Abstract
Background:
The axillary lymph node status is critical for breast cancer staging and individualized treatment planning.
Purpose:
To assess the effect of determining axillary lymph node (ALN) metastasis by breast MRI-derived radiomic signatures, and compare the discriminating abilities of different MR sequences.
Study Type:
Retrospective.
Population:
In all, 120 breast cancer patients, 59 with ALN metastasis and 61 without metastasis, all confirmed by pathology.
Field Strength/Sequence:
3 .0T scanner with T1-weighted imaging, T2-weighted imaging, diffusion-weighted imaging, and dynamic contrast-enhanced (DCE) sequences.
Assessment:
Typical morphological and texture features of the segmented tumor were extracted from four sequences, ie, T1WI, T2WI, DWI, and the second postcontrast phase (CE2) of the dynamic contrast-enhanced sequences. Additional contrast enhancement kinetic features were extracted from all DCE sequences (one pre- and seven postcontrast phases). Linear discriminant analysis classifiers were built and compared when using features from an individual sequence or the combination of the sequences in differentiating the ALN metastasis status.
Statistical Tests:
Mann–Whitney U-test, Fisher’s exact test, least absolute shrinkage selection operator (LASSO) regression, and receiver operating characteristic analysis were performed.
Results:
The accuracy/AUC of the four sequences was 79%/0.87, 77%/0.85, 74%/0.79, and 79%/0.85 for the T1WI, CE2, T2WI, and DWI, respectively. When CE2 was augmented by adding kinetic features, the model achieved the highest performance (accuracy = 0.86 and AUC = 0.91). When all features from the four sequences and the kinetics were combined, it did not lead to a further increase in the performance (P = 0.48).
Data Conclusion:
Breast tumor’s radiomic signatures from preoperative breast MRI sequences are associated with the ALN metastasis status, where CE2 phase and the contrast enhancement kinetic features lead to the highest classification effect.
In breast cancer patients, axillary lymph node metastasis is an important prognostic factor, and it is also critical for determining whether an adjuvant systemic chemotherapy or postoperative radiation is needed.1–3 Axillary lymph node (ALN) metastasis may be clinically confirmed by ultrasound-guided fine-needle aspiration (US-FNA) or lymph node dissection. For patients with confirmed ALN metastasis, axillary lymph node dissection (ALND) is the standard method for axillary lymph node staging. Sentinel lymph node dissection (SLND) is recommended to deal with nonpalpable axillary lymph nodes.4 However, both ALND and SLND are invasive, with potential complications of lymphedema, seroma, infection, hematoma, and arm pain.4 It would be beneficial to determine axillary lymph node status pre-operatively to reduce unnecessary lymph node dissection1–4 and patient distress.
Tumor heterogeneity reflects characteristics on tumor growth and biology and it has important clinical implications.5,6 The heterogeneity shown in images may be depicted by a set of quantitative radiomic image features.7,8 Quantitative radiomic features extracted from breast dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) have been shown to be associated with genetic expression profiles in breast cancer.9,10 Radiomic breast MRI features have also been shown to be predictive of the response of neoadjuvant chemotherapy, recurrence risk, and recurrence-free survival for breast cancer patients.11–15
Typical breast MRI protocols include multiple different sequences, including T1-weighted imaging (T1WI), T2-weighted imaging (T2WI), and diffusion-weighted imaging (DWI), as well as often DCE. The DCE sequence may reflect angiogenesis of tumor through the pattern of enhancement,16 and has been extensively investigated in many radiomic studies.9,15,17 The precontrast T1WI is mainly useful to evaluate the morphological information. T2W images are often clinically used to detect lesions and to reveal the cystic architecture within a lesion16; radiomic signatures derived by T2WI have been shown to be predictive of pathological response to neoadjuvant chemotherapy in breast cancer patients.11 DWI is used less in radiomic studies, possibly due to its low spatial resolution and image quality.18,19
Thus, the purpose of this study was to assess the feasibility of differentiating ALN metastasis using the radiomic signatures of primary breast cancer in breast MRI, and more important, to compare the potential discriminative effects of several multiparametric breast MR sequences, including precontrast T1WI, DCE, T2WI, and DWI, separately and combined.
Materials and Methods
Patients
The Institutional Review Board of our university hospital approved this study and the requirement of patients’ informed consent was waived. We retrospectively reviewed medical records to identify patients with invasive breast cancer confirmed by pathology between April 2013 and May 2017. A patient with a breast MRI examination performed within 10 days of treatment is considered eligible for enrollment. Patients were excluded if they had undergone core biopsy, operation, or neoadjuvant chemotherapy before imaging. Each enrolled patient was categorized as positive or negative for ALN metastasis status. The positive cases were confirmed by either lymph node dissection or US-FNA, and the negative cases were all confirmed by lymph node dissection.
