Abstract
Aim: In response to the COVID-19 pandemic, Regeneron developed the anti-SARS-CoV-2 monoclonal antibody cocktail, REGEN-COV® (RONAPREVE® outside the USA). Drug concentration data was important for determination of dose, so a two-part bioanalytical strategy was implemented to ensure the therapy was rapidly available for use. Results & methodology: Initially, a liquid chromatography-multiple reaction monitoring-mass spectrometry (LC-MRM-MS) assay, was used to analyze early-phase study samples. Subsequently, a validated electrochemiluminescence (ECL) immunoassay was implemented for high throughput sample analysis for all samples. A comparison of drug concentration data from the methods was performed which identified strong linear correlations and for Bland-Altman, small bias. In addition, pharmacokinetic data from both methods produced similar profiles and parameters. Discussion & conclusion: This novel bioanalytical strategy successfully supported swift development of a critical targeted therapy during the COVID-19 public health emergency.
Keywords: : bioanalytical, Bland-Altman, drug concentration assay, monoclonal antibody therapeutic, SARS-CoV-2
The SARS-CoV-2 virus, which causes COVID-19, was first identified in patients hospitalized with pneumonia-like symptoms in December 2019 [1]. The novel virus quickly spread around the globe and resulted in declaration by WHO of a global pandemic in March 2020. In June 2020, Regeneron began the first clinical trials of REGEN-COV for the prevention and treatment of COVID-19. REGEN-COV is a cocktail (1:1) of casirivimab and imdevimab: two human, IgG1 subclass, monoclonal antibodies that bind simultaneously to non-overlapping epitopes on the receptor binding domain of the SARS-CoV-2 spike protein, blocking normal virus entry via the angiotensin-converting enzyme 2 (ACE2) receptor [2,3].
In a public health emergency like the COVID-19 pandemic, Emergency Use Authorization (EUA) is a mechanism for the US FDA to enable rapid availability of an unapproved medicinal products for use as medicinal countermeasures. Pharmacokinetic data for the two mAbs was an important requirement to obtain an EUA for REGEN-COV, within 5 months after clinical program initiation [4,5]. Thus, a novel bioanalytical strategy was needed for swift development of an analytical method to measure the concentrations of casirivimab and imdevimab in serum from patients or subjects.
Immunoassays to detect human mAb therapeutics require highly specific anti-idiotypic Abs to distinguish between the drug and unrelated IgG in human serum. In typical development programs, drug candidates are selected and reagents are prepared prior to the commencement of clinical studies. However, given the rapid timeline for clinical development of REGEN-COV during the pandemic, these reagents were not available to develop and validate immunoassays to assess pharmacokinetics in patients at study initiation. Therefore, to support an EUA and trial progression, a two-pronged bioanalytical strategy was employed, consisting of an initial mass spectrometry-based assay and subsequent implementation of validated immunoassays.
The LC-MRM-MS assay was developed and qualified as a fit-for purpose assay in 2 months [6]. This rapid timeline was feasible because the method did not require specialized antibody reagents; the disadvantage was that it is a lower throughput method without a large capital investment. In contrast, the ECL immunoassays required lead-time to identify, scale-up and label anti-idiotypic mAbs needed for capture and detection reagents. However, once the immunoassays were developed and validated, these highly sensitive methods allowed higher throughput analysis of approximately 1500 samples per week on average for the REGEN-COV clinical program.
To support the initial EUA and early stages of the clinical trials, a subset of samples from early-phase patients were evaluated for drug concentrations using the LC-MRM-MS assay. Once the validated immunoassays were available, they were used to analyze all study samples (including re-analysis of the samples evaluated using the LC-MRM-MS method) to support the BLA [7–9]. This dual bioanalytical strategy presented a unique opportunity to directly evaluate two sets of data generated from the same samples using two orthogonal methods.
The objectives of this manuscript are to describe this strategy and to assess the agreement between the two methods employed for the measurement of casirivimab and imdevimab concentrations in serum to describe the pharmacokinetic parameters of these two mAbs in patients infected with SARS-CoV-2. Linear regression and Bland-Altman analysis [10], as well as comparison of pharmacokinetic profiles in serum and pharmacokinetic parameters (e.g., Cmax and AUC) demonstrated excellent agreement between the two methods of measurement.
Methods & materials
Clinical samples
Clinical trial serum samples were obtained from patients in Phases I, II and III of three different studies (ClinicalTrials.gov numbers NCT04426695, NT04425629 and NCT04452318). The serum samples were collected from adult patients or subjects administered a single dose of REGEN-COV (casirivimab and imdevimab) by intravenous infusion or subcutaneous injection at timepoints specified in the study protocols and ranged from baseline (prior to dosing of study drug) to 28 days post dosing. Samples were handled according to Biosafety Level 2 requirements.
Bioanalytical methods
LC-MRM-MS assay
Serum samples were denatured, reduced, alkylated and then underwent protease digestion with trypsin (Promega, WI, USA) and rAsp-N (Promega) prior to LC-MRM-MS analysis. The ratios of the peak areas from extracted ion chromatograms of the surrogate peptides derived from the variable complementarity-determining regions (CDRs) of antibody drugs and those from the corresponding internal standards were used to determine total casirivimab and total imdevimab concentrations [6]. Performance characteristics of the LC-MRM-MS method are shown in Table 1.
