Abstract
In Saccharomyces cerevisiae, the family of ATF/CREB transcriptional regulators consists of a repressor, Acr1 (Sko1), and two activators, Aca1 and Aca2. The AP-1 factor Gen4 does not activate transcription through ATF/CREB sites in vivo even though it binds these sites in vitro. Unlike ATF/CREB activators in other species, Aca1- and Aca2-dependent transcription is not affected by protein kinase A or by stress, and Aca1 and Aca2 are not required for Hog1-dependent salt induction of transcription through an optimal ATF/CREB site. Aca2 is important for a variety of biological functions including growth on nonoptimal carbon sources, and Aca2-dependent activation is modestly regulated by carbon source. Strains lacking Aca1 are phenotypically normal, but overexpression of Aca1 suppresses some defects associated with the loss of Aca2, indicating a functional overlap between Aca1 and Aca2. Acr1 represses transcription both by recruiting the Cyc8-Tup1 corepressor and by directly competing with Aca1 and Aca2 for target sites. Acr1 does not fully account for osmotic regulation through ATF/CREB sites, and a novel Hog1-dependent activator(s) that is not a bZIP protein is required for ATF/CREB site activation in response to high salt. In addition, Acr1 does not affect a number of phenotypes that arise from loss of Aca2. Thus, members of the S. cerevisiae ATF/CREB family have overlapping, but distinct, biological functions and target genes.
Eukaryotic organisms from yeast to human contain multiple ATF/CREB family proteins that activate or repress the expression of specific genes (14, 18, 19, 21). ATF/CREB proteins bind as homodimers or heterodimers to specific DNA sequences (consensus TGACGTCA) via a bZIP structural motif, which consists of a leucine zipper that mediates dimerization and an adjacent basic region that contacts DNA. ATF/CREB proteins can also form DNA-binding heterodimers with AP-1 proteins (17), members of a structurally related family that bind similar half-sites but differ in the requirement for half-site spacing (29, 50).
ATF/CREB proteins often serve as the ultimate targets of signal transduction pathways, and they play critical roles in many biological processes. In multicellular organisms, CREB stimulates transcription in response to cyclic AMP and calcium in a phosphorylation-dependent manner (41), and ATF-2 is the target of stress-activated mitogen-activated (MAP) kinases (16, 35, 63). ATF/CREB proteins control diverse biological functions such as memory (1, 53), opiate tolerance (37), spermatogenesis (10), circadian rhythms (9), and skeletal and neural development (47).
In the fission yeast Schizosaccharomyces pombe, ATF/CREB proteins are important for sexual development, entry into stationary phase, response to osmotic and oxidative stress, and activation of a hot spot for meiotic recombination (32, 51, 57, 66, 67). Atf1 is a direct target of the Spc1(Sty1) stress-activated MAP kinase (51, 67), an observation that is remarkably similar to the situation in mammalian cells. Although Atf1 is sufficient to mediate this stress response, activation of meiotic recombination requires an Atf1-Pcr1 heterodimer, whose activity also depends on Spc1 kinase (32, 33). Pcr1 is required for the nuclear localization of Atf1, and phosphorylation and association of Atf1 with Spc1 kinase is required for nuclear retention of Spc1 (12).
In the baker's yeast Saccharomyces cerevisiae, two ATF/CREB proteins have been identified. Acr1 (Sko1) is a repressor (43, 65), although the DNA-binding domain is not sufficient for repression (65). Overexpression of Acr1 suppresses the toxicity caused by high levels of protein kinase A (PKA) (43) or high levels of Rap1, a transcriptional regulator that also binds to telomeres (11), but the molecular bases for these effects are unknown. More recently, Acr1 has been implicated as a downstream effector of the HOG signal transduction pathway that responds to osmotic stress (46). Hac1 (44) is a transcriptional activator that induces a variety of genes in response to unfolded proteins in the endoplasmic reticulum (4, 42); for reasons discussed below, we believe that Hac1 should not be classified as an ATF/CREB protein.
Previously, we provided genetic and biochemical evidence for an ATF/CREB activator(s) in S. cerevisiae (65). Specifically, strains lacking the Acr1 repressor confer transcriptional activation through ATF/CREB sites in vivo, and they possess ATF/CREB-binding activities in vitro. Hac1 is not the putative ATF/CREB activator(s), because these transcriptional and DNA-binding activities are observed under normal growth conditions where Hac1 is not produced (52). Here, we show that two previously uncharacterized proteins, Aca1 and Aca2, are the ATF/CREB activators in S. cerevisiae. Phenotypic analysis indicates that Aca2 is important for growth on nonoptimal carbon sources as well as resistance to a variety of drugs, and that the individual ATF/CREB proteins play distinct biological roles. In addition, we show that Acr1 does not fully account for osmotic regulation through ATF/CREB sites, and that a novel activator(s) distinct from Aca1 and Aca2 supports ATF/CREB site-dependent activation in response to high salt.
MATERIALS AND METHODS
DNA molecules.
Plasmids used for synthesizing 35S-proteins in vitro were created by cloning PCR-generated fragments encoding the bZIP domains of Aca1 (C-terminal 300 or 138 residues), Aca2 (C-terminal 334 or 167 residues), or Hac1 (full-length 231 or N-terminal 108 residues) downstream of the SP6 promoter of Ycp88 (23). The comparable plasmids used to produce Gcn4 (23) or Acr1 (65) have been described previously. The plasmids expressing LexA fusion proteins were obtained by cloning SmaI-NcoI fragments encoding full-length ATF/CREB proteins into the YCp91-LexA vector (61); LexA-Acr1 has been previously described (26). Plasmids overexpressing the various proteins contain the following chromosomal fragments cloned into Yeplac195: Aca1, PvuII-XhoI; Aca2, SphI-SpeI; Acr1, NheI-XmaI; Hac1, PvuII-HindIII. YIp56-Sc3674, which contains a gcn4 deletion allele (7), and plasmids containing bcy1::URA3 (59), hog1::TRP1 (obtained from Haruo Saito), tup1::LEU2 (obtained from Joe Geisberg), and cyc8::LEU2 (5) were used to introduce null mutations into KY898 derivatives.
DNA-binding assays.
35S-labeled proteins were synthesized by transcription and translation in vitro using SP6 RNA polymerase and wheat germ extract (Promega), and the translation products were analyzed on denaturing polyacrylamide gels as described previously (55). For experiments to determine whether proteins bind as homodimers or heterodimers, proteins of different sizes were cotranslated. Equimolar amounts of the resultant proteins were incubated at 25°C for 30 min in buffer containing 20 mM Tris-HCl (pH 7.4), 0.1 mg of gelatin/ml 1 mM EDTA, 12.5% glycerol, 3 mM MgCl2, 50 mM KCl, 500 ng of poly(dI)-poly(dC), and 2 ng of a 32P-labeled 50-bp oligonucleotide probe containing a centrally positioned DNA-binding site. Protein-DNA complexes were analyzed on a 5% native polyacrylamide gel as described previously (55). Under the conditions used in these experiments, the intensities of the bands representing the protein-DNA complexes are roughly proportional to the binding constants (20, 22, 55).
