels was also observed in Bullet after pathogen challenge. However, their higher activities were not maintained consistently and a sharp decline in the levels was observed.
Nitric oxide estimation
NO levels in the pathogen-challenged sets of Bullet cultivar peaked at 1 DPI after which the levels started decreasing and at 3 DPI the levels were lower than that of the control set. NO levels in the pathogen-challenged sets of Beldanga cultivar started increasing from 1 DPI and continued to increase till 3 DPI (Fig. 6).
Fig. 6.
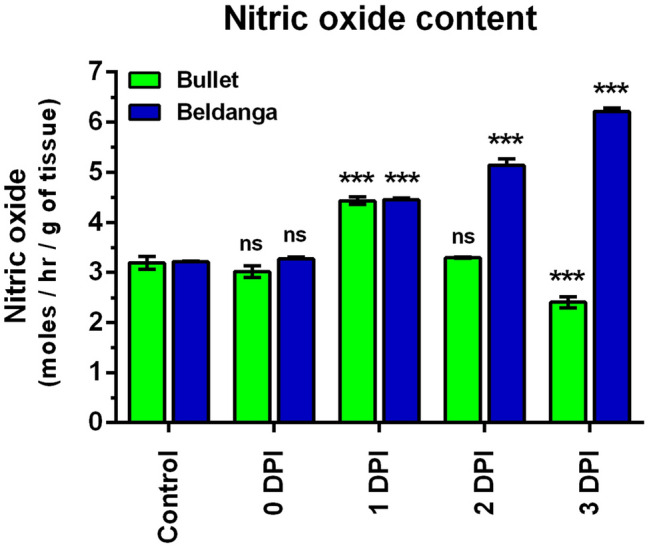
Nitric oxide (NO) estimation in pathogen-challenged sets of Bullet and Beldanga cultivars at different time intervals, i.e., 0 DPI, 1 DPI, 2 DPI, and 3 DPI. Non-infected plants were used as a control for the experiment. Values represent mean ± SD of three independent experiments each in triplicate. Comparison was performed for the infected plants with respect to the corresponding control of their respective varieties. ns = not significant, *p < 0.05, **p < 0.01 and ***p < 0.001 representing statistically significant differences with control
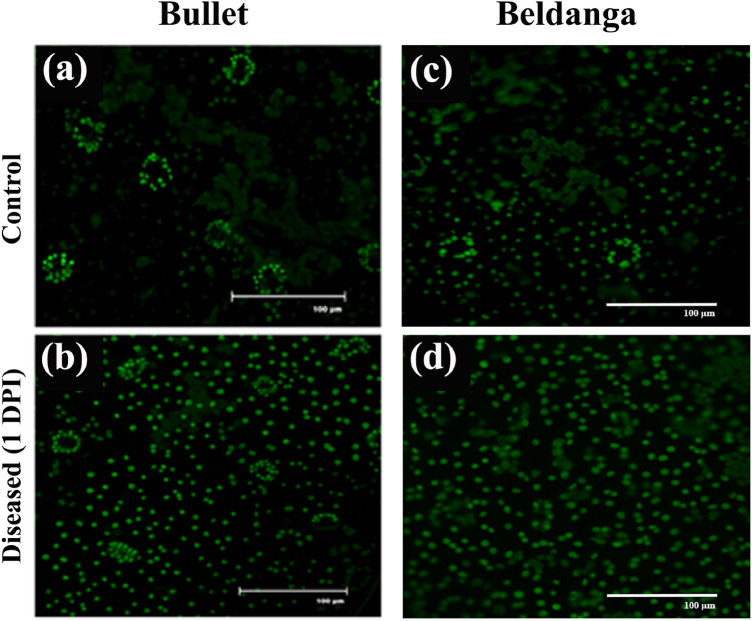
A similar pattern of a greater increase in NO levels was observed in pathogen-challenged sets of Beldanga cultivar than that of Bullet sets in real-time imaging of NO levels in the two cultivars (Fig. 7).
Fig. 7.