Table 1. Analytical parameters of the LC-MRM-MS and ECL immunoassay platforms.
| Method | Analyte | LLOQ (μg/ml) | Accuracy (%) | Precision (%) | Range (μg/ml) | Development time | Analysis throughput |
|---|---|---|---|---|---|---|---|
|
LC-MRM-MS |
Casirivimab | 20 | 95–103 | 3–13 | 20–2000 |
Shorter |
Lower |
| Imdevimab | 10 | 96–106 | 3–9 | 10–2000 | |||
|
ECL immunoassay |
Casirivimab | 0.156 | 93–103 | 7–11 | 0.156–10 |
Longer |
Higher |
| Imdevimab | 0.156 | 94–103 | 8–10 | 0.156–10 |
Reagents for ECL immunoassays
Mouse anti-idiotype monoclonal antibodies to casirivimab or to imdevimab were generated in-house (Regeneron Pharmaceuticals). As capture reagents, anti-casirivimab and anti-imdevimab mAbs were biotinylated according to manufacturer's instructions, using EZ-link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific). For detection, different, noncompeting anti-casirivimab and anti-imdevimab mAbs were ruthenylated according to manufacturer's instructions, using ruthenium NHS ester (MSD). A combination of casirivimab and imdevimab, in equal concentrations, is used as a standard. Serum samples with a known concentration of casirivimab and imdevimab in combination are used as high, mid and low quality controls.
ECL immunoassays
Two ECL immunoassays were developed to determine the concentrations of total casirivimab and total imdevimab in human serum samples as previously described [6–9]. Briefly, the assay procedures employ streptavidin microplates coated with either biotinylated mouse anti-casirivimab monoclonal antibody, or biotinylated mouse anti-imdevimab monoclonal antibody. Casirivimab and imdevimab captured on plates specific for each molecule are detected using two ruthenylated, noncompeting mouse monoclonal antibodies that are specific to either casirivimab or imdevimab. An electrochemiluminescent signal is generated by the ruthenium label when voltage is applied to the plate by the MSD reader. The measured electrochemiluminescence (i.e., counts) is proportional to the concentration of total casirivimab or total imdevimab in the serum samples. The performance characteristics of these bioanalytical methods were evaluated separately, for each anti-SARS-CoV-2 antibody, using multiple independent validation experiments, with respect to linearity, accuracy, precision, specificity, selectivity, dilution linearity, robustness and analyte stability [11,12]. Performance characteristics of the ECL immunoassays are shown in Table 1.
Statistical analysis
Linear regression was performed for sample analysis results from LC-MRM-MS and ECL immunoassay platforms for casirivimab and imdevimab (Figure 1). The 95% prediction intervals, coefficient of determination (r2), and slope of the line of best fit were determined. For casirivimab and imdevimab sample analysis results, normality of the differences between measures was determined (Figure 2) and then a Bland-Altman analysis was performed to plot the mean of methods against the difference of methods (Figure 3A & B) and against the percent difference of methods (Figure 3C & D). The bias and 95% limits of agreement were determined for each analysis.
Figure 1. The linear regression between the ECL immunoassay and the LC-MRM-MS assay results.
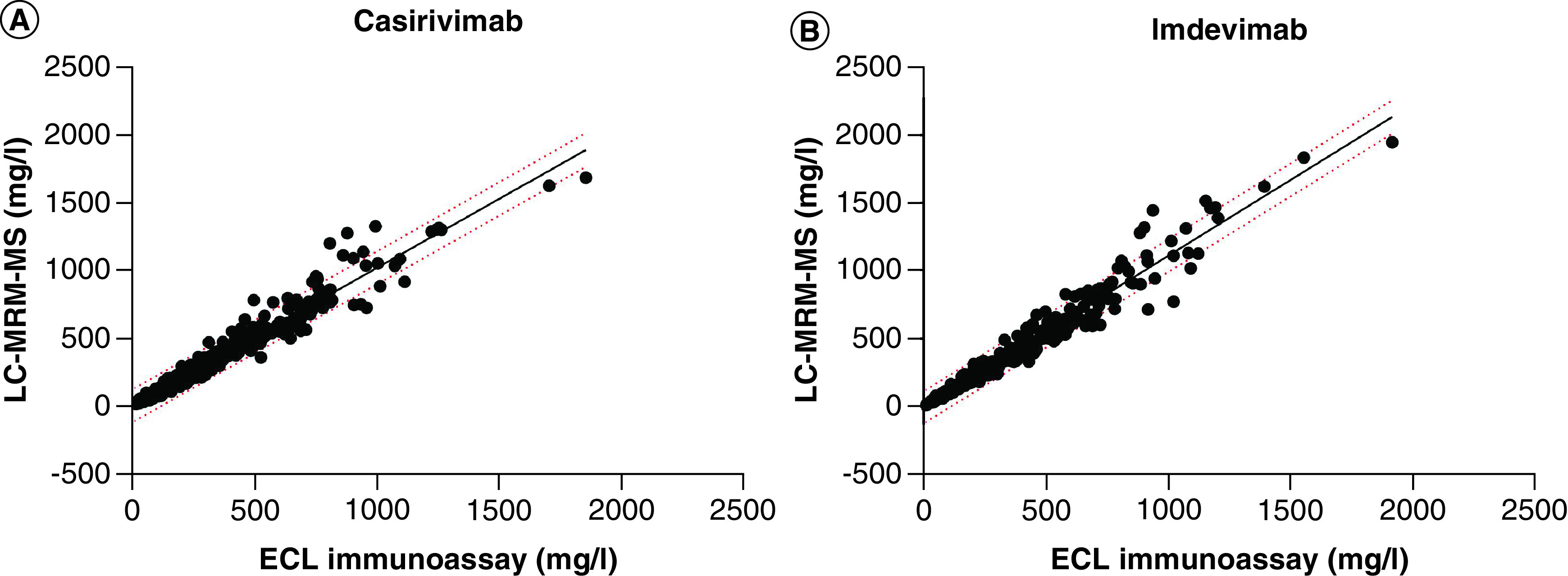
(A) For casirivimab, the correlation coefficient between the two methods is r2 = 0.9568, p < 0.0001 and slope = 1.014. (B) For imdevimab, the correlation coefficient between the two methods is r2 = 0.9562, p < 0.0001, and slope = 1.117. The 95% prediction intervals are shown with dotted lines.
