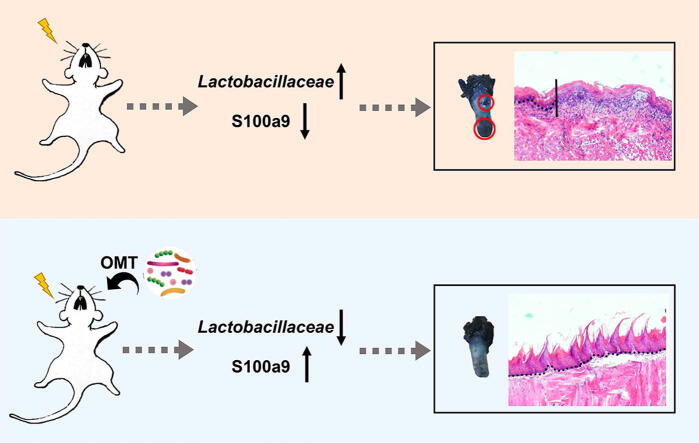
Graphical abstract
Keywords: Nasal, Oral and laryngeal cancer, Radiotherapy, Radiation-induced oral mucositis, Oral and gut microbiota, Lactobacillaceae, S100a9
Abstract
Oral mucositis is a common radiotherapy-induced complication among nasal, oral and laryngeal cancer (NOALC) patients. This complication leads to decreased quality of life and has few treatments. Here, fractionated radiation was performed to mimic radiotherapy for NOALCs in mouse models. Oral microbiota transplantation (OMT) mitigated oral mucositis, as judged by reconstructed epithelium and tongue papillae, fewer infiltrated leukocytes and more proliferative cells in the oral epithelium. The gut microbiota impacted oral mucositis progression, and OMT restructured oral and gut bacteria configurations and reprogrammed the gene expression profile of tongue tissues. In vivo silencing of glossal S100 calcium binding protein A9 debilitated the radioprotection of OMT. In light of clinical samples, we identified that patients with different alteration trends of Lactobacillaceae frequency presented different primary lesions and prognoses of NOALC following radiotherapy. Together, our findings provide new insights into the oral-gut microbiota axis and underpin the suggestion that OMT might be harnessed as a novel remedy to fight against oral mucositis in NOALC patients following radiotherapy in preclinical settings. Of note, oral microorganisms, such as Lactobacillaceae, might be employed as biomarkers to predict the prognosis of NOALC with radiotherapy.
1. Introduction
Head and neck squamous cell carcinomas (HNSCCs) have a yearly incidence of more than 600,000 cases worldwide, with 40–50% mortality [1]. HNSCC is a heterogeneous epithelial tumor that carries vastly diverse biological and clinical characteristics [2]. The classical risk factors for HNSCC are smoking and alcohol abuse. Currently, high-risk human papillomavirus (HPV) infection causes a substantial and rising proportion of these tumors, originating mainly in the oropharynx and occurring particularly in the Western world [3]. Surgery is the oldest and most traditional treatment for head and neck cancers, while modern radiotherapy has been proven to have similar success in treating head and neck cancers when compared to surgery if the lesion is at an early stage [4]. Although technological advancements in imaging and treatment delivery enable a more precise radiation region, therapeutic radiation to the head and neck region precipitates immediate and long-term sequelae covering cellulitis, mucositis, dysphagia, dysgeusia, severe pain of varying intensities, rampant caries, trismus, xerostomia and osteoradionecrosis [5]. Oral mucositis, marked by severe oral ulcerations, represents a common adverse side effect of large-dose radiation used before bone marrow transplantation or craniofacial radiotherapy. Radiation-induced loss of stem cells from the basal layer and subsequent denuding of the epithelium results in mucositis. As a result, patients with mucositis lesions frequently complain of mild to severe pain and pharyngeal dysphagia, which leads to the need for feeding tubes and a premature halt to radiotherapy [6].
The underlying molecular and cellular pathobiology of oral mucositis is characterized in five phases: initiation, the primary damage response, signaling and amplification, ulceration, and healing. Reactive oxygen species and transcription pathways, signaling and functional mediators, and importantly, microorganisms are involved in the development and resolution of mucositis [7]. Studies focusing on indigenous microbes residing within and throughout the human body, collectively termed the microbiota, have drawn a lot of attention during the past decade [8]. As a connection channel between outside environments and the respiratory/digestive tract, the oral cavity provides an appropriate temperature, humidity and nutrition for microorganism colonization [9]. The oral cavity carries distinct microenvironments, including the hard nonshedding surfaces of the teeth and the epithelial surfaces of the mucosal membranes [10]. Thus, the human mouth harbors one of the most diverse microbiomes throughout the whole body, including viruses, protozoa, fungi, archaea and bacteria, embedded within an extracellular matrix forming sophisticated biofilms [11]. The ecological community of commensal, symbiotic, and pathogenic microorganisms found in the oral cavity constitutes the oral microbiota. With hundreds of projects underway globally, investigations on gut microbiota experience a renaissance. However, its oral counterpart has not received the same level of attention for a long time. Oral microbial communities play pivotal roles in maintaining oral homeostasis, protecting the oral cavity and preventing disease development [12]. The symbiotic oral microbiota inhibits colonization by pathogens and is related to nitrate metabolism and cardiovascular health. Recently, the oral microbiota has been reported to correlate with the progression and aggravation of radiotherapy-induced mucositis in patients with nasopharyngeal carcinoma [13], indicating that oral microbiota analysis can be used in the prediction of mucositis during radiotherapy for nasal, oral and laryngeal cancer (NOALC). Our researches and studies from others report that radiation challenge shapes the gut microbiota configurations, and gut microbiota as well as microbial metabolites are promising therapeutic avenues to mitigate acute radiation syndrome covering hematopoietic and gastrointestinal (GI) toxicity [14], [15], [16], [17]. However, whether the oral microbiome can be utilized to battle against oral diseases, especially radiation-induced oral mucositis, deserves further investigation.
In the present study, we aimed to assess the effects of oral commensal microbiota on the development of radiation-induced mucositis and uncover the underlying mechanism. On the basis of mouse models and clinical samples, we found that transplantation of oral microbiota from healthy donors overtly mitigated radiation-induced oral mucositis by modulating S100 calcium binding protein A9 (S100a9). Importantly, the alteration paradigm of Lactobacillaceae abundance on the posterior pharyngeal wall predicted the prognosis of NOALC patients following radiotherapy. Thus, our findings provide novel insights into the relationship between the oral-gut microbiota axis and radiation-induced oral mucositis and underpin the predictive and therapeutic functions of the oral microbiota in radiation-induced oral mucositis in preclinical settings.
2. Materials and methods
2.1. Patients
Oral microbiota from posterior pharyngeal wall was collected from 44 nasal, oral and laryngeal cancer patients (30 men and 14 women; age range 19–70 years; average age 50.6) pre (before radiotherapy), during (after about 30 Gy performed) and post radiotherapy (after radiotherapy) from Nanfang Hospital, Southern Medical University (Guangzhou, China) using Copan CLASSIQSwabs (Copan Flock Technologier S.r.l., Brescia, Italy). Plasma samples were obtained from 27 patients pre- and post-radiotherapy, tumor volume information which calculated by nuclear magnetic resonance was obtained from 20 patients pre- and post-radiotherapy. Radiation therapy was administered in daily 2.0 Gy fractions, Monday through Friday, to a cumulative tumor dose of 50 to 60 Gy. For oral microbiota collection, microbial samples were taken from these patients prior to irradiation and after the course of treatment. The information of patients was listed in Supplementary Table 1. Written consents approving the use of clinical samples for research purposes were obtained from patients. The study protocol was approved by the ethics committee of Southern Medical University.
2.2. Mice
Six to eight-week-old male C57BL/6J mice were purchased from Beijing Huafukang Bioscience Co. Inc (Beijing, China). Mice were housed in the Specific Pathogen Free (SPF) level animal facility to avoid pathogenic bacteria infection at the Institute of Radiation Medicine (IRM), the Chinese Academy of Medical Sciences (CAMS). Mice were kept under standard conditions (ambient temperature 22 ± 2 ℃, air humidity 40–70% and a 12/12-h light/dark cycle) and continuous access to a standard diet (9% fat, 22% protein and 69% carbohydrate) and water. Animal experiments were performed according to the institutional guidelines approved by the Animal Care and Ethics Committee of IRM-PUMC, which complied with the Guide for the Care and Use of Laboratory Animals and the National Institutes of Health guide for the Care and Use of Laboratory Animals.
2.3. Experimental design
To assess the relationship between oral microbiota and radiation-induced oral mucositis, the mice were divided into two groups randomly based on body weight. One cohort was housed in normal cages (NC, n = 15), and the other cohort was housed in tailor-made cages (TC, n = 15). After 10 days, the oral microbes were collected and analyzed by 16S rRNA gene sequencing (data was shown in Fig. 1).
