ABSTRACT
Laboratory surveillance for poliovirus (PV) relies on virus isolation by cell culture to identify PV in stool specimens from acute flaccid paralysis (AFP) cases. Although this method successfully identifies PV, it is time-consuming and necessitates the additional biorisk of growing live virus in an increasingly polio-free world. To reduce the risk of culturing PV, the Global Polio Laboratory Network (GPLN) must switch to culture-independent diagnostic methods with sensitivity at least equivalent to that of cell culture procedures. Five commercial nucleic acid extraction kits and one enrichment method were tested for PV extraction efficiency. RNA yield was measured using real-time reverse transcription (RT)-PCR. Based on greater RNA yield, compared with the other kits, the Quick-RNA viral kit was selected for further testing and was optimized using an RNA extraction procedure for stool suspensions. RNA extraction was retrospectively tested with 182 stool samples that had previously tested positive for PVs, in parallel with the standard GPLN virus isolation algorithm. After virus isolation or RNA extraction, real-time RT-PCR assays were performed. RNA extraction was significantly more sensitive than virus isolation (McNemar’s test, P < 0.001). Thereafter, the RNA extraction method was tested in parallel for 202 prospective samples; RNA extraction and virus isolation were not significantly different from each other (McNemar’s test, P = 0.13). Direct RNA extraction was noninferior to current cell culture methods for detecting PV in stool samples. Our results show that direct RNA extraction can make downstream manipulation safer and can reduce the risk of accidental posteradication viral release. The method is amenable to implementation in a wide variety of polio laboratories.
IMPORTANCE Successfully identifying poliovirus from acute flaccid paralysis (AFP) cases is a vital role of the Global Polio Laboratory Network to achieve the goals of the Global Polio Eradication Initiative. Currently, laboratory surveillance relies on virus isolation by cell culture to test for PV present in stool samples. Although this method can identify polioviruses, laboratories must switch to culture-independent methods to reduce the risk associated with growing live viruses in a soon-to-be polio-free world. By implementing this streamlined method, in combination with real-time RT-PCR, laboratories can quickly screen for and type polioviruses of programmatic importance to support the final stages of global polio eradication.
KEYWORDS: poliovirus, culture-independent diagnostics, polio eradication, rRT-PCR, RNA extraction, direct detection
INTRODUCTION
The Global Polio Laboratory Network (GPLN) is a global surveillance system composed of 146 laboratories in 92 countries, in each of the six World Health Organization (WHO) regions (1, 2). Virus isolation, the “gold standard” of poliovirus (PV) diagnostics, is the first step in identifying PVs, followed by molecular screening using real-time reverse transcription-PCR (rRT-PCR) and finally sequencing to identify PVs of programmatic importance (e.g., wild, vaccine-derived, or type 2 PVs) (3, 4). PV isolation involves inoculating a 10% stool suspension into human rhabdomyosarcoma (RD) cells and mouse fibroblast L-cells genetically engineered to express the human PV receptor (L20B) (5, 6), followed by a series of cell passages from the R-arm (RD cells) or the L-arm (L20B cells), which can select for PVs in acute flaccid paralysis (AFP) specimens. Cytopathic effect (CPE)-positive specimens within the R-arm or the L-arm are then screened by a series of rRT-PCR assays to identify PVs for sequencing. PV rRT-PCR is a suite of six assays with nine targets for intratypic differentiation (ITD) of PVs according to serotype and vaccine or wild-type status (7). If a virus isolate is identified as programmatically important, then the VP1 capsid gene is sequenced for molecular identification (8–10).
Cell culture is affordable and sensitive; its use has contributed to the estimated >99% reduction in global PV cases (11). However, the PV isolation procedure can be time-consuming, and growing PV after eradication is undesirable for maintaining a polio-free world (12). The virus isolation algorithm can take up to 14 days for negative samples before characterization is completed (13). A simpler, faster method to detect PV is needed. As a result, cell culture–independent detection of PV, termed “direct detection,” is a major goal of the GPLN (14). Detection of PV directly from RNA extracted from stool specimens using rRT-PCR is faster than cell culture and removes the complication of maintaining cell lines and growing PV. Because the GPLN includes laboratories in low-and middle-income countries, procedures must be robust; any method should use internationally accessible supplies and reagents and be reproducible in all laboratories. The purpose of this study was to evaluate methods using commercial RNA extraction kits to determine the feasibility of a cell culture-independent method for detecting PV from stool by rRT-PCR and to assess the sensitivity, compared to virus isolation in culture.
