Abstract
In the present study, we investigated the role of p53 in G2 checkpoint function by determining the mechanism by which p53 prevents premature exit from G2 arrest after genotoxic stress. Using three cell model systems, each isogenic, we showed that either ectopic or endogenous p53 sustained a G2 arrest activated by ionizing radiation or adriamycin. The mechanism was p21 and retinoblastoma protein (pRB) dependent and involved an initial inhibition of cyclin B1-Cdc2 activity and a secondary decrease in cyclin B1 and Cdc2 levels. Abrogation of p21 or pRB function in cells containing wild-type p53 blocked the down-regulation of cyclin B1 and Cdc2 expression and led to an accelerated exit from G2 after genotoxic stress. Thus, similar to what occurs in p21 and p53 deficiency, pRB loss can uncouple S phase and mitosis after genotoxic stress in tumor cells. These results indicate that similar molecular mechanisms are required for p53 regulation of G1 and G2 checkpoints.
Most human tumors arise from multiple genetic changes which gradually transform growth-limited cells into highly invasive cells that are unresponsive to normal growth controls. The genetic evolution of normal cells into cancer cells is largely determined by the fidelity of DNA replication, repair, and division (40). Cell cycle arrest in response to DNA damage is an important mechanism for maintaining genomic integrity. The control mechanisms that restrain cell cycle transition after DNA damage are comprised of multiple signaling pathways and are known as cell cycle checkpoints (17). In normal tissue homeostasis, cells arrest after dissipation of essential growth factors, hormones, or nutrients or if differentiation is induced. After stress, cells arrest at checkpoints either to stall the initiation of DNA synthesis and cell division until cellular damage can be repaired or to activate pathways that lead to apoptosis.
It has become increasingly evident that loss of G1/S checkpoint function is a hallmark of human cancers. If one considers the frequency of alterations in p53, retinoblastoma protein (pRB), and their upstream regulatory pathways in cancer cells, the majority of human carcinomas have defective G1/S checkpoint function. Of all the genetic alterations identified in human tumors that lead to deregulation of G1/S checkpoint function, p53 gene mutation is the most common (24). p53 is a short-lived protein present at very low levels in the nuclei of normal cells; however, a variety of cellular insults, including DNA damage (27), hypoxia (15), aberrant oncogenic signaling (32), and inhibition of microtubule dynamics (51), result in elevated levels of the protein. p53 is a sequence-specific transcription factor (12, 29, 41) that induces expression of several target genes, including p21, Bax, and MDM2 (reviewed in reference 2). Through transactivation of p21, p53 is one of the major regulators of the G1/S checkpoint in response to cellular stress. p21 binds to G1 cyclin–cyclin-dependent kinase (Cdk) complexes and inhibits their ability to phosphorylate pRB (16). pRB acts as a transcriptional repressor in its hypophosphorylated state when it is bound to the E2F family of transcription factors (48). The E2F family mediates transcription of genes required for DNA synthesis, including cyclin E, cyclin A, dihydrofolate reductase, and thymidine kinase (reviewed in reference 57). The binding of hypophosphorylated pRB to E2F has been shown to inhibit E2F-dependent transcription of S-phase genes and arrest cells at the G1/S transition (22, 47).
Relative to p53's role in G1 checkpoint responses, a role for p53 in G2 cell cycle arrest is less well defined. Cells that do not contain wild-type p53 are deficient for G1/S checkpoint response (33, 49), although the ability of these cells to undergo a G2 arrest remains intact (31, 33). However, ectopic expression of p53 in the absence of damage is sufficient to induce cell cycle arrest at both the G1/S and G2/M transitions (1, 50) and is accompanied by reduced levels and/or differential subcellular localization of cyclin B1 protein (25, 60). Recent studies suggest that p53, p21, and 14-3-3ς are necessary to maintain a G2 arrest following DNA damage, since tumor cells lacking these proteins enter mitosis with accelerated kinetics (5, 7). Taken together, these results suggest that p53 is not required for the activation of a G2 arrest in response to genotoxic stress but that it may dictate the duration of G2 arrest through p21 transactivation.
The goal of this study was to further investigate the role of p53 in G2 checkpoint function by determining the mechanism by which p53 sustains G2 arrest after genotoxic stress. Three cell culture model systems, each isogenic, were used to show that p53 sustains the duration of G2 arrest after treatment of cells with ionizing radiation (IR) or adriamycin (ADR). The p53-mediated maintenance of G2 arrest appears to be dependent on an initial inhibition of the cyclin B1-Cdc2 activity by p21 and a secondary decrease in cyclin B1 and Cdc2 transcription that is p21 and pRB dependent.
MATERIALS AND METHODS
p53-inducible system.
The ecdysone-inducible expression system (Invitrogen, Carlsbad, Calif.) was used to generate cell lines that conditionally express p53. A human hemagglutinin-tagged p53 cDNA was ligated into the pIND vector. The resulting vector, pIND-p53, was cotransfected with the pVgRXR vector into the human large cell lung carcinoma cell line H1299, which is null for p53 expression. Stable clones were selected by limiting dilution in F-12 medium containing 10% fetal calf serum (FCS) (Gemini Bio-Products, Inc., Calabasas, Calif.), 1% penicillin-streptomycin (Sigma, St. Louis, Mo.), 600 μg of G418 (Mediatech, Herndon, Va.) per ml, and 400 μg of Zeocin (Cayla, Toulouse, France) per ml, and resulting cell lines were named H1299-inducible p53 (HIp53). Optimal p53 expression was observed after treatment of cells with the ecdysone analog ponasterone A (PonA) (Invitrogen) at a 10 μM concentration. In all of the experiments shown, PonA was readministered in fresh medium every 24 h to ensure that gene expression was maintained for the duration of the experiment.
Cell culture and treatment.
The RKO-E7 and RKO-NEO cell lines, kindly provided by K. Cho (University of Michigan, Ann Arbor), were cultured in McCoy's 5A medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% FCS 1% penicillin-streptomycin, and 500 μg of G418 per ml. The human colorectal carcinoma cell line HCT116 and the isogenic derivative lines HCT116 p53−/− and HCT116 p21−/− were kindly provided by B. Vogelstein (Johns Hopkins Oncology Center, Baltimore, Md.) and cultured in McCoy's 5A medium supplemented with 10% FCS and 1% penicillin-streptomycin. Cells were treated with ADR or IR as indicated in the figures. IR was delivered at room temperature with a 137Cs irradiator (J. L. Shepherd and Associates). All cells were cultured at 37°C with 5% CO2.
Fluorescence-activated cell sorter analysis.
Approximately 106 cells were incubated with 20 μg of propidium iodide (Sigma) per ml, and DNA content was determined using a FACS Caliber (Becton Dickinson). Data were plotted using CellQuest software, and axis scales were optimized using the control sample and were maintained at that value throughout each experiment. Fifteen thousand events were analyzed for each sample. Bromodeoxyuridine (BrdU) incorporation was performed and analyzed as previously described (13).
Western blot analysis and immunoprecipitations.
Cells were trypsinized, washed twice with ice-cold phosphate-buffered saline, and harvested in kinase lysis buffer (KLB) (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Triton X-100, 0.1% Nonidet P-40, 4 mM EDTA, 50 mM NaF, 0.2 mM Na vanadate) containing the protease inhibitors antipain (10 μg/ml), leupeptin (10 μg/ml), pepstatin A (10 μg/ml), chymostatin (10 μg/ml) (Sigma), and 4-(2-aminoethyl)-benzenesulfonylfluoride (200 μg/ml) (Calbiochem-Novabiochem Corp., La Jolla, Calif.). Cells were lysed by passage through a 23-gauge needle, and the protein supernatant was clarified by centrifugation at 13,000 × g for 10 min at 4°C. Protein concentration was determined by the Bradford protein assay (Bio-Rad Laboratories, Inc., Hercules, Calif.). Western blot analysis was performed as previously described (13) using the following primary antibodies: anti-p53 polyclonal antibody 1801 (Oncogene Research Products, Calbiochem, Cambridge, Mass.), anti-p21 antibody Waf1/Cip1 EA10 (Oncogene Research Products), anti-MDM2 antibody SMP14 (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.), anti-Cdc2 antibody 17 (Santa Cruz Biotechnology Inc.), anti-cyclin B1 antibody GNS1 (Santa Cruz Biotechnology Inc.), anti-pRb antibody LM95.1 (Oncogene Research Products), and anti-Bax antibody N-20 and anti-Actin antibody I-19 (Santa Cruz Biotechnology). In all Western blot analyses, uniform protein loading was confirmed by fast green staining. For immunoprecipitation-based experiments, anti-cyclin B1 GNS1, anti-Cdc2 17, and anti-p21 EA10 antibodies were cross-linked to protein G-Sepharose (Pharmacia Biotech Products, Piscataway, N.J.) as previously described (52). HIp53 cells were treated as indicated in the figure legends and harvested in KLB as described above. Cell lysates (500 μg) were immunoprecipitated with cross-linked antibody for 2 h at 4°C with rocking. Immunoprecipitated proteins were washed three times in KLB, resuspended in 1× Laemmli sample loading buffer, heated at 85°C for 10 min, and analyzed by Western blotting.
Kinase assays.
Kinase assays were performed as previously described (13). Cell lysates (250 μg) were immunoprecipitated with anti-Cdk2 antibody M2 (Santa Cruz Biotechnology Inc.) or anti-cyclin B1 antibody GNS1 (Santa Cruz Biotechnology Inc.). The kinase assay was initiated by adding 2 μCi of [γ-32P]ATP (3,000 Ci/mmol) (New England Nuclear Laboratories) and incubated at 30°C for 10 min using 7 μg of histone H1 (Boehringer Mannheim, Indianapolis, Ind.) as a substrate. The reaction was terminated by the addition of 25 μl of 2× Laemmli sample loading buffer. Reaction mixtures were heated at 85°C for 10 min and subjected to sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis. 32P-labeled histone H1 was quantified on an Instant Imager (Packard, Meriden, Conn.).