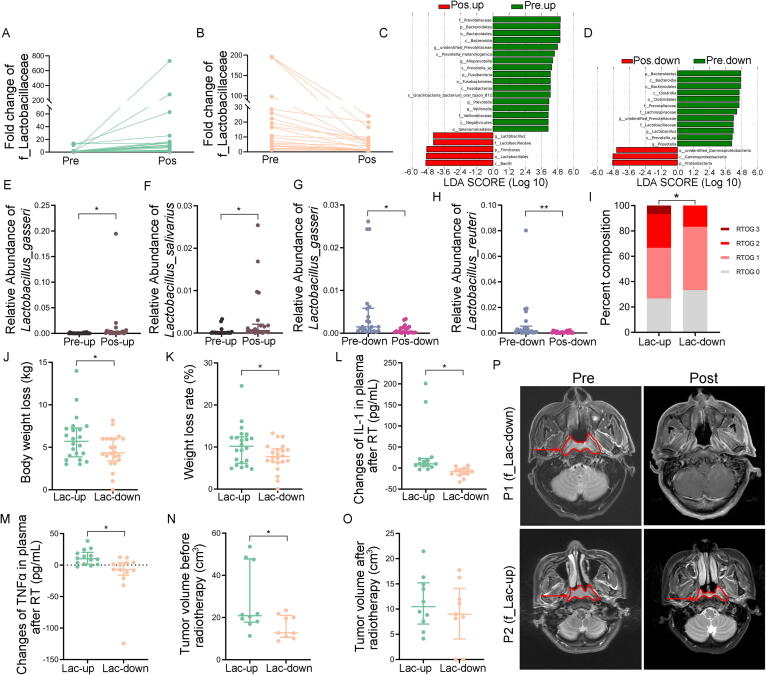
Fig. 1.
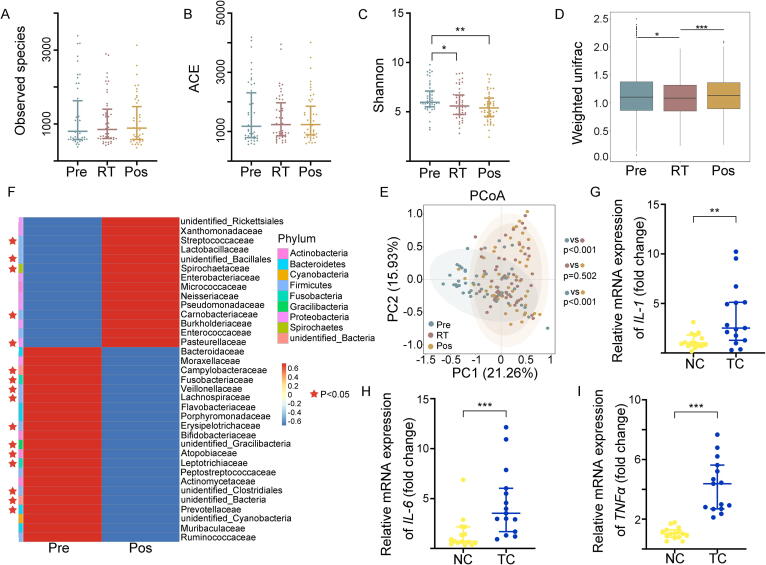
Radiotherapy shapes the oral microbiota composition. Figure A-F represented the data from 44 nasal, oral and laryngeal cancer patients pre-, during and post-radiotherapy respectively. Figure G-I represented the data from experimental mice grouped in normal cages (NC) and tailor-made cages (TC) respectively. (A) The observed species number of oral bacteria was examined by 16S rRNA high-throughput sequencing. (B, C) ACE and Shannon diversity index of oral bacteria was assessed by 16S rRNA high-throughput sequencing. (D) The β diversity of oral bacteria was compared by the weighted UniFrac analysis. The top and bottom boundaries of each box indicate the 75th and 25th quartile values, respectively, and lines within each box represent the 50th quartile (median) values. Ends of whiskers mark the lowest and highest diversity values in each instance. (E) Weighted principal coordinate analysis (PCoA) was used to measure the shift in oral bacterial composition profile. (F) The alteration of oral bacterial strains with high relative abundance at the family level was assessed before and after radiotherapy by 16S rRNA gene sequencing. The red star represented bacteria with significant differences (P < 0.05). (G-I) The expression levels of IL-1, IL-6 and TNFɑ were examined in tongue tissues by quantitative PCR from irradiated mice in normal and tailor-made cages. Mean ± SEM. Significant differences are indicated: * P < 0.05, ** P < 0.01, *** P < 0.005; Student’s t-test, n = 15 per group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
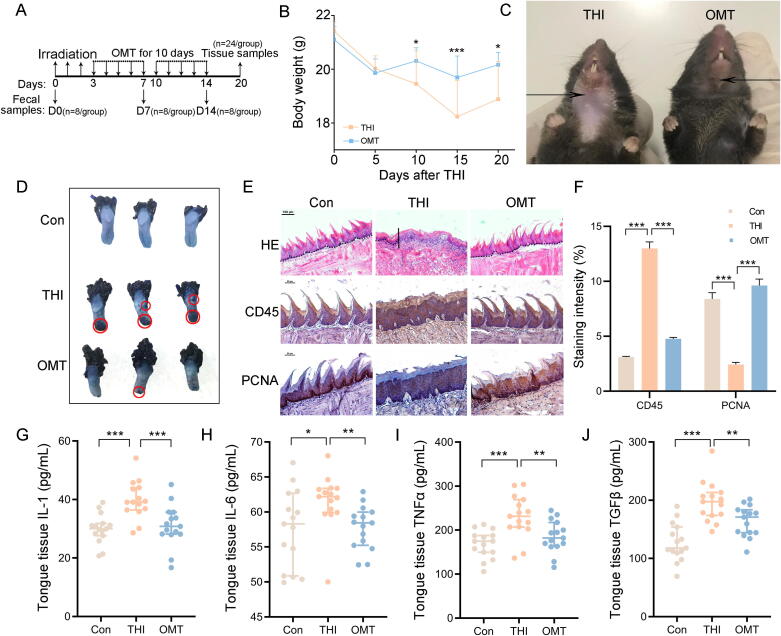
For OMT relative experiments: (1) Control group (n = 24): healthy 6- to 8-week-old male C57BL/6 mice were housed in the tailor-made cages with sham irradiation and sham OMT. (2) THI group (n = 24): Mice treated with head and neck irradiation (THI) were exposed to 8 Gy for three days. Then mice were subsequently orally administered with 150 µl of 2% methylcellulose (Sigma, Saint Louis, Missouri) for 10 days. The mice were housed in tailor-made cages avoid them eating feces. (3) OMT group (n = 24): Mice treated with THI were exposed to 8 Gy for three days. Then mice were subsequently performed with oral microbiota transplantation. The mice were housed in tailor-made cages avoid them eating feces. (4) THI + OMT + sh-S100a9 group (n = 18): Mice treated with THI were exposed to 8 Gy for three days. Then mice were injected with sh-S100a9 plasmid solution within 10 min after irradiation on the day before OMT. The mice were housed in tailor-made cages avoid them eating feces. The number of mice used here was based on previous study [18]. The tailor-made cage was showed in Fig. S7. Mice were sacrificed and tissue samples were collected after the 21 day course of the experiment.
2.4. Irradiation study
A Gammacell-40 137Cs irradiator (Atomic Energy of Canada Limited, Chalk River, ON, Canada) at a dose rate of 0.88 Gy per minute was used for all experiments. Mice treated with head and neck irradiation (THI) were exposed to 8 Gy for three days [6]. Mice were anesthetized with 3.5% chloral hydrate intraperitoneal injection (around 200 µl per mouse) using a lead shielding so that whole head and neck area was irradiated and the other parts of the mouse were shielded. Control mice were sham-irradiated.
2.5. Oral microbiota transplantation
Oral microbiota transplantation was performed based on previous study [19]. In detail, the healthy sex and age-matched C57BL/6 mice housed in normal cages were used as donors to collect oral microbiota and fresh oral microbiota used to transplantation was prepared every day. To ensure that recipient mice received similar inoculations, microbiota collected from donor mice of the same group were pooled. The donor’s oral microbiota was collected with oral swabs (Copan Flock Technologier S.r.l., Brescia, Italy) under SPF conditions. Oral swabs were taken of the whole oral cavity including tongue, maxilla, teeth etc. Then storage of swabs into saline and vortex. After centrifugation at 12000 rpm for 5 min, bacterial plaque was collected and resuspended in 2% methylcellulose. Each recipient mouse was inoculated in oral cavity using a gavage needle with 150 ul of the bacterial sample (∼5 × 106 CFU/mouse) in 2% methylcellulose from donor samples. 2% methylcellulose was used as control.
2.6. Antibiotics test
ABX-challenged mice were administrated with 200 μl solution of Ciprofloxacin (125 mg/L), Metronidazole (100 mg/L), Vancomycin (50 mg/L), Streptomycin (100U/L) and Penicillin (100U/L) intragastrically to avoid impairing oral microbiota [20], [21]. ABX treatment began on two days before irradiation for 20 days. The fresh antibiotic solution was prepared every day to promise its activity.
