Abstract
The RanGTP-binding protein RanBP1, which is located in the cytoplasm, has been implicated in release of nuclear export complexes from the cytoplasmic side of the nuclear pore complex. Here we show that Yrb1 (the yeast homolog of RanBP1) shuttles between the nucleus and the cytoplasm. Nuclear import of Yrb1 is a facilitated process that requires a short basic sequence within the Ran-binding domain (RBD). By contrast, nuclear export of Yrb1 requires an intact RBD, which forms a ternary complex with the Xpo1 (Crm1) NES receptor in the presence of RanGTP. Nuclear export of Yrb1, however, is insensitive towards leptomycin B, suggesting a novel type of substrate recognition between Yrb1 and Xpo1. Taken together, these data suggest that ongoing nuclear import and export is an important feature of Yrb1 function in vivo.
Exchange of macromolecules between the cytoplasmic and nuclear compartments is a hallmark of eukaryotic cells. This nucleocytoplasmic transport occurs through the nuclear pores (for reviews, see references 24, 58, and 76) and is generally a signal-mediated and energy-dependent process (for reviews, see references 25, 31, and 81). Only ions and smaller molecules seem to be able to traverse the pores by passive diffusion.
The small Ras-like GTPase Ran plays a crucial role in nucleocytoplasmic transport in that it provides the identity of the two compartments and thus ensures directionality of transport (for reviews, see references 20, 50, and 51). In the nucleoplasm, this small Ras-like GTPase is thought to exist mainly in the GTP-bound form, due to the exclusive nuclear localization of the only known guanine nucleotide exchange factor (GEF) for Ran, RCC1, or Prp20 in Saccharomyces cerevisiae (2). By contrast, the RanGTP concentration in the cytoplasm is supposed to be low since the only known GTPase-activating protein (GAP) for Ran, Rna1 in yeast, is a cytoplasmic protein (5, 21). This would create a steep gradient of RanGTP across the nuclear envelope.
Like in other processes governed by small Ras-like GTPases, effector proteins, which bind specifically to the GTP-bound form of Ran, play a key role in nucleocytoplasmic transport. An important group of Ran effectors is represented by the importin β-type nuclear transport receptors (karyopherin β) (for reviews, see references 1, 63, 86, and 89). Members of this protein family are characterized by a conserved RanGTP-binding domain at the N terminus. The effects of RanGTP on the binding of cargo determine whether a given importin or karyopherin β is an import or export receptor. In the case of import receptors or importins, the receptor-cargo complex is dissociated (in the nucleus) upon binding of RanGTP, whereas in the case of export receptors or exportins, a ternary complex between cargo, receptor, and RanGTP is formed and exported as an entity into the cytoplasm. In S. cerevisiae, 14 members of the karyopherin β family have been proposed to exist, and for many of them, cargoes were already identified. So far, two exportins, Cse1 and Xpo1, were found to be indispensable for yeast cell viability; Cse1 is the yeast homolog of CAS that exports importin α (Srp1) from the nucleus (38, 46, 74) and Xpo1, the yeast homolog of CRM1, is the export receptor for leucine-rich nuclear export signals (NES) (54, 75). In contrast to CRM1 of higher eukaryotes, Xpo1 is not sensitive towards the Streptomyces metabolite leptomycin B (LMB), which interferes directly with substrate recognition by CRM1 (4, 28). Recently, an LMB-sensitive variant of Xpo1 was engineered by a single amino acid substitution (53).
A second family of RanGTP-binding proteins is characterized by a conserved RanGTP-binding motif, also called a Ran-binding domain (RBD) (6, 15, 32). All members of this protein family have been implicated in nucleocytoplasmic transport, but their exact role in this process remains to be clarified. A prominent member of this protein family is the giant mammalian nucleoporin NUP358 (RanBP2), which contains four of these domains (90, 93) and was localized to the cytoplasmic filaments protruding from the nuclear pore complex (NPC) into the cytoplasm (60, 87, 90). Another member of this family is RanBP3 and its Schizosaccharomyces pombe homolog, Hba1, which were localized to the nucleoplasm, suggesting that proteins of this family exist on both sides of the nuclear envelope (52, 84). Besides the RanBP1-homolog Yrb1 (see below), S. cerevisiae contains two RBD-containing proteins, Yrb2 and Nup2 (23, 32). Whereas Yrb1 is essential for cell viability, null mutants of YRB2 and NUP2 exhibit only a cold sensitivity or no apparent growth defect, respectively (48, 55, 59, 70, 83). In agreement with its structural similarity to RanBP3 and Hba1, Yrb2 was localized to the nucleoplasm (55, 82), whereas Nup2 was localized to the NPC (48). All three RBD-containing proteins of yeast have been implicated in nucleocytoplasmic transport. Nup2 was found to interact genetically and physically with Nup1 and Srp1 (7), suggesting that it might participate in nuclear import of proteins with classical nuclear localization signals (NLSs). More recently, Nup2 was shown to play a role in the reexport of Srp1 from the nucleus (13). Consistent with this role, Nup2 was found in a two-hybrid screen with the nuclear tRNA export receptor, Los1, indicating a possible function in nuclear export of tRNA (36). Similarly, Yrb2 was recently shown to be a cofactor of Xpo1-mediated protein export (26, 56, 82).
The founding member of the protein family of RBD-containing proteins is RanBP1 (Yrb1). The protein has been implicated in both nuclear protein import and export in vivo (70) and in vitro (19, 44). Mechanistically, RanBP1 and other members of this protein family increase, via their conserved RBDs, the rate of RanGAP1-mediated GTP hydrolysis on Ran (6, 10, 55) and overcome the inhibition of this reaction by members of the karyopherin β family (9, 27, 49). Consistent with the cytoplasmic steady-state localization of RanBP1 and RanGAP1 (Rna1) (39, 66, 70, 95), it was proposed that these proteins are required for the release of karyopherin β from RanGTP and thus for the recycling of import receptors and for terminal steps of nuclear export (3, 26, 44). Such an exclusive cytoplasmic function of RanBP1, however, was recently questioned by the identification of nuclear pools of RanBP1 both in higher eukaryotes and in yeast (62, 66, 69, 95). RanBP1 from higher organisms differs from the S. cerevisiae and S. pombe counterparts in the sequence and the location of an extension outside of the conserved RBD. The C-terminal extension of metazoan RanBP1 harbors a sequence resembling that of a leucine-rich NES. This sequence was shown to be necessary but not sufficient for the cytoplasmic localization of a functional RBD (66, 95) and to function as an export signal in a heterologous context (65, 66). In agreement with these results, RanBP1 was shown to accumulate in the nucleus upon injection of peptides containing NES sequences from protein kinase A inhibitor protein or human immunodeficiency virus (HIV) Rev protein into Xenopus oocyte nuclei, suggesting that RanBP1 shuttles between the nucleus and cytoplasm and is exported via the pathway for leucine-rich NES (62). Yeast RanBP1 sequences exhibit an N-terminal extension outside of the RBD which shares no sequence similarity to the C-terminal extension of metazoan RanBP1 and also lacks the leucine-rich NES. Nevertheless, recent reports of nuclear pools of RanBP1 in yeast (36, 57, 69) indicated that the dynamic localization of RanBP1 might be conserved throughout the eukaryotic kingdom.
Here, we present evidence that Yrb1 shuttles between the nucleus and cytoplasm. The conserved RBD is necessary and sufficient for the essential function and nucleocytoplasmic shuttling of Yrb1. Yrb1 follows a facilitated transport route into the nucleus which requires a small basic sequence within the RBD. Despite the absence of an apparent leucine-rich NES, nuclear export of Yrb1 depends on Crm1 (Xpo1). Accordingly, a ternary complex between Yrb1, Xpo1, and RanGTP could be isolated, which forms both in vitro and in vivo. These data suggest that Yrb1 shuttles continuously between the cytoplasm and nucleus.
MATERIALS AND METHODS
Strains and growth conditions.
Yeast strains used in this study are listed in Table 1. Most of the strains were derived from laboratory strain W303 by standard yeast genetic techniques (42). The CSE1 and XPO1 shuffle strains were derived from strains M1702 (91) and LDY880 (92), respectively. DNA-mediated transformation of yeast cells was performed using a modified version of the lithium acetate method (29). Unless indicated otherwise, yeast cells were propagated at 30°C. YP (rich medium) and SC (synthetic complete medium) were prepared as previously described (42). Dextrose (D) or raffinose (R) was added as a carbon source at a final concentration of 2% (wt/vol) after autoclaving; induction with galactose (G) was performed by adding galactose (final concentration, 2%) to raffinose-grown cells. LMB treatment of CRM1T539C cultures was done as previously described (53). Drop-out media (SC lacking the appropriate nutrients) were used to maintain selection for plasmids. Agar plates containing 5-fluoroorotic acid were prepared as described by Boeke et al. (12). Escherichia coli DH5α (34) was used for propagation of all plasmid DNAs. Bacteria were cultivated using standard methods (68).
TABLE 1.