Northern blot analysis.
Cells were harvested in RNA lysis buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 2 mM EDTA, 1% SDS) and lysed by passage through a 23-gauge needle eight times. Proteinase K was added to the lysate to a final concentration of 100 μg/ml, and the lysate was incubated at 37°C for 1 h. Following proteinase K digestion, the NaCl concentration was adjusted to 400 mM. The samples were heated at 65°C for 5 min with constant agitation, followed by immediate cooling in ice water for 30 s. mRNA was isolated by incubation with oligo(dT) cellulose (Ambion Inc., Austin, Tex.) with rocking at room temperature for 2 h. The RNA-oligo(dT) cellulose mixture was washed twice with high-salt buffer (10 mM Tris [pH 7.5], 400 mM NaCl, 1 mM EDTA, 0.2% SDS) and packed with high-salt buffer on a poly prep chromatography column (Bio-Rad Laboratories, Inc.). The column was washed once with high-salt buffer and once with low-salt buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.2% SDS). The mRNA was eluted from the column with 55°C elution buffer (5 mM Tris [pH 7.5], 1 mM EDTA, 0.2% SDS). mRNA was precipitated at −20°C overnight with the addition of sodium acetate (pH 5.2) to a final concentration of 220 mM and two volumes of 95% ethyl alcohol. After precipitation, mRNA was recovered by centrifugation at 12,000 × g for 30 min and the pellet was rinsed once with 70% ethyl alcohol. The pellet was dried by inversion at room temperature and resuspended in sterile H2O. RNA (5 μg) was lyophilized, resuspended in sample buffer {1× morpholinepropanesulfonic acid (MOPS; 0.1 M MOPS [pH 7.0], 40 mM sodium acetate, 5 mM EDTA [pH 8.0]), 50% formamide, 6.5% formaldehyde} and heated at 55°C for 15 min. A 10× loading buffer (50% glycerol, 1 mM EDTA, 0.25% bromophenol blue, 0.25% xylene cyanol, 0.3 mg of ethidium bromide per ml) was added to the sample at a 1× concentration, and mRNA was resolved by gel electrophoresis on a 1% agarose gel containing 2% formaldehyde and 1× MOPS. The gel was washed twice in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer, and mRNA was transferred to a supported nitrocellulose membrane (Gibco BRL). Cyclin B1 and Cdc2 cDNAs were labeled with [α-32P]dCTP using Rediprime II (Amersham). Cyclin B1 cDNA was kindly provided by E. Nishida (Kyoto University, Kyoto, Japan), and Cdc2 cDNA was kindly provided by H. Piwnica-Worms (Washington University, St. Louis, Mo.). After a 2-h prehybridization in Express Hyb (Clontech Laboratories, Inc., Palo Alto, Calif.), membranes were incubated with 2 × 106 cpm of labeled cDNA per ml in Express Hyb at 42°C overnight. Membranes were washed twice at room temperature in 2× SSC–0.1% SDS, followed by two washed in 0.2× SSC–0.1% SDS at 42°C.
Electrophoretic mobility shift assays (EMSA).
After treatment as indicated above, either HIp53, HCT116, HCT116 p53−/−, or HCT116 p21−/− cells were harvested in microextraction buffer (20 mM HEPES [pH 7.8], 450 mM NaCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 25% glycerol) and sonicated. Protein supernatants were clarified by centrifugation, frozen in a methanol–dry-ice bath, and stored at −80°C until processed. An oligonucleotide duplex containing a consensus E2F binding site from the human c-myc promoter (23) was labeled with the Klenow fragment using [α-32P]dATP for 30 min at 37°C. Labeled oligonucleotide was purified by phenol-chloroform extraction and ethanol precipitation and resuspended in H2O. Gel shift reactions were performed by incubating 15 μg of protein lysate in 1× binding buffer (100 mM HEPES [pH 7.9], 5 mM MgCl2, 0.4 mM EGTA, 2 mM dithiothreitol, 200 mM KCl), 5% Ficoll, 200 ng of salmon sperm DNA, and 100,000 cpm of labeled oligonucleotide for 20 min at room temperature. For supershift assays, 1 μg of anti-Rb antibody IF8 (Santa Cruz Biotechnology, Inc.) was added per reaction mixture 5 min after the start of the 20-min incubation. For competition reactions, either a 25-, 50-, or 100-fold excess of unlabeled oligonucleotide was added to the reaction mixtures prior to the addition of protein lysate. Binding was resolved on a 4% acrylamide gel in 0.25× Tris-borate-EDTA.
CAT assays.
HIp53 cells were transiently transfected with pCAT-cyclin B1 or a pCAT vector control (25) using Lipofectamine (Gibco BRL) and treated with ADR 24 h after transfection. The chloramphenicol acetyltransferase (CAT) vectors were kindly provided by J. Lee (Hamilton Regional Cancer Center, Hamilton, Ontario, Canada). After 17 h of ADR treatment, the ADR was removed and the cells were washed twice with phosphate-buffered saline and cultured in the presence or absence of 10 μM PonA for 24 and 48 h. Cells were harvested in 0.25 M Tris, pH 7.8, and lysed by three cycles of freezing and thawing. Supernatants were clarified by centrifugation at 12,000 × g and stored at −80°C until assayed. Protein concentration was determined by the Bradford assay, and 75 μg of protein was used to determine CAT activity using acetyl coenzyme A as a substrate for chloramphenicol-d-threo-[dichloroacetyl-1,2-14C] ([14C]CAP) incorporation. Protein lysate was incubated with 5 μl of [14C]CAP (0.05 μCi/ml) and 450 μM acetyl coenzyme A for 40 min at 37°C. The reaction was terminated by the addition of 4°C ethyl acetate. Reaction mixtures were centrifuged at 12,000 × g for 2 min, and the upper layer was removed and dried under vacuum. Samples were resuspended in ethyl acetate, spotted onto thin-layer chromatography plates (J. T. Baker, Inc., Phillipsburg, N.J.), and resolved by ascending thin-layer chromatography in chloroform-methanol (95:5) until the solvent front was 1 cm from the top of the plate.
RESULTS
p53 expression sustains genotoxic stress-induced G2 growth arrest.
To determine the mechanism by which p53 regulates the G2 checkpoint response, we used three cell culture model systems: (i) a cell line that conditionally expresses ectopic p53; (ii) an isogenic set of HCT116 colorectal carcinoma cell lines that consists of the parental line, which endogenously expresses wild-type p53, and two derivative lines, HCT116 p53−/− (5) and HCT116 p21−/− (54), which are null for p53 and p21, respectively; and (iii) a pair of RKO colorectal carcinoma cell lines, the parental line, and an E7-expressing line, RKO-E7 (49).
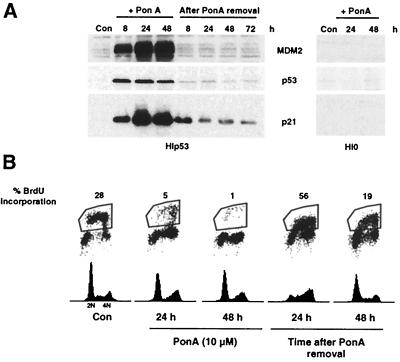
For the first line of experimentation, we developed a cell line that would conditionally express p53. A human lung carcinoma cell line (H1299) that is null for the endogenous expression of p53 was stably transfected with an ecdysone-inducible p53 expression vector. The resulting cell line, HIp53, was derived. After treatment of HIp53 cells with the ecdysone analog PonA, p53 protein levels increased in a dose-dependent manner. Optimal induction of p53 was achieved with a PonA dose of 10 μM (data not shown), and this concentration was used in the experiments whose results are shown. By 8 h after PonA treatment, p53, MDM2, and p21 proteins were detectable. Both MDM2 and p21 protein levels remained elevated through 48 h, while a decrease in p53 protein levels occurred by 48 h (Fig. 1A). MDM2 has been shown to mediate the degradation of p53 (18, 30), and thus the decline in p53 levels by 48 h of PonA treatment may be due in part to the continual induction of MDM2 (Fig. 1A). Treatment of a vector control cell line (H10) with PonA did not result in an increase in p21 or MDM2 protein in the absence of p53 (Fig. 1A).
FIG. 1.
Ectopic expression of p53 leads to reversible G1 and G2 growth arrest. HIp53 cells were treated with PonA for 48 h, the analog was removed, and cells were grown for an additional 72 h. (A) HIp53 cells were harvested at 8, 24, and 48 h following PonA addition and after PonA removal. Protein lysates were analyzed by Western blotting for the indicated proteins. (B) Simultaneous flow cytometric analyses for DNA synthesis (BrdU incorporation) and DNA content (propidium iodide staining) were performed at the indicated times. Results are representative of three independent experiments. Con, control.