Real-time NO detection in control and pathogen-challenged sets of the two cultivars. 1–3 DPI of Bullet control sets (a–c) and pathogen-challenged sets (d–f); 1–3 DPI of Beldanga control sets (i–k) and pathogen-challenged sets (l–n)
Gene expression studies
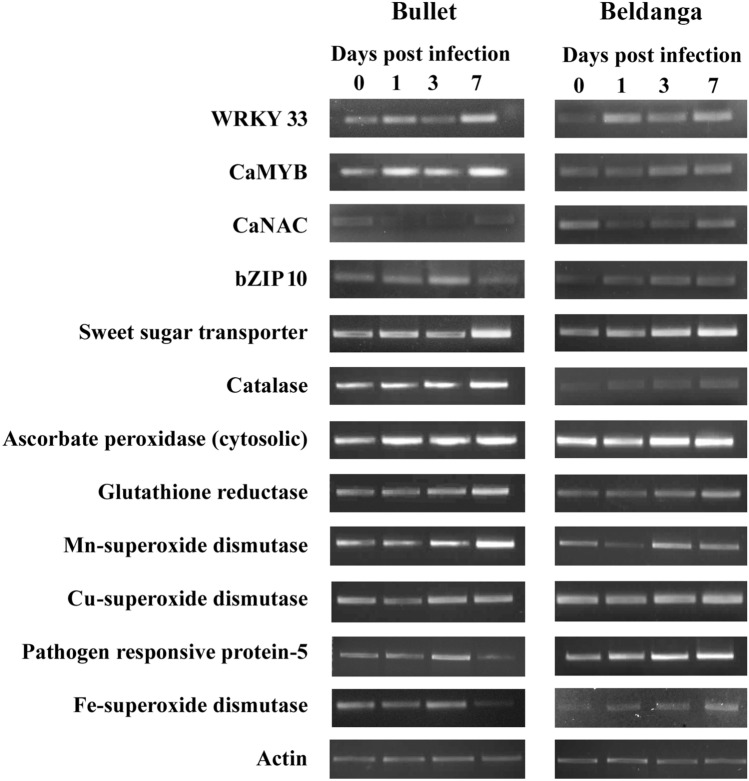
The semi-qRT-PCR results for 12 genes following infection in the chili cultivars Beldanga and Bullet depicted a disease resistance response in Beldanga cultivar. Gene expression analysis of important transcription factors such as WRKY33, CaMYB, CaNAC, and bZIP were carried out. The expression of WRKY33 increased post-infection, expression dropped at 3DPI in both the Bullet and Beldanga cultivars, and then rose again at 7DPI. The expression levels of CaMYB also increase post-infection in both. The basal expression of CaNAC is higher in Beldanga than in Bullet. Gene expression levels drop post-infection and rise again at 7 DPI being similar to the basal level in both the cultivars. Expression levels of bZIP 10 showed a steady rise in the cultivars of Beldanga and Bullet. However, expression fold dropped at 7 DPI for the susceptible cultivar of Bullet.
The sweet sugar transporter gene expression levels showed a sharp increase in fold expression post-3 DPI in Beldanga and at 7 DPI in Bullet. The basal level expression of CAT gene is lower in Beldanga as compared to Bullet. It shows increased gene expression at 7 DPI for both. The expression profile of ascorbate peroxidase increased several fold from 3DPI in Beldanga, whereas in Bullet, it increased and remained more or less constant till 7 DPI. Glutathione reductase showed increased gene expression profile post-3 DPI in both the cultivars. SODs such as Cu/Zn SODs and FeSOD showed higher expression at 7 DPI in Beldanga, whereas in Bullet, FeSOD expression declined at 7 DPI and there is no significant change in expression levels of Cu/Zn SOD. MnSOD showed an increase in gene expression at 3DPI for Beldanga, whereas at 7 DPI for Bullet. PR 5 showed several-fold upregulation in expression from 3DPI in Beldanga, whereas its expression dropped post-3 DPI in Bullet (Fig. 8).
Fig. 8.
Relative expression level analysis of several genes in Colletotrichum capsici-challenged sets of Bullet and Beldanga cultivars through semi-quantitative real-time PCR. Gene expression was analyzed at different time intervals, i.e., 0 DPI, 1 DPI, 3 DPI, and 7 DPI. Actin was used as an internal control
Discussion
Our results showed that, among the two cultivars of Capsicum annuum, the overall defense response was more pronounced and consistent in the Beldanga cultivar. The phenotypic disease expression in this cultivar showed discrete necrotic spots in the infection site without much effect on the whole leaf area. These necrotic spots pointed to an acute hypersensitive response in this cultivar leading to programmed cell death at the infection site containing the pathogen within and severing its connections with the rest of the leaf tissue. To destroy the invading pathogen, immediately after detection, the host resorts to ROS production (Heller et al., 2011; Prasad et al., 2020); the resultant oxidative burst causes damage in the cell membranes and membrane-bound structures, eventually aiding the process of programmed cell death (PCD) (Agrios, 2005; Prasad et al., 2020). Thus, the percent disease area and corresponding disease severity index (DSI) were much lower in the Beldanga cultivar which clearly points to the tolerance nature of Beldanga cultivar against anthracnose. In the Bullet cultivar, a rapid and massive pathogen spread caused yellowing of leaves and loss of tissue integrity, as evidenced by heavy pathogen localization in the leaf tissue. The might of this cultivar was not sufficient enough to completely restrict the infection within the infection site only, which points to more susceptible nature of this cultivar. Hence, greater percent disease area and disease severity indices (DSI) were recorded in Bullet.
Our study was intended to explain the differences in morphological disease symptoms of the two chili cultivars Bullet and Beldanga and to better characterize the underlying host–pathogen interaction. Evidence till date supports elevated levels of defense molecules which include defense enzymes, antioxidative enzymes, PR proteins as well as phenolics behind an overall improved disease resistance (Majid, 2017). Previous studies have reported enhanced activities of these defense molecules in a number of crop plants such as rice (Suthin et al. 2019), tomato (Anand et al. 2007), and tea (Chandra et al., 2015) resistant to specific diseases. In the present study, we found higher levels of all defense molecules post-inoculation in both the cultivars, the enhancement being more pronounced in Beldanga than in Bullet. Higher PO, PPO, and catalase activities were reported in chili fruits upon pathogen inoculation with C. capsici (Koppad and Mesta, 2017). The researchers further reported higher levels of these enzymes in infected resistant genotypes than in susceptible ones. These are antioxidant enzymes that are required to maintain the delicate balance between ROS production and scavenging (Singla et al. 2019). In the present study, both PO and catalase activity increased in Bullet as well as Beldanga cultivar following inoculation, which certainly testifies to ROS-mediated triggering of the downstream defense signaling pathways.
