Abstract
Neurodegenerative disorders like Alzheimer’s disease and Parkinson’s disease are characterized by progressive degeneration of synapses and neurons. Accumulation of misfolded/aggregated proteins represents a pathological hallmark of most neurodegenerative diseases, potentially contributing to synapse loss and neuronal damage. Emerging evidence suggests that misfolded proteins accumulate in the diseased brain at least in part as a consequence of excessively generated reactive oxygen species (ROS) and reactive nitrogen species (RNS). Mechanistically, not only disease-linked genetic mutations but also known risk factors for neurodegenerative diseases, such as aging and exposure to environmental toxins, can accelerate production of ROS/RNS, which contribute to protein misfolding – in many cases mimicking the effect of rare genetic mutations known to be linked to the disease. This review will focus on the role of RNS-dependent post-translational modifications, such as S-nitrosylation and tyrosine nitration, in protein misfolding and aggregation. Specifically, we will discuss molecular mechanisms whereby RNS disrupt the activity of the cellular protein quality control machinery, including molecular chaperones, autophagy/lysosomal pathways, and the ubiquitin-proteasome system (UPS). Because chronic accumulation of misfolded proteins can trigger mitochondrial dysfunction, synaptic damage, and neuronal demise, further characterization of RNS-mediated protein misfolding may establish these molecular events as therapeutic targets for intervention in neurodegenerative diseases.
Keywords: Protein S-nitrosylation, Tyrosine nitration, Protein misfolding, Autophagy, Molecular chaperones, Ubiquitin-proteasome system
Graphical Abstract
1. Introduction
A key pathological event in the majority of neurodegenerative diseases involves excessive generation of misfolded proteins that can contribute to progressive loss of synapses and neurons [1, 2, 3]. For example, aggregated/misfolded α-synuclein (αSyn) is mainly found in patients with synucleinopathies such as Parkinson’s disease (PD) and Lewy body dementia (LBD). Additionally, in Alzheimer’s disease (AD), the most common form of dementia in the elderly, extracellular amyloid-β (Aβ) and intracellular phosphor-tau form amyloid plaques and neurofibrillary tangles, respectively; interestingly, however, over 50% of AD brains examined postmortem also exhibit αSyn deposits [4]. Accordingly, in the past few decades numerous studies have investigated the toxic effects of soluble misfolded proteins on neuronal function and survival as well as the molecular mechanisms leading to this protein aggregation. Emerging evidence suggests that increased production of reactive oxygen species (ROS) and reactive nitrogen species (RNS), representing another key pathological feature of neurodegenerative diseases, is a critical driver of protein misfolding [5, 6]. In this review article, we provide a current overview of pathological events leading to excessive reactive nitrogen and oxygen species (RNS/ROS), including nitric oxide (NO)-related species, that trigger protein misfolding and aggregation, thus contributing to synaptic damage and neuronal injury. Here, we focus on NO-dependent signaling pathways mediated by post-translational modification (PTM) of critical cysteine thiols in a reaction known as S-nitros(yl)ation, which has been shown to be on a par with O-phosphorylation in terms of abundance and importance to cell physiology [7]. We will discuss specific examples showing that aberrant protein S-nitrosylation can compromise the quality control (QC) machinery of protein folding, including molecular chaperones, autophagy, and the ubiquitin-proteasome system (UPS) (Table 1). In addition, we delineate possible pathological roles of RNS-mediated nitration of tyrosine residues (see Section 2.3 for details).
Table 1.
List of major S-nitrosylated or nitrated proteins discussed in this review article
| Category/Pathway | Target protein | Type of NO-dependent PTM | Consequence of NO-dependent PTM | Key Reference |
|---|---|---|---|---|
| Molecular chaperon | PDI | S-Nitrosylation | Decrease in chaperone and isomerase activity | [131] |
| HSP90 | S-Nitrosylation and nitration | Decrease in chaperone activity | [122, 140] | |
| UPS | Parkin | S-Nitrosylation | Dysregulation in ubiquitination | [154–156] |
| Uch-L1 | S-Nitrosylation | Decrease in deubiquitinating activity; transnitrosylation | [69, 166] | |
| Autophagy | JNK1/IKKβ | S-Nitrosylation | Inhibition of activity, leading to decreased autophagy flux | [192] |
| PINK1/Parkin | S-Nitrosylation | inhibition of activity, leading to mitophagy dysfunction | [193] | |
| Conformational change | TDP-43 | S-Nitrosylation | Enhanced aggregation | [76] |
| α-Synuclein | Nitration | Enhanced aggregation | [109] |
1.1. Protein misfolding and aggregation in neurodegenerative diseases
Protein misfolding and aggregation serve as hallmarks of many neurodegenerative diseases including AD, PD, LBD, amyotrophic lateral sclerosis (ALS), Frontotemporal dementia (FTD), prion disease, and polyQ disorders (e.g., Huntington’s disease [HD]) [8]. Protein misfolding often causes abnormal oligomers of soluble protein followed by sequestration into fibrils or aggregates. The histopathological appearance of large aggregates in these neurodegenerative diseases has long been considered as pathological, leading to synaptic and neuronal malfunction. However, more recent data suggest that soluble oligomers are the most harmful species of misfolded protein [9]. Accordingly, the levels of large, aggregated Aβ plaques or tau tangles do not correlate with the severity of cognitive impairment in AD. Rather, soluble oligomeric species are thought to contribute to synaptic damage, representing the major pathological correlate of cognitive decline [10]. These oligomers often contain hydrophobic residues on their surface, facilitating binding to other cellular constituents such as proteins and lipid membranes. Through these aberrant interactions, soluble oligomers of misfolded proteins are thought to activate pathological signaling pathways, leading to synaptic and neuronal toxicity [11]. In contrast, the formation of large aggregates may be a protective response, at least in some cases, by sequestrating toxic oligomers into inclusion bodies [12, 13].
To counteract misfolded proteins, cells are equipped with QC machinery, including at least three components: Molecular chaperones, the UPS, and autophagy [1, 8, 14]. These QC systems not only assist proper folding of the proteins, thus preventing aggregation (e.g., via molecular chaperones), but target misfolded proteins for cellular degradation, eliminating toxicity associated with protein misfolding (via the UPS and autophagy mechanisms). Specifically, molecular chaperones (such as heat shock proteins [HSPs] and protein disulfide isomerase [PDI]) repair protein misfolding, and the UPS and autophagy are responsible for degradation of misfolded/aggregated proteins via the proteasome or lysosomal pathways. In general, the UPS participates in the degradation of short-lived, small-sized misfolded proteins that are tagged with polyubiquitin chains, whereas autophagy (generally meaning macroautophagy) is capable of removing larger insoluble substrates or even damaged organelles. Moreover, recent studies revealed extensive crosstalk between the UPS and autophagy-lysosomal degradation pathways, as exemplified by identification of key molecules common in both pathways, such as ubiquitin, parkin, p62, and HDAC6 [15]. There is also crosstalk between the macroautophagy pathway and the chaperone-mediated autophagy pathway (see section 6 for details) [16].
Supporting the view that protein aggregation represents an etiological feature of neurodegeneration, genetic mutations found in QC components (e.g., parkin, PINK1, Uch-L1, and ATP13A2) or aggregation-prone proteins (TDP-43 and αSyn) have been linked to rare forms of familial neurodegenerative diseases. Additionally, the activity of QC systems significantly declines with age or upon exposure to environmental risk factors, likely contributing to accumulation of misfolded proteins in sporadic neurodegenerative disorders. For example, increased oxidative and nitrosative stress in aged brains can facilitate accumulation of misfolded/aggreged proteins, in part via redox-based PTMs of key components of the QC machinery. In this review, we discuss signaling pathways in which excessive generation of NO and NO-related species contribute to protein misfolding and neurodegeneration via S-nitrosylation of critical cysteine thiols or nitration of tyrosine residues.
1.2. RNS in neurodegenerative diseases
There are three isoforms of NOsynthases (NOS) in mammalian cells, namely: NOS1 (or neuronal NOS [nNOS]), constitutively expressed in neural cells; NOS2 (or inducible NOS [iNOS]), expressed in various cell types upon inflammatory stimulation; and NOS3 (or endothelial NOS [eNOS]), mainly found in endothelium [17]. All three subtypes of NOS are homodimers, consume L-arginine and molecular oxygen as substrates, and utilize nicotinamide-adenine-dinucleotide phosphate (NADPH) as the electron donor. Each monomer of NOS has two domains, the C-terminal reductase domain and N-terminal oxygenase domain. The C-terminal domain contains the co-factor Flavin adenine dinucleotide (FAD) and Flavin mononucleotide (FMN), which can transfer electrons from NADPH to the heme site in the N-terminal domain, where the electrons are used to produce NO, probably in the form of •NO, with one free electron in its outer pi molecular orbital. The production of NO also requires calmodulin binding to NOS, which causes a conformational change that facilitates electron transfer from NADPH to heme. In the case of nNOS and eNOS, the binding of calmodulin relies on intracellular calcium/calmodulin levels. By contrast, the binding affinity of calmodulin to iNOS is much higher, allowing formation of the iNOS-calmodulin complex even in an environment with very low Ca2+ levels [17, 18]. When sufficient substrateL-arginine are not available, NOS has also been shown to generate ROS, specifically, superoxide anion (O2•−) [19–21]. More recent studies have suggested that even at adequate L-arginine levels, the reduction of oxygen to superoxide anion (O2•−) is an obligatory step in NO synthesis, but O2•− is not released under these conditions due to the action of co-factor tetrahydrobiopterin [21].
Endogenously produced RNS from the various NOS isoforms demonstrate Janus-faced effects, ranging from cytoprotective to neurotoxic effects. For example, when produced at low-to-moderate levels in vivo, RNS can activate signaling pathways maintaining physiological functions of the brain, including brain development, synaptic plasticity, and neuronal survival [22]. Initial studies on physiological functions of RNS revealed that the interaction of •NO with the heme moiety of soluble guanylyl cyclase (GC) leads to formation of cyclic guanosine monophosphate (cGMP) [23]. cGMP and the downstream signaling molecule, cGMP-dependent protein kinase I (cGKI), participate in the regulation of not only neuronal transmission in the nervous system but also smooth muscle relaxation [18, 24]. However, ever increasing evidence suggests that cGMP-independent mechanisms for RNS bioactivity play even more critical roles in brain function. One critical mechanism, termed protein S-nitros(yl)ation, is a redox-mediated PTM resulting from a covalent reaction of an NO-related group (likely in the form of nitrosonium cation, NO+) with a nucleophilic cysteine thiol group (more precisely, thiolate anion, RS−) on the target protein [25, 26]. It should be noted that, because NO+ is extremely unstable in solution, reacting immediately with water to generate nitrite (NO2−), transition metals, such as copper or iron, for example, in metalloproteins, are thought to catalyze transient oxidation of •NO to NO+, facilitating the in vivo formation of S-nitrosothiols (R-SNOs) [24, 27–29]. Alternative mechanisms of S-nitrosothiol formation have been proposed[29]. For instance, direct reaction between thiyl radical (RS•) and •NO radical can lead to S-nitrosothiol formation [30]. Mechanism notwithstanding, protein S-nitrosylation participates in both physiological and pathophysiological actions of RNS [6, 31–38]. Another important pathological PTM involving RNS is nitration of tyrosine residues, producing 3-nitrotyrosine. In the current review article, we will focus on the effects of these NO-dependent PTMs on protein misfolding and neuronal cell death [26].
2. RNS-mediated physiological and aberrant signaling in the CNS
In the central nervous system (CNS), nNOS is predominantly expressed in neuronal cells and is often associated with N-methyl-D-aspartate (NMDA)-type glutamate receptors (NMDARs) through a mutual interaction with the postsynaptic density protein 95 (PSD-95) [39]. Activation of NMDARs by glutamate triggers Ca2+ influx into the neuron and subsequent nNOS activation. Under physiological condition, the activation of the synaptic NMDARs leads to moderate NO generation and can contribute to neuroprotective effects. For instance, S-nitrosylation of NMDARs themselves (as a negative-feedback reaction) and caspases inhibits their activity, providing a neuroprotective mechanism [26, 40, 41]. However, activation of extrasynaptic NMDARs due to Aβ oligomers or other stimulatory pathways, can generate excessive amounts of RNS that contribute to neurodegenerative phenotypes [42, 43]. Moreover, Aβ oligomers, αSyn, and toxins such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) can induce inflammatory iNOS expression in other types of brain cells, such as astrocytes, macrophages, and microglia, further increasing NO production in degenerating brains [44, 45]. Additionally, dysregulation of the antioxidative machinery, resulting from environmental toxins such as paraquat and maneb, or age-associated low-grade chronic inflammation (termed inflammaging), can also cause an imbalance of pro- and antioxidant systems, and eventually contribute to protein misfolding and neurodegeneration [6]. For example, signs of compromised antioxidative machineries, such as a decrease in reduced/oxidized glutathione ratio and diminished levels of nuclear factor erythroid 2-related factor 2 (Nrf2), are evident in brains with advanced aging or early AD, and these changes engender oxidative and nitrosative stress [46]. Notably, occurring particularly with nitrosative stress, protein S-nitrosylation is now a well-recognized mediator of the biological activity of RNS [6, 47, 48]. Mechanistically, protein S-nitrosylation can affect cell signaling pathways and neuronal function by altering protein activity, causing conformational changes, modifying protein folding, changing protein-protein interactions, and influencing protein subcellular localization [6, 27, 49]. In this review article, we focus on aberrant S-nitrosylation and nitration of proteins due to high levels of RNS that contribute to protein misfolding and aggregation, in part due to diminished cellular activity of the protein QC machinery.
2.1. Specificity of protein S-nitrosylation
Despite the fact that many proteins contain multiple cysteine residues, S-nitrosylation typically occurs on specific free thiols (or more properly thiolates) [6, 26, 50–52]. Multiple mechanisms of selective S-nitrosylation have been proposed [6, 28, 29]. The first determinant of selective S-nitrosylation entails the close proximity of the target proteins to the source of NO production, exposing the target proteins to a high concentration of S-nitrosylating NO-related groups [6, 29, 52]. The most well-characterized example of this paradigm is the presence of nNOS, NMDARs, and PSD-95 in the same protein complex. Localization of NMDARs and PSD-95 adjacent to nNOS facilitates S-nitrosylation of these two proteins. The second biochemical mechanism that increases the selectivity of S-nitrosothiol formation is the compartmentalization of the target protein into a hydrophobic environment. In this case, hydrophobicity, as seen in lipid membranes or within a certain protein structure, stabilizes S-nitrosylating species, thereby promoting the efficiency of SNO-protein formation [52]. Thirdly, nucleophilic amino acid residues are often located near the target cysteine, potentially constituting a ‘SNO motif’ that facilitates reaction of the target cysteine residues with NO-related groups (particularly nitrosonium cation) [50, 53]. Along these lines, recent structural studies have revealed that the charged amino acid residue(s) is typically present within 8 Å of a SNO target site [54]. Fourthly, transnitrosylation between proteins is responsible for S-nitrosylation of one protein and removal of the SNO-group from the other (for a recent review, see [55] and section 2.2, below). Lastly, the stability of S-nitrosothiols can be counterbalanced by cellular de-nitrosylating enzymes, such as class III alcohol dehydrogenase (also known as ADH5 or GSNO reductase [GSNOR]), thioredoxin (Trx) and Trx-related proteins including PDI, and SNO-CoA reductase. These proteins catalyze reduction of a SNO group from the target cysteine thiol, removing the biological effects of S-nitrosylation [52]. S-Nitrosylation and denitrosylation are not necessarily enzymatically mediated [69]. Note that in most cases the proposed enzymatic reactions that catalyze S-nitrosylation (or transnitrosylation) and denitrosylation activity have not been deeply characterized. For example, apart from some studies by Marletta and colleagues on Trx-mediated transnitrosylation [29, 55–58], biochemical analyses to determine Michaelis-Menten kinetics as well as associated kcat/Km have not been conducted for the proposed S-nitrosylase/transnitrosylase or denitrosylase catalytic activities.