RESULTS
Quick-RNA viral kit yields the most viral RNA.
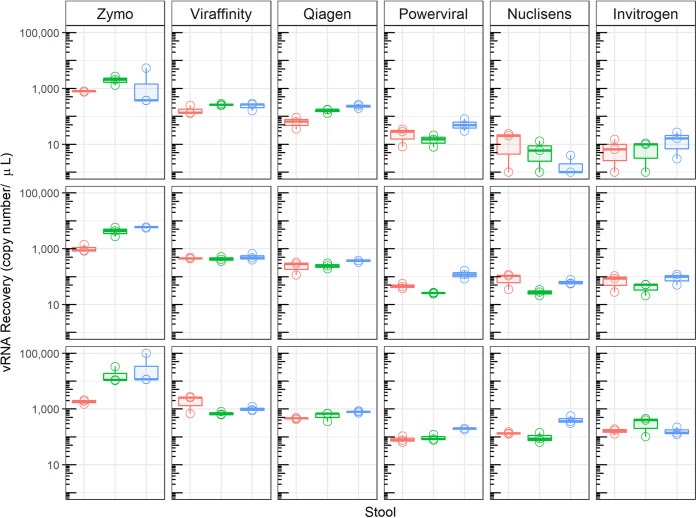
Three Sabin type 1 PV-containing stool samples were tested with five different RNA extraction kits and one bead enrichment step (Table 1). All extractions were eluted into 100 μl nuclease-free water. The Zymo Quick-RNA viral kit yielded significantly more viral RNA (Mann-Whitney test, P < 0.001) than the other RNA extraction kits for all three Sabin type 1 PV-containing stool samples (Fig. 1).
TABLE 1.
Characteristics of viral NA extraction kits
| Kit | Manufacturer | Extraction method | Additional reagents | Additional equipment |
|---|---|---|---|---|
| QIAamp viral RNA mini kit | Qiagen | Column | Ethanol | None |
| Dynabeads SILANE viral NA kit | Invitrogen | Magnetic beads | Isopropanol, ethanol, proteinase K | Magnet |
| PowerViral environmental isolation kit | Qiagen | Bead beating, columna | Ethanol, β-mercaptoethanola | Bead beatera |
| NucliSENS | bioMérieux | Magnetic beadsb | None | easyMAG system |
| Quick-RNA viral kit | Zymo Research | Column | Ethanol, β-mercaptoethanol | None |
| Viraffinity | Biotech Support Group | Virus enrichment | MES, lysis bufferc | None |
Optional.
Automated extraction.
Any commonly used lysis buffer (provided in the RNA extraction kit of choice).
FIG 1.
Assessment of viral RNA recovery from three previously reported Sabin type 1 PV-positive stool suspensions, comparing five RNA extraction kits and one virus enrichment method. Extracted RNA was tested with rRT-PCR assays using Sabin type 1 PV-specific primers and probe. Stool sample 1 (red), stool sample 2 (blue), and stool sample 3 (green) are represented. Each stool sample was extracted in triplicate and tested in rRT-PCR assays in triplicate. Each row represents a stool extraction replicate. The box plots show the median, lower quartile, and upper quartile of the data points for each stool.
The Quick-RNA viral kit was selected, and the final extraction procedure and rRT-PCR volumes were determined. A double extraction procedure was chosen based on ease of use and increased RNA yield. This method uses 400 μl of stool suspension and elution with 20 μl of nuclease-free water. Each extraction was performed in duplicate, and the eluates were combined for a final eluate of 40 μl to provide adequate volume for each ITD rRT-PCR assay. Samples were analyzed with rRT-PCR, and 5 μl of extracted RNA was used for each reaction.
RNA extraction is more sensitive than virus isolation for detection of PV in retrospective specimens.
Stool suspensions from PV-positive stool samples (n = 182) that had been previously identified through virus isolation were tested with the standard virus isolation procedure and the direct RNA extraction method in parallel. Virus isolates and extracted RNA were tested using the ITD rRT-PCR procedure and testing algorithm (7). Most samples (122/131 samples) that were positive by virus isolation were also positive by RNA extraction in the pan-PV (PanPV) assay. The direct RNA extraction method identified 31 samples that were negative by virus isolation; the differences between the RNA extraction procedure and virus isolation were significant (McNemar’s test, P < 0.001) (Table 2). Nine samples were identified by virus isolation but were missed with RNA extraction.
