Abstract
We have assayed the oncogenic, proliferative, and apoptotic activities of the frequent mutations that occur in the c-myc gene in Burkitt's lymphomas. Some alleles have a modest (50 to 60%) increase in transforming activity; however, the most frequent Burkitt's lymphoma allele (T58I) had an unexpected substantial decrease in transforming activity (85%). All alleles restored the proliferation function of c-Myc in cells that grow slowly due to a c-myc knockout. There was discordance for some alleles between apoptotic and oncogenic activities, but only the T58A allele had elevated transforming activity with a concomitant reduced apoptotic potential. We discovered a novel missense mutation, MycS71F, that had a very low apoptotic activity compared to wild-type Myc, yet this mutation has never been found in lymphomas, suggesting that there is no strong selection for antiapoptotic c-Myc alleles. MycS71F also induced very low levels of cytochrome c release from mitochondria, suggesting a mechanism of action for this mutation. Phosphopeptide mapping provided a biochemical basis for the dramatically different biological activities of the transformation-defective T58I and transformation-enhanced T58A c-Myc alleles. Furthermore, the antiapoptotic survival factor insulin-like growth factor 1 was found to suppress phosphorylation of T58, suggesting that the c-Myc transactivation domain is a direct target of survival signals.
Chromosomal translocations involving the c-myc gene occur in virtually all Burkitt's lymphomas (BL) and the related mouse plasmacytomas. Although these chromosomal abnormalities have been recognized for many years, it has recently become apparent that missense mutations may also play a role in the oncogenic activity of c-myc. More than 60% of BL and AIDS-associated lymphomas have mutations that alter the protein structure of the translocated c-myc gene (1, 5, 6, 8, 12, 28, 63). Similar mutations were recognized much earlier in three acutely oncogenic avian retroviruses harboring the myc gene, indicating that mutations can alter c-myc activity in many organisms. An understanding of the role of these mutations in tumor formation may provide important insight into the diverse functions of c-Myc in cell cycle progression, oncogenic transformation, apoptosis, and transcriptional regulation. Of broader interest is how c-Myc regulation and functional activities are interwoven with other signaling pathways.
Deletion mapping of c-Myc transforming activity consistently identifies the C-terminal DNA binding domain and an N-terminal region called Myc box II (amino acids 129 to 145) as essential for almost all biological activities. However, a second domain within the N terminus (amino acids 1 to 64) is also important in cell transformation. This region contains Myc homology box I (MbI, amino acids 45 to 64), an N-terminal domain that is nearly identical among all three Myc family proteins (c-, N-, and L-Myc). Early deletion mapping indicated that N-terminal mutants had variable transforming activity, but large deletions or those that target the conserved MbI were severely defective for oncogenic transformation (57). The importance of the c-Myc N terminus is accentuated by recent studies of BL that identified consistent mutations within MbI in >60% of the tumors (1, 5, 6, 8, 12, 28, 63). The finding of consistent, clustered mutations argues that they are not the result of benign genetic drift but rather represent selection for the outgrowth of tumor cells with specific c-Myc mutations. The frequency of the predominant naturally occurring c-Myc mutations from lymphomas or lymphoma cell lines is compiled in Table 1, showing that the two most common mutations are P57S and T58I. The largest cluster of mutations encompasses a proline-rich domain (amino acids 57 to 64), and it is striking that the mutation frequency drops just outside of the boundary of the proline cluster. Since this domain is a known site of phosphorylation in vivo (25, 37), the finding that most of the mutations remove or add phospho-acceptor sites suggests a simple model that phosphorylation of this domain significantly alters c-Myc function. Although the proline domain is one hot spot, two other sites of frequent mutation are E39 and A44, which are immediately N terminal to the proline domain. The E39 and A44 mutations are likely to be additive to those in the proline domain since tumors can have a mutation at only one of these sites or at all three positions simultaneously (1, 5, 63). Curiously, all of the mutations at E39 (glutamic acid) are to D (aspartic acid), which normally would be considered to be a conservative substitution. However, the same E39D mutation occurs in both human tumors and mouse plasmacytomas (the latter derived from a genetically defined inbred strain), indicating that they are not due to polymorphism. It was originally reported that many mutations in BL are homozygous and affect the nontranslocated allele as well as the translocated allele (5). However, reevaluation of the data suggests that the mutations affect only the translocated allele in both BL and plasmacytomas (K. Bhatia, personal communication). The mechanism of mutation at the c-myc locus is unknown.
TABLE 1.
c-Myc mutations in lymphomas
| Residue | Mutation | Occurrence | Other alleles (occurrence) |
|---|---|---|---|
| E39 | D | 11 | None |
| A44 | V | 6 | T (1), G (1) |
| P57 | S | 8 | T (5), A (1) |
| T58 | A | 2 | |
| T58 | I | 11 | N (2), D (1) |
| S62 | A* | 0 | |
| S62 | P | 3 | F (2) |
| S71 | E* | 0 | |
| S71 | F* | 0 | |
| S71 | W | 1 | None |
The frequency of predominant mutations within the Mb1 domain of c-Myc was tabulated from the literature. Other mutations not found in lymphomas (*) but which affect c-Myc phosphorylation were generated for functional analysis in this study.
Functional comparison of avian v-Myc to c-Myc proteins suggested that enhanced oncogenic activity in fibroblast and myeloid cell transformation is contributed by v-Myc mutations (20, 58). However, the functional analysis of mutations in mammalian c-myc genes is less clear. The T58A mutant has been reported to be fourfold more active than wild-type (wt) Myc in the transformation of rat embryo cells, as measured by focus formation in cooperation with the H-ras oncogene (49). However, the same mutation was indistinguishable from wt in the transformation of Rat1A fibroblasts (25), and this specific allele has only rarely been found in tumors (Table 1). Some complex mutant alleles derived from BL were found to have an increased size of anchorage-independent colonies but wt apoptotic activity in the same assay (26). It has been proposed that mutations in the c-Myc N terminus alter an interaction with the retinoblastoma protein-related p107 protein (23, 26); however, there has been only one report of an in vivo association of these proteins. Other studies reported an interaction between c-Myc and p107 only when both proteins were overexpressed in transient transfection assays (4, 55).
The ability to induce both tumor formation and apoptosis illustrates the extreme diversity in c-Myc biological activities (reviewed in reference 48). Initial studies demonstrated that c-Myc could induce apoptosis when constitutively expressed in growth factor-deprived cells (3, 17). This suggested a simple model in which unbalanced mitogenic signals induce a cell death program rather than growth. However, other forms of growth inhibition such as isoleucine deprivation or a thymidine block also promote Myc-induced apoptosis (17). Furthermore, some cytokines act as survival factors to block Myc-induced apoptosis even at stages of the cell cycle where they serve no mitogenic function (24). These observations led to a dual-signal model in which c-Myc can induce different downstream pathways depending on a complex integration of intracellular and extracellular cues (16, 24). A multiple-effector model has also been proposed in which c-Myc activates many targets that overlap in their contributions to both mitogenesis and apoptosis (46). Bcl-2 overexpression can block Myc-induced apoptosis in cell culture (7, 18, 60), and the latter two oncoproteins can cooperate in tumor formation in vivo (41). In contrast to the proapoptotic function usually ascribed to c-Myc, c-Myc serves to block apoptosis in a B-cell line (61). These contrasting observations indicate that cell background plays an important part in the activation of growth versus death pathways.
This present study addresses the function of c-Myc through an analysis of missense mutations within the N-terminal transactivation domain. Specific mutations that are associated with lymphomas and/or phosphorylation are shown to uncouple the oncogenic and apoptotic activities of c-Myc. Furthermore, the survival factor insulin-like growth factor 1 (IGF-1) is shown to exhibit allele-specific protection of Myc-induced apoptosis and to suppress phosphorylation at T58. These results suggest that subtle differences in the conformation or modification of the c-Myc transactivation domain can determine the fate of cells that harbor this oncogenic transcription factor.