Figure 2. The frequency distribution of logarithmic transformed (Log10) differences between the ECL immunoassay and the LC-MRM-MS assay for casirivimab and imdevimab.
The D'Agostino and Pearson test for normal distribution (A) for casirivimab (p = 0.2394) and (B) for imdevimab (p = 0.2099).
Figure 3. Bland-Altman analyses for the ECL immunoassay and the LC-MRM-MS assay.
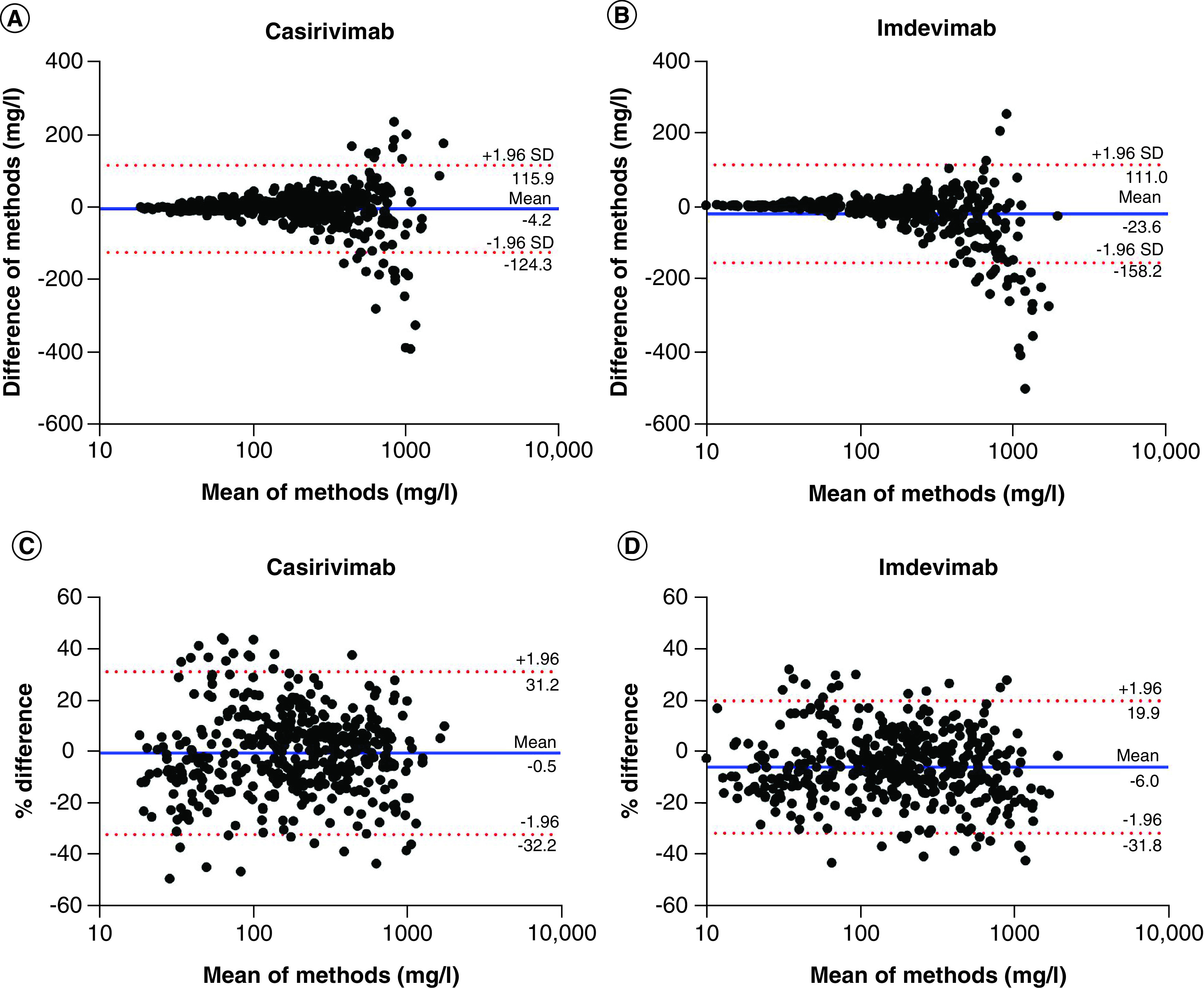
(A & B) The x-axis in each plot is the mean of the two measures and the y-axis is the difference between methods (ECL immunoassay result minus LC-MRM-MS assay result). (A) For casirivimab (n = 424), the bias from immunoassay to LC-MRM-MS (±SD) is -4.2 (±61.3; 95% limits of agreement -124.3 to 115.9). (B) For imdevimab (n = 444), the bias from immunoassay to LC-MRM-MS (±SD) is -23.6 (±68.7; 95% limits of agreement -158.2 to 111.0). (C & D) The x-axis in each plot is the average of the two measures and the y-axis is the percentage of the values on the axis ([ECL immunoassay result minus LC-MRM-MS assay result], divided by mean and expressed as a percentage). (C) For casirivimab, the bias from immunoassay to LC-MRM-MS (±SD) is -0.5% (±16.2; 95% limits of agreement -32.2 to 31.2). (D) For imdevimab, the bias from immunoassay to LC-MRM-MS (±SD) is -6.0% (±13.17; 95% limits of agreement -31.8 to 19.9).
