Abstract
Herpesviruses are ubiquitous, double-stranded DNA, enveloped viruses that establish lifelong infections and cause a range of diseases. Entry into host cells requires viral binding to specific receptors followed by the coordinated action of multiple viral entry glycoproteins to trigger membrane fusion. Although the core fusion machinery is conserved for all herpesviruses, each species uses distinct receptors and receptor-binding glycoproteins. Structural studies of the prototypical herpesviruses herpes simplex virus 1 (HSV-1), HSV-2, human cytomegalovirus (HCMV), and Epstein–Barr virus (EBV) entry glycoproteins have defined the interaction sites for glycoprotein complexes and receptors, as well as revealed conformational changes that occur upon receptor binding. Recent crystallography and electron microscopy studies have refined our model of herpesvirus entry into cells, clarifying both the conserved and unique features. In this Review, we discuss recent insights into herpesvirus entry by analyzing the structures of entry glycoproteins, including the diverse receptor-binding glycoproteins (HSV-1 gD, EBV gp42, and HCMV gH–gL–gO trimer and gH–gL–UL128–UL130–UL131A pentamer, as well gH–gL and the fusion protein gB, which are conserved in all herpesviruses.
Table of contents blurb
Recent crystallography and electron microscopy studies have refined our model of herpesvirus entry into cells. In this Review, Connolly, Jardetzky and Longnecker discuss recent insights into herpesvirus entry by analyzing the structures of entry glycoproteins, including the diverse receptor-binding glycoproteins and conserved fusion proteins.
Introduction
The Herpesviridae are a family of large, double-stranded DNA, enveloped viruses that cause a range of diseases. The nine human herpesviruses include herpes simplex virus 1 (HSV-1), HSV-2, varicella zoster virus (VZV), human cytomegalovirus (HCMV), human herpesvirus 6A (HHV6A), HHV6B, Epstein–Barr virus (EBV), HHV7, and Kaposi’s sarcoma herpesvirus (KSHV). Herpesvirus infections are ubiquitous and establish lifelong latency in infected hosts.
Despite infecting a variety of cell types, entry into host cells occurs through a conserved mechanism. This Review will focus on structural studies of entry for prototypical viruses from each subfamily: HSV-1 and HSV-2 from the Alphaherpesvirinae, HCMV from the Betaherpesvirinae, and EBV from the Gammaherpesvirinae. Extensive structural studies of these three viruses have been performed and the structures of multiple entry glycoproteins have been resolved for each virus allowing general features of herpesviruses to be discerned as well as virus specific features.
All three herpesviruses infect a large percentage of the human population. HSV-1 and HSV-2 most commonly cause mucocutaneous lesions in the oral or genital regions, but infection also can cause encephalitis or meningitis under rare circumstances. HCMV infection during childhood usually is asymptomatic; however, primary infection later in life can result in serious complications for fetal development in pregnant women, for transplant recipients, and for immune-compromised people. EBV typically causes infectious mononucleosis, but infection also can result in malignancies, including Burkitt’s and Hodgkin’s lymphoma. An understanding of the entry mechanisms for these viruses may provide a basis for the design of antiviral drug candidates and/or subunit vaccines.
This Review explains the mechanism of herpesvirus entry into cells, as outlined in multiple structural studies of the entry glycoproteins required by each representative virus. The structures of the distinct receptor-binding proteins from each virus will be compared, including HSV-1 glycoprotein D (gD), EBV glycoprotein 42 (gp42), and the HCMV gH–gL–gO trimer and gH–gL–UL128–UL130–UL131A pentamer. Structures of the conserved entry glycoproteins gH–gL and the fusion protein gB, examined using electron microscopy (EM) and crystallography, also are described.
These structural studies are linked to our understanding of the conformational changes and glycoprotein interactions required for entry to provide insight into how the fusion machinery drives virus entry into cells. These studies have broad implications because they provide targets to develop efficacious vaccines to prevent herpesvirus infections and associated disease.
Entry mechanism
Herpesvirus entry into cells requires the coordinated interaction of multiple glycoproteins on the surface of the virion. The initial attachment of a virus to a host cell tethers the virus to the cell, but does not trigger entry. This attachment is mediated by multiple viral glycoproteins and a variety of binding receptors. This Review focuses on the required entry events that occur after this attachment, including interactions among the glycoproteins and entry receptors that trigger membrane fusion and virus entry.
For herpesviruses, entry receptor binding and membrane fusion functions are performed by multifunctional viral glycoproteins1-3. The heterodimer gH–gL and the viral fusion protein gB represent a core set of entry glycoproteins that is required for all herpesviruses. By contrast, different herpesvirus subfamilies use distinct viral glycoprotein combinations to bind to various entry receptors. Even within a single virus species, different receptor-binding glycoprotein complexes may be required to mediate entry into different cell types.
In the current model of entry, binding to an entry receptor triggers conformational changes in the viral glycoproteins that signal to gB, the fusion protein, to execute membrane fusion (Fig. 1). Interactions among some of the entry glycoproteins, especially interactions with gB, have been difficult to capture, potentially because they are transient and/or low affinity.
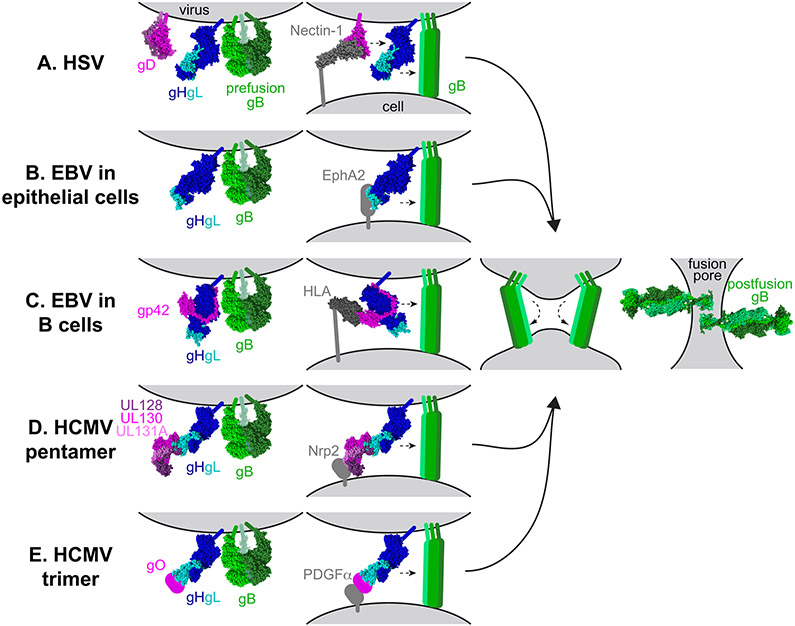
Fig. 1. Model of the herpesvirus entry mechanism.
a∣ Herpes simplex viruses 1 and 2 (HSV-1 and HSV-2) fuse with a host cell at the plasma or endosomal membrane. The glycoprotein D (gD) dimer (pink), gH–gL heterodimer (dark and light blue), and gB trimer (green) are necessary and sufficient for entry (column 1). gD binds to one of several entry receptors, including nectin-1 (grey, column 2). Receptor binding displaces the carboxyl-terminus of the gD ectodomain and transmits a signal to gH–gL (small arrow). gH–gL activates the fusion protein gB (small arrow) to insert hydrophobic fusion loops into the cell membrane. b∣ Epstein–Barr virus (EBV) fuses with the plasma membrane of an epithelial cell. gH–gL (blue) and gB (green) are sufficient for fusion (column 1). The binding of gH–gL to EphA2 (grey) triggers gB to insert into the host cell (column 2). c∣ EBV fusion with B cells occurs in the endosome. A complex of gp42 (pink) and gH–gL (blue) binds to human leukocyte antigen (HLA) class II (grey). The binding triggers gB and may impact membrane orientation (column 2). d∣ Human cytomegalovirus (HCMV) entry into epithelial and endothelial cells occurs after endocytosis and requires a pentamer complex of gH–gL (blue) bound to UL128–UL130–UL131A (shades of pink) (column 1). Pentamer binding to Neuropilin-2 (Nrp2) triggers gB (column 2). e∣ HCMV entry into all cells requires a trimer complex comprised of gO and gH–gL (column 1). In fibroblasts, the trimer binds to platelet-derived growth factor receptor α (PDGFRα) and triggers gB at the plasma membrane (column 2). The reason that the trimer is required for entry into epithelial and endothelial cells is unclear currently. After inserting into the target cell membrane, gB folds back on itself (column 3). Fusion most likely requires more than one gB trimer to be triggered. The other entry glycoproteins are removed for the figure for clarity. As gB refolds into its postfusion conformation, the viral and cell membranes are fused, creating a fusion pore, through which the viral capsid can enter the cell (column 4).
For HSV-1, gD serves the receptor-binding function, as described below (Table 1). gD binding to receptor prompts an interaction between gD and gH–gL. gH–gL serves as a regulator of fusion that transmits a signal to gB4. Upon triggering, gB undergoes a conformational change that results in insertion into the host cell membrane followed by refolding to bring the cell and viral membranes together. The refolding of multiple gB trimers creates a pore in the membrane, allowing the viral capsid to enter the cellular cytoplasm and to be transported to the nucleus.