Breast MRI Protocol
All patients were scanned in the prone position with breast immobilized using a 3 .0T scanner (Magnetom Verio Syngo, Siemens, Erlangen, Germany) and an 8-channel breast-specific phased-array coil. The imaging protocol consisted of the following sequences acquired in order: an axial 3D T1-weighted imaging (repetition time / echo time [TR/TE], 6.04/2.45 msec, field of view [FOV], 340 × 340 mm, slice thickness, 1.3 mm, flip angle, 20°, matrix size, 448 × 336), an axial fat-suppressed spoiled spin-echo T2WI imaging (TR/TE, 3600/61 msec, FOV, 340 × 340 mm, slice thickness, 4 mm, matrix size = 320 × 256), an axial DWI sequence, and axial DCE sequence. The diffusion-weighted images were obtained using the single-shot spin-echo echo-planar technique, with TR 9300 msec, TE 76 msec, 320 × 145 mm FOV, 4-mm slice thickness without gap, matrix of 168 × 168, three b values of 50 s/mm2, 400 s/mm2, and 1000 s/mm2, three diffusion directions, and echo planar imaging (EPI) factor of 76. The dynamic contrast-enhanced images were obtained using the 3D volumetric interpolated breath-hold examination (VIBE) technique, with TR 4.67 msec, TE 1.66 msec, 360 × 360 mm FOV, 1.2-mm slice thickness without a gap, flip angle of 10°, and matrix of 384 × 296. The dynamic-enhanced images included one precontrast and seven sequential postcontrast phases with a temporal resolution of 60 seconds. Magnevist (gadopentetate dimeglumine, Bayer Healthcare, Berlin, Germany) was injected immediately at the end of the precontrast phase acquisition, in a 0.2 ml/kg (or 0.1 mmol/kg body weight) through the elbow vein at a rate of 3.0 ml/s, followed by a flush of equal volume saline solution at the same rate.
Tumor Segmentation and Radiomic Feature Extraction
A single most representative slice was selected by a radiologist (R.C., with 11 years experience of breast MRI) for analysis. The tumor was segmented manually and a set of radiomic features were computed from the segmented tumor regions. Tumor segmentation was performed by the same radiologist who was blinded to the clinical and pathological information of the patients and by using the Mazda software (MaZda v. 4.6, Technical University of Lodz, Institute of Electronics, Poland). For radiomic feature extraction, we used the precontrast T1WI, T2WI, DWI (images of b = 1000 s/mm2), and the second postcontrast phase of the DCE sequence (denoted CE2) for comparative analyses. The reason for choosing the CE2 sequence here is because it is acquired at about 60–120 seconds after the contrast administration, a timepoint when the contrast between tumor and background is at its peak.16 The tumor region of interest (ROI) was firstly manually delineated on the CE2 image, and then was copied to the other three sequences (ie, T1WI, T2WI, and DWI), followed by a manual adjustment of the segmentation contours on these sequences as needed.
At preprocessing, all images were rescaled to 6 bits/pixel and gray-level normalization was carried out by the MaZda software by rescaling the histogram data to fit within μ ± 3σ (μ represents mean of gray-level, σ represents standard deviation of gray-level) to minimize the effects of the variations on imaging brightness and contrast.
A set of 475 quantitative radiomic imaging features was extracted from a segmented tumor ROI in each of the four MRI sequences (ie, T1WI, T2WI, DWI, and CE2) by using the Mazda software. These features included morphological features (eg, size and shape), histogram features, texture features (eg, gray-level co-occurrence matrix [GLCM], run length matrix [RLM]), and texture features extracted from some filtered images (eg, Wavelet transforms). GLCM-based features were calculated on four directions (θ = 0°, 45°, 90°, and 135°) with interpixel distances of n = 1, 2, 3, 4, and 5, respectively. RLM-based features were also calculated in the four directions.
In view of the kinetic features of DCE-MRI can reflect clinically important contrast enhancement characteristics of breast tumor, we computed an additional set of 109 kinetic features from the DCE sequence (ie, one pre- and seven postcontrast phases) using an in-house developed software. These features included those depicting time–intensity curve (TIC) (ie, washin slope [WIS], washout slope [WOS], peak enhancement [PE]) and features extracted from a time-to-peak (TTP) parametric map. The TTP parametric map is generated as follows. We first categorize all the pixels in a segmented tumor in terms of three classes/groups based on their TTP values (range: 1–7, corresponding to the seven postcontrast sequences). The three groups represent quick (Group 1), intermediate (Group 2), and slow (Group 3) arrival to the peak enhancement. Let Pi represent the ith pixel in the segmented tumor area, we use the following criteria to group the pixels:
| (1) |
Then a TTP parametric map was created as the pixel’s membership determined by Eq. 1 for each pixel in the tumor area. A pixel is labeled as red, green, or blue if it is a member of Group 1, Group 2, or Group 3, respectively. Note that while we chose to use three classes (for the sake of simplicity) to characterize the pixel’s heterogeneity on the arrival of the peak enhancement, one can use different number of classes as well. Next, the morphological and texture features were extracted from the TTP parametric map. The features extracted from the TTP parametric map may capture additional heterogeneity information of the contrast enhancement. Figure 1 shows an example of tumor segmentation in different MR sequences and the TTP parametric map.