Yeast strains.
All yeast strains were derived from KY898 (a ura3-52 lys2-801 ade2-101 leu2::PET56 trp1-Δ1 his3-303) (65), with the exception of FT4/pLF98 (8) and L9FT4 (61), which were used to analyze his3 transcription dependent on an optimal Yap site and a LexA operator, respectively. Deletions of the various ATF/CREB and other proteins were generated by standard two-step gene replacement. Structures of the deletion alleles are as follows: Δaca1, which lacks a BglII-HpaI fragment, removes nearly the entire protein-coding region but retains 16 amino acids (aa) at the C terminus; Δaca2, which lacks an MscI-XhoI fragment, removes most of the protein-coding region but retains 163 aa at the N terminus; Δacr1, which lacks an NheI-AflII fragment, is deleted for the entire protein-coding region as well as 482 bp upstream and 235 bp downstream; Δhac1, which lacks an SpeI-BssHII fragment, is deleted for the entire coding region as well as 28 bp upstream and 442 bp downstream. bcy1 disruption strains were generated by one-step gene replacement and were used immediately after construction to prevent accumulation of suppressor mutations (30).
Phenotypic analyses.
Growth phenotypes of the various strains were determined by spotting 105, 104, 103, and 102 cells on appropriate media and scored as follows: +++, grows better than wild type; ++, grows comparably to wild type; +, grows more poorly than wild type; ±, barely detectable growth; −, no growth. Transcriptional activation by LexA fusion proteins was assayed in cells containing JK103, a multicopy URA3 plasmid containing a lacZ reporter driven by a promoter with four LexA operators upstream of the GAL1 TATA and initiator elements (27). Transcriptional repression by LexA-Acr1 was assayed on reporters that either contain four (JK1621) or zero (pLGΔ312S) LexA operators upstream of the intact CYC1 promoter (28). β-Galactosidase assays were performed on permeabilized cells as described previously (61). Values were normalized to A600 and represent the average of at least four independent transformants; they are accurate to ±20%. To measure RNA levels, total RNA (40 μg as quantitated by A260) was hybridized to completion with an excess of the appropriate 32P-labeled oligonucleotide probes and treated with S1 nuclease as described previously (25). RNA levels were quantitated with respect to DED1 or tRNAw internal controls by PhosphorImager (Molecular Dynamics) analysis.
RESULTS
Identification of Aca1 and Aca2 as new members of the S. cerevisiae ATF/CREB family.
Previously, we identified the complete set of 14 bZIP proteins in S. cerevisiae by searching the complete genome with a degenerate motif based on the sequences of a large number of basic regions within bZIP domain (8). Twelve of these bZIP proteins have been characterized: the AP-1 factor Gcn4 (22); Met28 (34); Yap1 through 8, a novel and fungus-specific family of bZIP proteins (8); Hac1 (44); and the ATF/CREB repressor Acr1 (Sko1) (43, 65). Here, we describe the remaining two bZIP proteins, which for reasons discussed below are termed Aca1 and Aca2 (for ATF/CREB activators).
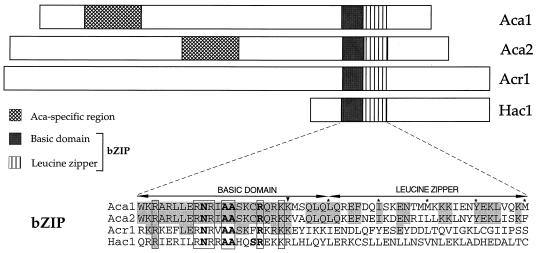
Aca1 (489 residues) and Aca2 (587 residues) have identical basic regions over the critical 17-aa region (Fig. 1), and they contain a lysine residue characteristic of ATF/CREB but not AP-1 proteins (29). The basic regions of Aca1 and Aca2 are most closely related to Acr1 and many ATF/CREB proteins in other eukaryotic species (averaging 80% identity and 90% similarity); their similarity to basic regions of AP-1 and other types of bZIP proteins is less pronounced. The Hac1 basic region is very divergent from those of ATF/CREB proteins, and we believe that Hac1 should not be categorized as an ATF/CREB protein even though it can bind the consensus ATF/CREB site in vitro (44) (see below). The Aca1 and Aca2 leucine zippers are similar to each other (54% identical, 75% similar) but are essentially unrelated in sequence to other leucine zippers. Aca1 and Aca2 contain a stretch of 80 aa with 85% similarity that is not present in known proteins. Thus, Aca1 and Aca2 are highly related proteins with an ATF/CREB-like bZIP domain.
FIG. 1.
Structures of S. cerevisiae ATF/CREB proteins. For each protein, the relative size, locations of the bZIP domains (striped and shaded boxes), and the Aca-specific region (hatched boxes) are indicated. Sequences of the bZIP domains of the ATF/CREB proteins and Hac1 are compared; residues in darker boxes are identical in Aca1 and Aca2, residues in lighter boxes are identical in Aca1, Aca2, and Acr1, and open boxes indicate residues common to all four proteins. Conserved residues in the basic region that contact DNA (bold), the lysine characteristic of ATF/CREB proteins (arrowhead), and the leucines or other residues defining position d of the leucine zipper (asterisks) are indicated.
Aca1 and Aca2 bind ATF/CREB sites as homodimers and heterodimers.
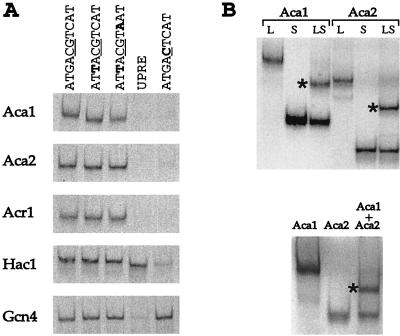
35S-labeled derivatives of Aca1 and Aca2 bind efficiently to the ATF/CREB consensus site, with affinities that are roughly comparable to those of Acr1, Hac1, and the AP-1 factor Gcn4 (Fig. 2A). However, Aca1 and Aca2, like other ATF/CREB proteins, do not bind the AP-1 consensus, in contrast to Gcn4, which prefers this site over the ATF/CREB site (50). The Aca proteins bind with comparable affinity to related ATF/CREB sites with single or double substitutions at positions ±2 that are tolerated by other ATF/CREB proteins. Thus, Aca1 and Aca2 display the DNA-binding specificity of ATF/CREB proteins.
FIG. 2.