While S-nitrosylation is typically a labile modification, the same cysteine residue can often undergo further oxidation. For instance, an S-nitrosylated cysteine residue can facilitate formation of a stable disulfide bond with a nearby (vicinal) cysteine [59, 60]. Additionally, SNO-induced changes in the 3D structure of the target protein can precipitate further oxidation of the same thiol group [26, 61, 62]. Namely, such conformational alterations make the remaining thiol group more susceptible to reaction with ROS, generating sulfenic acid (-SOH) or even more stable oxidation products such as sulfinic acid (-SO2H) or sulfonic acid (-SO3H) [59]. Hence, in this manner, pathological effects of SNO on the target protein can be prolonged through additional oxidation steps.
2.2. Protein-protein transnitrosylation
Low-molecular weight S-nitrosothiols, e.g., S-nitrosocysteine and S-nitrosoglutathione, serve as endogenous (S)NO donors, when kinetically and thermodynamically favorable, to produce S-nitrosylated proteins via transnitrosylation [29, 50]. In this reaction, mechanistically a cysteine thiol group (or, more precisely thiolate anion) most likely acts as a nucleophile, performing a reversible nucleophilic attack on the nitroso nitrogen to form an SNO-protein adduct. Additionally, proteins can transnitrosylate one another by a similar mechanism [47, 57, 63–69]. In protein-protein transnitrosylation, one protein effectively donates an NO-related group to another; one protein serving as the donor, while the other protein is the acceptor, forming a new S-nitrosothiol. Being in close contact during the reaction, the two proteins are typically found in a protein complex. Redox potential, kinetic and thermodynamic considerations can drive the reaction in one direction over another, i.e., with one of the proteins becoming tranS-nitrosylated while the other is denitrosylated [69]. However, these reactions can also be reversible depending on the exact kinetic and thermodynamic details [29]. Mounting evidence suggests that entire networks of transnitrosylating proteins may represent the predominant mechanism mediating the biological function of protein S-nitrosylation [47, 63, 68]. For example, we recently identified a transnitrosylation network that involves transfer of ‘SNO’ from Uch-L1 to Cdk5 to Drp1, contributing to mitochondrial impairment and synaptic loss in models of neurodegenerative diseases [69]. Whether these transnitrosylation reactions can be catalytically triggered as an enzymatic process, however, will require further studies of their biochemical characteristics, including Michaelis-Menten kinetics, kcat/Km, and increase in reaction rate over spontaneous reaction rate, as demonstrated in one example by Marletta and colleagues [29, 56–58].
2.3. Tyrosine nitration
Both in vitro and in vivo, •NO is also known to react extremely rapidly with superoxide anion (O2•−) to generate peroxynitrite (ONOO−). Peroxynitrite can produce a pathophysiologically relevant PTM called tyrosine nitration [70, 71], although peroxynitrite-independent mechanisms (e.g., possibly involving H2O2-mediated oxidation of NO2−) for nitration may also exist [72]. Tyrosine nitration arises from the addition of a nitro group (-NO2) to the 3’ position of the tyrosine aromatic ring. Similar to protein S-nitrosylation, this type of NO-dependent PTM can also affect protein folding and function, thus ultimately negatively influencing neuronal survival as discussed in detail below.
3. Effects of RNS on protein aggregation, oligomer and fibril formation, and aggregated protein spread
Compelling evidence suggests that abnormal accumulation of protein aggregates caused by misfolding and fibrilization of proteins contributes to cellular dysfunction, neuronal damage, and synaptic loss in many neurodegenerative diseases, including AD, PD, and ALS/FTD. Moreover, misfolded proteins can often form self-templating, soluble oligomeric assemblies (which are likely the most toxic forms to the cells) that engage additional misfolded proteins or even recruit correctly folded proteins to induce their conversion to a misfolded form, yielding protofibrils and ultimately fibrillar aggregates. Aggregated proteins or misfolded oligomers can spread to adjacent cells and seed further aggregation, contributing to neuronal damage in widespread brain areas [2, 73]. Among aggregated proteins reported to propagate along neural networks are tau in AD, αSyn in PD or LBD, TDP-43 in ALS or FTD, and PrPsc in prion disease. While genetic mutations in these genes can cause aggregation, aberrant PTMs (e.g., ubiquitination, phosphorylation, S-nitrosylation, or nitration) of non-mutant proteins can occur due to diverse environmental stimuli, thus triggering misfolding and aggregation, and hence simulating the more rare genetic mutations [74–76]. Here, we will discuss the evidence for RNS-mediated PTMs inducing aberrant protein aggregation and cell-to-cell spreading in neurodegenerative disorders.
3.1. S-Nitrosylation of TDP-43 contributes to its aggregation
FTD is the second most common form of pre-senile dementia after AD. FTD results in focal degeneration of the frontal and temporal lobes of the brain. ALS is a progressive neurodegenerative disorder characterized by loss of upper and lower motor neurons in the brain and spinal cord. These two neurodegenerative disorders are intrinsically linked by overlapping genetic, pathological, and clinical signatures, and may in fact represent two ends of a disease spectrum termed ALS/FTD spectrum disorder [77]. For example, mutations in the gene encoding TAR DNA Binding Protein (TARDBP; encoding the RNA-binding protein TDP-43) are associated with ALS/FTD spectrum disorder, although TARDBP mutations typically lead to ALS and represent rare familial cases (<1% of total ALS/FTD)[78]. Nonetheless, misfolded and aggregated TDP-43 has been identified as a major pathological protein accumulating in affected brain regions and motor neurons in 97% of sporadic ALS and 45% of FTD cases [79–82]. This high prevalence of TDP-43 proteinopathy in sporadic ALS/FTD prompted us to propose the hypothesis that the majority of TDP-43 aggregation in ALS/FTD spectrum disorder results from aging or various environmental factors associated with redox stress [83].
TDP-43 is a highly conserved and ubiquitously expressed DNA/RNA binding protein belonging to the heterogenous ribonucleoprotein family [84–86]. Under un-stressed conditions, TDP-43 is predominantly localized in the nucleus, regulating mRNA processing and stabilization, but it can form aggregates in the cytoplasm and possibly mitochondria under disease conditions [87, 88–91]. There are several key features that are commonly associated with aggregation of TDP-43, including TDP-43 ubiquitination, hyperphosphorylation, and S-nitrosylation [76, 81, 82, 92, 93].
In stressed cells, emerging evidence shows that TDP-43 is localized in cytoplasmic stress granules (SGs) and plays an important role in regulating the dynamics of SG formation and disassembly [94–97], although the biological function of SGs remain largely unknown. SGs are membraneless RNA-protein granules that form in the cytoplasm via liquid-liquid phase separation (LLPS) and often contain mRNA translation machineries [98]. Aberrant phase transition of cytoplasmic TDP-43 is associated with neurotoxicity, and cytoplasmic compartmentalization of the misfolded protein is thought to accumulate and result in the TDP-43 inclusions observed in ALS/FTD [99, 100]. Additionally, a number of studies have revealed that aggregated TDP-43 can spread to neighboring cells via cell-to-cell transmission and act as a seed to induce cytoplasmic aggregation of additional TDP-43 in the recipient cells, possibly contributing to the progression of TDP-43 proteinopathy [101, 102].
Advanced age and potential environmental toxins are thought to represent major risk factors for many neurodegenerative disorders, including ALS/FTD. Both of these risk factors can engender overproduction of intracellular RNS, likely contributing to protein misfolding and aggregation in ALS/FTD and other neurological diseases [6, 103]. Moreover, the FDA-approved drug edaravone (MCI-186), which scavenges RNS/ROS, delays progression of ALS if treatment begins at early stages [104–106]. This finding is consistent with the notion that oxidative and nitrosative stress represent contributors to ALS pathology.
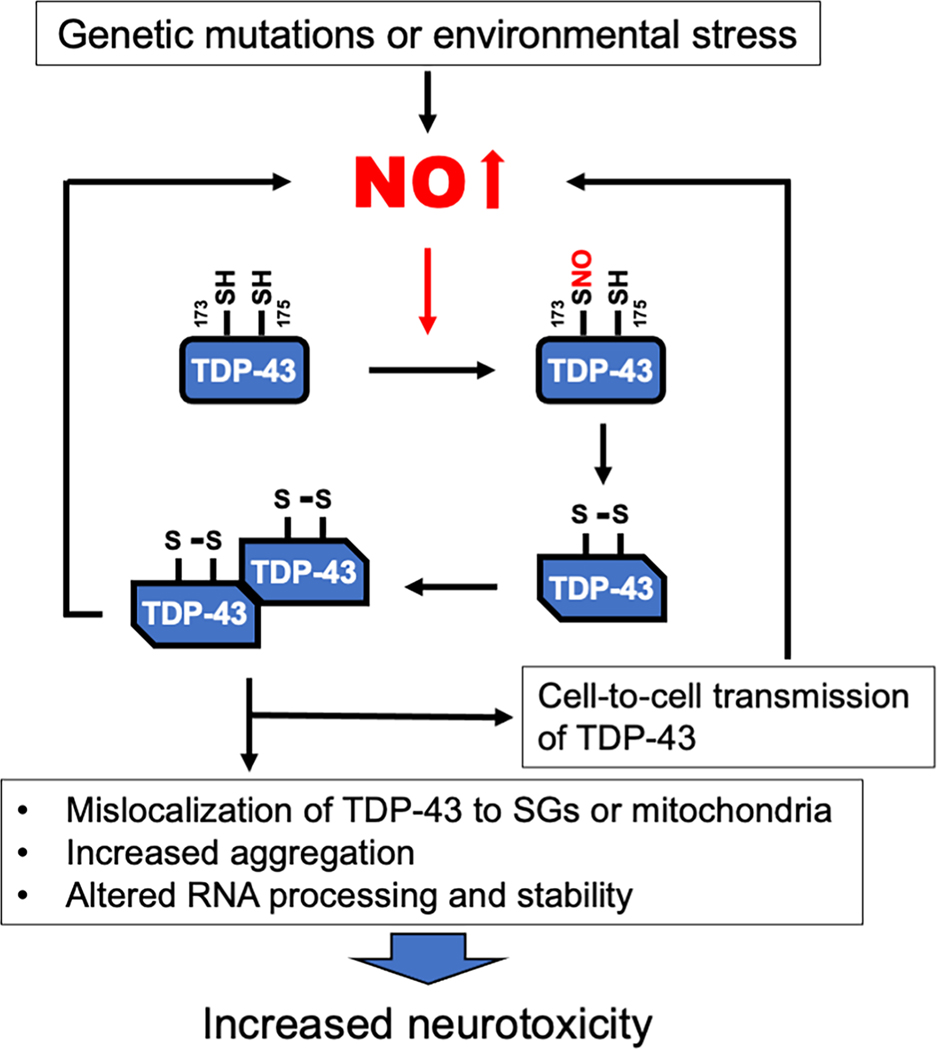
Along these lines, in a recent study we reported that S-nitrosylation of TDP-43 fosters its localization in cytosolic SGs. S-Nitrosylation of TDP-43 was associated with its aggregation in in vitro ALS/FTD models (including human induced pluripotent stem cell (hiPSC)-derived motoneurons) and in postmortem human brain from FTD patient [76]. TDP-43 contains a total of six cysteine residues [53], among which Cys173 and Cys175 are surrounded by a partial SNO motif; these cysteine residues have been identified as SNO sites [76], and are located in the RRM1 (RNA recognition motif) domain. S-Nitrosylation of at least one these thiol groups facilitates the formation of an intra-molecular disulfide linkage, causing a conformational change in the TDP-43 structure and leading to aggregation (Fig. 1). Consistent with this notion, non-nitrosylatable mutant TDP-43 (C173A/C175A) shows a decrease in insoluble TDP-43 aggregation [76].
Fig. 1.
Proposed mechanisms of SNO-TDP-43-mediated neurodegeneration. Genetic mutations or various environmental stimuli can cause an elevation in intracellular NO levels that increase S-nitrosylated TDP-43. SNO-TDP43 triggers intra-molecular disulfide bond formation that alters its protein conformation, contributing to aggregation of TDP-43. SNO-TDP-43 can spread to adjoining cells, leading to neurotoxicity.
Moreover, S-nitrosylation-induced oligomerized or aggregated TDP-43 is able to spread not only to adjoining cells but also induce the generation of additional intracellular NO, promoting cell death of recipient cells [76, 107]. Moreover, in a critical in vivo experiment, we showed that the NOS inhibitor L-NAME prevented neuronal loss from TDP-43 toxicity, consistent with the premise that NO-related species contribute to pathological aggregation, cell-to-cell propagation, and neurotoxicity associated with TDP-43 proteinopathy. Taken together, these findings suggest that SNO-TDP-43 formation is an important factor that contributes to the development of ALS/FTD spectrum disorders and possibly other neurodegenerative diseases [76, 108].
3.2. Tyrosine nitration of αSyn leads to its aggregation
Evidence suggests that the formation of nitrated proteins in biological systems can involve free radical reactions. For example, peroxynitrite-derived radical (e.g., •NO2) has been reported to react with a tyrosyl radical intermediate (Tyr•) to nitrate tyrosine residues, producing 3-nitrotyrosine [72]. Reminiscent of the effects of S-nitrosylation on protein aggregation, tyrosine nitration can precipitate protein misfolding [71]. Perhaps one of the most well characterized nitrated proteins associated with neurodegenerative disorders is αSyn [109, 110]. Using antibodies that recognize 3-nitrotyrosine, an early study found extensive accumulation of nitrated proteins in the core of LBs in human PD patient brains [111]; however, the identity of these nitrated proteins remain unknown. Subsequent studies by Ischiropoulos, Trojanowski, and Lee’s groups identified αSyn as a major molecular target of tyrosine nitration in LBs and demonstrated that nitration promotes aberrant aggregation of αSyn [109, 110]. Note that αSyn does not contain any cysteine residue for S-nitrosylation, whereas all four of its tyrosine residues (i.e., Tyr39, Tyr125, Tyr133 and Tyr136) are susceptible to nitration [112]. These findings are consistent with the notion that, unlike many cases of protein S-nitrosylation, nitration of tyrosine residues can occur more indiscriminately. Interestingly, nitration of αSyn enhances is oligomerization either via inhibition of fibril formation or stabilization of oligomers [110, 113]. In addition, nitration at residue Tyr39 hinders the ability of αSyn to bind lipids [113]. These studies suggest that αSyn nitration affects lipid binding and protein aggregation, thus contributing to α-synucleinopathies. However, many unanswered questions regarding the exact pathological role of nitrated αSyn still exist. For example, additional studies are needed to determine if nitration of αSyn starts in the early stages (or even pre-symptomatic stage) of the disease, thereby potentially also serving as a biomarker for PD or LBD [114]. Moreover, the impact of nitrated αSyn on neurotoxicity in vivo remains unclear.
3.3. Effects of tyrosine nitration on aggregation of Aβ, tau, and other proteins
Sequential cleavage of amyloid precursor protein (APP) by β- and γ-secretases results in production of Aβ peptides, in several forms including Aβ1–40 and Aβ1–42 [115]. Interestingly, human Aβ contains one tyrosine residue (Tyr10), which is replaced by a phenylalanine in mouse/rat Aβ. Additionally, RNS have been reported to trigger nitration of Aβ at Tyr10 in human AD [116]. An initial study demonstrated that nitration of Aβ accelerates its aggregation both in vitro and in transgenic mouse brains expressing human Aβ, possibly contributing to β-amyloidosis and cognitive impairment in AD. Moreover, a subsequent study revealed that Aβ nitration stabilizes toxic Aβ oligomers and impairs fibril formation, thus enhancing synaptotoxicity [117]. However, using a synthesized form of nitrated Aβ, other studies have shown that tyrosine nitration of Aβ prevents aggregation and enhances cell survival [118]. Thus, future studies are needed to resolve these discrepancies.
Tyrosine nitration of tau protein could alter its conformation, promoting tau aggregation. Along these lines, nitrated and aggregated tau was evident in human patient brains [119, 120]. In fact, tau nitration may be an early event in AD pathology, since nitrated tau appears before the mature neurofibrillary tangles [119]. Additionally, an S-nitrosoproteome analysis identified S-nitrosylated tau in a mouse model of AD [33]. Collectively, these findings are consistent with the notion that both S-nitrosylation and tyrosine nitration may modulate tau aggregation. Further studies will be required to determine possible molecular mechanisms of S-nitrosylation- and tyrosine nitration-induced tau aggregation.