FIGURE 1:
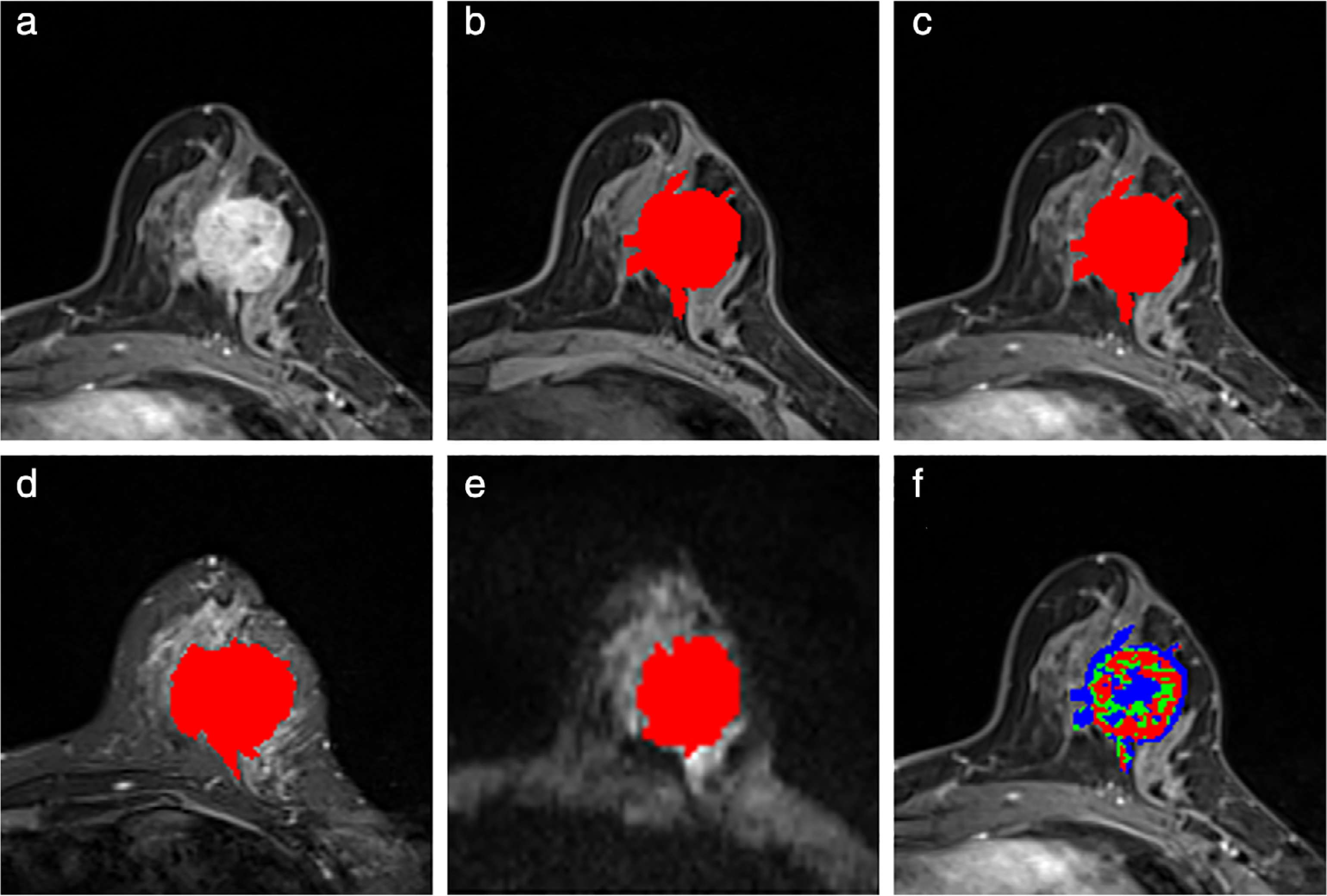
A 45-year-old woman diagnosed with invasive ductal carcinoma. (a) The second postcontrast image (ie, CE2). The malignant mass is seen in the left breast, with a size of 3.4 cm × 3.3 × 2.8 cm, histologic grade II (intermediate), axillary lymph node metastasis positive. (b–e) Segmentation (red masks) of the tumor overlaid on the original images in the precontrast T1 image (b), second postcontrast image (c), T2-weighted image (d), and diffusion-weighted image (e). (f) Example of a time-to-peak (TTP) parametric map (superposed on the second postcontrast image), where the red regions represent pixels that have arrived at their peaks in either the first or second postcontrast phase; the green regions represent pixels arrived at their peaks in either the third or fourth postcontrast phase; the blue regions represent pixels arrived at their peaks in the fifth, sixth, or seventh postcontrast phase.
