Abstract
Background
The global prevalence of nonalcoholic fatty liver disease (NAFLD) is increasing. The pathogenesis of NAFLD is multifaceted, and the underlying mechanisms are elusive. We conducted data mining analysis to gain a better insight into the disease and to identify the hub genes associated with the progression of NAFLD.
Methods
The dataset GSE49541, containing the profile of 40 samples representing mild stages of NAFLD and 32 samples representing advanced stages of NAFLD, was acquired from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were identified using the R programming language. The Database for Annotation, Visualization and Integrated Discovery (DAVID) online tool and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database were used to perform the enrichment analysis and construct protein–protein interaction (PPI) networks, respectively. Subsequently, transcription factor networks and key modules were identified. The hub genes were validated in a mice model of high fat diet (HFD)-induced NAFLD and in cultured HepG2 cells by real-time quantitative PCR.
Results
Based on the GSE49541 dataset, 57 DEGs were selected and enriched in chemokine activity and cellular component, including the extracellular region. Twelve transcription factors associated with DEGs were indicated from PPI analysis. Upregulated expression of five hub genes (SOX9, CCL20, CXCL1, CD24, and CHST4), which were identified from the dataset, was also observed in the livers of HFD-induced NAFLD mice and in HepG2 cells exposed to palmitic acid or advanced glycation end products.
Conclusion
The hub genes SOX9, CCL20, CXCL1, CD24, and CHST4 are involved in the aggravation of NAFLD. Our results offer new insights into the underlying mechanism of NAFLD progression.
Keywords: Nonalcoholic fatty liver disease, Fatty liver, Computational biology
Introduction
The incidence of nonalcoholic fatty liver disease (NAFLD) and its advanced subtypes has been rising rapidly, leading to health and economic burden on the patients.1,2 NAFLD is the leading cause of liver diseases globally and is associated with several metabolic disorders, such as type 2 diabetes.3,4 NAFLD includes a series of conditions from early steatosis to nonalcoholic steatohepatitis (NASH), and even hepatic carcinoma.5 However, the exact mechanisms of the development and progression of NAFLD are still not completely elucidated.6
Nowadays, microarray technology is a widely used method in discovery-based biomedical research.7 The pathogenesis of NAFLD involves a myriad of distinct molecular pathways and cellular changes. Several studies have reported the molecular mechanisms of NAFLD pathogenesis in the liver.8, 9, 10 However, the key genes associated with the disease progression and the underlying functional pathways remain obscure, and whether the differentially expressed genes (DEGs) are involved in hepatic lipid metabolism is still unclear.
In the present study, we have integrated the available microarray datasets of human NAFLD liver tissues to perform comprehensive bioinformatic analysis of DEGs. Moreover, we have verified the expression changes of the liver hub genes of high fat diet (HFD)-induced NAFLD mice, as well as in the HepG2 cells exposed to glucolipotoxicity. Our results might elucidate potential biomarkers and targets for the diagnosis and treatment of NAFLD.
Methods
Animal experiments ethics
The animal experiments in this study were approved by the Animal Care and Use Committee of Peking University (No. LA2018316). All ethical principles involved in the care and usage of laboratory animals were carried out.
Microarray data collection
The Gene Expression Omnibus dataset GSE49541, which was contributed by Moylan et al.,11 was downloaded from the National Center for Biotechnology Information website. The dataset contained a total of 72 RNA profiles from liver samples, including 40 belonging to mild NAFLD (fibrosis stage 0–1) and 32 belonging to advanced NAFLD (fibrosis stage 3–4). The dataset was generated using the GPL570 platform (Affymetrix Human Genome U133A Array).
Data preprocessing and DEG screening
The R language (Affy package, version 1.64.0) was used to manipulate the raw data. Based on annotation files, the probe IDs were converted into gene symbols, following background correction, standardization, and expression value calculation processes, as previously described.12 Subsequently, DEG screening was performed using the R language Limma package (version 3.42.2). The statistically significant screening criteria for the identification of DEGs were defined at |log 2 (fold change)| > 1 and P < 0.05.
Enrichment analysis of DEGs
To evaluate the functions of a cluster of DEGs, the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (https://david.ncifcrf.gov/) was used to perform the Gene Ontology (GO) analysis. Moreover, the Kyoto Encyclopedia of Genes and Genomes (KEGG) was used for the pathway analysis of DEGs. The enrichment analysis of DEGs was regarded as statistically significant at P < 0.05. The GO enrichment package of the R language, gplot, was used to list all the enriched pathways according to the P value.
Protein–protein interaction and module analyses
To investigate the connections among the proteins encoded by the identified DEGs, the Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/) was used to establish the protein–protein interaction (PPI) network, with a confidence score >0.4 as the threshold. Cytoscape software (version 3.8.2x; The Cytoscape Consortium, New York City, NY, USA) was used to visualize the PPI network. Molecular Complex Detection (MCODE) algorithm was used to identify the key modules of the PPI network within the set criteria of significance, which were defined at degree = 5, node score = 0.2, k-core = 2, and max depth = 100. According to this algorithm of Cytoscape, the obtained modules were ranked and scored. The top 2 modules with the highest score were considered to be significant. To identify hub genes, the top 16 genes were ranked. The genes with common diagrams ≥10 in every topological algorithm were considered hub genes.