TABLE 2.
Retrospective testing results with the direct RNA extraction method and virus isolation for the PanPV assay
| RNA extraction result | No. with virus isolation result of:a |
Total no. | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 122 | 31 | |
| Negative | 9 | 20 | |
| Total | 182 | ||
McNemar’s test, P < 0.001.
Results were parsed by individual target and were analyzed for statistical significance. RNA extraction was significantly more sensitive than virus isolation when tested with the Sabin type 1 assay (McNemar’s test, P = 0.003). There were 21 concordant-positive samples and 7 concordant-negative samples for RNA extraction and virus isolation with the Sabin type 1 assay. One sample was positive by virus isolation and negative by RNA extraction with the Sabin type 1 assay. Virus isolation failed to detect 13 samples that were positive by the direct RNA extraction method with the Sabin type 1 assay. There were no significant differences among all other ITD rRT-PCR assays (McNemar’s test, P > 0.4) (see Table S1 in the supplemental material).
RNA extraction is similarly sensitive, compared to virus isolation, for prospective samples.
A total of 202 stool samples were prospectively tested by RNA extraction and virus isolation to confirm the procedure with unknown clinical samples. Of the 202 prospectively tested specimens, 195 gave concordant results with both methods in the PanPV assay (13 concordant-positive samples and 182 concordant-negative samples). Virus isolation detected one more Sabin type 1 PV than did RNA extraction, and six specimens were positive by RNA extraction but negative by virus isolation; however, the differences were not significant (McNemar’s test, P = 0.13) (Table 3).
TABLE 3.
Prospective testing results with the direct RNA extraction method and virus isolation for the PanPV assay
| RNA extraction result | No. with virus isolation result of:a |
Total no. | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 13 | 6 | |
| Negative | 1 | 182 | |
| Total | 202 | ||
McNemar’s test, P = 0.13.
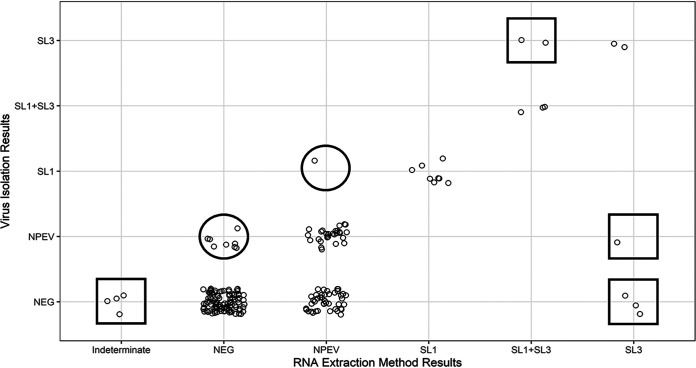
The six samples that were positive by RNA extraction but negative by virus isolation were investigated by cloning and sequencing of the PCR amplicons. The analysis verified two Sabin type 1 PVs and four Sabin type 3 PVs detected by RNA extraction. Four of the 182 negative samples were identified as indeterminate (pan-enterovirus [PanEV] positive, PanPV positive, and all other assays negative) with RNA extraction and negative with virus isolation. No PVs were detected by sequencing; however, sequencing results and BLAST analysis identified species C enteroviruses (nonpolio enterovirus [NPEVs]), i.e., three coxsackieviruses A13 (CVA13s) and one coxsackievirus A24 (CVA24), which are close relatives of PV in the same species. Additionally, the direct RNA extraction method identified 43 NPEVs that were not detected using the virus isolation method. Virus isolation identified eight NPEVs that the direct method did not detect. (Fig. 2).
FIG 2.
Virus isolation identified PV-positive samples that RNA extraction did not (indicated by circles). The direct RNA extraction method identified PV-positive samples that virus isolation did not (indicated by squares). RNA extraction method results are shown on the x axis, and virus isolation results are shown on the y axis. Each point represents a separate result. SL, Sabin-like; NEG, negative. Virus mixtures are indicated as SL1+SL3.
