ABSTRACT
Despite the importance of biofilm formation in the contamination of meat by pathogenic Escherichia coli at slaughter plants, drivers for biofilm remain unclear. To identify selection pressures for biofilm, we evaluated 745 isolates from cattle and 700 generic E. coli isolates from two beef slaughter plants for motility, the expression of curli and cellulose, and biofilm-forming potential. Cattle isolates were also screened for serogroup, stx1, stx2, eae, and rpoS. Generic E. coli isolates were compared by source (hide of carcass, hide-off carcass, and processing equipment) before and after the implementation of antimicrobial hurdles. The proportion of E. coli isolates capable of forming biofilms was lowest (7.1%; P < 0.05) for cattle isolates and highest (87.3%; P < 0.05) from equipment. Only one enterohemorrhagic E. coli (EHEC) isolate was an extremely strong biofilm former, in contrast to 73.4% of E. coli isolates from equipment. Isolates from equipment after sanitation had a greater biofilm-forming capacity (P < 0.001) than those before sanitation. Most cattle isolates were motile and expressed curli, although these traits along with the expression of cellulose and the detection of rpoS were not necessary for biofilm formation. In contrast, isolates capable of forming biofilms on equipment were almost exclusively motile and able to express curli. The results of the present study indicate that cattle rarely carry EHEC capable of making strong biofilms in slaughter plants. However, if biofilm-forming EHEC contaminates equipment, current sanitation procedures may not eliminate the most robust biofilm-forming strains. Accordingly, new and effective antibiofilm hurdles for meat-processing equipment are required to reduce future instances of foodborne disease.
IMPORTANCE As the majority of enterohemorrhagic E. coli (EHEC) isolates are not capable of forming biofilms, sources were undetermined for biofilm-forming EHEC isolated from “high-event periods” in beef slaughter plants. This study demonstrated that sanitation procedures used on beef-processing equipment may inadvertently lead to the survival of robust biofilm-forming strains of E. coli. Cattle only rarely carry EHEC capable of forming strong biofilms (1/745 isolates evaluated), but isolates with greater biofilm-forming capacity were more likely (P < 0.001) to survive equipment sanitation. In contrast, chilling carcasses for 3 days at 0°C reduced (P < 0.05) the proportion of biofilm-forming E. coli. Consequently, an additional antibiofilm hurdle for meat-processing equipment, perhaps involving cold exposure, is necessary to further reduce the risk of foodborne disease.
KEYWORDS: Escherichia coli, biofilm, microbial interventions, D value, locus of heat resistance, cattle, beef-packing plant, biofilms
INTRODUCTION
High-event periods (HEPs) occur sporadically in beef-processing plants when a higher-than-expected proportion of trim samples are positive for enterohemorrhagic Escherichia coli (EHEC) (1). Isolates of O157:H7 from HEPs have a strong ability to form mature biofilms (2), in contrast to the estimated 95% of O157:H7 isolates that lack individual biofilm-forming capacity (3), although EHEC may also become integrated into mixed-species biofilms (4). Among non-O157 E. coli isolates, biofilm formation is thought to be more common than in O157:H7 isolates (5), but with the exception of the O111 and O145 serogroups, carriage of Shiga toxins is also less frequent among non-O157 E. coli isolates (6). Impaired biofilm formation in all E. coli isolates has previously been linked to three factors: (i) an stx1 prophage insertion in mlrA preventing the expression of curli fimbriae, (ii) a mutation in rpoS reducing the expression of both cellulose and curli, or (iii) a lack of motility negatively impacting both the expression of curli and initial reversible biofilm attachment (7). Biofilm formation among E. coli isolates has been thought to be extremely variable among strains (8), but excluding O157, relatively few strains have been evaluated per serogroup (5, 8, 9).
Although isolates from HEPs have been characterized, the source of the biofilm-forming E. coli isolates causing HEPs has not yet been determined. One theory is that pathogenic E. coli isolates on the hides of cattle contaminate meat products after high bacterial loads overwhelm antimicrobial interventions at one or multiple stages within the processing facility (10). However, as Arthur et al. (1) found little genetic diversity in E. coli strains isolated from HEPs, they proposed that failures in sanitation might occur, transferring a single prevalent strain of EHEC to multiple sites within the slaughter plant. A third possibility, also proposed by Arthur et al. (1), would be that antimicrobial interventions within the slaughter plant would inadvertently select for robust biofilm-forming strains causing HEPs. Besides the enhanced formation of biofilm, strains of E. coli from HEPs have shown increased tolerance to sanitizers (2), although the relationship is unclear for biofilm-forming ability and attributes such as heat resistance, which may also allow E. coli to survive microbial interventions. Consequently, the present study was undertaken to determine the relationships among the biofilm-forming abilities of E. coli strains isolated along the production chain, including live cattle, hides of carcasses, hide-off carcasses, meat products, and processing equipment. For isolates collected in slaughter plants, the impacts of antimicrobial interventions (hide wash, carcass chilling, and equipment sanitation) were determined. Factors possibly influencing the biofilm-forming ability, including seasonality, the expression of curli and cellulose, and motility, of the isolates were evaluated, along with the relationship between biofilm-forming capacity and heat resistance. For cattle isolates, impacts of serogroup (O26, O45, O103, O111, O121, O145, and O157 ["top 7"]) on biofilm formation and the presence of stx1, stx2, eae, and rpoS were determined.
RESULTS
PCR characterization of top 7 E. coli isolates.
Each serogroup differed in the predominant combinations of Shiga toxin genes, eae, and rpoS that were detected (Table 1). For O26, the majority of isolates were eae positive and split between isolates lacking Shiga toxin genes and conserving rpoS and EHEC isolates positive for stx1 and eae. In contrast, the largest group of O45 isolates lacked eae, stx1, and stx2 and conserved rpoS. Only 13.7% of O45 isolates were EHEC, in comparison to 39.2% of O26 isolates. Isolates of O103 were evenly split between eae-positive and -negative isolates, with 35.7% being EHEC. However, in contrast to O26 and O45, 9.5% of O103 isolates were positive for rpoS, compared to only 1% of O45 and 2.9% of O26 isolates. Little diversity was present in O111, with 83% being EHEC carrying stx1 and none being positive for rpoS, although isolate numbers for this serogroup were limited (n = 12). Isolates of O121, O145, and O157 were distinguished from other serogroups by more carriage of stx2, with 40.2% of O121 isolates being EHEC and 16.7% being positive for rpoS. All isolates of O145 were eae and rpoS positive, with 46.7% being EHEC. By far the largest proportion of EHEC was for O157 (91.2%), with the majority of isolates carrying rpoS, stx1, and stx2.
TABLE 1.
