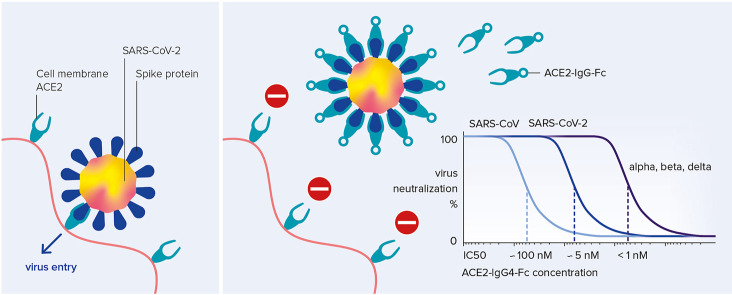
Abstract
SARS-CoV-2 enters host cells after binding through its spike glycoprotein to the angiotensin-converting enzyme 2 (ACE2) receptor. Soluble ACE2 ectodomains bind and neutralize the virus, yet their short in vivo half-live limits their therapeutic use. This limitation can be overcome by fusing the fragment crystallizable (Fc) part of human immunoglobulin G (IgG) to the ACE2 ectodomain, but this bears the risk of Fc-receptor activation and antibody-dependent cellular cytotoxicity. Here, we describe optimized ACE2-IgG4-Fc fusion constructs that avoid Fc-receptor activation, preserve the desired ACE2 enzymatic activity and show promising pharmaceutical properties. The engineered ACE2-IgG4-Fc fusion proteins neutralize the original SARS-CoV, pandemic SARS-CoV-2 as well as the rapidly spreading SARS-CoV-2 alpha, beta and delta variants of concern. Importantly, these variants of concern are inhibited at picomolar concentrations proving that ACE2-IgG4 maintains – in contrast to therapeutic antibodies - its full antiviral potential. Thus, ACE2-IgG4-Fc fusion proteins are promising candidate anti-antivirals to combat the current and future pandemics.
Keywords: COVID-19, Antiviral drug, Antiviral therapy, Entry inhibitor, Receptor trap
Graphical abstract
1. Introduction
Angiotensin-converting enzyme 2 (ACE2) serves as a common entry receptor for the human coronavirus (CoV)-NL63, the original SARS-CoV from 2003 and the novel, pandemic SARS-CoV-2 into their host cells by binding the spike protein on the virus surface (Hoffmann et al., 2020; Hofmann et al., 2005; Wrapp et al., 2020). Membrane fusion of SARS-CoV-2 is enabled by proteolytic activation of the spike protein by cell surface protease TMPRSS2 (Hoffmann et al., 2020). In addition, neuropilin-1 has recently been identified to bind a furin-cleaved spike protein and by this to significantly potentiate SARS-CoV-2 infectivity (Cantuti-Castelvetri et al., 2020). ACE2 is expressed on the plasma membrane of epithelial cells in the respiratory tract and lung (Wrapp et al., 2020), but also in other tissues like intestine, testes, liver, kidney, brain, and in the cardiovascular system (Crackower et al., 2002; Ding et al., 2004; Hamming et al., 2004).
ACE2 is an 805 amino acid type-I transmembrane protein consisting of extracellular, transmembrane and cytosolic domains (Jiang et al., 2014). The extracellular domain is a zinc metalloprotease, which enzymatically functions as a carboxypeptidase (Donoghue et al., 2000; Tipnis et al., 2000) suppressing the renin-angiotensin-aldosterone system (RAAS) by cleaving angiotensin II into heptapeptide angiotensin-(1–7) and cleaving other vasoactive peptides (Lu et al., 2016; Santos et al., 2018). Angiotensin-(1–7) lowers the diastolic blood pressure, has anti-inflammatory, anti-proliferative and anti-fibrotic effects (Burrell et al., 2004) and thereby protects lung, heart, kidney and other organs from injury (Crackower et al., 2002; Ding et al., 2004; Hamming et al., 2004). The potential contribution of angiotensin II to the COVID-19 pathophysiology has been indicated by reports that angiotensin II levels in plasma samples from COVID-19 patients were markedly elevated and correlated with viral load and severity of the disease (Liu et al., 2020a; Wu et al., 2020).
ACE2-Fc fusion proteins, composed of an IgG Fc domain fused to the extracellular domain of ACE2, have been suggested as a high-priority treatment option for COVID-19 (Batlle et al., 2020; Kruse, 2020). In addition to neutralizing SARS-CoV and SARS-CoV-2, the ACE2's enzymatic activity affecting the RAAS provides a second mode of action, potentially alleviating the pathophysiology of acute respiratory distress syndrome. Therapeutic use of a soluble human recombinant ACE2 dimer (APN01) with a half-life of 10 h is currently investigated in patients with COVID-19 (Haschke et al., 2013; Khan et al., 2017; Zoufaly et al., 2020). Strong in vitro SARS-CoV-2 neutralizing activity has been described for sequence variants of ACE2-IgG1-Fc fusion proteins (Glasgow et al., 2020; Higuchi Y et al., 2021; Huang et al., 2020; Iwanaga et al., 2020; Lei et al., 2020; Liu et al., 2020b; Lui et al., 2020), and ACE2-IgG1-Fc (HLX71) has entered phase 1 clinical studies.
A theoretical concern at the beginning of the pandemic was antibody-mediated enhancement (ADE) of SARS-CoV-2 infection, or disease-enhancing pathophysiology of sub-neutralizing or cross-reactive non-neutralizing antibodies by Fc-mediated complement activation or antibody-dependent cellular cytotoxicity (Bournazos et al., 2020). For the SARS-CoV-2 relative Middle East respiratory syndrome (MERS)-CoV, binding of Fc receptor gamma III (CD16) has led to infection of CD16 positive cells (Jafarzadeh et al., 2020; Manickam et al., 2020). While IgG1-Fc strongly binds to CD16 and induces pronounced cytotoxicity, Fc-related effector functions are minimal for IgG4-Fc (de Taeye et al., 2020). In this regard, the IgG4-Fc fragment appeared to be a preferred fusion partner for ACE2. However, it is well known that naturally occurring IgG4 antibodies are less stable than IgG1 variants due to the formation of half antibodies, which limits their use in pharmaceutical preparations (Aalberse and Schuurman, 2002; Correia, 2010; Dumet et al., 2019; Handlogten et al., 2020). To generate a stable ACE2-IgG4-Fc fusion protein, we have therefore chosen the immunoglobulin Fc region of an IgG4/kappa isotype with an S228P sequence alteration in the hinge region (Aalberse and Schuurman, 2002). For the ACE2 domain, two different truncations (Q18-G732 and Q18-S740) were used, and point mutations were introduced in the ACE2 domain to abrogate its enzymatic activity.
Here we show that an ACE2-IgG4-Fc fusion protein does not only have favorable pharmaceutical features but also efficiently neutralizes the 2003 SARS-CoV as well as circulating strains of SARS-CoV-2, including the variants of concern (VoC) alpha, beta and delta.
2. Materials and methods
2.1. Construct design
ACE2 amino acid sequence modifications were designed by computer-aided modelling. ACE2 ectodomains of different length, Q18-G732 and Q18-S740, with or without mutation of the catalytic site (wild type or H374N/H378N mutant) (Moore et al., 2004) were combined with the Fc fragment of IgG4 bearing a stabilizing S228P mutation in the hinge region (Aalberse and Schuurman, 2002). For comparison, the same ACE2 sequence variants were fused to the Fc fragment of IgG1 with a truncated hinge region (DKTHTCPPCPA).
2.2. Generation of expression plasmids
Plasmids encoding the Fc fusion proteins were generated at ThermoFisher. Genes of interest were subcloned into pcDNA3.1 Zeocin expression plasmids (Invitrogen V860-20, Life Technologies, Carlsbad, CA, USA) with an elongated CMV promoter using HindIII/XhoI restriction sites. Following amplification in Escherichia coli, expression plasmids were isolated and analyzed by restriction analysis as well as DNA sequencing.
2.3. Protein expression
Using the FreeStyle 293 Expression System (Thermo Fisher Scientific, Waltham, MA, USA), the different ACE2-Fc fusion proteins were transiently expressed in 3 x 240 mL culture media. On day six, samples were analyzed for cell viability as well as cell density and supernatants were harvested by centrifugation followed by sterile filtration (Walls et al., 2020). The material was either stored at −80 °C until purification or subjected directly to purification. Small samples were taken from the pools to determine expression yields by bio-layer interferometry (BLI).
2.4. Protein purification
Purification of the fusion proteins secreted into the culture medium was performed by protein A column chromatography followed by preparative Size Exclusion Chromatography (SEC). For protein A purification, after loading the sample, the Atmosphere A3 column (JSR Life Sciences, Sunnyvale, CA, USA) was washed and the ACE2-Fc fusion proteins were eluted using 40 mM sodium acetate buffer, pH 3.0. Following elution, samples were first neutralized to pH 7.5 using 1 M Tris, pH 9.0, subsequently diluted 1:1 with 50 mM Tris, pH 7.5, 300 mM NaCl and concentrated to 10 mg/mL using spin filters. Concentrated proteins were further purified with a Superdex 200 increase (GE Healthcare-Cytiva, Chicago, IL, USA) column equilibrated with 50 mM Tris, pH 7.5, 150 mM NaCl. The main peak was pooled, the protein concentration was determined by slope spectrometry (Lehr et al., 2015) and adjusted to 1 mg/mL. The protein solution was passed through a sterilizing filter and stored at 4 °C until further usage.