2.7. Bacterial diversity analysis
All samples were freshly collected from two independent experiments and stored at −80 °C until use (including clinical samples). Bacterial genomic DNA was extracted from mucosal samples using the DNA MAGNETICS and EXTRACT kit (Shenzhen BioEAsy Biotechnologies Co., Ltd., China) according to the manufacturer's instruction. DNA was extracted from the stool using the Power Fecal® DNA Isolation Kit (MoBio Carlsbad, CA USA). The DNA was recovered with 30 ml of buffer in the kit. PCR products were mixed in equidensity ratios. Then, mixture PCR products were purified with Qiagen Gel Extraction Kit (Qiagen, Germany). The 16S ribosomal RNA (rRNA) V4 was amplified used specific primer. All PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an IlluminaHiSeq2500 platform and 250 bp paired-end reads were generated. Paired-end reads was assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. Paired-end reads were merged using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/), a very fast and accurate analysis tool, which was designed to merge paired-end reads when at least some of the reads overlap the read generated from the opposite end of the same DNA fragment, and the splicing sequences were called raw tags. Quality filtering on the raw tags was performed under specific filtering conditions to obtain the high-quality clean tags according to the QIIME (V1.7.0, http://qiime.org/index.html) quality-controlled process. The tags were compared with the reference database (Gold database, http://drive5.com/uchime/uchime_download.html) using UCHIME algorithm (UCHIME Algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html) to detect chimera sequences, and then the chimera sequences were removed. Then the Effective Tags finally obtained. (Novogene Bioinformatics Technology Co., Ltd.). Sequence analysis was performed by Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/). Sequences with ≥ 97% similarity were assigned to the same OTUs. Representative sequence for each OTU was screened for further annotation. For each representative sequence, the Silva123 Database was used based on RDP classifier (Version 2.2, http://sourceforge.net/projects/rdpclassifier/) algorithm to annotate taxonomic information. Observed species, ACE index, Simpson index, Shannon index and chao1 index were applied to evaluate alpha diversity and the UniFrac distance was used to analyze the beta-diversity. Alpha and beta diversity calculation were performed using QIIME (V1.7.0). Statistical analysis of the relative abundance of the genera and the diversity indices and estimators were performed using R (v4.0.2). We collected 8 samples from each group. The primers are listed in Supplementary Table 2.
2.8. Transcriptome sequencing
Ribo-Zero rRNA removal kit (Epicentre, Madison, WI, USA) was used to remove ribosomal RNA. RNA libraries were prepared using the rRNA-depleted RNA with NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina (NEB, USA). Library sequencing was performed on an Illumina HiSeq X Ten platform (Illumina Inc., San Diego, CA) in ShenZhen Realomics Inc.
2.9. Statistical analysis
The data are presented as the means ± SEM with respect to the number of samples (n) in each group. Significance was assessed by comparing the mean values using Student’s t-test and Wilcoxon rank sum test for independent groups as follows: * P < 0.05, ** P < 0.01, *** P < 0.005. Two-way ANOVA and Chi-square test for body weight analysis and distribution of different grades of oral mucositis severity were performed in GraphPad Prism. Results with P < 0.05 were considered statistically significant. Experiments through the study have been performed at least three times.
3. Results
3.1. Radiotherapy shapes the oral microbiota configuration of NOALC patients
To address the effects of radiotherapy on the oral microbiota, we collected the oral microbiota from the posterior pharyngeal wall of 44 NOALC patients pre-, during and postradiotherapy. 16S rRNA high-throughput sequencing analysis showed no significant changes in the observed species of oral bacteria following radiotherapy (Fig. 1A). Statistical analysis further revealed that although the bacterial community richness ACE and Chao1 diversity indices (estimates the total number of species included in the community sample) displayed no alterations in oral microbiota species abundance (Fig. 1B and Fig. S1A), the Shannon and Simpson diversity indices (accounting for both the abundance and evenness of the species present) introduced a cumulative decrease following radiotherapy (Fig. 1C and Fig. S1B). β-diversity (representing phylogenetic differences in bacterial community structure) was estimated based on unweighted (taking into account species presence only) and weighted UniFrac distances (taking into account both species presence and species abundance), which compared intervariation of oral microbiota in NOALC patients pre-, during and postradiotherapy, respectively. The results showed that radiotherapy samples exhibited significantly lower average UniFrac values (representing phylogenetic differences in bacterial community structure) than those of patients preradiotherapy (Fig. 1D and Fig. S1C). Although unweighted principal coordinates analysis (PCoA) showed overlap of the oral microbiota (Fig. S1D), weighted PCoA represented their separation following radiotherapy (Fig. 1E). For example, radiotherapy elevated the frequency of Streptococcaceae and Lactobacillaceae at family (Fig. 1F) and genus (Fig. S1E) levels among the oral bacterial strains with high relative abundance. Together, our observations show that radiotherapy to NOALC patients influence oral microorganisms.
Next, to further investigate the effects of the oral microbiota on radiation-induced mucositis, mouse models were exposed to fractionated head and neck local irradiation to mimic radiotherapy for head and neck cancers. The mice were randomly divided into two groups based on body weight and housed in tailor-made cages to prevent coprophagy impacting the oral microbiota (the bottom of the cage was equipped with a metal grid, through which feces could fall into the padding. The details and photos of tailor-made cages are shown in Fig. S7) or normal cages, respectively. First, we analyzed the oral microorganism configuration after 10 days of preconditioning before irradiation. 16S rRNA gene sequencing identified the genus evolutionary tree information and discrepancy of oral microbiota between the two cohorts (Fig. S1F-J), suggesting that coprophagy impacts the oral microbiome. Then, the mice were exposed to local head and neck irradiation. qRT–PCR analysis showed that the irradiated mice in tailor-made cages carried higher levels of IL-1, IL-6 and TNFɑ in tongue tissues than those from normal cages after irradiation (Fig. 1G-I), further indicating that the resident microbial communities in oral indeed relate to the radiation-elevated inflammatory status of the oral cavity.
3.2. Oral microbiota transplantation protects against radiation-induced oral mucositis
First, we conducted oral bacterial taxonomic profiling analysis of healthy mouse donors, in which Streptococcus and Rodentibacter occupied the highest abundance at the genus level (Fig. S2A). Next, we transplanted oral microbes from healthy mouse donors to mice exposed to local head and neck irradiation (Fig. 2A). Intriguingly, oral microbiota transplantation (OMT) reduced the weight loss of the irradiated mice (Fig. 2B) and ameliorated radiation-intertwined alopecia and oral mucositis (Fig. 2C and D, Fig. S2B). Hematoxylin and eosin staining (H&E staining) further revealed thinning epithelium and flattened tongue papillae after radiation exposure; however, OMT erased the injuries (Fig. 2E, first line). Immunohistochemical staining showed that OMT led to fewer infiltrated leukocytes (Fig. 2E, second line and Fig. 2F) and higher numbers of proliferative cells in the oral epithelium (Fig. 2E, third line and Fig. 2F) of irradiated mice. ELISA analysis further validated that head and neck irradiation upregulated the levels of IL-1, IL-6, TNFɑ and TGFβ in tongue tissues and plasma. OMT eliminated the elevations (Fig. 2G-J and Fig. S2C-F), indicating that OMT alleviates systemic inflammation stimulated by head and neck irradiation. Together, our observations suggest that OMT contribute to battle against radiation-induced oral mucositis in mouse models.
Fig. 2.
