Summary
Spatial organization of molecules and cells in complex tissue microenvironments provides essential organizational cues in health and disease. A significant need exists for improved visualization of these spatial relationships. Here, we describe a multiplex immunofluorescence imaging method, termed SeqStain, that uses fluorescent-DNA-labeled antibodies for immunofluorescent staining and nuclease treatment for de-staining that allows selective enzymatic removal of the fluorescent signal. SeqStain can be used with primary antibodies, secondary antibodies, and antibody fragments to efficiently analyze complex cells and tissues. Additionally, incorporation of specific endonuclease restriction sites in antibody labels allows for selective removal of fluorescent signals while retaining other signals that can serve as marks for subsequent analyses. The application of SeqStain on human kidney tissue provided a spatialomic profile of the organization of >25 markers in the kidney, highlighting it as a versatile, easy-to-use, and gentle new technique for spatialomic analyses of complex microenvironments.
Keywords: Spatialomics, multiplex immunofluorescence, cellular neighborhoods, spatial pathology, relationship maps, antibody DNA conjugates, sequential staining
Graphical abstract
Highlights
-
•
A multiplex immunofluorescence staining platform for spatialomic analyses
-
•
Uses antibodies and fragments labeled with fluorescent double-stranded DNA
-
•
Uses nucleases for gentle removal of fluorescent signal
-
•
Can be readily adapted to the imaging setups commonly available
Motivation
Spatial profiling of molecules in complex microenvironments requires techniques for multiplex measurements. Immunofluorescence-based imaging has significant advantages for such spatialomic analyses. Yet newer techniques are needed that are easy to use, utilize off-the-shelf reagents, can be applied by using readily available instrumentation, and provide data with good signal-to-noise ratio and are gentle enough to not harm the tissues.
Multiplex imaging provides important insights into spatial organization of molecules and cells in complex tissues. In this issue of Cell Reports Methods, Rajagopalan et al. describe a method using sequential steps of staining with fluorescent-DNA-labeled antibodies and de-staining with nucleases to profile multiple molecules and cells in single tissue sections.
Introduction
Understanding the molecular and cellular composition of tissues and their relative organization in three-dimensional space of a complex tissue microenvironment (spatialomic organization) is essential for obtaining fundamental insights in tissue biology and interplay between various molecules and cells, and for more accurately determining changes due to disease or treatments. This is especially true in the case of cancer, where insights into the complex tumor microenvironment help guide therapeutic choices (Binnewies et al., 2018). Similar profiling of the cells infiltrating a transplanted tissue helps predict graft survival (Sablik et al., 2020). Techniques to quantify multiple molecules in a single sample, such as transcriptomics, proteomics, flow cytometry, and mass cytometry, have revolutionized the field by providing deep characterization of tissue composition, yet they lack information about spatial organization of the various molecules and cells (Bendall et al., 2012; Bodenmiller et al., 2012). Immunohistochemical (IHC)-based and immunofluorescence (IF)-based imaging methods provide information about spatial distribution of molecules and cells in tissues, yet were limited to detecting only 3–6 analytes at a time in the past (Cappi et al., 2019; Frampton et al., 2015; Gannot et al., 2007; Peng et al., 2011; Tóth and Mezey, 2007). Thus, a great deal of recent effort has focused on increasing the multiplexing capabilities of such imaging-based techniques, and several methodologies for detecting multiple antigens in a single tissue section are now being developed. For example, brightfield IHC-based techniques have been developed by using cycles of IHC staining and de-staining using organic solvents (Akturk et al., 2020; Dixon et al., 2015; Fountaine et al., 2006; Gerdes et al., 2013; Glass et al., 2009; Huang et al., 2013; Remark et al., 2016; Stack et al., 2014) (Dixon et al., 2015; Fountaine et al., 2006; Gerdes et al., 2013; Remark et al., 2016; Stack et al., 2014). Similarly, methodologies using IF-based imaging have been developed, such as CycIF that achieves multiplex imaging via de-staining using fluorescence bleaching (Lin et al., 2015, 2018), and the 4i method that achieves the same by eluting antibodies after each round of staining (Gut et al., 2018). These methods, although providing high multiplex IF capabilities, use harsh protocols to remove the stain in each cycle, which has the potential to harm sensitive tissues.
DNA-tagged antibodies are also used widely in imaging and offer the combined benefits of the specificity of antibodies and the versatility of DNA oligonucleotides (Kazane et al., 2012; Sano et al., 1992). Indeed, such reagents have redefined single-cell genomics by multiplexing cells from different samples (Gaublomme et al., 2019; Stoeckius et al., 2018). DNA-tagged antibodies have also recently been used very elegantly for multiplexed IF imaging (Agasti et al., 2017; Beechem, 2020; Goltsev et al., 2018; Jungmann et al., 2014; Saka et al., 2019; Schnitzbauer et al., 2017). The CODEX method combines DNA-tagged antibodies and in situ primer extension with fluorescently tagged nucleotides to achieve high-level multiplexing (Goltsev et al., 2018). Although CODEX aids in deeper understanding of the tissue architecture, its complex experimental setup and design impedes wider accessibility of the technique. Another technique, immuno-SABER, uses cycles of in situ DNA hybridization and removal (primer exchange reaction) for multiplexed IF imaging with DNA-tagged antibodies (Saka et al., 2019). However, it also requires a complex design of DNA oligonucleotides and setup for the exchange of DNA strands and concatemers. Thus, a need exists for a rapid, mild, and easy-to-use method for multiplexed immunofluorescence imaging.
Antibodies pre-labeled with a fluorophore provide significant advantages and reduce the number of steps needed in such processes, yet have not been routinely utilized in multiplex methods because of the difficulty in removing the fluorescent label after imaging. Endonucleases are enzymes that selectively cleave oligonucleotides either non-specifically (such as DNase I) or in a sequence-specific fashion (such as restriction endonucleases). These enzymes are routinely used in molecular and cell biology laboratories (Roberts, 2005) to cleave oligonucleotide sequences rapidly and under mild conditions that do not harm the cells and tissues. Here, we describe a multiplex immunofluorescence imaging technique, termed SeqStain (for Sequential Staining), whereby we use fluorescent DNA-labeled antibodies to selectively stain antigens on cells and tissues, and use nucleases to rapidly cleave fluorescent signal off the antibodies in the de-staining step. We show that the method is versatile, easy to use, gentle, and highly effective in a multiplex environment and can be rapidly adapted for use on a variety of cells and tissues.
Results
A sequential combination of staining with fluorescent DNA conjugated antibodies and de-staining with nuclease provides an easy-to-use, multiplexed SeqStain imaging platform
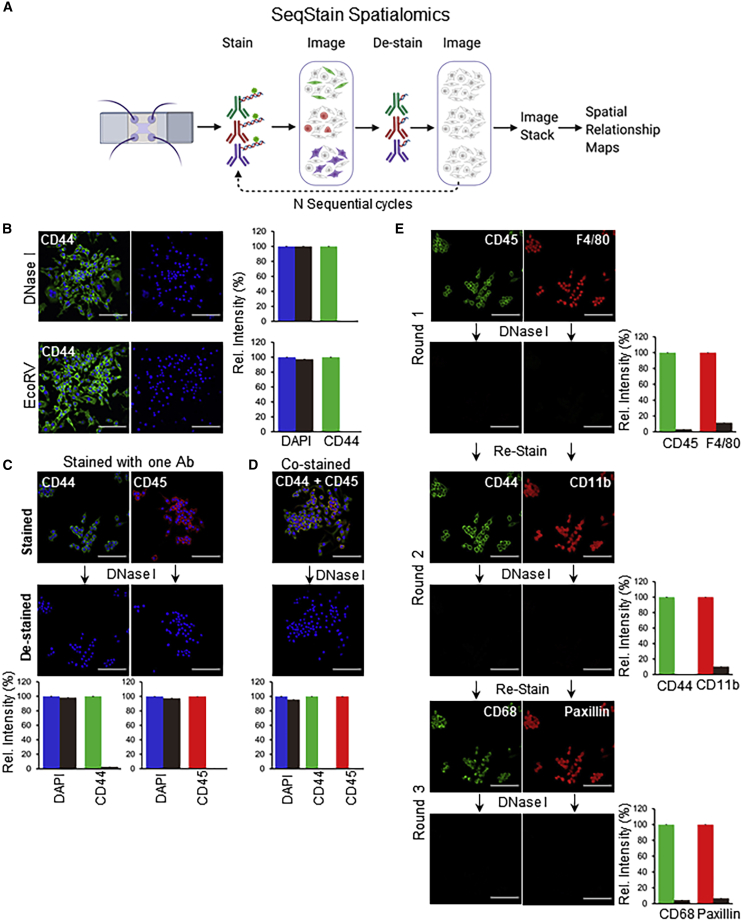
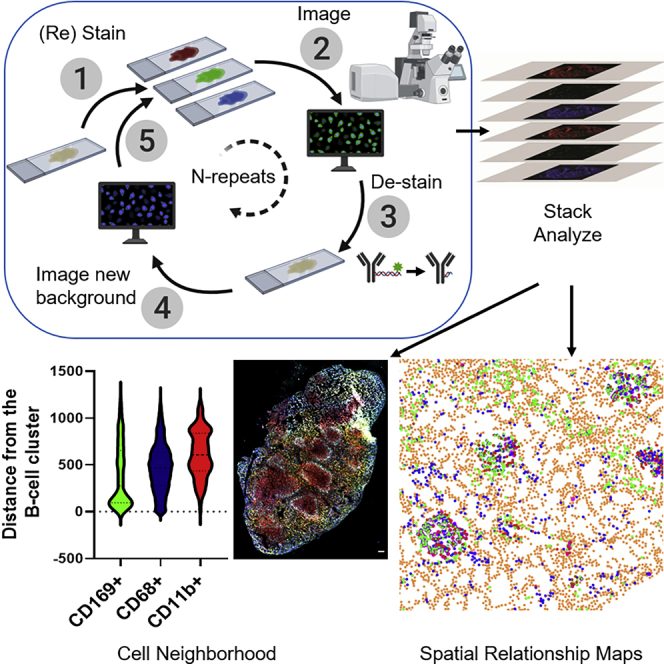
SeqStain uses antibodies conjugated with fluorescently labeled DNA oligonucleotides (termed “SeqStain antibodies”). The method relies on sequential steps of immunofluorescent labeling with a set of such antibodies and gentle removal of the fluorescent labels post imaging, followed by another round of labeling with a new set of fluorescently labeled antibodies (Figure 1A). Importantly, the SeqStain methodology utilizes efficient and selective enzymatic processes, such as treatment with DNase I or restriction enzymes, for rapidly cleaving fluorescently labeled oligonucleotides (oligos) off the antibodies, thereby removing the fluorescent labels from immunofluorescently labeled substrates in the de-staining steps. Subsequently, the cleaved labels are washed off prior to initiation of the next round of staining. The enzymatic removal of fluorescent oligonucleotides also offers flexibility in the oligo sequence design, the length and complexity of the oligos, and the types of oligos that can be used. Thus, cyclic steps of staining tissues with SeqStain antibodies and de-staining with nuclease treatment allow efficient and rapid multiplex immunofluorescent imaging of cells and tissues.
Figure 1.
SeqStain-based multiplex immunofluorescence imaging
(A) SeqStain methodology schematic. Immobilized cells and tissue sections are processed in multiple, sequential cycles of immunostaining with fluorescent DNA-labeled antibodies, imaging, gentle de-staining using a nuclease, and re-imaging. Post imaging, the data are analyzed by computational stacking and alignment of the images to generate spatial relationship maps. The schematic was generated using Biorender.
(B) Immunofluorescence images of RAW264.7 cells after staining with anti-CD44 SeqStain antibody and after de-staining with either DNase I (top panels) or the endonuclease EcoRV (bottom panels). All images are representative of at least three replicates. A bar graph showing quantification of fluorescence intensity for each panel is presented on the right.
(C) Immunofluorescence images of RAW264.7 cells after staining with anti-CD44 (fluorescently labeled with AF488 fluorophore) or anti-CD45 (labeled with Cy3 fluorophore) SeqStain antibodies (top panels) and after de-staining with DNase I for 1 min (bottom panels). Nuclei were labeled using DAPI. All images are representative of at least three replicates. A graph showing quantification of fluorescence intensity in each panel is presented on the bottom.
(D) Immunofluorescent images of RAW264.7 cells co-stained with anti-CD44 and anti-CD45 SeqStain antibodies (top panel) and 1 min after the addition of DNase I (bottom panel). All images are representative of at least three replicates. A graph showing quantification of fluorescence intensity in each panel is presented on the bottom.
(E) Immunofluorescence images of RAW264.7 cells after each of the three cycles of staining with two unique SeqStain antibodies and de-staining with DNase I. The antibodies used in each round are indicated in the panel, with SeqStain antibodies labeled using the AF488 fluorophore shown in green and the antibodies labeled using the Cy3 fluorophore shown in red. All images are representative of at least three replicates. A graph showing quantification of fluorescence intensity after staining (green and red bars) and de-staining (brown bars) in each panel is presented on the right.
Graphs show the mean ± standard deviation (SD). Scale bars, 100 μm.
SeqStain antibodies are highly efficient in multiplexed staining of mammalian cells
SeqStain antibodies were designed such that they could carry multiple fluorophores on each of the conjugated oligonucleotides (oligos) on the antibody (Figure S1A). Additionally, the oligo chains were designed so that the fluorescent dye molecules on the DNA were spaced apart by >5 nm dye-to-dye distance (∼15 nt apart) to prevent any unwanted dye-dye interactions (Woehrstein et al., 2017). SeqStain antibodies were generated in a two-step process (Figure S1B). In step 1, a 5′-amine modified 29-nt-long single-stranded DNA chain (linker oligo), containing a pre-specified 15-mer sequence for hybridization with a complementary docking oligo, was covalently attached to the antibody. SDS-PAGE showed an average of 2–5 linker oligos chemically conjugated to an antibody during a typical labeling experiment (Figure S1C). In step 2, a fluorescently labeled double-stranded DNA (dsDNA) complex, containing a docking oligo and multiple fluorescently labeled oligos, was hybridized to the antibody-DNA conjugate. This design of the dsDNA also allows for using many copies of fluorescently labeled DNA oligos to tune the signal intensity on the antibody. Subsequently, SeqStain antibodies were purified by using the 100-kDa size-exclusion column. Formation of the fluorescent DNA-antibody complex was confirmed by using 4% agarose gels (Figure S1D). This method of using fluorophores on the complementary oligonucleotides also avoids chemical quenching of the fluorophores during the antibody-DNA conjugation step. Analyses of fluorescence intensity after repeated cycles of heating and slow cooling during dsDNA hybridization (steps that are utilized to prepare fluorescent dsDNA-labeled SeqStain antibodies) showed no loss in fluorescence during this step (Figure S1E), confirming that this method of preparing SeqStain antibodies does not lead to any loss of fluorescence signal.