PO also plays important role in cell wall fortification (Saxena et al. 2019) which occurs to provide a mechanical barrier to the spread of the pathogen from the infection spot to the adjacent cells as well as to prevent additional invasion from the cell surface. Deposition of wall thickening materials such as lignin, suberin, and callose takes place with the aid of PO, PPO, PAL as well as phenolics (Saxena et al., 2019). Increased lignin deposition along with enhanced PO activity has been reported in C. annuum against C. capsici (Abhyashree et al. 2017; Saxena et al. 2019). In chili fruits, previous reports have shown induction of PO, PPO, PAL, and other enzymes in response to Alternaria alternata (Anand et al., 2009) and Pseudomonas (Ramamoorthy and Samiyappan, 2001). In the present study, an increase in the activity of PO, PPO, and PAL was observed in the pathogen-challenged sets of both Bullet and Beldanga cultivar post-inoculation. The activities peaked off and decreased in Bullet cultivar which clearly explains the extensive disease manifestation in this cultivar, while in the pathogen-challenged sets of Beldanga cultivar, PO, PPO, and PAL activities increased post-inoculation and maintained the enhanced levels till 3 DPI. The differences in the activity patterns of the enzymes PO, PPO, and PAL in the two cultivars further substantiates their differential response toward the pathogen. Studies with chili fruits infected with C. capsici have previously reported higher total content of orthodideoxy phenols as well as PO, PPO, and PAL levels in resistant genotypes and moderately resistant hybrids (Prasath and Ponnuswami, 2008). The researchers screened a total of 17 genotypes of chili against anthracnose pathogen and recorded the activity levels of the enzymes PO, PPO, PAL, total phenol, and orthodihydroxyphenol. The researchers further demonstrated that the least disease incidence in the resistant genotype Capsicum baccatum was concomitant with the highest levels of these enzymes and the highest accumulation of total phenol and orthodihydroxyphenol in this genotype. Our results thus are seeded well into the existing understanding of the role of these defense molecules in eliciting an immune response.
Phenol is the precursor of lignin and is an important signal molecule that activates a number of defense-related genes and pathways including systemic acquired resistance or SARS (Madhavan et al., 2010). Enhanced level of phenols in response to pathogen invasion has been reported owing to its role in the deposition of secondary cell wall materials around the infection spot (Saxena et al., 2019). In the present study, phenol levels were found to be elevated in both the cultivars post-inoculation. Previous reports have shown an improved level of phenolics in chili, upon pathogen ingress by C. truncatum (Madhavan et al., 2010; Saxena et al., 2019). Flavonoids are important phytoalexins that have antimicrobial properties (Facchini et al., 2002). In complete agreement with the previous reports, our results have shown enhanced levels of total phenol and flavonoids after pathogen invasion. The effect was more pronounced in Beldanga, owing to its severe fight-back mechanism that contributes to its resistance response. Thus total phenol and flavonoid levels were observed to be continuously increasing till 3 DPI in this cultivar. The levels dropped after 1 DPI in Bullet cultivar due to its eventual surrender to the disease as was substantiated by the disease progress studies. Our results are further validated by previous reports on an increased level of phenol against pathogen ingress in C. annum (Anand et al., 2009; Madhavan et al., 2010).
Another way of combating is to secrete extracellular enzymes that degrade fungal cell wall components. β-Glucanase and chitinase are two important PR proteins that are bestowed with this function. The former hydrolyzes glucan and the latter degrades chitin, thus diluting important cell wall constituents of the pathogen (Tian et al., 2006). Moreover, the interaction of these enzymes with the fungal cell wall induces the release of elicitor molecules that interact with the plant cell receptors and initiates downstream signal transduction (Ham et al., 1991). Accumulation of these PR proteins eventually leads to SAR (Ryals et al., 1996). Increased β-glucanase activity was reported in cucumber and tobacco against powdery mildew and in pear fruits against Alternaria alternata, in response to elicitor-mediated resistance (Herger and Klingauf, 1990; Tian 2006). Separate reports on increased activity of β-glucanase and chitinase have been correlated with improved resistance response in both tomato (Chaktaborty et al., 2017), and chili (Banu et al., 2019). In the current study, we report enhanced β-glucanase and chitinase activity in Beldanga cultivar in lieu of pathogen invasion, and in Bullet the activity levels have shown a gradual decrease, which is commensurate with the phenotypic characteristics. Thus a simulated defense response is touched up by close coordination of these enzymes PO, PPO, PAL, β-glucanase, chitinase, catalase as well as phenols and flavonoids to shape into a composite and more coordinated defense response.
Various signaling molecules have been identified as mediators in between the defense signaling cascades, of which NO has been recognized as an important modulator (Nasir et al., 2020). The role of NO in ROS scavenging (Mur et al., 2013) has been well elucidated. In the present study, NO levels as found in the quantitative estimation, and real time NO detection are in congruence with these findings. Both the cultivars showed an upsurge of NO levels post-pathogen inoculation and a prolonged elevated level was maintained in the Beldanga cultivar. Studies with both NO-enriching systems and NO-scavenging systems have shown a direct correlation of NO levels with plant defense (Chandra et al., 2015; Chakraborty et al., 2017).