Oral microbiota transplantation protects against radiation-associated oral mucositis. All mice in the following experiments were housed in tailor-made cages. 24 mice per group was used. (A) Sampling time points and scheme for oral microbiota transplantation combined with irradiation (THI). (B) Body weights were compared between THI and OMT groups, n = 24 per group; Significant differences are shown relative to the “THI” group using two-way group ANOVA (* P < 0.05; *** P < 0.005). (C) Photograph of mice in the two groups. The arrows point to the radiation-induced alopecia. (D) Photograph of tongue tissues from tailor-control, THI and OMT groups. The red circles point to mucositis. (E) The morphology of the tongue tissue was shown by H&E (first line) and immunohistochemistry staining (CD45 for second line; PCNA for third line), bar = 100 μm for first line, bar = 50 μm for second and third line. The solid diagonal line indicates the ulcer boundary, and dotted lines indicate the basement membrane. (F) The staining intensity of immunohistochemistry staining (CD45 and PCNA). (G-J) The content of IL-1, IL-6, TNFɑ and TGFβ in tongue tissues were examined by ELISA. Mean ± SEM. Significant differences are indicated: * P < 0.05, ** P < 0.01, *** P < 0.005 by Student’s t-test between each two cohort. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. OMT educates irradiation-shifted oral microbiota composition
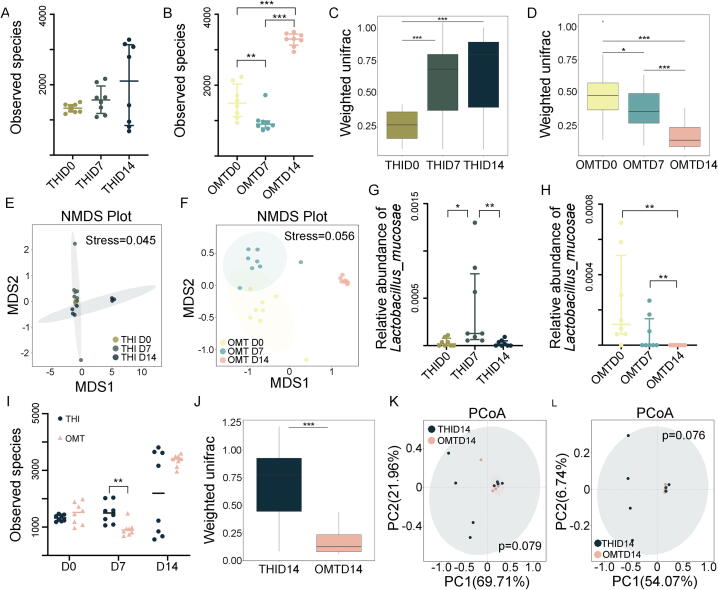
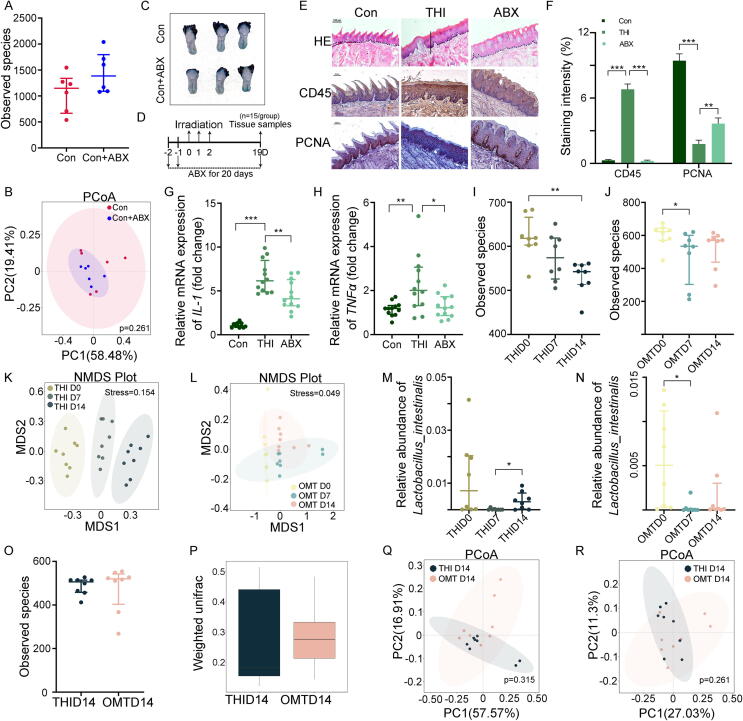
To tease out the underlying mechanism by which OMT mitigates radiation-induced oral mucositis, we scrutinized the oral bacteria taxonomic proportions with or without OMT from mice following head and neck irradiation. In parallel with the aforementioned clinical samples, irradiation unaltered the ɑ-diversity of oral bacteria (Fig. 3A and Fig. S3A); however, 10 days of OMT heightened the ɑ-diversity (Fig. 3B and Fig. S3B). Weighted UniFrac algorithm analysis showed that the β-diversity of oral bacteria was cumulative upregulated following irradiation, while OMT reversed the effects (Fig. 3C and D). Nonmetric multidimensional scaling (NMDS) and PCoA analysis all revealed more significant separation of oral microbial flora with OMT (Fig. 3E and F, Fig. S3C-F), indicating that OMT facilitates more significant migration of oral microbiota than irradiation alone. In accordance with the outcomes from clinical samples, head and neck irradiation elicited an elevatory frequency of Lactobacillaceae at the family level; however, OMT reversed the uptrend (Fig. S3G and H). The variation tendency of Lactobacillaceae was consistent in animal models and clinical samples following radiation, and OMT reversed the changes (Fig. S3G and H). Thus, we focused on Lactobacillaceae in the subsequent study. Metastat analysis further revealed that irradiation elevated the frequency of g_Lactobacillus_s_Lactobacillus_piscium and g_Lactobacillus_s_Lactobacillus_mucosae, while OMT lessened that of g_Lactobacillus_s_Lactobacillus_gasseri and g_Lactobacillus_s_Lactobacillus_mucosae (Fig. 3G and H, Fig. S3I and J). We also compared the oral bacterial configurations between the groups at 14 days after irradiation (10 days after OMT). Although the observed species number was unchanged at day 14, the oral bacteria in mice from the OMT cohort represented more homogeneous ɑ-diversity (Fig. 3I), and the microbial compositions between the two groups were quite different (Fig. 3J-L and Fig. S3K). As expected, irradiation-alone mice harbored higher Lactobacillaceae abundance than OMT mice at the genus level (Fig. S3L), implying that Lactobacillaceae might play a pivotal role in the development of radiation-induced oral mucositis. In addition, irradiation alone continued to increase the oral Proteobacteria phylum frequency, which was associated with ulcerative mucosal and/or skin lesions, but OMT reversed the elevation at day 14 (Fig. S3M and N). Together, our observations suggest that OMT remolds the oral bacteria taxonomic proportions to resist radiation-induced dysbiosis and mucositis in the oral cavity.
Fig. 3.
OMT educates irradiation-shifted oral microbiota composition. All mice in the following experiments were housed in tailor-made cages. (A, B) The observed species number of oral bacteria was examined by 16S rRNA high-throughput sequencing from mice at day 7 (THI D7) and day 14 (THI D14) after head and neck radiation exposure (A), day 7 (OMT D7) and day 14 (OMT D14) after oral microbiota transplantation (B). n = 8 per group. (C, D) The β diversity of oral bacteria was compared by the weighted UniFrac analysis from THI group (C) and OMT group (D). (E, F) Non-metric multidimensional scaling (NMDS) analysis was used to measure the shift in oral bacterial composition profile from THI group (E) and OMT group (F). (G, H) The abundance of g_Lactobacillus_s_ Lactobacillus_mucosae was assessed from THI group (G) and OMT group (H). (I) The observed species number of oral bacteria was examined from mice at day 0, 7, 14 after head and neck radiation exposure with or without oral microbiota transplantation. (J) The β diversity of oral bacteria was compared by the weighted UniFrac analysis between THI D14 and OMT D14. (K, L) Unweighted and weighted PCoA were used to measure the shift in oral bacterial composition profile from THI D14 and OMT D14. Significant differences for 16S rRNA high-throughput sequencing are indicated: Wilcoxon rank sum test.
3.4. Patients with the opposite alteration paradigm of Lactobacillaceae frequency present different primary lesions and prognoses of NOALC following radiotherapy
To further untangle the relationship between Lactobacillaceae frequency and the prognosis of nasal, oral and laryngeal carcinoma patients with radiotherapy, we returned to the clinical samples and divided the patients into two cohorts based on the variation tendency of Lactobacillaceae on the posterior pharyngeal wall at the family level (termed f_Lactobacillaceae). Demographic characteristics and cancer types factors of those two cohorts were summarized in Supplementary Table 3. At baseline, both groups were similar in terms of gender, age and cancer types. Fig. 4A-D and Fig. S4A-B represent the increase (Fig. 4A, C and Fig. S4A) and decrease (Fig. 4B, D and Fig. S4B) of f_Lactobacillaceae from the patients pre- and postradiotherapy (named as f_Lac-up and f_Lac-down, respectively). More specifically, metastat analysis showed elevations of g_Lactobacillus_s_Lactobacillus_gasseri and g_Lactobacillus_s_Lactobacillus_salivarius in the f_Lac-up group and reductions of g_Lactobacillus_s_Lactobacillus_gasseri and g_Lactobacillus_s_Lactobacillus_reuteri in the f_Lac-down group (Fig. 4E-H). The mucosa of patients’ oral cavity and oropharynx was scored clinically according to the Radiation Therapy Oncology Group (RTOG) criteria after radiotherapy. Approximately 33.3% of patients from the f_Lac-up group experienced a higher degree (RTOG2 + RTOG3) of mucositis, while only 16.67% of patients in the f_Lac-down group suffered from RTOG Grade 2 mucosa, and no patients developed RTOG3 mucosa (Fig. 4I). We counted the weight loss of patients in the two groups. Statistical analysis showed that the average weight reduction of patients in the f_Lac-up group was approximately 6.01 kg with a weight loss ratio of 10.30% and that of patients in the f_Lac-down group was 4.53 kg with a weight loss ratio of 7.69% (Fig. 4J and K). ELISA analysis measured the changes of inflammatory factors in blood plasma from the patients in the two cohorts, and revealed that radiotherapy facilitated more incremental of IL-1 and TNFɑ levels in patients from f_Lac-up group than those in patients from f_Lac-down group (Fig. 4L and M), indicating that alteration paradigm of f_Lactobacillaceae abundance relates to radiotherapy-mediated systemic inflammation of NOALC patients. We also calculated the tumor volume using nuclear magnetic resonance (NMR). Intriguingly, although the cure rate of radiotherapy showed no significance between the two cohorts (Fig. S4C), the primary tumor sizes were larger in patients from the f_Lac-up group than in patients from the f_Lac-down group (Fig. 4N-P), suggesting that patients with the opposite alteration paradigm of Lactobacillaceae frequency experience different primary lesions and prognoses of NOALC following radiotherapy.
Fig. 4.
