Abstract
Telomerase is a reverse transcriptase that adds single-stranded telomeric repeats to the ends of linear eukaryotic chromosomes. It consists of an RNA molecule including a template sequence, a protein subunit containing reverse transcriptase motifs, and auxiliary proteins. We have carried out an interference footprinting analysis of the Tetrahymena telomerase elongation complexes. In this study, single-stranded oligonucleotide primers containing telomeric sequences were modified with base-specific chemical reagents and extended with the telomerase by a single 32P-labeled dGMP or dTMP. Base modifications that interfered with the primer extension reactions were mapped by footprinting. Major functional interactions were detected between the telomerase and the six or seven 3′-terminal residues of the primers. These interactions occurred not only with the RNA template region, but also with another region in the enzyme ribonucleoprotein complex designated the telomerase DNA interacting surface (TDIS). This was indicated by footprints generated with dimethyl sulfate (that did not affect Watson-Crick hydrogen bonding) and by footprinting assays performed with mutant primers. In primers aligned at a distance of 2 nucleotides along the RNA template region, the footprints of the six or seven 3′-terminal residues were shifted by 2 nucleotides. This shift indicated that during the elongation reaction, TDIS moved in concert with the 3′ ends of the primers relative to the template region. Weak interactions occurred between the telomerase and residues located upstream of the seventh nucleotide. These interactions were stronger in primers that were impaired in the ability to align with the template.
Telomeres, the structures at the ends of linear eukaryotic chromosomes, consist of short DNA sequence repeats associated with specific DNA-binding proteins. In most eukaryotes, the telomeric DNA includes clusters of G residues and the complementary clusters of C residues that are interspersed with other short repeats (for reviews, see references 9 and 60). The G-rich strand, which is oriented in a 5′ → 3′ direction towards the ends of the chromosomes, protrudes as a single-stranded overhang that may interact with the double-stranded telomeric sequences (27, 35, 40, 43, 57, 58).
Telomerase, an enzyme found in most eukaryotes, is a reverse transcriptase (RT) that adds telomeric repeats to the G-rich extensions, thereby preventing chromosome shortening due to the inability of the normal replication apparatus to complete telomere synthesis (13, 23, 24). All telomerases characterized so far contain an RNA molecule and a catalytic protein subunit. The telomerase RNA of each organism includes a short sequence, complementary to the telomeric repeats of this organism, which serves as a template for synthesis of these repeats (10, 18, 26, 41, 54). The secondary structure of the RNAs of the ciliate telomerases has been conserved throughout evolution (41, 52). It is still not clear whether the telomerase RNA secondary structures in other groups of organisms have been similarly conserved. The catalytic protein subunit in all telomerases characterized so far includes reverse transcriptase motifs and was therefore termed telomerase RT (TERT) (17, 32, 42, 47, 49). The activities of ciliate and human telomerases can be reconstituted in vitro by mixing the TERT of each of the enzymes with the corresponding RNA subunit (1, 3, 5, 14). However, genetic and biochemical experiments have indicated that telomerases may also contain other auxiliary protein subunits (7, 16, 31, 34, 51).
The ciliate Tetrahymena thermophila telomerase was the first telomerase to be discovered (23, 24). The template sequence in the RNA of this telomerase was mapped and found to be 3′-CCCCAAC-5′, encoding the Tetrahymena telomeric repeat 5′-(GGGGTT)n-3′ (2, 19, 21, 26). In addition, A residues next to the 3′ end of the template region were found to be required for proper alignment of the primer along the template (19). Evidence was also provided for the involvement of other regions of the RNA in the primer extension process (8, 20, 39).
In vitro, primer extension by the Tetrahymena telomerase is processive (22). This processive mechanism requires that after completion of the synthesis of each repeat, the 3′ end of the primer be translocated to the beginning of the template region without complete dissociation of the primer-enzyme complex (15, 22, 37, 38). Therefore, it has been suggested that, in addition to the catalytic site, the telomerase must contain a second site that serves as an anchor to which the primer remains bound during the translocation step (15, 38). The existence of a second site was also suggested by experiments in which either the Tetrahymena telomerase or the human enzyme was used to extend primers that did not contain telomeric repeats at their 3′ termini but contained G-rich sequences at their 5′ termini. These primers were efficiently extended by the two telomerases, whereas primers that contained no G-rich sequences were rather poor substrates for the enzymes. Hence, it has been proposed that the second site preferentially binds G-rich sequences (33, 48; see also reference 7).
The interactions between the Tetrahymena enzyme and the DNA primers were also investigated by UV cross-linking techniques. In two studies, DNA primers substituted with the thymidine analog 5-[N-(p-azidobenzoyl)-3-aminoallyl]-deoxyuridine] (N3RdU) or with 5-iodouracil were found to specifically cross-link to 100- and 95-kDa proteins that copurified with the Tetrahymena telomerase activity (16, 30). In another study performed with the telomerase of the ciliate Euplotes aediculatus, DNA primers substituted with 5-iodopyrimidines were found to cross-link with a 130-kDa protein subunit and the RNA subunit of the enzyme (29). In this study, the cross-links were mapped to nucleotides located 20 to 22 bases upstream of the 3′ ends of the primers. From these data, it has been proposed that the upstream nucleotides were cross-linked to the anchor site of the enzyme (29).
Here we present an interference footprinting analysis designed to map functional interactions between the Tetrahymena telomerase and single-stranded DNA primers containing telomeric repeats during the elongation phase of primer extension reactions. In this study, major interactions were detected between the telomerase and the six or seven 3′-terminal nucleotide residues in the primers. The data indicated that these interactions occur not only with the RNA template region, but also with another region in the telomerase ribonucleoprotein complex (RNP). Furthermore, these two regions were found to move relative to each other during the elongation reaction. Only weak interactions were observed in most primers between the enzyme and nucleotide residues located upstream of the seventh nucleotide. These interactions were stronger in elongation complexes formed with mutant primers that were impaired in the ability to align with the template. We discuss the results in relation to previous studies of telomerases and other polymerases.
MATERIALS AND METHODS
Oligonucleotides.
DNA oligonucleotides were purchased from Biotechnology General (Nes Ziona, Israel) and purified by polyacrylamide gel electrophoresis, followed by Sephadex G-50 spun column chromatography (53). Prior to being modified or extended by telomerase, the oligonucleotide samples were heated for 5 min at 100°C and then fast cooled to 4°C.
Preparation of telomerase.
T. thermophila cells were grown to late logarithmic phase and starved without subsequent mating, as described before (25, 30). Partially purified telomerase was prepared as follows, using a published procedure that has been modified slightly (21). An S100 extract was prepared and bound to DEAE-Sepharose Fast Flow beads (Sigma) that had been washed with TMGI buffer (10 mM Tris-HCl [pH 8.0], 1 mM MgCl2, 10% glycerol, 5 mM mercaptoethanol, 0.10 mM phenylmethylsulfonyl fluoride, 0.25 μg of pepstatin per ml, 0.25 μg of chymostatin per ml) that also contained 0.20 M sodium acetate. Binding was accomplished by suspending the beads in the extract dissolved in TMGI buffer containing 0.20 M sodium acetate and 10 U of RNasin (Promega) per ml and by swirling the suspension for 30 min at 4°C. The mixture was centrifuged in a Heraeus centrifuge for 2 min at 500 × g at 4°C, and the beads were resuspended in TMGI buffer containing 0.20 M sodium acetate. The resuspended beads were washed and centrifuged again as described above, then resuspended in the same buffer, and poured into a column. The column was washed with 5 volumes of TMGI buffer containing 0.20 M sodium acetate, and telomerase activity was eluted with 2 volumes of TMGI buffer containing 0.50 M sodium acetate. This eluent was loaded on an Octyl-Sepharose CL-4B column (Pharmacia). The column was washed with 5 volumes of TMGI buffer containing 0.50 M sodium acetate and with 5 volumes of TMGI buffer. Telomerase was eluted with TMGI buffer containing 1% Triton X-100. Pooled fractions containing the enzyme were frozen and stored at −80°C.
Primer extension by telomerase.