Thus, the fundamental differences in disease response of the two cultivars of the same chili plant to the same pathogen could be explained due to differences in the activity levels of their important defense molecules. Enhanced levels of the defense molecules have been positively correlated to an enhanced degree of tolerance toward successful disease establishment (Ramamoorthy and Vellaiswamy, 2001; Banu et al., 2019). Previous reports have also suggested the role of sesquiterpenoids such as capsidiol and related genes active in pepper fruit in defense against Colletotrichum scovillei; however, their mechanism of biosynthesis remain to be understood (Baba et al., 2020). In our study, the elevated response of the defense molecules was maintained in Beldanga cultivar, but not in the Bullet cultivar which explains the susceptibility of the latter. In this context, the present study could contribute to the current understanding of successful disease establishment. To get a more complete picture of the differential disease response in the two cultivars, we studied the gene expression levels in whole plants.
The gene expression profile of 12 important genes remarkably substantiated the differences in morphological disease expression of the two cultivars. Till now, a total of 17 PR proteins have been identified in plants (Soh et al., 2012). Previous studies have linked increased expression levels of PR 2, 5, 9 to improved disease resistance in plants (Hong et al., 2018). Semi-q-RTPCR results showed a several-fold increase in the expression of pathogenesis responsive protein-5 in Beldanga cultivar from 3 DPI, whereas its expression dropped post-3DPI in the susceptible cultivar Bullet. Transcription factors (TFs) play a key role in transcriptional reprogramming, triggered by pathogen perception in plants, resulting in the differential regulation of downstream defense responsive genes and signal transduction (Peng et al., 2018). Transcription factor families of WRKY, NAC, MYB and bZIP are instrumental in plant immune response (Yuan et al., 2019). A large repertoire of the transcription factor family comprises WRKY, bZIP, bHLH, NAC, and AP2-ERF with a role in plant defense response (Seo et al., 2015). bZIPs play an important role in hormone and sugar signaling, oxidative stress and pathogen defense (Thurow et al., 2005). bZIP10 has been reported to play a role in the activation of pathogenesis-related genes and induce basal defense responses (Kaminaka et. al., 2006). Several bZIP genes are known to be involved upon biotic stimuli such as salicylic acid, methyl jasmonic acid, ethylene and pathogen defense (Gai et al., 2020).The gene expression analysis from semi-qRT-PCR results showed a steady rise in the expression of bZIP 10 in the resistant Beldanga cultivar post-infection. An increase in expression was also observed in the Bullet cultivar; however, its expression dropped below the basal level after 3DPI. Plant NAC TFs have also been known to play an important role in the development and biotic stress (Puranik et al., 2012). These TFs contain binding sites for W boxes, GCC boxes and MYB that mediates its interaction with other TFs such as WRKY and MYB in signal transduction (Pascual et al., 2015). An NAC domain containing transcription factor CaNAC1 was found to be involved in interactions between chili pepper and pathogens (Oh et al., 2005). Here, the expression levels of CaNAC in the resistant Beldanga cultivar were higher as compared to the Bullet cultivar. The expression dropped at 1 DP1 and 3 DPI following which it increased at 7 DPI and was the same as 0 DPI, suggesting an establishment of defense response after pathogen infection. The R2R3 type MYB transcription factors play a crucial role in plant defense, also regulating the expression of pathogen responsive proteins (Hong et al., 2018). Semi-qRT-PCR results show a steady rise in the expression level of CaMYB from 3DPI suggesting a basal response. WRKY33 is implicated in pathogen response in PTI (Hong et al., 2018) and via interactions with MAP kinase cascades (Mao et al., 2011). Earlier reports of a WRKY transcription factor, CaWRKY1 from chilli, have been shown to act as a negative regulator of defense for virus-induced gene silencing of the gene that decreased the growth of Xanthomonas, whereas overexpression resulted in a hypersensitive response and cell death in response to P. syringae and tobacco mosaic virus (Oh et al., 2008). Expression levels of WRKY33 in both the cultivars dropped at 3DPI and then increased again at 7 DPI suggesting a hypersensitive immune response.
The expression of sweet sugar transporters is often modified by extracellular pathogens so they can direct the sugars for their growth (Chen et al., 2010). AtSTP4 from Arabidopsis thaliana is overexpressed upon pathogen attack and invasion (Truernit et al., 1996). Sugar transporter proteins have been identified from Capsicum and implicated in growth, development and abiotic stress resistance (Wei et al., 2020). In the present study, upon pathogen attack, the semi qRT-PCR results showed a sharp rise in the expression level of the gene in both Beldanga cultivar (from 3 DPI) and Bullet cultivar (at 7 DPI) suggesting a faster response to pathogen attack in the resistant cultivar.