Visual comparison of concentration in serum by time was performed to assess the pharmacokinetic similarities in datasets generated from the same sample by each of the two analysis methods. Pharmacokinetic parameters of concern (Cmax, AUC0–28) were generated by noncompartmental analysis, as well as concentration in serum at 28 days post-dose (C28).
All statistical analyses were performed in GraphPad Prism 9 (GraphPad Software LLC). Concentration time data were plotted using ggplot 2 package in R [13]. Noncompartmental analysis was performed in Phoenix® WinNonlin®, v 8.3, Certara.
Results
Description of dataset for analysis
For the comparative analysis, sample data were combined from three studies where concentrations were available on both platforms (LC-MRM-MS and ECL immunoassay). From the casirivimab and imdevimab datasets, 6 and 1 analytical outliers (representing 1.4 and 0.22% of the data), respectively, were identified by ROUT method and were removed from the LC-MRM-MS and ECL immunoassay datasets; the ROUT method can detect any number of outliers and is based on the false discovery rate [14]. A total of 424 and 444 drug concentration results for casirivimab and imdevimab, respectively, from both methods were used for comparison.
Linear regression & Bland-Altman analysis
To determine the relationship between the datasets generated with the LC-MRM-MS method and ECL immunoassays, a linear regression analysis was performed (Figure 1). The correlation coefficient (r2) between the two different platforms for casirivimab and imdevimab was r2 = 0.957 and r2 = 0.956, respectively, and p < 0.0001 for both. As expected, a linear relationship was observed between the datasets, with a slope of ∼1 indicating similarities between measurements from two datasets.
Bland-Altman analysis is a procedure to compare the agreement between two different measures [15]. Correlation analysis assesses the relationship between two measurements, while the Bland-Altman method examines the mean differences and estimates an agreement interval. Using the 424 drug concentration results for casirivimab and 444 for imdevimab, the differences were calculated between measures obtained on the two different platforms. This type of analysis assumes a normal distribution of the mean differences. Therefore, a frequency distribution was plotted of the logarithmic transformed differences between the ECL immunoassay and LC-MRM-MS method datasets for casirivimab and for imdevimab (Figure 2). Using the differences between measures, the D'Agostino and Pearson test for normal distribution resulted in p = 0.239 for casirivimab data and p = 0.210 for imdevimab data, demonstrating the log-transformed differences were normally distributed.
In the Bland-Altman plots, the mean of the different measures (LC-MRM-MS method result and ECL immunoassay result) per sample was plotted against the difference of methods (i.e., ECL immunoassay result minus LC-MRM-MS result) (Figure 3A & B) or against the percentage of differences (i.e., ECL immunoassay result minus LC-MRM-MS result, divided by mean and expressed as a percentage) (Figure 3C & D) per sample for each mAb. The 95% limits of agreement were calculated for casirivimab and imdevimab: from -124.3 to 115.9 mg/l and -32.2 to 31.2% for casirivimab; from -158.2 to 111.0 mg/l and -31.8 to 19.9% for imdevimab. The mean of the differences and the standard deviation of the differences between the measurements acquired with the two different platforms were also calculated for each mAb, demonstrating a small negative bias from the immunoassay to the LC-MRM-MS method bias (±SD) of -4.2 (±61.28) mg/l and -0.5% (±16.2) for casirivimab and 23.6 (±68.7) mg/l and -6.0% (±13.2) for imdevimab.
