ABSTRACT
Campylobacter from contaminated poultry meat is a major source of human gastroenteritis worldwide. To date, attempts to control this zoonotic infection with on-farm biosecurity measures have been inconsistent in outcome. A cornerstone of these efforts has been the detection of chicken infection with microbiological culture, where Campylobacter is generally not detectable until birds are at least 21 days old. Using parallel sequence-based bacterial 16S profiling analysis and targeted sequencing of the porA gene, Campylobacter was identified at very low levels in all commercial flocks at less than 8 days old that were tested from the United Kingdom, Switzerland, and France. These young chicks exhibited a much greater diversity of porA types than older birds testing positive for Campylobacter by culture or quantitative PCR (qPCR). This suggests that as the bacteria multiply sufficiently to be detected by culture methods, one or two variants, as indicated by porA type, dominate the infection. The findings that (i) most young chicks carry some Campylobacter and (ii) not all flocks become Campylobacter positive by culture suggest that efforts to control infection, and therefore avoid contamination of poultry meat, should concentrate on how to limit Campylobacter to low levels by the prevention of the overgrowth of single strains.
IMPORTANCE Our results demonstrate the presence of Campylobacter DNA among fecal samples from a range of commercially reared meat chicks that are less than 8 days of age, consistent across 3 European countries. The recently developed, sensitive detection method indicates that infection occurs on commercial farms much earlier and more widely than previously thought, which opens up new opportunities to control Campylobacter contamination at the start of the food chain and reduce the unacceptably high levels of human disease.
KEYWORDS: broiler chickens, Campylobacter
INTRODUCTION
The consumption of raw or undercooked poultry meat contaminated with the bacterium Campylobacter is one of the major causes of gastroenteritis within human populations in high-income countries (1, 2). Current strategies for reducing infection in poultry have primarily focused on increasing on-farm biosecurity to prevent infection of chicken flocks during the growing period. These approaches have been largely ineffective as an intervention, and rates of infection have remained high with ∼70% of European Union flocks being contaminated (2, 3). Detecting the presence of Campylobacter within a commercially reared broiler flock currently relies on bacteriological culture or quantitative PCR (qPCR), which generally identify Campylobacter-positive flocks at >2 weeks of age (4–6). These findings have led to the widespread assumption that chickens are Campylobacter-free before this age, but it is not known whether they are genuinely free from infection or whether they are infected below levels detectable using these methods. To address this question, we applied different methods for detecting Campylobacter in chickens: (i) microbiological culture or qPCR, (ii) 16S bacterial profiling analysis, and (iii) parallel sequencing of the Campylobacter porA gene (7). The culture methods used standard growth media, and the 16S bacterial profiling analysis was conducted with an average sequencing depth of 65,000 reads per sample. The porA gene was chosen because it encodes a surface-exposed major outer membrane protein (MOMP) in Campylobacter and has been successfully included in typing schemes to assess genetic diversity (8). Parallel sequencing of a fragment of the short variable region of the gene (porAf2) was used since this efficiently identifies multiple strains within samples (7). Based upon extensive use of the porA locus in this context, it is known that while a porA variant shows general association with a Campylobacter species, clonal complex, or even sequence type, there is evidence that porA variants may also be exchanged between C. jejuni and C. coli. Since it is not possible to reliably distinguish between Campylobacter species, at least for C. jejuni and C. coli, which cause the majority of human disease, using fragments of this gene, we therefore refer to “Campylobacter” in this study. The species-specific qPCR results from flocks tested in this study showed that C. jejuni was detected in all of the Campylobacter-positive flocks at their endpoint, and C. coli was additionally detected in three flocks, indicative of mixed infection. The 34 commercial broiler (meat) flocks sampled for this study were from the United Kingdom, France, and Switzerland. The flocks were chosen to represent a range of different housing and management conditions in order to determine whether or not Campylobacter DNA could be detected from young chicks in a variety of circumstances. The Campylobacter status of each of the flocks shortly before slaughter was determined by standard culture or qPCR—henceforth referred to as Campylobacter endpoint culture/qPCR positive or negative for clarity. With a range of collaborating partners, and adhering to commercial practices across different farms, companies and countries, there was some variation in sample time points, for example, with some flock types being slaughtered later than others. We limited variation as much as possible by using centrally assembled sample kits, by limiting DNA storage and extraction to two methods, consistent by country, and by replicating each sample time point and flock type. Although we did not identify any substantive differences in DNA storage and extraction method, we make only limited comparisons between the different countries. Our aim was instead to identify the presence or absence of Campylobacter among a diverse set of samples from young chicks.
RESULTS
Detection of Campylobacter DNA among chicks <8 days of age.
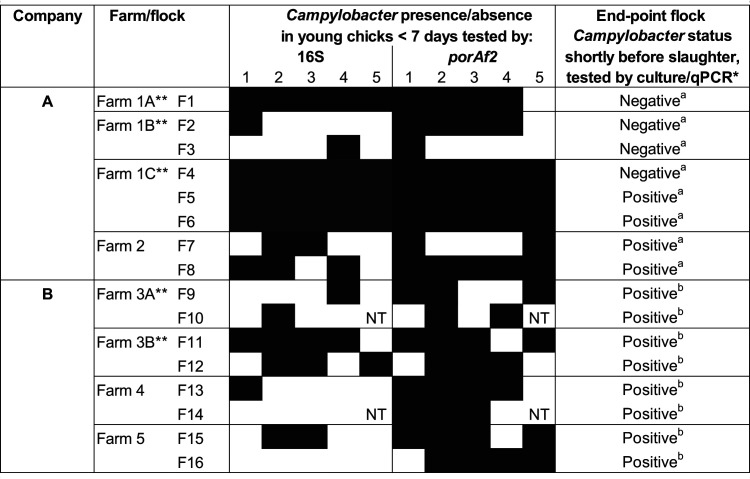
Detection of Campylobacter DNA, by both porAf2 and bacterial 16S rRNA gene (region 3 to 4) targets were compared among 78 fecal samples from chicks <7 days of age, for a subset of 16 U.K. flocks (Table 1). Campylobacter DNA was detected in 58/78 (74.4%) of fecal samples tested by porAf2 and from at least one bird in all of the flocks tested. Using the bacterial 16S rRNA gene sequencing approach, Campylobacter DNA was detected in 39/78 (50%) of fecal samples from at least one bird in 14/16 (87.5%) of the flocks tested.
TABLE 1.
Presence/absence of Campylobacter DNA among fecal samples from U.K. broiler chicks <7 days of age, detected by parallel sequencing of the 16S rRNA gene or porAf2 targetsa
Fresh fecal samples were collected from up to 5 birds selected at random at each time point. The 16S rRNA gene and porAf2 targets were amplified from DNA extracted directly from individual samples by PCR, and the number of variants was detected by parallel sequencing. 16S rRNA gene analysis at high depth was used to look for the presence of Campylobacter among the total bacterial profile, while amplification of the porAf2 target was used to enrich for Campylobacter-specific DNA relative to the other microbiota.
Flocks were tested for Campylobacter by conventional culture (a) or qPCR (b), using samples collected from the second sample date when birds were aged 28 to 35 days.
These flocks were present on farms 1 and 3 in separate rotations and distanced by time.
The remainder of the flocks tested in this study were subject to porAf2 parallel sequencing only. In total, porAf2 variants were identified among 139/169 (82.2%) fecal samples tested from chicks <8 days of age and from at least one chick among all of the 34 flocks tested in the study (Table 1 and Fig. 1a and b). Variants of porAf2 were detected among 9 of 10 samples from two flocks of 7-day-old chicks that were processed individually at the DNA extraction and PCR setup stages. Nontemplate controls for DNA extraction and PCRs were routinely negative. No Campylobacter DNA was detected among 60 samples tested from high-biosecurity specific-pathogen-free (SPF) chickens tested at 3 to 49 days of age.