2.5. Size exclusion chromatography with multi-angle light scattering (SEC-MALS)
A Shimadzu HPLC system with two concentration detectors (UV and refractive index) and a HELEOS II MALS detector were used for the measurements. The flow rate was 1 mL/min and the running buffer was 50 mM Tris, pH 7.5 and 150 mM NaCl. 50 μg of protein was injected on a Superdex 200 Increase 10/300 GL column (Cytiva). The chromatograms were evaluated with the Astra software.
2.6. Circular dichroism (CD)
All CD measurements were performed with a Jasco J-1500 spectropolarimeter at 20 °C. The sample buffer consisted of 50 mM Tris, pH 7.5 and 150 mM NaCl. The Far-UV CD spectra were obtained in a 1 mm quartz cuvette using a protein concentration of 0.1 mg/mL. The Near-UV CD spectra were measured in a 5 mm quartz cuvette using a protein concentration of 1 mg/mL.
2.7. ACE2 activity assay
An ACE2 activity assay kit from Abcam (Cat.No. ab273297) was used to measure the enzymatic activity of the constructs. The assay was performed according to the manufacturer's manual and is based on a synthetic peptidyl-4-methylcoumaryl-7-amide (MCA) that is cleaved by the ACE2 enzyme. Upon cleavage, free MCA is detected fluorometrically (Ex320nm/Em420nm) and quantified with a standard curve obtained with MCA standard solutions with known concentrations. Two commercially available ACE2-Fc proteins obtained from Genscript (Cat.No. Z03484-1) and Acrobiosystems (Cat.No. AC2-H5257) were used as reference.
2.8. Determination of binding affinity to spike protein RBD using surface plasmon resonance
The measurements were performed with a Biacore X-100 system and the Biotin CAPture kit (Cytiva). The running buffer was HBS-EP+ pH 7.4 (Cytiva). The ligand SARS-CoV-2 RBD with an AviTag (Acrobiosystems) was captured on the streptavidin chip to around 100 RU. Increasing concentrations of the analyte ACE2-Fc (0.32, 1.6, 8, 40 and 200 nM) were injected over the immobilized ligand in a single-cycle kinetic mode. The obtained sensorgrams were evaluated with the Biacore X-100 software to obtain a binding constant (KD).
2.9. Determination of binding affinity to Fc-receptors using surface plasmon resonance
A Biacore T200 was used for the Fc-receptor binding studies. For the FcγRI and the FcγRIIIa experiments, the his-tagged FcγRI and FcγRIIIa with a concentration of 1.5 nM were captured by injection of the solutions for 90 s with a flow rate of 5 μL/min over a covalently immobilized anti-his tag antibody on a CM5 chip. The running buffer was HBS-EP+ pH 7.4 (Cytiva). Five different concentrations of the ACE2-Fcs were injected in a single-cycle kinetic mode (3.7–300 nM for the experiment with FcγRI and 25–2000 nM for FcγRIIIa). The FcγRI-binding data was fit to a heterogeneous ligand model and the first binding constant was reported. The FcγRIIIa-binding data was fit to a two-state reaction model to derive the binding constant. For the FcRn experiments, the FcRn was covalently immobilized on a CM5 chip to around 50 RU. The sample buffer was HBS-EP+ pH 6.0 (Cytiva). The ACE2-Fcs were injected in five different concentrations from 205 to 8000 nM in a single-cycle kinetic mode. The FcRn binding was evaluated with a steady-state affinity fit.
2.10. SARS-CoV-2 spike S1 inhibition ELISA
Inhibition of binding of SARS-CoV-2 spike S1 protein to ACE2 was tested using the ACE2:SARS-CoV-2 Spike S1 Inhibitor Screening Assay Kit (BPS Bioscience, San Diego, CA, USA; Cat.No. 79945) according to the manufacturer's instructions with an adapted neutralization procedure. Briefly, biotinylated SARS-CoV-2 Spike S1 protein (25 nM) was incubated with serial dilutions of the ACE2-Fc fusion proteins in a 96-well neutralization plate at room temperature (RT) for 1 h with slow shaking (= neutralization mix).
ACE2 protein was added to a nickel-coated 96-well plate at a concentration of 1 μg/mL and incubated at RT for 1 h with slow shaking. Following a washing step to remove unbound ACE2, the plates were blocked at RT for 10 min with slow shaking. Subsequently, the neutralization mix was transferred to the ACE2 coated plate and the plate was incubated at RT for 1 h with slow shaking. Following a 10 min blocking step, the plate was incubated with Streptavidin-HRP at RT for 1 h with slow shaking. Following a washing and a 10 min blocking step, the HRP substrate was added and the plate was analyzed on a chemiluminescence reader.
2.11. Virus strains
SARS-CoV-2-GFP (kindly provided by Volker Thiel, University of Bern, Switzerland) is based on the original Wuhan SARS-CoV-2 isolate (GenBank accession MT108784) and was reconstituted from a synthetic construct derived from SARS-2 BetaCoV/Wuhan/IVDC-HB-01/2019 (Thi Nhu Thao et al., 2020).
SARS-CoV-2-Jan (SARS-CoV-2-Munich-TUM-1; EPI_ISL_582134), SARS-CoV-2-April (SARS-CoV-2 D614G; EPI_ISL_466888), SARS-CoV-2 alpha (EPI_ISL_755639), SARS-CoV-2 beta (EPI_ISL_1752394), SARS-CoV-2 delta (EPI_ISL_2772700) and SARS-CoV (AY291315.1) were isolated from patient material in Germany. Briefly, SARS-CoV-2-Jan was isolated from a COVID-19 infected patient who was infected during the earliest documented COVID-19 outbreak in Germany at the end of January 2020 with a virus imported from Wuhan via a single contact in Shanghai (Wolf et al., 2020). SARS-CoV-2-April was isolated during the first eminent wave of the pandemic in Europe in April 2020 from a patient in Munich, Germany. Both virus isolates as well as a control isolate from the early “Webasto” cluster outbreak (Wolfel et al., 2020) contain the S1 D614G mutation showing significantly higher infectious titers in vitro (Korber et al., 2020).
SARS-CoV-2-Jan, SARS-CoV-2 alpha, SARS-CoV-2 beta, SARS-CoV-2 delta, SARS-CoV and SARS-CoV-2-GFP (Thi Nhu Thao et al., 2020) were propagated and passaged in Vero E6 cells (derived from African green monkey kidney epithelial cells; ATCC-CRL-1586). SARS-CoV-2-April was isolated on Caco-2 cells followed by passaging in Vero E6 cells. All strains were cultured in DMEM medium (5% fetal calf serum (FCS), 1% penicillin/streptomycin (P/S), 200 mmol/L L-glutamine, 1% MEM-Non-Essential Amino Acids (NEAA), 1% sodium pyruvate (all from Gibco – Life Technologies). Virus stocks were sequenced after propagation to confirm their identity.
2.12. Plaque assay to determine SARS-CoV-2 titer
Viral titers were determined by plaque assay as described by Baer et al. (Baer and Kehn-Hall, 2014) with some modifications. All media and supplements were from Gibco – Life Technologies. Briefly, HepG2 or Vero E6 cells were plated in a 12-well plate at 5E05 cells/well in DMEM medium supplemented with 5% FCS, 1% P/S, 200 mmol/L L-glutamine, 1% MEM-NEAA, 1% sodium-pyruvate and incubated overnight at 37 °C and 5% CO2. Cells were infected with serial dilution of virus sample in cell culture medium at 37 °C for 1 h. After discarding the supernatant, 1 mL of 5% carboxymethylcellulose diluted in Minimum Essential Media was added per well and the plate was incubated at 37 °C until obvious plaques appeared. After removing the supernatant, cells were fixed with 10% paraformaldehyde at RT for 30 min. Next, a washing step with PBS was performed, followed by the addition of 1% crystal violet (diluted in 20% methanol and water). Following an incubation time of 15 min at RT, the solution was washed away with PBS and the plate was dried. The viral titer (PFU/mL) of the sample was determined by counting the average number of plaques for a dilution and the inverse of the total dilution factor.
2.13. In vitro infection assay using SARS-CoV-2-GFP
Vero-E6 cells were plated in a 96-well plate at 1.4E04 cells/well in DMEM medium (Gibco) supplemented with 5% FCS, 1% P/S, 200 mmol/L L-glutamine, 1% MEM-NEAA, 1% sodium-pyruvate (all from Gibco) and incubated overnight at 37 °C and 5% CO2. Serial dilutions of ACE2-Fc fusion proteins and SARS-CoV-2-GFP were mixed in fresh media and pre-incubated at 37 °C for 1 h. Afterwards, Vero E6 cells were infected with the neutralized virus solution at a multiplicity of infection (MOI) of 0.6 infectious viruses (IU) per cell at 37 °C. After 1 h, the neutralization mix was replaced by cell culture medium. Plates were placed in the IncuCyte S3 Live-Cell Analysis System (Essen BioScience, Newark, UK) and real-time images of uninfected mock cells (Phase channel) and infected (GFP and Phase channel) cells were captured every 4 h for 72 h. Virus control cells were infected with the same virus stock but without prior incubation with the ACE2-Fc fusion constructs using the identical protocol.