Flow-cytometric analyses with SeqStain antibodies showed comparable staining of cell surface proteins CD45 and CD11b on RAW murine macrophages as compared with staining using the conventional method (Figure S1F), confirming that antibody modification with DNA did not affect its ability to bind antigens. Staining of cells with the fluorescent oligo complex alone (no antibody) showed no staining. Additionally, imaging of cells with both SeqStain and conventional antibody labeling methods showed no difference between immunofluorescence labeling using DNA-modified primary antibody versus unmodified antibody (Figure S1G). Furthermore, counterstaining of non-fluorescent SeqStain antibody-labeled cells with a fluorescent secondary antibody showed cell staining similar to the unmodified primary antibody used via the conventional approach (Figure S1H), suggesting that the chemical conjugation of antibodies with oligonucleotides has minimal effect on their binding properties. Furthermore, oligos containing five repeats for complementary strand binding showed high fluorescence signal-to-background ratios (data not shown), and was, thus, utilized in all the assays presented here. Overall, this suggests that fluorescent dsDNA-labeled primary antibodies provide signal intensity similar to that of conventional approaches for use in immunofluorescence applications.
Next, RAW cells were immobilized on glass coverslips and labeled with anti-CD44 SeqStain antibody. Of note, we incorporated an EcoRV restriction site in the dsDNA sequence on the SeqStain antibodies. After fluorescence imaging, treatment of cells with DNase I resulted in rapid loss of the fluorescence signal (Figure 1B, top). Alternatively, treatment with EcoRV also resulted in rapid loss of fluorescence on the cells (Figure 1B, bottom), suggesting that both types of nucleases can be used. Treatment of cells stained with either a single SeqStain antibody (Figure 1C) or co-stained with two different SeqStain antibodies (Figure 1D) with DNase I rapidly reduced the fluorescence signal-to-background levels, suggesting that the fluorescence signal from multiple fluorophores can be simultaneously removed. Furthermore, DNase I removed the fluorescence on subsecond timescales from stained cells (Video S1). Keeping DNase I longer on the cells did not show any loss in sample integrity or increase in background signal (Figure S2A). SeqStain antibodies showed high fluorescence and low background that was similar or better than the cells immunostained by using the conventional methods (Figure S2B). Furthermore, staining of α-tubulin on RAW cells by using SeqStain antibody either prior to DNase I treatment or after two sequential DNase I treatments (Figure S2C) showed no loss in structural information or integrity of fine intracellular structures. Similarly, staining of α-tubulin on podocytes by using SeqStain antibody either prior to or after DNase I treatment (Figure S2D) showed no loss in structural integrity of fine intracellular structures, including F-actin, further suggesting that SeqStain is a gentle technique. Moreover, given that chemical conjugation of linker oligo to antibody can be performed by using a variety of methods, we tested two additional orthogonal chemistries to generate SeqStain antibodies (Figures S2E and S2F) (Gong et al., 2016; Stoeckius et al., 2017) and found that these methods had no effect on the staining and de-staining levels (Figures S2G and S2H), suggesting that antibody modification can be successfully accomplished by using multiple different methods.
RAW264.7 cells were co-stained with anti-CD44 AF488 and anti-CD45 Cy3 SeqStain antibodies. Subsequently, the cells were incubated with DNase I and the fluorescence images were acquired as a video every 3 s.
Finally, to evaluate SeqStain for multiplex staining, we used a 6-plex panel to iteratively stain and de-stain the immobilized RAW cells in three rounds of staining and de-staining (Figure 1E). Staining was performed with a set of two unique antibodies, each with a unique fluorophore-labeled DNA tag, per round. Cell nuclei were stained by using 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent signal from the antibodies was removed by using DNase I. Quantification showed complete removal of both fluorophores after each round, with no effect on DAPI staining, suggesting that this treatment does not affect the integrity of cells, nuclei, or nuclear materials. A concern for the antibody-based multiplex staining methods is that application of so many antibodies, either together or sequentially, could crowd the antigens, making multiplexing difficult. To address this, we stained and de-stained immobilized cells with the same SeqStain antibody for five cycles (Figure S3). Furthermore, the samples were further stained, de-stained, and re-stained with a second set of two antibodies for five more rounds. We found no loss in immunofluorescent labeling capability of these samples, likely because only a subset of epitopes were engaged during each cycle and that, although the fraction of available epitope for antibody binding decreases over time, it likely did not reach saturation during these experiments (only five cycles of binding) using the low antibody concentrations utilized here. It is quite possible that either use of much higher concentration of antibodies in each cycle or performing additional cycles would reach a point of full epitope occupancy or masking. Together, these data show that SeqStain offers a rapid and robust multiplex immunofluorescence imaging platform.
Primary antibodies pre-complexed with secondary antibodies and secondary antigen-binding fragments can also be used in SeqStain
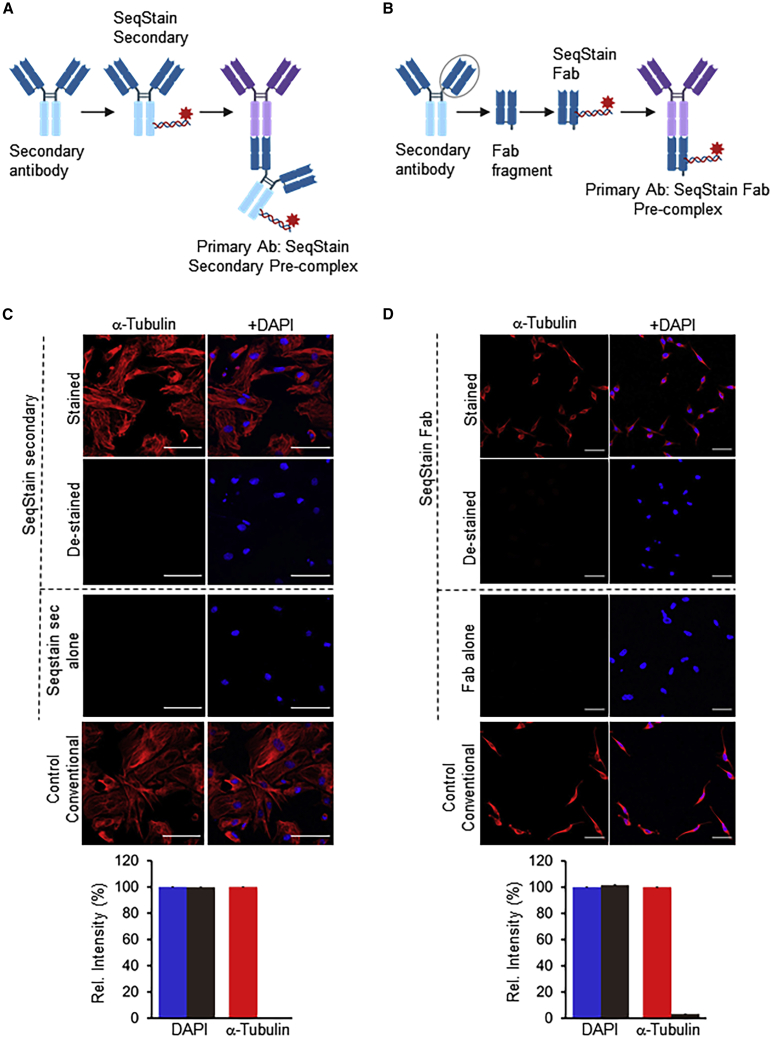
Conventional methods utilize an unlabeled primary antibody followed by a fluorescently labeled secondary antibody. However, extension of such methodology in a multiplex environment is limited by the number of species (such as mouse, rat, goat, or sheep) available to generate unique secondary antibodies. Pre-mixing primary antibodies with fluorescently labeled secondary antibodies or purified antigen-binding fragments (Fabs, the regions of antibodies with antigen-binding capacity; Coulter and Harris, 1983; Mariant et al., 1991) prior to their use in staining could potentially overcome some of these limitations. Here, as an alternative to using primary SeqStain antibodies for multiplex staining, we tested whether fluorescent-DNA-labeled secondary antibodies (SeqStain secondary Ab) or Fabs of secondary antibodies (SeqStain Fabs) (Figures 2A and 2B) could be applied in the SeqStain protocol. Such reagents have at least two major advantages: (1) by pre-complexing these reagents with primary antibodies, we can achieve signal amplification for targets with low level of expression; (2) these reagents might be quickly pre-complexed with many different primary antibodies, thus circumventing the need for modifying primary antibodies with fluorescent DNA.
Figure 2.
Application of SeqStain secondary antibodies and SeqStain Fabs in multiplex staining
(A) Schematic representation of SeqStain secondary antibody-based multiplex imaging method. Anti-Fc secondary antibodies are labeled with fluorescent DNA to develop SeqStain secondary antibodies. Subsequently, the SeqStain secondary antibodies are pre-complexed with appropriate primary antibodies and used in staining.
(B) Schematic representation of SeqStain Fab-based multiplex imaging method. Fab fragments generated by enzymatic digestion of antibodies are labeled with fluorescent DNA to develop SeqStain Fabs. Subsequently, SeqStain Fabs are pre-complexed with their corresponding primary antibodies for use in multiplex staining.
(C) Immunofluorescence images of murine podocyte cell line stained with anti-α-tubulin antibody pre-complexed with SeqStain secondary antibody (left panels) and with DAPI nuclear staining (right panels). Immunofluorescent images after de-staining with DNase I are shown below each panel. Control staining of these cells with SeqStain secondary alone and with conventional methodology is shown in the bottom panels. All images are representative of at least two replicates. A graph showing quantification of fluorescence intensity in each panel is also presented.
(D) Immunofluorescence images of HeLa cells stained with anti-α-tubulin antibody pre-complexed with SeqStain Fab (left panels) and with DAPI nuclear staining (right panels). Immunofluorescence images of these cells after de-staining with DNase I are shown below each panel. Control staining of these cells with SeqStain Fab alone (secondary Fab) and with conventional methodology is shown in the bottom panels. All images are representative of at least two replicates. A graph showing quantification of fluorescence intensity in each panel is also presented.
Graphs show the mean ± SD. Scale bars, 100 μm.
We prepared SeqStain Fabs by using commercially available secondary antibodies or with affinity-purified Fc-specific Fab fragments from secondary antibodies (mouse, rat, and rabbit) (Figure S4). Primary rabbit anti-mouse antibody against α-tubulin was pre-complexed with anti-rabbit SeqStain secondary Ab or with anti-rabbit secondary SeqStain Fab, respectively. Next, we stained immobilized mouse podocytes (Figure 2C) or HeLa cells (Figure 2D) with them. Imaging showed a high level of staining similar to the staining with conventional primary-secondary antibodies. As expected, treatment of these SeqStain secondary Ab or the SeqStain Fab-stained cells with DNase I resulted in rapid loss of the fluorescence signal. As a control, staining with SeqStain secondary Ab or SeqStain Fab alone, in the absence of the primary rabbit anti-mouse antibody against α-tubulin, did not show any staining, highlighting the specificity of this approach. Furthermore, use of a 6-plex panel of primary antibodies pre-complexed with SeqStain Fabs to iteratively stain and de-stain immobilized RAW cells also showed efficient labeling and clearing of fluorescent signal from these cells (Figure S4D), further suggesting that SeqStain methodology can be applied with a variety of reagents for rapid multiplex immunofluorescence imaging.