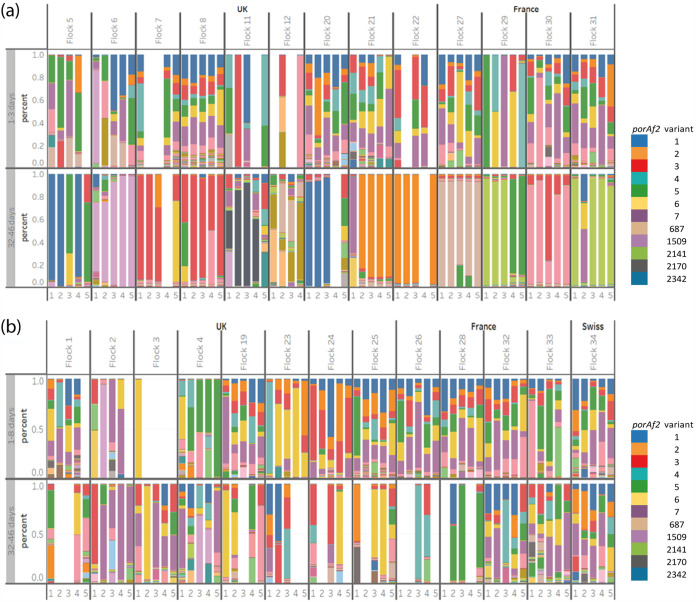
FIG 1.
The prevalence of Campylobacter porAf2 types among fecal samples collected from 34 commercially reared broiler flocks from the United Kingdom, France, and Switzerland (transformed data). (a and b) Results are shown for flocks that tested (a) Campylobacter positive and (b) Campylobacter negative at 32 to 46 days by culture/qPCR. Flocks that were tested at 28 to 31 days of age are not shown, together with flock 14, which had low sequencing depth. Each color represents a different porAf2 type; common variants are given in the legend.
Comparison of porAf2 type detected directly from fecal samples by parallel sequencing and cultured Campylobacter isolates.
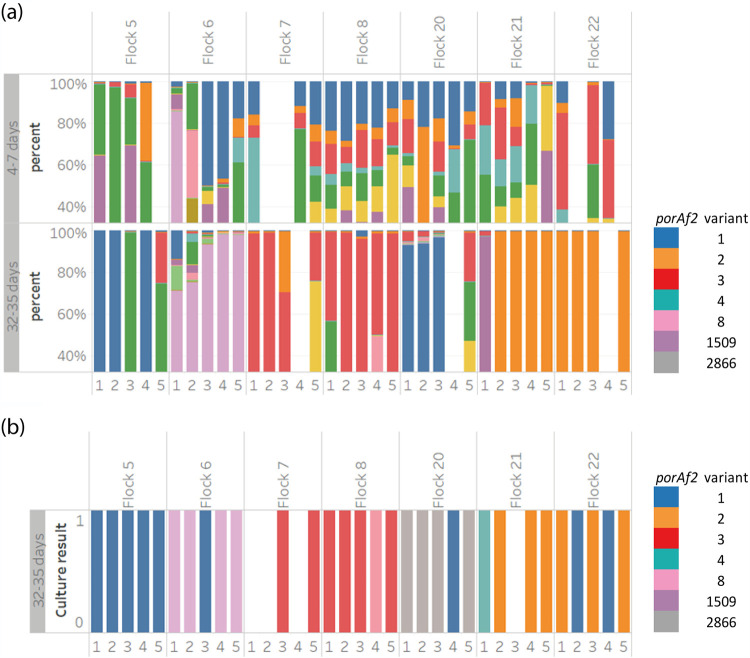
Campylobacter isolates were cultured from 28 samples within 7 flocks, from birds aged 25 to 32 days, from U.K. company A. The porAf2 type of a single colony pick per bird/sample was compared with the porAf2 variants detected by parallel sequencing from the same samples. The Campylobacter porAf2 type detected by conventional culture matched those that were predominant in the same sample and detected by parallel sequencing in 18/28 (64.3%) of samples tested (Fig. 2). For nine samples, the predominant porAf2 type detected by the parallel sequencing method was not the same as the porAf2 type detected by culture using a single colony pick. Almost all (27/28) of the cultured porAf2 types were detected in the same sample by parallel sequencing, at a percentage of the total Campylobacter porAf2 types recovered, ranging from 0.01% to 46.99%. For example, porAf2 type 2866 was identified among Campylobacter isolates cultured from four of five samples from flock 20 (Fig. 2), but it was present at ∼0.1% of porAf2 variants detected from the fecal samples by direct parallel sequencing (data not shown). The porAf2 variant 2866 was rarely identified among samples from the wider study, being identified on only four other occasions among single samples from four different flocks at <0.01% of the variants. The only exception where a cultured porAf2 variant was not detected by parallel sequencing from the same sample was also porAf2 type 2866 (Fig. 2), from sample 5/flock 20.
FIG 2.
(a and b) Comparison of Campylobacter porAf2 types identified among samples using (a) parallel sequencing and (b) conventional culture among a subset of flocks with a single colony pick per bird/sample. Each color represents a different porAf2 type; common variants are given in the legend.
Diversity of porAf2 variants recovered from the study.
A total of 1,098 porAf2 types (1,066 in samples from 25 U.K. flocks; 353 from 8 French flocks; 560 from 1 Swiss flock), corresponding to 746 peptide sequences, were identified among the flocks tested in the study (see Fig. S2 in the supplemental material). The porAf2 variants were manually checked on a sequencing alignment, with approximately 80% sequence similarity between the most disparate and up to 100% similarity to those identified among cultured Campylobacter isolates. Point mutations and indels were spread along the length of the short variable region fragment that was sequenced.
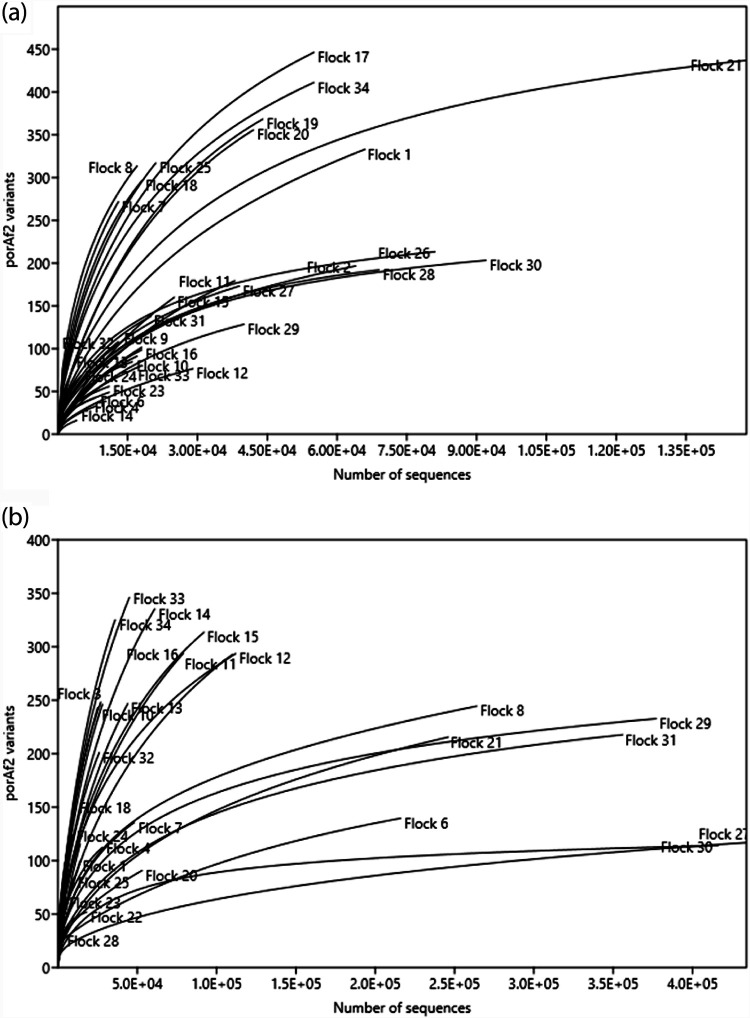
The number of different porAf2 types recovered from an individual fecal sample ranged from 9 to 358. Rarefaction curves demonstrated that the sequencing depth was sufficient to recover most of the flock diversity from samples from young chick <8 days of age, with a maximum of approximately 450 porAf2 variants recovered from both endpoint Campylobacter culture/qPCR negative and positive flocks (Fig. 3a). For birds >28 days of age, 100 to 300 porAf2 variants accounted for most of the flock diversity among endpoint Campylobacter culture/qPCR positive flocks, and approximately 200 porAf2 variants accounted for most of the diversity among endpoint culture/qPCR-negative flocks (Fig. 3b). There were four endpoint culture/qPCR positive U.K. flocks from company B where upwards of 350 porAf2 variants did not appear to capture the full diversity. Similarly, there were six more flocks that were endpoint culture/qPCR negative where either recovery of the porAf2 variants was relatively low, below n = 250, or in the case of one French and one Swiss flock, upward of 350 porAf2 variants did not appear to capture the full diversity in these samples. No systematic difference in porAf2 population structure related to DNA extraction method was noted (Fig. S3).