MATERIALS AND METHODS
Generation of mutant c-myc expression constructs.
Wild-type murine c-myc in a cytomegalovirus promoter-driven vector (42) was mutagenized by site-directed mutagenesis (33). The following antisense primers were used to generate various c-myc mutations: E39D/A44V (5′-CACTGGGC(A/G)CGGGCGGCTGCAG[C/G]TCGCTCTGC-3′), P57S (5′-GGCGGGGTGGAAAGCAGCTCGAA-3′), T58A (5′-GACAGGGGCGGGATGGGAAGCAGC-3′), T58I/S62P (5′-GGCTCGGGG[G/A]CAGGGGCGGG[A/G]TGGGAAGCAG-3′), S62A (5′-GGCTCGGGGCCAGGGGCGGG-3′), S71E (5′-ATAGGATGGTTCGCAGAGCCC-3′), and S71W (5′-ATAGGATGGCCAGCAGAGCCC-3′). S71F was generated fortuitously in the process of making another mutation. All mutant constructs were checked by restriction digests where appropriate and verified by dideoxy sequencing.
Cell lines and retroviral reconstitution of c-myc null fibroblasts.
TGR-1 (c-myc diploid) and HO15.19 (c-myc null) fibroblasts were gifts of J. Sedivy and cultured as described elsewhere (40). Retroviral reconstitution of HO cells was done as described previously (10). Briefly, wt or various mutant c-myc constructs were cloned into an LXSH retroviral vector (43). DNA (10 μg) was transfected by the calcium phosphate method into the ψ2 packaging cell line. Cells were washed with medium 24 h posttransfection and refed. Next day, the supernatant was harvested and filtered through a 0.4-μm-pore-size membrane. Polybrene was added at 8 μg/ml (final concentration) to the viral supernatant, which was used to infect HO cells at 50 to 60% confluence. Cells were incubated for 2 days and then split to low density. Cells were selected in hygromycin-containing medium (150 μg/ml), and two to seven randomly selected clones were picked and expanded.
Transformation assays.
Transformation of primary rat embryo fibroblasts was performed as described previously (9), using 1 μg of the cytomegalovirus promoter-driven c-myc (wt or mutant) expression vector and 1 μg of the activated H-rasG12V. Following transfections, the cells were maintained in 4% fetal bovine calf serum in Dulbecco modified Eagle medium, and foci were counted 12 to 16 days posttransfection.
Proliferation assays and growth curve analyses.
Wild-type or mutant c-myc reconstituted HO cells were plated in 6-cm-diameter plates at low density (∼104/dish). After each 24 h, cells were counted by hemacytometer for 4 or 5 days. Growth curves were plotted by CricketGraph III software, and doubling times were calculated by exponential curve fitting.
Flow cytometry.
Apoptosis was induced in c-myc reconstituted cell lines by serum deprivation (0.1% calf serum). After 24 h, both floating and attached cells were harvested by trypsinization and washed in phosphate-buffered saline–0.5% fetal calf serum. Cells were fixed in 70% ethanol overnight, and then DNA was extracted with phosphate-citrate buffer (0.1 M Na2HPO4, 2 mM citric acid [pH 7.8]). Cells were stained with propidium iodide (10 μg/ml), treated with RNase (50 μg/ml) at 37°C for 1 h, and analyzed by fluorescence-activated cell sorting (FACS) using a FACScan (Becton Dickinson). The relative extent of apoptosis measured by flow cytometry was independently confirmed by both DNA laddering and annexin binding assays (data available upon request).
Cytochrome c release assay.
Subcellular fractionation was performed as described previously (29) except with slight modifications. Cycling or apoptotic cells were resuspended in ice-cold sucrose cell extraction buffer (SCEB; 300 mM sucrose, 10 mM HEPES [pH 7.4], 50 mM KCl, 5 mM EGTA, 5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride). After 50 strokes of Dounce homogenization, the supernatant was collected, and the pellet was reextracted with SCEB plus 0.5% Triton X-100 and centrifuged at 2,000 × g for 10 min. Both the cytosolic supernatant and the mitochondrial pellet (heavy membrane fraction [HMF]) were frozen in liquid N2 and stored at −20°C prior to protein determination and Western blot analysis.
IGF-1 rescue experiment.
Wild-type or mutant c-myc reconstituted cell lines were serum starved in the presence or absence of IGF-1 (200 ng/ml; Sigma). After 18 to 20 h, both floating and attached cells were harvested and analyzed by FACS analysis.
Northern and Western blot analyses.
Total RNA was isolated from log-phase cells by a modification of the guanidium-thiocyanate method (Trizol; Gibco-BRL) and separated on a formaldehyde agarose gel (10 μg/lane). RNAs were transferred onto a nylon membrane (Nytran) by capillary action and hybridized at 65°C with various DNA gene probes labeled by random priming. Blots were washed at high stringency with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 50°C. For the Western blot assay, 3 mg of total cell lysate from cycling cells was immunoprecipitated with monoclonal anti-c-Myc antibody C33 (Santa Cruz Biotechnology) and subjected to polyacrylamide gel electrophoresis (SDS-PAGE). After transfer onto a nitrocellulose membrane (Protran), membranes were probed with polyclonal anti-c-Myc antibody N262 (Santa Cruz Biotechnology). For immunodetection of cytochrome c release, 100 μg of cytosolic supernatant and HMF proteins were separated by SDS-PAGE (15% gel), blotted onto a nitrocellulose membrane, and probed with monoclonal anti-cytochrome c and anti-cytochrome c oxidoreductase antibodies (Molecular Probes). For a loading control, the same blot was reprobed with a monoclonal anti-β-actin antibody (Amersham).
Two-dimensional (2D) phosphopeptide mapping.
c-Myc reconstituted cells were labeled with [32P]orthophosphate for 4 h in phosphate-free medium. Labeled c-Myc protein was immunoprecipitated from cells with anti-Myc (56), separated by SDS-PAGE, transferred to nitrocellulose, and digested from the membrane with 10 μg of thermolysin (Worthington Biochemicals) (38). Digestion was followed by performic acid oxidation (1 h at 0°C) and lyophilization. The digested fragments were separated in the first dimension by electrophoresis using a Hunter thin-layer electrophoresis chamber in pH 1.9 buffer (1.5 kV, 30 min) and then separated in the second dimension by ascending chromatography in the phosphochromatography buffer (38).
RESULTS
The initial goal of this study was to investigate the biological activities associated with c-Myc proteins containing missense mutations frequently found in BL and other Myc-induced cancers. A panel of single-site missense mutations was created in the mouse c-myc gene at positions corresponding to hot spots of mutation in the compiled tumor profile (Table 1). Although the majority of c-myc mutations have been described in human BL, the human and mouse c-Myc proteins are nearly identical and the same mutations are found in human, mouse, and chicken c-Myc proteins. The missense mutants were designed to match the predominant tumor alleles at each position (Table 1), although some other alleles not found in tumors were also analyzed.
Lymphoma-associated c-Myc mutants frequently have reduced oncogenic potential.