ALN Metastasis Status Classification Modeling and Statistical Analysis
Considering the large number of extracted radiomic features, we employed the least absolute shrinkage and selection operator (LASSO) technique to select/rank the most significant features prior to the modeling process for ALN metastasis status classification. After LASSO regression, a small set of selected features were used as the input of a linear discriminant analysis (LDA) classifier to differentiate the ALN metastasis status. In order to take advantage of the imaging data and maintain a robust test, we used 10-fold cross-validation and repeated 100 times of the training-testing loop to evaluate the model’s performance. The receiver operating characteristic (ROC) curve analysis was performed and the area under the ROC curve (AUC) and accuracy were used to assess the classification performance of the LDA models. The pipeline of feature extraction, LASSO-based feature selection, and LDA-based classification was applied separately to each of the four MRI sequences (T1WI, CE2, T2WI, and DWI). We also combined all the features extracted from the four sequences together as a single set of integrated features to evaluate the collective effects of the four sequences. Similarly, the LASSO feature selection and LDA classification on this integrated feature set were performed. At the end, we can compare the classification performance of the four sequences individually and combined.
Statistical analysis was performed by using the SPSS software (v. 25.0; IBM SPSS Statistics for Windows, Armonk, NY). Two-tailed statistical tests were used, and P < 0.05 was considered statistically significant. Continuous data were tested by a one-sample Kolmogorov–Smirnov test, then expressed as the mean ± standard deviation for data with a normal distribution, or expressed as median (interquartile range) in nonnormally distributed data. Corresponding to the distribution of the data, the Student’s t-test or Mann–Whitney U-test was used to compare the continuous data between patients with and without ALN metastasis, and the chi-square test or Fisher’s exact test was used to compare the categorical data.
Results
Patient Characteristics
A total of 120 breast cancer patients were enrolled in this study. The mean age of the full cohort was 45.4 ± 10.3 years old. There were 59 (49.2%) patients who were confirmed as ALN metastasis positive and the rest of the cohort (61 patients) as negative. Clinical and pathological characteristics of the patients are summarized in Table 1. Patient age, tumor size, and histologic grade were not significantly different between the positive and negative groups. The ALN metastasis positive group was mainly invasive ductal cancer (98.3%), while the negative group included 50 invasive ductal cancer (82.0%), eight invasive lobular cancer, two invasive cribriform cancer, and one invasive tubular cancer.
TABLE 1.
Patient Characteristics of the Study Cohort
| Total (n = 120) | ALN metastasis positive (n = 59) | ALN metastasis negative (n = 61) | P | |
|---|---|---|---|---|
| Age (mean ± SD), years old | 45.4 ± 10.3 | 47.7 ± 10.8 | 43.2 ± 9.3 | 0.081 |
| Tumor size (mean ± SD), cm | 3.2 ± 2.0 | 3.8 ± 2.2 | 2.6 ± 1.7 | 0.078 |
| Histological type, No (%) | 0.005 | |||
| Invasive ductal cancer | 108 (90.0) | 58 (98.3) | 50 (82.0) | |
| Other types | 12 (10.0) | 1 (1.7) | 11 (18.0) | |
| Histologic grade, No (%) | 0.066 | |||
| I | 14 (11.7) | 3 (5.1) | 11 (18.0) | |
| II | 97 (80.8) | 50 (84.8) | 47 (77.1) | |
| III | 9 (7.5) | 6 (10.2) | 3 (4.9) |
ALN: axillary lymph node.
Selected Features
Six different feature sets were selected by LASSO from the T1WI, CE2, T2WI, DWI, CE2 plus kinetic features, and the integrated features, respectively. The top-ranked five features from each feature set are listed in Table 2. As can be seen, for the T1WI, CE2, T2WI, and DWI sequences, the top features basically came from the GLCM measures, morphology, and Wavelet transforms, histogram measures, and RLM features. Interestingly, when combining the features from the CE2 and additional kinetics features, 14 kinetic features were selected and the top five also included kinetic features, indicating the importance of the kinetics features.
TABLE 2.