TABLE 1:
Herpesvirus receptor-binding glycoproteins and receptors
| Subfamily | Virus | Receptor-binding proteins |
Receptors |
|---|---|---|---|
| Alpha | Herpes simplex viruses 1 and 2 | gD | Herpesvirus entry mediator (HVEM), Nectin-1 and 3-O-sulfated heparan sulfate |
| Varicella zoster virus | gH–gL | Integrins | |
| Beta | Human cytomegalovirus | gH–gL–UL128–UL130–UL131A (pentamer) | Neuropilin-2 (Nrp2) |
| gH–gL–gO (trimer) | Platelet-derived growth factor receptor α (PDGFRα) | ||
| Human herpesviruses 6A and 6B | gH–gL–gQ1–gQ2 | CD46 (for HHV6A) CD134 (for HHV6B) | |
| gH–gL–gO | Unknown | ||
| Gamma | Epstein–Barr virus | gH–gL–gp42 | HLA class II |
| gH–gL | EphA2 | ||
| Kaposi’s sarcoma herpesvirus | gH–gL | EphA2 and EphA4 |
EBV enters cells in a similar manner, except that gp42 serves as the receptor-binding protein for EBV entry into B cells, instead of gD. gp42 forms a stable complex with gH–gL and binding of the gp42–gH–gL complex to receptor signals gB to mediate fusion. By contrast, for EBV entry into epithelial cells, gH–gL binds to receptor directly before signaling gB to trigger fusion.
For HCMV, receptor binding is mediated by two distinct complexes: a trimeric complex including gO and gH–gL or a pentameric complex including UL128, UL130, UL131A, and gH–gL. As for HSV-1 and EBV, binding of the HCMV trimer or pentamer to receptor transmits a signal to gB to trigger fusion.
Cell tropism
Herpesviruses exhibit broad cell tropism and the routes of entry can depend on the cell type and/or the viral determinants. During infection, HSV-1 and HSV-2 typically infect epithelial cells and neurons; however, the virus can infect a wide range of cells, including fibroblasts and lymphocytes. Entry into epithelial cells occurs by low pH-dependent fusion with the endosomal membrane, whereas entry into neurons occurs by fusion at the plasma membrane5. HCMV also infects a variety of cells, including epithelial cells, endothelial cells, fibroblasts, and leukocytes6. Entry into epithelial and endothelial cells requires endocytosis and low pH7, whereas fusion with fibroblasts occurs at the plasma membrane8. EBV mainly infects B cells and epithelial cells, but can infect other cells such as monocytes9. B cell entry occurs after endocytosis in a pH-independent manner, whereas epithelial cell entry occurs after fusion with the plasma membrane10. Despite adapting to infect a broad range of host cells, herpesviruses share a common entry mechanism with conserved core fusion machinery (gH–gL and gB) and divergent receptor-binding proteins.
Receptor-binding protein structures
Herpesviruses encode distinct receptor-binding proteins that influence cell tropism, but are not conserved among virus subfamilies, including gD for HSV-1 and HSV-2; gp42 for EBV; and gO and UL128–UL130–UL131A for HCMV (Table 1). Crystal structures of gD and gp42 have been resolved alone and in complex with their receptors11-15. A structure of gp42 bound to gH–gL has also been determined16. A complex of gH–gL with UL128–UL130–UL131A has been crystalized17 and a complex of gH–gL with gO has been visualized at low resolution by EM18. Receptors bound to both HCMV gH–gL complexes also have been observed by EM19,20. Although many binding receptors have been identified for these viruses and structures such as EBV gp350 (ref. 21) have been determined, this Review focuses on validated entry receptors that have been examined in structural studies.
HSV gD
gD serves as the required receptor-binding protein for most alphaherpesviruses, with the notable exception of VZV, which lacks gD22. gD binds to three classes of receptors, including nectins, herpesvirus entry mediator (HVEM), and a modified form of heparan sulfate. Nectin-1, a cell adhesion protein, functions as an entry receptor for most alphaherpesviruses and is expressed on a variety of tissues, including neurons, a crucial cell type for alphaherpesviruses23. Nectin-2 mediates entry of some HSV-1 and HSV-2 strains and the related poliovirus receptor can serve as an entry receptor for other alphaherpesviruses24,25. HVEM, a member of the tumor necrosis factor receptor (TNFR) family, is expressed primarily on immune cells and functions as a receptor for HSV-1 and HSV-2, but not other alphaherpesviruses26. 3-O-sulfonated derivatives of heparan sulfate (3-OST HS) also can serve as entry receptors for HSV-127.
Crystal structures of gD from HSV-1 (refs. 12,13), HSV-2 (ref. 28), and pseudorabies virus (PRV) (ref. 29) show that the gD core adopts an immunoglobulin (Ig) fold flanked by amino-terminal and carboxyl-terminal extensions (Fig. 2). Structures of gD bound to its receptor reveal that gD binds to the membrane-distal portion of both HVEM and nectin-111,13,29,30. HVEM binds to an N-terminal loop on HSV-1 gD that is stabilized only in the presence of HVEM13. In the absence of receptor, the gD N-terminus is disordered. This gD N-terminus is not conserved in bovine herpesvirus (BHV) and PRV, consistent with the inability of gD from these alphaherpesviruses to bind HVEM31. Nectin-1 binds to gD at a distinct but overlapping site, adjacent to the HVEM binding residues11,13,30,32,33. 3-OST HS also binds to gD and, although it has not been crystalized, mutational analysis suggests that the 3-OST HS binding site on gD also overlaps with that of HVEM27,34.
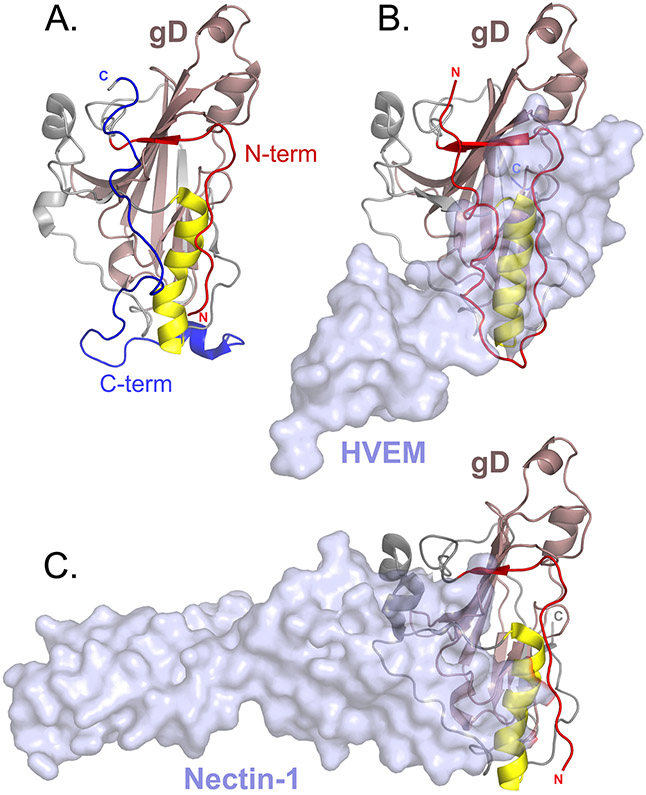
Fig. 2. Herpes simplex virus 1 glycoprotein D crystal structures.
a∣ A monomer from a dimeric form of glycoprotein D (gD) that was stabilized by introducing a disulfide bond at the carboxyl-terminus of the ectodomain (Protein Data Bank (PDB) ID 2C36)12. gD is oriented to show the receptor binding face. The core immunoglobulin fold of gD (pale pink) is flanked by amino-terminal (red) and C-terminal (blue) extensions. A α-helix (yellow) that supports the receptor binding site is shown and the N-termini and C-termini are marked. b∣ Herpesvirus entry mediator (HVEM) receptor (transparent surface rendering) bound to gD (PDB ID 1JMA)13. The N-terminus of gD (red) forms a loop that contains all of the contact residues for HVEM. c∣ Nectin-1 receptor (transparent surface rendering) bound to gD (PDB ID 3SKU)11. The HVEM and nectin-1 binding sites overlap. Binding of either HVEM or nectin-1 would displace the gD C-terminus.
The structure of a dimeric form of HSV-1 gD, artificially stabilized by an engineered intermolecular disulfide bond at the C-terminus of the gD ectodomain, shows that the C-terminal gD extension packs against the gD core in the absence of receptor and occludes both the nectin-1 and HVEM binding sites11,12,30 (Fig. 2). Thus, a conformational change must occur upon receptor binding to displace this C-terminal gD extension. In fact, truncation of the C-terminus enhances gD binding to both HVEM and nectin-1 (refs. 35,36). This conformational change may serve as a downstream signal for fusion. A panel of gD mutants designed to lock the C-terminal extension to the gD core at different sites successfully generated a mutant that retains the ability to bind to receptor but fails to trigger fusion, demonstrating that movement of the gD C-terminal extension contributes the fusion signal beyond simply permitting receptor binding37. The C-terminus of the gD ectodomain may serve as an interaction site for gH–gL or its movement may expose a gH–gL interaction site on gD (see below).
EBV gp42
Although EBV entry into epithelial cells requires only gH–gL and gB, gp42 serves as a required receptor-binding protein for EBV entry into B cells. gp42 binds to human leukocyte antigen type II (HLA class II), a member of the C-type lectin family. Crystal structures have been determined for gp42 alone, gp42 bound to HLA class II, and gp42 complexed with gH–gL plus an anti-gH antibody E1D1 (refs. 14-16) (Fig. 3). Using EM, the complex of gp42–gH–gL–HLA also has been visualized38.
