Abstract
Contribution of the renin-angiotensinogen system in the risk of COVID-19 and related complications have been assessed by several groups. However, the results are not consistent. We examined levels of ACE1 and ACE2 in the circulation of two groups of COVID-19 patients (ICU-admitted and general ward-admitted patients) compared with healthy controls. We also genotyped two polymorphisms in ACE1 gene (the ACE1-I/D polymorphism rs1799752 and rs4359) to appraise their association with expression levels of ACE1 and ACE2. Expression level of ACE1 was significantly higher in ICU patients compared with non-ICU patients (P value = 0.02). However, its expression was not significantly different between total COVID-19 patients and total controls (P value = 0.34). ACE2 expression was not different ether between two groups of COVID-19 patients (P value = 0.12) or between total COVID-19 patients and total controls (P value = 0.79). While distribution of rs1799752 and rs4359 alleles was similar between study groups, genotype frequencies of rs1799752 were differently distributed among total COVID-19 patients and controls (P value = 0.00001). Moreover, genotypes of the other polymorphism tended to be distinctively distributed among these two groups (P value = 0.06). In the total population of patients and controls, different ACE1 mRNA levels were observed among carriers of different rs1799752 genotypes; of note, ID genotype carriers showed a higher expression of ACE1 compared with II genotype carriers (P = 0.01). ACE1 polymorphisms might affect risk of COVID-19 and expression of ACE transcripts.
Keywords: ACE1, ACE2, COVID-19, Expression
Graphical abstract
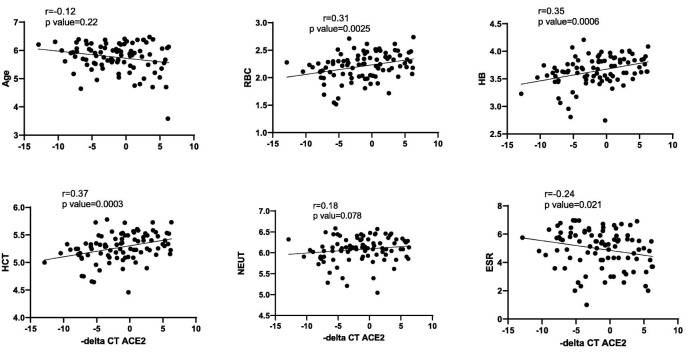
The correlation of ACE2 and ACE1 expression levels with age, blood cells counts, ESR and CRP was analyzed using nonparametric spearman correlation test. – delta CTs of ACE1 and ACE2 genes and log 2 of age and blood cells parameters were used for correlation tests. There was a significant positive correlation between ACE2 expression levels and RBC, HB and HCT levels in COVID-19 patients. However, ACE2 expression levels had a negative correlation with ESR in COVID-19 patients. In contrast to ACE2, ACE1 expressions had no correlations with assessed factors (except for HCT (r = 0.2, p = 0.04)).
1. Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has emerged as a global health problem since late 2019. This disorder has been associated with abnormal immune responses in some patients. Adult stem cells which have immunomodulatory and pro-reparative activities in the local environment [1,2], might be used as modulators of these response. Several research groups have aimed at identification of the underlying mechanism of susceptibility to this infection and development of severe forms of COVID-19 pointing to changes in the expression of certain genes or the presence of polymorphisms in genes mediating virus entry to target cells. Angiotensin-converting enzyme (ACE2) as the gene encoding the specific receptor of the SARS-CoV-2 has been at the center of attention. Expression level of ACE2 in the epithelial cells have been suggested as a factor for increasing susceptibility to COVID-19 [3]. Others have linked the lower risk of COVID-19 among children to the decreased levels of ACE2 receptor in them compared with adults [4].
Meanwhile, the presence of polymorphisms in renin-angiotensin system has been associated with risk of COVID-19 and disease course. For instance, ACE1-insertion/deletion (I/D) polymorphism has been shown to be strongly associated with COVID-19. Carriers of DD genotype have exhibited higher ACE1 levels and higher risk for development of acute respiratory distress syndrome and mortality [5,6]. In fact, D allele of this polymorphism has been found to be associated with progression of COVID-19 [7] and mortality rate from this infection [6].
Evidence suggests that SARS-CoV-2 interferes with normal balance of ACE1/ACE2 and induces the angiotensin II (Ang II)/Angiotensin II type 1 receptor (AT1R) pathway, resulting in severe COVID-19 consequences [8]. In addition, down-regulation of ACE2 and the imbalance between the renin-angiotensin system and ACE2/angiotensin- [[1], [2], [3], [4], [5], [6], [7]]/MAS following COVID-19 infection has been proposed to participate in the pathogenesis of multiple organ damage in this disorder [9].
In this study, we examined levels of ACE1 and ACE2 in the circulation of two groups of COVID-19 patients (ICU-admitted and general ward-admitted patients) compared with healthy controls. Moreover, we genotyped two polymorphisms in ACE1 gene (the ACE1-I/D polymorphism rs1799752 and rs4359) to appraise their association with expression levels of ACE1 and ACE2.