Moreover, RNS may contribute to accumulation of misfolded proteins through tyrosine nitration of additional proteins. For example, tyrosine nitration of HSP90 and Trx1 inhibits their chaperone and oxidoreductase activities, respectively [121, 122]. Additionally, nitration triggers aggregation of human islet amyloid polypeptide (hIAPP) [123]. Using models of neurodegenerative disorders, it will be important to determine if tyrosine nitration of these proteins contributes to protein misfolding, synaptic damage, and neurotoxicity.
4. RNS and molecular chaperones
Many proteins are typically synthesized in cells in a non-native state, requiring chaperones and other regulators for proper three-dimensional folding in order to become functionally active. Additionally, molecular chaperones can assist in re-folding of aggregation-prone proteins, such as αSyn and tau, to diminish their potential for aggregation and neurotoxicity. Along these lines, decreased availability of molecular chaperones has been linked to numerous diseases, ranging from neurodegenerative disorders to cancer, diabetes, cystic fibrosis, and cardiovascular disease [1]. Moreover, enhanced expression of molecular chaperons has been shown to delay progression of abnormal phenotypes in models of PD and other neurodegenerative disorders [124]. Molecular chaperone pathways are present in various cellular compartments, including, the cytosol, ER, mitochondria, and nucleus. Various members of the molecular chaperone family or proteins exist in mammalian cells and are often known as heat-shock proteins (HSPs) because they are highly expressed under conditions of temperature stress and other forms of cell stress. Additionally, classes of chaperones, such as protein disulfide isomerase (PDI) and glucose-regulated protein 78 (GRP78/BiP), are present in the endoplasmic reticulum (ER). However, the capacity of the molecular chaperones system, at least in model organisms, appears to decline with age, which represents a major risk factor for neurodegenerative diseases [125]. This decline in chaperone function likely contributes to the accumulation of misfolded proteins as we age. Here we discuss possible mechanisms of how aging-associated nitrosative stress disrupts molecular chaperone activity and contributes to protein aggregation.
4.1. S-Nitrosylation of PDI contributes to accumulation of misfolded proteins
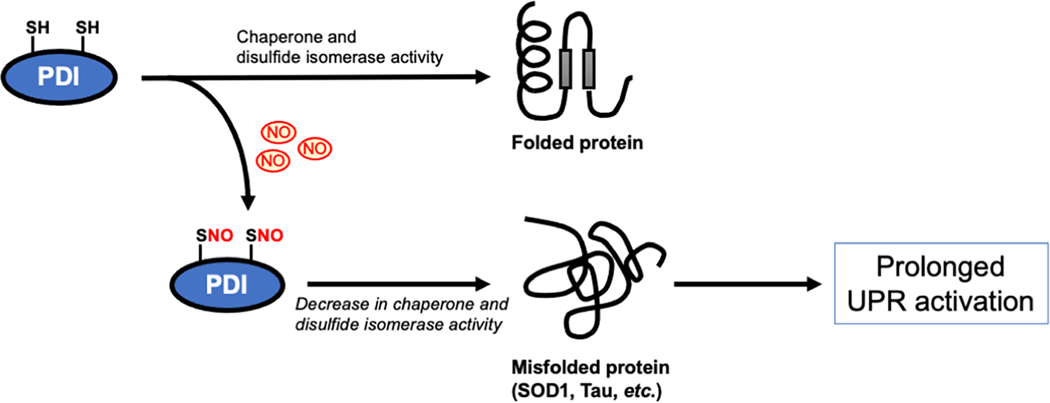
Protein processing and folding occur in the ER, particularly for secreted proteins. In neurodegenerative disorders misfolded proteins often accumulate in the ER lumen and trigger ER stress. Cells utilize two critical processes to relieve damage derived from the accumulation of misfolded proteins: The unfolded protein response (UPR) and ER-associated degradation (ERAD) [5, 126]. The UPR includes three downstream pathways: 1) activation of pancreatic ER kinase (PERK), leading to phosphorylation of eukaryotic initiation factor 2 (eIF2), which subsequently inhibits global protein synthesis and decreases the influx of nascent proteins into the ER; 2) activation of ATF6 (activating transcription factor 6), and 3) activation of the inositol-requiring enzyme 1 (IRE1) pathway, which up-regulates mRNA splicing of X box-binding protein 1 (Xbp1) and then stimulates transcription of UPR target genes, including ER chaperones such as PDI and GRP78/BiP. The UPR usually acts transiently to regain protein homeostasis and promote cell survival, while prolonged UPR activation results in cell damage and death. Emerging evidence suggests that elevated levels of NO-related species can induce ER stress through the disruption of ER Ca2+ homeostasis, possibly via inhibition of the Ca2+-ATPase activity of SERCA2a, contributing to prolonged activation of the UPR pathway [127–129]. Our group first reported that S-nitrosylation of PDI at the active site cysteines (Cys36 and Cys39) inhibits its protein isomerase and chaperone activities [130, 131]. Several PDI family members reside in the ER lumen and function as molecular chaperones as well as disulfide isomerases to assist protein folding. PDI belongs to the Trx-fold superfamily, featuring a common structural property named the Trx fold. All PDI isoforms possess at least one Trx domain with the conserved active site motif CXXC. Interestingly, we have found evidence for increased levels of SNO-PDI in the brains of human sporadic AD, PD and ALS. S-Nitrosylation-dependent inhibition of PDI activity results in increased accumulation of misfolded proteins in the ER, leading to prolonged activation of the UPR and subsequent neuronal cell death in models of AD, PD, and other neurological diseases (Fig. 2) [49, 130–134]. Moreover, we also observed that levels of S-nitrosylated PDI are increased in the spinal cord of the SOD1(G93A) mouse model of ALS, as well as in human patients with sporadic ALS [130, 135]. Because S-nitrosylation of PDI attenuates its ability to decrease aggregation of SOD1 and other aggregation-prone proteins in models of ALS [130], it is tempting to speculate that S-nitrosylation of PDI may be a critical reaction that contributes to accumulation of misfolded proteins in ALS as well as in AD, PD and possibly other neurodegenerative disorders associated with elevated levels of RNS. Interestingly, a more recent study found that PDI can act as a chaperone to physically interact with tau, thus preventing its hyperphosphorylation-mediated aggregation [133]. S-Nitrosylation of PDI prevents this protective effect against tau aggregation, consistent with the notion that SNO-PDI contributes to tau misfolding and aggregation. In addition to PDI, other UPR-related proteins, such as IRE1α and PERK, have also been found to be aberrantly S-nitrosylated in neurodegenerative and other diseases, thus altering their function [126, 136]. These findings suggest that protein S-nitrosylation compromises the neuroprotective functions of the UPR at multiple levels. Further studies will be required to examine if SNO-IRE1α or SNO-PERK can lead to dysregulated protein folding in neurodegenerative diseases.
Fig. 2.
S-Nitrosylation of PDI impairs its chaperone and disulfide isomerase activity that modulates protein folding. When an excessive amount of NO is present under pathological conditions, PDI is S-nitrosylated at its active site cysteines, thus inhibiting its protein folding and chaperone activity, resulting in accumulation of misfolded proteins with increased cell damage and death.
4.2. S-Nitrosylation of multiple molecular chaperones contributes to protein misfolding
In both neuronal and non-neuronal cells, S-nitrosylation appears to regulate the activity of other molecular chaperones and co-chaperones, including HSP90/70, mitochondrial chaperone TNF receptor–associated protein 1 (TRAP1), and valosin-containing protein (VCP). For example, HSP90 is a key regulator of protein homeostasis under both physiological and stress-induced damage with more than 200 substrate proteins identified to date [137–139]. HSP90 has three highly conserved domains, and each domain has its own function. The amino-terminal domain contains an ATP binding motif, the middle domain modulates ATPase activity and binding to co-chaperones and clients, and the carboxy-terminal domain (CTD) is important for both calmodulin binding and homodimerization. NO-related species can S-nitrosylate human HSP90 and thus inhibit its ATPase activity. Cys597 in the CTD, rather than in the ATPase domain, has been identified as the nitrosylation site, suggesting that a conformational change in the CTD inhibits the ATPase activity in an allosteric fashion at some distance from the active site [137, 140]. Intriguingly, SNO-HSP90 has been identified as a transnitrosylating agent towards the androgen receptor (AR), effectively transferring an NO group from HSP90 to AR(Cys601); S-nitrosylation of AR impairs its function as a transcription factor [141]. Further studies will be required to investigate if SNO-HSP90 contributes to protein aggregation in experimental models of neurodegenerative diseases.
Additionally, the mitochondrial-specific homolog of HSP90, TRAP1, has also been reported to be S-nitrosylated at Cys501 [142]. Acting as a molecular chaperone, TRAP1 has been reported to suppress the activity of mitochondrial electron transport chain complex II, which also represents succinate dehydrogenase (SDH) in the tricarboxylic acid (TCA) cycle. S-nitrosylation of TRAP1 accelerates its degradation, thus upregulating SDH, and sensitizing hepatocellular carcinoma cells to mitochondrially-targeted anti-cancer drugs such as SDH inhibitors. TRAP1 can also promote cell survival by decreasing ROS-dependent mitochondrial dysfunction [143, 144]. Additionally, the PD-linked kinase PINK1 (which has been shown to be S-nitrosylated in models of PD; see section 6.2 for details) opposes mitochondrial dysfunction and suppresses oxidative stress-mediated apoptosis, at least in part via phosphorylation of TRAP1 [145]. Further studies are needed to determine if aberrant S-nitrosylation modulates the PINK1-TRAP1 pathway to contribute to the etiology of PD and other neurological disorders.
Furthermore, S-nitrosylation of a co-chaperone of HSP70/HSP90, designated Hsp70/Hsp90 organizing protein (Hop; also known asstress-inducible phosphoprotein I [STI1 or STIP1]), causes the rapid degradation of Hop, and decreases its interaction with cystic fibrosis (CF) transmembrane conductance regulator (CFTR) [146]. The CFTR is an apical plasma membrane chloride channel, and the HSP70-HSP90-Hop complex is an essential component for trafficking CFTR to the cell surface. Considering the disease CF, which is caused by genetic mutations in the gene encoding CFTR, the most common mutation involves loss of phenylalanine 508 (ΔF508). This deletion results in misfolding of the CFTR and consequent degradation of virtually all of the protein prior to its reaching the cell surface. This is the cause of abnormal Cl− conductance in the lung and the resulting pulmonary disease phenotype. The decreased binding of SNO-Hop to ΔF508 CFTR assists maturation and cell surface expression of the mutant protein. Although ΔF508 CFTR is partly misfolded, it has the potential to be functional if expressed at the cell surface. Hence, an S-nitrosylating compound that promotes SNO-Hop formation has been proposed as an ancillary therapeutic approach for CF [146]. Moreover, because Hop has been shown to prevent Aβ and tau toxicity in models of AD[147, 148], it will be intriguing in future studies to determine if SNO-Hop affects Aβ or tau signaling.
Multiple proteomic studies have identified molecular chaperones as targets of S-nitrosylation in both neuronal and non-neuronal cell populations. For example, a proteomic study using neuroblastoma SH-SY5Y cells stimulated with rapamycin to challenge proteostasis identified more than 2,000 SNO-proteins, including a member of the Hsp70 family known as heat shock cognate (HSC70/HSPA8) [149]. In the case of HSC70/HSPA8, S-nitrosylation occurs at Cys17 located in the nucleotide binding region, presumably compromising the activity of HSC70/HSPA8 during protein folding, chaperone-mediated autophagy, and proteasomal degradation of substrate proteins. Other SNO-proteome analyses have also identified molecular chaperones in non-neuronal cells, including GRP78, Hsp70 protein 4, Hsc71, ORP150, calreticulin, and endoplasmin [150, 151]. In the future, additional S-nitroso-proteomic investigations using models of neurodegenerative disorders should help elucidate how S-nitrosylated chaperones may contribute to the pathophysiology of these diseases.
5. RNS and the UPS
The UPS is a key intracellular molecular machinery for degradation of ubiquitin(yl)ated proteins in eukaryotic cells. The sequential reaction of E1 (ubiquitin-activating enzymes), E2 (ubiquitin-conjugating enzymes), and E3 (ubiquitin-ligase enzymes) results in mono- or polyubiquitination of a substrate protein. In general, after polyubiquitinate, substrate proteins are selectively recognized by the proteasome for degradation into short peptides with subsequent reclamation of their amino acids [152]. In contrast, deubiquitinating enzymes in the UPS remove ubiquitin chains from substrates and prevent proteasomal degradation, reversing the effect of E3 ligases [153]. Under physiological conditions, misfolded proteins can be efficiently ubiquitinated and then degraded via the UPS. Accordingly, abnormalities in UPS function can result in the accumulation of misfolded proteins that subsequently form intra- and extracellular aggregates in the brain. As delineated below, protein S-nitrosylation has been shown to affect UPS function at multiple steps.
5.1. S-Nitrosylation of parkin results in protein misfolding and neurotoxicity
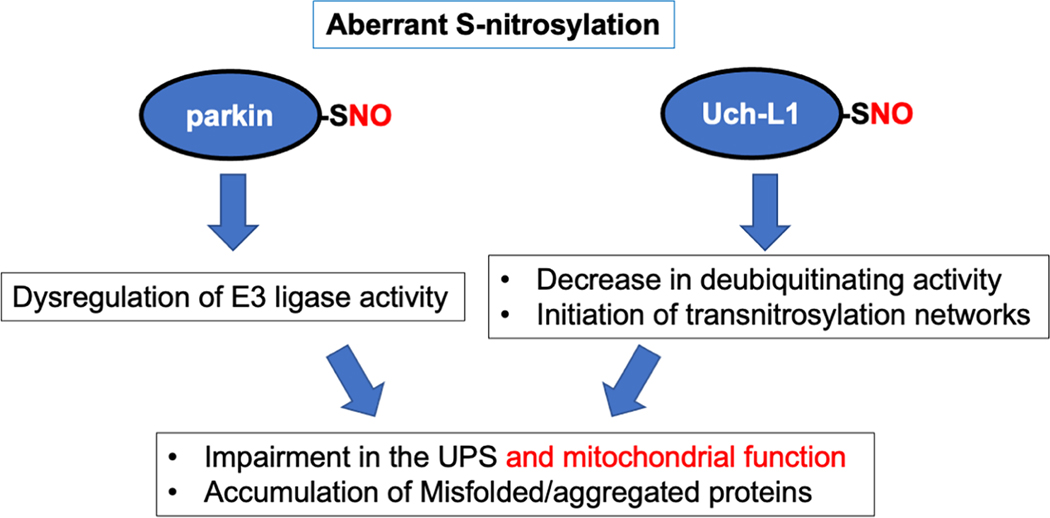
Rare mutations in the gene encoding parkin (PARK2) cause autosomal recessive juvenile PD and some very rare forms of adult PD. In contrast, dysfunctional parkin activity is thought to play an important role in virtually all cases of sporadic PD. Along these lines, we and others discovered that redox-mediated PTM of parkin largely account for its dysfunction in sporadic PD [154–157]. Parkin contains two RING domains as well as one in-between-RING (IBR) domain and belongs to the family of ubiquitin E3 ligase proteins. Our group and others demonstrated that S-nitrosylation occurs on multiple cysteine residues, mainly in the RING and IBR domains of parkin, disrupting its E3 ligase activity and leading to dysfunctional UPS activity (Fig. 3) [154–156, 158]. Moreover, S-nitrosylated parkin facilitates the formation of more stable oxidation products on the same cysteine residues, such as sulfinic acid (SO2H) and sulfonic acid (SO3H), via reaction with ROS, with the latter irreversibly inactivating the enzymatic activity of the protein [157]. Intriguingly, S-nitrosylation of parkin transiently stimulates its ubiquitin E3 ligase activity, followed by a decrease in activity [154, 155]. The initial increase in E3 ligase activity may reflect a neuroprotective aspect of SNO signaling, whereas downregulation of its activity results in UPS dysfunction, contributing to accumulation of misfolded proteins [154, 155, 159]. Additionally, parkin is an important mediator of mitophagy, a process by which the autophagy/lysosomal pathway removes damaged mitochondria (see also the section below on ‘RNS and autophagy’). Mitophagy is often impaired in many neurodegenerative diseases, including PD, AD, HD, ALS, and FTD [160]. Accordingly, recent studies have demonstrated that S-nitrosylation of parkin results in disruption of mitophagy activity, as described below [154–156, 161]. Additionally, our group and others found increased SNO-parkin levels in both animal models and human sporadic PD brains. Hence, dysfunctional parkin activity, occurring with either nitrosylation or rare genetic mutation, facilitates protein misfolding, defective mitophagy, and neuronal damage. These findings are consistent with the notion that aberrantly increased RNS/ROS production, as found in the vast majority of sporadic PD brains, phenocopies the effects of rare mutations in the gene encoding parkin via SNO-parkin-mediated protein misfolding, UPS dysfunction, and mitophagy impairment.