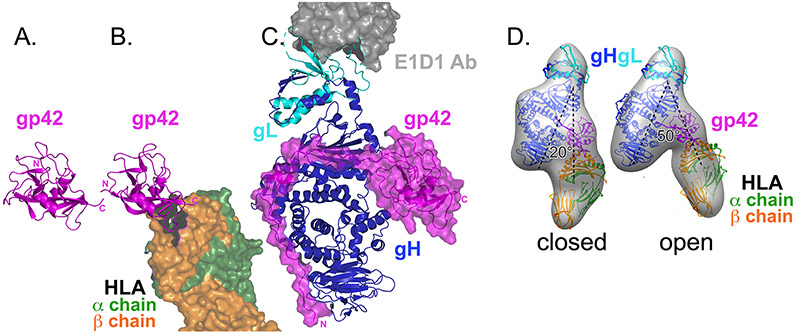
Fig. 3. Epstein–Barr virus glycoprotein 42 structures.
a∣ Crystal structure of glycoprotein 42 (gp42) alone (pink ribbons). The carboxyl-terminal C-type lectin domain (CTLD) is shown (Protein Data Bank (PDB) ID 3FD4)15. b∣ Crystal structure of gp42 (pink) bound to the human leukocyte antigen (HLA) class II receptor (green and orange surface rendering, partly shown) (PDB ID 1KG0)14. HLA binds to the gp42 CTLD and the gp42 amino-terminus is not resolved. HLA binding does not change the gp42 conformation drastically. c∣ Crystal structure of gp42–glycoprotein H (gH)–gL–E1D1 complex (PDB ID 5T1D)16. Although the CTLD of gp42 (pink ribbon with surface rendering) contacts gH–gL (blue and cyan ribbons, respectively), the majority of contacts lie in the gp42 N-terminal extension that extends down the length of gH–gL. Peptides from this gp42 N-terminus can inhibit entry into cells. E1D1 (grey surface rendering, partly shown) is a monoclonal antibody that binds to gL and partially neutralizes entry into epithelial cells but not B cells. d∣ Electron microscopy reconstructions of gp42–gH–gL–HLA complexes (grey surface). Fit inside the densities are the crystal structures of gH-gL (blue and cyan), gp42 (pink), and HLA (green and orange). Two conformations were observed: closed and open. In the closed conformation, HLA and gH–gL are arranged in a more parallel orientation than in the open conformation. Panel d is modified from ref. 38.
gp42 is a type II membrane protein that must be cleaved at the N-terminal transmembrane (TM) domain to generate a functional soluble form39. The C-terminal domain of gp42 adopts a C-type lectin domain (CTLD) fold and binds to the HLA class II receptor14. When complexed with gH–gL, the N-terminus of gp42 extends across gH, interacting with three gH domains16 (Fig. 3). Peptides derived from the gp42 N-terminal region can bind to gH–gL and inhibit entry into both B cells and epithelial cells40, suggesting that these peptides may occlude a binding site on gH–gL for the epithelial receptor (see below). The gp42 N-terminus anchors the gp42 C-terminal domain to gH–gL, however the interaction between the gp42 C-terminal domain and gH–gL is not extensive. gH–gL contacts three residues within a hydrophobic pocket (HP) in the gp42 C-terminal domain, located at the canonical CTLD ligand binding site. Mutations in this gp42 HP inhibit fusion without preventing gH–gL or HLA binding41, suggesting a functional role for the HP. The binding of gp42 to gH–gL positions the HLA binding site near the middle of the gH–gL–gp42 complex, in contrast to the arrangement seen for HCMV gH–gL complexes (see below).
EM reconstructions of gp42 simultaneously bound to HLA and gH–gL reveal multiple conformations consisting of open and closed forms38. gp42 bridges HLA and gH–gL, creating an approximately parallel orientation of gH–gL and HLA in the closed conformation and a highly variable orientation in open conformations. The TM anchors of gH (linked to the virus) and HLA (linked to the cell) would be located on the same side of this complex in the closed conformation, suggesting that binding to HLA may impact the arrangement of the juxtaposed membranes during fusion.
HCMV gO and UL128–UL130–UL131A
HCMV entry into all cells, including fibroblast, epithelial, and endothelial cells, requires gH–gL in a trimeric complex with gO20,42. Infection of epithelial and endothelial cells also requires gH–gL to form a pentameric complex with UL128, UL130, and UL131A7,43. Nearly all of the gH–gL in the virion is complexed with either gO or UL128–UL130–UL131A (ref. 44) and the formation of these complexes is mutually exclusive. Residue C144 in gL forms a disulfide bond with UL128 in the pentamer and with gO in the trimer18.
The gH–gL complexes bind to different receptors, although all of these have not been identified yet. The trimer binds to platelet-derived growth factor receptor α (PDGFRα) to mediate entry into fibroblasts, whereas the pentamer binds to neuropilin-2 (Nrp2) to mediate entry into epithelial and endothelial cells19,20,45. OR14I1 was identified recently as an additional epithelial cell receptor that mediates HCMV pentamer-dependent attachment and infection46 and CD147 also was found to promote pentamer-dependent entry, although direct binding did not occur47. In addition, the trimer is thought to interact with another, as yet unidentified receptor, on epithelial and endothelial cells48.
The crystal structure of pentamer bound to an antibody shows that UL128–UL130–UL131A assemble onto the N-terminus of gH–gL, docking at an N-terminal extension of gL that is conserved in betaherpesviruses but not present in other subfamilies17 (Fig. 4a). The limited interface between UL128–UL130–UL131A and gH–gL permits flexibility, which was observed experimentally17. Although the gH–gL–gO trimer has not been determined using crystallography, the trimer has been visualized using EM. Similar to UL128–UL130–UL131A, gO also docks at the tip of gH–gL (Fig. 4b) 18,20. This N-terminal gH–gL docking site differs from the gp42-docking site at the middle of gH–gL, suggesting that distinct arrangements of receptor complexes are capable of activating gH–gL between the subfamilies.
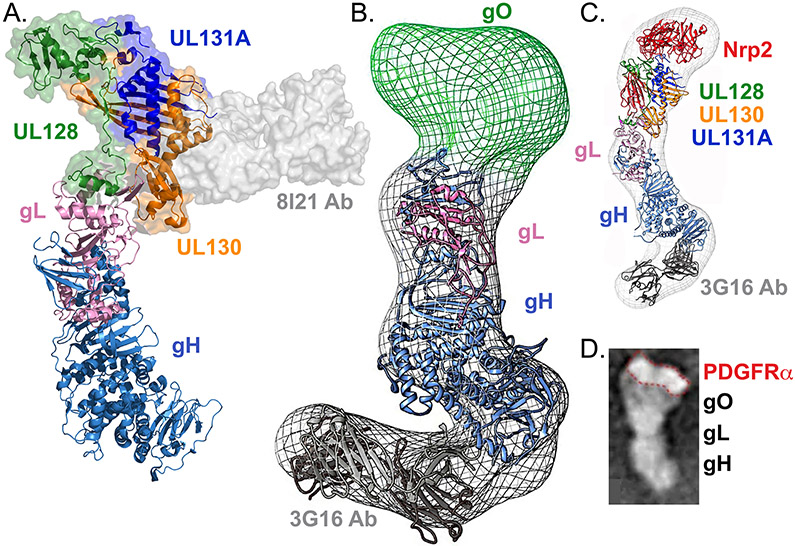
Fig. 4. Structures of human cytomegalovirus glycoprotein H–glycoprotein L complexes.
a∣ Crystal structure of pentamer bound to mAb 8I21 (grey surface rendering). UL128 (green), UL130 (orange), and UL131A (dark blue) assemble on the distal tip of glycoprotein H (gH)–gL (light blue and pink, respectively), contacting an amino-terminal extension of gL that is unique to betaherpesviruses (Protein Data Bank (PDB) ID 5V0B)17. b∣ Electron microscopy (EM) reconstruction of gH–gL–gO trimer bound to monoclonal antibody (mAb) 3G16 (Electron Microscopy Data Bank entry EMD-6431). The gH–gL (blue and pink) and mAb (grey) structures have been fit inside the density. gO (green) maps to the distal end of gH–gL, in a position analogous to UL128–UL130–UL131A. c∣ EM reconstruction of pentamer (colored as in A) bound to mAb 3G16 (grey) and Neuropilin-2 (Nrp2) receptor (red) (EMD-8884). Crystal structures have been fit inside the EM density and Nrp2 maps to the distal tip of the complex, contacting the UL128–UL130–UL131A–gL portion of the complex. d∣ EM image of trimer bound to the receptor platelet-derived growth factor receptor α (PDGFRα). A comparison of gH–gL–gO with and without receptor demonstrates that the receptor binds at the gO side of the complex, outlined in red. Panel b is modified from ref. 64 Panel c is modified from ref. 19. Panel d is modified from ref. 20.