To determine the cell cycle distribution and proliferative capacity of HIp53 cells in the absence of damage, simultaneous flow cytometric analyses for DNA synthesis (BrdU incorporation) and DNA content (propidium iodide staining) were performed (Fig. 1B). For these experiments, HIp53 cells were treated with PonA for 48 h, the analog was removed, and cells were grown for an additional 48 h. During each 24-h interval after PonA treatment and removal, cells were incubated with BrdU for 2 h prior to harvest (Fig. 1B). As evidenced by the flow cytometric histogram profile and the 28-fold decrease in BrdU incorporation, the HIp53 cells underwent cell cycle arrest at both G1 and G2 after 48 h of PonA treatment (Fig. 1B) while treatment of HI0 with PonA did not induce a cell cycle arrest or a decrease in BrdU incorporation (data not shown). After PonA removal, HIp53 cells reentered the cell cycle, as was confirmed by a 56-fold increase in BrdU incorporation at 24 h (Fig. 1B). Reentry of HIp53 cells into the cycle after removal of PonA was followed by a rapid decrease in p53 levels (Fig. 1A). Parallel decreases in MDM2 and p21 protein levels were also observed (Fig. 1A). Thus, in the absence of cellular stress, ectopic expression of p53 arrested HIp53 cells in the G1 and G2 phases of the cell cycle.
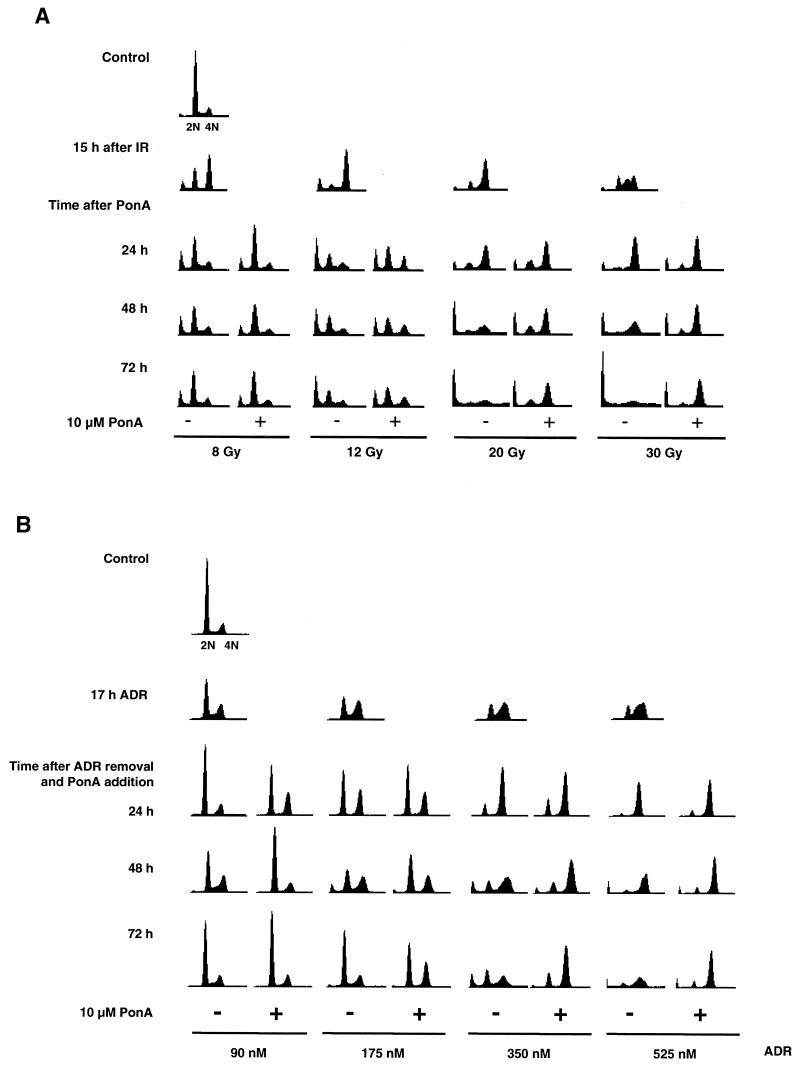
To determine if p53 expression affected the duration of G2 arrest after stress, HIp53 cells were treated with either IR or ADR prior to the induction of p53 with PonA. Treatment of HIp53 cells with IR (8, 12, 20, or 30 Gy) resulted in an accumulation of cells with a 4 N DNA content by 15 h (Fig. 2A and Table 1). After the IR-induced G2 arrest at 15 h, the HIp53 cells were cultured in the presence or absence of PonA for an additional 24, 48, or 72 h. The G2 arrest induced after exposure to 8 or 12 Gy of IR was transient, and p53 expression had only a minimal effect on the duration of the arrest. However, exposure of the cells to higher doses of IR (20 or 30 Gy) increased the length of G2 arrest and expression of p53 sustained the G2 arrest through the time course examined (Fig. 2A and Table 1). In the absence of p53 expression, cells did not maintain the cell cycle arrest; rather, a majority lost viability and detached from the plate by 48 and 72 h after exposure to IR, thus accounting for the diminished peaks in the histograms seen in Fig. 2A at those times.
FIG. 2.
p53 prolongs G2 cell cycle arrest after exposure to IR or ADR. Cells were harvested at the indicated times and analyzed by flow cytometric analysis. (A) Cells were treated with IR (8, 12, 20, and 30 Gy) and incubated for 15 h, after which time cells were cultured for an additional 24, 48, or 72 h in the presence or absence of PonA. (B) Cells were treated with ADR (90, 175, 350, or 525 nM) for 17 h, the drug was washed out, and the cells were cultured for an additional 24, 48, or 72 h in the presence or absence of PonA. Fifteen thousand events were analyzed for each condition, and all histograms were plotted by using the same scale for both axes. Results are representative of two independent experiments.
TABLE 1.
Cell cycle distribution of HIp53 cells after IR
| DNA content or cell cycle phase | Time after IR treatment (h) | % of cells after IRa at:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 8 Gy
|
12 Gy
|
20 Gy
|
30 Gy
|
||||||
| − | + | − | + | − | + | − | + | ||
| <2 N DNA | 15 | 13 | 0 | 2 | 1 | ||||
| G1 | 15 | 25 | 29 | 10 | 17 | ||||
| S | 15 | 8 | 20 | 13 | 46 | ||||
| G2/M | 15 | 54 | 51 | 75 | 36 | ||||
| <2 N DNA | 24 | 20 | 13 | 0 | 0 | 4 | 5 | 5 | 7 |
| G1 | 24 | 43 | 59 | 50 | 51 | 15 | 23 | 5 | 9 |
| S | 24 | 17 | 8 | 7 | 5 | 13 | 12 | 10 | 8 |
| G2/M | 24 | 20 | 20 | 43 | 44 | 68 | 60 | 80 | 76 |
| <2 N DNA | 48 | 21 | 13 | 11 | 4 | 31 | 9 | 16 | 10 |
| G1 | 48 | 41 | 60 | 34 | 50 | 10 | 18 | 6 | 9 |
| S | 48 | 17 | 8 | 15 | 6 | 18 | 10 | 20 | 9 |
| G2/M | 48 | 21 | 20 | 40 | 40 | 41b | 63 | 58b | 72 |
| <2 N DNA | 72 | 22 | 14 | 5 | 42 | 11 | 57 | 11 | |
| G1 | 72 | 44 | 57 | 54 | 53 | 10 | 16 | 5 | 6 |
| S | 72 | 15 | 10 | 12 | 7 | 12 | 10 | 15 | 7 |
| G2/M | 72 | 19 | 19 | 29 | 40 | 36b | 63 | 23b | 76 |
Results are representative of two independent experiments. Values for a control, untreated culture were as follows: <2 N, 0%; G1, 66%; S, 15%; G2/M, 19%. − and +, absence and presence of PonA (plus p53), respectively.
More than 75% of the cells underwent cell death and detached from the plate; the number is representative of the remaining cells (see Fig. 2A).
HIp53 cells were also treated with ADR (dose range, 90 to 525 nM) for 17 h, the drug was washed out, and the cells were cultured in the presence or absence of PonA for an additional 24, 48, or 72 h. ADR treatment induced a dose-dependent G2 arrest in HIp53 cells, with the highest percentage of cells arresting with a 4 N DNA content after treatment with 350 and 525 nM ADR (Fig. 2B and Table 2). At all of the ADR doses tested, the expression of p53 increased the duration of time that cells remained arrested in both the G1 and G2 phases of the cell cycle compared to arrest times for cells lacking p53 (Fig. 2B and Table 2). The most pronounced p53-mediated maintenance of G2 arrest occurred 48 and 72 h after treatment with 350 and 525 nM ADR (Fig. 2B and Table 2). In contrast, the majority of the cells lacking p53 expression did not maintain the cell cycle arrest, lost viability, and detached from the plate by 48 and 72 h after exposure to ADR, thus accounting for the diminished frequencies in the histograms seen in Fig. 2B at those times. Thus, after exposure of HIp53 cells to IR or ADR, p53 significantly extended the period of G2 arrest compared with that of cells deficient for p53.
TABLE 2.
Cell cycle distribution of HIp53 cells after ADR treatment
| DNA content or cell cycle phase | Time after ADR treatment (h) | % of cells after treatment with ADRa at:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 90 nM
|
175 nM
|
350 nM
|
525 nM
|
||||||
| − | + | − | + | − | + | − | + | ||
| <2 N | 17 | 0 | 0 | 0 | 0 | ||||
| G1 | 17 | 50 | 29 | 28 | 19 | ||||
| S | 17 | 19 | 20 | 19 | 39 | ||||
| G2/M | 17 | 31 | 51 | 53 | 42 | ||||
| <2 N | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| G1 | 24 | 70 | 71 | 50 | 51 | 13 | 23 | 7 | 10 |
| S | 24 | 11 | 8 | 7 | 5 | 8 | 5 | 6 | 4 |
| G2/M | 24 | 19 | 21 | 43 | 44 | 79 | 72 | 87 | 85 |
| <2 N | 48 | 0 | 0 | 11 | 4 | 13 | 3 | 5 | 2 |
| G1 | 48 | 44 | 73 | 34 | 50 | 18 | 15 | 4 | 11 |
| S | 48 | 17 | 6 | 15 | 6 | 16 | 7 | 7 | 4 |
| G2/M | 48 | 39 | 21 | 40 | 40 | 53b | 75 | 84b | 83 |
| <2 N | 72 | 0 | 0 | 5 | 27 | 2 | 9 | 4 | |
| G1 | 72 | 65 | 71 | 54 | 53 | 28 | 17 | 9 | 11 |
| S | 72 | 11 | 7 | 12 | 7 | 18 | 6 | 8 | 4 |
| G2/M | 72 | 24 | 22 | 29 | 40 | 27b | 75 | 74b | 81 |
Results are representative of two independent experiments. Values for a control, untreated culture were as follows: <2 N, 0%; G1, 71%; S, 12%; G2/M, 17%. − and +, absence and presence of PonA (plus p53), respectively.