2.14. Viral neutralization assay
Vero E6 cells were plated in a 96-well plate at 1.6E04 cells/well in DMEM medium (Gibco) supplemented with 5% FCS, 1% P/S, 200 mmol/L L-glumatine, 1% MEM-NEAA, 1% sodium-pyruvate (all from Gibco) and incubated overnight at 37 °C and 5% CO2. ACE2-Fc fusion proteins were serially diluted in fresh media, virus was added at an amount allowing to infect cells at a multiplicity of infection (MOI) of 0.06 IU/cell and pre-incubated at 37 °C for 1 h. Afterwards, Vero E6 cells were infected with the neutralized virus solution at 37 °C for 1 h. Next, the neutralization mix was removed, culture medium was added, and cells were incubated at 37 °C for 24 h. Mock cells represent uninfected Vero E6 cells, incubated with culture medium. After 24 h, cells were washed once with PBS and fixed with 4% paraformaldehyde at RT for 10 min. Following a washing step with PBS, fixed Vero E6 cells were permeabilized with 0.5% saponin (Carl Roth, Karlsruhe, Germany) in PBS at RT for 10 min to allow an in-cell ELISA. For this, cells were blocked with a mixture of 0.1% saponin and 10% goat serum (Gibco) in PBS with gentle shaking at RT for 1 h. Subsequently, Vero E6 cells were incubated with a 1:500 dilution of an anti-dsRNA J2 antibody (Jena Bioscience, Jena, Germany) in PBS supplemented with 1% FCS at 4 °C overnight with shaking. Following four washing steps with wash buffer (PBS supplemented with 0.05% Tween-20 (Carl Roth)). Next, the plates were incubated with a 1:2,000 dilution of a goat anti-mouse IgG2a-HRP antibody (Southern Biotech, Birmingham, AL, USA) in PBS supplemented with 1% FCS and incubated with gently shaking at RT for 1 h. Following four washing steps, 3,3′,5,5′-Tetramethylbenzidin (TMB) substrate (Invitrogen) was added to the wells and incubated in the dark for 10 min. Colorimetric detection on a Tecan infinite F200 pro plate reader (Tecan, Männedorf, Switzerland) at 450 nm and at 560 nm was performed after stopping the color reaction by the addition of 2N H2SO4 (Carl Roth).
2.15. Cell viability assay using SARS-CoV-2 variants of concern (VoC)
24 h before infection, human lung epithelial A549 cells (ATCC-CCL-185), engineered to overexpress the ACE2 (A549-hACE2), were plated at 1.5E04 cells/well in a 96-well white well half area plate with clear bottom (Corning, Corning, NY, USA) in DMEM containing 2% FCS, 1% P/S and 1% NEAA (all from Gibco) and incubated overnight at 37 °C and 5% CO2. A serial dilution of ACE2-IgG4-Fc construct 1 and 3 was performed and mixed with a defined volume of virus stocks of the indicated SARS-CoV-2 clinical isolates resulting in 80% cytotoxicity. After 1 h of pre-incubation at 37 °C, the neutralization mix of ACE2-IgG4-Fc construct and the respective SARS-CoV-2 isolates was added to A549-hACE2 cells. Virus-mediated killing of target cells was determined using a luminometric readout of virus-induced cytotoxicity. In brief, 72 h after infection cells were treated according to manufacturer's instructions: 15 μl CellTiter-Glo 2.0 reagent (Promega, Wisconsin, USA) were added to each well, incubated for 10 min in the dark at RT and luminescence was recorded (0.5 s integration time, no filter) using the Infinite F200 microplate reader (Tecan). Viability of cells and the corresponding infectious titer for each virus isolate was calculated by normalization of infected cells to untreated control cells (set to 100%).
3. Results
3.1. Engineering, expression and purification of ACE2-Fc proteins
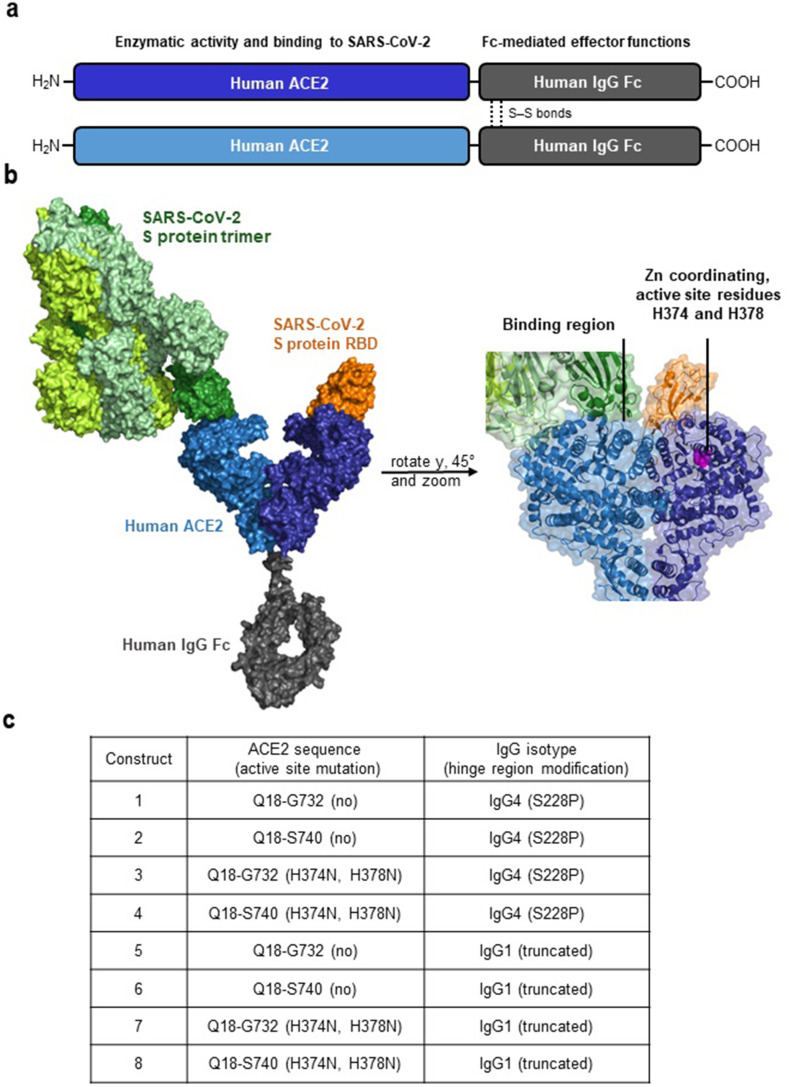
First, we wanted to create ACE2-Fc molecules with different biological properties using the PyMOL Molecular Graphics System (version 2.3.3., Schrödinger, LLC., 2020). ACE2-IgG4-Fc and ACE2-IgG1-Fc fusion proteins were designed based on crystal and EM structures of the ACE2 extracellular domain, the SARS-CoV-2 spike (S) protein and its receptor-binding domain (RBD), as well as the IgG4-Fc and IgG1-Fc domains (Fig. 1 a and b) (Lan et al., 2020; Scapin et al., 2015; Wrapp et al., 2020; Yan et al., 2020). Details of the ACE2 sequences fused to the Fc fragments of IgG4 and IgG1 are shown in Fig. 1c.
Fig. 1.
Structural elements in the ACE2-Fc constructs. a Schematic depiction of the main parts in an engineered ACE2-Fc molecule and their functional properties. b Design of the ACE2-Fc fusion protein; ACE2 parts in light and dark blue, IgG-Fc part in gray, spike (S) protein trimer in green and the receptor-binding domain (RBD) located at the tip of each spike protein in orange and dark green. The binding region as well as active site residues H374 and H378 important for the enzymatic activity of ACE2 are highlighted. Structures of the following Protein Data Bank (PBD) identifiers were used for modeling: 6M17, 6M0J, 6VSB, 5DK3. c Nomenclature and structural variations in the ACE2-Fc constructs.
The expression yields of all constructs were in a similar range, although slightly higher for Q18-G732 ACE2-Fc fusion proteins (Supplementary Table 1). The purity of the ACE2-Fc fusion proteins under investigation ranged between 95% and 98%, as evident from capillary electrophoresis sodium dodecyl sulfate (CE-SDS) and size exclusion chromatography (SEC) analysis (Supplementary Table 1). High molecular weight fractions (HMWS) measured after purification were slightly lower for Q18-G732 ACE2-Fc fusion proteins (Supplementary Table 1). Peptide mapping confirmed the presence of the modifications in the ACE2-Fc fusion proteins.