Top seven E. coli strains (n = 745) by serogroup and the presence of stx1, stx2, eae, and rpoSa
| Serogroup, eae status | No. of isolates with: |
Total no. of isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No stx1, stx2, or rpoS | stx1 only | stx2 only | stx1 and stx2 | rpoS and stx1 | rpoS and stx2 | rpoS, stx1, and stx2 | rpoS only | ||
| O26, eae positive | 45 | 36 | 2 | 1 | 1 | 0 | 0 | 1 | 86 |
| O26, eae negative | 5 | 5 | 5 | 0 | 1 | 0 | 0 | 0 | 16 |
| O45, eae positive | 19 | 13 | 0 | 1 | 0 | 0 | 0 | 0 | 33 |
| O45, eae negative | 43 | 20 | 2 | 3 | 0 | 0 | 0 | 1 | 69 |
| O103, eae positive | 25 | 33 | 6 | 7 | 2 | 4 | 4 | 6 | 87 |
| O103, eae negative | 39 | 13 | 0 | 3 | 3 | 0 | 0 | 12 | 70 |
| O111, eae positive | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| O111, eae negative | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| O121, eae positive | 7 | 13 | 15 | 6 | 7 | 0 | 0 | 1 | 49 |
| O121, eae negative | 23 | 7 | 0 | 14 | 2 | 0 | 0 | 7 | 53 |
| O145, eae positive | 2 | 0 | 0 | 0 | 13 | 0 | 3 | 17 | 35 |
| O145, eae negative | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| O157, eae positive | 1 | 9 | 0 | 6 | 29 | 8 | 162 | 1 | 216 |
| O157, eae negative | 7 | 8 | 0 | 0 | 2 | 0 | 0 | 2 | 19 |
| Total | 216 | 169 | 30 | 41 | 60 | 12 | 169 | 48 | 745 |
Predominant combinations for each serogroup are shaded in gray.
Biofilm-forming potential, motility, expression of curli and cellulose in top 7 isolates.
Few top 7 isolates were able to form biofilms, with 92.9% being classified as non-biofilm formers (Table 2). The proportion of EHEC isolates capable of forming biofilms was even smaller, at 2.1%. Only two EHEC isolates capable of forming biofilms also possessed the locus of heat resistance (LHR) and were classified as moderate and strong biofilm formers, respectively. As the biofilm-forming capacity increased, the numbers of top 7 isolates having these phenotypes decreased, with only 2 isolates being extremely strong biofilm formers and 1 of these being EHEC. However, none of the EHEC isolates that were biofilm formers were motile and expressed both curli and cellulose. Overall, motility and the expression of curli and cellulose were uncommon in top 7 isolates, with only 25.5% of the isolates having all three traits.
TABLE 2.
Biofilm-forming capacity, curli and cellulose expression, motility, and the presence of virulence genes and the locus of heat resistance in isolates of top seven E. coli strains (n = 745) collected from cattlea
| Cellulose, curli, and virulence attribute | No. of isolates with biofilm-forming capacity (no. with LHR)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Non-biofilm former |
Weak |
Moderate |
Strong |
Extreme |
||||||
| stx1 negative | stx1 positive | stx1 negative | stx1 positive | stx1 negative | stx1 positive | stx1 negative | stx1 positive | stx1 negative | stx1 positive | |
| Not CCMc | ||||||||||
| stx2 negative | ||||||||||
| eae negative | 72 (4 LHR) | 32 (1 LHR) | 4 | 3 | 3 (1 LHR) | 5 (2 LHR) | 1 | 1 LHR | 0 | 0 |
| eae positive | 94 (1 LHR) | 116 | 1 | 3 | 1 | 2 (1 LHR) | 0 | 4 (1 LHR) | 0 | 0 |
| stx2 positive | ||||||||||
| eae negative | 5 (3 LHR) | 11 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| eae positive | 22 (2 LHR) | 173 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 1 |
| CCMd | ||||||||||
| stx2 negative | ||||||||||
| eae negative | 53 | 20 | 3 | 1 | 2 | 2 | 1 | 1 | 1 | 0 |
| eae positive | 24 | 35 | 1 | 1 | 1 | 2 | 1 | 0 | 0 | 0 |
| stx2 positive | ||||||||||
| eae negative | 1 | 8 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| eae positive | 14 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 285 | 407 | 9 | 10 | 9 | 13 | 3 | 7 | 1 | 1 |
Shiga toxin-producing E. coli isolates are shaded in gray. Enterohemorrhagic E. coli isolates are in boldface type. LHR, locus of heat resistance.
Biofilm-forming capacity is as follows: non-biofilm former, x < ODc; weak, ODc < x < 2× ODc; moderate, 2× ODc < x < 4× ODc; strong, 4× ODc < x < 8× ODc; extreme, 16× ODc < x. No isolates were classified as very strong biofilm formers (8× ODc < x < 16× ODc).
Not CCM, the isolate does not have all three of the following characteristics: motility, expression of curli, and expression of cellulose.
CCM, the isolate is motile and expresses both curli and cellulose.
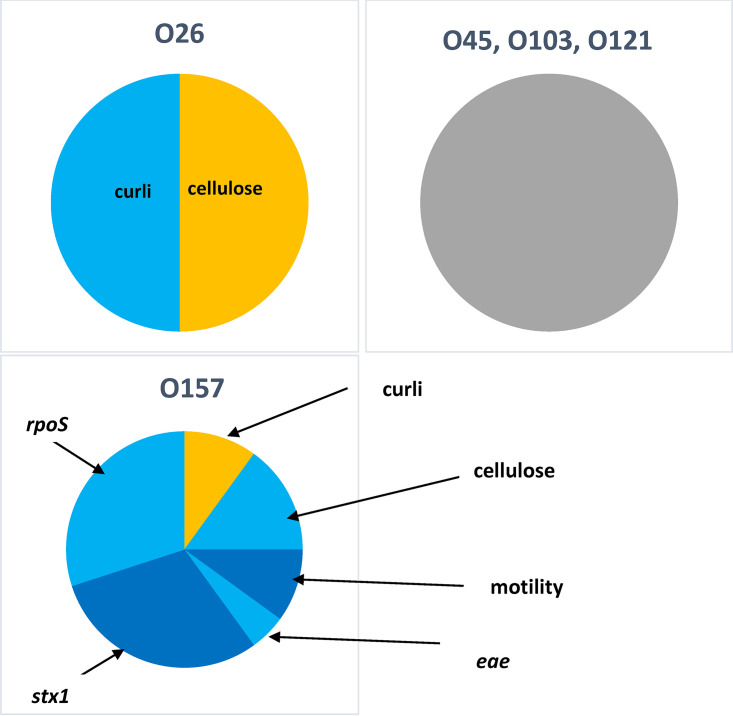
Although serogroup did not influence the proportion of top 7 isolates producing biofilms (Table 3), serogroup did influence motility, which was highest (P < 0.05) for O157 and lowest for O26, O111, and O145. Similarly, a higher proportion (P < 0.05) of O157 isolates expressed curli than all other serogroups with the exception of O26. Comparing isolates that both expressed curli and were positive for stx1, O157 again had the highest proportion (P < 0.05) compared to all serogroups with the exception of O111. However, evaluating the factors that influenced biofilm formation within a serogroup, the presence of eae and stx1 negatively influenced (P < 0.05) biofilm formation in O157 (Fig. 1), meaning that the small proportion of O157 isolates that were not EHEC were generally the strongest biofilm formers. For O157, the expression of curli increased (P < 0.05) biofilm formation, while the expression of cellulose, motility, and the presence of rpoS reduced (P < 0.05) biofilm formation. For O26, the expression of cellulose increased (P < 0.05) biofilm formation, while the expression of curli negatively impacted biofilm (P < 0.05). For other serogroups, no factors evaluated affected biofilm formation.
TABLE 3.