2. Material and methods
2.1. Cases and controls
The present genotyping and expression assay project was performed on 91 COVID-19 cases admitted to Nikan Hospital, Tehran, during 2020. COVID-19 was confirmed in all cases through RT-PCR method on nasopharyngeal swab samples. In addition, 91 control specimens were obtained from unaffected individuals without history of exposure to COVID-19 cases. The presence of any infectious or immune-related disorder was considered as criterion for exclusion of controls. The study protocol was approved by ethical committee of Shahid Beheshti University of Medical Sciences. Informed consent was obtained from all COVID-19 cases and controls. Laboratory parameters were gathered from all COVID-19 cases.
2.2. Genotyping
Tetra-primer amplification-refractory mutation system-PCR method was used for identification of rs4359 genotypes according to our former research [10]. Primers were designed using Primer1 software. The sequences of primers were as follow: Forward inner primer (T allele): GGGTCAGACAGAACTGGGTTCAATCT, Reverse inner primer (C allele): TTCTCTAGGAAACAAAGTAATGGAGACTGG, Forward outer primer: TGGCTAATGGTTACCTGACCTTGGTTAA and Reverse outer primer: TAGAGAGTGATGAATAGTGGGGTCCTGG. Annealing step was set at 62 °C.
Two rounds of PCR and electrophoresis were used for genotyping of the rs1799752 (I/D) polymorphism. First round of PCR was accomplished using the following primers: TGGAGAGCCACTCCCATCCTTTCT and GACGTGGCCATCACATTCGTCAGAT. Second round of PCR was performed using TGTAAGCCACTGCTGGAGAG and TGGCCATCACATTCGTCAGA as forward and reverse primers, respectively. The PCR program consisted an initial denaturing phase at 95 °C for 5 min; 35 cycles at 95 °C for 30 s, specific annealing temperature for 30 s and extension at 72 °C for 60 s. Finally, microtubes were incubated at 72 °C for 5 min. Genotyping results with confirmed with Sanger sequencing of a number of samples.
2.3. Expression assays
Blood samples were collected from COVID-19 patients and healthy controls in EDTA-containing tubes. Total RNA was extracted from all samples using GeneAll RNA extraction kit (Seoul, South Korea). Then, RNA was converted to complementary DNA using the BioFact™ kit (Seoul, South Korea). Levels of ACE1 and ACE2 genes were quantified in all samples in relation with expression of B2M gene. The RealQ Plus 2× Master Mix (Amplicon, Denmark) was used for preparation of reactions. Table 1 demonstrates the information about primers sequences and amplicons.
Table 1.
Sequence of primers used in expression assays.
| Gene | Primer sequence | Primer length | Product size | |
|---|---|---|---|---|
| ACE1 | Forward primer | ACGTGAGGATACAGCAAGGC | 20 | 75 |
| Reverse primer | AGAGTTCCTGCATGGTCTGG | 20 | ||
| ACE2 | Forward primer | ATCTACTCCACCGCCAAGGT | 20 | 187 |
| Reverse primer | TGCTGAGGGCAGTGAAATCC | 20 | ||
| B2M | Forward primer | AGATGAGTATGCCTGCCGTG | 20 | 105 |
| Reverse primer | GCGGCATCTTCAAACCTCCA | 20 | ||
2.4. Statistical methods
The Statistical Package for the Social Sciences (SPSS) v.22.0 (SPSS Inc., Chicago, IL) and SNP Analyzer 2.0 were used for statistical assessments. Graphics were created using GraphPad Prism version 9.0 for Windows (La Jolla California, USA). Expressions of ACE1 and ACE2 genes in each sample were calculated using the Efficiency adjusted Ct of normalizer gene (B2M) - Efficiency adjusted Ct of target genes (comparative –delta Ct method). Student t-test was used to compare expression levels of ACE1 and ACE2 between groups (COVID-19 patients vs. controls, and ICU patients vs. non-ICU patients). Mann Whitney's U test or student t-test was used to compare laboratory data between subgroups of COVID-19 patient (ICU patients vs. non-ICU patients).
Allele and genotype frequencies were compared between groups by the chi-squared test. Relative risk (odds ratio (OR)) for effect alleles and genotypes was calculated by logistic regression. Adjusted relative risks were calculated considering gender and age as covariates. Associations between genomic variants and COVID-19 risk were assessed in codominant, dominant, recessive and over-dominant models. The results of association analysis were described as OR and 95% confidence interval of OR (95% CI), P-value and FDR adjusted q-values. The FDR adjusted q-values were calculated through analyzing a stack of p values in column analyses by GraphPad Prism version 9.0. P-values less than 0.05 were considered as statistically significant. Estimation of accordance of genotype distributions with Hardy–Weinberg equilibrium, haplotype estimation, linkage disequilibrium (LD) blocking and were performed in SNP Analyzer 2.0.
The correlation of ACE2 and ACE1 expression levels with age, complete blood cells, ESR and CRP were analyzed using nonparametric spearman correlation test.
3. Results
3.1. General data of patients
First, we compared general laboratory data of ICU-admitted and general ward-admitted patients. This comparison showed higher levels of ESR and CRP in ICU-admitted group (P values = 0.011 and 0.000001, respectively). Moreover, ICU-admitted patients had lower lymphocyte count while higher neutrophil count (P values = 0.000492 and 0.000086, respectively) (Table 2 ).