Transcription factor analysis
The expression of genes is regulated by transcription factors. To predict and visualize the key transcription factors of the PPI network, the iRegulon plugin of Cytoscape (version 3.8.2x) was used, as previously described.13 Normalized enrichment score (NES) > 5 was defined to select transcription factors. According to the NES score, the top three modules were ranked and listed.
Establishment of the mouse NAFLD model
All mice were purchased from the Vital River Animal Center (Beijing, China). After one-week of acclimatization, twelve 8-week-old male C57BL/6N mice were divided into three groups. Two groups of mice were fed an HFD (dietary fat content of 60%) for 18 weeks (n = 6) and 24 weeks (n = 3), which represented the mild and advanced NAFLD models, respectively. One additional group of mice (n = 3) fed a normal diet (dietary fat content of 4%) served as the control. All the mice were reared in individually ventilated cages located in the same room. Food and water were accessed ad libitum. Before sacrificing the animals, magnetic resonance imaging (MRI; Siemens Prisma, Munich, Germany) was used to measure the body fat content.
Oil red O staining
The liver tissues were fixed overnight with 10% (v/v) neutral-buffered formalin at 4 °C and embedded in optimal cutting temperature compound. The 5 μm-thick sections were stained with oil red O solution (Servicebio, Wuhan, China) to assess the accumulation of hepatic fat. Images were obtained using a panoramic section scanner (3Dhistech, Pannoramic, Budapest, Hungary).
Establishment of liver cell glucolipotoxicity models in vitro
Human liver cell line, HepG2, (validated for gene expression and checked for mycoplasma contamination before use) were kindly gifted from the Medical Research Center, Peking University Third Hospital (Beijing, China).14 Palmitic acid (PA; Sigma, St. Louis-Aldrich, MO, USA) was used to establish the lipotoxicity-induced hepatic insulin resistance model, and advanced glycation end products (AGEs; Abcam, Cambridge, UK) were used to generate the glucotoxicity-induced hepatic damage model in vitro. HepG2 cells were incubated in Dulbecco's Modified Eagle's Medium (Gibco, Carlsbad, CA, USA) with 10% (v/v) fetal bovine serum (Gibco). PA (256 mg) was dissolved in 5 mL anhydrous ethanol, and then titrated with 5 mL sodium hydroxide (0.l mol/L). A total of 5 mL PA solution was slowly dripped into 95 mL 10% bovine serum albumin to obtain a complex with a concentration of 5 mmol/L as previously described.15 Subsequently, the HepG2 cells were incubated with PA (125, 250, 500, and 1000 nmol/L), or with AGEs (1, 10, and 100 μg/mL) for 24 h. Cells were then collected for RNA extraction.
Real-time quantitative PCR
RNA of liver tissues or HepG2 cells was extracted with Trizol (Thermo Fisher Scientific, Waltham, MA, USA) and reverse transcribed to cDNA using a Revert Aid First Strand cDNA Synthesis kit (Fermentas, Vilnius, Lithuania). The cDNA was subjected to quantitative analysis using the SYBR Green supermix (Bio-Rad Laboratories, Hercules, CA, USA) in a real-time quantitative PCR detection system (Bio-Rad Laboratories). The primer sequences synthesized by the Beijing AuGCT DNA-SYN Biotechnology Company (Beijing, China) are summarized in Supplementary Table S1. The housekeeping gene, GAPDH, was used to normalize the expression level of each gene.
Statistical analysis
All in vivo and in vitro studies were performed as three independent experiments. The experimental data are presented as the means ± standard deviation (SD). Statistical analysis was carried out using one-way ANOVA followed by the post-hoc Tukey–Kramer test. The statistical significance was defined at P < 0.05. All the analyses were performed using the Statistical Product and Service Solutions (SPSS) 22.0 software (IBM SPSS Inc, Chicago, IL, USA).
Results
Data preprocessing and DEG screening
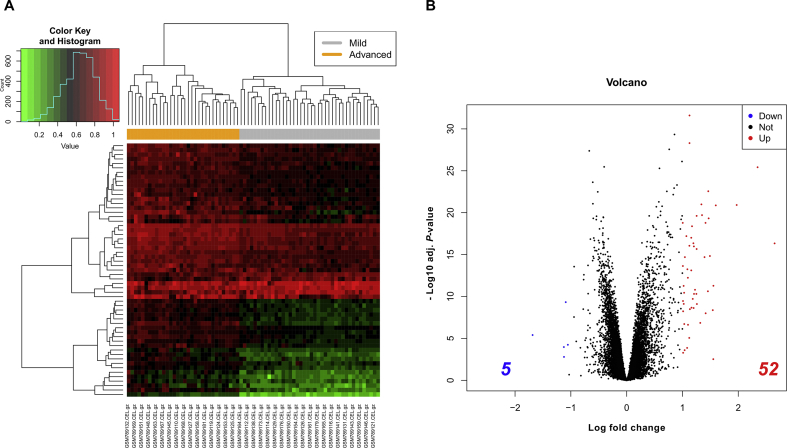
The dataset contained the microarray data of 40 patients with mild NAFLD (fibrosis stage 0–1) and 32 patients with advanced NAFLD (fibrosis stage 3–4). To identify the hub DEGs precisely, statistical significance was defined at |log 2 (fold change)| > 1 and P < 0.05. A total of 57 DEGs, including 52 upregulated DEGs and 5 downregulated DEGs, were selected (Supplementary Table S2), and displayed in form of a heat map and a volcano map (Fig. 1). The top 5 upregulated DEGs were EPCAM, STMN2, CTHRC1, EFEMP1 and CD24. The five downregulated DEGs were CYP2C19, DHRS2, MT1M, FITM1, and GNMT (Supplementary Table S2).