Low-resolution structures of the trimer bound to PDGFRα20 and the pentamer bound to Nrp2 receptor19 have been visualized using EM (Fig. 4c-d). The receptors appear to interact nearly exclusively with gO or UL128–UL130–UL131A. Crosslinking analysis confirms that Nrp2 contacts residues in gL, UL128, UL130, and UL131A19.
gH and gL
The gH–gL heterodimer is an essential component of the fusion machinery that is conserved among all herpesviruses. gH–gL is proposed to regulate fusion by interacting with gB. gH–gL serves a receptor-binding function for many herpesviruses; however, receptor-binding by gH–gL does not appear to be required for most alphaherpesviruses. Instead, gH–gL is proposed to relay a receptor-binding signal from gD to gB in alphaherpesviruses. For some herpesviruses, gH–gL alone acts directly as a receptor-binding complex, as is observed for EBV entry into epithelial cells and VZV entry (Table 1). In other instances, gH–gL forms a stable complex with other viral proteins to bind to entry receptors, as described above for EBV and HCMV.
EBV entry into epithelial cells is mediated by gH–gL binding directly to ephrin receptor tyrosine kinase A2 (EphA2)49. Similarly, KSHV entry is mediated by gH–gL binding to EphA2 or EphA4 (refs. 50-52). Soluble gp42 is able to block EphA2-mediated fusion49 and the ability of gp42 N-terminal peptides to block EBV entry into epithelial cells suggests that the epithelial receptor binding may overlap with the gp42 binding site on gH–gL or alternatively with a site that is crucial for gB activation in epithelial cell entry16,40. The E1D1 antibody, which binds exclusively to gL at a distinct site at the distal end of gH–gL, partially inhibits epithelial cell entry but not B cell entry, suggesting that E1D1 also inhibits epithelial receptor binding or post-receptor binding activation (Fig. 3c)16,53,54. These findings indicate that multiple regions of gH–gL are important for epithelial cell entry.
VZV gH–gL is proposed to interact with integrins, as cell–cell fusion can be inhibited by adding an anti-αV integrin antibody or knocking down αV integrin expression55. Interestingly, although not required for fusion, HSV gH–gL also binds to αvβ3, αvβ6, and αvβ8 integrins56,57.
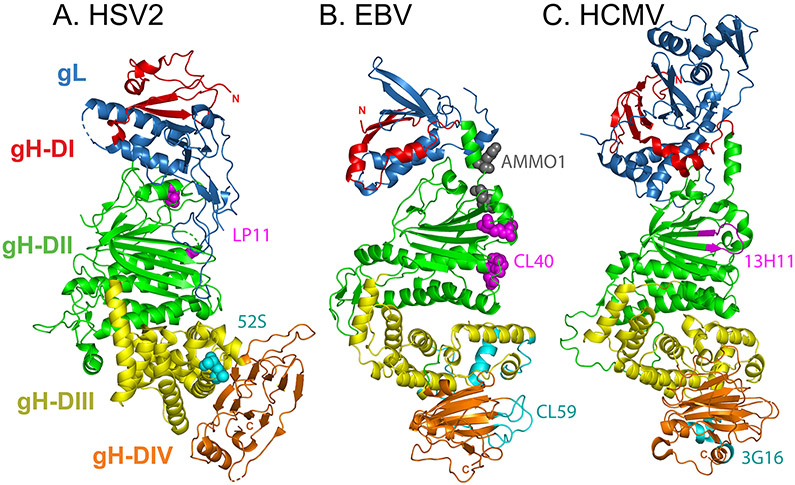
Crystal structures of gH–gL have been resolved for HSV-2 (ref. 58), VZV59, PRV60, EBV61, and HCMV17. The gH–gL ectodomain is comprised of four domains that adopt a boot-shaped organization for HSV-2 and VZV, and a more rod-like shape for PRV and EBV (Fig. 5). The overall architecture of HCMV gH–gL is intermediate to both forms.
Fig. 5. Crystal structures of the glycoprotein H–glycoprotein L complex.
a∣ Herpes simplex virus 2 (HSV-2) glycoprotein H (gH)–gL (Protein Data Bank (PDB) ID 3M1C)58. Four progressive gH domains (DI–DIV) and gL are shown. gH DI (red) is intimately associated with gL (blue). gL requires gH for proper folding and anchoring to the membrane. DII (green) includes parallel β-sheets and helices. DIII (yellow) is mostly helical and DIV (orange) includes a β-sandwich. Although domain designations between the gH–gL structures can differ, specifically for distinction between DII and DIII, this figure uses the domain designations identified for Epstein–Barr virus (EBV) gH–gL. The carboxyl-terminus would extent from DIV into the transmembrane (TM) domain. The overall complex adopts a boot-like shape. Substitutions at HSV-1 gH residues 168 or 329 (magenta spheres) prevent binding of the neutralizing monoclonal antibody (nAb) LP11. On the opposite face, substitution mutations at gH residues 536 or 537 (cyan spheres) prevent binding of the nAb 52S (ref. 63). b∣ EBV gH–gL (PDB 3PHF)61. The domains are colored as in part a and they adopt a more linear orientation. Co-crystallization revealed that nAb CL40 contacts gH residues 184, 239, 243, 284, and 286 (magenta spheres) and overlaps the binding site for the gp42 C-terminal domain. Electron microscopy reconstruction of bound nAb CL59 shows that this nAb binds to a distinct site within DIII and DIV (cyan ribbons; gH residues 406–415, 456–468, 494–503, 568–577, 623–626, and 645–656)65. The CL40 epitope partially overlaps with the nAb AMMO1 binding site (grey spheres; gH residues 73 and 76)66. c∣ Human cytomegalovirus (HCMV) gH–gL (PDB ID 5VOC) 17. Domains are colored as in part a. HCMV gL includes an amino-terminal extension that is absent from the other structures. Hydrogen deuterium exchange coupled to mass spectroscopy maps nAb 13H11 binding to gH residues 238–247 (magenta ribbons) and nAb 3G16 binding to gH residues 677–684 and 705–708 (cyan ribbons)64.
gH contains a C-terminal TM domain that anchors the complex to the membrane. gL lacks a TM anchor. The gH–gL structure can be described in four progressive domains, although specific domain designations between the gH–gL structures can differ62. Domain I is the most membrane distal region, comprised of gL and the N-terminus of gH. Domain II includes β-sheets and α-helices in a parallel arrangement. The N-terminal domains of gH are the least conserved in sequence, consistent with this region interacting with other species-specific viral proteins (see above). The two membrane proximal domains are more conserved, including the helices of domain III and β-sandwich of domain IV.
Neutralizing monoclonal antibodies (nAbs) map to several regions of gH–gL (Fig. 5), consistent with a requirement for gH–gL to interact with multiple partners during virus entry, including receptors, receptor-binding proteins, and gB. In HSV-1, selection of nAb resistance mutations indicates that the nAb LP11 binds to domain II of gH–gL, whereas the nAb 52S binds to domain III on the opposite face of the complex58,63. In HCMV, EM reconstructions show the nAbs 3G16 and 13H11 binding to distinct sites on gH–gL, neither of which interfere with the docking of gO or UL128–UL130–UL131A (refs. 20,64). 3G16 binds at the membrane proximal end of gH–gL, whereas 13H11 binds at the middle of gH–gL. In EBV, two nAbs (CL40 and CL59) inhibit entry into all cell types and also map to separate sites on gH–gL65. A crystal structure of CL40 bound to gH–gL shows this nAb at the middle of gH–gL, overlapping the site of the gp42 C-terminal domain, in a location analogous to that of 13H11. This CL40 binding site partially overlaps with the binding site for another nAb (AMMO1) which blocks entry into both B cells and epithelial cells66. By contrast, an EM reconstruction of CL59 bound to gH–gL places this nAb at domains III and IV, in a position analogous to 3G16 (ref. 65).
The gH cytoplasmic tail (CT) domain is short, ranging from 8 residues in EBV to 19 residues in PRV, but it contributes substantially to entry. Deletion or mutation of gH CT residues inhibits fusion for multiple species67-74 and enhances fusion for VZV72. When the gH–gL ectodomain is expressed in cells as a soluble protein or as a glycosylphosphatidylinositol-anchored form, cell–cell fusion either fails73,75,76 or occurs at only low levels4. Some mutations in the gH CT inhibit fusion without detectable changes to gH–gL ectodomain conformation or expression69,71,74, suggesting that an intracellular interaction, potentially between the CTs of gH and gB, may contribute to fusion regulation 71,77. Some EBV gH CT truncation mutants result in decreased gH–gL binding to gp42, suggesting that the gH CT may also impact the gH ectodomain74.
The fusion protein glycoprotein B
gB is the viral fusion protein, the glycoprotein responsible for inserting into the host cell membrane and refolding to drive fusion of the viral envelope and cell membrane. Viral fusion proteins are membrane-anchored glycoproteins that initially fold into a prefusion conformation78. Upon triggering by receptor binding and/or exposure to acidic pH, fusion proteins undergo a conformational change to insert hydrophobic residues into the host membrane and then fold back on themselves to bring the two membranes together.
Crystal structures of gB from all three herpesvirus subfamilies have been resolved, including from HSV-1 (ref. 77,79), PRV80,81, VZV82, HCMV83,84, and EBV85 (Fig. 6a-c). These gB homologs adopt similar structures. Unexpectedly, gB resembles the fusion proteins from rhabdovirus86,87, baculovirus88, and thogotovirus89, despite a lack of sequence homology between gB and the fusion proteins of these unrelated viruses. Together, these fusion proteins comprise the ‘class III’ fusion proteins90.