More than 75% of the cells underwent cell death and detached from the plate; the number is representative of the remaining cells (see Fig. 2B).
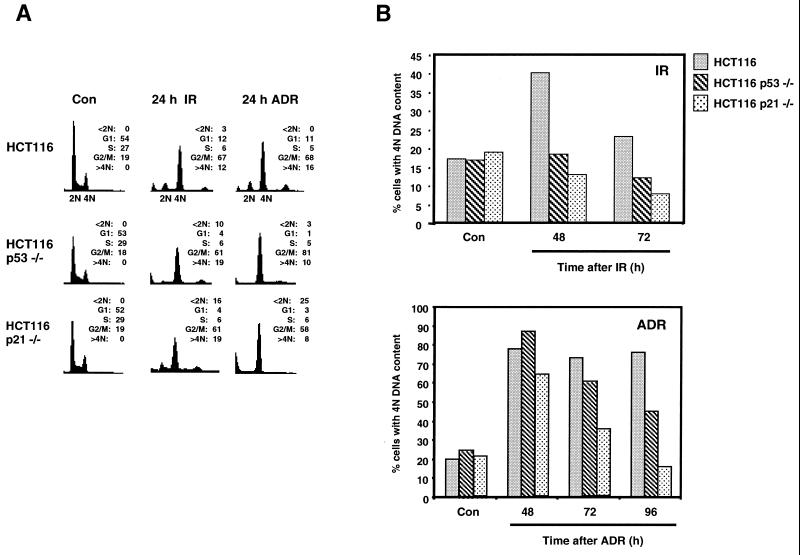
To verify and extend the observations described above, we also analyzed the role of p53 in G2 checkpoint response in an isogenic set of HCT116 colorectal carcinoma cell lines. HCT116, HCT116 p53−/−, and HCT116 p21−/− cells were treated with IR or ADR and analyzed for DNA content by flow cytometry at 24, 48, 72, and 96 h (in the case of ADR) after treatment. Twenty-four hours after treatment with IR or ADR, all of the HCT116-derived cell lines had an accumulation of cells with, predominantly, a 4 N DNA content (Fig. 3A). However, by 72 h after treatment with either genotoxic agent, the HCT116 p53−/− and the HCT116 p21−/− cells had reductions in the numbers of cells with a 4 N DNA content compared with the numbers for the parental HCT116 cells (Fig. 3B).
FIG. 3.
p21 is required for regulation of G2 checkpoint arrest. HCT116, HCT116 p53−/−, and HCT116 p21−/− cells were treated with IR (8 Gy) or ADR (350 nM) and analyzed 24, 48, and 72 h after treatment. (A and B) Cells were analyzed by flow cytometric analysis. (A) Propidium iodide fluorescence profile for control (Con) and 24-h time points; (B) quantifications from the flow histograms presented as percentages of cells with a 4 N DNA content after treatment with IR or ADR. Fifteen thousand events were analyzed for each condition, and all histograms were plotted by using the same scale for both axes. Results are representative of two independent experiments.
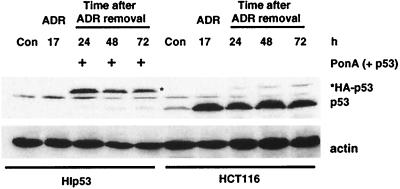
In order to compare the findings above and subsequent molecular results obtained with the two different cell model systems, we determined the relative levels of p53 in each cell line before and after ADR treatment. HIp53 and HCT116 cells were treated with ADR (350 nM) for 17 h, the drug was removed, and the cells were grown in fresh growth medium. With the HIp53 cells, PonA was added after 17 h to induce p53. For each line, the same number of cells (500,000) was harvested from control and treated cultures and protein lysates were prepared and analyzed on the same Western blot for p53 protein levels (Fig. 4). Relative to levels in the HIp53 cells, the HCT116 cells had higher levels of p53 protein at each time analyzed after ADR treatment. The slower migration of the p53 protein in the HIp53 cells was due to the inclusion of a hemagglutinin tag at the 5′ end of the p53 protein. The comparison of p53 levels in the two cell lines assured us that results obtained with the HIp53 cells were not due merely to the higher levels of overexpressed p53 protein, as is the concern with many ectopic expression systems.
FIG. 4.
Relative levels of p53 protein in HCT116 and HIp53 cells. HIp53 and HCT116 cells were treated with ADR (350 nM) for 17 h, the drug was removed, and the cells were grown in fresh growth medium. With the HIp53 cells, PonA was added after 17 h to induce p53. For each line, the same number of cells (500,000) was harvested from control (Con) and treated cultures and protein lysates were prepared and analyzed on the same Western blot for p53 protein levels. Actin analysis was included to assess protein loading and transfer. Results are representative of three independent experiments.
p53 expression results in loss of cyclin B1-Cdc2 activity.
To elucidate the mechanism underlying the prolonged stress-induced G2 arrest observed in the cells containing an intact p53 signaling pathway, we examined proteins known to play a role in the regulation of G2 transition. Progression of cells from G2 to mitosis requires the activity of the cyclin B1-Cdc2 complex (34). When cells undergo genotoxic stress, the activity of this complex is reduced through inhibitory phosphorylations of Cdc2 and cells arrest in G2 (36, 38). p21 is increased in a p53-dependent manner and can bind and inhibit the activities of several Cdks, including cyclin B1 and Cdc2 (16). We hypothesized that the observed p53 regulation of the G2 checkpoint was mediated through inhibition of cyclin B1-Cdc2 activity. To test this hypothesis, protein lysates were prepared from the cells described in the legends to Fig. 2 and 3 and processed using Western blot- and immunoprecipitation-based assays.
In the HIp53 cells, an increase in p21 protein occurred in cells that expressed p53 while only low-level p21 protein was detectable in IR- or ADR-treated cells lacking p53 (Fig. 5A and B). At all doses of IR or ADR tested, significant reductions in cyclin B1 and Cdc2 protein levels were observed by 48 and 72 h after induction of p53 (Fig. 5A and B). The cell cycle arrest pattern of the cells varied with the dose of the genotoxic agent (Fig. 2); however, the observed reduction of cyclin B1 and Cdc2 protein at 72 h was independent of cell cycle position. Of note, the kinetics of cyclin B1 protein loss were more rapid at lower doses and correlated with a greater number of cells arrested in G1. In contrast, cells treated with IR or ADR that lack p53 expression had an increase in cyclin B1 and Cdc2 protein levels at all doses examined.
FIG. 5.
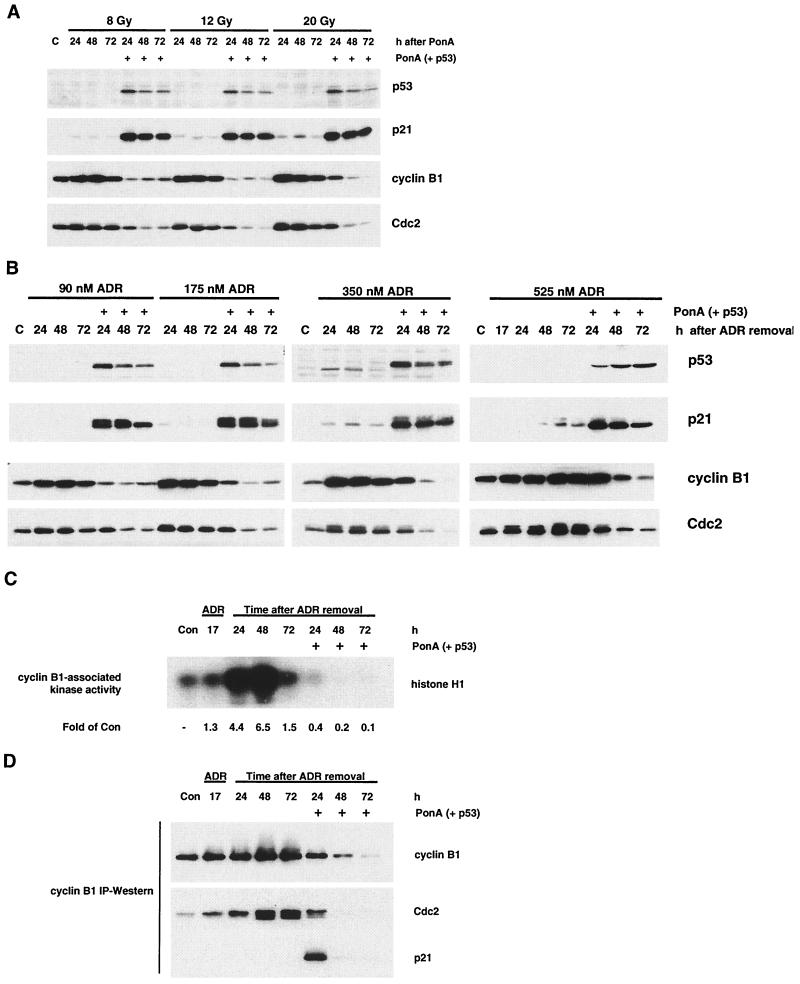
p53 expression after IR or ADR inhibits cyclin B1-Cdc2 kinase activity. (A) Cells were treated with IR (8, 12, and 20 Gy) and incubated for 15 h, after which time cells were cultured for an additional 24, 48, or 72 h in the presence or absence of PonA. Protein lysates were analyzed by Western blotting for the indicated proteins. (B) Cells were treated with ADR (90, 175, 350, or 525 nM) for 17 h, the drug was washed out, and the cells were cultured an additional 24, 48, or 72 h in the presence or absence of PonA. (C and D) Protein lysates were analyzed by cyclin B1 immunoprecipitation-based assays for Cdc2 kinase activity using histone H1 as a substrate (C) and for immunoprecipitable cyclin B1, Cdc2, and p21 proteins by Western blotting (D). HIp53 cells were treated with 525 nM ADR for the data presented in panels C and D. Results are representative of two independent experiments. Con, control; IP, immunoprecipitation.
