Abstract
β-Catenin and plakoglobin are highly homologous components of cell-cell adherens junctions linking cadherin receptors to the actin cytoskeleton. β-Catenin, in addition, activates transcription by forming a complex with LEF/TCF family transcription factors in the nucleus. Plakoglobin can also bind to LEF-1 and, when overexpressed in mammalian cells, enhances LEF-1-directed transcription. Plakoglobin overexpression, however, results in the elevation and nuclear translocation of endogenous β-catenin. We show here, by DNA mobility shift analysis, that the formation of a plakoglobin-LEF/TCF-DNA complex in vitro is very inefficient compared to a complex containing β-catenin-LEF-DNA. Moreover, in plakoglobin-transfected cells plakoglobin-LEF/TCF-DNA complexes were not formed; rather, the endogenous β-catenin, whose level is elevated by plakoglobin transfection, formed a β-catenin–LEF–DNA complex. Removal of the N- and C-terminal domains of both β-catenin and plakoglobin (leaving the armadillo repeat domain intact) induced plakoglobin-LEF-DNA complex formation and also enhanced β-catenin–LEF–DNA complexing, both with in vitro-translated components and in transfected cells. Transfection with these truncated catenins increased endogenous β-catenin levels, but the truncated catenins acted as dominant-negative inhibitors of β-catenin-driven transcription by forming transcriptionally inactive complexes with LEF-1. When these catenin mutants were prevented from entering the nucleus, by their fusion to the connexin transmembrane domain, they indirectly activated transcription by increasing endogenous β-catenin levels. These results suggest that overexpression of plakoglobin does not directly activate transcription and that formation of catenin-LEF-DNA complexes is negatively regulated by the catenin N- and C-terminal domains.
β-Catenin and plakoglobin (γ-catenin) are homologous proteins originally discovered as cytoplasmic components of cell-cell adherens junctions (52; for a review, see reference 6). Plakoglobin, in addition, is a component of the submembranal plaque of desmosomes (10, 18). In adherens junctions, β-catenin and plakoglobin independently bind to the cytoplasmic domain of cell-cell adhesion receptors of the cadherin family, linking them to the actin cytoskeleton by an association with α-catenin (reviewed in references 3, 6, 20, 34, 76, and 77).
In addition to their structural role in adherens junctions, β-catenin and its Drosophila homologue, armadillo, are part of the wg/wnt signaling pathway (56, 57, 85). Armadillo acts downstream of the secreted signaling molecule wg (the homologue of vertebrate wnt) to regulate wg-dependent developmental decisions (for example, specification of the anterior-posterior segment polarity [56]). During embryonic development in Xenopus laevis, β-catenin is required for the specification of the early dorsoventral axis (24). Ectopic elevation of β-catenin level (19), similar to wnt overexpression (73), results in the induction of a complete secondary embryonic axis. Signaling by β-catenin is believed to involve the nonjunctional pool of this protein, as recruitment of β-catenin to adherens junctions blocks its signaling function (16, 66, 70).
In the absence of a wnt signal, nonjunctional β-catenin is efficiently degraded by the ubiquitin-proteasome system (1, 51, 65). The targeting of β-catenin for degradation involves the phosphorylation of its N terminus by glycogen synthase kinase 3β (GSK) (1, 88), which requires engagement of β-catenin in a multiprotein complex that involves, in addition to β-catenin and GSK, the adenomatous polyposis coli tumor suppressor protein (47, 60), axin (32, 89) or conductin (5, 87), protein phosphatase 2A (27, 68), and dishevelled (17, 41, 72). This complex associates with the F-box/WD-repeat ubiquitin ligase β-TrCP that is responsible for the ubiquitination of β-catenin, leading to its degradation by the proteasome (22, 35, 64, 84).
The binding of wnt to its receptor frizzled results in the inhibition of GSK activity (9) and the subsequent elevation in β-catenin level (55). This is followed by the translocation of β-catenin into the nucleus and its complexing with LEF/TCF transcription factors (4, 29, 45). In this complex, LEF/TCF provides the DNA binding domain, while β-catenin contributes the transactivation domain, forming a bipartite transcription activator of LEF/TCF target genes (45, 80). Aberrant activation of β-catenin signaling, which may result from stabilization of β-catenin in cells carrying inactivating mutations in adenomatous polyposis coli, or mutations in the N terminus of β-catenin, is associated with tumors in a variety of tissues (8, 15, 40, 44, 46, 53, 61, 75, 81, 91). Increased β-catenin levels were shown elsewhere to enhance cell proliferation (50, 86, 90) and, in some cases, to induce neoplastic transformation (39). In line with these findings, the target genes of β-catenin–LEF/TCF, in mammalian cells, include oncogenes such as c-myc (23) and cyclin D1 (69, 78).
While the involvement of β-catenin in wnt signaling is well documented, it is still unclear whether its close homologue, plakoglobin, also has a signaling role. During early development in Xenopus, the reduction in plakoglobin level (38), unlike that of β-catenin (24), does not affect embryonic axis formation. The phenotypes of mice that are knockouts for β-catenin (21, 31) and plakoglobin (7, 62) are also different. While embryos lacking β-catenin die before gastrulation, plakoglobin-null mice undergo organogenesis and die owing to heart failure that results from compromised desmosome assembly in the absence of plakoglobin (7, 62). In addition, the level of plakoglobin is often reduced in cancer cells (2, 48, 74), and the human plakoglobin gene displays loss of heterozygosity in certain tumors (2). Finally, plakoglobin was shown previously to suppress tumorigenicity when overexpressed in various cell lines (71).
Despite these differences between β-catenin and plakoglobin, a transactivation domain was also identified in the plakoglobin molecule (70) and plakoglobin can bind to LEF-1, similarly to β-catenin (29, 70). An elevation in plakoglobin expression was shown to lead to ectopic axis formation in Xenopus (33) and activation of LEF/TCF-dependent transcription in mammalian cells (70). Plakoglobin can also activate LexA-responsive transcription when introduced into yeast cells together with a LEF-LexA fusion protein (25). The interpretation of these results regarding the signaling potential of plakoglobin in Xenopus (42) and mammalian cells is, however, complicated by the fact that increased levels of plakoglobin lead to compromised degradation and nuclear accumulation of the endogenous β-catenin (43, 70). This was suggested to account for the signaling effects reported in plakoglobin-overexpressing cells (43, 70).
In this study, we addressed the mechanism(s) underlying LEF/TCF-dependent transcriptional activation in mammalian cells expressing increased plakoglobin levels. We show that, while plakoglobin binds to LEF-1 with an affinity similar to that of β-catenin, it is inefficient in forming a ternary complex containing LEF-1 and the LEF-1 binding DNA sequence. This apparently results from an inhibitory action confined to the N- and C-terminal domains of the plakoglobin molecule and from the difference between the armadillo repeat domains of β-catenin and plakoglobin. In addition, we demonstrate that transactivation in plakoglobin-transfected cells results from elevated endogenous β-catenin that becomes engaged in a complex with LEF-1 and the LEF-1 binding DNA sequence.
MATERIALS AND METHODS
Cell culture and transfections.
293-T human embryonic kidney cells were cultured in Dulbecco modified Eagle medium and 10% calf serum (Gibco Laboratories), at 37°C, in the presence of 7% CO2. Cells were transiently transfected with the cDNA constructs described below, using Ca2+-phosphate, and the expression of transgenes was assessed 36 h after transfection.
Construction of plasmids.
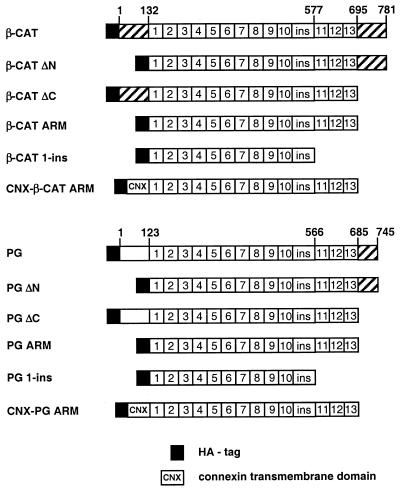
The hemagglutinin (HA)-tagged β-catenin (β-CAT), HA-tagged plakoglobin (PG), vesicular stomatitis virus (VSV)-tagged β-catenin (β-CAT-VSV-C), VSV-tagged plakoglobin (PG-VSV-C), β-CAT 1-ins, and PG 1-ins constructs were described previously (70). A β-catenin mutant containing armadillo repeats 1 to 13 (β-CAT ARM) was obtained by PCR amplification using oligonucleotides 5′-ACCTTCTAGATTGAAACATGCAGTTGTCAATTTG-3′ and 5′-ACCTGGATCCTGCAGTCTCATTCCAAGCC-3′. The amplified fragment was cloned into the XbaI/BamHI site of pCGN (70), resulting in pHA-β-CAT ARM. The PG ARM mutant was obtained by PCR amplification using oligonucleotides 5′-ACCTTCTAGACTCAAGTCGGCCATTGTGC-3′ and 5′-ACCTGGATCCTACTGGGCAGCCTCCCAGGC-3′. The amplified fragment was subcloned into the XbaI/BamHI site of the pCGN vector, resulting in pHA-PG ARM. To prepare fusion constructs of catenin armadillo repeat domains and the connexin transmembrane domain, the latter was obtained by PCR amplification from pCS7. CnxHPkg-UGP (42) using the following primers: 5′-ACCTTCTAGAAACTGGACAGGTCTATAC-3′ and 5′-ACCTACTAGTCGCACAGGCCCGGATGAT-3′. The PCR product was digested by XbaI/SpeI and subcloned into the XbaI site of pHA-β-CAT ARM and pHA-PG ARM. To obtain the β-CAT ARM, PG ARM, PG 1-ins, and β-catenin constructs containing the N-terminal VSV tag, pHA-β-CAT ARM, pHA-PG ARM, pHA PG 1-ins, and pHA-β-catenin were digested with XbaI/BamHI and the inserts were subcloned into pCGN-VSV bearing the N-terminal VSV tag (provided by D. Helfman, Cold Spring Harbor, N.Y.).
