Highlights
-
•
Adhesion and autophagy synergistically regulate one another.
-
•
These processes function at the intersection of protection and degeneration.
-
•
Understanding overlapping molecular functions may uncover therapeutic strategies.
Keywords: Adhesion, Autophagy, Extracellular matrix, Huntington’s disease, Integrin, Neurodegenerative disease
Abstract
Cellular adhesive connections directed by the extracellular matrix (ECM) and maintenance of cellular homeostasis by autophagy are seemingly disparate functions that are molecularly intertwined, each regulating the other. This is an emerging field in the brain where the interplay between adhesion and autophagy functions at the intersection of neuroprotection and neurodegeneration. The ECM and adhesion proteins regulate autophagic responses to direct protein clearance and guide regenerative programs that go awry in brain disorders. Concomitantly, autophagic flux acts to regulate adhesion dynamics to mediate neurite outgrowth and synaptic plasticity with functional disruption contributed by neurodegenerative disease. This review highlights the cooperative exchange between cellular adhesion and autophagy in the brain during health and disease. As the mechanistic alliance between adhesion and autophagy has been leveraged therapeutically for metastatic disease, understanding overlapping molecular functions that direct the interplay between adhesion and autophagy might uncover therapeutic strategies to correct or compensate for neurodegeneration.
Introduction
While early descriptions by Golgi and Cajal provided an elegant framework that formed the foundation of our understanding of neural cells within the brain [1], we now know that the cells that make up the brain don’t do so on their own, but are rather suspended within a dynamic extracellular matrix (ECM) [2]. This complex extracellular network guides critical cellular functions and is essential for supporting homeostasis, working in conjunction with autophagic machinery to maintain cellular balance. Autophagy is the process of trafficking cytoplasmic components to the lysosome for degradation or recycling to sustain energetic demands and remove cellular waste [3].
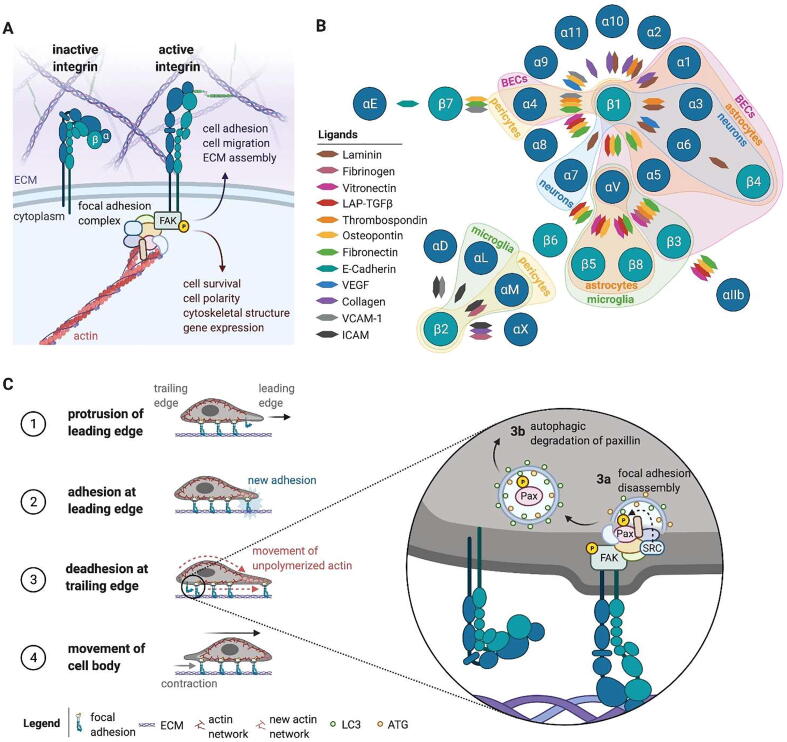
The interaction between cells and the ECM acts to direct critical cellular processes, such as cell migration, cell fate choice, and even cell survival. Cell-matrix anchorage is primarily mediated by integrins, a large family of transmembrane surface receptors named for their ability to integrate the intracellular milieu with the extracellular surroundings (for review [4]). α and β subunits heterodimerize to connect the extracellular environment to the actin cytoskeleton via adaptor molecules and kinases that form adhesive structures (Fig. 1A). Focal adhesions are the dynamic, large, sub-cellular protein complexes that form as a result of integrin-matrix adhesion that act to produce, transmit, and sense mechanical forces to potentiate signaling initiated from outside the cell [5]. Proteins are recruited in a hierarchical manner to the intracellular focal adhesion complex through a phosphorylation cascade mediated by the integrin-associated non-receptor kinases focal adhesion kinase (FAK) and integrin-linked kinase (ILK). Most integrins form focal adhesions through FAK association, where upon integrin activation the most immediate downstream molecular event following matrix adhesion is the recruitment of FAK and its autophosphorylation at tyrosine 397 [6]. Various integrins are expressed in different cell types and tissues throughout the brain [7] (Fig. 1B) and the ECM-integrin interaction regulates neurodevelopmental programs, including lumen formation [8], vascular patterning [9], and angiogenesis [10]. Additional reports indicate that integrin signaling and focal adhesions are dysregulated in neurodegenerative disease [11], [12], [13], suggesting a broader role for integrins, focal adhesion dynamics, and cell-matrix adhesion in disease that will be discussed in greater detail throughout this review.
Fig. 1.
Cell-type specific integrin-mediated adhesion directs intracellular and extracellular signaling, cell-matrix anchorage, and cell motility. (A) Integrins are a large family of α/β heterodimeric transmembrane receptors that are the primary mediators of cell-matrix anchorage. They exist in a conformational flux between inactive and active states, where dimers either adopt a bent/closed (left) or an extended/open (right) conformation, respectively. Only in their active, open states are integrins able to bind extracellular ligands that induce the formation of intracellular adhesion complexes at the cell surface, such as focal adhesion complexes. The most immediate downstream molecular event following matrix adhesion by most integrins is the recruitment of FAK and its autophosphorylation at tyrosine 397. FAK phosphorylation then instigates a cascade of hierarchical protein recruitments that form the focal adhesion complex, linking the focal adhesion to the cytoskeleton. This focal adhesion structure is stabilized by the mechanical forces from outside the cell, promoting outside-in signaling that relays signals necessary for cell survival, cell polarity, and activation of gene expression. In turn, the stability of the intracellular adhesion complex promotes inside-out signaling that creates the forces necessary for cell adhesion and cell migration. (B) Specific integrin α subunits form dimers with specific β subunits, with at least 24 combinations of heterodimeric pairs. This establishes receptors that are specific for various extracellular ligands. However, integrin-matrix adhesion is promiscuous, with most integrins recognizing several matrix proteins and an individual matrix protein serving as the ligand for multiple integrin dimers. Additionally, since the ECM environment is tissue specific, integrin expression is necessarily cell-type specific to bind the unique extracellular environments of cell types associated with those tissues. We have highlighted the known adhesion dynamics and integrin expression profiles associated with specific cell types of the brain. BEC = brain endothelial cells; LAP-TGFβ = latency-associated peptide-TGFβ, E-cadherin = epithelial cadherin, VEGF = vascular endothelial growth factor, VCAM-1 = vascular cell adhesion molecule-1, ICAM = intracellular adhesion molecule. (C) Dissolution of the intracellular adhesion complex, in part mediated by autophagic degradation, enables cellular motility. As a cell moves forward (1), integrin dimers at the cell’s leading edge make cell-matrix adhesions, promoting the formation of intracellular focal adhesion complexes and allowing for adhesion at the leading edge (2). To move forward, focal adhesion complexes at the trailing edge are disassembled to allow disengagement of integrin dimers with the ECM (3). Intracellularly, this can occur through selective autophagy, where the cytoskeletal protein paxillin (Pax) that links the focal adhesion complex to the cytoskeleton is targeted for removal by LC3-positive autophagosomes (3a). The removal of Pax from the adhesion complex breaks the mechanical forces promoting outside-in signaling that stabilize the adhesion complex, allowing integrins to disengage from the ECM and dissolving the adhesion complex (3b), allowing for forward movement of the cell body (4).
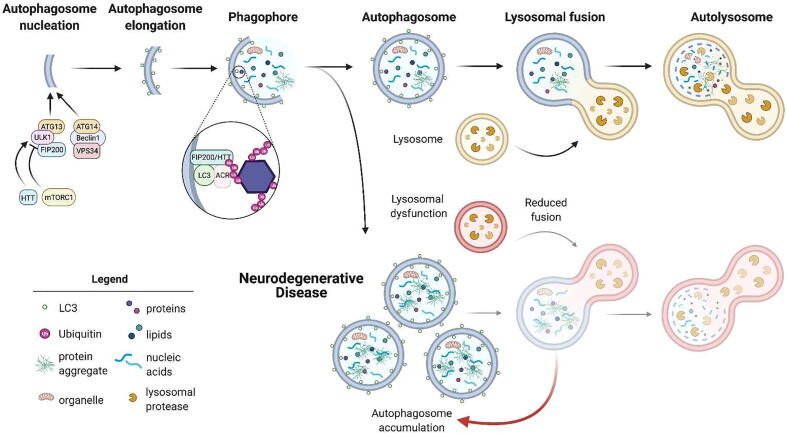
There are several mechanisms of autophagy (for review [14]). This review is focused primarily on macroautophagy, referred to simply as autophagy from here, which is a conserved catabolic process that maintains cellular homeostasis [14]. Autophagy entails enveloping cytoplasmic contents in a double-sided membrane, called an autophagosome, that is then transported to and fuses with the lysosome to deposit its contents [14] (Fig. 2). Fusion with the lysosome forms an autolysosome, which allows lysosomal proteases to degrade the autophagosome content for recycling of cellular components to form other proteins Alternatively, autophagic cargo modulated by Hsc70 can be directly targeted to the endosome/lysosome by direct invagination through microautophagy or by chaperone-mediated autophagy requiring the lysosomal membrane protein Lamp-2A [15]. Furthermore, autophagy can be non-selective or selective. Selective autophagy, under nutrient-rich or starvation conditions, can target specific proteins, such as misfolded, aggregated proteins and dysfunctional compromised organelles that characterize many neurodegenerative diseases, to the lysosome for degradation. The selection of specific autophagic targets can be facilitated by autophagy cargo receptors (ACRs), which often contain a ubiquitin-binding domain to bind ubiquitinated cargo, and an LC3-interacting region to connect these targets with the autophagosomal membrane [16], but may also function to cluster cargo in autophagosomes in an LC3- and ubiquitin-independent manner [17]. Fidelity of autophagic pathways is critical for maintaining cellular health, particularly in post-mitotic cell types of the brain that cannot dilute out cellular waste through division. As such, autophagy is the most significant mechanism that cell types of the brain have for removing compromised organelles and pathogenic, misfolded proteins that aggregate in neurodegenerative diseases. However, these same diseases have robust demonstrations of aberrant autophagic flux that contribute to pathophysiology [18]. Throughout disease progression, the balance of autophagic flux tips. While increased autophagy is beneficial in early disease, stress-related pathogenic mechanisms, including oxidative stress, hypoxic stress, and DNA damage, further increase the autophagic response despite reduced lysosomal function with aging, causing an accumulation of autophagosomes and a reduction of lysosomal fusion; late disease-stage therapies may therefore require inhibition of the autophagic response to maintain balance. This aberrant regulation of autophagic balance throughout disease progression demonstrates the requirement for understanding and maintaining balance of autophagic pathways in neurodegenerative disease [19]. Since autophagic stimulation has proven effective in clearance of aggregated disease-causing proteins in animal models [18], the discovery and development of biological processes that stimulate the autophagic response early in disease progression could serve as an effective therapeutic modality for these diseases, while inhibition of autophagy may be protective in late neurodegenerative disease and to slow some types of cancer cell proliferation. Throughout this review, we highlight how autophagy may be modulated for therapeutic gain in neurodegenerative disease by assessing the regulatory reciprocity between cellular adhesion and autophagy.
Fig. 2.
Macroautophagy in health and neurodegenerative disease Macroautophagy envelopes cytoplasmic contents in a double-membraned vesicle for transport to and fusion with the lysosome for degradation and recycling. Autophagosome formation begins with nucleation, followed by elongation, where the membrane becomes decorated with LC3 molecules. FIP200 scaffolds the autophagy initiation complex, facilitating Ulk1 kinase activity required for autophagosome nucleation. Sequence homology suggests that HTT may have a similar function. As the double-membraned vesicles elongates, it captures cellular components for degradation, either through selective or non-selective pathways. Selective autophagic pathways allow for the capture of compromised organelles and protein aggregates. Selective cargo slated for degradation by the autophagosome can be tagged with the post-translational modification ubiquitin and interact with LC3 at the autophagosome membrane, a process mediated by autophagy cargo receptors (ACR). Once fully formed, the autophagosome fuses with lysosomes to allow for degradation of the autophagosome contents by lysosomal proteases. These degradation products are then released back into the cell, where they are recycled for the formation of other proteins. In some neurodegenerative disease at later stages, protein aggregates accumulate, which may cause aberrant autophagic flux that leads to the accumulation of autophagosomes. This occurs because autophagosomes have reduced fusion with lysosomes because of disease-associated lysosomal dysfunction. Together, these alterations tip the balance of the autophagic pathway, contributing to a buildup of toxic, aggregated proteins.
Not only does the ECM and cell adhesion regulate the autophagic response, but autophagy in turn regulates adhesion dynamics, forming a synergistic partnership between these critical cellular functions in health and disease. While the interplay between cell adhesion and autophagy is better studied in non-CNS-related cell types and neoplastic diseases, it is emerging as an important regulator of biological functions that direct neuroprotection and neurodegeneration [20]. This review highlights the connection between cellular adhesion and autophagy and how each regulates the other in cell types of the brain, with a focus on neurodegenerative disease. An overview is provided of molecular mechanisms that regulate ECM- and adhesion-mediated autophagy, molecular events that govern autophagy-regulated cell adhesion, and intersecting molecular pathways that work cooperatively between adhesion and autophagy. Lastly, the merits and caveats of treating disease by modulating the autophagic response through the ECM are considered.
Adhesion-regulated autophagy
While autophagy primarily functions at a molecular level to recycle cellular components and provide balanced cellular homeostasis, autophagy induced by the ECM functions independently from nutrient demands or energetic requirements [21]. The extracellular regulation of the autophagic response can occur directly by ECM molecules or through mechanisms mediated by integrins that regulate cell-matrix anchorage. This section will detail the regulation of autophagy by the ECM and adhesive molecules.
Matrix-directed autophagy
Dysregulation of autophagic flux can contribute to neurodegenerative phenotypes [22], [23], and ECM remodeling that occurs either as a result of age or pathology appears to be a major determinant for developing dementias and neurodegenerative disease [24], [25]. Recent reports have shown that ECM dysregulation can not only directly contribute to neural dysfunction, but can also potentiate autophagic dysregulation in neurodegenerative disease, exacerbating the degenerative effect.
Both neurons and glia contribute to the ECM of the brain, which is encompassed by a structure called the perineuronal net (PNN) [26]. The PNN is primarily bound by proteoglycans, which promote and inhibit synaptic remodeling through neurite growth to protect against neurodegeneration under normal molecular programs [25], [27], [28]. In the striatum, the importance of the PNN for regulation of striatal function has been demonstrated through experiments that show enzymatic removal of the striatal PNN leads to abnormalities in mouse gait and Morris water maze performance [24]. CNS injury or disease causes a significant increase in chondroitin sulfate proteoglycans (CSPGs), which are a major component of the PNN [29]. In rodent models of spinal cord injury, increased CSPG signaling regulates autophagic flux at the axon growth cone by reducing autophagosome-lysosome fusion. This reduction in the autophagic response leads to axon regeneration failure, demonstrating matrix-directed contributions to autophagic function in the CNS that maintain a degenerative response following CNS injury [29].
Proteoglycan regulation of autophagy may also contribute to pathophysiology of neurodegenerative disease [30]; in Alzheimer’s mouse models with both tau and amyloid precursor protein mutations, enzymatic CSPG digestion by chondroitinase ABC ameliorated memory deficits by increasing neuronal plasticity [29]. In Alzheimer’s mouse primary cortical neurons treated with tau oligomers from Alzheimer’s disease (AD) patient brains, reduction of heparan sulfate proteoglycan synthesis or function prevents tau oligomer internalization, mediates AD-related autophagic alterations, and inhibits tau-related neuronal cytotoxicity [31]. Proteoglycans have also been connected to pathology in Huntington’s disease (HD). HD is a progressive, autosomal dominant, neurodegenerative disease caused by a genetic expansion of a CAG repeat within the huntingtin (HTT) gene, referred to as mutant HTT [32]. In addition to striking neuronal loss, HD also results in microglial activation that contributes to neuroinflammation. In the rapidly-progressing, late-stage R6/2 HD mouse model, a reduction in proteoglycan-containing PNNs contributes to volumetric brain loss. However, drug-induced microglial depletion in R6/2 mice globally increases CSPGs within the brain while partially restoring HD-associated aberrant protein clearance systems [33]. Pharmaceutical microglial depletion occurs through colony-stimulating factor 1 receptor (CSF1R) inhibition, and while the direct mechanistic relationship between CSF1R microglial depletion and global CSPG increases within this system was not assessed, the authors hypothesize it may be a direct or indirect mediation by microglia via astrocyte reactivity [33]. While these studies overall demonstrate that proteoglycans can be both harmful and beneficial and have differing effects on autophagy, these effects could vary based on disease, disease model, tissue/cell type, type of autophagy at play within the system, and/or proteoglycan family or specific proteoglycan, further highlighting the need for increased study in this area. However, taken together, these studies suggest that proteoglycans in the brain participate in regulating autophagic balance that may contribute to pathophysiology of both AD and HD.