Relationship between the expression of curli, the presence of stx1, and the production of biofilm by top 7 serogroup strainsa
| Serogroup | No. of isolates | % motile isolates | % of isolates positive for rpoS | % of isolates expressing curli | % of isolates expressing curli and positive for stx1 | % of isolates forming biofilms | % of isolates forming biofilms, expressing curli, and positive for stx1 |
|---|---|---|---|---|---|---|---|
| O26 | 102 | 73.1 A | 3.2 A | 82.3 AB | 31.4 A | 6.9 | 3.9 |
| O45 | 102 | 93.1 BC | 0.1 A | 78.4 A | 26.4 A | 7.8 | 1.0 |
| O103 | 157 | 86.6 AB | 19.8 B | 78.3 A | 34.4 A | 5.1 | 1.3 |
| O111 | 12 | 58.33 A | 0.0 A | 66.7 A | 66.7 AB | 16.7 | 0.0 |
| O121 | 102 | 91.0 ABC | 16.7 B | 73.5 A | 34.3 A | 5.9 | 1.0 |
| O145 | 35 | 69.7 A | 93.9 C | 68.6 A | 28.6 A | 0.0 | 0.0 |
| O157 | 235 | 95.7 C | 87.2 C | 92.8 B | 85.7 B | 7.6 | 5.4 |
Data in columns with different letters differ significantly (P < 0.05).
FIG 1.
Predictors of biofilm formation by serogroup of top 7 E. coli isolates, including the expression of curli and cellulose, motility, and the presence of eae, rpoS, and stx1. The presence of yellow-shaded attributes significantly increased the likelihood of biofilm formation. The presence of light blue- or dark blue-shaded attributes significantly reduced the likelihood of biofilm formation. The presence of gray shading indicates no significant predictors of biofilm formation. The proportion of the pie is equivalent to relative significance. O111 and O145 were excluded due to having two or fewer biofilm formers. The presence of stx2 did not influence biofilm formation by top 7 E. coli isolates.
Biofilm-forming potential, motility, and expression of curli and cellulose in generic E. coli isolates.
In contrast to top 7 isolates, 42.9% of generic E. coli isolates were capable of forming biofilms (Table 4). From processing equipment, 70% of isolates were strong biofilm formers, which were motile and expressed both curli and cellulose. At the opposite extreme, 73% of isolates from beef products were non-biofilm formers, with 40% of beef isolates being motile and expressing both curli and cellulose. Hide-off carcass isolates were 53.0% biofilm formers, and 49.8% were motile and expressed curli and cellulose. Similar to hide-off carcasses, isolates collected from the hides of carcasses were 44.6% biofilm formers, with 42.1% being motile and expressing both curli and cellulose. A total of seven isolates collected from hide-off carcasses classified as very strong or extremely strong biofilm formers also possessed the LHR, although no LHR-positive isolates were present in beef products or from processing equipment.
TABLE 4.
Biofilm-forming capacity, presence of the locus of heat resistance, motility, and curli and cellulose expression by stage of processing (n = 500) or processing equipment (n = 200) for generic E. coli isolates collected in two federally inspected beef slaughter plants in Alberta
| Biofilm-forming capacity categorya | No. of isolates (no. with LHR) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Hide of carcass |
Hide-off carcass |
Beef products |
Processing equipment |
|||||
| MCCb | Not MCCc | MCC | Not MCC | MCC | Not MCC | MCC | Not MCC | |
| Non-biofilm forming | 21 | 46 (1 LHR) | 54 | 77 | 34 | 39 | 24 | 5 |
| Weak | 1 | 3 | 13 | 10 | 1 | 5 | 4 | 0 |
| Moderate | 4 | 5 | 5 | 4 | 1 | 8 | 13 | 1 |
| Strong | 8 | 5 | 10 | 22 | 2 | 4 | 4 | 5 |
| Very strong | 5 | 3 | 25 | 19 (5 LHR) | 1 | 3 | 5 | 0 |
| Extreme | 12 | 8 | 32 | 8 (2 LHR) | 1 | 1 | 138 | 1 |
| Total | 51 | 70 | 139 | 140 | 40 | 60 | 188 | 12 |
Biofilm-forming capacity is as follows: non-biofilm forming, x < ODc; weak, ODc < x < 2× ODc; moderate, 2× ODc < x < 4× ODc; strong, 4× ODc < x < 8× ODc; very strong, 8× ODc < x < 16× ODc; extreme, 16× ODc < x.
The isolate is motile and expresses both curli and cellulose.