Drug concentration data & pharmacokinetic parameters
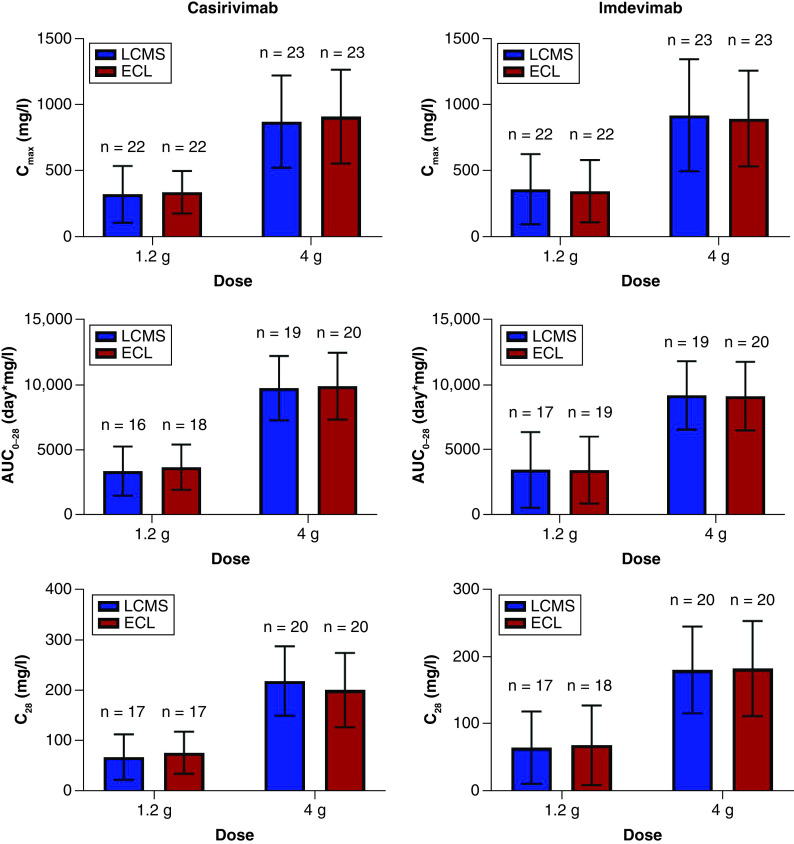
At all timepoints through 28 days after dosing, the concentrations in serum as measured by LC-MRM-MS method and ECL immunoassay are similar at both doses (Figure 4), suggesting that both methods will provide similar pharmacokinetic evaluation of the antibodies. These pharmacokinetic similarities were further confirmed by comparing the exposure of each antibody at both doses. Maximum concentration in serum (Cmax), area under concentration by time curve up to 28 days (AUC0–28), and concentration in serum at 28 days after dose (C28), determined by analysis of data generated by the LC-MRM-MS method and ECL immunoassay are not significantly different (Figure 5).
Figure 4. Comparison of drug concentration in serum by time for casirivimab (left) and imdevimab (right) in hospitalized (Study 2066, top) and non-hospitalized (Study 2067, bottom) patients with SARS-CoV-2.
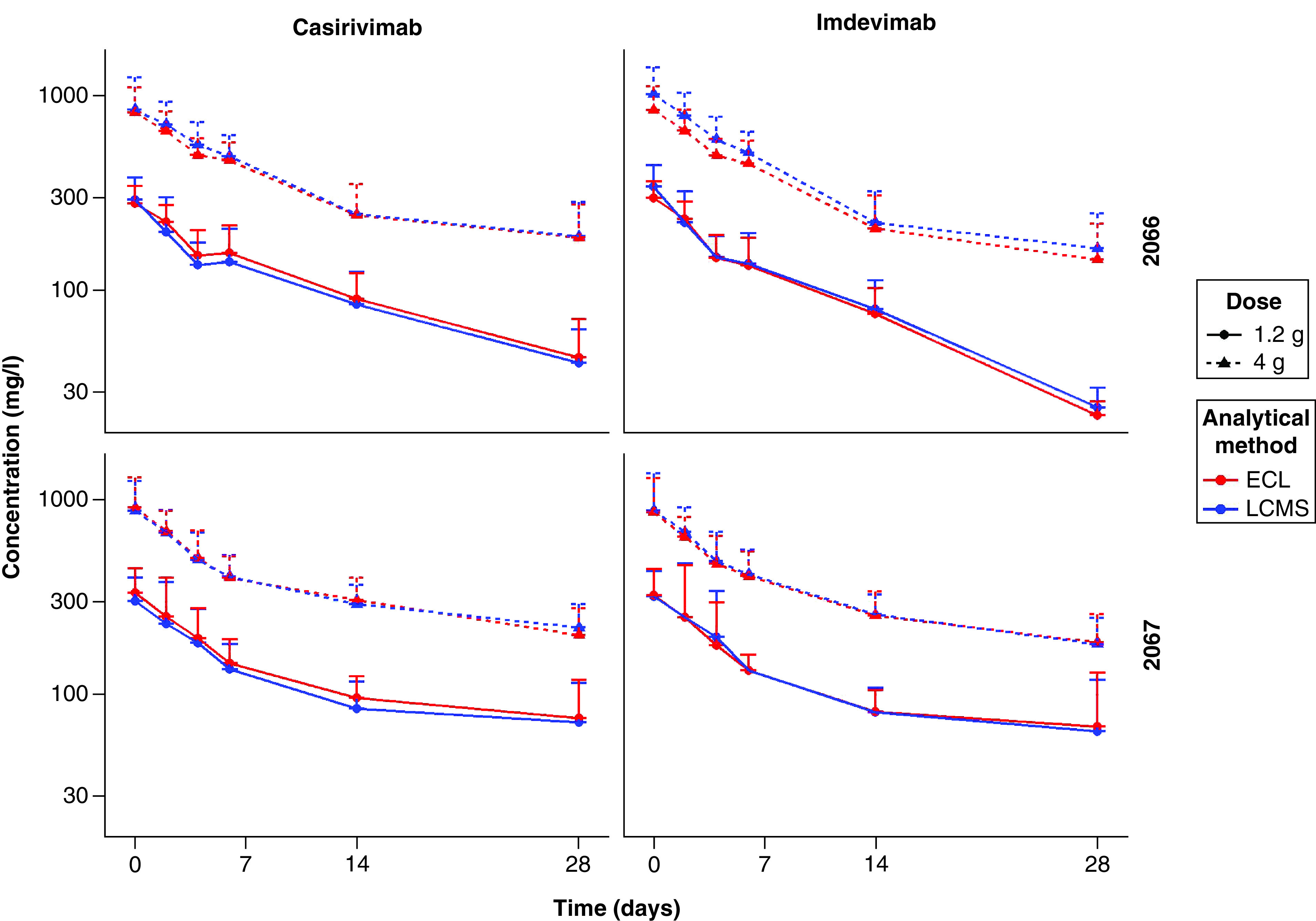
Error bars indicate standard deviation.