Matrix proteins, such as collagen, that act as ligands for transmembrane receptors also aid in autophagic regulation from outside the cell. While collagens are widely distributed in many tissues [34], [35], [36], [37], their expression in the brain is less common, with a few exceptions for collagens expressed by neurons. These neuronal collagens support functional roles rather than structural ones, with participation in axonal guidance, synaptogenesis, and differentiation [38]. Collagen VI serves a neuroprotective function in AD in both mice and patients [39]. A 2016 report demonstrates a connection between collagen VI, autophagy, and neurodegeneration – collagen VI null mice (Col6a1-/-) display motor and memory deficits and increased levels of p62 that co-localized with ubiquitin-positive aggregates in brain sections [40], similar to phenotypes observed in motor neuron disease patients [41], [42]. Col6a1-/- brain sections had increased LC3-II/actin ratio and Beclin 1 protein levels and increased signs of neurodegeneration, and primary cortical and hippocampal neurons from these Col6a1-/- mice display increased apoptosis and an impaired autophagic response with accumulation of the autophagic receptor p62 and decreased lipidated LC3, suggesting that ECM molecules can regulate the autophagy-lysosome system which in turn may contribute to neurodegenerative phenotypes [40].
Integrin activation and autophagy
Expression and activity of integrins have been shown to regulate autophagic function in neural models of neurodegenerative disease. Using SH-SY5Y 1-Methyl-4-phenylpyridinium ion (MPP+)-treated cells to model Parkinson’s disease, neurotoxicity occurred in conjunction with reduced expression of integrins αV and β3 and an increased LC3-II to LC3-I ratio, suggesting an increase in the autophagic response upon integrin reduction [43]. Previous work from this group showed that MPP+ treatment increases oxidative stress to induce cell death through changes in the insulin signaling pathway, depriving cells of this signaling molecule that is highly enriched in the brain and that maintains neural functions critical for brain health and promoting neuroprotection, such as glucose metabolism and neurotransmission [44]. In the MPP+-treated SH-SY5Y cells, pre-treatment with insulin had a neuroprotective effect that was observed concomitantly with increased expression of integrins αV and β3, ILK activation via AKT phosphorylation, and autophagic induction, implicating the cooperative effects of integrin activation and nutrient-rich autophagic induction under neuroprotective conditions [43].
Recently, a role for integrins in protein clearance of misfolded tau was described [45]. The integrin αV/β1 dimer is critical for endocytosis of recombinant extracellular tau aggregates by primary mouse astrocytes. Only in the presence of this integrin dimer can tau aggregates be phagocytosed by astrocytes, eliciting inflammatory signaling pathways that lead to pathogenic astrocytic activation [45]. While aggregate phagocytosis by astrocytes in neurodegenerative disease initially aids in facilitating clearance of misfolded proteins released by neurons [46], [47], the eventual inability to clear this toxic buildup in later disease stages leads to proteotoxic neurodegeneration through cytokine and chemokine release in various diseases [48], [49]. While the direct role of autophagy was not assessed as it relates to integrin-mediated astrocytic phagocytosis of tau aggregates, this work highlights the role that integrins play as receptors for protein clearance of pathogenic, misfolded proteins in neurodegenerative disease – an area that deserves further study given the contemporary interest in understanding how astrocytes convert to a neurotoxic state and the therapeutic potential of blocking integrin-mediated endocytosis to prevent astrocytic activation.
Deficits in the cerebrovascular system and blood–brain barrier (BBB) are a notable feature of many neurodegenerative diseases [50], [51], [52], [53], [54], [55], which precede and may even promote neurodegeneration [55], [56], [57], [58]. Induction of autophagy that prevents matrix detachment-induced cell death has been demonstrated in a rat model of cerebral ischemic stroke as well as human brain microvascular endothelial cells [59]. When human microvascular endothelial cells were treated with methylglyoxal, a chemical that induces endothelial cell injury and dysfunction, FAK phosphorylation increased and an autophagic response was mounted, as evidenced by increased LC3-II conversion [59]. Similar results were demonstrated in vivo. These results suggest that the protective effect of autophagy induced upon injury to brain endothelial cells is associated with FAK activation and thus integrin activation.
Anoikis resistance through autophagy
Most cells undergo apoptosis upon detachment from the ECM. The requirement of cell anchorage for survival was first described in 1993 using fibroblasts [60] and termed “anoikis” in 1994 [61], adopting the Greek term for homelessness to describe this unique form of detachment-induced apoptosis. Anoikis is critical for organ development, providing molecular cues that allow for the clearance of luminal spaces [62], [63]. Molecular evidence for anoikis has been described in a multitude of tissues and cell types, including cell types of the brain, with the demonstration that excitotoxic death is reduced in hippocampal neurons in vitro upon laminin adhesion [64], [65] and in vivo hippocampal neurons undergo anoikis upon laminin degradation [66]. Cellular detachment from the ECM results in programed cell death unless survival is molecularly promoted. One such mechanism that is robustly induced following cell-matrix detachment to promote survival is autophagy [67], [68].
The first report that an autophagic response was mounted following anoikis came in 2002 when studying lumen formation in mammary tissue [63]. Organoids were used to determine that neither apoptotic inhibition nor proliferation enhancement prevented lumen formation and microscopic analysis revealed many cytoplasmic vacuoles, suggesting the induction of autophagy contributes to lumen formation following anoikis. Subsequent reports described an inverse relationship between integrin activation and autophagic response by examining hypoosmotic swelling in rat livers [69], [70], specifically implicating loss of integrin-mediated adhesion in autophagic induction as a cellular survival strategy [71], [72]. In mammary epithelial cells, antibody inhibition of integrin β1 in attached cells is sufficient to induce autophagy, while providing suspended cells a laminin-rich ECM suppresses autophagy, suggesting that integrin-mediated adhesion inhibits autophagic function [71]. When these cells are detached from matrix, caspase-3 activation, and thus decreased survival, results from RNAi-mediated knockdown of the ATG proteins ATG5, ATG6 (Beclin 1), and ATG7, critical for an autophagic response. Together, these results suggest that disengagement of integrins from the ECM would lead to anoikis, however the cell’s response to induce autophagy sufficiently evades anoikis and promotes cell survival [71]. These findings highlight the necessary balance in autophagic flux necessary for cellular survival regulated by adhesion dynamics.
The role of anoikis resistance in neurodevelopment has only recently been described, examining the neurogenic niche in mice at the ventricular zone during cerebral cortex development [73]. Under a normal developmental paradigm, progenitor cells adhere to one another at the apical side of the ventricle and give rise to daughter neuroblasts that delaminate, altering their transcriptional profile as adhesion is lost to escape anoikis. When adhesion is disrupted at the apical surface, either through developmental errors or for pathological reasons, aberrant detachment and delamination occurs and developmental anoikis is initiated. This acts to prevent neurogenic defects that may result from progenitors with retained proliferative capacity residing outside of the neurogenic niche, which could lead to future brain malformations. Interestingly, aberrant neurodevelopment was recently described at the ventricular zone in HD using human embryonic, cortical tissue [74]. In HD, adhesion proteins are mislocalized in the ventricular zone and the number of progenitor cells is decreased while the pool of cells committed to a neuronal lineage is increased. It is hypothesized that these neurodevelopmental defects increase the vulnerability of the corticostriatal circuitry, contributing to the later neuronal dysfunctions in HD. Recent findings regarding developmental anoikis [73] suggest that aberrant adhesion dynamics as a result of mutant HTT expression may be regulating neurodevelopmental deficits in HD. HTT regulates cell–cell adhesion [75], and expression of mutant HTT may reduce cell–cell adhesion at the apical surface of the ventricle, leading to aberrant delamination and an increased propensity for neurogenesis. Uncovering and identifying molecular mechanisms that direct aberrant neurogenic programs in diseases like HD may help to identify mechanisms that can be leveraged for compensation later in life. While current work doesn’t define a link between developmental anoikis and autophagy, novel findings that either corroborate or invalidate anoikis resistance through autophagy in neurodegenerative disease could improve pathology not only by compensating for neurodevelopmental deficits, but also by stimulating clearance of protein aggregates that characterize many of these diseases.
Autophagic regulation of cell adhesion
The molecular players of focal adhesions form the complex that is primarily responsible for cellular motility by mediating intracellular connection to the extracellular matrix via integrins. These complexes are highly dynamic, continuously assembling at the leading edge and disassembling at the trailing edge to allow for forward movement of the cell (Fig. 1C) – a mechanism first described in 1971 by examining the leading edge of fibroblast motility with electron microscopy [76]. Since then, various pathways have been identified that disband adapter proteins that form integrin-mediated adhesion complexes to promote the destabilization of focal adhesions. While several mechanisms contribute to focal adhesion disassembly, including calpain-mediated proteolysis and microtubule-dependent transport [77], this section will focus on the role of autophagic turnover of focal adhesions.
Autophagic regulation of cell motility and adhesion dynamics
Not only does autophagy function downstream of the ECM through cells eliciting an autophagic response following cell-matrix detachment, but autophagy also functions upstream of integrin-mediated cell adhesion by regulating cellular migration via focal adhesion remodeling [78]. However, this relationship appears to be complex and various studies have elicited contradictory results. For example, some reports suggest that autophagic induction promotes cellular migration [79], [80], [81], [82] and others find that migration correlates with autophagic reduction [78], [83]. Reduction of Atg5 and Atg7 increases migration in HeLa cells, mouse embryonic fibroblasts [78], and endothelial progenitor cells [83]. In glioblastoma multiforme stem cells, the autophagy-associated proteins p62 and DRAM1 regulate cell motility and invasion of this aggressive brain cancer [81]. Additionally, autophagy functions to recycle focal adhesions to promote cellular migration, with autophagic inhibition leading to reduced migration [80] and focal adhesion enlargement [79]. In cancer cells, several reports have established autophagic regulation of adhesion dynamics through selective autophagy, which directly targets focal adhesion proteins, such as paxillin and Src kinase to LC3-positive autophagosomes [67], [79], [80]. Mechanistically, this breaks the intracellular focal adhesion connection with the actin cytoskeleton, allowing for cellular movement, as the adhesion complex protein paxillin acts to scaffold the adhesion complex to the cytoskeleton (Fig. 1C). Taken together, these reports suggest that autophagic regulation of cell adhesions appears to be cell type specific and context dependent [80], [84] or be differentially regulated by the type of autophagy that is induced or inhibited.
While much of the understanding of autophagic turnover of focal adhesion comes from metastatic cell types (for review [85]), focal adhesion dynamics are known to play critical roles for health and survival in cell types of the brain [86]. Cues that allow or prevent migration alter levels of autophagy-associated molecules in neuroblasts of tissue slices from adult mouse forebrain [87]. Compared to non-migrating cells, migrating neuroblasts expressed high levels of Atg5 and LC3-II, correlating the formation of autophagosomes with cell migration. The well-described inhibitor of autophagy, Bafilomycin A1 [88], as well as genetic depletion of Atg5 suppressed migration of neuroblasts in vivo in mice, implicating an autophagy-dependent mechanism for cell migration [87]. Similar to non-neuronal studies [79], [80], autophagic regulation of neuronal migration occurs via LC3-mediated recycling of paxillin, with reduced motility correlating with decreased paxillin recycling and an increased autophagosome to autolysosome ratio [87]. Interestingly, modulating neuroblast migration conversely influenced autophagy by inducing changes in LC3-II [87], demonstrating an inextricable link between the critical cellular processes of cellular motility and autophagy that allow them to act in concert to regulate and maintain cellular homeostasis.
FIP200 – An autophagy protein that regulates FAK-mediated adhesion
A major player in autophagic function is FIP200 (FAK family-interacting protein of 200 kDa). FIP200 is essential for the formation of autophagosomes as part of the autophagosome initiation complex (Fig. 2), which also includes Ulk1/Ulk2 and Atg13 [89], [90], [91]. In this initiation complex, FIP200 scaffolds the remodeling of portions of the endoplasmic reticulum (ER) into cup-like structures, which then recruit autophagosome elongation factors to generate an autophagosome [92]. FIP200 is also proposed to have sequence homology with the yeast autophagy scaffolds Atg17 and Atg11 [93]. Atg17 scaffolds non-selective, starvation-induced autophagy, whereas Atg11 scaffolds selective autophagy by binding to cargo-bound autophagy receptors. FIP200 appears to have a sequence more similar to Atg11 [94], and indeed, is required for the selective degradation of the ER in mammals as Atg11 is in yeast [95].
FIP200 is also required for normal neuronal development and function. FIP200 ablation in mouse radial glial cells resulted in postnatal progressive loss of neural stem cells and impairment of neuronal differentiation caused by apoptosis and cell cycle arrest [96]. The first report to link FIP200 to autophagic regulation of neuronal homeostasis in vivo deleted FIP200 selectively in neuronal cells of mice, demonstrating that loss of FIP200 leads to several degenerative phenotypes in the cerebellum, such as progressive neuronal loss, gliosis, neurite degeneration, increased apoptosis, and spongiosis [97], or intercellular edema, that may reflect extracellular matrix dysregulation. Additionally, ubiquitinated p62-positive protein aggregates accumulated in neurons while autophagosomes were not detected, suggesting aberrations in autophagic flux in neurons following FIP200 deletion [97].
Interestingly, prior to its reported contribution in autophagy, FIP200 was identified for its regulatory roles in FAK Inhibition [98]. FIP200 directly binds FAK in vitro and in vivo, inhibiting the phosphorylation event [98] that occurs following integrin-mediated adhesion [6]. Autophagy serves to inhibit focal adhesions by inhibiting FAK phosphorylation through the Atg13/Ulk1/FIP200 complex to promote a stationary cellular state in tumor cells [99]. The association between FIP200 and FAK is decreased following integrin-ligand association, allowing for FAK phosphorylation and progression of the downstream adhesion signaling cascade [6], [98], suggesting this key autophagy protein is critical in regulating FAK-mediated adhesion. While, to the best of our knowledge, no report has assessed the interplay between the adhesion and autophagic pathways as mediated by FIP200 in cell types of the brain, this relationship is well established in metastatic cell types [98], [99] and many of the degenerative cellular phenotypes described following FIP200 deletion in neurons have additionally been linked to adhesion-related deficits (anoikis) [61], reduced cell–cell adhesion [100], [101], and adhesion-mediated roles in axonal outgrowth and pathfinding [102], [103]), warranting future study of this area.
Huntingtin as an autophagic scaffold that modulates adhesion
An in vivo genome-wide single cell genetic screen in the CNS of the R6/2 exon 1 fragment and zQ175 full-length HTT HD mouse models was performed to identify genetic modifiers of mutant HTT toxicity [13]. Genes identified as being essential for neuronal viability in these models were assessed using KEGG and GO analysis, which identified genes associated with integrin-mediated signaling and focal adhesion terms as being protective against mutant HTT toxicity [13]. In an in vitro cell death assay using mouse striatal neurons expressing mutant HTT, knockdown of integrin αIIB reduced levels of a highly toxic small mutant HTT fragment [12]. These results suggest that integrin modulation may have therapeutic relevance for reducing mutant HTT toxicity. While the mechanism of mutant HTT toxicity in relation to integrin αIIB expression was not assessed, integrin reduction may function to stimulate an autophagic response that aids in clearance of toxic mutant HTT fragments. Together, these findings implicate a disease-specific role for integrins in HD pathophysiology and one may hypothesize that there may perhaps be an autophagic benefit for therapeutically targeting this large family of adhesion proteins.
In addition to FIP200, the HTT protein has been proposed to be a mammalian Atg11-like autophagy scaffold [94]. HTT is a large protein that interacts with many autophagy proteins, including autophagy receptor p62 and autophagosome initiation complex component ULK1 [104], [105]. Conditional knockout of HTT in the mouse CNS results in a significant accumulation of p62 and ubiquitin puncta [105], and in HTT loss-of-function Drosophila, p62 and ubiquitinated proteins accumulate in brain tissue from old animals [104]. There is some evidence that HTT plays a role in autophagosome cargo loading, and that HTT loss-of-function results in the formation of empty vesicles [104], [106]. HTT also interacts with the dynactin complex to facilitate transport of vesicles [107], [108], including autophagosomes. The association of HTT with cytoskeletal components may also be important for adhesion; HTT depletion in human fibroblasts reduces adhesion and alters cellular morphology. HTT staining localizes to adhesion contacts, membrane ruffles, and actin stress fibers [109]. Similarly, in Dictyostelium HTT is necessary for cell adhesion during starvation [110].
In the HdhQ111 full-length HTT knock-in mouse model, mutant HTT created a deficit of ATP, which reduced levels of energy-sensitive N-cadherin, a transmembrane protein that functions to mediate cell–cell adhesion, with age [111]. N-cadherin is ubiquitously expressed in the neural tube during development [112] and later functionally supports neurite outgrowth, axon guidance, dendritic arborization, and early synaptogenesis [113]. Later in development, N-cadherin localization is restricted to synapses [114], where it serves adhesive functions [115], [116], [117]. Unlike in cell-matrix detachment-induced anoikis, N-cadherin-mediated survival was not dependent on AKT activation. Rather the Erk1/2 MAP kinase pathway was engaged and the pro-apoptotic protein Bim was reduced [118]. Since Bim repression increases autophagosome production [119], this mechanism may serve to activate autophagy when N-cadherin adhesion is reduced. Under wild type conditions, HTT has an evolutionarily conserved role in adhesion through interaction with the metalloproteinase ADAM10 that cleaves N-cadherin, perhaps evolving as a regulatory mechanism to control neural adhesion for brain development [75]. In primary striatal neurons from HdhQ111 mice, reduced cell clustering and cell adhesion resulted from decreased N-cadherin mRNA expression and increased N-cadherin protein turnover, which in turn resulted in decreased neurite maturation [111]. In HD patient brains, ADAM10 aberrantly accumulates at the synapse where increased cleavage of N-cadherin is observed, ultimately leading to decreased extracellular adhesion and the loss of synaptic contacts [120]. These results suggest that the overlap between adhesion and autophagy participates in neurodegeneration in HD, perhaps through engagement of the ERK pathway and Bim repression to activate selective autophagy scaffolded by HTT that in turn affects adhesion mediated by N-cadherin, causing maturation abnormalities and structural defects at the synapse.
