ABSTRACT
Hemorrhagic pneumonia (HP) is a rare but highly lethal disease, mainly of dogs and cats, caused by hemolytic Escherichia coli strains that contain cnf1 (encoding cytotoxic necrotizing factor 1). After encountering fatal HP in two dogs, we used contemporary molecular methods, including multilocus sequence typing and whole-genome sequencing, to compare the corresponding case isolates with published HP clinical isolates and newly obtained fecal E. coli isolates from 20 humans and animals in the index HP case household. We also compared the aggregated HP clinical isolates, which represented 13 discrete strains, by pulsotype with a large, private pulsotype library of diverse-source E. coli. The HP clinical isolates represented a narrow range of phylogenetic group B2 lineages (mainly sequence types 12 and 127), O types (mainly O4 and O6), and H types (mainly H5 and H31), but diverse fimH alleles (type-1 fimbriae adhesin). Their extensive, highly conserved virulence genotypes, which qualified as extraintestinal pathogenic E. coli (ExPEC), encoded diverse adhesins, toxins, iron uptake systems, and protectins. Household surveillance identified multiple HP-like fecal strains, plus abundant between-host strain sharing, including of the household's index HP strain. The pulsotype library search identified, for five HP clinical strains, same-pulsotype human and animal fecal and clinical (predominantly urine) isolates, from diverse locales and time periods. Thus, E. coli strains that cause HP derive from a narrow range of ExPEC lineages within phylogroup B2, contain multiple virulence genes other than cnf1, are shared extensively between hosts, and likely function in nature mainly as intestinal colonizers and uropathogens.
IMPORTANCE This study clarifies the clonal background and extensive virulence genotypes of the E. coli strains that cause hemorrhagic pneumonia in domestic animals (mainly dogs and cats), shows that such strains circulate among animals and humans, identifies a substantial intestinal colonization component to their lifestyle, and extends their known clinical manifestations to include bacteremia and urinary tract infection. The findings place these strains better into context vis-à-vis current understandings of E. coli phylogeny, ecology, and pathogenesis; identify questions for future research; and may prove relevant for surveillance and prevention efforts.
KEYWORDS: Escherichia coli, ExPEC, cytotoxic necrotizing factor, hemorrhagic pneumonia, microbial ecology, molecular epidemiology, phylogenetic analysis, transmission, virulence determinants, whole-genome sequencing
INTRODUCTION
Escherichia coli is a ubiquitous gut colonizer and leading extraintestinal pathogen of mammals and birds. As an extraintestinal pathogen it causes, in humans, mainly urinary tract infection and bacteremia (1) and, in dogs and cats, mainly urinary tract infection (2). Rarely, in companion animals, it causes a dramatic illness characterized by fatal necrotizing hemorrhagic pneumonia (HP), with multiorgan involvement and overwhelming sepsis (3–8).
Collectively, the E. coli strains that cause extraintestinal infections at any site differ in multiple ways from those that colonize only the gut (i.e., commensal strains) or cause diarrhea, which has led to their designation as extraintestinal pathogenic E. coli (ExPEC) (9–11). Typical ExPEC strains derive from a limited set of lineages or sequence types (STs), mainly within E. coli phylogroup B2, whereas commensal and diarrheagenic strains are mainly from other phylogroups. ExPEC strains also express a limited range of (ST-associated) O, K, and H antigens that differ from those of commensal and diarrheagenic strains and contain distinctive virulence genes that encode the adhesins, toxins, siderophores, protectins, etc., needed for extraintestinal pathogenicity.
The E. coli HP case isolates reported to date all contain the virulence gene cnf1 (cytotoxic necrotizing factor 1 [CNF1]). CNF1 permanently activates the regulatory Rho, Rac, and Cdc42 GTPases in eukaryotic cells by deamidation of a glutamine residue, thereby promoting new activities in cells, including gene transcription and cell proliferation, and promotes survival of the bacteria (12). This feature, plus the necrohemorrhagic nature of the associated pneumonia, inspired the label necrotoxigenic E. coli (NTEC) for such strains, analogous to the label Shiga-toxigenic E. coli (STEC) for Shiga toxin-producing diarrheagenic E. coli strains (4, 13). However, the reported HP isolates have also contained genes for other toxins—including alpha hemolysin (hly) and secreted autotransporter toxin (sat)—plus multiple other virulence factors, leaving in question the true contribution of cnf1 to pathogenesis (3–8).
The occurrence of fatal E. coli HP in two epidemiologically unrelated dogs that presented to the University of Minnesota (UM) Veterinary Medical Center (VMC) approximately 8 years apart motivated the present study, which had three main goals. First, we sought to use contemporary molecular methods to characterize the present HP case isolates, in comparison with historical HP isolates. Second, to clarify possible transmission pathways and reservoirs, we sought to identify within-household sharing and persistence of such strains. Third, we sought for such strains more broadly, to clarify their overall ecology, host range, and pathogenic capabilities.
RESULTS
HP case 1.
In September 2009, a 6-month-old male Golden Retriever presented to the UM VMC after 1 day of vomiting, lethargy, and labored breathing. Clinical signs progressed over 2 h to hematemesis, epistaxis, and cardiorespiratory arrest, and the dog was euthanized.
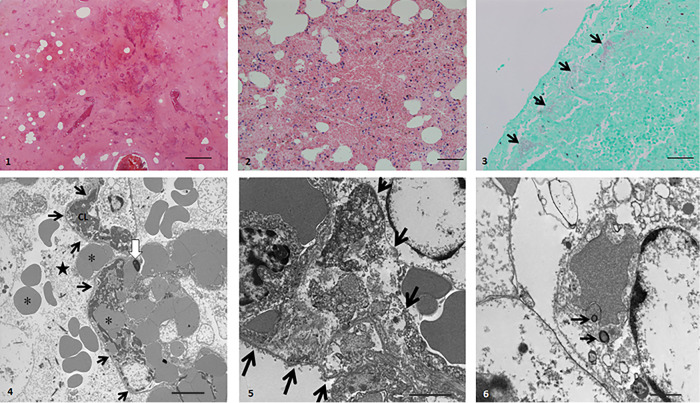
Postmortem examination revealed hemothorax, severe hemorrhagic pneumonia, and hemorrhagic enteritis. Microscopically, all sections of the affected lung regions were almost completely effaced by large amounts of hemorrhage, edema, and multifocal to confluent necrosis of the pulmonary parenchyma, with admixed necrotic mononuclear cells and numerous intra-alveolar colonies of short bacillary bacteria (Fig. 1). Transmission electron microscopy, which was used to further characterize the lung changes, showed loss of the alveolar capillary endothelium, apparently continuous basal membrane, and numerous microvascular thrombi (Fig. 1). In addition, there were segmental necrotic alveolar walls, segmental loss of the alveolar epithelium, extravasation of erythrocytes and fibrin, and bacteria within mononuclear cells.
FIG 1.