SeqStain allows rapid, multiplexed immunofluorescence imaging of complex tissues
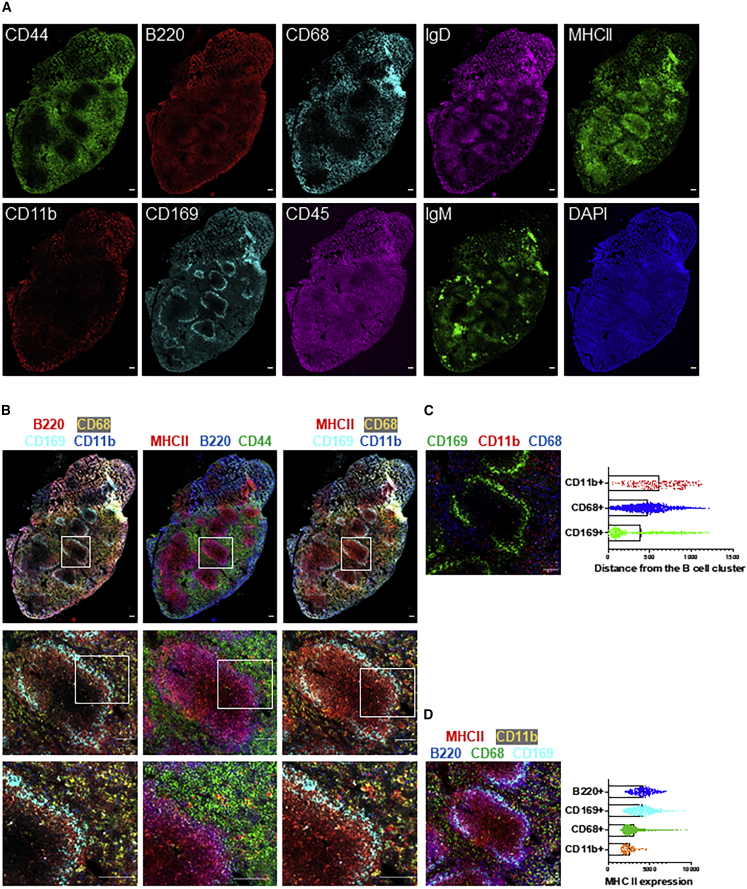
Next, we applied SeqStain to murine (spleen) and human tissues (kidney and tonsil). We selected a set of commercially available, well-characterized antibodies (see key resources table) that recognize several different cell types for these assays. First, we validated the prepared SeqStain antibodies. Figure S5 shows that the developed SeqStain reagents provided expected antigen-staining patterns on tissues and was comparable with the conventional IF staining. For example, staining of mouse spleen sections (Figure S5A) showed that anti-immunoglobulin D (IgD) and anti-IgM SeqStain antibodies expectedly stained B cell clusters, and the anti-CD169 SeqStain antibody stained marginal zone macrophages around the B cell cluster in ring-like pattern similar to the staining observed by using conventional methods (Goltsev et al., 2018; Sarvaria et al., 2017; Sic et al., 2014). Staining using anti-CD68 and anti-CD11b SeqStain antibodies, which stain macrophages in the spleen, similarly provided expected patterns. Remarkably, we also found that staining of serial sections of a murine spleen tissue for an antigen (MHC II) by using two different approaches—SeqStain and conventional immunostaining, respectively—yielded a similar pattern of MHC-II-positive B cell clusters throughout the tissue section (Kuwano et al., 2007) (Figure S5B), suggesting that SeqStain antibodies provide results similar to those by conventional approaches. Similarly, staining of human kidney tissue sections (Figure S5C) showed that anti-human SeqStain antibodies against cytokeratin-8, CD31, CD45, synaptopodin, podocin, Na+, K+-ATPase, EpCAM, and collagen IV provided expected and comparable staining patterns in comparison with the unmodified antibodies used in the conventional approach. Not surprisingly, use of SeqStain Fab pre-complexed with primary antibodies also provided staining-pattern results comparable with the staining patterns observed by using conventional indirect immunofluorescence, as exemplified on human kidney and tonsil tissue sections in Figures S5D and S5E. Furthermore, we found that the type of fluorophore used to label the dsDNA complex on SeqStain antibodies did not have any material effect on their performance, suggesting that most commonly available fluorophores can be used to fluorescently tag the DNA on the antibodies (Figure S6A). Additionally, as with the SeqStain-stained cells, fluorescent DNA-labeled SeqStain antibodies showed high fluorescence signal and low background signal, similar to or better than the tissues immunostained by using the conventional methods (Figure S6B). Together, these data suggest that SeqStain antibodies and Fabs are highly efficient in immunostaining a variety of complex tissues and provide staining results comparable with those of conventional methods.
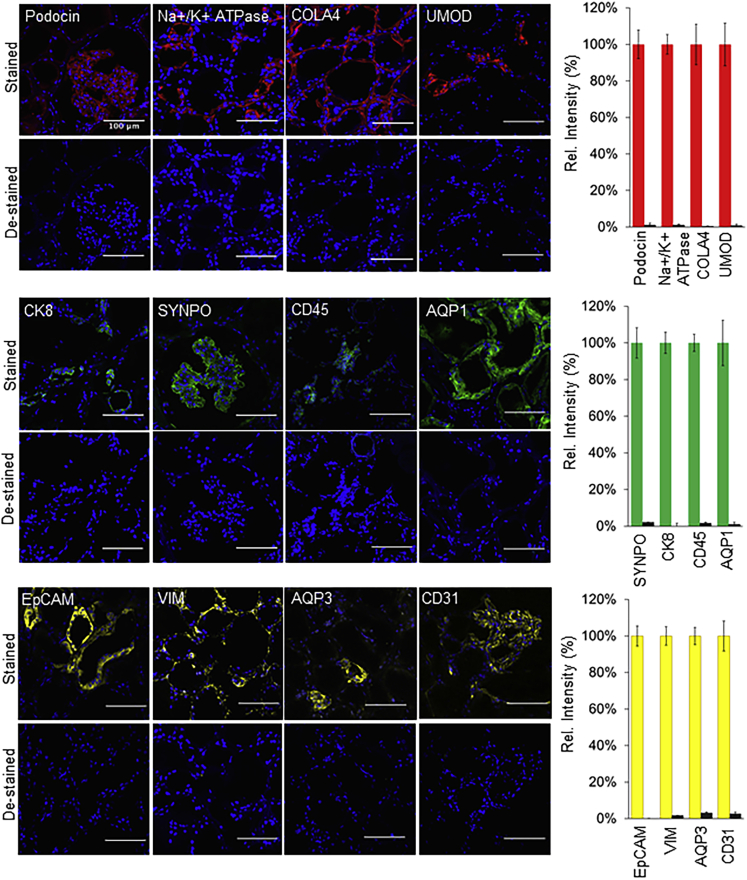
Next, we examined whether nucleases were equally efficient in removing the fluorescent signal from stained tissue sections as they were with cells, even though the tissues provide a highly complex microenvironment. Human kidney tissue (Figure 3) stained with a variety of SeqStain antibodies showed complete removal of fluorescence signal after a brief treatment with DNase I, and removed the fluorescence on subsecond timescales (Videos S2, S3, and S4). Notably, DNase I treatment of tissue sections stained by using the fluorescent antibodies via the conventional method (whereby the fluorophores do not contain a linker DNA) did not result in any loss of signal (Figures S6C and S6D). More importantly, DNase I treatment did not harm the integrity of the tissues. To probe this further, especially to make sure that the extensive DNase I treatment does not remove DNA-binding proteins from the nuclear DNA, we treated human kidney sections with increasing amounts of DNase I (by repeatedly staining the sections with DNase I either five times or ten times) and subsequently stained the cells with conventional anti-histone H1 antibody. Results showed no loss of histone levels or chromosomal DNA, suggesting that these treatments do not reduce sample quality (Figure S7A). To examine whether the technique is amenable to nuclear proteins, we stained tissue sections with anti-histone H1 SeqStain antibody. Results showed successful nuclear histone H1 staining by the antibody and, upon treatment with DNase I, efficient removal of the antibody-associated fluorescent signal, suggesting that SeqStain is amenable to staining such subcellular compartments as well (Figures S7B and S7C). Finally, when tissue sections stained with SeqStain antibodies were co-stained with conventional fluorescent secondary antibody, using spectrally non-overlapping fluorophores, it showed an overlapping staining pattern (Figures S7D and S7E), suggesting that the chemical modification of primary antibody with fluorescent dsDNA sequences does not harm the antibody properties and does not hamper its recognition by a secondary antibody. Again, treatment of the doubly labeled tissue section with DNase I showed selective removal of the SeqStain fluorophore without negatively affecting the conventionally conjugated fluorophore, the staining pattern, or the tissue integrity. DNase I treatment also completely removed the fluorophore signal from the SeqStain secondary antibodies (Figure S7F), Together, these data comprehensively show that SeqStain methodology is equally efficient for immunofluorescence staining of complex tissues.
Figure 3.
Enzymatic de-staining of tissues stained using the SeqStain antibodies
Immunofluorescence images showing human kidney tissue sections stained using SeqStain antibodies (as indicated in the panel). The antibodies were labeled using either the AF488 fluorophore (shown in green), the Cy3 fluorophore (shown in red), or the Cy5 fluorophore (shown in yellow). Immunofluorescence images of these tissue sections after de-staining with DNase I treatment are shown below each panel. All images are representative of at least three replicates. Graphs showing quantification of fluorescence intensity after staining (red bars, green bars, or yellow bars) and de-staining (brown bars) in each panel is also presented on the right. Graphs show the mean ± SD. Scale bar, 100 μm.
Normal human kidney tissue sections were stained with anti-cytokeratin 8 AF488 antibody and de-stained with DNase I, and the fluorescence images acquired as a video every 20 s.
Normal human kidney tissue sections were stained with anti-collagen IV Cy3 antibody and de-stained with DNase I, and the fluorescence images acquired as a video every 20 s.
Normal human kidney tissue sections were stained with anti-EpCAM Cy5 antibody and de-stained with DNase I, and the fluorescence images acquired as a video every 20 s.
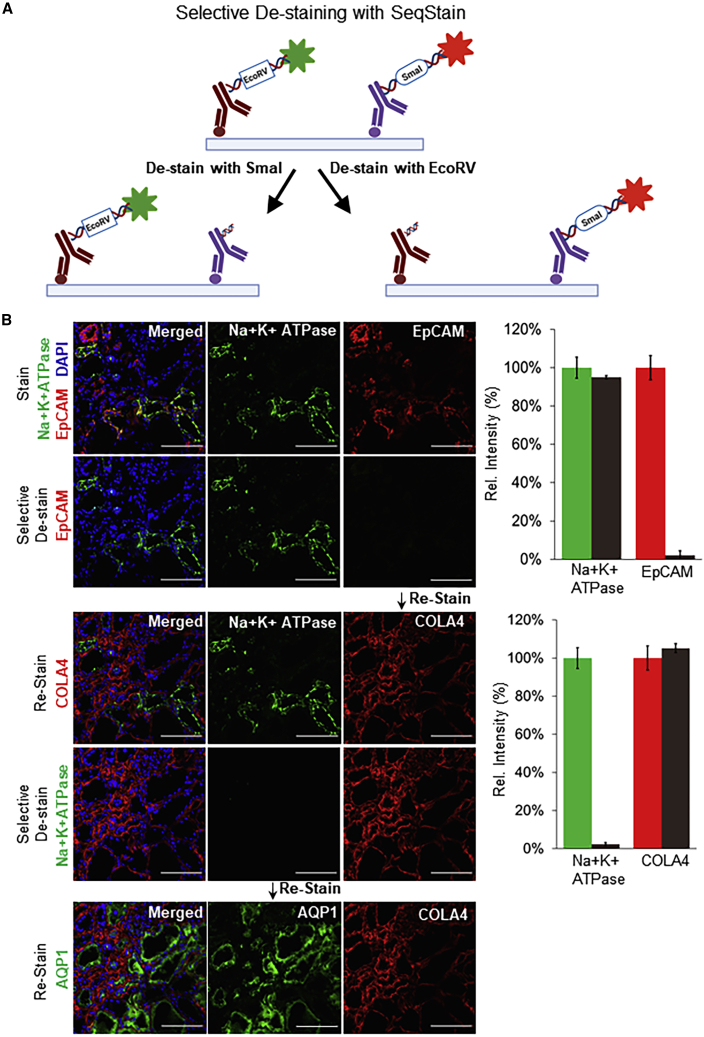
Fluorescent labels from SeqStain antibodies on tissues can be selectively removed by using pre-determined choice of nucleases
It is often desirable to maintain immunofluorescent labeling on an antigen while staining and de-staining other antigens on the same sample. However, such applications are difficult to incorporate in the current multiplex imaging techniques. Given that SeqStain relies on nuclease-based enzymatic removal of fluorescent signals, SeqStain methodology offers the possibility of selective removal of fluorophores. To test this, we engineered two unique restriction sites (EcoRV and SmaI) in the linker oligo sequences and used them to prepare the following SeqStain antibodies (Figure 4A): anti-EpCAM and anti-collagen IV SeqStain antibodies containing EcoRV restriction site in dsDNA linkers, and anti-Na+, K+-ATPase, and anti-aquaporin 1 SeqStain antibodies containing SmaI restriction site. Subsequently, we labeled human kidney cryosections with a combination of anti-Na+, K+-ATPase, and anti-EpCAM SeqStain antibodies bearing AF488 and Cy3 fluorochrome tag, respectively (Figure 4B). Imaging results showed expected labeling of the distinct tubular segments with the two antibodies (Borges Da Silva et al., 2015; Goltsev et al., 2018). Next, to selectively remove only one of the two fluorescent labels, we treated the tissue samples with EcoRV restriction endonuclease, which resulted in selective removal of the signal from the anti-EpCAM but did not affect the fluorescent signal from the anti-NA+, K+-ATPase antibody (Figure 4B). Subsequently, to determine whether this would have any negative consequences on staining with antibody labeled with DNA containing the same EcoRV restriction site and to study the utility of this method in a multiplexed setting, in the next round of staining we applied anti-collagen IV SeqStain antibody containing EcoRV restriction site and the Cy3 fluorescent tag. Imaging results show tubular basement membrane stained by the collagen IV antibody along with the Na+, K+-ATPase-positive transport channels in the tubules that were labeled in the previous round of staining (Figure 4B). Next, for selective cleavage of the other signal, we treated the tissue sample with SmaI restriction endonuclease, which selectively removed the signal from anti-Na+,K+-ATPase antibody. Finally, to determine whether this would have any negative consequences on staining with antibody labeled with DNA containing an SmaI site, in the next round we applied anti-aquaporin 1 antibody containing SmaI restriction site and the AF488 fluorophore, with expected staining of the cortical proximal tubules upon imaging (Figure 4B). The results clearly show that the SeqStain methodology allows selective de-staining of fluorescent markers in a multiplex setting, offering significant advantages where maintaining staining of specific markers or antigens is needed. Such retained stains can also serve as structural markers for imaging in subsequent rounds and help better orient the stacks of the whole tissue during image analyses.
Figure 4.
Selective de-staining of complex tissues using SeqStain antibodies and restriction endonucleases
(A) Schematic representation of the technique for selective removal of fluorophores from immunofluorescently labeled tissues. Tissue can be stained with a combination of SeqStain antibodies labeled using fluorescent DNA that contains specific recognition sites for restriction endonuclease(s) (such as EcoRV and SmaI). Treatment of samples with the specific restriction nuclease selectively removes fluorophores only from the antibodies carrying the respective DNA sequence, leaving all others undiminished.