FIG 3.
Rarefaction curves for Campylobacter porAf2 types isolated from commercial broiler flocks from the United Kingdom, France, and Switzerland. Results represent pooled sequence counts for up to 5 birds/flock and are grouped by bird age. (a) Young birds, 1 to 8 days; (b) older birds, 28 to 46 days.
Comparison of porAf2 population structure between endpoint Campylobacter culture/qPCR negative and positive flocks.
There was no significant difference in the average number of porAf2 types recovered from young broiler flocks that tested Campylobacter endpoint culture/qPCR negative at 28 to 46 days, compared to flocks that tested Campylobacter endpoint culture/qPCR positive by standard methods (transformed data, n = 33 flocks, Mann-Whitney U test, P = 0.6713) or among flocks tested at early and later time points (transformed data, Wilcoxon rank-sum exact test, n = 33 flocks, P = 0.618). For porAf2 types that became predominant within flocks, 76.1% (16/21) were identified among younger birds from the same flock, and often (11/21, 52.4%) at a higher proportion of the total porAf2 sequenced compared to young birds from other flocks (Wilcoxon rank-sum exact test, n = 13 flocks, P = 0.001 within countries and P = 0.002 between countries (Table 2).
TABLE 2.
The predominant porAf2 types identified within a flock among birds aged 28 to 46 days, compared to their prevalence among birds 1 to 8 days of agea
| Flock | Predominant porAf2 type(s)b (% of birds) | Average prevalence in young birds (1–7 days): |
||
|---|---|---|---|---|
| Same flock (% of birds) | Different flock: U.K. (% of birds) | Different flock: France (% of birds) | ||
| U.K. flocks | ||||
| Flock 5 | 1 (75) | 0.122 (100) | 0.124 (100) | 0.376 (100) |
| 5 (50) | 7.606 (100) | 0.109 (96) | 0.351 (100) | |
| Flock 6 | 1509 (100) | 6.877 (100) | <0.001 (75) | Not detected |
| Flock 7 | 3 (100) | 1.532 (100) | 0.112 (100) | 0.245 (100) |
| 6 (33) | 1.500 (100) | 0.119 (100) | 0.328 (100) | |
| Flock 8 | 3 (100) | 2.209 (100) | 0.111 (100) | 0.245 (100) |
| Flock 11 | 2170 (75) | Not detected | <0.001 (8) | <0.001 (14) |
| Flock 12 | 2366 (33) | Not detected | <0.001 (8) | <0.001 (14) |
| 687 (33) | <0.001 (33) | 0.003 (67) | 0.001 (86) | |
| 2328 (33) | Not detected | <0.001 (33) | <0.001 (72) | |
| Flock 20 | 2342 (60) | Not detected | <0.001 (13) | <0.001 (14) |
| Flock 21 | 7 (20) | 3.59 (100) | 0.190 (100) | 0.581 (100) |
| 2 (80) | 1.407 (100) | 0.040 (96) | 0.638 (86) | |
| Flock 22 | 2 (80) | 1.471 (100) | 0.055 (96) | 0.638 (86) |
| French flocks | ||||
| Flock 27 | 687 (100) | 0.007 (100) | 0.003 (67) | 0.001 (86) |
| Flock 29 | 2141 (60) | 0.043 (80) | 0.001 (38) | 0.001 (86) |
| 4.031 (80) | 0.107 (96) | 0.34 (100) | ||
| Flock 30 | 8 (80) | 6.64 (100) | 0.048 (92) | 0.127 (100) |
| 3 (20) | 1.479 (100) | 0.108 (100) | 0.228 (100) | |
| Flock 31 | 2141 (80) | Not detected | 0.001 (38) | 0.002 (86) |
| 7 (20) | 7.105 (100) | 0.218 (100) | 0.608 (100) | |
The prevalence of a porAf2 type was compared between young birds from the same flock and young birds from other flocks, based upon total sequence data recovered per sample.
Predominant porAf2 types were defined as being greater than 20% or, most often, greater than 75% of the total sequences recovered from a sample. The percentage of birds within a flock from which the specific porAf2 type was identified is shown in parentheses.
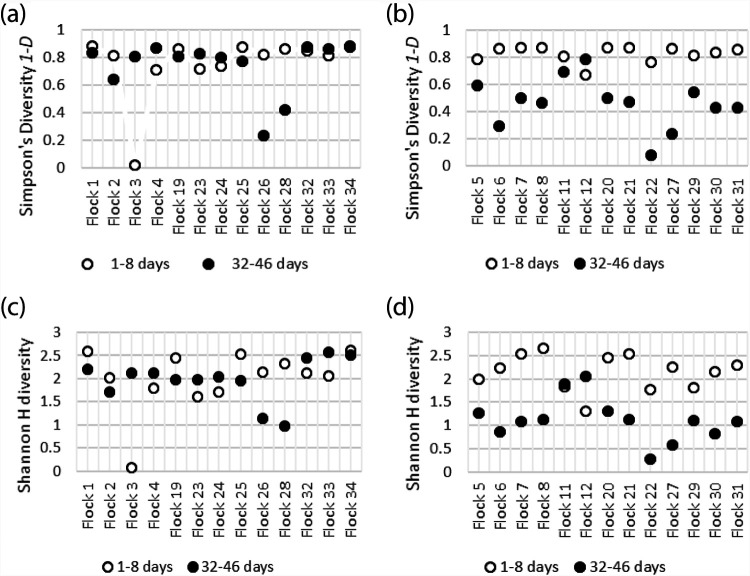
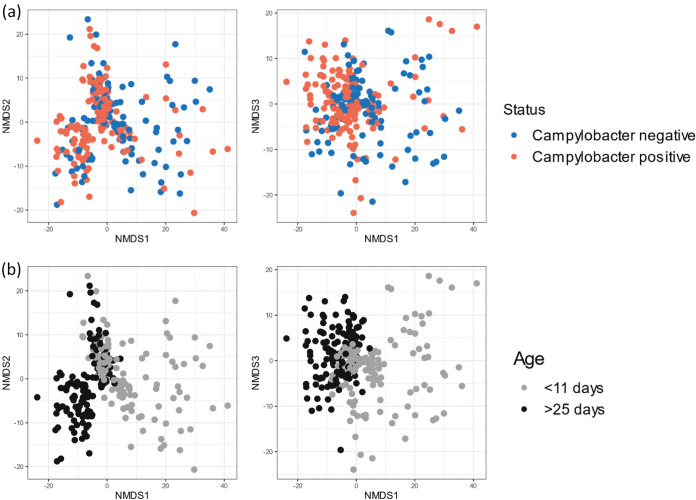
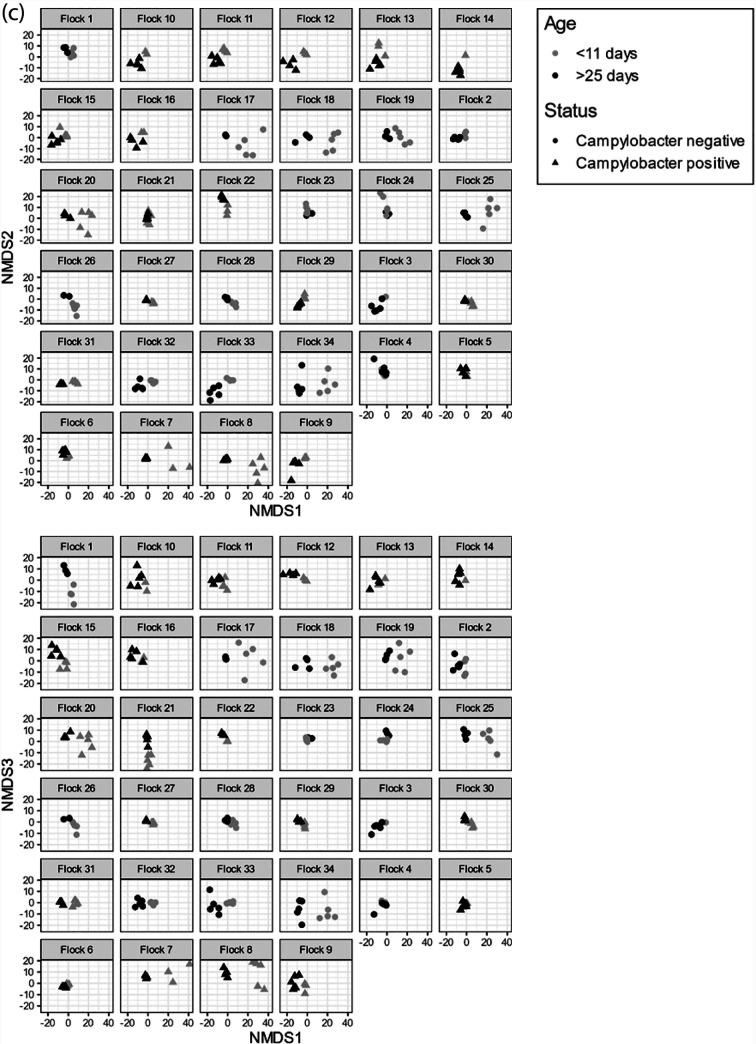
Simpson’s and Shannon’s diversity indices were used to compare the patterns of diversity within porAf2 of flocks sampled at earlier and later time points (Fig. 4). For Campylobacter endpoint culture/qPCR positive flocks, greater diversity was evident for samples from young birds, <8 days of age, compared to birds tested at 35 to 46 days of age from the same flock (n = 13 flocks, Simpson’s diversity P < 0.001, Shannon’s H diversity P = 0.002), but this was not true for endpoint culture/qPCR negative flocks (n = 13 flocks, Simpson’s diversity P = 0.787, Shannon’s H diversity P = 0.68). Aitchison distances, calculated between pairs of samples, demonstrated that there was no apparent relationship between porAf2 type population and whether or not the flock became Campylobacter endpoint culture/qPCR positive (Fig. 5a). The pattern of porAf2 sequences was, however, clearly correlated with age at time of sampling (Fig. 5b) and flock (Fig. 5c).