Primer extension was carried out in 20-μl reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 0.1 M sodium acetate, 1 mM spermidine, 6 mM mercaptoethanol, 0.4 U of RNasin per ml, 1 μM [α-32P]dGTP (specific radioactivity, 300 Ci/mmol; Dupont NEN, Boston, Mass.), 100 μM dTTP, 2.4 μM primer, and 5 to 10 μl of telomerase. The reaction mixtures were incubated for 15 min at 30°C. Primer extension by a single G residue was carried out in the same buffer except that dTTP was omitted from the reaction mixtures and the concentration of [α-32P]dGTP was 0.1 μM (specific radioactivity, 3,000 Ci/mmol). These reaction mixtures were incubated for 15 min at 10°C. Primer extension by a single T residue was performed similarly except that the 0.1 μM [α-32P]dGTP was replaced with 0.3 μM [α-32P]dTTP (specific radioactivity, 3,000 Ci/mmol). All reactions were terminated by addition of EDTA, sodium dodecyl sulfate (SDS), and proteinase K at final concentrations of 15 mM, 0.08%, and 0.05 mg/ml, respectively, and the mixtures were incubated for 45 min at 45°C. Next, ammonium acetate and yeast tRNA were added at final concentrations of 2.5 M and 15 μg/ml, respectively, and the reaction products were precipitated with ethanol and washed with 70% ethanol, as previously described (4). Samples were electrophoresed either in 10% polyacrylamide sequencing gels (products obtained in reactions performed in the presence of both dGTP and dTTP) or in 12% polyacrylamide sequencing gels (products obtained in reactions performed in the presence of either dGTP or dTTP). The gels were dried and exposed to phosphorimaging screens that were subsequently scanned with a Molecular Dynamics PhosphorImager.
Modification of DNA by chemical reagents. (i) Modification by DMS.
DNA at a concentration of 15 μg/ml was exposed to dimethyl sulfate (DMS) for 10 min at 10°C at the concentrations specified in Fig. 3. The reactions were terminated by the addition of mercaptoethanol at a final concentration of 1.0 M.
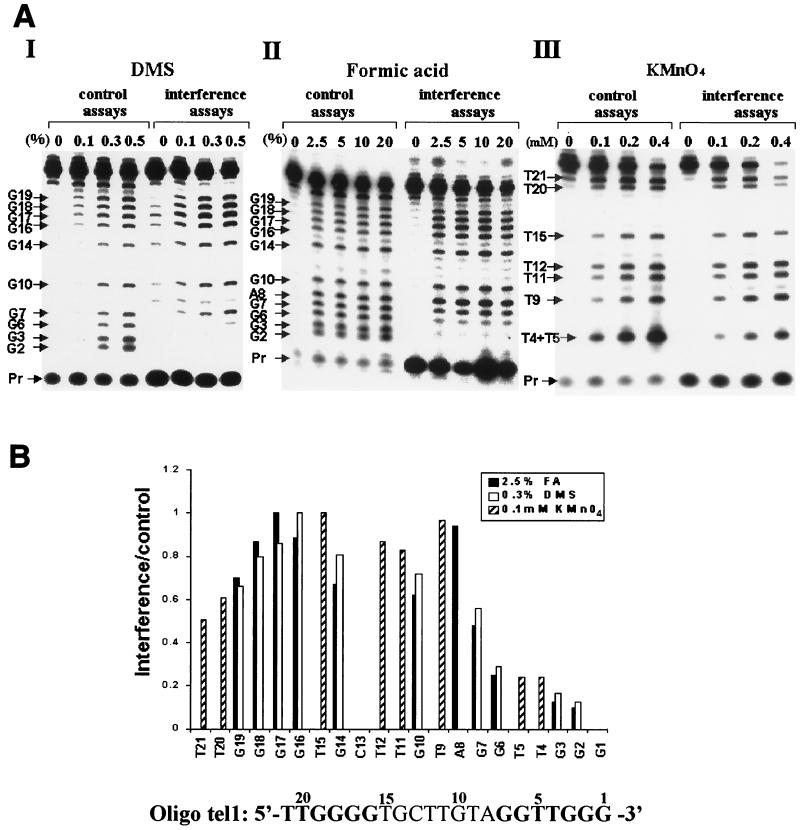
FIG. 3.
Interference footprinting performed with oligo tel1, which contains telomeric sequences at the 3′ and 5′ ends. (A) Interference footprinting was performed with oligo tel1, whose sequence is shown at the bottom of the figure, using the procedure described in Materials and Methods. The telomeric repeats in the oligo tel1 sequence are indicated with boldface letters. The concentrations of the reagents used for these assays, DMS (panel I), formic acid (panel II), and KMnO4 (panel III), are indicated. Pr, unincorporated 32P-labeled dATP. (B) Quantitative analysis of the data shown in A was performed as described in Materials and Methods. The histograms present data obtained in the analysis of one lane from each interference assay and of a corresponding control lane, namely the 0.3% DMS, 2.5% formic acid, and 0.1 mM KMnO4 lanes. The relative interference values are shown as bars. For calculations of the interference resulting from modification of T4 or T5, the molecules modified at T4 and T5 were assumed to be equally labeled (see Results).
(ii) Modification by formic acid.
DNA at a concentration of 50 μg/ml was exposed to formic acid for 30 min at 37°C at the concentrations specified in the figures. The reactions were terminated by the addition of sodium acetate and Trizma base at final concentrations of 0.8 M and 0.5 M, respectively.
(iii) Modification by KMnO4.
DNA at a concentration of 50 μg/ml was exposed to potassium permanganate (KMnO4) at the specified concentrations for 20 min at 37°C. The reactions were terminated by the addition of mercaptoethanol at a final concentration of 1.0 M. After completion of the reactions, all samples were precipitated with ethanol in the presence of 0.30 M sodium acetate and washed twice with 70% ethanol, as described above.
Interference footprinting.
Oligonucleotide primers were modified by one of the three reagents. For the modifications, samples of each oligonucleotide were exposed to several concentrations of the modifying reagent or not exposed at all. The modified DNAs were extended with telomerase by using a single 32P-labeled dGMP or dTMP, as described above. However, 60-μl reaction mixtures were used for these assays. The reactions were terminated by the addition of EDTA, SDS, and proteinase K and incubated as described above. Subsequently, the samples were extracted once with an equal volume of a mixture of phenol and chloroform (1:1), precipitated, and washed with 70% ethanol. In the control assays, the DNA was first labeled and then chemically modified. All the DNA samples were dried, exposed to piperidine (44), and analyzed by electrophoresis in 16% Long Ranger (FMC) gels containing 7 M urea, as described before (4). All footprinting assays were performed at least twice at several concentrations of the reagents (see the legends to the figures), and the results were reproducible.
Quantitative analysis of the footprinting data.
One control lane and the corresponding experimental lane, in which the modification apparently followed single-hit kinetics, were selected for quantitative analysis. Quantification of the bands observed in each gel was performed with the software supplied with the Molecular Dynamics PhosphorImager. Corrections for differential losses during the procedure were performed by first calculating the sum of the intensities of all the bands in each lane j, (ΣIi)j; then the observed intensity of each band, Ii, within a lane j was multiplied by the ratio (ΣIi)m/(ΣIi)j, where (ΣIi)m was the largest sum. The corrected intensity of each band in a sample that has not been exposed to the reagent (e.g., 0% formic acid), I0′, was subtracted from the corrected intensity of the corresponding band in a selected experimental lane, Ie′. Similarly, I0 was subtracted from the corresponding control lane value, Ic′. The differences were divided to give the relative interference values, (Ie′ − I0′)/(Ic′ − I0′). These ratios were normalized relative to the highest ratio, which was given the value 1.0, and plotted as histograms that are presented in Fig. 3 to 7.
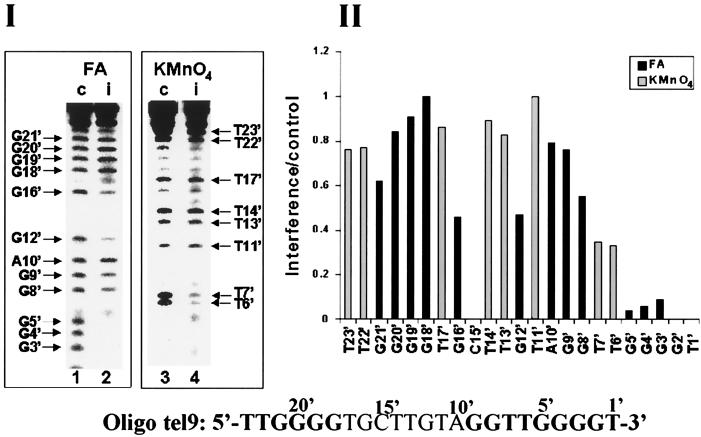
FIG. 7.