(B) Immunofluorescence images showing normal human kidney tissue sections after each of the cycles of staining with unique SeqStain antibodies (as indicated in the panel) and de-staining with a specific restriction endonuclease (as indicated). The antibodies were labeled using the AF488 fluorophore (shown in green) or the Cy3 fluorophore (shown in red). Merged images from the two fluorescence channels (along with images from DAPI-stained nuclear markers) are also shown. All images are representative of at least three replicates. Graph showing quantification of fluorescence intensity after staining (green and red bars) and de-staining (brown bars) in each panel is also presented on the right. Graphs show the mean ± SD. Scale bars, 100 μm.
SeqStain enables spatialomic profiling of multiple antigens on spleen and kidney tissues
To test multiplexed spatialomic profiling of a single tissue section by using SeqStain, we developed a panel of nine unique antibodies against various immune markers, along with pan-nuclear marker DAPI, and used it in five rounds of sequential staining and de-staining steps on mouse spleen tissue. Each round of staining used two unique SeqStain antibodies followed by imaging of the whole slide and de-staining with nuclease DNase I. Again, DAPI-stained nuclei provided a guidepost for aligning the various image sets at the end of the experiment. Results in Figure 5A show a high level of fluorescence staining by each SeqStain antibody, followed by complete and rapid removal of fluorescent label from both channels after DNase I treatment (Figure S8A). After completion of the imaging rounds, images were stacked and aligned by using DAPI-stained nuclei using ImageJ (Arganda-Carreras et al., 2006; Schneider et al., 2012). CellProfiler-based image analyses showed that the individual cells can be efficiently segmented computationally for cell-based fluorescent signal analyses of SeqStain-stained tissues (Figures S8B–S8D).
Figure 5.
Multiplex imaging of whole spleen tissue using a 9-plex SeqStain panel
(A) Immunofluorescence images of whole mouse spleen tissue sections after multiplex staining with multiple rounds of staining with unique SeqStain antibodies and DAPI (as indicated in the panel).
(B) Composite overlay of aligned immunofluorescence image stacks from whole mouse spleen tissue sections after multiplex staining with SeqStain antibodies and DAPI (as indicated on top of each panel). (Middle and bottom) Zoomed-in regions from each composite immunofluorescence image, showing co-localization of various markers based on SeqStain staining.
(C) Composite overlay showing the location of various myeloid cells (as indicated on the top of the panel) with respect to a selected B cell cluster. Boxed dot plot of cellular neighborhoods on the right shows the computed distance for each myeloid cell type on a per-cell basis from the B cell cluster.
(D) Composite overlay showing MHC II expression on various cell types (as indicated on the top of the panel) in a selected region. Boxed dot plot on the right shows computed co-expression of MHC II and the indicated markers on a per-cell basis.
Graphs show the mean ± SD. Scale bars, 100 μm.
Subsequently, select composites were generated from the aligned images to show the spatial organization of different markers in the spleen tissue (Figures 5B–5D). Images of the entire tissue section clearly showed no changes in overall tissue morphology or integrity due to the repeated cycles of staining and de-staining. Staining of different myeloid cells by using CD169, CD68, and CD11b revealed their spatial relationship with respect to B220+ B cells. Quantification of the inter-cell distances by using Fiji showed cells in distinct cellular neighborhoods (Figure 5C). As expected, the CD169+ macrophages were found in the marginal zone area lining B220+ B cell clusters, in close contact with the B cells as is typical for these cell populations, whereas the CD68+ macrophages were predominantly found in the red pulp region of the spleen. Additionally, the CD11b+ cells were found scattered outside the red pulp region (Borges Da Silva et al., 2015; McCulloch et al., 2018; Walzer et al., 2007). Thus, SeqStain can be used to profile the heterogeneous myeloid populations in the spleen whose distinct spatial location and relationship with other cell types help orchestrate the immune responses. Quantification of MHC II levels revealed that CD68+ and CD11b+ macrophages in the spleen had low levels of MHC II expression (Sheng et al., 2015) whereas CD169+ macrophages residing in the marginal zone area had higher expression of MHC II (Martinez-Pomares and Gordon, 2012; Veninga et al., 2015). As expected, B220+ B cells had the highest expression of MHC II among the cell types analyzed (Accolla et al., 2014; Murakami et al., 2007). Thus, by quantifying the relative spatial location of cells and their co-expression of markers, SeqStain offers a simple yet robust method to understand the spatial organization of various cell types in the tissue and an ability to generate spatial relationship maps (SRMs).
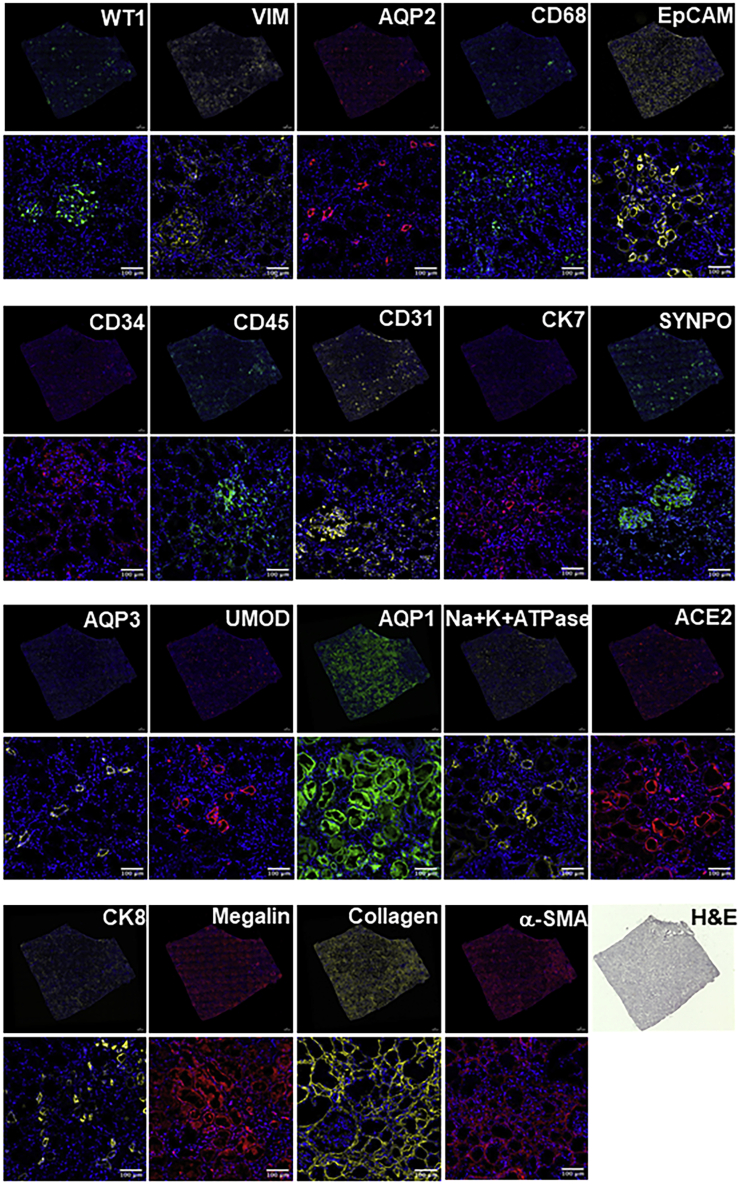
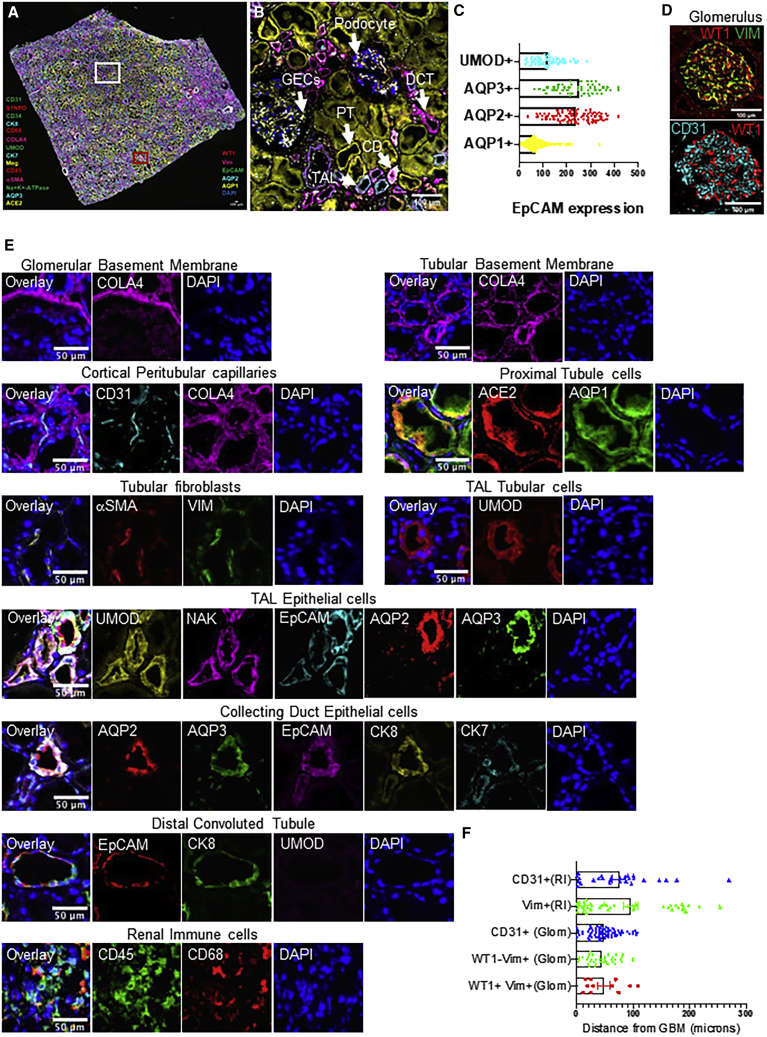
Next, we expanded the antibody set to profile human kidney tissue and also tested the feasibility of using three-color antibody mixtures for staining in each round. We developed a 20-color panel (19 unique antibodies + DAPI) and used it to stain a human kidney tissue section in nine cycles of staining and de-staining (Figures 6 and S9). Additionally, to confirm staining, we used a few of the antibodies twice to obtain a 25-plex image of the kidney tissue at high resolution. As above, images were stacked and aligned by using DAPI-stained nuclei for data quantification (Arganda-Carreras et al., 2006; Schneider et al., 2012). Results show a high level of immunofluorescence staining by each SeqStain antibody, followed by complete and rapid removal of fluorescent label from each of the three channels after DNase I treatment. Aligned images of the whole tissue section clearly showed no loss of integrity of the tissue or changes in tissue morphology due to the repeated cycles of staining and de-staining. This was further confirmed from the analyses of images after repeated staining with the same antibody in a different cycle, such as vimentin and aquaporin 1. Analyses of the stained images from two different cycles clearly showed high overlap of fluorescence signal without any loss in signal intensity or increase in background fluorescence (Figures S10A and S10B). Further analyses of various kidney tissue substructures by using 20-plex images from this experiment clearly showed the presence of expected immunophenotypes, spatial relationships, and distinct cellular neighborhoods in the various parts of the kidney (Figures 7A and 7E). Composites of pseudocolored images revealed a predominant presence of proximal tubules (AQP1) consistent with the histology of renal cortex (Figure 7B) (Nielsen et al., 2002; Singh et al., 2019). We were also able to identify the collecting duct (AQP2+ AQP3+ EpCAM+Cyto7+ Cyto8+) (Hatta et al., 1987; Schiano et al., 2019; Skinnider et al., 2005), Distal convoluted tubules (AQP2− AQP3− EpCAM+Cyto8+ UMOD−), thick ascending loop of Henle (EpCAM+ uromodulin+ Na+,K+-ATPase+ AQP2− AQP3−), and their spatial location with respect to one another (Figures 7B–7E) (Fissell and Miner, 2018). Similarly, in the glomerulus we were able to discern the three components of the glomerular capillary filter: podocytes (WT1+ and vimentin+), glomerular basement membrane (collagen IV+), and glomerular endothelial cells (CD31+) (Fissell and Miner, 2018). In addition, the specialized contractile mesangial cells of the glomerulus can be discerned by vimentin staining in the absence of WT1 staining (Johnson et al., 1991; Miner, 2012; Palmer et al., 2001; Qi et al., 2017). In the renal interstitium, collagen IV delineates the tubular basement membrane, forming a network throughout the entire tissue (Figures 6 and 7E) (Miner, 1999). Peritubular capillaries visualized by CD31 staining can be seen as a disconnected network of cells within this tubular basement membrane (Bábíčková et al., 2017). The renal interstitium also contains stromal cells that can be visualized by vimentin and α-smooth muscle actin staining (Figure 7E) (Boor and Floege, 2012). Angiotensin-converting enzyme 2 (ACE2), the functional receptor for the SARS coronaviruses, was found to be highly expressed in the proximal tubular cells identified by ACE2 co-expression with AQP1 (Hamming et al., 2004; Lely et al., 2004). The normal human kidney had resident immune cells that were identified by CD45 staining whereas the renal macrophages were identified by co-expression of CD45 and CD68 (Figure 7E) (Belliere et al., 2015). Image analyses using HALO and Fiji showed clear SRMs and the expected proximity of glomerular podocytes (WT1+ Vim+) with the glomerular endothelial cells (CD31+) and the glomerular mesangial cells (WT1- Vim+) as their cellular neighborhood in the glomerulus (Figures S10C and 7F). In summary, these experiments show that SeqStain is highly applicable for multiplexed spatial profiling of various types of tissues and allows for rapid generation of SRMs.
Figure 6.
SeqStain-based multiplex imaging of whole human kidney tissue provides a 20-plex image
Immunofluorescent images of whole kidney tissue sections after each round of staining with unique SeqStain antibodies (as indicated) and DAPI (as indicated in the panel). Zoomed-in sections of images are presented below each panel. A serial section stained with H&E is also presented. Scale bars, 100 μm.
Figure 7.
SeqStain multiplex imaged panels identify major substructures in the human kidney
(A) Image showing a composite overlay of aligned immunofluorescence image stack of 20 unique markers from the whole kidney tissue section. Scale bar, 100 μm.