Light and electron microscopy of lung tissue from case 1 of hemorrhagic pneumonia due to Escherichia coli. (Panel 1) (Hematoxylin and eosin [H&E] stain) Near-total effacement of normal architecture by hemorrhage, edema, fibrin, and necrotic debris. Bar, 200 μm. (Panel 2) (H&E stain) Alveolar walls are effaced. Alveolar spaces are filled with hemorrhage, edema, fibrin, and pyknotic mononuclear cells. Bar, 100 μm. (Panel 3) (Gram stain) Gram-negative bacilli (arrows) aggregate within alveolar spaces abutting the pleura. Bar, 50 μm. (Panel 4) (Transmission electron micrograph [TEM]) Alveolar space (star) contains erythrocytes (asterisks) and alveolar capillaries (black arrows). Capillary lumen (CL) contains fibrin thrombus. White arrow, pyknotic capillary endothelial cell. Bar, 5 μm. (Panel 5) (TEM) Alveolar wall (arrows) is expanded by fibrin strands and cell debris. Bar, 2 μm. (Panel 6) (TEM) Macrophage with bacteria in a phagolysosome (black arrows). Bar, 1 μm.
Escherichia coli O6:H31 was cultured from lung, spleen, liver, and small intestine. The liver and lung isolates were indistinguishable by pulsed-field gel electrophoresis (PFGE) (pulsotype 0064) and represented ST127 (Table 1). Virulence genotyping by PCR and whole-genome sequencing (WGS) analysis identified 24 extraintestinal virulence genes, qualifying the strain molecularly as ExPEC.
TABLE 1.
Characteristics of hemorrhagic pneumonia-associated Escherichia coli isolates, representing 13 epidemiologically unique strains, from the present study and previous reportsa
| Isolate(s) and yrb | Reference | Host | Tissue | Pulsotype | STc | O:H typed | Virulence genotypee |
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adhesins |
Iron | Protectins |
Miscellaneous | Colicins |
|||||||||||||||||||||||||||||
| P fimbriae |
S and F1C fimbriae | Capsule | |||||||||||||||||||||||||||||||
| papA allele | papG allele | sfaD | sfaE | sfaS | focG | focC | focI | fimH allele | ireA | sitA | K1 | K15 | kpsMII | kpsMTIII | traT | malX | fliC H7 | ibeA | tcpC | terC | usp | Cba (ColB) | Cea (ColE1) | celB (ColE2) | mchB (ColH) | Cma (ColM) | |||||||
| 4.0811 | 4 | Dog | L | 037 | 127 | O6:H31 | 7-2, 48 | III | + | + | + | − | − | − | 2 | − | + | − | − | + | − | − | + | − | − | − | + | + | − | − | − | − | − |
| 0.2025 | 3 | Dog A | L | 0064 | 127 | O6:H31 | 48 | III | + | + | + | − | − | − | 343 | − | + | − | − | + | − | + | + | − | − | + | + | + | + | − | − | − | + |
| 1.2922, 1.2924 | 3 | Dog B | L, T, B, S, J | 0445 | 127 | O6:H31 | 48 | III | + | + | + | − | − | − | 2 | − | + | − | − | + | − | + | + | − | − | + | + | + | + | − | − | − | + |
| 8.2906 | 8 | Tiger | L | 1421 | 127 | O6:H31 | 7-2, 48 | III | + | + | + | − | − | − | 2 | − | + | − | − | + | − | + | + | − | − | + | + | + | + | − | − | − | + |
| JJ2690, JJ2691 | This study (case 1) | Dog | L, Liv | 0064 | 127 | O6:H31 | 12 | III | + | + | + | − | − | − | 352 | − | + | − | − | + | − | + | + | − | − | + | + | + | − | − | − | − | − |
| 7.3597, 8.0112 | 7 | Cat B | L, I | 1422 | 625 | O6:H7 | 10, 12 | I, III | + | − | − | − | + | + | 115 | + | + | − | − | + | − | + | + | + | + | + | + | + | − | + | − | + | − |
| 1.2887 | 3 | Dog C | L | 1424 | 12 | O4:H5 | 13, 14 | I, III | − | − | − | + | − | + | 27 | + | + | − | − | − | + | − | + | − | − | + | + | + | − | + | − | + | − |
| 2.2945 | 3 | Dog D | L | 1425 | 12 | O4:H5 | 10 | III | + | − | − | − | + | − | 386 | − | + | − | − | + | − | − | + | − | − | − | + | + | − | − | − | + | − |
| 6.1636-8, 79-82 | 5 | Cat 1-7 | L | 1263 | 12 | O4:H5 | 10, 12 | I, III | + | − | − | − | + | − | 5 | + | + | − | − | + | − | − | + | − | − | + | + | + | − | + | − | + | − |
| 7.3596, 8.0201 | 7 | Cat A | L, C | 691 | 2604f | O4:H5 | 10, 12 | III | + | − | − | − | + | − | 511 | − | + | − | + | + | − | − | + | − | − | − | − | + | − | + | − | + | − |
| JJ3030 | This study (case 2) | Dog | L | 2320 | 2015 | O?:H14 | F13 | III | + | − | − | − | + | − | 197 | − | + | − | − | + | − | − | − | − | + | − | + | + | − | + | − | + | − |
| 7.3127 | 7 | Cat C | L | 1423 | 83 | O?:H5 | 10, 12 | III | + | − | − | − | − | + | 21 | − | + | − | − | + | − | + | + | − | + | − | + | + | − | + | + | + | − |
| 7.1451 | 6 | Horse | L | 2320 | 6210 | O2:H18 | 12 | 0 | + | + | − | + | − | + | 154 | − | − | + | − | + | − | + | − | − | + | − | + | − | + | + | − | + | + |
Boldface indicates isolates from the present study (cases 1 and 2). Abbreviations: B, blood; C, colon; I, intestine; J, jejunum; L, lung; Liv, liver; O?, O type indeterminate; S, stomach; ST, sequence type; T, trachea. +, present; −, absent.
All isolates were from phylogroup B2. Isolates are shown in sequence to match the phylogram (Fig. 2). Indistinguishable isolates (i.e., same strain) that are epidemiologically related (same host or temporospatial cluster) are shown in the same row. Years of isolation (or of publication, if isolation year unknown) are as follows: 4.0811, 2005; 0.2025, 2000; 1.2922 and 1.2924, 2001; 8.2906, 2010; JJ2690 and JJ2691, 2009; 7.3597 and 8.0112, 2007; 1.2887, 2001; 2.2945, 2002; 6.1636-8/6.1679-82, 2006; 7.3596/8.0201, 2007; JJ3030, 2017; 7.3127, 2007; 7.1451, 2008. U.S. states of origin are as follows: 4.0811, North Carolina(?), Maryland(?); 0.2025, 1.2922, 1.2924, 1.2887, 2.2945, and 7.1451, Pennsylvania; 8.2906, 6.1636-8, and 6.1679-82, Connecticut; JJ2690, JJ2691, and JJ3030, Minnesota; 7.3127, 7.3596, 7.3597, 8.0112, and 8.0201, Wisconsin(?), California(?).
STs (Achtman 7-locus system) were determined based on whole-genome sequence data.
O:H types were inferred from a combination of whole-genome sequence data, serotyping, and PCR-based typing.