FIG 4.
(a to d) Diversity of Campylobacter porAf2 populations in Campylobacter endpoint culture/qPCR-negative flocks (a and c) and Campylobacter endpoint culture/qPCR-positive flocks (b and d) using interpolated and extrapolated data. Inverse Simpson’s diversity 1-D (a and b) and Shannon H diversity (c and d).
FIG 5.
Ordination plots showing Aitchison distances between pairs of samples using nonmetric multidimensional scaling (NMDS). (a to c) The effects of (a) Campylobacter culture/qPCR endpoint status, (b) flock age at time of sampling, and (c) individual flock are shown.
Basic comparison of porAf2 variants recovered from countries.
Of the 1,098 porAf2 types identified from the study as a whole, 439 (40.0%) were unique to U.K. flocks and 32 (2.9%) were unique to French flocks. None of the 1,098 porAf2 types were unique to the Swiss flock, with all types identified among U.K. flocks and 254 types found in French flocks. Of the 1,098 7 (0.64%) porAf2 types (porAf2 types 1 to 7) were identified in all 34 flocks, accounting for 49.36% (2,315,725/4,691,464) of the total sequences recovered across the whole study.
Proportion of Campylobacter detected among the bacterial 16S profile.
Campylobacter 16S rRNA gene variants were recovered in samples from birds less than 8 days of age between 0 and 6 times (average 0.95), with an average sequencing depth of 65,000 reads per sample (Fig. S1). Campylobacter 16S rRNA gene variants were recovered in samples from birds 28 to 35 days of age between 0 and 543 times (average, 12.1), with an average sequencing depth of 49,000 reads per sample.
The proportion of Campylobacter detected among the bacterial 16S profile of young commercial bird samples was extremely small, ranging from 0.0001 to 0.012% of the total 16S rRNA gene reads (Table 3), and was similar, regardless of whether or not the flocks subsequently tested Campylobacter culture or qPCR positive at 28 to 35 days of age. However, a small subset of samples (5/151 samples; 3.3%) from 35-day-old chickens across three flocks that tested Campylobacter endpoint culture/qPCR positive at 35 days had greater than 0.1% Campylobacter abundance. This gave an average proportion of Campylobacter sequences among the bacterial 16S profile that was >100-fold larger, at 0.1377% of the sequencing reads (±0.490% standard deviation) (Wilcoxon rank-sum exact test, n = 6 flocks, P = 0.03). With flocks that never became endpoint culture/qPCR positive, the proportion of Campylobacter sequences/total 16S rRNA gene reads did not differ significantly between young and older birds from the same flock (Wilcoxon rank-sum exact text, n = 4 flocks, P = 0.375).
TABLE 3.
Detection of Campylobacter DNAa
| Flock |
Campylobacter negative |
Campylobacter positive |
|||
|---|---|---|---|---|---|
| 6–7 days (SD) | 34–35 days (SD) | 6–7 days (SD) | 28 days (SD) | 34–35 days (SD) | |
| Company A | |||||
| Flock 1 | 0.0022 (0.002) | 0.0012 (0.002) | |||
| Flock 2 | 0.0017 (0.004) | 0.0000 (0.000) | |||
| Flock 3 | 0.0003 (0.001) | 0.0016 (0.002) | |||
| Flock 4 | 0.0081 (0.003) | 0.0057 (0.004) | |||
| Flock 5 | 0.0120 (0.006) | 0.6856 (1.305) | |||
| Flock 6 | 0.0099 (0.010) | 0.1563 (0.162) | |||
| Flock 7 | 0.0005 (0.001) | 0.0012 (0.002) | |||
| Flock 8 | 0.0007 (0.001) | 0.0185 (0.028) | |||
| Company B | |||||
| Flock 9 | 0.0005 (0.001) | 0.0008 (0.001) | |||
| Flock 10 | 0.0001 (0.000) | 0.0002 (0.000) | |||
| Flock 11 | 0.0014 (0.001) | 0.0060 (0.008) | |||
| Flock 12 | 0.0018 (0.001) | 0.0507 (0.062) | |||
| Flock 13 | 0.0003 (0.002) | 0.0011 (0.001) | |||
| Flock 14 | 0.0000 (0.000) | 0.0008 (0.002) | |||
| Flock 15 | 0.0012 (0.002) | 0.0063 (0.012) | |||
| Flock 16 | 0.0000 (0.000) | 0.0146 (0.021) | |||
| Overall mean | 0.0031 | 0.0021 | 0.0024 | 0.0036 | 0.1377 |
Detection is defined as the percentage of the total number of 16S rRNA bacterial profile sequences recovered from fecal samples, with results pooled for each of the flocks. The figures shown are the average of samples from 4 to 5 birds, with sample standard deviation from the mean shown in parentheses. Results are shown for a subset of 16 U.K. flocks from 2 companies that were tested at early and late time points during rearing. The results are separated by flocks that were endpoint culture/qPCR negative and positive for Campylobacter at 28 to 35 days.