Yeast strains
| Strain | Characteristics | Source |
|---|---|---|
| W303-1A (CRY1) | MATa ura3-1 trp1-1 his3-11,15 leu2-3,112 ade2-1 can1-100 GAL+ | R. S. Fuller |
| W303-1B (CRY2) | MATα ura3-1 trp1-1 his3-11,15 leu2-3,112 ade2-1 can1-100 GAL+ | R. S. Fuller |
| W303-D(CRY1x2) | MATa/MATα (W303-1A × 1B) | R. S. Fuller |
| W303-1B xpo1Δ::LEU2 pRS313-XPO1 | 75 | |
| W303-1B xpo1Δ::LEU2 pRS313-xpo1-1 | 75 | |
| CRM1T539C | W303-1A xpo1Δ::KANR (pRS315-CRM1T539C) | 53 |
| HMK21 (YRB1-shuffle) | W303-1A yrb1Δ::HIS3 (pMK103) | Künzler et al., submitted |
| W303-1A yrb1Δ::HIS3 (pMK284-n) | This study | |
| XPO1-shuffle | W303-1A xpo1Δ::KANR (pRS316-XPO1) | This study |
| CSE1-shuffle | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 lys2-801 trp1-Δ101 cse1::LEU2 (pMK330) | This study |
| XPO1-shut off | W303-1A xpo1Δ::KANR (pGALPATG-XPO1) | This study |
| W303-1A xpo1Δ::LEU2 (pNOPPATA-TRP1-XPO1) | This study | |
| W303-1A xpo1Δ::LEU2 rna1-1 (pNOPPATA-TRP1-XPO1) | This study | |
| W303-1A cse1::LEU2 (pNOPPATA-TRP1-CSE1) | This study | |
| W303-1A cse1::LEU2 rna1-1 (pNOPPATA-TRP1-CSE1) | This study | |
| W303-1A xpo1Δ::LEU2 (pASZ11-xpo1-1) yrb1::HIS3 (pMK103) | This study | |
| W303-1A xpo1Δ::LEU2 (pASZ11-xpo1-1) yrb1::HIS3 (pMK284-n) | This study | |
| W303-1A xpo1Δ::KANR (pASZ11-xpo1-1) yrb1::HIS3 (pMK103) | This study | |
| W303-1A xpo1Δ::KANR (pASZ11-xpo1-1) yrb1::HIS3 (pMK346STOP-wt) | This study | |
| W303-1A xpo1Δ::KANR (pASZ11-xpo1-1) yrb1::HIS3 (pMK346STOP-ΔC) | This study |
Construction of plasmids.
Standard techniques were used for the manipulation of recombinant DNA (68). Plasmid DNA from E. coli was isolated as described previously (22). Unless specified otherwise, PCR amplifications were performed using standard conditions (67) and Vent DNA polymerase (New England Biolabs, Beverly, Mass.). The correct sequence of PCR-generated constructs was verified by nucleotide sequence analysis (80). Plasmids used in this study are listed in Table 2.
TABLE 2.
Plasmids used
| Plasmid | Characteristic(s) | Source |
|---|---|---|
| pEG202+PL | 2μm HIS3 ADH1p-LexA(DBD)-MCS-ADH1t | 33 |
| pJG4-5 | 2μm TRP1 GAL1p-B42(TAD)-MCS-ADH1t | 33 |
| pRS314 | CEN ARS TRP1 | 73 |
| pRS424 | 2μm TRP1 | 73 |
| pRS316 | CEN ARS URA3 | 73 |
| YCplac22 | CEN ARS TRP1 | 30 |
| YEplac112 | 2 μm TRP1 | 30 |
| pEMBLyex4 | 2 μm leu2d URA3 GAL1,10(UAS)-CYC1 | 17 |
| YEp352 | 2 μm URA3 | 37 |
| YEp351GAL | 2 μm LEU2 GAL1,10p | 8 |
| pASZ11 | CEN ARS ADE2 | 77 |
| pGEX4T3 | AmprT7-GST-MCS | Pharmacia |
| pET9d-His6-TEV | KanrT7-His6-TEV-MCS | G. Stier |
| pET8c | AmprT7-His6-MCS | S. Labeit |
| pRSETA | AmprT7-His6-MCS | Invitrogen |
| pTrcHisA | AmprTrc-His6-MCS | Invitrogen |
| pGALPATG | CEN ARS LEU2 GAL1p-ProtA-TEV-GAL4t | This study |
| pNOPPATA-TRP1-GSP2 | CEN ARS TRP1 NOP1p-ProtA-TEV-GSP2-ADH1t | This study |
| pEG-KG | 2μm leu2d URA3 GAL1,10(UAS)-CYC1-HA | I. Macara |
| pASZ11-xpo1-1 | H. Santos-Rosa | |
| pRS316-XPO1 | This study | |
| pMK330 | pRS316-CSE1 | 46 |
| pMK103 | YEp352-YRB1 | Künzler et al., submitted |
| pEMBLyex4-NUP2 | This study | |
| pEG-p36 | pEG-KG-YRB2 | I. Macara |
| pGEX-GSP1 | pGEX4T3-GSP1 | B. Senger |
| pLDB450-wt | pTrcHisA-GSP1 | This work |
| pMK104 | pRSETA-YRB1 | Künzler et al., submitted |
| pMK263 | pRSETA-RNA1 | This study |
| pGEX-YRB1 | pGEX4T3-YRB1(1-201) | This study |
| pGEX-YRB1-51 | pGEX4T3-yrb1-51(1-201) | This study |
| pGEX-YRB1-52 | pGEX4T3-yrb1-52(1-201) | This study |
| pGEX-YRB1RBD | pGEX4T3-YRB1(59-201) | This study |
| pGEX-YRB1N | pGEX4T3-YRB1(3-58) | This study |
| pGEX-YRB1RBDN | pGEX4T3-YRB1(59-131) | This study |
| pGEX-YRB1RBDC | pGEX4T3-YRB1(132-201) | This study |
| pGEX4T3-yrb1Δ(102-120) | This study | |
| pGEX4T3-yrb1-Δ(121-130) | This study | |
| pET9d-His6-TEV-XPO1 | This study | |
| pET9d-His6-TEV-CSE1 | This study | |
| pET8c-MTR10 | B. Senger | |
| pNOPPATA-TRP1-XPO1 | This study | |
| pNOPPATA-TRP1-CSE1 | This study | |
| pGALPATG-XPO1 | This study | |
| pMK275 | pRS314-YRB1 | Künzler et al., submitted |
| pMK277 | pRS314-yrb1-51 | Künzler et al., submitted |
| pMK278 | pRS314-yrb1-52 | Künzler et al., submitted |
| pRS314-yrb1-Δ(167-201) | This study | |
| pRS314-yrb1-Δ(132-201) | This study | |
| pRS314-yrb1-Δ(3-58) | This study | |
| pRS314-yrb1-Δ(59-131) | This study | |
| pRS314-yrb1-Δ(59-101) | This study | |
| pRS314-yrb1-Δ(102-131) | This study | |
| pRS314-yrb1-Δ(102-120) | This study | |
| pRS314-yrb1-Δ(121-130) | This study | |
| pRS314-yrb1-Δ(102-111) | This study | |
| pMK385-wt | pRS314-NLSSV40-YRB1 | This study |
| pMK385-51 | pRS314-NLSSV40-yrb1-51 | This study |
| pMK385-52 | pRS314-NLSSV40-yrb1-52 | This study |
| pRS314-NLSSV40-yrb1-Δ(121-131) | This study | |
| pMK284n | pRS314-YRB1-GFP(S65T) | 36 |
| pMK286n | pRS314-SRP1-GFP(S65T) | 46 |
| pMK346STOP-wt | YEp351GAL-muRanBP1-GFP(S65T) | This study |
| pMK346STOP-ΔC | YEp351GAL-muRanBP1ΔC-GFP(S65T) | This study |
| pMK291-wt | pRS424-YRB1-GFP(S65T) | Künzler et al., submitted |
| pMK291-51 | pRS424-yrb1-51-GFP(S65T) | Künzler et al., submitted |
| pMK291-52 | pRS424-yrb1-52-GFP(S65T) | Künzler et al., submitted |
| pRS424-yrb1-Δ(167-201)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(132-201)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(3-58)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(59-131)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(59-101)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(102-131)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(102-120)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(121-131)-GFP(S65T) | This study | |
| pRS424-yrb1-Δ(102-111)-GFP(S65T) | This study | |
| pRS424-YRB1-3xGFP(S65T) | This study | |
| pMK379-wt-n | pRS424-NLSSV40-YRB1-GFP(S65T) | This study |
| pMK379-51-n | pRS424-NLSSV40-yrb1-51-GFP(S65T) | This study |
| pMK379-52-n | pRS424-NLSSV40-yrb1-52-GFP(S65T) | This study |
| pRS424-NLSSV40-yrb1-Δ(121-131)-GFP(S65T) | This study | |
| pMK375 | YEplac112-YAP1-sGFP | This study |
| pEG202+PL-XPO1 | 54 | |
| pEG202+PL-MTR10 | 54 | |
| pEG202+PL-CSE1 | This study | |
| pJG4-5-Rev | 54 | |
| pJG4-5-GSP1 | This study | |
| pJG4-5-YRB1 | This study | |
| pJG4-5-SRP1 | This study |
Vector pGALPATG carrying a GAL1p-ProtA-TEV-GAL4t cassette was constructed based on pNOPPATA (pUN100-NOP1p-ProtA-TEV-ADH1t) (36) by replacing the ADH1 terminator by the GAL4 terminator and the NOP1 promoter by the GAL1 promoter. Plasmid pNOPPATA-TRP1-GSP2 was derived by recloning the NOP1p-ProtA-TEV-GSP2-ADH1t cassette from pNOPPATA-GSP2 (47) as a SacI-HindIII fragment into YCplac22 and subsequently destroying the BamHI restriction site (i.e., by cutting, filling in, and religating) in the polylinker of YCplac22. This plasmid served as the basis for plasmids pNOPPATA-TRP1-XPO1 and -CSE1. Plasmids pNOPPATA-TRP1-XPO1, pGALPATG-XPO1, and pET9d-His6-TEV-XPO1 all contain a PCR-generated NcoI-BamHI fragment comprising the entire coding region of XPO1 derived from pRS313-XPO1 (75). Plasmids pNOPPATA-TRP1-CSE1 and pET9d-His6-TEV-CSE1 were constructed analogously using a PCR-generated NcoI-BamHI fragment comprising the entire coding region of CSE1 derived from pMK330 (46). Plasmid pASZ11-xpo1-1 was made by recloning a SalI-BamHI fragment from pRS313-xpo1-1 (75; H. Santos-Rosa, unpublished data). Analogously, the XPO1 gene was recloned into pRS316 as a SalI-BamHI fragment from pRS313-XPO1 (75). A plasmid for galactose-induced overexpression of NUP2 was constructed by inserting a PCR-generated BamHI-HindIII NUP2 fragment comprising the entire coding region into pEMBLyex4 (17). Plasmid pMK375 was derived from plasmid pLDB419 (92) by recloning the SalI-SacI YAP1-sGFP fragment into YEplac112.