SODs play an important role in converting ROS, produced as a result of oxidative burst, into hydrogen peroxide and oxygen. The hydrogen peroxide is then broken down by enzymes such as catalase (CAT) and ascorbate peroxidase, thereby neutralizing the toxicity of ROS (Cox et al., 2003). Earlier studies have reported the induced expression of ascorbate peroxidase and thioredoxin peroxidase upon infection of Xanthomonas campestris triggering a hypersensitive response (Do et al., 2003) High expression of the SOD metallo-enzymes at 7 DPI in Beldanga suggested a pronounced defense response and mitigation of oxidative stress. In the Bullet cultivar, there was a decline in the expression of FeSOD at 7 DPI, whereas the CuSOD expression profile did not show a significant change. MnSOD showed an increased expression post 3 DPI in Beldanga cultivar but at 7DPI in the Bullet cultivar. The CAT gene expression also showed an increase at 7 DPI in both. The expression profile of ascorbate peroxidase increased post-infection in the Bullet cultivar and remained more or less constant till 7 DPI. However, comparably in the Beldanga cultivar, several-fold increase from 3DPI was observed suggesting a pronounced defense response to infection. Flavoproteins such as glutathione reductase (GR) play an important role in the conversion of glutathione disulfide to glutathione (GSSG/GSH cycle) and in the process eliminates excessive burst of H2O2 in the cell. The expression profile of glutathione reductase showed an increase in expression post-3 DPI in both the cultivars.
High-throughput phenotyping and gene expression analysis can also be complemented with the study of disease-associated marker analysis. There are very few marker-associated studies carried out. A previous study reported a new gene RCt1 for resistance against anthracnose and the same report identified few ISSR and AFLP markers showing polymorphism between resistant and susceptible cultivars (Mishra et al., 2019). Future studies related to such marker-assisted analysis can be helpful for the identification and breeding of disease-resistant varieties.
Conclusion
Anthracnose is a devastating disease of chili which causes huge economic loss. Thus studies contributing to the recognition of the basis of resistance in the tolerant cultivars can facilitate identification of genes responsible for the trait and, in turn, accelerate breeding programs to develop resistant cultivars. With this view, our study was aimed at better characterizing the defense response in the two host cultivars. While our disease progression and histological studies have shown discrete patterns of C. capsici colonization, the biochemical assay of defense enzymes and PR proteins have clearly pointed the differences in their activity levels. These results clearly explain their differential response. The results are further substantiated by the amount of total phenols, flavonoids and chlorophyll. Our characterization of the differentially expressed gene and proteins could facilitate the identification of important genetic markers in future studies and their application in successful breeding programs.
Author contribution statement
KA designed the experiments. JB and TH performed the experiments and analyzed the data. JB wrote the manuscript. All the authors have read and approved the manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1: Analysis of Disease Severity Index (DSI) and Disease area in Bullet and Beldanga cultivars at different time intervals i.e. 0 DPI, 1 DPI, 2 DPI and 3 DPI. (a) Disease Severity Index and (b) Disease area are represented in Percentage calculated with respect to their corresponding controls. Values represent Mean ± SD of three independent experiments each in triplicate. Comparison has been performed for the infected plants with respect to the corresponding control of their respective varieties. Where, ns=not significant, *p<0.05, **p<0.01 and ***p<0.001 representing statistically significant differences with control. (TIF 251 kb)
Supplementary Table 1: List of primers used for semi-quantitative gene expression studies (DOCX 13 kb)
Acknowledgements
The authors thank DST (WOS-A), UGC-UPE II, CAS Phase VII and DST (FIST), Govt. of India, for providing instrumental facility.
Abbreviations
- PR
Pathogenesis related
- PO
Peroxidase
- PPO
Polyphenol oxidase
- PAL
Phenylalanine ammonia lyase
- DPI
Days post-inoculation
- NO
Nitric oxide
- ROS
Reactive oxygen species
- SDW
Sterile distilled water
- SBM
Standard blotter method
- BAP
6-Benzylaminopurine
- PDA
Potato dextrose agar
- DSI
Disease severity index
- PVP
Polyvinylpyrrolidone
- PMSF
Phenylmethylsulfonyl fluoride
- DNS
Dinitrosalicylic acid
- EA
Enzyme activity
- BSA
Bovine serum albumin
- DAF-2DA
4,5-Diaminofluorescein diacetate
Funding
This research project was funded by Women Scientist Scheme–A (WOSA), Department of Science and Technology (DST), Government of India [Project sanction no. SR/WOS-A/LS-272/2017].