Patients with the opposite alteration paradigm of Lactobacillaceae frequency present different primary lesions and prognoses of NOALC following radiotherapy. The data was from nasal, oral and laryngeal cancer patients pre-, during and post-radiotherapy respectively. (A, B) Patients were divided into two cohorts based on the changes of their oral f_ Lactobacillaceae. Figures represented the increase (A) and decrease (B) of Lactobacillaceae at the family level from head and neck cancer patients pre- and post-radiotherapy named as f_Lac-up and f_Lac-down, respectively. (C, D) Linear discriminant analysis (LDA) effect size (LEfSe) results represented significantly shifts in abundance of oral bacteria in f_Lac-up groups (C) and f_Lac-down groups (D) following radiotherapy, and indicated the effect size of each differentially abundant bacterial taxon. (E-H) The abundance of most varied strain bacteria was assessed using 16S rRNA high-throughput sequencing from head and neck cancer patients pre- and post-radiotherapy. Significant differences for 16S rRNA high-throughput sequencing are indicated: Wilcoxon rank sum test. (I) The percentage distribution of different grades of oral mucositis severity in f_Lac-up group and f_Lac-down group based on RTOG criteria. * P < 0.05; Chi-square test. (J, K) Body weight loss (J) and weight loss rate (K) of patients were compared between f_Lac-up and f_Lac-down group. * P < 0.05; Student’s t-test. (L, M) The changes of IL-1 (L) and TNFɑ (M) in blood plasma from the patients after radiotherapy in the two groups. * P < 0.05; Student’s t-test. (N, O) Tumor sizes from patients pre- (N) and post-radiotherapy (O) were compared between f_Lac-up and f_Lac-down group. * P < 0.05; Student’s t-test. (P) The tumor volume was calculated using nuclear magnetic resonance (NMR). The arrows point to the tumor area.
3.5. OMT remodels enteric bacterial structure following head and neck irradiation
Given that oral bacteria are linked to various digestive systemic diseases [22], [23], we are curious about whether the gut microbiota is related to the development of oral diseases. Antibiotics can change the gut microbial composition, resulting in varied clinical effects [24], [25], [26]. Thus, we treated the mice with an antibiotic cocktail (ABX) intragastrically and assessed the effects of ABX on the oral cavity. 16S rRNA sequencing revealed that intragastrical treatment of antibiotics did not affect oral microbiota composition significantly (Fig. 5A and B). In addition, ABX alone did not impact the structure or inflammatory status of tongue tissues (Fig. 5C and Fig. S5A-D). Then, head- and neck-irradiated mice were treated with ABX intragastrically to abrogate gut microbes without impairing oral microbiota (Fig. 5D), and 16S rRNA gene sequencing validated ABX deleted the gut bacteria and altered microbial community taxonomic tree (Fig. S5E and F). H&E and immunohistochemical staining showed that ABX challenge introduced thicker epithelium, healthier tongue papillae, fewer infiltrated leukocytes and higher numbers of proliferative cells in irradiated mice (Fig. 5E and F), implying that gut microbiota contributes to the development of radiation-induced oral mucositis. qRT–PCR analysis further validated that ABX treatment erased radiation-elevated inflammatory factors such as IL-1, IL-6 and TNFɑ in tongue tissues (Fig. 5G and H, Fig. S5G). Accordingly, we investigated the effects of OMT on the gut bacterial composition of irradiated mice. Although the Shannon diversity index represented no significant changes (Fig. S5H and I), the observed species number of enteric bacteria continuously decreased following head and neck irradiation; however, OMT blocked the reduction (Fig. 5I and J). Whether OMT was performed or not, the intestinal bacterial pattern of irradiated mice revealed significant alterations by the unweighted UniFrac algorithm but no significant alterations by the weighted UniFrac algorithm (Fig. S5J-M). Unweighted PCoA and NMDS analysis both showed that head and neck irradiation caused sustained changes in the intestinal microbiota; however, OMT restrained the shift (Fig. 5K and L, Fig. S5N and O). Intriguingly, we also observed that the frequency of enteric Lactobacillus_intestinalis was maintained at a higher level at day 14 after irradiation alone compared with the irradiated mice with OMT (Fig. 5M and N). Comparing intestinal microbiota between the two cohorts showed that although ɑ- and β-diversity were unaltered at day 14 after head and neck irradiation (Fig. 5O and P, Fig. S5P), unweight and weight PCoA revealed a separation gut microbiota (Fig. 5Q and R). Intriguingly, irradiated mice with OMT also harbored a lower relative abundance of Lactobacillus at the genus level (Fig. S5Q).
Fig. 5.
OMT remodels enteric bacterial structure following head and neck irradiation. (A) The observed species number of intestinal bacteria was examined from healthy control with or without antibiotics treatment (ABX). (B) Weighted PCoA were used to measure the shift in intestinal bacterial composition profile from Con and Con + ABX groups. (C) Photograph of tongue tissues from Con and Con + ABX groups. (D) Tissue sampling time points and scheme for antibiotics treatment combined with irradiation. (E) The morphology of the tongue tissue was shown by H&E and immunohistochemistry staining among control, THI and ABX combined with THI group. Scale bar = 100 μm for first line, bar = 50 μm for second and third line. The solid diagonal line indicates the ulcer boundary, and dotted lines indicate the basement membrane. (F) The staining intensity of immunohistochemistry staining (CD45 and PCNA). (G, H) The expression levels of IL-1 (G) and TNFɑ (H) were examined in tongue tissues by quantitative PCR. Mean ± SEM. Significant differences are indicated: * P < 0.05, ** P < 0.01, *** P < 0.005 by Student’s t-test between each two cohort, n = 12 per group. (I, J) The observed species number of intestinal bacteria was examined from THI group (I) and OMT group (J). (K, L) NMDS analysis was used to measure the shift in the intestinal bacterial composition profile from THI group (K) and OMT group (L). (M, N) The abundance of Lactobacillus_intestinalis in small intestine was assessed from THI group (M) and OMT group (N). (O) The observed species number of intestinal bacteria was examined from THI D14 and OMT D14. (P) The β diversity of intestinal bacteria was compared by the weighted UniFrac analysis between THI D14 and OMT D14. (Q, R) Unweighted (Q) and weighted (R) PCoA were used to measure the shift in intestinal bacterial composition profile from THI D14 and OMT D14. Significant differences for 16S rRNA high-throughput sequencing are indicated: Wilcoxon rank sum test.
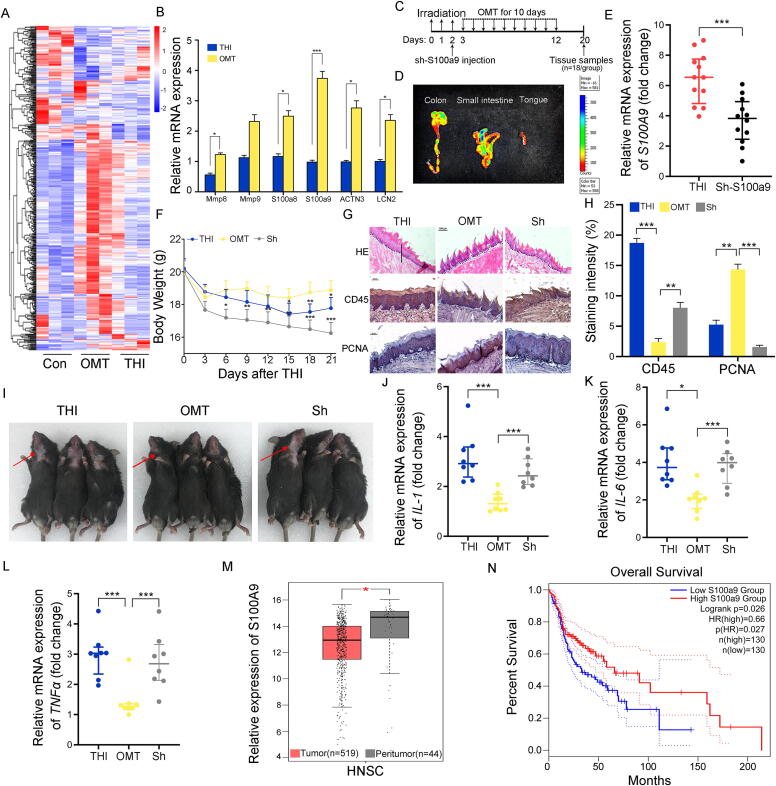
3.6. S100a9 is involved in the alleviation of OMT toward radiation-induced oral mucositis
To further address the underlying mechanism by which OMT protects against radiation-induced oral mucositis, we performed high-throughput sequencing to assess the gene expression profile in tongue tissues from mice with or without OMT after irradiation. A volcano plot exhibited the altered spectrum of mRNA expression driven by the stimuli (Fig. S6A-C). On the basis of the differentially expressed genes shown by the heatmap (Fig. 6A), we further validated the expression of target genes that were downregulated following irradiation exposure and upregulated after OMT by qRT–PCR. Among the target genes, S100a9 represented the most significant response to OMT (Fig. 6B). Thus, we performed hydrodynamic-based gene delivery to silence S100a9 expression in tongue tissues using pRNA-U6.1/Neo carrying shRNA targeting S100a9 within 10 min after irradiation (Fig. 6C). Fluorescence imaging analysis showed the accumulation of shRNA plasmid in the tongue and lower digestive tract (Fig. 6D), and qRT–PCR further validated that the shRNA attenuated the expression of S100a9 in tongue tissues (Fig. 6E). Then, the irradiated mice were treated with OMT with or without injection of shRNA. Importantly, S100a9 silencing abrogated OMT-reduced weight loss (Fig. 6F), exacerbated radiation-intertwined glottic toxicity, covering thinning epithelium and flattened tongue papillae (Fig. 6G, first line), more infiltrated leukocytes (Fig. 6G, second line and Fig. 6H), less proliferative cells (Fig. 6G, third line and Fig. 6H) and alopecia (Fig. 6I). qRT–PCR analysis further validated that S100a9 deletion elevated the expression of inflammatory factors (Fig. 6J-L), indicating that S100a9 might be implicated in the rehabilitation of radiation-induced oral mucositis mediated by OMT. Finally, we analyzed the overall survival rate of head and neck squamous carcinoma patients based on the expression of S100A9 (http://gepia.cancer-pku.cn/). The corresponding clinical information of the patients from whom the samples were obtained was downloaded from TCGA database (Supplementary Table 4). Notably, the peritumor tissues carried higher expression levels of S100A9 than tumor tissues (Fig. 6M), and patients with high levels of S100A9 had higher and longer overall survival rates (Fig. 6N), indicating that high levels of S100A9 might predict a good prognosis of patients with head and neck squamous cancers.
