Abstract
Myelodysplastic syndromes (MDS) are clonal neoplasms of the hematopoietic stem cell that result in aberrant differentiation of hematopoietic lineages caused by a wide range of underlying genetic, epigenetic, and other causes. Despite the myriad origins, a recognizable MDS phenotype has been associated with miRNA aberrant expression. A model of aberrant myeloid maturation that mimics MDS was generated using a stable knockdown of miR-378-3p. This model exhibited a transcriptional profile indicating aberrant maturation and function, immunophenotypic and morphologic dysplasia, and aberrant growth that characterizes MDS. Moreover, aberrant signal transduction in response to stimulation specific to the stage of myeloid maturation as indicated by CyTOF mass cytometry was similar to that found in samples from patients with MDS. The aberrant signaling, immunophenotypic changes, cellular growth, and colony formation ability seen in this myeloid model could be reversed with azacytidine, albeit without significant improvement of neutrophil function.
Myelodisplastic syndrome (MDS) is a group of clonal hematopoetic stem cell (HSC) disorders, predominantly of the elderly population, that carry an increased risk of transformation to an acute myeloid leukemia.1 MDS is characterized by cytopenias with dysplasia in one or more lineages of the bone marrow.1,2 MDS has been associated with a myriad of molecular genetic, epigenetic, and inflammatory lesions.2, 3, 4, 5, 6, 7, 8, 9 However, a recognizable phenotype of MDS used by pathologists and clinicians in the diagnosis of MDS includes granulocytic dysplasia (including hypogranulation and hypolobation), myeloid maturational dyssynchrony by flow cytometry, neutropenia, and aberrant function of neutrophils that manifest as increased risk of infections. miRNAs are critically important in the regulation of the cellular phenotype and are differentially expressed at different stages in hematopoietic maturation.10,11 Thus, the numerous causal lesions may funnel through a common pathway of dysregulated miRNAs in the pathogenesis of MDS.
miRNAs are small, noncoding RNAs of approximately 22 nucleotides that bind imperfectly to the 3′ untranslated region and other regions of target mRNAs to repress their translation and/or stability.10,11 Recent studies have confirmed disordered miRNA expression in patients with MDS, including a study from this research group that identified eight miRNAs that are highly discriminatory between MDS and normal control samples.12, 13, 14, 15, 16, 17
miR-378-3p was underexpressed in 80% of cases across MDS subtypes but only 10% of control normal bone marrows.12 miR-378-3p underexpression is associated with hypermethylation of its promoter, and targets of miR-378-3p are accordingly overexpressed.7,12 An antisense to miR-378-3p was introduced by lentiviral vector in HSCs and acute myeloid leukemia cell lines that can be induced to differentiate, creating a myeloid maturational model of MDS. This model demonstrated a transcriptional profile that indicated aberrant maturation and function and showed immunophenotypic, morphologic, growth, and functional defects that characterize MDS. Moreover, aberrant signal transduction in myeloid response to stimulation mimicked that seen in samples from patients with MDS. With the use of this model, azacytidine treatment reversed some of phenotypic alterations, albeit with no significant improvement of neutrophil function.
Materials and Methods
Cell Lines and Cell Cultures
HL60 cells were purchased from ATCC (Manassas, VA) and maintained in Iscove's modified Dulbecco's medium (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (Fisher Scientific, Grand Island, NY). Human bone marrow cells were obtained from patients diagnosed with MDS or normal controls under appropriate institutional review board approval [Brigham and Women's Hospital (BWH), Boston, MA]. Cells were cultured in RPMI 1640 medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum. CD34-positive primary human HSCs were isolated from patient bone marrow and mobilized peripheral blood samples under investigational review board approval (Vanderbilt University Medical Center) using EasySep Human CD34 Positive Selection Kit (catalog number 18056, StemCell Technologies Inc., Cambridge, MA).
ZIP Lentivirus Infection
miRZip-378a-3p (ZIP) and scramble (SCR) pGreenPur lentivectors (catalog number CS970A-1, System Biosciences, Palo Alto, CA) encode either an miRZip hairpin interfering RNA designed to generate the full-length antisense miR-378a-3p or an empty vector, respectively. HL60 or HSCs were seeded in 24-well plates and infected with a multiplicity of infection of 20 with ZIP or SCR using polybrene transduction reagent (8 μg/mL) (catalog number TR-1003-G, Millipore, Burlington, MA) by spin transduction (1500 × g, 2 hours). Initial transduction efficiency was measured by flow cytometry on day 3. After puromycin (catalog number P8833; Sigma-Aldrich, Burlington, MA) (2 μg/mL) treatment on day 3 through day 8, selection efficiency was measured by flow cytometry. Expression of the ZIP construct normalized to U6 was confirmed by real-time quantitative polymerase chain reaction using QuantiMir (catalog number RA420A-hU6, System Biosciences), using the ABI 7300 Real Time PCR System (Applied Biosystem, Foster City, CA). Expression of miR-378-3p normalized to U6 was assessed by real-time quantitative polymerase chain reaction using TaqMan miRNA assays (catalog number 4427975, Applied Biosystems). The presence of green fluorescent protein (GFP) in both ZIP and SCR vectors was confirmed by polymerase chain reaction and fluorescent microscopy (Eclipse TE2000-S Inverted Microscope, Nikon, Melville, NY).
Cell Maturation and Proliferation
HL60 cells stably transduced with ZIP, SCR, or untransduced (UNT) were plated at a density of 2.5 × 105 cells/mL in triplicate. The cells were treated with all-trans retinoic acid (ATRA, 1 μmol/L) (catalog number R2625; Sigma-Aldrich), azacytidine (0.5 μmol/L) (catalog number 2385; Sigma-Aldrich), or combined ATRA and azacytidine for 9 days. Cell number and viability were quantified by hemocytometer using Trypan blue staining (catalog number 25900CI, Corning, Manassas, VA). CD34-positive HSCs were induced to mature with granulocyte colony-stimulating factor (G-CSF; 100 ng/mL) (catalog number 300-23, Peprotech, Rocky Hill, NJ) and stem cell factor (20 ng/mL) (catalog number 300-07, Peprotech).
Morphologic Assessment
Cells were centrifuged at 91 × g (Shandon-Cytospin4, Thermo Electron Corporation, Waltham, MA) for 3 minutes, fixed in 100% methanol, and stained with Wright-Giemsa (catalog number 9380-16, Rica Chemical Company, Arlington, TX) for 15 minutes. Cells were morphologically binned as immature (myeloblasts and promyelocytes), intermediate (myelocytes and metamyelocytes), or mature myeloid cells (bands and segmented neutrophils) and scored for dysplasia on a per cell basis and represented as a percentage of total analyzed cells. Dysplastic features included hypolobation (bilobed or monolobed forms), hypogranulation, megaloblastoid change (nuclear/cytoplasmic dyssynchrony), and apoptotic bodies. Myeloid cells without these features were considered without significant atypia. Scoring was performed by two pathologists, one blinded to the conditions (Y.D.) and one not blinded to the conditions (A.S.K.). The blinded results are included in the table, but results were concordant between the pathologists.
Transcriptional Profiling by mRNA Sequencing
A library was constructed from total RNA using the TruSeq Small RNA sample preparation kit (Illumina, San Diego, CA) and sequenced on an Illumina HiSeq 2500. The raw sequencing reads in BCL format were processed through CASAVA-1.8.2 for FASTQ conversion and demultiplexing. The RTA chastity filter was used, and only the pass filter reads were retained for further analysis.
mRNA Sequencing Data Analysis
RNA-sequencing data went through multiperspective quality control procedures following established guidelines.18 Alignment was performed by TopHat 2 (Center for Computation Biology, Johns Hopkins University, Baltimore, MD).19 Gene quantification using both Cufflinks version 2.2.0 (University of Washington, Seattle, WA)20 and HTSeq version 0.13.5 (Python Software Foundtion, Fredericksburg, VA)21 was converted to differential expression analysis using Cuffdiff from the Cufflinks package and MultiRankSeq version 1.2.0 (Center for Quantitative Sciences, Vanderbilt University, Nashville, TN),22 respectively, which is a combination of DEGseq,23 edgeR,24 and baySeq.25 A list of the most differentially expressed genes was generated from all four methods based on a weighted flexible compound covariate method.26 Data (heatmaps and functional pathways) were visualized with permission using Ingenuity Pathway Analysis (Qiagen Inc., Germantown, MD).27
Colony-Forming Units Assay
HL60 cells stably expressing ZIP or SCR were seeded in 35-mm culture dishes in quadruplicate with a density of 2.2 × 105 cells/mL in methylcellulose (MethoCult H4535; StemCell Technologies). Cells were cultured in the presence or absence of azacytidine (0.5 μmol/L), ATRA (1 μmol/L), or combined azacytidine and ATRA for 7 days. Colonies were counted and imaged using an inverted microscope and gridded scoring dishes.