Integrin endocytosis and trafficking
Integrin trafficking (for review [121]), the process by which integrins are redistributed around and throughout a cell by endocytic/exocytic recycling [122], [123], [124], is critical for maintaining dynamic cell-ECM contact. Drosophila embryos were used to demonstrate that the primary mechanism of integrin trafficking for βPS, the primary β integrin in flies, is internalization rather than lateral diffusion within the membrane [125]. Integrins are initially internalized through several distinct mechanisms, classified under either clathrin-mediated or clathrin-independent endocytosis [121]. While the downstream molecular events involved in various routes of integrin internalization have been assessed and detailed, the discriminatory factors that determine initial internalization routes have not been fully elucidated. Several reports implicate the importance of extracellular cues by way of integrin mechanosensing for internalization and trafficking [126], [127] as well as the identity of the integrin dimer itself and available substratum [128]. As such, subsequent studies are required to determine the molecular events that direct initial integrin internalization. However, both integrin α and β subunits contain common endocytosis motifs [129], [130] that suggest the mechanisms involving integrin endocytosis are more ubiquitous than many of the adhesion-r elated mechanisms previously described.
Downstream from the initial internalization, clathrin or non-clathrin vesicles fuse with endosomes. Ultimately, integrin trafficking is dependent on the Rab family of small GTPases [122], [123], [131], [132]. Rab proteins are central to the elegant, organized eukaryotic endomembrane system that allows for exo- and endocytosis, bridging the exchange between the extra- and intracellular compartments [133]. With approximately 60 members in the Rab family, these proteins serve as mediators of the endocytic process [133]. Specific Rab proteins determine the recruitment of effector proteins, acting as endosomal zip codes that guide the shuttling of endosomes and their contents to specific locations throughout the cell [133]. Integrin-containing endosomes are shuttled by Rab proteins for sorting [122], [123], [131], [132]. Upon vesicle/endosome fusion, integrins are contained in Rab5-positive early endosomes [134] where the fate of endocytosed integrins is determined. They are either destined for recycling back to the plasma membrane or for lysosomal degradation. If integrins are fated for recycling, the contents transit to Rab11-positive or Rab4-positive recycling endosomes [135], [136], [137] that are preferentially exocytosed to the plasma membrane at sites of focal adhesions [138]. If integrins are fated for degradation, the endosomal contents transit to Rab-positive late endosomes which fuse with lysosomes [121] by way of autophagosomes [139], [140], [141], [142]. Recent work shows that integrins can also serve as receptors for a process called LC3-associated phagocytosis, a form of non-canonical autophagy. Integrin αM serves as a receptor to detect Listeria monocytogenes and initiate recruitment of LC3 to Listeria-containing phagosomes, which enhances degradation and prevents intracellular replication [143]. Similarly, αV or β3 integrin subunits in B cells direct TLR trafficking to the lysosome by recruiting LC3 to TLR-containing endosomes [144], [145].
While there is known overlap between cellular machinery that participates in the endosomal and autophagic pathways [146], such as Rab-mediated regulation of the merger of the autophagosome and the lysosome to form the autolysosme [147], evidence for autophagic contribution in endosomal trafficking is only recently coming to light. The intersection between the endosomal trafficking and autophagic pathways may converge on Rab11-positive recycling endosomes. In HEK293 cells, the autophagy proteins ULK1 and Atg9 co-localize with Rab11-positive endosomes [148]. Endosomal co-expression of TBC1D14 (TBC1 domain family member 14) blocks ULK1-mediated autophagic degradation, unless starvation is induced. Upon starvation, TCB1D14 dissociates and allows for ULK1-mediated autophagic induction.
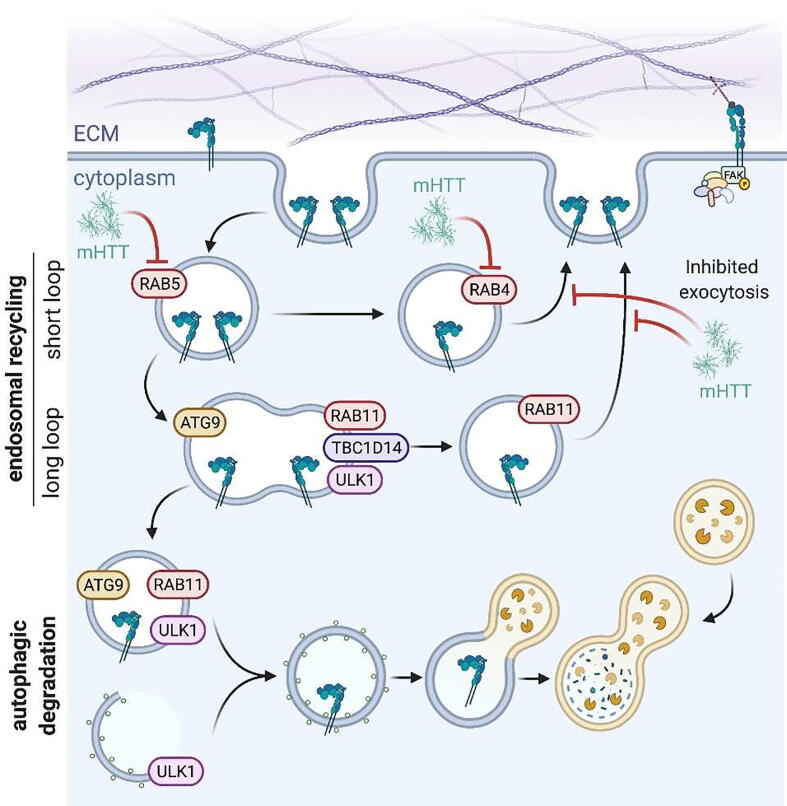
Autophagic participation in integrin trafficking has been demonstrated in vitro using embryonic neural crest cells [128] that give rise to several cell types, including neurons and glia of the CNS [149]. Integrin α6 is intracellularly trafficked in neural crest cells via Rab4-positive, Rab11-positive, and Rab4-/Rab11-positive vesicles for return to the cell surface to aid in rapid cellular movement [128]. When treated with Bafilomycin A1, cell migration is slowed and integrin trafficking is inhibited [128], suggesting contribution of the autophagic pathway in integrin trafficking in neuronal cell types. Rab11 also aids in integrin β1 trafficking in adult rat dorsal root ganglion neurons to support axon growth, perhaps indicating a neuroregenerative capacity for Rab11-directed integrin trafficking [150]. Indeed, in HD, there is a disruption in Rab11-mediated exocytosis in various models (Fig. 3), including Drosophila [151], [152], [153], HttQ74-expressing PC12 cells [153], HD patient-derived fibroblasts [154], HD140Q knock-in mouse model [154], and the transgenic, rapidly-progressing R6/2 mouse model [153]. Interestingly, in these models, Rab11 overexpression was sufficient to restore endosomal trafficking pathways and rescue HD-associate phenotypes. Specifically, phenotypes associated with integrin-mediated functions were restored, such as dendritic spine development in the hippocampus [153], [155], resistance to glutamate-induced neuronal death [64], [156], and amelioration of synaptic dysfunction [152], [157]. Furthermore, Rab11, HttQ47, and LC3 co-localized in amphisomes in dendrites of primary rat hippocampal neurons, connecting deficits in autophagy and Rab11-mediated exocytosis in HD [153]. Rab11 dysregulation also influences pathology in other neurodegenerative diseases, notably Alzheimer’s and Parkinson’s [158].
Fig. 3.
Rab-mediated integrin endocytosis and trafficking The Rab family of small GTPases directs integrin-containing endosomes for trafficking, where specific Rab proteins label endosomes for recycling back to the cell surface or for fusion with autophagosomes for degradation. In a healthy brain, immediately upon being endocytosed integrin-containing vesicles are labeled with Rab5. If they are to be recycled back to the cell surface, they either transit to Rab4- or Rab11-positive endosomes prior to returning to the surface at sites of focal adhesions. When co-labeled with ATG9, ULK1, and TBC1D14, endosomes are blocked from autophagic degradation. However, the removal of TBC1D14 from Rab11-positive endosomes slates them for fusion with autophagosomes for degradation. In a brain with neurodegenerative disease, Rab-mediated integrin trafficking is disrupted. Expression of mutant HTT (mHTT) disrupts endosomal transport of Rab5- and Rab4-positive vesicles. Additionally, Rab11-mediated exocytosis is disrupted in HD. However, endosomal trafficking pathways can be restored by Rab overexpression, rescuing pathological phenotypes at the dendrites and synapses.
Mutant HTT disrupts the functions of other Rab proteins via effector proteins [159], including Rab5 that is critical for integrin trafficking [160], [161]. More recently, Rab4 has been implicated in HD pathogenesis [162]. In patient-derived neurons differentiated from iPSCs and Drosophila larva, expression of mutant HTT perturbs the axonal motility of HTT-containing Rab4 vesicles, and HD Drosophila larva demonstrate aberrant movement, defects in synaptic morphology, and reduced lifespan, phenotypes all rescued by Rab4 overexpression [162]. These results suggest that expression of mutant HTT causes defects in HTT-mediated Rab4 vesicle transport to the synapses, which may alter integrin trafficking at these areas with susceptibility of early degeneration in HD. Previous knowledge about the intersection between Rab-mediated endocytosis/exocytosis, integrin trafficking, and autophagy makes it tempting to speculate that HD-related deficits involving Rab4 and Rab11 may be converging on the relevant, associated phenotypes directed by adhesion and autophagy in HD.
Intersecting pathways and cellular machinery
Key molecular pathways that intersect between integrin-mediated cell adhesion and autophagy have been nicely reviewed elsewhere, with a particular interest in the participation of these signaling events in cancer and the metastatic process [163]. Here, we review the intersection of selected pathways between autophagy and adhesion to highlight the relevance of this integration to the brain and neurodegenerative disorders. While a number of pathways have overlapping functions in adhesion and autophagy, few have been studied in cell types of the brain. As such, some seemingly obvious pathways are absent from our discussion, such as WNT and AMPK, since literature detailing the interplay between adhesion and autophagy in these pathways in cell types of the brain is lacking. Here we provide examples of pathways with a particular focus on the cooperative mechanisms that exist between adhesion and autophagy in cell types of the brain.
mTOR
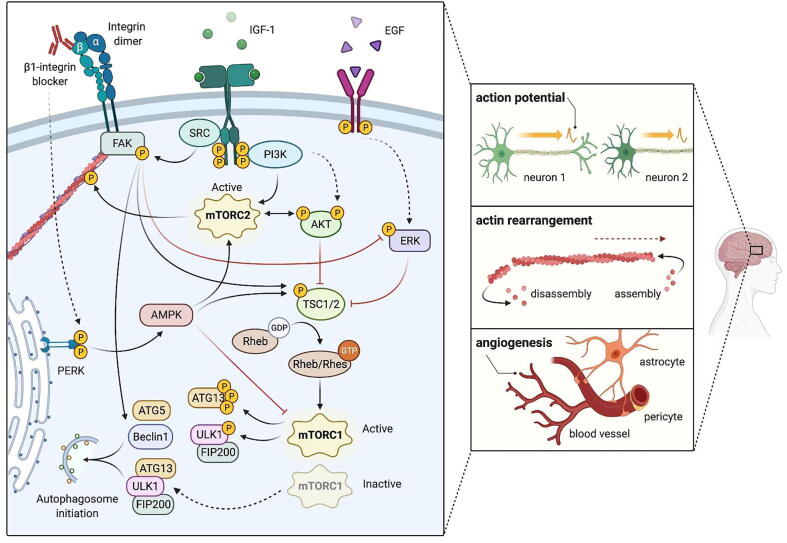
mTOR is a master regulator of growth and autophagy [164], [165], [166] and emerging evidence suggests that mTOR signaling can also regulate cellular adhesion, the intersection of which may mediate neuronal activity. mTOR is activated in conditions of nutrient and energy sufficiency, either by sensing abundance of intracellular nutrients, such as amino acids and cholesterol within lysosomes, or by extracellular ligands such as insulin or insulin-like growth factor-1 (IGF-1). In these nutrient-rich conditions, the autophagic starvation response is not necessary, therefore activated mTOR associates with the mTOR complex 1 (mTORC1). This complex suppresses autophagy at the level of the autophagosome initiation complex through two mechanisms (Fig. 2). First, it phosphorylates Atg13 to reduce its association with Ulk1. Second, it phosphorylates and thus represses the kinase activity of UlK1, which is required for the activity of the autophagosome initiation complex [167]. Functioning as an autophagic scaffold, wild type HTT may regulate the interaction of the mTORC1 complex with ULK1 to activate autophagy, a function that may become impaired with HTT mutation in HD(Rui et al., 2015). While HTT does not affect the kinase activity of mTORC1, it was found to compete with it for binding with ULK1. Starvation-independent stresses that induce selective autophagy like proteotoxicity, lipotoxicity, or mitotoxicity may result in an increased association of ULK1 with HTT at the expense of mTOR, thus freeing ULK1 from its inhibition by mTORC1 to activate autophagy under nutrient-rich conditions. mTOR may also associate with the mTORC2 complex, which may upregulate autophagy in some contexts, such as chaperone mediated autophagy in the neuroblastoma cell line SH-SY5Y and expanded autophagy in response to influenza A infection in MDCK cells [168], [169], [170]. A major function of mTORC2 is in the phosphorylation of the actin cytoskeleton [171]. Elimination of the mTORC2 complex causes thick, disconnected cytoplasmic actin fibers, similar to stress fibers, with reduced cortical actin. These cytoskeletal changes alter cellular morphology, with HeLa cells appearing flatter and square-like.
The mTOR complexes 1 and 2 (found as TOR complexes in yeast) are highly conserved [172] and can be found in neurons, where they regulate growth as in other types of cells [173], as well as contributing to neuronal-specific processes such as postsynaptic responsiveness (mTORC1) and presynaptic neurotransmitter release (mTORC2)[174]. Several studies have shown that mTOR signaling contributes to long-term potentiation, and mTORC2 in particular is required for long-term potentiation through its effect on actin polymerization and rearrangement in neurons [175], [176], [177]. mTOR is also expressed in astrocytes and microglia [178]. In neurodegenerative disease, clearance of protein aggregates and dead cells may stress the lysosomal system in glia [179], [180], making proper mTOR signaling critical. Dysregulation of mTOR signaling found in neurological disease has been previously studied and reviewed [181], [182], [183], [184]; for example, several studies have found that mTOR signaling is hyperactive in AD, which may suppress autophagy and lysosomal degradation of β-amyloid [185], [186], [187], [188], [189]. In HD, mTOR dysregulation is complex [190] and may depend on the stage of disease. Mutant HTT may promote mTOR signaling [191], and the mTOR inhibitor Rapamycin has been found to have therapeutic potential in models of HD [192]. An mTOR activating GTPase expressed specifically in the striatum, Rhes [193], binds to mutant HTT and induces its SUMOylation, which causes cytotoxicity [194], [195], again suggesting that upregulation of mTOR signaling may contribute to disease. However, mTOR may also become hypoactive late in disease as it is sequestered into mutant HTT aggregates [196], [197].
The mTOR signaling cascade has recently been shown to intersect with adhesion on several levels (4). IGF-1, an mTOR signaling pathway ligand, activates adhesion through both direct activation of β1-containing integrin dimers and through AKT signaling upstream of mTOR [198], [199]. Direct inhibition of either mTORC1 or mTORC2 reduces adhesion in a variety of cell types including hepatocellular carcinoma, cardiomyocytes, rhabdomyosarcoma cells, and monocytes [200], [201], [202], [203]. A proteomic screen of cells with reduced mTORC2 revealed that adhesion is significantly dysregulated [204], consistent with the role of mTORC2 in regulating the actin cytoskeleton. In mammary epithelial cells, the disruption of cell adhesion via a β1 integrin-blocking antibody leads to robust activation of AMPK, which activates mTORC2 and inhibits mTORC1 activity [205]. Finally, FAK phosphorylates a component of mTORC1 for its positive regulation [206].
Several studies have suggested that mTOR signaling may regulate adhesion specifically in neuronal cells. PC12 cells require mTOR/cadherin signaling for adhesion to extracellular matrix [207]. Reduction of mTOR complex activity results in reduction of neural cell adhesion molecule (NCAM) at neuromuscular junctions, resulting in muscle fiber denervation [208]. Finally, mTORC1 hyperactivation results in upregulation of genes associated with cellular adhesion in iPSC-derived neurons [209].
Erk
The ERK pathway intersects with the mTOR pathway (Fig. 4) and impacts both adhesion and autophagic function to participate in critical process for maintaining brain health, such as action potential, actin rearrangement, and angiogenesis, while regulating a response to neural insult. This is done through a signaling cascade initiated by extracellular signals, like Epidermal Growth Factor (EGF), that facilitates the communication of plasma membrane receptors with intracellular targets to regulate proliferation, differentiation, and survival through TSC1/2, an upstream inhibitor of mTORC1 [210]. In a rat subarachnoid hemorrhagic stroke model, nasal delivery of the ECM molecule osteopontin enhanced autophagy, resulting in reduced neuronal apoptosis and improved neurobehavior [211]. Mechanistically, osteopontin was shown to activate FAK phosphorylation, which in turn decreased the levels of ERK1/2 phosphorylation and increased expression of Beclin 1, ATG5, and the LC3-II to LC3-I ratio while decreasing p62 in neurons. These effects were abrogated with a FAK inhibitor, demonstrating a direct correlation between ECM-induced integrin activation through the FAK/ERK cascade to mediate autophagic flux, likely through disinhibition of TSC1/2 to reduce mTORC1 activity and allow for increased autophagy. However, given that different types of autophagy share core machinery, such as Beclin 1, ATG5, and LC3, no conclusions can be drawn about the type of autophagy at play in this system and autophagic balance that changes throughout disease progression to regulate neuroprotection and neurodegeneration must be considered. Though neurodegenerative diseases characterized by protein aggregation have documented autophagic flux aberrations [18] and the reports highlighted here show that exogenous addition of ECM molecules can be used to potentiate autophagy through the ERK pathway, the connection between integrin activation, ERK signaling, and autophagy has not been studied in neurodegenerative diseases to our knowledge. Nonetheless, the findings detailed above justify future studies exploring this topic in models of neurodegenerative disease.