Fig. 6.
S100a9 is involved in the alleviation of OMT toward radiation-induced oral mucositis. All mice in the following experiments were housed in tailor-made cages. (A) The mRNA expression profile in tongue tissues from mice with or without OMT after head and neck irradiation by high throughput sequencing. (B) The expression levels of target genes which down-regulated following irradiation exposure and up-regulated after OMT were validated by qRT-PCR. Mean ± SEM. n = 12 per group. (C) Tissue sampling time points and scheme for sh-S100a9 injection combined with irradiation and OMT. (D) Mice that received sh-S100a9 plasmid were sacrificed 24 h after the retro-orbital sinus injection, and the luciferase activity in various organs (colon, small intestine and tongue) was detected by bioluminescent imaging. (E) The expression level of S100a9 was examined in tongue tissues by qRT-PCR. n = 12 per group. (F) Body weights were compared among three group mice after THI. Significant differences are shown relative to the “THI + OMT” group using two-way group ANOVA (* P < 0.05, ** P < 0.01, *** P < 0.005). n = 18 per group. (G) The morphology of the tongue tissue was shown by H&E and immunohistochemistry staining among THI, THI + OMT and THI + OMT + sh-S100a9 group. Scale bar = 100 μm for first line, bar = 50 μm for second and third line. The solid diagonal line indicates the ulcer boundary, and dotted lines indicate the basement membrane. (H) The staining intensity of immunohistochemistry staining (CD45 and PCNA). (I) Photograph of mice in the three groups. The arrows point to the radiation-induced alopecia. (J-L) The expression levels of IL-1 (J), IL-6 (K) and TNFɑ (L) were examined in tongue tissues by quantitative PCR. Mean ± SEM. Significant differences are indicated: * P < 0.05, ** P < 0.01, *** P < 0.005 compared with THI + OMT; Student’s t-test, n = 8 per group. (M) The expression levels of S100A9 in peritumor tissues and tumor tissues of head and neck squamous carcinoma patients. (N) The overall survival rate of head and neck squamous carcinoma patients based on the expression of S100A9.
4. Discussion
Investigation on remedies for radiation-induced oral mucositis have yielded disappointing results; however, the possible role of oral-gut microbiota axis in the treatment of NOALC was rarely concerned. Accordingly, our work reveals that oral microbiota transplantation battles against radiation-induced oral mucositis by modulating S100a9.
The human oral cavity is heterogeneous with the spatial organization of microbial communities across sites, which governs the stability of microecology in facing environmental factors [27]. For example, cigarette smoking significantly shifts the microbiota of the buccal mucosa but not in other oral sites and nasal cavities [28]. Oral microbes maintain oral mesenchymal stem cell homeostasis [29], and oral microbiota imbalance relates to oral diseases, such as periodontitis and oral cancers [30], [31]. The oral microbiota is also related to the risk of other nonoral diseases. For instance, the presence of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans in the oral cavity is significantly associated with an increased risk of pancreatic cancer and that of the phylum Fusobacterium reduces the risk [23], [32]. In addition, the oral microbiome can be employed to predict the development of colorectal cancer [33]. Our previous studies identified that the gut microbiota and microbial metabolites are safe and effective remedies to battle against radiation-induced hematopoietic and GI tract toxicity. In this study, the mice without coprophagy carried higher levels of inflammatory cytokines which might partly due to the lack of protective and commensal bacteria in the oral cavity by eating fecal pellets. Thus, we reported that oral microbiota transplantation might be a promising option to ameliorate oral mucositis in mouse models following irradiation stimuli. The pathogenesis of radiation-induced oral mucositis contains two distinct pathways. The first pathway, referred to as the immediate injury pathway, generates radiation-induced DNA damage directly in basal epithelial cells; the second pathway, known as the indirect pathway, accumulates biological injury drivers such as free radicals and damaging cytokines in the microenvironment [34]. Owing to the bioactivity of microbial metabolites, compounds derived from commensal microbiota, such as short-chain fatty acids, have the potential to modulate immune cells to protect against radiation-induced oral mucositis [35], [36]. Although salivary gland is highly differentiated, slowly proliferating tissue, it is surprisingly sensitive to radiation. Radiation exposure precipitates salivary gland dysfunction, representing as hyposalivation, xerostomia, nutritional deficiencies, oral infections and functional changes, such as changes in salivary pH, dysphagia and taste loss [37]. The effects of OMT on salivary gland, the percentage of engraftment failure and how OMT affects the gut microbiome remain unclear. In addition, donor screening, collection of oral microorganisms and delivery method of transplantation need further investigation in clinical scenarios.
Intestinal microbial diversity relates to the development of severe mucositis [38]. In our study, irradiation alone or OMT altered both oral and enteric microbial diversity, implying that the symbiotic microbiome in the digestive tract might be involved in oral mucositis development. The relative abundance of several gram-negative bacteria belonging to the Proteobacteria phylum increased significantly as mucositis progressed to peak severity and was associated with ulcerative mucosal and/or skin lesions, which was also observed in our clinical samples (Fig. 1F) [13]. In mouse models, irradiation alone continued to increase the oral Proteobacteria phylum frequency, but OMT reversed the elevation at day 14 (Fig. S3M and N). The increase in these gram-negative bacteria may exacerbate mucosal inflammation by activating host pattern recognition receptors (e.g., TLR, NOD) by bacterial components (e.g., LPS, fimbriae, membrane vesicles, etc.) and releasing proinflammatory cytokines such as TNF-α, IL-6, PGE2 and MMPs [39]. Furthermore, we treated the irradiated mice with an antibiotic mixture intragastrically to deplete the gut microbiota, and the mice exhibited catabatic oral mucositis, indicating that gut flora contribute to the development of radiation-induced oral mucositis. Although antibiotics could clear gut microbes, there still has some limitations because its systemic effect. Manrique and colleagues [21] reports that oral ciprofloxacin did not affect overall community structure, but higher antibiotic dose could reduce the relative abundance of Streptococcus genus. Both in Manrique’s and our studies, antibiotic cocktail in drinking water or intragastrical treatment cleaned out the gut microbiota but the α-diversity of oral microbiota was slightly elevated without significant difference, which also evidenced the systemic effect of antibiotic. Multiple factors modulate the configurations and functions of the microbiome, but one of the major elements triggering gut microbiota establishment is diet. Long-term nutritional habits dominate the abundance and diversity of microbial populations in the GI tract [40]. Studies have identified that the nutritional value of food is partially affected by the gut microbiota; in turn, food molds the intestinal flora [41]. Thus, diet and nutritional supplies might impact the development and rehabilitation of radiation-induced oral mucositis. However, Zama D and colleagues report that patients with enteral nutrition or parenteral nutrition experience different incidence rates of acute graft-versus-host disease and improvement of gut eubiosis [42], hinting that the delivery methods of nutritional supplements require further investigation. In light of the evidence, the experiment mice in this study would experience different processes of oral mucositis when the diet supply was changed. The oral microbiota has been proven to overcome the physical and microbial barrier, colonize the gastrointestinal tract and profile the gut microbiota, especially in the small intestine [43]. Thus, we investigated the effects of OMT on the enteric bacteria taxonomic proportions in this study and found that OMT reprogrammed the gut bacterial composition shifted by head and neck irradiation. On the basis of the evidence, OMT ameliorates radiation-induced mucositis, which might partly depend on modulation of the oral-gut microbiome axis.