Cell Migration Assay
ZIP, SCR, or UNT HL60 cells were treated with ATRA (1 μmol/L) for 7 days. Viable cells were enriched by Ficoll-Paque purification (catalog number GE17-1440-02, Sigma-Aldrich). Cells (1 × 105) seeded in the upper wells of 96-well Transwell plates with 5-μm pores (catalog number 3388, Corning, Lowell, MA) were allowed to migrate into the lower wells that contained the mixture of chemotactic factors, N-formylmethionine-leucyl-phenylalanine (10 nmol/L; catalog number F3506, Sigma-Aldrich), complement component 5a (C5a, 1 nmol/L; catalog number 2037, R&D System, Minneapolis, MN), and lipopolysaccharides (10 μg/mL; catalog number L-8274, Sigma-Aldrich) for 120 minutes at 37°C. Cells from the lower wells were stained with alamarBlue HS (catalog number A50100, Invitrogen, Eugene, OR) for 120 minutes and fluorescence read at 570/610 nm.
Flow Cytometry
Cell surface markers were measured by flow cytometry in two laboratories. Cells were harvested by centrifugation, washed with phosphate-buffered saline, and incubated with antibodies for 20 to 30 minutes in the dark. In the Flow Cytometry Core of the Tennessee Valley Healthcare System, Veterans Affairs, two panels were used. For analysis of HL60 cells, the following antibodies were obtained from BD Biosciences (San Jose, CA): 7-aminoactinomycin D (catalog number 559925), CD11b-phycoerythrin (PE) (catalog number 340712), CD13-allophycocyanin (APC) (catatlog number 557454), CD16-V450 (catalog number 560474), and CD45-V500 (catalog number 560777). CD33–proprotein convertase 7 (PC7) was obtained from Beckman-Coulter (Indianapolis, IN) (catalog number A54824). For the analysis of CD34-positive HSCs, the following antibodies were obtained from BD Biosciences: CD11b-APC (catalog number 550019), CD13–Brilliant Violet (BV42) 1 (catalog number 562596), CD16-APC-cyanine 7 (catalog number 560195), and CD34–peridinin-chlorophyll-protein (catalog number 340666). CD33-PC7 was obtained from Beckman-Coulter (catalog number A54824). Samples were run on a BD FACSCanto and data analyzed on FACSDiva software version 6.1.3 (BD Biosciences). Co-expression of GFP with the myeloid cell surface marker confirmed analysis of transduced cells. The flow panel used in subsequent studies at BWH included the following antibodies: CD45-APC-H7 (catalog number 560178, BD Biosciences), CD13–fluorescein isothiocyanate (FITC) (catalog number 11-0138-42, eBioscience Inc., San Diego, CA), CD16-BV421 (catalog number 302038, Biolegend, San Diego, CA), and CD11b-PE (catalog number 301306, Biolegend). At the Dana Farber Cancer Institute Flow Cytometry Core, specimens were analyzed on a BD LSR Fortessa, and data were analyzed by FlowJo software version 10.2 through version 10.6.1 (BD Biosciences).
Cytometry Time of Flight (CyTOF) Mass Cytometry
Patient Sample Acquisition, Treatment, and Preparation
Bone marrow aspirates from patients with active myelodysplasia (n = 32) and controls (n = 32) were obtained under institutional review board approval from the BWH (Table 1 and Supplemental Table S1). After treatment with azacytidine (5 μmol/L) (catalog number A2385, Sigma-Aldrich) or dimethyl sulfoxide overnight at 37°C, erythrocytes were removed with ACK lysing buffer (catalog number A10492-01, Gibco). Cells were adjusted to 1 × 106 per sample and incubated with 1:1000 cisplatin (catalog number 201064, Fluidigm, San Francisco, CA) for 2 minutes.
Table 1.
Summary of Patient Characteristics
| Characteristic | Patients with MDS (n = 32) | Controls (n = 32) | P |
|---|---|---|---|
| Sex, n | |||
| Male | 26 | 20 | 0.098 |
| Female | 6 | 12 | |
| Age, mean (range), y | 65 (39–85) | 59 (28–76) | 0.055 |
| BM diagnoses, n | |||
| MDS-SLD | 1 | ||
| MDS-MLD | 14 | ||
| MDS-EB1 | 6 | ||
| MDS-EB2 | 8 | ||
| CMML-0∗ | 2 | ||
| MDS/MPN, NOS∗ | 1 | ||
| Negative for neoplasm | 26 | ||
| PCN (≤10% involved) | 5 | ||
| LPL (≤10% involved) | 1 | ||
| CBC counts, mean | |||
| WBCs, /μL (in thousands) | 3.49 | 6.49 | 0.0003 |
| ANC, μL (in thousands) | 1.56 | 4.43 | <0.0001 |
| HGB, g/dL | 10.00 | 12.93 | <0.0001 |
| MCV, fL | 92.13 | 93.02 | 0.6194 |
| Platelets, /μL (in thousands) | 110.78 | 211.34 | 0.0001 |
| PB blasts, % | 0.31 | 0.01 | 0.1098 |
| BM blasts, % | 6.34 | 0.91 | <0.0001 |
ANC, absolute neutrophil count; BM, bone marrow; CBC, complete blood cell; CMML, chronic myelomonocytic leukemia; EB, excess blasts; HGB, hemoglobin; LPL, lymphoplasmacytic lymphoma; MCV, mean cell volume; MDS, myelodysplastic syndrome; MDS/MPN, NOS, myelodysplastic/myeloproliferative neoplasms not otherwise specified; MLD, multilineage dysplasia; PB, peripheral blood; PCN, plasma cell neoplasm; SLD, single-lineage dysplasia; WBC, white blood cell.
The two cases of CMML and one case of MDS/MPN, NOS, are dysplastic-type MDS/MPNs and are accordingly included in this study.
Cell Surface Staining and Cytokine Stimulation
Cells (patient samples or cell lines) were incubated for 20 minutes at room temperature with a 100-μL cocktail of 16 metal conjugated cell surface marker antibodies obtained from the Lederer Laboratory of BWH (Table 2), followed by stimulation with granulocyte-monocyte colony stimulating factor (100 ng/mL) (catalog number 215-GM-010, R&D Systems, Minneapolis, MN), interferon-γ (100 ng/mL) (catalog number 285-IF-100, R&D Systems), and phorbol 12-myristate 13-acetate (50 nmol/L) (catalog number P8139, Sigma-Aldrich, St. Louis, MO) for 10 minutes at 37°C. Cells were fixed with 4% paraformaldehyde and permeabilized with methanol.
Table 2.
List of CyTOF Mass Cytometry Antibodies, Clone, and Isotope Label
| Target | Clone catalog number | Isotope |
|---|---|---|
| Cell surface markers | ||
| CD3 | UCHT1 | 115In |
| CD45 | HI30 | 141Pr |
| CD11b | M1/70 | 142Nd |
| CD34 | 581 | 143Nd |
| CD235a | HI264 | 144Nd |
| CD16 | 3G8 | 145Nd |
| CD8a | RPA T8 | 146Nd |
| CD13 | WM15 | 147Sm |
| CD20 | 2H7 | 148Nd |
| CD123 | 6H6 | 151Eu |
| CD14 | M5E2 | 152Sm |
| CD33 | WM53 | 154Sm |
| CD4 | RPA T4 | 155Gd |
| CD56 | HCB6 | 162Dy |
| CD117 cKit | 10422 | 158Gd |
| CD274/PD-L1 | 29E.2A3 | 170Er |
| Intracellular markers | ||
| Akt | Poly6034 | 150Nd |
| c-PARP | F21-852 | 164Dy |
| Cleaved caspase 3 (Asp175) | D3E9 | 156Gd |
| Ki-67 | 8D5 | 161Dy |
| Phospho-JAK2 (Y1008) | D4D8 | 153Eu |
| Phospho–NF-κB p65 (Ser536) | 93H1 | 159Tb |
| Phospho-STAT1 (Y701) | 58D6 | 160Gd |
| Phospho-PLCγ1 (Tyr783) | D6M9S | 165Ho |
| Phospho-p38 MAPK (Thr180/Tyr182) | D3F9 | 166Er |
| Phospho-p44/42 MAPK (T202/Y204)/Erk1/2 | D13.14.4E | 167Er |
| Phospho–cleaved CBL (Tyr700) | D1D67 | 169Tm |
| Phospho-STAT4 (Y693) | D2E4 | 171Yb |
| Phospho–4E-BP1 (T37/46) | 236B4 | 172Yb |
| Phospho-STAT5 (Y694) | D47E7 XP (R) | 174Yb |
| Phospho-S6 (p70RSK) S235/236 | D57.2.2E XP R | 175Lu |
| Phospho-CREB (S133) | 87G3 | 176Yb |
c-PARP, cleaved poly (ADP-ribose) polymerase; CREB, cAMP response element-binding protein; MAPK, mitogen-activated protein kinase; PLCγ1, phospholipase C γ1; STAT, signal transducer and activator of transcription.