Fig. 3.
S-Nitrosylation of parkin and Uch-L1 impairs UPS activity. Aberrant S-nitrosylation of parkin disrupts its ubiquitin E3 ligase activity. S-Nitrosylated Uch-L1 manifests decreased deubiquitinase activity and increased transnitrosylation activity. Aberrant protein S-nitrosylation eventually impairs the function of the UPS, contributing to the accumulation of misfolded and aggregated proteins.
In addition to proteasomal cleavage of polyubiquitin chains, non-degradative ubiquitin signaling is known to influence neuronal survival via receptor trafficking, mitochondrial homeostasis, and inflammatory responses [162, 163]. For example, parkin promotes cell survival through the epidermal growth factor (EGF)-Akt signaling pathway [164]. In this scenario, parkin monoubiquitinates Eps15, an adaptor protein involved in EGF receptor (EGFR) endocytosis, interfering with binding of Eps15 to the EGFR. This delays EGFR internalization and thus promotes Akt signaling for neuronal survival. Additionally, parkin-mediated monoubiquitination of PICK1 (protein interacting with C-kinase 1) decreases current fluxed by the acid-sensing ion channel (ASIC) [165]. Since ASIC-mediated current is implicated in synaptic plasticity and neuronal injury, it is tempting to speculate that parkin-mediated PICK1 monoubiquitination contributes to neurodegeneration in diseases such as PD. Future studies will be required to determine if S-nitrosylation of parkin affects neuronal viability via monoubiquitination of Eps15 or PICK1.
5.2. S-Nitrosylated Uch-L1 and protein misfolding
RNS can also regulate activities of deubiquitinating enzymes such as Uch-L1 and USP9X [69, 166, 167]. Of these deubiquitinating enzymes, Uch-L1 is most abundantly expressed in the brain, constituting about 2% of total soluble protein [168]. Additionally, dysfunctional Uch-L1 activity has been linked to the pathogenesis of several neurodegenerative diseases, including PD and AD [166, 169, 170]. Along these lines, recent studies from our group and others have shown that S-nitrosylation of Uch-L1 at Cys152 inhibits its binding to ubiquitin, thereby decreasing its deubiquitinating activity (Fig. 3) [69, 166]. We further demonstrated that substitution of an Ala for Cys152 (producing a non-nitrosylatable Uch-L1 mutant) abrogated the effect of RNS on Uch-L1 catalytic activity and ubiquitin binding [69]. Moreover, similar levels of SNO-Uch-L1 were found to be present in human AD brains, mouse models of AD, and cell-based AD models [69], consistent with the premise that pathologically-relevant levels of SNOUch-L1 exist in AD. Additionally, S-nitrosylation leads to structural instability of Uch-L1, potentially serving as a seed to accelerate aggregation of αSyn and possibly other aggregation-prone proteins [166].
In addition to the canonical function of Uch-L1 as a deubiquitinating enzyme, in multiple cell-based and animal models of AD we recently reported that SNO-Uch-L1 can transnitrosylate Cdk5, a kinase that influences neuronal development and survival [171]. Dysregulation of Cdk5 activity is known to contribute to the pathogenesis of AD and other neurodegenerative disorders such as PD, HD, and ALS [172]. Cdk5 is S-nitrosylated at Cys83 and Cys157 [67, 171, 173], leading to an aberrant increases in Cdk5 kinase activity, thus triggering loss of synapses and neurons [67]. A unique, previously unknown biochemical pathway mediates this damage, involving transnitrosylation from Uch-L1 to Cdk5 to Drp1, culminating in the formation of SNO-Drp1 [67, 69, 174]. Drp1 is a dynamin-like GTPase that normally facilitates mitochondrial fission. Upon S-nitrosylation, however, enhanced dimerization/multimerization of Drp1 increases its GTPase activity, resulting in excessive mitochondrial fragmentation, bioenergetic compromise, and synaptic impairment and loss in models of AD [174, 175]. We found that the concerted transnitrosylation from Uch-L1 to Cdk5 to Drp1 proceeds in a kinetically and thermodynamically favorable fashion and is present at pathological levels in human AD brains [67, 69, 174]. Importantly, evidence strongly suggests that this non-canonical transnitrosylation network from SNO-Uch-L1 to SNO-Cdk5 to SNO-Drp1 contributes to synapse loss and cognitive decline in vivo in mouse models of AD because lentiviral expression of non-nitrosylatable mutant Uch-L1(C152A) significantly protects synapses from damage due to toxic Aβ oligomers [69].
5.3. S-Nitrosylation of other UPS-associated enzymes: Potential effects on protein misfolding
In addition to parkin, other ubiquitin E3 ligases, particularly those bearing RING domains, have been shown to undergo S-nitrosylation [64, 176–180]. The list includes X-linked inhibitor of apoptosis (XIAP), cellular inhibitor of apoptosis 1 (cIAP1), ubiquitin protein ligase E3A (UBE3A), product of the von Hippel-Lindau gene (pVHL), carboxy-terminus of Hsc70 interacting protein (CHIP), and Ring Finger Proteins (RNFs). For example, XIAP is a potent anti-apoptotic protein that not only directly inhibits caspase activity but facilitates proteasomal degradation of caspases. S-Nitrosylation of XIAP occurs in the RING domain and inhibits its ubiquitin E3 ligase activity, thereby contributing to neuronal cell death. Additionally, S-nitrosylated caspase-3 can transnitrosylate XIAP. This reaction results in denitrosylation of caspase-3 (thus activating caspase activity) and S-nitrosylation of XIAP (consequently inhibiting its E3 ligase activity). The overall effect is to further activate the apoptotic pathway [64]. Concerning RNFs, Steve Tannenbaum’s group reported that levels of SNO-RNF213 are aberrantly increased in a mouse model of tauopathy [178]. Further studies are needed to delineate the pathological role of these SNO-E3 ligases on protein misfolding and synaptic damage in models of neurodegenerative diseases.
Additionally, protein S-nitrosylation appears to affect multiple processes of UPSdependent protein degradation, including proteasome components, ubiquitin-conjugating E2 enzymes, and the substrates of E3 ligases. Firstly, RNS can regulate UPS function via S-nitrosylation of proteasome components. Specifically, the 20S catalytic core of the 26S proteasome includes at least 10 cysteine residues that can undergo S-nitrosylation, leading to inhibition of proteasomal catalytic activity [181]. Secondly, S-nitrosylation of ubiquitin-conjugating enzyme E2 D1 (UBE2D1), known to be implicated in the ERAD pathway, decreases its E2 activity, potentially contributing to accumulation of misfolded proteins in the ER [182]. In addition, S-nitrosylation inhibits the activity of other ubiquitin-conjugating enzymes, such as E2D3 and E2D4, thereby decreasing p53 ubiquitination in neuroblastoma SH-SY5Y cells [149]. Thirdly, S-nitrosylation of many ubiquitination target proteins affects their protein turnover and thereby modulates the half-lives of these cellular proteins [183–189]. Examples include phosphatase and tensin homolog (PTEN), iron regulatory protein 2 (IRP2), latent TGF-β binding protein 1 (LTBP1), B-cell lymphoma 2 (Bcl-2), and FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein (FLIP). In the case of SNO-Bcl-2 and SNO-FLIP, S-nitrosylation inhibits their ubiquitination, thus increasing their cellular stability and suppressing apoptotic cell death. Given that S-nitrosylation regulates multiple UPS-related proteins and that transnitrosylation networks (e.g., SNO-Uch-L1/Cdk5/Drp1) have been shown to contribute to the pathogenesis of neurodegenerative disorders, it is anticipated that future studies will discover additional such networks that contribute to UPS dysfunction and protein misfolding.
6. RNS and autophagy
Autophagy is a lysosomal degradative process in which unfolded, aggregated proteins and damaged organelles are eliminated in order to maintain cellular homeostasis. Autophagic dysfunction disrupts the homeostatic state and influences cell survival, contributing to a variety of diseases including cancer, neurodegenerative disorders, cardiovascular disorders, and microbial infections [14]. Three major types of autophagy have been described: Macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). Macroautophagy (hereafter referred to simply as autophagy) is the most prevalent form of autophagy and is associated with the formation of cytosolic double-membrane vesicles that sequester and degrade damaged cytosolic contents [190]. Macroautophagy can also selectively degrade damaged mitochondria through a process known as mitophagy. In microautophagy, lysosome takes up cytosolic materials via invagination of the lysosomal membrane. CMA is another form of selective (and constitutive) autophagy, involving direct uptake of substrate proteins with the consensus KFERQ pentapeptide motif or a similar sequence into lysosomes; in this process, HSC70 recognizes and binds to the KFERQ motif and then delivers the substrate proteins to the lysosome. While excessive generation of ROS/RNS have been known to impair autophagy in many different disease states, including neurodegenerative disorders [191], mechanistic details for this phenomena have remained incompletely understood until recently [192, 193]. As summarized below, evidence now suggests that protein S-nitrosylation and tyrosine nitration of critical proteins compromise the autophagic machinery, thereby contributing to neurotoxicity associated with protein misfolding.
6.1. S-Nitrosylation of JNK and IKKβ inhibits autophagy
Many autophagy-related proteins are controlled by PTMs such as phosphorylation, O-GlcNAcylation, acetylation, and ubiquitination [194]. For example, starvation (a known inducer of autophagy) activates c-Jun N-terminal kinase 1 (JNK1), which in turn phosphorylates Bcl-2. Normally, Bcl-2 inhibits autophagy via interaction with the autophagy-related protein Beclin 1. Beclin 1 is a component of the class III phosphatidylinositol 3-kinase (PI3K)/hVps34 complex, which is critical for initiation of autophagosome formation [195]. Phosphorylation of Bcl-2, however, interrupts its binding to Beclin 1, allowing Beclin 1 to initiate autophagosome formation [196, 197]. Additionally, multiple stimuli favoring autophagy activate the IKK (IκB kinase) complex, containing IKKα and IKKβ [198]. Activated IKK stimulates autophagy signaling pathways through phosphorylation of AMP-activated protein kinase (AMPK). Activated AMPK induces autophagy through several signaling pathways. For example, AMPK stimulates autophagy by suppressing the inhibitory effect of mTORC1 [198].
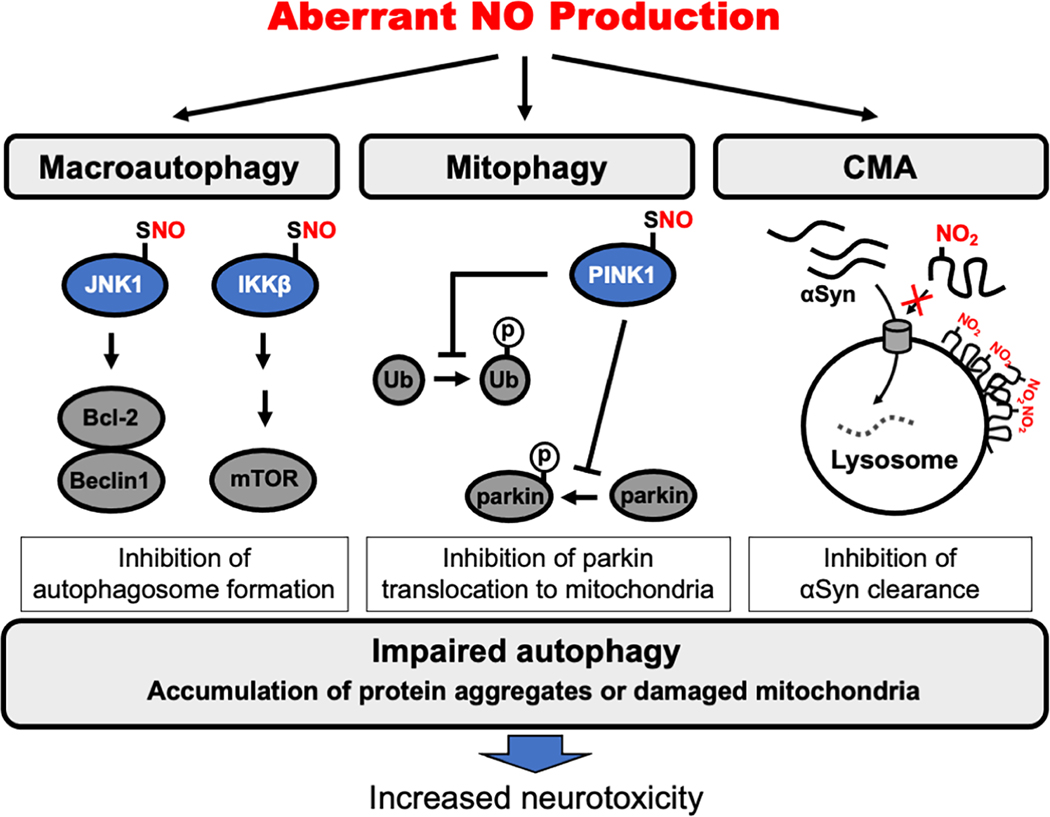
Initial studies on the effect of NO on autophagy include the demonstration by Rubinsztein that S-nitrosylation of JNK and IKKβ impairs induction of autophagy at an early stage (Fig. 4) [192]. Specifically, S-nitrosylation of JNK at Cys116 inhibits its kinase activity [199], leading to a decrease in Bcl-2 phosphorylation. This results in an increase in Bcl-2/Beclin 1 complex formation, thus inhibiting autophagy. While S-nitrosylation of Bcl-2 itself enhances anti-apoptotic activity as discussed above, SNO-Bcl-2 does not appear to influence autophagy [192, 200]. Like SNO-JNK, S-nitrosylation of IKKβ (at Cys179) suppresses autophagy by inhibiting its kinase activity and thus activating mTOR [192, 198, 201, 202]. S-Nitrosylation of PTEN also activates the mTOR signaling pathway to downregulate autophagy [203]. Thus, S-nitrosylation inhibits autophagy via multiple signaling pathways. In line with these observations, the NOS inhibitor, L-NAME, facilitates clearance of mutant huntingtin and αSyn by increasing autophagy [192], consistent with the notion that decreasing NO production promotes autophagy to protect cells from protein misfolding.
Fig. 4.
Regulation of autophagy by pathologically-elevated levels of NO-related species in neurodegenerative diseases. Pathologically increased levels of RNS inhibit autophagy under neuropathological conditions. Considering macroautophagy pathways, S-nitrosylation inhibits JNK1 and IKKβ kinase activity. As a consequence, downstream Bcl-2/Beclin 1 and mTOR pathways are altered, suppressing the initiation of autophagy. In mitophagy, S-nitrosylation of PINK1 decreases its kinase activity, thereby inhibiting phosphorylation of ubiquitin and parkin to suppress mitophagy. Considering CMA pathways, tyrosine nitration (denoted by the presence of NO2 and detected as 3-nitrotyrosine) triggers a conformational change in αSyn that decreases its clearance through CMA. Overall, NO-mediated impairment in autophagy pathways contributes to accumulation of toxic oligomers/aggregates and damaged mitochondria. The inability to clear these proteins and damaged organelles leads to formation of inclusion bodies and contributes to neurodegeneration.