Five Top-Ranked Features Selected by LASSO for Different Sequences
| Sequence | No. of selected features | Five top-ranked features (from rank 1 to rank 5) | Feature type |
|---|---|---|---|
| T1WI | 84 | Correlation (distance: [5, 5] pixels) | GLCM |
| Entropy (distance: [5, –5] pixels) | GLCM | ||
| Diameter range | Morphological | ||
| Number of skeletal loops | Morphological | ||
| Wavelet energy (Low frequency band at scale 1) | Wavelet | ||
| T2WI | 41 | Horizontal Feret’s diameter | Morphological |
| Number of profile contour pixels | Morphological | ||
| Sum of squares (distance: [4, –4] pixels) | GLCM | ||
| Oriental angle | Morphological | ||
| Angular second moment (distance: [5, 5] pixels) | GLCM | ||
| DWI | 90 | 90% percentile | Histogram |
| Area (distance: [5, 5] pixels) | GLCM | ||
| Horizontal long run emphasis | RLM | ||
| Width and length of the circumscribing rectangle of minimal area | Morphological | ||
| Sum of squares (distance: [0, 4] pixels) | GLCM | ||
| CE2 | 79 | Vertical Feret’s diameter | Morphological |
| Angular second moment (distance: [5, 5] pixels) | GLCM | ||
| Long run emphasis on the 45 degree | RLM | ||
| Gravity center to inscribed circle center horizontal distance | Morphological | ||
| Area of ARM | ARM | ||
| CE2 + Kinetics | 62 | Area of ARM | ARM |
| Entropy (distance: [5, 0] pixels) | GLCM | ||
| Skeleton length | Morphological | ||
| Long run emphasis on the 45 degree | RLM | ||
| Maximum peak enhancement | Kinetic | ||
| Union | 62 | Vertical Feret’s diameter (T2WI) | Morphological |
| Angular second moment (distance: [5, –5] pixels) (DWI) | GLCM | ||
| Sum of squares (distance: [5, 5] pixels) (T1WI) | GLCM | ||
| Difference variance (distance: [5, –5] pixels) (T1WI) | GLCM | ||
| 99% percentile (DWI) | Histogram |
CE2: the second postcontrast T1WI; Union: all features from four different sequences plus the kinetics features; GLCM: gray-level co-occurrence matrix; RLM: run length matrix; ARM: autoregressive model.
After we put all the features extracted from the four different sequences as well as the kinetics features (we called them the union), 57 features were selected, including two kinetic features, 15 from T2WI, 21 from DWI, 9 from T1WI, and 10 from CE2. The top-five features included a morphological feature from T2WI, a GLCM feature from DWI, two GLCM features from T1WI, and a histogram feature from DWI. Here, no kinetics features were among the five top-ranked features.
Classification Performance
The accuracy and ROC analysis results are shown and compared in Table 3 and Fig. 2. When comparing the four sequences individually, the highest accuracy (79.15%) comes from DWI and the highest AUC (0.8729) comes from T1WI. Overall, the performance measures are quite close between DWI and T1WI. It is obvious that the T2WI sequence exhibited the lowest overall performance compared with the other three sequences.
TABLE 3.
Performance of the Different Breast MRI Sequences, Separately or Combined, for Classifying the Axillary Lymph Node Metastasis Status
| Sequence | Accuracy (%, 95% CI) | AUC (95% CI) |
|---|---|---|
| T1WI | 78.67 (0.78, 0.79) | 0.8729 (0.8661, 0.8796) |
| CE2 | 77.32 (0.77, 0.78) | 0.8487 (0.8389, 0.8585) |
| T2WI | 74.16 (0.73, 0.75) | 0.7909 (0.7791, 0.8027) |
| DWI | 79.15 (0.78, 0.80) | 0.8508 (0.8409, 0.8607) |
| CE2 + kinetics | 86.37 (0.86, 0.87) | 0.9132 (0.9086, 0.9178) |
| Union | 84.43 (0.84, 0.85) | 0.9190 (0.9118, 0.9261) |
Note that we kept four digits after the decimal point in the AUC measures in order to show the subtle differences among the numbers. CI: confident interval; CE2: the second postcontrast T1WI; Union: all features from four different sequences plus the kinetics features.