Fig. 1.
Heat map (A) and volcano map (B) of the identified differentially expressed genes (DEGs) between mild (n = 40) and advanced (n = 32) NAFLD livers based on the GSE49541 dataset.
KEGG pathway and GO enrichment analyses of DEGs
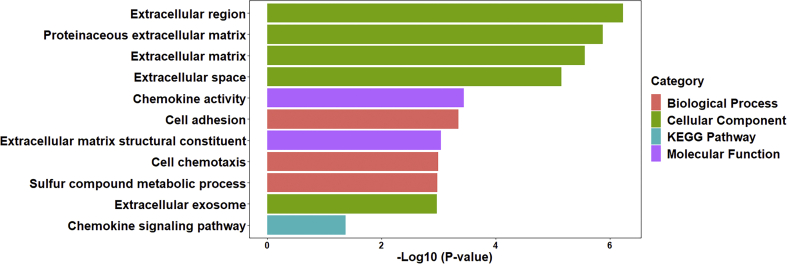
To determine the biological functions of the identified DEGs, enrichment analysis was performed using DAVID. As shown in Fig. 2, the upregulated DEGs were enriched in the extracellular region, proteinaceous extracellular matrix, extracellular matrix, extracellular space, and extracellular exosome in the cellular component GO term. In molecular function class, the DEGs were associated with chemokine activity and extracellular matrix structural constituent. In the biological process class, the DEGs were significantly associated with cell adhesion, cell chemotaxis, and sulfur compound metabolic process. In the KEGG pathway enrichment analysis, the DEGs were solely enriched in the chemokine signaling pathway (Fig. 2).
Fig. 2.
Top 11 pathways and biological functions enriched in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis related to DEGs.
PPI network analysis of DEGs
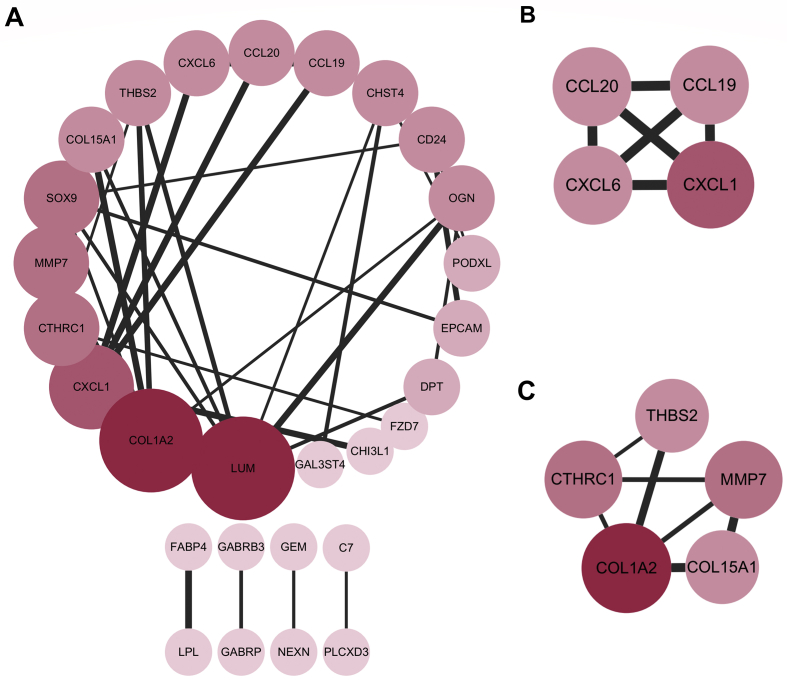
The identified DEGs were introduced into the online database, STRING. Subsequently, Cytoscape was used for network visualization analysis, and the isolated genes that showed no interactions were removed. As shown in Fig. 3A, there were 28 nodes and 36 edges in the PPI network. The MCODE plugin of Cytoscape software was further used to identify the densely connected significant modules that met the cutoff criteria. According to their score, two significant modules were identified from the PPI network. There were 4 nodes and 6 edges in module 1 (score: 4) (Fig. 3B), and 5 nodes and 7 edges in module 2 (score: 3.5) (Fig. 3C). The plugin CytoHubba was used to parse the PPI network. According to a network measures, including the degree, average shortest path length, eccentricity, betweenness centrality, radiality, neighborhood connectivity, stress, topological coefficient, closeness centrality, clustering coefficient, and the number of directed edges, the top 16 genes were regarded as important nodes in each topological analysis method, and the hub genes were selected with the frequency of occurrence ≥10 (Table 1). Based on the analysis of 11 topological algorithms, SOX9, CCL20, CXCL1, CD24, and CHST4 were considered as the hub genes (Table 1), which were used for the further validation studies.
Fig. 3.
Protein–protein interaction (PPI) and module analyses. (A) PPI network and module analyses of DEGs in GSE49541 dataset. (B–C) Significant modules, module 1 (B) and module 2 (C), selected from PPI network analysis. The color and size of node are relative to its degree (darker the color and larger the size, the greater is the degree). The strength of the confidence score is symbolized by the thickness of the line (the thicker the line, the higher is the confidence score).
Table 1.