The various plasmids containing internally deleted YRB1 genes were constructed by synthesizing by PCR, in a first step, various 5′ and 3′ portions of the gene as EcoRI-BamHI and BamHI-NotI fragments, respectively. The PCRs were performed using pMK275 as a template, T7 and T3 universal primers, and sequence-specific primers within the YRB1 open reading frame (ORF) which carried a BamHI site at their 5′ ends. In a second step, the PCR-generated 5′ and 3′ fragments were combined by cloning them sequentially into pRS314. The resulting deletion constructs were tagged with GFP(S65T) by exchanging the full-length ORF on pMK284n by the various internally deleted ORFs and recloning of the entire fusion genes on the multicopy vector pRS424 as EcoRI-NotI cassettes. The NLS of simian virus 40 (SV40) large T antigen was introduced into YRB1 by inserting a synthetic DNA fragment (coding strand, 5′-TCGAGCCACCAAAGAAGAAGCGTAAGGTTGAAC-3′; noncoding strand, 5′-TCGAGTTCAACCTTACGCTTCTTCTTTGGTGGC-3′; introduced cloning sites are underlined) into the SalI site at the 5′ end of the YRB1 ORF. Plasmids containing fusions of YRB1 to double and triple green fluorescent protein (GFP) were constructed by exchanging the BamHI single GFP cassette in pMK291-wt against tandem arrays of a PCR-generated BamHI-BglII GFP cassette (O. Gadal, unpublished data).
Plasmids pMK346STOP-wt and pMK346STOP-ΔC containing a GAL1-driven cDNA encoding full-length (amino acids 1 to 203) or C-terminally truncated (amino acids 1 to 160) GFP fusions of mouse RanBP1 (muRanBP1) were constructed by synthesizing respective fragments of the muRanBP1 ORF by PCR. The PCRs were performed using plasmid p12.6 lacking the first 12 amino acids (14) as template and 5′-CGGGATCCATGGCTGCAGCCAAGGACAG TCACGCTGACCATGATACTTCCACAGAGAATGCAGATGAGTCC-3′ combined with 5′-CCCCGGATCCGCGGCCGCCTTGTTTCTCCTCAGACTTCTCTTC-3′ (full length) or 5′-CCCCGGATCCGCGGCCGCCTTTCCTGCATTCTTCAAACTTT-3′ (ΔC), respectively, as primers. The PCR-generated fragments were digested with BamHI and inserted into the BamHI site of YEp351GAL (8). Finally, a PCR-generated GFP(S65T)-STOP NotI cassette was inserted into the NotI site at the C terminus of the above fragments.
Plasmids for expressing wild-type and various mutant forms of Yrb1 as glutathione S-transferase (GST) fusion proteins in E. coli cells were constructed by synthesizing respective fragments as EcoRI-NotI, BamHI-NotI, and BamHI by PCR and ligating them into pGEX4T3 (Pharmacia, Uppsala, Sweden). pGEX-GSP1 for expression of GST-Gsp1 in E. coli was made by recloning a BamHI-HindIII (HindIII 5′ overhang filled in using T4 DNA polymerase) fragment from plasmid pGPCNR1 (41) comprising the entire coding region of GSP1 into the BamHI-SmaI sites of vector pGEX4T3 (B. Senger, unpublished data). pMK450-wt was constructed from plasmid pLDB450 (92) by PCR amplification of the wild-type GSP1 ORF as an NheI-HindIII fragment and cloning it into pTrcHisA (Invitrogen, Carlsbad, Calif.). Plasmid pMK263, expressing a His6-tagged version of Rna1 in E. coli, contains the entire RNA1 ORF as a PCR-amplified BglII-PstI fragment.
Fusions of full-length HIV Rev protein to the B42 transcriptional activation domain (TAD) and full-length Mtr10 and Xpo1 to the E. coli LexA DNA-binding domain (DBD) were described previously (54, 79). A fusion of full-length Cse1 to the LexA DBD was generated by cloning a PCR-generated BamHI-SalI fragment comprising the entire CSE1 coding region into pEG202+PL (33). Fusions of the entire coding region of Yrb1 to the B42 TAD were obtained by insertion of respective PCR-generated EcoRI-XhoI fragments into vector pJG4-5 (33). Plasmids encoding fusions of full-length Gsp1 and Srp1 to the B42 TAD were constructed by inserting PCR-generated XhoI fragments comprising the entire coding regions of GSP1 and SRP1 into pJG4-5.
Preparation of rabbit polyclonal anti-Gsp1 antiserum.
A rabbit polyclonal antiserum against Gsp1 was raised using an affinity-purified maltose-binding protein fusion of the Gsp1(G21V) variant. A plasmid expressing the MBP-Gsp1(G21V) fusion protein was constructed in two steps. First, the BamHI-HindIII (HindIII 5′ overhang filled in using T4 DNA polymerase) fragment from YEp352GAL-GSP1(G21V) (36) comprising the entire ORF was introduced into the BamHI-SmaI sites of expression vector pGEX4T3 (Pharmacia). In a second step, the GSP1(G21V) ORF was released from this plasmid as a BamHI-SalI fragment and cloned into pMAL-c2 (New England Biolabs) opened with BamHI and SalI (B. Senger, unpublished data). The protein was expressed in BL21(DE3) cells (78) and affinity purified over amylose resin according to the manufacturer's recommendations (New England Biolabs). Immunization of two rabbits with this material was performed by a commercial antibody service (J. Pineda, Berlin, Germany). For detection of Gsp1 by immunoblotting, the resulting antisera were used as primary antibodies at a dilution of 1:3,000.
Two-hybrid assay.
Interactions between LexA(DBD) and B42(TAD) fusion proteins were assessed as described previously (54). β-Galactosidase activities were determined as described previously (46). Diploids were pregrown overnight in selective medium with 2% raffinose as the sole carbon source to mid-logarithmic phase (A600 = 0.5) and induced for 3 h by addition of 2% galactose before assaying.
Purification of ProtA fusion proteins from yeast.
Strains expressing ProtA-Xpo1 and ProtA-Cse1 in a null background for XPO1 and CSE1, respectively, were grown in yeast extract-peptone-dextrose (YPD) at 23°C to an A600 of ∼2.0. Purification of ProtA fusion proteins from these cells was performed as previously described (46) with the modification that cell lysis and washing of the immunoglobulin G (IgG)-Sepharose beads were done in Universal buffer without glycerol.
In vitro protein interaction assay (pull-down assay).
In vitro interaction between recombinant proteins was assayed as described previously (46). All proteins were expressed in BL21(DE3) (78) and purified as described previously (46). As a modification, His6-tagged proteins were purified over Talon beads (Clontech, Palo Alto, Calif.). The proteins were bound to this resin in the presence of 5 mM imidazole and eluted with 100 mM imidazole. Epitope-tagged (with GST or His6) Gsp1 from E. coli was purified accordingly except that, before ultracentrifugation, the lysate was incubated on ice for 30 min after addition of 1% Triton X-100 and 1% Tween 20. Affinity-purified Gsp1 was immediately loaded with GTP by adding 30 mM KPi (pH 7.5), 1 mM GTP, and 10 mM EDTA (pH 8), incubating at room temperature for 1 h, adding 20 mM Mg(OAc)2, incubating on ice for 30 min, and snap-freezing in liquid nitrogen. Purification and GTP loading of recombinant human Ran was described previously (11). Rna1-mediated GTP hydrolysis on Gsp1 was performed by incubating free Gsp1GTP or a GST-Yrb1–Xpo1–Gsp1GTP complex on glutathione-Sepharose beads in Universal buffer for 30 min at 30°C.
Quantification of RCC-mediated GTP exchange on Gsp1GTP.
Labeling of Gsp1 with [γ-32P]GTP and GTP exchange assays were performed as described for Ran (10). Briefly, 3 μM Gsp1GDP in 500 mM phosphate (pH 7.5), 1 mM 2-mercaptoethanol, and 10% glycerol was incubated for 30 min on ice with 20 mM EDTA and 6.7 μM [γ-32P]GTP (15 Ci/mmol; Du Pont, Wilmington, Del.). The buffer was changed in 20 mM HEPES–NaOH (pH 7.2), 50 mM NaOAc, 1 mM MgCl2, 0.5% hydrolyzed gelatin, and 0.4% NaN3 (incubation buffer) on a NAP 5 column (Pharmacia). A volume of 50 pM Gsp1-[γ-32P]GTP was incubated with indicated final concentrations of GST-Yrb1 in incubation buffer. After 30 min at 15°C, nucleotide exchange was induced for 30 s by addition of 20 nM human RCC1 and 200 μM GDP. Gsp1-bound radioactivity was determined after filtration of the samples through nitrocellulose. The dose dependence of GTP-exchange inhibition can be used to estimate the constant for dissociation of the Yrb1-Gsp1GTP complex.
Fluorescence microscopy.
Fluorescence microscopy of living yeast cells expressing GFP(S65T) fusion proteins was done as described previously (36). Cells were concentrated by a short spin and resuspended in the residual growth medium without any washing steps.
Miscellaneous.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were conducted as described previously (46).
RESULTS
Yrb1 is exported from the nucleus via a Crm1 (Xpo1)-dependent pathway.