FIGURE 2:
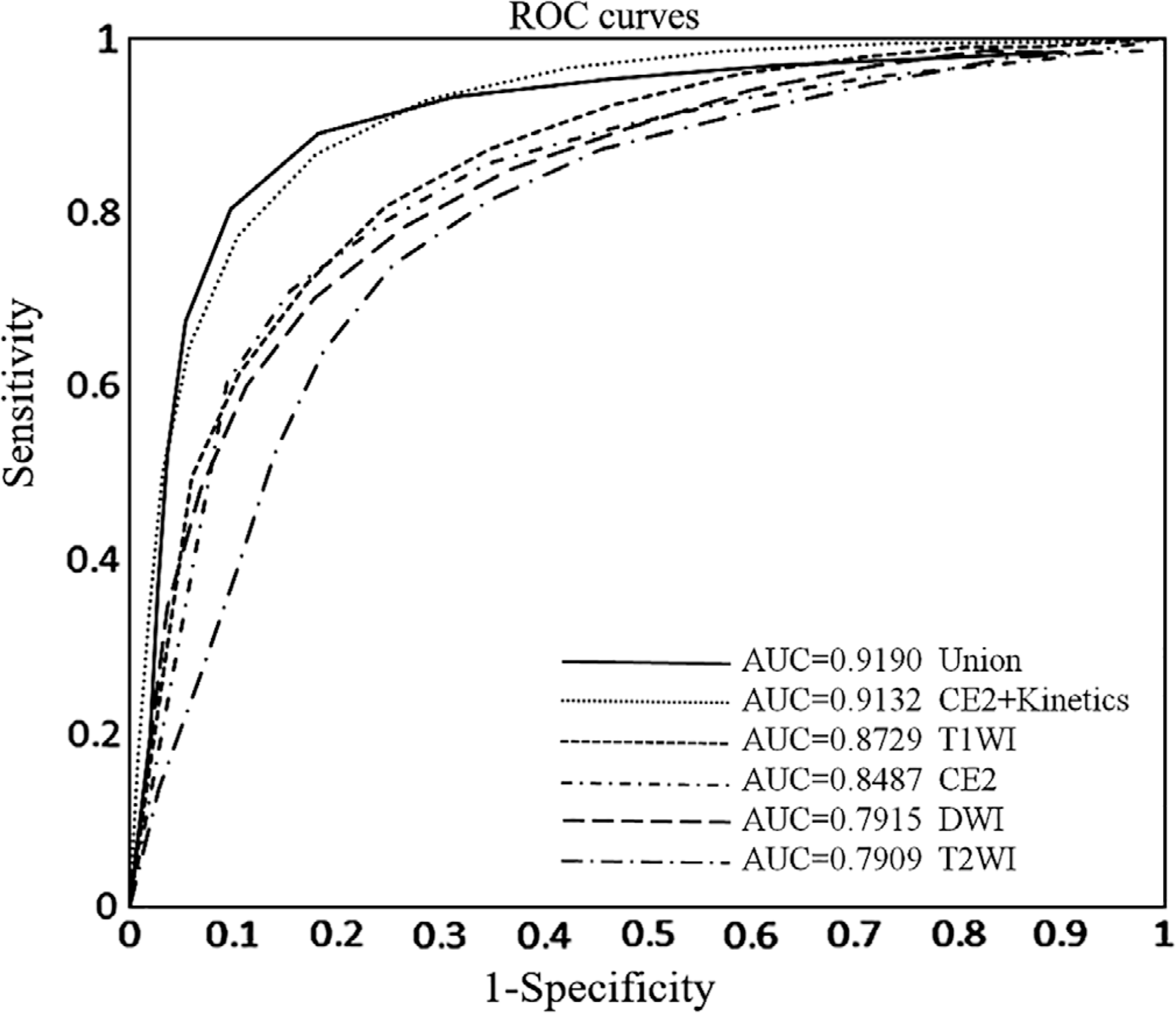
The ROC curves of using the different MRI sequences of precontrast T1WI, second postcontrast phase (CE2), T2WI, DWI, CE2 plus kinetic features (CE2 + Kinetics), and all of the features together (Union) for predicting axillary lymph node metastasis status.
However, when the kinetic features were added to the features from CE2, a substantial increase of the model performance was observed in terms of both accuracy (86.37%, P < 0.05) and AUC (0.9132, P < 0.05), obviously superior to the CE2 features alone. When all these features were mixed together, it did not lead to a performance improvement in comparison with the CE2 + kinetics scenario (AUC = 0.9190 for Union and AU = 0.9132 for CE2 + Kinetics; P = 0.48).
Discussion
In this study we showed that radiomic signatures extracted from preoperative breast MRIs are associated with the ALN metastasis status. The four MRI sequences: precontrast T1WI, CE2, T2WI, and DWI, all have features differential of the metastasis status. While the morphological and texture features from the four sequences exhibit a reasonable performance both in terms of accuracy and AUC, our results indicate that the kinetic features contribute substantially to the classification when combined with the features extracted from the CE2 phase, leading to the highest accuracy and AUC for this specific ALN metastasis status classification task.
Breast MRI is widely used in the preoperative evaluation of ALN status in breast cancer patients, showing superior performance than some other techniques such as ultrasound or positron emission tomography / computed tomography (PET-CT).21,22 While breast MRI is useful to diagnose ALN status via visual assessment, it suffers from some limitations. In most cases of breast MR scanning, the breast surface coil is mainly centered on the breast region,23–25 but some of the positive ALNs may locate at the border of the coil and some may even be out of the imaging FOV. Moreover, the air–axilla interfaces are often associated with magnetic sensitivity artifacts (or susceptible artifacts) that may affect the MRI of ALN.26 All these factors can restrict the visual assessment of breast MRI for diagnosing ALN metastasis status. In this study, we focus on the breast tumor itself in breast MRI and apply a radiomics-based approach to extract imaging features from the segmented tumor region to differentiate metastasis status of ALN. This may help avoid the aforementioned limits, and/or augment the direct visual assessment of ALN using breast MRI.