Genes shown were present in some but not all isolates, according to a combination of PCR and whole-genome sequencing (WGS). Definitions: cba (colicin B), cea (colicin E1), celB (colicin E2), cma (colicin M), fimH (type 1 fimbriae adhesin), fliC (flagellin), foc (F1C fimbriae), ibeA (invasion of brain endothelium A), ireA (siderophore receptor), K1 (capsule variant), K15 (capsule variant), kpsMII (group 2 capsules), kpsMTIII (group 3 capsules), malX (pathogenicity island marker), mchB (colicin H), papA (P fimbriae structural subunit), papG (P fimbriae adhesin), sfa (S fimbriae), sitA (iron transport), tcpC (sepsis-associated), terC (tellurite resistance), traT (serum resistance associated), usp (uropathogenic-specific protein). Genes present in all isolates: chuA (heme uptake), clbB/N (colibactin), cnf1 (cytotoxic necrotizing factor), fimH (type 1 fimbriae), fyuA (yersiniabactin), gad (glutamate decarboxylase), hlyD (alpha hemolysin), hra (heat-resistant agglutinin), iroN (salmochelin receptor), microcin M (part of colicin H), ompT (outer membrane protease), papAHC (P fimbriae structural subunit and assembly), sfa/focDE (S and F1C fimbriae), vat (vacuolating toxin), and yfcV (adhesin). papEFG (P fimbriae minor subunits, including adhesin) was present in all isolates except 7.1451. Genes sought but not detected: afa/draBC (Dr-binding adhesins), afaE8 (variant Dr-binding adhesin), astA (enteroaggregative E. coli toxin), bmaE (M fimbriae), cdtB (cytolethal distending toxin), clpG (adhesin), cvaC (colicin [microcin] V), F17 (adhesin), gafD (G fimbriae), hlyF (variant hemolysin), iha (adhesin-siderophore receptor), iss (increased serum survival), iutA (aerobactin receptor), K2/K100 (group 2 capsule variants), K5 (group 2 capsule variant), pic (protein associated with intestinal colonization), sat (secreted autotransporter toxin), and tsh (temperature-sensitive hemagglutinin).
ST2604 falls within ST complex 12 (STc12).
HP case 2.
In August 2017, an 8-month-old male Siberian Husky from a different Minnesota household developed a cough. The following morning it was found collapsed and unresponsive and so was taken to an animal hospital, where cardiopulmonary resuscitation was performed. Upon intubation, nonclotted blood came from the trachea. Resuscitation efforts were unsuccessful.
Postmortem examination showed severe hemothorax and extensive, bilateral, acute, necrohemorrhagic bronchopneumonia. Microscopically, affected lung sections exhibited abundant hemorrhage with fibrin accumulation and necrosis with moderate neutrophilic infiltration, accompanied by intra-alveolar colonies of short bacillary bacteria. Lung tissue grew 4+ beta-hemolytic E. coli and Streptococcus canis, as did multiple other organs, which in contrast lacked histopathological abnormalities. The E. coli lung isolate was typed as O?:H14, ST2015 (phylogroup B2), pulsotype 2320 (Table 1). It contained 19 extraintestinal virulence genes and qualified molecularly as ExPEC.
Comparison with published HP case isolates.
The two present HP case strains, both from dogs, were compared with all available isolates from published cases of E. coli HP, which, after deduplication, collectively represented 11 epidemiologically distinct strains, from dogs, cats, a tiger, and a horse (Table 1) (3–8). All 13 HP strains were from phylogroup B2. Eleven exhibited distinct pulsotypes, whereas the case 1 strain and archival HP isolate 0.2025 (also from a dog) both exhibited pulsotype 0064. The isolates represented eight different STs, predominantly ST127 (n = 5) and ST12 (n = 3); three defined O types, i.e., O6 (n = 6), O4 (n = 4), and O2 (n = 1); and five defined H types, i.e., H5 (n = 5), H31 (n = 5), and H7, H14, and H18 (n = 1 each). These variables corresponded extensively (Table 1).
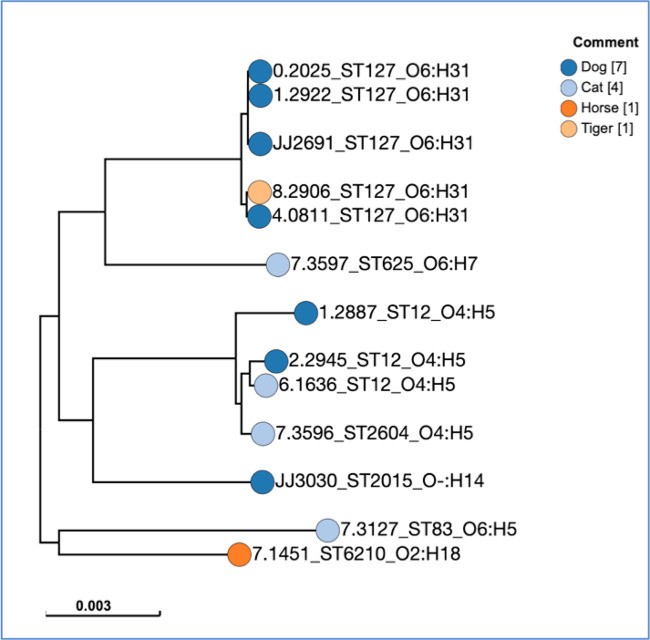
In an unrooted, single nucleotide polymorphism (SNP)-based maximum-likelihood core genome phylogeny, the 13 HP strains segregated by O:H type and ST into two main clusters, i.e., O6:H31–ST127 (n = 6) and O4:H5–ST12/ST2604 (n = 4), plus four singletons (Fig. 2). Archival HP isolate 7.3596, within the O4:H5 cluster, represented ST2604, a single-locus variant of ST12 (with mdh allele 244 instead of 16). Case 1 isolate JJ2026 fell within the O6:H31–ST127 cluster, whereas case 2 isolate JJ3030 was a singleton.
FIG 2.
Core-genome phylogram for 13 Escherichia coli isolates from animals with hemorrhagic pneumonia. The (unrooted) phylogram was inferred according to the maximum-likelihood method by using SNP Tree in Enterobase, based on 89,170 variant sites. Isolate 0.2025 (ST127–O6:H31) was the reference genome. Only variant sites present in all 13 isolates were included. Isolates JJ2691 and JJ3030 are from cases 1 and 2, respectively, presented here. Other isolates are archival (previously published) isolates.
Extended virulence genotyping of the 13 strains by PCR and WGS analysis identified 14 strictly conserved virulence genes or operons (Table 1 footnote e). These included genes for four adhesins, i.e., hra (heat-resistant agglutinin), papAHC (P fimbriae structural subunit and assembly), sfa/focDE (S and F1C fimbriae), and yfcV (variant adhesin); three toxins, i.e., cnf1 (cytotoxic necrotizing factor), hlyD (alpha hemolysin), and vat (vacuolating toxin); three iron uptake systems, i.e., chuA (heme uptake), fyuA (yersiniabactin), and iroN (salmochelin receptor); and four with miscellaneous functions, i.e., clbB/N (colibactin), gad (glutamate decarboxylase; acid tolerance), mchB (colicin H related), and ompT (outer membrane protease).
Six different F antigen-specific papA alleles occurred singly or in various pairwise combinations, suggesting multiple copies of the pap operon, and segregated partially by ST (Table 1). In contrast, 11 different fimH alleles occurred only once each, except for fimH2, which appeared in three O6:H31–ST127 isolates.