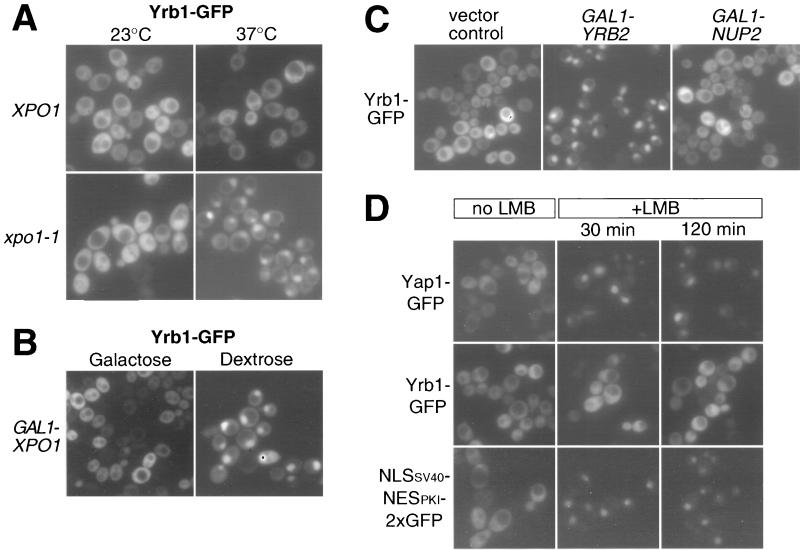
We reported previously that Yrb1-GFP accumulates in the nucleus when the N-terminally truncated nuclear tRNA export receptor Los1 is overexpressed (36). Since this observation indicated that Yrb1 may shuttle between the nucleus and cytoplasm, we studied the nuclear export of Yrb1 in various nucleocytoplasmic transport mutants. Strikingly, a dramatic and rapid nuclear accumulation of Yrb1-GFP was observed in the xpo1-1 mutant when shifted for only a few minutes to the restrictive temperature (Fig. 1A). Nuclear accumulation of Yrb1-GFP was also observed in mutants harboring the less severe alleles (crm1-1, crm1-2, and crm1-3) of XPO1 (92), albeit to a lesser extent (data not shown). In contrast, Yrb1-GFP remained cytoplasmic in the cse1-1 mutant, which is defective for nuclear export of another shuttling protein, Srp1-GFP (46). Conversely, shuttling of Srp1-GFP was not affected in the xpo1-1 mutant (data not shown). Yrb1-GFP also strongly accumulated inside the nucleus when XPO1 expression was repressed by using the regulatable GAL1 promoter (Fig. 1B; see also Table 1). As specificity controls, (i) Yrb1-GFP did not accumulate in an analogous strain harboring a glucose-repressible CSE1 gene and (ii) Srp1-GFP did not accumulate upon depletion of Xpo1 (data not shown). Finally, we examined the subcellular localization of Yrb1-GFP in cells overexpressing Yrb2 or Nup2, the two other yeast RBD-containing proteins, since it had been reported that overproduction of Yrb2 leads to nuclear accumulation of Xpo1 and NES-containing reporter proteins (82). Strikingly, Yrb2 overexpression, but not Nup2 overexpression, induces a strong nuclear accumulation of Yrb1-GFP (Fig. 1C). Consistent with a recently reported role of Nup2 in the nuclear export of Srp1 (13), we observed a strong nuclear accumulation of Srp1-GFP upon overexpression of Nup2, demonstrating the efficacy of our NUP2 construct (data not shown). This all shows that Yrb1 enters the nucleus and is exported from the nucleus by a mechanism that requires a functional Xpo1 and normal levels of Yrb2. Interestingly, nuclear export of Yrb1 was not inhibited by the fungicide LMB in an LMB-sensitive yeast strain (53) (Fig. 1D). This drug has been shown to interfere with the nuclear export and recognition of canonical leucine-rich NES by CRM1 (4, 28, 88). Accordingly, two established substrates of Xpo1 containing such canonical NES, the yeast transcription factor Yap1 (92), and an artificial reporter protein containing the NES of the cyclic AMP-dependent protein kinase inhibitor (75) revealed a rapid and efficient nuclear accumulation under the same conditions (Fig. 1D). This result suggests that recognition of Yrb1 by Xpo1 might be different from the recognition of a canonical NES (see Discussion).
FIG. 1.
Inhibition of nuclear export of Yrb1-GFP. (A) Yrb1-GFP accumulates in the nucleus of xpo1-1 cells. The double disruption xpo1Δ::LEU2 yrb1Δ::HIS3 strain complemented by xpo1-1 and YRB1-GFP on single-copy plasmids was transformed with either XPO1 on plasmid (XPO1) or empty vector (xpo1-1). Transformants were grown at 23°C and shifted for 10 min to 37°C. Shown are fluorescence micrographs depicting the localization of Yrb1-GFP. (B) Depletion of Xpo1 causes nuclear accumulation of Yrb1-GFP. A strain carrying a xpo1::KANR disruption complemented by GAL1-driven ProtA-XPO1 was transformed with YRB1-GFP on a single-copy plasmid. Transformants were grown in galactose-containing medium before transfer into dextrose-containing medium and repression for 18 h. (C) Overexpression of YRB2 causes nuclear accumulation of Yrb1-GFP. A strain harboring a yrb1Δ::HIS3 disruption complemented by single-copy YRB1-GFP was transformed with a multicopy plasmid carrying a galactose-inducible YRB2 or NUP2 gene. Transformants were grown in raffinose-containing medium before a 6-h induction by addition of 2% galactose. Shown is the fluorescence signal of Yrb1-GFP. (D) LMB has no effect on Yrb1 export. Transformants of LMB-sensitive yeast strain CRM1T539C containing plasmids encoding GFP fusion proteins of Yap1 or Yrb1 or an artificial NLS-NES-GFP reporter protein were cultivated in selective medium to mid-logarithmic phase. LMB was added to the cultures, and the cells were examined by fluorescence microscopy at the indicated time points.
Yrb1 interacts with Xpo1 in vivo.
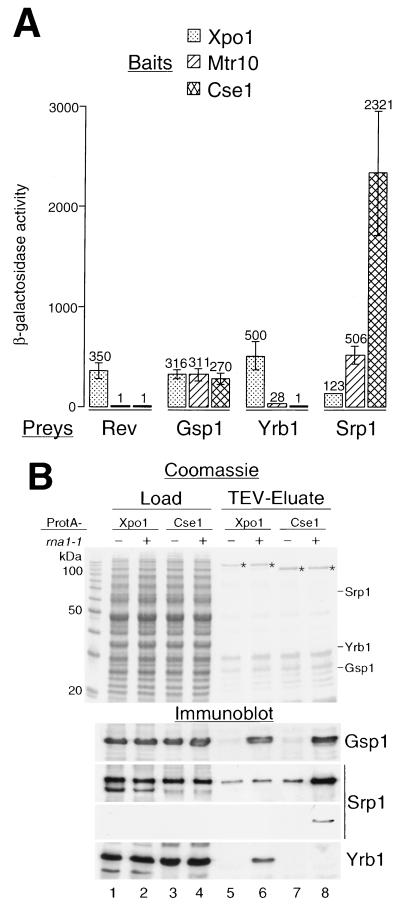
Since Xpo1 is required for the nuclear export of Yrb1, Yrb1 and Xpo1 may directly interact. To test for this, a two-hybrid analysis was performed. Indeed, an interaction was found between Yrb1 and Xpo1 (Fig. 2A). Yrb1 did not interact, however, with other members of the karyopherin β family, such as Mtr10 and Cse1. Conversely, Srp1 interacted strongly with Cse1 and weakly with Mtr10, but not with Xpo1, in this system. The strength of the two-hybrid interaction (as measured by the β-galactosidase activity) between Yrb1 and Xpo1 was as high as that of HIV Rev protein or Ran (Gsp1) with Xpo1. In addition to the two-hybrid assay, ProtA-tagged Xpo1 was affinity purified from yeast cells from both a wild-type and an rna1-1 mutant strain (Fig. 2B). We anticipated that a putative Yrb1-Xpo1-RanGTP complex would be stabilized in the rna1-1 strain, in which GTP hydrolysis is inhibited (47, 71). Indeed, Yrb1 and Gsp1 coisolate with ProtA-Xpo1, when purified from the rna1-1 strain (Fig. 2B, lane 6). In contrast, Srp1 is clearly enriched together with Gsp1 when ProtA-Cse1 is purified from the rna1-1 strain (Fig. 2B, lane 8). However, Srp1 is also seen, albeit weakly, associated with Cse1 in an RNA1 strain and with Xpo1 in both rna1-1 and RNA1 strains (Fig. 2B, lanes 5 to 7). This may be due to unspecific binding of Srp1 under our purification conditions, which was also observed with other, unrelated ProtA-fusion proteins (M. Künzler and E. Hurt, unpublished data). Overexpression of Yrb2 or Nup2 did not result in copurification of the export receptors with their respective cargoes and Gsp1, suggesting that the export complexes are not stable under these conditions (data not shown).
FIG. 2.
Yrb1 and Xpo1 interact in vivo. (A) Two-hybrid interaction. Full-length Yrb1 fused to the B42-TAD (Prey) was tested for interaction with full-length Xpo1, Mtr10, and Cse1 fused to the LexA DBD (Bait). B42-TAD fusion constructs of full-length HIV Rev, importin α (Srp1), and Gsp1 served as controls. β-Galactosidase activities are given in arbitrary units on top of the error bars. (B) Affinity purification of ProtA-Xpo1 from rna1-1 cells. xpo1::HIS3 cells complemented by a plasmid containing ProtA-XPO1 were grown in YPD at 23°C. As a control, cells deleted for CSE1 and rescued by ProtA-CSE1 were cultivated accordingly. The ProtA-fusion proteins were affinity purified on IgG-Sepharose and eluted using the TEV protease. The homogenate supernatants (Load) and TEV eluates were analyzed by SDS-PAGE, Coomassie blue staining, and immunoblotting using the indicated antibodies. In the case of the anti-Srp1 immunoblotting, both the enhanced chemiluminescence (upper panel) and the less-strong color reaction (4-chloro-1-naphthol, lower panel) are shown. The relative mobilities of Srp1, Yrb1, and Gsp1 in the Coomassie gel are also shown. Asterisks indicate the purified ProtA fusion proteins.