Figure 5. Pharmacokinetic parameters per platform for each casirivimab (left) and imdevimab (right) in non-hospitalized patients with SARS-CoV-2 diagnosis (Study 2067).
For the 1.2 g and 4 g doses per mAb, the following parameters are presented as mean ± standard deviation (SD) with number of subjects identified for each calculation (n): (top) maximum drug concentration in blood (Cmax), (middle) area under concentration by time curve up to 28 days post-dose (AUC0–28), and (bottom) drug concentration in serum at 28 days after dose (C28).
Discussion
During the clinical development of the REGEN-COV anti-SARS-CoV2 monoclonal antibody cocktail, a novel two-part bioanalytical approach was employed to measure drug concentrations. Initially a fit-for-purpose LC-MRM-MS method was used to analyze a subset of patient samples from early-phase studies to support advancement to later stages in the clinical studies and EUA submission. Subsequently, two validated immunoassays were used to analyze all samples (including re-analysis of samples evaluated by LC-MRM-MS) to support application for formal health authority approval. The determination of concentrations of two mAb therapeutics in clinical study samples with two analytical platforms permitted a comprehensive comparison of the two measurements in human samples.
The linear regressions for casirivimab and for imdevimab drug concentration data obtained by LC-MRM-MS method and ECL immunoassay identified strong, positive correlations that are highly significant. In addition, a wide range of sample concentrations was used for Bland-Altman comparison: from 17.6 to 1860.0 mg/l for casirivimab and from 9.94 to 1910.0 mg/l for imdevimab.
Furthermore, comparable concentrations in serum by time profile and exposure up to 28 days (AUC0–28) signifies that both analytical methods were able to characterize the pharmacokinetics of each antibody similarly, regardless of dose. Comparable Cmax as well as C28 signifies no bias in measurement of higher or lower concentrations.
Accurately and specifically quantitating human mAb drugs in complex biological matrices is challenging. Immunoassays use highly specific anti-idiotypic reagents to distinguish between each mAb therapeutic and unrelated human IgG in the sample, whereas LC-MRM-MS methods detect unique surrogate peptides derived from CDRs from the mAbs. Each method has advantages and disadvantages and when implemented in a coordinated manner, they can be used to complement each other [16].
Using the available variable domain amino acid sequence information to identify unique surrogate peptide candidates, LC-MRM-MS methods can be developed and qualified as fit-for-purpose assays in less time than is needed to produce highly specific anti-idiotype reagents needed for immunoassays. More specifically, for the REGEN-COV drug development program, the LC-MRM-MS assay was developed and qualified as a fit-for purpose assay in merely 2 months [6]. This rapid timeline was feasible because the method did not require specialized antibody reagents; the disadvantage was that it is a lower throughput method without a large capital investment. In contrast, the ECL immunoassays were slower to develop because of lead-time to identify, scale-up, and label anti-idiotypic mAbs needed for assay capture and detection reagents. However, the higher throughput potential for sample analysis via immunoassays is advantageous for long-term use due to greater throughput and wide use in the bioanalytical community. In order to further improve the throughput of ECL immunoassays, hybrid automation systems were employed, which allowed both mAbs to be analyzed simultaneously. Samples and controls containing both mAbs were diluted using the hybrid automation system; all subsequent assay steps were then completed manually. This allowed for an additional ∼50% increase on the number of samples analyzed per batch compared with what is typically run with a fully manual immunoassay. To date, the ECL immunoassays have been used to analyze more than 60,000 samples in 10 months for the REGEN-COV drug development program.
Chromatographic methods typically quantitate total drug levels, whereas immunoassays may detect different species (e.g., total, free, active) depending on how the assay is configured, the presence of soluble target or neutralizing anti-idiotype antibodies [17,18]. The good agreement between the two drug concentration methods presented here confirms the two platforms detect the same analyte. This suggests that there is an insufficient systemic concentration of target to impact drug clearance or to interfere in the assay. In addition, immunogenicity is low for both casirivimab and imdevimab with no apparent impact on PK [19]. Thus, for this therapeutic, neutralizing anti-idiotype antibodies are unlikely to interfere with the capture or detection step in the ECL assay. Therefore, both the LC-MRM-MS and the immunoassay methods detect total drug concentrations of casirivimab and imdevimab.
The sensitivity of the LC-MRM-MS and ECL immunoassay methods differed substantially: 10–20 mg/l for LC-MRM-MS and 0.156 mg/l for ECL immunoassay. However, the sensitivity of the LC-MRM-MS method was suitable for the early timepoints (up to day 28 post-dosing) analyzed in a subset of samples from early phases of the clinical studies. It is also possible to achieve greater sensitivity with LC-MS methods by employing a capture step with specific anti-idiotype antibodies or drug target (in this case the RBD of the spike protein) [20], however this would lengthen the development time needed for the LC-MRM-MS method. In this clinical program, the more sensitive immunoassays were used to analyze all study samples, including later timepoints important to understand additional PK parameters (e.g., terminal half-life), that were not analyzed with the LC-MRM-MS methods.