(B) Image of a zoomed-in region from the whole kidney tissue section in (A) (white square) showing various components of the kidney tissue, including the glomerular endothelial cells (GECs), proximal tubule (PT), collecting duct (CD), distal convoluted tubules (DCT), and podocytes. Scale bar, 100 μm.
(C) Boxed dot plot showing computed co-expression of EpCAM and the indicated markers on a per-cell basis (mean ± SD).
(D) Image of a zoomed-in region from the whole kidney tissue section in (A) (red square) showing one glomerulus (two panels), false colored for the indicated markers. Scale bar, 100 μm.
(E) Representative images showing zoomed-in regions of composite overlay of aligned immunofluorescence image stacks from (A) for the identification of various immunophenotypes of cells and tissue sections based on co-localization of various markers (as labeled). Scale bar, 50 μm.
(F) Boxed dot plot of cellular neighborhoods showing the computed distances of the indicated cells from glomerular basement membrane of a selected glomerulus in the kidney (mean ± SD). Glom refers to cells residing inside the glomerulus, whereas RI refers to cells residing outside the glomerulus, in the renal interstitium.
Discussion
Spatial profiling of cells in tissues provides critical insights into disease pathogenesis and can be diagnostic, especially for diseases such as cancer where spatial heterogeneity often leads to poor clinical outcomes (Binnewies et al., 2018; Dagogo-Jack and Shaw, 2018; Johnson et al., 2018). To address such needs, recent years have seen a surge of innovative multiplex staining techniques at both transcriptomic and proteomic levels (Akturk et al., 2020; Dixon et al., 2015; Fountaine et al., 2006; Gerdes et al., 2013; Goltsev et al., 2018; Gut et al., 2018; Halpern et al., 2017; Lin et al., 2015; Remark et al., 2016; Saka et al., 2019; Satija et al., 2015; Stack et al., 2014). However, the existing protein multiplexing methods either use harsh de-staining conditions or require complex experimental setup. The SeqStain methodology presented here is a gentle, easy-to-use, and efficient multiplex imaging technique that provides a unique platform for obtaining such spatialomic insights. The method combines antibodies that are pre-labeled with fluorescent dsDNA with a gentle, enzymatic method for removing fluorescence signal after each cycle of staining. In particular, we demonstrate multiplex staining of immobilized cells and tissue sections whereby de-staining gently removes the fluorescent signal to pre-staining levels on the whole slide. Strikingly, de-staining using the SeqStain method was also rapid and removed ∼99% of the signal in <1 min without affecting sample integrity or tissue morphology. The unexpected findings from our work are that the nuclease treatment did not seem to affect tissue integrity, as determined by repeated treatment of cells and tissues with DNase I. This is not entirely unexpected, as nucleases have also been used in the SeqFISH methodology (Lubeck et al., 2014). Additionally, by engineering specific restriction sites into DNA during antibody modification, we show that selective de-staining is possible with SeqStain. Retention of selective markers for subsequent rounds might be important for spatial alignment of tissues when it is not possible to perform whole-slide scanning or for measuring information about multiple neighboring antigens via fluorescence resonance energy transfer (FRET) or other such methods, whereby keeping a fluorophore fixed on one antigen might be helpful while changing the second or third fluorophores on different antigens. SeqStain is thus a highly configurable and rapid multiplex IF imaging method. Furthermore, the enzymatic approach for the removal of fluorescent labels offers flexibility in the design of oligo sequence used for conjugation, the length and complexity of the oligos, the types of fluorophores that can be included, and the types of oligo-based higher-order structures that can be used. Moreover, the method is easily scalable to tens of different markers, as exemplified by the data presented by using a 10-plex or a 25-plex SeqStain panel. Signal amplification, if necessary for markers expressed at low levels, can be done using a number of published techniques (Ali et al., 2014; Li et al., 2008; Mullis and Faloona, 1987), which include increasing the number of fluorophores on the oligos, increasing the number of fluorescent oligos on the docking oligos, using different DNA structures (such as branched DNA [Collins et al., 1997], origami folded plasmids [Jungmann et al., 2014], or Fluorocubes [Niekamp et al., 2020]), or pre-complexing primary antibodies with SeqStain Fabs and SeqStain secondary antibodies.
The technique offers significant new advantages for deeper understanding of complex tissue microenvironments. This methodology uses commercially available primary and secondary antibodies and Fab fragments that are chemically conjugated with fluorescently labeled dsDNA (SeqStain antibodies and Fabs) and that are easy to modify in any laboratory. We also show that such modifications do not affect their function in any way by testing in a variety of systems. To avoid cross-reactivity, pre-complexation of SeqStain Fabs or SeqStain secondary antibodies with primary antibodies can also be used during staining. This paves way to build a multiplex panel that combines SeqStain antibodies, SeqStain secondary antibodies, and SeqStain Fabs by opening up additional ways of staining tissues in multiplex analyses. Utilizing Fabs and secondary antibodies for SeqStain not only improves sensitivity of detection for markers expressed at low levels but also facilitates higher throughput by reducing the time it takes to prepare reagents for SeqStain. Furthermore, SeqStain is performed by using a simple perfusion setup with readily available components, which together forms an elegant staining platform. In fact, this setup transforms standard confocal microscopes into a multiplex imaging appliance which, when combined with a motorized stage, enables high-level data acquisition by this method.
SeqStain offers significant advantages over some of the other multiplex techniques that have recently been described in the literature, such as CODEX and ImmunoSABER. Unique, barcoded primers, each of different lengths that are attached to each antibody, are a key part of CODEX. Therefore, primer design, and keeping track of primers, barcodes, and primer pairs during iterative rendering becomes cumbersome. SeqStain does not require any complex primer design or barcodes for its application. Similarly, ImmunoSABER uses concatemer extension using the PER method for multiplexing. This requires a certain level of expertise and therefore cannot be readily employed. In contrast, our dsDNA design is very simple, allowing use of almost any sequence. Similarly, SeqStain has significant advantages in its simple de-staining step. CODEX uses TCEP to break the disulfide bonds for removing fluorophores attached to DNA, which could limit its applications and requires a reaction time of approximately 5–10 min. ImmunoSABER utilizes formamide treatment (50%) for 10 min followed by washes to destabilize the DNA duplex, which could also limit its applications to specific tissue types, whereas SeqStain does not have any such limitations. Additionally, de-staining in SeqStain is rapid, with almost complete removal of fluorescent signal within 1 min for most antigens. Such rapid de-staining also ensures that the integrity of nuclear DNA is preserved during the process. Moreover, SeqStain offers a simple solution for signal amplification, if needed, using SeqStain secondary antibodies and Fabs, whereas CODEX relies on tyramide signal-based amplification, which is incompatible for de-staining. Although ImmunoSABER offers signal amplification solutions, the PER method requires a certain level of expertise. Finally, unlike other methods, the SeqStain method utilizes publicly available and widely utilized image analysis programs such as Fiji (ImageJ) and CellProfiler for image registration, stacking, segmentation, and intensity measurements and for generating SRMs and cellular neighborhoods.
Limitations of the study
The data presented here show that SeqStain is a gentle, efficient, and rapid technique for multiplex immunofluorescent imaging of cells and tissues that has wide applicability. We also note some of the limitations in our current study, although we did not observe any of these shortcomings in our research. First, unlike CODEX and ImmunoSABER, where all the antibodies are applied to the tissue in the first cycle, SeqStain requires repeated rounds of staining with cocktails of antibodies, which might result in an overall longer time period for a multiplex imaging experiment or difficulty staining some of the molecules in later cycles of staining, due to instability or loss of antigen. Second, the de-staining step for some of the intracellular compartments, such as the nucleus, requires longer incubation with the nucleases to provide the enzyme enough time to diffuse into such intracellular compartments. Third, the hybridization step required to generate fluorescent dsDNA requires heating and cooling steps that might result in chemical or thermal instability of some of the fluorophores, making such fluorophores inappropriate for this method. Finally, the data presented here primarily concern fresh frozen tissue sections, and it is likely that further optimization will be required to adapt the protocols to fixed tissues and other types of tissue sections, although we believe that the method is fully adaptable to these other types of tissues.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD11b (clone M1/70) | Biolegend | Cat# 101248; RRID: AB_2561479 |
| α-Tubulin | Abcam | Cat# ab18251: RRID: AB_2210057 |
| Mouse CD45 (clone 30-F11) | Biolegend | Cat# 103164; RRID: AB_2819790 |
| Vinculin (clone EPR8185) | Abcam | Cat# ab129002; RRID: AB_11144129 |
| F4/80 (clone T45-2342) | BD Biosciences | Cat# 565409; RRID: AB_2739222 |
| Paxillin (clone Y113) | Abcam | Cat# ab32084; RRID: AB_779033 |
| CD45R/B220 (clone RA3-6B2) | BD Biosciences | Cat# 557390; RRID: AB_396673 |
| CD44 (clone IM7) | BD Biosciences | Cat# 553131; RRID: AB_394646 |
| MHC II (clone M5/114.15.2) | BD Biosciences | Cat# 556999; RRID: AB_396545 |
| IgD (clone 11-26c.2a) | BD Biosciences | Cat# 553438; RRID: AB_394858 |
| IgM (clone R6-60.2) | BD Biosciences | Cat# 553405; RRID: AB_394842 |
| CD169 (clone 3D6.112) | Biolegend | Cat# 142402; RRID: AB_10916523 |
| CD34 (clone QBEND/10) | Thermo Fisher Scientific | Cat# MA1-10202; RRID: AB_11156010 |
| Mouse CD68 (clone Y1/82A) | Biolegend | Cat# 333802; RRID: AB_1089058 |
| Human CD45 (clone H130) | BD Biosciences | Cat# 555480; RRID: AB_395872 |
| Human CD68 (KP1) | Thermo Fisher Scientific | Cat# 14-0688-80; RRID: AB_11151503 |
| CD8 (clone HIT8a) | BD Biosciences | Cat# 555631; RRID: AB_395995 |
| CD31 (clone WM59) | BD Biosciences | Cat# 555444; RRID: AB_395837 |
| Cytokeratin-8 (clone LP3K) | eBioscience | Cat# 14-9938-82; RRID: AB_10557242 |
| Cytokeratin-7 (clone RCK105) | Santa Cruz Biotechnology | Cat# sc-23876; RRID: AB_2265604 |
| Collagen IV (clone 1042) | Thermo Fisher Scientific | Cat# 14-9871-82; RRID: AB_10870985 |
| Histone (clone AE-4) | Santa Cruz Biotechnology | Cat# sc-8030; RRID: AB_675641 |
| Podocin | Sigma-Aldrich | Cat# P0372; RRID: AB_261982 |
| Synaptopodin (clone D9) | Santa Cruz Biotechnology | Cat# sc-515842 |
| EpCAM (clone HEA125) | Santa Cruz Biotechnolgy | Cat# sc-59906; RRID: AB_783252 |
| Aquaporin 1 | Millipore Sigma Santa Cruz Biotechnology |
Cat# SAB4501545; RRID: AB_10745542 and Cat# sc-25287; RRID: AB_626694 |
| Aquaporin 2 (clone E-2) | Santa Cruz Biotechnology | Cat# sc-515770; RRID: AB_2810957 |
| Aquaporin 3 (clone F-1) | Santa Cruz Biotechnology | Cat# sc-518001 |
| ACE2 (clone E-11) | Santa Cruz Biotechnology | Cat# sc-390851; RRID: AB_2861379 |
| WT1 (clone 6F-H2) | Thermo Fisher Scientific | Cat# MA1-46028; RRID: AB_962464 |
| Nephrin | Thermo Fisher Scientific | Cat# PA5-72826; RRID: AB_2718680 |
| Megalin (clone H-10) | Santa Cruz Biotechnology | Cat# sc-515772; RRID: AB_2783023 |
| Na+K+-ATPase (clone C464.6) | Santa Cruz Biotechnology | Cat# sc-21712; RRID: AB_626713 |
| Uromodulin (clone 877914) | R&D systems | Cat# MAB5144 |
| Vimentin (clone V9) | Santa Cruz Biotechnology | Cat# sc-6260; RRID: AB_628437 |
| α-SMA (clone 1A4) | Millipore Sigma | Cat# A5228; RRID: AB_262054 |
| Donkey anti-Mouse IgG Goat anti-mouse IgG |
Thermo Fisher Scientific | Cat# A21202; RRID: AB_141607 and Cat # A11301 |
| Donkey anti-Rabbit IgG | Thermo Fisher Scientific | Cat# A10042; RRID: AB_2534017 and Cat# A21206; RRID: AB_2535792 |
| Goat anti-Rat IgG | Thermo Fisher Scientific | Cat# A11081; RRID: AB_141738 and Cat# A11006; RRID: 2534074 |
| AffiniPure Fab fragment Goat Anit-Rabbit IgG, Fc fragment | Jackson ImmunoResearch | Cat# 111-007-008 |
| AffiniPure Fab fragment Goat Anit-Mouse IgG1, Fcy fragment | Jackson ImmunoResearch | Cat# 115-007-185 |
| AffiniPure Fab fragment Goat Anit-Rat IgG, Fcy fragment | Jackson ImmunoResearch | Cat# 112-007-008 |
| Biological samples | ||
| Murine spleen tissue | This paper | N/A |
| Normal human kidney | OriGene Technologies | AF1FR0002A0D3 AF3FR0002ED31 |
| Normal human tonsil tissue | OriGene Technologies | FR00001F19 |
| Chemicals, peptides, and recombinant proteins | ||
| Sulfo-SMCC (Sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate) | Thermo Fisher | A39268 |
| TCEP(Tris(2-carboxyethyl)phosphine hydrochloride) | Thermo Fisher | 77720 |
| DBCO-Sulfo-NHS ester | Click Chemistry Tools | A124 |
| EZ-Link™ Sulfo-NHS-LC-Biotin | Thermo Fisher | A39257 |
| Purified Streptavidin | BioLegend | 405150 |
| Traut’s reagent | Thermo Fisher | 26101 |
| Salmon sperm DNA | Thermo Fisher | AM9680 |
| DNase I | NEB | M0303 |
| EcoRV | NEB | R3195S |
| SmaI | NEB | R01041S |
| 3-Triethoxysilypropylamine (APES) | Millipore | A3648 |
| Experimental models: cell lines | ||
| RAW264.7 cells | ATCC | TIB-71 |
| K562 cells | ATCC | CCL-243 |
| Lewis Lung Carcinoma | ATCC | CRL-1642 |
| HeLa cells | ATCC | CCL-2 |
| K562 CD11b/CD18 cells | Laboratory of Vineet Gupta | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852245/ |
| Mouse podocytes | Laboratory of Jochen Reiser | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4625676/ |
| Oligonucleotides | ||
| mAb linker with EcoRV site; 5’-TTTTTTTTTTAGCAGATATCACAGC |
IDT | N/A |
| mAb linker with SmaI site; 5’-TTTTTTTTTTAGCACCCGGGACAGC |
IDT | N/A |
| mAb linker with Azide 5’; ACGGGATATCAGATACGGGATATCAGATACGGGA-TATCAGAT | IDT | N/A |
| mAb linker with Biotin 5’; ACGGGATATCAGATACGGGATATCAGATACGGGA-TATCAGAT | IDT | N/A |
| Bridging oligo with EcoRV site;5’; TTGACAGCTGCCGGATTGACAGCTGCCGGATTGA-CAGCTGCCGGATTGACAGCTGCCGGA- TTGACAGCTGCCGGA GCTGTGATATCTGCT | IDT | N/A |
| Bridging oligo with SmaI site; 5’-TTGACAGCTGCCGGATGACAGCTGCCGGA- TTGACAGCTGCCGGATTGACAGCTGCCGGA- TTGACAGCTGCCGGAGCTGTCCCGGGTGCT |
IDT | N/A |
| Fluorescent oligo (Maleimide-Sulfhydryl chemistry) 5’-TCCGGCAGCTGTCAA | IDT | N/A |
| Fluorescent oligo (DBCO-Azide chemistry) 5’-ATCTGATATCCCGT |
IDT | N/A |
| Fluorescent oligo (Biotin-Streptavidin chemistry) 5’-ATCTGATATCCCGT |
IDT | N/A |
| Single stranded blocking oligo 5’-TTTTCCCTCTTCTCTTCCTT |
IDT | N/A |
| Software and algorithms | ||
| Cell profiler version 3.1.9 | Carpenter et al., 2006 | https://cellprofiler.org |
| ImageJ | NIH | https://imagej.nih.gov/ij/download.html |
| Halo software version v3.2.1851.207 | Indica labs | https://indicalab.com/halo/ |
| Prism 8.2 software | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Vineet Gupta (vineet_gupta@rush.edu)
Materials availability
This study did not generate any unique materials. All materials needed to support the claims of the study are available commercially.
