Summary
Currently available SARS-CoV-2 therapeutics are targeted toward moderately to severely ill patients and require intravenous infusions, with limited options for exposed or infected patients with no or mild symptoms. Although vaccines have demonstrated protective efficacy, vaccine hesitancy and logistical distribution challenges will delay their ability to end the pandemic. Hence, there is a need for rapidly translatable, easy-to-administer-therapeutics that can prevent SARS-CoV-2 disease progression, when administered in the early stages of infection. We demonstrate that an orally bioavailable Hsp90 inhibitor, SNX-5422, currently in clinical trials as an anti-cancer therapeutic, inhibits SARS-CoV-2 replication in vitro at a high selectivity index. SNX-5422 treatment of human primary airway epithelial cells dampened expression of inflammatory pathways previously associated with poor SARS-CoV-2 disease outcomes. In addition, SNX-5422 interrupted expression of host factors demonstrated to be crucial for SARS-CoV-2 replication. Development of SNX-5422 as SARS-CoV-2-early-therapy will dampen disease severity, resulting in better clinical outcomes and reduced hospitalizations.
Subject areas: Therapeutics, Virology
Graphical abstract
Highlights
-
•
SNX-5422 inhibits SARS-CoV-2 replication in vitro at a high selectivity index
-
•
Transcriptomic profile of human airway epithelial cells treated with SNX-5422
-
•
SNX-5422 dampens proinflammatory pathways associated with poor COVID outcomes
-
•
SNX-5422 targets host factors crucial for SARS-CoV-2 replication
Therapeutics; Virology
Introduction
Since its emergence in 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly spread worldwide and the disease associated with SARS-CoV-2 infection, Coronavirus disease 2019 (COVID-19), was declared a pandemic by the WHO in March 2020 (Cucinotta and Vanelli, 2020). As of October 21st, 2021, there were more than 241 million confirmed infections and >4.9 million deaths, worldwide (WHO Coronavirus Disease COVID-19 Dashboard, 2020), making this ongoing disease the deadliest pandemic of the 21st century. Clinical manifestations of SARS-CoV-2 infection can range from asymptomatic (Nishiura et al., 2020) and mild upper airway symptoms to severe lower respiratory complications such as pneumonia, acute respiratory distress syndrome (ARDS), and multi-organ system dysfunction (Huang et al., 2020a), leading to respiratory compromise (Giamarellos-Bourboulis et al., 2020) and death (Mehraeen et al., 2020).
There is an ongoing effort to characterize SARS-CoV-2 pathogenesis and identify prophylactic and therapeutic approaches to suppress COVID-19 severity and improve clinical outcomes. Currently used treatment modalities for SARS-CoV-2 infection include repurposed small molecules such as remdesivir (Beigel et al., 2020), immunomodulatory agents such as corticosteroids (Prescott and Rice, 2020), and virus-specific monoclonal antibodies (Zhou et al., 2020b). However, most of these treatment strategies require intravenous infusions, and are administered in moderately to severely ill patients. Moreover, although prophylactic vaccines for SARS-CoV-2 infection have demonstrated robust protection, vaccine hesitancy and logistical distribution challenges, especially in low-to-middle income countries (LMIC), will restrict their ability to quickly end the global pandemic. Thus, there remains an urgent need for rapidly translatable post-exposure prophylactic interventions that can be administered during the early course of infection to prevent severe disease outcomes, hospitalizations, and reduce viral transmissions. Because such interventions will be administered to patients with no-to-mild symptoms, these therapies should be easy to store and administer, and have a wide margin of safety.
Cellular chaperone protein heat shock protein 90 (Hsp90), in addition to maintaining cellular homeostasis (Zhao and Houry, 2005; Taipale et al., 2010), also facilitates proper folding and functionality of virally encoded proteins (Gellezr et al., 2007). Hence, Hsp90 serves as host determinant for a wide diversity of viruses (Geller et al., 2012; Rathore et al., 2014; Smith and Haystead, 2017; Chase et al., 2008; Chen et al., 2012; Gao and Harhaj, 2013; Shatzer et al., 2017), including Coronaviruses (CoVs) such as severe acute respiratory syndrome coronavirus (SARS-CoV) (Li et al., 2004, 2020), Middle east respiratory syndrome coronavirus (MERS-CoV) (Li et al., 2020) and SARS-CoV-2 (Emanuel et al., 2020; Li et al., 2020). Recent in silico studies have proposed Hsp90 as a potential anti-SARS-CoV-2 target (Iyad et al., 2020). Furthermore, in a murine model of sepsis, Hsp90 inhibition has been associated with dampened systemic and pulmonary inflammation and acute lung injury (ALI) (Chatterjee et al., 2007), the key phenomena associated with poor SARS-CoV-2 clinical outcomes (Bussani et al., 2020; Pandolfi et al., 2020; Blanco-Melo et al., 2020). Hence, a trial was launched to test the efficacy of intravenously administered Hsp90 inhibitor, ganetespib (ADX-1612) against SARS-CoV-2 infection (Aldeyra Therapeutics Press Release, 2020).
SNX-5422 is an orally bioavailable Hsp90 inhibitor (Fadden et al., 2010; Huang et al., 2009), which is currently in clinical trials for solid state malignancies and lymphomas. The toxicity profile of this small molecule has been extensively characterized in humans, and found to be very well tolerated with no reported adverse effects or evidence of immunosuppression (Inafante et al., 2014; Rajan et al., 2011). Because Hsp90 can be exploited by viruses for their replication, here, we have evaluated the efficacy of SNX-5422 as a postexposure prophylactic treatment for SARS-CoV-2 infection and have mapped the transcriptional profile of the interaction of the drug with primary human airway epithelial cells from uninfected individuals with no known respiratory disease or smoking history. Our data reveal that SNX-5422, added shortly after infection, suppresses in vitro SARS-CoV-2 replication with a high selectivity index, and influences expression of cellular genes that have been previously associated with SARS-CoV-2 replication and poor disease outcomes, including airway inflammatory pathways. Our findings support further development of SNX-5422 as an oral post-exposure therapeutic for SARS-CoV-2 infection that will reduce the length of viral infectivity and dampen virus-associated inflammatory responses, initiated early in the course of infection. A postexposure therapy geared toward individuals in the initial phase of infection, will reduce COVID-19 disease severity and hospitalizations and will prevent additional global deaths, while the distribution of prophylactic vaccines ramps up.
Results and discussion
SNX-5422, an orally bioavailable Hsp90 inhibitor, is a potent suppressor of SARS-CoV-2 replication
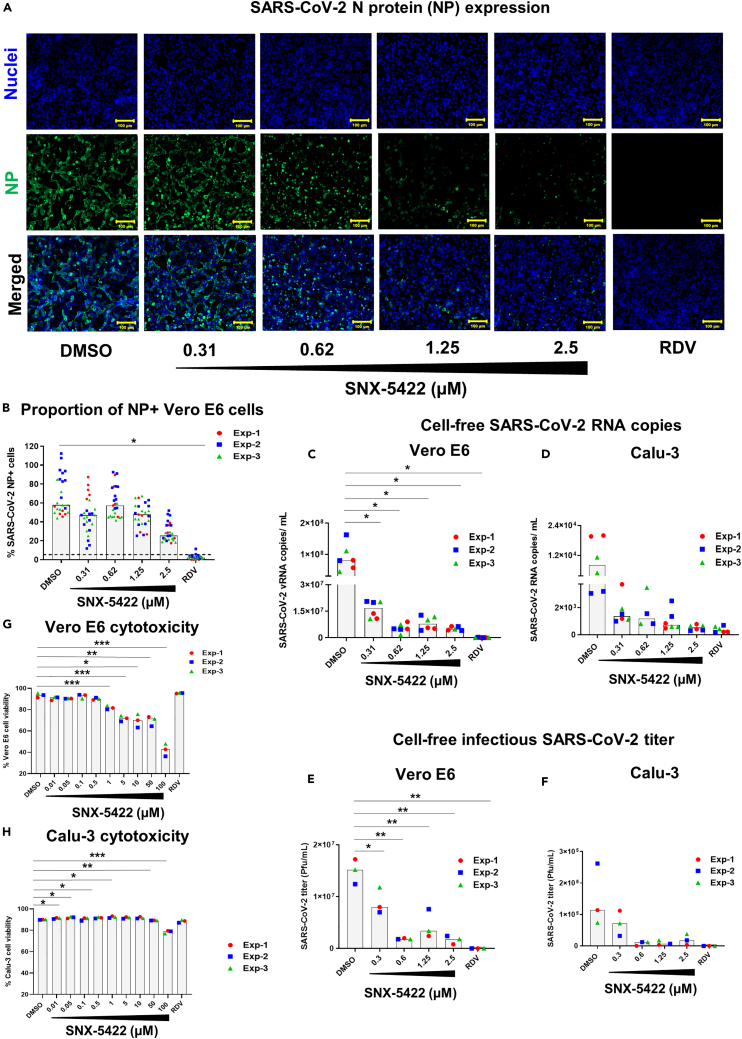
Recent in silico drug repositioning studies and single-cell transcriptomic analysis have identified Hsp90 as a host dependency factor for SARS-CoV-2 replication (Iyad et al., 2020; Emanuel et al., 2020). Inhibition of Hsp90 was associated with suppression of SARS-CoV-2 replication in vitro (Emanuel et al., 2020; Li et al., 2020; Kamel et al., 2020). Here, we sought to determine if an orally bioavailable, highly selective inhibitor of Hsp90 (SNX-5422) (Okawa et al., 2009), currently in clinical trials for cancer therapy (Inafante et al., 2014; Rajan et al., 2011), could serve as a post-exposure prophylactic or early therapeutic for SARS-CoV-2 infection. To mimic post-exposure prophylaxis in vitro, human lung epithelial cells (Calu-3), and African green monkey kidney cells (Vero E6) (Ogando et al., 2020) were incubated with SARS-CoV-2 strain USA-WA1/2020 for 1hr, and the virus-exposed cells were treated with increasing doses of SNX-5422 or DMSO (drug-vehicle). Using immunostaining and fluorescent imaging of virus-infected Vero E6 cells, we first assessed the antiviral activity of SNX-5422 by comparing proportion of viral nucleocapsid (NP) protein positive cells in drug-treated vs. vehicle-treated cells. Our data indicate that SNX-5422 attenuated intracellular NP expression (Figure 1A) and proportion of NP+ cells (Figure 1B) in a dose dependent manner, with a 50% inhibitory concentration (IC50) of 2.3μM (Table 1). In addition, we also confirmed that SNX-5422 reduced intracellular NP expression (Figure S1A) and proportion of NP+ cells (Figure S1B) in human lung epithelial cell line, Calu-3 (Harcourt et al., 2011), We further evaluated the antiviral activity of a dose range of SNX-5422 by comparing viral RNA shedding in cell supernatants of drug-treated vs. vehicle-treated Vero cells and human lung epithelial cells. SNX-5422 treatment resulted in a reduction of cell-free viral genomic copies (Figures 1C and 1D) with an IC50 of 0.2 μM in both cell types (Table 1). In addition, SNX-5422-treatment reduced cell-free infectious viral titers (Figures 1E and 1F) with an IC50 of 0.38 μM in Vero E6 cells and 0.4 μM in Calu-3 cells (Table 1). It is possible that differences in IC50s between each measure of viral replication in the same cell type are attributable to the difference in the antiviral mechanisms of SNX-5422 and the viral replication steps that the drug potentially interferes with. Interestingly, similar differences in IC50 measurements have been observed for a ribonucleoside analog β-D-N4-hydroxycytidine (NHC, EIDD-1931) (Sheahan et al., 2020).