Fig. 4.
mTOR and ERK signaling in adhesion and autophagy mTOR is a master autophagic regulator, activity of which is activated under nutrient-rich conditions. When energetic demands are sufficient, active mTOR associates with the mTORC1 complex to suppress autophagy by phosphorylating Atg13 and Ulk1 to reduce Atg13/Ulk1 association, which blocks the autophagosome initiation complex. Starvation inactivates mTORC1, preventing phosphorylation of Atg13 and Ulk1 and allowing for their association and initiation of autophagosome formation. mTOR also associates with the mTORC2 complex, which can upregulate chaperone-mediated autophagy. Active mTORC2 phosphorylates the actin cytoskeleton to aid in polymerization and rearrangement, which facilitates neuronal potentiation. Additionally, mTOR activity intersects with adhesion. IGF-1 is an upstream activator of mTOR signaling that activates adhesion through integrins and AKT signaling. In various cell types, inhibition of both mTORC1 and mTORC2 reduces adhesion, perhaps in part through the role mTORC2 plays in stabilization of the actin cytoskeleton. Additionally, integrin-targeting antibodies activate AMPK signaling that leads to mTORC1 inhibition and mTORC2 activation. ERK signaling, which can be initiated by extracellular signals such as Epidermal Growth Factor (EGF), intersects with the mTOR pathway by inhibiting TSC1/2, an upstream inhibitor of mTORC1. FAK phosphorylation inhibits ERK, disinhibiting TSC1/2, which reduces mTORC1 activity to increase autophagic activity. These signaling pathways aid in functions related to action potential, actin rearrangement, and angiogenesis within the brain. Solid arrows are indicative of a direct interaction, dashed arrows an indirect interaction.
Transglutaminase
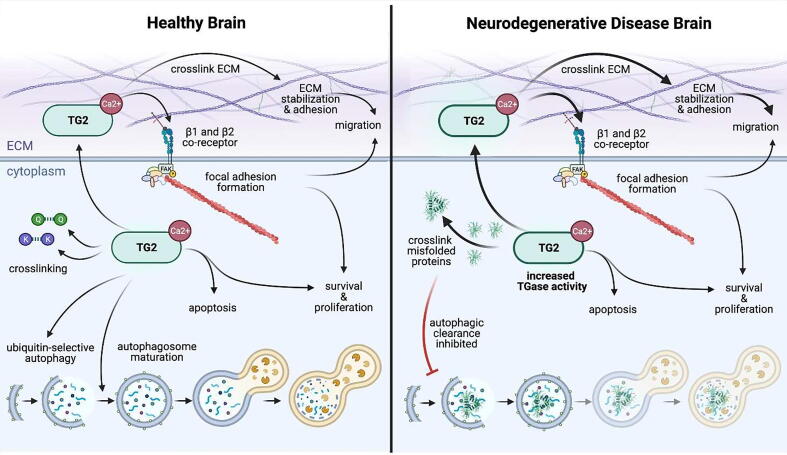
Signaling involving transglutaminase appears to regulate neuroinflammation through involvement with both autophagy and adhesion (Fig. 5). Transglutaminases are Ca2 + -dependent enzymes that cross-link glutamine and lysine residues [212]. Type 2 transglutaminase (TG2) separates itself from other transglutaminases in that it catalyzes additional biochemical reactions, including acting as a serine/threonine kinase [213]. While TG2 is localized both intra- and extracellularly, its localization dictates function [214]. Intracellularly, TG2 serves to modulate apoptosis, both positively and negatively [215], and is involved in autophagolysosome maturation [216]. In TG2 knockout mice, preautophagic vesicles accumulate, indicating an induction of the autophagic response [217], and ubiquinitated proteins accumulate, specifically implicating a role for TG2 in ubiquitin-mediated selective autophagy [218].
Fig. 5.
Intra- and extracellular signaling mediated by transglutaminase 2 in health and disease. Transglutaminases are calcium-dependent enzymes that cross-link glutamine (Q) and lysine (K) residues and are found both intra- and extracellularly, where localization, specifically of TG2, dictates function. In a healthy brain (left), intracellularly, TG2 modulates apoptosis and is involved in autophagolysosome maturation and ubiquitin-mediated selective autophagy. Extracellularly, TG2 cross-links ECM molecules to support cell-matrix stabilization and acts as an integrin co-receptor to mediate adhesion. Integrin-mediated adhesion potentiated by TG2 aids in focal adhesion formation, creating mechanical forces that allow for cellular migration and potentiating intracellular signaling related to cell survival. In neurodegenerative disease (right) where transglutaminase activity is increased (bold arrows), TG2 cross-links disease-causing protein aggregates. As a known autophagic inhibitor, TG2 may be contributing to pathology by cross-linking aggregation-prone proteins and reducing mechanisms for aggregate clearance. Solid arrows are indicative of a direct interaction.
The crossover between TG2 activity and autophagy has a significant role in protein aggregation and accumulation in neurodegenerative disease. TG2 cross-links various proteins associated with neurodegenerative disease that are prone to aggregation in the disease state, including β-amyloid, α-synuclin, tau, and HTT [219]. Additionally, transglutaminase activity is increased in patient brains associated with these diseases. These findings, combined with the TG2 function as an autophagic inhibitor [219], suggest that aberrant TG2 activity may be at least one of the mechanisms reducing autophagy in these diseases to contribute to pathogenesis – cross-linking aggregation-prone versions of disease-causing proteins and reducing autophagic mechanisms that would aid in aggregate clearance, further tipping protein homeostasis and autophagic balance in a pathogenic direction. Additionally, the disease-causing proteins themselves may contribute to aberrant TG2-mediated autophagy. In the case of HD, wild type HTT associates with TG2 in brain tissue, which together may target oligomerized, ubiquitinated proteins to the lysosome for degradation [220]. A reduction in this function by expression of mutant HTT may thus subsequently block autophagy. However, at least in HD, TG2 modulation for therapeutic purposes has variable effects in different mouse models, with some showing improvement upon TG2 knockout [221], [222] and others showing no effect [223], [224]. While not explored in these studies, this perhaps suggests the involvement of different kinds of autophagy and differences in autophagic balance requirements at various disease stages.
At the cell surface and extracellularly, TG2 cross-links ECM molecules to aid in cell-ECM stabilization [225] and adhesion [226]. It has also been reported to act as a co-receptor of β1 and β2 integrins through fibronectin to mediate adhesion and spreading and potentiate integrin signaling in fibroblasts [226], [227]. TG2 overexpression enhances adhesion by enlarging focal adhesions and increasing FAK phosphorylation, a process that is dependent on integrin association. Concordant results have been found in other cell types: in endothelial cells, reducing TG2 reduced adhesion and spreading on fibronectin [228], [229] and in melanoma cells, TG2 aids in stabilizing cell adhesion [230].
In cell types of the brain, TG2 has also been shown to interact with fibronectin and may contribute to neurodegenerative disease in a similar manner to its described extracellular function in endothelial and melanoma cells. In primary rat astrocytes, TG2 interacts with fibronectin to stimulate migration and the formation of focal adhesions following treatment with pro-inflammatory cytokines [231]. In neuroinflammatory disorders, such as multiple sclerosis (MS), glial scarring that results from astrocyte migration at demyelinated lesions may be the result of this mechanism. In fact, astrocytes associated with active and chronic MS lesions have increased TG2 positivity and localize with fibronectin [232], which demonstrates excessive accumulation at MS lesions [233], [234] and may therefore serve as a catalyst of elevated TG2 activation at these sites. Neuroinflammation activated in MS can normally be negatively regulated, in part, by autophagy [235]. However, this process is dysregulated in MS lesions where expression of Lamp2 and the LC3 ratio is decreased, suggesting the autophagic process, likely occurring through chaperone-mediated autophagy, at the site of MS lesions is aberrantly reduced [235]. Thus, aberrant ECM deposition at these lesions and concomitant deficits in autophagic flux may contribute to pathological hallmarks of neuroinflammation, highlighting a connection between TG2-regulated cell adhesion and autophagy in maintenance of brain health. However, to date, no study has explicitly examined the interplay between the intra- and extracellular functions of TG2 as it relates to adhesion and autophagy, respectively, to mediate pathological effects of neurodegenerative disease.
Primary cilia
The involvement of primary cilia in extracellular environment and autophagy [236], [237] as it relates to brain health and disease has been increasingly considered in the past several years [238], [239]. Primary cilia are non-motile microtubule-based sensory structures at the cell surface, acting as antennae that sense changes in the extracellular space to guide the cellular reaction to its environment.
The autophagic pathway has reciprocal regulation with primary cilia – each directing the biological function of the other. Primary cilia act to regulate the autophagic response by serving as a recruitment site for ATG proteins to mediate the formation of autophagosomes [236]. Mouse embryonic fibroblasts (MEFs) and kidney epithelial cells were used to show that functional primary cilia and active Hedgehog signaling are required for autophagosome formation under starvation conditions [240], suggesting that primary cilia participate in the regulation of autophagosome biogenesis. Conversely, autophagy controls ciliogenesis by degrading ciliary proteins; basal autophagy degrades proteins essential for ciliogenesis in MEFs and MCF7 cells, allowing autophagy to regulate primary cilia length in response to extracellular stimuli [240], [241].
While the regulatory crossover between primary cilia and autophagy is well documented, the overt connection between these processes in cell types of the brain has only recently been described. In the brain, primary cilia are found on progenitors, neurons, and astrocytes [242] where these structures receive input from neurotransmitters, neuromodulators, growth factors, and morphogens [238], [239]. In particular, primary cilia help to direct development of the cerebral cortex, participating in axon pathfinding, neuronal migration, neurogenesis, and circuit formation [238], [239]. Aberrant structure and/or function of primary cilia leads to a host of diseases referred to as ciliopathies that typically present with intellectual disability, brain malformations, and ataxia [238], [239]. Dysfunction of primary cilia has been most extensively studied in neurodevelopmental disorders [243] where disrupted ciliogenesis can be caused by autophagic perturbations resulting from genetic dysregulation of the mTOR pathway [244]. Primary cilia changes have also been described in neurodegenerative diseases [245], [246], [247]. In HdhQ111 mice and HD patient tissue, primary cilia length is increased in ependymal cells, altering the flow of cerebrospinal fluid [246]. Additionally, in human fetal tissue that carries the HD mutation, cilia length and number is increased in the developing cortex [74]. Primary cilia have also been implicated in Parkinson’s disease pathology in an MPP+-induced mouse model, where MPP+-treated animals have elongated cilia in neurons of the substantia nigra that fail to undergo autophagic induction and are more susceptible to apoptosis [248]. Together, these findings suggest that neurodegenerative disease-related changes in primary cilia regulated by autophagy directly lead to pathology.
Merits and caveats of treating disease via extracellular autophagy modulation
The interplay between cell adhesion and autophagy may represent a therapeutic interface for neurodegenerative disease, as neuropathological therapeutics may be feasibly designed to take advantage of ECM-regulated autophagic function. While autophagic flux has well-documented dysfunction in a variety of neurodegenerative diseases [18], modulating the extracellular environment to control the tunable matrix-integrin interaction could allow for autophagic calibration that may be leveraged to increase turnover of aggregated proteins for therapeutic benefit. The autophagic response to extracellular cues reinforces the idea that novel therapeutics for neurodegenerative pathologies could be delivered using matrix-based modalities [249]. An example of the therapeutic utility of this approach was cited earlier, where nasal delivery of the ECM molecule osteopontin activated autophagy to reduce neuronal apoptosis and improve neurobehavior in a rat stroke model [211]. As such, understanding ECM composition and adhesion dynamics in wild type and neurodegenerative disease conditions could be advantageous for treatment of neurodegenerative diseases, such as those mentioned here.
While the extracellular space is more accessible than plasma membrane-protected intracellular components, dosing of the neural matrix space requires passage across the BBB. Relevantly, deficits in the BBB are a significant feature of neurodegenerative diseases [50], [51], [52], [53]. Patients with Alzheimer’s and Parkinson’s have reduced glucose transport [57], [250], [251] and the reduced ability to remove pharmaceuticals and xenobiotics via P-glycoprotein transport, demonstrating BBB transport deficits [252], [253]. In HD patients, MRI shows that increased disease burden score positively correlates with BBB permeability [254], which is observed before atrophy and extensive neurodegeneration [55], [56], [57]. Additionally, post-mortem HD brains show capillary leakage [254], aberrant angiogenesis [51], [254], and disruption of tight junction protein expression in brain endothelial cells [254], suggesting autonomous deficits in the cell type primarily responsible for barrier fidelity. Studies such as these suggest that it may be feasible to treat neurological diseases with adhesion molecule-targeting drugs even if those drugs are known to not cross intact BBBs; studies specifically assessing drug penetrance across diseased BBB should be performed.
For neurodegenerative diseases characterized by large, intracellular protein aggregates, such as HD, AD, Parkinson’s disease, and amyotrophic lateral sclerosis, increasing autophagic function to clear aberrant misfolded proteins has had therapeutic success in various animal models [255]. Interestingly, certain autophagic modulators that have shown success also modulate adhesion. Verapamil, which induces autophagy through AMPK activation, has shown therapeutic benefit in neurodegenerative animal models and has roles in adhesion. In AD zebrafish, verapamil reduced insoluble tau levels and improved phenotypes related to morphology [256]. In HD PC12 cell, fly, and zebrafish models, verapamil reduced mutant HTT aggregates and decreased photoreceptor degeneration [257]. Additionally, verapamil inhibited adhesion of fibroblasts and reduced scar formation following peripheral nerve repair by inhibiting ECM secretion [258]. This secondary mechanism of action that has not been explored as it relates to neurodegenerative disease may also be contributing to the therapeutic benefit of verapamil in these models, particularly those related to morphological defects in AD zebrafish, as those results are suggestive of cellular remodeling that may involve changes in ECM deposition to aid in cellular migration. While other autophagy inducers that improve outcome in neurodegenerative models [259] also have roles in adhesion, namely rapamycin [260], resveratrol [261], lithium [262], nilotinib [263], bosutinib [264], and gemfibrozil [265], the link between these drugs and adhesion has primarily been explored in cancer or non-CNS-related cell types. However, adhesion-related functions for some, namely metformin [266], humanin [267], SB216763 [268], sodium valproate [269], and melatonin, have been explored in cell types of the brain. Melatonin, which induces autophagy through AMPK stimulation, increased autophagic flux and improved memory in AD mice [270]. Additionally, melatonin has regulatory roles in the expression of NCAMs, which are central for learning and memory [271]. Melatonin treatment in rats increased NCAM expression in the hippocampus and cortex, areas of the brain responsible for cognitive function [271]. Thus, the improvement of AD mice upon melatonin treatment may be the results of the dual roles for this compound in increasing autophagy to clear protein aggregates and restore protein homeostasis as well as increasing expression levels of adhesion molecules with known roles in learning and memory. The overlap in function for the drugs described here may be conferring benefit in neurodegenerative disease models by functionally targeting both autophagy and adhesion, compounding their therapeutic potential through the interplay between these critical biological functions using mechanisms and pathways described in this review. Future work is needed to explicitly determine the individual or dual roles of these drugs relating to their effects on adhesion and autophagy for therapeutic benefit of neurodegenerative disease.
Not only is the autophagic response amenable to modulation by various compounds, but integrins themselves are sensitive to pharmacologic manipulation. There are 117 integrin-targeting molecules in active or completed clinical trials (clinicaltrials.gov) and 6 with FDA approval [272], [273]. Natalizumab, an FDA approved integrin α4 antagonist that interferes with lymphocyte adhesion and migration to reduce inflammation, reduces neuroinflammation in MS [274] and improves MS-associated cognitive impairment [275]. Another drug that acts on adhesion-related molecules that recently completed trials for breast cancer metastasis is reparixin. Reparixin acts at the surface of cancer stem cells to block the activation of FAK phosphorylation that in turn activates the WNT pathway to regulate cell proliferation [276]. Recently published results from the reparixin trial demonstrated that the drug also acts to modulate autophagy, evidenced by decreased levels of p62 in tumors from reparixin-treated breast cancer patients [276]. Relevantly, reparixin was only found within the CNS of patients who experienced metastasis that compromised their BBB [277], perhaps suggesting a similar effect would be possible in patients with a compromised BBB due to neurodegeneration. While the clinical relevance of the interplay between adhesion and autophagy in this system is not yet clear, reparixin represents the only drug, to our knowledge, that conclusively targets both of these critical biological processes. Given the profound interconnection between adhesion and autophagy and their regulation of a vast number of molecular events with therapeutic potential for various diseases, including neurodegenerative pathologies, we predict that this is the first of many drugs that will target both adhesion and autophagy for therapeutic gain.