For in vitro translation assays, catenin inserts were excised from the pCGN expression vector by XbaI and BamHI and subcloned into pIND-tkUTR (14), which was digested by XbaI and BamHI. The resulting constructs were digested by PmeI, and the inserts containing catenins, with the 5′ untranslated region of herpes simplex virus thymidine kinase, were subcloned into the pCIneo vector, digested by SmaI, and treated with alkaline phosphatase.
Chimeras of β-catenin and plakoglobin were obtained by PCR and contained the N-terminal amino acids 1 to 132 of β-catenin fused to amino acids 123 to 687 of plakoglobin (β-N/PG ARM), amino acids 1 to 132 of β-catenin fused to amino acids 123 to 687 of plakoglobin and amino acids 698 to 781 of β-catenin (β-N/PG ARM/β-C), amino acids 133 to 695 of β-catenin fused to amino acids 686 to 745 of plakoglobin (β-ARM/PG C), and amino acids 123 to 685 of plakoglobin fused to amino acids 696 to 781 of β-catenin (PG ARM/β-C). β-N/PG ARM and β-N/PG ARM/β-C were kindly provided by M. Wheelock (University of Toledo, Toledo, Ohio) (82).
In vitro translation and DNA binding assays.
In vitro-translated proteins were synthesized from a T7 promoter with a coupled transcription and translation (TnT) kit (Promega). The efficiency of in vitro translation was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of parallel translation reactions performed in the presence of [35S]methionine (Amersham). To study the interaction between LEF-1 and catenins, in vitro-translated LEF-1 (0.4 μl) was incubated with a fivefold molar excess of in vitro-translated catenins in the presence of a 32P-labeled duplex oligonucleotide probe, containing a LEF-1 binding site from the cyclin D1 promoter (69). The molar ratio of catenins to LEF-1 or TCF-4 was determined by densitometric tracing of SDS-PAGE autoradiograms obtained from parallel translations in the presence of [35S]methionine. When in vitro-translated TCF-4 was used for the binding reactions, 0.2 μl of TCF-4 and a 10-fold molar excess of catenins were employed. The binding reaction and native gel electrophoresis in 4% acrylamide were performed as described previously (69).
Nuclear extracts.
293-T cells grown in 90-mm-diameter dishes were transfected with 2 μg of LEF-1 and 8 μg of catenin expression plasmids, or with the control pCIneo vector. Thirty-six hours after transfection, nuclear extracts were prepared as described previously (67). Briefly, cells were incubated for 15 min in low-salt buffer, then NP-40 was added, nuclei were pelleted by centrifugation, and nuclear proteins were extracted with high-salt buffer at 4°C. Protein concentrations were determined using the bicinchoninic acid protein assay reagent (Pierce) and bovine serum albumin as a standard. For DNA binding assays, 6 μg of nuclear extract was used. The polyclonal antibodies to β-catenin, the VSV-G epitope, and monoclonal antibody to the HA epitope were all previously described (70). One microgram of antibody was added to the binding reactions for analyzing the DNA mobility supershift. In competition experiments, a 1,000-fold excess of unlabeled oligonucleotides containing a mutant LEF-1 binding site from the cyclin D1 promoter was employed (69).
Immunoblotting.
Equal amounts of protein from nuclear extracts isolated from the transfected cells were separated by SDS-PAGE and subjected to Western blotting using a monoclonal anti-HA antibody (clone 12CA5; Boehringer Mannheim) and a polyclonal anti-VSV-G antibody (a gift from J. C. Perriard, Swiss Federal Institute of Technology, Zurich, Switzerland).
Coimmunoprecipitation.
293-T cells grown in 90-mm-diameter dishes were transfected with VSV-catenin and HA-LEF-1 plasmids and lysed 24 h after transfection in 600 μl of buffer containing 20 mM Tris-HCl, 140 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EGTA, 1.5 mM MgCl2, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride (pH 8.0) (61). The lysates were centrifuged at 15,000 rpm for 15 min at 4°C in an Eppendorf centrifuge and the supernatants were used for Western blotting and coprecipitation experiments. The lysates were precleared for 1 h at 4°C in the presence of 20 μl of protein A/G-coated agarose beads (Santa Cruz), centrifuged for 10 min at 15,000 rpm at 4°C, and incubated with 2 μg of anti-HA or anti-VSV antibodies at 4°C for 1 h, followed by another 1-h incubation with 20 μl of protein A/G-agarose bead suspension, after which the beads were pelleted; washed three times with 1 ml of a buffer containing 20 mM Tris-HCl, 150 mM NaCl, and 0.5% NP-40 (pH 8.0); and boiled for 5 min in 20 μl of SDS-PAGE sample buffer. The samples were subjected to SDS-PAGE and immunoblotted with anti-VSV and anti-HA antibodies.
Transactivation assays.
293-T cells, in 35-mm-diameter dishes, were transfected with 4 μg of catenin constructs, 0.5 μg of the TOPFLASH reporter (80), and 0.5 μg of the control simian virus 40 promoter-driven β-galactosidase, using Ca2+-phosphate, and luciferase activity was measured 36 h after transfection from duplicate plates (70). The inhibitory effects of catenin constructs on β-catenin-dependent transactivation were examined by transfecting 0.6 μg of β-catenin expression plasmid and 3.0 μg of catenin constructs, or control vector, together with 0.5 μg of the TOPFLASH or FOPFLASH reporter and 0.5 μg of LacZ, which served as a control for transfection efficiency. FOPFLASH activity was not affected by transfection of catenin expression plasmids.
Immunofluorescence microscopy.
Cells cultured on glass coverslips were fixed with 3.7% paraformaldehyde in phosphate-buffered saline and permeabilized with 0.5% Triton X-100 (Sigma). Monoclonal antibodies against the C terminus of β-catenin (6F9) and polyclonal antibodies against the HA epitope were described previously (70). The secondary antibody was fluorescein isothiocyanate- or Cy3-labeled goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, Pa.). The epifluorescence images were visualized with an Axiovert S100 TV microscope equipped with a cooled, scientific-grade, charge-coupled device camera (Photometrics, Tucson, Ariz.) and deconvoluted with the DeltaVision 2.10 software on a Silicon Graphics computer. Images were assembled using the Adobe PhotoShop 4.0 software.
For the quantification of β-catenin fluorescence intensity, images of transfected cells were collected with the DeltaVision 2.10 software and integrated cytoplasmic and nuclear fluorescence was measured using the Priism software. Values were normalized for the cell area, and the resulting intensity was divided by the corresponding values obtained with control untransfected cells (chosen within the same field of view). The resulting values were designated fold increase in fluorescence intensity.
RESULTS
Plakoglobin and β-catenin differ in their ability to form a ternary complex with LEF/TCF factors and DNA.
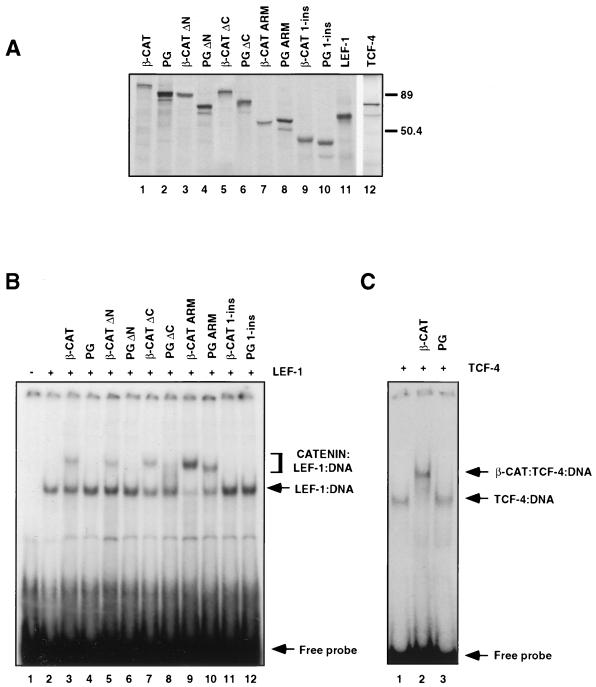
To compare the ability of β-catenin and plakoglobin to form ternary complexes with LEF/TCF and DNA, we used the DNA mobility shift analysis (electrophoretic mobility shift assay [EMSA]) with a radiolabeled LEF-1 DNA binding site as the DNA probe. First, in vitro-transcribed and -translated LEF-1, β-catenin, and plakoglobin and deletion mutants of the two catenins (Fig. 1) were prepared (Fig. 2A), and the interaction of LEF-1 with catenins and DNA was determined (Fig. 2B). Incubation of in vitro-translated LEF-1 with the DNA probe, in the absence of catenins, resulted in the formation of a single LEF-1–DNA band (Fig. 2B, lane 2). When LEF-1 was incubated with the DNA in the presence of β-catenin (at a fivefold molar excess of β-catenin over LEF-1), an additional slower-migrating band was formed (Fig. 2B, lane 3), corresponding to the β-catenin–LEF-1–DNA complex. In contrast to β-catenin, plakoglobin did not form a detectable complex with LEF-1 and DNA (Fig. 2B, lane 4). When β-catenin and plakoglobin were compared in their ability to form catenin–TCF-4–DNA ternary complexes, plakoglobin was again found unable to form a complex composed of plakoglobin–TCF-4–DNA (Fig. 2C, lane 3), while a β-catenin–TCF-4–DNA complex was efficiently formed under the same conditions (Fig. 2C, lane 2, compare to lanes 1 and 3). These results demonstrate that the observed difference between β-catenin and plakoglobin in the efficiency in forming a ternary complex with LEF/TCF factors and DNA was apparent with both LEF-1 and TCF-4 and was not unique to LEF-1.
FIG. 1.
Schematic representation of catenin constructs used in this study. Catenin domains that have transactivation capacity are hatched. The numbers 1 to 13 represent armadillo (ARM) repeats in β-catenin and plakoglobin with a nonrepeat region (ins) between repeats 10 and 11. HA designates the HA epitope tag; CNX marks the transmembrane domain of connexin-32.
FIG. 2.
