SUMMARY
In this review, we examine the fungal spore killers. These are meiotic drive elements that cheat during sexual reproduction to increase their transmission into the next generation. Spore killing has been detected in a number of ascomycete genera, including Podospora, Neurospora, Schizosaccharomyces, Bipolaris, and Fusarium. There have been major recent advances in spore killer research that have increased our understanding of the molecular identity, function, and evolutionary history of the known killers. The spore killers vary in the mechanism by which they kill and are divided into killer-target and poison-antidote drivers. In killer-target systems, the drive locus encodes an element that can be described as a killer, while the target is an allele found tightly linked to the drive locus but on the nondriving haplotype. The poison-antidote drive systems encode both a poison and an antidote element within the drive locus. The key to drive in this system is the restricted distribution of the antidote: only the spores that inherit the drive locus receive the antidote and are rescued from the toxicity of the poison. Spore killers also vary in their genome architecture and can consist of a single gene or multiple linked genes. Due to their ability to distort meiosis, spore killers gain a selective advantage at the gene level that allows them to increase in frequency in a population over time, even if they reduce host fitness, and they may have significant impact on genome architecture and macroevolutionary processes such as speciation.
KEYWORDS: genomic conflict, meiotic drive
INTRODUCTION
A foundational assumption in the study of genetics and evolution is that allele transmission between generations is fair. Specifically, Mendel’s law of segregation specifies that each allele comprising a heterozygous locus has a 50% chance of being passed to a given offspring (1). For natural selection, this equity serves an analogous role to that of fair tryouts for a sports team: it facilitates the process of choosing the best players (alleles) by giving each an unbiased opportunity to demonstrate their worth (fitness) (2).
Not all alleles, however, compete on an equal playing field. Some alleles, known as meiotic drivers, cheat during sexual reproduction to increase their transmission in the next generation (3, 4). For example, in a driver+/driver− heterozygote, the driver+ allele is passed to more than half, and, in some cases, all of the viable progeny. Because drivers spread at the expense of competing alleles, they can short-circuit natural selection’s ability to increase the frequency of the best-adapted alleles over generations. Drivers may even increase in spite of conferring a fitness cost to the host, and thereby driver alleles are usually considered selfish or parasitic (5–8).
Meiotic drive alleles are widespread in eukaryotes and can be divided into two major groups based on the stage in which they act. The first group of drivers is often called true meiotic drivers because they act during meiosis (9). The best-known examples of true meiotic drivers bias meiotic chromosome segregation so that they are preferentially transmitted to the gamete, as opposed to the polar body, during female (asymmetric) meiosis (10–12). The second group of drivers acts after the meiotic divisions and can be described as killer meiotic drivers because they gain a transmission advantage by destroying meiotic products that do not inherit the drive allele (13). Due to differences in their cheating mechanisms, these two groups differ in the level of cost they impose on a host’s ability to produce offspring. In true drive in female meiosis, the driver allele replaces the alternative allele without destroying meiotic products and thereby imposes little or no direct cost on host gamete formation. Killer meiotic drivers, on the other hand, kill the meiotic products carrying the alternative allele, which results in a reduction of the total number of viable meiotic products. The different mechanisms of drive also impact the evolutionary advantage of the driver allele. With true meiotic drive in female meiosis, the driver allele should increase in absolute number of copies each generation, while killer meiotic drive should result in only a relative increase of the driver allele because meiotic products carrying the alternative allele are killed and not replaced (6, 14).
Despite the identification of many drivers, studies of the mechanisms and evolutionary impacts of meiotic drive have been impeded by the notorious genetic intractability of most known drivers. However, in the last 10 years, meiotic drivers found in fungi of the Ascomycota phylum have emerged as important and tractable model systems. Although ascomycetes contain both true drivers (15–17) and killer meiotic drivers, we focus this review on the latter group, which are collectively known as spore killers. We first provide a general overview of what spore killing is. We then introduce the spore killers and describe what is known of their molecular mechanisms. Finally, we describe the population dynamics of spore killers and their potential impact on genome evolution and speciation.
WHAT IS SPORE KILLING?
Spore killing is a phenomenon found in several species of ascomycete fungi. Ascomycetes are a diverse group that includes both filamentous fungi and single-celled yeasts. A majority of ascomycetes spend the predominant part of their life cycle as haploids, but, during mating, nuclei fuse to form diploids, which subsequently undergo meiosis to generate haploid nuclei that are packaged in ascospores. The Ascomycota phylum is named for the common feature that the organisms package their sexual spores within sacs called asci (18). This feature has been a boon to genetics researchers for generations, because each ascus contains all of the products from a single meiosis, which allows for the direct assessment of gene segregation and independent assortment (Fig. 1) (19–21). When spore killing occurs, spores that do not carry the killer are destroyed (Fig. 2) (22). It is obvious that spore killing has a major impact on fungal fitness, especially given that the meiotic products of ascomycete fungi are offspring and not gametes, as in animals and plants (14).
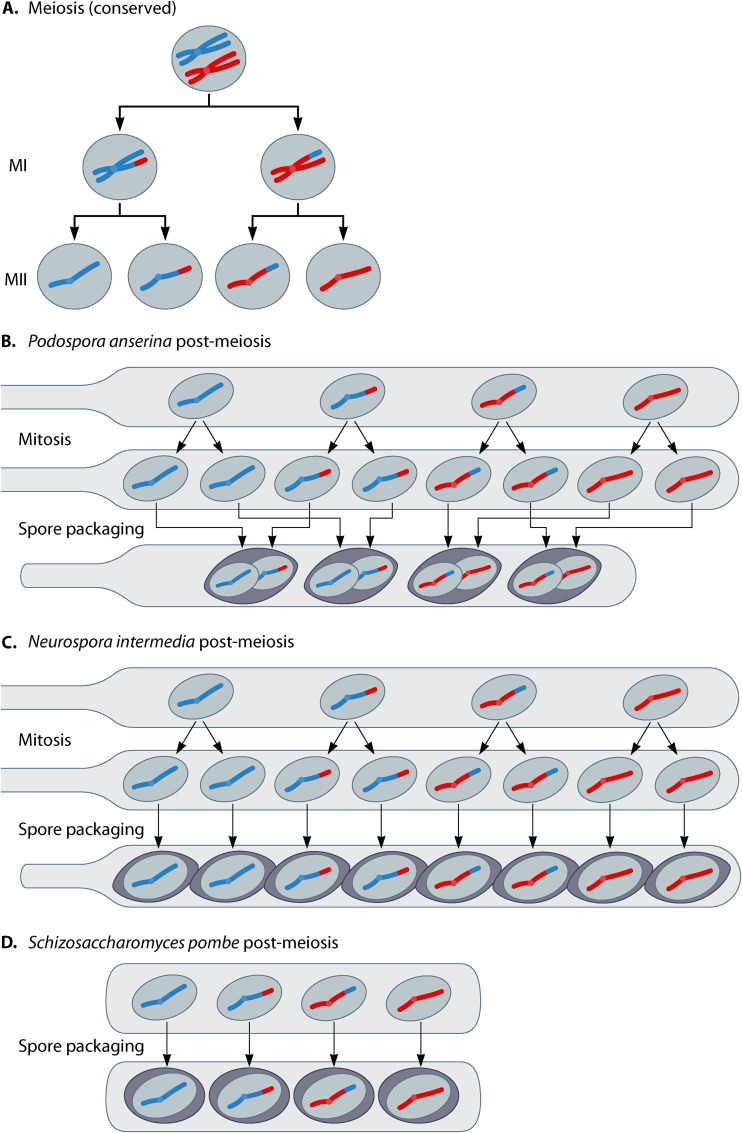
FIG 1.
Meiosis and spore packaging in select ascomycetes. (A) The basic steps of meiosis are shown for a hypothetical nucleus with one pair of homologous chromosomes. The products of the first and second meiotic divisions are abbreviated MI and MII, respectively. (B) In Podospora anserina, the products of MII undergo a mitotic division prior to packaging into four spores by following the indicated patterns. There are two segregation patterns for alleles that are particularly important for spore-killing phenotypes in Podospora. Heterozygous alleles undergo first-division segregation (FDS) if they are pulled to opposite poles during the first meiotic division. Alleles that undergo FDS are packaged into separate spores (e.g., in the diagram, chromosomes in each spore have either blue short arms or red short arms, not both). Heterozygous alleles undergo second-division segregation (SDS) if they are separated at the second meiotic division. After spore packaging, all alleles demonstrating SDS are found in all spores (e.g., in the diagram, each spore inherits both a blue and a red long-arm telomere allele). (C) In Neurospora intermedia, the meiotic products also undergo a mitotic division, but each of the resulting eight nuclei is packaged into individual spores. (D) In Schizosaccharomyces pombe, the four products of meiosis are directly packaged into spores.