DISCUSSION
The porAf2 and 16S rRNA gene sequencing approaches show that Campylobacter DNA is widely present in commercial broiler flocks in birds as young as 1 to 8 days of age, long before it is possible to detect Campylobacter using conventional culture or qPCR methods. The very low abundance (0.0001 to 0.012%) of the bacterial 16S profile attributable to Campylobacter provided a possible explanation for why these chicks are negative using these methods. With a developing microbiome, the already small proportion of Campylobacter may be vanishingly small in terms of biomass and thus below the threshold of detection by culture/qPCR, but not by the more sensitive parallel sequencing approach. That Campylobacter can be detected at an early age, however, is consistent with results from studies of cultured Campylobacter from 0.75% of 2,000 hatchery tray liners, representing pooled samples of 50 day-of-hatch chicks per tray liner (9). Other molecular studies have identified amplifiable Campylobacter DNA among hatchery samples and the reproductive tract of broiler breeders (10, 11). Vertical transfer of Campylobacter from parent flocks by direct or indirect routes provides a possible explanation for such early infection, but this idea remains controversial (6, 12–15).
Variants of porAf2 were identified from at least one fecal sample tested from young chicks <8 days old from each of the commercially reared flocks, irrespective of management regime or DNA extraction method and storage, but not from chickens reared under SPF conditions. A second sequencing-based method of detection using the 16S target also identified Campylobacter DNA among samples from young chicks, from all except two of the commercial flocks tested. Specific amplification of Campylobacter DNA using the porAf2 fragment giving added sensitivity of detection, compared to the broad 16S bacterial profile in which Campylobacter sequences were often rare, may explain the variation in results. Although 16S sequencing analysis pipelines typically include filters to remove very rare sequences, this was not appropriate in this case, where the purpose of the experiment was to detect the presence of predetermined rare sequences (i.e., Campylobacter). Instead, we treat these reads as informative because no samples had a high enough level of Campylobacter 16S sequence to plausibly suggest them as a significant source of cross-sample contamination, either in the laboratory or in data processing. It cannot be certain why so many porAf2 variants were detected by parallel sequencing among samples from older birds that tested negative by culture or qPCR. The correlation between low or high porAf2 variant diversity and culture/qPCR positive or negative result was consistent, however, with the same porAf2 variants predominant among individuals within a flock but different from those predominant in other flocks. Our results suggest that predominance of a single porAf2 type is linked with a higher proportional representation of Campylobacter among the microbiome in at least some birds from a culture/qPCR positive flock, potentially making it more easily detectable. Campylobacter has long proven to be difficult to culture in the laboratory and has been recognized to persist in a viable nonculturable state, with cells retaining an ability to resuscitate under favorable conditions (16). Even when Campylobacter can be cultured, there is evidence of different species or variants being preferentially selected, which could also be a contributing factor (7, 17).
While unable to determine the viability of all Campylobacter porAf2 variants, culture of isolates from endpoint samples for a subset of flocks demonstrated the viability of some at this stage. Each of the porA variants were manually checked by alignment with those derived by culture, and no other species were identified by nucleotide BLAST search using the NCBI database. Although it is possible that the diversity of porAf2 types may be overestimated if there is cross-reaction among other nonrelated species, or by cross-contamination during the sequencing process and technical pipeline, we have found no evidence of this (18).
Sensitive molecular techniques able to detect both low levels of Campylobacter and distinguish among species and strains will be essential in tracing the epidemiology of the organism, and techniques will no doubt be refined as the technology continues to develop (12).
A wide range of genotypes were present in all sample types, with rarefaction curves indicating that up to 450 porAf2 variants may be identified within pooled samples from a flock, irrespective of the endpoint culture/qPCR Campylobacter culture status of the flock. Many of the porAf2 variants were present at very low frequencies (less than 1% of the total). This level of resolution is unachievable by standard culture approaches. In addition, culture induces potential bias where Campylobacter porAf2 variants differ in their capacity for growth under standard laboratory conditions (5). This was evidenced by the recovery of porAf2 type 2866 from flock 20 by culture, where porAf2 type 1 was predominant by parallel sequencing direct from the fecal sample. There was ∼80% sequence homology between the most disparate porAf2 variant nucleotide sequences, which matches the sequence homology when comparing housekeeping loci between C. jejuni and C. coli species (19). The large number of porAf2 variants may reflect diversity derived from host immune selection, which is presumed to act on the outer membrane loop of the MOMP protein encoded by the porA locus (8). In this study, a subset of porAf2 variants (types 1 to 7) were common among all of the flocks tested. It is known that particular Campylobacter lineages are adapted to chickens and are widespread across the globe (20). However, while different porAf2 variants identify different Campylobacter genotypes, typing methods using more loci would give greater resolution to those Campylobacter strains with matching porAf2 variants. The proportion of each porAf2 variant among samples with saturated levels of detection were more evenly distributed in young birds, with Campylobacter DNA representing a very small amount of the total bacterial population. This contrasted markedly with older flocks that were Campylobacter endpoint culture/qPCR positive from 28 days of age, where there were one or two dominant genotypes (7). On average, there was an order of magnitude more Campylobacter 16S OTUs detectable among samples from endpoint culture/qPCR positive flocks, though this was dominated by a small number of birds within three flocks with up to 2.5% Campylobacter detectable. These birds may represent “super shedders,” or individuals that are more susceptible to infection within a flock, and may also be the reason that one or two porAf2 types become dominant among older birds within culture-positive broiler flocks (21). In 76% of cases, the porAf2 variant that became dominant among older birds was detected among samples from young chicks <8 days old from the same flock, despite only 5 samples being tested per flock at each time point.
The porAf2 analysis of the young bird samples showed no difference in either the abundance of Campylobacter present or in the range of genotypes between flocks that subsequently (i.e., from 28 days) became Campylobacter endpoint culture/qPCR positive and those that remained endpoint culture/qPCR negative. This suggests that all young flocks may have similar potential for becoming Campylobacter positive but that only some flocks experience the predominance of a limited number of Campylobacter variants that causes them to become culture or qPCR positive at slaughter. The observation that Campylobacter populations undergo a consistent change in composition with age (as revealed by porAf2 type structure) supports the view that these populations are subject to in vivo selective pressures. These selective pressures may relate to age-associated changes in gut physiology, immune processes, competition with other gut microbes, stress, or other factors.
The key implication of these observations is that they suggest the need for development in the focus of control methods in commercial poultry meat production. This change would expand the focus of interventions from biosecurity during the growing phase to prevention or mitigation of (i) colonization in the very youngest birds and (ii) Campylobacter outgrowth in growing birds. Defining the details of these two processes will be instrumental to reducing Campylobacter colonization or, more importantly, carriage at a high level, which represents the greater risk of transfer to humans. First, it is important to determine how so many chicken flocks become contaminated with Campylobacter at this young age. Second, given the widespread presence of Campylobacter in young flocks, it is important to understand why some maintain low levels of Campylobacter throughout their lives and others exhibit the overgrowth of single types that result in Campylobacter culture-positive flocks. In addition to the on-farm biosecurity measures that have formed the poultry industry’s main line of defense against contamination, it may also be necessary to consider upstream vertical sources of colonization in more detail, as well as the welfare and management of the birds themselves, as key to controlling absolute levels of Campylobacter contamination in foods.