Hub genes analyzed by different topological algorithms in the protein−protein interaction network.
| Different topological algorithms | Top genes |
|---|---|
| Degree | LUM, COL1A2, CXCL1, CTHRC1, SOX9, MMP7, CXCL6, CCL20, CCL19, CD24, CHST4, OGN, THBS2, COL15A1, PODXL, EPCAM |
| Average shortest path length | GAL3ST4, CHI3L1, CXCL6, CCL20, CCL19, PODXL, FZD7, EPCAM, CD24, DPT, CHST4, CXCL1, OGN, THBS2, CTHRC1, SOX9 |
| Eccentricity | CXCL6, CCL20, CCL19, PODXL, GAL3ST4, CHI3L1, CXCL1, CD24, CHST4, EPCAM, DPT, FZD7, LUM, CTHRC1, SOX9, MMP7 |
| Betweenness centrality | MMP7, COL1A2, CXCL1, LUM, SOX9, CHST4, CTHRC1, COL15A1, CD24, THBS2, OGN, PODXL, GAL3ST4, CHI3L1, CXCL6, CCL20 |
| Radiality | GAL3ST4, CHI3L1, CXCL6, CCL20, CCL19, PODXL, FZD7, EPCAM, DPT, CD24, CHST4, CXCL1, OGN, THBS2, CTHRC1, SOX9 |
| Neighborhood connectivity | COL15A1, THBS2, OGN, DPT, CHI3L1, MMP7, SOX9, COL1A2, FZD7, CTHRC1, CXCL6, CCL20, CCL19, LUM, EPCAM, CHST4 |
| Stress | MMP7, COL1A2, LUM, CXCL1, CHST4, SOX9, CTHRC1, COL15A1, CD24, THBS2, OGN, PODXL, DPT, CHI3L1, FZD7, CXCL6 |
| Topological coefficient | CXCL6, CCL20, CCL19, DPT, EPCAM, COL15A1, THBS2, OGN, PODXL, CD24, CTHRC1, CXCL1, SOX9, MMP7, CHST4, COL1A2 |
| Closeness centrality | GAL3ST4, CHI3L1, CXCL6, CCL20, CCL19, PODXL, FZD7, EPCAM, CD24, DPT, CHST4, CXCL1, OGN, THBS2, CTHRC1, SOX9 |
| Clustering coefficient | CXCL6, CCL20, CCL19, EPCAM, DPT, OGN, THBS2, COL15A1, CD24, CTHRC1, SOX9, MMP7, COL1A2, CXCL1, LUM, GAL3ST4 |
| Number of directed edges | COL1A2, LUM, CXCL1, CTHRC1, SOX9, MMP7, CXCL6, CCL20, CCL19, OGN, THBS2, COL15A1, CD24, CHST4, EPCAM, DPT |
| Common DEGs (≥10 diagrams) | CCL20, CXCL1, CD24, CHST4, SOX9 |
Modules of key transcription factors
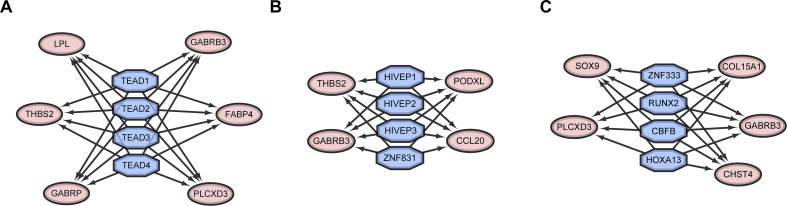
Transcription factors regulate gene expression and function by binding to a specific DNA sequence. Here, the iRegulon plugin was used to predict the transcription factors and the regulatory network of their target genes. All predicted transcription factor modules with NES >5 are listed in Supplementary Table S3. According to the NES score, the top 3 transcription factor modules are displayed in Fig. 4. In module 1, it was predicted that TEAD1, TEAD2, TEAD3, and TEAD4 transcription factors would regulate LPL, THBS2, GABRP, PLCXD3, FABP4, and GABRB3 (Fig. 4A). In module 2, the transcription factors HIVEP1, HIVEP2, HIVEP3, and ZNF831 would regulate THBS2, GABRB3, CCL20, and PODXL (Fig. 4B). In module 3, it was predicted that ZNF333, RUNX2, CBFB, and HOXA13 would regulate SOX9, PLCXD3, CHST4, GABRB3, and COL15A1 (Fig. 4C).
Fig. 4.
Transcription factor target networks in the top 3 modules using the iRegulon plugin. Blue octagon nodes represent the predicted transcription factors. Pink oval nodes represent the transcription factor-regulated genes.