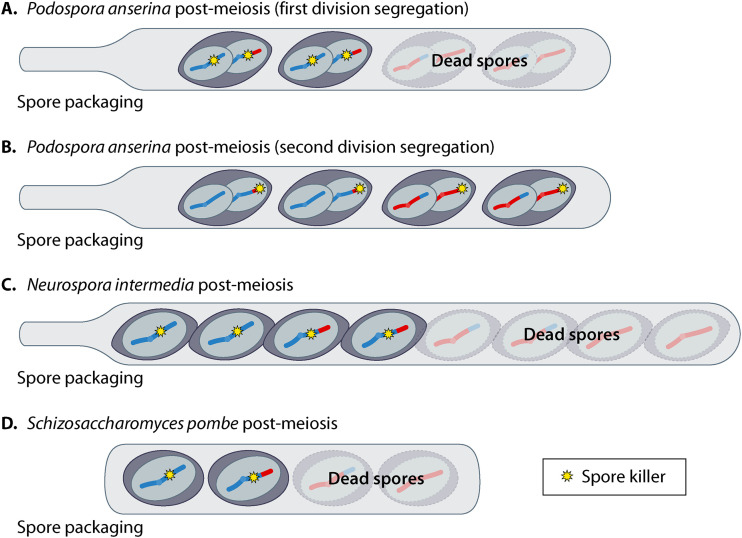
FIG 2.
Spore killing in select ascomycetes. In all images, the spore-killing haplotype is indicated with a yellow star. (A) A spore killer showing a first-division segregation pattern in Podospora anserina. This spore killer would destroy the two spores that do not inherit the killer haplotype regardless of the particular mechanism (i.e., killer-target or poison-antidote). (B) A spore killer showing a second-division segregation pattern in Podospora anserina. If this spore killer employs a killer-target mechanism, all spores would be destroyed (not shown) as each inherits the spore-killing haplotype (star) and the alternate allele that contains the target. If this spore killer employs a poison-antidote mechanism, all spores would survive (as shown) because each would receive the antidote. (C and D) Spore killing in Neurospora intermedia (C) and Schizosaccharomyces pombe (D), where half the spores are destroyed by a fully penetrant spore killer, regardless of mechanism.
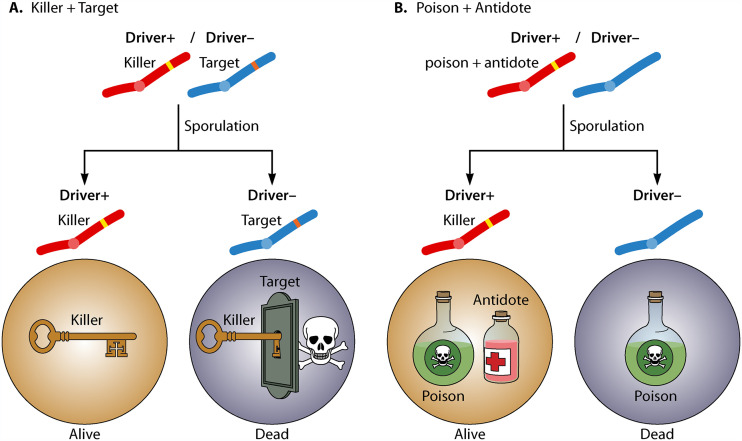
Like all killer meiotic drivers, spore killers must perform two key functions to cause drive (13). The first is that the driver must distinguish between spores that inherit the drive locus and those that do not. The second key function is that the driver must destroy spores that do not inherit the drive locus. Together, these two functions ensure that spores carrying the spore killer locus survive, while spores without the locus are destroyed. Spore killers employ one of two strategies to execute these two key functions, and they can be split into two mechanistic categories: killer-target drivers and poison-antidote drivers (Fig. 3). In killer-target drive systems, the drive locus encodes an element that can be described as a killer. All developing spores are exposed to the killer, but the killer is only detrimental to spores that inherit a second element, referred to as the target. In all known killer-target drive systems, the target is an allele found tightly linked to the drive locus but on the nondriving haplotype. In other words, an individual harboring the driving allele lacks the target allele, and the sensitive individual carries the competing allele that serves as the target. Killer-target drive therefore occurs in driver+ target−/driver− target+ heterozygotes. The spores that inherit the driver− target+ haplotype are destroyed, whereas those that inherit the driver+ target− haplotype are not (Fig. 3A). Close genetic linkage between the two factors helps ensure that the two elements do not become uncoupled by recombination, which would create a suicide genotype that includes both the killer and the target.
FIG 3.
Mechanisms of spore killing. (A) In the killer-target systems, all spores are exposed to the killer element encoded by the yellow locus. Only those spores that inherit the target locus (orange) are destroyed by the killer. (B) In the poison-antidote systems, the drive haplotype (yellow) encodes both a poison and an antidote. All spores are exposed to the poison, but only those that inherit the drive haplotype receive the antidote and are rescued.
As the name suggests, poison-antidote drive systems carry both a poison and an antidote element within the drive locus. Like the killer element from killer-target drivers, all developing spores are exposed to the poison of poison-antidote drive systems. Unlike killer-target drivers, however, all spores are also sensitive to the poison of poison-antidote drivers. The key to drive in this system is the restricted distribution of the antidote. Only the spores that inherit the drive locus receive the antidote and are rescued from the toxicity of the poison (Fig. 3B). Similar to the killer-target drivers, close genetic linkage between the two components of a poison-antidote drive system prevents their uncoupling by recombination to generate a suicide locus that encodes a poison without an antidote. However, unlike killer-target drivers, both functions are on the same haplotype: some of poison-antidote spore killers encode both functions within a single gene and others encode them in separate genes.
WHY STUDY SPORE KILLERS?
The study of spore killers is important in many ways. At the most basic cellular level, spore killers must carry out efficient, targeted cell destruction. Studying the molecular mechanisms of these killers therefore offers a unique opportunity to study cellular processes. For example, the het-s spore killer (described more below) is a great model for understanding prion proteins and programed cell death pathways, and the wtf (with Tf) spore killers are suitable models for addressing the biology of protein aggregates (23–25). Additionally, other killer meiotic drivers may act through broadly conserved cellular pathways. Thus, understanding their drive mechanisms may lead to a better understanding of factors limiting fertility and breeding potential in species, such as crop plants, that are important to human health and well-being.
Furthermore, the study of meiotic drivers is conceptually important for basic evolutionary biology. In particular, spore killers offer an opportunity to study natural selection acting at multiple levels in a biological hierarchy. The original view of the genome from the early twentieth century was that of a highly integrated and coordinated network that has evolved to produce a viable and reproductively successful individual. However, recently, genomes have been viewed as containing intrinsically conflicting parts that coevolve antagonistically (26, 27). As mentioned above, a spore killer imposes a clear fitness cost to the organism carrying it; hence, its activity is likely to result in intragenomic conflict. Conflicts caused by selfish genetic elements are not only common but are also expected to be drivers of evolutionary innovation, and, hence, are of fundamental importance for evolution (6, 27). Understanding the interaction between spore killers and their host genomes may also be applicable to other systems affected by conflicts. In cancer biology, for example, there is conflict because cells divide at the expense of the individuals carrying them. Similar conflicts likely occur during the evolution of multicellularity, as a new layer of biological organization is generated by the aggregation of smaller independent units. While theoretical work can predict the evolutionary outcomes of genetic conflicts, empirical data that support or refute predictions are scarce. Thus, spore killers are highly tractable examples in which both theoretical and empirical analyses can be used to study evolutionary conflict.
Finally, understanding spore killers could have a practical applied importance in designing effective artificial driver systems known as gene drives (28). These transgenic constructs wield the evolutionary power of meiotic drive to spread traits within a target population. Specifically, genetically modified organisms containing a desired trait linked to a drive element are released in a population. The drive element promotes the spread of the trait throughout the population. Gene drives have an enormous potential to improve human health, e.g., by suppressing mosquito populations that serve as vectors of human diseases. Gene drives also have the potential to improve crop yields by suppressing insect pests and populations of pathogenic fungi. For example, components of natural spore killers could be used as components in gene drives to help suppress pathogenic ascomycete fungi on crops, and, more broadly, spore killers could be used outside fungi to spread traits in plants or insect populations. Knowledge about the molecular function of the drive elements is, of course, critical to the development of artificial drive systems. Also crucial in this context is an understanding of spore killer evolution, which can help predict what characteristics of the host population will determine the fate of a drive element over generations, and, hence, how efficiently gene drives will spread within natural populations. Research about the evolutionary history of drive elements in nature is also important for assessing potential risks when developing them for synthetic drive. For example, understanding the likelihood of spread of spore killers between populations and species is important for predicting the possibility to control an introduced drive element after release. All this knowledge is essential for rational design of safe and effective gene drives.
WHO ARE THE KNOWN SPORE KILLERS?
Spore killing has been detected in a number of ascomycete species. The majority of these are model species used to study genetics and biochemistry, and, given the prevalence of spore killers found in these well-studied species, it is likely there are numerous spore killers in nature yet to be discovered. In each of the following sections we provide details on the known ascomycete spore killers, including what is known about the killer’s molecular mechanisms, and we provide information on the prevalence of driver alleles within the species.