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abhayashree MS, Murali M, Thriveni MC, Sindhu GM, Amruthesh KN. Crude oligosaccharides mediated resistance and histo-chemical changes in Capsicum annuum against anthracnose disease caused by Colletotrichum capsici. Plant Biosyst. 2017;151:221–233. [Google Scholar]
- Agrios GN. Plant pathology. 5. San Diego: Academic Press; 2005. p. 922. [Google Scholar]
- Anand T, Chandrasekaran A, Kuttalam S, Raguchander T, Prakasam V, Samiyappan T. Association of some plant defense enzyme activities with systemic resistance to early leaf blight and leaf spot induced in tomato plants by azoxystrobin and Pseudomonas fluorescens. J Plant Interact. 2007;2(4):233–244. doi: 10.1080/17429140701708985. [DOI] [Google Scholar]
- Anand T, Bhaskaran R, Raguchander T, Samiyappan R, Prakasham V, Gopalakrihnan C. Defence responses of chilli fruits to Colletotrichum capsici and Alternaria alternata. Biol Plant. 2009;53:553. doi: 10.1007/s10535-009-0100-5. [DOI] [Google Scholar]
- Anderson MD, Prasad TK, Stewart CR. Changes in Isozyme Profiles of catalase, peroxidase, and glutathione reductase during acclimation to chilling in mesocotyls of maize seedlings. Plant Physiol. 1995;109(4):1247–1257. doi: 10.1104/pp.109.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba VY, Powell AF, Ivamoto-Suzuki ST, et al. Capsidiol-related genes are highly expressed in response to Colletotrichum scovillei during Capsicum annuum fruit development stages. Sci Rep. 2020;10:12048. doi: 10.1038/s41598-020-68949-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansode Y, Bajekals S. Characterization of chitinase from microorganisms isolated from Lonar Lake. Indian J Biotechnol. 2006;5:357–363. [Google Scholar]
- Banu N, Mahadevamurthy M, Amruthesh K. Plant growth-promoting fungi (PGPF) instigate plant growth and induce disease resistance in Capsicum annuum L. upon Infection with Colletotrichum capsici (Syd.) Butler and Bisby. Biomolecules. 2019;10:41. doi: 10.3390/biom10010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartha B, Kolbert Z, Erdei L. Nitric oxide production induced by heavy metals in Brassica juncea L. Czern and Pisum sativum L. Acta Biol Szeged. 2005;49:9–12. [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chakraborty N, Chandra S, Acharya K. Biochemical basis of improvement of defense in tomato plant against Fusarium wilt by CaCl2. Physiol Mol Biol Plants Int J Funct Plant Biol. 2017;23(3):581–596. doi: 10.1007/s12298-017-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Chakraborty N, Dasgupta A, Sarkar J, Panda K, Acharya K. Chitosan nanoparticles: a positive modulator of innate immune responses in plants. Sci Rep. 2015;5:15195. doi: 10.1038/srep15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar S, Umesha S. Induction of antioxidant enzymes associated with bacterial spot pathogenesis in tomato. Int J Food Agric Veterin Sci. 2012;2:22–34. [Google Scholar]
- Chang CC, Yang MH, Wen HM, Chern JC. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J Food Drug Anal. 2002;10:178–182. [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, Harrison TS, McDade HC, et al. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun. 2003;71(1):173–180. doi: 10.1128/iai.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M. NO news is good news for plants. Curr Opin Plant Biol. 2005;8:390–396. doi: 10.1016/j.pbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Dickerson DP, Pascholati SF, Hagerman AE, et al. Phenylalanine ammonia-lyase and hydroxycinnamate: CoA ligase in maize mesocotyls inoculated with Helminthosporium maydis or Helminthosporium carbonum. Physiol Plant Pathol. 1984;25:111–123. doi: 10.1016/0048-4059(84)90050-X. [DOI] [Google Scholar]
- Do HM, Hong JK, Jung HW, Kim SH, Ham JH, Hwang BK. Expression of peroxidase-like genes, H2O2 production, and peroxidase activity during the hypersensitive response to Xanthomonas campestris pv. vesicatoria in Capsicum annuum. Mol Plant Microbe Interact. 2003;16(3):196–205. doi: 10.1094/MPMI.2003.16.3.196. [DOI] [PubMed] [Google Scholar]
- Du J, Verzaux E, Chaparro-Garcia A, Bijsterbosch G, Keizer LP, Zhou J, Liebrand TW, Xie C, Govers F, Robatzek S. Elicitin recognition confers enhanced resistance to phytophthora infestans in potato. Nat Plants. 2015;1:15034. doi: 10.1038/nplants.2015.34. [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG. Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot. 2002;80:577–589. [Google Scholar]
- Gai WX, Ma X, Qiao YM, et al. Characterization of the bZIP transcription factor family in pepper (Capsicum annuum L.): CabZIP25 positively modulates the salt tolerance. Front Plant Sci. 2020;11:139. doi: 10.3389/fpls.2020.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham K, Kauffmann SS, Albersheim P, Darvill AG. Host-pathogen interactions. A soybean pathogenesis-related protein with b-1,3- glucanase activity releases phytoalexin elicitor-active heat-stable fragments from fungal cell walls. Mol Pl Microbe Interact. 1991;4:545–552. [Google Scholar]
- Hameda HM, Klein BP. Effects of naturally occurring antioxidants on peroxidase activity of vegetable extracts. J Food Sci. 1990;55:184–185. [Google Scholar]
- Heller J, Tudzynski P. Reactive oxygen speciesin phytopathogenic fungi: signaling, development, and dis-ease. Annu Rev Phytopathol. 2011;49(1):369–390. doi: 10.1146/annurev-phyto-072910-095355. [DOI] [PubMed] [Google Scholar]
- Herger G, Klingauf F. Control of powdery mildew fungi with extracts of the giant knotweed, Reynoutria sachalinensis (Polygonaceae) Mededelingen Faculteit Landbouwwetenschappen Rijksuniversiteit Gent. 1990;55:1007–1014. [Google Scholar]
- Hong Z, Shikai L, Changyou W, Wanquan J. The role of transcription factor in wheat defense against pathogen and its prospect in breeding. J Plant Biol Crop Res. 2018;1:1005. [Google Scholar]
- Inglis DA, Haedorn DJ, Rand RE. Use of dry inoculum to evaluate beans for resistance to anthracnose and angular leaf spot. Plant Dis. 1988;72:771–774. [Google Scholar]
- Isaac S. Fungal plant interaction. London: Chapman and Hall Press; 1992. p. 115. [Google Scholar]
- ISTA (2005) International seed testing association. In: Proceedings of the international seed testing association. International rules of seed testing. Seed Sci Technol 15: 1–9.