The isolate does not have all three of the following characteristics: motility, expression of curli, and expression of cellulose.
Effects of source of isolates and seasonality on biofilm formation.
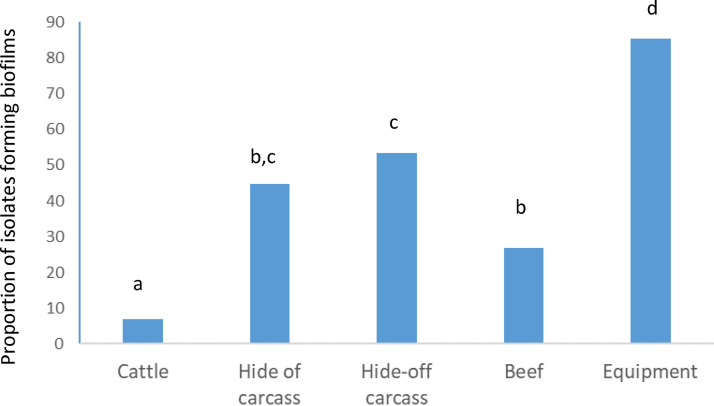
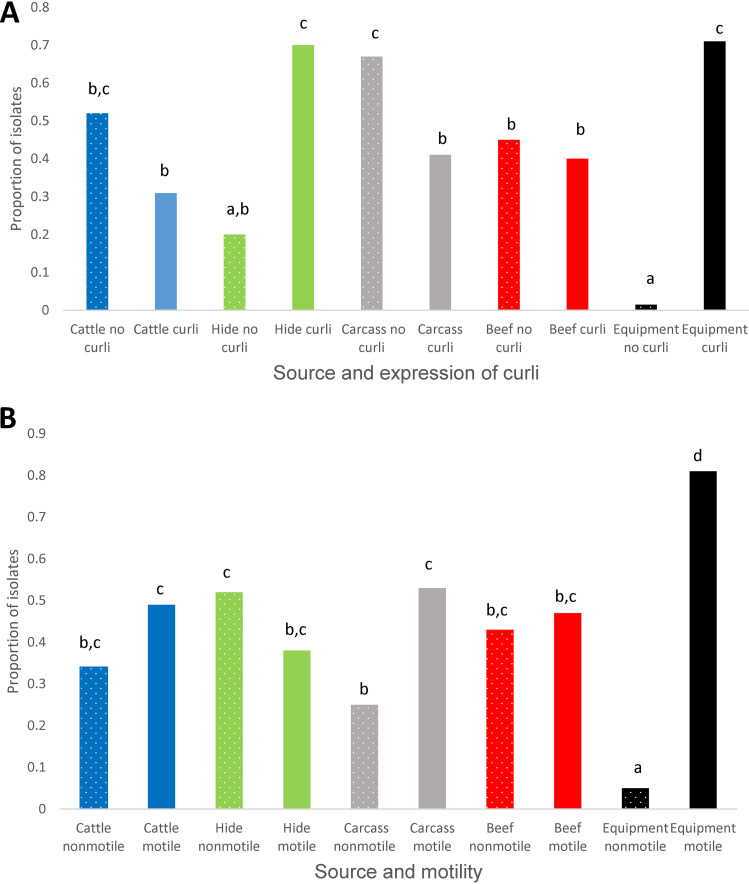
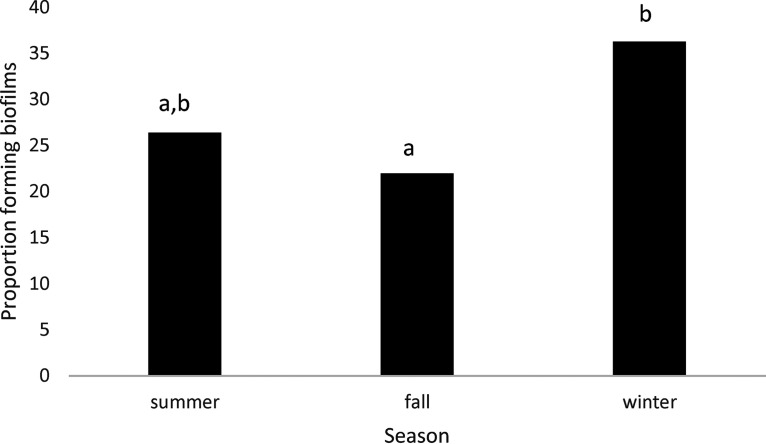
Comparing all sources, the proportion of isolates capable of forming biofilm was highest (P < 0.05) in those collected from processing equipment, lowest (P < 0.05) in those from live cattle, and intermediate in isolates from hides of carcasses, hide-off carcasses, and beef products (Fig. 2). Significant interactions between source of isolates and two biofilm-related phenotypes (expression of curli and motility) were present (Fig. 3). While the expression of curli had no influence on the proportion of live cattle or beef product isolates making biofilms, this phenotype increased (P < 0.05) the proportions of isolates from processing equipment and from hides of carcasses that formed biofilms. Unexpectedly, in hide-off carcasses, the expression of curli reduced (P < 0.05) the proportion of isolates forming biofilms. For both hide-off carcasses and processing equipment, motility increased (P < 0.05) the proportion of isolates forming biofilms but did not influence biofilm formation in isolates from live cattle, hides of carcasses, or beef products. For isolates from live cattle, hides of carcasses, and hide-off carcasses where the season of isolate collection was known, the biofilm-forming potential was increased (P < 0.05) in the winter compared to the fall, with summer months being intermediate (Fig. 4).
FIG 2.
Effect of source on the proportions of E. coli isolates capable of forming biofilms as determined by the optical density at 570 nm. Sources with different superscript letters differ (P < 0.05).
FIG 3.
Interactions between source and phenotypic attributes for the proportions of isolates of E. coli forming biofilms as determined by the optical density (OD) at 570 nm: expression of curli (A) and motility (B). Values with different superscript letters differ (P < 0.05).
FIG 4.
Proportion of isolates forming biofilms by season, including E. coli isolates from cattle, hides of carcasses, and hide-off carcasses (n = 658).
Impacts of antimicrobial interventions at slaughter plants on biofilm.
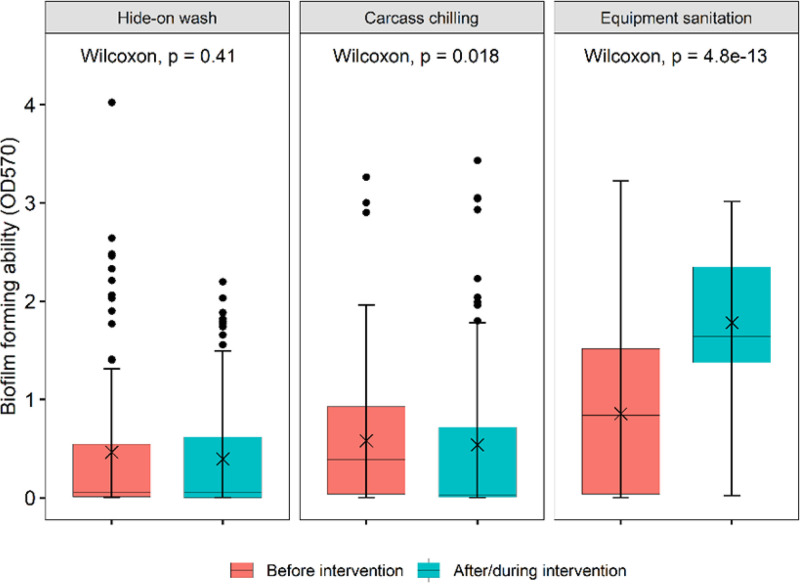
Of the three antimicrobial interventions evaluated, hide washing was the only one that did not significantly impact the subsequent biofilm-forming capacity of the isolates (Fig. 5). Chilling (carcasses were held at 0°C for 3 days) reduced (P = 0.018) the proportion of isolates forming biofilms. In contrast, the biofilm-forming capacity of isolates markedly increased in those collected after sanitation of equipment (P < 0.001).
FIG 5.
Effect of antimicrobial interventions at beef-processing plants on the biofilm-forming ability of E. coli isolates as determined by the optical density (OD) at 570 nm.
Relationship between biofilm formation and heat resistance.
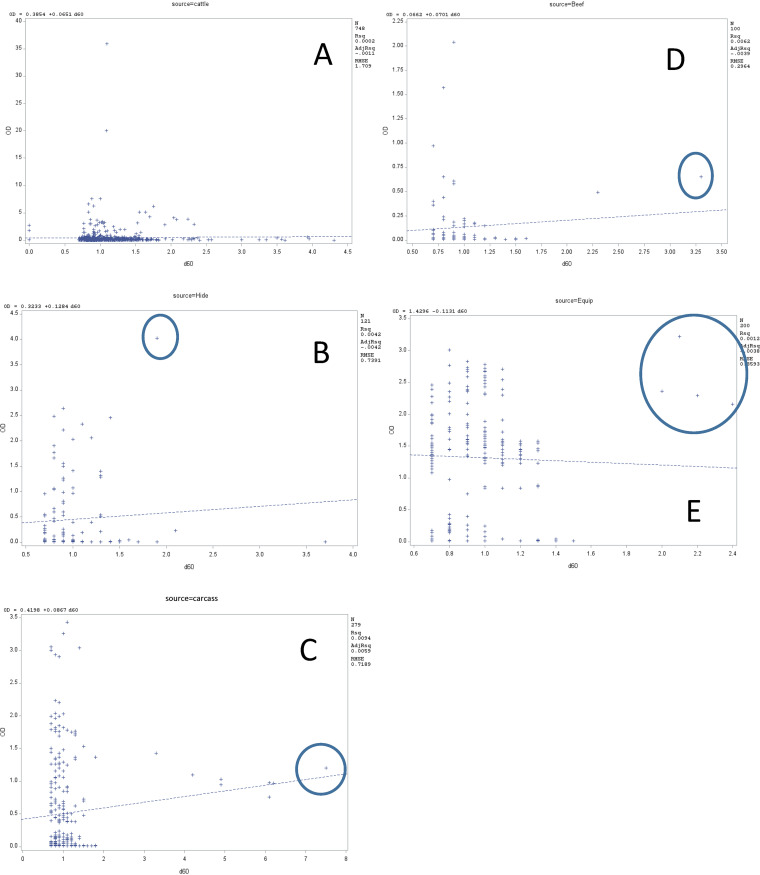
Although biofilm formation as measured by the optical density (OD) and heat resistance of isolates as determined by D60°C (time required at 60°C to kill 90% of cells) were not significantly related in regression analyses (Fig. 6), heightened heat resistance and biofilm-forming ability were not mutually exclusive. With the exception of live cattle, all sources had one or more isolates that were relatively heat-resistant biofilm formers. The presence of the LHR did not influence the heat resistance of these biofilm-forming isolates as no isolates from processing equipment were positive for the LHR, yet four of these were both heat resistant (D60°C of >2 min) and extremely strong biofilm formers. Similarly, although two strong biofilm formers isolated from live cattle were positive for the LHR, the D60°C values for these isolates were <1.2 min, similar to those of the majority of the cattle isolates.