Spore Killers in Podospora
Podospora anserina is a coprophilic filamentous fungus (29). As with other coprophilous fungi, P. anserina develops fruiting bodies (perithecia) in herbivore dung, from which it shoots ascospores onto the surrounding vegetation. There, the spores may get eaten by a herbivore, e.g., a horse or a rabbit. Going through the digestive tract of the animal activates the ascospores, which germinate after passing through the digestive tract and ending up in new dung. As P. anserina is pseudohomothallic, it produces ascospores that are dikaryotic (n+n) for mating type. This is achieved by the mating-type locus undergoing second-division segregation in the majority of meioses (Fig. 1). The germinating dikaryotic mycelium grows in dung until starvation, at which point it forms reproductive organs containing ascogenous hyphae that differentiate into sac-like structures (asci) where nuclear fusion and meiosis take place. The diploid zygotes undergo a canonical meiosis (Fig. 1A), and following the second meiotic division (MII), the nuclei proceed through one round of mitosis. After that, the resulting eight haploid nuclei are packaged into four ascospores. Each ascospore contains two haploid nuclei, one derived from each set of sister chromatids from a single MII division (Fig. 1B). The two nuclei within a given spore therefore inherit the same set of centromeres but are generally genetically distinct due to meiotic recombination. Each ascus, however, contains spores representing only two overall genotypes (Fig. 1B) (29).
Because of the spore packaging system, spore killing in Podospora is affected by the meiotic segregation pattern of the locus. For example, killer-target drive systems should be centromere-linked to ensure the killer and the target are separated by the first meiotic division and not packaged in the same spore. In poison-antidote drivers, spore killing occurs only when the spore killer segregates from the competing allele at the first meiotic division (Fig. 2A). Otherwise, all spores inherit the antidote and survive (Fig. 2B). There are two known types of spore killers in Podospora, and we will describe them in order of their discovery.
het-s, a single-gene killer-target driver.
In 1965, Bernet was the first to describe phenotypes caused by the het-s fungal meiotic drive locus, although these phenotypes were not conclusively demonstrated to result from meiotic drive until later work in 2003 by Dalstra et al. (described below) (30–32). The het-s gene is now well-studied and is today arguably the best understood meiotic drive system at the molecular level. Interestingly, many important insights came from investigations of asexual cell fusion rather than meiotic studies (33). We will therefore first explain the role of het-s in asexual cell fusion and then revisit how this gene acts as a selfish spore killer in light of these mechanistic studies.
Cells of filamentous fungi like P. anserina can fuse hyphae and exchange cytoplasm, leading to the formation of vegetative (asexual) cells, known as heterokaryons, that contain different types of nuclei. Heterokaryon incompatibility systems are thought to help protect fungi from a variety of parasitic elements that could be transferred between individuals via cytoplasmic mixing (33–36). Because of these incompatibility systems, heterokaryons can be established only if two fusing hyphae have the same genotype at a series of het loci. When fusions occur between incompatible hyphae, the cells at the fusion junction either die or fail to grow due to the actions of HET proteins (37). The het-s meiotic drive locus was named for its role in heterokaryon incompatibility.
There are two different alleles of the het-s gene: het-S and het-s. The het-s allele encodes HET-s protein, which can adopt either a soluble conformation or an insoluble prion conformation, neither of which is toxic. The prion conformation of HET-s can arise spontaneously and can then convert the soluble form of the protein to the prion form on contact. The het-S allele encodes the HET-S protein, which does not form a prion (38–42). When hyphae carrying HET-s prions fuse (asexually) with hyphae carrying HET-S proteins, the prions induce a deadly conformational change in the HET-S proteins. Specifically, the HET-s–HET-S interaction exposes a previously buried HET-S transmembrane domain (43). The altered HET-S proteins oligomerize, perforate cell membranes, and cause death of the fused cells (23, 44). However, when the HET-s proteins are not in the prion conformation, het-s and het-S cells are compatible to form heterokaryons (33).
Although asexual fusion of het-S and het-s strains leads to cell death when the HET-s protein is in the prion conformation, the two strain types are sexually compatible. Bernet initially observed that at low temperature (18°C), crosses in which the female parents carry a het-s allele and the male parents carry a het-S allele yield a high fraction of asci containing two normally developed spores and two aborted spores. The viable spores from the two-spored asci have the phenotype of het-s strains. The reciprocal cross, and crosses at higher temperature, yield normal asci (30, 31).
Dalstra et al. repeated and expanded these studies to show that het-s can act as a killer-target type spore killer when crossed to het-S (32). For example, when a female carrying HET-s prions mates with a het-S male, the prions act as a killer element that is transmitted to all developing spores. The HET-S proteins are the targets, likely because the prions turn them into deadly membrane-disrupting complexes (Fig. 3A) (23, 44). Centromere linkage causes ∼90% of asci to contain two spores with het-S in both nuclei, while the other two spores carry the het-s allele in both nuclei (Fig. 2A). The het-S spores encode the target protein and are destroyed, whereas the het-s spores do not contain the target, so they survive (Fig. 3A). Like heterokaryon incompatibility, the het-s locus only drives when its HET-s proteins are in the prion conformation. In addition, spore killing likely does not occur in the reciprocal cross because the male parent transfers too little cytoplasm to ensure all developing spores contain HET-s prions (32). It remains unclear why the zygotes (and their heterokaryotic precursors) generated by mating het-s females containing prions to het-S males are not killed. Dalstra et al. suggest that the level of HET-S protein expressed in these cells could be too low to induce killing, with HET-S levels reaching a lethal level only in developing spores (32).
The het-s locus offers an illustrative example of the potential fitness costs of meiotic drive. When the locus drives, it causes the death of half the spores. Moreover, in ∼10% of meioses, a crossover between the het-s locus and the centromere generates an ascus in which the spores will each contain one het-s nucleus and one het-S nucleus. In other words, the drive mechanism backfires because all four spores carry the killer and the target and, thus, are destroyed (32).
The het-S locus is not essential for life or sexual reproduction, as strains lacking the gene are viable and make asci normally (32, 39). het-S orthologs are found in other fungi, and their similarity to het-S suggests that the het-s killer allele arose via mutation of a het-S-like ancestor (38, 45). Minimally, it seems that both the het-s and het-S alleles have persisted in P. anserina populations since their discovery in 1965 (30, 31). For example, in a survey of 112 strains collected in The Netherlands in 1991, 72 strains contained the het-s allele, 66 of which contained HET-s prions (38).
The overrepresentation of the het-s allele relative to het-S could be due to meiotic drive, but why has het-s not gone to fixation? Debets et al. propose a model in which the meiotic drive of het-s is counteracted by selection acting on the HET-s/HET-S heterokaryon incompatibility system (38). As heterokaryon incompatibility can prevent the spread of cytoplasmic parasitic elements, cells bearing the rarer variant at a given het locus could have a selective advantage due to a decreased ability to exchange cytoplasm with infected individuals. In the absence of other selective pressures, this scenario causes balancing selection to maintain alternate het alleles in roughly equal frequencies in a population. In their model, the ability of het-s to drive in meiosis shifts the allele frequency in its favor, but the het-S allele is maintained because rare variants are less likely to acquire parasites. Consistent with this idea, they found that 51% of het-s strains were infected with a harmful senescence plasmid, compared to only 20% of the rarer het-S strains (38). Still, there may be other factors involved in preventing het-s fixation. For example, it has been suggested that HET-S has a role in nonheterokaryon incompatibility-based defense against unidentified parasites or pathogens (46). This role also may provide selective incentive for maintaining the het-S allele in P. anserina populations (24, 32, 45).
The Spok gene family constitutes multiple single-gene poison-antidote killers.
In the early 2000s, van der Gaag et al. crossed P. anserina strains from France and the Netherlands and observed the presence, absence, and frequency of killed spores (47). The underlying spore killers were designated Psk, for Podospora spore killing. With this classical genetic approach, the authors identified seven spore killer types (Psk-1 through Psk-7) that interact in a hierarchical way. Although the genetic basis of the Psk genes was not clarified (47), a correlation was observed between vegetative incompatibility (different from het-s) and the Psk phenotype, suggesting that they are connected either physically or mechanistically (48). However, it was later discovered that the genes of the Spok gene family underlie the Psk spore killers (49), and the relationship between the Spok gene family and vegetative incompatibility remains unclear.
In 2014, Grognet et al. first identified and characterized two genes of the Spok gene family, Spok1 and Spok2, as novel spore-killing elements in Podospora (50). They showed that a single Spok gene can act as both the poison and antidote (Fig. 3A). By crossing reference strains of P. anserina and P. comata, the authors revealed that spore killing observed in the wild-type cross was due to a gene on chromosome 5 of P. comata: Spok1. Interestingly, when deleting Spok1 from the P. comata strain, they discovered that the P. anserina strain has its own killer gene in the same chromosome but at a different location. They named this gene Spok2. Grognet et al. reported a dominant epistatic relationship between the two genes in that Spok1 is resistant and dominant to Spok2. While Spok1 is capable of killing in the presence of Spok2, Spok2 cannot kill in the presence of Spok1 (50). As this relationship between the Spok genes is reminiscent of the hierarchy of killing among the Psk genes, Vogan et al. set out to investigate the connection between the activity of Spok genes and the Psk genes in P. anserina (49).