Twenty-five additional virulence genes or operons were variably present, of which seven (papCEFG, sfaD [S and F1C fimbriae], sitA [iron uptake], kpsMII [group 2 capsules], malX [pathogenicity island marker], terC [tellurite resistance], and usp [uropathogenic-specific protein]) were nearly canonical, i.e., they occurred in all but one or two of the 13 strains (Table 1). All 12 strains with papG (P fimbriae adhesin) had papG allele III; three additionally had allele I (two O4:H5–ST12, one O6:H7–ST625). The remaining variably present genes either tracked with ST127, with or without ST625 (e.g., sfaE [S and F1C fimbriae], sfaS [S fimbriae], and traT [serum resistance associated]); occurred only in non-ST127 strains (e.g., cea [colicin E1] and mchB [colicin H]); or were distributed sporadically by ST (e.g., focG [F1C fimbriae], ireA [siderophore receptor], and K1 [group 2 capsule variant]). Conspicuously absent (in contrast to their appreciable prevalence among ExPEC isolates generally) were afa/draBC (Dr-binding adhesins), cdtB (cytolethal distending toxin), cvaC (colicin [microcin] V), iha (adhesin-siderophore receptor), iss (increased serum survival), iutA (aerobactin receptor), K2/K100 (group 2 capsule variants), K5 (group 2 capsule variant), papG allele II, and sat (secreted autotransporter toxin).
Fecal surveillance (case 1 household).
Nine months after the dog in case 1 died, fecal samples were collected from the 20 surviving hosts associated with that household (4 humans, 6 dogs, 9 cats, 1 horse). Selective plating and molecular deduplication yielded 27 unique E. coli strains (Table 2).
TABLE 2.
Characteristics of fecal Escherichia coli isolates from 20 human and animal household members associated with present case 1a
| Isolateb | Pulsotype | Phylogroup | STc | O or O:H typed | No. of strains in host |
Virulence genotypee |
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adhesins |
Toxins |
Iron uptake |
Protectins |
Miscellaneous |
ExPEC | ||||||||||||||||||||||||||||||||||||
| Human (n = 4) | Cat (n = 9) | Dog (n = 7) | Horse (n = 1) | papAHCEFG | papG allele I | papG allele III | sfa/focDE | sfaS | focG | afaE8 | hra | hlyA | hlyF | cnf1 | pic | vat | astA | iroN | fyuA | ireA | iutA | kpsMII | kpsMTIII | K1 | K5 | K15 | traT | iss | cvaC | usp | ibeA | ompT | H7 | malX | clbB/clbN | ||||||
| CU838f | 964 | A | 10 | O4 | 0 | 0 | 0 | 1 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| CU839 | 1385 | A | O−:H+ | 0 | 0 | 0 | 1 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| CU840 | 1386 | A | O−:H16 | 0 | 0 | 0 | 1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| CU842 | 1388 | A | O−:H19 | 0 | 2 | 0 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | |
| CU853 | 983 | B1 | O−:H7 | 0 | 1 | 0 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | |
| CU857 | 988 | B1 | O−:H16 | 0 | 1 | 0 | 0 | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| CU870 | 974 | B1 | O−:H21 | 0 | 1 | 0 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | |
| CU841 | 1387 | B1 | O−:H41,44 | 0 | 0 | 1 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| CU843 | 1389 | B1 | O8:H16 | 0 | 0 | 1 | 0 | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | |
| CU844 | 998 | B1 | O8:H51 | 0 | 0 | 1 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| CU849 | 965 | B1 | O113:H21 | 0 | 1 | 0 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | − | − | − | |
| CU832 | 949 | B1 | O150:H8 | 0 | 0 | 2 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | − | − | − | |
| CU829 | 1072 | B1 | O153:H7 | 1 | 0 | 0 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | |
| CU880 | 1323 | B1 | O161:H16 | 0 | 1 | 0 | 0 | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| CU827 | 948 | B1 | O169:H8 | 1 | 0 | 0 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | |
| CU846 f | 1006 | B2 | 12 | O4 | 0 | 3 | 0 | 0 | + | − | + | + | + | + | − | + | + | − | + | − | + | − | + | + | + | − | − | + | − | − | − | − | − | − | + | − | + | − | + | + | + |
| CU866 f | 1390 | B2 | 12 | O4 | 0 | 0 | 1 | 0 | + | − | + | + | − | − | − | + | + | − | + | − | + | − | + | + | − | − | − | − | − | − | − | − | − | − | + | − | + | − | + | + | + |
| CU850 f | 977 | B2 | 12 | O4 | 0 | 3 | 1 | 0 | + | + | + | − | − | − | − | + | + | − | + | − | + | − | − | + | + | − | + | − | − | − | − | + | − | − | + | − | − | − | + | + | + |
| CU836 | 1384 | B2 | 73 | O2:H1 | 0 | 1 | 0 | 0 | + | − | + | + | − | + | − | + | + | − | + | + | + | − | + | + | − | − | + | − | − | − | − | − | − | − | + | − | + | − | + | + | + |
| CU858 f | 986 | B2 | 73 | O25a | 0 | 1 | 0 | 0 | + | − | + | + | − | + | − | + | + | − | + | + | + | − | + | + | − | − | + | − | − | − | − | + | − | − | + | − | + | − | + | + | + |
| CU864 | 914 | B2 | 127 | O6:H31 | 0 | 0 | 2 | 0 | + | − | + | + | + | − | − | + | + | − | + | − | + | − | + | + | − | − | + | − | − | − | − | + | − | − | + | − | + | − | + | + | + |
| CU845 | 1005 | B2 | 372 | O83:H31 | 0 | 0 | 1 | 0 | − | − | − | + | − | + | − | − | − | − | − | − | + | − | + | + | − | − | − | − | − | − | − | − | − | − | + | + | + | − | + | − | − |
| CU825 | 921 | B2 | 429 | O83w:H4 | 3 | 0 | 0 | 0 | − | − | − | − | − | − | − | − | − | + | − | − | + | − | + | + | − | + | + | − | + | − | − | + | + | + | + | + | + | − | + | − | + |
| CU851 | 980 | B2 | 929 | O21:H14 | 0 | 1 | 0 | 0 | + e | − | + | + | + | + | − | + | + | − | + | − | + | − | + | + | + | − | + | − | − | − | − | + | − | − | + | + | + | − | + | − | + |
| CU834 | 1295 | B2 | 998 | O2:H6,41 | 0 | 0 | 2 | 0 | + | − | + | + | + | − | − | + | + | − | + | − | + | − | + | + | + | − | + | − | + | − | − | − | − | − | + | + | + | − | + | − | + |
| CU835 | 1296 | B2 | 1056 | O−:H7 | 0 | 0 | 1 | 0 | − | − | − | − | − | − | + | − | − | − | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | + | − | − | − |
| CU848 | 1016 | D1 | ND | 0 | 1 | 0 | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
Boldface type indicates cnf1-positive isolates; italic type indicates the strain shared by case 1 (lung, liver) and a healthy dog (feces). ExPEC, extraintestinal pathogenic E. coli; ND, not done; ST, sequence type. +, present; −, absent.
One representative isolate is shown per unique strain (i.e., pulsotype), regardless of how many hosts the strain was isolated from.
ST was determined if a strain was from phylogroup B2 or exhibited an ExPEC-associated O type (O2, O4, O6, O7, O15, O16, O18, O25, O75).
For strains with no O type result after a PCR-based screen, O:H serotyping was done. Isolates with footnote symbol: PCR-based O type.
All strains had fimH (type 1 fimbriae). None had afa/draBC (Dr-binding adhesins), bmaE (M fimbriae), cdtB (cytolethal distending toxin), clpG (variant adhesin), F17 (variant adhesin), gafD (G fimbriae), iha (adhesin-siderophore), K2/K100 (group 2 capsule variants), K5 (group 2 capsule variant), K15 (group 2 capsule variant), papG allele II (variant P fimbriae adhesin), sat (secreted autotransporter toxin), or tsh (temperature-sensitive hemagglutinin).
This strain lacked papAH.