Data and code availability
The article includes all data generated or analyzed during this study. Original source data for Figures in the paper are available upon request to the Lead Contact author. No proprietary software was used in the data analysis.
Experimental model and subject details
Cell lines: RAW264.7 cells (mouse), Lewis Lung Carcinoma (LLC) cells (mouse), K562 cells (human) and HeLa cells (human) were obtained from ATCC (Manassas, VA) and were cultured as per manufacturer’s recommendations. K562 CD11b/CD18 cells and mouse podocytes have been previously described (Maiguel et al., 2011; Shankland et al., 2007). All cell lines were maintained according to the published procedures at 37°C with 5% CO2.
All animal studies were performed in compliance with the Institutional Animal Care and Use Committee (IACUC) at Rush University Medical Center. Wild type C57Bl/6 mice (6-8 week old) were obtained from The Jackson Laboratory. Murine spleen tissues were collected from mice bearing orthotopically transplanted LLC cells, as previously described (Schmid et al., 2018). Briefly, LLC cells were passaged at least 3 times before inoculating into mice. Cells were routinely checked for mycoplasma using the MycoAlert assay (Lonza) and found negative prior to their use in animals. LLC cells (1.0 x 106) in 100μL cold phosphate-buffered saline (PBS) were subcutaneously inoculated into the mouse rear flank. Once the tumors grew to 1000mm3 in volume, the animals were sacrificed, and the harvested spleen tissue was immediately embedded in Tissue Tex OCT. Embedded tissue was cryopreserved in liquid nitrogen and transferred to -80˚C for long term storage. Cryopreserved normal human kidney and tonsil tissue blocks were purchased from Origene (Rockville, MD).
Method details
Preparation of fluorescent-DNA conjugated antibodies (SeqStain antibodies)
Preparation of linker oligos for conjugation to antibodies
Synthetic oligonucleotides (oligos) were obtained from IDT (Coralville, Iowa). Linker oligos were activated and conjugated to antibodies using published protocols (Williams and Chaput, 2010). Briefly, linker oligos with a 5’-terminal amine modification were activated using sulfo-SMCC (Sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate) reagent (Thermo Fisher, #A39268) according to manufacturer’s instructions. This SMCC-modified oligo was desalted to remove excess sulfo-SMCC using the 7KDa molecular weight cut-off Zeba columns (Thermo Fisher, # 89882) and used in conjugation reaction with the reduced antibodies.
Conjugation of antibodies to the linker oligos
Primary antibodies, secondary antibodies, and affinity purified Fc-specific Fab fragments were obtained from commercial sources. Any preservative, such as sodium azide or glycerol, was removed from the commercially obtained antibodies using 50 kDa molecular weight cut-off Amicon ultra filtration columns (Millipore, # UFC505096) and buffer exchanged into phosphate-buffered saline (PBS) buffer pH 7.2. Antibodies were chemically conjugated to SMCC-modified linker oligos using published protocols (Dovgan et al., 2019; Gong et al., 2016; Kazane et al., 2012; Winkler, 2013). Briefly, antibodies were reduced using TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) (Thermo Fisher, # 77720) according to manufacturer’s instructions, purified using 50 kDa molecular weight cut-off Amicon ultra filtration columns (Millipore, # UFC505096) and buffer exchanged into phosphate-buffered saline (PBS) buffer pH 7.2. Next, SMCC-modified oligo was added to the reaction mixture (at a 1:25 molar ratio), incubated at 4°C for 1 hour and purified using Amicon ultra filtration columns. Conjugation efficiency was determined using SDS-PAGE gels, which showed an average of 2-5 linker oligos chemically conjugated to an antibody during a typical labelling experiment. Subsequently, the conjugated antibody was washed, quantified by measuring the intensity of bands on an SDS-PAGE gel, and stored in PBS containing 0.5M NaCl.
Hybridization of oligos to create fluorescent-DNA labelled antibodies
The linker oligo conjugated antibody was mixed with fluorescent-DNA complex to prepare fluorescent-DNA labelled antibody. First, the fluorescent-DNA complex was prepared in a separate tube by mixing a docking oligo (Figure S1A) that contains a sequence complementary to the linker oligo and a set of sequence repeats for binding fluorescently labelled oligos. The fluorescent oligos were designed to contain a fluorophore at its 3’-end. The mixture was annealed by heating at 85°C for 5 minutes and slow cooling to generate the complex. All the oligos were obtained from IDT (Coralville, Iowa), with the fluorescent oligos containing a single fluorescent label at its 3’ end. Next, the complex was mixed with linker oligo conjugated antibody at 45°C and slow cooled to 37°C. The reaction mix was kept at 37°C for 30 minutes, slow cooled to room temperature and the completion of reaction was monitored by running it on a 4% agarose gel. Subsequently, the fluorescent-DNA labelled antibody complex was purified using 100 kDa molecular weight cut-off Amicon ultra filtration columns (Millipore #UFC510096) and stored in PBS buffer containing 0.5M NaCl at 4°C for use in SeqStain assays.
Preparation of SeqStain antibodies using DBCO-Azide chemistry
Antibodies were covalently conjugated to the linker oligo using the DBCO-Azide click chemistry using published protocols (31). Briefly, purified linker oligos containing 3’-terminal Azide were purchased from IDT (Coralville, Iowa) and resuspended in PBS, pH 7.2. Antibody was modified with DBCO-Sulfo-NHS ester (Click Chemistry Tools, #A124) according to manufacturer’s instructions. The DBCO activated antibodies were prepared and purified according to published protocols using 50 kDa molecular weight cut-off Amicon ultra filtration columns (Millipore, # UFC505096) and buffer exchanged into PBS, pH 7.2. DBCO-activated antibodies were combined with 20-molar fold excess of azide-modified linker oligo and incubated at 4°C overnight. Conjugation efficiency was estimated by running the mixture on a SDS-PAGE gel. The linker conjugated antibody was hybridized with fluorescent-DNA complex, as above, to prepare SeqStain antibodies.
Preparation of SeqStain antibodies using biotin-streptavidin methodology
Antibodies were biotin labelled in-house using a biotin labelling kit (Thermo Fisher, #A39257) by following manufacturer’s instructions. Linker oligo with 3’-terminal biotin was purchased from IDT (Coralville, Iowa) and purified streptavidin was purchased from BioLegend (Biolegend, #405150). Biotinylated antibody, streptavidin and biotinylated linker oligo were combined in a 1:1:3 molar ratio and incubated at room temperature for 30 minutes. Subsequently, the linker conjugated antibody was hybridized with fluorescent-DNA complex, as above, to prepare SeqStain antibodies.
Preparation of fluorescent-DNA conjugated Fabs (SeqStain Fabs)
Affinity-purified Fc-specific Fab fragments were modified according to literature protocols using Traut’s reagent, which adds sulfhydryl groups by reacting with the primary amines in Fab (Jue et al., 1978; Newton et al., 2001). Briefly, Traut’s reagent (Thermo Fisher, #26101) was added at 20-fold molar excess to the Fab fragments according to manufacturer’s instructions. This thiolation reaction proceeded for an hour at room temperature. Subsequently, excess reagent was quenched by adding 20mM glycine for 5 minutes. Thiolated Fab fragments were purified using 30 kDa molecular weight Amicon ultra filtration columns (Millipore, #UFC503096) and the concentration was estimated by A280 absorbance using Nanodrop 2000 (Thermo Fisher, Waltham, Massachusetts). Conjugation of the Fab with linker oligos and hybridization with fluorescent-DNA complex was performed as described for the antibodies above.
Preparation of pre-complex between unlabelled primary antibody and SeqStain Fabs
SeqStain Fabs were mixed with unlabelled primary antibodies at equal molar ratio (3:1 weight ratio) in PBS, pH 7.2, and incubated at room temperature for 2 hours. Any unbound, excess Fab was removed by filtration using 100 kDa molecular weight cut-off size exclusion Amicon ultra filtration columns (Millipore, # UFC510096). Subsequently, the complex was used for staining of cells and tissue sections.
Preparation of pre-complex between primary antibody and SeqStain secondary antibodies
Secondary antibodies were conjugated to the linker oligo using the maleimide-sulfhydryl chemistry and hybridized to the fluorescent oligo complex as detailed for primary antibodies. SeqStain secondary antibodies were mixed with unlabelled primary antibodies at 2:1 molar ratio at 0.1uM concentration with respect to the primary antibody to avoid formation of superclusters of primary and secondary antibodies. This reaction was incubated at room temperature for 15 minutes. Excess secondary antibody was then blocked by adding the corresponding normal serum at 1:20 v/v ratio and the mixture was incubated for 5 minutes at room temperature (Thermo Fisher Scientific, #10410 and #10510). The complex was used immediately for staining of cells and tissue sections.
Gel analysis of SeqStain antibodies and Fabs
To determine linker oligo conjugation efficiency of antibodies, the reaction mixture was analysed using 10% SDS PAGE gel (Thermo Fisher Scientific, NW04122BOX) following manufacturer’s instructions. Controls, including unmodified antibodies and a protein ladder (Bio-rad, #1610375) were also used. Subsequently, the gel was Coomassie stained (Thermo Fisher Scientific, #24617) to visualize the protein bands. To determine efficiency of hybridization reaction, the annealing reaction mixture was analysed using a 4% agarose gel containing a DNA stain (Thermo Fisher Scientific, #S33102). The bands were visualized using a UV transilluminator (Bio-Rad laboratories, Hercules, California).
Thermostability assay
In order to assess the thermostability of the fluorescent oligos, serial dilutions of the fluorescence oligos containing either the AF488 or cy3 fluorochrome were prepared in PBS. The oligos rwere cyclically heated to 85°C for 5 minutes and slow cooled to ambient temperature and the fluorescence was measured at ambient temperature at the end of each cycle using CFX connect real time system (Bio-Rad, #1855201).
Flow cytometry assays
Single cell suspensions of cells in culture were prepared in PBS containing 1% BSA. Non-specific antibody recognition sites on cells were blocked by adding Fc Block (Biolegend, #101301) to the suspension for ten minutes at 4°C. Next, cells were stained with either unlabelled primary antibodies (conventional staining) or with SeqStain antibodies for 20 min at 4°C. Subsequently, cells were washed with PBS containing 1% BSA and analysed using LSR-Fortessa flow cytometer. Data was analysed using FlowJo software version 10.2.
Staining of cells and tissue sections using SeqStain
Buffers and reagents
The following buffers and reagents were used in the assays. Block-1 solution (PBS containing 1% Bovine Serum Albumin (Sigma-Aldrich, #B4287-1G)), Block-2 solution (PBS containing 200ng/ml salmon sperm DNA (Thermo Fisher, #AM9680) and 3 nanomoles/ml single-stranded DNA and 0.5M NaCl), Wash buffer (PBS containing 0.1% Tween-20 (Sigma-Aldrich, #P1379), DNase I de-staining buffer (PBS containing 1X DNase buffer and 20 units of DNase I (NEB, #M0303) in 500ul), EcoRV de-staining buffer (deionized water containing 1X cut-smart buffer and 200 units of EcoRV (NEB, #R3195S) in 500ul), SmaI de-staining buffer (de-ionized water containing 1X cut smart buffer and 200 units of SmaI (NEB, #R01041S) in 500ul).