Validation of key genes associated with NAFLD in vivo and in vitro
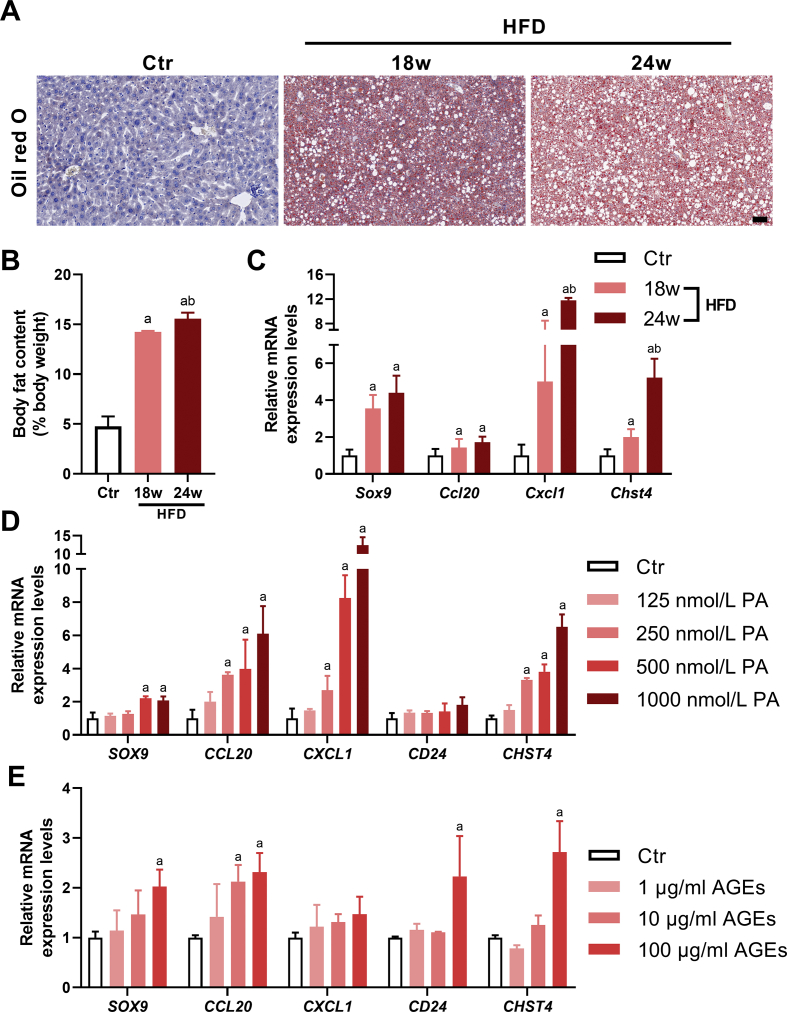
We screened the possible hub genes of NAFLD based on highly correlated topological algorithms from the PPI networks. The 5 hub genes, SOX9, CCL20, CXCL1, CD24, and CHST4, were identified (Table 1). To confirm the role of these hub genes in the liver during different stages of NAFLD, mice were fed an HFD with different periods. First, we detected deposition of adipose in the liver of HFD-fed mice. In addition, all the HFD-fed mice developed some form of hepatic steatosis. The oil red O staining showed more fat droplets and hepatocyte ballooning in the liver of the HFD-fed mice compared with the control group. These changes were more severe in mice fed HFD for 24 weeks than in mice fed HFD for 18 weeks (Fig. 5A). The percentage of total adipose tissue (as detected by the MRI scan) to body weight was higher in the 18-week HFD-fed mice than in the control mice [(14.20 ± 0.11) % vs. (4.72 ± 0.99) %, t = 15.95, P < 0.01], and much higher in the 24-week HFD-fed mice than in the 18-week HFD-fed mice [(15.60 ± 0.60) % vs. (14.20 ± 0.11) %, t = 3.791, P < 0.05)] (Fig. 5B). These results indicate that HFD could successfully induce adipose accumulation and lead to the development and progression of NAFLD in mice. Next, we determined the expression of the key genes in the liver of the HFD-induced NAFLD mice. The mRNA levels of Sox 9, Ccl20, Cxcl1, and Chst4 in the liver were higher in mice fed an 18-week HFD than in the control mice, and their levels were further increased after 24-week HFD (Fig. 5C).
Fig. 5.
Validation of the potential key genes in the livers of NAFLD mice and in cultured HepG2 cells exposed to glucolipotoxicity. C57BL/6N mice were fed a high fat diet (HFD) for 18 (n = 6) and 24 (n = 3) weeks. Age-matched C57BL/6N mice fed a normal diet (n = 3) were used as the control. (A) Oil red O staining of liver tissues. Scale bar = 50 μm. (B) The percentage of total adipose tissue (as detected by magnetic resonance imaging scan) to body weight. (C) Relative mRNA levels of hub genes in mouse liver tissues detected by real-time quantitative PCR (qPCR). (D) Relative mRNA levels of hub genes determined by qPCR in HepG2 cells cultured with palmitic acid (PA) or vehicle for 24 h (n = 3). (E) Relative mRNA levels of hub genes detected by qPCR in HepG2 cells cultured with advanced glycation end products (AGEs) or vehicle for 24 h (n = 3). Data are expressed as the means ± standard deviation. Statistical analysis was conducted using one-way ANOVA followed by the post-hoc Tukey–Kramer test. aP < 0.05 (vs. control). bP < 0.05 (vs.18-week HFD exposure).
Given that NAFLD is strongly associated with an abnormal metabolism of lipids and glucose, we further explored the expression levels of the hub genes in cultured liver cell line, HepG2, exposed to lipotoxic or glucotoxic conditions, induced by the application of PA or AGEs, respectively. The mRNA levels of SOX9, CCL20, CXCL1, and CHST4 were upregulated by high concentrations (500 and 1000 nmol/L) of PA (Fig. 5D). Furthermore, the mRNA levels of SOX9, CCL20, CD24 and CHST4 were upregulated by 100 μg/mL of AGEs (Fig. 5E). These results indicate that the suggested hub genes might be highly relevant to the development of NAFLD. Interestingly, CD24 and CCL20, the key genes involved in the progression of NAFLD, were also upregulated in the livers of patients with type 2 diabetes, when the GSE15653 database (including 5 normal liver tissues and 9 liver samples from diabetic patients) was used for validation analysis (Supplementary Fig. S1).