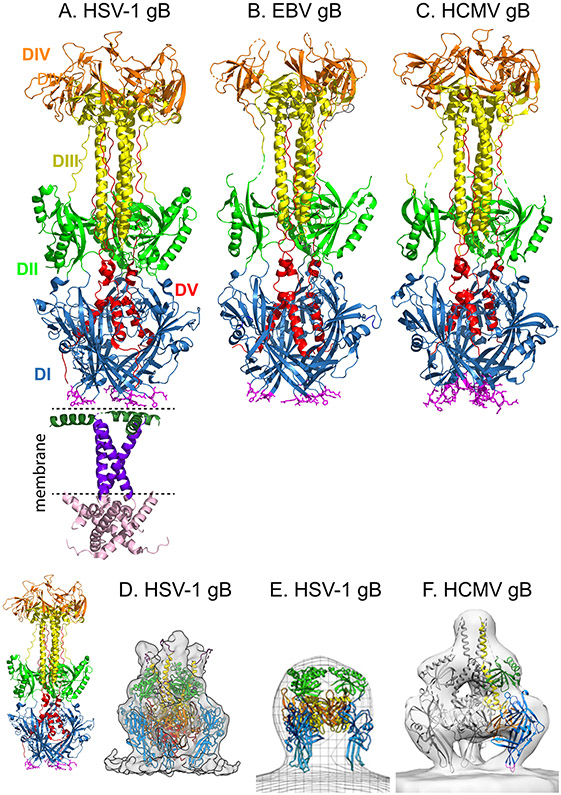
Fig. 6. Glycoprotein B structures.
a∣ Crystal structure of full-length postfusion herpes simplex virus 1 (HSV-1) glycoprotein B (gB) (Protein Data Bank (PDB) ID 5V2S)77. The trimeric ectodomain is comprised of five domains (DI–DV). DI (blue) contains hydrophobic fusion loops (magenta sticks) that insert into the host cell membrane. DIII (yellow) includes an extended central trimeric coiled-coil, against which DV (red) packs in an anti-parallel orientation, as DV proceeds through DI, towards the transmembrane (TM) domain. The membrane proximal region (dark green), TM (dark purple), and cytoplasmic (pink) domains are shown. b∣ Crystal structure of postfusion Epstein–Barr (EBV) gB ectodomain (PDB 3FVC) 85. c∣ Crystal structure of postfusion human cytomegalovirus (HCMV) gB ectodomain (PDB ID 5CXF)84. For parts b and c, the domains are colored as in part a. For crystallization, residues in the fusion loops of EBV and HCMV gB were replaced with HSV-1 residues. d∣ Cryoelectron tomography (cryoET) reconstruction of the compact conformation of a mutant form of HSV-1 gB, 12 nm in height. Density fitting of the gB domains from the postfusion gB structure is shown. Domains are colored as in part a. The fusion loops of DI (light blue) are oriented towards the membrane. For comparison, the ectodomain of postfusion HSV-1 gB is shown at a similar scale on the left, at 16 nm in height. e∣ Alternative cryoET reconstruction of a compact form of HSV-1 gB, 9 nm in height. A pseudoatomic model of prefusion gB121, based on the prefusion structure of vesicular stomatitis virus protein G (VSV G) (PDB ID 5I2S)100, was fit into the 3D electron microscopy (EM) reconstruction. Domains are colored as in part a. DV is excluded from the model. f∣ CryoET reconstruction of human cytomegalovirus (HCMV) gB (Electron Microscopy Data Bank entry EMD-9328), 13 nm in height. Domains from a single protomer are colored as in part a. Domain fitting using the postfusion HCMV structure placed DI (blue) near the membrane, with the fusion loops (magenta) oriented towards the membrane. As the EM reconstruction cannot accommodate the DIII postfusion conformation, the DIII fit was modeled on VSV G. DV is excluded from the model. Part d is modified from ref. 103. Part e is modified from ref. 123. Part f is modified from ref. 124.
gB is a trimeric single-pass transmembrane protein with five extracellular domains. nAbs map to multiple sites in the gB ectodomain83,91-93 and mutations in each of the gB domains can prevent fusion94,95, suggesting that multiple regions of gB participate in the conformational changes required for fusion and/or interactions with the proteins that trigger gB. The formation of a fusion pore likely requires multiple gB trimers. Lateral interactions among gB trimers have been observed by EM and may contribute to the expansion of the fusion pore96.
Receptors that bind to gB and mediate entry have been reported, such as paired immunoglobulin-like type 2 receptor-α97, myelin-associated glycoprotein98, and myosin-999. Triggering fusion by binding to gB directly departs from the current model of virus entry and future structural studies of these receptors may explain how they enhance entry.
Postfusion gB structure
Crystal structures of the rhabdovirus vesicular stomatitis virus (VSV) fusion protein G have been resolved in both the prefusion100 and postfusion86 conformations. All of the gB crystal structures resolved thus far resemble the postfusion form of VSV G. Removal of the gB TM and CT regions appears to destabilize gB, preventing the crystallization of a prefusion form77,79. In fact, gB folds into a postfusion form even when mutations that reduce fusion are introduced101-103, when the TM is substituted101,102, or when the membrane is disrupted with detergent77,79. Antibodies specific for the prefusion conformation that might facilitate stabilization of a prefusion form have not been identified. All the antibodies that have been characterized, including neutralizing antibodies, appear to be able to bind to the postfusion form91,93.
Domain I lies at the base of the molecule in the postfusion structure and contains two fusion loops with hydrophobic residues (Fig. 6a-c). These fusion loops are thought to insert into the host cell membrane during fusion104 and mutations within the loops demonstrate that they are required for fusion function80,105-107. Domain II contains a pleckstrin-homology domain. Extended linker regions that lead into and out of domains I and II may allow for large-scale conformational rearrangements during fusion. Domain III forms an extended trimer of helices that comprises the core of the structure. Domain IV forms a crown at the top of the structure. Domain V extends down the length of the molecule, packing against the helices of domain III in an antiparallel orientation that is reminiscent of the hallmark ‘six-helix bundle’ (6HB) structure present in the postfusion form of class I fusion proteins108. The formation of a stable 6HB structure during fusion is thought to contribute energy for fusion pore formation109. Mutations in domain V of gB designed to weaken its packing against domain III result in reduced fusion and a small plaque phenotype110,111, suggesting that the interaction of domain III and V may also contribute to the energetics of fusion.
As seen for class I fusion proteins112,113, the CT domain of gB regulates fusion. The mechanism for this regulation is unclear; however, inter-protomer interactions among the CTs or between the CTs and the membrane may stabilize the prefusion state of the gB ectodomain, preventing premature activation77. Mutations in the gB CT can inhibit or enhance fusion, depending on the specific mutation114-119. The crystal structure of HSV-1 gB including its TM and CT shows that the CT forms a trimer that interacts with the membrane (Fig. 6a)77. The gB CT also may interact with the gH CT (see below)71. Among class III fusion proteins, fusion regulation by the CT may be unique to the herpesviruses because the VSV and baculovirus fusion protein CT domains are substantially shorter than the gB CT120.
Prefusion gB models
Computational homology models of the prefusion gB of HSV and EBV have been created based on the prefusion VSV G crystal structure85,121. These models suggest that gB undergoes substantial refolding to transition to postfusion.
More recently, alternate conformations of full-length gB anchored in a membrane have been studied using cryoelectron tomography (cryoET) (Fig. 6d-f)103,122-124. These cryoET reconstructions reveal compact trimers that are shorter than postfusion gB (8–12 nm versus 16–18 nm in height). These structures may represent prefusion or intermediate conformations of gB. When gB domains from the crystal structures were fit into the cryoET reconstructions, the resulting models orient the fusion loops towards the membrane103,123,124. The postfusion conformation of domain III cannot be fit into the cryoET reconstructions, suggesting that transition from this alternate compact form of gB to the postfusion form requires substantial structural rearrangement.
Glycoprotein interactions
Interaction between HSV-1 and HSV-2 gD and gH–gL
Although complexes of gH–gL and the receptor-binding proteins of HCMV and EBV have been examined structurally, a complex between the alphaherpesvirus gD and gH–gL has not been visualized. Biochemical and genetic studies suggest that they interact, perhaps transiently and/or with low affinity. gD and gH–gL can be crosslinked in the virion125 and they are reported to co-precipitate126,127. An interaction between gD and gH–gL has been observed using bimolecular fluorescence complementation (BiFC)128,129 and stabilization of the gD–gH–gL complex by the BiFC tags inhibits fusion, suggesting that the interaction between gD and gH–gL may need to be transient128,129. A species-specific functional interaction between gD and gH–gL was shown by coexpressing combinations of entry glycoproteins from the non-complementing species HSV-1 and saimiriine herpesvirus 1 (SaHV1)130. gH–gL chimeras including segments of HSV-1 and SaHV1 mapped the gD interaction site to gH domains DI and DII (Fig. 5)131. This membrane-distal site on gH–gL coincides with the location of the receptor-binding proteins for EBV and HCMV, suggesting gD may trigger a conformational change in the N-terminus of gH–gL. In fact, an N-terminally truncated form of HSV gH–gL can mediate cell–cell fusion in the absence of gD132.
Most recently, the binding of HSV-2 gD to gH–gL was demonstrated by surface plasmon resonance (SPR) using purified soluble forms of gD and gH–gL133. Presentation of gD by some antibodies allowed gH–gL binding, whereas other antibodies occluded the gH–gL interaction site. These data defined the gH–gL interaction site on one face of gD, distinct from the receptor-binding face. The ability of an anti-gD nAb (MC2) to block fusion without blocking receptor binding or gH–gL binding indicates that an additional site on gD is required for function. The fast on-rate and off-rate observed suggests that the interaction between gD and gH–gL is transient.