Preparation of cells for SeqStain
25 mm round cover glass (Fisher Brand, #12-545-102 25CIR-1) was coated with 3-Triethoxysilypropylamine APES (Millipore, #A3648) following manufacturer’s instructions. RAW264.7 cells were cultured as a monolayer on the coated cover glass in a 6-well plate. When the cells were about 50% confluent, they were removed from culture and fixed by treating with 4% paraformaldehyde at room temperature for 15 minutes. Once fixed, the cells were washed three times with PBS pH 7.4, permeabilized with 0.5% Triton-X at room temperature for 15 minutes. Subsequently, the cells were washed twice with PBS pH7.4 and blocked with the block-1 solution at room temperature for 1 hour. During this time, the cover glass was mounted onto the perfusion chamber.
Preparation of tissue sections for SeqStain
Tissue sections of 5μm thickness were mounted on the APES coated cover glass and fixed with acetone at room temperature for 5 minutes. Sections were further fixed by treating with 1.6% paraformaldehyde at room temperature for 15 minutes. The fixed sections were washed thrice with PBS pH7.4 and blocked with block-1 solution at room temperature for 1 hour. During incubation, the cover glass was mounted onto the perfusion chamber.
SeqStain immunofluorescence staining
The cover glass containing cells or tissue sections were blocked with Block-2 solution at room temperature for 1 hour. Subsequently, these samples were stained with either SeqStain antibodies or primary antibodies pre-complexed with SeqStain Fabs diluted in Block-2 solution at room temperature for 1 hour. Next, the chamber was perfused with the wash buffer continuously for 5 minutes to remove non-specific staining. Nuclei were counterstained by adding DAPI (Sigma-Aldrich, #D9542). The cells were washed again by perfusing wash buffer for 5 minutes and imaged. To de-stain, the chamber was perfused with 500ul of the de-staining buffer (NEB, #M0303) and imaged at the indicated time points. The de-staining buffer was replaced after 10 minutes for an additional round of de-staining. The de-stained samples were blocked again with block-2 solution for 20 minutes before adding the next round of SeqStain antibodies or primary antibodies pre-complexed with SeqStain Fabs. For the selective de-staining experiment, de-staining was performed by perfusing the appropriate de-staining buffers. Nuclear staining with DAPI was repeated every round. For staining by indirect immunofluorescence (conventional), the cover glass containing the cells or tissue sections were prepared as described before and mounted onto the perfusion chamber. The mounted samples were stained with primary antibodies after blocking with block-1 solution for 1 hour at room temperature. The chamber was perfused with wash buffer for 5 minutes to remove non-specific staining and then incubated with the corresponding secondary antibodies and stained at room temperature for 30 minutes. The samples were washed by perfusing wash buffer for 5 minutes and imaged after nuclear staining with DAPI.
Perfusion set-up
Cover glass was mounted on the closed bath chamber (Warner instruments, #RC-43C) following manufacturer’s instructions. This, in-turn, was mounted on the quick exchange platform (Warner instruments, #64-0375 (QE-1)) with built-in perfusion and suction holders. Buffers and solutions were perfused in and suctioned out via the inlet and outlet ports, respectively. A stage adapter (Warner instruments, #64-2415) was used to place the platform on the microscope to perform the iterative rounds of staining and de-staining.
Image acquisition
Immunofluorescence images were acquired using the Zeiss 700 LSM confocal microscope and Zen software (Carl Zeiss Group, Hartford, Connecticut). The destaining videos were acquired using the time-lapse image acquisition option in the Zen software. DNase I was added after the first image was acquired (-20 sec), following which images were acquired every 20 seconds for 5 minutes to monitor the rate of destaining. For the whole slide imaging, the images were acquired using the Nikon Eclipse TI2-E inverted confocal microscope (Nikon Corporation, Tokyo, Japan) with a motorized stage. The image tiles were acquired with the perfect focus system (PFS) to correct for any focal drifts between the tiles and then processed with the NIS Elements software.
Quantification and statistical analysis
Image analysis using cell profiler
Cell profiler version 3.1.9 was used to identify individual cells and their nuclei for both immunofluorescent staining of cells and tissue sections according to published protocols (Carpenter et al., 2006; Lamprecht et al., 2007; McQuin et al., 2018). Briefly, a working pipeline was created to run the required analysis. A set of modules were designed to identify objects and their relative intensities. For each image set, nuclei were considered as starting points and was identified using identify primary objects module. Identify secondary objects module was used to identify objects such as cells based on objects identified in previous module. Measure object intensity module was used to measure the intensity of each identified object. Overlay Object module created a color-coded label of the previously identified objects. Overlay Outline was used to place outlines of an object on a desired image as to create segmentation in the cellular region. Intensity measured was exported to excel files for further analysis.
Image analysis using Fiji
For the whole slide images obtained from the multiplex imaging experiment, to align the images, DAPI images from each round of staining and de-staining were rigid registered (translation + rotation) to each other using the Register Virtual Stack Slices plugin in Fiji (Arganda-Carreras et al., 2006). For the other channels in each round, the same transformation as the corresponding DAPI channel was applied with the Transform Virtual Stack Slices plugin in Fiji. To measure distances of various cell types from the B-cell cluster, a region of interest was selected, and the cluster was traced manually using the signal from the DAPI and B220 channels. Euclidean Distance Map to the boarder of the cluster was calculated with Fiji. To measure signal intensities of each channel, the nuclei were first segmented using the DAPI channel. Segmentation was performed using the StarDist plugin, which uses an already trained model for convolution neural network (DSB 2018 dataset) (Schmidt et al., 2018). Once segmented, intensities for each channel were calculated on a per cell basis. Alongside, the EDM measurements for each segmented cell was calculated. In order to generate the plot profiles of the same markers from distinct rounds, first a stack of the corresponding images was generated. From the stack, a random region was cropped, and stack was converted to individual images. Plot profiles were calculated by drawing an arbitrary yellow line at the same position in both the images using Fiji. The overlay plots were generated using GraphPAD PRISM 9. To compute the signal to noise ratio for the immunofluorescence images, a region of interest (ROI) was defined using Fiji. The defined ROI was positioned in a region with positive staining to calculate the integrated density (signal), this was repeated for four more regions with positive staining and the average is shown. Subsequently, the defined ROI was positioned in a region without cell or tissue staining to calculate the integrated density (noise) and repeated for four additional times as before. Between the SeqStain and conventional immunofluorescence images for each set, the same ROI was used by utilizing the selection tool in Fiji.
Image analysis using HALO
To generate the spatial relationship maps, HALO spatial analysis software was used. First the images containing markers of interest were aligned using the registration module in HALO. Next, positive signal for each marker was identified using the HALO High-plex module. The identified cell types were plotted as a spatial plot using the Spatial analysis module to generate the spatial maps and distances were computed using the proximity analysis option in the software.
Statistics
Statistical analyses were performed using Excel (Microsoft, Bothell, WA) and Prism 8.2 software (GraphPad Software). Student’s t-test were utilized when the data were normally distributed. Mann-Whitney test was used for comparison between two groups when data was not normally distributed.
Acknowledgments
We thank Liudmila Romanova and Diptaman Chatterjee in Kordower laboratory at Rush University Medical Center and the Center for Advanced Microscopy at Northwestern University for help with tissue staining and image analysis, and all past and current members of the Gupta laboratory for their technical help and suggestions. This work was supported in part by funding from the Oppenheimer Family Foundation, Bears Care, the Department of Internal Medicine at Rush University Medical Center, and by the National Institutes of Health (grants R01DK107984, R01DK084195, and R01CA244938 to V.G.).
Author contributions
Conceptualization, V.G., A.R., and I.V.; methodology, A.R., I.V., R.A., D.K., J.H.K., and V.G.; investigation, A.R., I.V., R.A., and S.K.; software, A.R., S.K., and D.K.; supervision, V.G.; writing – original draft, A.R., I.V., R.A., D.K., S.K., D.C., and V.G.; writing – review & editing, A.R., I.V., J.K., and V.G.; funding acquisition, V.G.
Declaration of interests
A.R. and V.G. are inventors on a patent application filed by Rush University Medical Center related to these studies. V.G. is a co-founder of 149 Bio, LLC and Spatialomyx, LLC that is licensing this intellectual property and has significant financial interest in it. V.G. is also an Officer and a Board member of 149 Bio, LLC and Spatialomyx, LLC. The remaining authors declare no competing interests.
Published: May 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100006.
Supplemental information
References
- Accolla R.S., Lombardo L., Abdallah R., Raval G., Forlani G., Tosi G. Boosting the MHC class II-restricted tumor antigen presentation to CD4+ T helper cells: a critical issue for triggering protective immunity and re-orienting the tumor microenvironment toward an anti-tumor state. Front. Oncol. 2014;4:32. doi: 10.3389/fonc.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agasti S.S., Wang Y., Schueder F., Sukumar A., Jungmann R., Yin P. DNA-barcoded labeling probes for highly multiplexed Exchange-PAINT imaging. Chem. Sci. 2017;8:3080–3091. doi: 10.1039/c6sc05420j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akturk G., Sweeney R., Remark R., Merad M., Gnjatic S. Multiplexed immunohistochemical consecutive staining on single slide (MICSSS): multiplexed chromogenic IHC assay for high-dimensional tissue analysis. Methods Mol. Biol. 2020;2055:497–519. doi: 10.1007/978-1-4939-9773-2_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.M., Li F., Zhang Z., Zhang K., Kang D.K., Ankrum J.A., Le X.C., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- Arganda-Carreras, I., Sorzano, C.O.S., Marabini, R., Carazo, J.M., Ortiz-De-Solorzano, C., and Kybic, J. (2006). Consistent and elastic registration of histological sections using vector-spline regularization. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) vol. 4241 (Springer), pp. 85–95.
- Bábíčková J., Klinkhammer B.M., Buhl E.M., Djudjaj S., Hoss M., Heymann F., Tacke F., Floege J., Becker J.U., Boor P. Regardless of etiology, progressive renal disease causes ultrastructural and functional alterations of peritubular capillaries. Kidney Int. 2017;91:70–85. doi: 10.1016/j.kint.2016.07.038. [DOI] [PubMed] [Google Scholar]
- Beechem J.M. High-plex spatially resolved RNA and protein detection using digital spatial profiling: a technology designed for immuno-oncology biomarker discovery and translational research. Methods Mol. Biol. 2020;2055:563–583. doi: 10.1007/978-1-4939-9773-2_25. [DOI] [PubMed] [Google Scholar]
- Belliere J., Casemayou A., Ducasse L., Zakaroff-Girard A., Martins F., Iacovoni J.S., Guilbeau-Frugier C., Buffin-Meyer B., Pipy B., Chauveau D., et al. Specific macrophage subtypes influence the progression of rhabdomyolysis-induced kidney injury. J. Am. Soc. Nephrol. 2015;26:1363–1377. doi: 10.1681/ASN.2014040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall S.C., Nolan G.P., Roederer M., Chattopadhyay P.K. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Zunder E.R., Finck R., Chen T.J., Savig E.S., Bruggner R.V., Simonds E.F., Bendall S.C., Sachs K., Krutzik P.O., et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boor P., Floege J. The renal (myo-)fibroblast: a heterogeneous group of cells. Nephrol. Dial. Transplant. 2012;27:3027–3036. doi: 10.1093/ndt/gfs296. [DOI] [PubMed] [Google Scholar]
- Borges Da Silva H., Fonseca R., Pereira R.M., Cassado A.A., Álvarez J.M., D’Império Lima M.R. Splenic macrophage subsets and their function during blood-borne infections. Front. Immunol. 2015;6:480. doi: 10.3389/fimmu.2015.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappi G., Dupouy D.G., Comino M.A., Ciftlik A.T. Ultra-fast and automated immunohistofluorescent multistaining using a microfluidic tissue processor. Sci. Rep. 2019;9:4489. doi: 10.1038/s41598-019-41119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M.L., Irvine B., Tyner D., Fine E., Zayati C., Chang C.A., Horn T., Ahle D., Detmer J., Shen L.P., et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter A., Harris R. Simplified preparation of rabbit fab fragments. J. Immunol. Methods. 1983;59:199–203. doi: 10.1016/0022-1759(83)90031-5. [DOI] [PubMed] [Google Scholar]
- Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- Dixon A.R., Bathany C., Tsuei M., White J., Barald K.F., Takayama S. Recent developments in multiplexing techniques for immunohistochemistry. Expert Rev. Mol. Diagn. 2015;15:1171–1186. doi: 10.1586/14737159.2015.1069182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovgan I., Koniev O., Kolodych S., Wagner A. Antibody-oligonucleotide conjugates as therapeutic, imaging, and detection agents. Bioconjug. Chem. 2019;30:2483–2501. doi: 10.1021/acs.bioconjchem.9b00306. [DOI] [PubMed] [Google Scholar]
- Fissell W.H., Miner J.H. What is the glomerular ultrafiltration barrier? J. Am. Soc. Nephrol. 2018;29:2262–2264. doi: 10.1681/ASN.2018050490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountaine T.J., Wincovitch S.M., Geho D.H., Garfield S.H., Pittaluga S. Multispectral imaging of clinically relevant cellular targets in tonsil and lymphoid tissue using semiconductor quantum dots. Mod. Pathol. 2006;19:1181–1191. doi: 10.1038/modpathol.3800628. [DOI] [PubMed] [Google Scholar]