Table 2.
General laboratory data of COVID-19 patients' subgroups.
| Parameters | ICU (N = 37) | Non-ICU (N = 54) | pa |
|---|---|---|---|
| WBCs | 10,316.48 ± 1980.19 | 6613.3 ± 598.21 | 0.027 |
| RBC | 4.6 ± 0.14 | 4.73 ± 0.091 | 0.29 |
| HB | 12.29 ± 0.41 | 12.98 ± 0.26 | 0.133 |
| HCT | 38.21 ± 1.25 | 39.98 ± 0.78 | 0.23 |
| MCV | 83.08 ± 0.9 | 84.6 ± 0.79 | 0.32 |
| MCH | 26.76 ± 0.39 | 27.41 ± 0.31 | 0.106 |
| MCHC | 32.24 ± 0.25 | 32.42 ± 0.16 | 0.317 |
| PLT | 225,718.91 ± 17,222.78 | 199,827.77 ± 12,044.75 | 0.17 |
| LYM | 16.35 ± 1.68 | 24.25 ± 1.5 | 0.000492 |
| NEU | 75.21 ± 1.93 | 64.90 ± 1.69 | 0.000086 |
| ESR | 54.21 ± 5.56 | 37.22 ± 4.16 | 0.011 |
| CRP | 115.29 ± 12.21 | 44.45 ± 6.76 | 0.000001 |
Mann Whitney's U test or student t-test was used to analyze data between subgroups of COVID-19 patients.
3.2. Genotyping and expression assays
Expression level of ACE1 was significantly higher in ICU patients compared with non-ICU patients (P value = 0.02). However, its expression was not significantly different between total COVID-19 patients and total controls (P value = 0.34). ACE2 expression was not different ether between two groups of COVID-19 patients (P value = 0.12) or between total COVID-19 patients and total controls (P value = 0.79). While distribution of rs1799752 and rs4359 alleles was similar between study groups, genotype frequencies of rs1799752 were differently distributed among total COVID-19 patients and controls (P value = 0.00001). Moreover, genotypes of the other polymorphism tended to be distinctively distributed among these two groups (P value = 0.06). Table 3 shows demographic data, relative expressions of ACE1 and ACE2 and distribution of ACE1 variants in COVID-19 patients and matched controls.
Table 3.
Demographic data, ACE1 and ACE2 relative expressions and distribution of ACE1 variants in COVID-19 patients and matched controls.
| Parameters | ICU patients (N = 37) | Non-ICU patients (N = 54) | pa | All COVID-19 patients (N = 91) | Controls (N = 91) |
pa |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male | 24 (64.9) | 29 (53.7) | 0.28 | 53 (58.2) | 52 (57.1) | Adjusted |
| Female | 13 (35.1) | 25 (46.3) | 38 (41.8) | 39 (42.9) | variable | |
| Age (year) | 58.62 ± 2.66 | 56.18 ± 2.39 | 0.50 | 57.17 ± 1.78 | 51.82 ± 1.5 | Adjusted variable |
| ACE1 Expression (−delta CT) | −3.63 ± 1.08 | −6.43 ± 0.65 | 0.02 | −5.29 ± 0.6 | - 4.85 ± 0.5 | 0.34 |
| ACE2 Expression (−delta CT) | −0.61 ± 0.57 | −2.06 ± 0.66 | 0.12 | −1.47 ± 0.46 | −1.28 ± 0.58 | 0.79 |
| Allele (n %) | ||||||
| rs1799752 | ||||||
| I* D* |
31 (41.9) 43 (58.1) |
47 (43.5) 61 (56.5) |
0.82 | 78 (42.9) 104 (57.1) |
79 (43.4) 103 (56.6) |
0.91 |
| rs4359 | ||||||
| T* | 33 (44.6) | 51 (47.2) | 0.72 | 84 (46.2) | 73 (40.1) | 0.24 |
| C* | 41 (55.4) | 57 (52.8) | 98 (53.8) | 109 (59.9) | ||
| ACE1 Genotypes (n %) | ||||||
| rs1799752 | ||||||
| II | 0 (0) | 4 (7.4) | 0.09 | 4 (4.4) | 21 (23.1) | <0.00001 |
| ID | 31 (83.8) | 39 (72.2) | 70 (76.9) | 37 (40.7) | ||
| DD | 6 (16.2) | 11 (20.4) | 17 (18.7) | 33 (36.6) | ||
| rs4359 | ||||||
| TT | 5 (13.5) | 7 (13) | 0.78 | 12 (13.2) | 14 (15.4) | |
| TC | 23 (62.2) | 37 (68.5) | 60 (65.9) | 45 (49.5) | 0.06 | |
| CC | 9 (24.3) | 10 (18.5) | 19 (20.9) | 32 (35.5) |
Data are shown as mean ± standard error of mean or percentage. Alleles*, total number of chromosomes.
student's t-test, Mann Whitney's U test or Chi square test as appropriate.