The 27 fecal strains were from phylogroups B2 (n = 11), B1 (n = 11), A (n = 4), and D (n = 1) (Table 2). Nine strains, all from phylogroup B2, qualified molecularly as ExPEC, and eight strains, all ExPEC, contained cnf1 (Tables 2 and 3). The cnf1-positive strains included fecal isolate CU864, from one surviving dog; it corresponded molecularly with the case 1 HP clinical strain.
TABLE 3.
Characteristics of 27 fecal Escherichia coli strains from 20 human and animal household members associated with present case 1 of hemorrhagic pneumonia due to E. coli
| Characteristic |
Prevalence of characteristic (no. [column %]) |
P value | Prevalence of characteristic (no. [column %]) |
P value | ||||
|---|---|---|---|---|---|---|---|---|
| Category | Specific variablea | Total (n = 27) | Non-B2 (n = 16) | B2 (n = 11) | cnf1 negative (n = 19) | cnf1 positive (n = 8) | ||
| Sequence type (ST) | STc12 or ST127 | 4 (15) | 0 (0) | 4 (36) | 0.02 | 0 (0) | 4 (50) | 0.004 |
| O type | O2, O4, or O6 | 7 (26) | 1 (6) | 6 (55) | 0.02 | 1 (5) | 6 (75) | <0.001 |
| Pathotype | ExPEC | 9 (33) | 0 (0) | 9 (82) | <0.001 | 1 (5) | 7 (88) | <0.001 |
| Adhesins | papAHCEFG, papG III | 8 (30) | 0 (0) | 8 (73) | <0.001 | 0 (0) | 8 (100) | <0.001 |
| sfa/focDE | 8 (30) | 0 (0) | 8 (73) | <0.001 | 1 (5) | 7 (88) | <0.001 | |
| hra | 10 (37) | 2 (13) | 8 (73) | 0.003 | 2 (11) | 8 (100) | <0.001 | |
| Toxins | hlyA | 8 (30) | 0 (0) | 8 (73) | <0.001 | 0 (0) | 8 (100) | <0.001 |
| cnf1 | 8 (30) | 0 (0) | 8 (73) | <0.001 | NAb | NA | NA | |
| vat | 10 (37) | 0 (0) | 10 (91) | <0.001 | 2 (11) | 8 (100) | <0.001 | |
| Iron acquisition | iroN | 9 (33) | 0 (0) | 9 (82) | <0.001 | 2 (11) | 7 (88) | <0.001 |
| fyuA | 12 (44) | 2 (13) | 10 (91) | <0.001 | 4 (21) | 8 (100) | <0.001 | |
| ireA | 4 (15) | 0 (0) | 4 (36) | 0.02 | 0 (0) | 4 (50) | 0.004 | |
| Protectins | kpsMII or kpsMTIII | 8 (30) | 0 (0) | 8 (73) | <0.001 | 1 (5) | 7 (88) | <0.001 |
| Miscellaneous | usp | 10 (37) | 0 (0) | 10 (91) | <0.001 | 2 (11) | 8 (100) | <0.001 |
| ibeA | 4 (15) | 0 (0) | 4 (36) | 0.02 | 2 (11) | 2 (25) | 0.56 | |
| ompT | 16 (59) | 6 (38) | 10 (91) | 0.008 | 9 (47) | 7 (88) | 0.09 | |
| malX | 10 (37) | 0 (0) | 10 (91) | <0.001 | 2 (11) | 8 (100) | <0.001 | |
| clbB/N | 6 (22) | 0 (0) | 6 (55) | 0.002 | 0 (0) | 6 (75) | <0.001 | |
Variables shown are those that yielded P < 0.05 for at least one comparison. Definitions: clb/N (colibactin synthesis), cnf1 (cytotoxic necrotizing factor), ExPEC (extraintestinal pathogenic E. coli), fyuA (yersiniabactin), hlyA (alpha hemolysin), hra (heat-resistant agglutinin), ibeA (invasion of brain endothelium A), ireA (siderophore receptor), iroN (salmochelin receptor), kpsMII (group 2 capsules), kpsMTIII (group 3 capsules), malX (pathogenicity island marker), ompT (outer membrane protein T), papAHCEFG (P fimbriae); papG allele III (P adhesin variant), sfa/focDE (S and F1C fimbriae), STc (ST complex); usp (uropathogenic-specific protein), vat (vacuolating toxin).
NA, not applicable (cnf1 status was invariant within each group).
The phylogroup B2 fecal strains, especially those with cnf1, were significantly enriched with HP-associated STs, O antigens, and virulence genes (Table 3). Notably, all eight cnf1-positive strains contained a consensus set of eight virulence genes or operons (including cnf1), four were from ST12 or ST127, and six were O2, O4, or O6, features suggesting the potential to cause HP.
Including case 1, the household's 27 fecal strains each occurred in a single host (19 strains) or in two hosts (5 strains, including the case 1 clinical strain), three hosts (2 strains), or four hosts (1 strain) (Table 2). Of the eight multiple-host strains, one occurred only in humans, four (including the case 1 strain) only in dogs, two only in cats, and one in a dog and three cats. Conversely, of the 21 household members, 17 shared one or more strains (including the case 1 strain) with another household member. Shared-strain status was associated with phylogroup B2 (6/11, versus 2/16 for other phylogroups: P = 0.03), ExPEC (6/9, versus 2/18 non-ExPEC isolates; P = 0.006), and cnf1 (5/8, versus 3/19 cnf1-negative isolates; P = 0.03).
Death of a second dog in the case 1 household.
In January 2011, 16 months after the death of the dog in case 1, an 8-year-old male German Shepherd from the case 1 household also fell ill. Notably, this dog was the only household member in which the earlier fecal surveillance had detected the case 1 HP strain (Table 2). After illness onset the dog stopped eating, became severely dehydrated, and 2 days later died.
Formalin-fixed tissues (lung, heart, liver, pancreas, spleen, duodenum, stomach, urinary bladder; not brain or kidney) were received and processed for microscopic analysis, which identified moderate to advanced postmortem autolysis, but no apparent cause of death, including specifically no evidence of infectious agents, inflammation, or neoplasia. Cultures of the corresponding unfixed tissues and a rectal swab yielded Enterococcus and eight distinct pulsotypes of E. coli. However, none of the E. coli strains were hemolytic on blood agar or corresponded by PFGE to the case 1 HP strain (not shown).
Comparison with a large PFGE library.
A search of a large private PFGE profile library identified one or more same-pulsotype isolates for five of the 13 present HP strains (Table 4). Pulsotype 0064 (O6:H31; ST127), which accounted for two of the present dog-derived HP strains, also included archival dog, cat, and human isolates, as recovered from feces (two dogs, one cat, and three humans), urine (one dog and seven humans), blood (one human), and lung (two dogs, one reportedly with HP and one without clinical data), from across the United States (1985 through 2018). Pulsotype 0691 (O4:H5; ST12 complex), which accounted for a present cat-derived HP strain, also included three archival human urine isolates from Japan (1997) and Minnesota (2018). Pulsotype 1263 (O4:H5, ST12), which accounted for the present HP strain that caused an outbreak among multiple cats at a Connecticut facility, also included archival kidney, spleen, and uterus isolates from one or more cats in Florida (1996). Finally, pulsotype 1424, which accounted for a present dog-source HP strain, also included a human urine isolate from Michigan (1998) and a retail ground turkey meat isolate from Indiana (2001).
TABLE 4.