Taken together, the results of the two-hybrid and biochemical analyses suggest that Yrb1, Xpo1, and RanGTP can be found in complex in vivo.
Yrb1 forms a complex with Xpo1 and RanGTP in vitro.
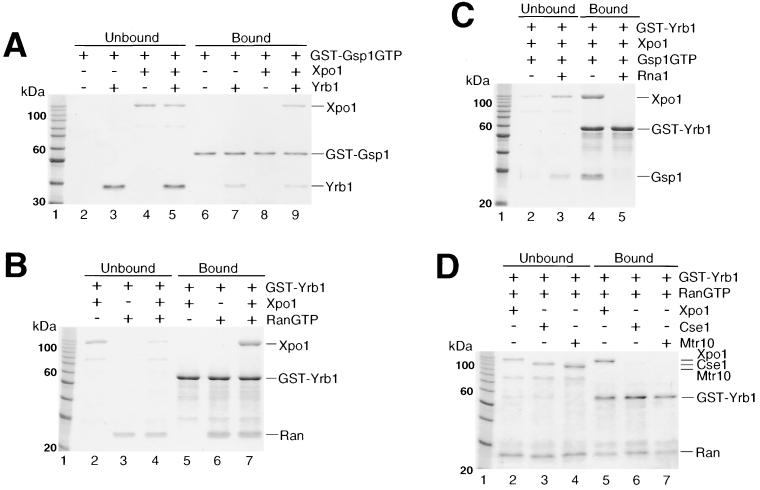
To test whether Yrb1 can form a complex with Xpo1 and RanGTP in vitro, we performed pull-down assays using recombinant proteins. First, we immobilized GST-Gsp1GTP on glutathione-Sepharose beads and analyzed binding of His6-tagged Xpo1 and Yrb1, separately or in combination, to these beads as described previously (46) (Fig. 3A). As expected for an exportin-type receptor, Xpo1 alone did not reveal a significant affinity for Gsp1GTP in this assay (Fig. 3A, lane 8), whereas Yrb1 readily associated with Gsp1GTP in the absence of Xpo1 due to its RBD (Fig. 3A, lane 7). In contrast, Xpo1 bound to the GST-Gsp1GTP beads in the presence of Yrb1 (Fig. 3A, lane 9). In a second experiment, we tested under the same conditions immobilized GST-Yrb1 for binding to His6-tagged Xpo1 and human RanGTP (Fig. 3B). As anticipated, Yrb1 formed a complex with RanGTP alone (Fig. 3B, lane 6), whereas no complex formation was observed with Xpo1 alone (Fig. 3B, lane 5). In the presence of RanGTP, however, Xpo1 can be significantly bound to Yrb1 (Fig. 3B, lane 7). The same result was obtained using His6-tagged Gsp1GTP instead of RanGTP (data not shown and Fig. 3C). These experiments suggest that a ternary complex between Yrb1, Xpo1, and RanGTP is formed in a cooperative way in vitro. This complex (like the dimeric complex between Yrb1 and RanGTP) requires the GTP-bound state of Ran, since conversion of Gsp1GTP to Gsp1-GDP by His6-tagged Rna1 (RanGAP) prior to the binding assay abolished its formation (Fig. 3C). A similar result was obtained by incubation of a preformed GST-Yrb1–Xpo1–Gsp1GTP complex with Rna1, demonstrating that the complex can be disassembled by RanGAP-mediated GTP hydrolysis on Gsp1 (data not shown). To test whether Yrb1 forms a complex with other members of the karyopherin β family, the nuclear import receptor Mtr10, which has a low affinity to RanGTP alone (72), and the nuclear export receptor Cse1 were tested in the GST-Yrb1 in vitro binding assay. No significant binding of Yrb1 to these proteins, even in the presence of RanGTP, could be detected (Fig. 3D). Taken together, these data suggest that a specific complex forms between Xpo1, Yrb1, and RanGTP, which could represent the nuclear export complex of Yrb1.
FIG. 3.
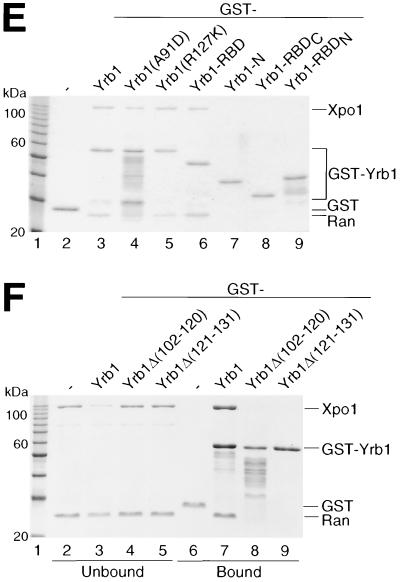
In vitro complex formation between Yrb1, Xpo1, and RanGTP (Gsp1GTP). (A) Cooperative binding of Yrb1 and Xpo1 to GST-Gsp1GTP. Recombinant GST-Gsp1GTP was bound to glutathione-Sepharose and incubated with recombinant His6-Xpo1 and His6-Yrb1 as indicated. Unbound and bound fractions were analyzed by SDS-PAGE and Coomassie blue staining. (B) Cooperative binding of Xpo1 and RanGTP to GST-Yrb1. Recombinant GST-Yrb1 was bound to glutathione-Sepharose and incubated with recombinant His6-Xpo1 and RanGTP as indicated. (C) Complex formation is dependent on the GTP-bound form of Gsp1. GST-Yrb1 was bound to glutathione-Sepharose and incubated with His6-Xpo1 together with His6-Gsp1GTP or His6-Gsp1GDP that had been produced by preincubation of His6-Gsp1GTP with recombinant RanGAP (His6-Rna1) (see Materials and Methods). (D) Yrb1 does not interact with other importins or exportins in the presence of RanGTP. Recombinant His6-tagged karyopherins, Cse1 and Mtr10, were tested, in comparison with His6-Xpo1, for interaction with GST-Yrb1 in the presence of RanGTP as described above for panel B. (E and F) RanGTP binding by the RBD of Yrb1 is necessary and sufficient for in vitro formation of the Yrb1-Xpo1-RanGTP complex. Indicated GST-Yrb1 fusion proteins were affinity purified and analyzed for binding to His6-Xpo1 in the presence of RanGTP as described above for panel B. The boundaries of the Yrb1-N, -RBD, -RBDN, and -RBDC fragments are amino acids 3 to 58, 59 to 201, 59 to 131, and 132 to 201, respectively. (E) For the pull-down assays, only the bound fractions are shown.
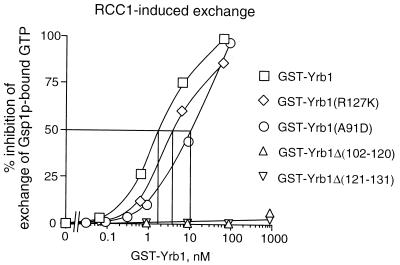
In order to identify the sequence within Yrb1 responsible for the formation of this ternary complex, various mutant forms of Yrb1 were expressed as GST fusion proteins in E. coli and examined in pull-down assays for interaction with RanGTP alone (data not shown) and with Xpo1 in the presence of RanGTP (Fig. 3E and F). This revealed that an intact RBD comprising residues 59 to 201 of Yrb1 is necessary and sufficient both for RanGTP binding (data not shown) and for the formation of the Yrb1-Xpo1-RanGTP ternary complex. Any deletion of the RBD, either truncations from the N terminus (RBDC, comprising residues 132 to 201) or the C terminus (RBDN, comprising residues 59 to 131) or small internal deletions which led to distinct changes in the localization of Yrb1 in vivo (see Fig. 5), abolished binding of RanGTP or Gsp1GTP (data not shown; Fig. 4) and formation of the ternary complex (Fig. 3E and F). The N-terminal extension outside of the RBD, however, can be deleted without significant loss of RanGTP binding and complex formation and, conversely, did not reveal any significant binding activity on its own (Yrb1-N, comprising residues 3 to 58) (Fig. 3E). In the pull-down assay, since two point mutations within the RBD, A91D and R127K (which lead to temperature-sensitive growth and differential localization of Yrb1 in vivo) (M. Künzler et al., submitted for publication) did not detectably affect complex formation, we quantified their affinity to Gsp1GTP by determining their inhibitory effect on RCC1-mediated nucleotide exchange on Gsp1GTP (Fig. 4). The small internal deletions in the Yrb1 RBD were included as controls in this assay. In agreement with the results of the pull-down assay, the two point mutations, although located in conserved residues of the RBD, reduced the affinity of Yrb1 for Gsp1GTP only moderately, from approximately 2 nM for the wild-type protein to 4 nM (R127K) and 10 nM (A91D) for the altered proteins. The determined affinity of the A91D mutant might even be underestimated because of reduced stability of the altered protein both in vivo (Künzler et al., submitted) and in vitro. An affinity of 10 nM is thereby sufficient for a stable interaction in the pull-down assay, as could be confirmed by another RanGTP-binding protein (A. Braunwarth, E. Hurt, and M. Künzler, unpublished data).