Accompanying the reduction in cyclin B1 and Cdc2 protein levels in the HIp53 cells, we observed a 60% decrease in cyclin B1-Cdc2 activity by 24 h after p53 induction and a further decline to 10% of control levels by 72 h (Fig. 5C). Conversely, in the absence of p53 expression there was an increase in cyclin B1-Cdc2 activity through 48 h, with levels elevated 6.5-fold higher than that of the control (Fig. 5C). At 24 h after ADR removal, the decrease in cyclin B1-Cdc2 kinase activity in HIp53 cells expressing p53 (Fig. 5C) preceded any marked decrease in cyclin B1 and Cdc2 protein levels (Fig. 5B). In addition, the loss of cyclin B1-Cdc2 kinase activity 24 h after p53 induction did not correlate with inhibitory phosphorylation of Cdc2, as the predominant form of the protein was in the faster-migrating, hypophosphorylated state (Fig. 5B).
We hypothesized that the initial down-regulation of cyclin B1-Cdc2 kinase activity 24 h after p53 induction was due to the direct association of p21 with the cyclin B1-Cdc2 complex. To test this hypothesis, protein lysates were immunoprecipitated with cyclin B1 antibodies and analyzed by Western blotting for cyclin B1, Cdc2, and p21. Both p21 and Cdc2 coimmunoprecipitated with cyclin B1 24 h after p53 expression in ADR-treated HIp53 cells (Fig. 5D). Consistent with the decrease in cyclin B1 levels seen in Fig. 5B, the levels of immunoprecipitable cyclin B1, as well as those of any coimmunoprecipitable Cdc2 and p21, were reduced by 72 h after p53 expression (Fig. 5D). p21 was not coimmunoprecipitated with cyclin B1 in ADR-treated cells that lacked p53 expression; however, increasing amounts of Cdc2 coprecipitated with cyclin B1 at 24, 48, and 72 h in the absence of p53 expression. Thus, the maintenance of G2 arrest in cells expressing p53 after genotoxic stress appears to involve a two-step mechanism, including an initial inhibition of cyclin B1-Cdc2 activity through p21 association with the complex, followed by a marked decrease in cyclin B1 and Cdc2 protein levels.
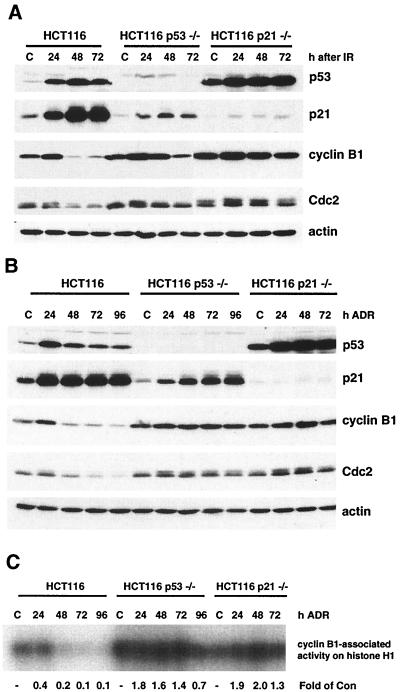
In both HCT116 and HCT116 p21−/− cells, p53 levels increased and remained elevated through 72 h after IR (Fig. 6A). Similar changes in p53 levels were observed after ADR treatment (Fig. 6B). p21 protein levels were significantly elevated only in HCT116 cells after IR and ADR treatment (Fig. 6A and B). p53-independent elevation of p21 protein was also observed in the HCT116 p53−/− cells. There was a decrease in cyclin B1 and Cdc2 protein levels by 24 h in HCT116 cells exposed to both IR and ADR, while cyclin B1 and Cdc2 protein levels remained elevated in HCT116 p53−/− and HCT116 p21−/− cells (Fig. 6A and B). Similar to the results for the HIp53 cells, we observed a 60% decrease in cyclin B1-Cdc2 activity by 24 h after ADR treatment and a further decline to 10% of control levels by 72 h in the HCT116 cells (Fig. 6C). Conversely, in the absence of a functional p53 signaling pathway in the HCT116 p53−/− and HCT116 p21−/− cells, there was an increase in cyclin B1-Cdc2 activity through 72 h, with levels elevated 1.3- to 2-fold higher than that of the control (Fig. 6C). These data are consistent with the results obtained with the HIp53 cells and support the hypothesis that p53 regulation of the G2 checkpoint occurs through a p21-dependent mechanism that involves a reduction of cyclin B1 and Cdc2 protein levels.
FIG. 6.
p21 is required for p53-mediated loss of cyclin B1 and Cdc2 protein levels. Cells were treated as described in the legend to Fig. 3 with IR (A) or ADR (B), harvested, and analyzed by Western blotting for the indicated proteins. Actin analysis was included to assess protein loading and transfer. (C) Protein lysates were analyzed by cyclin B1 immunoprecipitation-based assays for Cdc2 kinase activity using histone H1 as a substrate. Results are representative of three independent experiments. C and Con, control.
Loss of cyclin B1 and Cdc2 after p53 expression is due to a reduction in cyclin B1 and Cdc2 mRNA.
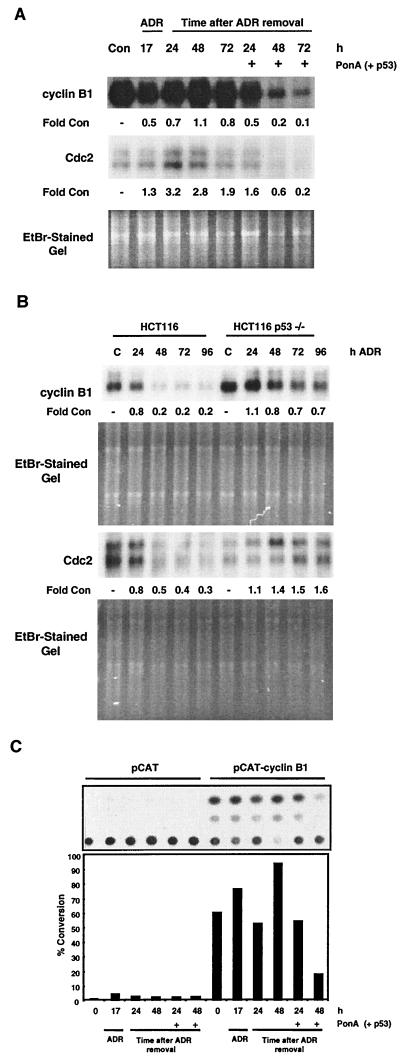
To determine whether the p53-dependent decrease in cyclin B1 and Cdc2 protein levels was due to a reduction in cyclin B1 and Cdc2 mRNA, Northern blot analyses were performed. A time-dependent loss of cyclin B1 and Cdc2 mRNA was observed in ADR-treated HIp53 cells and in HCT116 cells that expressed p53 (Fig. 7A and B). In both cell lines, the levels of cyclin B1 and Cdc2 mRNA were reduced by 70 to 90% by 72 h (Fig. 7A and B). These results are consistent with the observed decrease in cyclin B1 and Cdc2 protein levels (Fig. 5B and 6B). In contrast, cyclin B1 and Cdc2 mRNA levels were elevated at 24 and 48 h after ADR treatment in cells that lacked an intact p53 signaling pathway (Fig. 7A and B). To further extend these results, a cyclin B1 promoter-CAT reporter vector (pCAT-cyclin B1) was transfected into the HIp53 cells. The cells were treated with ADR for 17 h, the drug was washed out, and the cells were cultured for an additional 24 or 48 h in the presence or absence of PonA. Induction of p53 after ADR treatment resulted in a 4.5-fold reduction in cyclin B1 promoter-CAT activity by 48 h compared with the CAT activity in cells lacking p53 (Fig. 7C). Taken together, these data suggest that the loss of cyclin B1 and Cdc2 protein observed during p53-mediated sustained G2 arrest was due to decreases in cyclin B1 and Cdc2 mRNA levels and that the regulation of cyclin B1 occurred at the transcriptional level.
FIG. 7.