Interference footprinting performed with oligo tel9, which ends with the sequence GGGGT-3′ instead of GGG-3′. Formic acid (FA) and KMnO4 interference assays were carried out as described in the legends to Fig. 3 and 4 except that [32P]dTTP was used for the primer extension reactions instead of [32P]dGTP. The data presented are those obtained in 5% formic acid assays and 0.15 mM KMnO4 assays of oligo tel9.
RESULTS
Primer extension by telomerase.
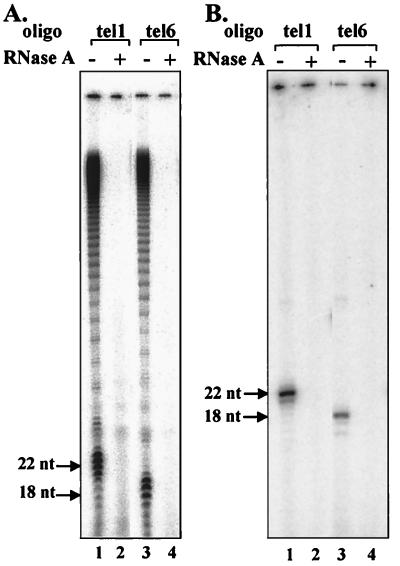
Table 1 shows nine of the oligonucleotide primers used for our footprinting analysis. Eight of these oligonucleotides (oligos tel1 to tel8) ended with the sequence GGG-3′. As Fig. 1A shows, these oligonucleotides were expected to align with the telomerase RNA template region so that G would be the first nucleotide added to them by the enzyme. The ninth (oligo tel9) ended with the sequence GGGGT-3′ and was expected to align with the template as shown in Fig. 1B, so that T would be the first residue added to it by the enzyme. Figure 2A shows assays in which two of the primers that ended with GGG-3′, oligo tel1 and oligo tel6, were extended in the presence of 32P-labeled dGTP and unlabeled dTTP. It can be seen that typical telomerase extension products containing many Tetrahymena telomeric repeats were obtained (lanes 1 and 3). Furthermore, RNase inhibited these reactions, indicating that the extension was catalyzed by telomerase and not by contaminating polymerases (lanes 2 and 4) (24, 25). Figure 2B (lanes 1 and 3) shows experiments in which the same primers were extended in the presence of 32P-labeled dGTP alone. Unlike the assays shown in Fig. 2A that were performed at 30°C, these assays were carried out at 10°C. At this lower temperature, the nuclease activity of the telomerase (15, 45) or the activities of other possible contaminating nucleases were found to be minimized (H. Wang and E. H. Blackburn, personal communication; S. Benjamin, N. Baran, and H. Manor, unpublished data). As expected, the major products obtained in these reactions were primers extended by a single 32P-labeled dGMP residue; only relatively small amounts of shorter products were generated by nucleolytic cleavage followed by reinsertion of radioactively labeled dGMP residues. These reactions were also inhibited by exposure of the enzyme to RNase (Fig. 2B, lanes 2 and 4), an indication that the extended primers, like the products of the reactions shown in Fig. 2A, were authentic telomerase extension products. Similar assays were performed with all the other primers and gave similar results. It should be noted that in primer extension reactions performed at 10°C in the presence of both 32P-labeled dGTP and unlabeled dTTP, the primers were extended by many telomeric repeats, like those in the reactions performed at 30°C (not shown).
TABLE 1.
Oligonucleotide primers used for the interference assays and relative incorporation of a single [32P]dGMP residue into oligos tel1 through tel8
| Oligo | Sequencea | No. of nucleotides | Relative incorporationb ± SD |
|---|---|---|---|
| tel1 | TTGGGGTGCTTGTAGGTTGGG | 21 | 100 ± 9 |
| tel2 | TTGGGGTGCTCGAGGTAGGG | 20 | 5.3 ± 2 |
| tel3 | TTGGGGTGCTCGAGGATGGG | 20 | 49 ± 6 |
| tel4 | TTGGGGTGCTTGTAGATTGGG | 21 | 93 ± 32 |
| tel5 | TTGGGGTGCTTGTAGGTTTTGGG | 23 | 173 ± 54 |
| tel6 | TTGGGGTTGGGGTTGGG | 17 | 29 ± 1.7 |
| tel7 | TAGTCGTGCTTGTACATTGGG | 20 | 118 ± 11 |
| tel8 | TTCCGTTAGTCGAGTTGGGGCGCTTGCAGGTTGGG | 35 | 104 ± 12 |
| tel9 | TTGGGGTGCTTGTAGGTTGGGGT | 23 |
Telomeric repeats are indicated by bold letters. Substitutions and insertions are underlined.
The incorporation of a single [32P]dGMP residue into oligos tel1 to tel8 was determined in assays of the type shown in Fig. 2B. A labeled oligonucleotide was added to each sample before the ethanol precipitation step, and the radioactivity recovered in this oligonucleotide was used to correct the data for losses that occurred during the procedure. The data are expressed relative to the incorporation into oligo tel1, which was set at 100. Each assay was repeated three times. Average values and standard deviations are shown here.
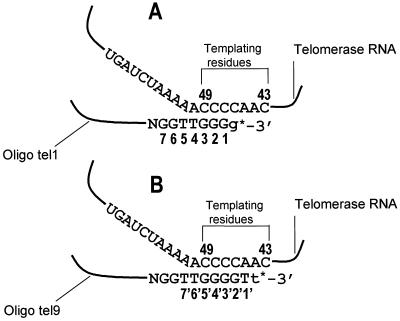
FIG. 1.
Schematic representation of the alignment of oligonucleotide primers with the Tetrahymena telomerase RNA template region and extension of these primers by a single nucleotide residue. (A) Scheme showing the alignment of primers ending with the sequence GGG-3′ along the telomerase RNA template region and their extension by a single 32P-labeled dGMP residue (g*). (B) Scheme showing the alignment of primers ending with the sequence GGGGT-3′ along the telomerase RNA template region and their extension by a single 32P-labeled dTMP residue (t*).
FIG. 2.
Elongation of oligonucleotide primers by the telomerase. (A) Primer extension reactions were carried out with the telomerase at 30°C in the presence of 32P-labeled dGTP and unlabeled dTTP, as described in Materials and Methods (lanes 1 and 3). Similar reactions were performed with telomerase that had been treated with 0.50 U of RNase A per ml for 15 min at 22°C (lanes 2 and 4). The products were electrophoresed in denaturing polyacrylamide gels, and the gels were dried and visualized in a Molecular Dynamics PhosphorImager. nt, nucleotides. (B) Same as panel A except that the reactions were performed at 10°C in the absence of dTTP.
Table 1 also shows a quantitative analysis of the incorporation of 32P-labeled dGTP into the oligos tel1 to tel8, based on data of the type shown in Fig. 2B. The data were normalized relative to the incorporation into oligo tel1, which was set at 100. It can be seen that the normalized incorporation into the oligonucleotides varied between 5.3 ± 2 and 173 ± 54. The significance of these variations will be discussed below.
Interference footprinting of elongation complexes generated with oligo tel1, which contains telomeric repeats at the 3′ and 5′ ends.
Interference footprinting was used to study the interactions between individual primer residues and the telomerase in elongation complexes during extension of the primers by a single nucleotide. Footprinting assays were initially performed with the 21-residue oligo tel1, which consisted of telomeric repeats at the 3′ and 5′ ends separated by a nontelomeric sequence (Table 1). These assays were designed to assess the interactions of the enzyme with the telomeric repeats at the two separate positions. The procedure was as follows. Oligo tel1 molecules were first modified with each of the three base-specific chemical reagents DMS, formic acid, and KMnO4. The modified primers were incubated with telomerase in the presence of [32P]dGTP so that the chains were extended by a single 32P-labeled dGMP residue. Thus, the primers were end labeled at the 3′ termini. DNA was subsequently purified, cleaved at the modified sites by treatment with piperidine, and electrophoresed in denaturing gels. In control assays that were performed in parallel, the primer molecules were first extended by a single 32P-labeled dGMP residue, then chemically modified, treated with piperidine, and electrophoresed in the same gels. The gels were exposed to PhosphorImager screens.