On the basis of 16S rRNA gene sequencing, we found that the frequency of Lactobacillaceae in the oral cavity from clinical samples and animal models shifted following a certain paradigm after irradiation exposure. Radiotherapy to NOALC patients or total head irradiation to mice increased the abundance of Lactobacillaceae at the family level; however, OMT erased the change. From the oral cavity to feces, Lactobacillus species inhabit the entirety of the human GI tract and dominate vaginal microbes [44]. Lactobacilli has its pros and cons. On the one hand, Lactobacilli governs host immune systems, promotes digestive tract metabolic capacities and maintains homeostasis of the enteric microbiome. Lactobacilli colonization, especially Lactobacillus murinus, recruits Treg cells in the colon and ameliorates colitis [45]. Moderate high-salt challenge in the diet impairs intestinal survival of Lactobacillus spp., resulting in an increase in TH17 cells and blood pressure [46]. Probiotics containing Lactobacilli and Bifidobacteria prevent radiation-induced diarrhea [47], [48], [49], and Lactobacillus brevis CD2 lozenges reduce chemoradiotherapy-induced grade III and IV mucositis in patients with head and neck cancer [50]. On the other hand, Lactobacillus reuteri colonization worsens autoimmune manifestations and elicits systemic lupus erythematosus in a mouse model [51]. Overgrowth of Lactobacillus murinus dampens metabolism of the GI tract and prompts spurs the development of alopecia [52]. Here, we focused on the variation tendency of bacteria propelled by radiotherapy and divided the cancer patients into two cohorts according to the alteration paradigm of Lactobacillaceae at the family level, termed the f_Lac-up group and f_Lac-down group. Of note, the patients in the f_Lac-up group had a larger primary tumor volume and poorer prognosis than the patients in the f_Lac-down group. This suggests that the shift paradigm of specific oral microorganisms, such as Lactobacillaceae, can be employed as a biomarker to predict the prognosis of head and neck cancer, especially for nasal, oral and laryngeal carcinoma. We also observed specific strains of Lactobacillaceae shifts following radiation stimuli and OMT, such as Lactobacillus_mucosae in the oral cavity and Lactobacillaceae_intestinalis in the intestine. The specific strains might contribute to the progress or mitigation of radiation-induced oral mucositis, but in our opinion, the whole transplanted microbiome, including the microbiota and metabolites, works together to fight against oral mucositis. The key bioactive components in the microbiome require further study.
The molecular responses of animal models to OMT were assessed by high-throughput sequencing. Irradiation exposure reprogrammed the gene expression profile of tongue tissues, but OMT educated and tuned the shift. S100a9 was one of the altered genes that decreased following irradiation and increased after OMT. S100A9 is a calcium binding protein with multiple ligands and posttranslational modifications that is implicated in inflammatory processes and spurs the development of primary carcinoma to metastatic carcinoma [53]. Thus, we assessed the relationship between S100A9 expression and the prognosis of head and neck cancer sufferers and found that a low level of S100A9 was observed in tumor tissues and corresponded to poor prognosis. In addition, recombinant S100A9 protein administration promotes acute myeloid leukemia (AML) cell maturation, elicits growth arrest, and prolongs survival in AML mouse models [54]. S100A9 has been reported to participate in injury development. For instance, S100A9 is a key element necessary for neutrophil recruitment in acute and chronic liver injury [55] and reduces acute lung injury [56]. We silenced S100a9 in tongue tissues of mouse models; interestingly, the S100a9-depleted mice were unresponsive to OMT and exhibited serious oral mucositis after irradiation exposure. These observations suggest that S100A9 is involved in the radioprotection of buccal microbiota toward radiation-induced oral mucositis. Intracellular S100A9 mediates the interactions between the cytoskeleton and plasma membrane in a calcium-dependent manner and facilitates the resistance of epithelial cells to bacterial infections [57]. In line with the evidence, our findings further identify the protective function of S100A9 against external pressures, especially radiation challenge.
In conclusion, our work shows that oral microbiota transplantation contributes to protect against intractable radiation-induced oral mucositis in mouse models. Specifically, OMT reduces weight loss, mitigates glossal and systemic inflammation, and restructures the histrionic disorganization of the tongue following local head and neck irradiation. Mechanistically, oral microbiota transplantation educates oral and intestinal bacteria taxonomic proportions and reprograms the mRNA expression profile of tongue tissues disturbed by radiation challenge. Importantly, hydrodynamic-based gene delivery assays further validated that S100a9 contributes to the radioprotective processes of OMT in vivo. Patients with opposite alteration paradigms of Lactobacillaceae frequency presented different primary lesions and prognoses of nasal, oral and laryngeal cancer following radiotherapy. Together, our findings provide new insights into the oral-gut microbiota axis and corroborate the suggestion that oral microbiota transplantation might be employed as a novel therapeutic avenue for radiation-induced oral mucositis of patients with head and neck cancer after radiotherapy in preclinical settings. Specific oral microorganisms, such as Lactobacillaceae, can be harnessed as biomarkers to predict the prognosis of cancer.
CRediT authorship contribution statement
Huiwen Xiao: Conceptualization, Investigation, Methodology, Data curation, Formal analysis, Validation, Visualization, Writing – original draft. Yao Fan: Resources. Yuan Li: Investigation, Methodology. Jiali Dong: Investigation, Methodology. Shuqin Zhang: Investigation, Methodology. Bin Wang: Investigation, Methodology. Jia Liu: Investigation, Methodology. Xingzhong Liu: Funding acquisition. Saijun Fan: Funding acquisition. Jian Guan: Funding acquisition. Ming Cui: Conceptualization, Methodology, Data curation, Formal analysis, Validation, Visualization, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee. The studies involving human participants were reviewed and approved by the Ethics Committee of the Nanfang Hospital of Southern Medical University (LC2016PY015, LC2019ZD008, 2018CR021 and 2020CR025).
Acknowledgments
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 32100087, 81872555, 82003399, and 82173467), the Science Foundation for Distinguished Young Scholars of Tianjin (20JCJQJC00100).
Author contributions
All authors made substantive intellectual contributions to the present study and approved the final manuscript. M.C. elaborated the study design; H.W.X., M.C., Y.F., Y.L., J.L.D., S.Q.Z., B.W., and J.L. collected the data; M.C. and H.W.X. contributed to data analysis and interpretation; M.C. and H.W.X. drafted the article; M.C., J.G., X.Z.L. and S.J.F. provided experimental funding support. All the authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.10.028.
Contributor Information
Jian Guan, Email: guanj@smu.edu.cn.
Ming Cui, Email: cuiming0403@bjmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.M.L. Read B. Modasia A. Fletcher R.J. Thompson K. Brookes P.C. Rae et al. PTTG and PBF functionally interact with p53 and predict overall survival in head and neck cancer canres.0855.2018 10.1158/0008-5472.CAN-18-0855 [DOI] [PMC free article] [PubMed]
- 2.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171(7):1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leemans C.R., Snijders P.J.F., Brakenhoff R.H. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18(5):269–282. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 4.Ray-Chaudhuri A., Shah K., Porter R.J. The oral management of patients who have received radiotherapy to the head and neck region. Br Dent J. 2013;214(8):387–393. doi: 10.1038/sj.bdj.2013.380. [DOI] [PubMed] [Google Scholar]
- 5.Jawad H., Hodson N.A., Nixon P.J. review of dental treatment of head and neck cancer patients, before, during and after radiotherapy: part 1. Br Dent J. 2015;218:65–68. doi: 10.1038/sj.bdj.2015.28. [DOI] [PubMed] [Google Scholar]
- 6.Han G., Bian L.i., Li F., Cotrim A., Wang D., Lu J., et al. Preventive and therapeutic effects of Smad7 on radiation-induced oral mucositis. Nat Med. 2013;19(4):421–428. doi: 10.1038/nm.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonis S.T. Pathobiology of oral mucositis: novel insights and opportunities. J Support Oncol. 2007;5:3–11. [PubMed] [Google Scholar]
- 8.Zmora N., Suez J., Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(1):35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 9.Jia G., Zhi A., Lai P.F.H., Wang G., Xia Y., Xiong Z., et al. The oral microbiota - a mechanistic role for systemic diseases. Br Dent J. 2018;224(6):447–455. doi: 10.1038/sj.bdj.2018.217. [DOI] [PubMed] [Google Scholar]
- 10.Lamont R.J., Koo H., Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16(12):745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade W.G. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Belda-Ferre P., Alcaraz L.D., Cabrera-Rubio R., Romero H., Simón-Soro A., Pignatelli M., et al. The oral metagenome in health and disease. ISME J. 2012;6(1):46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X.-X., Yang X.-J., Chao Y.-L., Zheng H.-M., Sheng H.-F., Liu H.-Y., et al. The potential effect of oral microbiota in the prediction of mucositis during radiotherapy for nasopharyngeal carcinoma. EBioMedicine. 2017;18:23–31. doi: 10.1016/j.ebiom.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonneau M., Elkrief A., Pasquier D., Paz Del Socorro T., Chamaillard M., Bahig H., et al. The role of the gut microbiome on radiation therapy efficacy and gastrointestinal complications: A systematic review. Radiother Oncol. 2021;156:1–9. doi: 10.1016/j.radonc.2020.10.033. [DOI] [PubMed] [Google Scholar]