Loss of cyclin B1 and Cdc2 expression after p53 expression. (A) HIp53 cells were treated with 525 nM ADR for 17 h, the drug was washed out, and the cells were cultured in the presence or absence of PonA for 24, 48, or 72 h. Cells were harvested, mRNA was isolated, and Northern blot analyses were performed for cyclin B1 or Cdc2 mRNA. (B) HCT116 and HCT116 p53−/− cells were treated with 350 nM ADR for the times indicated, and cells were harvested, mRNA was isolated, and Northern blot analyses were performed for cyclin B1 and Cdc2. (C) HIp53 cells were transfected with either pCAT-cyclin B1 or with the pCAT vector. Twenty-four hours after transfection, the cells were treated with 525 nM ADR. The drug was washed out 17 h after treatment, and the cells were cultured an additional 24 or 48 h in the presence or absence of PonA. Cells were harvested and analyzed for CAT activity. Results are representative of two independent experiments. Con, control; EtBr, ethidium bromide.
Cyclin B1 and Cdc2 transcription is dependent on the activity of cyclin A-Cdk2 complexes in late S phase (35). Furthermore, the expression of Cdc2 and cyclin A are regulated by the E2F family of transcription factors (8, 9, 14, 46). We hypothesized that the p53-mediated inhibition of cyclin B1 transcription was dependent on p21 inhibition of cyclin-Cdk complexes, the resulting hypophosphorylation of pRB, and subsequent inhibition of an E2F-dependent transcriptional cascade. To test this hypothesis, we analyzed ADR-treated HIp53 and HCT116 cells for Cdk2 activity and pRB phosphorylation state. Cdk2 activity was significantly inhibited by 24 h in the HIp53 and HCT116 cell lines and completely inhibited by 72 h in the HIp53 cells and by 90% in the HCT116 cells (Fig. 8). In contrast, cells without an intact p53 signaling pathway displayed a 1.5- to 3-fold increase in Cdk2 activity over the time course (Fig. 8). Analysis of pRB phosphorylation revealed that pRB was predominantly in the hyperphosphorylated, inactive state in HIp53 cells lacking p53 expression as well as the HCT116 p21−/− cells but was predominantly in the hypophosphorylated, active state in ADR-treated HIp53 and HCT116 cells expressing p53 and p21 (Fig. 8). Faster-migrating forms of pRB were detectable in HCT116 p53−/− cells and were likely due to the p53-independent elevation of p21 protein (Fig. 6B) that occurred after ADR treatment; however, these forms were not sufficient to inhibit cyclin B1-Cdc2 or Cdk2 activity (Fig. 6C and 8B). These results led to the hypothesis that in the presence of p53, there is a pRB-dependent inhibition of E2F transcriptional activity in G2-arrested cells.
FIG. 8.
Dephosphorylation of pRB during p53 regulation of the G2 checkpoint. (A) HIp53 cells were treated with 525 nM ADR for 17 h, the drug was washed out, and the cells were cultured for the indicated times in the presence or absence of PonA. (A) Cells were harvested and analyzed by Western blot analysis for pRB protein levels and phosphorylation state (upper blot) and for Cdk2 kinase activity using histone H1 as a substrate (lower blot). (B) HCT116, HCT116 p53−/−, and HCT116 p21−/− cells were treated with 350 nM ADR for the times indicated, and the cells were harvested and analyzed by Western blot analysis for pRB protein levels and phosphorylation state (upper blot) and for Cdk2 kinase activity using histone H1 as a substrate (lower blot). Results are representative of three independent experiments. C and Con, control.
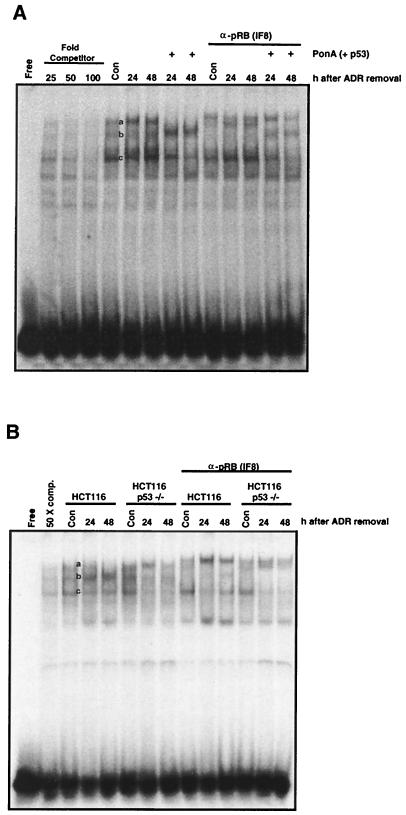
To assess the interaction between pRB and E2F transcription factors, EMSA were performed using an E2F binding element from the human c-myc gene (23). Gel shift analyses showed that three different E2F complexes were formed with proteins harvested from rapidly cycling populations of HIp53, HCT116, and HCT116 p53−/− cells (Fig. 9, complexes a, b, and c). The formation of three protein-DNA complexes with this E2F binding element is consistent with previous observations (23). These three complexes could be efficiently competed with excess unlabeled binding site DNA (Fig. 9). In ADR-treated cells that lacked p53 expression, there were increases in levels of the slower-migrating complex at 24 and 48 h (Fig. 9, complex a). In ADR-treated cells expressing p53, slower-migrating complex a was undetectable by 48 h (Fig. 9). The decrease in complex a formation was accompanied by an increase in the intensity of complex b in cells expressing p53. To determine which of these complexes contained pRB, supershift assays were performed with an antibody specific for pRB. The supershift analyses revealed that complex b contained pRB (Fig. 9). The results indicate that a significant fraction of the E2F protein is in complex with pRB in G2-arrested cells that express p53.
FIG. 9.
pRB interaction with E2F transcription factors in HIp53, HCT116, and HCT116 p53−/− cells after genotoxic stress. An EMSA was performed to analyze pRB and E2F interaction using an E2F binding element derived from the human c-myc promoter. For competition assays, either a 25-, 50-, or 100-fold excess of unlabeled oligonucleotide was added. For supershift assays, 1 μg of anti-pRB antibody was added to reaction mixtures 5 min after the incubation was initiated. Free, oligonucleotide duplex alone. (A) HIp53 cells were treated with 525 nM ADR for 17 h, the drug was washed out, and the cells were cultured for the indicated times in the presence or absence of PonA. Cells were harvested and analyzed. (B) HCT116 and HCT116 p53−/− cells were treated with 350 nM ADR for the times indicated, and the cells were harvested and analyzed. Results are representative of three independent experiments. Con, control; comp., competitor.
Disruption of pRB signaling abrogates the maintenance of G2 arrest.
Based on the results described above, we hypothesized that p53 regulation of G2 arrest was pRB dependent. To test this hypothesis, we used a set of RKO cells developed by Slebos and colleagues that stably express either the human papillomavirus (HPV) type 16 E7 protein (RKO-E7) or the vector alone (RKO-NEO) (49). Expression of the HPV type 16 E7 viral protein abrogates pRB function in these cells (11, 49, 59). RKO-NEO and RKO-E7 cells were treated with IR or ADR, and DNA contents were assessed by flow cytometric analysis at 24, 48, and 72 h after exposure to the agents. In both RKO-NEO and RKO-E7 cultures, there was an accumulation of cells with a predominant 4 N DNA content at 24 h after IR and 17 h after ADR treatment (Fig. 10A). There was also an arrest of the RKO-NEO cells in G1 that was abrogated by E7 expression (Fig. 10A) as previously reported (49). RKO-NEO cells sustained the G2 arrest after both IR and ADR treatment through 72 h (Fig. 10B), whereas a significant fraction of RKO-E7 cells exited from G2 and endoreduplicated as evidenced by the accumulation of cells with a 8 N DNA content (Fig. 10A, ADR treatment).
FIG. 10.
pRB is required for prolonged G2 arrest and loss of cyclin B1 and Cdc2 protein levels. RKO-NEO and RKO-E7 cells were treated with IR (10 Gy) or ADR (350 nM), the drug was washed out at 17 h, and cells were harvested at 24, 48, and 72 h. (A and B) Cells were analyzed by flow cytometric analysis. (A) Propidium iodide fluorescence profile for control (Con) and treated cells; (B) quantifications from the flow histograms presented as percentages of cells with a 4 N DNA content after treatment with IR or ADR. Fifteen thousand events were analyzed for each condition, and all histograms were plotted by using the same scale for both axes. Protein lysates from cells exposed to IR (C) and ADR (D) were analyzed by Western blotting for the indicated proteins. (E) Protein lysates were analyzed for Cdc2 and Cdk2 kinase activities using histone H1 as a substrate. Results are representative of two independent experiments.
Analysis of proteins from the RKO cell lines revealed that p53 and p21 levels increased in similar manners in both RKO-NEO and RKO-E7 lines after both treatments (Fig. 10C and D). Cyclin B1 and Cdc2 protein levels were reduced in RKO-NEO cells 72 h after IR and 48 h after ADR treatment, while RKO-E7 cells maintained control or higher levels of cyclin B1 and Cdc2 after treatment with IR and ADR (Fig. 10C and D). pRB was in the hypophosphorylated form in RKO-NEO cells between 48 and 72 h after IR and ADR treatment, whereas pRB remained predominantly in the phosphorylated state in RKO-E7 cells (Fig. 10C and D). Dephosphorylation of pRB in RKO-NEO cells occurred with kinetics similar to those seen with the decrease in cyclin B1 and Cdc2 protein levels. Consistent with the reduction in cyclin B1 and Cdc2 proteins levels, we observed a decrease in cyclin B1-Cdc2 and Cdk2 activities in RKO-NEO cells by 72 h after IR and ADR exposure (Fig. 10E). In contrast, there was an increase in cyclin B1-Cdc2 and Cdk2 kinase activities after IR and ADR treatment in RKO-E7 cells (Fig. 10E). These biochemical changes mirror those observed in the HIp53 and the HCT116 cell systems and indicate that pRB plays an integral role in p53-mediated maintenance of the G2 arrest after genotoxic stress.