Discussion
The prevalence of NAFLD, one of the most common chronic liver diseases, is increasing at an alarming pace globally.16 However, the pathogenesis of NAFLD is not completely understood.17 It has been suggested that NAFLD is strongly correlated with genetic components.18 In this study, we downloaded the GSE49541 dataset to obtain gene expression data of the advanced NAFLD liver tissues and compared them with mild NAFLD liver tissues. A total of 57 DEGs, 52 upregulated genes and 5 downregulated genes, were selected. Functional and enrichment analyses indicated that the DEGs were mainly enriched in the extracellular region, chemokine activity, and cell adhesion. KEGG pathway analysis demonstrated that the DEGs were only enriched in the chemokine signaling pathway. We identified SOX9, CCL20, CXCL1, CD24, and CHST4 as hub genes based on the PPI network analysis. Furthermore, we validated the upregulated expression of these hub genes in the livers of HFD-induced NAFLD mice and in cultured HepG2 cells exposed to glucolipotoxicity.
A total of 57 DEGs were chosen in this study. As the expression of a single gene is not sufficient to explain the entire biological process, and the changes in biological phenotype, it is necessary to study the interaction of a series of genes and proteins. Enrichment analysis is fundamental for biological interpretation of experimental “omics” data.19 Our enrichment analysis revealed that DEGs were significantly enriched in extracellular process and cell adhesion in the cellular component and biological process classes, respectively. Extracellular processes such as neutrophil extracellular traps,20 have been reported to participate in the inflammation associated with NASH.21 Some adhesion molecules promote leukocyte recruitment in the liver and exacerbate the NAFLD.22 These results suggest that the extracellular region is the main pathological site for the aggravation of fatty liver phenotype and that cell adhesion, especially the adhesion of inflammatory factors, is the main biological process of the disease.
Next, we screened the hub genes associated with the progression of NAFLD. Through the PPI network analysis, SOX9, CCL20, CXCL1, CD24, and CHST4 were selected as the most common genes in 11 topological algorithms. SRY-box transcription factor 9 (SOX9) is mainly expressed in bile duct cells under physiological conditions.23 During the process of chronic liver injury, SOX9-positive cells act as facultative liver stem cells and are involved in liver regeneration.24 SOX9 is also highly expressed in hepatocellular carcinoma tissues, which is related to poor prognosis in the patients.25,26 In the present study, SOX9 was upregulated in the livers of HFD-induced NAFLD mice and in HepG2 cells exposed to PA or AGEs. These results suggest that SOX9 is involved in metabolic liver diseases and may serve as a potential biomarker to diagnose and assess the severity of NAFLD.
Liver steatosis is associated with a presence of many chemokines and active inflammatory cells, which is a sign of chronic inflammation.27 Our study showed that the DEGs were enriched in the chemokine pathway and activity in both the KEGG pathway and molecular function analyses. Moreover, C–C motif chemokine ligand 20 (CCL20) and C-X-C motif chemokine ligand 1 (CXCL1) were predicted as the hub genes from the PPI network analysis. Furthermore, we found that the expression levels of Ccl20 and Cxcl1 were higher in the livers of HFD-induced NAFLD mice than in the control mice, and the mRNA levels of CCL20 and CXCL1 were upregulated by PA in HepG2 cells. Many studies in rodent models indicate that chemokines play a crucial role in NAFLD.28,29 The levels of CCL20 were increased in the animal models of liver injury, especially with the acute-on-chronic condition.30 Results from a network meta-analysis showed that the concentrations of chemokines, including CCL20, in the NASH group were higher than those in the control group.29 Additionally, the CCL20 gene is one of the most upregulated transcripts observed in fibrosis associated with NAFLD, in comparison to normal conditions, which was further validated in a replication group.31 These results suggest that the CCL20 chemokine is a potential therapeutic target, and can be regarded as one of the most important chemokines involved in the mechanisms underlying NAFLD.
The cluster of differentiation 24 (CD24) and carbohydrate sulfotransferase 4 (CHST4) were the two other DEGs that we identified and validated in the livers of HFD-induced NAFLD mice and in HepG2 cells exposed to PA or AGEs. A previous study imported three GEO datasets of NAFLD samples (GSE66676, GSE49541, and GSE834521), and found that CD24 was the only gene co-expressed in all three datasets.10 In a cross-sectional study, liver tissue-transcriptome differences were evaluated in a subset of 25 mild-NAFLD and 20 NASH biopsies. Five identified DEGs, including CD24, were positively associated with disease severity and were found to be important classifiers of mild NAFLD and severe NAFLD.32 CD24-positive cells isolated from hepatocellular carcinoma cell lines exhibited stemness properties, such as self-renewal, chemotherapy resistance, metastasis, and tumorigenicity.33 These results indicate that CD24 may play a role in hepatocyte injury and promote regeneration during the development and progression of NAFLD. Another hub gene, CHST4, encodes sulfotransferase, an enzyme which utilizes 3′-phospho-5′-adenylyl sulfate to catalyze the transfer of sulfate, ultimately serving as ligand for L-selectins (SELL, Selectin L, a lymphocyte homing receptor). SELL ligands are highly expressed in endothelial cells and play a central role in lymphocyte homing at sites of inflammation.34 Therefore, our findings suggest that CHST4 may participate in the inflammation associated with NAFLD. Up till now, the precise functions and the underlying mechanisms of CD24 and CHST4 in NAFLD progression remain unclear.10
All the 12 transcription factors identified in the present study using transcription factor analysis were likely to be implicated in the progression of NAFLD. The transcription factors of transcriptional enhanced associate (TEA) domain DNA-binding family (TEAD1, TEAD2, TEAD3, and TEAD4) regulate gene expression primarily through interaction with transcriptional co-activators with PDZ-motif (TAZ).35 A previous study demonstrated that inhibiting liver TAZ in murine NASH models prevented or even reversed hepatic inflammation, hepatocyte death and hepatic fibrosis, but not liver steatosis.36 Upregulation of Runt-related transcription factor 2 (Runx 2) in activated murine hepatic stellate cells promoted hepatic infiltration of macrophages by increasing the expression of monocyte chemotactic protein 1.37 The involvement of other transcription factors, including HIVEP, ZNF, CBFB, and HOXA13, identified in this study has not been reported in liver diseases. The specific function of these transcription factors in NAFLD, especially in hepatic fibrosis, requires further research.