MATERIALS AND METHODS
Fecal sample collection.
Fresh fecal samples were collected from 2015 to 2018 from 34 commercially grown broiler chicken flocks, representing 15 farms, 5 companies, and 3 countries (Table 4). Samples from company A (United Kingdom) were cultured for Campylobacter within 48 h of collection and then stored at −80°C. Samples from company B (United Kingdom) were frozen at −80°C within a day of collection and later tested by qPCR. Samples from companies C, D, and E (France and Switzerland) were immediately added to RNAlater (Thermo Fisher Scientific, UK) and stored at −20°C before long-term storage at −80°C. Fecal samples collected from 10 SPF chickens at 3, 7, 14, 21, 35, and 49 days of age and stored at −80°C were employed as negative controls.
TABLE 4.
Details of the commercial broiler flocks from which fecal samples were testeda
| Sample source | Flock | Year | 1st age (days) | 2nd age (days) | Campylobacter statusb (culture/qPCR) |
|---|---|---|---|---|---|
| U.K. | |||||
| Company A | |||||
| Farm 1Ac | Flock 1 | 2018 | 7 | 35 | Negative |
| Flock 25 | 2018 | 7 | 35 | Negative | |
| Farm 1Bc | Flock 2 | 2018 | 7 | 35 | Negative |
| Flock 3 | 2018 | 7 | 35 | Negative | |
| Farm 1Cc | Flock 4 | 2017 | 6 | 35 | Negative |
| Flock 5 | 2017 | 6 | 35 | Positive | |
| Flock 6 | 2017 | 6 | 35 | Positive | |
| Farm 2 | Flock 7 | 2016 | 7 | 35 | Positive |
| Flock 8 | 2016 | 7 | 35 | Positive | |
| Farm 6 | Flock 17 | 2016 | 3 | 31 | Negative |
| Flock 18 | 2016 | 3 | 31 | Negative | |
| Farm 7 | Flock 19 | 2017 | 4 | 32 | Negative |
| Flock 20 | 2017 | 4 | 32 | Positive | |
| Farm 8 | Flock 21 | 2017 | 7 | 35 | Positive |
| Flock 22 | 2017 | 7 | 35 | Positive | |
| Farm 9 | Flock 23 | 2017 | 7 | 35 | Negative |
| Flock 24 | 2017 | 7 | 35 | Negative | |
| Company B | |||||
| Farm 3A | Flock 9 | 2015 | 7 | 28 | Positive |
| Flock 10 | 2015 | 7 | 28 | Positive | |
| Farm 3B | Flock 11 | 2015 | 7 | 35 | Positive |
| Flock 12 | 2015 | 7 | 35 | Positive | |
| Farm 4 | Flock 13 | 2015 | 7 | 28 | Positive |
| Flock 14 | 2015 | 7 | 28 | Positive | |
| Farm 5 | Flock 15 | 2015 | 7 | 28 | Positive |
| Flock 16 | 2015 | 7 | 28 | Positive | |
| France | |||||
| Company C | |||||
| Farm 10 | Flock 26 | 2017 | 1 | 42 | Negative |
| Flock 27 | 2017 | 1 | 46 | Positive | |
| Farm 11 | Flock 28 | 2016/7 | 1 | 43 | Negative |
| Flock 29 | 2017 | 3 | 42 | Positive | |
| Flock 30 | 2017 | 1 | 42 | Positive | |
| Farm 12 | Flock 31 | 2017 | 3 | 46 | Positive |
| Company D | |||||
| Farm 13 | Flock 32 | 2016 | 3 | 32 | Negative |
| Farm 14 | Flock 33 | 2016/7 | 3 | 32 | Negative |
| Switzerland | |||||
| Company E | |||||
| Farm 15 | Flock 34 | 2016 | 8 | 36 | Negative |
A total of 34 broiler flocks were tested from 3 countries and 5 companies, sampled from 2015 to 2018. Up to 5 fresh fecal samples were tested from different birds at early (<8 days of age) and late (28 to 46 days of age) time points for each flock.
Flocks were tested for Campylobacter by conventional culture or qPCR, using samples collected from the second sample date when birds were aged 28 to 46 days.
Flocks from farms 1 and 3 were placed in rotation and separated by time.
Detection of Campylobacter using culture or qPCR.
Samples from company A were cultured on charcoal cefoperazone deoxycholate agar (CCDA) (PO0119; Oxoid, UK) and incubated in a microaerobic atmosphere using the GenBox Microaer system (bioMérieux Ltd., UK) at 42°C for 48 h. Preliminary work for this study, as well as that previously published, has shown that Campylobacter culture-positive fecal samples from our flocks are typically dominated by a single porAf2 variant, and hundreds of colonies would need to be subcultured per sample before rarer porAf2 variants could be identified (7). Given the volume of samples, a pragmatic approach was therefore taken, with a single presumptive Campylobacter colony from each sample subcultured onto Columbia blood agar (CBA) (PB0122; Oxoid, UK) and incubated for another 48 h at 42°C. Identity was confirmed by characteristic appearance and Gram stain, by catalase and oxidase tests, and by DNA sequencing of the porAf1 fragment (previously referred to as the porA fragment) using the Sanger sequencing method (7).
Fecal samples from companies B, C, D, and E were tested for the presence of Campylobacter using qPCR. Between 10 and 20 samples were pooled for each flock at each time point, and the DNA was extracted using the protocols described below. Primers and probes detecting portions of the mapA (C. jejuni) and ceuE (C. coli) genes, published by Best et al. (22), were used to detect the presence of C. jejuni and/or C. coli. Positive results were recorded for threshold cycle (CT) values between 20 and 32.
DNA extraction from fecal samples.
Equipment and surfaces were decontaminated using 2% Virkon and DNAZap PCR DNA degradation solutions (AM9890; Thermo Fisher Scientific, UK) and 70% ethanol before use and between batches. DNA was extracted from fecal samples not stored in RNAlater using the Maxwell 16 LEV blood DNA kit (AS1290; Promega, UK) and automated DNA extraction system. Briefly, 250 mg of sample was added to a tube containing one-quarter volume 0.5-mm zirconia/silica beads (Thistle Scientific, UK) and 600 μl lysis buffer. Samples were subjected to a bead beater for 1 min at high speed and then heated at 95°C for 10 min in a water bath. They were centrifuged in a microcentrifuge at 13,200 rpm for 5 min before the supernatant was transferred to fresh tubes containing proteinase K and incubated at 56°C for 20 min. The samples were then loaded onto the Maxwell 16 LEV blood kit cartridges, and DNA extraction was performed using the manufacturer’s “blood” protocol.
Fecal samples stored in RNAlater were centrifuged at 13,200 rpm for 10 min, and the supernatant was removed prior to DNA extraction procedures in order to concentrate and retrieve the original sample material. They were then washed in excess cold phosphate-buffered saline (PBS) and centrifuged for another 10 min at 13,200 rpm, and the supernatant was discarded. DNA was then extracted using the Qiagen DNeasy PowerSoil kit (no. 12888-100; Qiagen, UK) following the manufacturer’s instructions. DNA prepared using the Maxwell system was diluted 1:10 before use to remove the effects of PCR inhibitors but was used neat for Qiagen extractions
Parallel sequencing for 16S (bacterial profiling) and porAf2 (Campylobacter).
Using a standard approach, 25-μl PCRs were prepared in triplicate in a PCR UV cabinet in a designated clean room. Equipment was decontaminated before use and between batches using DNAZap (AM98902; Thermo Fisher Scientific, UK), 70% ethanol, and UV light for a minimum of 15 min. Fresh plastic ware and reagents/aliquots and nontemplate controls containing molecular water were used for every set of PCRs.