One important aspect of our study is to compare the discriminative abilities of multiparametric breast MR sequences. The majority of the features used are morphological and texture features and the performance margins are not wide among these sequences. It is a bit surprising that the postcontrast sequence CE2 does not outperform T1WI, but note that here no kinetic features were included for the CE2 sequence alone. This may also explain why the precontrast T1WI and postcontrast CE2 are close to each other, since only morphological and texture features were computed. But the additional kinetic features from the time–intensity curve and the TTP map are able to show a difference. This informs the importance of the heterogeneity of tumor contrast enhancements and the multiphase DCE sequences in breast MRI. In fact, the utilities of multiple temporal postcontrast sequence and kinetics-related measures have been shown in previous work.27–29 Our study on differentiating the ALN metastasis status adds an example showing the value of kinetic features extracted from multiple postcontrast sequences.
Interestingly, when we combined all the radiomic features of the four sequences and the kinetic features (ie, the union), it did not lead to improved performance than using CE2 plus the kinetic features. This implies that those other sequences may contribute minimally in the ALN metastasis differentiation task, if we have already had the CE2 sequence and kinetic features computed from the multiple temporal DCE phase. We do notice that in terms of the selected features, the “union” setting identified fewer kinetic features but more DWI features. This observation merits further investigation in future work. Considering the relatively low spatial resolution of DWI, it may benefit from the complement of other MR sequences that are more appropriate to compute morphology and texture features. We plan to further look into the comparisons of DWI to other sequences when high-resolution DWI data become available in the future.
Our study has several aspects worth pointing out. The dataset for this study was acquired from a single MR scanner in a medical center with a consistent scanning protocol, which may minimize potential bias and confounding factors in radiomic feature extraction and analysis. The segmentation and feature extraction were performed by using free commercial software that has been validated in many previous studies.11,30 In addition, we compared the effects of different MR sequences for our specific classification task. Such comparative studies are valuable to the community because it helps us gain insight and understanding of the overall utilities of these different MR sequences in radiomic-based clinical outcome differentiation.
Our study has several limitations. This is a retrospective study and our sample size was relatively small. Since the entire study cohort came from a single medical center, it lacks an evaluation of the generalization and robustness of our results. A larger patient cohort collected potentially from multiple institutions is planned for a more rigorous analysis. Second, tumor ROIs were drawn manually by radiologists, and thus is time-consuming and prone to error and user variability. An automatic, reliable, and validated breast tumor segmentation method is ideal but not available to us at this stage of our study. Third, we selected a single representative slice to analyze in this preliminary study. Although this is a common approach used in numerous previous studies,11,13,15 we acknowledge that it may lose some important spatial information compared with using the whole tumor volume. We are planning to explore a 3D segmentation method and then volumetric-based study in the next-step research. In addition, when computing the TTP maps we did not perform intersequence coregistration that may not be optimal.
In conclusion, this study showed that the radiomic signatures of primary breast cancer extracted from preoperative breast MR sequences are associated with the ALN metastasis status. The precontrast T1WI, CE2, T2WI, and DWI sequences all showed a level of discriminative performance, where the combination of CE2 features and the kinetic features from multiple temporal postcontrast sequence performs the best. A larger study is warranted to further evaluate our findings.
Acknowledgment
Contract grant sponsor: National Key R&D Program of China; Contract grant sponsor: Fundamental Research Funds for the Central Universities; Contract grant numbers: 2017YFC 1309100, N172008008; Contract grant sponsor: National Institutes of Health (NIH) / National Cancer Institute (NCI); Contract grant numbers: 1R01CA193603, 3R01CA193603–03S1, 1R01CA218405; Contract grant sponsor: University of Pittsburgh Physicians (UPP) Academic Foundation Award.