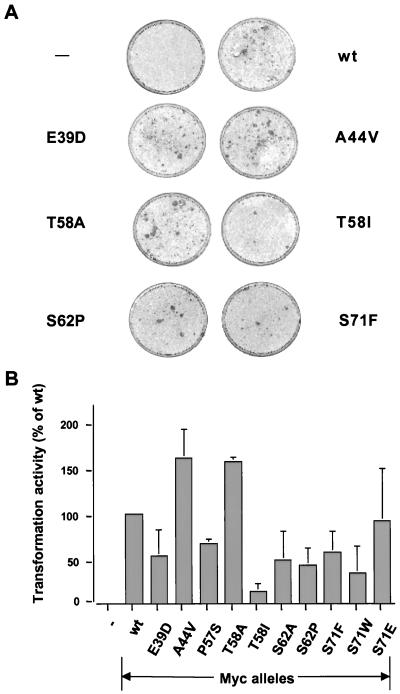
The simplest hypothesis to account for the high frequency of mutations within the c-Myc N terminus was that these mutations cause an increase in oncogenic potential that might accelerate tumor growth. Each c-Myc allele was assayed for oncogenic transformation of early-passage rat embryo cells in cooperation with the H-rasG12V oncogene (Fig. 1) (34). One common mutant allele, MycA44V, was reproducibly more active (160%) than wt Myc in this assay. The uncommon MycT58A allele was also more active (160%) than wt Myc. In contrast, the most common mutation in BL (T58I) had dramatically reduced oncogenic activity (13% of wt Myc activity). Thus, relatively similar alternate mutations at a single position in Myc (T58I versus T58A) can have strikingly different consequences for oncogenic activity. Other frequent lymphoma-associated mutations (E39D, P57S, and S62P) had reproducibly lower oncogenic activity (40 to 80%) than wt Myc. A transformation defect was also observed with the S62A mutation (50% of wt Myc), in agreement with a previous report (49), but this mutation has never been observed in lymphomas. From this analysis, we conclude that the frequent mutations in lymphomas do not potentiate a consistent enhancement of oncogenic potential, at least as assessed by the oncogene cooperation assay.
FIG. 1.
Transformation activity of wt and mutant c-myc genes in the rat embryo fibroblast focus assay. (A) Representative pictures of the rat embryo fibroblast monolayers with H-rasG12V only (−) or H-rasG12V with wt or mutant c-myc. (B) Relative transformation activities of various mutant c-myc alleles as a percentage of wt c-myc activity. Data shown are averages of two to three independent experiments. Differences in transforming activity are statistically significant based on P values of <0.05 using the Student t test. Protein expression was equal for all alleles after transient transfection into rat embryo fibroblasts (data not shown).
Three additional mutant c-Myc alleles were also analyzed for oncogenic potential, S71F (60% of wt activity), S71W (35%), and S71E (90%) (Fig. 1). The S71W mutation has been found in only one AIDS-associated lymphoma (12), whereas the S71F allele has not been found in tumors but was created fortuitously in the course of this study. These alleles were studied because they were mutations in a known c-Myc phosphorylation site (S71), and they proved to have unique biological properties as described below.
Rare lymphoma-associated Myc mutants uncouple apoptosis from oncogenic transformation.
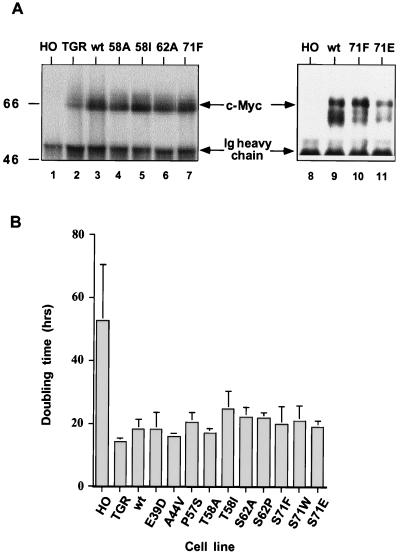
A second hypothesis that could account for the expansion of tumor cells with c-Myc mutations is that the mutations might reduce Myc-induced apoptosis. Since net tumor growth is a balance of cell division and cell death, any mutation that diminished the rate of cell death could be clonally selected. Misregulation of c-Myc expression through chromosomal translocation leads to an autosuppression of the normal, untranslocated c-myc gene in the tumor cell. Since mutations are found exclusively on the translocated alleles, only the mutant protein is produced in these tumors. To approximate this in a controlled experimental setting, we took advantage of a cell line lacking all endogenous Myc expression after knocking out both c-myc genes by homologous recombination (40). Wild-type and mutant c-myc genes were retrovirally transduced into the c-myc null cells, and multiple randomly selected clones for each mutation were studied for a range of parameters, including protein expression, growth rate, apoptosis, and endogenous gene expression. The levels of transduced wt and mutant c-Myc proteins (Fig. 2A) and RNA (not shown) were virtually identical between cell lines, and this level was two- to fourfold higher than the level of endogenous c-Myc protein in the parental diploid Rat1 fibroblasts (TGR). Two to seven clones for each allele were analyzed with similar results. The finding of equivalent levels of wt Myc and of MycT58A and other mutant proteins in these stably reconstituted cell lines conflicts with recent reports from ectopic expression that mutant Myc proteins have a prolonged half-life and accumulate to higher levels than wt c-Myc (50, 52). All of the mutant c-Myc proteins restored the growth rate of the null cells to a level similar to that of cells reconstituted with wt c-Myc and of the parental c-myc diploid cell line (Fig. 2B). Some variation in growth rate was observed, but the doubling time for all of the reconstituted alleles (∼20 to 24 h) was very close to that of the parental line (18 h), compared to 50 to 60 h for the slow-growing c-myc null cells. Interestingly, cells reconstituted with the T58I allele had a reproducible 20 to 30% longer doubling time, consistent with the weaker transforming activity in the H-rasG12V cooperation assay. However, this minor defect in proliferative activity was much less severe than the defect in the H-ras cooperation assay.
FIG. 2.
Reconstitution of c-Myc protein expression and cell proliferation in c-myc null cells. (A) Wild-type or mutant c-myc reconstituted cell lines have similar levels of c-Myc protein, but the level is slightly higher than that in the c-myc diploid TGR cells. c-Myc proteins were immunoprecipitated from equal amounts of cycling cell lysate and detected by Western blotting. Ig, immunoglobulin. Sizes are given in kilodaltons. (B) Average doubling time of fibroblast cell lines expressing either no c-Myc (HO), endogenous diploid levels of c-Myc (TGR), or wt or mutant c-Myc in reconstituted cell lines. Bars represent doubling times ± standard error for three to five experiments, and two to four clones were assayed for each reconstituted allele. The difference in doubling time between wt Myc and T58I or S62A reconstituted lines is statistically significant based on P values of <0.02 using the Student t test.
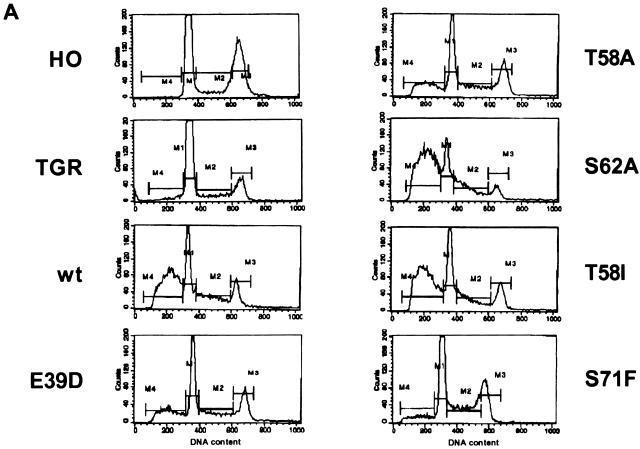
The apoptotic activity of each c-Myc mutant was assessed by serum deprivation of the reconstituted cell lines. Apoptosis was measured as the accumulation of cells with a sub-G1 DNA content by flow cytometry (Fig. 3A). In the parental diploid line, there is very little apoptosis because the endogenous c-Myc level is low and suppressed by growth factor withdrawal. The c-myc null cells have virtually no detectable apoptosis, even with prolonged incubation in low serum. In contrast to these cell lines with low or no c-Myc, cell lines in which wt c-Myc is expressed from the retroviral long terminal repeat exhibited high levels of apoptosis within 24 h of serum starvation. Apoptosis was independently confirmed by the appearance of DNA ladders and annexin staining (data not shown). The extent of apoptosis induced by each mutant c-Myc protein was compared to that induced by wt c-Myc in the same experiment (Fig. 3B). For several of the lymphoma-associated mutants, the apoptotic activity directly paralleled the transforming activity as measured in the H-rasG12V cooperation assay. The A44V mutant had reproducibly 25% more apoptotic cells than wt Myc, consistent with its increased oncogenic activity. Similarly, the E39D, S62P, S71W, and S71E mutants had less than wt apoptotic activities that paralleled their reduced oncogenic activities. We stress that the differential apoptosis described here and below was not the result of the kinetics of the process, since the relative extent of apoptosis was the same at shorter (18 h) and longer (48 h) times (data not shown).