Distribution of genotype frequencies of both polymorphisms were in accordance with Hardy-Weinberg equilibrium in control subjects (P values = 0.77 and 0.1, respectively), but not in patients (P values = 0.0018 and < 0.0001, respectively) (Table 4 ).
Table 4.
The results of exact-test for Hardy-Weinberg equilibrium (P values and genotype distributions are shown).
| Variants | rs4359 |
rs1799752 |
||||||
|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | Hardy-Weinberg P-value | DD | ID | II | Hardy-Weinberg P-value | |
| COVID-19 patients | 19 | 60 | 12 | 0.0018 | 17 | 70 | 4 | <0.0001 |
| Normal controls | 32 | 45 | 14 | 0.77 | 33 | 37 | 21 | 0.1 |
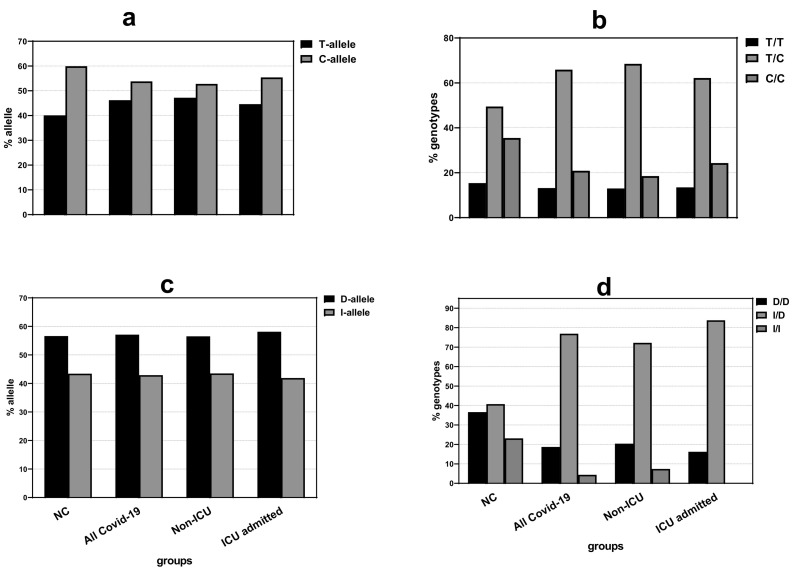
Fig. 1 shows distributions of alleles and genotypes of rs4359 and rs1799752 variants among normal controls, all COVID-19 patients, non-ICU and ICU-admitted cases.
Fig. 1.
Distributions of alleles (a, c) and genotypes (b, d) of rs4359 and rs1799752 among normal controls (NC), COVID-19 patients, non-ICU and ICU-admitted cases. There was significant difference between genotypes distribution of rs1799752 among COVID-19 patients compared to matched controls.
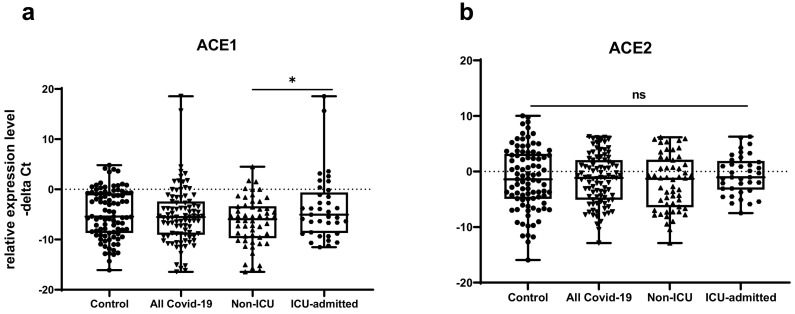
Fig. 2 demonstrates relative expressions of ACE1 and ACE2 in COVID-19 patients compared with healthy controls.
Fig. 2.
Relative expressions of ACE1 and ACE2 in COVID-19 patients (n = 91) compared with healthy controls (n = 91). There was no significant difference in expression of ACE1 or ACE2 between COVID-19 patients and healthy controls. However, a moderate increase in ACE1 expression was observed in ICU-admitted COVID-19 patients compared to other group of COVID-19 patients. – delta Ct values in the figures were plotted as box and whisker plots (showing the median [line], interquartile range [box], and minimum and maximum values).