Cells were barcoded using Cell-ID 20-Plex Pd Barcoding Kit (catalog number S00114, Fluidigm Corporation, South San Francisco, CA). Samples were incubated in BarCode Reagent for 15 minutes at room temperature and washed with cell staining buffer. Barcoded samples were then pooled and rinsed.
Cells were incubated at room temperature for 60 minutes with a cocktail of 16 metal conjugated intracellular antibodies (1:100 dilution of each antibody) (Table 2). Cells were fixed with paraformaldehyde, resuspended in cell staining buffer, and stored at 4°C overnight.
Data Acquisition by Mass Cytometry
On day 2, cells were incubated with intercalator Ir (191 and 193) (1:4000 with phosphate-buffered saline) for 20 minutes at room temperature. Cells were resuspended with EQ beads and analyzed on the mass cytometry (Helios, Fluidigm). Events were gated using Cytobank (Cytobank Inc., Santa Clara, CA)28 and binned as immature, intermediate, or mature myeloid cells based on expression of CD33 and the variable expression of CD45, CD34, CD117, CD123, CD16, CD11b, and CD13. For patient samples, mature monocytes were also gated by expression of CD14. T, B, and natural killer lymphoid cells were excluded using CD3, CD4, CD8, CD20, CD16, and CD56. Erythroid cells were excluded using CD235a. Gating assignments were confirmed by SPADE analysis in Cytobank software version 7.2.1 (Cytobank Inc., Santa Clara, CA).28 Expression of all markers was normalized to the unstimulated state.
Statistical Analysis
For cell line–based experiments, data are expressed as means ± SD or SE of triplicates. Experiments were performed at least three times. Statistical analyses were performed by analysis of variance followed by Tukey's multiple comparison test or by t-test as appropriate. P < 0.05 was considered to be significant. Replicates of mass cytometry CyTOF data were averaged across experiments per experimental condition for each intracellular marker expression level and analyzed by the t-test. Expression levels were all normalized to the unstimulated paired samples, and Δ expression levels were calculated (condition 1 – condition 2). For patient CyTOF mass cytometry data, mean expression levels for the control and myeloid samples were obtained and analyzed by the t-test. After normalization, delta expression levels were calculated (control-myeloid).
Results
Transduction by ZIP Results in Reduced Expression of miR-378-3p
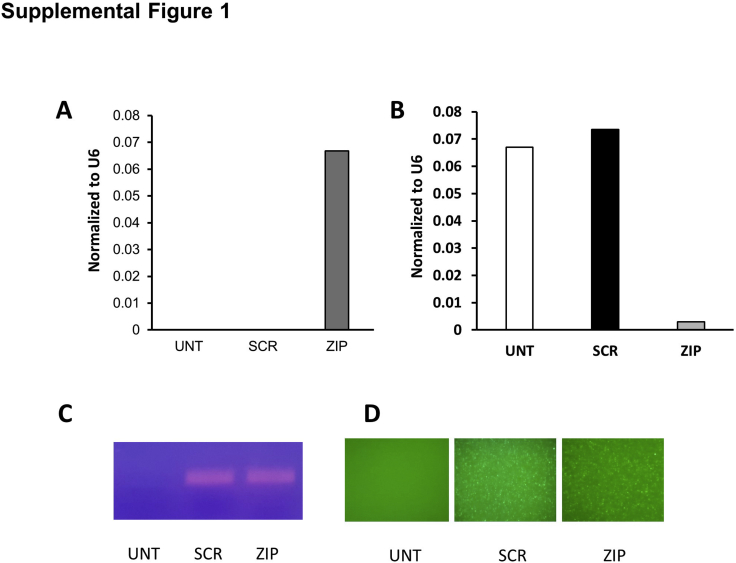
HL60 cells and CD34 HSCs were transduced with ZIP or SCR lentiviral vectors. After puromycin selection, ZIP and SCR cells were confirmed to express GFP by fluorescent microscopy, flow cytometry, and PCR, whereas only ZIP cells expressed the miRZIP-378-3p by real-time quantitative polymerase chain reaction. Compared with UNT and SCR, ZIP cells demonstrated reduced expression of miR-378a-3p (Supplemental Figure S1). Initially, several other miRNAs, including combinations, were targeted (data not shown); however, eventually miR-378-3p was selected for full analysis detailed in this article because it had the most robust phenotype, as detailed below.
ZIP Induces a Transcriptional Profile of Aberrant Cellular Function and Growth
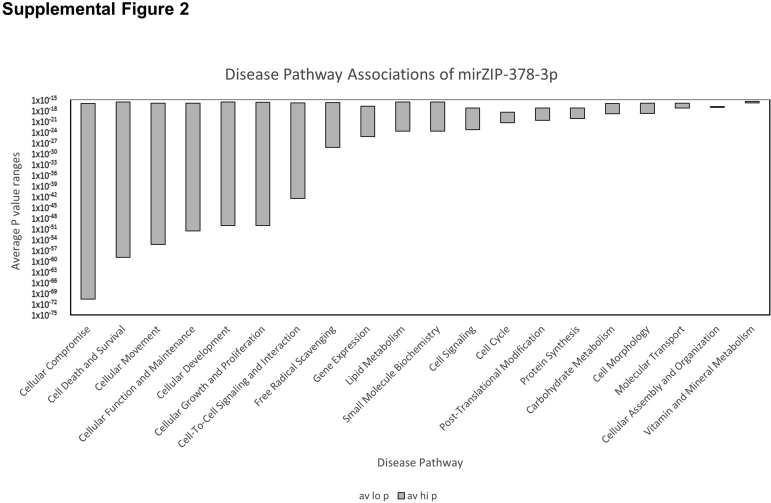
mRNA sequencing of duplicate HL60 transductions was followed by comparisons using a combination ranking derived from DEGseq, edgeR, and baySeq methods. The top 2000 discriminatory genes for the comparisons were input into Ingenuity Pathway Analysis with DEGseq, edgeR, and baySeq data. P values and fold change data were provided separately. All three algorithms identified the same most impacted cellular functions in ZIP compared with SCR cells with significant deactivation of cell movement, signaling, and phagocytosis (Figure 1, Supplemental Table S2, and Supplemental Figure S2). DEGseq and baySeq provided the identical six pathways with a score ≥35, whereas edgeR provided seven pathways that met the criteria. These top-scoring pathways encompassed a total of 328 key proteins involved in cellular maturation, growth and proliferation, cellular movement, DNA replication, and cell death and survival (Supplemental Table S3).
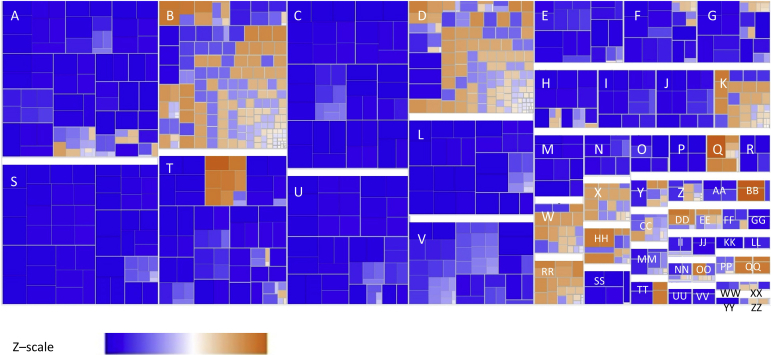
Figure 1.
Snapshot from Ingenuity Pathway Analysis, demonstrating key diseases and functions affected by knockdown of miR-378-3p by mirZIP-378-3p. Most highly affected pathways include those that involve cell movement and function. Key pathways are predominantly underexpressed as indicated by the blue end of the color scale of z scores that represent decrease of implicated biologic function (blue) (versus increased function in orange). A: Hematologic system development and function; B: organismal injury and abnormalities; C: cell-to-cell signaling and interaction; D: cancer; E: tissue development; F: cellular development; G: cell growth and function; H: lymphoid tissue structure; I: lipid metabolism; J: small molecule biochemistry; K: gastrointestinal disease; L: cell function and maintenance; M: hematopoiesis; N: cellular compromise; O: organismal death; P: cardiovascular disease; Q: infectious disease; R: free radical scavenging; S: cellular movement; T: inflammatory response; U: immune cell trafficking; V: cell death and survival; W: immunologic disease; X: reproductive system disease; Y: skeletal and muscular disorders; Z: endocrine system disorders; AA: embryonic development; BB: organismal development; CC: tissue morphology; DD: respiratory disease; EE: connective tissue development; FF: metabolic disease; GG: antimicrobial response; HH: inflammatory disease; II: renal and urologic disease; JJ: cell signaling; KK: cellular growth and proliferation; LL: molecular transport; MM: neurologic disease; NN: connective tissue development; OO: hepatic system disease; PP: dermatologic disease; QQ: hepatic and digestive system disease; RR: hematologic disease; SS: gene expression; TT: organ development; UU: cell migration; VV: posttranslational modification; WW: vitamin and mineral metabolism; XX: humoral immune response and hypersensitivity response; YY: carbohydrate metabolism; ZZ: protein synthesis and cellular function and maintenance.