Identification of β-catenin and plakoglobin domains governing the formation of catenin–LEF-1–DNA complexes. (A) The catenin constructs and LEF/TCF factors were transcribed and translated in vitro in the presence of [35S]methionine, resolved by SDS-PAGE, and visualized by autoradiography. The positions of molecular weight markers (in thousands) are indicated on the right. (B) The in vitro-translated LEF-1 was incubated with 32P-labeled DNA, containing a LEF/TCF binding site, in the absence or presence of a fivefold molar excess of the various in vitro-translated catenin constructs. Protein-DNA complexes were separated by native 4% PAGE and visualized by autoradiography. (C) In vitro-translated TCF-4 was incubated with a 32P-labeled DNA probe containing the LEF/TCF binding site in the presence or absence of a 10-fold molar excess of β-catenin or plakoglobin, and the EMSA was performed as described for panel B.
Since the armadillo repeat domain of β-catenin was implied to contain the binding site for LEF/TCF (4, 80), we next compared the abilities of deletion mutants of β-catenin and plakoglobin comprised of the armadillo repeat domains of these proteins (Fig. 1, β-CAT ARM and PG ARM) to those of full-length catenins in forming ternary complexes by EMSA. The results shown in Fig. 2B demonstrate that there was a significant difference in catenin–LEF-1–DNA complex formation between these constructs: both β-CAT ARM and PG ARM were substantially more efficient in forming catenin–LEF-1–DNA complexes than were the respective full-length molecules (Fig. 2B, lanes 9 and 10, compare to lanes 3 and 4). To determine whether the N or the C termini of catenins were responsible for conferring the inefficient capacity to form ternary complexes by the full-length catenins, we prepared ΔN and ΔC deletion mutants of both β-catenin and plakoglobin (Fig. 1, β-CAT ΔC, β-CAT ΔN, PG ΔC, and PG ΔN) and tested their ability to form complexes with LEF-1 and DNA. Both ΔN and ΔC β-catenin mutants interacted with LEF-1 with an efficiency that was similar to that of the full-length protein but which was much lower than that of the armadillo repeat domain (Fig. 2B, lanes 5 and 7, compare to lanes 3 and 9). ΔN- and ΔC-plakoglobin constructs were also significantly less effective in their interaction with LEF-1 than was PG ARM (Fig. 2B, lanes 6 and 8, compare to lane 10). PG ΔC was slightly more efficient than full-length plakoglobin in forming such a complex (Fig. 2B, compare lane 4 to 8, which displays a smear). Thus, both the N- and C-terminal domains of β-catenin and plakoglobin could apparently act as negative regulators of catenin–LEF-1–DNA ternary complex formation. Removal of the last three armadillo repeats of both β-catenin and plakoglobin (Fig. 1, β-CAT 1-ins and PG 1-ins) completely abolished the interaction with LEF-1 and DNA (Fig. 2B, lanes 11 and 12), indicating that the integrity of the armadillo repeat domain is necessary for binding to LEF-1 (see below).
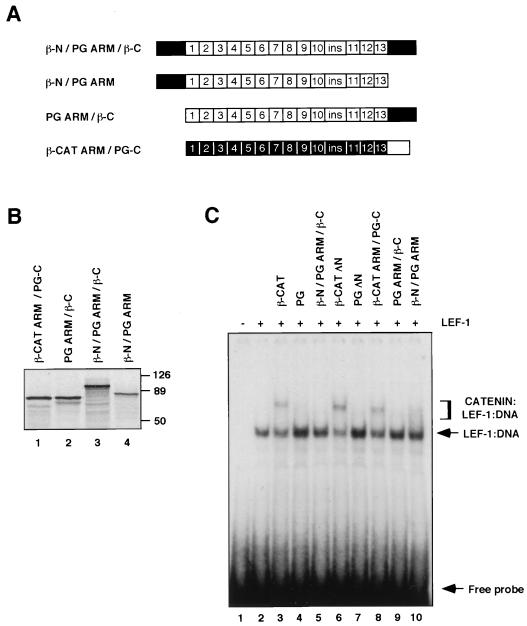
To determine the contribution of the catenin terminal domains and the armadillo repeats to the difference in ternary complex formation between the two catenins, chimeras of β-catenin and plakoglobin were also prepared (Fig. 3A). These constructs were translated in vitro (Fig. 3B) and tested for the ability to form ternary complexes with LEF-1 (Fig. 3C). When the terminal domains of plakoglobin were replaced by those of β-catenin, the resulting chimera (β-N/PG ARM/β-C), similarly to full-length plakoglobin (Fig. 3C, lane 4), was unable to form a detectable complex with LEF-1 and the DNA (Fig. 3C, lane 5), thus being significantly different from β-catenin (Fig. 3C, lane 3). This implies that the terminal domains of either β-catenin or plakoglobin exert a similar inhibitory effect on ternary complex formation by PG ARM.
FIG. 3.
Formation of catenin–LEF–DNA complexes by a chimera of β-catenin and plakoglobin. (A) Schematic representation of β-catenin and plakoglobin chimeric molecules. The β-catenin domains are shown in black, and those of plakoglobin are shown in white. (B) The catenin constructs were translated in vitro in the presence of [35S]methionine, resolved by SDS-PAGE, and visualized by autoradiography. Numbers at right of panel B represent molecular masses in kilodaltons. (C) In vitro-translated LEF-1 was incubated with a 32P-labeled DNA probe that contained a LEF/TCF binding site in the presence or absence of a fivefold molar excess of the in vitro-translated catenin mutants over LEF-1, and the EMSA was performed as described in the legend to Fig. 2.
To determine the inhibitory properties of the C-terminal domains, in the absence of the N terminus, we tested chimeric molecules consisting of the ARM repeats of plakoglobin and β-catenin linked to the C-terminal domain of the other molecule (Fig. 3A, PG ARM/β-C and β-CAT ARM/PG-C). Comparison of these chimeras to β-catenin ΔN and plakoglobin ΔN showed that the β-CAT ARM/PG-C chimeric molecule behaved similarly to β-catenin ΔN (Fig. 3C, lanes 6 and 8) and that PG ARM/β-C was similar to plakoglobin ΔN (Fig. 3C, lanes 7 and 9), in their efficiency in forming ternary complexes. The β-N/PG ARM chimeric molecule formed a smear (Fig. 3C, lane 10) indicative of weak complexing, similar to that observed with plakoglobin ΔC (Fig. 2C, lane 8). Taken together, these experiments demonstrate that the inhibitory properties of the N- and C-terminal domains of β-catenin and plakoglobin are similar, and the major difference between the catenins in ternary complex formation is apparently determined by the ARM domains of these molecules (Fig. 2B, compare lanes 9 and 10).
β-Catenin–LEF-1–DNA but not plakoglobin–LEF-1–DNA complexes are formed in cells transfected with plakoglobin.
We have previously demonstrated that the transfection of plakoglobin into 293-T cells results in LEF-1-dependent transcriptional activation (70). Since plakoglobin was inefficient in the formation of a complex with LEF-1 and DNA in vitro (Fig. 2B and C and Fig. 3C), we next asked whether it could form such a complex in the nuclei of cells overexpressing plakoglobin. 293-T cells were transfected with HA-tagged LEF-1 either alone or together with VSV-tagged plakoglobin, or β-catenin (Fig. 4A), and EMSAs with nuclear extracts from the transfected cells were conducted (Fig. 4B). Nuclear extracts from cells transfected with a control vector formed bands (Fig. 4B, lane 1, N. S.) that were competed by excess unlabeled DNA comprised of the mutant LEF-1 binding site and were thus considered nonspecific (results not shown).
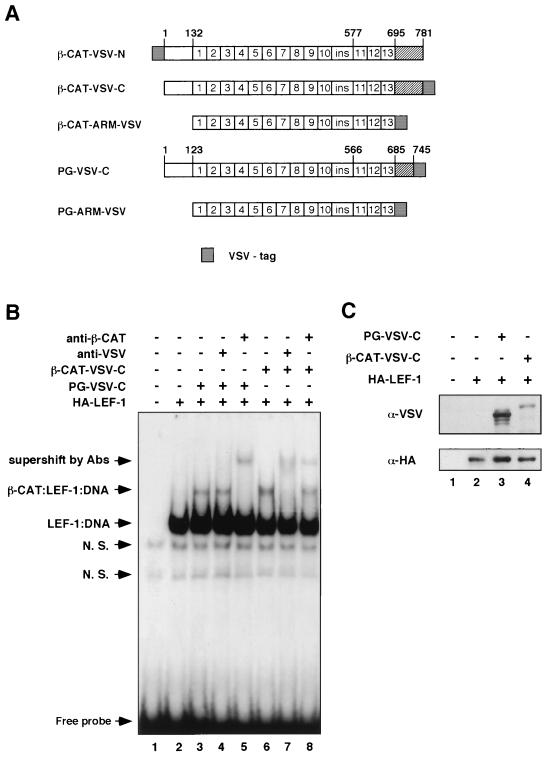
FIG. 4.
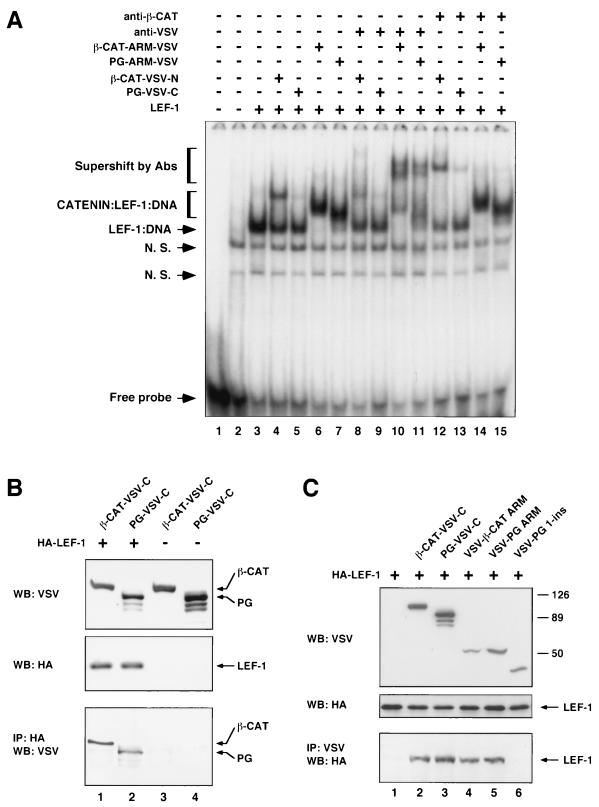
β-Catenin–LEF-1–DNA complex formation in plakoglobin-transfected cells. (A) Schematic representation of N- and C-terminally VSV-tagged catenin constructs. (B) Catenin–LEF-1–DNA complex formation was analyzed with nuclear extracts from 293-T cells transfected with LEF-1 and VSV-plakoglobin, or VSV–β-catenin. Gel EMSAs were performed with 6 μg of nuclear extract from 293-T cells transfected with control vector (lane 1) or with LEF-1 and the indicated catenin mutants. Nuclear extracts were incubated with a 32P-labeled DNA probe containing the LEF-1 binding site, in the absence or presence of the indicated antibodies. DNA-protein complexes were analyzed as described in the legend to Fig. 2. N. S., nonspecific. (C) The nuclear extracts used in panel B were analyzed, by immunoblotting, for levels of catenins (with anti-VSV antibody [upper panel]) and LEF-1 (with anti-HA antibody [lower panel]).