DISCUSSION
The results presented provide a mechanism for how p53 sustains G2 arrest after genotoxic stress. Treatment of p53-deficient cells with genotoxic agents induced a transient G2 arrest that was followed by an increase in cyclin B1-Cdc2 kinase activity and premature progression of cells into mitosis, whereas in cells expressing p53, G2 arrest was sustained through an initial inhibition of cyclin B1-Cdc2 activity, followed by a marked decrease in cyclin B1 and Cdc2 levels. Of significance, p53 maintenance of G2 arrest was p21 and pRB dependent. Induction of p53 after cell stress resulted in a marked elevation of p21, conversion of pRB to the hypophosphorylated, active form, and increased pRB-E2F complex formation. The abrogation of pRB activity by E7 in the RKO cells resulted in a premature G2 exit in response to genotoxic stress and subsequent endoreduplication. Thus, our study not only provides insight into how p53 sustains G2 arrest after stress, it also shows that pRB loss can uncouple S phase and mitosis after genotoxic stress in tumor cells.
Agarwal et al. first provided evidence that p53 could mediate a G2 growth arrest (1). The observed p53-mediated G2 arrest in the HIp53 cells, in the absence of genotoxic stress, is consistent with the results of this previous study. Our findings support previous studies demonstrating that Cdc2 is down-regulated in a p53-dependent manner after IR (3), cyclin B1 and Cdc2 levels decrease after ectopic p53 expression (25), and p53 expression inhibits cyclin B1 and Cdc2 transcription (53). In agreement with the study of Bunz et al. (5), we show that HCT116 p53−/− and HCT116 p21−/− cells are unable to maintain a G2 arrest after exposure of cells to IR. Our results support and provide insight into previous findings that pRB overexpression mediates G2 growth arrest independently of p53 (26) and that hypophosphorylation of pRB occurs during stress-induced G2 arrest (43, 62). Further, Park et al. show that constitutive activation of cyclin B1-Cdc2 overrides p53-mediated G2 arrest (37).
This study shows that similar molecular mechanisms are involved in p53 regulation of G1 and G2 checkpoints. The comparable mechanisms are exemplified by the results obtained after treatment of HIp53 cells with a dose range of genotoxic agents. Exposure of HIp53 cells to lower doses of either IR or ADR, followed by p53 expression, led to a predominant G1 cell cycle arrest, whereas higher doses of the agents caused an accumulation of cells at G2. Analyses of cyclin B1 and Cdc2 proteins showed that a reduction in levels occurred regardless of whether a G1 or a G2 arrest was initiated if p53 was present; however, the kinetics of cyclin B1 and Cdc2 protein reduction varied and appeared to correlate with the phase of cell cycle arrest. Exposure of cells to doses of genotoxic agent that resulted in a predominant accumulation of cells in the G1 phase of the cell cycle resulted in a more rapid decrease in cyclin B1 and Cdc2 protein levels compared to levels seen after doses that resulted in a more pronounced G2 arrest. The difference in the kinetics of cyclin B1 and Cdc2 protein loss can be explained by cell cycle-dependent gene expression. Cells in G1 have not activated the transcription of genes that encode G2-phase-specific proteins, and thus cyclin B1 and Cdc2 proteins are absent. In contrast, cells in G2 have elevated levels of cyclin B1 and Cdc2 protein. In the presence of hypophosphorylated pRB, transcription of cyclin B1 and Cdc2 is inhibited regardless of cell cycle phase; however, the time required for cyclin B1 and Cdc2 protein degradation in a culture of cells that is predominantly in G2 would account for the apparent difference in kinetics of protein loss. These data are consistent with those of a recent study by de Toledo et al. which showed that down-regulation of E2F-responsive genes, including those for Cdc2, cyclin A, cyclin B1, thymidine kinase, and topoisomerase II, occur in a p53-dependent manner after treatment of cells with IR (10). Also, our results depicting the integral role of pRB in p53-mediated maintenance of the G2 checkpoint response are consistent with those of a previous study by Hickman et al. showing that HPV E7 abrogates p53-induced growth arrest and inhibition of Cdk activities (21).
An obvious question from the results presented is does the E2F family of transcription factors play a direct role in regulation of the cyclin B1 promoter? The promoters of Cdc2 and cyclin A have been shown to be regulated by E2F transcription factors directly (8, 9). Previous studies have demonstrated that cyclin B1 expression is dependent on the kinase activity of cyclin A-Cdk2 (35) and that deregulated expression of Cdk2 abrogates IR-induced G2 arrest (56). One possibility is that E2F regulates cyclin B1 transcription indirectly by affecting the expression of cyclin A. Alternatively, E2F transcription factors may regulate cyclin B1 transcription through direct promoter binding. In our preliminary analyses of the cyclin B1 promoter, we have located a putative E2F binding element proximal to the transcriptional start site (P. M. Flatt and J. A. Pietenpol, unpublished data).
Several cell model systems were used in this study to show that both the ectopic expression of p53 and the activation of endogenous p53 were sufficient to sustain a G2 arrest after stress. Our combined results are in contrast with the report of Passalaris et al. indicating that p53 does not affect the duration of G2 arrest in response to DNA damage (39). In the previous study, a decrease in cyclin B1 and Cdc2 protein levels in ADR-treated normal fibroblasts was observed; however, the change in protein levels did not affect the length of G2 arrest (39). Normal human fibroblasts were used as a model system in the previous study, whereas all of the cell types used in our study were of epithelial tumor origin. We have previously shown that primary cultures of normal human keratinocytes (epithelial) and fibroblasts (mesenchymal) have marked differences in cell cycle checkpoint response, duration of growth arrest, and cell fate after exposure to genotoxic agents (13). Thus, the contribution of p53 at the G2 checkpoint may be cell type specific. In addition, Passalaris et al. used low-passage-number cultures of normal fibroblasts for their study and noted that increased passage number resulted in the inability of E6-containing cells to maintain a G2 arrest compared with fibroblasts that retained an intact p53 signaling pathway (39). In fact, Kaufmann et al. demonstrated that E6 expression in normal human fibroblasts correlated with inactivation of the G2 checkpoint and acquisition of chromosomal abnormalities (28).
A different mechanism by which p53 regulates the duration of the G2 checkpoint is thought to be dependent on the transactivation of another p53 downstream target gene, 14-3-3ς (20). Similar to the results with HCT116 p53−/− and HCT116 p21−/− cells, HCT116 cells null for 14-3-3ς prematurely exit G2 and undergo mitotic catastrophe after genotoxic stress (7). The p53-dependent transactivation of the 14-3-3ς gene product has recently been shown to play an integral role in the cytoplasmic localization of the cyclin B1-Cdc2 complex after genotoxic stress (7). 14-3-3ς can form a complex with Cdc2 and wee1, and the complex of proteins can be identified in the cytoplasms of cells during G2 arrest (7). However, as stated above, the contribution of p53 to the G2 checkpoint response may be cell type specific since 14-3-3ς has not been detected in rapidly growing or irradiated human diploid fibroblasts (20). Thus, inhibition of cyclin B1-Cdc2 activity in response to cellular stress involves redundant biochemical pathways that work in concert to regulate phosphorylation, subcellular localization, activity, and expression of cyclin B1 and Cdc2 proteins. The interplay of multiple pathways is likely required to achieve the maximal activation and maintenance of G2 cell cycle arrest in response to cellular stress in different cell types.
A hallmark of human tumors is deficiency of checkpoint function. Several preclinical studies have correlated loss of specific cell cycle regulatory gene products with enhanced vulnerability to anticancer agents (6, 19, 51, 55). There is growing evidence that ablation of G2 arrest in tumor cell lines alters sensitivity to several anticancer agents (4, 42, 44, 45, 58, 61). As we expand our understanding of how cell cycle regulatory pathways interplay to constitute a checkpoint, our ability to design rational therapies that exploit the molecular defects in tumor cells will increase.
ACKNOWLEDGMENTS
Caroline D. Scatena and Luo Jia Tang contributed equally to this work.
We thank K. Cho for the RKO-NEO and RKO-E7 cells lines, B. Vogelstein for the isogenic set of HCT116 cells, J. Lee for the cyclin B1 promoter-CAT construct, the staff of the S. Hiebert laboratory for assistance with the EMSA, and E. Nishida and H. Piwnica-Worms for cyclin B1 and Cdc2 cDNAs, respectively. We thank the members of the Pietenpol laboratory for critical reading of the manuscript.
This work was supported by a Burroughs Wellcome New Investigator in Toxicology award (J.A.P.), NIH grant CA70856 (J.A.P.), NIEHS institutional training grant ES07028 (C.D.S.), NIH grant CA68485 (core services), and NIEHS grants ES07028 and ES00267 (core services).