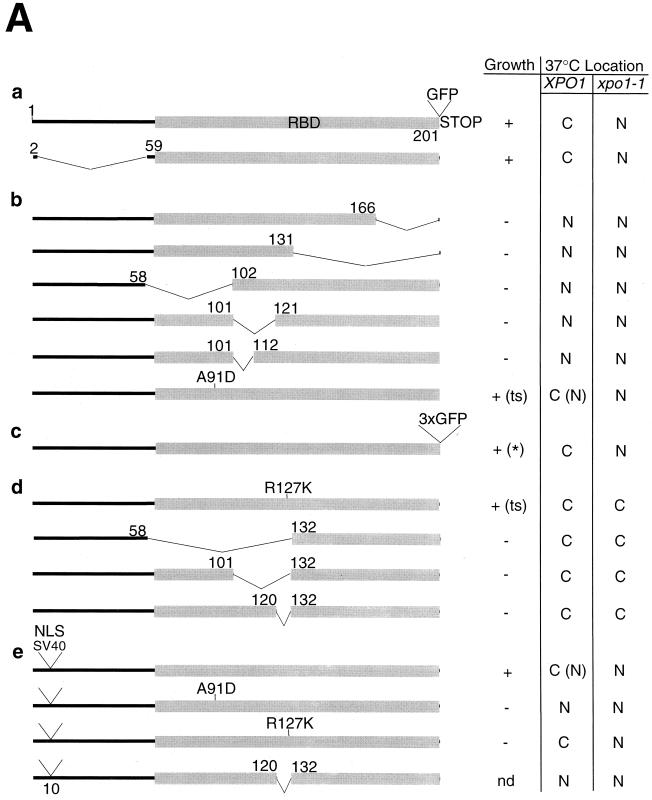
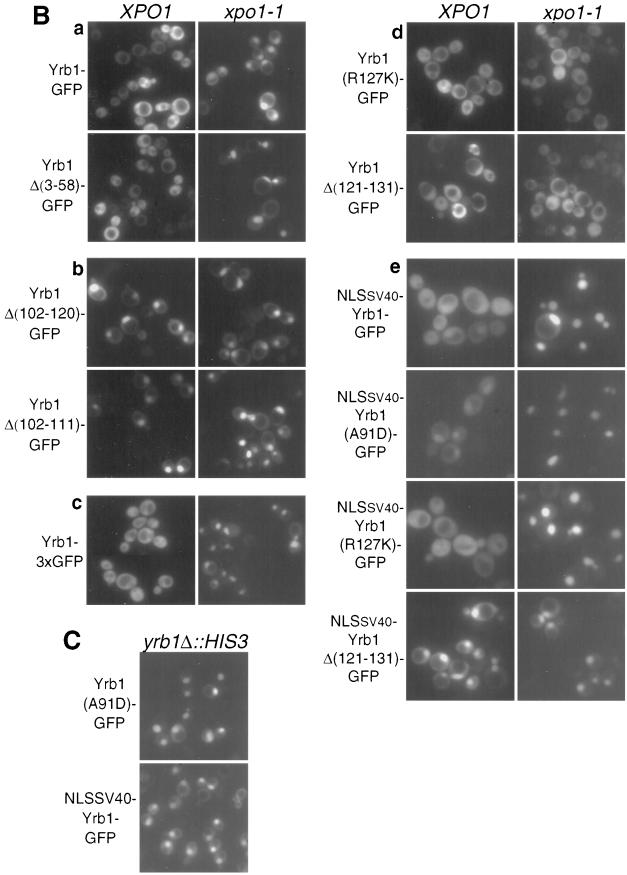
FIG. 5.
Analysis of nuclear export and import signals within Yrb1. (A) YRB1 and muRanBP1 deletion constructs with indicated complementation of the otherwise nonviable yrb1Δ::HIS3 strain (nd, not determined) and subcellular localization of derived GFP-tagged fusion proteins in XPO1 wild-type and xpo1-1 cells. For complementing constructs whose localization in a genetic YRB1 wild-type background differs from the one in a genetic yrb1 null background, the latter localization is indicated in brackets. Complementation tests were performed using untagged constructs in low-copy-number plasmids, except for the Yrb1-3xGFP construct (c), which was tested in a high-copy-number plasmid (indicated by an asterisk). The two single amino acid changes (A91D and R127K) both lead to thermosensitive (ts) growth. For the subcellular localization studies, corresponding fusion genes to single GFP(S65T) were cloned in a multicopy plasmid. (B) Subcellular localization of GFP-tagged Yrb1 mutant constructs in yeast. The constructs described for panel A were transformed in xpo1-1 and isogenic wild-type strains. Transformants were grown at 23°C and examined by fluorescence microscopy upon shift for 2 h to 37°C. Representatives of each category are shown. (C) Localization of Yrb1(A91D)-GFP and NLSSV40-Yrb1-GFP in yrb1Δ::HIS3 cells. YRB1 shuffle strain HMK21 was transformed with multicopy plasmids encoding the respective Yrb1-GFP fusions. Upon shuffling out of the YRB1 wild-type plasmid, the strains were cultivated in YPD at 30°C to mid-logarithmic phase and examined by fluorescence microscopy.
FIG. 4.
Inhibition of RCC1-mediated GTP exchange on Gsp1GTP. The graph shows the affinity of the indicated GST-Yrb1 fusion proteins for Gsp1GTP as measured by the inhibition of RCC1-induced GTP exchange (see Materials and Methods for details).
Taken together, these results show that the formation of the Yrb1-Xpo1-RanGTP complex requires the functional RBD of Yrb1.
The RBD of Yrb1 harbors both nuclear export and import signals.
In order to determine the domains required for proper function and subcellular localization of Yrb1, we performed a mutational analysis of YRB1, including a comprehensive set of deletions and two previously isolated point mutations, yrb1-51 (A91D) and yrb1-52 (R127K) (Künzler et al., submitted) (Fig. 5A). These mutants were examined (i) for their ability to complement the otherwise nonviable yrb1::HIS3 mutant and (ii) for their localization in wild-type and xpo1-1 mutant strains (Fig. 5B). This series of experiments reconfirmed the finding that the integrity of the RBD of Yrb1 is essential for Yrb1 function and location, whereas the N-terminal extension is dispensable (Fig. 5A). Accordingly, full-length and Δ(3-58) Yrb1 behave very similarly except that the nuclear accumulation of Yrb1Δ(3-58) in the xpo1-1 strain occurs already at permissive temperature (data not shown; Fig. 5Aa). Different truncations of the RBD, i.e., Δ(167-201), Δ(132-201), Δ(59-101), Δ(102-120), and Δ(102-111), were made, all of which abolish recognition by Xpo1 and lead to a nuclear accumulation of the respective GFP fusion proteins (Fig. 5Ab). Strikingly, even a single amino acid substitution (yrb1-51; A91D) in a conserved residue of this domain caused nuclear accumulation in a yrb1Δ::HIS3 strain (Künzler et al., submitted; Fig. 5C). This accumulation was evident already at the semipermissive temperature (30°C). In summary, mutations and/or deletions scattered throughout the Yrb1 RBD sequence cause nuclear accumulation.
Nuclear accumulation of Yrb1 could, in principle, due to the small size of protein, occur by passive diffusion and nuclear retention. To exclude this possibility, fusion constructs of Yrb1 to double or triple GFP were made and tested for nuclear accumulation in the xpo1-1 mutant strain (Fig. 5Ac). Even the Yrb1-3xGFP construct, which has a calculated molecular mass of 100 kDa and an apparent molecular mass of 120 kDa (data not shown) (which is clearly above the exclusion limit for passive diffusion), accumulated to the same extent and with the same kinetics as Yrb1-GFP in xpo1-1 cells when shifted to 37°C (Fig. 5Bc). This suggests that nuclear import of Yrb1 is not due to passive diffusion and may be a facilitated process. In searching for an NLS in Yrb1, we noticed that another yrb1 allele (yrb1-52; R127K) affected nucleocytoplasmic transport of Yrb1 in a different way. GFP-tagged Yrb1(R127K) remained cytoplasmic even in the xpo1-1 mutant, suggesting that it cannot be imported into the nucleus (Fig. 5Ad). This observation prompted us to test whether the region around residue 127 represents a nuclear import signal. Despite the fact that any deletion in the RBD tested so far abolished complementation and nuclear export by Xpo1, we found that any deletion that removed residues 121 to 131 of the RBD prevented nuclear import in the xpo1-1 mutant (Fig. 5Ad). Consistent with these results, both the deletion mutants and Yrb1(R127K) remained cytoplasmic in cells overproducing Yrb2 (data not shown). In order to exclude the possibility that these mutants were defective in nuclear accumulation rather than nuclear import, we inserted the NLS of SV40 large T antigen near the N terminus of Yrb1, Yrb1(R127K), Yrb1(A91D), and Yrb1Δ(121-131) and their respective GFP fusion proteins and examined the subcellular localization of the resulting fusion proteins (Fig. 5Ae). The heterologous NLS is active within the context of Yrb1, as shown by nuclear accumulation of NLSSV40-Yrb1-GFP in yrb1Δ::HIS3 cells (Fig. 5C) and NLSSV40-Yrb1(A91D)-GFP in wild-type cells (Fig. 5Be). Consistent with a nuclear import defect rather than a problem in nuclear accumulation of Yrb1(R127K) and Yrb1Δ(121-131), these mutant forms of Yrb1 accumulated in xpo1-1 cells, or even in XPO1 cells, respectively, when fused to the NLS (Fig. 5Be). The steady-state localization of these fusion proteins is thereby in agreement with the in vitro binding experiments (Fig. 3E and F) in which, in contrast to the Δ(121-131) mutant, both point mutants of Yrb1 are still able to form a ternary complex with Xpo1 and RanGTP. The R127K mutant, which has the highest affinity for RanGTP among the mutants, is thereby exported most efficiently and accumulated least efficiently. Therefore, the nuclear import of Yrb1 depends on a short sequence (121 KVRILMRRDKT 131) within the RBD surrounding R127 (underlined). However, neither this sequence alone or bigger parts of the RBD comprising this sequence (residues 102 to 166) were able to target GFP into the nucleus (data not shown; see also Discussion), suggesting that the NLS might be more complex. Since the growth defect of the yrb1-52 mutant (R127K) (Künzler et al., submitted) could have been (partly) due to impaired nuclear import of Yrb1, we tested complementation of the otherwise lethal yrb1Δ::HIS3 deletion by the NLSSV40-fusion proteins of Yrb1, Yrb1(A91D), and Yrb1(R127K). In contrast to the untagged alleles, however, neither NLSSV40-yrb1-51 nor NLSSV40-yrb1-52 was able to complement (data not shown). Interestingly, however, NLSSV40-YRB1 (as well as NLSSV40-YRB1-GFP) was functional and did not show any obvious growth defect, suggesting that yeast can live even if Yrb1 is largely nuclear at steady state (data not shown; see Fig. 5C for steady-state distribution of NLSSV40-Yrb1-GFP).