3.3. Association between ACE1 genotypes and ACE1 and ACE2 expressions
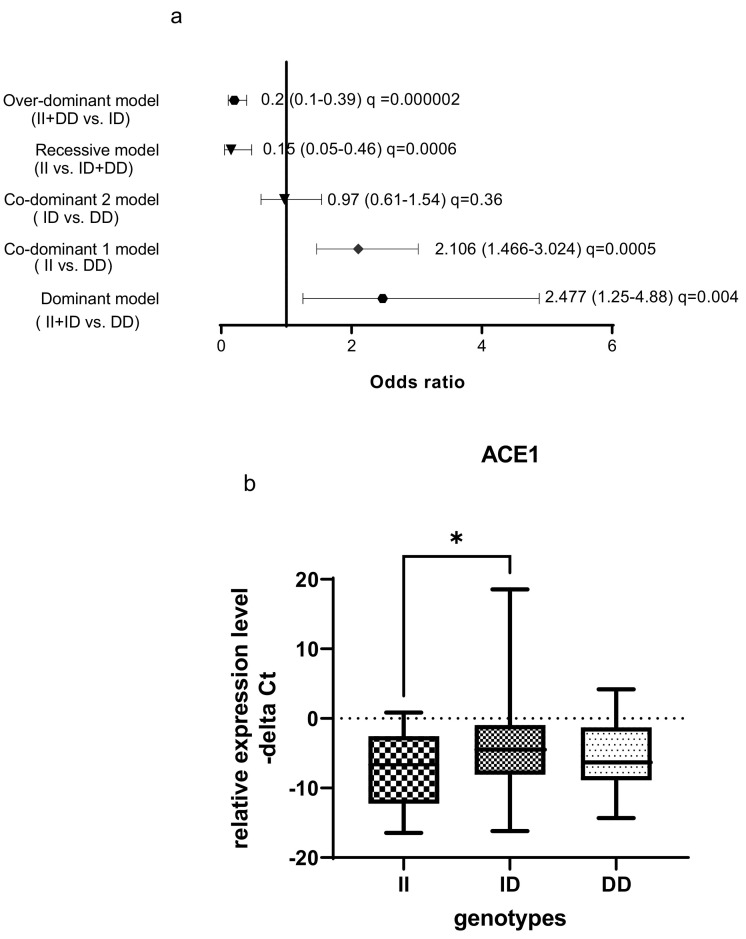
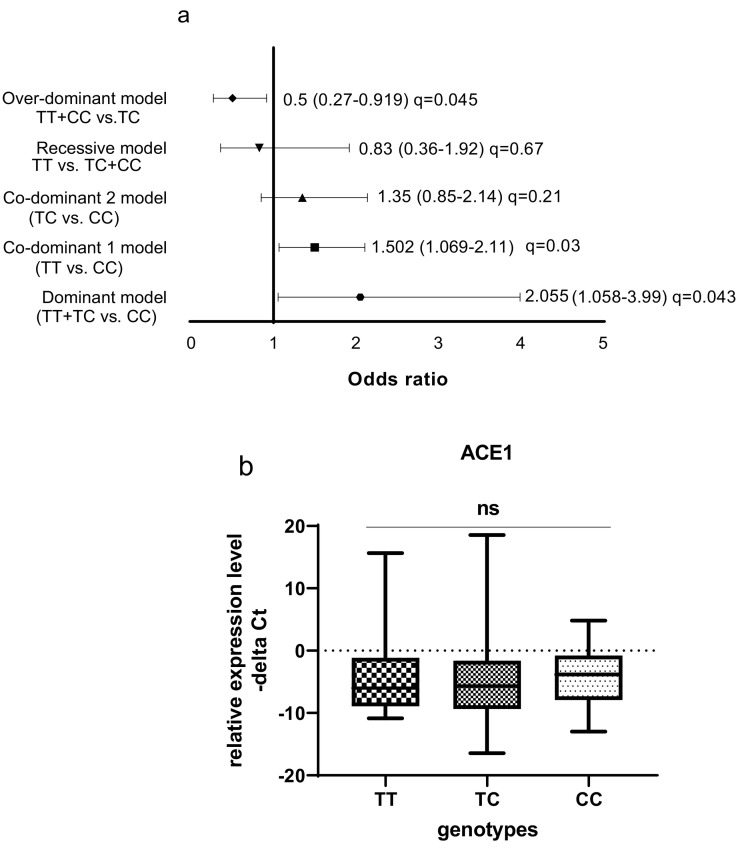
The ACE1 rs1799752 polymorphism was associated with a high risk of COVID-19 in dominant and co-dominant models (Fig. 3 ). In the dominant model, the presence of at least one mutated (−) allele was tested against the homozygous wildtype genotype (wt/wt). The ACE1 rs1799752 polymorphism showed a significant protective effect against COVID-19 risk in over-dominant model. In the total population of patients and controls, different ACE1 mRNA levels were observed among carriers of different rs1799752 genotypes; of note, ID genotype carriers showed a higher expression of ACE1 compared with II genotype carriers (P = 0.01).
Fig. 3.
Association between ACE1 rs1799752 polymorphism and COVID-19 infection as well as ACE1 mRNA levels by rs1799752 genotypes. (a) The results of association tests under five different inheritance models are shown. The Odds Ratios (plus Confidence Intervals) are reported on the X axis in a linear scale. Data on the right of Y axis indicates causative effects toward the risk and the data on the left indicates protective effects. The ACE1 rs1799752 polymorphism was associated with a high risk of COVID-19 in dominant and co-dominant models. In the dominant model, the presence of at least one mutated (−) allele was tested against the homozygous wildtype genotype (wt/wt). The ACE1 rs1799752 polymorphism showed a significant protective effect against COVID-19 risk in over-dominant model. (b) In the total population of patients and controls, different ACE1 mRNA levels were observed among carriers of different rs1799752 genotypes; of note, ID genotype carriers showed a higher expression of ACE1 compared with II genotype carriers (P = 0.01). The level of Odds Ratios was showed as FDR adjusted q-values. MAF: minor allele frequency.