3.2. Variations in the ACE2-Fc sequences allow preserving structural properties and engineering enzymatic activity of ACE2
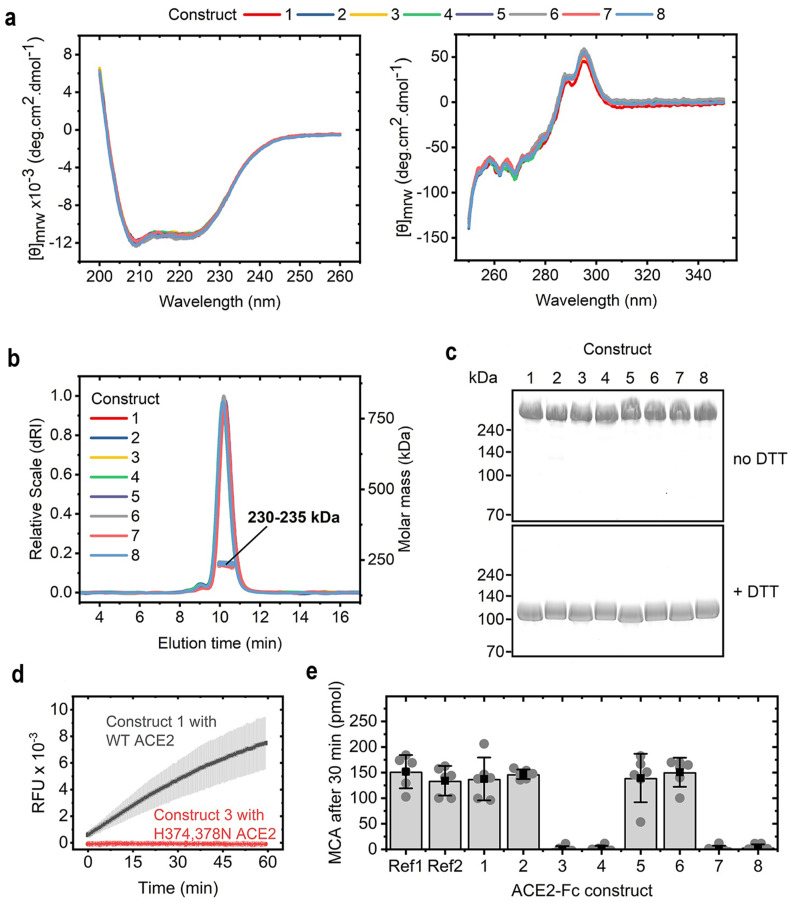
To study the secondary structure to the ACE2-Fc fusion construct, we used protein circular dichroism (CD). The Far-UV (200–260 nm) CD spectra of the fusion proteins could be superimposed indicating that the secondary structures are preserved among all constructs, regardless of the sequence variations (Fig. 2 a). The same held true for the Near-UV (250–350 nm) CD spectra, which indicated that the overall tertiary structure is also highly similar in all ACE2-Fc proteins investigated (Fig. 2a).
Fig. 2.
Structural and functional characteristics of the ACE2-Fc proteins. a Far-UV CD spectra (left) and Near-UV CD spectra (right) of ACE2-Fc constructs indicating that all proteins exhibit similar secondary and tertiary structures. b Chromatograms and molecular mass from size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) indicating that the ACE2-Fc molecules form homodimers. c Non-reducing (top) and reducing (bottom) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis showing that intermolecular disulfide bonds in the homodimers are formed. d Comparison of fluorescence signals over time obtained in an assay testing the cleavage of a fluorescent peptidyl-4-methylcoumaryl-7-amide (MCA). Relative fluorescent units (RFU) are given. e Amount of MCA cleaved after 30 min of incubation with the ACE2-Fc constructs. Ref1 and Ref2 are two different commercially available ACE2-Fc proteins from Genscript and Acrobiosystems, respectively. Bars are mean values; error bars depict the 95% confidence interval of six independent experiments shown as circles.
Size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) was used to investigate the oligomeric state of the fusion proteins. All ACE2-Fc fusion molecules tested exhibit similar elution times and molecular masses of 230–235 kDa (Fig. 2b). These values agreed well with the theoretical mass of a homodimer, which is 216–218 kDa based on calculations from the primary sequence. The slightly higher molecular mass measured in solution was due to the glycosylation of the proteins. Reducing and non-reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2c) revealed that all ACE2-Fc fusion proteins analyzed under reducing conditions with dithiothreitol (DTT) run at about 110–120 kDa, which correlated to the theoretical mass of a monomeric ACE2-Fc molecule. The non-reduced ACE2-Fc fusion proteins exhibited a much higher apparent molecular weight, indicating that the intermolecular disulfide bonds in the ACE2-Fc homodimers are formed, which agrees with results from capillary electrophoresis sodium dodecyl sulfate (CE-SDS) under reducing and non-reducing conditions.
The enzymatic activity of the engineered ACE2-Fc fusions was evaluated by their ability to cleave synthetic ACE2 substrate peptidyl-4-methylcoumaryl-7-amide (MCA), releasing a fluorescent product. All ACE2-Fc constructs with wildtype (WT) ACE2 sequences cleave the same amount of MCA during a 30-min incubation period, while mutations in the active ACE2 site resulted in a complete loss of enzymatic activity (Fig. 2d and e).
3.3. ACE2-Fc fusion constructs strongly interact with the receptor-binding domain (RBD) of SARS-CoV-2
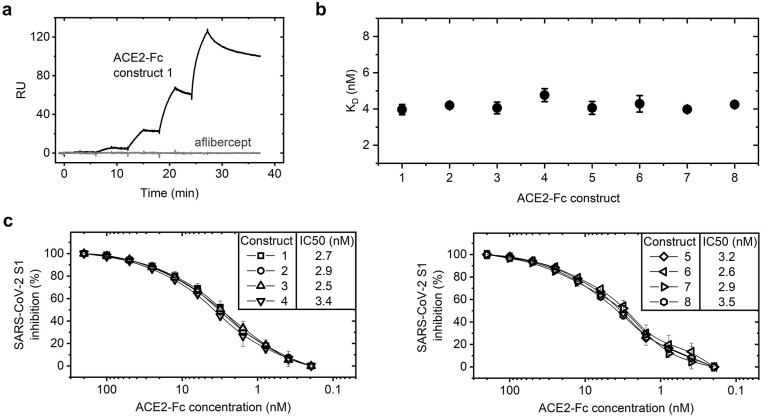
Surface plasmon resonance (SPR) allowed us to determine the binding affinity of our ACE2-Fc constructs to the RBD of the spike protein of SARS-CoV-2 that was recombinantly expressed and immobilized. The ACE2-Fc fusion proteins bound in a concentration-dependent manner to the viral protein domain, while an unrelated Fc fusion protein (aflibercept) used as a control showed no interaction with the ligand (Fig. 3 a). The binding constants revealed that all constructs analyzed bind to the immobilized SARS-CoV-2 RBD with an equilibrium dissociation constant (KD) of around 4 nM (Fig. 3b). This indicated that the structural variations do not influence the interaction between the ACE2-Fc fusion proteins and the RBD of SARS-CoV-2.
Fig. 3.
Interaction of the ACE2-Fc constructs with the SARS-CoV-2 RBD. a Surface plasmon resonance (SPR) was performed to obtain binding curves of ACE2-Fc fusion constructs and an unrelated Fc fusion protein (aflibercept) to an immobilized RBD from SARS-CoV-2. An exemplary binding curve is shown (RU = Response Units). b Binding constants of the ACE2-Fc constructs towards the RBD of SARS-CoV-2 (Mean ± SD of triplicate measurements). c ACE2-Fc fusion proteins were pre-incubated with the SARS-CoV-2 spike S1 protein and tested in a competition ELISA for their ability to neutralize S1 binding to immobilized ACE2 protein. Potent inhibition of SARS-CoV-2 spike S1 protein by ACE2-IgG4-Fc constructs (left) and ACE2-IgG1-Fc constructs (right). Data are represented as means ± SD of at least two independent experiments.
Neutralizing activities of the ACE2-Fc fusion proteins against the SARS-CoV-2 spike protein were tested in a competition enzyme-linked immunoassay (ELISA). All ACE2-Fc constructs tested potently inhibited the binding of spike S1 protein of SARS-CoV-2 to ACE2 (Fig. 3c). Consistent with their affinities to the SARS-CoV-2 RBD, there were no significant differences between the different fusion proteins. The half-maximal inhibitory concentrations (IC50 values) for the SARS-CoV-2 spike S1 protein ranged from 2.5 to 3.5 nM.
3.4. Effect of the Fc part on ACE2-IgG-Fc binding to FcγRI, FcγRIIIa and FcRn
The Fc-part can be important for the interaction of the molecules with Fc-receptors. We therefore used SPR to test the interaction of two ACE2-IgG4-Fc and two ACE2-IgG1-Fc proteins with FcγRI and FcγRIIIa. The ACE2-IgG4-Fc fusion proteins showed slightly lower affinity to FcγRI when compared to the IgG1 counterparts (Table 1 ). The ACE2-IgG4-Fc showed no binding to FcγRIIIa, which contrasts with the ACE2-IgG1-Fc molecules (Table 1).
Table 1.