Figure 1.
SNX-5422 potently suppresses SARS-CoV-2 replication at non-toxic concentrations
Vero E6 and Calu-3 cells were infected with SARS-CoV-2 strain USA-WA1/2020 and treated with either 0.1% DMSO (drug-vehicle) or 0.31–2.5 μM SNX-5422, or 5μM remdesivir (RDV).
(A) Both SNX-5422 and RDV were reconstituted in DMSO. After 48 h post-infection (pi), (A) Vero cells were immunostained for SARS-CoV-2 nucleocapsid protein (NP) (green). Nuclei were counterstained (blue) before imaging. (B) Immunofluorescent images were quantified to evaluate proportions of SARS-CoV-2-NP+ cells. Each symbol and color represent an independent experiment and each data point represents an independent quantified image field. Medians of data points are reported as gray bars. Dotted line represents detection cut off, which is the mean + 3 standard deviation (SD) of the percentage of NP+ cells in uninfected controls. Image quantification was performed using Fiji as described in the STAR methods section.
(C and D) Cell-free viral RNA in the supernatant of infected and treated (C) Vero-E6 and (D) Calu-3 cells were measured by qRT-PCR of the viral N gene.
(E and F) The infectious viral titers of the supernatant of (E) Vero E6 and (F) Calu-3 were determined by plaque assay.
(G and H) (G) Vero E6 and (H) Calu-3 cells were treated with either 0.1% DMSO (drug-vehicle) or 0.01–100 μM SNX-5422, or 5 μM RDV. After 48 h, the proportion of viable cells was evaluated by flow cytometry. Each symbol and color represent an independent experiment and medians of the data points are reported as gray bars. Two-sided p value was computed using Welch’s two-sample t test and statistical significance was evaluated at the 0.05 alpha level. ∗: p ≤ 0.05; ∗∗: p ≤ 0.01; ∗∗∗p ≤ 0.001.
Table 1.
Cell-specific SNX-5422 potency and selectivity
| Cytotoxic concentration (CC) | ||||
| Cell line | Assay | CC50a (μM) | ||
| Vero E6 | Cellular viability | 91.97 | ||
| Calu-3 | Cellular viability | >100 | ||
| Inhibitory concentration (IC) | ||||
| Cell line | Assay | IC50b (μM) | SIc | |
| Vero E6 | Cell-free SARS-CoV-2 RNA | 0.2 | 459.85 | |
| Cell-free infectious SARS-CoV-2 titer | 0.38 | 242.03 | ||
| Intracellular SARS-CoV-2 N protein expression | 2.3 | 39.99 | ||
| Calu-3 | Cell-free SARS-CoV-2 RNA | 0.2 | >500 | |
| Cell-free infectious SARS-CoV-2 titer | 0.4 | >250 | ||
| Intracellular SARS-CoV-2 N protein expression | NEd | NEd | ||
CC50: 50% cytotoxic concentration.
IC50: 50% inhibitory concentration.
SI: Selectivity index = CC50/IC50.
NE: Not estimated.
Because SNX-5422 inhibits the functionality of Hsp90 (Fadden et al., 2010; Huang et al., 2009), a host chaperone necessary for maintaining cellular homeostasis (Zhao and Houry, 2005; Taipale et al., 2010), we determined the selectivity index of the drug in inhibiting SARS-CoV-2 replication. Uninfected Vero E6 and Calu-3 cells were treated with increasing concentrations of SNX-5422 and the frequency of live cells was monitored by flow cytometry (Figures 1G and 1H). The 50% cellular cytotoxicity (CC50) of SNX-5422 was computed to be 91.97 μM and >100 μM in Vero E6 and Calu-3 cells, respectively, highlighting the minimal cytotoxic effects of the drug on these two cell types. Consequently, the selectivity index (SI = CC50/IC50) of SNX-5422 in suppressing viral replication was evaluated to be high in both cell types (Table 1). Collectively, these data suggest that treatment with an oral inhibitor of Hsp90, SNX5422, upon SARS-CoV-2 exposure, can potently attenuate viral replication in vitro, at a high selectivity index.
SNX-5422 treatment of primary human tracheobronchial epithelial (TBE) cells alters expression of cellular inflammatory and metabolic pathways
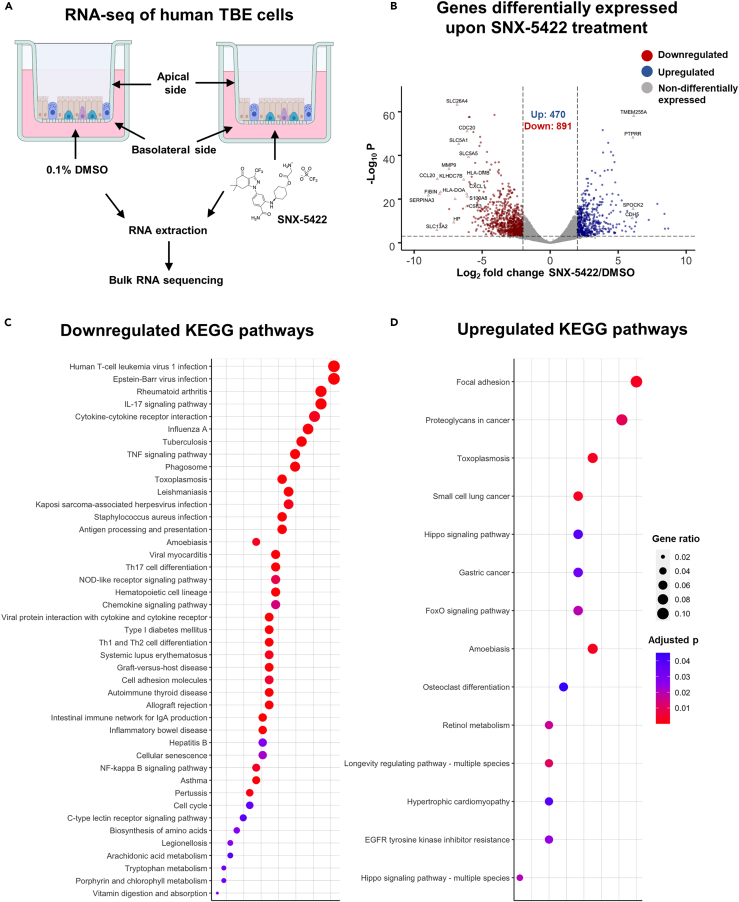
In addition to developing antivirals targeting specific viral proteins, there has been a growing effort in developing therapeutics targeting host proteins exploited by viruses for their replication or implicated in viral pathogenesis (Nitulescu et al., 2020). As such therapeutics can impair normal functioning of host proteins, a clear understanding of their impact on cellular mechanisms is critically important. We therefore characterized the interactions of SNX-5422 with the cellular machinery using a physiologically relevant ex vivo human primary tracheobronchial epithelial (TBE) cell model (Mason, 2020). Fully differentiated human TBE cells from three independent donors (Table S1) without known respiratory disease or smoking history were cultured ex vivo at the air-liquid interface. The basolateral side of the cell culture was treated with either 0.1% DMSO (drug-vehicle) or 1 μM SNX-5422 for 48 h, followed by bulk RNA sequencing of the total cellular RNA (Figure 2A). Principal component analysis of the read counts revealed that treatment with SNX-5422 formed distinct clusters compared to the control group (Figure S2), highlighting differential expression of cellular genes upon drug treatment. Differentially expressed genes (DEGs) between the control group and the SNX-5422-treated group were defined as protein coding genes with a Log2 fold change (FC) ratio >2 and adjusted p value ≤0.001. A volcano plot of the protein coding genes indicated that 1361 genes were differentially expressed between the two groups, with 470 cellular genes being upregulated and 891 cellular genes being downregulated upon SNX-5422 treatment (Figure 2B). To identify cellular genes whose expression was highly regulated upon SNX-5422 treatment of TBE cells, we collated DEGs with a Log2 FC > 6, for which the average normalized read count across donors and experimental conditions was ≥300 (Table S2). Using this stringent threshold, we identified 16 cellular genes that were downregulated and 4 cellular genes that were upregulated upon drug treatment (Figure 2B). Further categorization of those genes based on functions revealed that SNX-5422-treatment of TBE cells downregulated cytokines and chemokines that serve as chemoattractant for lymphocytes and neutrophils (CCL20, CXCL1), regulators of cellular inflammation (CSF3, S100A8) and proliferation (CDC20), and antigen presentation pathway-associated genes (HLA-DOA, HLA-DMB) and genes functioning as solute transporters (SLC13A2, SLC26A4, SLC5A1, SLC5A5) (Figure 2B).