Aca1 and Aca2 bind ATF/CREB sites as homodimers and heterodimers. (A) The indicated 35S-labeled proteins were incubated with an optimal ATF/CREB site (TGACGTCA), derivatives with one or two substitutions (bold) at the ±2position, the UPRE (GGAACTGGACAGCGTGTCGAAA), and the optimal AP-1 site (TGACTCA). (B) 35S-labeled derivatives of the indicated proteins (L, large; S, short; LS, cosynthesized mixture) were incubated with the ATF/CREB optimal binding site. The intermediate-sized products indicative of large-small and Aca1-Aca2 heterodimers are marked with asterisks.
bZIP proteins bind their DNA targets as dimers, and certain combinations of bZIP domains can interact with each other and bind as heterodimers. A mixture of two differently sized versions of Aca1 or Aca2 yields a protein-DNA complex of intermediate mobility (Fig. 2B), indicating that these proteins bind as homodimers. Examination of all pairwise combinations of ATF/CREB proteins indicates that Aca1 and Aca2 can bind ATF/CREB sites as a heterodimer with an affinity comparable to that of the corresponding homodimers. However, DNA-binding heterodimers were not observed for any other combination (data not shown).
In addition to binding ATF/CREB sites (44), Hac1 binds the UPRE (unfolded protein response element), a 22-bp sequence in KAR2 and other promoters that are activated in response to unfolded proteins in the endoplasmic reticulum (4, 42). Levels of Hac1 binding to the ATF/CREB and UPRE sequences are roughly comparable, whereas Aca1, Aca2, and Acr1 do not bind the UPRE (Fig. 2A). Thus, ATF/CREB proteins are not involved in the unfolded protein response, and Hac1 and ATF/CREB proteins have distinct DNA-binding specificities.
Aca1 and Aca2 are transcriptional activators.
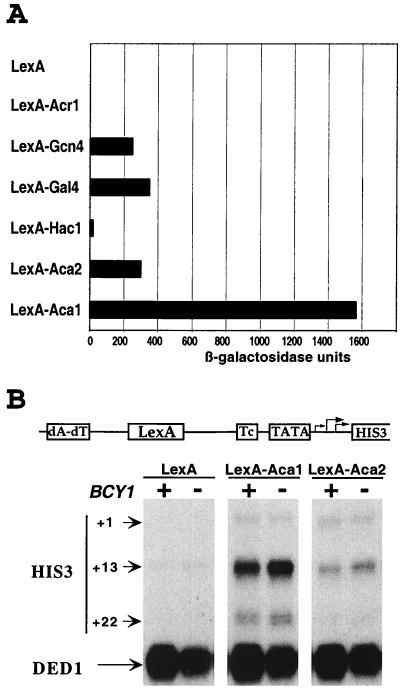
As an initial test to determine whether Aca1 and Aca2 are transcriptional activators, we fused their full-length coding sequences to the LexA DNA-binding domain. When assayed on a GAL1 promoter derivative with four LexA operators, activation by LexA-Aca2 occurs at a level comparable to that achieved by LexA-Gcn4 or LexA-Gal4, whereas activation by LexA-Aca1 is approximately fivefold more efficient (Fig. 3A). As expected from the fact that LexA-Acr1 functions as a repressor (26), no activation was observed in strains containing LexA-Acr1. LexA-Aca1 and LexA-Aca2 also stimulate transcription from a his3 promoter derivative with LexA sites in a manner that is unaffected by a bcy1 mutation and hence high levels of PKA (Fig. 3B). Thus, Aca1 and Aca2 contain functional activation domains.
FIG. 3.
Aca1 and Aca2 contain transcriptional activation domains. (A) β-Galactosidase activities (average from six independent transformants; values accurate to ±15%) of cells carrying the indicated LexA fusion proteins and a lacZ reporter with four LexA operators upstream of the GAL1 TATA element. (B) RNAs from strain XY and its bcy1 derivatives containing the indicated LexA fusion protein were analyzed for levels of his3 (+1, +13, and +22 transcripts) and ded1 (internal control) RNAs by quantitative S1 analysis.
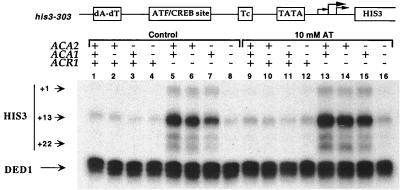
Using a modified his3 promoter (his3-303) in which the AP-1 site is converted to an ATF/CREB site, we previously showed that ATF/CREB sites can stimulate transcription in a manner repressed by Acr1 (65). To address whether Aca1 and/or Aca2 are responsible for ATF/CREB site-dependent transcription from the his3-303 promoter, we analyzed his3 mRNA levels in aca1, aca2, or double-mutant strains. When Acr1 is present, his3 transcription occurs at a low level (Fig. 4, lanes 1 to 4) that appears to be independent of Aca1 and Aca2. In the absence of Acr1, his3 transcription occurs a high level that is essentially unaffected by loss of Aca1 and is only slightly reduced in the absence of Aca2 (lanes 5 to 7). However, the loss of both Aca1 and Aca2 reduced his3 transcription in the acr1 deletion strain to the background level (lane 8), demonstrating that Aca1 and Aca2 are the activators responsible for transcription through the ATF/CREB site. Both proteins contribute to ATF/CREB site-dependent activation, with Aca2 appearing to be more important.
FIG. 4.
Aca1 and Aca2 activate transcription through ATF/CREB sites. RNAs from KY898 derivatives (contain the his3-303 promoter; functional elements indicated) with the indicated genotypes were analyzed for levels of his3 (+1, +13, and +22 transcripts) and ded1 (internal control) RNAs by quantitative S1 analysis. Strains were grown in minimal medium lacking histidine in the absence (lanes 1 to 8) or presence (lanes 9 to 16) of 10 mM AT.
Gcn4 does not activate transcription from ATF/CREB sites in vivo.
In vitro, the AP-1 factor Gcn4 binds with comparable affinity to its natural target sites and the consensus ATF/CREB site (50). It has been suggested that Gcn4 might not activate transcription through ATF/CREB sites (56), but this study was complicated by the presence of Aca1 and Aca2. Treatment of the strains described above with aminotriazole (AT), which induces Gcn4-dependent activation, yields results similar to those obtained under normal growth conditions (Fig. 4, lanes 9 to 16). In particular, AT does not induce his3 transcription in the acr1 aca1 aca2 strain, conditions in which there should be no other ATF/CREB proteins to compete for binding the ATF/CREB site. As the Gcn4 activation domain functions when fused to heterologous DNA-binding domains (24, 54), it is likely that the transcriptional defect reflects the inability of Gcn4 to bind ATF/CREB sites in vivo. This striking discrepancy between in vitro and in vivo activity might involve some feature of chromatin structure (e.g., rotational positioning of the target site on nucleosomes) or the interaction of proteins such as Mbf1 that increase Gcn4-dependent binding in vitro and transcription in vivo (58).
Physiological functions of Aca1 and Aca2.