Interactions between gB and gH–gL
Using cryoET, an interaction between gB and a linear structure, presumed to be gH–gL, was visualized on HCMV virions124. The linear structure was frequently associated with a compact conformation of gB, which may represent prefusion gB, but not with the postfusion gB present in the membrane. Exclusive association with the prefusion form of gB is consistent with gH–gL stabilizing gB prior to the receptor-binding step that triggers fusion. Whether this gB complex forms with gH–gL trimers or pentamers, and how these proteins interact to coordinate viral entry, remains to be determined. This low-resolution structure suggests that gH–gL and gB may interact at a region adjacent to the membrane. A close association of the TM domains is consistent with a proposed interaction between the gH and gB CT domains71,77. The gB CT may function as a clamp, preventing premature triggering of gB. In an alternate model, gH is proposed to act as a wedge, potentially triggering gB upon receptor-binding by disrupting gB CT interprotomer interactions and/or gB CT interactions with the membrane77. The proposed stabilizing and wedge functions of gH–gL are not mutually exclusive. In additional support for a functional interaction between the gH and gB CT domains, the substitution of EBV gB CT residues with those of rhesus lymphocryptovirus (rhLCV) confers partial fusion function with rhLCV gH–gL74.
The ectodomains of gB and gH–gL also are proposed to interact. An interaction between gB and gH–gL detected using BiFC was blocked by nAbs that bind to the gB and gH–gL ectodomains58,134, including nAb LP11 (Fig. 5A). Furthermore, gB and gH–gL from HSV-1 or HCMV can mediate low levels of entry when expressed on opposing membranes, as can a soluble form of the HSV-2 gH–gL ectodomain4,135.
Conclusions and perspectives
The complexity of herpesvirus entry into cells results from the broad range of receptor usage and cell tropism, and the requirement for multiple glycoproteins to mediate membrane fusion. The extensive structural studies described here clarify our understanding of the stepwise mechanism of virus entry and lay the groundwork for the development of antiviral interventions. The only herpesviruses vaccines currently available include a live attenuated VZV vaccine136 and a VZV gE subunit vaccine137. Current antiviral medications for herpesviruses, such as acyclovir, target virus replication, rather than the viral entry glycoproteins. Understanding the structural details of herpesvirus entry may aid the design of better vaccines, inhibitors, or neutralizing antibodies that target receptor binding, interactions among the entry glycoproteins, and/or the refolding of gB. The requirement of gH–gL and gB for all herpesviruses means that discovery of an inhibitor of one species could serve as a model for analogous inhibitors for other species. The entry glycoproteins generate neutralizing antibodies and the structural determination of the binding sites for these antibodies also provides candidate sites for the design of inhibitors.
Advances in EM imaging have provided 3D reconstructions of prefusion or intermediate conformations of gB103,122-124, as well gB interacting with gH–gL124. For all of the herpesviruses, atomic resolution of the prefusion gB conformation and the gH–gL–gB complex remains to be determined. For HSV-1, the interaction between gD and gH–gL has not been visualized using any structural approach. For EBV, the structure of gH–gL bound to EphA2 and the complex of gp42–HLA–gH–gL at atomic resolution remain unknown. For HCMV, the structures of the gH–gL–gO trimer and the gH–gL complexes bound to PDGFRα or Nrp2 receptors at atomic resolution have not been determined.
Although biochemical and genetic approaches have provided models for the functional interactions among entry glycoproteins, future crystallography and cryoET studies will continue to identify the interaction sites and conformational changes required for virus-induced membrane fusion. These studies may be assisted by the use of antibodies and/or mutations that stabilize the complexes and/or transient conformations. Additional receptors for these viruses may be identified and characterized using structural methods as well.
Acknowledgements
This work was supported by grant AI-137267 and AI-148478 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. All crystal structures in this manuscript were rendered using MacPymol.
Glossary terms
- Enveloped viruses
Viruses with an outer layer consisting of a lipid bilayer, in which the viral glycoproteins responsible for mediating virus entry into cells are embedded.
- Conformational change
A change in protein structure made possible by the intrinsic flexibility of the protein that can be triggered by environmental factors, such as binding to a receptor or another glycoprotein.
- Cell tropism
The specific cell type(s) that support the replication of different viruses
- Entry receptors
Molecules present in host cells that bind directly to viruses and mediate virus entry into the cell.
- Neutralizing monoclonal antibody (nAb)
An antibody that binds to a virus particle and prevents infection, typically by preventing virus entry into the cell.
- Fusion loops
Short stretches of hydrophobic residues within a fusion protein that insert into the host cell membrane during the fusion event.
- Electron tomography
Method to produce high-resolution 3D models of molecules by reconstructing a series of 2D electron microscopy images taken from multiple angles.
- Crystal structure
Structural model based on x-ray diffraction of a crystal that often permits atomic resolution for protein structures.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Vallbracht M, Backovic M, Klupp BG, Rey FA & Mettenleiter TC Common characteristics and unique features: A comparison of the fusion machinery of the alphaherpesviruses Pseudorabies virus and Herpes simplex virus. Advances in virus research 104, 225–281, doi: 10.1016/bs.aivir.2019.05.007 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Mohl BS, Chen J & Longnecker R Gammaherpesvirus entry and fusion: A tale how two human pathogenic viruses enter their host cells. Advances in virus research 104, 313–343, doi: 10.1016/bs.aivir.2019.05.006 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Nishimura M & Mori Y Entry of betaherpesviruses. Advances in virus research 104, 283–312, doi: 10.1016/bs.aivir.2019.05.005 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Atanasiu D, Saw WT, Cohen GH & Eisenberg RJ Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol 84, 12292–12299, doi: 10.1128/JVI.01700-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicola AV Herpesvirus Entry into Host Cells Mediated by Endosomal Low pH. Traffic 17, 965–975, doi: 10.1111/tra.12408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerna G, Baldanti F & Revello MG Pathogenesis of human cytomegalovirus infection and cellular targets. Human immunology 65, 381–386, doi: 10.1016/j.humimm.2004.02.009 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA & Johnson DC Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80, 710–722, doi: 10.1128/JVI.80.2.710-722.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compton T, Nepomuceno RR & Nowlin DM Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191, 387–395 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Hutt-Fletcher LM Epstein-Barr virus entry. J Virol 81, 7825–7832, doi: 10.1128/JVI.00445-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller N & Hutt-Fletcher LM Epstein-Barr virus enters B cells and epithelial cells by different routes. J Virol 66, 3409–3414 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Giovine P et al. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog 7, e1002277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krummenacher C et al. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. The EMBO journal 24, 4144–4153 (2005). Compares the crystal structure of unbound HSV-1 gD to the previously determined gD–HVEM complex, revealing a conformational change that occurs upon receptor-binding.
- 13.Carfi A et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Molecular cell 8, 169–179 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Mullen MM, Haan KM, Longnecker R & Jardetzky TS Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Molecular cell 9, 375–385 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Kirschner AN, Sorem J, Longnecker R & Jardetzky TS Structure of Epstein-Barr virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure 17, 223–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sathiyamoorthy K et al. Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat Commun 7, 13557, doi: 10.1038/ncomms13557 (2016). Reports the crystal structure of the EBV gp42–gH–gL complex, demonstrating that the amino-terminus of gp42 spans the length of gH.
- 17. Chandramouli S et al. Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Science immunology 2, doi: 10.1126/sciimmunol.aan1457 (2017). Reports the crystal structure of the HCMV pentamer, demonstrating that HCMV gH-gL folds similarly to EBV and UL128–UL130–UL131A binds to an extension in gL.
- 18. Ciferri C et al. Structural and biochemical studies of HCMV gH/gL/gO and Pentamer reveal mutually exclusive cell entry complexes. PNAS 112, 1767–1772, doi: 10.1073/pnas.1424818112 (2015). EM reconstructions of the HCMV trimer and pentamer complexes demonstrate that both gO and UL128–UL130–UL131A dock at the tip of gH–gL.
- 19. Martinez-Martin N et al. An Unbiased Screen for Human Cytomegalovirus Identifies Neuropilin-2 as a Central Viral Receptor. Cell 174, 1158–1171 e1119, doi: 10.1016/j.cell.2018.06.028 (2018). Identifies Nrp2 as an HCMV receptor and shows that this receptor binds to distal end of the pentamer using EM reconstruction.
- 20. Kabanova A et al. Platelet-derived growth factor-alpha receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat Microbiol 1, 16082, doi: 10.1038/nmicrobiol.2016.82 (2016). Identifies PDGFRα as an HCMV receptor and shows that this receptor binds to distal end of the trimer using EM.