Occurrence of hemorrhagic pneumonia (HP)-associated Escherichia coli pulsotypes within a reference pulsed-field gel electrophoresis library
| Pulsotype | HP isolate(s) from this report (new and published) | Matching isolates from private PFGE profile library |
||||||
|---|---|---|---|---|---|---|---|---|
| Source | No. of isolates from specimen type |
Yr(s), range | Location | |||||
| Feces | Urine | Blood | Other | |||||
| 0064 | 0.0205,a JJ2690,b JJ2691b | Dog | 2 | 1 | Lung, 2c | 2005–2013 | NY, OH (USA) | |
| Cat | 1 | 1998 | MN (USA) | |||||
| Human | 3 | 7d | 1 | 1985–2017 | MN, OH, TX, and WA (USA) | |||
| 0691 | 7.3596,e 8.0201e | Human | 3d | 1997–2018 | Japan; MN (USA) | |||
| 1263 | 6.136–6.138,f 6.1679–6.1682f | Cat | Kidney, 1g; spleen, 1g; uterus, 1g | 1996 | FL (USA) | |||
| 1424 | 1.2887 | Human | 1 | 1998 | MI (USA) | |||
| Food | Ground turkey, 1 | 2001 | IN (USA) | |||||
From a unique case (3).
Both from present case 1 (lung, liver).
One dog (from same vendor that provided the canine host for HP isolate 0.0205) had hemorrhagic pneumonia. The other dog lacked clinical data.
From males and females.
Both from the same case (lung, colon) (7).
All from the same outbreak (multiple cats at one facility) (5).
Possibly from the same case.
DISCUSSION
The present HP cases, which both involved previously healthy, epidemiologically unrelated, community-dwelling household dogs, were similar in nearly all respects to previously reported cases, which involved multiple dogs and cats, a tiger, and a horse (3–8). Specifically, like previous cases, the present cases were rapidly fatal, presented with overwhelming sepsis, and demonstrated multiorgan involvement (by gross and microscopic pathological examination and culture), with a predominant finding of necrohemorrhagic pneumonia.
According to the extensive molecular and phenotypic testing done here, the new HP E. coli isolates shared many characteristics with the previously reported HP case isolates, which were studied in parallel. Collectively, the HP-causing strains represented a narrow range of clonal lineages within phylogroup B2 (mainly the ST12 complex and ST127), contained cnf1 plus multiple other virulence genes (some strictly conserved or quasi-conserved, consistent with genetic linkage or coselection), and exhibited a narrow range of clonally distributed O antigens (mainly O4 and O6) and H antigens (mainly H5 and H31). The WGS-based virulence gene screen newly identified sitA (iron uptake) and terC (tellurite resistance) as potential contributors to these strains' ability to cause HP. One of the new case isolates corresponded by pulsotype with a temporally and geographically unrelated historical case isolate, suggesting the existence of a widely disseminated, endemic strain with the ability to cause HP.
A possible mechanism for such dissemination was suggested by the study's fecal surveillance component. It documented that multiple members of the case 1 household shared either the household's index HP strain (two dogs) or several other E. coli strains, some from the same STs and with the same O:H serotypes as and virulence genotypes similar to those of the HP case isolates. These findings suggest that a fecal reservoir and extensive host-host transmission may underlie sporadic HP cases, with HP being only the proverbial tip of the iceberg. Within-household sharing of E. coli strains, likely from host-to-host transmission, has been documented extensively for other types of E. coli (14, 15); the present findings extend this phenomenon to HP-causing strains.
Regarding endemicity, recovery of the case 1 HP strain from a second dog in the same household 9 months after it caused fatal sepsis in case 1 suggests that it persisted in the household at least that long. Notably, however, it was not detected in the second dog 8 months later, when that dog died of unknown (non-HP) causes. Whether in the meantime the strain had passed to other household members, and so persisted within the household even after the death of the two known-to-be-colonized dogs, is unknown. However, prolonged within-household persistence is not unusual for ExPEC strains (15–17).
The search of the private reference PFGE profile library identified multiple archival isolates with close genomic similarity to the HP clinical strains, implying the potential to cause HP. In nature, nearly all of these archival isolates had displayed diverse non-HP phenotypes; i.e., they were intestinal colonizers of humans and companion animals, versus human urinary and bloodstream pathogens, canine and feline urinary pathogens, or a contaminant of retail turkey meat. This broad ecological range is consistent with both zoonotic potential and, conceivably, foodborne transmission, although this is speculative.
Despite the fact that the study’s opportunistic sampling approach permits no firm conclusions, the finding of abundant HP-like archival urine isolates, plus the known much higher incidence of urinary tract infection (UTI) than HP (1), suggests that HP likely represents a comparatively rare clinical presentation even for the distinctive strains that are able to cause it. Such strains, like other ExPEC strains (18), therefore probably exist mainly as gut colonizers, and on the rare occasions when they do cause extraintestinal infection, usually cause a more common syndrome, such as UTI.
These considerations, plus the fact that the HP-causing strains contained multiple canonical virulence genes other than cnf1, detract from the appeal of labeling them as NTEC, a term that gives what is probably unwarranted emphasis to the contribution of a necrotizing process like HP to their lifestyle and of cnf1 to their pathogenic capabilities. That said, such strains do seem to be uniquely capable of causing HP, which, curiously, has been reported almost exclusively in dogs and cats (3–8), despite humans sometimes carrying such strains and developing other types of infection caused by them.
This seeming host specificity clouds the interpretation of experimental data from mouse and rat pneumonia models and observational data from humans with ventilator-associated pneumonia. These findings include the observation that, in a rat pneumonia model with ST12 challenge strain CP9, hly contributed to both necrosis and apoptosis but cnf1 contributed only to apoptosis (19), whereas in a mouse pneumonia model with ST127 challenge strain 536, neither hly nor cnf1 contributed to virulence (20). Likewise, among human clinical respiratory E. coli isolates from mechanically ventilated patients with or without pneumonia and all-comer bloodstream isolates, characteristics significantly associated with pneumonia included ST127, sfa/foc, papG allele III, and iroN (21), which also typify HP isolates. Thus, many questions remain regarding the HP syndrome, the distinctive causative strains, and the relevant ecological and host-pathogen relationships.
Study limitations include the opportunistic sampling approach, limited clinical and epidemiological data, and absence of genome sequence data for the fecal and archival PFGE library isolates. Study strengths and novel features include the analysis of all available published HP case isolates, addition of two new HP cases, household fecal surveillance surrounding one of these cases, extensive virulence genotyping, leveraging of a large private PFGE library, and genomic sequencing of the HP isolates.
In summary, E. coli strains that cause HP derive from a narrow range of ExPEC lineages within phylogroup B2, contain multiple virulence genes other than cnf1, are shared extensively between hosts, and likely function in nature mainly as intestinal colonizers and uropathogens. The findings identify questions for future research and may prove relevant for surveillance and prevention efforts.
MATERIALS AND METHODS
Clinical HP case isolates.
Clinical isolates from the two present HP cases and necropsy tissues from a dog from the same household as case 1 that died 2 years after case 1 were obtained from the UM Veterinary Diagnostic Laboratory (VDL). Clinical isolates from published cases of HP (from dogs, cats, a tiger, and a horse), submitted previously to the Escherichia coli Reference Center (ECRC; The Pennsylvania State University) for serotyping, were obtained from the ECRC. For the present HP cases, histopathologic analysis was done by the VDL using standard methods, and electron microscopy of lung tissue from case 1 was done as described elsewhere (22). Briefly, formalin-fixed lung tissues (1-mm3 fragments) were postfixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, followed by a second postfixation with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer. Samples were dehydrated and embedded in EMbed 812 resin. Sixty- to 70-nm thin sections were contrasted with 5% uranyl acetate and Santos’ lead citrate. (All reagents were from Electron Microscopy Sciences.) These samples were visualized using a transmission electron microscope (JEOL 1400; JEOL, Ltd., Tokyo, Japan).