Figure 2.
SNX-5422 treatment alters gene expression of human tracheobronchial epithelial (TBE) cells
(A) Schematic diagram illustrating treatment of human TBE cells. TBE cells from 3 independent healthy donors were cultured in air–liquid interface and treated with either 0.1% DMSO (drug-vehicle) or 1 μM SNX-5422 reconstituted in DMSO, for 48 h. RNA was then extracted from the cells and bulk RNA sequencing was performed.
(B) Volcano plot demonstrating protein coding genes altered upon SNX-5422 treatment of human TBE cells. Red, blue, and gray indicate downregulated, upregulated, and non-differentially expressed genes, respectively. Differentially expressed genes (DEGs) were defined as protein coding genes with a Log2 fold change >2 between the drug-treated and control groups and adjusted p value < 0.001. Open symbols represent genes with Log2 fold change >6 between the drug-treated and control groups. Cellular pathway overrepresentation analysis was performed with DEGs with a normalized average read count across donors and experimental conditions ≥300.
(C and D) (C) Downregulated and (D) Upregulated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with an adjusted p value < 0.05 were reported. Dot size represents gene ratio and color schema represents adjusted p values.
Next, we mapped the cellular signaling pathways that were significantly altered upon SNX-5422 treatment of human TBE cells, by performing a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway overrepresentation analysis on the DEGs (Figures 2C and 2D). Notably, cellular inflammatory pathways such as IL-17 signaling pathway, cytokine-cytokine receptor interaction, TNF-signaling pathway, Th17 cell differentiation, NOD-like receptor signaling and chemokine signaling pathways were among the top 20 identified pathways that were downregulated by SNX-5422 treatment. In addition, signaling pathways associated with regulation of cell cycle and cellular senescence, and metabolic pathways such as biosynthesis of amino acids and tryptophan metabolism, were also downregulated by SNX-5422 (Figure 2C). Furthermore, treatment with this oral Hsp90 inhibitor resulted in upregulation of key cellular pathways involved in physiological functions such as cellular-adhesion, regulation of apoptosis and cellular proliferation (Figure 2D). Taken together, our data suggest that SNX-5422 alters TBE cell inflammation, cellular proliferation, and metabolism.
SNX-5422 dampens cellular inflammatory pathways previously associated with SARS-CoV-2 replication and poor disease prognosis
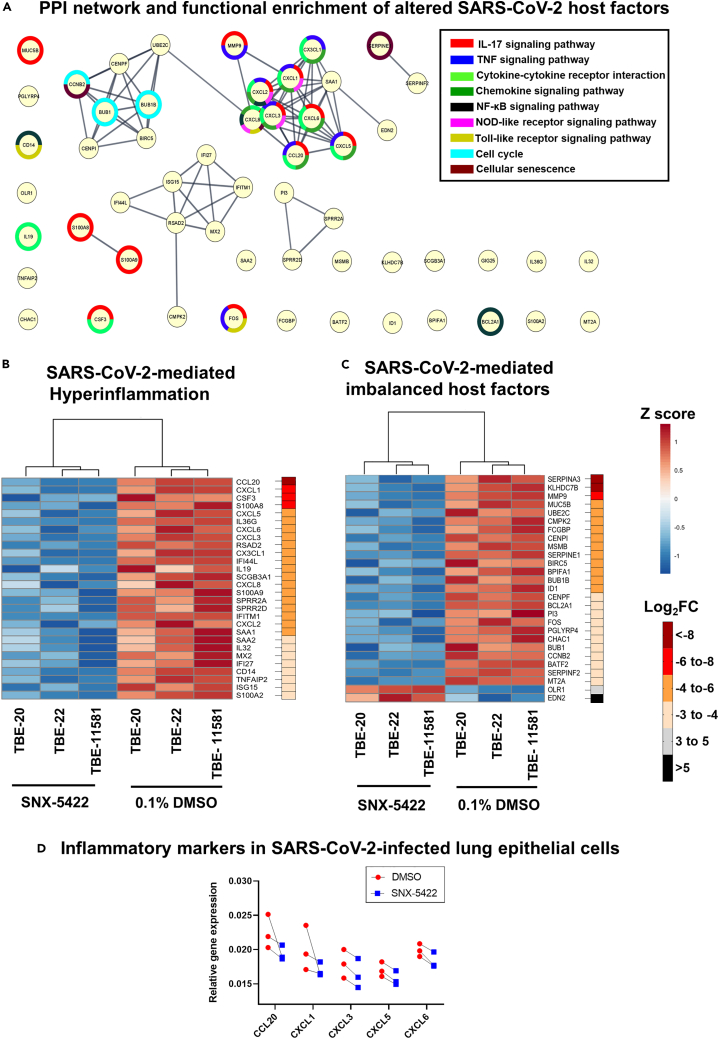
SARS-CoV-2 infection has been associated with hyper-induction of pro-inflammatory cytokines resulting in multiple organ failure, ALI and ARDS (Song et al., 2020). Interestingly, Hsp90 inhibition dampened systemic and pulmonary inflammation and lung injury in a murine model of sepsis (Chatterjee et al., 2007). Because SNX-5422 exhibited downregulated inflammatory transcriptomic pathways in human TBE cells, we interrogated whether these pathways have been previously associated with SARS-CoV-2 disease progression. To this end, we first selected a gene set whose expression was stringently regulated upon SNX-5422 treatment of human TBE cells (Log2 FC > 3, total 287 genes) (Table S2). We then performed an extensive literature search in PubMed and non-peer reviewed prepublication repositories to compare our identified gene set with the cellular genes and pathways previously associated with SARS-CoV-2 replication. To maintain physiological relevance, we restricted our literature search to genes and pathways reported in ex vivo SARS-CoV-2-infected primary airway epithelial cells or those identified in blood, bronchoalveolar lavage fluid (BALF), and lung biopsies of SARS-CoV-2 positive patients. Our search identified 55 genes regulated by SNX-5422, which have been associated with SARS-CoV-2 pathogenesis and disease progression. A protein-protein interaction (PPI) network and functional enrichment analysis mapped several of these 55 genes to cellular inflammatory pathways including IL-17 signaling pathway, TNF signaling pathway, cytokine-cytokine receptor interaction, and chemokine signaling pathway (Figure 3A). SNX-5422 dampened expression of cellular genes, previously shown to be associated with SARS-CoV-2 -mediated proinflammatory response and hyper-cytokinemia (Figure 3B). Specifically, these inflammatory genes could be broadly categorized into chemokines (CXCL1 (Coperchini et al., 2020), CXCL5 (Coperchini et al., 2020), CXCL6 (Blanco-Melo et al., 2020), CXCL3 (Blanco-Melo et al., 2020) and CXCL2 (Zhou et al., 2020a)), chemokine-receptor ligands (CCL20 (Zhou et al., 2020a), CX3CL1 (Nienhold et al., 2020)), cytokines (IL36G (Xiong et al., 2020)), interleukins (IL-19 (Vastrad et al., 2020), CXCL8 (Zhou et al., 2020a), IL-32 (Blanco-Melo et al., 2020)), tumor necrosis factor alpha-induced protein (TNFAIP2 (Fagone et al., 2020)), regulators of inflammatory responses (CSF3 (Nunnari et al., 2020), S100A8 (Zhou et al., 2020a), S100A9 (Zhou et al., 2020a), SCGB3A1 (Mick et al., 2020), S100A2 (Xu et al., 2020)) and interferon-induced proteins (RSAD2 (Zhou et al., 2020a), IFI44L (Mick et al., 2020), IFITM1 (Zhou et al., 2020a), MX2 (Lieberman et al., 2020), IFI27 (Mick et al., 2020), and ISG15 (Zhou et al., 2020a)). Collectively, our analysis suggests that early treatment with SNX-5422 may mitigate SARS-CoV-2-mediated hyperinflammation, thereby improving clinical outcomes of COVID-19.