Additionally, recent proteomic studies identified the lysosomal protease cathepsin D (CTSD) as a target of S-nitrosylation [204, 205]. S-Nitrosylation of CTSD at Cys329 was reported to inhibit processing of CTSD, which is required for its maturation, thus decreasing cellular CTSD activity [204]. However, the potential pathophysiological role of S-nitrosylated CTSD in neurodegenerative disorders remains unclear and requires future investigation. Further studies will also be needed to determine if other endosome/lysosome-resident proteins are regulated via S-nitrosylation.
6.2. S-Nitrosylation of PINK1 inhibits parkin-mediated mitophagy
In contrast to the effect of RNS in inhibiting autophagy, as just discussed, RNS can also activate the autophagic process. For example, moderately low concentrations of RNS/ROS can stimulate autophagy in part via suppression of mTOR expression [206]. Notably, the dual actions of RNS on autophagy-related processes are even more evident during mitophagy.
Mitophagy represents a form of (macro)autophagy that selectively removes damaged, defective mitochondria. The process of mitophagy is tightly regulated by two PD-related proteins: parkin and PINK1 (phosphatase and tensin homolog-induced putative kinase protein 1) [207]. While PINK1 is unstable and rapidly degraded via the UPS under normal conditions, once mitochondria are damaged, PINK1 is stabilized on the outer mitochondrial membrane (OMM) where it recruits parkin. Additionally, at the damaged mitochondria membrane, PINK1 phosphorylates parkin, which activates its E3 ubiquitin ligase activity toinitiate mitophagy [208–210]. Moreover, as discussed above, at moderately low levels of RNS, S-nitrosylation of parkin itself in the early stages of PD may initially enhance its E3 ligase activity [154, 155]. Accordingly, activated parkin conjugates a polyubiquitin chain onto the OMM proteins VDAC1, MFN1 and MFN2, leading to the recruitment of autophagy adaptor proteins, including optineurin, nuclear dot protein 52 and p62/SQSTM1, among others, to stimulate mitophagy as a protective mechanism. Autophagy adaptor proteins bind directly to microtubule-associated protein 1A/1B-light chain 3 (LC3), sequestering defective mitochondria into autophagosomes for subsequent removal of the organelle [211].
Recent evidence suggests that depending on concentration, NO-related species can either upregulate or downregulate the mitophagic process [212]. For example, as just discussed, moderately low levels of NO, as occur under physiological conditions or in the early stages of neurodegeneration, can enhance parkin-dependent ubiquitination, mitophagy, and neuronal protection [154, 155, 213]. In contrast, we recently demonstrated that high levels of RNS, as seen under pathological conditions, can impair mitophagy via S-nitrosylation of PINK1 [193]. In this case, S-nitrosylation of PINK1 at Cys568, which is localized at the C-terminal region of the protein, allosterically decreases its kinase activity. Formation of SNO-PINK1 thus results in decreased phosphorylation of parkin as well as ubiquitin, with a consequent decrease in ubiquitin polymerization. This decrease in parkin phosphorylation inhibits its translocation to the OMM, leading to inhibition of mitophagy (Fig. 4) [193].
The mitochondrial uncoupler, valinomycin is known to stabilize PINK1 on the OMM and induce mitophagy. As expected, therefore, co-exposure of hiPSC-derived dopaminergic (DA) neurons to valinomycin and the NOS inhibitor L-NAME decreases SNO-PINK1 formation and increases mitophagy [193]. These findings are consistent with the notion that RNS negatively regulate mitophagy via SNO-PINK1-dependent pathways. As mentioned earlier, S-nitrosylation of parkin at Cys323 transiently increases its activity and promotes mitophagy, possibly reflecting a neuroprotective effect of lower concentrations of RNS at the early stages of a neurodegenerative disease [161]. However, under pathological situations in which NO production is dramatically increased for prolonged periods, (i) S-nitrosylation of PINK1 can counteract the transient effects of SNO on parkin activity, and (ii) S-nitrosylation of parkin eventually diminishes its E3 ligase activity, as described above, further compromising mitophagy [155, 156, 193]. In the future, additional studies will undoubtedly elucidate additional SNO-proteins that influence autophagy in general and mitophagy in particular.
6.3. Tyrosine nitration of αSyn and autophagy
Misfolded αSyn can be degraded by both the UPS and autophagy, in particular CMA [214–216]. As delineated above, nitrated αSyn demonstrates an increased propensity to aggregate and phase separate into cytoplasmic inclusion bodies in PD and LBD brains [217]. Additionally nitrated αSyn appears to be neurotoxic to DA neurons in models of α-synucleinopathy [218]. Monomeric and dimerized forms of αSyn can be degraded via CMA. However, oligomerized or nitrated αSyn are not efficient substrates for CMA [113, 219]. Therefore, it has been postulated that nitration can cause a conformational change in αSyn that interferes with its targeting to the CMA machinery [113, 220]. Future studies are needed to support this premise.
7. Conclusions
Over the last few decades, numerous studies have identified protein misfolding and aggregation as key features in a myriad of neurodegenerative diseases. In this review, we highlighted pathological roles of RNS in accumulation of misfolded/aggregated proteins (Table 1). Specifically, we summarized recent evidence from both in vitro and in vivo studies that aberrant protein S-nitrosylation can compromise activity of the cell’s protein QC machinery, thus contributing to abnormal protein accumulation and neurotoxicity. Further studies will be needed to uncover additional mechanisms underlying the proteotoxic effects of aberrant S-nitrosylation and should aid in development of new therapeutic approaches to neurodegenerative diseases. The utilization of novel S-nitrosoproteomic approaches with enhanced sensitivity (e.g., SNO-TRAP and organomercury enrichment [33, 221]) are expected to identify additional aberrantly S-nitrosylated proteins that disrupt molecular chaperones, the UPS and autophagy, and should provide new molecular insights into the role of NO-related species in neurodegenerative disorders.
Highlights.
RNS contribute to protein misfolding and aggregation via aberrant S-nitrosylation
Aberrant protein S-nitrosylation affects molecular chaperones
Aberrant protein S-nitrosylation compromises the ubiquitin-proteasome system
Aberrant protein S-nitrosylation impairs the autophagy-lysosomal pathway
S-Nitrosylation is a potential therapeutic target for neurodegenerative disorders
Acknowledgments
The authors thank Dr. Scott R. McKercher (The Scripps Research Institute) for critically reading and evaluating the manuscript.
Funding
This work was supported in part by NIH grants R01 NS086890, R01 DA048882, DP1 DA041722, RF1 AG057409, and R01 AG056259 (to S.A.L.), and R01 AG061845 (to T.N.).
Abbreviations
- ADH5
Alcohol dehydrogenase class III
- ALS
Amyotrophic lateral sclerosis
- AMPK
AMP-activated protein kinase
- APP
Amyloid precursor protein
- ATF6
Activating transcription factor 6
- ATP13A2
ATPase cation transporting 13A2
- Bcl-2
B-cell lymphoma 2
- CF
Cystic fibrosis
- CFTR
Cystic fibrosis transmembrane conductance regulator
- cGKI
cGMP-dependent protein kinase I
- cGMP
Cyclic guanosine monophosphate
- CHIP
Carboxy-terminus of Hsc70 interacting protein
- CMA
Chaperon-mediated autophagy
- CNS
Central nervous system
- cIAP1
Cellular inhibitor of apoptosis protein-1
- CTD
Carboxy-terminal domain
- DA
Dopaminergic
- eIF2
Eukaryotic initiation factor 2
- ER
Endoplasmic reticulum
- ERAD
ER-associated degradation
- FAD
Flavin adenine dinucleotide
- FLIP
FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein
- FMN
Flavin mononucleotide
- FTD
Frontotemporal dementia
- GC
Guanylyl cyclase
- GRP78
Glucose-regulated protein 78
- GSH
Glutathione
- GSNO
S-Nitrosoglutathione
- GSNOR
GSNO reductase
- HD
Huntington’s disease
- HDAC6
Histone deacetylase 6
- HSC
Heat shock cognate
- HSP
Heat shock protein
- hIAPP
Human islet amyloid polypeptide
- IBR
In-between-RING
- IKK
IκB kinase
- IRE1
Inositol-requiring enzyme 1
- IRP2
Iron regulatory protein 2
- JNK1
c-Jun N-terminal kinase 1
- LBD
Lewy body dementia
- LC3
Microtubule-associated protein 1A/1B-light chain 3
- LIR
LC3 interacting region
- LLPS
Liquid-liquid phase separation
- LTBP1
Latent TGF-β binding protein 1
- MFN
Mitofusin
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- NMDAR
N-methyl-D-aspartate (NMDA)-type glutamate receptor
- NOS
Nitric oxide synthase
- OMM
Outer mitochondrial membrane
- ORP150
Oxygen-regulated protein 150
- PDI
Protein disulfide isomerase
- PERK
Pancreatic ER kinase
- PI3K
Class III phosphatidylinositol 3-kinase
- PINK1
Phosphatase and tensin homolog-induced putative kinase protein 1
- PSD-95
Postsynaptic density protein-95
- PTEN
Phosphatase and tensin homolog
- PTM
Post-translational modification
- pVHL
von Hippel–Lindau protein
- QC
Quality control
- RNF
RING finger protein
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- RRM1
RNA recognition motif 1
- SDH
Succinate dehydrogenase
- SG
Stress granule
- SNOC
S-Nitrosocysteine
- SOD1
Superoxide dismutase 1
- SQSTM1
Sequestosome 1
- αSyn
α-Synuclein
- TARDBP
Transactivation response (TAR) DNA binding protein
- TDP-43
TAR DNA binding protein-43
- mTORC
Mammalian target of rapamycin complex
- TRAP1
TNF receptor–associated protein 1
- UBE2D1
Ubiquitin-conjugating enzyme E2 D1
- UBE3A
Ubiquitin protein ligase E3A
- Uch-L1
Ubiquitin C-terminal hydrolase L1
- UPR
Unfolded protein response
- UPS
Ubiquitin-proteasome system
- USP9X
Ubiquitin specific peptidase 9 X-linked
- VCP
Valosin-containing protein
- VDAC1
Voltage dependent anion channel 1
- XIAP
X-linked inhibitor of apoptosis
Footnotes
Declaration of competing interest
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Muchowski PJ, Wacker JL, Modulation of neurodegeneration by molecular chaperones, Nat. Rev. Neurosci 6 (1) (2005) 11–22. [DOI] [PubMed] [Google Scholar]
- [2].Soto C, Unfolding the role of protein misfolding in neurodegenerative diseases, Nat. Rev. Neurosci 4 (1) (2003) 49–60. [DOI] [PubMed] [Google Scholar]
- [3].Soto C, Pritzkow S, Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases, Nat. Neurosci 21 (10) (2018) 1332–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hamilton RL, Lewy bodies in Alzheimer’s disease: a neuropathological review of 145 cases using α-synuclein immunohistochemistry, Brain Pathol. 10 (3) (2000) 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nakamura T, Lipton SA, S-Nitrosylation of critical protein thiols mediates protein misfolding and mitochondrial dysfunction in neurodegenerative diseases, Antioxid. Redox Signal 14 (8) (2011) 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nakamura T, Tu S, Akhtar MW, Sunico CR, Okamoto S, Lipton SA, Aberrant protein S-nitrosylation in neurodegenerative diseases, Neuron 78 (4) (2013) 596–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lane P, Hao G, Gross SS, S-Nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation, Sci. STKE 2001 (86) (2001) re1. [DOI] [PubMed] [Google Scholar]
- [8].Ciechanover A, Brundin P, The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg, Neuron 40 (2) (2003) 427–446. [DOI] [PubMed] [Google Scholar]
- [9].Shankar GM, Bloodgood BL, Townsend M, Walsh DM, Selkoe DJ, Sabatini BL, Natural oligomers of the Alzheimer amyloid-β protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway, J. Neurosci 27 (11) (2007) 2866–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Masliah E, Mallory M, Alford M, DeTeresa R, Hansen LA, McKeel DW Jr., Morris JC, Altered expression of synaptic proteins occurs early during progression of Alzheimer’s disease, Neurology 56 (1) (2001) 127–129. [DOI] [PubMed] [Google Scholar]
- [11].Campioni S, Mannini B, Zampagni M, Pensalfini A, Parrini C, Evangelisti E, Relini A, Stefani M, Dobson CM, Cecchi C, Chiti F, A causative link between the structure of aberrant protein oligomers and their toxicity, Nat. Chem. Biol 6 (2) (2010) 140–147. [DOI] [PubMed] [Google Scholar]
- [12].Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S, Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death, Nature 431 (7010) (2004) 805–810. [DOI] [PubMed] [Google Scholar]
- [13].Benilova I, Karran E, De Strooper B, The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes, Nat. Neurosci 15 (3) (2012) 349–357. [DOI] [PubMed] [Google Scholar]
- [14].Nixon RA, The role of autophagy in neurodegenerative disease, Nat. Med 19 (8) (2013) 983–997. [DOI] [PubMed] [Google Scholar]
- [15].Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP, HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS, Nature 447 (7146) (2007) 859–863. [DOI] [PubMed] [Google Scholar]
- [16].Wu H, Chen S, Ammar AB, Xu J, Wu Q, Pan K, Zhang J, Hong Y, Crosstalk Between Macroautophagy and Chaperone-Mediated Autophagy: Implications for the Treatment of Neurological Diseases, Mol. Neurobiol 52 (3) (2015) 1284–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Forstermann U, Sessa WC, Nitric oxide synthases: regulation and function, Eur. Heart J 33 (7) (2012) 829–837, 837a-837d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hobbs AJ, Ignarro LJ, Nitric oxide-cyclic GMP signal transduction system, Methods Enzymol. 269 (1996) 134–148. [DOI] [PubMed] [Google Scholar]
- [19].Heinzel B, John M, Klatt P, Bohme E, Mayer B, Ca2+/calmodulin-dependent formation of hydrogen peroxide by brain nitric oxide synthase, Biochem. J 281 (Pt 3) (1992) 627–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pou S, Pou WS, Bredt DS, Snyder SH, Rosen GM, Generation of superoxide by purified brain nitric oxide synthase, J. Biol. Chem 267 (34) (1992) 24173–24176. [PubMed] [Google Scholar]
- [21].Xia Y, Superoxide generation from nitric oxide synthases, Antioxid. Redox Signal 9 (10) (2007) 1773–1778. [DOI] [PubMed] [Google Scholar]
- [22].Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH, Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures, Proc. Natl. Acad. Sci. U. S. A 88 (14) (1991) 6368–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO, Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide, Science 299 (5608) (2003) 896–899. [DOI] [PubMed] [Google Scholar]
- [24].Heinrich TA, da Silva RS, Miranda KM, Switzer CH, Wink DA, Fukuto JM, Biological nitric oxide signalling: chemistry and terminology, Br. J. Pharmacol 169 (7) (2013) 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Garthwaite J, Charles SL, Chess-Williams R, Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain, Nature 336 (6197) (1988) 385–388. [DOI] [PubMed] [Google Scholar]
- [26].Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS, A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds, Nature 364 (6438) (1993) 626–632. [DOI] [PubMed] [Google Scholar]
- [27].Gaston BM, Carver J, Doctor A, Palmer LA, S-Nitrosylation signaling in cell biology, Mol. Interv 3 (5) (2003) 253–263. [DOI] [PubMed] [Google Scholar]