FIG 6.
Relationship between the biofilm optical density and heat tolerance (D60°C) of E. coli by source of isolates: live cattle (A), hide of carcass (B), hide-off carcass (C), beef products (D), and processing equipment (E). Isolates having relatively heightened heat resistance and biofilm-forming capacity are circled. Rsq, coefficient of determination; AdjRsq, adjusted coefficient of determination; RMSE, mean squared error of the coefficient of determination.
DISCUSSION
Biofilm formation by top 7 strains.
Although it has been reported that 95% of O157 strains are unable to express curli (11), this was not the case in the present study, and the majority of the top 7 strains shared this capability. Prophages carrying stx1 have been found to insert within and inactivate mlrA, which is a positive regulator of CsgD and necessary for the transcription of curli operons (3). However, in reverse of expectations, the expression of curli and carriage of stx1 were higher in O157 than in most serogroups evaluated. Some curli-positive stx1-positive O157 strains have a mutation in the csgD promoter that leads to the overexpression of curli, although other curli-positive O157 strains have been stx1 negative after the loss of the mlrA-embedded prophage (12). Whole-genome sequence analyses of curli-expressing stx1-positive O157 isolates are in progress and may better explain our results. Potentially, prophages carrying stx1 may integrate at sites other than within mlrA, or mutations in the csgD promoter may be more common than previously thought, as curli-expressing stx1-positive isolates were more than 25% of those evaluated for all serogroups of top 7 strains.
Even though curli was expressed by the majority of top 7 strains, few were capable of forming biofilms. Strains of EHEC that do not produce curli or cellulose and are nonmotile are thought to have less biofilm-forming capacity than strains with a combination of these traits (9). Flagella have been shown to be important in the initial attachment of E. coli to a surface (13), but in the present study, the expression of cellulose was the only trait positively influencing biofilm formation by O26, while the expression of curli was the only trait positively influencing biofilm formation by O157 (Fig. 1). Motility and the expression of cellulose were actually negatively associated with biofilm formation by O157, but these and others such as carriage of conserved rpoS and eae may have been negatively associated only due to their predominance in EHEC. Even though non-EHEC O157 strains were uncommon, they constituted 29% of O157 biofilm formers. Although not all isolates were serotyped, some of the strongest-biofilm-forming O157 isolates were O157:H12, which has been used as a nonpathogenic surrogate for E. coli O157:H7 (14). Even though EHEC isolates forming biofilms were rare in the present study, and similar to previously reported O157:H7 biofilm-forming prevalence (3), they were not unknown. One EHEC isolate was an extremely strong biofilm former (Table 2), as has been reported for EHEC responsible for HEPs at abattoirs (2).
All top 7 isolates were evaluated for the presence of rpoS due to its role in improved tolerance to stress by O157:H7 as well as its influence on biofilm formation as the master regulator of the “curli/cellulose control cascade” (13). Results of the present study, however, did not demonstrate any positive role of rpoS in biofilm formation, possibly due to the incubation conditions used. Although biofilms were evaluated in stationary phase, rpoS is expressed during periods of stress and starvation (15), which may not have occurred in incubations in a nutrient-dense medium such as Luria-Bertani (LB) medium. No biofilm was formed by isolates of O145, although it, along with O157, had the highest (P < 0.05) proportions of rpoS detected (Table 3). rpoS was detected in 38.8% of top 7 isolates, and in the remainder of the isolates, mutations in rpoS that interfered with PCR detection may have been present. Genomic evaluations currently in progress will determine if mutations in rpoS may lead to a lack of function. Both motility and the expression of cellulose were lower (P < 0.05) in isolates of O145 than in isolates of O157, which perhaps contributed to less biofilm formation by O145, although motility was also not positively linked to biofilm formation by top 7 strains in the present study.
Picozzi et al. (8) concluded that biofilm formation among EHEC isolates was extremely variable and could not be predicted by serogroup or by the presence of virulence genes, although they evaluated only 45 isolates across six serogroups. Our results demonstrate that other than by directly evaluating biofilm formation, it was not possible to predict which top 7 isolates would be biofilm formers based on motility, the expression of curli and cellulose, and the presence of conserved rpoS, in agreement with a recent study by Ma et al. (16), who concluded that genetic or phenotypic characterizations of curli, cellulose, and motility did not correspond with the biofilm-forming potential of E. coli. Those authors also evaluated an extensive list of 21 biofilm-related genes, the presence or absence of which was also not able to predict biofilm formation. Based on our results, none of the three factors identified by Chen et al. (7) as reasons for impaired biofilm formation could be confirmed in the top 7 isolates evaluated, as motility and the expression of curli and cellulose were common compared to their capacity to form biofilms. Methods to quantify the expression of curli and cellulose may provide additional insight into biofilm formation by EHEC.
Effects of source of isolates and antimicrobial interventions on biofilm formation.
To our knowledge, this is the first study to compare the biofilm-forming capacities of E. coli strains isolated from the complete beef production chain. In contrast to our previous study of the same isolates, which for heat tolerance found no differences among sources of isolates or evidence of antimicrobial interventions exerting selection pressure for increased heat tolerance (17), both the source of the isolates and antimicrobial interventions affected the biofilm-forming capacity of the isolates. Comparing sources, a heightened biofilm-forming capacity was found in isolates from processing equipment compared to all others evaluated. Previous work raised this possibility as biofilm-forming ability was found to improve the persistence of E. coli on meat fabrication equipment (18). In the present study, only strong biofilm formers were likely able to remain on equipment and be recovered after sanitation. As air chilling of carcasses at 0°C reduced the fraction of isolates forming biofilms, perhaps a chilled water wash or another chilling procedure might be a beneficial additional antibiofilm hurdle after the completion of equipment sanitation, although additional studies would be necessary to develop and evaluate the efficacy of such a process, and Dourou et al. (19) found that biofilms of O157:H7 could form on beef-processing surfaces at 4°C.