Conclusions
Data in non-CNS-related cell types robustly demonstrates biological synergy between cellular adhesion and autophagy to maintain homeostasis, and mounting evidence suggests that these processes may also work cooperatively in cell types of the brain. The interplay between adhesion and autophagy governs many critical biological functions in the brain that have roles in both neuroprotection and neurodegeneration. In neural cell types, there is a matrix-directed response to engage autophagic flux following cellular detachment to promote cell survival. Concomitantly, autophagy aids in controlling adhesion, by directly regulating focal adhesion turnover to mediate cell motility. This autophagic regulation of adhesion dynamics to control movement is critical for neurodevelopment, directing the migration of cells as the brain forms to maintain a normal developmental paradigm. Additionally, autophagy of focal adhesions molecularly contributes to programs essential for neurite outgrowth and synaptic plasticity, which are disrupted in the neurodegenerative diseases described here. Thus, known disease-mediated aberrations in autophagy and adhesion may directly contribute to neurodegenerative pathologies. Further, the molecular responses to adhesion and autophagy converge on the same signaling pathways to elicit differing biological responses with the same molecules. While the individual contributions of adhesion and autophagy for each of the pathways highlighted here is evident, the direct intersection of both of these biological processes in each of these pathways has not been fully elucidated. Gaining a deeper understanding of this intersection in neurodegenerative diseases and how adhesion and autophagy cooperatively contribute to molecular signaling pathways may help to elucidate disease pathology. Understanding molecular mechanisms that exist at this synergistic junction – between adhesion and autophagy – may offer novel therapeutic modalities for neurodegenerative disease. The extracellular space is significantly more accessible than intracellular components, since bypassing the plasma membrane is no longer a challenge. Additionally, adhesion-mediating proteins are highly-druggable. Thus, leveraging the extracellular response to stimulate autophagy in cell types of the brain may be a feasible mechanism for clearing protein aggregates that characterize many neurodegenerative diseases to improve or compensate for pathophysiology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by a Huntington’s Disease Society of America Berman/Topper Career Development Fellowship to S.J.H., CHDI to J.S.S., the Hereditary Disease Foundation to J.S.S., and grants from the National Institute of Health (R35 NS116872 and R01 NS089076 to L.M.T., NS072453 to J.S.S., National Research Service Award 1F30AG060704-01A1 to G.F.).
Author Contributions
S.J.H. conceptually designed the manuscript and contributed to all sections. G.F. contributed expertise related to autophagy and designed figures. A.R. designed figures. J.S.S. and L.M.T. revised the manuscript along lines of expertise.
Contributor Information
Sarah J. Hernandez, Email: sjherna2@uci.edu.
Leslie M. Thompson, Email: lmthomps@uci.edu.
References
- 1.Glickstein M. Golgi and Cajal: The neuron doctrine and the 100th anniversary of the 1906 Nobel Prize. Curr. Biol. 2006;16(5):R147–R151. doi: 10.1016/j.cub.2006.02.053. [DOI] [PubMed] [Google Scholar]
- 2.Celio M.R., Spreafico R., De Biasi S., Vitellaro-Zuccarello L. Perineuronal nets: past and present. Trends Neurosci. 1998;21(12):510–515. doi: 10.1016/s0166-2236(98)01298-3. [DOI] [PubMed] [Google Scholar]
- 3.Simon H.-U., Tschan M., Bütikofer P. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmann M., Kukkurainen S., Hytönen V.P., Wehrle-Haller B. Cell adhesion by integrins. Physiol. Rev. 2019;99(4):1655–1699. doi: 10.1152/physrev.00036.2018. [DOI] [PubMed] [Google Scholar]
- 5.Burridge K., Guilluy C. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 2016;343(1):14–20. doi: 10.1016/j.yexcr.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaller M.D., Hildebrand J.D., Shannon J.D., Fox J.W., Vines R.R., Parsons J.T. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol. Cell. Biol. 1994;14(3):1680–1688. doi: 10.1128/mcb.14.3.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan C.-S., Weeber E.J., Kurup S., Sweatt J.D., Davis R.L. Integrin requirement for hippocampal synaptic plasticity and spatial memory. J. Neurosci. 2003;23(18):7107–7116. doi: 10.1523/JNEUROSCI.23-18-07107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zovein A.C., Luque A., Turlo K.A., Hofmann J.J., Yee K.M., Becker M.S., Fassler R., Mellman I., Lane T.F., Iruela-Arispe M.L. β1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3-dependent mechanism. Dev. Cell. 2010;18(1):39–51. doi: 10.1016/j.devcel.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei L., Liu D., Huang Y., Jovin I., Shai S.-Y., Kyriakides T., Ross R.S., Giordano F.J. Endothelial expression of β1 integrin is required for embryonic vascular patterning and postnatal vascular remodeling. Mol. Cell. Biol. 2008;28(2):794–802. doi: 10.1128/MCB.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stupack D.G., Cheresh D.A. ECM remodeling regulates angiogenesis: endothelial integrins look for new ligands. Sci. STKE. 2002;2002(119):pe7-pe7. doi: 10.1126/stke.2002.119.pe7. [DOI] [PubMed] [Google Scholar]
- 11.Caltagarone J., Jing Z., Bowser R. Focal adhesions regulate Aβ signaling and cell death in Alzheimer's disease. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2007;1772(4):438–445. doi: 10.1016/j.bbadis.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J.P., Holcomb J., Al-Ramahi I., De Haro M., Gafni J., Zhang N., Kim E., Sanhueza M., Torcassi C., Kwak S. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron. 2010;67(2):199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wertz M.H., Mitchem M.R., Pineda S.S., Hachigian L.J., Lee H., Lau V., Powers A., Kulicke R., Madan G.K., Colic M. Genome-wide in vivo CNS screening identifies genes that modify CNS neuronal survival and mHTT toxicity. Neuron. 2020 doi: 10.1016/j.neuron.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018;19(6):349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 15.Yim W.-W.-Y., Mizushima N. Lysosome biology in autophagy. Cell Disc. 2020;6(1):1–12. doi: 10.1038/s41421-020-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018;20(3):233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohnstad A.E., Delgado J.M., North B.J., Nasa I., Kettenbach A.N., Schultz S.W., Shoemaker C.J. Receptor-mediated clustering of FIP200 bypasses the role of LC3 lipidation in autophagy. EMBO J. 2020;39(24) doi: 10.15252/embj.2020104948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkbeiner S. The autophagy lysosomal pathway and neurodegeneration. Cold Spring Harbor Perspect. Biol. 2020;12(3) doi: 10.1101/cshperspect.a033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochaba J., Fote G., Kachemov M., Thein S., Yeung S.Y., Lau A.L., Hernandez S., Lim R.G., Casale M., Neel M.J. IKKβ slows Huntington’s disease progression in R6/1 mice. Proc. Natl. Acad. Sci. 2019;116(22):10952–10961. doi: 10.1073/pnas.1814246116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer L., Dikic I. Matrix Biology; 2021. Autophagy: Instructions from the Extracellular Matrix. [DOI] [PubMed] [Google Scholar]
- 21.Neill T., Schaefer L., Iozzo R.V. Instructive roles of extracellular matrix on autophagy. Am. J. Pathol. 2014;184(8):2146–2153. doi: 10.1016/j.ajpath.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J.-I., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 24.Lee H., Leamey C.A., Sawatari A. Perineuronal nets play a role in regulating striatal function in the mouse. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0032747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morawski M., Filippov M., Tzinia A., Tsilibary E., Vargova L. ECM in brain aging and dementia. Prog. Brain Res. 2014;214:207–227. doi: 10.1016/B978-0-444-63486-3.00010-4. [DOI] [PubMed] [Google Scholar]
- 26.Dityatev A., Schachner M. Extracellular matrix molecules and synaptic plasticity. Nat. Rev. Neurosci. 2003;4(6):456–468. doi: 10.1038/nrn1115. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi Y. Lecticans: organizers of the brain extracellular matrix. Cell. Mol. Life Sci. CMLS. 2000;57(2):276–289. doi: 10.1007/PL00000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandtlow C.E., Zimmermann D.R. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol. Rev. 2000;80(4):1267–1290. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 29.Tran A.P., Warren P.M., Silver J. Regulation of autophagy by inhibitory CSPG interactions with receptor PTPσ and its impact on plasticity and regeneration after spinal cord injury. Exp. Neurol. 2020 doi: 10.1016/j.expneurol.2020.113276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultheis N., Jiang M., Selleck S.B. Putting the brakes on autophagy: the role of heparan sulfate modified proteins in the balance of anabolic and catabolic pathways and intracellular quality control. Matrix Biol. 2021 doi: 10.1016/j.matbio.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Puangmalai N., Bhatt N., Montalbano M., Sengupta U., Gaikwad S., Ventura F., McAllen S., Ellsworth A., Garcia S., Kayed R. Internalization mechanisms of brain-derived tau oligomers from patients with Alzheimer’s disease, progressive supranuclear palsy and dementia with Lewy bodies. Cell Death Dis. 2020;11(5):1–16. doi: 10.1038/s41419-020-2503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Group, T. H. s. d. C. R. (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72(6) 971-983. [DOI] [PubMed]
- 33.Crapser J.D., Ochaba J., Soni N., Reidling J.C., Thompson L.M., Green K.N. Microglial depletion prevents extracellular matrix changes and striatal volume reduction in a model of Huntington's disease. Brain. 2020;143(1):266–288. doi: 10.1093/brain/awz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braghetta P., Fabbro C., Piccolo S., Marvulli D., Bonaldo P., Volpin D., Bressan G.M. Distinct regions control transcriptional activation of the alpha1 (VI) collagen promoter in different tissues of transgenic mice. J. Cell Biol. 1996;135(4):1163–1177. doi: 10.1083/jcb.135.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keene D.R., Engvall E., Glanville R.W. Ultrastructure of type VI collagen in human skin and cartilage suggests an anchoring function for this filamentous network. J. Cell Biol. 1988;107(5):1995–2006. doi: 10.1083/jcb.107.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quarto R., Dozin B., Bonaldo P., Cancedda R., Colombatti A. Type VI collagen expression is upregulated in the early events of chondrocyte differentiation. Development. 1993;117(1):245–251. doi: 10.1242/dev.117.1.245. [DOI] [PubMed] [Google Scholar]
- 37.Chen P., Cescon M., Megighian A., Bonaldo P. Collagen VI regulates peripheral nerve myelination and function. FASEB J. 2014;28(3):1145–1156. doi: 10.1096/fj.13-239533. [DOI] [PubMed] [Google Scholar]
- 38.Hubert T., Grimal S., Carroll P., Fichard-Carroll A. Collagens in the developing and diseased nervous system. Cell. Mol. Life Sci. 2009;66(7):1223–1238. doi: 10.1007/s00018-008-8561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng J.S., Dubal D.B., Kim D.H., Legleiter J., Cheng I.H., Yu G.-Q., Tesseur I., Wyss-Coray T., Bonaldo P., Mucke L. Collagen VI protects neurons against Aβ toxicity. Nat. Neurosci. 2009;12(2):119. doi: 10.1038/nn.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cescon M., Chen P., Castagnaro S., Gregorio I., Bonaldo P. Lack of collagen VI promotes neurodegeneration by impairing autophagy and inducing apoptosis during aging. Aging (Albany NY) 2016;8(5):1083. doi: 10.18632/aging.100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soontornniyomkij V., Risbrough V.B., Young J.W., Soontornniyomkij B., Jeste D.V., Achim C.L. Increased hippocampal accumulation of autophagosomes predicts short-term recognition memory impairment in aged mice. Age. 2012;34(2):305–316. doi: 10.1007/s11357-011-9234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seppänen A., Pikkarainen M., Hartikainen P., Hofmann S.C., Majamaa K., Alafuzoff I. Expression of collagen XVII and ubiquitin-binding protein p62 in motor neuron disease. Brain Res. 2009;1247:171–177. doi: 10.1016/j.brainres.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 43.Ramalingam M., Kim S.-J. Insulin on activation of autophagy with integrins and syndecans against MPP+-induced α-synuclein neurotoxicity. Neurosci. Lett. 2016;633:94–100. doi: 10.1016/j.neulet.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Ramalingam M., Kim S.J. The neuroprotective role of insulin against MPP+-induced parkinson's disease in differentiated SH-SY5Y Cells. J. Cell. Biochem. 2016;117(4):917–926. doi: 10.1002/jcb.25376. [DOI] [PubMed] [Google Scholar]
- 45.Wang P., Ye Y. Filamentous recombinant human Tau activates primary astrocytes via an integrin receptor complex. Nat. Commun. 2021;12(1):1–14. doi: 10.1038/s41467-020-20322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Burchett J.M., Schuler D.R., Cirrito J.R. Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 2014;34(29):9607–9620. doi: 10.1523/JNEUROSCI.3788-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye Y. Regulation of protein homeostasis by unconventional protein secretion in mammalian cells Seminars in cell & developmental biology. Elsevier. 2018:29–35. doi: 10.1016/j.semcdb.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sofroniew M.V. Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 2015;16(5):249–263. doi: 10.1038/nrn3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ransohoff R.M. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 50.Hua J., Unschuld P.G., Margolis R.L., Van Zijl P.C., Ross C.A. Elevated arteriolar cerebral blood volume in prodromal Huntington's disease. Mov. Disord. 2014;29(3):396–401. doi: 10.1002/mds.25591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin C.-Y., Hsu Y.-H., Lin M.-H., Yang T.-H., Chen H.-M., Chen Y.-C., Hsiao H.-Y., Chen C.-C., Chern Y., Chang C. Neurovascular abnormalities in humans and mice with Huntington's disease. Exp. Neurol. 2013;250:20–30. doi: 10.1016/j.expneurol.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 52.Kook S.-Y., Hong H.S., Moon M., Ha C.M., Chang S., Mook-Jung I. Aβ1–42-RAGE interaction disrupts tight junctions of the blood–brain barrier via Ca2+-calcineurin signaling. J. Neurosci. 2012;32(26):8845–8854. doi: 10.1523/JNEUROSCI.6102-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zlokovic B.V. Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nat. Rev. Neurosci. 2011;12(12):723. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim R.G., Quan C., Reyes-Ortiz A.M., Lutz S.E., Kedaigle A.J., Gipson T.A., Wu J., Vatine G.D., Stocksdale J., Casale M.S. Huntington’s disease iPSC-derived brain microvascular endothelial cells reveal WNT-mediated angiogenic and blood-brain barrier deficits. Cell Rep. 2017;19(7):1365–1377. doi: 10.1016/j.celrep.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montagne A., Barnes S.R., Sweeney M.D., Halliday M.R., Sagare A.P., Zhao Z., Toga A.W., Jacobs R.E., Liu C.Y., Amezcua L. Blood-brain barrier breakdown in the aging human hippocampus. Neuron. 2015;85(2):296–302. doi: 10.1016/j.neuron.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takechi R., Lam V., Brook E., Giles C., Fimognari N., Mooranian A., Al-Salami H., Coulson S.H., Nesbit M., Mamo J.C. Blood-brain barrier dysfunction precedes cognitive decline and neurodegeneration in diabetic insulin resistant mouse model: an implication for causal link. Front. Aging Neurosci. 2017;9:399. doi: 10.3389/fnagi.2017.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simpson I.A., Chundu K.R., Davies-Hill T., Honer W.G., Davies P. Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer's disease. Ann. Neurol. Offi. J. Am. Neurol. Assoc. Child Neurol. Soc. 1994;35(5):546–551. doi: 10.1002/ana.410350507. [DOI] [PubMed] [Google Scholar]
- 58.Winkler E.A., Sengillo J.D., Sagare A.P., Zhao Z., Ma Q., Zuniga E., Wang Y., Zhong Z., Sullivan J.S., Griffin J.H. Blood–spinal cord barrier disruption contributes to early motor-neuron degeneration in ALS-model mice. Proc. Natl. Acad. Sci. U.S.A. 2014;111(11):E1035–E1042. doi: 10.1073/pnas.1401595111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang L., Li X., Zhong Y., Yu J., Yu L., Dai H., Yan M. Autophagy protects human brain microvascular endothelial cells against methylglyoxal-induced injuries, reproducible in a cerebral ischemic model in diabetic rats. J. Neurochem. 2015;135(2):431–440. doi: 10.1111/jnc.13277. [DOI] [PubMed] [Google Scholar]
- 60.Meredith J., Jr, Fazeli B., Schwartz M. The extracellular matrix as a cell survival factor. Mol. Biol. Cell. 1993;4(9):953–961. doi: 10.1091/mbc.4.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frisch S.M., Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994;124(4):619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy. 2008;4(3):351–353. doi: 10.4161/auto.5523. [DOI] [PubMed] [Google Scholar]
- 63.Debnath J., Mills K.R., Collins N.L., Reginato M.J., Muthuswamy S.K., Brugge J.S. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111(1):29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 64.Gary D.S., Mattson M.P. Integrin signaling via the PI3-kinase–Akt pathway increases neuronal resistance to glutamate-induced apoptosis. J. Neurochem. 2001;76(5):1485–1496. doi: 10.1046/j.1471-4159.2001.00173.x. [DOI] [PubMed] [Google Scholar]