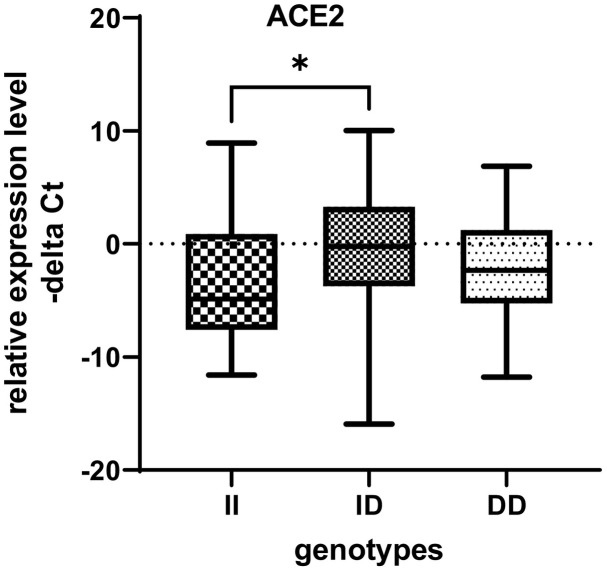
ACE2 expression was significantly higher in ID genotype carriers than the II genotype carriers in total population. In fact, increase in ACE1 expression was associated with increase in the ACE2 expression in ID genotype carriers (Fig. 4 ).
Fig. 4.
ACE2 expression was significantly higher in ID genotype carriers than the II genotype carriers in total population.
The ACE rs4359 polymorphism was associated with a higher risk of COVID-19 in dominant and co-dominant models. The ACE1 rs4359 polymorphism showed a significant protective effect against the risk for COVID-19 in over-dominant model. However, in contrast to the rs1799752 genotypes, there was no significant difference in the ACE1 expression level between carriers of different rs4359 genotypes in the total population of patients and controls (Fig. 5 ).
Fig. 5.
Association between ACE1 rs4359 gene polymorphism and COVID-19 risk as well as ACE1 mRNA levels by rs4359 genotypes. (a) The results of association tests under five different inheritance models are shown. The Odds Ratios (plus Confidence Intervals) are reported on the X axis in a linear scale. Data on the right of Y axis indicates causative effects toward the risk and the data on the left indicates protective effects. The ACE1 rs4359 polymorphism was associated with a higher risk of COVID-19 in dominant and co-dominant models. The ACE1 rs4359 polymorphism showed a significant protective effect against the risk for COVID-19 in over-dominant model. (b) There was no significant difference in the ACE1 expression level between rs4359 genotypes in the total population of patients and controls. Odds Ratios are shown as FDR adjusted q-values. MAF: minor allele frequency.
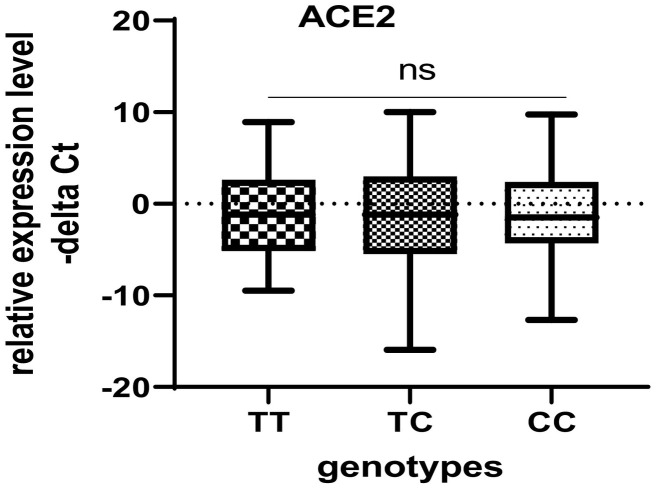
There was no significant difference in ACE2 expression level among the rs4359 genotype carriers which was consistent with ACE1 expression levels among the rs4359 genotype carriers (Fig. 6 ).
Fig. 6.
ACE2 expression in carriers of different rs4359 genotypes in total population.
There was no significant difference in distribution of rs4359 and rs1799752 variants between CIVID-19 cases and control in allelic model.
Then, we appraised association between estimated haplotypes and COVID-19 risk. The results showed similar distribution of ACE1 haplotypes between COVID-19 cases and healthy controls (Table 5 ).
Table 5.
The results of haplotype analysis in COVID-19 patients and controls.
| rs4359 | rs1799752 | Case | Control | Total | OR (95% CI) | P-value | FDR q-Value |
|---|---|---|---|---|---|---|---|
| C | D | 0.22 | 0.35 | 0.32 | 1.14 (0.75–1.74) | 0.52 | 0.525 |
| C | I | 0.31 | 0.24 | 0.23 | 0.48 (0.26–0.87) | 0.014 | 0.057 |
| T | D | 0.34 | 0.21 | 0.23 | 0.81 (0.46–1.43) | 0.47 | 0.525 |
| T | I | 0.11 | 0.19 | 0.19 | 1.55 (0.97–2.48) | 0.06 | 0.121 |
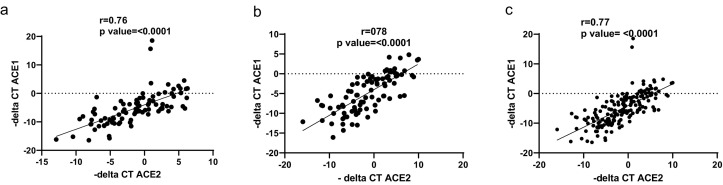
A strong positive correlation has been detected between ACE1 and ACE2 expression in COVID-19 patients, healthy controls as well as total population (Fig. 7 ).