DISCUSSION
As polio eradication nears, polio diagnostic laboratories need molecular methods to replace cell culture-dependent diagnostics (3, 13, 15, 16). Culture-independent detection of PV must be noninferior to virus isolation, as well as robust, rapid, and widely accessible. The RNA extraction procedure uses internationally available materials and reagents. Parallel testing indicated that the direct RNA extraction method was more sensitive than virus isolation when tested with the Sabin type 1 assay. This is critical to detect low-titer PVs in stool. Prospective testing of clinical samples showed that the RNA extraction procedure was similarly sensitive, compared to virus isolation, and detected PV-positive samples that virus isolation did not identify. Sequencing showed that potential false-positive samples with the direct RNA extraction method were true-positive samples. Identification of PV through molecular methods means faster results and obviates the need to grow PV to high titers, which poses a risk for laboratory-acquired infections or unintended release of an eradicated agent.
PV can be reliably detected independent of cell culture through RNA extraction by using the standard ITD rRT-PCR kit. Preliminary testing with Sabin type 1 PV-positive stool samples showed that the Zymo Quick-RNA viral kit recovered more viral RNA than the other five commercial RNA extraction methods tested.
Retrospective results showed that the Zymo Quick-RNA viral kit direct extraction method is more sensitive than virus isolation for detection of PV. The discrepancy between historical and parallel testing results is expected, since those samples were from 2014 and before and the virus could have lost viability. Virus isolation results from parallel testing were considered the true results for our analysis. Prospective testing showed equivalent results for RNA extraction and virus isolation. Preliminary pilot testing results show advantages of the new method, such as quicker turnaround time, compared to virus isolation (C. Harrington, E. Vega, & N. Gerloff, unpublished observations).
Using the commercially available Zymo Quick-RNA viral kit has advantages in terms of quality control, compared to preparing in-house extraction reagents, which is important for reproducibility. Commercial reagents are prepared in controlled environments and distributed worldwide, as opposed to in-house reagents that may vary from laboratory to laboratory. Additionally, a desirable characteristic of an RNA extraction kit is the ability to concentrate samples. This is important to improve the limit of detection of the molecular assays, since one limitation of molecular assays, compared to cell culture, is the amount of sample that can be analyzed.
A challenge to broad implementation of culture-independent detection will be converting established PV isolation laboratories to molecular laboratories. In many laboratories, personnel may need to be recruited or retrained, and laboratory space may need to be renovated or identified, since cell culture laboratories may not have the appropriate work areas for molecular procedures such as RNA extraction and rRT-PCR (17, 18). Additionally, a working algorithm and threshold cycle (CT) cutoff value would have to be implemented in case the integrity of the RNA from the direct method is not as optimal as a virus isolate. The limitation of both the retrospective analysis and the prospective analysis was the small number of PV-positive samples. Each analysis was performed with the most recent result of virus isolation and the direct RNA extraction method for a head-to-head comparison. The NPEVs that were missed by virus isolation were likely viruses that do not grow well in RD or L20B cells; however, this is difficult to confirm, since these samples were not sequenced and the isolation algorithm is not designed to detect NPEVs. During prospective testing, the four samples with indeterminate results were species C enterovirus by the direct RNA extraction method and sequencing but were negative by virus isolation. In addition, there is a limit on the number of stool suspensions that can be processed at one time with the direct RNA extraction method. Because the RNA extraction procedure is column based, the number of samples that can be centrifuged at one time may create a bottleneck. The current recommendation is to process 10 stool suspensions at one time. The RNA extraction procedure increases hands-on time and, if samples are processed incorrectly, also increases the possibility of cross contamination. Automated procedures are being validated for high-workload laboratories to mitigate these issues.
This study is not the first to attempt to increase the sensitivity of molecular methods, compared to virus isolation, for PV detection (3, 19). A bead-based approach developed by Arita et al. uses an efficient method to amplify the entire PV capsid region with virus concentrated from stool extracts using PV receptor linked to magnetic beads (3). The method is sensitive but not commercially available and would require in-house production of reagents. Production of receptor-coupled magnetic beads would be costly and time-consuming and would require extensive quality assurance and quality control procedures to minimize lot-to-lot variation. Nevertheless, the capture procedure is efficient and sensitive; although both procedures are viable options for detection of PV, the double extraction was selected because of simplicity and cost.