Figure 3A, panel I, shows PhosphorImager traces of footprints obtained after modification of oligo tel1 molecules with three concentrations of DMS, a reagent that methylates the N-7 atoms of guanine residues and also the N-1 and N-3 atoms of adenine residues at much lower efficiencies. A comparison between the experimental and the control lanes indicated that modification of residues G2, G3, and G6 strongly interfered with the primer extension by telomerase. These footprints also indicated that modification of the G residues upstream of G6, including the residues constituting the 5′ telomeric repeat, did not substantially interfere with the primer extension. It should be noted that in these assays and in most subsequent assays, the G1 band was not resolved well from the band containing the unincorporated dGTP and hence could not be analyzed. However, in some assays (not shown), the G1 cleavage product was better resolved, and modification at this residue was also found to strongly interfere with the extension of the primer. The primer residues G1, G2, and G3 must form Watson-Crick hydrogen bonds with the telomerase RNA-templating residues C47, C48, and C49 (Fig. 1). However, these interactions should not have been affected by modification with DMS. Hence, the interference patterns described above are indicative of additional interactions of G1, G2, and G3 with other components of the telomerase ribonucleoprotein (RNP).
A second reagent used for these experiments, formic acid, causes removal of both guanine and adenine bases from the DNA backbone. Figure 3A, panel II, shows the PhosphorImager traces of assays performed with formic acid at four concentrations. These assays too indicated that modification of residues G2, G3, and G6 strongly interfered with primer extension by the telomerase. As already found in the assays performed with DMS, modifications of other upstream residues did not appear to substantially affect the extension of the modified primers.
A third reagent, KMnO4, oxidizes the double bond between the atoms C-5 and C-6 of thymine and causes disruption of the ring structure. Figure 3A, panel III, shows interference footprinting assays performed with KMnO4 at three different concentrations. In these footprints and in other footprints shown below, residues T4 and T5 were not resolved. Clearly, the intensity of the combined T4 plus T5 band in the interference assays is considerably lower than that of the corresponding band in the control assays. The KMnO4 interference patterns are discussed further below.
Of the data shown in Fig. 3A, the footprints obtained with one concentration of each reagent were quantitated and used to calculate the relative interference due to modification of each residue in the primer, as described in Materials and Methods. Figure 3B shows a graphic representation of these data. Each bar represents the normalized ratio of the intensity of a band in the experimental lane and the intensity of the corresponding band in the control lane. The lower the bar, the higher the degree of interference caused by modification of the nucleotide corresponding to this band. It can be seen that modifications of each of residues G2, G3, and G6 by formic acid, interfered with the elongation of oligo tel1 to an extent of ≥75% relative to the elongation of molecules in which residue G17 was modified. In other experiments performed with oligo tel1 and various derivatives of this oligonucleotide, the extents of interference caused by modifications of nucleotides G2 and G3 were as high as 98% (see below). Figure 3B also shows that moderate interference was caused by modification of residue G7 with formic acid. Modifications of the residues upstream of G7 did not substantially affect the extension of the modified primer. Modifications of residues G2, G3, G6, and G7 by DMS caused a somewhat reduced interference. This could be due to the difference in the modification products generated by exposure to these two reagents. Otherwise, the DMS and formic acid interference profiles were similar.
In the quantitation of the KMnO4 data, the interference due to T4 modification was assumed to be equal to the interference due to T5 modification. In another experiment (not shown), oligo tel1 was 5′-end labeled, modified with KMnO4, and cleaved with piperidine. In gel electrophoresis of these cleavage products, T4 and T5 were resolved and found to be equally modified. Hence, the interference due to modification of either residue T4 or residue T5 was determined by dividing the intensity of the combined T4 plus T5 band in the interference lane by that of the corresponding band in the control lane. It is apparent that modification of either T4 or T5 strongly inhibited primer extension.
It should be noted that even though the interference profiles obtained in the assays performed with the three modifying reagents are correlated, each of the profiles should be assessed separately; for the various modifications caused by the three reagents could affect the reaction in different ways and might cause different degrees of interference.
Interference footprinting of complexes generated with oligo tel1 variants containing substitutions at the 3′ telomeric repeat. (i) Interference footprinting of complexes generated with primers in which residues facing nucleotides 50 and 51 of the RNA subunit were replaced.
As Figure 1A shows, of the residues 5′-TTGGG-3′ in oligo tel1, nucleotides G1, G2, and G3 must form Watson-Crick hydrogen bonds with the telomerase RNA nucleotides C47, C48, and C49, which serve as templating residues (2, 19, 21). Nucleotides T4 and T5 may also form Watson-Crick hydrogen bonds with telomerase RNA nucleotides A50 and A51. These RNA nucleotides were found to be required for alignment of telomeric primers along the template, even though they do not serve as templating residues (19). The following experiments were designed to examine the relative contributions of the Watson-Crick pairings between T4 and A50 and between T5 and A51 vis-à-vis other interactions required for a proper alignment of the primers. For this purpose, interference footprinting analyses were performed with primers in which the T4 and T5 residues were replaced with other nucleotides.
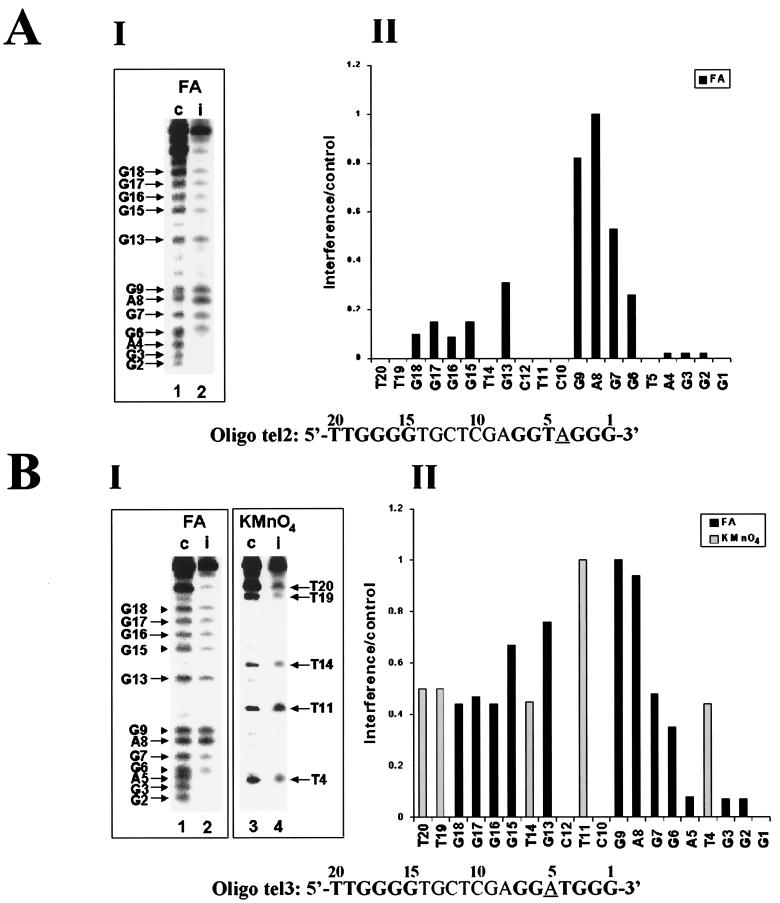
We first used for these assays oligo tel2, which contains a telomeric repeat at the 5′ terminus and a 3′ telomeric sequence that has been mutated by replacing residue T4 with A. As Table 1 shows, only about 5% as much radioactively labeled dGMP was incorporated into this primer as into oligo tel1. This result supports the notion that Watson-Crick pairing with RNA residue A50 plays a major role in the primer extension reaction (19). Figure 4A shows a footprinting analysis of this low-efficiency reaction. Formic acid rather than DMS was used for these and subsequent assays reported below because it gave more reproducible results and provided reliable interference patterns for A as well as G residues. Panel I shows one control lane (c) and the corresponding experimental (i) lane that were selected from a whole experiment of the type shown in Fig. 3. Panel II shows a histogram of the relative interference values calculated from the data shown in panel I. It can be seen that modification of residues G2, G3, and A4 strongly interfered with the extension of this primer and that modification of G6 also resulted in a rather strong interference. The strong relative interference resulting from modification of nucleotide A4, which cannot form a Watson-Crick hydrogen bond with A50 of the telomerase RNA, showed that the extension of oligo tel2 depended on interactions other than Watson-Crick base-pairing with A50. It should also be noted that, unlike the footprints of oligo tel1, modification of the G residues in the upstream telomeric repeat of oligo tel2 did cause a rather strong interference with its extension. These data indicated that the interactions of the upstream G's with the telomerase were functionally significant for this inefficient reaction (see Discussion).