Indeed, Vogan et al. demonstrated that three genes of the Spok gene family underlie the Psk genes identified by van der Gaag et al. (47, 49). Vogan et al. identified two novel Spok homologs (Spok3 and Spok4) and confirmed that the individual SPOK proteins perform both poison and antidote functions. In addition, the authors showed that the poison and antidote functions are dependent on distinct domains, a predicted nuclease and kinase domain, respectively. Strikingly, the combination of Spok2, Spok3, and Spok4 genes at different chromosomal locations was found to create a killing hierarchy and represent the genetic basis of the Psk spore killers in P. anserina (49). It is noteworthy that Vogan et al. made use of knock-in strains to assay pairwise interactions among the Spok genes and revealed that, in contrast to Spok1 of P. comata and Spok2 of P. anserina, which were shown by Grognet et al. to interact epistatically, Spok2, Spok3, and Spok4 of P. anserina do not interact (49, 50). Instead, the Spok genes kill independently of each other, and the spore-killing hierarchy observed in P. anserina is simply an emergent property of the presence and absence of the various Spok homologs in the different genomes (49).
Genomic and phylogenetic analyses across ascomycetes suggest that the Spok genes disperse via cross-species transfer and evolve by duplication and diversification within lineages (49, 51). This finding was in accordance with the finding presented by Grognet et al. in that the Spok family is widespread across ascomycetes (50). Whether Spok genes function as meiotic drivers in other species is, however, unknown and needs further investigation.
In P. anserina, Spok2 is found at high population frequencies, while two other genes of the Spok family, Spok3 and Spok4, are found at low to intermediate frequencies (49). In the study by Vogan et al., it was revealed that, unlike Spok2, Spok3 and Spok4 occur in a large (74 to 167 kbp) region (the Spok block) that can be found in different genomic locations in different strains. Notably, the block is never found at more than one genomic locus within natural strains. The Spok block is novel to P. anserina and represents an interesting genomic feature given the high genome synteny among strains of P. anserina, P. comata, and P. pauciseta (49). In a recent follow-up study, Vogan et al. found evidence supporting the Spok block to be a long DNA transposon that has captured the Spok genes and subsequently grown to a massive size through the gradual accumulation of DNA sequence. This DNA transposon is named Enterprise and is expected to be mobilized by a tyrosine recombinase (51). The authors provided experimental evidence that the Spok block has detrimental effects on spore production in strains that carry it, suggesting that it reduces fitness, which may be the reason for the lower frequencies of the Spok block in the population relative to Spok2.
Spore Killers in Neurospora
Neurospora is a genus of filamentous fungi that are often found on burned vegetation. The name was derived from nerve-like grooves that are found on the surface of their ascospores. Neurospora enjoy a process of sexual reproduction similar to that of Podospora anserina; however, most of the well-studied species (including N. crassa, N. sitophila, and N. intermedia, discussed below) are heterothallic, meaning that they grow primarily as haploids and are sexually deficient at the haploid stage. For mating and meiosis to take place, individuals of different mating types must meet. After the meiotic divisions, the nuclei of several Neurospora species undergo one round of mitosis to yield eight nuclei. Unlike in Podospora, however, the nuclei are each packaged into individual spores (Fig. 1C) (20, 52). In crosses between a completely penetrant spore killer and a sensitive strain, four of the eight spores are killed (Fig. 2C) (53).
Sk-1: a single gene causing a poison-antidote killing.
The Sk-1 killer in N. sitophila was the first spore killer element to be identified in Neurospora. Turner and Perkins reported a consistent pattern of four viable spores and four inviable spores per ascus when crossing natural isolates of N. sitophila from Nigeria and the United States (53). When the surviving offspring were backcrossed to the parents, the pattern was only observed in crosses to Nigerian strains, and in crosses within each population dead spores could not be observed. Based on these results, the authors determined that the U.S. strains were carrying Sk-1, a spore killer with close to 100% killing efficiency. The authors did not identify the locus responsible for the killing, but they found it was centromere-linked. The scarcity of genetic markers in N. sitophila and the difficulty of introgressing genomic regions from N. sitophila into the model species N. crassa made further genetic characterization difficult (53).
Over 40 years later, Svedberg et al. reported the identification of the single gene (Spk-1) responsible for spore killing in N. sitophila (54). Spk-1 shows no homology to any other known spore-killer gene. The gene was identified by whole-genome sequencing of 56 N. sitophila strains, a sample that captured the global diversity of N. sitophila and included a balanced representation of both killers and sensitive strains, which made it possible to use a genome-wide association test to associate the spore-killer phenotype with genotype. Using this approach, Svedberg et al. identified a 2-kb region on chromosome 6 that was highly associated with the spore-killer phenotype. With confirmatory crosses, deletion mutants, and insertion of the candidate locus in a nonkilling strain, the authors confirmed that this locus is sufficient for both spore killing and for resistance, as expected for a spore killer that kills with a poison-antidote mechanism. Transcriptomic analyses and molecular dissection further revealed that Sk-1 spore killing is caused by a single gene (Spk-1) that encodes a protein product of no more than 134 amino acids on a 1,450-bp transcript. This single gene is capable of both killing sensitive sibling spores and of protecting the spores producing the protein from self-killing. The mechanism behind these phenotypes is unknown, but the protein has predicted transmembrane domains, presenting the possibility that the poison could disrupt membrane integrity, as has been observed with the het-s meiotic driver (23) and in some bacterial toxin-antitoxin systems (55). A phylogenetic analysis of alleles from several different species of Neurospora suggests that Spk-1 in N. sitophila originated in a different Neurospora species, potentially N. hispaniola (54).
A noteworthy finding of Svedberg et al. was that spore killing can be suppressed by an RNA interference-based genome defense mechanism known as meiotic silencing by unpaired DNA (MSUD). This makes Spk-1 currently unique among the spore killers in that suppression by a host genome defense mechanism has been identified (54).
Today, over 700 natural isolates of N. sitophila have been tested for spore killing, and roughly 15% of these natural isolates display the phenotype (56, 57). A single resistant strain that can neither kill nor be killed has also been identified. Killer strains have been found all over the world, but there are notable differences in regional distribution. Some geographic regions appear to be fixed for Sk-1, whereas it is completely absent from others. In Italy and Tahiti, dense sampling has revealed an even ratio between killers and sensitives (56, 57). In Svedberg et al., it was shown that the population structure of N. sitophila is divided into three subclades, one of which is fixed for Sk-1, one where Sk-1 is absent, and a third where killers and sensitives intermix. The variation in Sk-1 distribution is still unknown and could be explained by multiple factors, such as time since introduction or association with fitness costs. However, as the interaction with the genome defense mechanism (MSUD) showed geographic confinement, it is particularly tempting to speculate that the presence of MSUD determines the likelihood with which the spore killer will invade and go to fixation in a population (54).
Sk-2 and Sk-3: multigene poison-antidote drivers kept together in large haplotypes.
A second spore-killer element of Neurospora, Sk-2, was later found in an N. intermedia strain from Borneo (53). The extensive sampling and phenotyping of Neurospora from natural populations revealed a total of four Sk-2 strains, which all produce only four spores when crossed to sensitive strains and eight spores when crossed to each other. Importantly, though, an additional spore killer strain of N. intermedia did not show this pattern: while it killed when crossed to a sensitive strain, it produced no viable spores when crossed to strains classified as Sk-2. It was subsequently determined that the spore death phenotype was due to the presence of two distinct incompatible killer systems that are capable of killing each other. Based on these results, this last spore killer strain of N. intermedia was classified as a new spore killer, Sk-3 (56).
Both Sk-2 and Sk-3 were successfully introgressed into N. crassa to facilitate genetic analysis. Campbell and Turner were able to use the rich set of genetic markers established in N. crassa to map both of these killers to a 30-cM region surrounding the centromere of chromosome 3 (58). In this region, recombination is suppressed between killer and sensitive strains, which prevented more refined mapping (58). Later, Hammond et al. took advantage of the nonkiller but resistant strains (rSk-2 and rSk-3) that had been found in nature to identify the factors conferring resistance to killing (59). Their work revealed that Sk-2 and Sk-3 were poison-antidote type drivers and that the two types of killing strains, in addition to the resistant strains, all use alleles of the same gene (rsk) for resistance (i.e., the antidote function). The rsk gene is found at the left flank of the 30-cM haplotype, and, depending on the allelic variant of the resistance gene (which differ by specific indel patterns), it confers resistance to Sk-2, resistance to Sk-3, or sensitivity to killing through unknown mechanisms (59).