Conclusion
This study examined a coordinated dual-platform bioanalytical approach that aided the rapid clinical development of a mAb therapeutic, which was crucial during the COVID-19 healthcare emergency. The drug concentration data obtained with the LC-MRM-MS and immunoassay platforms were comparable and there was good agreement between the measures. The pros and cons of the respective methods complemented each other and permitted the strategic choice to use both analytical techniques. This approach contributed to the overall success of the clinical development of the therapeutic, REGEN-COV.
As the SARS-CoV-2 virus persists in our communities and continues to mutate, the need for effective therapies such as REGEN-COV will remain [21]. During future pandemics when mAb therapies are needed to combat infectious pathogens, or when the need arises to investigate new anti-SARS-CoV-2 spike antibodies, we have successfully demonstrated that a strategy where two orthogonal methods can be implemented sequentially to measure drug concentrations. Data obtained from LC-MRM-MS and from ECL immunoassays can yield highly comparable data and can be used successfully to understand the pharmacokinetic properties of biotherapeutics.
Future perspective
Historically, small and large molecule bioanalysis was performed on different analytical platforms, LC-MS for small molecules and ligand binding assays (LBA) for biologics. However, immunoassay and chromatographic methods are being used increasingly in new ways for analysis of both small and large molecules, analytes from drug programs with novel modalities and even immunogenicity assessment [22–24].
In some bioanalytical scenarios, unlike the assays described for REGEN-COV, sample analysis results may differ between assay platform. These differences may be beneficial for the pharmacokineticist if they require total drug concentrations (LC-MS) as compared with free/active drug (LBA) that is not bound to target or to neutralizing anti-idiotype antibodies (or vice versa). Again, these different analytical approaches can be complementary and provide a richer understanding of the drug disposition [25,26].
The work presented here is further evidence that these two platforms, and also other techniques such flow cytometry and quantitative PCR, are not competing methodologies but rather components of a suite of bioanalytical tools that can be implemented in a complementary fashion [16]. Neither approach is inherently better than the other. Instead, each technique has strengths and weaknesses and the role of the bioanalytical scientist is to select the most appropriate approach to satisfy the needs at each point of the drug development program.
Summary points.
Background
In response to the COVID-19 pandemic, Regeneron developed the anti-SARS-CoV-2 monoclonal antibody cocktail REGEN-COV® (RONAPREVE® outside of USA) which consists of two neutralizing noncompeting antibodies, casirivimab and imdevimab, that bind the SARS-CoV-2 spike protein.
Drug concentration data was important for determination of dose and dosing regimens, so a two-part bioanalytical strategy was implemented to ensure the therapy was rapidly available for use.
Initially, a liquid chromatography-multiple reaction monitoring-mass spectrometry (LC-MRM-MS) assay was developed and qualified as a fit-for-purpose assay to analyze early-phase clinical study samples. Subsequently, a validated electrochemiluminescence (ECL) immunoassay was implemented for high throughput analysis for all samples.
Results & methodology
A statistical comparison was performed for drug concentration data using serum samples analyzed in both methods.
The comparison identified strong linear correlations and minimal bias by Bland-Altman analysis.
In addition, similar drug concentration versus time profiles and pharmacokinetic parameters were obtained using data from the different analytical methods.
Discussion & conclusion
This bioanalytical strategy successfully supported swift development of a critical targeted therapy during the COVID-19 public health emergency and demonstrates that orthogonal methods may be successfully implemented to aid rapid clinical drug development.
Acknowledgments
The authors thank the patients who participated in these trials and the investigators at the individual study sites. We thank the other members of the Bioanalytical Sciences COVID-19 Team members including A Denizard, A Fodera, A Vallejos, B Capparelli, A Patel, B Moran, I Perez, J Rodriguez, J Sutherland, K Kameron, L Colligan, M Lorenz and Y Liu for their essential contributions to all of the COVID-19 studies.
Disclaimer
The views expressed in this article are those of the authors and do not reflect official policy of Regeneron Pharmaceuticals, Inc.
Ethical conduct of research
For investigations involving human subjects, informed consent has been obtained from the participants involved.
Data sharing statement
The authors certify that this manuscript reports original clinical trial data (NCT04426695, NT04425629, and NCT04452318). No individual participant data that underlies the results reported in the article will be available, and nor will information regarding the study protocol or statistical analysis, beyond what is reported in the text, and what is available on ClinicalTrials.gov.
Footnotes
Financial & competing interests disclosure
This research was funded by Regeneron Pharmaceuticals, Inc. Two clinical trials were funded by the Biomedical and Advanced Research and Development Authority of the Department of Health and Human Services; ClinicalTrials.gov numbers, NCT04426695 and NT04425629. One clinical trial was funded by Regeneron and NIAID; ClinicalTrials.gov number NCT04452318. All authors are employees of Regeneron Pharmaceuticals, Inc. and may hold stock and/or stock options in the company. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest
- 1.Zhu N, Zhang D, Wang W et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382(8), 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum A, Fulton BO, Wloga E et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369(6506), 1014–1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen J, Baum A, Pascal KE et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369(6506), 1010–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) OF REGEN-COV (casirivimab and imdevimab). US FDA; (2021). [Google Scholar]
- 5.Weinreich DM, Sivapalasingam S, Norton T et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 384(3), 238–251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; • The drug concentration data generated with the LC-MRM-MS method is presented in this manuscript.