Fecal isolates.
For HP case 1, fecal swabs from all surviving human (n = 4) and animal (n = 16: 6 dogs, 9 cats, 1 horse) hosts associated with the household, collected by the owner 9 months after the dog's death, plus (separately) necropsy tissue specimens from one of these household dogs, which died 2 years after case 1, underwent selective plating to recover generic and hemolytic E. coli (14). From each plate, up to five putative E. coli colonies (as inferred from colony appearance and lactose, indole, and citrate phenotypes) underwent PCR-based genomic profiling to resolve putative unique genotypes (23).
Pulsed-field gel electrophoresis.
All clinical HP isolates and a representative of each putatively unique fecal genotype per host underwent XbaI pulsed-field gel electrophoresis (PFGE) according to the PulseNet protocol (24). With computer assistance (BioNumerics; Applied Maths), profiles were assigned to an existing versus novel pulsotype, i.e., clone, based on a 94% similarity threshold (according to Dice coefficients) in comparison with a large private PFGE profile library (2,573 pulsotypes) (25). The PFGE library, which included profiles for 9,376 isolates from diverse locales, clinical/epidemiological contexts, specimen types, and time periods, was searched for archival isolates that corresponded by pulsotype with the present HP clinical isolates.
Clonal characterization.
For all fecal and HP study isolates, after deduplication by genomic profiling, a representative of each unique clone per host (or outbreak, for published isolates) was further characterized. If not already done, O:H typing was done by the ECRC, using serological detection for (somatic) O antigens (26) and PCR-based detection for (flagellar) H types (27). For all selected representative isolates, established PCR-based assays were used to determine major E. coli phylogenetic group, O type (if unclear from serotyping), and extended virulence genotypes (52 extraintestinal virulence genes and variants), including, for the clinical isolates, F types (papA alleles) (23, 28). Fecal clones from phylogroup B2 underwent conventional Achtman multilocus sequence typing (MLST) (29, 30). Isolates were classified presumptively as ExPEC according to established molecular criteria, i.e., if positive for ≥2 of papAH or papC (counted as one; P fimbriae), sfa/focDE (S and F1C fimbriae), afa/draBC (Dr-binding adhesins), iutA (aerobactin receptor), and kpsMII (group 2 capsule) (31).
Whole-genome sequence analysis.
A representative of each new and published HP clinical strain underwent whole-genome sequencing (WGS). DNA was extracted from cultures using the DNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. Genomic DNA libraries were created using the Nextera XT library preparation kit and Nextera XT index kit v2 (Illumina, San Diego, CA), and sequencing was performed using 2 × 250-bp dual-index runs on an Illumina MiSeq instrument at the University of Minnesota Mid-Central Research and Outreach Center (Willmar, MN). By using applications available via Enterobase (https://enterobase.warwick.ac.uk/) (32) and the Center for Genomic Epidemiology (https://www.genomicepidemiology.org/), the genome sequence data were used to infer STs, O:H types, and extraintestinal virulence genotypes, including alleles of papA and fimH (33–36). Additionally, a unrooted, single-nucleotide-polymorphism (SNP)-based maximum-likelihood core genome phylogram was inferred by using SNP Tree, via Enterobase (32).
Statistical methods.
Two-group comparisons involving dichotomous variables were tested using Fisher's exact text. The significance criterion was a P value of <0.05.
Data availability.
Raw reads from sequenced isolates have been deposited in the NCBI database, with BioProject accession number PRJNA762997 (isolate 0.2025, SRX12173624; isolate 1.2887, SRX12173625; isolate 1.2922, SRX12173629; isolate 2.2945, SRX12173630; isolate 4.0811, SRX12173631; isolate 6.1636, SRX12173632; isolate 7.1451, SRX12173633; isolate 7.3127, SRX12173634; isolate 7.3596, SRX12173635; isolate 7.3597, SRX12173636; isolate 8.2906, SRX12173626; isolate JJ2691, SRX12173627; isolate JJ3030, SRX12173628).
ACKNOWLEDGMENTS
This work was supported in part by the Office of Research and Development, U.S. Department of Veterans Affairs. The opinions expressed are strictly those of the authors and do not reflect those of the authors' respective institutions.
Contributor Information
James R. Johnson, Email: JOHNS007@UMN.EDU.
Christopher A. Elkins, Centers for Disease Control and Prevention
REFERENCES
- 1.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect 5:449–456. 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 2.Ling GV, Biberstein EL, Hirsh DC. 1979. Bacterial pathogens associated with urinary tract infections. Vet Clin North Am Small Anim Pract 9:617–630. 10.1016/S0195-5616(79)50077-1. [DOI] [PubMed] [Google Scholar]
- 3.Handt LK, Stoffregen DA, Prescott JS, Pouch WJ, Ngai DTW, Anderson CA, Gatto NT, DebRoy C, Fairbrother JM, Motzel SL, Klein HJ. 2003. Clinical and microbiologic characterization of hemorrhagic pneumonia due to extraintestinal pathogenic Escherichia coli in four young dogs. Comp Med 53:663–670. [PubMed] [Google Scholar]
- 4.Breitschwerdt E, Debroy C, Mexas A, Brown T, Remick A. 2005. Isolation of necrotoxigenic Escherichia coli from a dog with hemorrhagic pneumonia. J Am Vet Med Assoc 226:2016–2019. 10.2460/javma.2005.226.2016. [DOI] [PubMed] [Google Scholar]
- 5.Sura R, Van Kruiningen HJ, DebRoy C, Hinckley LS, Greenberg KJ, Gordon Z, French RA. 2007. Extraintestinal pathogenic Escherichia coli-induced acute necrotizing pneumonia in cats. Zoonoses Public Health 54:307–313. 10.1111/j.1863-2378.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 6.Debroy C, Roberts E, Jayarao BM, Brooks JW. 2008. Bronchopneumonia associated with extraintestinal pathogenic Escherichia coli in a horse. J Vet Diagn Invest 20:661–664. 10.1177/104063870802000524. [DOI] [PubMed] [Google Scholar]
- 7.Highland MA, Byrne BA, Debroy C, Samitz EM, Peterson TS, Oslund KL. 2009. Extraintestinal pathogenic Escherichia coli-induced pneumonia in three kittens and fecal prevalence in a clinically healthy cohort population. J Vet Diagn Invest 21:609–615. 10.1177/104063870902100504. [DOI] [PubMed] [Google Scholar]
- 8.Carvallo F, Debroy C, Baeza E, Hinckley L, Gilbert K, Choi SJ, Risatti G, Smyth JA. 2010. Necrotizing pneumonia and pleuritis associated with extraintestinal pathogenic Escherichia coli in a tiger (Panthera tigris) cub. J Vet Diagn Invest 22:136–140. 10.1177/104063871002200130. [DOI] [PubMed] [Google Scholar]