FIG. 4.
Interference footprinting assays performed with oligo tel1 variants containing substitutions of residues N4 and N5. Formic acid (FA) and KMnO4 interference assays were carried out as described in the legend to Fig. 3. For each reagent, one control lane (c) and one experimental lane (i) are shown in panel I, and the corresponding histograms are presented in panel II. (A) Formic acid (10%) assays of oligo tel2. (B) Formic acid (10%) assays and 0.15 mM KMnO4 assays of oligo tel3.
We next studied the extension of oligo tel3, in which residue T5 rather than T4 was replaced with A. As Table 1 shows, the efficiency of extension of this primer was only about twofold lower than that of oligo tel1. This result is compatible with a previous observation that formation of a Watson-Crick base pair between residue N5 in the primer and residue A51 in the RNA template is less essential for the extension reaction than formation of a base pair between residue N4 in the primer and A50 in the template (19). Figure 4B shows interference footprinting assays performed with this oligonucleotide, using formic acid and KMnO4. It can be seen that modifications of residues G2, G3, and A5 strongly interfered with the extension of the primer, whereas the interference due to modifications of residues G6 and G7 was more moderate. The strong interference caused by modification of A5 indicates that interactions other than Watson-Crick base-pairing between the N5 nucleotide in the primer and RNA residue A51 played a major role in extension of this primer by the enzyme. It should also be noted that modifications of the G residues in the upstream telomeric repeat of this primer moderately interfered with primer extension. A similar degree of interference caused by modification of the upstream G residues was also observed in assays performed with another variant of oligo tel1 in which the T5 residue facing A51 in the RNA was replaced with G (not shown). It should also be noted that modification of residue G5 in this oligonucleotide strongly interfered with the elongation of this primer.
Inspection of the KMnO4 footprint of oligo tel3 reveals that the interference caused by modification of residue T4 was much less substantial than that observed in the KMnO4 footprint of oligo tel1. A similar observation was made in a KMnO4 footprinting assay of an additional variant of oligo tel1 in which residue T5 was deleted (not shown). Apparently, the absence of T5 affected the interactions of these primers with the enzyme so that the role of T4 in the reaction became less significant.
(ii) Interference footprinting of complexes generated with primers in which residues facing nucleotides 52 and 53 of the RNA subunit were replaced.
It has been reported that nucleotides 52 and 53 in the RNA template may also play a role in the alignment of telomeric primers with the Tetrahymena telomerase (19). It was therefore interesting to determine whether replacement of the oligo tel1 residues G6 and G7, which face these RNA nucleotides, with other bases would affect the interference profiles of the substituted primers. We first used for these assays oligo tel4, a variant of oligo tel1 in which G6 was replaced with A. This substitution did not affect the efficiency of extension of the unmodified oligo tel4 compared with that of oligo tel1 (Table 1). Figure 5A shows formic acid and KMnO4 interference assays performed with oligo tel4. It can be seen that the interference profiles obtained in the assays of this primer did not substantially differ from the profiles obtained in the corresponding assays of oligo tel1. In particular, modification of residue A6 caused a strong interference with the extension of oligo tel4. In addition, modification of G7 in this oligonucleotide also caused substantial interference with its elongation. Similar interference profiles were obtained in assays of another oligonucleotide in which G6 has been replaced with T and in assays of two other oligonucleotides in which G7 has been replaced with either A or T (not shown).
FIG. 5.
Interference footprinting assays performed with oligo tel1 variants containing substitutions of residues N6 and N7. Formic acid (FA) and KMnO4 interference assays were carried out as described in the legends to Fig. 3 and 4. (A) Formic acid (2.5%) assays and 0.10 mM KMnO4 assays of oligo tel4. (B) Formic acid (5%) assays and 0.10 mM KMnO4 assays of oligo tel5.
We have also studied the interactions of the telomerase with oligo tel5, another variant of oligo tel1 in which two T residues have been inserted at positions 6 and 7 of the primer. As Table 1 shows, the efficiency of extension of oligo tel5 was significantly higher than that of oligo tel1. This result could be due to the ability of all four residues T4, T5, T6, and T7 in this oligonucleotide to form Watson-Crick hydrogen bonds with residues A50, A51, A52, and A53 in the telomerase RNA, unlike oligo tel1 (see Fig. 1). Figure 5B shows formic acid and KMnO4 interference assays performed with oligo tel5. It can be seen that, as in the case of oligo tel1, modification of residues 2 to 6 strongly interfered with the extension of oligo tel5. Modification of T7, G8, and G9 caused a slight interference compared with modification of other upstream residues. Thus, the ability of primer residues T6 and T7 to form two additional Watson-Crick hydrogen bonds with RNA residues A52 and A53 did not substantially affect the interference profile of this primer.
Evidence that the presence or distribution of upstream telomeric repeats did not substantially affect the interference patterns.
The data presented so far indicated that upstream telomeric repeats do not play a major role in the elongation of primers that are properly aligned with the RNA template region of the telomerase. Here we present footprinting assays designed to further investigate the role of upstream telomeric sequences in the elongation reaction.