ZIP Causes Dysplasia and Apoptosis
UNT, SCR, and ZIP HSCs were induced to mature by G-CSF treatment over 9 days. Wright-Giemsa–stained cytospins of the cultured cells were scored for cell maturation and dysplasia (Table 3 and Figure 2, A–C). Although there was some effect of the transduction on both ZIP and SCR cells, of mature neutrophils, 100% of ZIP cells demonstrated abnormal bilobed (8% of neutrophils) and monolobated neutrophils (92% of neutrophils) (bilobed: P = 0.050 versus SCR and P = 0.027 versus UNT; monolobated: P = 0.015 versus SCR and P = 0.0034 versus UNT). By contrast, 71% of SCR cells were abnormally lobated, including 49% monolobated and 22% bilobed neutrophils, and only a few UNT cells were abnormally lobated. ZIP cells also demonstrated significantly increased apoptosis (P = 0.0021 versus SCR and P = 1.15 × 10−6 versus UNT). Similarly, the HL60 cells were studied under the analogous conditions (Table 3) after treatment with ATRA for 7 days. Despite a higher background of cellular atypia (as defined in Materials and Methods) with the HL60 UNT, SCR, and ZIP cells compared with the primary HSCs, the ZIP cells were significantly left-shifted compared with both UNT and SCR cells with increased immature cells (P = 0.0148 versus UNT), increased intermediate myeloid cells (P = 0.045 versus UNT, P = 0.0255 versus SCR), and decreased mature forms (P = 0.003 versus UNT, P = 0.029 versus SCR). Similar results were identified in NB4 cells (data not shown). Morphologic atypia was not statistically different between the ZIP cells and either set of control cells.
Table 3.
Manual Differential of Morphologic Stages of Myeloid Maturation and Dysplasia in UNT, SCR, and ZIP Cells in Primary HSCs (Mean of 4 Replicate Experiments, with Goal of 100 Cell Counts per Experiment) and the HL60 Cell Line (Mean of 4 Replicate Experiments, with Goal of 200 Cell Counts per Experiment)
| Morphologic feature (mean lobes) | UNT, % | SCR, % | ZIP, % |
P value |
||
|---|---|---|---|---|---|---|
| SCR versus ZIP | UNT versus ZIP | UNT versus SCR | ||||
| HSCs | ||||||
| Neutrophils (3–5 lobes) | 30.50 | 7.00 | 0.00 | 0.0500 | 0.0001 | 0.0009 |
| Neutrophils (2 lobes) | 6.50 | 5.25 | 0.50 | 0.0500 | 0.0267 | 0.4918 |
| Neutrophils (1 lobe) | 0.25 | 12.00 | 5.75 | 0.0154 | 0.0034 | 0.0026 |
| Intermediate myeloid | 34.25 | 23.75 | 9.25 | 0.0344 | 0.0099 | 0.2209 |
| Immature myeloid | 25.25 | 13.75 | 3.00 | 0.0481 | 0.0036 | 0.0904 |
| Apoptotic body | 3.25 | 38.25 | 81.50 | 0.0021 | <0.0001 | 0.0033 |
| HL60s | ||||||
| Mature myeloid without significant atypia | 32.12 | 26.25 | 4.41 | 0.0290 | 0.0027 | 0.5951 |
| Mature myeloid and dysplastic | 8.68 | 4.50 | 1.13 | 0.2319 | 0.0796 | 0.2599 |
| Intermediate myeloid without significant atypia | 32.07 | 28.75 | 53.92 | 0.0255 | 0.0450 | 0.7353 |
| Intermediate myeloid and dysplastic | 2.67 | 1.00 | 1.68 | 0.5664 | 0.7692 | 0.3198 |
| Immature myeloid without significant atypia | 12.22 | 29.25 | 27.36 | 0.8691 | 0.0148 | 0.3353 |
| Immature myeloid and dysplastic | 0.67 | 0.50 | 2.59 | 0.2129 | 0.3133 | 0.9929 |
| Apoptotic body | 11.56 | 9.75 | 8.92 | 0.7429 | 0.4120 | 0.1961 |
| HL60s, azacytidine treated | ||||||
| Mature myeloid without significant atypia | 36.79 | 34.92 | 26.59 | 0.7848 | 0.0628 | 0.2641 |
| Mature myeloid and dysplastic | 6.68 | 8.54 | 1.80 | 0.2722 | 0.1013 | 0.5838 |
| Intermediate myeloid without significant atypia | 39.25 | 21.83 | 35.58 | 0.4003 | 0.6572 | 0.0100 |
| Intermediate myeloid and dysplastic | 3.71 | 0.13 | 1.14 | 0.0622 | 0.1305 | 0.0263 |
| Immature myeloid without significant atypia | 5.84 | 19.27 | 22.01 | 0.8391 | 0.0619 | 0.2413 |
| Immature myeloid and dysplastic | 0.39 | 1.02 | 0.88 | 0.8811 | 0.5935 | 0.9601 |
| Apoptotic body | 7.33 | 14.29 | 12.01 | 0.7937 | 0.2318 | 0.4792 |
HSCs, human hematopoietic stem cells; SCR, scramble; UNT, untransduced; ZIP, miRZIP-378-3p.
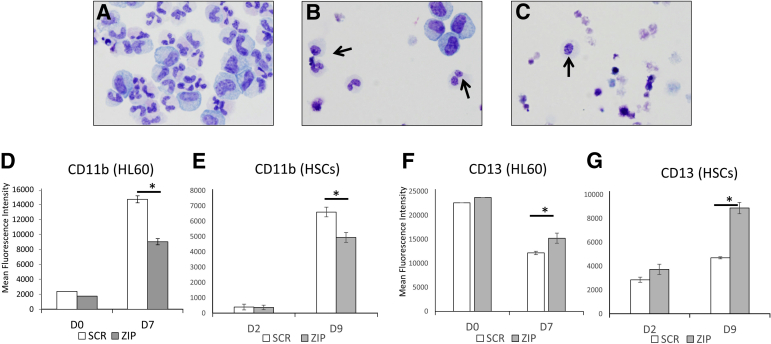
Figure 2.
Morphologic and immunophenotypic dysplasia of miRZip-378a-3p (ZIP). A–C: Human CD34-positive hematopoietic stem cells (HSCs) were transfected with ZIP or scramble (SCR) vector and induced to mature by treatment with granulocyte colony-stimulating factor (G-CSF) for 9 days. Arrows indicate hypolobated neutrophils. A: untransduced human HSCs. B: HSCs transduced with SCR. C: HSCs transduced with ZIP. D–G: Myeloid markers of maturation on ZIP and SCR cell on days 0 (D0), Day 7 (D7) of all-trans retinoic acid treatment for HL60 cells and day 2 (D2) and day 9 (D9) of G-CSF treatment for HSCs. D and E: CD11b expression on HL60 cells (D) and HSCs (E). F and G: CD13 expression on HL60 cells (F) and HSCs (G). ∗P < 0.05. Original magnification, ×40 (Wright-Giemsa stain) (A–C).
ZIP Induces Immunophenotypic Dyssynchronous Maturation
Both HL60 and CD34 HSCs were induced to mature by treatment with ATRA for 7 days (HL60) or G-CSF for 9 days (CD34 HSCs), respectively. Flow cytometry was then performed using a panel of myeloid maturation markers (Figure 2, D–G). ZIP cells exhibited decreased CD11b (HL60 cells: P = 0.0035; CD34 HSCs: P = 0.0019) but increased CD13 expression over SCR (HL60 cells: P = 0.0090; CD34 HSCs: P = 0.0055). Because CD11b typically increases with maturation and CD13 typically increases in the later stages of maturation (after decreasing early in maturation), these findings indicate true dyssynchrony. Similar dyssynchrony was seen with other markers of maturation (CD16, CD33, CD34, and CD45) (Supplemental Table S4).