- 21.Szakonyi G et al. Structure of the Epstein-Barr virus major envelope glycoprotein. Nature structural & molecular biology 13, 996–1001 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Oliver SL, Yang E & Arvin AM Varicella-Zoster Virus Glycoproteins: Entry, Replication, and Pathogenesis. Curr Clin Microbiol Rep 3, 204–215, doi: 10.1007/s40588-016-0044-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ & Spear PG Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280, 1618–1620 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Krummenacher C et al. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 322, 286–299 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Warner MS et al. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246, 179–189 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Montgomery RI, Warner MS, Lum BJ & Spear PG Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87, 427–436 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Shukla D et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99, 13–22 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Lee CC et al. Structural basis for the antibody neutralization of herpes simplex virus. Acta Crystallogr D Bio Crystallogr 69, 1935–1945, doi: 10.1107/S0907444913016776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A et al. Structural basis of nectin-1 recognition by pseudorabies virus glycoprotein D. PLoS Pathog 13, e1006314, doi: 10.1371/journal.ppat.1006314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu G et al. Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition. J Virol 88, 13678–13688, doi: 10.1128/JVI.01906-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connolly SA et al. Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC(nectin-1) with different affinities. Virology 280, 7–18, doi: 10.1006/viro.2000.0747 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Krummenacher C et al. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol 72, 7064–7074 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitbeck JC et al. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J Virol 73, 9879–9890 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon M, Zago A, Shukla D & Spear PG Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1. J Virol 77, 9221–9231 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willis SH et al. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol 72, 5937–5947 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krummenacher C et al. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J Virol 73, 8127–8137 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazear E et al. Engineered disulfide bonds in herpes simplex virus type 1 gD separate receptor binding from fusion initiation and viral entry. J Virol 82, 700–709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sathiyamoorthy K et al. Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog 10, e1004309, doi: 10.1371/journal.ppat.1004309 (2014). EM reconstructions of the gp42–HLA–gH–gL complex reveal open and closed conformations.
- 39.Sorem J, Jardetzky TS & Longnecker R Cleavage and secretion of Epstein-Barr virus glycoprotein 42 promote membrane fusion with B lymphocytes. J Virol 83, 6664–6672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirschner AN, Lowrey AS, Longnecker R & Jardetzky TS Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J Virol 81, 9216–9229 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva AL, Omerovic J, Jardetzky TS & Longnecker R Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J Virol 78, 5946–5956 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou M, Lanchy JM & Ryckman BJ Human Cytomegalovirus gH/gL/gO Promotes the Fusion Step of Entry into All Cell Types, whereas gH/gL/UL128-131 Broadens Virus Tropism through a Distinct Mechanism. J Virol 89, 8999–9009, doi: 10.1128/JVI.01325-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D & Shenk T Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol 79, 10330–10338, doi: 10.1128/JVI.79.16.10330-10338.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou M, Yu Q, Wechsler A & Ryckman BJ Comparative analysis of gO isoforms reveals that strains of human cytomegalovirus differ in the ratio of gH/gL/gO and gH/gL/UL128-131 in the virion envelope. J Virol 87, 9680–9690, doi: 10.1128/JVI.01167-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y et al. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-alpha as a key for entry. PLoS Pathog 13, e1006281, doi: 10.1371/journal.ppat.1006281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.E X et al. OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism. PNAS 116, 7043–7052, doi: 10.1073/pnas.1814850116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanarsdall AL et al. CD147 Promotes Entry of Pentamer-Expressing Human Cytomegalovirus into Epithelial and Endothelial Cells. mBio 9, doi: 10.1128/mBio.00781-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Jardetzky TS, Chin AL, Johnson DC & Vanarsdall AL The Human Cytomegalovirus Trimer and Pentamer Promote Sequential Steps in Entry into Epithelial and Endothelial Cells at Cell Surfaces and Endosomes. J Virol 92, doi: 10.1128/JVI.01336-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J et al. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol 3, 172–180, doi: 10.1038/s41564-017-0081-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.TerBush AA, Hafkamp F, Lee HJ & Coscoy L A Kaposi's Sarcoma-Associated Herpesvirus Infection Mechanism Is Independent of Integrins alpha3beta1, alphaVbeta3, and alphaVbeta5. J Virol 92, doi: 10.1128/JVI.00803-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Zhang X, Schaller S, Jardetzky TS & Longnecker R Ephrin Receptor A4 is a New Kaposi's Sarcoma-Associated Herpesvirus Virus Entry Receptor. mBio 10, doi: 10.1128/mBio.02892-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hahn AS et al. The ephrin receptor tyrosine kinase A2 is a cellular receptor for Kaposi's sarcoma-associated herpesvirus. Nat Med 18, 961–966, doi: 10.1038/nm.2805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chesnokova LS & Hutt-Fletcher LM Fusion of Epstein-Barr virus with epithelial cells can be triggered by alphavbeta5 in addition to alphavbeta6 and alphavbeta8, and integrin binding triggers a conformational change in glycoproteins gHgL. J Virol 85, 13214–13223, doi: 10.1128/JVI.05580-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q, Turk SM & Hutt-Fletcher LM The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol 69, 3987–3994 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang E, Arvin AM & Oliver SL Role for the alphaV Integrin Subunit in Varicella-Zoster Virus-Mediated Fusion and Infection. J Virol 90, 7567–7578, doi: 10.1128/JVI.00792-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parry C, Bell S, Minson T & Browne H Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J Gen Virol 86, 7–10 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Gianni T, Salvioli S, Chesnokova LS, Hutt-Fletcher LM & Campadelli-Fiume G alphavbeta6- and alphavbeta8-integrins serve as interchangeable receptors for HSV gH/gL to promote endocytosis and activation of membrane fusion. PLoS Pathog 9, e1003806, doi: 10.1371/journal.ppat.1003806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chowdary TK et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Strut Mol Biol 17, 882–888 (2010). Reports the crystal structure of HSV-2 gH–gL, exhibiting extensive interactions between gL and the amino-terminal domain of gH.
- 59.Xing Y et al. A site of varicella-zoster virus vulnerability identified by structural studies of neutralizing antibodies bound to the glycoprotein complex gHgL. PNAS 112, 6056–6061, doi: 10.1073/pnas.1501176112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Backovic M et al. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. PNAS 107, 22635–22640 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsuura H, Kirschner AN, Longnecker R & Jardetzky TS Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. PNAS 107, 22641–22646, doi: 10.1073/pnas.1011806108 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Connolly SA, Jackson JO, Jardetzky TS & Longnecker R Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol 9, 369–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gompels UA et al. Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J Virol 65, 2393–2401 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ciferri C et al. Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies. PLoS Pathog 11, e1005230, doi: 10.1371/journal.ppat.1005230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sathiyamoorthy K et al. Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies. PNAS 114, doi: 10.1073/pnas.1704661114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Snijder J et al. An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity 48, 799–811 e799, doi: 10.1016/j.immuni.2018.03.026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson DW, Davis-Poynter N & Minson AC Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J Virol 68, 6985–6993 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Browne HM, Bruun BC & Minson AC Characterization of herpes simplex virus type 1 recombinants with mutations in the cytoplasmic tail of glycoprotein H. J Gen Virol 77, 2569–2573 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Jackson JO, Lin E, Spear PG & Longnecker R Insertion mutations in herpes simplex virus 1 glycoprotein H reduce cell surface expression, slow the rate of cell fusion, or abrogate functions in cell fusion and viral entry. J Virol 84, 2038–2046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silverman JL & Heldwein EE Mutations in the cytoplasmic tail of herpes simplex virus 1 gH reduce the fusogenicity of gB in transfected cells. J Virol 87, 10139–10147, doi: 10.1128/JVI.01760-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rogalin HB & Heldwein EE Interplay between the Herpes Simplex Virus 1 gB Cytodomain and the gH Cytotail during Cell-Cell Fusion. J Virol 89, 12262–12272, doi: 10.1128/JVI.02391-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang E, Arvin AM & Oliver SL The cytoplasmic domain of varicella-zoster virus glycoprotein H regulates syncytia formation and skin pathogenesis. PLoS Pathog 10, e1004173, doi: 10.1371/journal.ppat.1004173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vallbracht M, Fuchs W, Klupp BG & Mettenleiter TC Functional Relevance of the Transmembrane Domain and Cytoplasmic Tail of the Pseudorabies Virus Glycoprotein H for Membrane Fusion. J Virol 92, e00376–00318, doi: 10.1128/JVI.00376-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J, Jardetzky TS & Longnecker R The Cytoplasmic Tail Domain of Epstein-Barr Virus gH Regulates Membrane Fusion Activity through Altering gH Binding to gp42 and Epithelial Cell Attachment. mBio 7, doi: 10.1128/mBio.01871-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rowe CL, Connolly SA, Chen J, Jardetzky TS & Longnecker R A soluble form of Epstein-Barr virus gH/gL inhibits EBV-induced membrane fusion and does not function in fusion. Virology 436, 118–126, doi: 10.1016/j.virol.2012.10.039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones NA & Geraghty RJ Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology 324, 213–228 (2004). [DOI] [PubMed] [Google Scholar]
- 77. Cooper RS, Georgieva ER, Borbat PP, Freed JH & Heldwein EE Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB. Nat Struct Mol Biol 25, 416–424, doi: 10.1038/s41594-018-0060-6 (2018). Reports the crystal structure of full-length HSV-1 gB, including the membrane proximal, transmembrane, and cytoplasmic tail domains.
- 78.White JM, Delos SE, Brecher M & Schornberg K Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Bio 43, 189–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heldwein EE et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313, 217–220 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Vallbracht M et al. Structure-function dissection of the Pseudorabies virus glycoprotein B fusion loops. J Virol 92, e01203–01201, doi: 10.1128/JVI.01203-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li X et al. Two classes of protective antibodies against Pseudorabies virus variant glycoprotein B: Implications for vaccine design. PLoS Pathog 13, e1006777, doi: 10.1371/journal.ppat.1006777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oliver SL et al. A glycoprotein B-neutralizing antibody structure at 2.8 Å uncovers a critical domain for herpesvirus fusion initiation. Nature communications 11, 1–15, doi: 10.1038/s41467-020-17911-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chandramouli S et al. Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat Commun 6, 8176, doi: 10.1038/ncomms9176 (2015). Reports the crystal structure of the postfusion conformation of HCMV gB bound to a nAb.