When nuclear extracts from cells transfected with HA-tagged LEF-1 were analyzed, an additional strong band was detected (Fig. 4B, lane 2), which corresponded to the LEF-1–DNA complex and displayed a lower mobility in the presence of anti-HA antibodies (results not shown). When extracts from cells transfected with LEF-1 and β-catenin, or plakoglobin, were analyzed, an additional band (β-CAT–LEF–DNA) whose position corresponded to the expected position of a catenin–LEF-1 complex was apparent (Fig. 4B, lanes 3 and 6). To identify the molecular composition of this band, nuclear extracts were incubated with antibodies to the VSV tag, or to β-catenin (Fig. 4B, lanes 4 and 7 and 5 and 8, respectively). As expected, in extracts from β-catenin-transfected cells this band was supershifted by anti-VSV and anti-β-catenin antibodies (Fig. 4B, lanes 7 and 8, compare to lane 6), confirming that it contained the β-catenin–LEF-1–DNA complex. In contrast, with extracts from cells transfected with VSV-plakoglobin, only the anti-β-catenin antibody induced a shift in this band (Fig. 4B, lane 5), while the anti-VSV antibody did not alter the migration of this band (Fig. 4B, lane 4). Since the level of transfected plakoglobin in the nuclear extracts was higher than that of transfected β-catenin (Fig. 4C), these results imply that plakoglobin did not form a detectable complex with LEF-1–DNA in the VSV-plakoglobin-transfected cells (Fig. 4B, lane 4) but that, rather, the endogenous β-catenin became engaged in a complex with LEF-1 and the DNA.
Removal of the N- and C-terminal domains of β-catenin and plakoglobin enhances catenin–LEF-1–DNA complex formation in transfected cells.
To analyze the ability of the terminal domains of β-catenin and plakoglobin to regulate catenin–LEF-1–DNA complex formation in cells, nuclear extracts from cells transfected with LEF-1 and with VSV-tagged armadillo repeat domains of β-catenin and plakoglobin (Fig. 4A) were subjected to DNA mobility shift analysis. In these experiments, the strong band corresponding to the LEF-1–DNA complex (Fig. 5A, lane 3) was almost completely supershifted in extracts containing the armadillo repeat domains of β-catenin (Fig. 5A, lane 6) and plakoglobin (Fig. 5A, lane 7) compared to full-length catenins (Fig. 4B, lanes 3 and 6). This demonstrates the very efficient formation of DNA-bound complexes comprised of LEF-1–DNA and the armadillo domains. To confirm that these complexes indeed contained β-CAT-ARM-VSV and PG-ARM-VSV, the anti-VSV antibody was added to the nuclear extracts. This resulted in the induction of a supershift in the migration of these bands (Fig. 5A, lanes 10 and 11, compare to lanes 6 and 7). The addition of antibodies to β-catenin to such lysates did not induce an efficient supershift (Fig. 5A, lanes 14, 15), indicating that the nuclear extracts from cells cotransfected with LEF-1 and the armadillo repeat domains of catenins did not contain significant amounts of endogenous β-catenin complexed with LEF-1. Since the polyclonal antibody to β-catenin that was used here did not react with the armadillo repeat domain of β-catenin (data not shown), we attributed the very weak supershift observed with this antibody (Fig. 5A, lanes 14 and 15) to a ternary complex formed by the endogenous β-catenin.
FIG. 5.
The armadillo repeats of β-catenin and plakoglobin efficiently form ternary complexes with LEF-1 and DNA in transfected cells. (A) EMSA was conducted with nuclear extracts from 293-T cells transfected with control vector (lane 2) or with LEF-1 and the indicated catenin constructs (lanes 3 to 15, illustrated in Fig. 4A). Nuclear extracts were incubated with a 32P-labeled DNA probe containing the LEF/TCF binding site, in the absence or presence of antibodies, as indicated. In lane 1, the radiolabeled DNA was incubated without the nuclear extract. DNA-protein complexes were analyzed by native gel electrophoresis as described in Fig. 2. N. S., nonspecific bands that could be competed with wild-type or mutant LEF-1 binding DNA sequences. Abs, antibodies. (B and C) Coimmunoprecipitation of VSV-tagged catenins (B) and their deletion mutants (C) with HA-tagged LEF-1 using cell lysates of transfected 293 cells. (B) The amount of catenins coprecipitated by LEF-1 was determined (lower panel); (C) the amount of LEF-1 coprecipitated by catenins (lower panel) is shown. The levels of transfected proteins in the cell lysates were determined by Western blotting (upper and middle panels). Numbers to the right of panel C are molecular masses in kilodaltons. WB, Western blot; IP, immunoprecipitation.
The VSV-tagged β-catenin used in this experiment (Fig. 5A, lane 4) contained an N-terminal VSV tag (Fig. 4A), in contrast to the C-terminal VSV tag used in the experiments described for Fig. 4B. This resulted in a different efficiency in the supershifts induced by the anti-VSV and anti-β-catenin antibodies in the two experiments (compare Fig. 4B, lanes 7 and 8, to Fig. 5A, lanes 8 and 12). It is noteworthy that in nuclear extracts from transfected cells, PG-ARM-VSV was an efficient partner for LEF-1–DNA, while full-length plakoglobin did not form a detectable complex with LEF-1–DNA (Fig. 5A, lanes 7 and 11, compare to lanes 5 and 9), although the level of plakoglobin in such extracts was higher than that of the PG ARM, as determined by Western blotting (results not shown). These results demonstrate that the increase in catenin–LEF-1–DNA complex formation observed after removal of the N- and C-terminal domains using in vitro-translated components (Fig. 2B) was also apparent with nuclear lysates from cells transfected with these constructs.
To examine whether the differences observed in ternary complex formation resulted from different affinities of the various catenin constructs for LEF-1, we determined the interaction of catenins and their deletion mutants with LEF-1 by coimmunoprecipitation (Fig. 5B and C). VSV-tagged catenins were cotransfected with HA-tagged LEF-1, and immunoprecipitation was performed with either monoclonal anti-HA antibody (Fig. 5B) or polyclonal anti-VSV antibodies (Fig. 5C). In cells expressing similar levels of catenins and LEF-1 (Fig. 5B), the efficiencies of coprecipitation of full-length β-catenin and plakoglobin with LEF-1, by an antibody recognizing the transfected LEF-1 (HA), were very similar (Fig. 5B, lower panel, lanes 1 and 2). Similarly, the coprecipitations of LEF-1 with both catenins by anti-VSV that recognized the catenins were also similar (Fig. 5C, lower panel, lanes 2 and 3). These results, together with previously published data (29), strongly suggest that the differences observed between β-catenin and plakoglobin in ternary complex formation with LEF-1 and DNA apparently do not result from different affinities of the catenins for LEF-1 but most probably are due to the weaker capacity of the plakoglobin–LEF-1 complex to interact with DNA.
We have also compared the abilities of full-length catenins to those of their deletion mutants (by coimmunoprecipitation) in the interaction with LEF-1. The results shown in Fig. 5C (lower panel, lanes 4 and 5) demonstrate that both ARM repeats efficiently bind LEF-1 while PG 1-ins does not display a detectable binding to LEF-1 (Fig. 5C, lane 6), in agreement with its inability to shift the LEF-1–DNA complex in EMSA (Fig. 2B, lanes 11 and 12). The low expression level of the VSV-tagged β-CAT 1-ins precluded us from using it in this experiment (results not shown).
Differential ability of catenin deletion mutants to elevate and translocate endogenous β-catenin into the nucleus.
The formation of a complex between endogenous β-catenin and LEF-1 and DNA, using extracts from cells transfected with catenin mutants (β-CAT-ARM-VSV and PG-ARM-VSV), suggested that these mutants could elevate endogenous β-catenin, similar to observations made with cells transfected with full-length plakoglobin (70). To examine this possibility, we determined, by immunofluorescence microscopy, the abilities of the catenin deletion mutants to elevate and consequently alter the localization of endogenous β-catenin.
293-T cells were transiently transfected with deletion mutants of catenins and doubly stained for the endogenous β-catenin and for the transfected protein (Fig. 6 and 7). In general, the various deletion mutants displayed similar subcellular localizations: they were mainly localized in the nuclei of the transfected cells (Fig. 6C and G and 7A and E) and only sometimes also localized at cell-cell junctions (arrows in Fig. 6 and 7). In addition, PG 1-ins staining was mainly diffuse, while the other constructs were mostly confined to the nucleus.
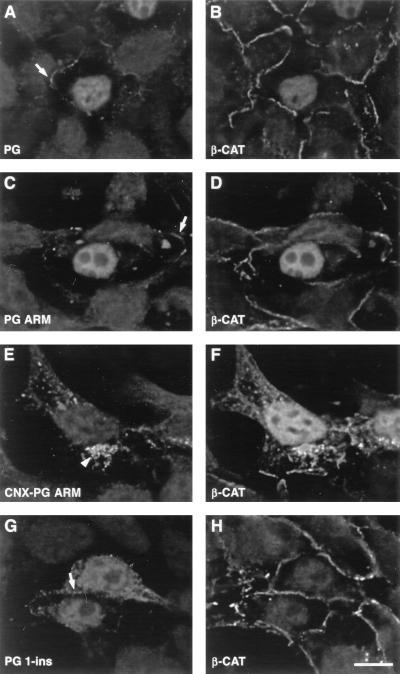
FIG. 6.