- [28].Lancaster JR Jr., How are nitrosothiols formed de novo in vivo?, Arch. Biochem. Biophys 617 (2017) 137–144. [DOI] [PubMed] [Google Scholar]
- [29].Smith BC, Marletta MA, Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling, Curr. Opin. Chem. Biol 16 (5–6) (2012) 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tannenbaum SR, White FM, Regulation and specificity of S-nitrosylation and denitrosylation, ACS Chem. Biol 1 (10) (2006) 615–618. [DOI] [PubMed] [Google Scholar]
- [31].Ahern GP, Klyachko VA, Jackson MB, cGMP and S-nitrosylation: two routes for modulation of neuronal excitability by NO, Trends Neurosci. 25 (10) (2002) 510–517. [DOI] [PubMed] [Google Scholar]
- [32].Stamler JS, Lamas S, Fang FC, Nitrosylation. the prototypic redox-based signaling mechanism, Cell 106 (6) (2001) 675–683. [DOI] [PubMed] [Google Scholar]
- [33].Seneviratne U, Nott A, Bhat VB, Ravindra KC, Wishnok JS, Tsai LH, Tannenbaum SR, S-Nitrosation of proteins relevant to Alzheimer’s disease during early stages of neurodegeneration, Proc. Natl. Acad. Sci. U. S. A 113 (15) (2016) 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Amal H, Gong G, Yang H, Joughin BA, Wang X, Knutson CG, Kartawy M, Khaliulin I, Wishnok JS, Tannenbaum SR, Low Doses of Arsenic in a Mouse Model of Human Exposure and in Neuronal Culture Lead to S-Nitrosylation of Synaptic Proteins and Apoptosis via Nitric Oxide, Int. J. Mol. Sci 21 (11) (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rizza S, Cardaci S, Montagna C, Di Giacomo G, De Zio D, Bordi M, Maiani E, Campello S, Borreca A, Puca AA, Stamler JS, Cecconi F, Filomeni G, S-Nitrosylation drives cell senescence and aging in mammals by controlling mitochondrial dynamics and mitophagy, Proc. Natl. Acad. Sci. U.S.A 115 (15) (2018) E3388–e3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sen T, Saha P, Sen N, Nitrosylation of GAPDH augments pathological tau acetylation upon exposure to amyloid-β, Sci Signal 11 (522) (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shin MK, Vazquez-Rosa E, Koh Y, Dhar M, Chaubey K, Cintron-Perez CJ, Barker S, Miller E, Franke K, Noterman MF, Seth D, Allen RS, Motz CT, Rao SR, Skelton LA, Pardue MT, Fliesler SJ, Wang C, Tracy TE, Gan L, Liebl DJ, Savarraj JPJ, Torres GL, Ahnstedt H, McCullough LD, Kitagawa RS, Choi HA, Zhang P, Hou Y, Chiang CW, Li L, Ortiz F, Kilgore JA, Williams NS, Whitehair VC, Gefen T, Flanagan ME, Stamler JS, Jain MK, Kraus A, Cheng F, Reynolds JD, Pieper AA, Reducing acetylated tau is neuroprotective in brain injury, Cell 184 (10) (2021) 2715–2732 e2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yi W, Zhang Y, Liu B, Zhou Y, Liao D, Qiao X, Gao D, Xie T, Yao Q, Zhang Y, Qiu Y, Huang G, Chen Z, Chen C, Ju Z, Protein S-nitrosylation regulates proteostasis and viability of hematopoietic stem cell during regeneration, Cell Rep. 34 (13) (2021) 108922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sattler R, Xiong Z, Lu WY, Hafner M, MacDonald JF, Tymianski M, Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein, Science 284 (5421) (1999) 1845–1848. [DOI] [PubMed] [Google Scholar]
- [40].Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS, Fas-induced caspase denitrosylation, Science 284 (5414) (1999) 651–654. [DOI] [PubMed] [Google Scholar]
- [41].Tenneti L, D’Emilia DM, Lipton SA, Suppression of neuronal apoptosis by S-nitrosylation of caspases, Neurosci. Lett 236 (3) (1997) 139–142. [DOI] [PubMed] [Google Scholar]
- [42].Talantova M, Sanz-Blasco S, Zhang X, Xia P, Akhtar MW, Okamoto S, Dziewczapolski G, Nakamura T, Cao G, Pratt AE, Kang YJ, Tu S, Molokanova E, McKercher SR, Hires SA, Sason H, Stouffer DG, Buczynski MW, Solomon JP, Michael S, Powers ET, Kelly JW, Roberts A, Tong G, Fang-Newmeyer T, Parker J, Holland EA, Zhang D, Nakanishi N, Chen HS, Wolosker H, Wang Y, Parsons LH, Ambasudhan R, Masliah D, Heinemann SF, Pina-Crespo JC, Lipton SA, Aβ induces astrocytic glutamate release, extrasynaptic NMDA receptor activation, and synaptic loss, Proc. Natl. Acad. Sci. U. S. A 110 (27) (2013) E2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Molokanova E, Akhtar MW, Sanz-Blasco S, Tu S, Pina-Crespo JC, McKercher SR, Lipton SA, Differential effects of synaptic and extrasynaptic NMDA receptors on Aβ-induced nitric oxide production in cerebrocortical neurons, J. Neurosci 34 (14) (2014) 5023–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liberatore GT, Jackson-Lewis V, Vukosavic S, Mandir AS, Vila M, McAuliffe WG, Dawson VL, Dawson TM, Przedborski S, Inducible nitric oxide synthase stimulates dopaminergic neurodegeneration in the MPTP model of Parkinson disease, Nat. Med 5 (12) (1999) 1403–1409. [DOI] [PubMed] [Google Scholar]
- [45].Medeiros R, Prediger RD, Passos GF, Pandolfo P, Duarte FS, Franco JL, Dafre AL, Di Giunta G, Figueiredo CP, Takahashi RN, Campos MM, Calixto JB, Connecting TNF-α signaling pathways to iNOS expression in a mouse model of Alzheimer’s disease: relevance for the behavioral and synaptic deficits induced by amyloid β protein, J. Neurosci 27 (20) (2007) 5394–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wojsiat J, Zoltowska KM, Laskowska-Kaszub K, Wojda U, Oxidant/antioxidant imbalance in Alzheimer’s disease: therapeutic and diagnostic prospects, Oxid. Med. Cell Longev 2018 (2018) 6435861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Seth D, Hess DT, Hausladen A, Wang L, Wang YJ, Stamler JS, A multiplex enzymatic machinery for cellular protein S-nitrosylation, Mol. Cell 69 (3) (2018) 451–464 e456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mnatsakanyan R, Markoutsa S, Walbrunn K, Roos A, Verhelst SHL, Zahedi RP, Proteome-wide detection of S-nitrosylation targets and motifs using bioorthogonal cleavable-linker-based enrichment and switch technique, Nat. Commun 10 (1) (2019) 2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen X, Guan T, Li C, Shang H, Cui L, Li XM, Kong J, SOD1 aggregation in astrocytes following ischemia/reperfusion injury: a role of NO-mediated S-nitrosylation of protein disulfide isomerase (PDI), J. Neuroinflammation 9 (1) (2012) 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS, Protein S-nitrosylation: purview and parameters, Nat. Rev. Mol. Cell Biol 6 (2) (2005) 150–166. [DOI] [PubMed] [Google Scholar]
- [51].Lei SZ, Pan ZH, Aggarwal SK, Chen HS, Hartman J, Sucher NJ, Lipton SA, Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex, Neuron 8 (6) (1992) 1087–1099. [DOI] [PubMed] [Google Scholar]
- [52].Stomberski CT, Hess DT, Stamler JS, Protein S-nitrosylation: determinants of specificity and enzymatic regulation of S-nitrosothiol-based signaling, Antioxid. Redox Signal 30 (10) (2019) 1331–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stamler JS, Toone EJ, Lipton SA, Sucher NJ, (S)NO signals: translocation, regulation, and a consensus motif, Neuron 18 (5) (1997) 691–696. [DOI] [PubMed] [Google Scholar]
- [54].Marino SM, Gladyshev VN, Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation, J. Mol. Biol 395 (4) (2010) 844–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nakamura T, Oh CK, Zhang X, Tannenbaum SR, Lipton SA, Protein transnitrosylation signaling networks contribute to inflammaging and neurodegenerative disorders, Antioxid. Redox Signal (2021) [DOI] [PMC free article] [PubMed]
- [56].Barglow KT, Knutson CG, Wishnok JS, Tannenbaum SR, Marletta MA, Site-specific and redox-controlled S-nitrosation of thioredoxin, Proc. Natl. Acad. Sci. U. S. A 108 (35) (2011) E600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mitchell DA, Marletta MA, Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine, Nat. Chem. Biol 1 (3) (2005) 154–158. [DOI] [PubMed] [Google Scholar]
- [58].Mitchell DA, Morton SU, Fernhoff NB, Marletta MA, Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells, Proc. Natl. Acad. Sci. U. S. A 104 (28) (2007) 11609–11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA, S-Nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death, Science 297 (5584) (2002) 1186–1190. [DOI] [PubMed] [Google Scholar]
- [60].Chang TS, Jeong W, Woo HA, Lee SM, Park S, Rhee SG, Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine, J. Biol. Chem 279 (49) (2004) 50994–51001. [DOI] [PubMed] [Google Scholar]
- [61].Arnelle DR, Stamler JS, NO+, NO, and NO- donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation, Arch. Biochem. Biophys 318 (2) (1995) 279–285. [DOI] [PubMed] [Google Scholar]
- [62].Wolhuter K, Whitwell HJ, Switzer CH, Burgoyne JR, Timms JF, Eaton P, Evidence against stable protein S-nitrosylation as a widespread mechanism of post-translational regulation, Mol. Cell 69 (3) (2018) 438–450 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH, GAPDH mediates nitrosylation of nuclear proteins, Nat. Cell Biol 12 (11) (2010) 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, Clemente AT, Okamoto S, Salvesen GS, Riek R, Yates JR 3rd, Lipton SA, Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death, Mol. Cell 39 (2) (2010) 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chen L, Wu R, Feng J, Feng T, Wang C, Hu J, Zhan N, Li Y, Ma X, Ren B, Zhang J, Song CP, Li J, Zhou JM, Zuo J, Transnitrosylation mediated by the non-canonical catalase ROG1 regulates nitric oxide signaling in plants, Dev. Cell 53 (4) (2020) 444–457 e445. [DOI] [PubMed] [Google Scholar]
- [66].Choi MS, Nakamura T, Cho SJ, Han X, Holland EA, Qu J, Petsko GA, Yates JR 3rd, Liddington RC, Lipton SA, Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in Parkinson’s disease models, J. Neurosci 34 (45) (2014) 15123–15131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Qu J, Nakamura T, Cao G, Holland EA, McKercher SR, Lipton SA, S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide, Proc. Natl. Acad. Sci. U. S. A 108 (34) (2011) 14330–14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jia J, Arif A, Terenzi F, Willard B, Plow EF, Hazen SL, Fox PL, Target-selective protein S-nitrosylation by sequence motif recognition, Cell 159 (3) (2014) 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nakamura T, Oh CK, Liao L, Zhang X, Lopez KM, Gibbs D, Deal AK, Scott HR, Spencer B, Masliah E, Rissman RA, Yates JR 3rd, Lipton SA, Non-canonical transnitrosylation network contributes to synapse loss in Alzheimer’s disease, Science (2021) eaaw0843. [DOI] [PMC free article] [PubMed]
- [70].Ferrer-Sueta G, Campolo N, Trujillo M, Bartesaghi S, Carballal S, Romero N, Alvarez B, Radi R, Biochemistry of Peroxynitrite and Protein Tyrosine Nitration, Chem. Rev 118 (3) (2018) 1338–1408. [DOI] [PubMed] [Google Scholar]
- [71].Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS, Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase, Arch. Biochem. Biophys 298 (2) (1992) 431–437. [DOI] [PubMed] [Google Scholar]
- [72].Radi R, Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects, Acc. Chem. Res 46 (2) (2013) 550–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ross CA, Poirier MA, Protein aggregation and neurodegenerative disease, Nat. Med 10 Suppl (2004) S10–17. [DOI] [PubMed] [Google Scholar]
- [74].Davis AA, Leyns CEG, Holtzman DM, Intercellular Spread of Protein Aggregates in Neurodegenerative Disease, Annu. Rev. Cell Dev. Biol 34 (2018) 545–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ischiropoulos H, Beckman JS, Oxidative stress and nitration in neurodegeneration: cause, effect, or association?, J. Clin. Invest 111 (2) (2003) 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Pirie E, Oh CK, Zhang X, Han X, Cieplak P, Scott HR, Deal AK, Ghatak S, Martinez FJ, Yeo GW, Yates JR 3rd, Nakamura T, Lipton SA, S-Nitrosylated TDP-43 triggers aggregation, cell-to-cell spread, and neurotoxicity in hiPSCs and in vivo models of ALS/FTD, Proc. Natl. Acad. Sci. U. S. A 118 (11) (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gao FB, Almeida S, Lopez-Gonzalez R, Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder, EMBO J. 36 (20) (2017) 2931–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Nguyen HP, Van Broeckhoven C, van der Zee J, ALS genes in the genomic era and their implications for FTD, Trends Genet. 34 (6) (2018) 404–423. [DOI] [PubMed] [Google Scholar]
- [79].Ling SC, Polymenidou M, Cleveland DW, Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis, Neuron 79 (3) (2013) 416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mackenzie IR, Neumann M, Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies, J. Neurochem 138 Suppl 1 (2016) 54–70. [DOI] [PubMed] [Google Scholar]
- [81].Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T, TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Biochem. Biophys. Res. Commun 351 (3) (2006) 602–611. [DOI] [PubMed] [Google Scholar]
- [82].Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM, Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Science 314 (5796) (2006) 130–133. [DOI] [PubMed] [Google Scholar]
- [83].Ash PEA, Stanford EA, Al Abdulatif A, Ramirez-Cardenas A, Ballance HI, Boudeau S, Jeh A, Murithi JM, Tripodis Y, Murphy GJ, Sherr DH, Wolozin B, Dioxins and related environmental contaminants increase TDP-43 levels, Mol. Neurodegener 12 (1) (2017) 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG, hnRNP proteins and the biogenesis of mRNA, Annu. Rev. Biochem 62 (1993) 289–321. [DOI] [PubMed] [Google Scholar]
- [85].Wang HY, Wang IF, Bose J, Shen CK, Structural diversity and functional implications of the eukaryotic TDP gene family, Genomics 83 (1) (2004) 130–139. [DOI] [PubMed] [Google Scholar]
- [86].Buratti E, Baralle FE, Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9, J. Biol. Chem 276 (39) (2001) 36337–36343. [DOI] [PubMed] [Google Scholar]
- [87].Wang W, Wang L, Lu J, Siedlak SL, Fujioka H, Liang J, Jiang S, Ma X, Jiang Z, da Rocha EL, Sheng M, Choi H, Lerou PH, Li H, Wang X, The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity, Nat. Med 22 (8) (2016) 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM, Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation, J. Biol. Chem 283 (19) (2008) 13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Rabdano SO, Izmailov SA, Luzik DA, Groves A, Podkorytov IS, Skrynnikov NR, Onset of disorder and protein aggregation due to oxidation-induced intermolecular disulfide bonds: case study of RRM2 domain from TDP-43, Sci. Rep 7 (1) (2017) 11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Herzog JJ, Xu W, Deshpande M, Rahman R, Suib H, Rodal AA, Rosbash M, Paradis S, TDP-43 dysfunction restricts dendritic complexity by inhibiting CREB activation and altering gene expression, Proc. Natl. Acad. Sci. U.S.A 117 (21) (2020) 11760–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Vivoli-Vega M, Guri P, Chiti F, Bemporad F, Insight into the folding and dimerization mechanisms of the N-terminal domain from human TDP-43, Int. J. Mol. Sci 21 (17) (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, Beach TG, Buratti E, Baralle F, Morita M, Nakano I, Oda T, Tsuchiya K, Akiyama H, Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Ann. Neurol 64 (1) (2008) 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bargsted L, Medinas DB, Martinez Traub F, Rozas P, Munoz N, Nassif M, Jerez C, Catenaccio A, Court FA, Hetz C, Matus S, Disulfide cross-linked multimers of TDP-43 and spinal motoneuron loss in a TDP-43A315T ALS/FTD mouse model, Sci. Rep 7 (1) (2017) 14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A, TDP-43 is recruited to stress granules in conditions of oxidative insult, J. Neurochem 111 (4) (2009) 1051–1061. [DOI] [PubMed] [Google Scholar]