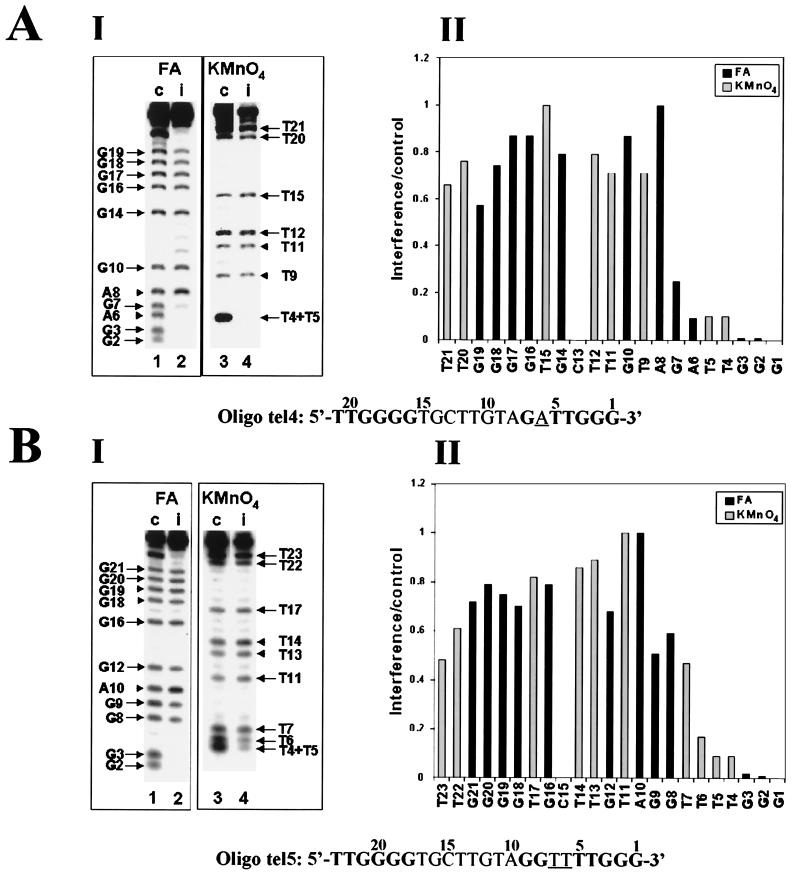
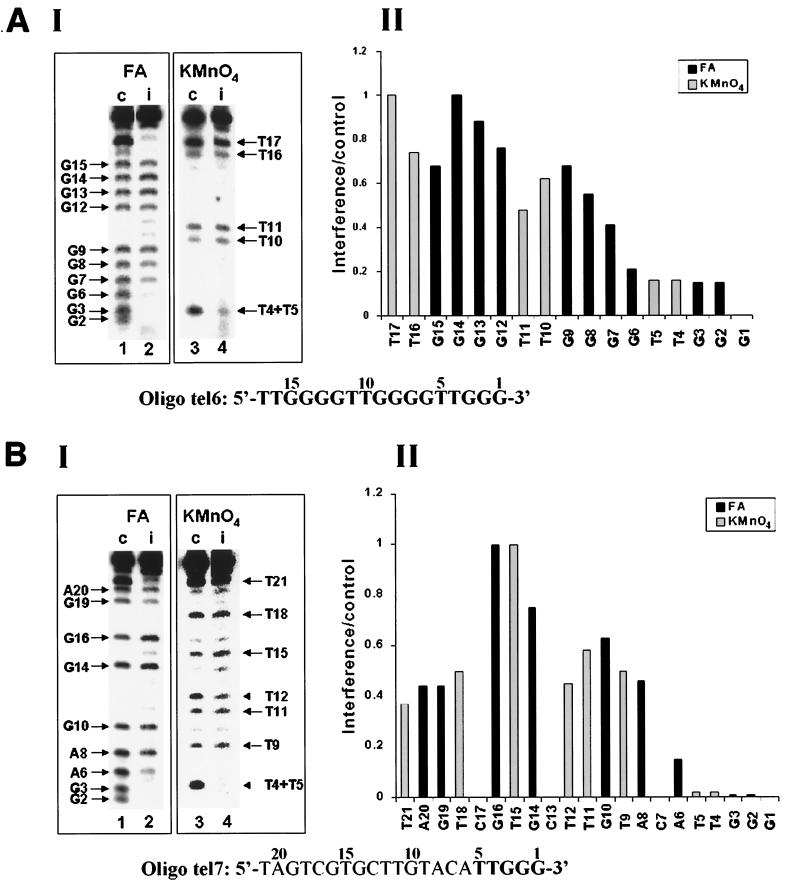
First, we studied the elongation of primers consisting of telomeric repeats exclusively. Attempts to extend a 23-nucleotide-long primer containing four telomeric repeats (and ending with GGG-3′) by a single [32P]dGMP residue revealed that the reaction was very strongly inhibited. The inhibition was probably due to formation of intramolecular four-stranded DNA structures (59). Therefore, we used for these assays a 17-base primer designated oligo tel6, which consisted of three telomeric repeats. It can be seen (Table 1) that the efficiency of elongation of this primer by the telomerase was threefold lower than that of oligo tel1. This result could be due to the formation of unusual DNA structures other than four-stranded DNA by oligo tel6. Alternatively, it could result from the difference in length between the two oligonucleotides, oligo tel6 (17 residues) being shorter than oligo tel1 (21 residues). Figure 6A shows formic acid and KMnO4 interference assays performed with oligo tel6. It can be seen that modifications of residues N2 to N6 caused strong interference with the elongation of this primer. The formic acid data also showed a gradual decrease in the degree of interference resulting from modifications of residues G7, G8, and G9. Overall, these data and the data obtained in the assays performed with KMnO4 indicated that only moderate to slight interference was caused by modifications of G and T residues located upstream of residue G6 in oligo tel6.
FIG. 6.
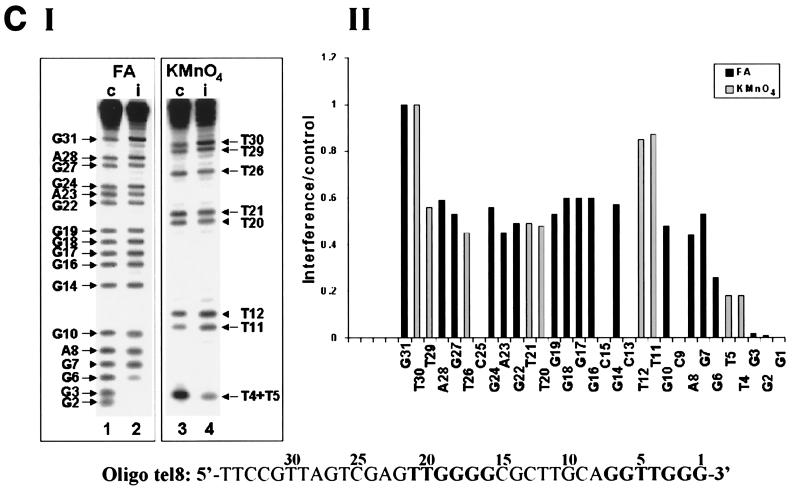
Interference footprinting assays performed with primers in which the number and distribution of telomeric repeats have been varied. Formic acid (FA) and KMnO4 interference assays were carried out as described in the legends to Fig. 3 and 4. (A) Formic acid (2.5%) assays and 0.10 mM KMnO4 assays of oligo tel6. (B) Formic acid (4%) assays and 0.12 mM KMnO4 assays of oligo tel7. (C) Formic acid (3%) assays and 0.15 mM KMnO4 assays of oligo tel8. In these assays, bands corresponding to residues 32 to 35 could not be resolved and hence were not quantitated.
We next studied the elongation of oligo tel7, which lacks upstream telomeric sequences. As Table 1 shows, the efficiency of elongation of oligo tel7 by a single G residue was about the same as that of oligo tel1. Figure 6B shows interference assays carried out with oligo tel7. It can be seen that strong interference was only observed when residues 2 to 6 were modified. Only slight to moderate interference was caused by modification of G, A, or T residues located further upstream in the nontelomeric sequences.
We have also studied the interference patterns obtained with a considerably longer oligonucleotide, oligo tel8, which contained two telomeric sequences at similar positions as in oligo tel1 and also contained a 14-base nontelomeric sequence at the 5′ end (Table 1). Figure 6C shows formic acid and KMnO4 interference profiles obtained in assays of oligo tel8 and the corresponding histogram. It can be seen that modification of each of the nucleotides G2, G3, T4, T5, and G6 strongly interfered with the elongation of oligo tel8 by a single G residue. Modification of other residues, including G7 and nucleotides in the upstream telomeric repeat, only caused a moderate to a slight interference with the reaction. These data and the data reported in the previous sections indicate that the upstream telomeric sequences do not play a major role in the elongation of properly aligned primers by a single dGMP residue.
Evidence that the interference footprint is shifted relative to the template during primer elongation by the telomerase.
We next performed formic acid and KMnO4 interference footprinting analyses of the elongation of oligo tel9, which consisted of the oligo tel1 sequence and two additional residues, GT, at the 3′ terminus (Table 1). For this analysis, oligo tel9, in which the residues were numbered 1′, 2′, 3′, etc., was extended by a single radioactively labeled dTMP residue, as indicated in Fig. 1B. Figure 7 (panel I) presents the footprints obtained in these assays. In these gels, the bands corresponding to residues T1′ and G2′ were not resolved well from the radioactively labeled dTTP precursor and could not be analyzed. Panel II shows the histogram of the relative interference values of the bands that were identified in panel I. It can be seen that modification of the residues included in the sequence extending between G3′ and T7′ caused substantial interference with the extension of oligo tel9. However, the interference that resulted from modification of T6′ and T7′ was not as strong as that caused by modification of G3′, G4′, and G5′. Slight to moderate interference was caused by modification of some residues located further upstream.