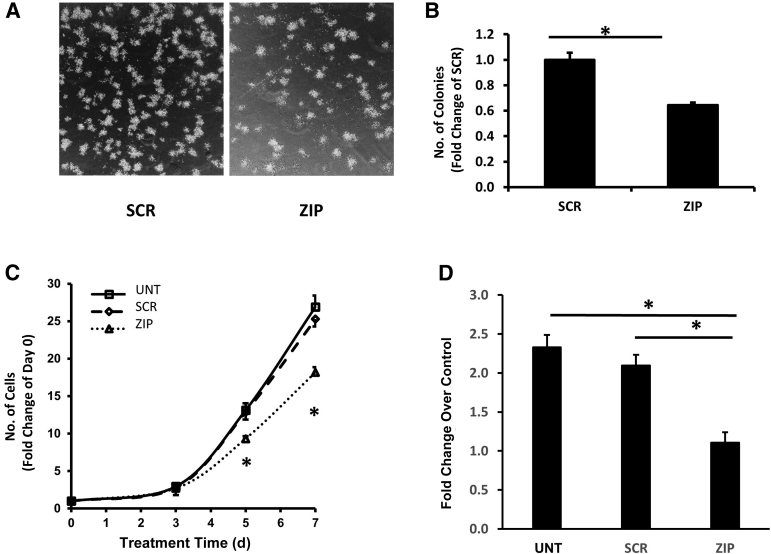
ZIP Inhibits Colony Formation and Cell Proliferation
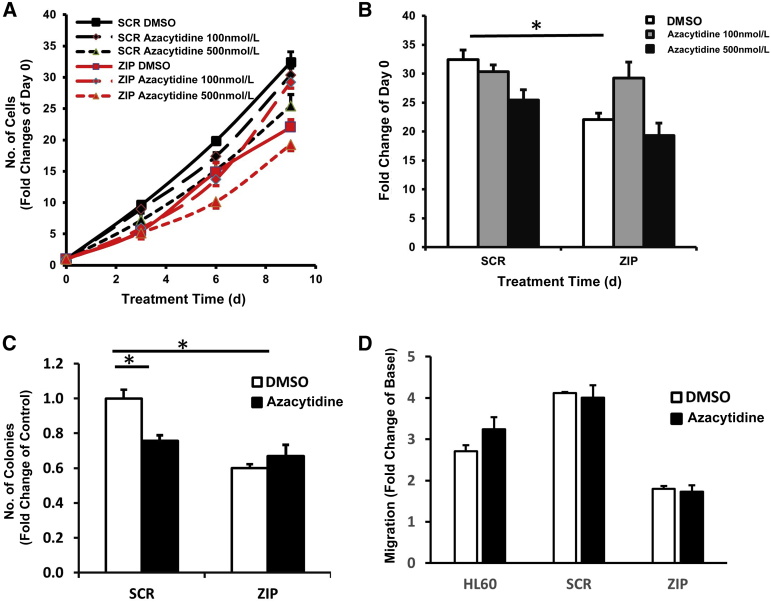
UNT, SCR, and ZIP HL60 cells were assessed for the ability to form methylcellulose colonies. ZIP cells exhibited diminished colony formation ability compared with SCR cells (P = 0.005) (Figure 3, A and B). Cell proliferation studies were performed by viable cell enumeration on days 0, 3, 5, and 7. No significant difference was found between the growth of UNT and SCR cells (P = 0.49), whereas ZIP cells were significantly less proliferative (P = 0.017 versus UNT and P = 0.026 versus SCR) (Figure 3, C and D).
Figure 3.
Colony formation and proliferation of untransduced (UNT), scramble (SCR), and miRZip-378a-3p (ZIP) cells. A and B: Colony formation in methylcellulose of SCR and ZIP cells. C: Time course of cell proliferation of UNT, SCR, and ZIP cells in triplicate. D: Neutrophil migration experiments were performed using the Transwell system assay and UNT, SCR, or ZIP cells treated with all-trans retinoic acid (1 μmol/L) for 7 days. Data are expressed as the fold change compared with basal in each group. ∗P < 0.05. Original magnification (inverted microscope), ×10 (A).
ZIP Inhibits Cell Migration
Because of the need to sufficiently expand cells for maturational studies, all subsequent studies were performed on HL60 cells alone. The HL60 cell line was chosen because of its ease of expansion compared with HSCs,29 its superior transduction efficiency compared with NB4 cells, and its well-known ability to mature into the full spectrum of myeloid maturation morphologically and immunphenotypically.30 To investigate the effects of ZIP on cell migration, UNT, SCR, and ZIP HL60 cells were treated with ATRA (1 μmol/L) for 7 days. In response to a cocktail of neutrophil chemotactic factors (N-formylmethionine-leucyl-phenylalanine, C5a, and lipopolysaccharides), ZIP cells exhibited significantly reduced migration compared with UNT and SCR cells (P = 0.0044 and 0.0063, respectively) (Figure 3D).
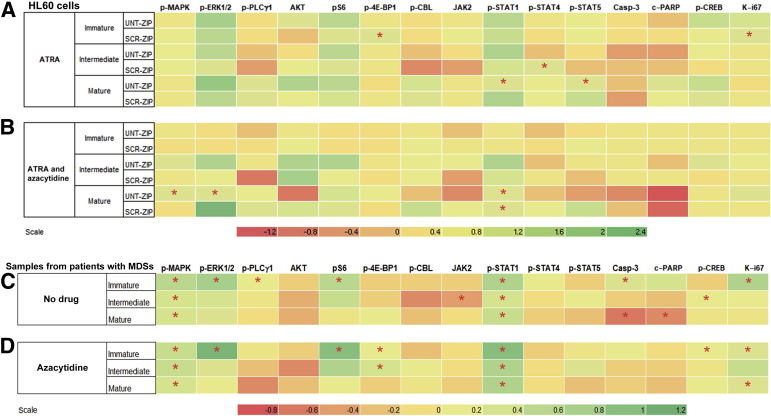
ZIP Induces Immunophenotypic Evidence of Aberrant Signaling
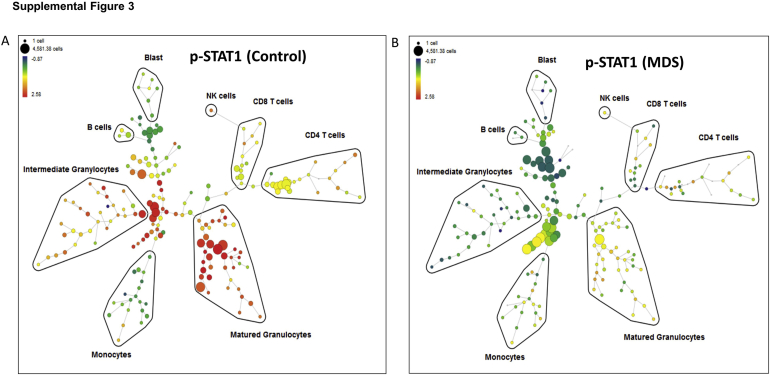
Single-cell phenotyping by CyTOF mass cytometry was performed on UNT, SCR, and ZIP HL60 cells in response to stimulation by a cocktail of known neutrophilic stimulants (granulocyte-monocyte colony stimulating factor, interferon-γ, and phorbol 12-myristate 13-acetate). Cells were induced to mature for 7 days with ATRA before stimulation and CyTOF mass cytometry assessment. The myeloid cells were binned into three stages of maturation by cell surface markers followed by assessment of intracellular signaling proteins (Supplemental Figure S3). As shown in the heatmap in Figure 4A and in Supplemental Table S5, differences were found in pathway use by ZIP cells compared with both SCR and UNT cells, including underutilization of phospho (p)–ERK, pS6, and p-STAT1 across all stages of maturation. This underutilization of p-STAT1 was especially prominent in the most mature ZIP cells (P = 0.014 versus UNT). Interestingly, ZIP cells had decreased Ki-67 in the most immature cells (P = 0.020 versus SCR cells).
Figure 4.
Heatmaps of relative use of intracellular signaling pathways by CyTOF mass cytometry. Expression levels are normalized to the unstimulated paired condition, and the difference between two test arms after normalization represented on the heat map scale. The myeloid cells are binned into three stages of maturation (immature, intermediate, and mature). A and B: miRZip-378a-3p (ZIP) expression levels shown relative to either untransduced (UNT) or scramble (SCR) cells with all-trans retinoic acid (ATRA) (A) or ATRA plus azacytidine (B). Green indicates lower expression in the ZIP cells, whereas red indicates higher expression in the ZIP cells. C and D: Expression levels from samples of patients with myelodysplasia compared with control samples treated with dimethyl sulfoxide vehicle control (C) or azacytidine (D). Green indicates lower expression in the patient cells, whereas red indicates higher expression in the patient cells compared with controls. ∗P < 0.05. Casp-3, caspase-3; CREB, cAMP response element-binding protein; c-PARP, cleaved poly (ADP-ribose) polymerase; MAPK, mitogen-activated protein kinase; MDS, myelodysplastic syndromes; p-, phospho; PLCγ1, phospholipase C γ1.
ZIP Signaling Aberrations Mimic Those Seen in Patient Samples
The optimal validation for any human model system is the concordance with primary patient samples. To ascertain whether the signaling aberrations seen in the ZIP myeloid model were representative of patients with MDS, bone marrow aspirate samples were obtained from 32 patients with myelodysplasia and 32 controls (Table 1 and Supplemental Table S1) and were analyzed by CyTOF mass cytometry. As shown in Figure 4C, the MAPK pathway and p-STAT1 were significantly underresponsive to granulocytic stimulation in patients with MDS compared with controls at all stages of myeloid maturation, mirroring that seen in the ZIP cells (Supplemental Table S6). By contrast, p-CBL and JAK2 were more highly responsive in MDS myeloid cells, similar to that seen in the ZIP cells. Mature MDS myeloid cells had significant activation of caspase-3 and c-PARP (P = 0.033 and 0.012, respectively), whereas Ki-67 was underexpressed in immature MDS cells (P = 0.002), as was seen in the ZIP cells.