There are certain limitations in this study. First, the duration of HFD exposure in our animal model may not be long enough to induce severe NAFLD and to successfully compare the different lengths of HFD treatment in mice. Second, the sample size is relatively small. Larger sample sizes obtained from animal studies and prospective clinical cohort studies are warranted to verify the function of these hub genes.
In summary, we used bioinformatics analyses to identify 57 DEGs in mild and advanced NAFLD liver tissues. We identified SOX9, CCL20, CXCL1, CD24, and CHST4 as hub genes, and identified intersecting pathways involved in extracellular space, cell adhesion, and inflammation. Notably, we verified the upregulated expression of these hub genes in the livers of HFD-induced NAFLD mice and in HepG2 cells exposed to PA or AGEs. These hub genes may serve as biomarkers for advanced NAFLD stages and offer new insights into drug discovery. Nevertheless, further studies are required to clarify the detailed function and specific mechanisms of these hub genes in the development and progression of NAFLD.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81830022 and 81970671).
Data supplied
Microarray data is available at NCBI GEO, accession numbers: GSE49541; Raw codes are available at Github (https://github.com/JinFeng-bio/NAFLD).
Conflict of interest
None.
Acknowledgements
We sincerely thank Professor Lixiang Xue (Peking University Third Hospital, Beijing, China) for the kind gifting of the HepG2 cells. We also thank Yangpeng Zhang (Shanghai Jiaotong University, Shanghai, China) for his technical assistance.
Edited by Yi Cui
Footnotes
Peer review under responsibility of Chinese Medical Association.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cdtm.2021.08.002.
Contributor Information
Rui Wei, Email: weirui@bjmu.edu.cn.
Tianpei Hong, Email: tpho66@bjmu.edu.cn.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Younossi Z.M., Henry L., Bush H., Mishra A. Clinical and economic burden of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Clin Liver Dis. 2018;22:1–10. doi: 10.1016/j.cld.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Hardy T., Oakley F., Anstee Q.M., Day C.P. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu Rev Pathol. 2016;11:451–496. doi: 10.1146/annurev-pathol-012615-044224. [DOI] [PubMed] [Google Scholar]
- 3.Kotronen A., Seppala-Lindroos A., Bergholm R., Yki-Jarvinen H. Tissue specificity of insulin resistance in humans: fat in the liver rather than muscle is associated with features of the metabolic syndrome. Diabetologia. 2008;51:130–138. doi: 10.1007/s00125-007-0867-x. [DOI] [PubMed] [Google Scholar]
- 4.Perry R.J., Kim T., Zhang X.M., et al. Reversal of hypertriglyceridemia, fatty liver disease, and insulin resistance by a liver-targeted mitochondrial uncoupler. Cell Metabol. 2013;18:740–748. doi: 10.1016/j.cmet.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Association for the Study of the Liver, European Association for the Study of Diabetes, European Association for the Study of Obesity. EASL-EASD-EASO clinical practice guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388–1402. doi: 10.1007/s00125-016-3902-y. [DOI] [PubMed] [Google Scholar]
- 6.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Govindarajan R., Duraiyan J., Kaliyappan K., Palanisamy M. Microarray and its applications. J Pharm BioAllied Sci. 2012;4:S310–S312. doi: 10.4103/0975-7406.100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pihlajamaki J., Boes T., Kim E.Y., et al. Thyroid hormone-related regulation of gene expression in human fatty liver. J Clin Endocrinol Metab. 2009;94:3521–3529. doi: 10.1210/jc.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchner H., Sinha I., Gao H., et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol Metab. 2016;5:171–183. doi: 10.1016/j.molmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S., Sun C., Hou Y., et al. A comprehensive bioinformatics analysis on multiple Gene Expression Omnibus datasets of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Sci Rep. 2018;8:7630. doi: 10.1038/s41598-018-25658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moylan C.A., Pang H., Dellinger A., et al. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2014;59:471–482. doi: 10.1002/hep.26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu W., Li N., Liao F. Identification of key genes and pathways in pancreatic cancer gene expression profile by integrative analysis. Genes. 2019;10:612. doi: 10.3390/genes10080612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L., Pan Z., Yang X. Key genes and co-expression network analysis in the livers of type 2 diabetes patients. J Diabetes Investig. 2019;10:951–962. doi: 10.1111/jdi.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao S.B., Li K.L., Qiu H., et al. Enhancing chemotherapy sensitivity by targeting PcG via the ATM/p53 pathway. Am J Canc Res. 2017;7:1874–1883. [PMC free article] [PubMed] [Google Scholar]