The ATF/CREB family is not essential for cell growth, because strains lacking any combination of Aca1, Aca2, Acr1, Hac1, and Gcn4 are viable. However, Aca2 is important for a variety of physiological functions. Strains lacking Aca2 have no apparent defects in mating, sporulation, or cell wall function (assayed by flocculence and sensitivity to calcofluor white), but their growth is slightly reduced in standard rich and minimal media and severely affected at 15°C. In addition, aca2 deletion strains are sensitive to a variety of unrelated drugs such as cycloheximide, hygromycin B, formamide, rapamycin, and oligomycin (Table 1), although they behave normally in response to staurosporine, canavanine, tunicamycin, and 5-fluoro-orotic acid (data not shown). Finally, aca2 strains grow extremely poorly or not at all on a variety of alternative carbon sources (Table 2), whereas they grow normally on medium containing poor nitrogen sources (1% proline or glutamate).
TABLE 1.
Temperature and drug sensitivities of wild-type and mutant strains
| Strain or genotype | Growth
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| YPD
|
Cycloheximide, 0.4 μg/μl | Hygromycin B, 25 μg/μl | Caffeine, 7.5 mM | Formamide, 3% | Rapamycin, 5 μg/μl | Oligomycin, 1.5 μg/μl | |||
| 16°C | 30°C | 37°C | |||||||
| KY898 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Δaca1 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Δaca2 | − | + | + | − | ± | ++ | − | + | ± |
| Δaca1 Δaca2 | − | + | + | − | ± | ++ | − | + | ± |
| Δacr1 | ++ | ++ | ++ | ++ | ++ | − | + | ++ | ++ |
| Δacr1 Δaca1 | ++ | ++ | ++ | ++ | ++ | + | + | ++ | ++ |
| Δacr1 Δaca2 | − | + | + | − | ± | + | − | + | ± |
| Δacr1 Δaca1 Δaca2 | − | + | + | − | ++ | ++ | − | + | ± |
| 2μm-Aca1 in Δaca2 | ++ | ++ | NDa | ND | ND | ND | ND | ND | ND |
ND, not determined.
TABLE 2.
Effects of carbon source on growth of wild-type and mutant strains
| Strain or genotype | Growth
|
||||||
|---|---|---|---|---|---|---|---|
| Glucose, 2% | Glycerol, 3% | Ethanol, 3% | Maltose, 2% | Galactose, 2% | Sucinate + 2-diacylglycerol 2% + 0.2 mg/ml | Raffinose, 2% | |
| KY898 | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Δaca1 | ++ | +++ | ++ | ++ | ++ | ++ | ++ |
| Δaca2 | + | − | − | − | ± | − | ± |
| Δaca1 Δaca2 | + | − | − | − | ± | − | ± |
| Δacr1 | ++ | +++ | ++ | ++ | ++ | +++ | ++ |
| Δacr1 Δaca1 | ++ | +++ | ++ | ++ | ++ | +++ | ++ |
| Δacr1 Δaca2 | + | − | − | − | ± | − | ± |
| Δacr1 Δaca1 Δaca2 | + | ± | ± | ± | + | ± | + |
| 2μm-Aca1 in Δaca2 | ++ | + | + | ++ | + | ++ | NDa |
ND, not determined.
In contrast, Aca1 plays a minor physiological role. Loss of Aca1 does not cause or exacerbate any of the phenotypes associated with the loss of Aca2. However, overexpression of Aca1 can suppress the cold sensitivity and poor growth in nonoptimal carbon sources caused by an aca2 deletion. This suggests that the Aca1 proteins are functionally related and that, in many respects, Aca1 is a less active or less abundant version of Aca2. Curiously, loss of Aca1 results in increased growth of an acr1 aca2 double-mutant strain on all nonoptimal carbon sources tested (Table 2), indicating that Aca1 and Aca2 functions do not completely overlap.
Consistent with the result that Acr1 and the Aca proteins have opposing transcriptional effects at the his3-303 promoter, the caffeine sensitivity conferred by an acr1 mutation is partially suppressed by either an aca1 or aca2 mutation and completely suppressed by the combination of aca1 and aca2 (Table 1). However, an acr1 mutation does not suppress any of the phenotypes conferred by the aca2 deletion. Conversely, overexpression of Acr1 does not cause or exacerbate any of the phenotypes associated with aca2 deletions (data not shown). These results suggest that Acr1 and Aca2 affect distinct biological functions and target genes.
In contrast to the ATF/CREB activators in the fission yeast S. pombe (51, 67), Aca1 and Aca2 do not seem to be involved in the response to osmotic or oxidative stress. Specifically, the various aca mutant strains are comparable to the wild-type strain when grown in the presence of hydrogen peroxide, NaCl, KCl, LiCl, or sorbitol. Aca1 and Aca2 are functionally distinct from Hac1. Unlike hac1 strains, aca1 or aca2 strains are insensitive to tunicamycin (an inducer of the unfolded protein response), capable of growing in medium without inositol, and unable to activate UPRE-dependent transcription of KAR2 (Table 1 and data not shown).
Aca2-dependent transcription is modestly regulated by carbon source but not by PKA.
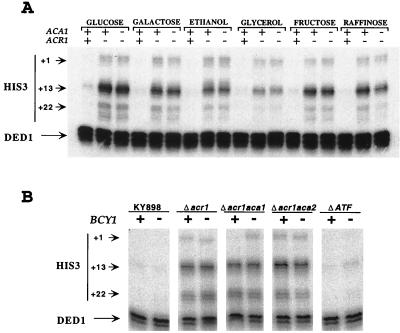
Because aca2 mutant strains grow poorly in nonoptimal carbon sources, we examined whether ATF/CREB site-dependent transcription is regulated by carbon source (Fig. 5A). In the wild-type strain, levels of transcription from the his3-303 promoter are comparably low in all media tested, suggesting that repression by Acr1 is unaffected by carbon source. However, in the acr1 deletion strain, transcription from the his3-303 promoter is reduced when cells are grown in glycerol (3.5-fold), ethanol (2-fold), and galactose (1.5-fold) but not in fructose or raffinose. Indistinguishable results are observed in the acr1 aca1 double-mutant strain, indicating that Aca2-dependent transcription is modestly regulated by carbon source. These results also suggest that Aca1-dependent transcription is not significantly regulated by carbon source, but this cannot be examined directly because the growth of aca2 strains is severely impaired in nonoptimal carbon sources. The modest reduction in Aca2-dependent transcription in certain nonoptimal carbon sources is unexpected given the importance of Aca2 for growth in these conditions. While growth in nonoptimal carbon sources could conceivably result in Aca2 being a more efficient activator or repressor of the relevant natural target genes, we suspect that the reduction in Aca2-dependent transcription is simply too modest to have an appreciable effect on the growth phenotype. In this regard, diploids heterozygous for ACA2 grow normally in nonoptimal carbon sources.
FIG. 5.