- Kamble P, Giri S, Mane R, Tiwana A. Estimation of chlorophyll content in young and adult leaves of some selected plants. Univ J Environ Technol. 2015;5:306–310. [Google Scholar]
- Kaminaka H, et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25:4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppad S, Mesta RK. Role of enzymes in resistance against fruit rot of chilli caused by Colletotrichum capsici (Syd.) Butler and Bisby. Int J Chem Stud. 2017;5(4):536–540. [Google Scholar]
- Kumar K, Khan P. Peroxidase and polyphenol oxidase in excised ragi (Eleusine corocana cv PR 202) leaves during senescence. Indian J Exp Biol. 1982;20:412–416. [PubMed] [Google Scholar]
- Madhavan S, Paranidharan V, Velazhahan R. Foliar application of Burkholderia sp. strain TNAU-1 leads to activation of defense responses in chili (Capsicum annuum L.) Braz J Plant Physiol. 2010;23:261–266. [Google Scholar]
- Majid MU, Awan MF, Fatima K, et al. Genetic resources of chili pepper (Capsicum annuum L.) against Phytophthora capsici and their induction through various biotic and abiotic factors. Cytol Genet. 2017;51:296–304. doi: 10.3103/S009545271704003X. [DOI] [Google Scholar]
- Malik NA, Kumar IS, Nadarajah K. Elicitor and receptor molecules: orchestrators of plant defense and immunity. Int J Mol Sci. 2020;21(3):963. doi: 10.3390/ijms21030963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, et al. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23:1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos RM, Jimenez A, Roman P, Romojaro F, Bacaeizo S, Leterrier M, Gomez M, Sevilla F, del Rio LA, Corpas FJ, Palma JM. Antioxidant systems from pepper (Capsicum annuum L.): involvement in the response to temperature changes in ripe fruits. Int J Mol Sci. 2013;14:9556–9580. doi: 10.3390/ijms14059556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M, Navaud O, Peyraud R, Barascud M, Badet T, Vincent R, Barbacci A, Raffael S. Emerging trends in molecular interactions between plants and the broad host range fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum. Front Plant Sci. 2016;7:422. doi: 10.3389/fpls.2016.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R, Nanda S, Rout E, Chand SK, Mohanty JN, Joshi RK. Differential expression of defense-related genes in chilli pepper infected with anthracnose pathogen Colletotrichum truncatum. Physiol Mol Plant Pathol. 2017;97:1–10. [Google Scholar]
- Mishra R, Rout E, Mohanty JN, Joshi RK. Sequence-tagged site-based diagnostic markers linked to a novel anthracnose resistance gene RCt1 in chili pepper (Capsicum annuum L) 3 Biotech. 2019;9(1):9. doi: 10.1007/s13205-018-1552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15(4):349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, Hall MA, Harren FJM, Hebelstrup KH, Gupta KJ. Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants. 2013;5:52. doi: 10.1093/aobpla/pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir NNM, Ho CL, Lamasudin DU, Saidi NB. Nitric oxide improves tolerance to Fusarium oxysporum f. sp. cubense tropical race 4 in banana. Physiol Mol Plant Pathol. 2020 doi: 10.1016/J.PMPP.2020.101503. [DOI] [Google Scholar]
- Oh SK, Lee S, Yu SH, Choi D. Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta. 2005;222(5):876–887. doi: 10.1007/s00425-005-0030-1. [DOI] [PubMed] [Google Scholar]
- Oh SK, Baek KH, Park JM, Yi SY, Yu SH, Kamoun S, Choi D. Capsicum annuum WRKY protein CaWRKY1 is a negative regulator of pathogen defense. New Phytol. 2008;177(4):977–989. doi: 10.1111/j.1469-8137.2007.02310.x. [DOI] [PubMed] [Google Scholar]
- Pan SQ, Ye XS, Kuć J. Association of β-1,3-glucanase activity and isoform pattern with systemic resistance to blue mould in tobacco induced by stem injection with Peronospora tabacina or leaf inoculation with tobacco mosaic virus. Physiol Mol Plant Pathol. 1991;39:25–39. doi: 10.1016/0885-5765(91)90029-H. [DOI] [Google Scholar]
- Pascual MB, Canovas FM, Avila C. The NAC transcription factor family in maritime pine (Pinus pinaster): molecular regulation of two genes involved in stress responses. BMC Plant Biol. 2015;15:254. doi: 10.1186/s12870-015-0640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, van Wersch R, Zhang Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant Microbe Interact. 2018;31:403–409. doi: 10.1094/MPMI-06-17-0145-CR. [DOI] [PubMed] [Google Scholar]
- Prasad A, Sedlářová M, Balukova A, Rác M, Pospíšil P. Reactive oxygen species as a response to wounding. Vivo imaging in Arabidopsis thaliana. Front Plant Sci. 2020;10:1660. doi: 10.3389/fpls.2019.01660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasath D, Ponnuswami V. Screening of chilli (Capsicum annuum L.) genotypes against Colletotrichum capsici and analysis of biochemical and enzymatic activities in inducing resistance. Indian J Genet Plant Breed. 2008;68:344–346. [Google Scholar]