All this shows that the RBD of Yrb1 mediates both nuclear import and export of this RanGTP-binding protein.
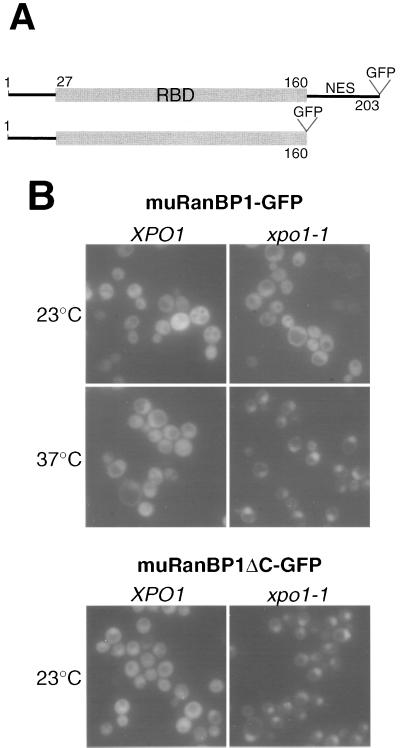
Nuclear export of mouse RanBP1 in yeast does not require the leucine-rich NES.
To test whether the import and export signals identified within Yrb1 are also present in higher eukaryotic homologs, we analyzed RanBP1 from mouse and its derived RBD in yeast (lacking the carboxy-terminally located leucine-rich NES) (Fig. 6A). Interestingly, both RanBP1-GFP and the RBD-GFP driven from the galactose-inducible GAL1 promoter complement the yrb1Δ::HIS3 null strain, indicating that the RBD of muRanBP1 is functional in yeast (data not shown). In addition, the constructs complement the same mutation also in a xpo1-1 background and accumulate, like Yrb1-GFP, in the nucleus of these mutant cells (Fig. 6B). This suggests that the RBD is sufficient for the essential function and the dynamic subcellular localization of mouse RanBP1 in yeast.
FIG. 6.
Mouse RanBP1 complements the yrb1Δ::HIS3 mutant and exhibits a shuttling behavior like that of Yrb1. (A) Schematic representation of the muRanBP1 constructs. Expression was under the control of the galactose-inducible GAL1 promoter from a multicopy plasmid. (B) Localization of muRanBP1-GFP and muRanBP1ΔC-GFP. Double disruption xpo1Δ::KAN yrb1Δ::HIS3 strain complemented by pADE2-xpo1-1 and muRanBP1-GFP or muRanBP1-ΔC-GFP were transformed with a single-copy plasmid harboring XPO1 or empty vector, respectively. Transformants were grown at 23°C and examined by fluorescence microscopy before and after a 5-min shift to 37°C.
DISCUSSION
Nucleocytoplasmic transport factors of the karyopherin β family and Ran require for their function continuous shuttling between the nucleus and cytoplasm. In contrast, other factors appear to be resident and restricted to either the nuclear or the cytoplasmic compartment, e.g., RanGAP in the cytoplasm and RanGEF in the nucleus.
The data presented here suggest that the RanGTP-binding protein RanBP1(Yrb1) is efficiently shuttling between the nucleus and cytoplasm in yeast. It is imported into the nucleus by a yet unknown mechanism and exported via the CRM1 (Xpo1)-dependent nuclear export pathway. Our data also show that Yrb1 does not enter the nucleus simply by passive diffusion but rather by a facilitated transport mechanism. This finding is surprising in view of its suggested role in the cytoplasm, where it should release RanGTP from karyopherin β-type receptors in conjunction with Rna1 (RanGAP) (see the introduction). The identification of Xpo1 as the nuclear export receptor for Yrb1 raises the question of the nature of the NES in Yrb1. The absence of a leucine-rich sequence in Yrb1 suggested that the signals recognized by Xpo1 are different from a canonical NES. Interestingly, the entire and intact RBD of both Yrb1 and muRanBP1 is necessary and sufficient for Xpo1-mediated nuclear export in yeast. Accordingly, RanGTP binding by Yrb1 is necessary for interaction with Xpo1 in vitro. Based on the low affinity of Xpo1 to RanGTP in the absence of export cargo, we interpret the in vitro-isolated Yrb1-Xpo1-RanGTP ternary complex as a true export complex. Thus, the domain of Yrb1 recognized by Xpo1 appears to be complex and not short and continuous like the canonical leucine-rich NES. Recently, it was shown that snurportin, another shuttling nucleocytoplasmic transport factor, is exported from the nucleus by CRM1 and requires an extended NES (61). This suggests that CRM1 may be able to recognize different types of NES. The insensitivity of Yrb1 export to LMB in an engineered S. cerevisiae strain which is sensitive to this drug might indicate that the residues on CRM1 involved in the recognition of noncanonical NES are different from the ones responsible for the binding of canonical NES.
The aspects of nuclear import of Yrb1 in respect to shuttling are less clear. Our results suggest that Yrb1 import is a facilitated process dependent on cis- and trans-acting factors. By deletion analysis, we identified a short sequence of 11 amino acid residues within the RBD which is necessary for nuclear import of Yrb1. The sequence is relatively rich in basic residues, but does not seem to be a classical NLS (see below). Consistent with the result of the deletion analysis, a point mutation, which leads to an import-defective Yrb1, was mapped to a conserved Arg residue within this sequence. Interestingly, this residue makes direct contact with the C terminus of RanGTP according to the crystal structure of a complex between the first RBD of RanBP2 and Ran bound to a GTP analogue (85). This finding suggests that the putative nuclear import mediator and RanGTP use the same binding site on Yrb1. If so, this would offer a possible release mechanism of Yrb1 from its import mediator in the nucleus by binding to RanGTP. Despite this knowledge, we were not yet able to delineate the minimal NLS sufficient for nuclear import of Yrb1. This suggests that the NLS of Yrb1 may be more complex than anticipated or is only formed within the folded Yrb1.
With regard to trans-acting factors involved in Yrb1 nuclear import, we found that the prp20-1 mutation in RanGEF, which affects a number of different nuclear import pathways (45), also inhibits nuclear import of Yrb1 (M. Künzler and E. Hurt, unpublished data). In contrast, no inhibition of Yrb1 import was observed in the karyopherin mutants that were tested so far (pse1-1, yrb4Δ::HIS3, nmd5Δ::HIS3, kap104Δ::HIS3, mtr10Δ::HIS3, pdr6Δ::HIS3, and sxm1Δ::HIS3) (M. Künzler and E. Hurt, unpublished data). The classical import pathway involving importin α and β is also unlikely to be used, since Yrb1 did not show any affinity for Srp1 or Kap95, either alone or in combination, in vitro (M. Künzler and E. Hurt, unpublished data). Therefore, it is possible that Yrb1 uses a nonconventional route to enter the nucleus. A possible candidate for such a nonconventional import complex is a previously reported ternary complex between RanBP1, importin β, and RanGDP (18, 19), whose physiological role is still unclear (see also below). However, so far we have no experimental evidence that this complex represents an import complex.
What could be the physiological significance of Yrb1 shuttling? Several scenarios can be discussed. One possibility is that Yrb1 somehow gets into the nucleus and has to be continuously exported into the cytoplasm by Xpo1, because nuclear Yrb1 is not compatible with its assumed function in dissociating nuclear export complexes on the cytoplasmic side of the NPC. Indeed, it has been shown that high levels of RanBP1 microinjected into the nucleus are detrimental for nuclear export in Xenopus oocytes (40). How, in this model, would Yrb1 enter the nucleus? It may do so as a result of its proposed role during nuclear import (18, 19). In agreement with such a role, mutations in YRB1 inhibit nuclear protein import in vivo (70) (Künzler et al., submitted). However, this inhibition could also be the consequence of the impaired recycling of nuclear import receptors. Alternatively, it was proposed in higher eukaryotic cells that a nuclear export mechanism for RanBP1 might be necessary to reestablish the “normal” subcellular distribution of RanBP1 following nuclear envelope assembly after mitosis. However, in yeast, no nuclear envelope breakdown occurs during mitosis. As an alternative explanation for the shuttling of Yrb1, our finding that yeast tolerates a significant pool of Yrb1 in the nucleus at steady state (when Yrb1 is targeted to the nucleus by the NLS of SV40 large T antigen) could mean that Yrb1, in principle, could fulfill a function in the nucleus. As one possibility for such a nuclear function, it may play a role in the early steps of Xpo1-mediated nuclear export. Such a role was suggested for the nuclear RBD-containing Yrb2 protein (82). Alternatively, a nuclear pool of Yrb1 may be required for nuclear processes different from nucleocytoplasmic transport. In this regard, it has recently been demonstrated that RanBP1 affects (Ran-dependent) microtubule organization in vitro (16, 43, 94). Such a nuclear function of RanBP1 would provide physiological significance to the described in vitro activity of RanBP1 as a guanine nucleotide dissociation inhibitor of guanine nucleotide exchange by nuclear RanGEF (RCC1) (10) and a reported physical interaction between RanBP1 (Yrb1) and RCC1 (Prp20) (35, 64).
ACKNOWLEDGMENTS
Special thanks go to D. Lau for technical support, D. Zenklusen (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) for sharing unpublished results, B. Wolff (Novartis Research Institute, Vienna, Austria) for leptomycin B, and J. Thorner (University of California, Berkeley), for generous support. We thank K. Weis (University of California, Berkeley), I. Macara (Markey Center for Cell Signaling, Charlottesville, Va.), M. Rosbash (M. Neville) and L. Davis (Brandeis University, Waltham, Mass.), M. Nomura (University of California, Irvine), M. Fitzgerald-Hayes (University of Massachusetts, Amherst), and G. Stier and S. Labeit (EMBL, Heidelberg, Germany) for providing us with strains and plasmids. We are grateful to G. Simos and O. Gadal for critical reading of the manuscript.