FIG. 3.
Apoptotic versus transforming activity with differing c-Myc mutations. (A) Representative flow cytometry profiles of HO, TGR, and various reconstituted cell lines following serum starvation (0.1%) for 24 h. DNA content was measured by fluorescence with propidium iodide staining. M1, M2, M3, and M4 represent gating for G1, S, G2, and sub-G1 populations, respectively. (B) Histogram of average percent apoptosis of the various cell lines in low serum as calculated by FACS gating for the sub-G1 population (M4). Data represent averages of three to five independent determinations. The oncogenic activity of each mutant c-Myc protein from Fig. 1B is shown for comparison.
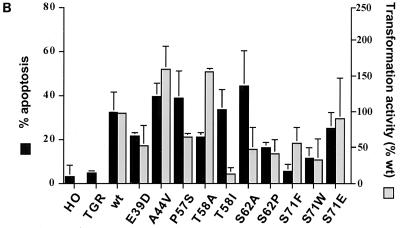
Several of the c-Myc mutants had a notable uncoupling of apoptosis from oncogenic activity, although the pattern was not as one would predict if tumor formation was linked to reduced apoptosis. Only one mutant, MycT58A, had a reduced rate of apoptosis that was concomitant with elevated oncogenic activity (Fig. 3B). However, this mutation is only rarely observed in lymphomas, although it is found in two chicken v-Myc proteins (reviewed in reference 35). While T58 is the most common site of mutation in c-Myc, the predominant allele in lymphomas is T58I rather than T58A (Table 1). Surprisingly, cells reconstituted with the T58I allele had an apoptotic rate that was similar to the wt rate (Fig. 3B), even though the oncogenic activity was severely reduced in the H-rasG12V cooperation assay (Fig. 1B) and the doubling time of the MycT58I reconstituted lines was consistently longer than for cells expressing wt Myc (Fig. 2B). Thus, subtle differences in the specific missense mutation at T58 (A versus I) produce drastic differences in biological activity. The origin of this difference will be discussed below. Similar to the T58I mutant, the MycS62A mutant induced reproducibly higher levels of apoptosis even though it had reduced oncogenic activity. As with the different T58 mutations, the alternate S62P allele was distinct from S62A and had parallel apoptotic and oncogenic activities. The frequent lymphoma-associated mutation P57S was similar to S62A, with high levels of apoptosis despite a reduced oncogenic activity (Fig. 3B). Thus, the apoptotic activity of c-Myc can be sustained in mutant proteins, even though the oncogenic and proliferative activity is curtailed. This finding was unexpected if a major contribution of c-Myc mutation is to reduce apoptosis.
The most interesting c-Myc mutant studied was S71F. Multiple reconstituted clonal lines expressing the MycS71F protein had dramatically lower levels of apoptosis compared to wt c-Myc and other mutants, as assayed by both sub-G1 DNA content with flow cytometry (Fig. 3B) and DNA laddering (not shown). A low level of apoptosis was still detected, but apoptotic activity was only 15% of the wt level. This reduced apoptosis contrasts with the much smaller reduction in oncogenic activity of the MycS71F mutant (Fig. 1B), as well as its ability to restore the wt doubling time of reconstituted c-myc null cells. Interestingly, as discussed in the introduction, no S71F mutant alleles have ever been found in tumor cells. The only mutation at this position is one example of S71W (12). To determine if a lymphoma-associated mutation had the same phenotype as MycS71F, the c-myc null cells were reconstituted with MycS71W. As with MycS71F, serum starvation of MycS71W-expressing cells induced a low level of apoptosis (Fig. 3B), although the level paralleled the reduced oncogenic activity when assayed for focus formation in rat embryo fibroblasts. MycS71F and MycS71W proteins were produced at levels equivalent to those of other mutants and to the wt protein in reconstituted cell lines.
The two MycS71 alleles with very low apoptotic potential contained substitutions of hydrophobic amino acids in place of a phospho-acceptor site. It was interesting to consider if another mutation at this site would have the same activity, especially a mutation (S71E) that could potentially mimic phosphorylation. In contrast to the hydrophobic missense mutations above, cells reconstituted with a MycS71E mutant protein had wt rates of proliferation and apoptosis (Fig. 3B), as well as wt oncogenic activity in cooperation with H-rasG12V (Fig. 1B). This high level of apoptosis was induced even though the level of MycS71E protein expression was lower than the wt level (Fig. 2A). Thus, the presence of a negative charge on S71, through either phosphorylation or the amino acid side group, may be critical for apoptotic activity but less important for oncogenic activity. It is also apparent that there is no selection for reduced apoptosis by mutation of S71 to F or W in tumor outgrowth.
Phosphorylation correlates with c-Myc biological activity.
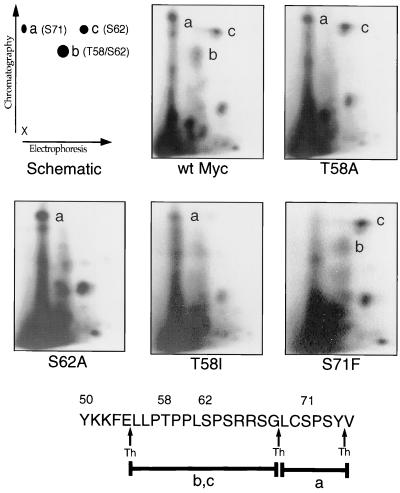
The lymphoma-associated mutations in c-Myc are tightly clustered around the sites of growth factor-induced phosphorylation, suggesting that altered phosphorylation could be important for the different biological activities of specific c-Myc alleles. We were particularly interested in understanding the opposite effects on c-Myc biological activity caused by the T58I versus T58A mutations. Previous studies have shown that T58 and S62 phosphorylation are growth factor regulated and that T58 phosphorylation is dependent on prior phosphorylation of S62 (25, 37). In growth factor-stimulated cells, a large fraction of the c-Myc protein is phosphorylated at both T58 and S62, whereas S71 is phosphorylated both in serum-starved cells and after growth factor stimulation (37).
To determine if mutations affected c-Myc phosphorylation, each of the proteins used in this study was phosphopeptide mapped from log-phase Myc-reconstituted cells (Fig. 4). Compared to wt Myc, the MycS62A mutant lacked both S62 and T58 phosphorylation, but S71 phosphorylation was unaffected. In contrast to the interdependence of the T58 and S62 phosphorylation sites, the MycS71F mutant lacked only phosphorylation of the mutated S71 site; T58/S62 phosphorylation appeared unaltered. The E39D and A44V mutants had wt phosphopeptide maps (data not shown), indicating that the effects on phosphorylation were localized to mutations that affect the proline domain.
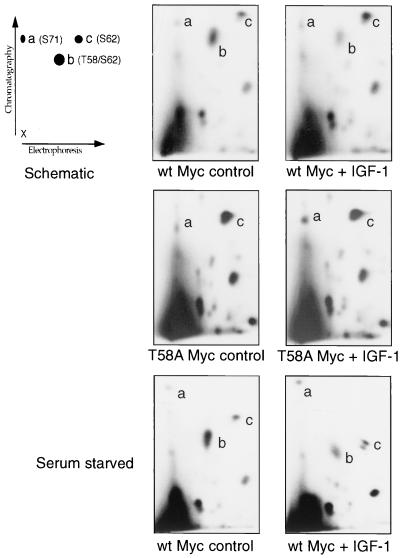
FIG. 4.