- Frampton J.P., Tsuei M., White J.B., Abraham A.T., Takayama S. Aqueous two-phase system-mediated antibody micropatterning enables multiplexed immunostaining of cell monolayers and tissues. Biotechnol. J. 2015;10:121–125. doi: 10.1002/biot.201400271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannot G., Tangrea M.A., Erickson H.S., Pinto P.A., Hewitt S.M., Chuaqui R.F., Gillespie J.W., Emmert-Buck M.R. Layered peptide array for multiplex immunohistochemistry. J. Mol. Diagn. 2007;9:297–304. doi: 10.2353/jmoldx.2007.060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaublomme J.T., Li B., McCabe C., Knecht A., Yang Y., Drokhlyansky E., Van Wittenberghe N., Waldman J., Dionne D., Nguyen L., et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat. Commun. 2019;10:2907. doi: 10.1038/s41467-019-10756-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes M.J., Sevinsky C.J., Sood A., Adak S., Bello M.O., Bordwell A., Can A., Corwin A., Dinn S., Filkins R.J., et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. U S A. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass G., Papin J.A., Mandell J.W. SIMPLE: a sequential immunoperoxidase labeling and erasing method. J. Histochem. Cytochem. 2009;57:899–905. doi: 10.1369/jhc.2009.953612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G.P. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H., Holcomb I., Ooi A., Wang X., Majonis D., Unger M.A., Ramakrishnan R. Simple method to prepare oligonucleotide-conjugated antibodies and its application in multiplex protein detection in single cells. Bioconjug. Chem. 2016;27:217–225. doi: 10.1021/acs.bioconjchem.5b00613. [DOI] [PubMed] [Google Scholar]
- Gut G., Herrmann M.D., Pelkmans L. Multiplexed protein maps link subcellular organization to cellular states. Science. 2018;361:eaar7042. doi: 10.1126/science.aar7042. [DOI] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Tóth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E., et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K., Takagi S., Fujisawa H., Takeichi M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Dev. Biol. 1987;120:215–227. doi: 10.1016/0012-1606(87)90119-9. [DOI] [PubMed] [Google Scholar]
- Huang W., Hennrick K., Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum. Pathol. 2013;44:29–38. doi: 10.1016/j.humpath.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Johnson D.B., Bordeaux J., Kim J.Y., Vaupel C., Rimm D.L., Ho T.H., Joseph R.W., Daud A.I., Conry R.M., Gaughan E.M., et al. Quantitative spatial profiling of PD-1/PD-L1 interaction and HLA-DR/Ido-1 predicts improved outcomes of anti-PD-1 therapies in metastatic melanoma. Clin. Cancer Res. 2018;24:5250–5260. doi: 10.1158/1078-0432.CCR-18-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.J., Iida H., Alpers C.E., Majesky M.W., Schwartz S.M., Pritzl P., Gordon K., Gown A.M. Expression of smooth muscle cell phenotype by rat mesangial cells in immune complex nephritis: α-Smooth muscle actin is a marker of mesangial cell proliferation. J. Clin. Invest. 1991;87:847–858. doi: 10.1172/JCI115089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jue R., Lambert J.M., Leland P.L., Pierce R., Traut R.R. Addition of sulfhydryl groups to Escherichia coli ribosomes by protein modification with 2-iminothiolane (methyl 4-mercaptobutyrimidate) Biochemistry. 1978;17:5399–5406. doi: 10.1021/bi00618a013. [DOI] [PubMed] [Google Scholar]
- Jungmann R., Avendaño M.S., Woehrstein J.B., Dai M., Shih W.M., Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazane S.A., Sok D., Cho E.H., Uson M.L., Kuhn P., Schultz P.G., Smider V.V. Site-specific DNA-antibody conjugates for specific and sensitive immuno-PCR. Proc. Natl. Acad. Sci. U S A. 2012;109:3731–3736. doi: 10.1073/pnas.1120682109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano Y., Prazma C.M., Yazawa N., Watanabe R., Ishiura N., Kumanogoh A., Okochi H., Tamaki K., Fujimoto M., Tedder T.F. CD83 influences cell-surface MHC class II expression on B cells and other antigen-presenting cells. Int. Immunol. 2007;19:977–992. doi: 10.1093/intimm/dxm067. [DOI] [PubMed] [Google Scholar]
- Lamprecht M.R., Sabatini D.M., Carpenter A.E. CellProfiler™: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Lely A.T., Hamming I., van Goor H., Navis G.J. Renal ACE2 expression in human kidney disease. J. Pathol. 2004;204:587–593. doi: 10.1002/path.1670. [DOI] [PubMed] [Google Scholar]
- Li J.J., Chu Y., Lee B.Y.H., Xie X.S. Enzymatic signal amplification of molecular beacons for sensitive DNA detection. Nucleic Acids Res. 2008;36:e36. doi: 10.1093/nar/gkn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-R., Fallahi-Sichani M., Sorger P.K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 2015;6:8390. doi: 10.1038/ncomms9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-R., Izar B., Wang S., Yapp C., Mei S., Shah P.M., Santagata S., Sorger P.K. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife. 2018;7:e31657. doi: 10.7554/eLife.31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiguel D., Faridi M.H., Wei C., Kuwano Y., Balla K.M., Hernandez D., Barth C.J., Lugo G., Donnelly M., Nayer A., et al. Small molecule-mediated activation of the integrin CD11b/CD18 reduces inflammatory disease. Sci. Signal. 2011;4:ra57. doi: 10.1126/scisignal.2001811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariant M., Camagna M., Tarditi L., Seccamani E. A new enzymatic method to obtain high-yield F(ab)2 suitable for clinical use from mouse IgGl. Mol. Immunol. 1991;28:69–77. doi: 10.1016/0161-5890(91)90088-2. [DOI] [PubMed] [Google Scholar]
- Martinez-Pomares L., Gordon S. CD169+ macrophages at the crossroads of antigen presentation. Trends Immunol. 2012;33:66–70. doi: 10.1016/j.it.2011.11.001. [DOI] [PubMed] [Google Scholar]
- McCulloch L., Alfieri A., McColl B.W. Experimental stroke differentially affects discrete subpopulations of splenic macrophages. Front. Immunol. 2018;9:1108. doi: 10.3389/fimmu.2018.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D., et al. CellProfiler 3.0: next-generation image processing for biology. Plos Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner J.H. Renal basement membrane components. Kidney Int. 1999;56:2016–2024. doi: 10.1046/j.1523-1755.1999.00785.x. [DOI] [PubMed] [Google Scholar]
- Miner J.H. The glomerular basement membrane. Exp. Cell Res. 2012;318:973–978. doi: 10.1016/j.yexcr.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K.B., Faloona F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Murakami T., Chen X., Hase K., Sakamoto A., Nishigaki C., Ohno H. Splenic CD19-CD35+B220+ cells function as an inducer of follicular dendritic cell network formation. Blood. 2007;110:1215–1224. doi: 10.1182/blood-2007-01-068387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton D.L., Hansen H.J., Mikulski S.M., Goldenberg D.M., Rybak S.M. Potent and specific antitumor effects of an anti-CD22-targeted cytotoxic ribonuclease: potential for the treatment of non-Hodgkin lymphoma. Blood. 2001;97:528–535. doi: 10.1182/blood.v97.2.528. [DOI] [PubMed] [Google Scholar]
- Niekamp S., Stuurman N., Vale R.D. A 6-nm ultra-photostable DNA FluoroCube for fluorescence imaging. Nat. Methods. 2020;17:437–441. doi: 10.1038/s41592-020-0782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S., Frøkiær J., Marples D., Kwon T.H., Agre P., Knepper M.A. Aquaporins in the kidney: from molecules to medicine. Physiol. Rev. 2002;82:205–244. doi: 10.1152/physrev.00024.2001. [DOI] [PubMed] [Google Scholar]
- Palmer R.E., Kotsianti A., Cadman B., Boyd T., Gerald W., Haber D.A. WT1 regulates the expression of the major glomerular podocyte membrane protein Podocalyxin. Curr. Biol. 2001;11:1805–1809. doi: 10.1016/s0960-9822(01)00560-7. [DOI] [PubMed] [Google Scholar]
- Peng C.W., Liu X.L., Chen C., Liu X., Yang X.Q., Pang D.W., Zhu X.B., Li Y. Patterns of cancer invasion revealed by QDs-based quantitative multiplexed imaging of tumor microenvironment. Biomaterials. 2011;32:2907–2917. doi: 10.1016/j.biomaterials.2010.12.053. [DOI] [PubMed] [Google Scholar]
- Qi H., Casalena G., Shi S., Yu L., Ebefors K., Sun Y., Zhang W., D’Agati V., Schlondorff D., Haraldsson B., et al. Glomerular endothelial mitochondrial dysfunction is essential and characteristic of diabetic kidney disease susceptibility. Diabetes. 2017;66:763–778. doi: 10.2337/db16-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remark R., Merghoub T., Grabe N., Litjens G., Damotte D., Wolchok J.D., Merad M., Gnjatic S. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci. Immunol. 2016;1:aaf6925. doi: 10.1126/sciimmunol.aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R.J. How restriction enzymes became the workhorses of molecular biology. Proc. Natl. Acad. Sci. U S A. 2005;102:5905–5908. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablik K.A., Jordanova E.S., Pocorni N., Clahsen-van Groningen M.C., Betjes M.G.H. Immune cell infiltrate in chronic-active antibody-mediated rejection. Front. Immunol. 2020;10:3106. doi: 10.3389/fimmu.2019.03106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka S.K., Wang Y., Kishi J.Y., Zhu A., Zeng Y., Xie W., Kirli K., Yapp C., Cicconet M., Beliveau B.J., et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 2019;37:1080–1090. doi: 10.1038/s41587-019-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T., Smith C.L., Cantor C.R. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- Sarvaria A., Madrigal J.A., Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell. Mol. Immunol. 2017;14:662–674. doi: 10.1038/cmi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiano G., Glaudemans B., Olinger E., Goelz N., Müller M., Loffing-Cueni D., Deschenes G., Loffing J., Devuyst O. The urinary excretion of uromodulin is regulated by the potassium channel ROMK. Sci. Rep. 2019;9:19517. doi: 10.1038/s41598-019-55771-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M.C., Khan S.Q., Kaneda M.M., Pathria P., Shepard R., Louis T.L., Anand S., Woo G., Leem C., Faridi M.H., et al. Integrin CD11b activation drives anti-tumor innate immunity. Nat. Commun. 2018;9:5379. doi: 10.1038/s41467-018-07387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, U., Weigert, M., Broaddus, C., and Myers, G. (2018). Cell detection with star-convex polygons. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) vol. 11071 (Springer), pp. 265–273.
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzbauer J., Strauss M.T., Schlichthaerle T., Schueder F., Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- Shankland S.J., Pippin J.W., Reiser J., Mundel P. Podocytes in culture: past, present, and future. Kidney Int. 2007;72:26–36. doi: 10.1038/sj.ki.5002291. [DOI] [PubMed] [Google Scholar]
- Sheng J., Ruedl C., Karjalainen K. Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity. 2015;43:382–393. doi: 10.1016/j.immuni.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Sic H., Kraus H., Madl J., Flittner K.A., Von Münchow A.L., Pieper K., Rizzi M., Kienzler A.K., Ayata K., Rauer S., et al. Sphingosine-1-phosphate receptors control B-cell migration through signaling components associated with primary immunodeficiencies, chronic lymphocytic leukemia, and multiple sclerosis. J. Allergy Clin. Immunol. 2014;134:420–428. doi: 10.1016/j.jaci.2014.01.037. [DOI] [PubMed] [Google Scholar]
- Singh N., Avigan Z.M., Kliegel J.A., Shuch B.M., Montgomery R.R., Moeckel G.W., Cantley L.G. Development of a 2-dimensional atlas of the human kidney with imaging mass cytometry. JCI Insight. 2019;4:e129477. doi: 10.1172/jci.insight.129477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider B.F., Folpe A.L., Hennigar R.A., Lim S.D., Cohen C., Tamboli P., Young A., De Peralta-Venturina M., Amin M.B. Distribution of cytokeratins and vimentin in adult renal neoplasms and normal renal tissue: potential utility of a cytokeratin antibody panel in the differential diagnosis of renal tumors. Am. J. Surg. Pathol. 2005;29:747–754. doi: 10.1097/01.pas.0000163362.78475.63. [DOI] [PubMed] [Google Scholar]
- Stack E.C., Wang C., Roman K.A., Hoyt C.C. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M., Zheng S., Houck-Loomis B., Hao S., Yeung B.Z., Mauck W.M., Smibert P., Satija R. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth Z.E., Mezey É. Simultaneous visualization of multiple antigens with tyramide signal amplification using antibodies from the same species. J. Histochem. Cytochem. 2007;97:873–885. doi: 10.1369/jhc.6A7134.2007. [DOI] [PubMed] [Google Scholar]
- Veninga H., Borg E.G.F., Vreeman K., Taylor P.R., Kalay H., van Kooyk Y., Kraal G., Martinez-Pomares L., den Haan J.M.M. Antigen targeting reveals splenic CD169+ macrophages as promoters of germinal center B-cell responses. Eur. J. Immunol. 2015;45:747–757. doi: 10.1002/eji.201444983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T., Bléry M., Chaix J., Fuseri N., Chasson L., Robbins S.H., Jaeger S., André P., Gauthier L., Daniel L., et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl. Acad. Sci. U S A. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.A.R., Chaput J.C. Synthesis of peptide-oligonucleotide conjugates using a heterobifunctional crosslinker. Curr. Protoc. Nucleic Acid Chem. 2010;Chapter 4:Unit4.41. doi: 10.1002/0471142700.nc0441s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J. Oligonucleotide conjugates for therapeutic applications. Ther. Deliv. 2013;4:791–809. doi: 10.4155/tde.13.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehrstein J.B., Strauss M.T., Ong L.L., Wei B., Zhang D.Y., Jungmann R., Yin P. Sub-100-nm metafluorophores with digitally tunable optical properties self-assembled from DNA. Sci. Adv. 2017;3:e1602128. doi: 10.1126/sciadv.1602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RAW264.7 cells were co-stained with anti-CD44 AF488 and anti-CD45 Cy3 SeqStain antibodies. Subsequently, the cells were incubated with DNase I and the fluorescence images were acquired as a video every 3 s.
Normal human kidney tissue sections were stained with anti-cytokeratin 8 AF488 antibody and de-stained with DNase I, and the fluorescence images acquired as a video every 20 s.
Normal human kidney tissue sections were stained with anti-collagen IV Cy3 antibody and de-stained with DNase I, and the fluorescence images acquired as a video every 20 s.
Normal human kidney tissue sections were stained with anti-EpCAM Cy5 antibody and de-stained with DNase I, and the fluorescence images acquired as a video every 20 s.
Data Availability Statement
The article includes all data generated or analyzed during this study. Original source data for Figures in the paper are available upon request to the Lead Contact author. No proprietary software was used in the data analysis.