Binding affinities of ACE2-Fc constructs to Fc-receptors.
| Construct | FcγRI |
FcγRIIIa |
FcRn |
|---|---|---|---|
| KD (nM) | KD (μM) | KD (μM) | |
| 1 | 45 ± 0.2 | No binding | 2.9 ± 0.68 |
| 3 | 43 ± 5.2 | No binding | 3.9 ± 0.62 |
| 5 | 10 ± 2.2 | 0.6 ± 0.45 | 3.4 ± 0.05 |
| 7 | 11 ± 0.8 | 0.8 ± 0.54 | 3.5 ± 0.62 |
Mean ± SD of duplicate measurements.
IgG half-life is primarily regulated by binding to the neonatal Fc Receptor, FcRn, through the IgG's constant region, Fc. Therefore, we were interested in whether the ACE2-IgG-Fc constructs still bind to FcRn and used SPR to test the binding of two ACE2-IgG4-Fc and two ACE2-IgG1-Fc proteins to FcRn. All four constructs had similar affinity to the FcRn (Table 1) indicating that usage of the IgG4 Fc domain allows to preserve this feature of the Fc-fusions.
3.5. Neutralization of SARS-CoV-2-GFP by ACE2-IgG4-Fc
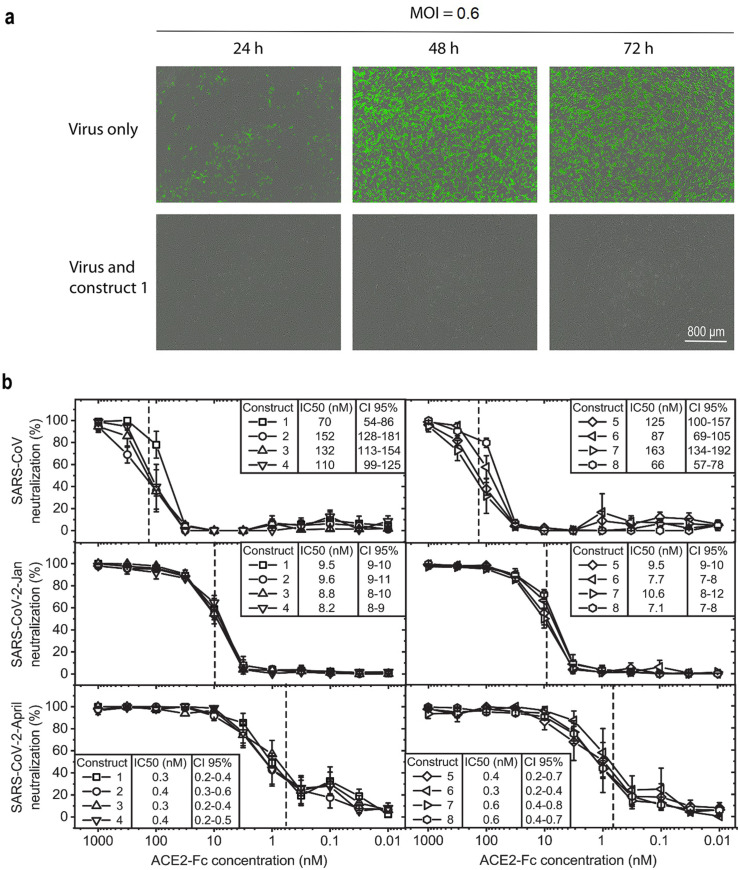
We next wanted to test whether the competition of our ACE2-Fc fusion constructs with binding of the SARS-CoV-2 via S1 to its receptor ACE2 translates into neutralization of infectious virus. Because of the expected favorable in vivo characteristics, we chose two IgG4-based ACE2-Fc fusion proteins, construct 1 and construct 3, for virus neutralization tested by live-cell imaging using the IncuCyte S3 platform in a biosafety level 3 laboratory. When Vero E6 cells were infected with SARS-CoV-2-GFP (Thi Nhu Thao et al., 2020) pre-incubated with serial dilutions of the two ACE2-IgG4-Fc fusion proteins (ACE2 Q18-G732 wildtype and ACE2 Q18-G732 H374N/H378N fused to IgG4-Fc S228P) the virus was neutralized in a concentration-dependent manner and no green fluorescent protein (GFP) expression was detected in contrast to cell layers showing increasing GFP expression when infected with the non-treated virus. (Fig. 4 a and Supplementary Movies). This demonstrates that construct 1 and construct 3 effectively inhibit SARS-CoV-2-GFP infection in vitro.
Fig. 4.
Effect of ACE2-Fc on SARS-CoV-2-GFP infection and inhibition of SARS-CoV-2 primary isolates. a ACE2-IgG4-Fc reduces SARS-CoV-2-GFP replication. Representative fluorescent images of Vero E6 cells infected with SARS-CoV-2-GFP (multiplicity of infection (MOI) = 0.6 IU/cell) pre-incubated with ACE2-IgG4-Fc fusion construct 1 (632 nM). b ACE2-Fc fusion proteins potently neutralize coronaviruses. Serial dilutions of ACE2-Fc fusion proteins were pre-incubated with different coronaviruses and tested for their ability to neutralize the virus before infection of Vero E6 cells. Neutralization of SARS-CoV (top), SARS-CoV-2-Jan (middle) and SARS-CoV-2-April (bottom) by ACE2-IgG4-Fc constructs (left) and ACE2-IgG1-Fc constructs (right) is shown. Data given are means ± SEM of three independent experiments each. 50% inhibitory concentrations (IC50) determined as well as the 95% confidence interval (CI 95%) are given for each construct. The dashed lines indicate the IC50 values on the corresponding curves.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.antiviral.2021.105197
The following is/are the supplementary data related to this article:
3.6. Efficient neutralization of primary SARS-CoV-2 isolates including major variants ofconcern
To determine whether the ACE2-Fc constructs neutralize different isolates of SARS-CoV-2, we compared the neutralization capacity of each of the eight ACE2-Fc fusion constructs against a range of primary isolates from patients. Hereby, we also included the original SARS-CoV isolate from 2003 (Drosten et al., 2003) as well as two different SARS-CoV-2 strains isolated from patients in January and April 2020 carrying the D614G mutation in the viral spike protein. First, all eight ACE2-IgG-Fc fusion proteins developed against the pandemic SARS-CoV-2 also neutralized the first human pathogenic SARS-CoV circulating in 2003 (Drosten et al., 2003) with 50% inhibitory concentration (IC50 values) in the range of 90–250 nM (Fig. 4b).
Second, to test the ACE2-Fc constructs for their ability to neutralize SARS-CoV-2, we evaluated the antiviral activity against SARS-CoV-2-Jan isolated from the earliest documented COVID-19 patient in Germany (Bohmer et al., 2020; Rothe et al., 2020), which was directly connected to the initial outbreak in Wuhan, China. All ACE2-Fc fusion proteins displayed strong neutralizing potential against SARS-CoV-2-Jan (Fig. 4b) with IC50 values in the range of 7–11 nM.
Third, the ACE2-Fc fusion protein variants were also compared for their ability to neutralize a second SARS-CoV-2 isolate, SARS-CoV-2-April, which was isolated when the virus was massively spreading in Europe. SARS-CoV-2-April displayed a different plaque-forming phenotype than SARS-CoV-2-Jan (Supplementary Fig. 1). All ACE2-Fc constructs displayed significantly increased neutralizing potential against SARS-CoV-2-April (Fig. 4b) with IC50 in the picomolar range. The comparison of IC50 values showed an increasing neutralizing potential of ACE2-Fc fusion proteins, the more the SARS coronavirus evolved to allow for more efficient spread in the community.
3.7. Increasing antiviral potential of ACE2-IgG4-Fc against emerging SARS-CoV-2 variants of concern
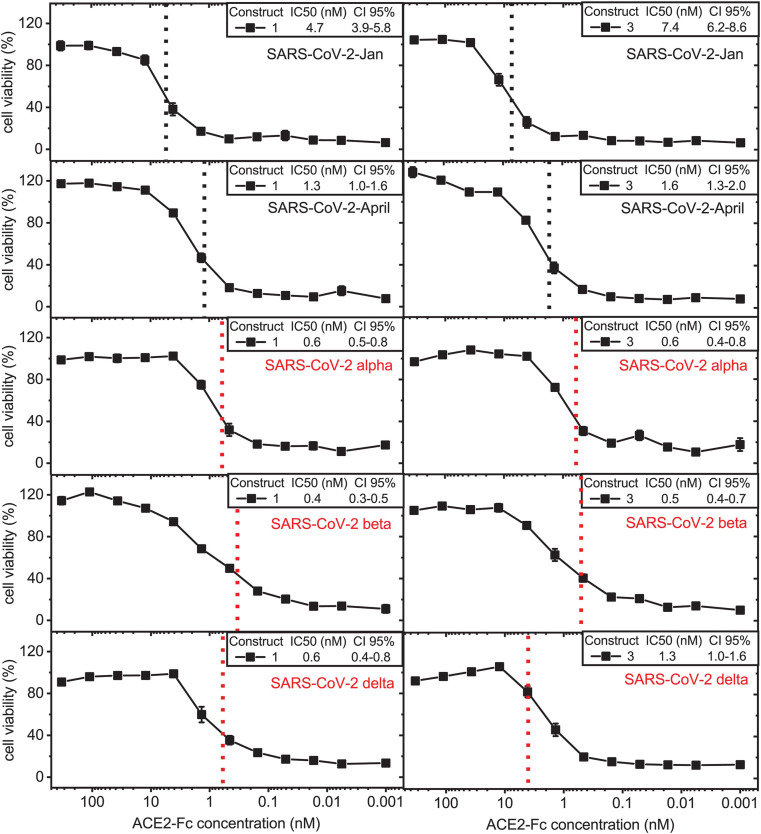
To confirm our results obtained in VeroE6 cells, we determine whether the ACE2-Fc fusion proteins can prevent cell toxicity resulting from infection with SARS-CoV-2 VoC in human alveolar basal epithelium-derived A549-hACE2 cells. A549-hACE2 cells were challenged with the different ACE2-Fc fusion protein-pre-treated VoCs. Because of their expected favorable in vivo features, we chose the two IgG4-based ACE2-Fc fusion proteins, construct 1 and construct 3 (Fig. 5 ). Both ACE2-IgG4-Fc constructs entirely prevented SARS-CoV-2-induced cytotoxicity and, therefore, potently neutralized SARS-CoV-2-Jan (Fig. 5) and SARS-CoV-2-April (Fig. 5) with IC50 values in the range of 1–7 nM confirming previous results (Fig. 4b).