- 84. Burke HG & Heldwein EE Crystal Structure of the Human Cytomegalovirus Glycoprotein B. PLoS Pathog 11, e1005227, doi: 10.1371/journal.ppat.1005227 (2015). Reports the crystal structure of the postfusion conformation of HCMV gB.
- 85.Backovic M, Longnecker R & Jardetzky TS Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. PNAS 106, 2880–2885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roche S, Bressanelli S, Rey FA & Gaudin Y Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313, 187–191 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Yang F et al. Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody. Cell Host Microbe, doi: 10.1016/j.chom.2019.12.012 (2020). [DOI] [PubMed] [Google Scholar]
- 88.Kadlec J, Loureiro S, Abrescia NG, Stuart DI & Jones IM The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol 15, 1024–1030 (2008). [DOI] [PubMed] [Google Scholar]
- 89.Peng R et al. Structures of human-infecting Thogotovirus fusogens support a common ancestor with insect baculovirus. PNAS 114, E8905–E8912, doi: 10.1073/pnas.1706125114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Backovic M & Jardetzky TS Class III viral membrane fusion proteins. Curr Opin Struc Biol 19, 189–196, doi: 10.1016/j.sbi.2009.02.012 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bender FC et al. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol 81, 3827–3841 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bootz A et al. Protective capacity of neutralizing and non-neutralizing antibodies against glycoprotein B of cytomegalovirus. PLoS Pathog 13, e1006601, doi: 10.1371/journal.ppat.1006601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cairns TM et al. Mechanism of neutralization of herpes simplex virus by antibodies directed at the fusion domain of glycoprotein B. J Virol 88, 2677–2689, doi: 10.1128/JVI.03200-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin E & Spear PG Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. PNAS 104, 13140–13145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reimer JJ, Backovic M, Deshpande CG, Jardetzky T & Longnecker R Analysis of Epstein-Barr virus glycoprotein B functional domains via linker insertion mutagenesis. J Virol 83, 734–747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maurer UE et al. The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein-membrane and lateral protein-protein interaction. Structure 21, 1396–1405, doi: 10.1016/j.str.2013.05.018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Satoh T et al. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132, 935–944 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suenaga T et al. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. PNAS 107, 866–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arii J et al. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature 467, 859–862 (2010). [DOI] [PubMed] [Google Scholar]
- 100.Roche S, Rey FA, Gaudin Y & Bressanelli S Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315, 843–848 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Silverman JL, Sharma S, Cairns TM & Heldwein EE Fusion-deficient insertion mutants of herpes simplex virus 1 glycoprotein B adopt the trimeric postfusion conformation. J Virol 84, 2001–2012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vitu E, Sharma S, Stampfer SD & Heldwein EE Extensive mutagenesis of the HSV-1 gB ectodomain reveals remarkable stability of its postfusion form. J Mol Biol 425, 2056–2071, doi: 10.1016/j.jmb.2013.03.001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Vollmer B et al. The pre-fusion structure of herpes simplex virus glycoprotein B. Sci Adv 6, eabc1726, doi: 10.1126/sciadv.abc1726 (2020). EM reconstructions of a membrane-anchored HSV-1 gB mutant designed to trap the prefusion form reveal a compact structure at an overall resolution of 9 Å.
- 104.Falanga A et al. Biophysical characterization and membrane interaction of the two fusion loops of glycoprotein B from herpes simplex type I virus. PLoS One 7, e32186, doi: 10.1371/journal.pone.0032186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oliver SL et al. Mutagenesis of varicella-zoster virus glycoprotein B: putative fusion loop residues are essential for viral replication, and the furin cleavage motif contributes to pathogenesis in skin tissue in vivo. J Virol 83, 7495–7506, doi: 10.1128/JVI.00400-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Backovic M, Jardetzky TS & Longnecker R Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J Virol 81, 9596–9600 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hannah BP et al. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J Virol 83, 6825–6836 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Harrison SC Viral membrane fusion. Virology 479-480, 498–507, doi: 10.1016/j.virol.2015.03.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Melikyan GB et al. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151, 413–423 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Connolly SA & Longnecker R Residues within the C-terminal arm of the herpes simplex virus 1 glycoprotein B ectodomain contribute to its refolding during the fusion step of virus entry. J Virol 86, 6386–6393, doi: 10.1128/JVI.00104-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fan Q, Kopp SJ, Connolly SA & Longnecker R Structure-Based Mutations in the Herpes Simplex Virus 1 Glycoprotein B Ectodomain Arm Impart a Slow-Entry Phenotype. mBio 8, doi: 10.1128/mBio.00614-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Waning DL, Russell CJ, Jardetzky TS & Lamb RA Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. PNAS 101, 9217–9222 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wyss S et al. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain on the gp41 cytoplasmic tail. JVirol 79, 12231–12241 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garcia NJ, Chen J & Longnecker R Modulation of Epstein-Barr virus glycoprotein B (gB) fusion activity by the gB cytoplasmic tail domain. mBio 4, e00571–00512, doi: 10.1128/mBio.00571-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nixdorf R, Klupp BG, Karger A & Mettenleiter TC Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J Virol 74, 7137–7145 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fan Z et al. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J Virol 76, 9271–9283 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Muggeridge MI, Grantham ML & Johnson FB Identification of syncytial mutations in a clinical isolate of herpes simplex virus 2. Virology 328, 244–253, doi: 10.1016/j.virol.2004.07.027 (2004). [DOI] [PubMed] [Google Scholar]
- 118.Silverman JL, Greene NG, King DS & Heldwein EE Membrane requirement for folding of the herpes simplex virus 1 gB cytodomain suggests a unique mechanism of fusion regulation. J Virol 86, 8171–8184, doi: 10.1128/JVI.00932-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oliver SL et al. An immunoreceptor tyrosine-based inhibition motif in varicella-zoster virus glycoprotein B regulates cell fusion and skin pathogenesis. PNAS 110, 1911–1916, doi: 10.1073/pnas.1216985110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cooper RS & Heldwein EE Herpesvirus gB: A Finely Tuned Fusion Machine. Viruses 7, 6552–6569, doi: 10.3390/v7122957 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gallagher JR et al. Functional fluorescent protein insertions in herpes simplex virus gB report on gB conformation before and after execution of membrane fusion. PLoS Pathog 10, e1004373, doi: 10.1371/journal.ppat.1004373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zeev-Ben-Mordehai T et al. Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. PNAS 113, 4176–4181, doi: 10.1073/pnas.1523234113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fontana J et al. The Fusion Loops of the Initial Prefusion Conformation of Herpes Simplex Virus 1 Fusion Protein Point Toward the Membrane. mBio 8, doi: 10.1128/mBio.01268-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Si Z et al. Different functional states of fusion protein gB revealed on human cytomegalovirus by cryo electron tomography with Volta phase plate. PLoS Pathog 14, e1007452, doi: 10.1371/journal.ppat.1007452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Handler CG, Cohen GH & Eisenberg RJ Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol 70, 6076–6082 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gianni T, Amasio M & Campadelli-Fiume G Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J Biol Chem 284, 17370–17382, doi: 10.1074/jbc.M109.005728 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Perez-Romero P, Perez A, Capul A, Montgomery R & Fuller AO Herpes simplex virus entry mediator associates in infected cells in a complex with viral proteins gD and at least gH. J Virol 79, 4540–4544 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Avitabile E, Forghieri C & Campadelli-Fiume G Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol 81, 11532–11537 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Atanasiu D et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. PNAS 104, 18718–18723 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fan Q, Longnecker R & Connolly SA Substitution of herpes simplex virus 1 entry glycoproteins with those of saimiriine herpesvirus 1 reveals a gD-gH/gL functional interaction and a region within the gD profusion domain that is critical for fusion. J Virol 88, 6470–6482, doi: 10.1128/JVI.00465-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fan Q, Longnecker R & Connolly SA A Functional Interaction between Herpes Simplex Virus 1 Glycoprotein gH/gL Domains I and II and gD Is Defined by Using Alphaherpesvirus gH and gL Chimeras. J Virol 89, 7159–7169, doi: 10.1128/JVI.00740-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Atanasiu D et al. Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors. mBio 4, doi: 10.1128/mBio.00046-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Cairns TM et al. Surface Plasmon Resonance Reveals Direct Binding of Herpes Simplex Virus Glycoproteins gH/gL to gD and Locates a gH/gL Binding Site on gD. J Virol 93, doi: 10.1128/JVI.00289-19 (2019). Demonstrates direct interaction between purified forms of the HSV-2 gD and gH-gL ectodomains.
- 134.Atanasiu D et al. Bimolecular complementation defines functional regions of Herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol 84, 3825–3834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vanarsdall AL, Ryckman BJ, Chase MC & Johnson DC Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed in cis or in trans. J Virol 82, 11837–11850 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Oxman MN et al. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N Engl J Med 352, 2271–2284, doi: 10.1056/NEJMoa051016 (2005). [DOI] [PubMed] [Google Scholar]
- 137.Cunningham AL et al. Efficacy of the Herpes Zoster Subunit Vaccine in Adults 70 Years of Age or Older. N Engl J Med 375, 1019–1032, doi: 10.1056/NEJMoa1603800 (2016). [DOI] [PubMed] [Google Scholar]