Other than being numerous, biofilm-forming isolates from equipment were also notable as they were almost exclusively motile (Fig. 3), although motility also increased (P < 0.05) the proportion of biofilm formers on carcasses. Motility of isolates has often been linked to the ability of E. coli to form biofilms (7, 16, 20), and while not associated in the present study with biofilm formation in isolates from cattle, the hides of carcasses, or meat products, motility may be more necessary to form biofilms on processing equipment and hide-off carcasses. Perhaps, movement to locations more protected from sanitation procedures affecting carcasses and processing equipment is a requirement for biofilm initiation and the overall survival of E. coli from these sources. Similarly, Chitlapilly Dass et al. (20) theorized that motile, biofilm-forming EHEC isolates swim upstream after sanitation and accumulate in protected locations such as floor drains of slaughter facilities.
Along with requirements for motility, biofilm-forming isolates from processing equipment also almost exclusively expressed curli, a phenotype shared by the majority of biofilm-forming isolates from the hides of carcasses. Both hides and processing equipment are routinely subjected to high-pressure water washes, in contrast to other sources of E. coli evaluated, with the exception of hide-off carcasses. That the proportion of isolates from hide-off carcasses that formed biofilm was reduced in those expressing curli was unexpected, but a number of adhesins contribute to biofilm formation (16), and alternatives to curli may be more important for the adherence of E. coli to the layer of subcutaneous fat on the surface of the hide-off carcass.
Seasonality of biofilm formation.
The seasonality of E. coli biofilm formation in the beef production chain has been little examined, although several studies have evaluated the seasonality of multispecies biofilms in rivers or water systems (21, 22). Although the seasonality results of the present study are preliminary as data were not available for all four seasons, the results were consistent for isolates from cattle, hides of carcasses, and hide-off carcasses, the only isolates where the season of collection was known. Seasonal differences in isolates would be most expected in those collected from cattle and their environment as many were collected from fecal pats or the hides of cattle exposed to ambient temperatures.
The biofilm-forming ability was heightened in the winter when temperatures in Alberta are commonly <0°C, and cattle hides commonly carry a continuous layer of hardened tag, i.e., a mixture of mud, bedding, and manure, particularly on the belly and brisket (23). The genetic diversity of E. coli isolates collected from cattle in winter months has been shown to be reduced (6), likely as only a smaller, more-related population is able to survive. With surviving populations in the winter likely being cold stressed, and stress being linked to the expression of rpoS and the production of curli and cellulose (15), perhaps the increased proportion of biofilm formers in the winter is related to increased stress to E. coli or the condition of hides. The close relationship among top 7 isolates from cattle and generic E. coli isolates collected at slaughter plants has been previously demonstrated (17), with the increased biofilm-forming capacity of cattle isolates entering the slaughter plant during the winter likely being passed to other steps of the slaughter chain. Although the seasonality of biofilm formation on processing equipment is as yet unknown, any increased biofilm-forming capacity in the winter would be balanced by a reduced population of viable E. coli cells (6), potentially reducing the risk of the transfer of EHEC to processing equipment.
Relationship between biofilm-forming ability and heat resistance.
The biofilm-forming capacity of E. coli is known to be temperature dependent (8, 19), but it was only recently that the relationship between heat resistance and biofilm formation was investigated by Ma et al. (16). Those authors found that the six heat-resistant (LHR-positive) generic E. coli isolates that they investigated were all biofilm formers. While we agree with those authors that heat-resistant biofilm-forming strains may be a “serious food safety and public health risk,” in evaluating larger numbers of E. coli isolates, we found no relationship between the LHR and biofilm-forming capacity or between heat resistance as determined by the D60°C and the OD of the biofilms formed. Of 24 isolates evaluated with the LHR, 54% were capable of forming biofilms, with the majority of these being generic E. coli isolates. Instead, the results of the present study support that even on processing equipment where biofilm formers were most common, heat-resistant biofilm-forming isolates (n = 4 [none with the LHR]) were rare.
Most likely mechanisms for HEPs.
Arthur et al. (1) postulated that failures in hygiene may spread a single dominant strain of EHEC throughout a slaughter plant or that sanitation procedures within the slaughter plant were perhaps inadvertently selecting for strains of EHEC capable of causing HEPs. The results of the present study support roles for both of these mechanisms in the establishment of biofilm-forming EHEC capable of causing a HEP. Biofilm-forming EHEC isolates were rare, and only 1/745 E. coli isolates from cattle was EHEC and an extremely strong biofilm former. As a strong biofilm former, potentially this or other similar EHEC isolates would have the highest likelihood of surviving sanitation procedures and contaminating processing equipment, perhaps being transported through wash water pipes, as proposed previously by Chitlapilly Dass et al. (20), to multiple areas of the slaughter plant. If EHEC biofilm formers were to contaminate processing equipment, the results of the present study demonstrate that current equipment sanitation procedures may not eliminate the most robust biofilm formers. Accordingly, a new and effective antibiofilm hurdle is required for processing equipment if we are ever to reduce HEPs and increase the safety of beef products.
MATERIALS AND METHODS
Bacterial isolates, antimicrobial hurdles in slaughter plants, and culture conditions.
Escherichia coli isolates (n = 1,445), included 745 isolates of the “top 7” serogroups O26, O45, O103, O111, O121, O145, and O157 recovered from live cattle or their environment and 700 generic E. coli isolates recovered from two federally inspected beef slaughter plants (A and B) as described previously by Zhang et al. (17). At plant A, antimicrobial treatments were minimal for carcasses and included carcass trimming, cold-water washing, and air chilling of dressed carcasses. Processing equipment sanitation occurred at the end of each day and included physical removal of detritus, prerinsing with pressurized water at 40°C to 50°C, spraying with a chlorine-based alkaline foaming agent, and sanitization with a quaternary sanitizer (24). In contrast, plant B also employed a hide-on carcass wash with 1.5% sodium hydroxide at 55°C, spraying of skinned carcasses with 5% lactic acid, and pasteurization of carcasses at the end of the dressing process with steam at >90°C (25).
Top 7 isolates were screened for serogroup and the presence of stx1, stx2, and eae using primers and PCR conditions described previously by Conrad et al. (26). Isolates were classified as EHEC if they were positive for eae and stx1 and/or stx2. The presence of RNA polymerase sigma factor S (rpoS) was detected by PCR as described previously by Uhlich et al. (3) for O157:H7, with this assay also being used to identify isolates with possible rpoS mutations for future whole-genome sequence analyses. The presence of the locus of heat resistance (LHR) was determined as described previously by Mercer et al. (27). For generic E. coli isolates, their species was verified by PCR using the primers for uidA described previously by Bej et al. (28). After characterization by PCR, each E. coli isolate was streaked onto MacConkey agar (BD Bioscience, Canada) and incubated at 35°C for 24 ± 2 h. A single colony was then subcultured in 5 ml Luria-Bertani (LB) medium (BD Bioscience) and incubated for 16 to 18 h at 35°C, with shaking at 80 to 100 rpm.
Biofilm-forming potential.
A culture grown overnight was diluted by taking 50 μl of the culture and adding it to 5 ml of fresh LB medium, and 160 μl of the diluted inoculum was then added to duplicate wells of a round-bottom 96-well microtiter plate (Thermo Scientific, Canada). Each microtiter plate also included duplicate blank wells of LB medium as a negative control, while positive controls contained an isolate of O121:H23 known to be a strong biofilm former (29). Microtiter plates were gently covered with a Nunc-TSP 96-peg lid (Fisher Scientific, Canada) to avoid splashing and cross-contamination and incubated at 15°C for 4 days. After incubation, the pegged lid was removed and washed with gentle agitation for 1 min in two new Nunc plates, each containing 180 μl of phosphate buffer solution (PBS; Millipore Sigma, Canada). The pegged lid was then transferred to a new plate containing 180 μl of 0.1% crystal violet (Millipore Sigma, Canada) and stained for 20 min at room temperature with gentle agitation, followed by washing the lid twice in 180 μl PBS with gentle agitation as described above. The washed pegged lid was then placed into a new plate containing 180 μl 80% ethanol for 20 min at room temperature with slight agitation.