References
- 1.Jatoi I, Hilsenbeck SG, Clark GM, Osborne CK. Significance of axillary lymph node metastasis in primary breast cancer. J Clin Oncol 1999;17: 2334–2340. [DOI] [PubMed] [Google Scholar]
- 2.Lee MC, Joh JE, Chau A. Axillary staging prior to neoadjuvant chemotherapy: The roles of sentinel lymph node biopsy and axillary ultrasonography. Cancer Control 2012;19:277–285. [DOI] [PubMed] [Google Scholar]
- 3.van Roozendaal LM, Schipper RJ, Van de Vijver KK, et al. The impact of the pathological lymph node status on adjuvant systemic treatment recommendations in clinically node negative breast cancer patients. Breast Cancer Res Treat 2014;143:469–476. [DOI] [PubMed] [Google Scholar]
- 4.Giuliano AE, Ballman KV, McCall L, et al. Effect of axillary dissection vs no axillary dissection on 10-year overall survival among women with invasive breast cancer and sentinel node metastasis: The ACOSOG Z0011 (Alliance). JAMA 2017;318:918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer 2012;12:323–334. [DOI] [PubMed] [Google Scholar]
- 7.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol 2016;2:1636–1642. [DOI] [PubMed] [Google Scholar]
- 9.Ashraf AB, Daye D, Gavenonis S, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: Preliminary associations with gene expression profiles. Radiology 2014;272:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Zhu Y, Burnside ES, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 2016;281:382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson S, Purdie C, Michie C, et al. Interim heterogeneity changes measured using entropy texture features on T2-weighted MRI at 3.0 T are associated with pathological response to neoadjuvant chemotherapy in primary breast cancer. Eur Radiol 2017;27:4602–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parikh J, Selmi M, Charles-Edwards G, et al. Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology 2014;272:100–112. [DOI] [PubMed] [Google Scholar]
- 13.Chamming’s F, Ueno Y, Ferre R, et al. Features from computerized texture analysis of breast cancers at pretreatment MR imaging are associated with response to neoadjuvant chemotherapy. Radiology 2018; 286:412–420. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Ko ES, Lim Y, et al. Breast cancer heterogeneity MR imaging texture analysis and survival outcomes. Radiology 2017;282:665–675. [DOI] [PubMed] [Google Scholar]
- 15.Michoux N, Van den Broeck S, Lacoste L, et al. Texture analysis on MR images helps predicting non-response to NAC in breast cancer. BMC Cancer 2015;15:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterji M, Mercado CL, Moy L. Optimizing 1.5-Tesla and 3-Tesla dynamic contrast-enhanced magnetic resonance imaging of the breasts. Magn Reson Imaging Clin N Am 2010;18:207–224, viii. [DOI] [PubMed] [Google Scholar]
- 17.Agner SC, Rosen MA, Englander S, et al. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images. Radiology 2014;272:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidic I, Egnell L, Jerome NP, et al. Support vector machine for breast cancer classification using diffusion-weighted MRI histogram features: Preliminary study. J Magn Reson Imaging 2018;47:1205–1216. [DOI] [PubMed] [Google Scholar]
- 19.Schipper RJ, Paiman ML, Beets-Tan RG, et al. Diagnostic performance of dedicated axillary T2- and diffusion-weighted MR imaging for nodal staging in breast cancer. Radiology 2015;275:345–355. [DOI] [PubMed] [Google Scholar]
- 20.Szczypinski PM, Strzelecki M, Materka A, Klepaczko A. MaZda—A software package for image texture analysis. Comput Methods Programs Biomed 2009;94:66–76. [DOI] [PubMed] [Google Scholar]
- 21.Valente SA, Levine GM, Silverstein MJ, et al. Accuracy of predicting axillary lymph node positivity by physical examination, mammography, ultrasonography, and magnetic resonance imaging. Ann Surg Oncol 2012;19:1825–1830. [DOI] [PubMed] [Google Scholar]
- 22.Liang X, Yu J, Wen B, Xie J, Cai Q, Yang Q. MRI and FDG-PET/CT based assessment of axillary lymph node metastasis in early breast cancer: A meta-analysis. Clin Radiol 2017;72:295–301. [DOI] [PubMed] [Google Scholar]
- 23.Yun SJ, Sohn YM, Seo M. Differentiation of benign and metastatic axillary lymph nodes in breast cancer: Additive value of MRI computer-aided evaluation. Clin Radiol 2016;71:403.e1–7. [DOI] [PubMed] [Google Scholar]
- 24.Hyun SJ, Kim EK, Moon HJ, Yoon JH, Kim MJ. Preoperative axillary lymph node evaluation in breast cancer patients by breast magnetic resonance imaging (MRI): Can breast MRI exclude advanced nodal disease? Eur Radiol 2016;26:3865–3873. [DOI] [PubMed] [Google Scholar]
- 25.Kim EJ, Kim SH, Kang BJ, Choi BG, Song BJ, Choi JJ. Diagnostic value of breast MRI for predicting metastatic axillary lymph nodes in breast cancer patients: Diffusion-weighted MRI and conventional MRI. Magn Reson Imaging 2014;32:1230–1236. [DOI] [PubMed] [Google Scholar]
- 26.Huang SY, Seethamraju RT, Patel P, Hahn PF, Kirsch JE, Guimaraes AR. Body MR imaging: Artifacts, k-space, and solutions. Radiographics 2015;35:1439–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Cao G, Sun X, et al. Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology 2018;288: 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karahaliou A, Vassiou K, Arikidis NS, Skiadopoulos S, Kanavou T, Costaridou L. Assessing heterogeneity of lesion enhancement kinetics in dynamic contrast-enhanced MRI for breast cancer diagnosis. Br J Radiol 2010;83:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golden DI, Lipson JA, Telli ML, Ford JM, Rubin DL. Dynamic contrast-enhanced MRI-based biomarkers of therapeutic response in triple-negative breast cancer. J Am Med Inform Assoc 2013;20:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waugh SA, Purdie CA, Jordan LB, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol 2016; 26:322–330. [DOI] [PubMed] [Google Scholar]