The modification of the Quick-RNA viral kit for cell culture-independent detection of PV is a major but incremental step in the transition from cell culture to complete molecular detection. Identification of additional manual extraction kits and an automated procedure is necessary to implement direct detection across the entire GPLN. It is necessary to have multiple validated kits, since not all countries have access to the same RNA extraction reagents/kits, due to import restrictions or the lack of vendor support within the country. An additional manual procedure will ensure that all laboratories have access to a validated procedure for detecting PV from stool specimens. An automatic extraction method is desirable for high-workload laboratories or those that already use automated methods in their workflows. Additionally, the Quick-RNA viral kit does not completely inactivate PV during the 1-min incubation period. A 5-min incubation period is necessary to inactivate the virus prior to extraction of RNA, to avoid any risk of exposure or viral release (20). The Zymo Quick-RNA viral kit direct extraction method, in combination with the WHO-recommended ITD rRT-PCR kit, can rapidly screen and type PV in support of the final stages of global polio eradication.
MATERIALS AND METHODS
Specimen selection and preparation.
Three Sabin type 1 PV-confirmed stool samples were selected for initial evaluation of commercial RNA extraction kits and method optimization; 10% stool suspensions were prepared following the GPLN protocol (21, 22). Briefly, ∼1 g of stool was added to 0.5 g of glass beads, 0.25 ml of chloroform, and 5 ml of minimum essential medium (MEM) in a conical tube. The suspension was shaken for 30 min in a vertical shaker and then centrifuged at 3,000 × g for 30 min at 4°C. The clarified supernatant was then transferred to a 2-ml cryovial and stored at −70°C for subsequent direct RNA extraction independent of virus isolation.
Evaluation of commercial RNA extraction kits.
Sabin type 1 PV RNA transcripts were quantitated on a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and used as RNA standards (10). Briefly, a 10-fold standard RNA ladder, in 5-step serial dilutions in triplicate, were set up for each rRT-PCR run to create a standard curve. The extracted RNA was tested in rRT-PCR assays and compared to the RNA ladder on the same run. Copy number was determined based on the linear relationship between the dilution factor and the CT value. Five nucleic acid (NA) extraction kits were selected for evaluation, namely, QIAamp viral RNA mini kit (catalogue number 52906; Qiagen, Germantown, MD), Dynabeads MyOne SILANE viral NA kit (catalogue number 37011D; Thermo Fisher Scientific, Waltham, MA), PowerViral environmental isolation kit (catalogue number 28000; Qiagen), NucliSENS easyMAG system (catalogue number 280130; bioMérieux, Marcy l’Etoile, France), and Quick-RNA viral kit (catalogue number R1035; Zymo Research, Irvine, CA). The QIAamp viral RNA mini kit is routinely used for RNA extraction for sequencing in the GPLN and was used as the baseline for all results. In order to directly compare all kits, sample volume was adjusted to 200 μl, with elution into 100 μl using nuclease-free water. Extracted RNA was quantitated by rRT-PCR with the ITD rRT-PCR kit, using 1 μl of sample for each assay (10). Briefly, the ITD rRT-PCR kit is made of six assays with nine targets that work as a single algorithm to screen for PVs. The nine targets are PanEV, PanPV, PanPV Type 2, Sabin 1, Sabin 2, Sabin 3, wild PV 1 (WPV1), WPV3-I, and WPV3-II. Isolates of programmatic importance are then referred for sequencing as part of the standard GPLN procedure.
Viraffinity (catalogue number V1062; Biotech Support Group, Monmouth, NJ), a virus enrichment method, was tested in conjunction with the QIAamp viral RNA mini kit to enrich virus prior to RNA extraction. Briefly, 1 volume (400 μl) of 60 mM 2-(N-morpholino)ethanesulfonic acid (MES), 150 mM NaCl (pH 6.5), was added to 400 μl stool suspension. A 1:4 volume ratio of Viraffinity sample, as recommended by the manufacturer for enterovirus-picornavirus samples, was mixed and incubated at room temperature for 5 min (23). After incubation, the beads were pelleted at 1,000 × g for 10 min. The supernatant was discarded, and the pellet was washed three times with 800 μl of 60 mM MES, 150 mM NaCl (pH 6.5). To recover viral NAs, the pellet was resuspended in 200 μl of lysis buffer, and viral RNA was extracted with the QIAamp viral RNA mini kit and eluted into 100 μl water. All experiments were performed in triplicate, and viral RNA was quantitated by rRT-PCR with the EV-Sabin rRT-PCR assay (10).