M.K. is a recipient of grants from the Deutsche Forschungsgemeinschaft (Ku 1235/1-1) and is supported by a fellowship provided by the Swiss National Research Foundation.
REFERENCES
- 1.Adam S A. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- 2.Amberg D C, Fleischmann M, Stagljar I, Cole C N, Aebi M. Nuclear PRP20 protein is required for mRNA export. EMBO J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askjaer P, Bachi A, Wilm M, Bischoff F R, Weeks D L, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj I W, Fornerod M. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 5.Becker J, Melchior F, Gerke V, Bischoff F R, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- 6.Beddow A L, Richards S A, Orem N R, Macara I G. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belanger K D, Kenna M A, Wei S, Davis L I. Genetic and physical interactions between Srp1p and nuclear pore complex proteins Nup1p and Nup2p. J Cell Biol. 1994;126:619–630. doi: 10.1083/jcb.126.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benton B, Zang J H, Thorner J. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae as a proline rotamase localized to the nucleolus. J Cell Biol. 1994;127:623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff F R, Görlich D. RanBP1 is crucial for the release of RanGTP from importin b-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 10.Bischoff F R, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischoff F R, Ponstingl H. Catalysis of guanine nucleotide exchange of Ran by RCC1 and stimulation of hydrolysis of Ran-bound GTP by Ran-GAP1. Methods Enzymol. 1995;257:135–144. doi: 10.1016/s0076-6879(95)57019-5. [DOI] [PubMed] [Google Scholar]
- 12.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 13.Booth J W, Belanger K D, Sannella M I, Davis L I. The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J Biol Chem. 1999;274:32360–32367. doi: 10.1074/jbc.274.45.32360. [DOI] [PubMed] [Google Scholar]
- 14.Bressan G, Somma M P, Lewis J, Santolamezza C, Copeland N G, Gilbert D J, Jenkins N A, Lavia P. Characterization of the opposite-strand genes from the mouse bidirectionally transcribed HTF9 locus. Gene. 1991;103:201–209. doi: 10.1016/0378-1119(91)90274-f. [DOI] [PubMed] [Google Scholar]
- 15.Butler G, Wolfe K H. Yeast homologue of mammalian Ran binding protein 1. Biochim Biophys Acta. 1994;1219:711–712. doi: 10.1016/0167-4781(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 16.Carazo-Salas R E, Guarguaglini G, Gruss O J, Segref A, Karsenti E, Mattaj I W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 17.Cesareni G, Murray J A H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages in genetic engineering. Genet Eng. 1987;9:135–154. [Google Scholar]
- 18.Chi N C, Adam E J H, Adam S A. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- 19.Chi N C, Adam E J H, Visser G D, Adam S A. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole C N, Hammell M. Driving and directing transport. Curr Biol. 1998;8:368–372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- 21.Corbett A H, Koepp D M, Schlenstedt G, Lee M S, Hopper A K, Silver P A. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Sal G, Manfioletti G, Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988;16:9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dingwall C, Kandels-Lewis S, Séraphin B. Targeting Ran to the nuclear pore: a family of Ran binding proteins that includes nucleoporins. Proc Natl Acad Sci USA. 1995;92:7525–7529. doi: 10.1073/pnas.92.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doye V, Hurt E C. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 25.Englmeier L, Mattaj I. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 26.Floer M, Blobel G. Putative reaction intermediates in Crm1-mediated nuclear protein export. J Biol Chem. 1999;274:16279–16286. doi: 10.1074/jbc.274.23.16279. [DOI] [PubMed] [Google Scholar]
- 27.Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- 28.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 29.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base-pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 31.Görlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Görlich D, Hartmann E. A Ran-binding motif in nuclear pore proteins. Trends Cell Biol. 1995;5:192–193. doi: 10.1016/s0962-8924(00)88992-8. [DOI] [PubMed] [Google Scholar]
- 33.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi N, Yokoyama N, Seki T, Azuma Y, Ohba T, Nishimoto T. RanBP1, a Ras-like nuclear G protein binding to Ran/TC4, inhibits RCC1 via Ran/TC4. Mol Gen Genet. 1995;247:661–669. doi: 10.1007/BF00290397. [DOI] [PubMed] [Google Scholar]
- 36.Hellmuth K, Lau D, Bischoff R, Künzler M, Simos G, Hurt E. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 38.Hood J K, Silver P A. Cse1p is required for export of Srp1p/importin-α from the nucleus in Saccharomyces cerevisiae. J Biol Chem. 1998;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- 39.Hopper A K, Traglia H M, Dunst R W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadowaki T, Goldfarb D, Spitz L M, Tartakoff A M, Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 43.Kalab P, Pu R T, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 44.Kehlenbach R H, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koepp D M, Wong D H, Corbett A H, Silver P A. Dynamic localization nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Künzler M, Hurt E C. Cse1p functions as the nuclear export receptor for importin a in yeast. FEBS Lett. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- 47.Lau D, Künzler M, Braunwarth A, Hellmuth K, Podtelejnikov A, Mann M, Hurt E. Purification of protein A-tagged yeast Ran reveals association with a novel karyopherin β family member Pdr6p. J Biol Chem. 2000;274:467–471. doi: 10.1074/jbc.275.1.467. [DOI] [PubMed] [Google Scholar]
- 48.Loeb J D J, Davis L I, Fink G R. NUP2, a novel yeast nucleoporin, has functional overlap with other proteins of the nuclear pore complex. Mol Biol Cell. 1993;4:209–222. doi: 10.1091/mbc.4.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lounsbury K M, Macara I G. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- 50.Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 51.Moore M S. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 52.Mueller L, Cordes V C, Bischoff R, Ponstingl H. Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett. 1998;427:330–336. doi: 10.1016/s0014-5793(98)00459-1. [DOI] [PubMed] [Google Scholar]
- 53.Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 55.Noguchi E, Hayashi N, Nakashima N, Nishimoto T. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-GTPase-activating protein. Mol Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noguchi E, Saitoh Y-h, Sazer S, Nishimoto T. Disruption of the YRB2 gene retards nuclear protein export, causing a profound mitotic delay, and can be rescued by overexpression of XPO1/CRM1. J Biochem. 1999;125:574–585. doi: 10.1093/oxfordjournals.jbchem.a022323. [DOI] [PubMed] [Google Scholar]
- 57.Novoa I, Rush M G, D'Eustachio P. Isolated mammalian and Schizosaccharomyces pombe Ran-binding domains rescue S. pombe spb1 (RanBP1) genomic mutants. Mol Biol Cell. 1999;10:2175–2190. doi: 10.1091/mbc.10.7.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 59.Ouspenski I I, Mueller U W, Matynia A, Sazer S, Elledge S J, Brinkley B R. Ran-binding protein-1 is an essential component of the Ran/RCC1 molecular switch system in budding yeast. J Biol Chem. 1995;270:1975–1978. doi: 10.1074/jbc.270.5.1975. [DOI] [PubMed] [Google Scholar]
- 60.Panté N, Aebi U. Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science. 1996;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- 61.Paraskeva E, Izaurralde E, Bischoff F R, Huber J, Kutay U, Hartmann E, Lührmann R, Görlich D. CRM1-mediated recycling of Snurportin 1 to the cytoplasm. J Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasquinelli A E, Powers M A, Lund E, Forbes D J, Dahlberg J E. Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc Natl Acad Sci USA. 1997;94:13394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pemberton L F, Blobel G, Rosenblum J. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 64.Pu R T, Dasso M. The balance of RanBP1 and RCC1 is critical for nuclear assembly and nuclear transport. Mol Biol Cell. 1997;8:1955–1970. doi: 10.1091/mbc.8.10.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richards S A, Carey K L, Macara I G. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 66.Richards S A, Lounsbury K M, Carey K L, Macara I G. A nuclear export signal is essential for the cytosolic localization of the ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–494. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 68.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 69.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff F R. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schlenstedt G, Wong D H, Koepp D M, Silver P A. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seedorf M, Damelin M, Kahana J, Taura T, Silver P A. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Senger B, Simos G, Bischoff F R, Podtelejnikov A V, Mann M, Hurt E C. Mtr10p functions as a nuclear import receptor for the mRNA binding protein Np13p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sikorski R S, Hieter R. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Solsbacher J, Maurer P, Bischoff F R, Schlenstedt G. Cse1p is involved in export of yeast importin α from the nucleus. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 76.Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- 77.Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 78.Studier F W, Rosemberg H A, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:62–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 79.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 80.Tabor S, Richardson C. DNA sequence analysis with a modified T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Talcott B, Moore M S. Getting across the nuclear pore complex. Trends Cell Biol. 1999;9:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- 82.Taura T, Krebber H, Silver P A. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taura T, Schlenstedt G, Silver P A. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- 84.Turi T G, Mueller U W, Sazer S, Rose J. Characterization of a nuclear protein conferring Brefeldin A resistance in Schizosaccharomyces pombe. J Biol Chem. 1996;271:9166–9171. doi: 10.1074/jbc.271.15.9166. [DOI] [PubMed] [Google Scholar]
- 85.Vetter I, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 86.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 87.Wilken N, Senécal J L, Scheer U, Dabauvalle M C. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur J Cell Biol. 1995;68:211–219. [PubMed] [Google Scholar]
- 88.Wolff B, Sanglier J-J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent RNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 89.Wozniak R W, Rout M P, Aitchison J D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 90.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 91.Xiao Z, McGrew J T, Schroeder A J, Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yan C, Lee L H, Davis L I. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, Fukui M, Nishimoto T. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 94.Zhang C, Hughes M, Clarke P R. Ran-GTP stabilizes microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci. 1999;112:2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]
- 95.Zolotukhin A S, Felber B K. Mutations in the nuclear export signal of human Ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Biol Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]