- 15.Cui M., Xiao H., Li Y., Zhou L., Zhao S., Luo D., et al. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol Med. 2017;9(4):448–461. doi: 10.15252/emmm.201606932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao H.-W., Cui M., Li Y., Dong J.-l., Zhang S.-Q., Zhu C.-C., et al. Gut microbiota-derived indole 3-propionic acid protects against radiation toxicity via retaining acyl-CoA-binding protein. Microbiome. 2020;8(1) doi: 10.1186/s40168-020-00845-610.21203/rs.3.rs-100316/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Dong J., Xiao H., Zhang S., Wang B., Cui M., et al. Gut commensal derived-valeric acid protects against radiation injuries. Gut microbes. 2020;11(4):789–806. doi: 10.1080/19490976.2019.1709387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui M., Xiao H., Li Y., Zhang S., Dong J., Wang B., et al. Sexual dimorphism of gut microbiota dictates therapeutics efficacy of radiation injuries. Adv Sci. 2019;6(21):1901048. doi: 10.1002/advs.v6.2110.1002/advs.201901048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao E., Mattos M., Vieira G.H.A., Chen S., Corrêa J.D., Wu Y., et al. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe. 2017;22(1):120–128.e4. doi: 10.1016/j.chom.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H., Ma Y., Peng X., Qiu W., Kong L., Ren B., et al. Antibiotic-induced dysbiosis of the rat oral and gut microbiota and resistance to Salmonella. Arch Oral Biol. 2020;114:104730. doi: 10.1016/j.archoralbio.2020.104730. [DOI] [PubMed] [Google Scholar]
- 21.Manrique P., Freire M.O., Chen C., Zadeh H.H., Young M., Suci P. Perturbation of the indigenous rat oral microbiome by ciprofloxacin dosing. Mol Oral Microbiol. 2013;28(5):404–414. doi: 10.1111/omi.2013.28.issue-510.1111/omi.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj J.S., Betrapally N.S., Hylemon P.B., Heuman D.M., Daita K., White M.B., et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62(4):1260–1271. doi: 10.1002/hep.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan X., Alekseyenko A.V., Wu J., Peters B.A., Jacobs E.J., Gapstur S.M., et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67(1):120–127. doi: 10.1136/gutjnl-2016-31258010.1136/gutjnl-2016-312580.supp110.1136/gutjnl-2016-312580.supp2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mu C., Zhu W. Antibiotic effects on gut microbiota, metabolism, and beyond. Appl Microbiol Biotechnol. 2019;103(23-24):9277–9285. doi: 10.1007/s00253-019-10165-x. [DOI] [PubMed] [Google Scholar]
- 25.Masetti R., Zama D., Leardini D., Muratore E., Turroni S., Prete A., et al. The gut microbiome in pediatric patients undergoing allogeneic hematopoietic stem cell transplantation. Pediatr Blood Cancer. 2020;67(12) doi: 10.1002/pbc.v67.1210.1002/pbc.28711. [DOI] [PubMed] [Google Scholar]
- 26.Becattini S., Taur Y., Pamer E.G. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016;22(6):458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proctor D.M., Fukuyama J.A., Loomer P.M., Armitage G.C., Lee S.A., Davis N.M., et al. A spatial gradient of bacterial diversity in the human oral cavity shaped by salivary flow. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-02900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G., Phillips S., Gail M.H., Goedert J.J., Humphrys M.S., Ravel J., et al. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome. 2017;5(1) doi: 10.1186/s40168-016-0226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su Y., Chen C., Guo L., Du J., Li X., Liu Y.i. Ecological balance of oral microbiota is required to maintain oral mesenchymal stem cell homeostasis. Stem Cells. 2018;36(4):551–561. doi: 10.1002/stem.v36.410.1002/stem.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C., Hemme C., Beleno J., Shi Z.J., Ning D., Qin Y., et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 2018;12(5):1210–1224. doi: 10.1038/s41396-017-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim Y., Totsika M., Morrison M., Punyadeera C. Oral microbiome: A new biomarker reservoir for oral and oropharyngeal cancers. Theranostics. 2017;7(17):4313–4321. doi: 10.7150/thno.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy S., Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 33.Flemer B., Warren R.D., Barrett M.P., Cisek K., Das A., Jeffery I.B., et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonis S.T. A hypothesis for the pathogenesis of radiation-induced oral mucositis: when biological challenges exceed physiologic protective mechanisms. Implications for pharmacological prevention and treatment. Support Care Cancer. 2021;29(9):4939–4947. doi: 10.1007/s00520-021-06108-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W., Yu T., Huang X., Bilotta A.J., Xu L., Lu Y., et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-18262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parada Venegas D., De la Fuente M.K., Landskron G., González M.J., Quera R., Dijkstra G., et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jasmer K., Gilman K., Forti K., Weisman G., Limesand K. Radiation-induced salivary gland dysfunction: mechanisms, therapeutics and future directions. J Clin Med. 2020;9:4095. doi: 10.3390/jcm9124095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Vliet M.J., Harmsen H.J.M., de Bont E.S.J.M., Tissing W.J.E., Manchester M. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog. 2010;6(5):e1000879. doi: 10.1371/journal.ppat.100087910.1371/journal.ppat.1000879.g00110.1371/journal.ppat.1000879.g002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasconcelos R.M., Sanfilippo N., Paster B.J., Kerr A.R., Li Y., Ramalho L., et al. Host-microbiome cross-talk in oral mucositis. J Dent Res. 2016;95(7):725–733. doi: 10.1177/0022034516641890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moszak M., Szulińska M., Bogdański P. You are what you eat-the relationship between diet, microbiota, and metabolic disorders-a review. Nutrients. 2020;12:1096. doi: 10.3390/nu12041096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illiano P., Brambilla R., Parolini C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020;287(5):833–855. doi: 10.1111/febs.v287.510.1111/febs.15217. [DOI] [PubMed] [Google Scholar]
- 42.Zama D., Gori D., Muratore E., Leardini D., Rallo F., Turroni S., et al. Enteral versus parenteral nutrition as nutritional support after allogeneic hematopoietic stem cell transplantation: a systematic review and meta-analysis. Transplant Cell Ther. 2021;27(2):180.e1–180.e8. doi: 10.1016/j.jtct.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Li B., Ge Y., Cheng L., Zeng B., Yu J., Peng X., et al. Oral bacteria colonize and compete with gut microbiota in gnotobiotic mice. Int J Oral Sci. 2019;11(1) doi: 10.1038/s41368-018-0043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heeney D.D., Gareau M.G., Marco M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr Opin Biotechnol. 2018;49:140–147. doi: 10.1016/j.copbio.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang C.e., Kamiya T., Liu Y., Kadoki M., Kakuta S., Oshima K., et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe. 2015;18(2):183–197. doi: 10.1016/j.chom.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Wilck N., Matus M.G., Kearney S.M., Olesen S.W., Forslund K., Bartolomaeus H., et al. Salt-responsive gut commensal modulates T(H)17 axis and disease. Nature. 2017;551(7682):585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delia P., Sansotta G., Donato V., Frosina P., Messina G., De Renzis C., et al. Use of probiotics for prevention of radiation-induced diarrhea. World J Gastroenterol. 2007;13:912–915. doi: 10.3748/wjg.v13.i6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demers M., Dagnault A., Desjardins J. A randomized double-blind controlled trial: impact of probiotics on diarrhea in patients treated with pelvic radiation. Clin Nutr. 2014;33(5):761–767. doi: 10.1016/j.clnu.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 49.Linn Y.H., Thu K.K., Win N.H.H. Effect of Probiotics for the prevention of acute radiation-induced diarrhoea among cervical cancer patients: a randomized double-blind placebo-controlled study. Probiotics Antimicrob Proteins. 2019;11(2):638–647. doi: 10.1007/s12602-018-9408-9. [DOI] [PubMed] [Google Scholar]
- 50.Sharma A., Rath G.K., Chaudhary S.P., Thakar A., Mohanti B.K., Bahadur S. Lactobacillus brevis CD2 lozenges reduce radiation- and chemotherapy-induced mucositis in patients with head and neck cancer: a randomized double-blind placebo-controlled study. Eur J Cancer. 2012;48(6):875–881. doi: 10.1016/j.ejca.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Zegarra-Ruiz D.F., El Beidaq A., Iñiguez A.J., Lubrano Di Ricco M., Manfredo Vieira S., Ruff W.E., et al. A diet-sensitive commensal Lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. 2019;25(1):113–127.e6. doi: 10.1016/j.chom.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayashi A., Mikami Y., Miyamoto K., Kamada N., Sato T., Mizuno S., et al. Intestinal dysbiosis and biotin deprivation induce alopecia through overgrowth of Lactobacillus murinus in Mice. Cell Rep. 2017;20(7):1513–1524. doi: 10.1016/j.celrep.2017.07.057. [DOI] [PubMed] [Google Scholar]
- 53.Markowitz J., Carson W.E. Review of S100A9 biology and its role in cancer. Biochim Biophys Acta. 2013;1835(1):100–109. doi: 10.1016/j.bbcan.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Laouedj M.R. Tardif L. Gil M.-A. Raquil A. Lachhab M. Pelletier et al. S100A9 induces differentiation of acute myeloid leukemia cells through TLR4 129 14 2017 1980 1990 10.1182/blood-2016-09-738005 [DOI] [PubMed]
- 55.Moles A., Murphy L., Wilson C.L., Chakraborty J.B., Fox C., Park E.J., et al. A TLR2/S100A9/CXCL-2 signaling network is necessary for neutrophil recruitment in acute and chronic liver injury in the mouse. J Hepatol. 2014;60(4):782–791. doi: 10.1016/j.jhep.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hiroshima Y., Hsu K., Tedla N., Wong S.W., Chow S., Kawaguchi N., et al. S100A8/A9 and S100A9 reduce acute lung injury. Immunol Cell Biol. 2017;95(5):461–472. doi: 10.1038/icb.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Austermann J., Spiekermann C., Roth J. S100 proteins in rheumatic diseases. Nat Rev Rheumatol. 2018;14(9):528–541. doi: 10.1038/s41584-018-0058-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.