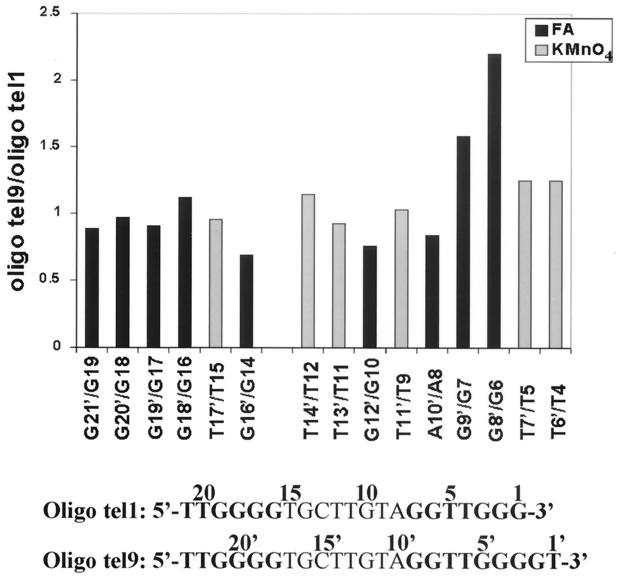
It was interesting to compare these data with the data obtained in the assays of oligo tel1. As Fig. 1 shows, oligo tel9 and oligo tel1 were aligned with the RNA template so that residues 1′, 2′, 3′, etc., in oligo tel9 were shifted downstream by 2 nucleotides relative to residues 1, 2, 3, etc., in oligo tel1. For example, G5′ in oligo tel9 corresponded to G3 in oligo tel1. We note that modification of residues G4′ and G5′ in oligo tel9 (Fig. 7) and of the corresponding residues G2 and G3 in oligo tel1 (Fig. 3) caused strong interference with the extension of oligo tel9 and oligo tel1, respectively. Also, the interference patterns resulting from modification of all the residues upstream of G9′ in oligo tel9 were similar to the interference patterns resulting from modification of the corresponding residues upstream of G7 in oligo tel1. Notwithstanding these similarities, it is evident that modification of residue G6 in oligo tel1 caused strong interference with the extension of this oligonucleotide, while modification of the corresponding residue G8′ in oligo tel9 caused a considerably smaller interference with its extension. This difference is clearly illustrated in Fig. 8, which displays the ratios of the interference values of corresponding residues in oligo tel9 and oligo tel1. The ratio G8′/G6 was significantly higher than the other ratios displayed in Fig. 8. The ratios T6′/T4, T7′/T5, and G9′/G7 were also slightly higher than the other ratios. It should be noted that two other independent assays performed with both oligonucleotides gave similar results. These data indicated that the footprint of the six or seven residues at the 3′ terminus was shifted downstream by 2 nucleotides in oligo tel9 relative to the corresponding footprint of oligo tel1. The significance of this observation will be discussed below.
FIG. 8.
Ratios of the relative interference values at corresponding positions along oligo tel9 and oligo tel1. The sequences of the two oligonucleotides are shown at the bottom. The formic acid (FA) and KMnO4 interference values observed at the indicated positions in oligo tel9 were divided by the corresponding values in oligo tel1.
DISCUSSION
We have presented in this paper an interference footprinting analysis of interactions of the Tetrahymena telomerase with single-stranded DNA primers in active enzyme-primer elongation complexes. Specifically, this analysis provided data on the degree to which chemical modifications of each individual nucleotide in the primers interfered with their extension by a single dGMP or dTMP residue. We surmise that the degree of interference is a measure of the functional significance of the interactions of each nucleotide in the primers with the telomerase RNP. It should be noted, however, that these assays did not reveal whether the modifications interfered with the binding of the primers to the enzyme or with the catalysis of phosphodiester bond formation.
Our experiments have shown that the six 3′-terminal nucleotides of all the primers that were used in this study strongly interacted with the telomerase in the elongation complexes. Substantial interactions were also observed with the seventh primer residue, but these interactions varied in the various primers. In primers that ended with the sequence TGGG-3′, the interactions detected by our footprinting assays presumably included Watson-Crick hydrogen bonds formed between these four nucleotides and the telomerase RNA residues CCCA at positions 47 to 50 (see Fig. 1A). This inference is based on previous studies of telomerase RNA mutants, which indicated that the RNA residues CCC (47 to 49) served as templating nucleotides and the A50 residue was needed for alignment of the primers at the active site (19, 21). However, those studies have also indicated that, in addition to the interactions with the RNA template region, the same primer residues also interact with the telomerase protein(s) or with other regions of the RNA. This notion is now supported by our observation that modifications of the three 3′-terminal G residues by DMS caused strong interference with the extension of the primers, even though these modifications (methylation of N7 in guanine residues) should not have affected the ability of the primers to form Watson-Crick hydrogen bonds with the templating nucleotides in the RNA. Interactions other than Watson-Crick base-pairing with residue 4 in the primers were also demonstrated by the footprinting assays of oligo tel2, in which the T4 residue has been replaced with A (Fig. 4A). In line with these observations, kinetic studies indicated that the binding of the E. aediculatus telomerase to primers involves limited base-pairing (4 to 10 bp) with the RNA template region and substantial interactions with a protein component of the enzyme (28).
We have also determined that the predominant interactions of the telomerase with residues 5, 6, and 7, found upstream of the 3′-terminal TGGG in these primers, were not hydrogen bonds of the Watson-Crick type (see Fig. 1). This conclusion was based on the observation that similar extents of interference occurred in assays of primers containing either T, G, or A residues at these positions, while only T's could form Watson-Crick hydrogen bonds with the A residues at the corresponding positions in the telomerase RNA.
Only weak interactions were observed between the telomerase and residues located upstream of nucleotide 7 in the primers containing a 3′ canonical Tetrahymena telomeric repeat. Furthermore, these interactions were not substantially different in a primer containing additional upstream telomeric repeats and primers containing nontelomeric sequences at the upstream positions. However, the interactions between the enzyme and the upstream sequences were considerably enhanced in oligonucleotides containing a mutated 3′ telomeric repeat. The enhancement was found to be most prominent in the assays performed with oligo tel2, but some enhancement was also observed in the assays of oligo tel3 and another related mutant oligo (see Fig. 4 and Results). These primers were apparently impaired in the ability to align with the RNA template region, and hence the overall efficiency of their extension was lower than the efficiency of extension of properly aligned primers (Table 1). We suggest that the enhanced interactions with the upstream sequences compensated for the impaired alignment of the 3′-terminal sequences. These data appear to be compatible with previous studies of the extension of primers which did not contain telomeric repeats at the 3′-termini that could anneal with the RNA template region before the first elongation cycle. The presence of upstream telomeric sequences increased the efficiency of extension of such primers by the Tetrahymena telomerase and by two other telomerases (33, 45, 48). Studies on the length dependence of the extension of entirely nontelomeric primers also indicated that interactions of the telomerase with upstream sequences play a significant role in their extension (55; for a review, see reference 13).
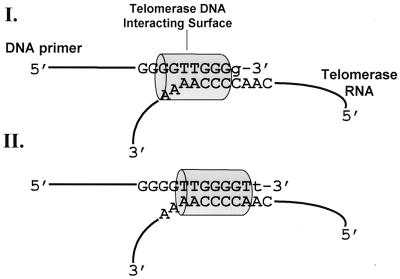
Another significant observation made in the present study was the shift in the footprint generated with oligo tel9 relative to the footprint generated with oligo tel1 (Fig. 1, 3, 7, and 8). We suggest the scheme shown in Fig. 9 to account for these data. According to this scheme, at stage I, a structural element in the telomerase RNP, which is illustrated as a cylinder and designated the telomerase DNA-interacting surface (TDIS), strongly interacts with the six 3′-terminal nucleotides (and to a lesser extent with the seventh nucleotide) in the primers. During elongation, TDIS moves along the primers and the RNA template region and attains stage II. At this stage, TDIS is also associated with the six or seven 3′-terminal nucleotides in the primers.
FIG. 9.
Scheme for primer elongation by telomerase based on the footprinting data. A structural element in the telomerase, which is schematically illustrated as a cylinder and designated the TDIS, strongly interacts with the six or seven 3′-terminal nucleotides in the primers. As the primers are elongated, TDIS moves in concert with their 3′ termini along the template region in the telomerase RNA (see the Discussion). g and t are newly added oligonucleotides (Fig. 1).