The effect of plakoglobin mutants on the localization of endogenous β-catenin. 293-T cells were transiently transfected with the HA-tagged plakoglobin constructs described in Fig. 1 and doubly stained for the HA tag (A, C, E, and G) and the endogenous β-catenin (B, D, F, and H). Cells were transfected with PG (A and B), PG ARM (C and D), CNX-PG ARM (E and F), and PG 1-ins (G and H). Note that PG ARM is efficient in translocating the endogenous β-catenin into the nucleus while PG 1-ins is not. The localization of the transfected proteins to cell-cell contacts is marked by arrows, while localization of CNX-PG ARM to cytoplasmic vesicles is marked by an arrowhead. Bar, 10 μm.
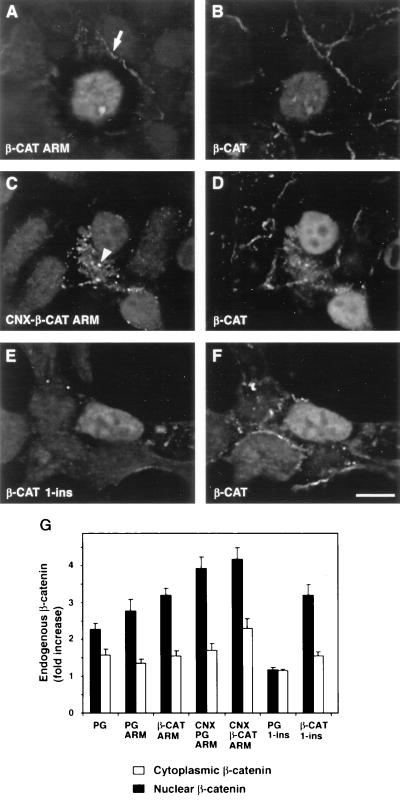
FIG. 7.
Transfection of deletion mutants or membrane-tethered forms of β-catenin increases the level and translocates endogenous β-catenin to the nucleus. 293-T cells were transiently transfected with plasmids encoding the armadillo repeat of β-catenin (β-CAT ARM) (A and B), a connexin-fused derivative (CNX-β-CAT ARM) (C and D), and β-CAT 1-ins (E and F). Cells were stained for the HA tag to detect the transfected proteins (A, C, and E) and doubly stained for endogenous β-catenin with the 6F9 antibody (B, D, and F), which does not recognize the truncated β-catenin mutants. Note that both CNX-β-CAT ARM and β-CAT ARM are efficient in translocating the endogenous β-catenin into the nucleus. Localization of the transfected proteins to cell-cell contact sites (arrow) and to cytoplasmic vesicles (arrowhead) is indicated. Bar, 10 μm. (G) Levels of nuclear and cytoplasmic β-catenin in control and transfected cells were determined by computerized quantitation of the fluorescence intensity as described in Materials and Methods. The values were derived from 30 cells and represent the means ± standard errors of the means. Note that transfection of the catenin constructs induced an increase in the levels of the endogenous β-catenin in both the cytoplasm and the nucleus of the transfected cells.
Double staining for the endogenous β-catenin showed that transfection with both β-CAT ARM (Fig. 7A) and PG ARM (Fig. 6C) resulted in nuclear translocation of the endogenous β-catenin (Fig. 7B and 6D, respectively), similar to the effect of full-length plakoglobin (Fig. 6A and B). This result is attributed to the compromised degradation of β-catenin in these cells (43). The shorter truncation mutants, PG 1-ins and β-CAT 1-ins (Fig. 1), differed in their effect on endogenous β-catenin (Fig. 6H and 7F), as previously described (70). While transfection with β-CAT 1-ins (Fig. 7E) resulted in nuclear localization of the endogenous β-catenin (Fig. 7F), transfection with PG 1-ins (Fig. 6G) did not display a detectable effect on the endogenous β-catenin (Fig. 6H). This implies that the regions within the armadillo repeat domains that determine the binding of catenins to components of the degradation machinery are different in β-catenin and in plakoglobin.
To examine whether nuclear localization of the armadillo repeat domains of both catenins was necessary for nuclear translocation of the endogenous β-catenin, we also prepared fusion constructs of these molecules to the transmembrane domain of connexin (Fig. 1). Such membrane-tethered armadillo repeats of β-catenin and plakoglobin were excluded from the nucleus and localized to cytoplasmic vesicles and to cell borders (arrowheads in Fig. 6E and 7C). The ability to affect the level and localization of endogenous β-catenin, however, was preserved in such constructs, and cells overexpressing these cytoplasm-anchored mutants displayed nuclear staining of the endogenous β-catenin (Fig. 6F and 7D).
We have also determined the fluorescence intensity of endogenous β-catenin in the nuclei and cytoplasm of cells transfected with plakoglobin and with various deletion mutants of catenins (Fig. 7G) and found that, except for PG 1-ins, which did not affect the level of endogenous β-catenin, transfection with the other catenin mutants moderately elevated the cytoplasmic pool of β-catenin (between 1.5- and 2.5-fold) and induced a two- to fourfold increase in the nuclear level of β-catenin (Fig. 7G). The connexin-anchored catenin mutants were the most efficient in their effect on endogenous β-catenin, most probably owing to their exclusive cytoplasmic localization. Interestingly, the level of junctional β-catenin was not significantly affected by transfection with the catenin constructs (Fig. 6 and 7 and data not shown).
Taken together, these results suggest that the β-catenin and plakoglobin mutants examined (except PG 1-ins), were capable of increasing the level and induced the translocation into the nucleus of the endogenous β-catenin, and their cytoplasmic sequestration did not affect this ability.
Transcriptional capacities of deletion mutants of β-catenin and plakoglobin.
The ability of the various catenin deletion mutants to increase the level and translocate endogenous β-catenin into the nucleus did not always correlate with their ability to form ternary complexes with LEF-1–DNA (Fig. 2B, compare to Fig. 6 and 7). For example, while both β-CAT ARM and β-CAT 1-ins could translocate the endogenous β-catenin into the nucleus (Fig. 7B and F), only β-CAT ARM, not β-CAT 1-ins, formed a complex with LEF-1 and DNA (Fig. 2B, lanes 9 and 11). We have therefore analyzed the transactivation capacity of these constructs. We expected that constructs lacking the transactivation domain, but which efficiently formed a complex with LEF-1 and the DNA, would act as dominant-negative inhibitors of transactivation by β-catenin, irrespective of their influence on endogenous β-catenin. We also expected that the transactivation capacity of catenin deletion mutants that do not form complexes with LEF-1 and DNA (β-CAT 1-ins and PG 1-ins) would depend on their capacity to elevate endogenous β-catenin (70).
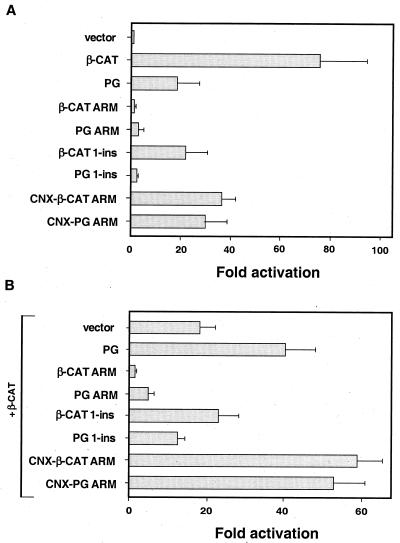
To test these predictions, 293-T cells were transfected with the various constructs, together with a reporter plasmid that detected LEF-1-responsive transcription, and the activity of this reporter was determined (Fig. 8A). As expected, neither β-CAT ARM nor PG ARM and PG 1-ins induced significant transcriptional activation of the reporter, while transfection of β-CAT 1-ins led to a 20-fold activation of the reporter (Fig. 8A). The transactivation potential of β-CAT 1-ins was similar to that of PG (Fig. 8A) (70), in line with their similar capacities to elevate the level of endogenous β-catenin (Fig. 6 and 7). PG ARM and PG 1-ins were both weak in transactivation (Fig. 8A), while their abilities to form complexes with LEF-1–DNA were very different (Fig. 2B, lanes 10 and 12).
FIG. 8.
Transcriptional properties of various β-catenin and plakoglobin constructs. 293-T cells were transfected with different catenin constructs together with a LEF-1-responsive reporter and the LacZ plasmid that served as control for transfection efficiency and lysed 36 h after transfection, and luciferase activity was determined. The values (fold activation) were normalized for transfection efficiency by analyzing β-galactosidase activity. (A) Note that transfection with the armadillo repeat constructs PG ARM and β-CAT ARM, which lack the transactivation domain, was inefficient in the induction of reporter activation, while the connexin fusion of these constructs (CNX-PG ARM and CNX-β-CAT ARM) effectively induced transactivation. (B) The inhibitory effect of catenin constructs on transcriptional activation driven by full-length β-catenin was determined by transfecting 0.6 μg of β-catenin and 3 μg of control plasmid, or the indicated catenin constructs, together with the reporter plasmid and the LacZ control. Note the inhibitory effect of β-CAT ARM and PG ARM (probably by a dominant-negative action in the nucleus). In contrast, CNX-β-CAT ARM and CNX-PG ARM, which are sequestered in the cytoplasm, elevated transcription similarly to plakoglobin. In addition, β-CAT 1-ins and PG 1-ins, which could not complex with LEF-DNA (Fig. 2B), had no significant effect on β-catenin-directed transactivation.
To examine whether an efficient complex formation by these mutants with LEF-1–DNA is reflected in a dominant-negative effect on β-catenin-directed transactivation, β-catenin was transfected together with the expression plasmids coding for β-CAT ARM, PG ARM, β-CAT 1-ins, and PG 1-ins (Fig. 8B). In agreement with the in vitro binding studies, both β-CAT ARM and PG ARM inhibited β-catenin-driven transactivation, while β-CAT 1-ins and PG 1-ins had no effect (Fig. 8B; see Fig. 9C for the proposed mechanism).