- 9.Russo TA, Johnson JR. 2000. A proposal for an inclusive designation for extraintestinal pathogenic Escherichia coli: ExPEC. J Infect Dis 181:1753–1754. 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JR, Russo TA. 2002. Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. J Infect Dis 186:859–864. 10.1086/342490. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JR, Gajewski A, Lesse AJ, Russo TA. 2003. Extraintestinal pathogenic Escherichia coli as a cause of invasive non-urinary infections. J Clin Microbiol 41:5798–5802. 10.1128/JCM.41.12.5798-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabbri A, Travaglione S, Fiorentini C. 2010. Escherichia coli cytotoxic necrotizing factor 1 (CNF1): toxin biology, in vivo applications and therapeutic potential. Toxins (Basel) 2:283–296. 10.3390/toxins2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borriello G, Lucibelli M, De Carlo E, Auriemma C, Cozza D, Ascione G, Scognamiglio F, Iovane G, Galiero G. 2012. Characterization of enterotoxigenic E. coli (ETEC), Shiga-toxin producing E. coli (STEC) and necrotoxigenic E. coli (NTEC) isolated from diarrhoeic Mediterranean water buffalo calves (Bubalus bubalis). Res Vet Sci 93:18–22. 10.1016/j.rvsc.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox T, Clabots C, Porter S, Bender T, Thuras P, Colpan A, Boettcher J, Johnson JR. 2020. Bacterial “virulence” traits and host demographics predict Escherichia coli colonization behaviors within households. Open Forum Infect Dis 7:ofaa495. 10.1093/ofid/ofaa495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JR, Davis G, Clabots C, Johnston BD, Porter S, Debroy C, Pomputius W, Ender PT, Cooperstock M, Slater BS, Banerjee R, Miller S, Kisiela D, Sokurenko E, Aziz M, Price LB. 2016. Household clustering of Escherichia coli sequence type 131 clinical and fecal isolates according to whole genome sequence analysis. Open Forum Infect Dis 3:ofw129. 10.1093/ofid/ofw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JR, Clabots C, Kuskowski MA. 2008. Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. J Clin Microbiol 46:4078–4082. 10.1128/JCM.00980-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray AC, Kuskowski MA, Johnson JR. 2004. Virulence factors predict Escherichia coli colonization patterns among human and animal household members. Ann Intern Med 140:848–849. 10.7326/0003-4819-140-10-200405180-00032. [DOI] [PubMed] [Google Scholar]
- 18.Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, Tenaillon O. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol Biol Evol 24:2373–2384. 10.1093/molbev/msm172. [DOI] [PubMed] [Google Scholar]
- 19.Russo T, Davidson BA, Genagon SA, Warholic N, MacDonald U, Pawlicki P, Beanan JM, Olson R, Holm B, Knight IIP. 2005. E. coli virulence factor hemolysin induces neutrophil apoptosis and necrosis/lysis in vitro and necrosis/lysis and lung injury in a rat pneumonia model. Am J Physiol Lung Cell Mol Physiol 289:L207–L216. 10.1152/ajplung.00482.2004. [DOI] [PubMed] [Google Scholar]
- 20.Phillips-Houlbracq M, Ricard J-D, Foucrier A, Yoder-Himes D, Gaudry S, Bex J, Messika J, Margetis D, Chatel J, Dobrindt U, Denamur E, Roux D. 2018. Pathophysiology of Escherichia coli pneumonia: respective contribution of pathogenicity islands to virulence. Int J Med Microbiol 308:290–296. 10.1016/j.ijmm.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 21.La Combe B, Clermont O, Messika J, Eveillard M, Kouatchet A, Lasocki S, Corvec S, Lakhal K, Billard-Pomares T, Fernandes R, Armand-Lefevre L, Bourdon S, Reignier j, Fihman V, de Prost N, Bador J, Goret J, Wallet F, Denamur E, Ricard J-D, COLOCOLI Group. 2019. Pneumonia-specific Escherichia coli with distinct phylogenetic and virulence profiles, France, 2012–2014. Emerg Infect Dis 25:710–718. 10.3201/eid2504.180944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armién A, Wolf T, Mor S, Ng T, Bracht A, Goyal S, Rasmussen J. 2020. Molecular and biological characterization of a cervidpoxvirus isolated from moose with necrotizing dermatitis. Vet Pathol 57:296–310. 10.1177/0300985819891240. [DOI] [PubMed] [Google Scholar]
- 23.Mohamed M, Clabots C, Porter SB, Bender T, Thuras P, Johnson JR. 2020. Large fecal reservoir of Escherichia coli sequence type 131-H30 subclone strains that are shared within households and resemble clinical ST131-H30 isolates. J Infect Dis 221:1659–1668. 10.1093/infdis/jiz669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 25.Johnson JR, Nicolas-Chanoine M, Debroy C, Castanheira M, Robicsek A, Hansen G, Weissman SJ, Urban C, Platell JL, Trott DJ, Zhanel GG, Clabots C, Johnston BD, Kuskowski MA, the MASTER Investigators. 2012. Comparison of Escherichia coli sequence type ST131 pulsotypes by epidemiologic traits, 1967 – 2009. Emerg Infect Dis 18:598–607. 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orskov F, Orskov I. 1984. Serotyping of Escherichia coli. Methods Microbiol 14:43–111. 10.1016/S0580-9517(08)70447-1. [DOI] [Google Scholar]
- 27.Machado J, Grimont F, Grimont PA. 2000. Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res Microbiol 151:535–546. 10.1016/s0923-2508(00)00223-0. [DOI] [PubMed] [Google Scholar]
- 28.Johnson JR, Stell AL, Scheutz F, O'Bryan TT, Russo TA, Carlino UB, Fasching CC, Kavle J, van Dijk L, Gaastra W. 2000. Analysis of F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect Immun 68:1587–1599. 10.1128/IAI.68.3.1587-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon DM, Clermont O, Tolley H, Denamur E. 2008. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol 10:2484–2496. 10.1111/j.1462-2920.2008.01669.x. [DOI] [PubMed] [Google Scholar]
- 30.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 95:3140–3145. 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 47:2161–2168. 10.1128/AAC.47.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Alikhan N-F, Mohamed K, Fan Y, Achtman M, Agama Study Group. 2020. The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res 30:138–152. 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen M, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig R, Jelsbak L, Sicheritz-Pontén T, Ussery D, Aarestrup F, Lund O. 2012. Multilocus sequence typing of total genome sequenced bacteria. J Clin Microbiol 50:1355–1361. 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joensen KG, Tetzschner A, Iguchi A, Aarestrup F, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J Clin Microbiol 53:2410–2426. 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roer L, Tchesnokova V, Allesoe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, Thomsen M, Lund O, Hansen F, Hammerum A, Sokurenko E, Hasman H. 2017. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol 55:2538–2543. 10.1128/JCM.00737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tetzschner AMM, Johnson J, Johnson B, Lund O, Scheutz F. 2020. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol 58:e01269-20. 10.1128/JCM.01269-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw reads from sequenced isolates have been deposited in the NCBI database, with BioProject accession number PRJNA762997 (isolate 0.2025, SRX12173624; isolate 1.2887, SRX12173625; isolate 1.2922, SRX12173629; isolate 2.2945, SRX12173630; isolate 4.0811, SRX12173631; isolate 6.1636, SRX12173632; isolate 7.1451, SRX12173633; isolate 7.3127, SRX12173634; isolate 7.3596, SRX12173635; isolate 7.3597, SRX12173636; isolate 8.2906, SRX12173626; isolate JJ2691, SRX12173627; isolate JJ3030, SRX12173628).