Although our footprinting assays did not reveal which part of the telomerase RNP interacts with the 3′-terminal nucleotides in the primers, it appears likely that the TERT subunit is involved in these interactions and that TDIS is a part of TERT or contains a part of TERT. The Tetrahymena TERT contains RT motifs, like the TERTs of other organisms (11, 14). Thus, it belongs to the RT subfamily of polymerases (12, 42). It is therefore interesting that in human immunodeficiency virus type 1 (HIV1) RT template-primer elongation complexes, the seven 3′-terminal nucleotides of the primers were found by footprinting assays to be protected against hydroxyl radical cleavage (46). A more detailed structural analysis revealed that a subdomain of this enzyme termed the minor groove-binding track interacts with the minor groove of the primer-template duplex along the second through the sixth base pairs from the 3′ primer terminus. It has been proposed that the minor groove-binding track may slide along the minor groove as the nascent strand is elongated (6). The similarity in the number and positions of strongly interacting primer residues in the elongation complexes of the telomerase and the HIV RT provides support for the notion that the two enzymes are structurally related.
However, there are apparent differences in the topography of the elongation complexes of the two enzymes. In the RT complexes, primer residues N2 to N6 and all the residues located further upstream form a duplex with the template. As Fig. 9 shows, of the corresponding N2 to N6 primer residues in the stage I telomerase complexes, which strongly interact with TDIS, only N2 to N5 can form duplexes with the template. N6 is not included in the duplex region. Furthermore, in some of the primers analyzed in the present study, N5 too is in a single-stranded region. This situation changes as TDIS moves along the template and attains stage II, in which all seven 3′ residues in the primers may form a duplex with the RNA. Clearly, in both stages I and II, the primer residues located upstream of N7 do not form a duplex with the template. Thus, unlike the minor groove-binding track in RT, the interactions of TDIS with the primers may vary as the latter are extended. These observations are consistent with the data of Wang et al. (56), which also indicated that the telomerase-primer interface may vary as the primers are extended and form a longer duplex region with the template.
The scheme shown in Fig. 9 implies that during the elongation phase of the primer extension process, two parts of the same telomerase molecule, the RNA template region and TDIS, move relative to each other. We suggest that a similar movement may also occur during the translocation phase, in which the telomerase active site retreats from the 5′ to the 3′ end of the template region before the onset of a new round of elongation (13, 23). This suggestion implies that while the Watson-Crick hydrogen bonds between the primer and the RNA template region are disrupted during the translocation, the primers remain anchored to the RNP through binding of the TDIS to some or all of the six or seven 3′-terminal nucleotide residues. Thus, the translocation mechanism suggested here may not require binding of a second site in the telomerase to primer residues located upstream of nucleotide N7. This type of movement would be analogous to the coordinated retreat of the Escherichia coli RNA polymerase and the transcription bubble that occurs at certain transcription pause sites (36, 50). Clearly, further work is required to test this hypothesis.
ACKNOWLEDGMENTS
Sima Benjamin and Nava Baran made equal contributions to the work reported in this article.
This study was supported by grant 96-417 from the United States-Israel Binational Science Foundation and by grant 980003-B from the Israel Cancer Association through the Ber-Lehmsdorf Memorial Fund.
We thank Elizabeth H. Blackburn, David Gilley, and He Wang for advice and helpful comments. We also thank James A. Borowiec for critical reading of the manuscript.
REFERENCES
- 1.Autexier C, Greider C W. Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev. 1994;8:563–575. doi: 10.1101/gad.8.5.563. [DOI] [PubMed] [Google Scholar]
- 2.Autexier C, Greider C W. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 3.Autexier C, Pruzan R, Funk W D, Greider C W. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 4.Baran N, Pucshansky L, Marco Y, Benjamin S, Manor H. The SV40 large T-antigen helicase can unwind four stranded DNA structures linked by G-quartets. Nucleic Acids Res. 1997;25:297–303. doi: 10.1093/nar/25.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 6.Bebenek K, Beard W A, Darden T A, Li L, Prasad R, Luton B A, Gorenstein D G, Wilson S H, Kunkel T A. A minor groove binding track in reverse transcriptase. Nat Struct Biol. 1997;4:194–197. doi: 10.1038/nsb0397-194. [DOI] [PubMed] [Google Scholar]
- 7.Bednenko J, Melek M, Greene E C, Shippen D E. Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J. 1997;16:2507–2518. doi: 10.1093/emboj/16.9.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya A, Blackburn E H. A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc Natl Acad Sci USA. 1997;94:2823–2827. doi: 10.1073/pnas.94.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn E H, Greider C W. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 10.Blasco M A, Funk W, Villeponteau B, Greider C W. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- 11.Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cech T R, Nakamura T M, Lingner J. Telomerase is a true reverse transcriptase: a review. Biochemistry (Engl Transl Biokhimiya) 1997;62:1202–1205. [PubMed] [Google Scholar]
- 13.Collins K. Ciliate telomerase biochemistry. Annu Rev Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- 14.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins K, Greider C W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 16.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 17.Counter C M, Meyerson M, Eaton E N, Weinberg R A. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng J, Funk W D, Wang S S, Weinrich S L, Avilion A A, Chiu C P, Adams R R, Chang E, Allsopp R C, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 19.Gilley D, Blackburn E H. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilley D, Blackburn E H. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Natl Acad Sci USA. 1999;96:6621–6625. doi: 10.1073/pnas.96.12.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilley D, Lee M S, Blackburn E H. Altering specific telomerase RNA template residues affects active site function. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 22.Greider C W. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greider C W. Telomerase biochemistry and regulation. In: Blackburn E H, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 35–68. [Google Scholar]
- 24.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 25.Greider C W, Blackburn E H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 26.Greider C W, Blackburn E H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 27.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 28.Hammond P W, Cech T R. Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry. 1998;37:5162–5172. doi: 10.1021/bi972988o. [DOI] [PubMed] [Google Scholar]
- 29.Hammond P W, Lively T N, Cech T R. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrington L, Hull C, Crittenden J, Greider C. Gel shift and UV cross-linking analysis of Tetrahymena telomerase. J Biol Chem. 1995;270:8893–8901. doi: 10.1074/jbc.270.15.8893. [DOI] [PubMed] [Google Scholar]
- 31.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 32.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D K, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrington L A, Greider C W. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 34.Holt S E, Aisner D L, Baur J, Tesmer V M, Dy M, Ouellette M, Trager J B, Morin G B, Toft D O, Shay J W, Wright W E, White M A. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klobutcher L A, Swanton M T, Donini P, Prescott D M. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komissarova N, Kashlev M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc Natl Acad Sci USA. 1998;95:14699–14704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee M S, Blackburn E H. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee M S, Gallagher R C, Bradley J, Blackburn E H. In vivo and in vitro studies of telomeres and telomerase. Cold Spring Harbor Symp Quant Biol. 1993;58:707–718. doi: 10.1101/sqb.1993.058.01.078. [DOI] [PubMed] [Google Scholar]
- 39.Licht J D, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingner J, Cooper J P, Cech T R. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 41.Lingner J, Hendrick L L, Cech T R. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 42.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 43.Makarov V L, Hirose Y, Langmore J P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 44.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 45.Melek M, Greene E C, Shippen D E. Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metzger W, Hermann T, Schatz O, Le Grice S F, Heumann H. Hydroxyl radical footprint analysis of human immunodeficiency virus reverse transcriptase-template primer complexes. Proc Natl Acad Sci USA. 1993;90:5909–5913. doi: 10.1073/pnas.90.13.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 48.Morin G B. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 50.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 51.Ramakrishnan S, Sharma H W, Farris A D, Kaufman K M, Harley J B, Collins K, Pruijn G J M, Vanvenrooij W J, Martin M L, Narayanan R. Characterization of human telomerase complex. Proc Natl Acad Sci USA. 1997;94:10075–10079. doi: 10.1073/pnas.94.19.10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romero D P, Blackburn E H. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 55.Wang H, Blackburn E H. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–879. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Gilley D, Blackburn E H. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–1160. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wellinger R J, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 58.Wright W E, Tesmer V M, Huffman K E, Levene S D, Shay J W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zahler A M, Williamson J R, Cech T R, Prescott D M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 60.Zakian V A. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]