Aca2-dependent transcription is regulated by carbon source but not PKA. (A) RNAs from KY898 derivatives with the indicated genotypes were grown in various carbon sources and analyzed for levels of his3 (+1, +13, and +22 transcripts) and ded1 (internal control) RNAs by quantitative S1 analysis. (B) Effect of bcy1 mutation, which causes high levels of PKA, on transcription from the his3-303 promoter.
CREB proteins in various species activate transcription in a manner dependent on phosphorylation by PKA (41). However, in strains lacking various combinations of ATF/CREB proteins, transcription from the his3-303 promoter is essentially unaffected by loss of Bcy1, which causes very high PKA levels (Fig. 5B). This observation suggests that Aca1 and Aca2, unlike CREB, are not activated by cyclic AMP and PKA.
Transcriptional activation through ATF/CREB sites in response to high salt is independent of Aca1 and Aca2.
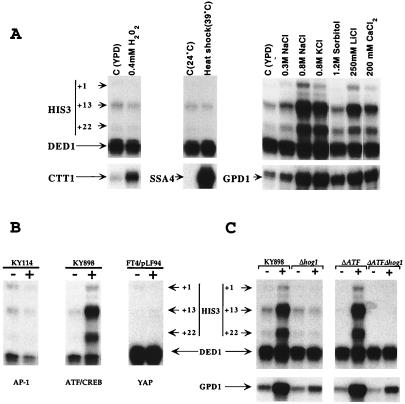
ATF/CREB activators in S. pombe and mammalian cells are targets of stress-inducible MAP kinases and play an important role in the response to several kind of stress (16, 35, 51, 63, 67). Although aca1 and aca2 mutant strains grow well in response to all stress agents tested, we directly examined ATF/CREB site-dependent transcription in response to stress (Fig. 6A). Transcription from the his3-303 promoter is unaffected by heat shock and oxygen stress (0.4 mM hydrogen peroxide), but it is strongly induced by high salt (15- to 20-fold in 0.8 M NaCl or KCl). Transcription from this promoter is also significantly induced by 250 mM LiCl (10-fold) or 200 mM CaCl2 (5-fold) but is only slightly induced by 0.3 M NaCl or 1.2 M sorbitol (2-fold). This pattern of induction resembles that of the osmosensing HOG signal transduction pathway kinase (2, 36), although it is unclear if the response of the his3-303 promoter is related to ionic strength, osmolarity, or the distinct effects of particular ions. For this reason, we will use the term “salt induction” to describe the behavior of the his3-303 promoter.
FIG. 6.
ATF/CREB site-dependent transcription is induced by salt in a Hog1-dependent manner. (A) RNAs from KY898 cells (contains ATF/CREB site) grown in YPD medium that were subjected to the indicated compounds for 1 h. (B) RNAs from KY898, KY114 (contains AP-1 site of the natural his3 promoter), and FT4/pLF94 (contains the optimal Yap-binding site) cells that were (+) or were not (−) treated for 1 h with 0.8 M NaCl. (C) RNAs from KY898 and the acr1 aca1 aca2 (ΔATF) derivative as well as their isogenic hog1 deletion derivatives were (+) or were not (−) treated for 1 h with 0.8 M NaCl. In all cases, levels of his3 (+1, +13, and +22 transcripts) and ded1 (internal control) RNAs were analyzed by quantitative S1 analysis.
Salt induction depends on the ATF/CREB site in the his3-303 promoter, because comparable promoters containing the natural AP-1 site or an optimal Yap site (8) are uninducible by NaCl (Fig. 6B), and ATF/CREB sites upstream of the cyc1 TATA element are sufficient to confer salt regulation of a lacZ reporter construct (data not shown). Induction of his3-303 transcription is detectable after 5 min of treatment with 0.8 M NaCl (data not shown) and is completely eliminated in a hog1 deletion strain (Fig. 6C), indicating the involvement of the HOG pathway and the Hog1 MAP kinase (2, 36). The complete Hog1 dependence of the his3-303 promoter on high salt differs from the situation with native genes (e.g., GPD1) whose salt induction is only partially Hog1 dependent. Salt induction of ATF/CREB site-dependent transcription via the HOG pathway has been observed previously, and it has been suggested that it reflects inactivation of Acr1 in response to osmotic stress (46).
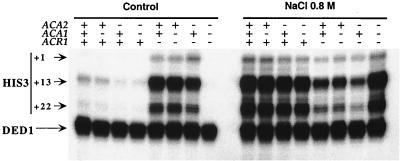
Whether or not high salt inactivates Acr1, there must be an activator(s) that binds the ATF/CREB site to account for the high level of transcription from the his3-303 promoter under these conditions. Surprisingly, Aca1 and Aca2 are not required for activation in response to high salt, because the aca1 aca2 acr1 triple-mutant strain is indistinguishable from the wild-type strain for transcription from the his3-303 promoter (Fig. 7). Moreover, salt induction of the his3-303 promoter in the triple-mutant strain depends on Hog1 (Fig. 6C). Thus, there must be a salt-regulated activator(s) that stimulates transcription through an optimal ATF/CREB site but is not a member of the ATF/CREB family. Analysis of mutant strains indicates that Hac1 and Gcn4 are not responsible for salt regulation of the his3-303 promoter, and Gcn4- and Hac1-regulated genes are not activated in response to salt (data not shown). Similarly, transcription dependent on the optimal Yap-binding site is unaffected by salt stress (Fig. 6B), suggesting that the eight Yap proteins are not involved. Although overexpression of Yap4 (Hal6) or Yap6 (Hal7) causes increased resistance to high salt (40), a strain lacking Yap4, Yap6, and all ATF/CREB proteins is fully capable of induction of the his3-303 promoter in response to 0.8 M NaCl (data not shown). Finally, the his3-303 promoter is fully salt inducible in strains lacking all ATF/CREB proteins as well as Met28 or Hot1, a nuclear protein involved in osmotic stress-inducible regulation (48).
FIG. 7.
Aca1 and Aca2 are not involved in the salt response of the his3-303 promoter. RNAs from KY898 derivatives with the indicated genotypes were or were not treated for 1 h with 0.8 M NaCl and analyzed for levels of his3 (+1, +13, and +22 transcripts) and ded1 (internal control) RNAs by quantitative S1 analysis.
Curiously, acr1 mutants actually have threefold-lower levels of transcription (Fig. 7), indicating that Acr1 contributes positively to transcription under these conditions. Decreased transcription in the acr1 mutant strain is reversed when both Aca1 and Aca2 are also eliminated, suggesting that the ATF/CREB activators function negatively. The simplest explanation for these observations is that the ATF/CREB proteins regulate the transcription of an inhibitor of the salt-regulated activator(s) in a manner similar to the his3-303 promoter.
The ATF/CREB family regulates GRE2 transcription but is not responsible for salt regulation of native yeast promoters containing ATF/CREB sites.