Thermolytic phosphopeptide mapping analysis of Myc null cells stably expressing wt c-Myc or c-Myc proteins with phosphorylation point mutations, as indicated. Cells were labeled for 4 h with [32P]orthophosphate. Myc proteins were immunoprecipitated and processed for 2D phosphopeptide mapping as described in Materials and Methods. The schematic representation of the 2D thermolytic peptide map (top left) indicates major sites of phosphorylation at S71 (peptide a), T58/S62 (peptide b), and S62 (peptide c) following thermolytic digestion. The diagram at the bottom represents predicted thermolytic peptides a, b, and c. Arrows indicate thermolysin (Th) digestion sites.
Phosphopeptide mapping of the T58 mutants provided a critical insight into their strikingly different biological activities. The phosphopeptide map of the MycT58A protein showed normal phosphorylation of S62 and a wt level of S71 phosphorylation. In contrast, the MycT58I protein lacked not only T58 phosphorylation but also virtually all phosphorylation of S62 (Fig. 4). Phosphorylation of S71 was unaffected in the T58I mutant. Presumably, the increased hydrophobicity of the isoleucine compared to alanine at amino acid 58 either blocks the recognition of S62 by the c-Myc kinase or increases recognition by an undefined phosphatase. Since the MycS62A mutant has reduced transforming activity in several studies, blocking S62 phosphorylation indirectly by a nearby T58I mutation may have the same effect or induce an even more unfavorable conformation for oncogenic transformation. The most likely explanation of these observations is that the conformation of the c-Myc N-terminal transactivation domain (and hence biological activity) is highly sensitive to phosphorylation or specific missense mutations in the conserved MbI region.
Regulation of known target genes is indistinguishable between wt and mutant c-Myc proteins.
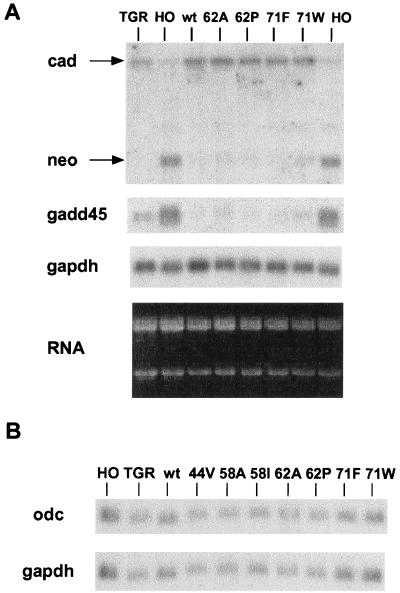
The c-Myc protein can both transactivate and repress several cellular target promoters (13, 14). The distinct biological activities of the mutant c-Myc proteins described above are presumably due to either quantitative or qualitative differences in target gene regulation. To assess the transcription factor activity of each c-Myc allele, the reconstituted cell lines were studied for the expression of several target genes that are highly dependent on basal levels of c-Myc. The most unique c-Myc target gene is cad, which is the only proposed transactivation target that is dependent on endogenous c-Myc expression in log-phase fibroblasts (10). The level of cad mRNA is three- to fourfold lower in c-myc null fibroblasts than in the diploid c-myc parental line or null cells reconstituted with wt c-Myc under a viral promoter. Reconstitution of cells with all of the c-Myc mutant proteins reactivated the expression of the endogenous cad gene to the same extent as with wt c-Myc (Fig. 5A). The odc (ornithine decarboxylase) gene has been proposed as a critical target gene for c-Myc-induced apoptosis in myeloid cells (45), but there were no differences in odc mRNA levels between the diploid parental cells and c-myc null cells, and odc mRNA levels were unaffected in the c-Myc-reconstituted lines, regardless of the apoptotic potential of each c-Myc mutant (Fig. 5B). No differences in cdc25A levels were observed in the presence or absence of c-Myc as previously reported (10) (data not shown).
FIG. 5.
c-Myc proteins induce equivalent activation and repression of target genes. (A) Northern blot of the parental and reconstituted cell line RNAs sequentially probed for cad, neo, gadd45, and gapdh RNAs. The ethidium bromide-stained gel is shown in the lower panel. (B) Northern blot of odc and gapdh expression as above.
The repression of cellular promoters has been linked to functionally critical domains of c-Myc, although the mechanism of repression is unknown. Two well-characterized targets of c-Myc repression were studied in the reconstituted cell lines. In the c-myc null cells, the c-myc promoter drives the production of neo mRNA as the result of gene targeting, and the exogenous c-Myc produced by retroviral transduction represses the endogenous c-myc promoter and hence neo RNA. All of the c-Myc mutants repressed the c-myc promoter to the same extent as wt Myc (Fig. 5A). Repression of the gadd45 gene is also dependent on basal levels of c-Myc since the c-myc null cells have elevated levels compared to the parental diploid line (10). As with the c-myc promoter, gadd45 is repressed equally by both wt and mutant c-Myc proteins (Fig. 5A). Thus, all of the c-Myc proteins with lymphoma-associated mutations are indistinguishable in both transactivation and repression of the cellular target genes that were examined. We presume that differences in c-Myc-dependent gene expression exist between the reconstituted cell lines, but this will require a more systematic analysis.
Allele-specific cytochrome c release correlates with c-Myc apoptotic activity.
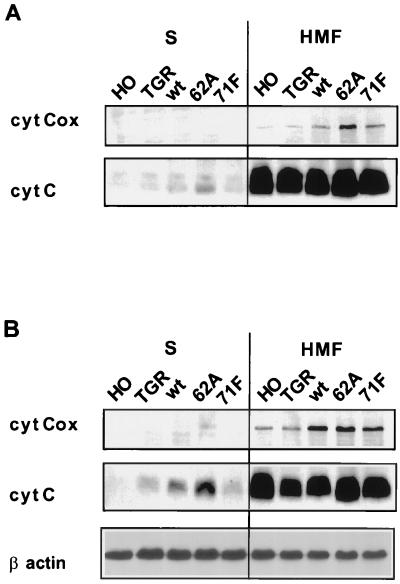
A recent study indicates that c-Myc overexpression in the context of an apoptotic signal such as growth factor deprivation promotes the release of cytochrome c from mitochondria (29, 31), which complexes with Apaf-1 to initiate a caspase cascade and cell death (reviewed in reference 22). The reconstituted cell lines with differing apoptotic potential were assayed for the extent of cytochrome c release into the cytoplasm before and after serum starvation. The mitochondria fractionate to the heavy membrane pellet after gentle cell lysis, and their integrity can be followed with the mitochondrial enzyme cytochrome c oxidase. The cytochrome c remained within the mitochondria (pellet fraction) in all cell lines cultured in 10% serum, regardless of the expressed c-Myc protein or in the complete absence of c-Myc in the null cell line (Fig. 6A). In contrast, cytochrome c release was readily detectable after serum starvation in all cell lines expressing c-Myc (Fig. 6B). The amount of cytochrome c released was highest in cell lines with constitutive wt Myc or MycS62A, which have the highest rates of apoptosis. Cytochrome c release was substantially lower in the parental diploid c-myc cell line with endogenous c-Myc levels that are suppressed upon serum starvation. Notably, the cell line reconstituted with MycS71F had the same low level of cytochrome c release as the diploid c-myc parental line, both of which have very low levels of apoptosis (Fig. 6B). It was also noteworthy that the c-myc null cells, which have no detectable apoptosis when serum starved, have no detectable cytochrome c release (Fig. 6B). The cytochrome c oxidase remained in the pellet fraction in all lysates, verifying that the mitochondria remained intact.
FIG. 6.
c-Myc-induced apoptotic activity correlates with cytochrome c release from mitochondria. (A) Cycling c-myc null cells (HO), c-myc diploid cells (TGR), and null cells reconstituted with wt c-Myc, MycS62A, and MycS71F were lysed in an isotonic sucrose buffer by Dounce homogenization and fractionated by centrifugation. Equivalent amounts of protein from the resultant supernatant and pellet (HMF) were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with anti-cytochrome c (cyt C) or anti-cytochrome c oxidoreductase (cyt Cox). The latter protein serves as a control for mitochondrial integrity. (B) Cytochrome c release from the above cells in 0.1% serum following the same lysis protocol. After transfer, the blot was probed with antibodies for cytochrome c and cytochrome c oxidoreductase. To verify equal protein loading, the same blot was reprobed with anti-β-actin (bottom panel).
c-Myc mutants are differentially sensitive to apoptotic survival factors.