With the resistance gene identified, the Hammond group was able to further screen for mutations that disrupt spore killing by Sk-2 (i.e., the poison function) (60). Specifically, they performed mutagenesis on suicidal genotypes containing the killer but not the resistance gene, reasoning that only a mutation in a gene required for killing (rfk) would allow viable ascospores to be produced. With this method, they located rfk-1 to a 45-kb region within Sk-2 on chromosome 3 (60). In a follow-up study, the rfk-1 gene was identified and characterized (61). The rfk-1 gene is located next to the right border of the Sk-2 haplotype, a location that allows it to escape silencing caused by the MSUD genome defense process (62). The rfk-1 gene contains four exons, three introns, and two stop codons, the first of which undergoes RNA editing to a tryptophan codon during sexual development. As a consequence, translation of an unedited rfk-1 transcript in vegetative tissue is expected to produce a 102-amino-acid protein, whereas translation of an edited rfk-1 transcript in sexual tissue is expected to produce a protein with 130 amino acids. The mechanism underlying rfk-1 killing has so far remained unknown, but it is possible that the unedited and edited rfk-1 transcripts have different roles with respect to the mechanism of meiotic drive by spore killing (61). The region of the Sk-3 locus responsible for killing has not yet been identified.
As described above, the two components of the Sk-2 spore killer (rsk and rfk-1) flank a region of suppressed recombination (58–60). Such suppressed recombination between components of multilocus drive systems is common, likely because a crossover that separates the two components would generate suicidal haplotypes. Svedberg et al. formally investigated the consequences of these multilocus meiotic drives on genome architecture in N. intermedia (63). Their study shows that Sk-2 and Sk-3 have induced independent and convergent structural changes in the same genomic region. Evidence for this hypothesis was found by generating short- and long-read-based genome assemblies for all five available spore killer strains and one sensitive strain of N. intermedia. The authors also used short-read sequencing for phylogenetic and population genetic analysis of a large collection of N. intermedia strains from natural populations. Using this large and diverse data set, they showed that even though Sk-2 and Sk-3 are located in the same chromosomal region, their respective haplotypes do not cluster in phylogenetic analyses, suggesting separate origins. Both the Sk-2 and Sk-3 haplotypes have accumulated a dense set of inversions that are interspersed with transposable elements (TEs). The inversions are unique for each killer type, further supporting an ancient evolutionary split. In the nonrecombining region of Sk-2, for which multiple genomes allowed substitution analyses, the authors identified signs of relaxed selection. For example, TEs have spread in the nonrecombining regions of both Sk-2 and Sk-3 despite what appears to be a set of fully functional mechanisms to limit their spread. This is consistent with the hypothesis that recombination suppression reduces the efficacy of selection in this region (63).
It is noteworthy that, in spite of extensive sampling and phenotyping, only a few killers have been found in N. intermedia. Instead, resistant strains are common and both killers and resistant strains are only found together in southeast Asia (56). As for the spore killers, the factors affecting their distributions are unknown, but one explanation for their low incidence is that the high frequency of resistant strains can prevent the invasion of a killer in a population. However, one can also envision a fitness cost of Sk-2 and Sk-3, as they are located within a region of suppressed recombination that could accumulate deleterious mutations that lower their ability to drive or the general fitness of any individual that carries them (54).
Spore Killers in Schizosaccharomyces pombe
Schizosaccharomyces pombe is commonly known as fission yeast, because the pill-shaped single-celled organism grows clonally by fission. The ecology of S. pombe is not well known, but it has been collected from fruits and a wide variety of fermented beverages (64). S. pombe cells generally grow as haploids, but when starved, cells of opposite mating types can fuse to form diploids. The diploid zygotes usually enter meiosis immediately, and the four products of meiosis are packaged as spores (Fig. 1D) (65).
Crosses between distinct natural isolates of S. pombe produce very few viable spores (66–69). Studies aimed at uncovering the cause of this infertility discovered the existence of spore killers affecting all chromosomes. Genes from the wtf gene family were subsequently demonstrated to contribute to spore killing (66, 70, 71). In addition, there is at least one additional coarsely mapped locus that causes weak drive in S. pombe that does not contain a wtf gene (72).
wtf genes are a family of single-gene poison-antidote killers.
The wtf genes comprise a large gene family, originally identified during the assembly of the S. pombe genome and named based on the genes’ tendency to be flanked with sequences derived from the Tf transposons (73, 74). There are a variable number of wtf genes and pseudogenes present in different isolates of S. pombe, ranging from 25 to 38 (70, 71, 74, 75). The driving wtf genes cause meiotic drive using a poison-antidote mechanism. Like the Spok drivers, both the poison and antidote functions are encoded by one gene. Unlike the Spok genes, however, the poison and antidote functions of wtf drivers are carried out by separate proteins, Wtfpoison and Wtfantidote. The two proteins are encoded by separate but largely overlapping transcripts. The transcript for the Wtfantidote protein is longer and contains an additional exon, leading the Wtfantidote protein to contain an additional ∼45 N-terminal amino acids not found in the Wtfpoison protein. During spore development, all spores are exposed to a lethal dose of the Wtfpoison protein of a given driver. Those spores that inherit the wtf drive gene, however, are rescued from death by the Wtfantidote. In S. pombe, this leads wtf driver+/− heterozygotes to generally produce two viable driver+ spores and two dead driver− spores (Fig. 2D) (70, 71).
A given S. pombe genome can contain up to 14 distinct wtf predicted meiotic drive genes (75). Interestingly, the Wtfantidote proteins act specifically against Wtfpoison proteins that are highly similar or identical in sequence outside the ∼45 antidote-specific amino acids. In other words, the antidote of wtfA generally cannot protect a spore from the poison produced by wtfB (71, 76). Moreover, due to rapid evolution of the gene family, the suite of alleles carried by a given natural isolate is largely unique (71, 75). This means that in a cross between a S. pombe haploid isolate predicted to contain 14 wtf drivers and another predicted to contain 10, there could be 24 distinct spore killers acting. In such cases, the only spores likely to survive are those that inherit all wtf drive alleles by inheriting two copies of chromosome 3, which houses all the wtf drivers in most isolates (72). It is not, however, clear how often this extreme situation occurs in nature, as the ecology of S. pombe, including outcrossing rates, is not well understood (64, 77, 78).
Not all wtf genes, however, can autonomously cause meiotic drive. Some wtf genes encode only a Wtfantidote protein. These antidote-only genes act as drive suppressors in that they can rescue spores from Wtfpoisons produced by wtf driver genes that share a highly similar coding sequence (72, 79). In addition, there are four other wtf genes found in all sequenced isolates of S. pombe that are highly diverged from the wtf genes involved in drive and from each other. The functions of these four genes are unclear, although they do not appear to cause meiotic drive (72).
The wtf drive genes are incredibly diverse, sharing as little as 30% pairwise amino acid identity. Despite this, the phenotypes caused by these drivers are highly similar (72). Mechanistic analyses of one wtf gene, wtf4 (from the S. kambucha isolate of S. pombe), revealed a potential explanation for how dramatically different Wtf proteins cause the same phenotype. The Wtf4 proteins use homotypic protein interactions to assemble into aggregates. Both the Wtf4poison and Wtf4antidote proteins can self-assemble, but the similarity between the two proteins also allows them to coassemble into aggregates. Aggregates comprised exclusively of Wtf4poison proteins remain distributed in the cytoplasm, where they are toxic. Aggregates that contain Wtf4antidote proteins are recognized by aggregate management pathways and are trafficked to the vacuole (fungal lysosome), where they are sequestered or destroyed (25).
Originally, Wtfpoison proteins, which contain predicted transmembrane domains, were proposed to cause cell death by assembling into pores and disrupting membrane integrity (70, 71). This pore model is still viable, but an alternative has also been proposed based on the aggregation propensity of Wtf proteins. This alternative model posits that the toxicity of the Wtf4poison aggregates is due to many nonspecific interactions that disrupt cellular protein homoeostasis. In simpler terms, the aggregates could nonspecifically disrupt the folding of many cytoplasmic proteins, which could overwhelm the cells’ ability to refold or destroy misfolded proteins. Under this model, diverse Wtf proteins could have the same phenotypic effects if they maintain the homotypic assembly properties of Wtf4 (25). As this model proposes no specific interactions are required for Wtfpoison toxicity, the proteins could diverge widely and maintain functionality. The Wtfantidote would similarly be free to diverge as long as it could maintain the ability to recruit aggregate management systems. Consistent with this constraint, the antidote-specific first exon of the wtf coding sequences is the best conserved (75).
Unmapped Spore Killer in Bipolaris maydis
Bipolaris maydis (also known as Cochliobolus heterostrophus) is a filamentous fungus known for causing southern leaf blight in maize. B. maydis sexual reproduction is similar to that of N. crassa (52). Bronson et al. fortuitously discovered spore killers in this system in a genetic study designed to search for genes required for the production of T-toxin, a metabolite involved in virulence of the fungus to maize (80). The authors found no new genes affecting T-toxin production, but they did detect the effects of a spore killer linked to a previously known gene, TOX1. The authors did not image asci, but in 5 out of 11 crosses between field isolates lacking the ability to produce T-toxin (Tox−) and TOX1 (Tox+) lab strains, the authors noticed reduced spore viability and an excess of progeny (up to 95%) that inherited the Tox− phenotype. The surviving spores (Tox−) inherited the ability to bias allele transmission in their favor in backcrosses to the sensitive parental (Tox+) strain (80). The locus responsible for the spore killing is unknown beyond its presumed linkage to the TOX1 locus. Identifying this driver and deciphering its mechanism could lead to control mechanisms to limit the impact of B. maydis.