- 65.Gary D.S., Milhavet O., Camandola S., Mattson M.P. Essential role for integrin linked kinase in Akt-mediated integrin survival signaling in hippocampal neurons. J. Neurochem. 2003;84(4):878–890. doi: 10.1046/j.1471-4159.2003.01579.x. [DOI] [PubMed] [Google Scholar]
- 66.Chen Z.-L., Strickland S. Neuronal death in the hippocampus is promoted by plasmin-catalyzed degradation of laminin. Cell. 1997;91(7):917–925. doi: 10.1016/s0092-8674(00)80483-3. [DOI] [PubMed] [Google Scholar]
- 67.Sandilands E., Serrels B., McEwan D.G., Morton J.P., Macagno J.P., McLeod K., Stevens C., Brunton V.G., Langdon W.Y., Vidal M. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat. Cell Biol. 2012;14(1):51–60. doi: 10.1038/ncb2386. [DOI] [PubMed] [Google Scholar]
- 68.Lock R., Debnath J. Extracellular matrix regulation of autophagy. Curr. Opin. Cell Biol. 2008;20(5):583–588. doi: 10.1016/j.ceb.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.vom Dahl S., Schliess F., Reissmann R., Görg B., Weiergräber O., Kocalkova M., Dombrowski F., Häussinger D. Involvement of integrins in osmosensing and signaling toward autophagic proteolysis in rat liver. J. Biol. Chem. 2003;278(29):27088–27095. doi: 10.1074/jbc.M210699200. [DOI] [PubMed] [Google Scholar]
- 70.Schliess F., Reissmann R., Reinehr R., vom Dahl S., Häussinger D. Involvement of integrins and Src in insulin signaling toward autophagic proteolysis in rat liver. J. Biol. Chem. 2004;279(20):21294–21301. doi: 10.1074/jbc.M313901200. [DOI] [PubMed] [Google Scholar]
- 71.Fung C., Lock R., Gao S., Salas E., Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol. Biol. Cell. 2008;19(3):797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen N., Debnath J. IκB kinase complex (IKK) triggers detachment-induced autophagy in mammary epithelial cells independently of the PI3K-AKT-MTORC1 pathway. Autophagy. 2013;9(8):1214–1227. doi: 10.4161/auto.24870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.László Z.I., Lele Z., Zöldi M., Miczán V., Mógor F., Simon G.M., Mackie K., Kacskovics I., Cravatt B.F., Katona I. ABHD4-dependent developmental anoikis safeguards the embryonic brain. Nat. Commun. 2020;11(1):1–16. doi: 10.1038/s41467-020-18175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barnat M., Capizzi M., Aparicio E., Boluda S., Wennagel D., Kacher R., Kassem R., Lenoir S., Agasse F., Braz B.Y. Huntington’s disease alters human neurodevelopment. Science. 2020;369(6505):787–793. doi: 10.1126/science.aax3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sardo V.L., Zuccato C., Gaudenzi G., Vitali B., Ramos C., Tartari M., Myre M.A., Walker J.A., Pistocchi A., Conti L. An evolutionary recent neuroepithelial cell adhesion function of huntingtin implicates ADAM10-Ncadherin. Nat. Neurosci. 2012;15(5):713. doi: 10.1038/nn.3080. [DOI] [PubMed] [Google Scholar]
- 76.Abercrombie M., Heaysman J.E., Pegrum S.M. The locomotion of fibroblasts in culture: IV. Electron microscopy of the leading lamella. Exp. Cell Res. 1971;67(2):359–367. doi: 10.1016/0014-4827(71)90420-4. [DOI] [PubMed] [Google Scholar]
- 77.Stehbens S., Wittmann T. Targeting and transport: how microtubules control focal adhesion dynamics. J. Cell Biol. 2012;198(4):481–489. doi: 10.1083/jcb.201206050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tuloup-Minguez V., Hamai A., Greffard A., Nicolas V., Codogno P., Botti J. Autophagy modulates cell migration and β1 integrin membrane recycling. Cell Cycle. 2013;12(20):3317–3328. doi: 10.4161/cc.26298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kenific C.M., Stehbens S.J., Goldsmith J., Leidal A.M., Faure N., Ye J., Wittmann T., Debnath J. NBR1 enables autophagy-dependent focal adhesion turnover. J. Cell Biol. 2016;212(5):577–590. doi: 10.1083/jcb.201503075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharifi M.N., Mowers E.E., Drake L.E., Collier C., Chen H., Zamora M., Mui S., Macleod K.F. Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of paxillin with LC3. Cell Rep. 2016;15(8):1660–1672. doi: 10.1016/j.celrep.2016.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galavotti S., Bartesaghi S., Faccenda D., Shaked-Rabi M., Sanzone S., McEvoy A., Dinsdale D., Condorelli F., Brandner S., Campanella M. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32(6):699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 82.Lock R., Kenific C.M., Leidal A.M., Salas E., Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4(4):466–479. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li W.-D., Hu N., Lei F.-R., Wei S., Rong J.-J., Zhuang H., Li X.-Q. Autophagy inhibits endothelial progenitor cells migration via the regulation of MMP2, MMP9 and uPA under normoxia condition. Biochem. Biophys. Res. Commun. 2015;466(3):376–380. doi: 10.1016/j.bbrc.2015.09.031. [DOI] [PubMed] [Google Scholar]
- 84.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dower C.M., Wills C.A., Frisch S.M., Wang H.-G. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy. 2018;14(7):1110–1128. doi: 10.1080/15548627.2018.1450020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kerstein P.C., Patel K.M., Gomez T.M. Calpain-mediated proteolysis of talin and FAK regulates adhesion dynamics necessary for axon guidance. J. Neurosci. 2017;37(6):1568–1580. doi: 10.1523/JNEUROSCI.2769-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bressan, C., Pecora, A., Gagnon, D., Snapyan, M., Labrecque, S., De Koninck, P., Parent, M., Saghatelyan, A. (2020). The dynamic interplay between ATP/ADP levels and autophagy sustain neuronal migration in vivo. bioRxiv. [DOI] [PMC free article] [PubMed]
- 88.Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23(1):33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 89.Hara T., Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5(1):85–87. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 90.Hara T., Takamura A., Kishi C., Iemura S.-I., Natsume T., Guan J.-L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ganley I.G., Lam D.H., Wang J., Ding X., Chen S., Jiang X. ULK1· ATG13· FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kenny, S. J., Chen, X. S., Ge, L., Xu, K. (2019). Super-resolution microscopy unveils FIP200-scaffolded, cup-shaped organization of mammalian autophagic initiation machinery. bioRxiv 712828.
- 93.Zientara-Rytter K., Subramani S. Mechanistic insights into the role of Atg11 in selective autophagy. J. Mol. Biol. 2020;432(1):104–122. doi: 10.1016/j.jmb.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Steffan J.S. Does Huntingtin play a role in selective macroautophagy? Cell Cycle. 2010;9(17):3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith M.D., Harley M.E., Kemp A.J., Wills J., Lee M., Arends M., von Kriegsheim A., Behrends C., Wilkinson S. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell. 2018;44(2) doi: 10.1016/j.devcel.2017.11.024. 217-232.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang C., Liang C.-C., Bian Z.C., Zhu Y., Guan J.-L. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat. Neurosci. 2013;16(5):532. doi: 10.1038/nn.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liang C.-C., Wang C., Peng X., Gan B., Guan J.-L. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 2010;285(5):3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abbi S., Ueda H., Zheng C., Cooper L.A., Zhao J., Christopher R., Guan J.-L. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol. Biol. Cell. 2002;13(9):3178–3191. doi: 10.1091/mbc.E02-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Caino M.C., Chae Y.C., Vaira V., Ferrero S., Nosotti M., Martin N.M., Weeraratna A., O’Connell M., Jernigan D., Fatatis A. Metabolic stress regulates cytoskeletal dynamics and metastasis of cancer cells. J. Clin. Investig. 2013;123(7):2907–2920. doi: 10.1172/JCI67841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirano S., Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol. Rev. 2012;92(2):597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- 101.Redies C., Hertel N., Hübner C.A. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 102.Robles E., Gomez T.M. Focal adhesion kinase signaling at sites of integrin-mediated adhesion controls axon pathfinding. Nat. Neurosci. 2006;9(10):1274–1283. doi: 10.1038/nn1762. [DOI] [PubMed] [Google Scholar]
- 103.Myers J.P., Santiago-Medina M., Gomez T.M. Regulation of axonal outgrowth and pathfinding by integrin–ECM interactions. Devel. Neurobiol. 2011;71(11):901–923. doi: 10.1002/dneu.20931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rui Y.-N., Xu Z., Patel B., Chen Z., Chen D., Tito A., David G., Sun Y., Stimming E.F., Bellen H.J. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015;17(3):262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ochaba J., Lukacsovich T., Csikos G., Zheng S., Margulis J., Salazar L., Mao K., Lau A.L., Yeung S.Y., Humbert S. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. U.S.A. 2014;111(47):16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., De Vries R., Arias E., Harris S., Sulzer D. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13(5):567. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Caviston J.P., Holzbaur E.L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19(4):147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caviston J.P., Ross J.L., Antony S.M., Tokito M., Holzbaur E.L. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. 2007;104(24):10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tousley A., Iuliano M., Weisman E., Sapp E., Richardson H., Vodicka P., Alexander J., Aronin N., DiFiglia M., Kegel-Gleason K.B. Huntingtin associates with the actin cytoskeleton and α-actinin isoforms to influence stimulus dependent morphology changes. PLoS ONE. 2019;14(2) doi: 10.1371/journal.pone.0212337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thompson M.N., MacDonald M.E., Gusella J.F., Myre M.A. Huntingtin supplies a csaA-independent function essential for EDTA-resistant homotypic cell adhesion in Dictyostelium discoideum. J. Huntington's Dis. 2014;3(3):261–271. doi: 10.3233/JHD-140112. [DOI] [PubMed] [Google Scholar]
- 111.Reis S.A., Thompson M.N., Lee J.-M., Fossale E., Kim H.-H., Liao J.K., Moskowitz M.A., Shaw S.Y., Dong L., Haggarty S.J. Striatal neurons expressing full-length mutant huntingtin exhibit decreased N-cadherin and altered neuritogenesis. Hum. Mol. Genet. 2011;20(12):2344–2355. doi: 10.1093/hmg/ddr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takeichi M., Matsunami H., Inoue T., Kimura Y., Suzuki S., Tanaka T. Roles for cadherins in patterning of the developing brain. Dev. Neurosci. 1997;19(1):86–87. doi: 10.1159/000111189. [DOI] [PubMed] [Google Scholar]
- 113.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 2007;8(1):11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 114.Uchida N., Honjo Y., Johnson K.R., Wheelock M.J., Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 1996;135(3):767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Benson D.L., Colman D.R., Huntley G.W. Molecules, maps and synapse specificity. Nat. Rev. Neurosci. 2001;2(12):899–909. doi: 10.1038/35104078. [DOI] [PubMed] [Google Scholar]
- 116.Benson D.L., Schnapp L.M., Shapiro L., Huntley G.W. Making memories stick: cell-adhesion molecules in synaptic plasticity. Trends Cell Biol. 2000;10(11):473–482. doi: 10.1016/s0962-8924(00)01838-9. [DOI] [PubMed] [Google Scholar]
- 117.Benson D.L., Tanaka H. N-cadherin redistribution during synaptogenesis in hippocampal neurons. J. Neurosci. 1998;18(17):6892–6904. doi: 10.1523/JNEUROSCI.18-17-06892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lelièvre E.C., Plestant C., Boscher C., Wolff E., Mège R.-M., Birbes H. N-cadherin mediates neuronal cell survival through Bim down-regulation. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0033206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luo S., Garcia-Arencibia M., Zhao R., Puri C., Toh P.P., Sadiq O., Rubinsztein D.C. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol. Cell. 2012;47(3):359–370. doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vezzoli E., Caron I., Talpo F., Besusso D., Conforti P., Battaglia E., Sogne E., Falqui A., Petricca L., Verani M. Inhibiting pathologically active ADAM10 rescues synaptic and cognitive decline in Huntington’s disease. J. Clin. Investig. 2019;129(6) doi: 10.1172/JCI120616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moreno-Layseca P., Icha J., Hamidi H., Ivaska J. Integrin trafficking in cells and tissues. Nat. Cell Biol. 2019;21(2):122–132. doi: 10.1038/s41556-018-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.De Franceschi N., Hamidi H., Alanko J., Sahgal P., Ivaska J. Integrin traffic–the update. J. Cell Sci. 2015;128(5):839–852. doi: 10.1242/jcs.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Paul N.R., Jacquemet G., Caswell P.T. Endocytic trafficking of integrins in cell migration. Curr. Biol. 2015;25(22):R1092–R1105. doi: 10.1016/j.cub.2015.09.049. [DOI] [PubMed] [Google Scholar]
- 124.Valdembri D., Serini G. Regulation of adhesion site dynamics by integrin traffic. Curr. Opin. Cell Biol. 2012;24(5):582–591. doi: 10.1016/j.ceb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 125.Yuan L., Fairchild M.J., Perkins A.D., Tanentzapf G. Analysis of integrin turnover in fly myotendinous junctions. J. Cell Sci. 2010;123(6):939–946. doi: 10.1242/jcs.063040. [DOI] [PubMed] [Google Scholar]
- 126.Yu C.-H., Rafiq N.B.M., Cao F., Zhou Y., Krishnasamy A., Biswas K.H., Ravasio A., Chen Z., Wang Y.-H., Kawauchi K. Integrin-beta3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat. Commun. 2015;6(1):1–12. doi: 10.1038/ncomms9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pines M., Das R., Ellis S.J., Morin A., Czerniecki S., Yuan L., Klose M., Coombs D., Tanentzapf G. Mechanical force regulates integrin turnover in Drosophila in vivo. Nat. Cell Biol. 2012;14(9):935–943. doi: 10.1038/ncb2555. [DOI] [PubMed] [Google Scholar]
- 128.Strachan L.R., Condic M.L. Cranial neural crest recycle surface integrins in a substratum-dependent manner to promote rapid motility. J. Cell Biol. 2004;167(3):545–554. doi: 10.1083/jcb.200405024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Calderwood D.A., Fujioka Y., de Pereda J.M., García-Alvarez B., Nakamoto T., Margolis B., McGlade C.J., Liddington R.C., Ginsberg M.H. Integrin β cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. 2003;100(5):2272–2277. doi: 10.1073/pnas.262791999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.De Franceschi N., Arjonen A., Elkhatib N., Denessiouk K., Wrobel A.G., Wilson T.A., Pouwels J., Montagnac G., Owen D.J., Ivaska J. Selective integrin endocytosis is driven by interactions between the integrin α-chain and AP2. Nat. Struct. Mol. Biol. 2016;23(2):172. doi: 10.1038/nsmb.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhen Y., Stenmark H. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 2015;128(17):3171–3176. doi: 10.1242/jcs.166074. [DOI] [PubMed] [Google Scholar]
- 132.Dozynkiewicz M.A., Jamieson N.B., MacPherson I., Grindlay J., van den Berghe P.V., von Thun A., Morton J.P., Gourley C., Timpson P., Nixon C. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell. 2012;22(1):131–145. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Homma Y., Hiragi S., Fukuda M. Rab family of small GTPases: an updated view on their regulation and functions. FEBS J. 2021;288(1):36–55. doi: 10.1111/febs.15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sandri C., Caccavari F., Valdembri D., Camillo C., Veltel S., Santambrogio M., Lanzetti L., Bussolino F., Ivaska J., Serini G. The R-Ras/RIN2/Rab5 complex controls endothelial cell adhesion and morphogenesis via active integrin endocytosis and Rac signaling. Cell Res. 2012;22(10):1479–1501. doi: 10.1038/cr.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nader G.P., Ezratty E.J., Gundersen G.G. FAK, talin and PIPKIγ regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat. Cell Biol. 2016;18(5):491–503. doi: 10.1038/ncb3333. [DOI] [PubMed] [Google Scholar]
- 136.Roberts M., Barry S., Woods A., van der Sluijs P., Norman J. PDGF-regulated rab4-dependent recycling of αvβ3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 2001;11(18):1392–1402. doi: 10.1016/s0960-9822(01)00442-0. [DOI] [PubMed] [Google Scholar]
- 137.Atherton P., Lausecker F., Harrison A., Ballestrem C. Low-intensity pulsed ultrasound promotes cell motility through vinculin-controlled Rac1 GTPase activity. J. Cell Sci. 2017;130(14):2277–2291. doi: 10.1242/jcs.192781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huet-Calderwood C., Rivera-Molina F., Iwamoto D.V., Kromann E.B., Toomre D., Calderwood D.A. Novel ecto-tagged integrins reveal their trafficking in live cells. Nat. Commun. 2017;8(1):1–13. doi: 10.1038/s41467-017-00646-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lobert V.H., Brech A., Pedersen N.M., Wesche J., Oppelt A., Malerød L., Stenmark H. Ubiquitination of α5β1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev. Cell. 2010;19(1):148–159. doi: 10.1016/j.devcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 140.Fader C., Colombo M.I. Multivesicular bodies and autophagy in erythrocyte maturation. Autophagy. 2006;2(2):122–125. doi: 10.4161/auto.2.2.2350. [DOI] [PubMed] [Google Scholar]