Fig. 5.
Neutralization potency of ACE2-IgG4-Fc fusion proteins increases with evolution of pandemic SARS-CoV-2 variants. Serial dilutions of ACE2-IgG4-Fc fusion constructs 1 and 3 were pre-incubated with the indicated SARS-CoV-2 primary isolates or VoCs and tested for their ability to prevent cytotoxicity following infection of A549-hACE2 cells. Neutralization of SARS-CoV-2-Jan, SARS-CoV-2-April and SARS-CoV-2 VoCs alpha, beta and delta by enzymatically active ACE2-IgG4-Fc construct 1 (left) and enzymatically inactive ACE2-IgG4-Fc construct 3 (right). Data given are means ± SEM of three independent experiments each. 50% inhibitory concentrations (IC50 values) determined as well as the 95% confidence interval (CI 95%) are shown for each construct. The dashed lines indicate the IC50 values on the corresponding curves.
In addition, ACE2-IgG4-Fc constructs neutralized the SARS-CoV-2 VoC alpha showing significantly increased neutralizing potential in comparison to SARS-CoV-2-Jan and April reaching IC50 values in the picomolar range (Fig. 5, middle panel). Excitingly, ACE2-IgG4-Fc fusion constructs displayed an even higher antiviral potential against the SARS-CoV-2 VoC beta, which is known to exhibit the strongest immune escape potential of all SARS-CoV-2 strains known so far (Fig. 5, second lowest panel) and also proved resistant to neutralization by an anti-RBD monoclonal antibody in our experiments (Supplementary Fig. 2). Finally, ACE2-IgG4-Fc also potently neutralized the SARS-CoV-2 VoC delta variant (Fig. 5, lowest panel). In contrast, an unrelated IgG4 antibody did not show any virus neutralization in our assays (Supplementary Fig. 3).
Taken together, ACE2-IgG4-Fc constructs show an increasing capacity to neutralize SARS-CoV-2 with increasing adaptation of the virus to the human population including the major VoCs.
4. Discussion
In this study, we designed, expressed and evaluated different ACE2-Fc fusion proteins with favorable biochemical features and demonstrate that they elicit a broad antiviral activity in the nano-molar or even picomolar range against human pathogenic beta-coronaviruses including SARS-CoV-2 VoCs as well as the 2003 SARS-CoV. Importantly, the more the SARS-CoV-2 spike protein evolved and diversified in the pandemic, and the more problematic immune escape became, the more efficient these virus isolates were inhibited by our ACE2-Fc fusion proteins.
All ACE2-IgG4-Fc and ACE2-IgG1-Fc fusion proteins designed inhibited infection of Vero E6 cells with the original SARS-CoV from 2003 with IC50 values in the range of 90–250 nM. A SARS-CoV-2 isolate from January 2020, which was obtained from the earliest documented COVID-19 cases in Germany, and thus is closely related to the original Wuhan strain (Bohmer et al., 2020; Rothe et al., 2020; Wolf et al., 2020), was inhibited with IC50 values of around 10 nM. Infection by the SARS-CoV-2-April variant predominantly circulating worldwide was prevented even more efficiently. Most importantly, our ACE2-IgG4-Fc constructs prevented infection and cell toxicity of the most infectious SARS-CoV-2 variant known so far, the alpha strain, and the variants of highest concern world-wide, the beta and delta variants reaching picomolar IC50. This demonstrates that, in contrast to therapeutic antibodies targeting the viral spike protein, the virus cannot escape neutralization by or ACE2-IgG4-Fc fusion construct.
Surveillance of SARS-CoV-2 variants emerging over time identified genomic regions with increasing genetic variation (Islam et al., 2020; Khan et al., 2020; Mercatelli and Giorgi, 2020). A D614G substitution in the C-terminal region stabilizing the spike protein is associated with an improved ability of the virus to bind its receptor, ACE2, and rapidly became the most prevalent variant worldwide (Fernandez, 2020; Korber et al., 2020). Over time, novel and even more infectious SARS-CoV-2 strains emerged, the alpha variant being the prime example (Santos and Passos, 2021). Our results indicate that both the ACE2-IgG4-Fc fusion proteins and the ACE2-IGg1-Fc fusion proteins neutralize these pandemic SARS-CoV-2 variants even more potently than the early SARS-CoV-2, which originated from Wuhan. The lower efficacy against the original SARS-CoV from 2003 is in line with the fact that the affinity of the SARS-CoV-2 spike protein for ACE2 is significantly higher than the affinity of the SARS-CoV spike protein (Wrapp et al., 2020). It, however, also indicates, that the VoCs evolving because they can bind more efficiently to ACE2 and therefore become more infectious can be targeted with our ACE2-IgG4 fusion protein even more efficiently. This helps to prepare for new potentially pandemic viruses stemming from animal reservoirs, as it happened recently with “cluster 5″, a SARS-CoV-2 variant with a combination of mutations still investigated for their impact on disease severity and resistance to vaccination and therapeutic antibodies (Mallapaty, 2020; Oude Munnink et al., 2021).
Our findings are highly relevant in the context of the emerging SARS-CoV-2 variants that at least partially escape neutralization by therapeutic antibodies (Hu et al., 2021; Wang et al., 2021). The use of one of the first antibodies used in COVID-19 therapy, bamlanivimab, was even halted by the US-FDA only four months after it had gained authorization for emergency use. The FDA is now recommending to develop and use antibodies for combination therapies targeting different epitopes due to viral resistance concerns. However, it is probably only a matter of time until we see SARS-CoV-2 variants emerging that escape multiple therapeutic antibodies. Unlike antibodies, our ACE2-IgG4-Fc fusion protein will retain its potent virus neutralization efficiency as it is vital for the virus to retain its receptor binding capacity. Therefore, coronavirus variants that mutate to escape ACE2-Fc-binding will lose fitness and become less infectious.
In addition to the alpha VoC, the emergence of SARS-CoV-2 VoCs which are able to at least partially escape immunity raises additional concerns, namely those which carry a mutation of amino acid 484 such as the beta, the gamma and the most recent delta variant detected in India. SARS-CoV-2 VoC alpha has been shown to be refractory to neutralization by most monoclonal antibodies against the N-terminal domain of the spike protein and is relatively resistant to a few monoclonal antibodies against the receptor-binding domain (Wang et al., 2021) while it is still sensitive to plasma from individuals who have recovered from COVID-19 or sera from individuals who have been vaccinated against SARS-CoV-2. The beta and delta VoC are not only refractory to neutralization by most monoclonal antibodies directed against the N-terminal domain of the spike protein but also by multiple individual monoclonal antibodies against the receptor-binding motif. In addition, both variants show a worrisome escape from neutralization by convalescent plasma and sera from individuals who have been vaccinated (Garcia-Beltran et al., 2021; Planas et al., 2021; Wang et al., 2021). Our data strongly indicate that all these variants can still be targeted by the engineered ACE2-IgG4 fusion constructs.
ACE2 plays a central role in the homeostatic control of cardio-renal actions and has been shown to protect against severe acute lung injury and acute angiotensin II-induced hypertension (Tikellis and Thomas, 2012; Wysocki et al., 2010). A soluble dimer (APN019) has been safely tested in a clinical phase I study in healthy volunteers and in a phase II study in patients with an acute respiratory distress syndrome (Haschke et al., 2013; Khan et al., 2017). It is currently being tested for its therapeutic effect in a phase II study in COVID-19 patients (Zoufaly et al., 2020). We used the full length ACE2 ectodomain sequence Q18-S740 and a shortened version comprising Q18-G732. Most importantly, binding to the SARS-CoV-2 viral spike protein to both variants was comparable with a KD of 4 nM.