- [95].Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B, Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue, PLoS One 5 (10) (2010) e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C, TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1, Hum. Mol. Genet 20 (7) (2011) 1400–1410. [DOI] [PubMed] [Google Scholar]
- [97].Khalfallah Y, Kuta R, Grasmuck C, Prat A, Durham HD, Vande Velde C, TDP-43 regulation of stress granule dynamics in neurodegenerative disease-relevant cell types, Sci. Rep 8 (1) (2018) 7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wolozin B, Ivanov P, Stress granules and neurodegeneration, Nat. Rev. Neurosci 20 (11) (2019) 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhang P, Fan B, Yang P, Temirov J, Messing J, Kim HJ, Taylor JP, Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology, Elife 8 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, Copley KE, Hurtle B, Portz B, Pyles NJ, Guo L, Calder CB, Wills ZP, Pandey UB, Kofler JK, Brodsky JL, Thathiah A, Shorter J, Donnelly CJ, RNA binding antagonizes neurotoxic phase transitions of TDP-43, Neuron 102 (2) (2019) 321–338 e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, Yoshida M, Murayama S, Mann DM, Akiyama H, Hasegawa M, Prion-like properties of pathological TDP-43 aggregates from diseased brains, Cell Rep. 4 (1) (2013) 124–134. [DOI] [PubMed] [Google Scholar]
- [102].Feiler MS, Strobel B, Freischmidt A, Helferich AM, Kappel J, Brewer BM, Li D, Thal DR, Walther P, Ludolph AC, Danzer KM, Weishaupt JH, TDP-43 is intercellularly transmitted across axon terminals, J. Cell Biol 211 (4) (2015) 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Urushitani M, Shimohama S, The role of nitric oxide in amyotrophic lateral sclerosis, Amyotroph. Lateral Scler. Other Motor Neuron Disord 2 (2) (2001) 71–81. [DOI] [PubMed] [Google Scholar]
- [104].Writing G, Edaravone ALSSG, Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial, Lancet Neurol. 16 (7) (2017) 505–512. [DOI] [PubMed] [Google Scholar]
- [105].Satoh K, Ikeda Y, Shioda S, Tobe T, Yoshikawa T, Edarabone scavenges nitric oxide, Redox Rep. 7 (4) (2002) 219–222. [DOI] [PubMed] [Google Scholar]
- [106].Watanabe T, Yuki S, Egawa M, Nishi H, Protective effects of MCI-186 on cerebral ischemia: possible involvement of free radical scavenging and antioxidant actions, J. Pharmacol. Exp. Ther 268 (3) (1994) 1597–1604. [PubMed] [Google Scholar]
- [107].Porta S, Xu Y, Restrepo CR, Kwong LK, Zhang B, Brown HJ, Lee EB, Trojanowski JQ, Lee VM, Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo, Nat. Commun 9 (1) (2018) 4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Al-Chalabi A, Perspective: Don’t keep it in the family, Nature 550 (7676) (2017) S112. [DOI] [PubMed] [Google Scholar]
- [109].Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM, Oxidative damage linked to neurodegeneration by selective α-synuclein nitration in synucleinopathy lesions, Science 290 (5493) (2000) 985–989. [DOI] [PubMed] [Google Scholar]
- [110].Souza JM, Giasson BI, Chen Q, Lee VM, Ischiropoulos H, Dityrosine cross-linking promotes formation of stable α-synuclein polymers. Implication of nitrative and oxidative stress in the pathogenesis of neurodegenerative synucleinopathies, J. Biol. Chem 275 (24) (2000) 18344–18349. [DOI] [PubMed] [Google Scholar]
- [111].Good PF, Hsu A, Werner P, Perl DP, Olanow CW, Protein nitration in Parkinson’s disease, J. Neuropathol. Exp. Neurol 57 (4) (1998) 338–342. [DOI] [PubMed] [Google Scholar]
- [112].Barrett PJ, Timothy Greenamyre J, Post-translational modification of α-synuclein in Parkinson’s disease, Brain Res. 1628 (Pt B) (2015) 247–253. [DOI] [PubMed] [Google Scholar]
- [113].Hodara R, Norris EH, Giasson BI, Mishizen-Eberz AJ, Lynch DR, Lee VM, Ischiropoulos H, Functional consequences of α-synuclein tyrosine nitration: diminished binding to lipid vesicles and increased fibril formation, J. Biol. Chem 279 (46) (2004) 47746–47753. [DOI] [PubMed] [Google Scholar]
- [114].Prigione A, Piazza F, Brighina L, Begni B, Galbussera A, Difrancesco JC, Andreoni S, Piolti R, Ferrarese C, α-Synuclein nitration and autophagy response are induced in peripheral blood cells from patients with Parkinson disease, Neurosci. Lett 477 (1) (2010) 6–10. [DOI] [PubMed] [Google Scholar]
- [115].LaFerla FM, Green KN, Oddo S, Intracellular amyloid-β in Alzheimer’s disease, Nat. Rev. Neurosci 8 (7) (2007) 499–509. [DOI] [PubMed] [Google Scholar]
- [116].Kummer MP, Hermes M, Delekarte A, Hammerschmidt T, Kumar S, Terwel D, Walter J, Pape HC, Konig S, Roeber S, Jessen F, Klockgether T, Korte M, Heneka MT, Nitration of tyrosine 10 critically enhances amyloid β aggregation and plaque formation, Neuron 71 (5) (2011) 833–844. [DOI] [PubMed] [Google Scholar]
- [117].Guivernau B, Bonet J, Valls-Comamala V, Bosch-Morato M, Godoy JA, Inestrosa NC, Peralvarez-Marin A, Fernandez-Busquets X, Andreu D, Oliva B, Munoz FJ, Amyloid-β peptide nitrotyrosination stabilizes oligomers and enhances NMDAR-mediated toxicity, J. Neurosci 36 (46) (2016) 11693–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhao J, Shi Q, Zheng Y, Liu Q, He Z, Gao Z, Liu Q, Insights Into the mechanism of tyrosine nitration in preventing β-amyloid aggregation in Alzheimer’s disease, Front. Mol. Neurosci 14 (2021) 619836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Horiguchi T, Uryu K, Giasson BI, Ischiropoulos H, LightFoot R, Bellmann C, Richter-Landsberg C, Lee VM, Trojanowski JQ, Nitration of tau protein is linked to neurodegeneration in tauopathies, Am. J. Pathol 163 (3) (2003) 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Reyes JF, Fu Y, Vana L, Kanaan NM, Binder LI, Tyrosine nitration within the proline-rich region of Tau in Alzheimer’s disease, Am. J. Pathol 178 (5) (2011) 2275–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Tao L, Jiao X, Gao E, Lau WB, Yuan Y, Lopez B, Christopher T, RamachandraRao SP, Williams W, Southan G, Sharma K, Koch W, Ma XL, Nitrative inactivation of thioredoxin-1 and its role in postischemic myocardial apoptosis, Circulation 114 (13) (2006) 1395–1402. [DOI] [PubMed] [Google Scholar]
- [122].Franco MC, Ye Y, Refakis CA, Feldman JL, Stokes AL, Basso M, Melero Fernandez de Mera RM, Sparrow NA, Calingasan NY, Kiaei M, Rhoads TW, Ma TC, Grumet M, Barnes S, Beal MF, Beckman JS, Mehl R, Estevez AG, Nitration of Hsp90 induces cell death, Proc. Natl. Acad. Sci. U. S. A 110 (12) (2013) E1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zhao J, Wu J, Yang Z, Ouyang L, Zhu L, Gao Z, Li H, Nitration of hIAPP promotes its toxic oligomer formation and exacerbates its toxicity towards INS-1cells, Nitric Oxide 87 (2019) 23–30. [DOI] [PubMed] [Google Scholar]
- [124].Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM, Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease, Science 295 (5556) (2002) 865–868. [DOI] [PubMed] [Google Scholar]
- [125].Balch WE, Morimoto RI, Dillin A, Kelly JW, Adapting proteostasis for disease intervention, Science 319 (5865) (2008) 916–919. [DOI] [PubMed] [Google Scholar]
- [126].Nakato R, Ohkubo Y, Konishi A, Shibata M, Kaneko Y, Iwawaki T, Nakamura T, Lipton SA, Uehara T, Regulation of the unfolded protein response via S-nitrosylation of sensors of endoplasmic reticulum stress, Sci. Rep 5 (2015) 14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Gotoh T, Oyadomari S, Mori K, Mori M, Nitric oxide-induced apoptosis in RAW 264.7 macrophages is mediated by endoplasmic reticulum stress pathway involving ATF6 and CHOP, J. Biol. Chem 277 (14) (2002) 12343–12350. [DOI] [PubMed] [Google Scholar]
- [128].Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, Wada I, Akira S, Araki E, Mori M, Nitric oxide-induced apoptosis in pancreatic β cells is mediated by the endoplasmic reticulum stress pathway, Proc. Natl. Acad. Sci. U. S. A 98 (19) (2001) 10845–10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Viner RI, Williams TD, Schoneich C, Peroxynitrite modification of protein thiols: oxidation, nitrosylation, and S-glutathiolation of functionally important cysteine residue(s) in the sarcoplasmic reticulum Ca-ATPase, Biochemistry 38 (38) (1999) 12408–12415. [DOI] [PubMed] [Google Scholar]
- [130].Jeon GS, Nakamura T, Lee JS, Choi WJ, Ahn SW, Lee KW, Sung JJ, Lipton SA, Potential effect of S-nitrosylated protein disulfide isomerase on mutant SOD1 aggregation and neuronal cell death in amyotrophic lateral sclerosis, Mol. Neurobiol 49 (2) (2014) 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA, S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration, Nature 441 (7092) (2006) 513–517. [DOI] [PubMed] [Google Scholar]
- [132].Kabiraj P, Marin JE, Varela-Ramirez A, Zubia ES, Narayan M, Ellagic acid mitigates SNO-PDI induced aggregation of Parkinsonian biomarkers, ACS Chem. Neurosci (2014) [DOI] [PubMed]
- [133].Wang K, Liu JQ, Zhong T, Liu XL, Zeng Y, Qiao X, Xie T, Chen Y, Gao YY, Tang B, Li J, Zhou J, Pang DW, Chen J, Chen C, Liang Y, Phase Separation and Cytotoxicity of Tau are Modulated by Protein Disulfide Isomerase and S-nitrosylation of this Molecular Chaperone, J. Mol. Biol 432 (7) (2020) 2141–2163. [DOI] [PubMed] [Google Scholar]
- [134].Wang J, Yuan Y, Zhang P, Zhang H, Liu X, Zhang Y, Neohesperidin prevents Aβ25–35-induced apoptosis in primary cultured hippocampal neurons by blocking the S-nitrosylation of protein-disulphide isomerase, Neurochem. Res 43 (9) (2018) 1736–1744. [DOI] [PubMed] [Google Scholar]
- [135].Walker AK, Farg MA, Bye CR, McLean CA, Horne MK, Atkin JD, Protein disulphide isomerase protects against protein aggregation and is S-nitrosylated in amyotrophic lateral sclerosis, Brain 133 (Pt 1) (2010) 105–116. [DOI] [PubMed] [Google Scholar]
- [136].Yang L, Calay ES, Fan J, Arduini A, Kunz RC, Gygi SP, Yalcin A, Fu S, Hotamisligil GS, METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction, Science 349 (6247) (2015) 500–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Retzlaff M, Stahl M, Eberl HC, Lagleder S, Beck J, Kessler H, Buchner J, Hsp90 is regulated by a switch point in the C-terminal domain, EMBO Rep. 10 (10) (2009) 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Schopf FH, Biebl MM, Buchner J, The HSP90 chaperone machinery, Nat. Rev. Mol. Cell Biol 18 (6) (2017) 345–360. [DOI] [PubMed] [Google Scholar]
- [139].Hoter A, El-Sabban ME, Naim HY, The HSP90 family: structure, regulation, function, and implications in health and disease, Int. J. Mol. Sci 19 (9) (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, Lopez-Ferrer D, Higueras MA, Tarin C, Rodriguez-Crespo I, Vazquez J, Lamas S, S-Nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities, Proc. Natl. Acad. Sci. U. S. A 102 (24) (2005) 8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Qin Y, Dey A, Purayil HT, Daaka Y, Maintenance of androgen receptor inactivation by S-nitrosylation, Cancer research 73 (22) (2013) 6690–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Rizza S, Montagna C, Cardaci S, Maiani E, Di Giacomo G, Sanchez-Quiles V, Blagoev B, Rasola A, De Zio D, Stamler JS, Cecconi F, Filomeni G, S-Nitrosylation of the Mitochondrial Chaperone TRAP1 Sensitizes Hepatocellular Carcinoma Cells to Inhibitors of Succinate Dehydrogenase, Cancer research 76 (14) (2016) 4170–4182. [DOI] [PubMed] [Google Scholar]
- [143].Zhang X, Dong Y, Gao M, Hao M, Ren H, Guo L, Guo H, Knockdown of TRAP1 promotes cisplatin-induced apoptosis by promoting the ROS-dependent mitochondrial dysfunction in lung cancer cells, Mol. Cell Biochem 476 (2) (2021) 1075–1082. [DOI] [PubMed] [Google Scholar]
- [144].Altieri DC, Stein GS, Lian JB, Languino LR, TRAP-1, the mitochondrial Hsp90, Biochim. Biophys. Acta 1823 (3) (2012) 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Pridgeon JW, Olzmann JA, Chin LS, Li L, PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1, PLoS Biol. 5 (7) (2007) e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Marozkina NV, Yemen S, Borowitz M, Liu L, Plapp M, Sun F, Islam R, Erdmann-Gilmore P, Townsend RR, Lichti CF, Mantri S, Clapp PW, Randell SH, Gaston B, Zaman K, Hsp 70/Hsp 90 organizing protein as a nitrosylation target in cystic fibrosis therapy, Proc. Natl. Acad. Sci. U. S. A 107 (25) (2010) 11393–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Ostapchenko VG, Beraldo FH, Mohammad AH, Xie YF, Hirata PH, Magalhaes AC, Lamour G, Li H, Maciejewski A, Belrose JC, Teixeira BL, Fahnestock M, Ferreira ST, Cashman NR, Hajj GN, Jackson MF, Choy WY, MacDonald JF, Martins VR, Prado VF, Prado MA, The prion protein ligand, stress-inducible phosphoprotein 1, regulates amyloid-beta oligomer toxicity, J. Neurosci 33 (42) (2013) 16552–16564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Ambegaokar SS, Jackson GR, Functional genomic screen and network analysis reveal novel modifiers of tauopathy dissociated from tau phosphorylation, Hum. Mol. Genet 20 (24) (2011) 4947–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Valek L, Heidler J, Scheving R, Wittig I, Tegeder I, Nitric oxide contributes to protein homeostasis by S-nitrosylations of the chaperone HSPA8 and the ubiquitin ligase UBE2D, Redox Biol. 20 (2019) 217–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Dall’Agnol M, Bernstein C, Bernstein H, Garewal H, Payne CM, Identification of S-nitrosylated proteins after chronic exposure of colon epithelial cells to deoxycholate, Proteomics 6 (5) (2006) 1654–1662. [DOI] [PubMed] [Google Scholar]
- [151].Huang B, Li FA, Wu CH, Wang DL, The role of nitric oxide on rosuvastatin-mediated S-nitrosylation and translational proteomes in human umbilical vein endothelial cells, Proteome Sci. 10 (1) (2012) 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Ross CA, Pickart CM, The ubiquitin-proteasome pathway in Parkinson’s disease and other neurodegenerative diseases, Trends Cell Biol. 14 (12) (2004) 703–711. [DOI] [PubMed] [Google Scholar]
- [153].Harrigan JA, Jacq X, Martin NM, Jackson SP, Deubiquitylating enzymes and drug discovery: emerging opportunities, Nat. Rev. Drug Discov 17 (1) (2018) 57–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lipton SA, Nakamura T, Yao D, Shi ZQ, Uehara T, Gu Z, Comment on “S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function”, Science 308 (5730) (2005) 1870; author reply 1870. [DOI] [PubMed] [Google Scholar]
- [155].Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA, Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity, Proc. Natl. Acad. Sci. U. S. A 101 (29) (2004) 10810–10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM, S-Nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function, Science 304 (5675) (2004) 1328–1331. [DOI] [PubMed] [Google Scholar]