- 141.Fader C.M., Sánchez D., Furlán M., Colombo M.I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9(2):230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 142.Morvan J., Köchl R., Watson R., Collinson L.M., Jefferies H.B., Tooze S.A. In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy. 2009;5(5):676–689. doi: 10.4161/auto.5.5.8378. [DOI] [PubMed] [Google Scholar]
- 143.Gluschko A., Herb M., Wiegmann K., Krut O., Neiss W.F., Utermöhlen O., Krönke M., Schramm M. The β2 integrin Mac-1 induces protective LC3-associated phagocytosis of Listeria monocytogenes. Cell Host Microbe. 2018;23(3) doi: 10.1016/j.chom.2018.01.018. 324–337.e5. [DOI] [PubMed] [Google Scholar]
- 144.Acharya M., Sokolovska A., Tam J.M., Conway K.L., Stefani C., Raso F., Mukhopadhyay S., Feliu M., Paul E., Savill J. αv Integrins combine with LC3 and atg5 to regulate Toll-like receptor signalling in B cells. Nat. Commun. 2016;7(1):1–15. doi: 10.1038/ncomms10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Raso F., Sagadiev S., Du S., Gage E., Arkatkar T., Metzler G., Stuart L.M., Orr M.T., Rawlings D.J., Jackson S.W. α v Integrins regulate germinal center B cell responses through noncanonical autophagy. J. Clin. Investig. 2018;128(9):4163–4178. doi: 10.1172/JCI99597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lamb C.A., Dooley H.C., Tooze S.A. Endocytosis and autophagy: shared machinery for degradation. BioEssays. 2013;35(1):34–45. doi: 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- 147.Ao X., Zou L., Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21(3):348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Longatti A., Lamb C.A., Razi M., Yoshimura S.-I., Barr F.A., Tooze S.A. TBC1D14 regulates autophagosome formation via Rab11-and ULK1-positive recycling endosomesRecycling endosomes contribute to autophagosomes. J. Cell Biol. 2012;197(5):659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Anderson D.J. Cellular and molecular biology of neural crest cell lineage determination. Trends Genet. 1997;13(7):276–280. doi: 10.1016/s0168-9525(97)01187-6. [DOI] [PubMed] [Google Scholar]
- 150.Eva R., Dassie E., Caswell P.T., Dick G., Norman J.C., Fawcett J.W. Rab11 and its effector Rab coupling protein contribute to the trafficking of β1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J. Neurosci. 2010;30(35):11654–11669. doi: 10.1523/JNEUROSCI.2425-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Akbergenova Y., Littleton J.T. Pathogenic huntington alters BMP signaling and synaptic growth through local disruptions of endosomal compartments. J. Neurosci. 2017;37(12):3425–3439. doi: 10.1523/JNEUROSCI.2752-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Steinert J.R., Campesan S., Richards P., Kyriacou C.P., Forsythe I.D., Giorgini F. Rab11 rescues synaptic dysfunction and behavioural deficits in a Drosophila model of Huntington's disease. Hum. Mol. Genet. 2012;21(13):2912–2922. doi: 10.1093/hmg/dds117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Richards P., Didszun C., Campesan S., Simpson A., Horley B., Young K., Glynn P., Cain K., Kyriacou C., Giorgini F. Dendritic spine loss and neurodegeneration is rescued by Rab11 in models of Huntington's disease. Cell Death Differ. 2011;18(2):191–200. doi: 10.1038/cdd.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li X., Standley C., Sapp E., Valencia A., Qin Z.-H., Kegel K.B., Yoder J., Comer-Tierney L.A., Esteves M., Chase K. Mutant huntingtin impairs vesicle formation from recycling endosomes by interfering with Rab11 activity. Mol. Cell. Biol. 2009;29(22):6106–6116. doi: 10.1128/MCB.00420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shi Y., Ethell I.M. Integrins control dendritic spine plasticity in hippocampal neurons through NMDA receptor and Ca2+/calmodulin-dependent protein kinase II-mediated actin reorganization. J. Neurosci. 2006;26(6):1813–1822. doi: 10.1523/JNEUROSCI.4091-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li X., Sapp E., Chase K., Comer-Tierney L.A., Masso N., Alexander J., Reeves P., Kegel K.B., Valencia A., Esteves M. Disruption of Rab11 activity in a knock-in mouse model of Huntington's disease. Neurobiol. Dis. 2009;36(2):374–383. doi: 10.1016/j.nbd.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McGeachie A.B., Cingolani L.A., Goda Y. A stabilising influence: integrins in regulation of synaptic plasticity. Neurosci. Res. 2011;70(1):24–29. doi: 10.1016/j.neures.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang, J., Su, G., Wu, Q., Liu, J., Tian, Y., Liu, X., Zhou, J., Gao, J., Chen, W., Chen, D. (2020). Rab11-mediated recycling endosome role in nervous system development and neurodegenerative diseases. International Journal of Neuroscience (just-accepted) 1-12. [DOI] [PubMed]
- 159.Saudou F., Humbert S. The biology of huntingtin. Neuron. 2016;89(5):910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 160.Pal A., Severin F., Lommer B., Shevchenko A., Zerial M. Huntingtin–HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington's disease. J. Cell Biol. 2006;172(4):605–618. doi: 10.1083/jcb.200509091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hattula K., Peränen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000;10(24):1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- 162.White J.A., Krzystek T.J., Hoffmar-Glennon H., Thant C., Zimmerman K., Iacobucci G., Vail J., Thurston L., Rahman S., Gunawardena S. Excess Rab4 rescues synaptic and behavioral dysfunction caused by defective HTT-Rab4 axonal transport in Huntington’s disease. Acta Neuropathol. Commun. 2020;8(1):1–22. doi: 10.1186/s40478-020-00964-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vlahakis A., Debnath J. The interconnections between autophagy and integrin-mediated cell adhesion. J. Mol. Biol. 2017;429(4):515–530. doi: 10.1016/j.jmb.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hahn-Windgassen A., Nogueira V., Chen C.-C., Skeen J.E., Sonenberg N., Hay N. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J. Biol. Chem. 2005;280(37):32081–32089. doi: 10.1074/jbc.M502876200. [DOI] [PubMed] [Google Scholar]
- 165.Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 166.Laplante M., Sabatini D.M. mTOR signaling at a glance. J. Cell Sci. 2009;122(20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dunlop E., Tee A. mTOR and autophagy: a dynamic relationship governed by nutrients and energy Seminars in cell & developmental biology. Elsevier. 2014:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 168.Datan E., Shirazian A., Benjamin S., Matassov D., Tinari A., Malorni W., Lockshin R.A., Garcia-Sastre A., Zakeri Z. mTOR/p70S6K signaling distinguishes routine, maintenance-level autophagy from autophagic cell death during influenza A infection. Virology. 2014;452:175–190. doi: 10.1016/j.virol.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Arias E., Koga H., Diaz A., Mocholi E., Patel B., Cuervo A.M. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol. Cell. 2015;59(2):270–284. doi: 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chang K., Kang P., Liu Y., Huang K., Miao T., Sagona A.P., Nezis I.P., Bodmer R., Ocorr K., Bai H. TGFB-INHB/activin signaling regulates age-dependent autophagy and cardiac health through inhibition of MTORC2. Autophagy. 2019:1–16. doi: 10.1080/15548627.2019.1704117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sarbassov D.D., Ali S.M., Kim D.-H., Guertin D.A., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 172.Tatebe H., Shiozaki K. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules. 2017;7(4):77. doi: 10.3390/biom7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.LiCausi F., Hartman N.W. Role of mTOR complexes in neurogenesis. Int. J. Mol. Sci. 2018;19(5):1544. doi: 10.3390/ijms19051544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.McCabe M.P., Cullen E.R., Barrows C.M., Shore A.N., Tooke K.I., Laprade K.A., Stafford J.M., Weston M.C. Genetic inactivation of mTORC1 or mTORC2 in neurons reveals distinct functions in glutamatergic synaptic transmission. Elife. 2020;9 doi: 10.7554/eLife.51440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang W., Zhu P.J., Zhang S., Zhou H., Stoica L., Galiano M., Krnjević K., Roman G., Costa-Mattioli M. mTORC2 controls actin polymerization required for consolidation of long-term memory. Nat. Neurosci. 2013;16(4):441. doi: 10.1038/nn.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tang S.J., Reis G., Kang H., Gingras A.-C., Sonenberg N., Schuman E.M. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc. Natl. Acad. Sci. 2002;99(1):467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cammalleri M., Lütjens R., Berton F., King A.R., Simpson C., Francesconi W., Sanna P.P. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc. Natl. Acad. Sci. 2003;100(24):14368–14373. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li C.-Y., Li X., Liu S.-F., Qu W.-S., Wang W., Tian D.-S. Inhibition of mTOR pathway restrains astrocyte proliferation, migration and production of inflammatory mediators after oxygen–glucose deprivation and reoxygenation. Neurochem. Int. 2015;83:9–18. doi: 10.1016/j.neuint.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 179.Sung K., Jimenez-Sanchez M. Autophagy in astrocytes and its implications in neurodegeneration. J. Mol. Biol. 2020;432(8):2605–2621. doi: 10.1016/j.jmb.2019.12.041. [DOI] [PubMed] [Google Scholar]
- 180.Plaza-Zabala A., Sierra-Torre V., Sierra A. Autophagy and microglia: novel partners in neurodegeneration and aging. Int. J. Mol. Sci. 2017;18(3):598. doi: 10.3390/ijms18030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Francois A., Verite J., Bilan A.R., Janet T., Calon F., Fauconneau B., Paccalin M., Page G. Elsevier; Molecules to Medicine with mTOR: 2016. The mTOR Signaling Pathway in Neurodegenerative Diseases; pp. 85–104. [Google Scholar]
- 182.Talboom J.S., Velazquez R., Oddo S. The mammalian target of rapamycin at the crossroad between cognitive aging and Alzheimer’s disease. NPJ Aging Mech. Dis. 2015;1(1):1–7. doi: 10.1038/npjamd.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.B Jahrling, J., Laberge, R.-M. (2015). Age-related neurodegeneration prevention through mTOR inhibition: potential mechanisms and remaining questions. Current topics in medicinal chemistry 15(21) 2139-2151. [DOI] [PMC free article] [PubMed]
- 184.Perluigi M., Di Domenico F., Butterfield D.A. mTOR signaling in aging and neurodegeneration: At the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol. Dis. 2015;84:39–49. doi: 10.1016/j.nbd.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 185.An W.-L., Cowburn R.F., Li L., Braak H., Alafuzoff I., Iqbal K., Iqbal I.-G., Winblad B., Pei J.-J. Up-regulation of phosphorylated/activated p70 S6 kinase and its relationship to neurofibrillary pathology in Alzheimer's disease. Am. J. Pathol. 2003;163(2):591–607. doi: 10.1016/S0002-9440(10)63687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pei J.J., Hugon J. mTOR-dependent signalling in Alzheimer's disease. J. Cell Mol. Med. 2008;12(6b):2525–2532. doi: 10.1111/j.1582-4934.2008.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sun Y.-X., Ji X., Mao X., Xie L., Jia J., Galvan V., Greenberg D.A., Jin K. Differential activation of mTOR complex 1 signaling in human brain with mild to severe Alzheimer's disease. J. Alzheimers Dis. 2014;38(2):437–444. doi: 10.3233/JAD-131124. [DOI] [PubMed] [Google Scholar]
- 188.Griffin R.J., Moloney A., Kelliher M., Johnston J.A., Ravid R., Dockery P., O'Connor R., O'Neill C. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer's disease pathology. J. Neurochem. 2005;93(1):105–117. doi: 10.1111/j.1471-4159.2004.02949.x. [DOI] [PubMed] [Google Scholar]
- 189.Li X., Alafuzoff I., Soininen H., Winblad B., Pei J.J. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer's disease brain. FEBS J. 2005;272(16):4211–4220. doi: 10.1111/j.1742-4658.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 190.Ferrarelli L.K. Is mTOR a good guy or bad guy in Huntington’s disease? Sci. Signaling. 2015;8(362):ec26-ec26. [Google Scholar]
- 191.Pryor, W. M., Biagioli, M., Shahani, N., Swarnkar, S., Huang, W.-C., Page, D. T., MacDonald, M. E., Subramaniam, S. (2014). Huntingtin promotes mTORC1 signaling in the pathogenesis of Huntington’s disease. Science signaling 7(349) ra103-ra103. [DOI] [PubMed]
- 192.Sarkar S., Krishna G., Imarisio S., Saiki S., O'Kane C.J., Rubinsztein D.C. A rational mechanism for combination treatment of Huntington's disease using lithium and rapamycin. Hum. Mol. Genet. 2008;17(2):170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- 193.Subramaniam S., Napolitano F., Mealer R.G., Kim S., Errico F., Barrow R., Shahani N., Tyagi R., Snyder S.H., Usiello A. Rhes, a striatal-enriched small G protein, mediates mTOR signaling and L-DOPA–induced dyskinesia. Nat. Neurosci. 2012;15(2):191–193. doi: 10.1038/nn.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mealer R.G., Subramaniam S., Snyder S.H. Rhes deletion is neuroprotective in the 3-nitropropionic acid model of Huntington's disease. J. Neurosci. 2013;33(9):4206–4210. doi: 10.1523/JNEUROSCI.3730-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Subramaniam S., Sixt K.M., Barrow R., Snyder S.H. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324(5932):1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36(6):585. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 197.Lee J.H., Tecedor L., Chen Y.H., Monteys A.M., Sowada M.J., Thompson L.M., Davidson B.L. Reinstating aberrant mTORC1 activity in Huntington’s disease mice improves disease phenotypes. Neuron. 2015;85(2):303–315. doi: 10.1016/j.neuron.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schiaffino, S., Mammucari, C. (2011). Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skeletal muscle 1(1) 4. [DOI] [PMC free article] [PubMed]
- 199.Tai Y.-T., Podar K., Catley L., Tseng Y.-H., Akiyama M., Shringarpure R., Burger R., Hideshima T., Chauhan D., Mitsiades N. Insulin-like growth factor-1 induces adhesion and migration in human multiple myeloma cells via activation of β1-integrin and phosphatidylinositol 3′-kinase/AKT signaling. Cancer Res. 2003;63(18):5850–5858. [PubMed] [Google Scholar]
- 200.Chen L., Xu B., Liu L., Liu C., Luo Y., Chen X., Barzegar M., Chung J., Huang S. Both mTORC1 and mTORC2 are involved in the regulation of cell adhesion. Oncotarget. 2015;6(9):7136. doi: 10.18632/oncotarget.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Engl T., Rutz J., Maxeiner S., Juengel E., Roos F., Khoder W., Bechstein W.O., Nelson K., Tsaur I., Haferkamp A. mTOR inhibition reduces growth and adhesion of hepatocellular carcinoma cells in vitro. Mol. Med. Rep. 2017;16(5):7064–7071. doi: 10.3892/mmr.2017.7401. [DOI] [PubMed] [Google Scholar]
- 202.Ackermann M.A., King B., Lieberman N.A., Bobbili P.J., Rudloff M., Berndsen C.E., Wright N.T., Hecker P.A., Kontrogianni-Konstantopoulos A. Novel obscurins mediate cardiomyocyte adhesion and size via the PI3K/AKT/mTOR signaling pathway. J. Mol. Cell. Cardiol. 2017;111:27–39. doi: 10.1016/j.yjmcc.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ribeiro, M. C., Peruchetti, D. B., Silva, L. S., Silva-Filho, J. L., Souza, M. C., Henriques, M. d. G., Caruso-Neves, C., Pinheiro, A. A. S. (2018). LPS induces mTORC1 and mTORC2 activation during monocyte adhesion. Frontiers in molecular biosciences 5 67. [DOI] [PMC free article] [PubMed]
- 204.Wang H., Shao X., He Q., Wang C., Xia L., Yue D., Qin G., Jia C., Chen R. Quantitative proteomics implicates Rictor/mTORC2 in cell adhesion. J. Proteome Res. 2018;17(10):3360–3369. doi: 10.1021/acs.jproteome.8b00218. [DOI] [PubMed] [Google Scholar]
- 205.Avivar-Valderas A., Bobrovnikova-Marjon E., Diehl J.A., Bardeesy N., Debnath J., Aguirre-Ghiso J. Regulation of autophagy during ECM detachment is linked to a selective inhibition of mTORC1 by PERK. Oncogene. 2013;32(41):4932–4940. doi: 10.1038/onc.2012.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gan B., Yoo Y., Guan J.-L. Association of focal adhesion kinase with tuberous sclerosis complex 2 in the regulation of s6 kinase activation and cell growth. J. Biol. Chem. 2006;281(49):37321–37329. doi: 10.1074/jbc.M605241200. [DOI] [PubMed] [Google Scholar]
- 207.Wei G., Wang L., Dong D., Teng Z., Shi Z., Wang K., An G., Guan Y., Han B., Yao M. Promotion of cell growth and adhesion of a peptide hydrogel scaffold via mTOR/cadherin signaling. J. Cell. Physiol. 2018;233(2):822–829. doi: 10.1002/jcp.25864. [DOI] [PubMed] [Google Scholar]
- 208.Baraldo M., Geremia A., Pirazzini M., Nogara L., Solagna F., Türk C., Nolte H., Romanello V., Megighian A., Boncompagni S. Skeletal muscle mTORC1 regulates neuromuscular junction stability. J. Cachexia Sarcopenia Muscle. 2020;11(1):208–225. doi: 10.1002/jcsm.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Winden K.D., Sundberg M., Yang C., Wafa S.M., Dwyer S., Chen P.-F., Buttermore E.D., Sahin M. Biallelic mutations in TSC2 lead to abnormalities associated with cortical tubers in human iPSC-derived neurons. J. Neurosci. 2019;39(47):9294–9305. doi: 10.1523/JNEUROSCI.0642-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wortzel I., Seger R. The ERK cascade: distinct functions within various subcellular organelles. Genes Cancer. 2011;2(3):195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sun C., Enkhjargal B., Reis C., Zhang T., Zhu Q., Zhou K., Xie Z., Wu L., Tang J., Jiang X. Osteopontin-enhanced autophagy attenuates early brain injury via FAK–ERK pathway and improves long-term outcome after subarachnoid hemorrhage in rats. Cells. 2019;8(9):980. doi: 10.3390/cells8090980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Folk, J., Finlayson, J., The ɛ-(γ-glutamyl) lysine crosslink and the catalytic role of transglutaminases, Advances in protein chemistry, Elsevier1977, pp. 1-133. [DOI] [PubMed]