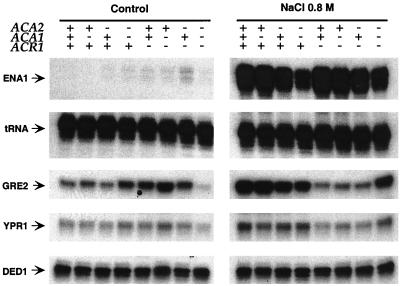
We also examined whether the ATF/CREB proteins affect transcription of ENA1, YPR1, and GRE2 (Fig. 8), genes that are induced by osmotic stress (13, 38, 45) and that contain ATF/CREB sites in their promoters. Regulation of ENA1 by high salt is mediated through the ATF/CREB site in the promoter (46), whereas this has not been demonstrated for GRE2 and YPR1. In contrast to a previous study using an ENA1-LacZ reporter (46), the very low level of ENA1 expression in YPD medium is minimally affected by loss of Acr1 function. Similarly, YPR1 transcription is essentially unaffected by the ATF/CREB proteins under these growth conditions. However, GRE2 transcription is increased approximately threefold in an acr1 deletion strain and is virtually eliminated in the acr1 aca1 aca2 triple-mutant strain. The transcriptional profile of GRE2 resembles (but is not identical to) that of the his3-303 promoter, indicating that GRE2 is a physiological target of Acr1, Aca1, and Aca2.
FIG. 8.
ATF/CREB proteins are not significantly involved in salt regulation of native yeast promoters containing ATF/CREB sites. RNAs from KY898 derivatives with the indicated genotypes were or were not treated for 1 h with 0.8 M NaCl and analyzed for levels of the indicated RNAs by quantitative S1 analysis.
As observed with the artificial his3-303 promoter, transcriptional induction of ENA1 (approximately 100-fold), GRE2 (6-fold), and YPR1 (2-fold) in response to salt stress is observed in strains lacking Acr1, Aca1, and Aca2 (Fig. 8). Furthermore, GRE2 and YPR1 resemble the his3-303 promoter in that osmotic induction is reduced in strains lacking Acr1 except in the situation where Aca1 and Aca2 are also absent. In the case of ENA1, transcription in 0.8 M NaCl is reduced approximately 1.5-fold in aca2 strains, suggesting that Aca2 makes a minor contribution to expression of this gene. Thus, salt regulation of ENA1, and perhaps GRE2 and YPR1, is mediated primarily by an activator(s) that binds ATF/CREB sites but is not a member of the ATF/CREB family.
Acr1 represses through the Cyc8-Tup1 corepressor complex.
Several lines of evidence suggest that Acr1 does not repress transcription solely by competing with Aca1 and Aca2 for ATF/CREB sites. First, Acr1 represses the his3-303 promoter below the level observed in the corresponding promoter lacking the ATF/CREB site (50). Second, deletion of the C-terminal 15 residues of Acr1 abolishes repression in vivo but does not affect ATF/CREB binding in vitro (65). Third, Acr1 represses transcription even when tethered upstream of a heterologous promoter via the LexA DNA-binding domain (26). These observations suggest that Acr1 may repress transcription by recruiting a corepressor, although repression by Acr1 does not involve the Sin3-Rpd3 histone deacetylase corepressor complex (26).
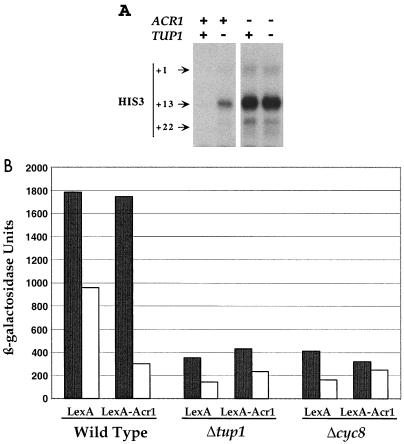
The Cyc8-Tup1 corepressor complex is required for repression by a variety of pathway-specific DNA-binding proteins such as α2, Mig1, and Rox1 (6, 28, 31, 60, 61, 62). To examine whether Acr1-dependent repression requires Cyc8-Tup1, we examined transcription from the his3-303 promoter in a tup1 deletion strain. As shown in Fig. 9A, his3 transcription is significantly increased in a tup1 strain, although the level is approximately threefold less than observed in the isogenic acr1 strain. Furthermore, repression of a heterologous promoter by LexA-Acr1 is abolished by cyc8 and tup1 strains (Fig. 9B). These observations suggest that Acr1-dependent repression is mediated by the Cyc8-Tup1 corepressor complex. However, the difference in transcription from the his3-303 promoter in tup1 and acr1 strains suggests that part of the repression involves ATF/CREB site competition between Acr1 and the Aca proteins.
FIG. 9.
Acr1 repression is mediated by the Cyc8-Tup1 corepressor complex. (A) RNAs (40 μg) from KY898 derivatives with the indicated genotypes were analyzed for levels of his3 (+1, +13, and +22 transcripts) and ded1 (internal control) RNAs by quantitative S1 analysis. (B) β-Galactosidase activities (average from three to four independent transformants) of wild-type and indicated mutant strains expressing LexA or LexA-Acr1 and lacZ reporter plasmids that either lack (shaded boxes) or contain (open bars) four LexA operators upstream of the CYC1 promoter. Values are accurate to ±15%.
DISCUSSION
The ATF/CREB family of S. cerevisiae consists of two activators and a repressor.
ATF/CREB proteins are defined by three criteria: DNA-binding specificity, amino acid sequence of the basic region within the bZIP domain, and transcriptional regulation of promoters containing ATF/CREB sites. By these criteria, Aca1 and Aca2 and the previously described Acr1 (Sko1) represent the complete set of ATF/CREB proteins in S. cerevisiae. Although Hac1 can bind the consensus ATF/CREB site and was originally designated an ATF/CREB protein (44), we believe that Hac1 should not be classified as an ATF/CREB protein because its basic region is highly diverged from those of other members of the family. Moreover, unlike other ATF/CREB proteins, Hac1 efficiently binds an unrelated sequence (UPRE) through which it mediates the response to unfolded proteins, its major physiological role (4, 42). Within the S. cerevisiae ATF/CREB family, Aca1 and Aca2 are more closely related, as they share a conserved domain and can form a heterodimeric complex that binds ATF/CREB sites.
Aca1 and Aca2 can independently stimulate transcription through ATF/CREB sites, and each protein contains a functionally autonomous activation domain. Conversely, Acr1 represses transcription through ATF/CREB sites, and it presumably contains a repression domain at the extreme C terminus (65) that recruits the Cyc8-Tup1 complex to target promoters. Taken together, these observations suggest that Aca1 homodimers, Aca2 homodimers, and Aca1-Aca2 heterodimers function as activators, whereas the Acr1 homodimer functions as a repressor. However, it remains possible that these ATF/CREB proteins might associate with other proteins (either by heterodimerization through the leucine zipper or by some other interaction) and hence affect transcription in more complex ways.