The absorbance of the microplate at 570 nm was then measured using a microplate reader (Synergy HT; Bio-Tek Instruments Inc., USA). Optical densities (ODs) were then grouped into classes from 0 (non-biofilm former) to 5 (extremely strong biofilm former) (Table 5) based on the mean OD from two independent replicates for each isolate compared to the cutoff OD (ODc), which was equal to three times the standard deviation of the OD of the negative control plus the average OD of the negative control.
TABLE 5.
Classification of the biofilm-forming abilities of isolates based on the mean optical density at 570 nm from two independent replicates in duplicate
| OD rangea | Biofilm-forming ability category | Biofilm class |
|---|---|---|
| x < ODc | Non-biofilm former | 0 |
| ODc < x < 2× ODc | Weak biofilm former | 1 |
| 2× ODc < x < 4× ODc | Moderate biofilm former | 2 |
| 4× ODc < x < 8× ODc | Strong biofilm former | 3 |
| 8× ODc < x < 16× ODc | Very strong biofilm former | 4 |
| 16× ODc < x | Extremely strong biofilm former | 5 |
ODc, cutoff optical density (OD) equal to three times the standard deviation of the OD of the negative control plus the average OD of the negative control.
Motility and expression of curli and cellulose.
The motility of isolates was determined using the soft-agar method as described previously by Visvalingam et al. (30). E. coli cultures grown overnight in LB medium were diluted 100-fold, and a 1-μl aliquot was point inoculated into the center of TYE agar (tryptone at 10 g/liter, yeast extract at 5 g/liter, and Bacto agar at 3.5 g/liter) by stabbing with a micropipette tip halfway through the depth of the agar. Plates were incubated at 15°C for 48 h. The diameter of each motility halo produced was then measured, and an isolate was classified as motile if its halo diameter measured ≥4 mm.
The Congo red indicator (CRI) agar method was used to assess the expression of extracellular matrix components (30). Cultures grown overnight in LB medium were streaked onto CRI agar (10 g/liter Casamino Acids, 1 g/liter yeast extract, 20 mg/liter Congo red, 10 mg/liter brilliant blue, and 20 g/liter Bacto agar) and incubated at 15°C for 4 days. The color of the resulting colonies determined the phenotype. Colonies were red, brown, pink, or white and were considered positive for the expression of both cellulose and curli, curli only, cellulose only, or neither, respectively (31).
Phenotypic characterization of heat resistance.
Heat inactivation experiments for isolates were performed as described previously by Zhang et al. (17). Briefly, 1.5 ml of cultures grown overnight in LB medium in the stationary phase of growth was added to 2-ml microcentrifuge tubes (Eppendorf, Canada) and incubated in a 60°C water bath (model FSGPD28; Fisher Scientific, Canada) for up to 30 min. The time required for medium to reach 60°C (come-up time [T0]) was measured by a long needle probe attached to a thermometer (Thermapen Mk4; Thermoworks, USA) stabbed into the medium through the lids of the tubes. After removal from the water bath, the tubes were immediately placed in an ice water bath for rapid cooling. Cultures were then serially diluted in 0.1% (wt/vol) peptone water, and 1 ml of the appropriate dilutions was plated onto Petrifilm aerobic count plates (3M Corp., St. Paul, MN, USA), according to the manufacturer’s instructions. The plates were incubated at 35°C for 18 to 24 h, and CFU were enumerated. Log-transformed counts were plotted against the incubation time, and the regression of the plot was used to calculate the D60°C for each isolate (32).
Statistical analyses.
Statistical analyses were performed using SAS version 9.4 (SAS Institute Inc., Cary, NC). As OD values did not follow normal distributions as determined by Shapiro-Wilk tests, nonparametric analyses (Wilcoxon rank sum test and Kruskal-Wallis test) were used within the NPAR1way procedure to compare the biofilm-forming abilities of isolates collected before and after sanitation on the OD of E. coli sourced from meat processors. For presence/absence values such as the formation of biofilm, the expression of curli or cellulose, motility, and PCR detection of virulence- or biofilm-related factors, isolates were compared using generalized linear mixed models (Proc Glimmix) with a binomial distribution. Model-adjusted means (back-transformed to the original scale) and 95% confidence intervals were determined, with isolate being the experimental unit and source of isolate and season being fixed effects. Analyses of top 7 isolates also included serogroup as a fixed effect. To determine the relationships between D60°C and OD values, regression analyses were conducted within Proc Reg for each source of isolates. In all analyses, P values of <0.05 were deemed significant.
ACKNOWLEDGMENTS
We thank the Beef Cattle Research Council for project funding (FOS.01.17) and Results Driven Agriculture for additional funding (2021r014R).
We thank Susanne Trapp, Allison McNaughton, and Rebecca Lohmann for sterling technical assistance.
Contributor Information
Kim Stanford, Email: kim.stanford@uleth.ca.