Cell culture-independent detection of PV.
The Quick-RNA viral kit was selected for optimization based on viral RNA recovery from the three Sabin type 1 PV-containing stool samples (Fig. 1). To optimize workflow and RNA recovery, two RNA extractions were run from the same sample in parallel (two extractions per sample). Briefly, 400 μl of stool suspension was added to a microcentrifuge tube. Viral RNA buffer (1,200 μl) was added to the aliquoted stool suspension in the microcentrifuge tube, briefly vortex-mixed, incubated at room temperature for 1 min, and then centrifuged at 10,000 × g for 1 min. Then, 800 μl of stool suspension and viral RNA buffer mixture was removed from the tube and loaded onto a column, which was centrifuged at 10,000 × g for 1 min. The column matrix was reloaded until the entire sample-viral RNA buffer suspension (1,600 μl) was centrifuged through the matrix. Five hundred microliters of viral wash buffer was added to the column, the column was centrifuged 10,000 × g for 2 min, and the flowthrough fraction was discarded. Nuclease-free water (20 μl) was added to each column for final elution. After a 1-min incubation at room temperature, the column was centrifuged for 30 s to elute the RNA. The spent column was then discarded and replaced with the second column used for RNA extraction. This column was recentrifuged. The method yields a final volume of 40 μl viral RNA, i.e., 20 μl from each column.
Retrospective parallel testing.
Stool suspensions were prepared as described above, divided into two equal aliquots, and stored at −70°C until needed. The panel consisted of 182 previously reported PV-positive stool specimens from the African, Eastern Mediterranean, and European WHO regions. Virus isolates following the WHO standard procedures with CPE were harvested and assayed using the ITD rRT-PCR with 1 μl clarified cell culture supernatant per reaction (5, 22, 24, 25). The RNA extracted via the Quick-RNA viral kit, as described above, was tested using standard ITD rRT-PCR assays and conditions (7) with 5 μl RNA. Virus isolation by cell culture and PV detection by direct RNA extraction followed by rRT-PCR were compared.
Prospective parallel testing.
Stool suspensions were prepared, as described above, from 202 stool samples received from AFP cases in Yemen and were divided into two aliquots for RNA extraction and virus isolation, as described above.
Resolution of discordant results.
Discordant results between virus isolation and RNA extraction were resolved by sequencing the ∼900-nucleotide VP1 capsid region using the standard GPLN protocol (26). If standard sequencing was unsuccessful, then amplicons were generated with serotype-targeted primer and probe mixes from the ITD rRT-PCRs. Briefly, the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) was used to clone ITD rRT-PCR amplicons into the TOPO TA plasmid using the manufacturer’s recommended procedure, and M13 primers were used to sequence the insert using the Sanger method (27, 28). Contigs were assembled using Sequencher v5.4.6, and their identities were determined by BLAST (29).
Data management and statistical and visual analyses.
CT values were recorded for each sample and target. Results were compiled, edited, and merged using R (30). RNA yields for different RNA extraction kits were compared by using the Mann-Whitney U test (Wilcoxon rank sum test) (31). McNemar’s chi-square test with continuity correction was used for parallel testing analysis using the gmodels package (32). Data visualizations were made using the ggplot2 package in R (33).
Ethical considerations.
The CDC internal program for research determination deemed that this study is categorized as public health nonresearch.
ACKNOWLEDGMENTS
We thank our CDC colleagues Jane Iber and Naomi Dybdahl-Sissoko for input on viral inactivation methods, Kimberly Wong for RNA extraction support, Shannon Rogers for assistance with the NucliSENS easyMAG automated system, Elizabeth Henderson for sample and database management, and Qi Chen for her sequencing expertise. We thank Humayun Asghar, Ousmane Diop, and the members of the WHO Ad Hoc Small Working Group for suggestions and valuable discussions on practical conditions in GPLN laboratories.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The use of trade names is for identification only and does not imply endorsement by the CDC or the U.S. government.
Footnotes
Supplemental material is available online only.