REFERENCES
- 1.Agarwal M L, Agarwal A, Taylor W R, Stark G R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. The p53 network. J Biol Chem. 1999;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Azzam E I, De Toledo S M, Pykett M J, Nagasawa H, Little J B. CDC2 is down-regulated by ionizing radiation in a p53-dependent manner. Cell Growth Differ. 1997;8:1161–1169. [PubMed] [Google Scholar]
- 4.Blume E. Researchers gain footholds in the cell cycle. J Natl Cancer Inst. 1995;87:1504–1505. doi: 10.1093/jnci/87.20.1504. [DOI] [PubMed] [Google Scholar]
- 5.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 6.Bunz F, Hwang P M, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler K W, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Investig. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan T A, Hermeking H, Lengauer C, Kinzler K W, Vogelstein B. 14-3-3 sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 8.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1999;15:5846–5847. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Toledo S, Azzam E, Keng P, Laffrenier S, Little J. Regulation by ionizing radiation of CDC2, cyclin A, cyclin B, thymidine kinase, topoisomerase IIa, and RAD51 expression in normal human diploid fibroblasts is dependent on p53/p21. Cell Growth Differ. 1998;9:887–896. [PubMed] [Google Scholar]
- 11.Demers G W, Foster S A, Halbert C L, Galloway D A. Growth arrest by induction of p53 in DNA damaged keratinocytes is bypassed by human papillomavirus 16 E7. Proc Natl Acad Sci USA. 1994;91:4382–4386. doi: 10.1073/pnas.91.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 13.Flatt P M, Price J O, Shaw A, Pietenpol J A. Differential cell cycle checkpoint response in normal human keratinocytes and fibroblasts. Cell Growth Differ. 1998;9:535–543. [PubMed] [Google Scholar]
- 14.Furukawa Y, Terui Y, Sakoe K, Ohta M, Saito M. The role of cellular transcription factor E2F in the regulation of cdc2 mRNA expression and cell cycle control of human hematopoietic cells. J Biol Chem. 1994;269:26249–26258. [PubMed] [Google Scholar]
- 15.Graeber T G, Peterson J F, Tsai M, Monica K, Fornance A J, Jr, Giaccia A J. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 17.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 18.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 19.Hawkins D S, Demers G W, Galloway D A. Inactivation of p53 enhances sensitivity to multiple chemotherapeutic agents. Cancer Res. 1996;56:892–898. [PubMed] [Google Scholar]
- 20.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1998;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 21.Hickman E S, Bates S, Vousden K H. Perturbation of the p53 response by human papillomavirus type 16 E7. J Virol. 1997;71:3710–3718. doi: 10.1128/jvi.71.5.3710-3718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiebert S W, Chellappan S, Horowitz J M, Nevins J R. The interaction of RB with E2F coincides with inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 23.Hiebert S W, Packham G, Strom D K, Haffner R, Oren M, Zambetti G, Cleveland J L. E2F-1:DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol Cell Biol. 1995;15:6864–6874. doi: 10.1128/mcb.15.12.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain S P, Harris C C. p53 mutation spectrum and load: the generation of hypotheses linking the exposure of endogenous or exogenous carcinogens to human cancer. Mutat Res. 1999;428:23–32. doi: 10.1016/s1383-5742(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 25.Innocente S A, Abrahamson J L A, Cogswell J P, Lee J M. p53 regulates a G2 checkpoint through cyclin B1. Proc Natl Acad Sci USA. 1999;96:2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karantza V, Maroo A, Fay D, Sedivy J M. Overproduction of Rb protein after the G1/S boundary causes G2 arrest. Mol Cell Biol. 1993;13:6640–6652. doi: 10.1128/mcb.13.11.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 28.Kaufmann W K, Schwartz J L, Hurt J C, Byrd L L, Galloway D A, Levedakou E, Paules R S. Inactivation of G2 checkpoint function and chromosomal destabilization are linked in human fibroblasts expressing human papillomavirus type 16 E6. Cell Growth Differ. 1997;8:1105–1114. [PubMed] [Google Scholar]
- 29.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 30.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 31.Li C Y, Nagasawa H, Dahlberg W K, Little J B. Diminished capacity for p53 in mediating a radiation-induced G1 arrest in established human tumor cell lines. Oncogene. 1995;11:1885–1892. [PubMed] [Google Scholar]
- 32.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 33.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 34.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 35.Moro A, Zerfass K, Joswig S, Jansen-Duerr P. Effect of cyclins and Cdks on the cyclin B1 promoter activation. Biochem Mol Biol Int. 1997;41:919–924. doi: 10.1080/15216549700201971. [DOI] [PubMed] [Google Scholar]
- 36.Mueller P R, Coleman T R, Kumagai A, Dunphy A. Myt1: a membrane-associated inhibitory kinase that phosphorylates cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–93. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 37.Park M, Chae H D, Yun J, Jung M, Kim Y S, Kim S H, Han M H, Shin D Y. Constitutive activation of cyclin B1-associated cdc2 kinase overrides p53-mediated G2-M arrest. Cancer Res. 2000;60:542–545. [PubMed] [Google Scholar]
- 38.Parker L L, Piwnica-Worms H. Inactivation of p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 39.Passalaris T M, Benanti J A, Gewin L, Kiyono T, Galloway D A. The G2 checkpoint is maintained by redundant pathways. Mol Cell Biol. 1999;19:5872–5881. doi: 10.1128/mcb.19.9.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 41.Pietenpol J A, Tokino T, El-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell S N, DeFrank J S, Connell P, Eogan M, Preffer F, Dombkowski D, Tang W, Friend S. Differential sensitivity of p53(−) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995;55:1643–1648. [PubMed] [Google Scholar]
- 43.Rigberg D A, Kim F S, Sebastian J L, Kazanjian K K, McFadden D W. Hypophosphorylated retinoblastoma protein is associated with G2 arrest in esophageal squamous cell carcinoma. J Surg Res. 1999;84:101–105. doi: 10.1006/jsre.1999.5617. [DOI] [PubMed] [Google Scholar]
- 44.Roberge M, Berlinck R G S, Xu L, Anderson H J, Lim L Y, Curman D, Stringer C M, Friend S H, Davies P, Vincent I, Haggarty S J, Kelly M T, Britton R, Piers E, Andersen R J. High-throughput assay for G2 checkpoint inhibitors and identification of the structurally novel compound isogranulatimide. Cancer Res. 1998;58:5701–5706. [PubMed] [Google Scholar]
- 45.Russell K J, Wiens L W, Demers W, Galloway D A, Plon S E, Groudine M. Abrogation of the G2 checkpoint results in differential radiosensitization of G1 checkpoint-deficient and G1-checkpoint competent cells. Cancer Res. 1995;55:1639–1642. [PubMed] [Google Scholar]
- 46.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwarz J K, Bassing C H, Kovesdi I, Datto M B, Blazing M, George S, Wang X-F, Nevins J R. Expression of the E2F1 transcription factor overcomes type β transforming growth factor-mediated growth suppression. Proc Natl Acad Sci USA. 1995;92:483–487. doi: 10.1073/pnas.92.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellers W R, Kaelin W G. pRB as a modulator of transcription. Biochim Biophys Acta. 1996;1288:M1–M5. doi: 10.1016/0304-419x(96)00014-5. [DOI] [PubMed] [Google Scholar]
- 49.Slebos R J, Lee M H, Plunkett B S, Kessis T D, Williams B O, Jacks T, Hedrick L, Kastan M B, Cho K R. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1994;91:5320–5324. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart N, Hicks G G, Paraskevas F, Mowat M. Evidence for a second cell cycle block at G2/M by p53. Oncogene. 1995;10:109–115. [PubMed] [Google Scholar]
- 51.Stewart Z A, Mays D, Pietenpol J A. Defective G1-S cell cycle checkpoint function sensitizes cells to microtubule inhibitor-induced apoptosis. Cancer Res. 1999;59:3831–3837. [PubMed] [Google Scholar]
- 52.Szak S T, Pietenpol J A. High affinity insertion/deletion lesion binding by p53—evidence for a role of the p53 central domain. J Biol Chem. 1999;274:3904–3909. doi: 10.1074/jbc.274.6.3904. [DOI] [PubMed] [Google Scholar]
- 53.Taylor W R, DePrimo S E, Agarwal A, Agarwal M L, Schönthal A H, Katula K S, Stark G R. Mechanisms of G2 arrest in response to overexpression of p53. Mol Biol Cell. 1999;10:3607–3622. doi: 10.1091/mbc.10.11.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 55.Waldman T, Zhang Y, Dillehay L, Yu J, Kinzler K, Vogelstein B, Williams J. Cell-cycle arrest versus cell death in cancer therapy. Nat Med. 1997;3:1034–1036. doi: 10.1038/nm0997-1034. [DOI] [PubMed] [Google Scholar]
- 56.Walker D H, Adami G R, Dold K M, Babiss L E. Misregulated expression of the cyclin dependent kinase 2 protein in human fibroblasts is accompanied by the inability to maintain a G2 arrest following DNA damage. Cell Growth Differ. 1995;6:1053–1061. [PubMed] [Google Scholar]
- 57.Wang J Y. Retinoblastoma protein in growth suppression and death protection. Curr Opin Genet Dev. 1997;7:39–45. doi: 10.1016/s0959-437x(97)80107-4. [DOI] [PubMed] [Google Scholar]
- 58.Wang Q, Fan S, Eastman A, Worland P J, Sausville E A, O'Connor P M. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88:956–965. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 59.White A E, Livanos E M, Tlsty T D. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 60.Winters Z E, Ongkeko W M, Harris A L, Norbury C J. p53 regulates Cdc2 independently of inhibitory phosphorylation to reinforce radiation-induced G2 arrest in human cells. Oncogene. 1998;17:673–684. doi: 10.1038/sj.onc.1201991. [DOI] [PubMed] [Google Scholar]
- 61.Yao S L, Akhtar A J, McKenna K A, Bedi G C, Sidransky D, Mabry M, Ravi R, Collector M I, Jones R J, Sharkis S J, Fuchs E J, Bedi A. Selective radiosensitization of p53-deficient cells by caffeine-mediated activation of p34cdc2 kinase. Nat Med. 1996;2:1140–1143. doi: 10.1038/nm1096-1140. [DOI] [PubMed] [Google Scholar]
- 62.Yen A, Sturgill R. Hypophosphorylation of the Rb protein in S and G2 as well as G1 during growth arrest. Exp Cell Res. 1998;241:324–331. doi: 10.1006/excr.1998.4007. [DOI] [PubMed] [Google Scholar]