Figure 3.
SNX-5422 treatment targets cellular pathways and genes involved in SARS-CoV-2 replication and disease progression
(A) Protein–protein interaction (PPI) network and functional enrichment of genes altered by SNX-5422 that are involved in SARS-CoV-2 replication and COVID-19 progression. Each node represents a protein and interactions with confidence score >0.9 are presented.
(B and C) Heat maps of DEGs following SNX-5422 treatment of human TBE cells, that have been identified to be (B) associated with SARS-CoV-2-mediated hyperinflammation and (C) imbalanced upon viral infection. Red represents relative upregulation of gene expression and blue represents relative downregulation of gene expression. Gene expression counts were transformed, and Z score normalized for heatmap generation. Donors were clustered with complete linkage. Genes are arranged based on fold changes, as indicated by the fold change key.
(D) Calu-3 cells were infected with SARS-CoV-2 strain USA-WA1/2020 and treated with either 0.1% DMSO (drug-vehicle) or 2.5 μM SNX-5422 reconstituted in DMSO. After 48 h post-infection (pi), total RNA was extracted, and relative expression of selected inflammatory genes were assessed using RT-qPCR. Two-sided p value was computed using Welch’s two-sample t test and statistical significance was evaluated at the 0.05 alpha level.
SNX-5422 regulates expression of host factors known to be imbalanced upon SARS-CoV-2 infection
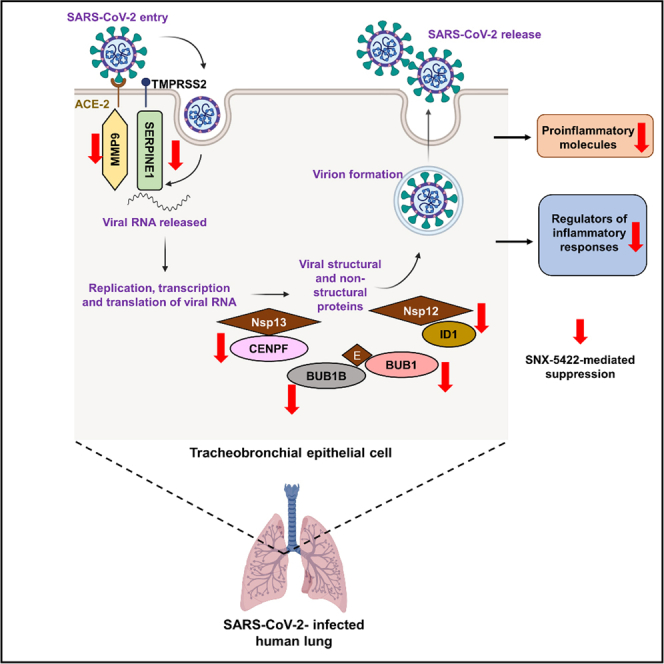
Recently, several host factors of SARS-CoV-2 replication have been identified (Gordon et al., 2020; Wang et al., 2021; Daniloski et al., 2021). In addition, SARS-CoV-2 infection and COVID-19 progression have been associated with imbalanced expression of cellular pathways (Blanco-Melo et al., 2020) and heightened innate immune responses (Zhou et al., 2020a). Here, we aimed to investigate whether SNX-5422 treatment regulated expression of those previously identified SARS-CoV-2 host factors and cellular mechanisms imbalanced upon infection. Our data indicate that of the 55 genes altered by SNX-5422 treatment that were previously reported in the context of SARS-CoV-2 replication (Figure 3A), 27 genes had an imbalanced cellular expression, in virally infected cells (Figure 3C). Further categorization of these 55 genes revealed that the majority of these genes fall into 4 functional categories,- namely, regulators of innate immunity (SERPINA3 (D'Alessandro et al., 2020), BPIFA1 (Liu et al., 2020), and PGLYRP4 (Chandrashekar et al., 2020)), regulators of cell cycle (UBE2C (Bock and Ortea, 2020), CENPI (Venkatakrishnan et al., 2020), BIRC5 (Xiong et al., 2020), CCNB2 (Xiong et al., 2020), BUB1B (Ahmed et al., 2020), BUB1 (Ahmed et al., 2020) and CENPF (Gordon et al., 2020)), mucosal structural proteins (MMP9 (Hazra et al., 2020) and FCGBP (Liu et al., 2020)) and regulators of blood coagulation (SERPINA3 (D'Alessandro et al., 2020), and SERPINE1 (Mccord et al., 2020) and SERPINF2 (D'Alessandro et al., 2020)). Furthermore, cellular genes whose expression was impacted by SNX-5422 treatment, encoded for proteins that were previously shown to be associated with SARS-CoV-2 replication phases (MMP9 (Hazra et al., 2020) and SERPINE1 (Mccord et al., 2020)) or those interacting with SARS-CoV-2 structural and non-structural proteins (BUB1B (Ahmed et al., 2020), BUB1 (Ahmed et al., 2020), CENPF (Gordon et al., 2020) and ID1 (Islam and Khan, 2020)). Taken together, our analysis indicates that SNX-5422 treatment interrupted cellular pathways and genes associated with replication of SARS-CoV-2.
SNX-5422 treatment downregulates expression of proinflammatory genes in SARS-CoV-2-infected human lung epithelial cells
Our data indicated that SNX-5422 treatment of uninfected human airway epithelial cells dampened expression of pro-inflammatory genes that have been associated with SARS-CoV-2 replication and disease progression in previous literature (Figure 3B). Hence, next, we sought to validate whether this anti-inflammatory mechanism of SNX-5422 was also observed in SARS-CoV-2-infected human lung epithelial cells. To this end, Calu-3 cells were incubated with SARS-CoV-2 strain USA-WA1/2020 for 1hr and the virus-exposed cells were treated with 2.5μM SNX-5422 or DMSO (drug-vehicle) for 48 h. After 48 h, total cellular RNA was extracted, and relative gene expression of 5 pro-inflammatory genes (CCL20, CXCL1, CXCL3, CXCL5, and CXCL6) identified to be downregulated by SNX-5422 in uninfected condition (Figure 3B) was assessed. SNX-5422 treatment downregulated expression of all 5 genes compared to DMSO-treated controls in infected cell populations (Figure 3D). Based on this finding, we postulate that early administration of SNX-5422 will dampen SARS-CoV-2-mediated inflammation, although further validation of this hypothesis in physiologically relevant ex vivo and in vivo models of infection will be necessary.
Viruses rely on the biosynthetic machinery of the host for their replication. Therefore, an attractive strategy to attenuate viral propagation and mitigate poor disease outcomes is to target viral host-dependency factors. Although the advantage of such an antiviral strategy is their efficacy against newly evolving viral variants and a high barrier toward development of drug resistant viral strains, such therapies might interfere with the normal cellular functioning. Hence, development of host-targeted therapeutics with a very high selectivity index for suppressing viral replication yet having minimal off-target effect is crucial. Moreover, as post-exposure prophylactics are administered to individuals with no-to-mild symptoms, the safety bar for such a therapeutic modality will be higher than those administered in acutely infected patients. Hence, although host-directed drug candidates are currently fast-tracked into clinical trials as anti-SARS-CoV-2 therapeutics, characterizing the interactions of the drug candidates with host cell mechanisms will be absolutely critical to minimize adverse outcomes.