Contributor Information
Chelsea Harrington, Email: lur1@cdc.gov.
Heba H. Mostafa, Johns Hopkins Hospital
REFERENCES
- 1.Diop OM, Kew OM, de Gourville EM, Pallansch MA. 2017. The Global Polio Laboratory Network as a platform for the Viral Vaccine-Preventable and Emerging Diseases Laboratory Networks. J Infect Dis 216(Suppl 1):S299–S307. doi: 10.1093/infdis/jix092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutter PD, Hinman AR, Hegg L, King D, Sosler S, Swezy V, Hussey AL, Cochi SL. 2017. Transition planning for after polio eradication. J Infect Dis 216(Suppl 1):S287–S292. doi: 10.1093/infdis/jix026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arita M, Kilpatrick DR, Nakamura T, Burns CC, Bukbuk D, Oderinde SB, Oberste MS, Kew OM, Pallansch MA, Shimizu H. 2015. Development of an efficient entire-capsid-coding-region amplification method for direct detection of poliovirus from stool extracts. J Clin Microbiol 53:73–78. doi: 10.1128/JCM.02384-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morales M, Tangermann RH, Wassilak SG. 2016. Progress toward polio eradication: worldwide, 2015–2016. MMWR Morb Mortal Wkly Rep 65:470–473. doi: 10.15585/mmwr.mm6518a4. [DOI] [PubMed] [Google Scholar]
- 5.Wood DJ, Hull B. 1999. L20B cells simplify culture of polioviruses from clinical samples. J Med Virol 58:188–192. doi:. [DOI] [PubMed] [Google Scholar]
- 6.Xu W, Zhang Y. 2016. Isolation and characterization of vaccine-derived polioviruses: relevance for the Global Polio Eradication Initiative, p 213–226. In Martín J (ed), Poliovirus: methods and protocols. Springer Science+Business Media, New York, NY. doi: 10.1007/978-1-4939-3292-4_10. [DOI] [PubMed] [Google Scholar]
- 7.Gerloff N, Sun H, Mandelbaum M, Maher C, Nix WA, Zaidi S, Shaukat S, Seakamela L, Nalavade UP, Sharma DK, Oberste MS, Vega E. 2018. Diagnostic assay development for poliovirus eradication. J Clin Microbiol 56:e01624-17. doi: 10.1128/JCM.01624-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu HM, Zheng DP, Zhang LB, Oberste MS, Kew OM, Pallansch MA. 2003. Serial recombination during circulation of type 1 wild-vaccine recombinant polioviruses in China. J Virol 77:10994–11005. doi: 10.1128/jvi.77.20.10994-11005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minor P. 2009. Vaccine-derived poliovirus (VDPV): impact on poliomyelitis eradication. Vaccine 27:2649–2652. doi: 10.1016/j.vaccine.2009.02.071. [DOI] [PubMed] [Google Scholar]
- 10.Wassilak S, Pate MA, Wannemuehler K, Jenks J, Burns C, Chenoweth P, Abanida EA, Adu F, Baba M, Gasasira A, Iber J, Mkanda P, Williams AJ, Shaw J, Pallansch M, Kew O. 2011. Outbreak of type 2 vaccine-derived poliovirus in Nigeria: emergence and widespread circulation in an underimmunized population. J Infect Dis 203:898–909. doi: 10.1093/infdis/jiq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorley BR, Roberts JA. 2016. Isolation and characterization of poliovirus in cell culture systems. Methods Mol Biol 1387:29–53. doi: 10.1007/978-1-4939-3292-4_4. [DOI] [PubMed] [Google Scholar]
- 12.Aylward B, Yamada T. 2011. The polio endgame. N Engl J Med 364:2273–2275. doi: 10.1056/NEJMp1104329. [DOI] [PubMed] [Google Scholar]
- 13.Buonagurio DA, Coleman JW, Patibandla SA, Prabhakar BS, Tatem JM. 1999. Direct detection of Sabin poliovirus vaccine strains in stool specimens of first-dose vaccinees by a sensitive reverse transcription-PCR method. J Clin Microbiol 37:283–289. doi: 10.1128/JCM.37.2.283-289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Previsani N, Tangermann RH, Tallis G, Jafari HS. 2015. World Health Organization guidelines for containment of poliovirus following type-specific polio eradication: worldwide, 2015. MMWR Morb Mortal Wkly Rep 64:913–917. doi: 10.15585/mmwr.mm6433a5. [DOI] [PubMed] [Google Scholar]
- 15.Old MO, Martinez CV, Kwock D, Garcia J, Martin G, Chan C, Maldonado YA. 2003. Direct extraction of Sabin poliovirus genomes from human fecal samples using a guanidine thiocyanate extraction method. J Virol Methods 110:193–200. doi: 10.1016/S0166-0934(03)00133-2. [DOI] [PubMed] [Google Scholar]