For a subset of samples, variable regions 3 and 4 of the 16S rRNA gene were sequenced using the following primers: E338 5′-ACTCCTACGGGAGGCAGCAGT-3′ and R806 5′-GGACTACHVGGGTWTCTAAT-3′. For all samples, a short 405- to 473-bp region of the porA gene was sequenced using the following primers: MOMP B 5′-CCA CAA TTA TGG TTA GCT TA-3′ and MOMP 2R 5′-TGA GAA GTT AAG TTT TGG AGA G-3′ (7). The E338 forward primer was tagged with 10 different nucleotide barcodes for each reaction, and the MOMP2R reverse primer was tagged with different 7 nucleotide barcodes for each reaction, enabling reactions to be multiplexed within the same sequencing library. For both sets of PCR, the high-fidelity Phusion Hot Start Flex DNA polymerase enzyme and 5X Phusion HF buffer (New England Biolabs, UK; M0535) were used according to the manufacturer’s recommendations. The following thermocycling conditions were used: (i) for 16S, initial denaturation at 98°C for 5 min, 25 cycles of 98°C for 15 s, 50°C for 15 s, 72°C for 90 s, and final extension of 72°C for 10 min; (ii) for porAf2, initial denaturation at 98°C for 30 s, 35 cycles of 98°C for 10 s, 58°C for 30 s, 72°C for 30 s, and final extension of 72°C for 10 min. Library preparation was performed in a third room, separate from the DNA extraction and PCR setup, using protocols described previously (7). PCR products were sequenced using the Illumina MiSeq platform and 600-cycle MiSeq reagent kit v3 (Illumina, UK; MS-102-3003), giving 300 paired nucleotide reads. Samples were loaded with 10% phiX.
Quality control.
The processes of DNA extraction, PCR set-up, and sequencing library preparation were performed in separate rooms. All surfaces, including equipment, were decontaminated with DNAZap (Invitrogen, UK) between batches of samples. Samples from young chicks containing small amounts of DNA were handled before samples from older chicks, apart from a pilot study where DNA was extracted and sequenced from both sample types in a mixed arrangement. PCR was performed in a dedicated PCR room, within separate PCR hoods, reserved for master mix preparation and addition of sample DNA. The sequencing reactions were performed across 10 nonsuccessive MiSeq runs and were set up by three members of staff.
A subset of 10 samples, from flocks 1 and 21, aged 7 days, were prepared individually through DNA extraction and PCR processes to control for potential cross-contamination between samples prepared in the same batch. At each stage, only one of the samples was handled in a room at a time, and the process of DNA extraction was performed, or PCR was completed for a particular sample, before the next sample was started. DNA extraction was performed for three samples from flock 1 using a Maxwell 16 robot (a single cartridge used at a time). DNA was extracted for two samples from flock 1 and five samples from flock 21 using the Qiagen DNeasy PowerSoil kit in enclosed tubes.
Processing of MiSeq data.
Barcoded reads were demultiplexed and terminal primer sequences were trimmed using custom Python scripts. Any reads still containing a copy of either primer sequence after this trimming were identified using cutadapt v1.15 and excluded from further analyses on the assumption that they represented PCR artifacts (e.g., concatemers). The fastq_eestats2 command in USEARCH v10.0.240 was used to assess the optimal 3′ trimming length to remove low-quality sequences, which were 270 bp for our data (23). The DADA2 3.10 software package was used to trim reads, filter to ≤1 expected errors/read, assign sequence variants (using pooled sequences), merge read pairs, and remove chimeras (using the removeBimeraDenovo function with method = consensus) (24). The counts of Campylobacter 16S operational taxonomic units (OTUs) per sample were calculated from a phyloseq object using the subset_taxa function (25). The porAf2 nucleotide sequences were aligned in MEGA X to confirm that they were coding sequences for correct fragments from Campylobacter (26). A small number of sequences containing stop codons were removed from further analyses. Allele numbers for nucleotide and peptide sequences for porAf2 were assigned using the PubMLST Campylobacter database (https://pubmlst.org/organisms/campylobacter-jejunicoli) (27). The assigned porAf2 alleles are publicly available and can be found by downloading allele sequences that are not within assigned typing schemes, on the typing section of the database. The raw data reads have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB45083.
Data analyses.
Data were transformed to an even sampling depth, giving proportional frequency of each porAf2 type, using the phyloseq package in R (25). Data from flock 14, aged 7 days, were removed from the analyses at this point, as the sequencing depth was low, between 127 and 281. These data were used for all subsequent analyses, with the exception of Simpson’s and Shannon’s diversity indices, which were performed on raw and interpolated/extrapolated data (28) and rarefaction analyses performed on the raw data. Bar charts and Table 3 calculations were prepared using Tableau v2019.4 software. Paired Wilcoxon rank-sum exact tests were used to compare (i) the difference in the percentage of bacterial 16S profile represented by Campylobacter 16S rRNA gene variants between flocks tested at young and older time points, (ii) the prevalence of a porAf2 type among young birds in a flock before it became predominant, with the average prevalence among young birds from other flocks, (iii) the number of porAf2 types identified among flocks tested at young and older times points, and (iv) Simpson’s diversity index and Shannon’s H diversity index between Campylobacter culture-positive and -negative flocks. A Mann-Whitney U test was used to compare the number of porAf2 types identified among Campylobacter culture-positive and -negative flocks. The tests were performed using Paleontological Statistics (PAST) v4.0 software (29).
PAST v4.0 software was also used to calculate rarefaction curves for samples pooled by flock/age (total sequencing data). The iNEXT (interpolation/extrapolation) R package was used to calculate Simpson’s and Shannon’s diversity indices on raw data, as well as to give interpolated/extrapolated diversity estimates for each sample, allowing them to be compared to each other in a standardized way (28). For Simpson’s diversity index, a 1-D value of 1.0 indicated that all members of a population could be distinguished from each other, and a 1-D value of 0 indicated that all members of a population were identical (30). Shannon’s diversity index was included, as it is considered to give more weight to rare species (pofAf2 variants) (31). An H value of 0 indicated that all species were the same. H increases with increasing number of species.
As parallel sequencing data are by nature compositional, differences between Campylobacter porAf2 populations was assessed using a nonmetric multidimensional scaling (NMDS) visualization of the Aitchison distance between samples (32). DADA2-processed MiSeq data (see above) were further analyzed using various R packages, as noted below, with the use of the data.table package (33) for data handling and the ggplot2 package (34) for plotting. First, the data were filtered to remove samples with <500 reads and sequences which occurred in only a single sample. To permit the subsequent (log-based) transform, low nonzero values were then input for zero values using zCompositions::multRepl (35) with the CZM method. The zero-input data were transformed by centered log-ratio (clr) using compositions::clr (36), and the Aitchison distances between samples were calculated (Aitchison distance being the Euclidean distance following clr-transform). NMDS was then performed in three dimensions with vegan::metaMDS (37) using the monoMDS engine.
The Bray-Curtis dissimilarity index was used to assess the effects of using two different DNA extraction kits (38). The ordination function with the NMDS method, together with the phyloseq, ggPlot2, and Plyr packages were used to produce the Bray-Curtis ordination plots in R (25, 34, 39).
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Science Research Council (grant numbers BB/N023803/1 and BB/K004468/1) including part of the Animal Health and Welfare ERA-net call, and also the BBSRC DTP Integrated Biosciences (grant number BB/M011224/1). The views expressed are those of the author(s) and not necessarily those of the BBSRC. We gratefully acknowledge in-kind support from Avara Foods, and thank all companies for providing access for samples. We also thank Benjamin Schusser from the Technical University of Munich for providing fecal samples from SPF birds.
This study used broiler flocks that were reared commercially by industry partners, in line with standard industry practice, and involved noninvasive sampling of feces. All prevailing local, national, and international regulations and conventions and normal scientific ethical practices were respected. The Swiss samples were approved by the Canton of Bern (BE97/16) and met all cantonal and federal regulations for the treatment of animals.