SARS-CoV-2 infection results in highly activated T cells (Chen and John Wherry, 2020), Th17 skewing (de Biasi et al., 2020), activation of the NF-κB signaling pathway (Huang et al., 2020b), and abnormal elevation of inflammatory cytokines (de Biasi et al., 2020), leading to hypercytokinemia and an imbalanced host immune response (Zhou et al., 2020a; Blanco-Melo et al., 2020). Hyperinflammation is a key determinant of the severity of COVID-19 outcomes (Mehta et al., 2020), where patients with higher levels of inflammatory cytokines have been associated with higher risk of lung injury and ARDS (Chen et al., 2020; Torres Acosta and Singer, 2020). Hence, targeted treatments, early during the course of infection, to prevent such cytokine storms have been proposed (Gallelli et al., 2020). We demonstrate that SNX-5422-treatment of human TBE cells downregulated key inflammatory pathways and genes already associated with poor COVID-19 outcomes (Zhou et al., 2020a; Nienhold et al., 2020). Importantly, SNX-5422 treatment also altered expression of host responses reported to be imbalanced upon SARS-CoV-2 infection such as innate immune responses (Blanco-Melo et al., 2020), cell cycle regulation (Yuan et al., 2006), biosynthesis, and metabolism of amino acids (Blasco et al., 2020). Moreover, SNX-5422 interrupted expression of cellular factors that directly or indirectly interact with the crucial steps of SARS-CoV-2 replication (Mccord et al., 2020; Hazra et al., 2020) and with viral structural and non-structural proteins (Ahmed et al., 2020; Gordon et al., 2020; Islam and Khan, 2020).
In summary, our data support further testing and development of SNX-5422 as an oral post-exposure prophylactic or early-phase therapeutic for SARS-CoV-2 infection. Based on our findings, we hypothesize that early administration of SNX-5422 will interfere with SARS-CoV-2 replication machinery and dampen virus-induced inflammation and severe disease outcomes. Our observations of the antiviral properties of SNX-5422 was primarily limited to African green monkey kidney cells (Vero E6) and human lung epithelial (Calu-3) cell lines, hence, experimental validation of such mechanisms in physiologically relevant ex vivo human primary tissue sections, in animal models and finally, in human clinical trials remains necessary. The fact that SNX-5422 is orally bioavailable, and is clinically tested, makes this drug a quality candidate for an easy-to-administer and rapidly translatable post-exposure prophylactic therapy for SARS-CoV-2 infections. Of note, the in vitro drug levels necessary for achieving SARS-CoV-2 suppression was very comparable to the anti-tumor drug levels suggested in human phase II clinical trials (100 mg/m2 every other day, which is equivalent to ∼2.3 μM plasma drug levels), although in vivo pharmacokinetic and pharmacodynamic validation in SARS-CoV-2 infected conditions remains necessary. Finally, because SNX-5422 targets dependency factors that are utilized by multiple CoVs (Aldeyra Therapeutics Press Release, 2020, Emanuel et al., 2020; Iyad et al., 2020; Li et al., 2004, 2020) this drug may have the potential to serve as a broad-spectrum ready-to-deploy therapeutic, which might be efficacious in not only addressing the current pandemic, but also facilitating future pandemic preparedness against newly emerging, genetically diverse, CoV strains.
Limitations of the study
Our observations of the antiviral properties of SNX-5422 was primarily limited to cell lines, hence, experimental validation of such mechanisms in physiologically relevant ex vivo human primary tissue sections, in animal models and finally, in human clinical trials remains necessary.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS-CoV-2 (2019-nCoV) Nucleocapsid Antibody, Rabbit MAb | SinoBiological | Cat # 40143-R019 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher | Cat # A-21206 |
| Bacterial and virus strains | ||
| SARS-CoV-2 USA-WA1/2020 | BEI Resources | NR-52281 |
| Biological samples | ||
| Fully differentiated human tracheobronchial epithelial (TBE) cells | MATTEK | EpiAirwayTM |
| Chemicals, peptides, and recombinant proteins | ||
| Remdesivir | MedChemExpress | CAS # 1809249-37-3 |
| SNX-5422 | Fadden et al. (2010) and Huang et al. (2009) | N/A |
| Critical commercial assays | ||
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Thermo Fisher | Cat # L34957 |
| QIAamp viral RNA mini kit | Qiagen | Cat # 52906 |
| SuperScript™ III First-Strand Synthesis System | Thermo Fisher | Cat # 18080051 |
| TaqMan™ Gene Expression Master Mix | Thermo Fisher | Cat # 4369542 |
| RNeasy Mini Kit | Qiagen | Cat # 74106 |
| TruSeq RNA library Prep Kit v2 | Illumina | Cat # RS-122-2001 and RS-122-2002 |
| Deposited data | ||
| Raw and processed RNA Seq data | This paper | GEO: GSE166397 |
| Experimental models: Cell lines | ||
| African Green Monkey: Vero E6 cells | ATCC | CRL-1586 |
| Human: Calu-3 | ATCC | HTB-55 |
| Oligonucleotides | ||
| SARS-CoV-2 N fwd primer: 5′-CACATTGGCACCCGCAATC-3′ | IDT | N/A |
| SARS-CoV-2 N rev primer: 5′-GAGGAACGAGAAGAGGCTTG-3′ | IDT | N/A |
| SARS-CoV-2 N probe: 5′-FAM-ACTTCCTCAAGGAACAACATT GCCA-QSY-3′ |
IDT | N/A |
| TaqMan gene expression assay- CCL20 | Thermo Fisher | Cat # Hs00355476_m1 |
| TaqMan gene expression assay- CXCL1 | Thermo Fisher | Cat # Hs00236937_m1 |
| TaqMan gene expression assay- CXCL3 | Thermo Fisher | Cat # Hs00171061_m1 |
| TaqMan gene expression assay- CXCL5 | Thermo Fisher | Cat # Hs00607029_g1 |
| TaqMan gene expression assay- CXCL6 | Thermo Fisher | Cat # Hs06632196_S1 |
| Recombinant DNA | ||
| pCDNA3.1(+)-N-eGFP | Molecular Cloud/GenScript Biotech | Cat # MC_0101137 |
| Software and algorithms | ||
| Fiji | Schindelin et al. (2012) | https://imagej.net/software/fiji/ |
| Zeiss Zen Black | Zeiss | https://www.micro-shop.zeiss.com/en/us/softwarefinder/software-categories/zen-black/ |
| Graph Pad Prism | Graph Pad | https://www.graphpad.com/ |
| FlowJo™ Vs.10.1 | BD Life Sciences | https://www.flowjo.com/ |
| R Vs.4.0.5 | N/A | https://cran.r-project.org/src/base/R-4/ |
| STRING Vs. 11.5 | Szklarczyk et al. (2021) | https://string-db.org/ |
| Cytoscape Vs 3.8.2 | Shannon et al. (2003) | https://cytoscape.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sallie R. Permar (sap4017@med.cornell.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject detail
Fully differentiated human TBE cells (EpiAirwayTM) from 3 independent donors with no reported respiratory disease or smoking history (Table S1) were obtained from MATTEK (Ashland, MA). The cells were cultured at the air-liquid interface in 1 ml of AIR-100-MM culture medium (MatTek) in 6 well plates at 37°C in 5% CO2. Upon receipt of cells, the cultures were acclimated for 16–24 hr prior to the start of experiments. In vitro efficacy studies for SNX-5422 were performed in African Green Monkey kidney epithelial cells (Vero E6) and human lung epithelial cells, Calu-3. Vero E6 cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1X Penicillin/Streptomycin (Gibco), and 1X non-essential amino acid (NEAA) mixture (Gibco) and maintained at 37°C in 5% CO2. Calu-3 cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (FBS), 25 mM HEPES, 1X Penicillin/Streptomycin (Gibco), and maintained at 37°C in 5% CO2.
Method details
Compounds
The small molecular inhibitor of Hsp90, SNX-5422, has been previously described (Fadden et al., 2010; Huang et al., 2009). The compound was synthesized in-house (Duan et al., 2012) and was characterized by proton nuclear magnetic resonance (NMR) and liquid chromatography/mass-spectrometry (LC/MS). The compound was solubilized in 100% DMSO to a 10 mM stock concentration. Remdesivir (MedChemExpress LLC, USA) was reconstituted in 100% DMSO to a concentration of 10 mM.
SARS-CoV-2 propagation and titering
SARS-CoV-2 USA-WA1/2020 (BEI Resources; NR-52281) was propagated on Vero E6 cells at a multiplicity of infection (MOI) of 0.001 in virus diluent (DMEM supplemented with 2% FBS, 1X Penicillin/Streptomycin (Gibco), 1mM sodium pyruvate (Gibco) and 1X NEAA (Gibco)) at 37°C in 5% CO2. At day 4 post-infection (pi), cell supernatant containing the released virus was harvested, spun at 1500 rpm for 5 minutes, filtered through a 0.22 μM sterile vacuum filtration system, aliquoted and stored at −80°C until further use.
Stock viral titer was determined by plaque assay. Briefly, 0.72 × 106 Vero E6 cells were seeded in 6 well plates. The stock virus was serially diluted and incubated on cellular monolayer at 37°C in 5% CO2. After 1 hr, virus was aspirated, and cells were overlayed with carboxy-methyl cellulose (CMC) containing media (0.6% CMC, MEM supplemented with 1X Penicillin/Streptomycin (Gibco), 2% FBS, 1mM sodium pyruvate (Gibco), 1X NEAA (Gibco), 0.3% sodium bicarbonate (Gibco), and 1X GlutaMAX (Gibco). After 4 days of incubation at 37°C in 5% CO2, plaque assays stained with 1% crystal violet in 10% neutral buffered formalin (NBF), and plaque forming unit/mL (Pfu/mL) was determined.