FIG. 9.
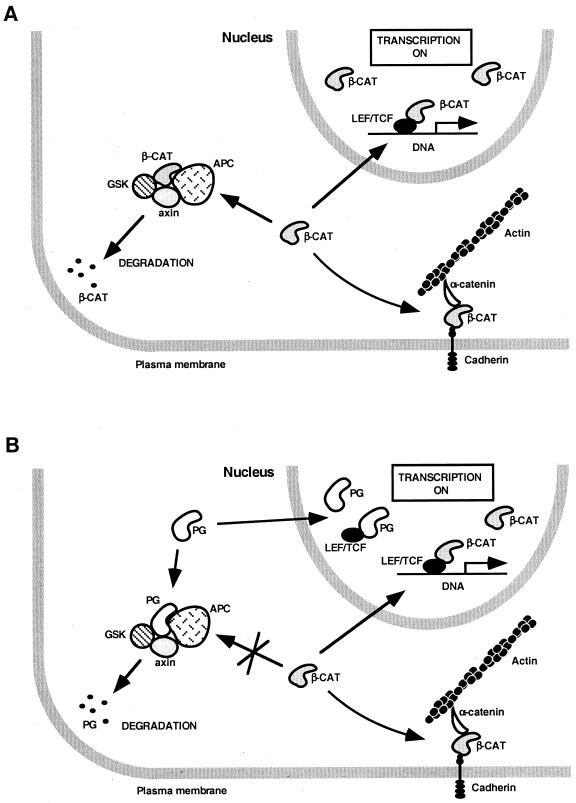
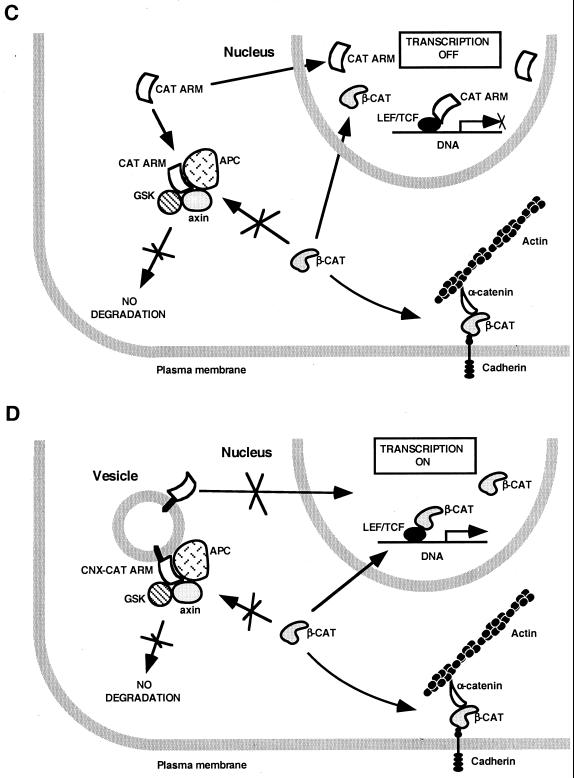
Schematic representation of the proposed effect of plakoglobin and catenin mutants on endogenous β-catenin and LEF/TCF-dependent transcription. (A) When β-catenin levels are elevated, β-catenin activates transcription directly by forming a β-catenin–LEF/TCF complex. (B) Plakoglobin blocks degradation of endogenous β-catenin in the cytoplasm and also localizes to the nucleus but does not form a plakoglobin-LEF/TCF-DNA complex; rather, it induces transactivation indirectly by elevating the level of endogenous β-catenin. (C) The armadillo repeat domains of β-catenin and plakoglobin (CAT ARM) elevate endogenous β-catenin but act as dominant-negative inhibitors of transcription by forming a transcriptionally inactive complex with LEF/TCF and DNA. (D) The membrane-tethered armadillo repeats of β-catenin and plakoglobin (CNX-CAT ARM) act as transcriptional activators by elevating endogenous β-catenin similarly to plakoglobin.
To demonstrate that this inhibition of transcription requires nuclear localization of the β-CAT ARM and PG ARM constructs, we have also used fusion proteins of these catenin mutants with the transmembrane domain of connexin (Fig. 1). With these membrane-tethered forms of β-CAT ARM and PG ARM (CNX-PG ARM and CNX-β-CAT ARM), there was no inhibition of transactivation, but rather a significant induction of transcription (Fig. 8B). This was expected, as these fusion proteins were localized in the cytoplasm (Fig. 7C and 6E), elevated endogenous β-catenin levels, and efficiently activated transcription (Fig. 8A), most probably by the endogenous β-catenin (Fig. 9D).
Taken together, the transactivation studies demonstrated that the differences observed in vitro in LEF-1–DNA complex formation with the various β-catenin and plakoglobin mutants were also reflected in their different transactivation capacities in transfected cells.
DISCUSSION
β-Catenin and plakoglobin display a high degree of homology and have similar functions in the assembly of cell-cell adherens junctions, but the two proteins are clearly different in various aspects (for a review, see reference 6). This was most convincingly demonstrated by the different phenotypes observed in mice that were knockouts for plakoglobin (7, 62) and β-catenin (21, 31) and by the different effects of the interference with the function of maternal β-catenin (24) and plakoglobin (38) in Xenopus embryos. Mice lacking plakoglobin did not display abnormalities that could be attributed to compromised wnt signaling in early development, and a decrease in maternal plakoglobin level in Xenopus did not affect embryonic axis formation, which, in contrast, is critically dependent on β-catenin–LEF/TCF-driven transcription. Furthermore, activating mutations in β-catenin were reported to lead to oncogenic transformation (reviewed in references 6 and 58), and oncogenes such as c-myc and cyclin D1 (23, 69, 78) and invasion-promoting secreted proteases including matrilysin (13) are direct transcriptional targets of the β-catenin–LEF/TCF complex. In contrast, the levels of plakoglobin are often reduced in cancer cells (2, 48, 71, 74), and overexpression of plakoglobin was shown to suppress tumorigenicity (71).
In spite of these differences, when several functional tests were applied to β-catenin and plakoglobin, they revealed a high degree of similarity in the effects of the two catenins. For example, ectopic overexpression of either β-catenin (19, 24) or plakoglobin (33) leads to axis duplication in Xenopus, and both proteins can induce transcriptional activation of a LEF/TCF reporter plasmid when transfected into mammalian cells (46, 70). To address this apparent contradiction between the similarities and the differences in β-catenin and plakoglobin functions, we studied the mechanism of transcriptional activation in plakoglobin-overexpressing cells and compared the abilities of β-catenin and plakoglobin to form complexes with LEF/TCF and DNA.
The difference between β-catenin and plakoglobin in the formation of catenin–LEF/TCF–DNA complexes.
By applying the DNA EMSA, we demonstrated that plakoglobin differs from β-catenin in its ability to form a catenin–LEF/TCF–DNA complex. While β-catenin–LEF/TCF–DNA complexes were efficiently formed using this experimental approach, plakoglobin–LEF/TCF–DNA complex formation was undetectable. However, we (this study) and others (29) found that transfected β-catenin and plakoglobin were both effective in coimmunoprecipitating LEF-1, and vice versa, and the catenin-binding domain of LEF-1 is equally potent in recruiting β-catenin and plakoglobin to a LexA-driven promoter (25). In addition, overexpression of either β-catenin or plakoglobin results in its colocalization with LEF-1 in the nuclei of the transfected cells (4, 63, 70). Taken together with the present study, these results suggest that, while a plakoglobin–LEF-1 complex can form, this complex is probably very inefficient in binding to DNA. This difference in DNA binding of the catenin complexes was detected with nuclear extracts from transfected cells and also with in vitro-translated proteins, using either LEF-1 or TCF-4. Such a difference, therefore, likely results from inherent structural variations between β-catenin and plakoglobin and not from differential regulation of catenins by their interaction with other regulatory proteins.
Since cells transfected with plakoglobin display LEF/TCF-dependent transactivation, while plakoglobin–LEF-1–DNA complexes were not detected in the nuclear extracts of these cells, we examined the reason for LEF-directed transcription in these cells. We demonstrated that the catenin–LEF-1–DNA complex detected by DNA mobility shift analysis with nuclear extracts from plakoglobin-transfected cells is comprised, by and large, of the endogenous β-catenin whose level was increased in these cells (43, 70). It appears, therefore, that transactivation in cells transfected with plakoglobin is most likely mediated by the elevated endogenous β-catenin and not by plakoglobin (Fig. 9A and B). In addition, the apparent similarity in the effects of β-catenin and plakoglobin on transcriptional activation does not reflect similar mechanisms of action of the two proteins (Fig. 9). While the overexpression of β-catenin probably mimics the elevation in β-catenin following activation of the wnt pathway, which leads to β-catenin-driven transactivation after complexing with LEF/TCF, overexpression of plakoglobin results in compromised degradation of the endogenous β-catenin (by the interaction of plakoglobin with components of the degradation machinery [30, 37, 43, 59, 64]), thereby indirectly leading to transactivation (Fig. 9A and B). This plakoglobin-induced transcription masks the apparent inability of plakoglobin to act as coactivator of LEF/TCF factors as shown in the present study. These findings support the hypothesis that the function of plakoglobin in Xenopus axis specification is mediated by the elevation of the endogenous β-catenin (43) and are also in agreement with the phenotype of plakoglobin knockout mice (7, 62), which display normal wnt signaling. These results are also consistent with the inability of plakoglobin to rescue the signaling function of armadillo in arm−/− Drosophila embryos (83).
Taking these results together with studies demonstrating the ability of both plakoglobin and β-catenin to complex with LEF-1 in transfected cells (this study and references 29 and 70), we suggest a model (Fig. 9B) in which plakoglobin can form a complex with LEF-1 in the nuclei of cells transfected with plakoglobin but this complex is not bound to DNA, while the elevated endogenous β-catenin in such cells forms transcriptionally active β-catenin–LEF/TCF–DNA complexes.