Xianqin Yang, Email: xianqin.yang@canada.ca.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Arthur TM, Bono JL, Kalchayanand N. 2014. Characterization of Escherichia coli O157:H7 strains from contaminated raw beef trim during “high event periods.” Appl Environ Microbiol 80:506–514. doi: 10.1128/AEM.03192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R, Kalchayanand N, King DA, Luedtke BE, Bosilevac JM, Arthur TM. 2014. Biofilm formation and sanitizer resistance of Escherichia coli O157:H7 strains isolated from “high event period” meat contamination. J Food Prot 77:1982–1987. doi: 10.4315/0362-028X.JFP-14-253. [DOI] [PubMed] [Google Scholar]
- 3.Uhlich GA, Chen C-Y, Cottrell BJ, Hofmann CS, Dudley EG, Strobaugh TP, Nguyen L-H. 2013. Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli O157:H7. Microbiology (Reading) 159:1586–1596. doi: 10.1099/mic.0.066118-0. [DOI] [PubMed] [Google Scholar]
- 4.Visvalingam J, Wang H, Ells TC, Yang X. 2019. Facultative anaerobes shape multispecies biofilms composed of meat processing surface bacteria and Escherichia coli O157:H7 or Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 85:e01123-19. doi: 10.1128/AEM.01123-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cookson AL, Cooley WA, Woodward MJ. 2002. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int J Med Microbiol 292:195–205. doi: 10.1078/1438-4221-00203. [DOI] [PubMed] [Google Scholar]
- 6.Stanford K, Johnson RP, Alexander T, McAllister TA, Reuter T. 2016. Influence of season and feedlot location on prevalence and virulence of Escherichia coli from feces of western-Canadian slaughter cattle. PLoS One 11:e0159866. doi: 10.1371/journal.pone.0159866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C-Y, Hofmann CS, Cottrell BJ, Strobaugh TP, Paoli GC, Nguyen L-H, Yan X, Uhlich GA. 2013. Phenotypic and genotypic characterization of biofilm-forming capacities in non-O157 Shiga toxin-producing Escherichia coli strains. PLoS One 8:e84863. doi: 10.1371/journal.pone.0084863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picozzi C, Antoniani D, Vigentini I, Foschino R, Kneifel W. 2017. Genotypic characterization and biofilm formation of Shiga toxin-producing Escherichia coli. FEMS Microbiol Lett 364:fnw291. doi: 10.1093/femsle/fnw291. [DOI] [PubMed] [Google Scholar]
- 9.Lajhar SA, Brownlie J, Barlow R. 2018. Characterization of biofilm-forming capacity and resistance to sanitizers of a range of E. coli O26 pathotypes from clinical cases and cattle in Australia. BMC Microbiol 18:41. doi: 10.1186/s12866-018-1182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vosough Ahmadi B, Velthuis AGJ, Hogeveen H, Huirne RBM. 2006. Simulating Escherichia coli O157:H7 transmission to assess effectiveness of interventions in Dutch dairy-beef slaughterhouses. Prev Vet Med 77:15–30. doi: 10.1016/j.prevetmed.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Sheng H, Xue Y, Zhao W, Hovde CJ, Minnich SA. 2020. Escherichia coli O157:H7 curli fimbriae promotes biofilm formation, epithelial cell invasion and persistence in cattle. Microorganisms 8:580. doi: 10.3390/microorganisms8040580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlich GA, Chen C-Y, Cottrell BJ, Hofmann CS, Yan X, Nguyen L. 2016. Stx1 prophage excision in Escherichia coli strain PA20 confers strong curli and biofilm formation by restoring native mlrA. FEMS Microbiol Lett 363:fnw123. doi: 10.1093/femsle/fnw123. [DOI] [PubMed] [Google Scholar]
- 13.Mika F, Hengge R. 2014. Small RNAs in the control of RpoS, CsfD and biofilm architecture of Escherichia coli. RNA Biol 11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H-B, Chen C-H, Boomer A, Pradhan A, Patel J. 2020. Persistence of Escherichia coli O157:H12 and Escherichia coli K12 as non-pathogenic surrogates for O157:H7 on lettuce cultivars irrigated with secondary-treated wastewater and roof-collected rain water in the field. Front Sustain Food Syst 4:555459. doi: 10.3389/fsufs.2020.555459. [DOI] [Google Scholar]
- 15.Sheldon JR, Yim M-S, Saliba JH, Chung W-H, Wong K-Y, Leung KT. 2012. Role of rpoS in Escherichia coli O157:H7 strain H32 biofilm development and survival. Appl Environ Microbiol 78:8331–8339. doi: 10.1128/AEM.02149-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma A, Neumann N, Chui L. 2021. Phenotypic and genetic determination of biofilm formation in heat-resistant Escherichia coli possessing the locus of heat resistance. Microorganisms 9:403. doi: 10.3390/microorganisms9020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, Tran F, Stanford K, Yang X. 2020. Are antimicrobial interventions associated with heat-resistant Escherichia coli on meat? Appl Environ Microbiol 86:e00512-20. doi: 10.1128/AEM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X, Wang H, He A, Tran F. 2018. Biofilm formation and susceptibility to biocides of recurring and transient Escherichia coli isolated from meat fabrication equipment. Food Control 90:205–211. doi: 10.1016/j.foodcont.2018.02.050. [DOI] [Google Scholar]
- 19.Dourou D, Beauchamp CS, Yoon Y, Geornaras I, Belk KE, Smith GC, Nychas G-JE, Sofos JN. 2011. Attachment and biofilm formation by Escherichia coli O157:H7 at different temperatures on various food-contact surfaces encountered in beef processing. Int J Food Microbiol 149:262–268. doi: 10.1016/j.ijfoodmicro.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Chitlapilly Dass S, Bosilevac JM, Weinroth M, Elowsky CG, Zhou Y, Anandappa A, Wang R. 2020. Impact of mixed biofilm formation with environmental microorganisms on E. coli O157:H7 survival against sanitation. NPJ Sci Food 4:16. doi: 10.1038/s41538-020-00076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wey JH, Jurgens K, Weitere M. 2012. Seasonal and successional influences on bacterial community composition exceed that of protozoan grazing in river biofilms. Appl Environ Microbiol 78:2013–2024. doi: 10.1128/AEM.06517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling F, Hwang C, LeChevallier MW, Andersen GL, Liu W-T. 2016. Core-satellite populations and seasonality of water meter biofilms in a metropolitan drinking water distribution system. ISME J 10:582–595. doi: 10.1038/ismej.2015.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Environment Canada. 2021. Alberta weather forecasts by locations and historical data. https://weather.gc.ca/forecast/canada/index_e.html?id=ab. Accessed 22 February 2021.
- 24.Yang X, Wang H, He A, Tran F. 2017. Microbial efficacy and impact on the population of Escherichia coli of a routine sanitation process for the fabrication facility of a beef packing plant. Food Control 71:353–357. doi: 10.1016/j.foodcont.2016.07.016. [DOI] [Google Scholar]
- 25.Yang X, Badoni M, Tran F, Gill CO. 2015. Microbiological effects of a routine treatment for decontaminating hide-on carcasses at a large beef packing plant. J Food Prot 78:256–263. doi: 10.4315/0362-028X.JFP-14-226. [DOI] [PubMed] [Google Scholar]
- 26.Conrad CC, Stanford K, McAllister TA, Thomas J, Reuter T. 2014. Further development of sample preparation and detection methods for O157 and the top 6 non-O157 STEC serogroups in cattle feces. J Microbiol Methods 105:22–30. doi: 10.1016/j.mimet.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Mercer RG, Zheng J, Garcia-Hernandez R, Ruan L, Gänzle MG, McMullen LM. 2015. Genetic determination of heat resistance of E. coli. Front Microbiol 6:932. doi: 10.3389/fmicb.2015.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bej AK, DiCesare JL, Haff L, Atlas RM. 1991. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl Environ Microbiol 57:1013–1017. doi: 10.1128/aem.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bumunang EW, McAllister TA, Zaheer R, Ortega Polo R, Stanford K, King R, Niu YD, Ateba CN. 2019. Characterization of non-O157 Escherichia coli from cattle faecal samples in the North-West province of South Africa. Microorganisms 7:272. doi: 10.3390/microorganisms7080272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visvalingam J, Ells TC, Yang X. 2017. Impact of persistent and non-persistent generic Escherichia coli and Salmonella sp. recovered from a beef packing plant on biofilm formation by E. coli O157. J Appl Microbiol 123:1512–1521. doi: 10.1111/jam.13591. [DOI] [PubMed] [Google Scholar]
- 31.Uhlich GA, Chen C-Y, Cottrell BJ, Nguyen L-H. 2014. Growth media and temperature effects on biofilm formation by serotype O157:H7 and non-O157 Shiga toxin producing Escherichia coli. FEMS Microbiol Lett 354:133–141. doi: 10.1111/1574-6968.12439. [DOI] [PubMed] [Google Scholar]
- 32.van Asselt ED, Zwietering MH. 2006. A systematic approach to determine global thermal inactivation parameters for various food pathogens. Int J Food Microbiol 107:73–82. doi: 10.1016/j.ijfoodmicro.2005.08.014. [DOI] [PubMed] [Google Scholar]