- 6.Zhong X, Nayak S, Guo L et al. Liquid chromatography-multiple reaction monitoring-mass spectrometry assay for quantitative measurement of therapeutic antibody cocktail REGEN-COV concentrations in COVID-19 patient serum. Anal. Chem. 93(38), 12889–12898 (2021). [DOI] [PubMed] [Google Scholar]; • Details the method used to acquire drug concentration data for the REGEN-COV drug-development program and was subsequently used for the analyses presented here.
- 7.O'Brien MP, Forleo-Neto E, Sarkar N et al. Subcutaneous REGEN-COV antibody combination in early SARS-CoV-2 infection. medRxiv (2021). [Google Scholar]
- 8.O'Brien MP, Forleo-Neto E, Musser BJ et al. Subcutaneous REGEN-COV antibody combination to prevent Covid-19. N. Engl. J. Med. 385(13), 1184–1195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinreich DM, Sivapalasingam S, Norton T et al. REGEN-COV antibody combination and outcomes in outpatients with Covid-19. N. Engl. J. Med. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; • The drug concentration data generated with the ECL immunoassay is presented in this manuscript.
- 10.Giavarina D. Understanding Bland Altman analysis. Biochem Med (Zagreb) 25(2), 141–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides guidance on performing Bland-Altman analysis and on interpretation.
- 11.Bioanalytical Method Validation Guidance for Industry. U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER), CfVMC; (2018). [Google Scholar]
- 12.Booth B, Stevenson L, Pillutla R et al. 2019 White Paper On Recent Issues in Bioanalysis: FDA BMV Guidance, ICH M10 BMV Guideline and Regulatory Inputs (Part 2 - Recommendations on 2018 FDA BMV Guidance, 2019 ICH M10 BMV Draft Guideline and Regulatory Agencies' Input on Bioanalysis, Biomarkers and Immunogenicity). Bioanalysis 11(23), 2099–2132 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer International Publishing, NY, USA: (2016). [Google Scholar]
- 14.Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformat. 7, 123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. J. Royal Statistical Soc. 32(3), 307–317 (1983). [Google Scholar]
- 16.Papadimitriou A, Bansal S, Heinrich J, Staack RF. Can LC-MS/MS and ligand-binding assays live in harmony for large-molecule bioanalysis? Bioanalysis 6(13), 1735–1737 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Schick E, Staack RF, Haak M et al. Validation of a ligand-binding assay for active protein drug quantification following the ‘free analyte QC concept’. Bioanalysis 8(24), 2537–2549 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Staack RF, Jordan G, Viert M, Schafer M, Papadimitriou A, Heinrich J. Quantification of a bifunctional drug in the presence of an immune response: a ligand-binding assay specific for ‘active’ drug. Bioanalysis 7(24), 3097–3106 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Report on the Deliberation Results and Report on Special Approval for Emergency: ronapreve. Japan Pharmaceuticals and Medical Devices Agency Pharmaceutical Evaluation Division, PSaEHBMoH, Labour and Welfare; (2021). [Google Scholar]
- 20.Yuan M, Ismaiel OA, Mylott WRJ. Hybrid ligand binding immunoaffinity-liquid chromatography/mass spectrometry for biotherapeutics and biomarker quantitation: how to develop a hybrid LBA-LC-MS/MS method for a protein? Rev. Separation Sci. 1(1), 47–55 (2019). [Google Scholar]
- 21.Copin R, Baum A, Wloga E et al. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell 184(15), 3949–3961 e3911 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Liu L, Yu N et al. Immunoaffinity LC-MS/MS is more suitable than ELISA to quantify a PEGylated molecule in cynomolgus monkey serum. Bioanalysis 12(15), 1061–1069 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Huang X, Xu X, Partridge MA et al. Isotyping and semi-quantitation of monkey antiidiotype antibodies by immunocapture liquid chromatography-mass spectrometry. AAPS J. 23(1), 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neubert H, Alley SC, Lee A et al. 2020 White Paper on Recent Issues in Bioanalysis: BMV of Hybrid Assays, Acoustic MS, HRMS, Data Integrity, Endogenous Compounds, Microsampling and Microbiome (Part 1 – Recommendations on Industry/Regulators Consensus on BMV of Biotherapeutics by LCMS, Advanced Application in Hybrid Assays, Regulatory Challenges in Mass Spec, Innovation in Small Molecules, Peptides and Oligos). Bioanalysis. 13(4), 203–238 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Wang SJ, Wu ST, Gokemeijer J et al. Attribution of the discrepancy between ELISA and LC-MS/MS assay results of a PEGylated scaffold protein in post-dose monkey plasma samples due to the presence of antiidiotype antibodies. Anal Bioanal Chem. 402(3), 1229–1239 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Gu H, Liu A et al. Antibody-drug conjugate bioanalysis using LB-LC-MS/MS hybrid assays: strategies, methodology and correlation to ligand-binding assays. Bioanalysis. 8(13), 1383–1401 (2016). [DOI] [PubMed] [Google Scholar]