Azacytidine Reverses Differentiation Skewing of ZIP Cells
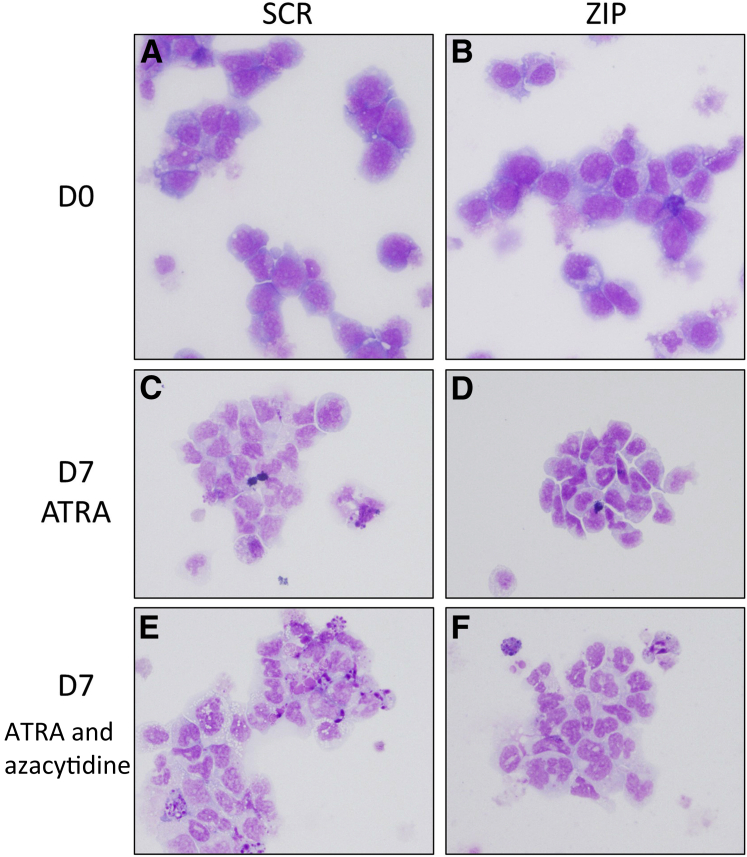
The DNA methyltransferase inhibitor azacytidine is used clinically in the treatment of MDS.31 HL60 cells having undergone ATRA-induced maturation with and without azacytidine were examined morphologically. Azacytidine markedly improved the differentiation of the ZIP cells, eliminating all significant differences in maturation from the UNT or SCR cells seen with ATRA alone (Table 3 and Figure 5).
Figure 5.
Azacytidine reverses the effects of miRZip-378a-3p (ZIP) on HL60 maturational morphologic features. ZIP and scramble (SCR) cells were assessed in presence of all-trans retinoic acid (ATRA) with dimethyl sulfoxide vehicle control or azacytidine. A–F: Morphologic assessment was performed on cytospins on SCR (A, C, and E) and ZIP cells (B, D, and F) at day 0 (D0) (A and B) and day 7 (D7) of ATRA (C and D) or ATRA and azacytidine (E and F). C and D: The untreated SCR cells demonstrate increased mature forms (C), whereas the ZIP cells show more intermediate forms (D). E and F: With ATRA treatment, there is an increase in the number of mature myeloid forms in the ZIP cells (F), such that there is no significant difference between SCR and ZIP (E versus F). Original magnification, ×40 (Wright-Giemsa stain).
Azacytidine Mitigates Cell Signaling Abnormalities of ZIP Cells
UNT, SCR, and ZIP HL60 cells were induced to mature with ATRA in the presence of azacytidine before stimulation and CyTOF mass cytometry analysis. As shown in the heatmap of signaling molecules (Figure 4B), azacytidine mitigated the expression of p-CBL in the mature ZIP cells and expression of caspase-3 and cPARP in immature cells compared with untreated cells (Figure 4A). Interestingly, there was marked activation of caspase-3 and c-PARP in mature cells with azacytidine treatment, potentially consistent with azacytidine toxicity in the mature cells. In addition, there was increased AKT expression relative to UNT in the mature myeloid cells treated with azacytidine in marked reversal of that seen in the untreated cells.
Effect of Azacytidine on Aberrant Signaling in Patient Samples
Patient samples were incubated overnight with azacytidine before stimulation and CyTOF mass cytometry analysis. As shown in Figure 4D, some reversal of the signaling transduction pathway use occurred with azacytidine compared with untreated cells (Figure 4C). Azacytidine appears to mitigate the abnormal use of p-CBL and JAK2 in intermediate and mature myeloid cells and of c-PARP and caspase-3 in mature myeloid cells, findings similar to those seen in the ZIP cell line.
Azacytidine Restores the Growth of ZIP Transduced Cells on Cell Proliferation and Colony Formation
SCR and ZIP HL60 cells were treated with azacytidine to test the effects of azacytidine on ZIP-induced cell growth inhibition. As previously shown (Figure 3C), ZIP markedly decreased cell proliferation compared with control (Figure 6, A and B). However, low-dose azacytidine (100 nmol/L) but not higher-dose (500 nmol/L) azacytidine reversed the cell growth defect such that the difference between the SCR and ZIP cells treated with low-dose azacytidine was not significant. ZIP cells also responded differently to azacytidine than SCR cells in the colony formation assay; although the SCR cells treated with azacytidine demonstrated further attenuation of colony formation, ZIP cells did not (Figure 6C and Supplemental Figure S4).
Figure 6.
Azacytidine reverses the effects of miRZip-378a-3p (ZIP) on HL60 cell proliferation, colony formation, and migration. A: Time course of cell proliferation with two different zzacytidine doses. B: Comparison of the different groups at the end of day of treatment (10 days). C: Quantification of cell colony formation in each group [dimethyl sulfoxide (DMSO) or azacytidine] after culture for 7 days. D: Effects of azacytidine on cell migration. ∗P < 0.05.
Azacytidine Has no Effect on Neutrophil Migration in ZIP Cells
HL60 cell maturation was induced by ATRA treatment for 7 days. Cells were then incubated with either azacytidine or vehicle control (dimethyl sulfoxide) overnight. The Transwell system assay (Fisher Scientific, Waltham, MA) was used to detect cell migration. As previously shown (Figure 3D), ZIP HL60 cells demonstrated reduced cell migration compared with UNT and SCR groups, but this effect was not modified by azacytidine treatment (Figure 6D).
Discussion
The lack of adequate model systems hampers the study of MDS. Primary MDS cells are challenging to culture because of their poor viability ex vivo.32 MDS models, historically created from a single genetic lesion, have been plagued by the assumption that the biology of this one lesion recapitulates the whole etiologic diversity of MDS. Recent advances in multiplex gene editing and patient-derived xenograft models have greatly increased the avenues for exploration of molecular diversity and clonal evolution but do not provide a single model of MDS but rather many genetically distinct models.33,34
Rather than targeting this molecular diversity, this study focused downstream on the miRNA alterations that regulate the broadly recognizable MDS phenotype. In particular, one miRNA, mir-378-3p, has been implicated in the pathogenesis of MDS12,35,36 and oncogenesis in general.37, 38, 39, 40, 41 Although other miRNAs that were significantly differentially expressed in MDS compared with the controls in the discovery and validation sets from the initial study in this area12 were initially examined with the an eye toward potential for combinatorial effects, mir-378-3p alone appeared to recapitulate much of the MDS phenotype and was the focus of this study.
The ZIP cells developed in this study demonstrated significant aberrations in cellular maturation, growth and proliferation, and cellular movement across several different methods. These features mimic the cytopenias and dysplasia seen in patients with MDS. Increased apoptosis of MDS cells occurs ex vivo and in this ZIP model.42 Criteria for the use of flow cytometry in the diagnosis of dysplasia associated with MDS include the asynchronous expression of CD13/CD11b or CD13/CD16, all markers that had aberrant dyssynchrony in this ZIP model.43 The diminished growth and viability of the ZIP cells under standard culture conditions and decreased colony formation ability are also seen in primary MDS cells ex vivo,32,44 whereas overexpression of miR-378-3p has been associated with increased tumor sphere formation.45 Finally, neutrophils from patients with MDS demonstrate decrease migration, as seen in the ZIP cells.46, 47, 48
A previous study from this group detailed some of the predicted targets of miR-378-3p and demonstrated the expression of differentiation of one of these targets, suppressor of fused, in the bone marrow biopsies of patients with MDS compared with controls. It discussed the possible function of suppressor of fused in MDS, particularly in the erythroid findings of the disease.12 A second potential target, transducer of ERBB2 (TOB2), was also implicated in this earlier report and could explain many of the cell growth/proliferation defects seen in this study. TOB2 is an antiproliferative tumor suppressor that inhibits cell cycle progression, reportedly through repression of cyclin D1.49 TOB2 is directly regulated by miR-378-3p through a luciferase reporter assay that uses the 3′ untranslated region of TOB2, demonstrating that overexpression of miR-378-3p results in decreased TOB2 expression.49 Derepression of TOB2 expression by knockdown of miR-378-3p might therefore result in the observed decreased cell proliferation. What role TOB2 or other targets of miR-378-3p might play in the function of the myeloid cells remains to be seen.