Apoptosis induced by c-Myc is blocked by serum growth factors, in particular by IGF-1 (24). Since signaling pathways culminate in the phosphorylation of numerous substrates, the uncoupling of apoptosis from oncogenic transformation by mutations around the primary sites of c-Myc phosphorylation suggested that the c-Myc protein itself could be a downstream target of survival signals. The major sites of serum-induced phosphorylation are T58 and S62, and T58 phosphorylation is dependent on prior phosphorylation of S62. Therefore, we focused the analysis on the MycS62A protein since this mutant induced the same high level of apoptosis as wt Myc.
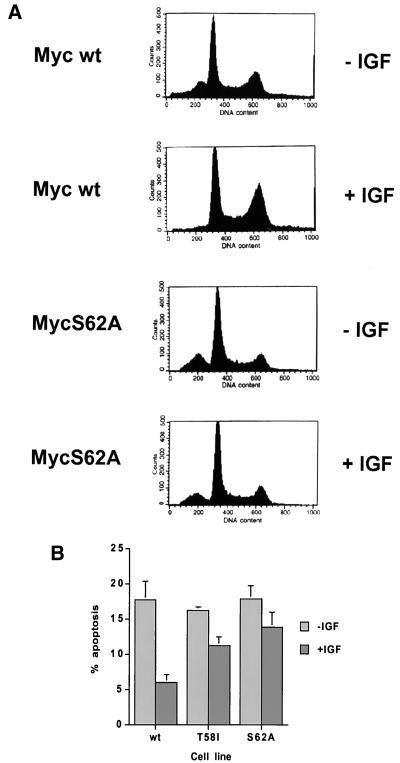
The wt and MycS62A reconstituted cell lines were either starved for serum-derived survival factors or starved in the presence of the survival factor IGF-1. Apoptosis was measured by the appearance of cells with a sub-G1 DNA content after 24 h. As described previously, IGF-1 protects the cells expressing wt Myc from apoptosis, reducing the number of apoptotic cells to 35% of level in the untreated control (Fig. 7). IGF-1 treatment simultaneously promoted an increase in the fraction of cells in the G2/M phase of the cell cycle. In contrast, IGF-1 provided very little protection from apoptosis for the cells expressing the MycS62A protein, in which the rate of apoptosis remained at 80% of that in the untreated cells. Comparison of serum-starved cells to starved cells in the presence of IGF-1 showed that the IGF-1-resistant apoptosis of the MycS62A reconstituted cells was accompanied by no change in the fraction of cells in G2/M phase. Thus, the S62 phosphorylation site is required for the majority of the IGF-1-mediated survival. Analysis of the MycT58I reconstituted cells, which also lack S62 phosphorylation and have high rates of apoptosis, gave generally similar results, although there was slightly more protection by IGF-1 compared to S62A. It should be noted that the S62A mutation does not completely block all IGF-1-mediated survival since there is still a 20% reduction in apoptosis with IGF-1 treatment of the MycS62A cells. IGF-1 is expected to induce a pleiotropic cellular response that will affect other pathways that influence the apoptotic cascade besides c-Myc.
FIG. 7.
IGF-1 rescue of c-Myc-induced apoptosis is mutation specific. (A) Flow cytometry of wt Myc and MycS62A reconstituted cells cultured for 18 h in 0.1% serum in the presence or absence of IGF-1 (200 ng/ml). Both floating and attached cells were harvested, fixed, and stained with propidium iodide before determination of the DNA content by flow cytometry. (B) Apoptotic activity of different c-Myc mutants after gating for the sub-G1 population in serum-starved cells with or without IGF-1. Data represent the averages of three independent experiments ± standard error.
IGF-1 induces altered c-Myc phosphorylation.
The altered response of the MycS62A cells to IGF-1-mediated survival suggested that the c-Myc protein might be a direct target of the IGF-1 receptor signal pathway. The wt Myc protein from reconstituted cells was phosphopeptide mapped from growing or serum-starved cells and from parallel cultures treated for 20 h with IGF-1 (Fig. 8). The phosphopeptide maps show that IGF-1 induced a substantial decrease in the abundance of the doubly phosphorylated T58/S62 peptide and an approximately equivalent increase in the signal for the same peptide phosphorylated on S62 alone. Decreased T58 phosphorylation was apparent in 10% serum (Fig. 8, top panels) and serum-starved cells (bottom panels). Other phosphopeptide spots including S71 retained the same signal intensity, both in absolute and in relative terms. This shift in c-Myc phosphorylation could be due to the induction of S62 phosphorylation by IGF-1, to a reduction in T58 phosphorylation, or to altered phosphatase activity affecting either site. To address these possibilities, the phosphorylation of the MycT58A protein was studied in response to IGF-1 treatment. Since this mutation prevents phosphorylation of T58, only a strong signal from the singly phosphorylated peptide containing S62 is present. IGF-1 treatment of cells harboring this mutant protein showed no induction of S62 phosphorylation (Fig. 8, middle panels). This finding strongly suggests that IGF-1 treatment either suppresses the activity of the kinase that phosphorylates T58 or induces a phosphatase that preferentially dephosphorylates T58 without affecting S62 phosphorylation.
FIG. 8.
IGF-1 suppresses the phosphorylation of c-Myc T58, as determined by thermolytic phosphopeptide mapping analysis of Myc null cells stably expressing either wt c-Myc or MycT58A. Cells were untreated or treated with IGF-1 (200 ng/ml) for 20 h prior to labeling for 4 h with [32P]orthophosphate. The top and middle panels were derived from log-phase cells in 10% serum, whereas the bottom panels were derived from cells starved in 0.1% serum for 20 h before labeling. Myc proteins were immunoprecipitated and processed for 2D phosphopeptide mapping as described in Materials and Methods. The schematic representation of the 2D thermolytic peptide map (top left) indicates major sites of phosphorylation at S71 (peptide a), T58/S62 (peptide b), and S62 (peptide c) following thermolytic digestion. Peptide c resolved into two closely spaced spots in the serum-starved IGF-1 treated cells, but this was not found in other experiments.
DISCUSSION
Complex signals govern the cellular pathways that culminate in the decision to progress through the cell cycle, exit into quiescence, or trigger programmed cell death. The c-Myc protein plays a role in all three of these pathways since it is essential for proper cell cycle progression, prevents exit into quiescence when constitutively expressed, and triggers apoptosis in the absence of appropriate survival signals. We demonstrate here that subtle modifications of the c-Myc transactivation domain can dramatically alter its activity in individual pathways and that survival factors signal directly to the c-Myc protein as one mediator of their antiapoptotic activity.
The ability to separate the oncogenic function of c-Myc from its proapoptotic activity has a number of implications for understanding the role of c-Myc in the outgrowth of tumor cells. First, for the majority of c-Myc mutations, there is good correlation between oncogenic and apoptotic activities. These include large deletions examined earlier (17) and several of the missense mutations analyzed in this study, such as E39D, A44V, S62P, and S71W. Both oncogenic and apoptotic activities are either enhanced (A44V) or suppressed (for example, E39D) in parallel. This finding is supported by a comparison of c-Myc and v-Myc, in which the latter has both increased transforming and apoptotic activity (reviewed in reference 35). On the other hand, only a single missense mutation (T58A) was found to enhance oncogenic activity while at the same time reducing the extent of apoptosis compared to wt Myc. This was surprising given that the most frequent mutation in BL is T58I, which has the opposite biological effects of reduced transforming activity and wt levels of apoptosis. Intuitively, one might have expected mutations such as T58A to be strongly selected within tumor cell populations, but only if Myc-induced apoptosis is a rate-limiting step in tumor formation. A trivial, but interesting, resolution of this apparent paradox is that the c-Myc missense mutants may have distinctly different biological activities in B cells than in the fibroblasts studied here. Resolving this will require the development of a suitable B-cell model system in which both oncogenic and apoptotic potential can be analyzed. It is interesting that c-Myc acts to prevent rather than induce apoptosis in the WEHI231 lymphoid cell line (61). Yet another possibility is that the apoptotic pathway is suppressed by other genetic lesions such as p53 or p19ARF mutation in the majority of Myc-induced tumors (15, 27, 51).