- [157].Meng F, Yao D, Shi Y, Kabakoff J, Wu W, Reicher J, Ma Y, Moosmann B, Masliah E, Lipton SA, Gu Z, Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation, Mol. Neurodegener 6 (2011) 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Wilkaniec A, Lenkiewicz AM, Czapski GA, Jesko HM, Hilgier W, Brodzik R, Gassowska-Dobrowolska M, Culmsee C, Adamczyk A, Extracellular α-synuclein oligomers induce parkin S-nitrosylation: relevance to sporadic parkinson’s disease etiopathology, Mol. Neurobiol 56 (1) (2019) 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Zhong Y, Li X, Du X, Bi M, Ma F, Xie J, Jiang H, The S-nitrosylation of parkin attenuated the ubiquitination of divalent metal transporter 1 in MPP(+)-treated SH-SY5Y cells, Sci. Rep 10 (1) (2020) 15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Lou G, Palikaras K, Lautrup S, Scheibye-Knudsen M, Tavernarakis N, Fang EF, Mitophagy and neuroprotection, Trends Mol. Med 26 (1) (2020) 8–20. [DOI] [PubMed] [Google Scholar]
- [161].Ozawa K, Komatsubara AT, Nishimura Y, Sawada T, Kawafune H, Tsumoto H, Tsuji Y, Zhao J, Kyotani Y, Tanaka T, Takahashi R, Yoshizumi M, S-Nitrosylation regulates mitochondrial quality control via activation of parkin, Sci. Rep 3 (2013) 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Schmidt MF, Gan ZY, Komander D, Dewson G, Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities, Cell Death Differ. 28 (2) (2021) 570–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Atkin G, Paulson H, Ubiquitin pathways in neurodegenerative disease, Front. Mol. Neurosci 7 (2014) 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, Brice A, van PM Bergen En Henegouwen, E.A. Fon, A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling, Nat. Cell Biol 8 (8) (2006) 834–842. [DOI] [PubMed] [Google Scholar]
- [165].Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, Corera AT, Brice A, Seguela P, Fon EA, Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels, Mol. Biol. Cell 18 (8) (2007) 3105–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Kumar R, Jangir DK, Verma G, Shekhar S, Hanpude P, Kumar S, Kumari R, Singh N, Sarovar Bhavesh N, Ranjan Jana N, Kanti Maiti T, S-Nitrosylation of UCHL1 induces its structural instability and promotes α-synuclein aggregation, Sci. Rep 7 (2017) 44558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Majumdar U, Manivannan S, Basu M, Ueyama Y, Blaser MC, Cameron E, McDermott MR, Lincoln J, Cole SE, Wood S, Aikawa E, Lilly B, Garg V, Nitric oxide prevents aortic valve calcification by S-nitrosylation of USP9X to activate NOTCH signaling, Sci. Adv 7 (6) (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Setsuie R, Wada K, The functions of UCH-L1 and its relation to neurodegenerative diseases, Neurochem. Int 51 (2–4) (2007) 105–111. [DOI] [PubMed] [Google Scholar]
- [169].Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L, Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases, J. Biol. Chem 279 (13) (2004) 13256–13264. [DOI] [PubMed] [Google Scholar]
- [170].Butterfield DA, Gnjec A, Poon HF, Castegna A, Pierce WM, Klein JB, Martins RN, Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer’s disease: an initial assessment, J. Alzheimers. Dis 10 (4) (2006) 391–397. [DOI] [PubMed] [Google Scholar]
- [171].Zhang P, Yu PC, Tsang AH, Chen Y, Fu AK, Fu WY, Chung KK, Ip NY, S-nitrosylation of cyclin-dependent kinase 5 (cdk5) regulates its kinase activity and dendrite growth during neuronal development, J. Neurosci 30 (43) (2010) 14366–14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH, Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration, Nature 402 (6762) (1999) 615–622. [DOI] [PubMed] [Google Scholar]
- [173].Foster MW, Forrester MT, Stamler JS, A protein microarray-based analysis of S-nitrosylation, Proc. Natl. Acad. Sci. U. S. A 106 (45) (2009) 18948–18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA, S-Nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury, Science 324 (5923) (2009) 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Lee DS, Kim JE, PDI-mediated S-nitrosylation of DRP1 facilitates DRP1-S616 phosphorylation and mitochondrial fission in CA1 neurons, Cell Death Dis. 9 (9) (2018) 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Tsui AK, Marsden PA, Mazer CD, Adamson SL, Henkelman RM, Ho JJ, Wilson DF, Heximer SP, Connelly KA, Bolz SS, Lidington D, El-Beheiry MH, Dattani ND, Chen KM, Hare GM, Priming of hypoxia-inducible factor by neuronal nitric oxide synthase is essential for adaptive responses to severe anemia, Proc. Natl. Acad. Sci. U. S. A 108 (42) (2011) 17544–17549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lee YI, Giovinazzo D, Kang HC, Lee Y, Jeong JS, Doulias PT, Xie Z, Hu J, Ghasemi M, Ischiropoulos H, Qian J, Zhu H, Blackshaw S, Dawson VL, Dawson TM, Protein microarray characterization of the S-nitrosoproteome, Mol. Cell. Proteomics 13 (1) (2014) 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Amal H, Gong G, Gjoneska E, Lewis SM, Wishnok JS, Tsai LH, Tannenbaum SR, S-Nitrosylation of E3 ubiquitin-protein ligase RNF213 alters non-canonical Wnt/Ca+2 signaling in the P301S mouse model of tauopathy, Transl. Psychiatry 9 (1) (2019) 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zaman K, Knight J, Hussain F, Cao R, Estabrooks SK, Altawallbeh G, Holloway K, Jafri A, Sawczak V, Li Y, Getsy P, Sun F, Raffay T, Cotton C, Brodsky JL, Periasamy A, Lewis SJ, Gaston B, S-Nitrosylation of CHIP enhances F508Del-CFTR maturation, Am. J. Respir. Cell Mol. Biol 61 (6) (2019) 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Romagny S, Bouaouiche S, Lucchi G, Ducoroy P, Bertoldo JB, Terenzi H, Bettaieb A, Plenchette S, S-Nitrosylation of cIAP1 switches cancer cell fate from TNFα/TNFR1-mediated cell survival to cell death, Cancer research 78 (8) (2018) 1948–1957. [DOI] [PubMed] [Google Scholar]
- [181].Kapadia MR, Eng JW, Jiang Q, Stoyanovsky DA, Kibbe MR, Nitric oxide regulates the 26S proteasome in vascular smooth muscle cells, Nitric Oxide 20 (4) (2009) 279–288. [DOI] [PubMed] [Google Scholar]
- [182].Fujikawa K, Nakahara K, Takasugi N, Nishiya T, Ito A, Uchida K, Uehara T, S-nitrosylation at the active site decreases the ubiquitin-conjugating activity of ubiquitin-conjugating enzyme E2 D1 (UBE2D1), an ERAD-associated protein, Biochem. Biophys. Res. Commun 524 (4) (2020) 910–915. [DOI] [PubMed] [Google Scholar]
- [183].Kwak YD, Ma T, Diao S, Zhang X, Chen Y, Hsu J, Lipton SA, Masliah E, Xu H, Liao FF, NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration, Mol. Neurodegener 5 (2010) 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Numajiri N, Takasawa K, Nishiya T, Tanaka H, Ohno K, Hayakawa W, Asada M, Matsuda H, Azumi K, Kamata H, Nakamura T, Hara H, Minami M, Lipton SA, Uehara T, On-off system for PI3-kinase-Akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN), Proc. Natl. Acad. Sci. U. S. A 108 (25) (2011) 10349–10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Kim S, Wing SS, Ponka P, S-Nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway, Mol. Cell Biol 24 (1) (2004) 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Chanvorachote P, Nimmannit U, Stehlik C, Wang L, Jiang BH, Ongpipatanakul B, Rojanasakul Y, Nitric oxide regulates cell sensitivity to cisplatin-induced apoptosis through S-nitrosylation and inhibition of Bcl-2 ubiquitination, Cancer research 66 (12) (2006) 6353–6360. [DOI] [PubMed] [Google Scholar]
- [187].Chanvorachote P, Nimmannit U, Wang L, Stehlik C, Lu B, Azad N, Rojanasakul Y, Nitric oxide negatively regulates Fas CD95-induced apoptosis through inhibition of ubiquitin-proteasome-mediated degradation of FLICE inhibitory protein, J. Biol. Chem 280 (51) (2005) 42044–42050. [DOI] [PubMed] [Google Scholar]
- [188].Gupta A, Anjomani-Virmouni S, Koundouros N, Dimitriadi M, Choo-Wing R, Valle A, Zheng Y, Chiu YH, Agnihotri S, Zadeh G, Asara JM, Anastasiou D, Arends MJ, Cantley LC, Poulogiannis G, PARK2 depletion connects energy and oxidative stress to PI3K/Akt activation via PTEN S-nitrosylation, Mol. Cell 65 (6) (2017) 999–1013 e1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Zhao Q, Zheng K, Ma C, Li J, Zhuo L, Huang W, Chen T, Jiang Y, PTPS facilitates compartmentalized LTBP1 S-nitrosylation and promotes tumor growth under hypoxia, Mol. Cell 77 (1) (2020) 95–107 e105. [DOI] [PubMed] [Google Scholar]
- [190].Schuck S, Microautophagy - distinct molecular mechanisms handle cargoes of many sizes, J. Cell Sci 133 (17) (2020) [DOI] [PubMed] [Google Scholar]
- [191].Chen Y, Azad MB, Gibson SB, Superoxide is the major reactive oxygen species regulating autophagy, Cell Death Differ. 16 (7) (2009) 1040–1052. [DOI] [PubMed] [Google Scholar]
- [192].Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, Garcia-Arencibia M, Rose C, Luo S, Underwood BR, Kroemer G, O’Kane CJ, Rubinsztein DC, Complex inhibitory effects of nitric oxide on autophagy, Mol. Cell 43 (1) (2011) 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Oh CK, Sultan A, Platzer J, Dolatabadi N, Soldner F, McClatchy DB, Diedrich JK, Yates JR 3rd, Ambasudhan R, Nakamura T, Jaenisch R, Lipton SA, S-Nitrosylation of PINK1 attenuates PINK1/parkin-dependent mitophagy in hiPSC-based Parkinson’s disease models, Cell Rep. 21 (8) (2017) 2171–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J, Regulation of autophagy by protein post-translational modification, Lab. Invest 95 (1) (2015) 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P, Regulation of macroautophagy by mTOR and Beclin 1 complexes, Biochimie 90 (2) (2008) 313–323. [DOI] [PubMed] [Google Scholar]
- [196].Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B, Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy, Cell 122 (6) (2005) 927–939. [DOI] [PubMed] [Google Scholar]
- [197].Wei Y, Pattingre S, Sinha S, Bassik M, Levine B, JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy, Mol. Cell 30 (6) (2008) 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, Tailler M, Delahaye N, Tesniere A, De Stefano D, Younes AB, Harper F, Pierron G, Lavandero S, Zitvogel L, Israel A, Baud V, Kroemer G, The IKK complex contributes to the induction of autophagy, EMBO J. 29 (3) (2010) 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Park HS, Huh SH, Kim MS, Lee SH, Choi EJ, Nitric oxide negatively regulates c-Jun N-terminal kinase/stress-activated protein kinase by means of S-nitrosylation, Proc. Natl. Acad. Sci. U. S. A 97 (26) (2000) 14382–14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, Rojanasakul Y, S-Nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis, J. Biol. Chem 281 (45) (2006) 34124–34134. [DOI] [PubMed] [Google Scholar]
- [201].Reynaert NL, Ckless K, Korn SH, Vos N, Guala AS, Wouters EF, van der Vliet A, Janssen-Heininger YM, Nitric oxide represses inhibitory κB kinase through S-nitrosylation, Proc. Natl. Acad. Sci. U. S. A 101 (24) (2004) 8945–8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, He X, Hung JY, Lai CC, Ding Q, Su JL, Yang JY, Sahin AA, Hortobagyi GN, Tsai FJ, Tsai CH, Hung MC, IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway, Cell 130 (3) (2007) 440–455. [DOI] [PubMed] [Google Scholar]
- [203].Zhu L, Li L, Zhang Q, Yang X, Zou Z, Hao B, Marincola FM, Liu Z, Zhong Z, Wang M, Li X, Wang Q, Li K, Gao W, Yao K, Liu Q, NOS1 S-nitrosylates PTEN and inhibits autophagy in nasopharyngeal carcinoma cells, Cell Death Discov. 3 (2017) 17011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Zhou Y, Wynia-Smith SL, Couvertier SM, Kalous KS, Marletta MA, Smith BC, Weerapana E, Chemoproteomic strategy to quantitatively monitor transnitrosation uncovers functionally relevant S-nitrosation sites on cathepsin D and HADH2, Cell Chem. Biol 23 (6) (2016) 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Kohr MJ, Aponte A, Sun J, Gucek M, Steenbergen C, Murphy E, Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: short communication, Circ. Res 111 (10) (2012) 1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].He H, Feng YS, Zang LH, Liu WW, Ding LQ, Chen LX, Kang N, Hayashi T, Tashiro S, Onodera S, Qiu F, Ikejima T, Nitric oxide induces apoptosis and autophagy; autophagy down-regulates NO synthesis in physalin A-treated A375-S2 human melanoma cells, Food Chem. Toxicol 71 (2014) 128–135. [DOI] [PubMed] [Google Scholar]
- [207].Pickrell AM, Youle RJ, The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease, Neuron 85 (2) (2015) 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N, Ubiquitin is phosphorylated by PINK1 to activate parkin, Nature 510 (7503) (2014) 162–166. [DOI] [PubMed] [Google Scholar]
- [209].Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ, PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity, J. Cell Biol 205 (2) (2014) 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J, PINK1 controls mitochondrial localization of parkin through direct phosphorylation, Biochem. Biophys. Res. Commun 377 (3) (2008) 975–980. [DOI] [PubMed] [Google Scholar]
- [211].Palikaras K, Lionaki E, Tavernarakis N, Mechanisms of mitophagy in cellular homeostasis, physiology and pathology, Nat. Cell Biol 20 (9) (2018) 1013–1022. [DOI] [PubMed] [Google Scholar]
- [212].Nakamura T, Lipton SA, Nitric oxide-dependent protein post-translational modifications impair mitochondrial function and metabolism to contribute to neurodegenerative diseases, Antioxid. Redox Signal 32 (12) (2020) 817–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Han JY, Kang MJ, Kim KH, Han PL, Kim HS, Ha JY, Son JH, Nitric oxide induction of parkin translocation in PTEN-induced putative kinase 1 (PINK1) deficiency: functional role of neuronal nitric oxide synthase during mitophagy, J. Biol. Chem 290 (16) (2015) 10325–10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC, α-Synuclein is degraded by both autophagy and the proteasome, J. Biol. Chem 278 (27) (2003) 25009–25013. [DOI] [PubMed] [Google Scholar]
- [215].Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA, Expression of A53T mutant but not wild-type α-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death, J. Neurosci 21 (24) (2001) 9549–9560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D, Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy, Science 305 (5688) (2004) 1292–1295. [DOI] [PubMed] [Google Scholar]
- [217].Gao HM, Kotzbauer PT, Uryu K, Leight S, Trojanowski JQ, Lee VM, Neuroinflammation and oxidation/nitration of α-synuclein linked to dopaminergic neurodegeneration, J. Neurosci 28 (30) (2008) 7687–7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Yu Z, Xu X, Xiang Z, Zhou J, Zhang Z, Hu C, He C, Nitrated α-synuclein induces the loss of dopaminergic neurons in the substantia nigra of rats, PLoS One 5 (4) (2010) e9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV, Hodara R, Fredenburg R, Wu DC, Follenzi A, Dauer W, Przedborski S, Ischiropoulos H, Lansbury PT, Sulzer D, Cuervo AM, Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy, J. Clin. Invest 118 (2) (2008) 777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Salvador N, Aguado C, Horst M, Knecht E, Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state, J. Biol. Chem 275 (35) (2000) 27447–27456. [DOI] [PubMed] [Google Scholar]
- [221].Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H, Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation, Proc. Natl. Acad. Sci. U. S. A 107 (39) (2010) 16958–16963. [DOI] [PMC free article] [PubMed] [Google Scholar]