- Puranik S, et al. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy V, Samiyappan R. Induction of defense-related genes in Pseudomonas fluorescens treated chilli plants in response to infection by Colletotrichum capsici. J Mycol Plant Pathol. 2001;31:146–155. [Google Scholar]
- Roy M, Harris J, Afreen S, Deak E, Gade L, Balajee SA, Park B, Chiller T, Luby S (2013) Aflatoxin contamination in food commodities in Bangladesh. Food Addit Contam B6(1):17–23 [DOI] [PubMed]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter M, Knowles GR. Assay of NOS activity by the measurement of conversion of oxyhemoglobin to methemoglobin by NO. In: Titheradge MA, editor. Nitric oxide protocols. Totowa: Humana Press; 1998. pp. 61–65. [DOI] [PubMed] [Google Scholar]
- Saxena A, Mishra S, Ray S, Raghuwanshi R, Singh H. Differential reprogramming of defense network in Capsicum annum L. plants against colletotrichum truncatum infection by phyllospheric and rhizospheric trichoderma strains. J Plant Growth Regul. 2019 doi: 10.1007/s00344-019-10017-y. [DOI] [Google Scholar]
- Seo E, Choi D. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief Funct Genom. 2015;14(4):260–267@@. doi: 10.1093/bfgp/elv011. [DOI] [PubMed] [Google Scholar]
- Sharma PN, Kaur M, Sharma OP, Sharma P, Pathania A. Morphological, pathological and molecular variability in Colletotrichum capsici, the cause of fruit rot of chillies in the subtropical region of north-western India. J Phytopathol. 2005;153:232–237. [Google Scholar]
- Singla P, Devi R, Kaur S, Kaur J. Stripe rust induced defence mechanisms in the leaves of contrasting barley genotypes (Hordeum vulgare L.) at the seedling stage. Protoplasma. 2019 doi: 10.1007/s00709-019-01428-5. [DOI] [PubMed] [Google Scholar]
- Soh HC, Park AR, Park S, Back K, Yoon JB, Park HG, Kim YS. Comparative analysis of pathogenesis-related protein 10 (PR10) genes between fungal resistant and susceptible peppers. Eur J Plant Pathol. 2012;132(1):37–48. [Google Scholar]
- Suthin T, Sudhagar GB, Rao S, Ann RS, Muthukumar A, Muthukumaran N, Rao S (2019) Induction of defence enzymes activities in rice plant treated by seaweed algae against Rhizoctonia solani Kuhn causing sheath blight of rice. 210–218.
- Taylor PWJ, Mongkolporn O, Than PP, et al (2007) Pathotypes of Colletotrichum spp. infecting chili peppers and mechanisms of resistance. In: Oh DG, Kim KT (eds) First International Symposium on Chilli Anthracnose, Abstracts, Seoul, South Korea. National Horticultural Research Institute. p. 29.
- Than PP, Jeewon R, Hyde KD, Pongsupasamit S, Mongkolporn O, Taylor PWJ. Characterization and pathogenicity of Colletotrichum species associated with anthracnose disease on chili (Capsicum spp.) in Thailand. Plant Pathol. 2008;57:562–572. [Google Scholar]
- Thurow C, et al. Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 2005;44:100–113. doi: 10.1111/j.1365-313X.2005.02513.x. [DOI] [PubMed] [Google Scholar]
- Tian S, Wan Y, Qin G, Xu Y. Induction of defense responses against Alternaria rot by different elicitors in harvested pear fruit. Appl Microbiol Biotechnol. 2006;70:729–734. doi: 10.1007/s00253-005-0125-4. [DOI] [PubMed] [Google Scholar]
- Truernit E, Schmid J, Epple P, Illig J, Sauer N. The sink-specific and stress regulated Arabidopsis gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell. 1996;8:2169–2182. doi: 10.1105/tpc.8.12.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Liu J, Zheng J, et al. Sugar transporter proteins in Capsicum: identification, characterization, evolution and expression patterns. Biotechnol Biotechnol Equip. 2020;34(1):341–353. [Google Scholar]
- Yadav V, Wang Z, Wei C, Amo A, Ahmed B, Yang X, Zhang X. Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens. 2020;9(4):312. doi: 10.3390/pathogens9040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Wang H, Cai J, et al. NAC transcription factors in plant immunity. Phytopathol Res. 2019;1:3. doi: 10.1186/s42483-018-0008-0. [DOI] [Google Scholar]
- Zieslin N, Ben Zaken R. Peroxidase activity and presence of phenolic substances in peduncles of rose flowers. Plant Physiol Biochem. 1993;31:333–339. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Analysis of Disease Severity Index (DSI) and Disease area in Bullet and Beldanga cultivars at different time intervals i.e. 0 DPI, 1 DPI, 2 DPI and 3 DPI. (a) Disease Severity Index and (b) Disease area are represented in Percentage calculated with respect to their corresponding controls. Values represent Mean ± SD of three independent experiments each in triplicate. Comparison has been performed for the infected plants with respect to the corresponding control of their respective varieties. Where, ns=not significant, *p<0.05, **p<0.01 and ***p<0.001 representing statistically significant differences with control. (TIF 251 kb)
Supplementary Table 1: List of primers used for semi-quantitative gene expression studies (DOCX 13 kb)