Since both the DNA binding domain and the N-terminal transactivation domain are required for apoptosis, it is assumed that c-Myc activates or represses cellular genes that potentiate the cell death pathway. However, different genes have been proposed to mediate Myc-induced apoptosis in different cell backgrounds. In myeloid cells, the odc gene is misregulated by ectopic c-Myc expression and the ODC protein appears to be necessary and at least partially sufficient to mediate Myc-induced cell death (45, 46). In fibroblasts, it has been proposed that cdc25A and ldh-a are directly regulated by c-Myc and that both can participate in apoptotic signals (21, 53). However, ectopic expression of c-Myc does not hyperactivate any of these genes in fibroblasts but rather leads to sustained expression only with growth factor deprivation or under anchorage-independent culture conditions (21, 54). A loss-of-function analysis suggests that c-Myc may play only a minor role in the regulation of all three proposed proapoptotic targets, with no effect in log-phase cells (10). An important consideration for studies of candidate target genes is the observation that cycloheximide itself can induce apoptosis in cells that overexpress c-Myc (17) and cycloheximide does not block Myc-dependent apoptosis with p53 (59), indicating that de novo protein synthesis (and by inference gene misregulation) is not required subsequent to an apoptotic signal. The most straightforward interpretation of this observation is that c-Myc overexpression establishes a state of sensitization to apoptotic triggers, which include growth-inhibitory conditions besides serum starvation, such as disruption of protein synthesis and nucleotide or amino acid metabolism. This predicts that the target genes mediating the induction of apoptosis should be constitutively misregulated by the relatively modest (two- to fourfold) overexpression of c-Myc that can potentiate an apoptotic response. This level of c-Myc overexpression can induce a slight constitutive elevation in one Myc target gene (cad) in fibroblasts but no elevation in the expression of odc, ldh-a, or cdc25A (10; A. Bush, unpublished observations).
In addition to gene activation, c-Myc also induces gene repression such as autosuppression of the c-myc promoter itself and the suppression of gadd45 among others (39, 47). The mechanism of Myc-induced gene repression remains unresolved, but it does not seem to involve direct Myc binding to responsive promoters (reviewed in reference 11). Nevertheless, it is noteworthy that rough mapping of the c-Myc domains required for apoptosis shows a concordance with gene repression rather than transactivation (17). Furthermore, studies of a naturally occurring c-Myc protein form called MycS supports a link between apoptosis and gene repression. MycS lacks transactivation activity yet retains the ability to promote cell proliferation, apoptosis, and transcriptional repression (62). The fact that the reconstitution of c-myc null cells with constitutive c-Myc expression leads to a suppression of both the c-myc promoter and gadd45 is direct evidence for the stable reprogramming of gene expression in cells that accompanies an increased apoptotic potential. On the other hand, both the c-myc promoter and gadd45 are suppressed by all of the c-myc alleles assayed in this study, regardless of their high or low rates of apoptosis. Thus, the suppressor activity of c-Myc on known targets can be largely uncoupled from apoptosis with the S71F mutation and from oncogenic transformation with the T58I mutation. Since the c-Myc transactivation domain recruits cofactors to the chromosomal sites recognized by the DNA binding domain, subtle conformational changes due to missense mutation or posttranslational modification may induce qualitative or quantitative differences in cofactor binding. It is interesting that the MycS protein lacks the entire N-terminal domain that is mutated in lymphomas (56). A simple resolution of this apparent paradox is that the primary purpose of the N terminus is to modulate c-Myc activity and that the complete absence of this domain sustains activity.
A crucial outcome of the Myc-induced sensitization to apoptotic triggers is the enhanced release of cytochrome c from the mitochondria (29, 31), which is sufficient to initiate caspase activation and programmed cell death (22). The observation that c-myc null cells have no detectable cytochrome c release provides further support for a link between the level of c-Myc expression and mitochondrial integrity. Furthermore, these data suggest that even a low level of endogenous c-Myc expression may promote some cytochrome c release, which could account for the low but measurable level of apoptosis in the parental diploid fibroblasts compared to the c-myc null cells. The most striking finding was the reduced cytochrome c release with the MycS71F mutation which correlates with apoptotic potential. We presume that the MycS71F protein promotes qualitative or quantitative changes in target gene expression compared to the wt protein or other mutants.
The allele-specific protection of cells reconstituted with mutant c-Myc proteins provided the first hint that the c-Myc transactivation domain might be a direct target of antiapoptotic survival signals. Mapping of the in vivo phosphorylation sites demonstrated that IGF-1 selectively suppressed the phosphorylation of T58 without affecting the extent of phosphorylation at nearby sites. Coupling this observation with the reduced apoptosis of the MycT58A protein provides compelling evidence that the suppression of T58 phosphorylation is an important mediator of antiapoptotic survival factors. Interestingly, the MycT58I protein lacks both T58 and S62 phosphorylation through the apparent interference with phosphorylation by the isoleucine 58 side chain. It was previously shown that T58 phosphorylation is dependent on prior phosphorylation of S62 (37). Since both MycT58I and MycS62A are apoptotic and relatively resistant to IGF-1 protection, the sustained phosphorylation of S62 may be important to suppress apoptotic activity. However, it is important to stress that any mutation in the MbI domain of c-Myc may induce conformational changes that extend beyond the loss of phosphorylation.
The differential biological activities associated with T58, S62, and S71 alleles adds a greater impetus to define the kinase(s) responsible for phosphorylation of these adjacent sites. Predominantly in vitro studies have suggested several candidates for T58 and S62, such as MAPK/ERK, GSK-3, JNK, and cdc2 (2, 25, 37, 44), but the actual kinases that are responsible for the in vivo phosphorylation of the c-Myc proline-rich domain remain poorly defined (36). In addition to the importance of S71 for starvation-induced apoptosis, S71 has also recently been implicated in UV-induced apoptosis (44). Of particular relevance to the present study is the possible role of GSK-3 in the phosphorylation of T58, which has previously been demonstrated to function at least in vitro (25, 37). GSK-3 activity is suppressed through phosphorylation by Akt/protein kinase B, which is in turn activated by the phosphatidylinositol 3-kinase pathway (reviewed in reference 19). Previous studies have implicated this pathway in the survival signals that suppress c-Myc-induced apoptosis (30, 32), and the data presented here suggest that at least one downstream effector of the survival signal may be the suppression of GSK-3-dependent c-Myc phosphorylation. Survival signals undoubtedly act through multiple downstream effectors, but the identification of the c-Myc protein itself in this pathway provides a novel and functionally important intersection of proapoptotic and antiapoptotic signals.
ACKNOWLEDGMENTS
We thank Kerri Dugan and Andy Beavis for help with retroviral transduction and flow cytometry. We thank Heather Van Buskirk for critical reading of the manuscript.
This work was supported by research grants from the National Institutes of Health to M.D.C. and S.R.H. This work was also supported by a predoctoral fellowship from the New Jersey Commission on Cancer Research to D.W.C.