Unmapped Spore Killers in Fusarium verticillioides
Fusarium verticillioides (also known as Fusarium fujikuroi, Gibberella fujikuroi, and Fusarium moniliforme) is a filamentous plant pathogen that grows on many important crops (81). Sexual reproduction in F. verticillioides is similar to that of N. crassa (52). Spore killing was first reported to occur in a number of crosses between different F. verticillioides isolates carried out by Kathariou and Speith in 1982 (82). This conclusion was supported by the work of Xu and Leslie, who mapped the spore killing locus, Sk, to a chromosome (83). Recently, the location of the Sk locus was further refined by Pyle et al., who narrowed down the causative locus to a 102-kb region containing 42 genes (84).
It is possible that spore killers are common in Fusarium species. In their early work, Kathariou and Speith tested the phenotypes of 225 natural isolates of F. verticillioides for the presence of spore killers by crossing them to only one of two tester strains: a sensitive strain or a killing strain. The authors reported finding the killer phenotype in 80% of strains, while they classified the remaining strains as sensitive or intermediate (82). Sidhu also reported similar observations using strains of F. subglutinans (85). However, given our current knowledge about the complexities that arise in the presence of multiple spore killer loci, the conclusions of these studies require further investigation. For example, Kathariou and Speith classified some strains as sensitive to killing because they yielded predominantly (≥95%) 8-spored asci when crossed to the sensitive tester strain, although sensitivity to a killer was not demonstrated (82). In addition, Kathariou and Speith, as well as Sidhu, classified strains as killers because they produced 8-spored asci when crossed to a known killer tester strain, although their ability to kill was not demonstrated (82, 85).
It will be exciting to explore if Fusarium species house multiple spore killers like Podospora and Schizosaccharomyces. In particular, Spok gene homologs are found in Fusarium species, so it is possible these genes could cause spore killing in some strains or genetic contexts (49). As with B. maydis, deciphering the drive mechanism(s) in Fusarium species could reveal control mechanisms for this and other related pathogenic ascomycetes.
THE EVOLUTIONARY DYNAMICS OF SPORE KILLING
As mentioned above, because of their ability to distort meiosis, spore killers gain a selective advantage at the gene level that allows them to increase in frequency at the population level over time. This increase occurs even though spore killing imposes a fitness cost on the organism. Under the proper conditions, spore killers can even spread if they impose fitness costs beyond those imposed directly by the spore killing. For example, a spore killer could spread along with deleterious alleles accumulated within an associated region of suppressed recombination (e.g., see reference 63). Because of this, spore killers could spread deleterious alleles to fixation and leave a population less fit even after spore killing has ceased.
Spore killers are not, however, predicted to always be able to invade or spread to fixation in a population. Spore killers can also go extinct or coexist with alternate alleles as stable polymorphisms. The evolutionary dynamics of a spore killer can be affected by factors such as the strength of drive, the prevalence of resistance, fitness costs of carrying the spore killer, features of the life cycle, behavior, and population structure of their host organism. In a recent study by Martinossi-Allibert et al., a single-locus population genetics model of spore killing in an ascomycete fungal host was developed (14). The model is adaptable to fungi with different life cycles, such as Neurospora and Podospora. Interestingly, the model shows that, in spite of the fungal life cycle being fundamentally different from those of animals and plants (meiotic products are progeny not gametes), the population dynamics show many similarities between fungi and animals/plants. For example, the model by Martinossi-Allibert et al. predicts that a stable coexistence of spore killers and sensitive strains in populations can be explained by the fitness costs of carrying the killer, a scenario also expected for drive in animal and plants (9). Furthermore, the model identifies characteristics of the killer locus as well as life cycle characteristics that may determine the fate of a spore killer upon entry into a population. These are, in addition to fitness costs, killing advantage (when killing of sensitive meiotic products results in either the production of additional meiotic products or higher absolute fitness of surviving ones), host population size, and mating system. In general, the work by Martinossi-Allibert et al. shows that a spore killer without costs or a killing advantage is more likely to invade a population than a neutral allele, but the presence of killing advantage makes invasion considerably more likely. In contrast, selfing of the host and fitness costs associated with the killer can hinder its invasion or stop its spread at intermediate frequencies (14). López Hernández et al. employed a population genetics model to explore the evolutionary dynamics of wtf spore killers in S. pombe. That work also focused on how inbreeding and linked fitness costs impede the spread of a spore killer, and, notably, the predictions of the model matched well with experimental evolution analyses exploring the same parameters (78).
Upon establishment in a population, meiotic drive can shape genetic architecture in other important ways (12, 86–89). As mentioned above, in multilocus drive systems, such as Sk-2 and Sk-3, a large region full of inversions can link the drive components. Furthermore, in ascomycetes carrying spore killers, the molecular machinery required for sporulation is under selection to not only carry out meiosis and spore formation but also to mitigate the impact of the spore killers. This could lead to evolutionary tradeoffs where variants that are nonideal for sporulation are selected due to their ability to offset the costs of spore killers. For example, many natural isolates of S. pombe produce spores disomic for chromosome 3 at high frequency. This disomy comes at a cost, as it likely reduces the number of overall spores produced. The disomy is adaptive, however, when S. pombe outcrosses as spores that inherit both parental copies of chromosome 3, which houses the wtf drivers, produce more wtf antidotes, and, thus, are more likely to survive (76).
Finally, the evolutionary impact of spore killers can extend beyond the haplotype housing the driver and competing alternate alleles. For example, in S. pombe, mutual killing by wtf drivers contributes to the considerable reduction in fertility observed between closely related isolates (66, 67, 70, 71, 76, 90). Interactions between spore killers (or linked variants in the haplotype) and the background genome have also been implicated in contributing to reproductive isolation between N. intermedia and N. metzenbergii (91). Reproductive isolation may also in theory stem from the genetic conflicts that arise between spore killers and their hosts (6, 27). Specifically, rapid coevolution between spore killers and counteracting genes, called suppressors, could accelerate speciation by creating genetic incompatibilities between recently separated populations, as shown in other systems of meiotic drive (92, 93).
CONCLUDING REMARKS
In recent years, major advances have been made in Podospora, Neurospora, and Schizosaccharomyces spore killer research. The commonality of spore killers in these model systems and the enormous diversity in form and function (both in the way they function [killer-target versus toxin-antidote] and in their genetic architecture [multiple versus single gene]) suggest that we would find countless more spore killers by simply looking for them. The ascus may be a good target for the evolution of spore killing, as the meiotic products share cytoplasm before spore delineation, which is key to killing in at least Neurospora and Schizosaccharomyces (59, 61, 70, 71).
In the future, it will be important to map the genes responsible for spore killing in Bipolaris and Fusarium, as well as to search for spore killers in other fungi, such as the Basidiomycetes, that package spores differently (18). Identifying the genes, however, is insufficient. Ideally, the field should strive to understand the origin, evolutionary history, and molecular mechanisms of each driver. Such studies have the potential to help unravel commonalities of meiotic drive in fungi, including their molecular mechanisms and population dynamics. Examples of future research directions include the origin of spore killers to unravel commonalities in their association with transposition and introgression. The future of research on spore killers should also focus on identifying interactions with the host genome, e.g., defense reactions and incompatibilities. With this research, we will learn about basic cell biology, be better able to judge the importance of intragenomic conflict in driving genome evolution, and also better understand macroevolutionary processes such as speciation. Ideally, we will also be able to apply the increased knowledge of drive to the development of synthetic gene drives.
ACKNOWLEDGMENTS
We thank Mark Miller and Patrick Lane for help in generating the figures and the reviewers for useful suggestions to the initially submitted manuscript.
Contributor Information
Sarah Zanders, Email: sez@stowers.org.
Hanna Johannesson, Email: Hanna.Johannesson@ebc.uu.se.