Aca1 and Aca2 have different biological functions than ATF/CREB activators in other eukaryotic organisms.
ATF/CREB activator proteins in fission yeast and mammalian cells play an important role in mediating the response to activation of PKA and to a wide variety of environmental stresses (16, 51, 67). The molecular basis for the stress response is remarkably conserved in that these ATF/CREB proteins are directly phosphorylated by stress-responsive MAP kinases, whereupon they activate target genes containing ATF/CREB sites. In contrast, the ATF/CREB family in S. cerevisiae does not appear to mediate the responses to PKA or to stress. In response to thermal, osmotic, or oxidative stress, S. cerevisiae strains lacking any combination of ATF/CREB protein are essentially normal. Furthermore, transcriptional activity through ATF/CREB sites is not induced by PKA, heat shock, or oxidative stress, and the strong response to high salt is independent of Aca1 and Aca2. These observations are consistent with the idea that the biological functions of homologous transcriptional regulators can differ considerably between budding and fission yeasts (64). In this regard, the general stress response in S. cerevisiae is mediated primarily by the Msn2 and Msn4 activators (15, 39, 49), whose activity is negatively regulated by PKA. Thus, these results provide another example supporting the view that mammals and many other multicellular eukaryotes are more evolutionarily related to S. pombe than to S. cerevisiae.
Aca2 is important for carbon source utilization and regulation.
Although the ATF/CREB proteins do not appear to be involved in stress responses, Aca2 is important for a variety of physiological functions, suggesting that Aca2 affects distinct classes of target genes. Most interestingly, Aca2 is important for cells to grow on nonoptimal carbon sources. It is unclear whether the poor growth on nonoptimal carbon sources reflects a common function related to glucose repression or a set of analogous functions that are more specific to individual or particular types of carbon sources. Although aca1 mutant strains are phenotypically normal, Aca1 probably contributes to carbon source regulation, because overexpression of Aca1 suppresses the inability of aca2 mutant strains to grow on nonoptimal carbon sources.
Some growth defects of aca2 deletion strains are similar to those observed in strains lacking Snf1 kinase, which plays a key role in glucose repression and utilization of alternative carbon sources (3). This suggests the possibility that Aca2 and Snf1 act in a common pathway of glucose repression and that Aca2 might be a substrate or transcriptional regulator of Snf1. Poor growth in nonoptimal carbon sources is also observed in strains lacking Bcy1, the regulatory subunit of PKA (59), although our results do not suggest a connection between PKA and the ATF/CREB family.
A salt-inducible activator(s) that functions through ATF/CREB sites but is distinct from the ATF/CREB family and is not a bZIP protein.
The ATF/CREB site in the ENA1 promoter is a physiological target for Acr1, and it is responsible for salt regulation via the HOG pathway (46). In addition, repression by artificial recruitment of Acr1 (via the Gal4 DNA-binding domain) is alleviated by high salt, suggesting that Acr1 is functionally inactivated in response to salt stress (46). However, our results indicate that salt regulation through ATF/CREB sites is not simply due to inactivation of Acr1. First, ENA1 transcription is minimally affected by loss of Acr1, whereas it is dramatically stimulated by treatment with 0.8 M NaCl. Second, transcription of GRE2, YPR1, and the artificial his3-303 promoter under conditions of high salt is actually reduced in the acr1 deletion strain. Third, and most important, salt induction of the his3-303, ENA1, GRE2, and YPR1 promoters occurs in the acr1 aca1 aca2 strain, which lacks all members of the ATF/CREB family. In the case of the his3-303 promoter, transcriptional activation is completely dependent on the ATF/CREB site, thereby defining a novel, Hog1-dependent activator(s) that functions through an optimal ATF/CREB site in response to high salt. This novel activator(s) is likely to contribute to salt regulation of the native ENA1, GRE2, and YPR1 promoters, but this remains to be demonstrated. Although this salt-inducible activator(s) has yet to be identified, our results have essentially excluded all bZIP proteins and Hot1.
Evidence that the ATF/CREB activators and the repressor have related but distinct biological functions and target genes.
The simplest model for transcriptional regulation by the S. cerevisiae ATF/CREB family is that Acr1 opposes the action of the functionally redundant activators Aca1 and Aca2 at a common set of promoters. Acr1 inhibits transcription both by competing with Aca1 and Aca2 for ATF/CREB sites and by an active repression mechanism involving recruitment of the Cyc8-Tup1 corepressor. This model accounts for the activity of the artificial his3-303 and natural GRE2 promoters, and it can explain why the caffeine sensitivity conferred by an acr1 mutation is partially suppressed by either an aca1 or aca2 mutation and completely suppressed by the combination of aca1 and aca2. In this view, caffeine sensitivity is caused by high Aca1- and Aca2-dependent activation through ATF/CREB sites, and the natural target promoters responsible for this phenotype are functionally analogous to the his3-303 promoter. For promoters subject to this competition model, transcriptional regulation in response to a signal could occur by affecting the activities or levels of one or more of the ATF/CREB proteins.
However, several observations suggest that individual ATF/CREB proteins have distinct biological functions. First, many phenotypes of an aca2 strain are not suppressed by loss of Acr1, nor are they caused or enhanced by overexpression of Acr1. This suggests that Acr1 does not play a significant role in many biological processes affected by the ATF/CREB activators. Second, although Aca1 and Aca2 activate the his3-303 and GRE2 promoters to near comparable levels, aca1 mutations do not cause or enhance various aca2 phenotypes, indicating that Aca1 and Aca2 make unequal contributions to a variety of biological functions. This inequality could be explained by proposing that Aca1 is a weakened version of Aca2 and that the overall level of ATF/CREB activation differentially affects specific biological functions, but this suggestion does not account for why loss of Aca1 increases growth of an acr1 aca2 strain on nonoptimal carbon sources. Thus, despite the shared properties of Aca1 and Aca2, these ATF/CREB activators may not be functionally redundant in all respects.
The distinct biological functions of individual ATF/CREB proteins imply that these proteins have distinct promoter specificities and target genes. Although the basic regions of Aca1 and Aca2 are identical, the Acr1 basic region has a few differences, including one at a residue that makes base-specific contacts with the ATF/CREB site. Hence, differential recognition of native yeast promoters might be involved in the functional distinction between Acr1 and the ATF/CREB activators. In addition, differences in protein-protein interactions that mediate cooperative binding to promoters or modulate transcriptional activity are likely to contribute to promoter specificity. Finally, the various ATF/CREB proteins may be differentially affected by signal transduction pathways, thereby resulting in distinct transcriptional outputs depending on the physiological conditions.