- 16.Tambini G, Andrus JK, Marques E, Boshell J, Pallansch M, de Quadros CA, Kew O. 1993. Direct detection of wild poliovirus circulation by stool surveys of healthy children and analysis of community wastewater. J Infect Dis 168:1510–1514. doi: 10.1093/infdis/168.6.1510. [DOI] [PubMed] [Google Scholar]
- 17.Gardner TJ, Diop OM, Jorba J, Chavan S, Ahmed J, Anand A. 2018. Surveillance to track progress toward polio eradication: worldwide, 2016–2017. MMWR Morb Mortal Wkly Rep 67:418–423. doi: 10.15585/mmwr.mm6714a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snider CJ, Diop OM, Burns CC, Tangermann RH, Wassilak SG. 2016. Surveillance systems to track progress toward polio eradication: worldwide, 2014–2015. MMWR Morb Mortal Wkly Rep 65:346–351. doi: 10.15585/mmwr.mm6513a3. [DOI] [PubMed] [Google Scholar]
- 19.Arita M, Masujima S, Wakita T, Shimizu H. 2010. Development of a particle agglutination method with soluble virus receptor for identification of poliovirus. J Clin Microbiol 48:2698–2702. doi: 10.1128/JCM.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honeywood MJ, Jeffries-Miles S, Wong K, Harrington C, Burns CC, Oberste MS, Bowen MD, Vega E. 2021. Use of guanidine thiocyanate-based nucleic acid extraction buffers to inactivate poliovirus in potentially infectious materials. J Virol Methods 297:114262. doi: 10.1016/j.jviromet.2021.114262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adeniji JA, Oragwa AO, George UE, Ibok UI, Faleye TOC, Adewumi MO. 2017. Preponderance of enterovirus C in RD-L20B-cell-culture-negative stool samples from children diagnosed with acute flaccid paralysis in Nigeria. Arch Virol 162:3089–3101. doi: 10.1007/s00705-017-3466-2. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization. 2004. Polio laboratory manual, 4th ed. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 23.Leggitt PR, Jaykus LA. 2000. Detection methods for human enteric viruses in representative foods. J Food Prot 63:1738–1744. doi: 10.4315/0362-028x-63.12.1738. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. 2007. S1. Supplement to the WHO Polio Laboratory Manual: an alternative test algorithm for poliovirus isolation and characterization. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 25.World Health Organization. 2015. The 21st Informal Consultation on the Global Polio Laboratory Network 25. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 26.Liu HM, Zheng DP, Zhang LB, Oberste MS, Pallansch MA, Kew OM. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J Virol 74:11153–11161. doi: 10.1128/JVI.74.23.11153-11161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorba J. 2016. Phylogenetic analysis of poliovirus sequences. Methods Mol Biol 1387:227–237. doi: 10.1007/978-1-4939-3292-4_11. [DOI] [PubMed] [Google Scholar]
- 28.Jorba J, Campagnoli R, De L, Kew O. 2008. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J Virol 82:4429–4440. doi: 10.1128/JVI.02354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.R Core Team. 2016. R: a language and environment for statistical computing, v3.3.1. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 31.Fagerland MW, Lydersen S, Laake P. 2013. The McNemar test for binary matched-pairs data: mid-p and asymptotic are better than exact conditional. BMC Med Res Methodol 13:91. doi: 10.1186/1471-2288-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trajman A, Luiz RR. 2008. McNemar χ2 test revisited: comparing sensitivity and specificity of diagnostic examinations. Scand J Clin Lab Invest 68:77–80. doi: 10.1080/00365510701666031. [DOI] [PubMed] [Google Scholar]
- 33.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer Science+Business Media, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00668-21_Supp_1_seq6.pdf, PDF file, 0.05 MB (49.3KB, pdf)