This model does not rule out the possibility that plakoglobin may have a role in transactivation under other circumstances. This notion is based on the presence of a C-terminal transactivation domain in the plakoglobin molecule that is active in transcription when linked to a Gal4 DNA binding domain, similar to the C terminus of β-catenin and armadillo (70). In addition, the comparison of the ARM repeats of the two catenins in their ability to complex with LEF–DNA suggests a quantitative, rather than a qualitative, difference between the two proteins (Fig. 2, lanes 9 and 10). Moreover, a fusion protein consisting of a heterologous DNA binding domain with LEF-1 (LexA–LEF-1) could activate transcription with both β-catenin and plakoglobin in yeasts, which lack endogenous catenins (25). It is noteworthy, however, that the heterologous LexA DNA binding domain that mediated DNA binding in the plakoglobin–LEF-1–DNA complex (25) could have been differently affected by plakoglobin, compared to the binding of LEF-1 to DNA via its native high-mobility group domain (as used in the present study).
Regulation of catenin–LEF-1–DNA complex formation by the N- and C-terminal domains of catenins.
While plakoglobin was inefficient in the formation of a catenin–LEF-1–DNA complex, we found that its armadillo repeat domain was highly competent in this activity. This rather unexpected finding suggests that catenins may have built-in regulatory domains that affect catenin–LEF/TCF–DNA complex formation. Our attempts to map such regulatory domains led us to conclude that both the N- and C-terminal domains of the catenin molecules are required for such negative regulation of plakoglobin–LEF-1–DNA complex formation. Interestingly, a similar negative effect on β-catenin–LEF-1–DNA complex formation of the N- and C-terminal domains of β-catenin was also observed, suggesting a common regulatory mechanism for both catenins.
Our efforts to determine whether the difference between catenins in ternary complex formation results from different terminal domains or from differences in the ARM repeats lead us to conclude that this is most probably attributable to the ARM domains of the two proteins and is enhanced by the inhibitory effect of the terminal domains. Our finding that the terminal domains of one catenin could be replaced by those of the other, without a significant change in the efficiency of the molecules in forming a ternary complex, supports this notion.
A recent study using the yeast two-hybrid screen demonstrated that the C terminus of the Drosophila melanogaster β-catenin (armadillo) could bind to the armadillo repeat domain of this molecule (11). It is possible that the inhibitory effect of the terminal domains of catenins on catenin–LEF-1–DNA complex formation results from intramolecular interactions within the catenin molecule, a property conserved in evolution between Drosophila and mammals. In this respect, it is noteworthy that the C termini of β-catenin and plakoglobin are acidic, while the armadillo repeat domain forms a positively charged groove (28) that can potentially serve as a binding site for the terminal domains. Interestingly, deletion of the C terminus of plakoglobin was shown to increase its binding in vitro to the cytoplasmic domain of E-cadherin (79) and to enhance desmosome assembly in transfected cells (54). In addition, terminal domains of β-catenin were shown to negatively regulate its interaction with desmoglein, the effect that may arise from the intramolecular interactions within the catenin molecule (82). Together, these results strongly support the existence of a negative regulation of ternary complex formation with LEF/TCF–DNA and the interaction of catenins with other partners by their terminal domains.
The in vitro ternary complex formation studies were corroborated by transactivation experiments demonstrating that the armadillo repeat domains of both catenins (which lack the transactivation domains) can act as dominant-negative effectors of β-catenin-mediated transcription, by forming inactive complexes with LEF-1 on the DNA. Such inactive complexes on the DNA could prevent the complexing of endogenous β-catenin with LEF–DNA and thus inhibit transactivation (Fig. 9C).
Interestingly, the ARM domain of β-catenin was shown to act positively in the wnt signaling pathway of Xenopus (19), in contrast to the results of the present study. One possible explanation for this difference could be that the balance between the negative effect (owing to ARM complexing with LEF-1) and the positive effect (by elevating endogenous β-catenin) is different when comparing the injection studies in Xenopus and the transfection in mammalian cells, therefore leading to an activation rather than to inhibition in the Xenopus system.
As predicted, cytoplasmic anchorage of the armadillo domains, of either plakoglobin or β-catenin, by their fusion to the transmembrane domain of connexin abolished their inhibitory effect on transcription. In contrast, such membrane-tethered forms of the armadillo domains of both catenins acted as transcription activators, by elevating the endogenous β-catenin, similar to the effect of membrane-anchored β-catenin (12, 26, 42, 43, 49) (Fig. 9C and D) and plakoglobin (36, 43).
In conclusion, this study demonstrated significant differences between the abilities of β-catenin and plakoglobin to form DNA-bound complexes with LEF/TCF factors. In addition, transcriptional activation in cells overexpressing plakoglobin is apparently not directly regulated by plakoglobin but is mediated by the elevated endogenous β-catenin. These results are consistent with the differential roles that the two catenins may play in wnt signaling (7, 38, 62).
Nevertheless, since the armadillo repeat domain of plakoglobin can efficiently bind to the LEF-1–DNA complex and since there is a transactivation domain in the C terminus of plakoglobin (25, 70), plakoglobin may have a potential to act as a transcription activator under certain conditions. Current studies in our laboratory, employing DNA-microarray technology, are addressing the question of the possible capacity of plakoglobin to induce genes in cells lacking β-catenin.
ACKNOWLEDGMENTS
This study was supported by grants from the German-Israeli Foundation for Scientific Research and Development and the Cooperation Program in Cancer Research between DKFZ and IMOSA. A.B.Z. holds the Lunenfeld-Kunin Chair in Genetics and Cell Biology.
We are grateful to M. Wheelock, H. Clevers, M. van de Wetering, M. Klymkowsky, and D. Helfman for sending reagents; to D. Riveline for stimulating discussions; to E. Sadot and T. Gottlieb for critical reading of the manuscript; and to Inbal Simcha for continuous support.
J.Z. and M.S. contributed equally to this work.
REFERENCES
- 1.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aberle H, Bierkamp C, Torchard D, Serova O, Wagner T, Natt E, Wirsching J, Heidkamper C, Montagna M, Lynch H. The human plakoglobin gene localizes on chromosome 17q21 and is subjected to loss of heterozygosity in breast and ovarian cancers. Proc Natl Acad Sci USA. 1995;92:6384–6388. doi: 10.1073/pnas.92.14.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams C, Nelson J W. Cytomechanics of cadherin-mediated cell-cell adhesion. Curr Opin Cell Biol. 1998;10:572–577. doi: 10.1016/s0955-0674(98)80031-8. [DOI] [PubMed] [Google Scholar]
- 4.Behrens J, von Kries J, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 5.Behrens J, Jerchow B, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Ze'ev A, Geiger B. Differential molecular interactions of β-catenin and plakoglobin in adhesion, signaling and cancer. Curr Opin Cell Biol. 1998;10:629–639. doi: 10.1016/s0955-0674(98)80039-2. [DOI] [PubMed] [Google Scholar]
- 7.Bierkamp C, Mclaughlin K, Schwarz H, Huber O, Kemler R. Embryonic heart and skin defects in mice lacking plakoglobin. Dev Biol. 1996;180:780–785. doi: 10.1006/dbio.1996.0346. [DOI] [PubMed] [Google Scholar]
- 8.Chan E, Gat U, McNiff J, Fuchs E. A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 9.Cook D, Fry M, Hughes K, Sumathipala R, Woodgett J, Dale T. Wingless inactivates glycogen synthase kinase-3 via an intracellular signalling pathway which involves a protein kinase C. EMBO J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]
- 10.Cowin P, Kapprell H, Franke W, Tamkun J, Hynes R. Plakoglobin: a protein common to different kinds of intercellular adhering junctions. Cell. 1986;46:1063–1073. doi: 10.1016/0092-8674(86)90706-3. [DOI] [PubMed] [Google Scholar]
- 11.Cox R, Pai I, Kirkpatrick C, Stein J, Peifer M. Roles of the C terminus of armadillo in wingless signaling in Drosophila. Genetics. 1999;153:319–332. doi: 10.1093/genetics/153.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox R, Pai I, Miller J, Orsulic S, Stein J, McCormick C, Audeh Y, Wang W, Moon R, Peifer M. Membrane-tethered Drosophila Armadillo cannot transduce Wingless signal on its own. Development. 1999;126:1327–1335. doi: 10.1242/dev.126.6.1327. [DOI] [PubMed] [Google Scholar]
- 13.Crawford H, Fingleton B M, Rudolph-Owen L, Goss K, Rubinfeld B, Polakis P, Matrisian L. The metalloproteinase matrilysin is a target of β-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 14.Damalas A, Ben-Ze'ev A, Simcha I, Shtutman M, Leal J F, Zhurinsky J, Geiger B, Oren M. Excess β-catenin promotes accumulation of transcriptionally active p53. EMBO J. 1999;18:3054–3063. doi: 10.1093/emboj/18.11.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de La Coste A, Romagnolo B, Billuart P, Renard C, Buendia M, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, Perret C. Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagotto F, Funayama N, Gluck U, Gumbiner B. Binding to cadherins antagonizes the signaling activity of β-catenin during axis formation in Xenopus. J Cell Biol. 1996;132:1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fagotto F, Jho E, Zeng L, Kurth T, Joos T, Kaufmann C, Costantini F. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J Cell Biol. 1999;145:741–756. doi: 10.1083/jcb.145.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke W, Goldschmidt M, Zimbelmann R, Mueller H, Schiller D, Cowin P. Molecular cloning and amino acid sequence of human plakoglobin, the common junctional plaque protein. Proc Natl Acad Sci USA. 1989;86:4027–4031. doi: 10.1073/pnas.86.11.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funayama N, Fagotto F, McCrea P, Gumbiner B. Embryonic axis induction by the armadillo repeat domain of β-catenin: evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiger B, Ayalon O. Cadherins. Annu Rev Cell Biol. 1992;8:307–332. doi: 10.1146/annurev.cb.08.110192.001515. [DOI] [PubMed] [Google Scholar]