- 15.Le Y., Wei R., Yang K., et al. Liraglutide ameliorates palmitate-induced oxidative injury in islet microvascular endothelial cells through GLP-1 receptor/PKA and GTPCH1/eNOS signaling pathways. Peptides. 2020;124:170212. doi: 10.1016/j.peptides.2019.170212. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani A., Petracca G., Beatrice G., et al. Non-alcoholic fatty liver disease and risk of incident chronic kidney disease: an updated meta-analysis. Gut. 2020 doi: 10.1136/gutjnl-2020-323082. [DOI] [PubMed] [Google Scholar]
- 17.Sharpton S.R., Schnabl B., Knight R., Loomba R. Current concepts, opportunities, and challenges of gut microbiome-based personalized medicine in nonalcoholic fatty liver disease. Cell Metabol. 2020;33:21–32. doi: 10.1016/j.cmet.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams C.D., Stengel J., Asike M.I., et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 19.Huang Q., Wu L.Y., Wang Y., Zhang X.S. GOMA: functional enrichment analysis tool based on GO modules. Chin J Canc. 2013;32:195–204. doi: 10.5732/cjc.012.10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang F., Chen Q., Wang W., Ling Y., Yan Y., Xia P. Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. J Hepatol. 2020;72:156–166. doi: 10.1016/j.jhep.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Hirsova P., Ibrahim S.H., Verma V.K., et al. Extracellular vesicles in liver pathobiology: small particles with big impact. Hepatology. 2016;64:2219–2233. doi: 10.1002/hep.28814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weston C.J., Shepherd E.L., Claridge L.C., et al. Vascular adhesion protein-1 promotes liver inflammation and drives hepatic fibrosis. J Clin Invest. 2015;125:501–520. doi: 10.1172/JCI73722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Athwal V.S., Pritchett J., Martin K., et al. SOX9 regulated matrix proteins are increased in patients serum and correlate with severity of liver fibrosis. Sci Rep. 2018;8:17905. doi: 10.1038/s41598-018-36037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaguchi Y. Sox 9 and programming of liver and pancreatic progenitors. J Clin Invest. 2013;123:1881–1886. doi: 10.1172/JCI66022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X.L., Hu B., Tang W.G., et al. CD73 sustained cancer-stem-cell traits by promoting SOX9 expression and stability in hepatocellular carcinoma. J Hematol Oncol. 2020;13:11. doi: 10.1186/s13045-020-0845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nouri M., Massah S., Caradec J., et al. Transient Sox 9 expression facilitates resistance to androgen-targeted therapy in prostate cancer. Clin Cancer Res. 2020;26:1678–1689. doi: 10.1158/1078-0432.CCR-19-0098. [DOI] [PubMed] [Google Scholar]
- 27.Sun H.J., Wu Z.Y., Nie X.W., Wang X.Y., Bian J.S. Implications of hydrogen sulfide in liver pathophysiology: mechanistic insights and therapeutic potential. J Adv Res. 2021;27:127–135. doi: 10.1016/j.jare.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haberl E.M., Feder S., Pohl R., et al. Chemerin is induced in non-alcoholic fatty liver disease and hepatitis B-related hepatocellular carcinoma. Cancers. 2020;12:2967. doi: 10.3390/cancers12102967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X., Chiwanda Kaminga A., Liu A., Wen S.W., Chen J., Luo J. Chemokines in non-alcoholic fatty liver disease: a systematic review and network meta-analysis. Front Immunol. 2020;11:1802. doi: 10.3389/fimmu.2020.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Affo S., Morales-Ibanez O., Rodrigo-Torres D., et al. CCL20 mediates lipopolysaccharide induced liver injury and is a potential driver of inflammation and fibrosis in alcoholic hepatitis. Gut. 2014;63:1782–1792. doi: 10.1136/gutjnl-2013-306098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu X., Jin Q., Chen H., et al. CCL20 is up-regulated in non-alcoholic fatty liver disease fibrosis and is produced by hepatic stellate cells in response to fatty acid loading. J Transl Med. 2018;16:108. doi: 10.1186/s12967-018-1490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee A., Basu A., Das K., et al. Hepatic transcriptome signature correlated with HOMA-IR explains early nonalcoholic fatty liver disease pathogenesis. Ann Hepatol. 2020;19:472–481. doi: 10.1016/j.aohep.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Wang R., Xiong S., et al. Cancer-associated fibroblasts promote the stemness of CD24(+) liver cells via paracrine signaling. J Mol Med (Berl) 2019;97:243–255. doi: 10.1007/s00109-018-1731-9. [DOI] [PubMed] [Google Scholar]
- 34.Jinawath N., Chamgramol Y., Furukawa Y., et al. Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma. Hepatology. 2006;44:1025–1038. doi: 10.1002/hep.21330. [DOI] [PubMed] [Google Scholar]
- 35.Liu F., Lagares D., Choi K.M., et al. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2015;308:L344–L357. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin K.C., Moroishi T., Meng Z., et al. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat Cell Biol. 2017;19:996–1002. doi: 10.1038/ncb3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong L., Huang L., Xue Q., et al. Cell-specific elevation of Runx 2 promotes hepatic infiltration of macrophages by upregulating MCP-1 in high-fat diet-induced mice NAFLD. J Cell Biochem. 2019 doi: 10.1002/jcb.28456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.