REFERENCES
- 1.Albert T, Urlbauer B, Kohlhuber F, Hammersen B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt's lymphoma cell lines. Oncogene. 1994;9:759–763. [PubMed] [Google Scholar]
- 2.Alverez E, Northwood I, Gonzalez F, Latour D, Seth A, Abate C, Curran T, Davis R. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J Biol Chem. 1991;266:15277–15285. [PubMed] [Google Scholar]
- 3.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 4.Beijersbergen R L, Hijmans E M, Zhu L, Bernards R. Interaction of c-Myc with the pRb-related protein p107 results in inhibition of c-Myc-mediated transactivation. EMBO J. 1994;13:4080–4086. doi: 10.1002/j.1460-2075.1994.tb06725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia K, Spangler G, Gaidano G, Hamdy N, Dalla-Favera R, Magrath I. Mutations in the coding region of c-myc occur frequently in acquired immunodeficiency syndrome-associated lymphomas. Blood. 1994;84:883–888. [PubMed] [Google Scholar]
- 7.Bissonnette R P, Echeverri F, Mahboubi A, Green D R. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1993;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 8.Brennscheidt U, Eick D, Kunzmann R, Martens U, Kiehntopf M, Mertelsmann R, Herrmann F. Burkitt-like mutations in the c-myc gene locus in prolymphocytic leukemia. Leukemia. 1994;8:897–902. [PubMed] [Google Scholar]
- 9.Brough D E, Hofmann T J, Ellwood K B, Townley R A, Cole M D. An essential domain of the c-Myc protein interacts with a nuclear factor that is also required for E1A-mediated transformation. Mol Cell Biol. 1995;15:1536–1544. doi: 10.1128/mcb.15.3.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bush A, Mateyak M, Dugan K, Obaya A, Adiachi S, Sedivy J, Cole M D. c-myc null cells misregulate cad and gadd45 but not other proposed c-myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claassen G, Hann S. Myc-mediated transformation: the repression connection. Oncogene. 1999;18:2925–2933. doi: 10.1038/sj.onc.1202747. [DOI] [PubMed] [Google Scholar]
- 12.Clark H M, Yano T, Otsuki T, Jaffe E S, Shibata D, Raffeld M. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 1994;54:3383–3386. [PubMed] [Google Scholar]
- 13.Cole M D, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 14.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evan G, Littlewood T D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 17.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 18.Fanidi A, Harrington E A, Evan G I. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature. 1992;359:554–556. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- 19.Franke T F, Kaplan D R, Cantley L C. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 20.Frykberg L, Graf T, Vennstrom B. The transforming activity of the chicken c-myc gene can be potentiated by mutations. Oncogene. 1987;1:415–421. [PubMed] [Google Scholar]
- 21.Galaktionov K, Chen X, Beach D. CDC25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 22.Green D, Reed J. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Bhatia K, Magrath I T, Dang C V, Dalla-Favera R. Binding and suppression of the Myc transcriptional activation domain by p107. Science. 1994;264:251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- 24.Harrington E A, Bennett M R, Fanidi A, Evan G I. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriksson M, Bakardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- 26.Hoang A T, Lutterbach B, Lewis B C, Yano T, Chou T-Y, Barrett J F, Raffeld M, Hann S R, Dang C V. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol Cell Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs J J, Scheijen B, Voncken J W, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston J M, Yu M T, Carroll W L. c-myc hypermutation is ongoing in endemic, but not all Burkitt's lymphoma. Blood. 1991;78:2419–2425. [PubMed] [Google Scholar]
- 29.Juin P, Hueber A O, Littlewood T, Evan G I. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 1999;13:1367–1381. doi: 10.1101/gad.13.11.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy S G, Kandel E S, Cross T K, Hay N. Akt/protein kinase B inhibits cell death by preventing the release of cytochrome c from mitochondria. Mol Cell Biol. 1999;19:5800–5810. doi: 10.1128/mcb.19.8.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy S G, Wagner A J, Conzen S D, Jordan J, Bellacosa A, Tsichlis P N, Hay N. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 33.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 34.Land H, Parada L F, Weinberg R A. Tumorigenic conversion of primary embryo fibroblasts requires at least 2 cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 35.Lee C, Reddy E. The v-myc oncogene. Oncogene. 1999;18:2997–3003. doi: 10.1038/sj.onc.1202786. [DOI] [PubMed] [Google Scholar]
- 36.Lutterbach B, Hann S R. c-Myc transactivation domain-associated kinases: questionable role for map kinases in c-Myc phosphorylation. J Cell Biochem. 1999;72:483–491. doi: 10.1002/(sici)1097-4644(19990315)72:4<483::aid-jcb4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 37.Lutterbach B, Hann S R. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14:5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutterbach B, Hann S R. Overexpression of c-Myc and cell immortalization alters c-Myc phosphorylation. Oncogene. 1997;14:967–975. doi: 10.1038/sj.onc.1200920. [DOI] [PubMed] [Google Scholar]
- 39.Marhin W W, Chen S, Facchini L M, Fornace A J J, Penn L Z. Myc represses the growth arrest gene gadd45. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- 40.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Phenotypes of Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 41.McDonnell T J, Korsmeyer S J. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14;18) Nature. 1991;349:254–256. doi: 10.1038/349254a0. [DOI] [PubMed] [Google Scholar]
- 42.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 43.Miller A D, Miller D, Garcia J V, Lynch C. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 44.Noguchi K, Kitanaka C, Yamana H, Kokubu A, Mochizuki T, Kuchino Y. Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. J Biol Chem. 1999;274:32580–32587. doi: 10.1074/jbc.274.46.32580. [DOI] [PubMed] [Google Scholar]
- 45.Packham G, Cleveland J L. Ornithine decarboxylase is a mediator of c-Myc-induced apoptosis. Mol Cell Biol. 1994;14:5741–5747. doi: 10.1128/mcb.14.9.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Packham G, Porter C W, Cleveland J L. c-Myc induces apoptosis and cell cycle progression by separable, yet overlapping, pathways. Oncogene. 1996;13:461–469. [PubMed] [Google Scholar]
- 47.Penn L J Z, Brooks M W, Laufer E M, Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990;9:1113–1122. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prendergast G. Mechanisms of apoptosis by c-Myc. Oncogene. 1999;18:2967–87. doi: 10.1038/sj.onc.1202727. [DOI] [PubMed] [Google Scholar]
- 49.Pulverer B J, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett J R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 50.Salghetti S E, Kim S, Tansey W P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitt C A, McCurrach M E, de Stanchina E, Wallace-Brodeur R R, Lowe S W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sears R, Leone G, DeGregori J, Nevins J. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 53.Shim H, Chun Y S, Lewis B C, Dang C V. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci USA. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shim H, Dolde C, Lewis B C, Wu C S, Dang G, Jungmann R A, Dalla-Favera R, Dang C V. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith-Sorensen B, Hijmans E M, Beijersbergen R L, Bernards R. Functional analysis of Burkitt's lymphoma mutant c-Myc proteins. J Biol Chem. 1996;271:5513–5518. doi: 10.1074/jbc.271.10.5513. [DOI] [PubMed] [Google Scholar]
- 56.Spotts G D, Patel S V, Xiao Q, Hann S R. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stone J, De L T, Ramsay G, Jakobovits E, Bishop J M, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Symonds G, Hartshorn A, Kennewell A, O'Mara M, Bruskin A, Bishop M. Transformation of murine myelomonocytic cells by myc: point mutations in v-myc contribute synergistically to transforming potential. Oncogene. 1989;4:285–294. [PubMed] [Google Scholar]
- 59.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell-cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 60.Wagner A J, Small M B, Hay N. Myc-mediated apoptosis is blocked by ectopic expression of Bcl-2. Mol Cell Biol. 1993;13:2432–3440. doi: 10.1128/mcb.13.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu M, Arsura M, Bellas R E, FitzGerald M J, Lee H, Schauer S L, Sherr D H, Sonnenshein G E. Inhibition of c-myc expression induces apoptosis of WEHI-231 murine B cells. Mol Cell Biol. 1996;16:5015–5025. doi: 10.1128/mcb.16.9.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann S R. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yano T, Sander C A, Clark H M, Dolezal M V, Jaffe E S, Raffeld M. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt's lymphoma. Oncogene. 1993;8:2741–2748. [PubMed] [Google Scholar]