To explore the signal transduction alterations that might underlie these functional defects, signaling patterns of ZIP cells were compared with those of patients with myelodysplasia. The use of primary patient samples was selected as the optimal comparator rather than use of these cells in a secondary model system, such as a mouse model. The MAPK and STAT1 pathways were found to be significantly underresponsive to granulocyte stimulation in both patients with MDS and ZIP cells at all stages of maturation. Similar parallels were seen between the patient samples and the cell model with decreased Ki-67 expression of immature myeloid cells and increased caspase-3 activation in mature myeloid cells. Although preliminary associations between signaling pathway use and the specific genetic findings in these samples from patients with MDS were examined, the number of samples with any specific genetic lesion was too small for any robust associations to be made and will need to await larger studies.
Finally, the utility of this model was demonstrated by studying the effects of azacytidine on the various defects in MDS. Azacytidine was the first compound approved by the US Food and Drug Administration for the treatment of all types of MDS. Although cytotoxic at high doses, azacytidine at low doses has been previously reported to improve differentiation.31 In these studies, azacytidine increased maturation of ZIP cells and mitigated the expression changes in the signaling pathways seen in the ZIP cells. In addition, azacytidine was able to partially reverse the growth and colony-forming defects in the ZIP cells, although no effect on neutrophil function was observed. The effect of azacytidine is intriguing in these studies. It has been previously demonstrated that the promoter of miR-378-3p is hypermethylated in MDS.7 However, the decreased expression of miR-378-3p in this experimental model was not caused by hypermethylation but by the presence of an antisense oligonucleotide. Although the most cited effect of azacytidine is on DNA methylation, azacytidine may also be incorporated into RNA and affect tRNA methyltransferase levels, resulting in defective mRNAs and tRNAs.50,51 This alteration in the mRNA and tRNA complements in the cells caused by azacytidine administration may negate the loss of the translational repression of mir-378-3p targets by the antisense. It is well known that the effects of azacytidine are multifactorial. This study suggests that, at least at low doses, azacytidine may exert a beneficial normalizing effect on the myeloid cells in addition to the known cytotoxic effects at higher dosages.52,53
Admittedly, this study examined the effect of knocking down only a single miRNA, although several dysregulated miRNAs (both up- and down-regulated) have been reported in MDS.12, 13, 14, 15, 16, 17 It is possible that the combinatorial mix of these aberrancies is required to recapitulate fully the findings in patients with MDS. However, it is remarkable that a single knockdown of miR-378-3p is effective in mimicking so many of the myeloid findings in MDS. The extension of this model to the other hematopoietic lineages involved in the MDS and to interactions of those hematopoietic cells with their microenvironment should be examined in the future.
In conclusion, this study uses a miRNA implicated in MDS to create an miRNA-based cell line model of MDS. This myeloid model recapitulates many of the phenotype aberrations in MDS and can be used in the study of MDS biology and potentially in the identification of novel therapies for MDS.
Acknowledgment
We thank the Vanderbilt University Medical Center Vangard core facilities for technical support.
Footnotes
Supported by Vanderbilt-Ingram Cancer Center grant P30 CA068485) (Y.G., Vanderbilt Vision Center grant P30 EY08126 (Y.G.), Doris Duke Charitable Foundation Clinician Scientist Development Award 2011048 (A.S.K.), American Cancer Society Institutional Research Grant IRG-58-009-51 (A.S.K.), and National Center for Research Resources/NIH–funded Vanderbilt Clinical and Translational Science Award UL1 RR024975 (A.S.K.).
Disclosures: None declared.
Current addresses of D.W., Boston Children's Hospital, Boston, MA; of M.L., Icahn School of Medicine, New York, NY; of B.U., Medicortex, Turku, Finland; of Y.G., Comprehensive Cancer Center, University of New Mexico, Albuquerque, NM; of Y.D., Loma Linda University Medical Center, Loma Linda, CA; of S.A.N., Washington University, St. Louis, MI; of J.S., University of Michigan, Ann Arbor, MI; of E.Y.G., University of California, Davis, CA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2021.07.006.
Supplemental Data
Supplemental Figure S1.
Evidence of the successful transduction and knockdown of HL60 cells with Scramble (SCR) vector and mirZIP-378-3p (ZIP). A: Expression of the antisense to miR-378-3p by the lentiviral vector by quantitative real-time RT-PCR (RT-qPCR) showing expression of the antisense only the ZIP cells. B: Expression of miR-378-3p by RT-qPCR showing knockdown of the miRNA expression in ZIP cells. C: Expression of green fluorescent protein (GFP) by PCR and gel electrophoresis, showing successful transduction of the vector in SCR and ZIP cells. D: Fluorescent images cells demonstrating expression of GFP by SCR and ZIP cells. Original magnification, ×10 (D). UNT, untransduced.
Supplemental Figure S2.
Disease pathway associations of mirZIP-378-3p. Three transcriptional profiling analyses (DESeq, edgeR, and bayseq) were assessed using Ingenuity Pathway Analysis. The top associated diseases pathways, based on lowest P value ranges, were obtained, and the average high (av hi) and average low (av lo) P values from the three analysis methods were plotted. Key pathways involved cell survival, proliferation, and function.
Supplemental Figure S3.
Examples of SPADE analysis of patient samples in response to stimulation demonstrating binning of cell populations (based on expression of cell surface markers) and phospho (p)–signal transducer and activator of transcription 1 (p-STAT1) expression in control (A) and myelodysplastic syndrome (MDS) (B) samples. NK, natural killer.
Supplemental Figure S4.
Images of cell colony formation in each group [dimethyl sulfoxide (DMSO) or azacytidine (AZA)] after culture for 7 days (quantitated in Figure 6C). Original magnification (inverted microscope), ×10. SCR, scramble; ZIP, miRZip-378a-3p.