Pharmacokinetic studies in mice and humans revealed that recombinant human ACE2 exhibits fast clearance rates resulting in a short half-life of only a few hours (Haschke et al., 2013; Wysocki et al., 2010; Zoufaly et al., 2020). When the extracellular domain of murine ACE2 was fused to an Fc fragment of murine immunoglobulin IgG the ACE2-Fc fusion protein demonstrated a prolonged in vivo half-life and effective organ protection in murine models of both, acute and chronic angiotensin II-dependent hypertension (Liu et al., 2018).
ACE2-Fc fusion proteins are homodimers stabilized by disulfide bonds in the hinge region, similar to IgGs (Bernardi et al., 2020). Functional variants of ACE2-IgG1-Fc fusion proteins have been described in the literature (Glasgow et al., 2020; Huang et al., 2020; Iwanaga et al., 2020; Lei et al., 2020; Liu et al., 2020b; Lui et al., 2020), and a first fusion protein designed with an IgG1 Fc portion to prolong the circulating half-life is currently in a phase II clinical study by Hengenix Biotech Inc. In our study, novel ACE2-Fc fusion proteins were designed in which the ACE2 domain is fused to the Fc fragment of human IgG4 (ACE2-IgG4-Fc) containing a stabilizing S228P mutation in the hinge region. Although ACE2 domain and the Fc part most likely fold independently, both Fc domains could be used to obtain stable fusion proteins and neither the interaction of ACE2 with the virus spike protein nor its enzymatic function was affected. Thus, engineering general features of the fusion protein is possible without interfering with spike protein interaction.
The presence of the Fc domain could markedly increase the plasma half-life of ACE2-Fc due to the interaction of the Fc domain with the neonatal Fc-receptor (FcRn), and therefore a slower renal clearance of the fusion molecule. FcRn is broadly expressed on many cell types including endothelial cells and respiratory epithelial cells (Latvala et al., 2017; Sockolosky and Szoka, 2015). Binding to FcRn could extend the systemic half-life by chaperoning bound Fc fusion proteins away from lysosomal degradation. In addition, FcRn transports IgG and Fc fusion molecules across mucosal barriers into the lumen of the respiratory and intestinal tract thereby providing dynamic trafficking between circulating and luminal IgG molecules at mucosal sites (Sockolosky and Szoka, 2015; Tzaban et al., 2009). Here we showed that the ACE2-IgG4-Fc has preserved binding to FcRn with the same affinity as the ACE2-IgG1-Fc counterparts. On the other hand, the ACE2-IgG4-Fc did not bind to CD16 (FcγRIIIa) in contrast to ACE2-IgG1-Fc, although the constant heavy chain regions of the different IgG subclasses share over 95% sequence homology. IgG4 in particular has poor ability to engage C1q and Fc gamma receptors and has been associated with anti-inflammatory properties. Most importantly, using our design, ACE2-IgG4-Fc fusion constructs show equally high binding affinity to the SARS-CoV-2 RBD and spike neutralizing the virus with IC50 values in the picomolar range.
The SARS-CoV-2 pandemic has caused an unprecedented challenge to develop COVID-19 therapies. However, progress with antiviral drugs has been slow. Here we show that ACE2-IgG4-Fc fusion proteins have favorable biophysical and pharmaceutical characteristics and significant in vitro SARS-CoV-2 neutralizing potency. The Fc part from IgG4 could bring clinical benefits for ACE2-Fc proteins by avoiding potential antibody-dependent disease enhancement during treatment. In addition, our data showed that ACE2-IgG4-Fc fusion proteins displayed increasing neutralizing potential, the further the SARS-CoV variant adapted to its human host becoming more infectious or escaping immune responses. For the currently emerging SARS-CoV-2 VoCs, alternative therapies are urgently needed, and ACE2-Fc fusion proteins certainly are interesting candidates. Thus, our ACE2-IgG4-Fc fusion protein is a promising candidate not only for therapeutic use in the current SARS-CoV-2 pandemic including more infectious SARS-CoV-2 VoC but also for future coronavirus infectious diseases. Upcoming studies will reveal whether the promising results presented herein can be translated to a therapeutic efficiency in vivo.
Author contributions
J.B., C.B. and U.P. developed the concept and designed the experiments. H.S., J.S., L.W., M.S., designed and performed experiments and analyzed the data. A.R. designed the constructs, supervised construct manufacturing and supervised physicochemical and functional analysis. C.-C.C, V.G., M.F. and O.T.K. provided critical reagents and methods. F.-P. W., N.S., S.P. and F.W. gave conceptual support and supervised experiments. H.S., J.S., A.R., F.-P. W., J.B. C.B. and U.P. wrote and revised the manuscript.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: A patent application has been filed by Formycon AG for the content disclosed in this study. The authors A.R., F.-P. W., N.S., S.P., F.W., and C.B. are employees of Formycon AG. J.B. is advisory board member of Formycon AG. Remaining authors declare no conflict of interest. U.P. is shareholder and member of the board of SCG Cell Therapy seeking a license from Formycon to co-develop the product. Remaining authors declare no conflict of interest.
Acknowledgements
The authors like to thank Polpharma Biologics Utrecht B.V. for performing the transient transfections and providing the fusion molecules. SARS-CoV-2 Spike S1 Inhibition ELISA was performed in collaboration with TebuBio, France. Binding to Fc receptors was performed in collaboration with Vela Laboratories, Austria. We are grateful to Volker Thiel, University of Bern, Switzerland, for providing the SARS-CoV-2-GFP, to Friedemann Weber, University of Giessen, Germany, for providing SARS-CoV (Frankfurt 1), and to Dr. Joachim J. Bugert, Institut für Mikrobiologie der Bundeswehr, for the alpha (B.1.1.7) variant.
The study was supported by grant AZ-1433-20 of the Bayerische Forschungsstiftung (Formycon AG, TUM) and AZ-1459-20C (O.T. Keppler), by the President's fund of the Helmholtz Association (CoViPa) and by the FOR-COVID network financed by the Ministry of Science and Arts (StMWK) of the State of Bavaria. H.L.S. was supported by a postdoctoral fellowship from the Peter und Traudl Engelhorn Stiftung.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2021.105197.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aalberse R.C., Schuurman J. IgG4 breaking the rules. Immunology. 2002;105:9–19. doi: 10.1046/j.0019-2805.2001.01341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer A., Kehn-Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J Vis Exp. 2014 doi: 10.3791/52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci. (Lond.) 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Huang Y., Harris B., Xiong Y., Nandi S., McDonald K.A., Faller R. Development and simulation of fully glycosylated molecular models of ACE2-Fc fusion proteins and their interaction with the SARS-CoV-2 spike protein binding domain. PLoS One. 2020;15 doi: 10.1371/journal.pone.0237295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmer M.M., Buchholz U., Corman V.M., Hoch M., Katz K., Marosevic D.V., Bohm S., Woudenberg T., Ackermann N., Konrad R., Eberle U., Treis B., Dangel A., Bengs K., Fingerle V., Berger A., Hormansdorfer S., Ippisch S., Wicklein B., Grahl A., Portner K., Muller N., Zeitlmann N., Boender T.S., Cai W., Reich A., An der Heiden M., Rexroth U., Hamouda O., Schneider J., Veith T., Muhlemann B., Wolfel R., Antwerpen M., Walter M., Protzer U., Liebl B., Haas W., Sing A., Drosten C., Zapf A. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect. Dis. 2020;20:920–928. doi: 10.1016/S1473-3099(20)30314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell L.M., Johnston C.I., Tikellis C., Cooper M.E. ACE2, a new regulator of the renin-angiotensin system. Trends Endocrinol. Metabol. 2004;15:166–169. doi: 10.1016/j.tem.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Osterlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia I.R. Stability of IgG isotypes in serum. MAbs. 2010;2:221–232. doi: 10.4161/mabs.2.3.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y., Scholey J., Ferrario C.M., Manoukian A.S., Chappell M.C., Backx P.H., Yagil Y., Penninger J.M. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- de Taeye S.W., Bentlage A.E.H., Mebius M.M., Meesters J.I., Lissenberg-Thunnissen S., Falck D., Senard T., Salehi N., Wuhrer M., Schuurman J., Labrijn A.F., Rispens T., Vidarsson G. FcgammaR binding and ADCC activity of human IgG allotypes. Front. Immunol. 2020;11:740. doi: 10.3389/fimmu.2020.00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z., Geng J., Cai J., Han H., Li X., Kang W., Weng D., Liang P., Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Dumet C., Pottier J., Gouilleux-Gruart V., Watier H. Insights into the IgG heavy chain engineering patent landscape as applied to IgG4 antibody development. MAbs. 2019;11:1341–1350. doi: 10.1080/19420862.2019.1664365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. Structural impact of mutation D614G in SARS-CoV-2 spike protein: enhanced infectivity and therapeutic opportunity. ACS Med. Chem. Lett. 2020;11:1667–1670. doi: 10.1021/acsmedchemlett.0c00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., Murali M.R., Alter G., Charles R.C., Dighe A., Branda J.A., Lennerz J.K., Lingwood D., Schmidt A.G., Iafrate A.J., Balazs A.B. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488. doi: 10.1016/j.cell.2020.12.015. e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow A., Glasgow J., Limonta D., Solomon P., Lui I., Zhang Y., Nix M.A., Rettko N.J., Zha S., Yamin R., Kao K., Rosenberg O.S., Ravetch J.V., Wiita A.P., Leung K.K., Lim S.A., Zhou X.X., Hobman T.C., Kortemme T., Wells J.A. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28046–28055. doi: 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handlogten M.W., Peng L., Christian E.A., Xu W., Lin S., Venkat R., Dall'Acqua W., Ahuja S. Prevention of Fab-arm exchange and antibody reduction via stabilization of the IgG4 hinge region. MAbs. 2020;12 doi: 10.1080/19420862.2020.1779974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschke M., Schuster M., Poglitsch M., Loibner H., Salzberg M., Bruggisser M., Penninger J., Krahenbuhl S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin. Pharmacokinet. 2013;52:783–792. doi: 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- Higuchi Y S.T., Arimori T., Ikemura N., Mihara E., Kirita Y., Ohgitani E., Mazda O., Motooka D., Nakamura S., Sakai Y., Itoh Y., Sugihara F., Matsuura Y., Matoba S., Okamoto T., Takagi J., Hoshino A. Engineered ACE2 receptor therapy overcomes mutational escape of SARS-CoV-2. Nat. Commun. 2021;12:3802. doi: 10.1038/s41467-021-24013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Peng P., Wang K., Fang L., Luo F.-y., Jin A.-s., Liu B.-z., Tang N., Huang A.-l. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.Y., Lin M.S., Kuo T.C., Chen C.L., Lin C.C., Chou Y.C., Chao T.L., Pang Y.H., Kao H.C., Huang R.S., Lin S., Chang S.Y., Yang P.C. EMBO Mol Med n/a; 2020. Humanized COVID-19 Decoy Antibody Effectively Blocks Viral Entry and Prevents SARS-CoV-2 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., Alam A., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga N., Cooper L., Rong L., Beddingfield B., Crabtree J., Tripp R.A., Kolls J.K. Novel ACE2-IgG1 fusions with improved activity against SARS-CoV2. bioRxiv. 2020;2020 doi: 10.1016/j.isci.2021.103670. 2006.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarzadeh A., Chauhan P., Saha B., Jafarzadeh S., Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Yang J., Zhang Y., Dong M., Wang S., Zhang Q., Liu F.F., Zhang K., Zhang C. Angiotensin-converting enzyme 2 and angiotensin 1-7: novel therapeutic targets. Nat. Rev. Cardiol. 2014;11:413–426. doi: 10.1038/nrcardio.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Benthin C., Zeno B., Albertson T.E., Boyd J., Christie J.D., Hall R., Poirier G., Ronco J.J., Tidswell M., Hardes K., Powley W.M., Wright T.J., Siederer S.K., Fairman D.A., Lipson D.A., Bayliffe A.I., Lazaar A.L. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care. 2017;21:234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.I., Khan Z.A., Baig M.H., Ahmad I., Farouk A.E., Song Y.G., Dong J.J. Comparative genome analysis of novel coronavirus (SARS-CoV-2) from different geographical locations and the effect of mutations on major target proteins: an in silico insight. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse R.L. 2020. Therapeutic Strategies in an Outbreak Scenario to Treat the Novel Coronavirus Originating in Wuhan, China. F1000Res 9; p. 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Latvala S., Jacobsen B., Otteneder M.B., Herrmann A., Kronenberg S. Distribution of FcRn across species and tissues. J. Histochem. Cytochem. 2017;65:321–333. doi: 10.1369/0022155417705095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehr B., Robinson J., Burdick R., Tholudur A. Evaluation of a variable-pathlength spectrophotometer: a comparable instrument for determining protein concentration. BioProcess Int. 2015;13:46–51. [Google Scholar]
- Lei C., Qian K., Li T., Zhang S., Fu W., Ding M., Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020;11:2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.Y., Zheng B., Zhang Y., Li J.P. Role and mechanism of angiotensin-converting enzyme 2 in acute lung injury in coronavirus disease 2019. Chronic Dis Transl Med. 2020;6:98–105. doi: 10.1016/j.cdtm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Wysocki J., Souma T., Ye M., Ramirez V., Zhou B., Wilsbacher L.D., Quaggin S.E., Batlle D., Jin J. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 2018;94:114–125. doi: 10.1016/j.kint.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Liu P., Xie X., Gao L., Jin J. Designed variants of ACE2-Fc that decouple anti-SARS-CoV-2 activities from unwanted cardiovascular effects. Int. J. Biol. Macromol. 2020;165:1626–1633. doi: 10.1016/j.ijbiomac.2020.10.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Cassis L.A., Kooi C.W., Daugherty A. Structure and functions of angiotensinogen. Hypertens. Res. 2016;39:492–500. doi: 10.1038/hr.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui I., Zhou X.X., Lim S.A., Elledge S.K., Solomon P., Rettko N.J., Zha B.S., Kirkemo L.L., Gramespacher J.A., Liu J., Muecksch F., Lorenzi J.C.C., Schmidt F., Weisblum Y., Robbiani D.F., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D., Rosenburg O.S., Leung K.K., Wells J.A. Trimeric SARS-CoV-2 Spike interacts with dimeric ACE2 with limited intra-Spike avidity. bioRxiv. 2020;2020 2005.2021. [Google Scholar]
- Mallapaty S. COVID mink analysis shows mutations are not dangerous - yet. Nature. 2020;587:340–341. doi: 10.1038/d41586-020-03218-z. [DOI] [PubMed] [Google Scholar]
- Manickam C., Sugawara S., Reeves R.K. Friends or foes? The knowns and unknowns of natural killer cell biology in COVID-19 and other coronaviruses in July 2020. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., Bouwmeester-Vincken N., Harders F., Hakze-van der Honing R., Wegdam-Blans M.C.A., Bouwstra R.J., GeurtsvanKessel C., van der Eijk A.A., Velkers F.C., Smit L.A.M., Stegeman A., van der Poel W.H.M., Koopmans M.P.G. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Pere H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Loriere E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wolfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.C., Passos G.A. The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. bioRxiv. 2021;2020 2012.2029. [Google Scholar]
- Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., Campagnole-Santos M.J. The ACE2/angiotensin-(1-7)/MAS Axis of the renin-angiotensin system: focus on angiotensin-(1-7) Physiol. Rev. 2018;98:505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapin G., Yang X., Prosise W.W., McCoy M., Reichert P., Johnston J.M., Kashi R.S., Strickland C. Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab. Nat. Struct. Mol. Biol. 2015;22:953–958. doi: 10.1038/nsmb.3129. [DOI] [PubMed] [Google Scholar]
- Sockolosky J.T., Szoka F.C. The neonatal Fc receptor, FcRn, as a target for drug delivery and therapy. Adv. Drug Deliv. Rev. 2015;91:109–124. doi: 10.1016/j.addr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Nhu Thao T., Labroussaa F., Ebert N., V'Kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., Gultom M., Schmied K., Laloli L., Husser L., Wider M., Pfaender S., Hirt D., Cippa V., Crespo-Pomar S., Schroder S., Muth D., Niemeyer D., Corman V.M., Muller M.A., Drosten C., Dijkman R., Jores J., Thiel V. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Tikellis C., Thomas M.C. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int J Pept. 2012;2012 doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- Tzaban S., Massol R.H., Yen E., Hamman W., Frank S.R., Lapierre L.A., Hansen S.H., Goldenring J.R., Blumberg R.S., Lencer W.I. The recycling and transcytotic pathways for IgG transport by FcRn are distinct and display an inherent polarity. J. Cell Biol. 2009;185:673–684. doi: 10.1083/jcb.200809122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292 e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wolf G.K., Glueck T., Huebner J., Muenchhoff M., Hoffmann D., French L.E., Keppler O.T., Protzer U. Clinical and epidemiological features of a family cluster of symptomatic and asymptomatic severe acute respiratory syndrome coronavirus 2 infection. J Pediatr. Infect Dis Soc. 2020;9:362–365. doi: 10.1093/jpids/piaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Deng W., Li S., Yang X. Advances in research on ACE2 as a receptor for 2019-nCoV. Cell. Mol. Life Sci. 2020;78:531–544. doi: 10.1007/s00018-020-03611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki J., Ye M., Rodriguez E., Gonzalez-Pacheco F.R., Barrios C., Evora K., Schuster M., Loibner H., Brosnihan K.B., Ferrario C.M., Penninger J.M., Batlle D. Targeting the degradation of angiotensin II with recombinant angiotensin-converting enzyme 2: prevention of angiotensin II-dependent hypertension. Hypertension. 2010;55:90–98. doi: 10.1161/HYPERTENSIONAHA.109.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoufaly A., Poglitsch M., Aberle J.H., Hoepler W., Seitz T., Traugott M., Grieb A., Pawelka E., Laferl H., Wenisch C., Neuhold S., Haider D., Stiasny K., Bergthaler A., Puchhammer-Stoeckl E., Mirazimi A., Montserrat N., Zhang H., Slutsky A.S., Penninger J.M. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.