Fig. 7.
Correlation between ACE1 and ACE2 expression levels. There was a strong positive correlation between ACE1 and ACE2 expression in COVID-19 patients (a), healthy controls (b), and total population (c).
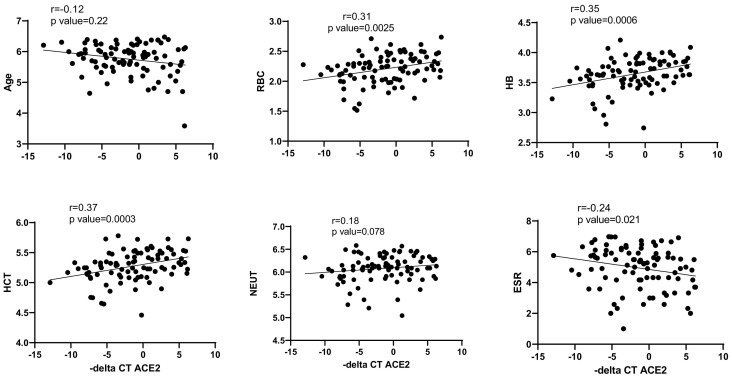
Finally, the correlation of ACE2 and ACE1 expression levels with age, blood cells counts, ESR and CRP was analyzed using nonparametric spearman correlation test. – delta CTs of ACE1 and ACE2 genes and log 2 of age and blood cells parameters were used for correlation tests. There was a significant positive correlation between ACE2 expression levels and RBC, HB and HCT levels in COVID-19 patients. However, ACE2 expression levels had a negative correlation with ESR in COVID-19 patients (Fig. 8 ). In contrast to ACE2, ACE1 expressions had no correlations with assessed factors (except for HCT (r = 0.2, p = 0.04)).
Fig. 8.
Correlation of ACE2 expression levels with age, RBC, HB, HCT, NEUT and ESR of COVID-19 patients. – delta CTs of ACE2 and log 2 of age and blood cells parameters were used for correlation tests.
4. Discussion
The importance of renin-angiotensin system in the pathogenesis of COVID-19 has been assessed by several studies. Over-activation of the renin-angiotensin-aldosterone has been suggested to participate in abnormal biochemical and clinical manifestations of SARS-CoV-2 infection [11]. The protective renin-angiotensin system medicated by ACE2 might be inhibited in COVID-19 [11]. ACE2 is regarded as a negative regulator of renin angiotensin system which converts Ang II to angiotensin 1–7 [12]. On the other hand, ACE1 catalyzes biogenesis of Ang II from Ang I [13]. Thus, ACE1/ACE2 level has a critical significance in the pathogenesis of disorders associated with renin-angiotensin system [14].
In the current study, we demonstrated higher levels of ACE1 in ICU patients compared with non-ICU patients. However, its expression was not significantly different between total COVID-19 patients and total controls. ACE2 expression was not different ether between two groups of COVID-19 patients or between total COVID-19 patients and total controls. These findings indicate imbalance in ACE1/ACE2 level in severely affected COVID-19 patients.
The ACE1 rs1799752 polymorphism was associated with a high risk of COVID-19 in dominant and co-dominant models. In the dominant model (II + ID versus DD), the presence of at least one mutated allele has been shown to increase risk of COVID-19. This polymorphism showed a significant protective effect against COVID-19 risk in over-dominant model (II + DD versus ID). Yet, due to small sample size, this result is not conclusive. In the total population of patients and controls, different ACE1 mRNA levels were observed among carriers of different rs1799752 genotypes. Most notably, ID genotype carriers showed a higher expression of ACE1 compared with II genotype carriers. Supposing the impact of ACE1 up-regulation in COVID-19, this finding is in accordance with the observed association between this polymorphism and risk of COVID-19 in dominant and over-dominant models in the current study. However, these findings are in contrast with the previously reported higher levels of ACE1 in carriers of DD genotype [5,6]. ACE2 expression was also higher in ID genotype carriers than the II genotype carriers in total population.
The ACE1 rs4359 polymorphism was associated with a higher risk of COVID-19 in dominant and co-dominant models. The ACE1 rs4359 polymorphism showed a significant protective effect against the risk for COVID-19 in over-dominant model. However, in contrast to the rs1799752 genotypes, there was no significant difference in the ACE1 or ACE2 expression level between carriers of different rs4359 genotypes in the total population of patients and controls. Thus, this polymorphism can be regarded as a non-functional polymorphism in this regard, although a previous study had reported the role of this polymorphism in modulation of response of patients to a certain ACE inhibitor medication [15].
A strong positive correlation has been detected between ACE1 and ACE2 expression in COVID-19 patients, healthy controls as well as total population. Thus, one can deduce that the balance between these two transcripts is only impaired in severely affected COVID-19 cases.