SARS-CoV-2 infection and treatment
1.4 × 105 Vero E6 or Calu-3 cells were seeded in 24 well plates. After 24 hrs, cells were incubated with the SARS-CoV-2 isolate at an MOI of 0.01–0.02 at 37°C and 5% CO2, with intermittent plate rocking. After 1 hr, the virus was aspirated, cells were rinsed twice with 1X PBS and fresh maintenance medium containing dilutions of SNX-5422 or Remdesivir or 0.1% DMSO was added. The cells were incubated for 48 hrs at 37°C and 5% CO2.
Cellular cytotoxicity using flow cytometry
Suspensions of Vero E6 and Calu-3 cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Thermo Fisher) according to manufacturer’s instructions. The stained cells were fixed using methanol-free 4% formaldehyde (Thermo Fisher) for 30 minutes and acquired on an LSRII flow cytometer (BD Biosciences) using BD FACS Diva software and analyzed with FlowJo software version 10.1 (BD Life Sciences). LIVE/DEAD marker negative viable cells were selected from total cells after sequential selection of forward scatter and side scatter singlets.
Infectious viral titer of supernatant
Cellular supernatant was collected from infected and drug-treated cells, 48 hrs after infection. The supernatant was clarified by spinning at 1500 rpm for 5 minutes and infectious viral titer was measured by plaque assay as described above.
qRT-PCR
SARS-CoV-2 RNA from cell supernatant was extracted using the QIAamp viral RNA mini kit (Qiagen). A two-step qRT-PCR was used to detect viral RNA released in the cell supernatant. In the first step, viral cDNA for the nucleocapsid (N) gene was generated using SuperScript™ III First-Strand Synthesis System (Thermo Fisher) and N-reverse primer (5′-GAGGAACGAGAAGAGGCTTG-3′), following manufacturer’s instructions. In the second step, 7uL cDNA from step-1 was amplified using N gene forward primer (5′-CACATTGGCACCCGCAATC-3′), N gene reverse primer (5′-GAGGAACGAGAAGAGGCTTG-3′) and probe (5′-FAM-ACTTCCTCAAGGAACAACATTGCCA-QSY-3′) using TaqMan gene expression mastermix (Thermo Fisher). The thermal cycling steps were: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min, and qPCR was performed on a Step-One-Plus real time PCR machine (Applied Biosystems) using the StepOne Software v2.3. Viral RNA copy number/mL supernatant was assessed using pCDNA3.1(+)-N-eGFP plasmid (Molecular Cloud/GenScript Biotech) as standard.
For extraction of total cellular RNA from SARS-CoV-2-infected Calu-3 cells, cells were resuspended in TRIzol reagent (Thermo Fisher) followed by phase separation with chloroform and cellular RNA was extracted subsequently using the RNeasy Mini Kit (Qiagen). A two-step qRT-PCR was used to detect expression of selected cellular genes. In the first step, total cDNA was generated using SuperScript™ III First-Strand Synthesis System (Thermo Fisher) and oligo dT following manufacturer’s instructions. In the second step, 8uL cDNA from step-1 was amplified using TaqMan™ Gene Expression Master Mix and TaqMan Gene Expression Assaysfor specific genes (Thermo Fisher). The thermal cycling steps were: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min, and qPCR was performed on a Step-One-Plus real time PCR machine (Applied Biosystems) using the StepOne Software v2.3. Relative expression of each target with respect to total input RNA quantity was computed.
Immunofluorescence assay
1.4 × 105 Vero E6 or Calu-3 cells were seeded in 4-well chamber slides (Corning). After 24 hrs, cells were incubated with SARS-CoV-2 USA-WA1/2020 strain at an MOI of 0.01–0.02 at 37°C and 5% CO2, with intermittent rocking. After 1 hr, the virus was aspirated, cells were rinsed twice with 1X PBS and fresh maintenance medium containing dilutions of SNX-5422 or Remdesivir or 0.1% DMSO was added. After 48 hrs, cells were fixed by submerging the chamber slides in 10% NBF for 2 hrs. Fixed cells were washed three times with DPBS (Sigma) + 0.02% Triton X-100. For immunostaining, cells were permeabilized with DPBS+ 0.1% Triton-X-100 for 20 minutes and blocked in DPBS supplemented with 5% BSA, 0.02% Tween-20 and 10% donkey sera (Sigma) for 1hr. The cells were then incubated with SARS-CoV-2 (2019-nCoV) Nucleocapsid Antibody, Rabbit Mab (Sinobiological; 1:100) for 2hrs at room temperature. After subsequent washing of the samples, the cells were treated with Donkey anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 (Thermo Fisher; 1:1000) for 1 hr at room temperature. After further washing, the coverglasses were mounted on glass slides with ProLong Glass Antifade with NucBlue (Invitrogen) and sealed. Coverglasses were allowed to cure overnight prior to imaging using Axio Imager fluorescent microscope (Carl Zeiss). Images were captured with 20× objective, processed using Zeiss Zen Black software and proportion of SARS-CoV-2-NP+ cells were counted using Fiji (Yin et al., 2021; Schindelin et al., 2012). To this end, a cut off threshold for both DAPI and AF-488 channel was computed in Fiji software based on uninfected cell images, and this threshold was applied to Carl Zeiss Image metadata files for DMSO (drug-vehicle) or SNX-5422-treated infected cells. The same threshold was maintained during quantification of each image from the same experiment day. After the threshold was adjusted, positive cell counts in both channels were documented using ‘analyze particle’ function in Fiji. Finally, percent NP+ cells were computed from the evaluated cell counts from 5-10 fields per treatment condition each experiment day.
CC50 and IC50 calculations
CC50 and IC50 were calculated using Fit spline/LOWESS analysis in GrahPad prism, where we used the “point-to-point” curve fitting approach for standard curve generation.
Bulk RNA-sequencing of human TBE cells
Human TBE cells from 3 independent donors (Table S1) were treated with 1 μM SNX-5422 or 0.1% DMSO added to the media on the basolateral side of the culture (Figure 2A), in three biological replicates. After 48 hrs, cells were resuspended in TRIzol reagent (Thermo Fisher) and total RNA from the cells was extracted by phase separation with chloroform and subsequently using the RNeasy Mini Kit (Qiagen). RNA-Seq libraries were prepared using TruSeq RNA library Prep Kit v2 (Illumina, Inc. USA). Before pooling and sequencing, fragment length distribution and library quality were assessed on a TapeStation 2200 (Agilent Technologies), and the libraries were validated by Qubit Fluorometers (Thermo Fisher). All libraries were then pooled in a concentration at 4nM and sequenced on a NextSeq 500 Illumina sequencing platform system using NextSeq 500/550 High Output Kit v2.0 (150 cycles) (Illumina, Inc. USA).
Analysis of the bulk RNA-seq data
RNA-Seq data was quality checked with FastQC (Andrews, 2010) and preprocessing was carried out using TrimGalore (Krueger, 2012) toolkit to trim low-quality bases and Illumina adapter sequences using default settings. Reads were aligned to the ENSEMBL Homo_sapiens.GRCh38.dna.primary_assembly genome using the ENSEMBL Homo_sapiens.GRCh38.100 transcript (Kersey et al., 2012) annotation file with STAR (Dobin et al., 2013) splice-aware RNA-seq alignment tool in paired mode allowing maximum multimapping of 3. Gene level counts were quantified using FeatureCounts (Liao et al., 2014) tool, counting unique features in non-stranded mode and retaining both gene ID, gene name, and gene biotype mapping from the ENSEMBL annotation file. Prior to differential expression analysis, count data was collapsed to donor level and genes for which mean raw count was at least 15 were kept. Normalization and differential expression were carried out with DESeq2 (Love et al., 2014) Bioconductor (Huber et al., 2015) package, utilizing the ‘apeglm’ Bioconductor package (Zhu et al., 2019) for log fold change shrinkage, in R statistical programming environment. The design formula was constructed to test for the effect of treatment while controlling for donor.
Principal component analysis (PCA) was performed with plotPCA in ‘DESeq2’ R package. PCA was done on variance stabilizing transformed (vst) count data and batch corrected with ‘limma’ package for genes with an average raw count of at least 15 across samples. Volcano plot with differentially expressed genes was generated using ‘EnhancedVolcano’ package in R, after filtering for described differential expression cutoffs. Heatmaps for differential expression of genes were generated using ‘pheatmap’ package in R. A vst was applied to count data, and batch corrected with ‘limma’ package, followed by Z-score normalization. Dot plots demonstrating upregulated and downregulated KEGG pathways were generated using ‘ClusterProfiler’ package in R using a “universe” of all human genes. Protein-protein interaction (PPI) network was constructed using STRING Vs. 11.5 (Szklarczyk et al., 2021) and Cytoscape (Vs 3.8.2) (Shannon et al., 2003).
Biocontainment and biosafety
Work with live SARS-CoV-2 isolate (USA-WA1/2020; BEI Resources NR-52281) was performed under Biosafety Level-3 (BSL-3) in the Duke Regional Biocontainment Laboratory.