References
- 1.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Hasserjian R.P. Myelodysplastic syndrome updated. Pathobiology. 2019;86:7–13. doi: 10.1159/000489702. [DOI] [PubMed] [Google Scholar]
- 3.Figueroa M.E., Skrabanek L., Li Y., Jiemjit A., Fandy T.E., Paietta E., Fernandez H., Tallman M.S., Greally J.M., Carraway H., Licht J.D., Gore S.D., Melnick A. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114:3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G., Schnittger S., Sanada M., Kon A., Alpermann T., Yoshida K., Roller A., Nadarajah N., Shiraishi Y., Shiozawa Y., Chiba K., Tanaka H., Koeffler H.P., Klein H.U., Dugas M., Aburatani H., Kohlmann A., Miyano S., Haferlach C., Kern W., Ogawa S. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinker E.B., Dueber J.C., Qualtieri J., Tedesco J., Erdogan B., Bosompem A., Kim A.S. Differential expression of ribosomal proteins in myelodysplastic syndromes. J Clin Pathol. 2016;69:176–180. doi: 10.1136/jclinpath-2015-203093. [DOI] [PubMed] [Google Scholar]
- 6.Narla A., Ebert B.L. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdogan B., Bosompem A., Peng D., Han L., Smith E., Kennedy M.E., Alford C.E., Wu H., Zhao Z., Mosse C.A., El-Rifai W., Kim A.S. Methylation of promoters of microRNAs and their host genes in myelodysplastic syndromes. Leuk Lymphoma. 2013;54:2720–2727. doi: 10.3109/10428194.2013.790542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muto T., Walker C.S., Choi K., Hueneman K., Smith M.A., Gul Z., Garcia-Manero G., Ma A., Zheng Y., Starczynowski D.T. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat Immunol. 2020;21:535–545. doi: 10.1038/s41590-020-0663-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reilly B., Tanaka T.N., Diep D., Yeerna H., Tamayo P., Zhang K., Bejar R. DNA methylation identifies genetically and prognostically distinct subtypes of myelodysplastic syndromes. Blood Adv. 2019;3:2845–2858. doi: 10.1182/bloodadvances.2019000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzon R., Croce C.M. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 11.Großhans H., Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 12.Erdogan B., Facey C., Qualtieri J., Tedesco J., Rinker E., Isett R.B., Tobias J., Baldwin D.A., Thompson J.E., Carroll M., Kim A.S. Diagnostic microRNAs in myelodysplastic syndrome. Exp Hematol. 2011;39:915–926.e2. doi: 10.1016/j.exphem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Fang J., Varney M., Starczynowski D.T. Implication of microRNAs in the pathogenesis of MDS. Curr Pharm Des. 2012;18:3170–3179. doi: 10.2174/1381612811209023170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussein K., Theophile K., Busche G., Schlegelberger B., Gohring G., Kreipe H., Bock O. Aberrant microRNA expression pattern in myelodysplastic bone marrow cells. Leuk Res. 2010;34:1169–1174. doi: 10.1016/j.leukres.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Sokol L., Caceres G., Volinia S., Alder H., Nuovo G.J., Liu C.G., McGraw K., Clark J.A., Sigua C.A., Chen D.T., Moscinski L., Croce C.M., List A.F. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br J Haematol. 2011;153:24–32. doi: 10.1111/j.1365-2141.2011.08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giudice V., Banaszak L.G., Gutierrez-Rodrigues F., Kajigaya S., Panjwani R., Ibanez M.D.P.F., Rios O., Bleck C.K., Stempinski E.S., Raffo D.Q., Townsley D.M., Young N.S. Circulating exosomal microRNAs in acquired aplastic anemia and myelodysplastic syndromes. Haematologica. 2018;103:1150–1159. doi: 10.3324/haematol.2017.182824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milunović V., Mandac Rogulj I., Planinc-Peraica A., Bulycheva E., Kolonić Ostojić S. The role of microRNA in myelodysplastic syndromes: beyond DNA methylation and histone modification. Eur J Haematol. 2016;96:553–563. doi: 10.1111/ejh.12735. [DOI] [PubMed] [Google Scholar]
- 18.Sheng Q., Vickers K., Zhao S., Wang J., Samuels D.C., Koues O., Shyr Y., Guo Y. Multi-perspective quality control of Illumina RNA sequencing data analysis. Brief Funct Genomics. 2017;16:194–204. doi: 10.1093/bfgp/elw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y., Zhao S., Ye F., Sheng Q., Shyr Y. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. Biomed Res Int. 2014;2014:248090. doi: 10.1155/2014/248090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 24.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardcastle T.J., Kelly K.A. BaySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shyr Y., Kim K. In: A Practical Approach to Microarray Data Analysis. Berrar D.P., Dublitzky W., Granzow M.A., editors. Springer; New York, NY: 2005. Weighted flexible compound covariate method for classifying microarray data; pp. 186–200. [Google Scholar]
- 27.Krämer A., Green J., Pollard J., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotecha N., Krutzik P.O., Irish J.M. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. Chapter 10:Unit10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S., Geiger H. HSC niche biology and HSC expansion ex vivo. Trends Mol Med. 2017;239:799–819. doi: 10.1016/j.molmed.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta D., Shah H.P., Malu K., Berliner N., Gaines P. Differentiation and characterization of myeloid cells. Curr Protoc Immunol. 2014;104:22F.5.1–22F.5.28. doi: 10.1002/0471142735.im22f05s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj K., Mufti G.J. Azacytidine (Vidaza®) in the treatment of myelodysplastic syndromes. Ther Clin Risk Manag. 2006;2:377–388. doi: 10.2147/tcrm.2006.2.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J.E., Conlon J.P., Yang X., Sanchez P.V., Carroll M. Enhanced growth of myelodysplastic colonies in hypoxic conditions. Exp Hematol. 2007;35:21–31. doi: 10.1016/j.exphem.2006.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tothova Z., Krill-Burger J.M., Popova K.D., Landers C.C., Sievers Q.L., Yudovich D., Belizaire R., Aster J.C., Morgan E.A., Tsherniak A., Ebert B.L. Multiplex CRISPR/Cas9-based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell. 2017;21:547–555.e8. doi: 10.1016/j.stem.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Y., Rongvaux A., Taylor A., Jiang T., Tebaldi T., Balasubramanian K., Bagale A., Terzi Y.K., Gbyli R., Wang X., Fu X., Gao Y., Zhao J., Podoltsev N., Xu M., Neparidze N., Wong E., Torres R., Bruscia E.M., Kluger Y., Manz M.G., Flavell R.A., Halene S. A highly efficient and faithful MDS patient-derived xenotransplantation model for pre-clinical studies. Nat Commun. 2019;10:366. doi: 10.1038/s41467-018-08166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuang X., Wei C., Zhang T., Yang Z., Chi J., Wang L. MiR-378 inhibits cell growth and enhances apoptosis in human myelodysplastic syndromes. Int J Oncol. 2016;49:1921–1930. doi: 10.3892/ijo.2016.3689. [DOI] [PubMed] [Google Scholar]
- 36.Wu D., Zhu X., Wen X., Zhang Y., Ma J., Yao D., Zhou J., Guo H., Wu P., Zhang X., Qiu H., Lin J., Qian J. Hypomethylation of MIR-378 5’-flanking region predicts poor survival in young patients with myelodysplastic syndrome. Mol Genet Genomic Med. 2020;8:e1067. doi: 10.1002/mgg3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee D.Y., Deng Z., Wang C.H., Yang B.B. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng H., Guo Y., Song H., Xiao B., Sun W., Liu Z., Yu X., Xia T., Cui L., Guo J. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene. 2013;518:351–359. doi: 10.1016/j.gene.2012.12.103. [DOI] [PubMed] [Google Scholar]
- 39.Zhang G., Zhou H., Xiao H., Li Y., Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109. doi: 10.1186/1471-2407-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z., Ma J. MiR-378 serves as a prognostic biomarker in cholangiocarcinoma and promotes tumor proliferation, migration, and invasion. Cancer Biomark. 2019;24:173–181. doi: 10.3233/CBM-181980. [DOI] [PubMed] [Google Scholar]
- 41.Ho C.S., Noor S.M., Nagoor N.H. MiR-378 and MiR-1827 regulate tumor invasion, migration and angiogenesis in human lung adenocarcinoma by targeting RBX1 and CRKL, respectively. J Cancer. 2018;9:331–345. doi: 10.7150/jca.18188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raza A., Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer. 2012;12:849–859. doi: 10.1038/nrc3321. [DOI] [PubMed] [Google Scholar]
- 43.Westers T.M., Ireland R., Kern W., Alhan C., Balleisen J.S., Bettelheim P., et al. Standardization of flow cytometry in myelodysplastic syndromes: a report from an International Consortium and the European LeukemiaNet Working Group. Leukemia. 2012;26:1730–1741. doi: 10.1038/leu.2012.30. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Z., Qianqiao Z., Qi H., Feng X., Chunkang C., Xiao L. In vitro deprivation of CD8+CD57+T cells promotes the malignant growth of bone marrow colony cells in patients with lower-risk myelodysplastic syndrome. Exp Hematol. 2010;38:677–684. doi: 10.1016/j.exphem.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Ma J., Wu D., Yi J., Yi Y., Zhu X., Qiu H., Kong R., Lin J., Qian J., Deng Z. MiR-378 promoted cell proliferation and inhibited apoptosis by enhanced stem cell properties in chronic myeloid leukemia K562 cells. Biomed Pharmacother. 2019;112:108623. doi: 10.1016/j.biopha.2019.108623. [DOI] [PubMed] [Google Scholar]
- 46.Ricevuti G., Mazzone A., Pasotti D., Fossati G., Mazzucchelli I., Notario A. The role of integrins in granulocyte dysfunction in myelodysplastic syndrome. Leuk Res. 1993;17:609–619. doi: 10.1016/0145-2126(93)90092-y. [DOI] [PubMed] [Google Scholar]
- 47.Pasotti D., Mazzone A., Fossati G., Mazzucchelli I., Pistone M.C., Montagna M., Parachini N., Labbate G., Corti C., Ricevuti G. Correlations between membrane integrins and granulocyte defects in myelodysplastic syndromes. Recenti Prog Med. 1993;84:742–749. [PubMed] [Google Scholar]
- 48.Fuhler G.M., Knol G.J., Drayer A.L., Vellenga E. Impaired interleukin-8- and GROα-induced phosphorylation of extracellular signal-regulated kinase result in decreased migration of neutrophils from patients with myelodysplasia. J Leukoc Biol. 2005;77:257–266. doi: 10.1189/jlb.0504306. [DOI] [PubMed] [Google Scholar]
- 49.Feng M., Li Z., Aau M., Wong C.H., Yang X., Yu Q. Myc/miR-378/TOB2/cyclin D1 functional module regulates oncogenic transformation. Oncogene. 2011;30:2242–2251. doi: 10.1038/onc.2010.602. [DOI] [PubMed] [Google Scholar]
- 50.Lu L., Randerath K. Mechanism of 5-azacytidine-induced transfer RNA cytosine-5-methyltransferase deficiency. Cancer Res. 1980;40:2701–2703. [PubMed] [Google Scholar]
- 51.Schaefer M., Hagemann S., Hanna K., Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 52.Christman J.K. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 53.Diesch J., Zwick A., Garz A.-K., Palau A., Buschbeck M., Götze K. A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin Epigenetics. 2016;8:71. doi: 10.1186/s13148-016-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.