Finally, ACE2 expression levels were positively correlated with RBC, HB and HCT levels in COVID-19 patients. However, ACE2 expression levels had a negative correlation with ESR in COVID-19 patients. In contrast, ACE1 expression was only correlated with HCT. A previous study has demonstrated the impact of Ang II on enhancement of erythropoiesis [16]. In fact, ACE1 knockout mice have exhibited anemia in spite of having normal kidney function [16]. Considering the opposite impacts of ACE1 and ACE2 on Ang II levels [13,14], ACE1 correlation with HCT can be contributed to the impact of this enzyme on erythropoiesis. However, the underlying mechanism for the observed correlation between ACE2 levels and the mentioned parameters should be clarified in future.
Taken together, ACE1 polymorphisms might affect risk of COVID-19 and expression of ACE transcripts.
Authors statement
SGF wrote the draft and revised it. MT designed and supervised the study. SE analyzed the data. MA, GA, NA and BMH performed the experiment and collected the clinical data and samples. All the authors read and approved the submitted version.
Declaration of competing interest
The authors declare they have no conflict of interest.
Acknowledgments
Acknowledgement
The current study was supported by a grant from Shahid Beheshti University of Medical Sciences.
References
- 1.Ballini A., Cantore S., Scacco S., Coletti D., Tatullo M. Mesenchymal stem cells as promoters, enhancers, and playmakers of the translational regenerative medicine 2018. Stem Cells Int. 2018;2018:6927401. doi: 10.1155/2018/6927401. PubMed PMID: 30510586. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatullo M., Codispoti B., Pacifici A., Palmieri F., Marrelli M., Pacifici L., et al. Potential use of human periapical cyst-mesenchymal stem cells (hPCy-MSCs) as a novel stem cell source for regenerative medicine applications. Front. Cell Develop. Biol. 2017;5:103. doi: 10.3389/fcell.2017.00103. PubMed PMID: 29259970. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. e00127–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunyavanich S., Do A., Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. Jama. 2020;323(23):2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H., Cao J.J. Angiotensin-converting enzyme gene polymorphism and severe lung injury in patients with coronavirus disease 2019. Am. J. Pathol. 2020 Oct;190(10):2013–2017. doi: 10.1016/j.ajpath.2020.07.009. 32735889 PMC7387924. (Epub 2020/08/01. eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pati A., Mahto H., Padhi S., Panda A.K. ACE deletion allele is associated with susceptibility to SARS-CoV-2 infection and mortality rate: an epidemiological study in the Asian population. Clin. Chim. Acta. 2020;510:455–458. doi: 10.1016/j.cca.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gómez J., Albaiceta G.M., García-Clemente M., López-Larrea C., Amado-Rodríguez L., Lopez-Alonso I., et al. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene. 2020 Dec 15;762:145102. doi: 10.1016/j.gene.2020.145102. 32882331 PMC7456966. (Epub 2020/09/04. eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyerstedt S., Casaro E.B., Rangel É.B. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40(5):905–919. doi: 10.1007/s10096-020-04138-6. PubMed PMID: 33389262. Epub 01/03. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24(1):1–10. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaeghmaie R., Ghafouri-Fard S., Noroozi R., Tavakoli F., Taheri M., Ayatollahi S.A. Polymorphisms in the angiotensin I converting enzyme (ACE) gene are associated with multiple sclerosis risk and response to interferon-β treatment. Int. Immunopharmacol. 2018;64:275–279. doi: 10.1016/j.intimp.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Wiese O.J., Allwood B.W., Zemlin A.E. COVID-19 and the renin-angiotensin system (RAS): A spark that sets the forest alight? Med. Hypotheses. 2020;144:110231. doi: 10.1016/j.mehy.2020.110231. PubMed PMID: 33254538. Epub 09/03. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275(43):33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 13.Dinh D.T., Frauman A.G., Johnston C.I., FABIANI M.E. Angiotensin receptors: distribution, signalling and function. Clin. Sci. 2001;100(5):481–492. [PubMed] [Google Scholar]
- 14.Chaudhary M. COVID-19 susceptibility: potential of ACE2 polymorphisms. Egyptian J. Med. Hum. Genet. 2020;21(1):1–8. doi: 10.1186/s43042-020-00099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatnagar V., O’Connor D.T., Schork N.J., Salem R.M., Nievergelt C.M., Rana B.K., et al. Angiotensin-converting enzyme gene polymorphism predicts the time-course of blood pressure response to angiotensin converting enzyme inhibition in the AASK trial. J. Hypertens. 2007 Oct;25(10):2082–2092. doi: 10.1097/HJH.0b013e3282b9720e. 17885551 PMC2792638. (Epub 2007/09/22. eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole J., Ertoy D., Lin H., Sutliff R.L., Ezan E., Guyene T.T., et al. Lack of angiotensin II-facilitated erythropoiesis causes anemia in angiotensin-converting enzyme-deficient mice. J. Clin. Invest. 2000;106(11):1391–1398. doi: 10.1172/JCI10557. PubMed PMID: 11104792. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]