- 213.Mishra S., Murphy L.J. Tissue transglutaminase has intrinsic kinase activity identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase. J. Biol. Chem. 2004;279(23):23863–23868. doi: 10.1074/jbc.M311919200. [DOI] [PubMed] [Google Scholar]
- 214.Mehta K., Fok J.Y., Mangala L.S. Tissue transglutaminase: from biological glue to cell survival cues. Front Biosci. 2006;11(3):173–185. doi: 10.2741/1789. [DOI] [PubMed] [Google Scholar]
- 215.Fésüs L., Szondy Z. Transglutaminase 2 in the balance of cell death and survival. FEBS Lett. 2005;579(15):3297–3302. doi: 10.1016/j.febslet.2005.03.063. [DOI] [PubMed] [Google Scholar]
- 216.D'Eletto M., Grazia Farrace M., Falasca L., Reali V., Oliverio S., Melino G., Griffin M., Fimia G.M., Piacentini M. Transglutaminase 2 is involved in autophagosome maturation. Autophagy. 2009;5(8):1145–1154. doi: 10.4161/auto.5.8.10040. [DOI] [PubMed] [Google Scholar]
- 217.Mastroberardino P.G., Piacentini M. Type 2 transglutaminase in Huntington’s disease: a double-edged sword with clinical potential. J. Intern. Med. 2010;268(5):419–431. doi: 10.1111/j.1365-2796.2010.02275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.D'eletto M., Farrace M., Rossin F., Strappazzon F., Di Giacomo G., Cecconi F., Melino G., Sepe S., Moreno S., Fimia G.M. Type 2 transglutaminase is involved in the autophagy-dependent clearance of ubiquitinated proteins. Cell Death Differ. 2012;19(7):1228–1238. doi: 10.1038/cdd.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Grosso H., Mouradian M.M. Transglutaminase 2: biology, relevance to neurodegenerative diseases and therapeutic implications. Pharmacol. Ther. 2012;133(3):392–410. doi: 10.1016/j.pharmthera.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 220.Franich N.R., Basso M., André E.A., Ochaba J., Kumar A., Thein S., Fote G., Kachemov M., Lau A.L., Yeung S.Y. Striatal mutant huntingtin protein levels decline with age in homozygous Huntington’s disease knock-in mouse models. J. Huntington's Dis. 2018;7(2):137–150. doi: 10.3233/JHD-170274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mastroberardino P., Iannicola C., Nardacci R., Bernassola F., De Laurenzi V., Melino G., Moreno S., Pavone F., Oliverio S., Fesus L. ‘Tissue’transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington's disease. Cell Death Differ. 2002;9(9):873–880. doi: 10.1038/sj.cdd.4401093. [DOI] [PubMed] [Google Scholar]
- 222.Karpuj M.V., Becher M.W., Springer J.E., Chabas D., Youssef S., Pedotti R., Mitchell D., Steinman L. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat. Med. 2002;8(2):143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 223.Menalled L.B., Kudwa A.E., Oakeshott S., Farrar A., Paterson N., Filippov I., Miller S., Kwan M., Olsen M., Beltran J. Genetic deletion of transglutaminase 2 does not rescue the phenotypic deficits observed in R6/2 and zQ175 mouse models of Huntington's disease. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0099520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kumar A., Kneynsberg A., Tucholski J., Perry G., van Groen T., Detloff P.J., Lesort M. Tissue transglutaminase overexpression does not modify the disease phenotype of the R6/2 mouse model of Huntington's disease. Exp. Neurol. 2012;237(1):78–89. doi: 10.1016/j.expneurol.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Martinez J., Chalupowicz D.G., Roush R.K., Sheth A., Barsigian C. Transglutaminase-mediated processing of fibronectin by endothelial cell monolayers. Biochemistry. 1994;33(9):2538–2545. doi: 10.1021/bi00175a024. [DOI] [PubMed] [Google Scholar]
- 226.Telci D., Wang Z., Li X., Verderio E.A., Humphries M.J., Baccarini M., Basaga H., Griffin M. Fibronectin-tissue transglutaminase matrix rescues RGD-impaired cell adhesion through syndecan-4 and β1 integrin co-signaling. J. Biol. Chem. 2008;283(30):20937–20947. doi: 10.1074/jbc.M801763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Akimov S.S., Krylov D., Fleischman L.F., Belkin A.M. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J. Cell Biol. 2000;148(4):825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jones R., Nicholas B., Mian S., Davies P., Griffin M. Reduced expression of tissue transglutaminase in a human endothelial cell line leads to changes in cell spreading, cell adhesion and reduced polymerisation of fibronectin. J. Cell Sci. 1997;110(19):2461–2472. doi: 10.1242/jcs.110.19.2461. [DOI] [PubMed] [Google Scholar]
- 229.Verderio E., Nicholas B., Gross S., Griffin M. Regulated expression of tissue transglutaminase in Swiss 3T3 fibroblasts: effects on the processing of fibronectin, cell attachment, and cell death. Exp. Cell Res. 1998;239(1):119–138. doi: 10.1006/excr.1997.3874. [DOI] [PubMed] [Google Scholar]
- 230.Menter D.G., Patton J., Updyke T., Kerbel R., Maamer M., McIntire L., Nicolson G. Transglutaminase stabilizes melanoma adhesion under laminar flow. Cell Biophys. 1991;18(2):123–143. doi: 10.1007/BF02989810. [DOI] [PubMed] [Google Scholar]
- 231.Van Strien M.E., Brevé J.J., Fratantoni S., Schreurs M.W., Bol J.G., Jongenelen C.A., Drukarch B., Van Dam A.-M. Astrocyte-derived tissue transglutaminase interacts with fibronectin: a role in astrocyte adhesion and migration? PLoS ONE. 2011;6(9) doi: 10.1371/journal.pone.0025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Van Strien M.E., Drukarch B., Bol J.G., Van Der Valk P., Van Horssen J., Gerritsen W.H., Breve J.J., Van Dam A.M. Appearance of tissue transglutaminase in astrocytes in multiple sclerosis lesions: a role in cell adhesion and migration? Brain Pathol. 2011;21(1):44–54. doi: 10.1111/j.1750-3639.2010.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sobel R.A., Mitchell M.E. Fibronectin in multiple sclerosis lesions. Am. J. Pathol. 1989;135(1):161. [PMC free article] [PubMed] [Google Scholar]
- 234.Van Horssen J., Dijkstra C.D., De Vries H.E. The extracellular matrix in multiple sclerosis pathology. J. Neurochem. 2007;103(4):1293–1301. doi: 10.1111/j.1471-4159.2007.04897.x. [DOI] [PubMed] [Google Scholar]
- 235.Liang P., Le W. Role of autophagy in the pathogenesis of multiple sclerosis. Neurosci. Bull. 2015;31(4):435–444. doi: 10.1007/s12264-015-1545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Orhon I., Dupont N., Pampliega O., Cuervo A., Codogno P. Autophagy and regulation of cilia function and assembly. Cell Death Differ. 2015;22(3):389–397. doi: 10.1038/cdd.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lai Y., Jiang Y. Reciprocal regulation between primary cilia and mTORC1. Genes. 2020;11(6):711. doi: 10.3390/genes11060711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Liu S., Trupiano M.X., Simon J., Guo J., Anton E. The essential role of primary cilia in cerebral cortical development and disorders. Curr. Top. Dev. Biol. 2021;142:99–146. doi: 10.1016/bs.ctdb.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hasenpusch-Theil K., Theil T. The multifaceted roles of primary cilia in the development of the cerebral cortex. Front. Cell Dev. Biol. 2021;9:86. doi: 10.3389/fcell.2021.630161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pampliega O., Orhon I., Patel B., Sridhar S., Díaz-Carretero A., Beau I., Codogno P., Satir B.H., Satir P., Cuervo A.M. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502(7470):194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tang Z., Lin M.G., Stowe T.R., Chen S., Zhu M., Stearns T., Franco B., Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502(7470):254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Guemez-Gamboa A., Coufal N.G., Gleeson J.G. Primary cilia in the developing and mature brain. Neuron. 2014;82(3):511–521. doi: 10.1016/j.neuron.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Valente E.M., Rosti R.O., Gibbs E., Gleeson J.G. Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 2014;10(1):27–36. doi: 10.1038/nrneurol.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Park, S. M., Lim, J. S., Ramakrishina, S., Kim, S. H., Kim, W. K., Lee, J., Kang, H.-C., Reiter, J. F., Kim, D. S., Kim, H. H. (2018). Brain somatic mutations in MTOR disrupt neuronal ciliogenesis, leading to focal cortical dyslamination. Neuron 99(1) 83-97.e7. [DOI] [PubMed]
- 245.Sotthibundhu A., Li Q.-X., Thangnipon W., Coulson E.J. Aβ1–42 stimulates adult SVZ neurogenesis through the p75 neurotrophin receptor. Neurobiol. Aging. 2009;30(12):1975–1985. doi: 10.1016/j.neurobiolaging.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 246.Keryer G., Pineda J.R., Liot G., Kim J., Dietrich P., Benstaali C., Smith K., Cordelières F.P., Spassky N., Ferrante R.J. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J. Clin. Investig. 2011;121(11):4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kaliszewski M., Knott A., Bossy-Wetzel E. Primary cilia and autophagic dysfunction in Huntington’s disease. Cell Death Differ. 2015;22(9):1413–1424. doi: 10.1038/cdd.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bae J.-E., Kang G.M., Min S.H., Jo D.S., Jung Y.-K., Kim K., Kim M.-S., Cho D.-H. Primary cilia mediate mitochondrial stress responses to promote dopamine neuron survival in a Parkinson’s disease model. Cell Death Dis. 2019;10(12):1–15. doi: 10.1038/s41419-019-2184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Casas C., Isus L., Herrando-Grabulosa M., Mancuso F.M., Borrás E., Sabidó E., Forés J., Aloy P. Network-based proteomic approaches reveal the neurodegenerative, neuroprotective and pain-related mechanisms involved after retrograde axonal damage. Sci. Rep. 2015;5:9185. doi: 10.1038/srep09185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mooradian A., Chung H., Shah G. GLUT-1 expression in the cerebra of patients with Alzheimer’s disease. Neurobiol. Aging. 1997;18(5):469–474. doi: 10.1016/s0197-4580(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 251.Hunt A., Schönknecht P., Henze M., Seidl U., Haberkorn U., Schröder J. Reduced cerebral glucose metabolism in patients at risk for Alzheimer's disease. Psychiatry Res. Neuroimag. 2007;155(2):147–154. doi: 10.1016/j.pscychresns.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 252.van Assema D.M., Lubberink M., Bauer M., van der Flier W.M., Schuit R.C., Windhorst A.D., Comans E.F., Hoetjes N.J., Tolboom N., Langer O. Blood–brain barrier P-glycoprotein function in Alzheimer's disease. Brain. 2012;135(1):181–189. doi: 10.1093/brain/awr298. [DOI] [PubMed] [Google Scholar]
- 253.Kortekaas R., Leenders K.L., Van Oostrom J.C., Vaalburg W., Bart J., Willemsen A.T., Hendrikse N.H. Blood–brain barrier dysfunction in parkinsonian midbrain in vivo. Ann. Neurol. 2005;57(2):176–179. doi: 10.1002/ana.20369. [DOI] [PubMed] [Google Scholar]
- 254.Drouin-Ouellet J., Sawiak S.J., Cisbani G., Lagacé M., Kuan W.L., Saint-Pierre M., Dury R.J., Alata W., St-Amour I., Mason S.L. Cerebrovascular and blood–brain barrier impairments in Huntington's disease: potential implications for its pathophysiology. Ann. Neurol. 2015;78(2):160–177. doi: 10.1002/ana.24406. [DOI] [PubMed] [Google Scholar]
- 255.Ciechanover, A., Kwon, Y. T. (2015). Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Experimental & molecular medicine 47(3) e147-e147. [DOI] [PMC free article] [PubMed]
- 256.Siddiqi F.H., Menzies F.M., Lopez A., Stamatakou E., Karabiyik C., Ureshino R., Ricketts T., Jimenez-Sanchez M., Esteban M.A., Lai L. Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing. Nat. Commun. 2019;10(1):1–14. doi: 10.1038/s41467-019-09494-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4(5):295. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Han A.-C., Deng J.-X., Huang Q.-S., Zheng H.-Y., Zhou P., Liu Z.-W., Chen Z.-B. Verapamil inhibits scar formation after peripheral nerve repair in vivo. Neural Regener. Res. 2016;11(3):508. doi: 10.4103/1673-5374.179075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Djajadikerta A., Keshri S., Pavel M., Prestil R., Ryan L., Rubinsztein D.C. Autophagy induction as a therapeutic strategy for neurodegenerative diseases. J. Mol. Biol. 2020;432(8):2799–2821. doi: 10.1016/j.jmb.2019.12.035. [DOI] [PubMed] [Google Scholar]
- 260.Liu L., Chen L., Chung J., Huang S. Rapamycin inhibits F-actin reorganization and phosphorylation of focal adhesion proteins. Oncogene. 2008;27(37):4998–5010. doi: 10.1038/onc.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Azios N.G., Dharmawardhane S.F. Resveratrol and estradiol exert disparate effects on cell migration, cell surface actin structures, and focal adhesion assembly in MDA-MB-231 human breast cancer cells. Neoplasia. 2005;7(2):128–140. doi: 10.1593/neo.04346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Xu W., Ge Y., Liu Z., Gong R. Glycogen synthase kinase 3β dictates podocyte motility and focal adhesion turnover by modulating paxillin activity: implications for the protective effect of low-dose lithium in podocytopathy. Am. J. Pathol. 2014;184(10):2742–2756. doi: 10.1016/j.ajpath.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hadzijusufovic E., Albrecht-Schgoer K., Huber K., Hoermann G., Grebien F., Eisenwort G., Schgoer W., Herndlhofer S., Kaun C., Theurl M. Nilotinib-induced vasculopathy: identification of vascular endothelial cells as a primary target site. Leukemia. 2017;31(11):2388–2397. doi: 10.1038/leu.2017.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Botros L., Pronk M.C., Juschten J., Liddle J., Morsing S.K., van Buul J.D., Bates R.H., Tuinman P.R., van Bezu J.S., Huveneers S. Bosutinib prevents vascular leakage by reducing focal adhesion turnover and reinforcing junctional integrity. J. Cell Sci. 2020;133(9) doi: 10.1242/jcs.240077. [DOI] [PubMed] [Google Scholar]
- 265.Roy A., Pahan K. Gemfibrozil, stretching arms beyond lipid lowering. Immunopharmacol. Immunotoxicol. 2009;31(3):339–351. doi: 10.1080/08923970902785253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Al Hassan, M., Fakhoury, I., El Masri, Z., Ghazale, N., Dennaoui, R., El Atat, O., Kanaan, A., El-Sibai, M. (2018). Metformin treatment inhibits motility and invasion of glioblastoma cancer cells. Analytical Cellular Pathology 2018. [DOI] [PMC free article] [PubMed]
- 267.Wang X., Wu Z., He Y., Zhang H., Tian L., Zheng C., Shang T., Zhu Q., Li D., He Y. Humanin prevents high glucose-induced monocyte adhesion to endothelial cells by targeting KLF2. Mol. Immunol. 2018;101:245–250. doi: 10.1016/j.molimm.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 268.Liu J., Zhu B., Zhang G., Wang J., Tian W., Ju G., Wei X., Song B. Electric signals regulate directional migration of ventral midbrain derived dopaminergic neural progenitor cells via Wnt/GSK3β signaling. Exp. Neurol. 2015;263:113–121. doi: 10.1016/j.expneurol.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 269.Kolozsi E., Mackenzie R., Roullet F., Decatanzaro D., Foster J. Prenatal exposure to valproic acid leads to reduced expression of synaptic adhesion molecule neuroligin 3 in mice. Neuroscience. 2009;163(4):1201–1210. doi: 10.1016/j.neuroscience.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 270.Luengo E., Buendia I., Fernández-Mendívil C., Trigo-Alonso P., Negredo P., Michalska P., Hernández-García B., Sánchez-Ramos C., Bernal J.A., Ikezu T. Pharmacological doses of melatonin impede cognitive decline in tau-related Alzheimer models, once tauopathy is initiated, by restoring the autophagic flux. J. Pineal Res. 2019;67(1) doi: 10.1111/jpi.12578. [DOI] [PubMed] [Google Scholar]
- 271.Baydas G., Nedzvetsky V., Nerush P., Kirichenko S., Demchenko H., Reiter R. A novel role for melatonin: regulation of the expression of cell adhesion molecules in the rat hippocampus and cortex. Neurosci. Lett. 2002;326(2):109–112. doi: 10.1016/s0304-3940(02)00328-2. [DOI] [PubMed] [Google Scholar]
- 272.Goodman S.L., Picard M. Integrins as therapeutic targets. Trends Pharmacol. Sci. 2012;33(7):405–412. doi: 10.1016/j.tips.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 273.Raab-Westphal S., Marshall J.F., Goodman S.L. Integrins as therapeutic targets: successes and cancers. Cancers. 2017;9(9):110. doi: 10.3390/cancers9090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Polman C.H., O'Connor P.W., Havrdova E., Hutchinson M., Kappos L., Miller D.H., Phillips J.T., Lublin F.D., Giovannoni G., Wajgt A. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 2006;354(9):899–910. doi: 10.1056/NEJMoa044397. [DOI] [PubMed] [Google Scholar]
- 275.Gudesblatt M., Wissemann K., Zarif M., Bumstead B., Fafard L., Wilken J., Blitz K., Buhse M., Santra S., Hotermans C. Improvement in cognitive function as measured by neurotrax in patients with relapsing multiple sclerosis treated with natalizumab: a 2-year retrospective analysis. CNS Drugs. 2018;32(12):1173–1181. doi: 10.1007/s40263-018-0553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Goldstein L.J., Perez R.P., Yardley D., Han L.K., Reuben J.M., Gao H., McCanna S., Butler B., Ruffini P.A., Liu Y. A window-of-opportunity trial of the CXCR1/2 inhibitor reparixin in operable HER-2-negative breast cancer. Breast Cancer Res. 2020;22(1):1–9. doi: 10.1186/s13058-019-1243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ruffini P.A. The CXCL8-CXCR1/2 axis as a therapeutic target in breast cancer stem-like cells. Front. Oncol. 2019;9:40. doi: 10.3389/fonc.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]