- 21.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of β-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 22.Hart M, Concordet J, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 23.He T, Sparks A, Rago C, Hermeking H, Zawel L, da Costa L, Morin P, Vogelstein B, Kinzler K. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 24.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro C, Wylie C. Overexpression of cadherins and underexpression of β-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 25.Hecht A, Litterst C, Huber O, Kemler R. Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- 26.Hsu S, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with β-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu W, Zeng L, Costantini F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem. 1999;274:3439–3445. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- 28.Huber A, Nelson J W, Weis W. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 29.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B, Kemler R. Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 30.Hulsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. Requirement for β-catenin in anterior-posterior axis formation in mice. J Cell Biol. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karnovsky A, Klymkowsky M. Anterior axis duplication in Xenopus induced by the over-expression of the cadherin-binding protein plakoglobin. Proc Natl Acad Sci USA. 1995;92:4522–4526. doi: 10.1073/pnas.92.10.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 35.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K, Nakayama R. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klymkowsky M, Williams B, Barish G, Varmus H, Vourgourakis Y. Membrane-anchored plakoglobins have multiple mechanisms of action in Wnt signaling. Mol Biol Cell. 1999;10:3151–3169. doi: 10.1091/mbc.10.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kodama S, Ikeda S, Asahara T, Kishida M, Kikuchi A. Axin directly interacts with plakoglobin and regulates its stability. J Biol Chem. 1999;274:27682–27688. doi: 10.1074/jbc.274.39.27682. [DOI] [PubMed] [Google Scholar]
- 38.Kofron M, Spagnuolo A, Klymkowsky M, Wylie C, Heasman J. The roles of maternal α-catenin and plakoglobin in the early Xenopus embryo. Development. 1997;124:1553–1560. doi: 10.1242/dev.124.8.1553. [DOI] [PubMed] [Google Scholar]
- 39.Kolligs F, Hu G, Dang C, Fearon E. Neoplastic transformation of RK3E by mutant β-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korinek V, Barker N, Morin P, van Wichen D, de Weger R, Kinzler K, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Yuan H, Weaver C, Mao J, Farr G, Sussman D, Jonkers J, Kimelman D, Wu D. Axin and frat1 interact with dvl and GSK, bridging dvl to GSK in wnt-mediated regulation of LEF-1. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merriam J, Rubenstein A, Klymkowsky M. Cytoplasmically anchored plakoglobin induces a WNT-like phenotype in Xenopus. Dev Biol. 1997;185:67–81. doi: 10.1006/dbio.1997.8550. [DOI] [PubMed] [Google Scholar]
- 43.Miller J, Moon R. Analysis of the signaling activities of localization mutants of β-catenin during axis specification in Xenopus. J Cell Biol. 1997;139:229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyoshi Y, Iwao K, Nawa G, Yoshikawa H, Ochi T, Nakamura Y. Frequent mutations in the β-catenin gene in desmoid tumors from patients without familial adenomatous polyposis. Oncol Res. 1998;10:591–594. [PubMed] [Google Scholar]
- 45.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 46.Morin P, Sparks A, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 47.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular β-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro P, Lozano E, Cano A. Expression of E- or P-cadherin is not sufficient to modify the morphology and the tumorigenic behavior of murine spindle carcinoma cells: possible involvement of plakoglobin. J Cell Sci. 1993;105:923–934. doi: 10.1242/jcs.105.4.923. [DOI] [PubMed] [Google Scholar]
- 49.Nelson R, Gumbiner B. β-Catenin directly induces expression of the Siamois gene, and can initiate signaling indirectly via a membrane-tethered form. Ann N Y Acad Sci. 1998;854:86–98. doi: 10.1111/j.1749-6632.1998.tb10109.x. [DOI] [PubMed] [Google Scholar]
- 50.Orford K, Orford C, Byers S. Exogenous expression of β-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J Cell Biol. 1999;146:855–868. doi: 10.1083/jcb.146.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orford K, Crockett C, Jensen J, Weissman A, Byers S. Serine phosphorylation-regulated ubiquitination and degradation of β-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 52.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palacios J, Gamallo C. Mutations in the β-catenin gene (CTNNB1) in endometrioid ovarian carcinomas. Cancer Res. 1998;58:1344–1347. [PubMed] [Google Scholar]
- 54.Palka H, Green K. Roles of plakoglobin end domains in desmosome assembly. J Cell Sci. 1997;110:2359–2371. doi: 10.1242/jcs.110.19.2359. [DOI] [PubMed] [Google Scholar]
- 55.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peifer M, Orsulic S, Pai L, Loureiro J. A model system for cell adhesion and signal transduction in Drosophila. Dev Suppl. 1993;1993:163–176. [PubMed] [Google Scholar]
- 57.Peifer M, Wieschaus E. The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell. 1990;63:1167–1176. doi: 10.1016/0092-8674(90)90413-9. [DOI] [PubMed] [Google Scholar]
- 58.Polakis P. The oncogenic activation of β-catenin. Curr Opin Genet Dev. 1999;9:15–21. doi: 10.1016/s0959-437x(99)80003-3. [DOI] [PubMed] [Google Scholar]
- 59.Rubinfeld B, Souza B, Albert I, Munemitsu S, Polakis P. The APC protein and E-cadherin form similar but independent complexes with α-catenin, β-catenin, and plakoglobin. J Biol Chem. 1995;270:5549–5555. doi: 10.1074/jbc.270.10.5549. [DOI] [PubMed] [Google Scholar]
- 60.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3β to the APC-β-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 61.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 62.Ruiz P, Brinkmann V, Ledermann B, Behrend M, Grund C, Thalhammer C, Vogel F, Birchmeier C, Gunthert U, Franke W, Birchmeier W. Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J Cell Biol. 1996;135:215–225. doi: 10.1083/jcb.135.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadot E, Simcha I, Shtutman M, Ben-Ze'ev A, Geiger B. Inhibition of β-catenin-mediated transactivation by cadherin derivatives. Proc Natl Acad Sci USA. 1998;95:15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadot E, Simcha I, Iwai K, Ciechanover A, Geiger B, Ben-Ze'ev A. Differential interaction of plakoglobin and β-catenin with the ubiquitin-proteasome system. Oncogene. 2000;19:1992–2001. doi: 10.1038/sj.onc.1203519. [DOI] [PubMed] [Google Scholar]
- 65.Salomon D, Sacco P, Roy S, Simcha I, Johnson K, Wheelock M, Ben-Ze'ev A. Regulation of β-catenin levels and localization by overexpression of plakoglobin and inhibition of the ubiquitin-proteasome system. J Cell Biol. 1997;139:1325–1335. doi: 10.1083/jcb.139.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanson B, White P, Vincent J. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- 67.Schreiber E, Matthias P, Muller M, Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seeling J, Miller J, Gil R, Moon R, White R, Virshup D. Regulation of β-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- 69.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, Ben-Ze'ev A. Differential nuclear translocation and transactivation potential of β-catenin and plakoglobin. J Cell Biol. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simcha I, Geiger B, Yehuda-Levenberg S, Salomon D, Ben-Ze'ev A. Suppression of tumorigenicity by plakoglobin: an augmenting effect of N-cadherin. J Cell Biol. 1996;133:199–209. doi: 10.1083/jcb.133.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smalley M, Sara E, Paterson H, Naylor S, Cook D, Jayatilake H, Fryer L, Hutchinson L, Fry M, Dale T. Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. EMBO J. 1999;18:2823–2835. doi: 10.1093/emboj/18.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sokol S, Christian J, Moon T, Melton D. Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell. 1991;67:741–752. doi: 10.1016/0092-8674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 74.Sommers C, Gelmann E, Kemler R, Cowin P, Byers S. Alterations in β-catenin phosphorylation and plakoglobin expression in human breast cancer cells. Cancer Res. 1994;54:3544–3552. [PubMed] [Google Scholar]
- 75.Sparks A, Morin P, Vogelstein B, Kinzler K. Mutational analysis of the APC/β-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 76.Takeichi M. Cadherins: a molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- 77.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 78.Tetsu O, McCormick F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 79.Troyanovsky R, Chitaev N, Troyanovsky S. Cadherin binding sites of plakoglobin: localization, specificity and role in targeting to adhering junctions. J Cell Sci. 1996;109:3069–3078. doi: 10.1242/jcs.109.13.3069. [DOI] [PubMed] [Google Scholar]
- 80.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma H A D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 81.Voeller H, Truica C, Gelmann E. β-Catenin mutations in human prostate cancer. Cancer Res. 1998;58:2520–2523. [PubMed] [Google Scholar]
- 82.Wahl J, III, Sacco-Bubulya P, Sadler T, Johnson K, Wheelock M. The amino and carboxyl-terminal tails of β-catenin reduce its affinity for desmoglein 2. J Cell Sci. 2000;113:1737–1745. doi: 10.1242/jcs.113.10.1737. [DOI] [PubMed] [Google Scholar]
- 83.White P, Aberle H, Vincent J. Signaling and adhesion activities of mammalian β-catenin and plakoglobin in Drosophila. J Cell Biol. 1998;140:183–195. doi: 10.1083/jcb.140.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Winston J, Strack P, Beer-Romero P, Chu C, Elledge S, Harper J. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 86.Wong M, Rubinfeld B, Gordon J. Effects of forced expression of an NH2-terminal truncated β-catenin on mouse intestinal epithelial homeostasis. J Cell Biol. 1998;141:765–777. doi: 10.1083/jcb.141.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto H, Kishida S, Uochi T, Ikeda S, Koyama S, Asashima M, Kikuchi A. Axil, a member of the Axin family, interacts with both glycogen synthase kinase 3β and β-catenin and inhibits axis formation of Xenopus embryos. Mol Cell Biol. 1998;18:2867–2875. doi: 10.1128/mcb.18.5.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yost C, Torres M, Miller J, Huang E, Kimelman D, Moon R. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 89.Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek T, Perry W, Lee J, Tilghman G, Costantini F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 90.Zhu A, Watt F. β-Catenin signalling modulates proliferative potential of human epidermal keratinocytes independently of intercellular adhesion. Development. 1999;126:2285–2298. doi: 10.1242/dev.126.10.2285. [DOI] [PubMed] [Google Scholar]
- 91.Zurawel R, Chiappa S, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic β-catenin mutations. Cancer Res. 1998;58:896–899. [PubMed] [Google Scholar]