Quantification and statistical analysis
To evaluate the dose-dependent effect of SNX-5422 treatment on SARS-CoV-2 replication outcomes and cellular cytotoxicity, the mean values of different replicates from individual experiments from cells treated with different concentrations of SNX-5422 were compared to DMSO (drug-vehicle)-treated cells by performing the Welch’s Two Sample t-test. All calculated p-values are two-sided with statistical significance evaluated at the 0.05 alpha level. P-values have not been corrected for multiple comparisons. All analyses were performed in R Version 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria). These data will serve as preliminary data for future confirmatory and mechanistic studies using physiologically relevant human airway epithelial cell and animal models of infection.
Acknowledgements
This work was supported by a grant from Open Philanthropy (R.G and S.R.P) and COVID-19 research startup funds (S.R.P). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The content is solely the responsibility of the authors. Work with live SARS-CoV-2 was performed under BSL-3 in the Duke Regional Biocontainment Laboratory (RBL), which received partial support for construction from the NIH/NIAID (UC6AI058607; G. D. Sempowski). Flow cytometry and fluorescent microscopy was performed at the DHVI Flow Cytometry Facility (Durham, NC) and Duke Light Microscopy Core facility (Durham, NC), respectively. Bulk-RNA sequencing was performed at the DHVI Viral Genetic Analysis core (Durham, NC). The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281, and was provided by Dr. Gregory Sempowski. We thank Dr. Stacy Horner, Duke Molecular Genetics and Microbiology, for kindly providing Calu-3 (ATCC) cells.
Author contributions
R.G., S.M.P., T.H., M.B. and S.R.P. designed the study and interpreted the data; R.G., J.J.T., F.K., S.N.L., J.S. and M.B. performed experiments and analyzed the data; V.R. performed the RNA-seq and pathway analysis; P.H. and T.H. provided SNX-5422 and consulted on the drug properties. S.M.P., T.H. and M.B. contributed important insights for the interpretation and discussion of the results. C. T. performed the statistical analysis for the manuscript. R.G. and S.R.P. drafted the manuscript. All authors read and approved the final manuscript.
Declarations of interests
The authors declare no competing interests. A patent has been filed with Duke University for use of SNX-5422 as an anti-SARS-CoV-2 therapeutic.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103412.
Contributor Information
Maria Blasi, Email: maria.blasi@duke.edu.
Sallie R. Permar, Email: sap4017@med.cornell.edu.
Supplemental information
Data and code availability
-
•
The raw and processed RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ahmed S., Paramasivam P., Raj K., Kumar V., Murugesan R., Ramakrishnan V. Regulatory cross talk between SARS-CoV-2 receptor binding and replication machinery in the human host. Front. Physiol. 2020;11:802. doi: 10.3389/fphys.2020.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldeyra Therapeutics Press Release Aldeyra therapeutics to advance ADX-1612, an investigational new HSP90 inhibitor with potential nanomolar potency against SARS-CoV-2, to Clinical Testing for COVID-19; ADX-629 Accepted for BARDA CoronaWatch Meeting. 2020. https://ir.aldeyra.com/news-releases/news-release-details/aldeyra-therapeutics-advance-adx-1612-investigational-new-hsp90
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco H., Bessy C., Plantier L., Lefevre A., Piver E., Bernard L., Marlet J., Stefic K., Benz-de Bretagne I., Cannet P., et al. The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 2020;10:16824. doi: 10.1038/s41598-020-73966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock J.O., Ortea I. Re-analysis of SARS-CoV-2-infected host cell proteomics time-course data by impact pathway analysis and network analysis: a potential link with inflammatory response. Aging. 2020;12:11277–11286. doi: 10.18632/aging.103524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., Volpe M.C., Colliva A., Zanconati F., Berlot G., et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61:103104. doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar D.S., Manne U., Varambally S. Comparative transcriptome analyses reveal genes associated with SARS-CoV-2 infection of human lung epithelial cells. bioRxiv. 2020 doi: 10.1101/2020.06.24.169268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase G., Deng T., Fodor E., Leung B.W., Mayer D., Schwemmle M., Brownlee G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology. 2008;377:431–439. doi: 10.1016/j.virol.2008.04.040. [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Dimitropoulou C., Drakopanayiotakis F., Antonova G., Snead C., Cannon J., Venema R.C., Catravas J.D. Heat shock protein 90 inhibitors prolong survival, attenuate inflammation, and reduce lung injury in murine sepsis. Am. J. Respir. Crit. Care Med. 2007;176:667–675. doi: 10.1164/rccm.200702-291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-D., Zhang Z.-Y., Wei X.-J., Cai Y.-Q., Yao W.-Z., Wang M.-H., Huang Q.-F., Zhang X.-B. Association between cytokine profiles and lung injury in COVID-19 pneumonia. Respir. Res. 2020;21:201. doi: 10.1186/s12931-020-01465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Sin S.H., Wen K.W., Damania B., Dittmer D.P. Hsp90 inhibitors are efficacious against Kaposi Sarcoma by enhancing the degradation of the essential viral gene LANA, of the viral co-receptor EphA2 as well as other client proteins. Plos Pathog. 2012;8:e1003048. doi: 10.1371/journal.ppat.1003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessandro A., Thomas T., Dzieciatkowska M., Hill R.C., Francis R.O., Hudson K.E., Zimring J.C., Hod E.A., Spitalnik S.L., Hansen K.C. Serum proteomics in COVID-19 patients: altered coagulation and complement status as a function of IL-6 level. J. Proteome Res. 2020;19:4417–4427. doi: 10.1021/acs.jproteome.0c00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M., et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S., Venkatraman S., Hong X., Huang K., Ulysse L., Mobele B.I., Smith A., Lawless L., Locke A., Garigipati R. Process development and scale-up of an Hsp90 inhibitor. Org. Process Res. Dev. 2012;16:1787–1793. [Google Scholar]
- Emanuel W., Kirstin M., Vedran F., Asija D., Theresa G.L., Roberto A., Filippos K., David K., Salah A., Christopher B., et al. Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention. bioRxiv. 2020 doi: 10.1101/2020.05.05.079194. [DOI] [Google Scholar]
- Fadden P., Huang K.H., Veal J.M., Steed P.M., Barabasz A.F., Foley B., Hu M., Partridge J.M., Rice J., Scott A., et al. Application of chemoproteomics to drug discovery: identification of a clinical candidate targeting hsp90. Chem. Biol. 2010;17:686–694. doi: 10.1016/j.chembiol.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Fagone P., Ciurleo R., Lombardo S.D., Iacobello C., Palermo C.I., Shoenfeld Y., Bendtzen K., Bramanti P., Nicoletti F. Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun. Rev. 2020;19:102571. doi: 10.1016/j.autrev.2020.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallelli L., Zhang L., Wang T., Fu F. Severe acute lung injury related to COVID-19 infection: a review and the possible role for escin. J. Clin. Pharmacol. 2020;60:815–825. doi: 10.1002/jcph.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Harhaj E.W. HSP90 protects the human T-cell leukemia virus type 1 (HTLV-1) tax oncoprotein from proteasomal degradation to support NF-kappaB activation and HTLV-1 replication. J. Virol. 2013;87:13640–13654. doi: 10.1128/JVI.02006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller R., Taguwa S., Frydman J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim. Biophys. Acta. 2012;1823:698–706. doi: 10.1016/j.bbamcr.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellezr R., Vignuzzi M., Andino R., Frydman J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 2007;21:195–205. doi: 10.1101/gad.1505307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J.L., Caidi H., Anderson L.J., Haynes L.M. Evaluation of the Calu-3 cell line as a model of in vitro respiratory syncytial virus infection. J. Virol. Methods. 2011;174:144–149. doi: 10.1016/j.jviromet.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra S., Chaudhuri A.G., Tiwary B.K., Chakrabarti N. Matrix metallopeptidase 9 as a host protein target of chloroquine and melatonin for immunoregulation in COVID-19: a network-based meta-analysis. Life Sci. 2020;257:118096. doi: 10.1016/j.lfs.2020.118096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J., et al. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. bioRxiv. 2020 doi: 10.1101/2020.06.30.175695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.H., Veal J.M., Fadden R.P., Rice J.W., Eaves J., Strachan J.P., Barabasz A.F., Foley B.E., barta T.E., Ma W., et al. Discovery of novel 2-aminobenzamide inhibitors of heat shock protein 90 as potent, selective and orally active antitumor agents. J. Med. Chem. 2009;52:4288–4305. doi: 10.1021/jm900230j. [DOI] [PubMed] [Google Scholar]
- Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inafante J.R., Weiss G.J., Jones S., Tibes R., Bauer T.M., Bendell J.C., Hinson J.M., Jr., von Hoff D.D., Burris H.A., 3rd, Orlemans E.O., Ramanathan R.K. Phase I dose-escalation studies of SNX-5422, an orally bioavailable heat shock protein 90 inhibitor, in patients with refractory solid tumours. Eur. J. Cancer. 2014;50:2897–2904. doi: 10.1016/j.ejca.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Islam A., Khan M.A. Lung transcriptome of a COVID-19 patient and systems biology predictions suggest impaired surfactant production which may be druggable by surfactant therapy. Sci. Rep. 2020;10:19395. doi: 10.1038/s41598-020-76404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyad S., Scott H., Abdelghani T. Drug repositioning suggests a role for the heat shock protein 90 inhibitor geldanamycin in treating COVID-19 infection. Preprint. 2020 doi: 10.21203/rs.3.rs-18714/v1. (Version 1) [DOI] [Google Scholar]
- Kamel W., Noerenberg M., Cerikan B., Chen H., Järvelin A., Kammoun M., Lee J., Shuai N., Garcia-Moreno M., Andrejeva A., et al. Global analysis of protein-RNA interactions in SARS-CoV-2 infected cells reveals key regulators of infection. bioRxiv. 2020 doi: 10.1101/2020.11.25.398008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J., Staines D.M., LAWSON D., Kulesha E., Derwent P., Humphrey J.C., Hughes D.S., Keenan S., Kerhornou A., Koscielny G., et al. Ensembl genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–D97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. 2012. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- Li C., Chu H., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Hou Y., Shuai H., Cai J., et al. Human coronavirus dependency on host heat shock protein 90 reveals an antiviral target. Emerg. Microbes Infect. 2020;9:2663–2672. doi: 10.1080/22221751.2020.1850183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., Tao P.Z., Liu Y.Z., Jiang J.D. Geldanamycin, a ligand of heat shock protein 90, inhibits the replication of herpes simplex virus type 1 in vitro. Antimicrob. Agents Chemother. 2004;48:867–872. doi: 10.1128/AAC.48.3.867-872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lieberman N.A.P., Peddu V., Xie H., Shrestha L., Huang M.-L., Mears M.C., Cajimat M.N., Bente D.A., Shi P.-Y., Bovier F., et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18:e3000849. doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Jia P., Fang B., Zhao Z. Differential expression of viral transcripts from single-cell RNA sequencing of moderate and severe COVID-19 patients and its implications for case severity. Front. Microbiol. 2020;11:603509. doi: 10.3389/fmicb.2020.603509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020;55:2000607. doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccord J.M., Hybertson B.M., Cota-Gomez A., Gao B. Nrf2 Activator PB125(R) as a potential therapeutic agent against COVID-19. bioRxiv. 2020 doi: 10.1101/2020.05.16.099788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehraeen E., Karimi A., Barzegary A., Vahedi F., Afsahi A.M., Dadras O., Moradmand-Badie B., Seyed Alinaghi S.A., Jahanfar S. Predictors of mortality in patients with COVID-19-a systematic review. Eur. J. Integr. Med. 2020;40:101226. doi: 10.1016/j.eujim.2020.101226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., Mcauley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., HLH Across Speciality Collaboration U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick E., Kamm J., Pisco A.O., Ratnasiri K., Babik J.M., Calfee C.S., Castaneda G., Derisi J.L., Detweiler A.M., Hao S., et al. Upper Airway gene expression differentiates COVID-19 from other acute respiratory illnesses and reveals suppression of innate immune responses by SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.05.18.20105171. [DOI] [Google Scholar]
- Nienhold R., Ciani Y., Koelzer V.H., Tzankov A., Haslbauer J.D., Menter T., Schwab N., Henkel M., Frank A., Zsikla V., et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat. Commun. 2020;11:5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Kobayashi T., Miyama T., Suzuki A., Jung S.M., Hayashi K., Kinoshita R., Yang Y., Yuan B., Akhmetzhanov A.R., Linton N.M. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitulescu G.M., Paunescu H., Moschos S.A., Petrakis D., Nitulescu G., Ion G.N.D., Spandidos D.A., Nikolouzakis T.K., Drakoulis N., Tsatsakis A. Comprehensive analysis of drugs to treat SARSCoV2 infection: mechanistic insights into current COVID19 therapies (Review) Int. J. Mol. Med. 2020;46:467–488. doi: 10.3892/ijmm.2020.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari G., Sanfilippo C., Castrogiovanni P., Imbesi R., Li Volti G., Barbagallo I., Musumeci G., di Rosa M. Network perturbation analysis in human bronchial epithelial cells following SARS-CoV2 infection. Exp. Cell Res. 2020;395:112204. doi: 10.1016/j.yexcr.2020.112204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-DOBBE J.C., Limpens R., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Barcena M., et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa Y., Hideshima T., Steed P., Vallet S., Hall S., Huang K., Rice J., Barabasz A., Foley B., Ikeda H., et al. SNX-2112, a selective Hsp90 inhibitor, potently inhibits tumor cell growth, angiogenesis, and osteoclastogenesis in multiple myeloma and other hematologic tumors by abrogating signaling via Akt and ERK. Blood. 2009;113:846–855. doi: 10.1182/blood-2008-04-151928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi L., Fossali T., Frangipane V., Bozzini S., Morosini M., D'Amato M., Lettieri S., Urtis M., Di Toro A., Saracino L., et al. Broncho-alveolar inflammation in COVID-19 patients: a correlation with clinical outcome. BMC Pulm. Med. 2020;20:301. doi: 10.1186/s12890-020-01343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott H.C., Rice T.W. Corticosteroids in COVID-19 ARDS: evidence and hope during the pandemic. JAMA. 2020;324:1292–1295. doi: 10.1001/jama.2020.16747. [DOI] [PubMed] [Google Scholar]
- Rajan A., Kelly R.J., Trepel J.B., Kim Y.S., Alarcon S.V., Kummar S., Gutierrez M., Crandon S., Zein W.M., Jain L., et al. A phase I study of PF-04929113 (SNX-5422), an orally bioavailable heat shock protein 90 inhibitor, in patients with refractory solid tumor malignancies and lymphomas. Clin. Cancer Res. 2011;17:6831–6839. doi: 10.1158/1078-0432.CCR-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore A.P., Haystead T., Das P.K., Merits A., Ng M.L., Vasudevan S.G. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antivir. Res. 2014;103:7–16. doi: 10.1016/j.antiviral.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatzer A., Ali M.A., Chavez M., Dowdell K., Lee M.J., Tomita Y., El-Hariry I., Trepel J.B., Proia D.A., Cohen J.I. Ganetespib, an HSP90 inhibitor, kills Epstein-Barr virus (EBV)-infected B and T cells and reduces the percentage of EBV-infected cells in the blood. Leuk. Lymphoma. 2017;58:923–931. doi: 10.1080/10428194.2016.1213823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.P., Haystead T.A.J. Hsp90: a key target in HIV infection. Future Virol. 2017;12:55–59. [Google Scholar]
- Song P., Li W., Xie J., Hou Y., YOU C. Cytokine storm induced by SARS-CoV-2. Clin. Chim. Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Jarosz D.F., lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Torres Acosta M.A., Singer B.D. Pathogenesis of COVID-19-induced ARDS: implications for an ageing population. Eur. Respir. J. 2020;56:2002049. doi: 10.1183/13993003.02049-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrad B., Vastrad C., Tengli A. Identification of potential mRNA panels for severe acute respiratory syndrome coronavirus 2 (COVID-19) diagnosis and treatment using microarray dataset and bioinformatics methods. 3 Biotech. 2020;10:422. doi: 10.1007/s13205-020-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A.J., Kayal N., Anand P., Badley A.D., Church G.M., Soundararajan V. Benchmarking evolutionary tinkering underlying human–viral molecular mimicry shows multiple host pulmonary–arterial peptides mimicked by SARS-CoV-2. Cell Death Discov. 2020;6:96. doi: 10.1038/s41420-020-00321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus Disease (COVID-19) Dashboard 2020. https://covid19.who.int/?gclid=Cj0KCQiAmL-ABhDFARIsAKywVafJxVkqo9vmNjGaiJEw5SEQU7xbiPnPU7_WwnfIc3xMAkp7QwF1CHMaAijZEALw_wcBhttps://covid19.who.int/?gclid=Cj0KCQiAmL-ABhDFARIsAKywVafJxVkqo9vmNjGaiJEw5SEQU7xbiPnPU7_WwnfIc3xMAkp7QwF1CHMaAijZEALw_wcB
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Qi F., Li H., Yang Q., Wang H., Wang X., Liu X., Zhao J., Liao X., Liu Y., et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., de Jesus P.D., et al. MDA5 Governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wu J., Shan Y., Yao Z., Dong B., Chen B., Zhao Z., Wang S., Chen J., Cong Y. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346:74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Houry W.A. Hsp90: a chaperone for protein folding and gene regulation. Biochem. Cell Biol. 2005;83:703–710. doi: 10.1139/o05-158. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890 e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Wang X., Fu Y., Zhang X., LIU C. Neutralizing antibodies for the treatment of COVID-19. Acta Pharm. Sin B. 2020;11:304–307. doi: 10.1016/j.apsb.2020.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A., Ibrahim J.G., Love M.I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35:2084–2092. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw and processed RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.