Footnotes
Supplemental material is available online only.
Contributor Information
F. M. Colles, Email: frances.colles@zoo.ox.ac.uk.
Charles M. Dozois, INRS—Institut Armand-Frappier
REFERENCES
- 1.Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, Ogden ID, Maiden MC, Forbes KJ. 2009. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis 48:1072–1078. 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Food Safety Authority. 2010. Analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Campylobacter and Salmonella on broiler carcasses in the EU, 1008. EFSA J 8:1503–1602. 10.2903/j.efsa.2010.1503. [DOI] [Google Scholar]
- 3.Jorgensen F, Charlett A, Swift C, Corcionivoschi N, Elviss NC. 2019. A microbiological survey of Campylobacter contamination in fresh whole UK-produced chilled chickens at retail sale. https://www.food.gov.uk/sites/default/files/media/document/campylobacter-contamination-uk-chickens-year-4-report.pdf.
- 4.Newell DG, Fearnley C. 2003. Sources of Campylobacter colonization in broiler chickens. Appl Environ Microbiol 69:4343–4351. 10.1128/AEM.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hermans D, Pasmans F, Messens W, Martel A, Van Immerseel F, Rasschaert G, Heyndrickx M, Van Deun K, Haesebrouck F. 2012. Poultry as a host for the zoonotic pathogen Campylobacter jejuni. Vector Borne Zoonotic Dis 12:89–98. 10.1089/vbz.2011.0676. [DOI] [PubMed] [Google Scholar]
- 6.Koutsoumanis K, Allende A, Alvarez-Ordonez A, Bolton D, Bover-Cid S, Davies R, De Cesare A, Herman L, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Alter T, Crotta M, Ellis-Iversen J, Hempen M, Messens W, Chemaly M, EFSA Panel on Biological Hazards (BIOH). 2020. Update and review of control options for Campylobacter in broilers at primary production. EFSA J 18:4145–4154. 10.2903/j.efsa.2020.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colles FM, Preston SG, Barfod KK, Flammer PG, Maiden MCJ, Smith AL. 2019. Parallel sequencing of porA reveals a complex pattern of Campylobacter genotypes that differs between broiler and broiler breeder chickens. Sci Rep 9:6204. 10.1038/s41598-019-42207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cody AJ, Maiden MJC, Dingle KE. 2009. Genetic diversity and stability of the porA allele as a genetic marker in human Campylobacter infection. Microbiology 155:4145–4154. 10.1099/mic.0.031047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd J, Bailey RH, Wills R, Nisbet D. 2007. Recovery of Campylobacter from commercial broiler hatchery trayliners. Poult Sci 86:26–29. 10.1093/ps/86.1.26. [DOI] [PubMed] [Google Scholar]
- 10.Rothrock MJ, Jr, Locatelli A, Feye KM, Caudill AJ, Guard J, Hiett K, Ricke SC. 2019. A microbiomic analysis of a pasture-raised broiler flock elucidates foodborne pathogen ecology along the farm-to-fork continuum. Front Vet Sci 6:260. 10.3389/fvets.2019.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox NA, Richardson LJ, Maurer JJ, Berrang ME, Fedorka-Cray PJ, Buhr RJ, Byrd JA, Lee MD, Hofacre CL, O'Kane PM, Lammerding AM, Clark AG, Thayer SG, Doyle MP. 2012. Evidence for horizontal and vertical transmission in Campylobacter passage from hen to her progeny. J Food Prot 75:1896–1902. 10.4315/0362-028.JFP-11-322. [DOI] [PubMed] [Google Scholar]
- 12.Ingresa-Capaccioni S, Jimenez-Trigos E, Marco-Jimenez F, Catala P, Vega S, Marin C. 2016. Campylobacter epidemiology from breeders to their progeny in eastern Spain. Poult Sci 95:676–683. 10.3382/ps/pev338. [DOI] [PubMed] [Google Scholar]
- 13.Battersby T, Whyte P, Bolton DJ. 2016. The pattern of Campylobacter contamination on broiler farms; external and internal sources. J Appl Microbiol 120:1108–1118. 10.1111/jam.13066. [DOI] [PubMed] [Google Scholar]
- 14.Tangkham W, Janes M, LeMieux F. 2016. Prevalence and distribution of Campylobacter jejuni in small-scale broiler operations. J Food Prot 79:75–81. 10.4315/0362-028X.JFP-15-331. [DOI] [PubMed] [Google Scholar]
- 15.Callicott KA, Friethriksdottir V, Reiersen J, Lowman R, Bisaillon JR, Gunnarsson E, Berndtson E, Hiett KL, Needleman DS, Stern NJ. 2006. Lack of evidence for vertical transmission of Campylobacter spp. in chickens. Appl Environ Microbiol 72:5794–5798. 10.1128/AEM.02991-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv R, Wang K, Feng J, Heeney DD, Liu D, Lu X. 2019. Detection and quantification of viable but non-culturable Campylobacter jejuni. Front Microbiol 10:2920. 10.3389/fmicb.2019.02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams LK, Sait LC, Cogan TA, Jorgensen F, Grogono-Thomas R, Humphrey TJ. 2012. Enrichment culture can bias the isolation of Campylobacter subtypes. Epidemiol Infect 140:1227–1235. 10.1017/S0950268811001877. [DOI] [PubMed] [Google Scholar]
- 18.Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS. 2019. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol 27:105–117. 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Dingle KE, Colles FM, Falush D, Maiden MC. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol 43:340–347. 10.1128/JCM.43.1.340-347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheppard SK, Colles F, Richardson J, Cody AJ, Elson R, Lawson A, Brick G, Meldrum R, Little CL, Owen RJ, Maiden MC, McCarthy ND. 2010. Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol 76:5269–5277. 10.1128/AEM.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawson T, Paton RS, Colles FM, Maiden MCJ, Dawkins MS, Bonsall MB. 2020. A Mathematical modeling approach to uncover factors influencing the spread of Campylobacter in a flock of broiler-breeder chickens. Front Microbiol 11:576646. 10.3389/fmicb.2020.576646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best EL, Powell EJ, Swift C, Grant KA, Frost JA. 2003. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol Lett 229:237–241. 10.1016/S0378-1097(03)00845-0. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh TC, Ma KA, Chao A. 2016. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7:1451–1456. 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 29.Hammer O, Harper DAT, Ryan PD. 2001. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electronica 4:1903–1905. [Google Scholar]
- 30.Hunter PR. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol 28:1903–1905. 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon CE. 1948. A mathematical theory of communication. AT&T Tech J 27. [PubMed] [Google Scholar]
- 32.Quinn TP, Erb I, Richardson MF, Crowley TM. 2018. Understanding sequencing data as compositions: an outlook and review. Bioinformatics 34:2870–2878. 10.1093/bioinformatics/bty175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowle F, Srinivasan A. 2020. data.table: Extension of ‘data frame’. R package 1.13.0. https://CRAN.R-project.org/package=data.table.
- 34.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 35.Palarea-Albaladejo J, Martin-Fernandez JA. 2015. zCompositions: R package for multivariate imputation of left-censored data under a compositional approach. Chemom Intell Lab Syst 143:85–96. 10.1016/j.chemolab.2015.02.019. [DOI] [Google Scholar]
- 36.Gerald van der Boogart K, Tolosana-Delgado R, Bren M. 2020. Compositions: compositional data analysis. R package version 2.0-0. https://CRAN.R-project.org/package=compositions. Accessed
- 37.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan.
- 38.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. 10.2307/1942268. [DOI] [Google Scholar]
- 39.Wickham H. 2011. The split-apply-combine strategy for data analysis. J Stat Softw 40:1–29. 10.18637/jss.v040.i01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3. Download aem.01060-21-s0001.pdf, PDF file, 0.3 MB (325.4KB, pdf)