REFERENCES
- 1.Abbott S, Fairbanks DJ. 2016. Experiments on plant hybrids by Gregor Mendel. Genetics 204:407–422. 10.1534/genetics.116.195198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crow JF. 1991. Why is Mendelian segregation so exact? Bioessays 13:305–312. 10.1002/bies.950130609. [DOI] [PubMed] [Google Scholar]
- 3.Sandler L, Novitski E. 1957. Meiotic drive as an evolutionary force. Am Nat 91:105–110. 10.1086/281969. [DOI] [Google Scholar]
- 4.Zimmering S, Sandler L, Nicoletti B. 1970. Mechanisms of meiotic drive. Annu Rev Genet 4:409–436. 10.1146/annurev.ge.04.120170.002205. [DOI] [PubMed] [Google Scholar]
- 5.Crow JF. 1988. The ultraselfish gene. Genetics 118:389–391. 10.1093/genetics/118.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burt A, Trivers R. 2006. Genes in conflict: the biology of selfish genetic elements. Belknap Press of Harvard University Press, Cambridge, MA. [Google Scholar]
- 7.Price TA, Wedell N. 2008. Selfish genetic elements and sexual selection: their impact on male fertility. Genetica 134:99–111. 10.1007/s10709-008-9253-y. [DOI] [PubMed] [Google Scholar]
- 8.Zanders SE, Unckless RL. 2019. Fertility costs of meiotic drivers. Curr Biol 29:R512–R520. 10.1016/j.cub.2019.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L, Johannesson H, Knief U, Kokko H, Larracuente AM, Manser A, Montchamp-Moreau C, Petrosyan VG, Pomiankowski A, Presgraves DC, Safronova LD, Sutter A, Unckless RL, Verspoor RL, Wedell N, Wilkinson GS, Price TA. 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol Evol 31:315–326. 10.1016/j.tree.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, Higgins DM, Ross-Ibarra J, Wallace JG, Kanizay LB, Alabady M, Qiu W, Tseng KF, Wang N, Gao Z, Birchler JA, Harkess AE, Hodges AL, Hiatt EN. 2018. A kinesin-14 motor activates neocentromeres to promote meiotic drive in Maize. Cell 173:839–850. 10.1016/j.cell.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Akera T, Chmatal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, Lampson MA. 2017. Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358:668–672. 10.1126/science.aan0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akera T, Trimm E, Lampson MA. 2019. Molecular strategies of meiotic cheating by selfish centromeres. Cell 178:1132–1144. 10.1016/j.cell.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bravo Nunez MA, Nuckolls NL, Zanders SE. 2018. Genetic villains: killer meiotic drivers. Trends Genet 34:424–433. 10.1016/j.tig.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinossi-Allibert I, Veller C, Ament-Velasquez SL, Vogan AA, Rueffler C, Johannesson H. 2021. Invasion and maintenance of meiotic drivers in populations of ascomycete fungi. Evolution 75:1150–1169. 10.1111/evo.14214. [DOI] [PubMed] [Google Scholar]
- 15.Boulton A, Myers RS, Redfield RJ. 1997. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci USA 94:8058–8063. 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habig M, Kema GH, Holtgrewe Stukenbrock E. 2018. Meiotic drive of female-inherited supernumerary chromosomes in a pathogenic fungus. Elife 7:e40251. 10.7554/eLife.40251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolas A, Treco D, Schultes NP, Szostak JW. 1989. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338:35–39. 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 18.Alexopoulos CJ, Mims CW, Blackwell M. 1996. Introductory mycology, 4th ed. Wiley, New York, NY. [Google Scholar]
- 19.Zickler D. 2009. Observing meiosis in filamentous fungi: Sordaria and Neurospora. Methods Mol Biol 558:91–114. 10.1007/978-1-60761-103-5_7. [DOI] [PubMed] [Google Scholar]
- 20.Raju NB. 2009. Neurospora as a model fungus for studies in cytogenetics and sexual biology at Stanford. J Biosci 34:139–159. 10.1007/s12038-009-0015-5. [DOI] [PubMed] [Google Scholar]
- 21.Phadnis N, Hyppa RW, Smith GR. 2011. New and old ways to control meiotic recombination. Trends Genet 27:411–421. 10.1016/j.tig.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner BC, Perkins DD. 1991. Meiotic drive in Neurospora and other fungi. Am Nat 137:416–429. 10.1086/285174. [DOI] [Google Scholar]
- 23.Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R. 2012. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol 10:e1001451. 10.1371/journal.pbio.1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. 2014. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156:1207–1222. 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuckolls NL, Mok AC, Lange JJ, Yi K, Kandola TS, Hunn AM, McCroskey S, Snyder JL, Bravo Nunez MA, McClain M, McKinney SA, Wood C, Halfmann R, Zanders SE. 2020. The wtf4 meiotic driver utilizes controlled protein aggregation to generate selective cell death. Elife 9:e55694. 10.7554/eLife.55694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawkins R. 1976. The selfish gene. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 27.Rice WR. 2013. Nothing in genetics makes sense except in light of genomic conflict. Annu Rev Ecol Evol Syst 44:217–237. 10.1146/annurev-ecolsys-110411-160242. [DOI] [Google Scholar]
- 28.Collins JP. 2018. Gene drives in our future: challenges of and opportunities for using a self-sustaining technology in pest and vector management. BMC Proc 12:9. 10.1186/s12919-018-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silar P. 2020. Podospora anserina. hal-02475488. https://hal.archives-ouvertes.fr/hal-02475488
- 30.Bernet J. 1965. Mode d'action des gènes de “barrage” et relation entre l'incompatibilité cellulaire et l'incompatibilité sexuelle chez “Podospora anserina”, vol Série A, No d'ordre: 115. Masson, Paris, France. [Google Scholar]
- 31.Padieu E, Bernet J. 1967. Mode of action of the genes responsible for abortion of certain products of meiosis in the Ascomycete, Podospora anserina. C R Acad Hebd Seances Acad Sci D 264:2300–2303. [PubMed] [Google Scholar]
- 32.Dalstra HJ, Swart K, Debets AJ, Saupe SJ, Hoekstra RF. 2003. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci USA 100:6616–6621. 10.1073/pnas.1030058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riek R, Saupe SJ. 2016. The HET-S/s prion motif in the control of programmed cell death. Cold Spring Harb Perspect Biol 8:a023515. 10.1101/cshperspect.a023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caten CE. 1972. Vegetative incompatibility and cytoplasmic infection in fungi. J Gen Microbiol 72:221–229. 10.1099/00221287-72-2-221. [DOI] [PubMed] [Google Scholar]
- 35.Debets F, Yang X, Griffiths AJ. 1994. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet 26:113–119. 10.1007/BF00313797. [DOI] [PubMed] [Google Scholar]
- 36.Hartl DL, Dempster ER, Brown SW. 1975. Adaptive significance of vegetative incompatibility in Neurospora crassa. Genetics 81:553–569. 10.1093/genetics/81.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goncalves AP, Heller J, Daskalov A, Videira A, Glass NL. 2017. Regulated forms of cell death in fungi. Front Microbiol 8:1837. 10.3389/fmicb.2017.01837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debets AJ, Dalstra HJ, Slakhorst M, Koopmanschap B, Hoekstra RF, Saupe SJ. 2012. High natural prevalence of a fungal prion. Proc Natl Acad Sci USA 109:10432–10437. 10.1073/pnas.1205333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turcq B, Deleu C, Denayrolles M, Begueret J. 1991. Two allelic genes responsible for vegetative incompatibility in the fungus Podospora anserina are not essential for cell viability. Mol Gen Genet 228:265–269. 10.1007/BF00282475. [DOI] [PubMed] [Google Scholar]
- 40.Coustou V, Deleu C, Saupe S, Begueret J. 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA 94:9773–9778. 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein ML, Ness F, Cescau S, Soragni A, Leitz D, Saupe SJ, Riek R. 2010. The mechanism of prion inhibition by HET-S. Mol Cell 38:889–899. 10.1016/j.molcel.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. 2008. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science 319:1523–1526. 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 43.Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, Bathany K, Lascu I, Schmitter JM, Riek R, Saupe SJ. 2003. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J 22:2071–2081. 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathur V, Seuring C, Riek R, Saupe SJ, Liebman SW. 2012. Localization of HET-S to the cell periphery, not to [Het-s] aggregates, is associated with [Het-s]-HET-S toxicity. Mol Cell Biol 32:139–153. 10.1128/MCB.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daskalov A, Paoletti M, Ness F, Saupe SJ. 2012. Genomic clustering and homology between HET-S and the NWD2 STAND protein in various fungal genomes. PLoS One 7:e34854. 10.1371/journal.pone.0034854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daskalov A, Saupe SJ. 2021. The expanding scope of amyloid signalling. Prion 15:21–28. 10.1080/19336896.2021.1874791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Gaag M, Debets AJ, Oosterhof J, Slakhorst M, Thijssen JA, Hoekstra RF. 2000. Spore-killing meiotic drive factors in a natural population of the fungus Podospora anserina. Genetics 156:593–605. 10.1093/genetics/156.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Gaag M, Debets AJ, Hoekstra RF. 2003. Spore killing in the fungus Podospora anserina: a connection between meiotic drive and vegetative incompatibility? Genetica 117:59–65. 10.1023/a:1022364632611. [DOI] [PubMed] [Google Scholar]
- 49.Vogan AA, Ament-Velasquez SL, Granger-Farbos A, Svedberg J, Bastiaans E, Debets AJ, Coustou V, Yvanne H, Clave C, Saupe SJ, Johannesson H. 2019. Combinations of Spok genes create multiple meiotic drivers in Podospora. Elife 8:e46454. 10.7554/eLife.46454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grognet P, Lalucque H, Malagnac F, Silar P. 2014. Genes that bias Mendelian segregation. PLoS Genet 10:e1004387. 10.1371/journal.pgen.1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogan AA, Ament-Velasquez SL, Bastiaans E, Wallerman O, Saupe SJ, Suh A, Johannesson H. 2021. The Enterprise, a massive transposon carrying Spok meiotic drive genes. Genome Res 31:789–798. 10.1101/gr.267609.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coppin E, Debuchy R, Arnaise S, Picard M. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev 61:411–428. 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner BC, Perkins DD. 1979. Spore killer, a chromosomal factor in Neurospora that kills meiotic products not containing it. Genetics 93:587–606. 10.1093/genetics/93.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svedberg J, Vogan AA, Rhoades NA, Sarmarajeewa D, Jacobson DJ, Lascoux M, Hammond TM, Johannesson H. 2021. An introgressed gene causes meiotic drive in Neurospora sitophila. Proc Natl Acad Sci USA 118:e2026605118. 10.1073/pnas.2026605118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner BC. 2001. Geographic distribution of Neurospora spore killer strains and strains resistant to killing. Fungal Genet Biol 32:93–104. 10.1006/fgbi.2001.1253. [DOI] [PubMed] [Google Scholar]
- 57.Jacobson DJ, Dettman JR, Adams RI, Boesl C, Sultana S, Roenneberg T, Merrow M, Duarte M, Marques I, Ushakova A, Carneiro P, Videira A, Navarro-Sampedro L, Olmedo M, Corrochano LM, Taylor JW. 2006. New findings of Neurospora in Europe and comparisons of diversity in temperate climates on continental scales. Mycologia 98:550–559. 10.3852/mycologia.98.4.550. [DOI] [PubMed] [Google Scholar]
- 58.Campbell JL, Turner BC. 1987. Recombination block in the spore killer region of Neurospora. Genome 29:129–135. 10.1139/g87-022. [DOI] [PubMed] [Google Scholar]
- 59.Hammond TM, Rehard DG, Xiao H, Shiu PK. 2012. Molecular dissection of Neurospora spore killer meiotic drive elements. Proc Natl Acad Sci USA 109:12093–12098. 10.1073/pnas.1203267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harvey AM, Rehard DG, Groskreutz KM, Kuntz DR, Sharp KJ, Shiu PK, Hammond TM. 2014. A critical component of meiotic drive in Neurospora is located near a chromosome rearrangement. Genetics 197:1165–1174. 10.1534/genetics.114.167007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhoades NA, Harvey AM, Samarajeewa DA, Svedberg J, Yusifov A, Abusharekh A, Manitchotpisit P, Brown DW, Sharp KJ, Rehard DG, Peters J, Ostolaza-Maldonado X, Stephenson J, Shiu PKT, Johannesson H, Hammond TM. 2019. Identification of rfk-1, a meiotic driver undergoing RNA editing in Neurospora. Genetics 212:93–110. 10.1534/genetics.119.302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiu PK, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–916. 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 63.Svedberg J, Hosseini S, Chen J, Vogan AA, Mozgova I, Hennig L, Manitchotpisit P, Abusharekh A, Hammond TM, Lascoux M, Johannesson H. 2018. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nat Commun 9:4242. 10.1038/s41467-018-06562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeffares DC. 2018. The natural diversity and ecology of fission yeast. Yeast 35:253–260. 10.1002/yea.3293. [DOI] [PubMed] [Google Scholar]
- 65.Egel R. 2004. The molecular biology of Schizosaccaromyces pombe: genetics, genomics, and beyond. Springer, Berlin, Germany. [Google Scholar]
- 66.Zanders SE, Eickbush MT, Yu JS, Kang JW, Fowler KR, Smith GR, Malik HS. 2014. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. Elife 3:e02630. 10.7554/eLife.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Avelar AT, Perfeito L, Gordo I, Ferreira MG. 2013. Genome architecture is a selectable trait that can be maintained by antagonistic pleiotropy. Nat Commun 4:2235. 10.1038/ncomms3235. [DOI] [PubMed] [Google Scholar]
- 68.Hu W, Suo F, Du LL. 2015. Bulk segregant analysis reveals the genetic basis of a natural trait variation in fission yeast. Genome Biol Evol 7:3496–3510. 10.1093/gbe/evv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bahler J, Sedlazeck FJ. 2017. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8:14061. 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nuckolls NL, Bravo Nunez MA, Eickbush MT, Young JM, Lange JJ, Yu JS, Smith GR, Jaspersen SL, Malik HS, Zanders SE. 2017. wtf genes are prolific dual poison-antidote meiotic drivers. Elife 6:e26033. 10.7554/eLife.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu W, Jiang ZD, Suo F, Zheng JX, He WZ, Du LL. 2017. A large gene family in fission yeast encodes spore killers that subvert Mendel's law. Elife 6:e26057. 10.7554/eLife.26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bravo Nunez MA, Sabbarini IM, Eickbush MT, Liang Y, Lange JJ, Kent AM, Zanders SE. 2020. Dramatically diverse Schizosaccharomyces pombe wtf meiotic drivers all display high gamete-killing efficiency. PLoS Genet 16:e1008350. 10.1371/journal.pgen.1008350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, Quail MA, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 74.Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. 2003. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res 13:1984–1997. 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eickbush MT, Young JM, Zanders SE. 2019. Killer meiotic drive and dynamic evolution of the wtf gene family. Mol Biol Evol 36:1201–1214. 10.1093/molbev/msz052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bravo Nunez MA, Sabbarini IM, Eide LE, Unckless RL, Zanders SE. 2020. Atypical meiosis can be adaptive in outcrossed Schizosaccharomyces pombe due to wtf meiotic drivers. Elife 9:e57936. 10.7554/eLife.57936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tusso S, Nieuwenhuis BPS, Sedlazeck FJ, Davey JW, Jeffares DC, Wolf JBW. 2019. Ancestral admixture is the main determinant of global biodiversity in fission yeast. Mol Biol Evol 36:1975–1989. 10.1093/molbev/msz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.López Hernández JFH, Helston RM, Lange JJ, Billmyre RB, Schaffner SH, Eickbush MT, McCroskey S, Zanders SE, 2021. Diverse mating phenotypes impact the spread of wtf meiotic drivers in S. pombe. bioRxiv 10.1101/2021.05.28.446231. [DOI] [PMC free article] [PubMed]
- 79.Bravo Nunez MA, Lange JJ, Zanders SE. 2018. A suppressor of a wtf poison-antidote meiotic driver acts via mimicry of the driver's antidote. PLoS Genet 14:e1007836. 10.1371/journal.pgen.1007836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bronson CR, Taga M, Yoder OC. 1990. Genetic control and distorted segregation of T-toxin production in field isolates of Cochliobolus heterostrophus. Phytopathology 80:819–823. 10.1094/Phyto-80-819. [DOI] [Google Scholar]
- 81.Cen YK, Lin JG, Wang YL, Wang JY, Liu ZQ, Zheng YG. 2020. The gibberellin producer Fusarium fujikuroi: methods and technologies in the current toolkit. Front Bioeng Biotechnol 8:232. 10.3389/fbioe.2020.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kathariou S, Spieth PT. 1982. Spore killer polymorphism in Fusarium moniliforme. Genetics 102:19–24. 10.1093/genetics/102.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu JR, Leslie JF. 1996. A genetic map of Gibberella fujikuroi mating population A (Fusarium moniliforme). Genetics 143:175–189. 10.1017/s0016672300034066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pyle J, Patel T, Merrill B, Nsokoshi C, McCall M, Proctor RH, Brown DW, Hammond TM. 2016. A meiotic drive element in the Maize pathogen Fusarium verticillioides is located within a 102 kb region of chromosome V. G3 (Bethesda) 6:2543–2552. 10.1534/g3.116.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sidhu GS. 1984. Genetics of Gibberella fujikuroi V. spore killer alleles in G. fujikuroi. J Heredity 75:237–238. 10.1093/oxfordjournals.jhered.a109923. [DOI] [Google Scholar]
- 86.Henikoff S, Ahmad K, Malik HS. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102. 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 87.Hurst GD, Werren JH. 2001. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet 2:597–606. 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 88.Werren JH. 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci USA 108(Suppl 2):10863–10870. 10.1073/pnas.1102343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finseth FR, Nelson TC, Fishman L. 2021. Selfish chromosomal drive shapes recent centromeric histone evolution in monkeyflowers. PLoS Genet 17:e1009418. 10.1371/journal.pgen.1009418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jeffares DC, Rallis C, Rieux A, Speed D, Převorovský M, Mourier T, Marsellach FX, Iqbal Z, Lau W, Cheng TMK, Pracana R, Mülleder M, Lawson JLD, Chessel A, Bala S, Hellenthal G, O'Fallon B, Keane T, Simpson JT, Bischof L, Tomiczek B, Bitton DA, Sideri T, Codlin S, Hellberg JEEU, van Trigt L, Jeffery L, Li J-J, Atkinson S, Thodberg M, Febrer M, McLay K, Drou N, Brown W, Hayles J, Carazo Salas RE, Ralser M, Maniatis N, Balding DJ, Balloux F, Durbin R, Bähler J. 2015. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nat Genet 47:235–241. 10.1038/ng.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vogan Aas J, Grudzinska-Sterno M, Johannesson H. 2020. Meiotic drive is associated with sexual incompatibility in Neurospora. bioRxiv 10.1101/2020.11.27.400796. [DOI] [PMC free article] [PubMed]
- 92.Frank SA. 1991. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45:262–267. 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 93.Hurst LD, Pomiankowski A. 1991. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics 128:841–858. 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]