ACKNOWLEDGMENTS
We thank Martijn Rep for enjoyable discussions and open lines of communication about the functions of Aca1 and Aca2, and we recognize that he independently came to many of the same conclusions as presented in this paper. We thank Haruo Saito for conversations about the HOG signal transduction pathway, Jutta Deckert, Joe Geisberg, and Haruo Saito for plasmids, and Jutta Deckert, Lisete Fernandes, and Marie Keaveney for useful advice throughout the course of the project.
This work was supported by a postdoctoral fellowship to M.A.G-G. from Conselleria de Educacio I Ciencia and from Ministerio de Educacion y Cultura and by research grants GM30186 and GM53720 to K.S. from the National Institutes of Health.
REFERENCES
- 1.Bartsch D, Casadio A, Karl K A, Serodio P, Kandel E R. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- 2.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 3.Celenza J L, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 4.Cox J S, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 5.Deckert J, Perini R, Balasubramanian B, Zitomer R S. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics. 1995;139:1149–1158. doi: 10.1093/genetics/139.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deckert J, Rodriguez-Torres A M, Simon J T, Zitomer R S. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6109–6117. doi: 10.1128/mcb.15.11.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engleberg D, Klein C, Martinetto H, Struhl K, Karin M. The UV response involving the Ras signalling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell. 1994;77:381–390. doi: 10.1016/0092-8674(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foulkes N S, Duval G, Sassone-Corsi P. Adaptive inducibility of CREB as transcriptional memory of circadian rhythms. Nature. 1996;381:83–85. doi: 10.1038/381083a0. [DOI] [PubMed] [Google Scholar]
- 10.Foulkes N S, Mellstrom B, Benusiglio E, Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature. 1992;335:80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- 11.Freeman K, Gwadz M, Shore D. Molecular and genetic analysis of the toxic effect of RAP1 overexpression in yeast. Genetics. 1995;141:1253–1262. doi: 10.1093/genetics/141.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaits F, Degols G, Shiozaki K, Russell P. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev. 1998;12:1464–1473. doi: 10.1101/gad.12.10.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garay-Arroyo A, Covarrubias A A. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast. 1999;15:879–892. doi: 10.1002/(SICI)1097-0061(199907)15:10A<879::AID-YEA428>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez G A, Yamamoto K K, Fischer W H, Karr D, Menzel P, Biggs W, Vale W W, Montminy M R. A cluster of phosphorylation sites on the cyclic-AMP regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- 15.Gorner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p i regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta S D, Campbell B, Derijard B, Davies R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 17.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hai T, Liu F, Allegretto E A, Karin M, Green M R. A family of immunologically related transcription factors that includes multiple forms of ATF and AP-1. Genes Dev. 1988;2:1216–1226. doi: 10.1101/gad.2.10.1216. [DOI] [PubMed] [Google Scholar]
- 19.Hai T, Liu F, Coukos W J, Green M R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 20.Hill D E, Hope I A, Macke J P, Struhl K. Saturation mutagenesis of the yeast HIS3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986;234:451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 21.Hoeffler J P, Meyer T E, Yun Y, Jameson J L, Haebner J F. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science. 1988;242:1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- 22.Hope I A, Struhl K. GCN4 protein, synthesized in vitro, binds to HIS3 regulatory sequences: implications for the general control of amino acid biosynthetic genes in yeast. Cell. 1985;43:177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- 23.Hope I A, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 24.Hope I A, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 27.Kamens J, Richardson P, Mosialos G, Brent R, Gilmore T. Oncogenic transformation by vRel requires an amino-terminal activation domain. Mol Cell Biol. 1990;10:2840–2847. doi: 10.1128/mcb.10.6.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Struhl K. Determinants of half-site spacing preferences that distinguish AP-1 and ATF/CREB bZIP domains. Nucleic Acids Res. 1995;23:2531–2537. doi: 10.1093/nar/23.13.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komachi K, Redd M J, Johnson A D. The WD repeats of Tup1 interact with the homeo domain protein a2. Genes Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- 32.Kon N, Krawchuk M D, Warren B G, Smith G R, Wahls W P. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kon N, Schroeder S C, Krawchuk M D, Wahls W P. Regulation of the Mts1-Mts2-dependent ade6-M26 meiotic recombination hot spot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol Cell Biol. 1998;18:7575–7583. doi: 10.1128/mcb.18.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F, Green M R. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1A protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 36.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 37.Maldonado R, Blendy J A, Tzavara E, Gass P, Roques B P, Hanoune J, Schutz G. Reduction of morphine abstinence in mice with a mutation in the gene encoding CREB. Science. 1996;273:657–659. doi: 10.1126/science.273.5275.657. [DOI] [PubMed] [Google Scholar]
- 38.Marquez J A, Serrano R. Multiple transduction pathways regulate the sodium-extrusion gene PMR1/ENA1 during the salt stress in yeast. FEBS Lett. 1996;382:89–92. doi: 10.1016/0014-5793(96)00157-3. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 40.Mendizabal I, Rios G, Mulet J M, Serrano R, deLarrinoa I F. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 1998;425:323–328. doi: 10.1016/s0014-5793(98)00249-x. [DOI] [PubMed] [Google Scholar]
- 41.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 42.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 43.Nehlin J O, Carlberg M, Ronne H. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 1992;20:5271–5278. doi: 10.1093/nar/20.20.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nojima H, Leem S H, Araki H, Sakai A, Nakashima N, Kanaoka Y, Ono Y. Hac1: a novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994;22:5279–5288. doi: 10.1093/nar/22.24.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norbeck J, Blomberg A. Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 M NaCl. Evidence for osmotic induction of glycerol dissimilation via the dihydroxyacetone pathway. J Biol Chem. 1997;272:5544–5554. doi: 10.1074/jbc.272.9.5544. [DOI] [PubMed] [Google Scholar]
- 46.Proft M, Serrano R. Repressors and upstream repressing sequences of the stress-regulation ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reimold A M, Grusby M J, Kosaras B, Fries J W, Mori R, Maniwa S, Clauss I M, Collins T, Sidman R L, Glimcher M J, Glimcher L H. Chondrodysplasia and neurological abnormalities in ATF-2 deficient mice. Nature. 1996;379:262–265. doi: 10.1038/379262a0. [DOI] [PubMed] [Google Scholar]
- 48.Rep M, Reiser V, Gartner U, Thevelein J M, Hohmann S, Ammerer G, Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sellers J W, Vincent A C, Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990;10:5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 52.Sidrauski C, Cox J S, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 53.Silva A J, Kogan J H, Frankland P W, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 54.Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator are functionally homologous. Cell. 1987;50:841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- 55.Struhl K. Reverse biochemistry: methods and applications for synthesizing yeast proteins in vitro. Methods Enzymol. 1991;194:520–535. doi: 10.1016/0076-6879(91)94039-f. [DOI] [PubMed] [Google Scholar]
- 56.Suckow M, Hollenberg C P. The activation specificities of wild-type and mutant Gcn4p in vivo can be different from the DNA binding specificities of the corresponding bZIP peptides in vitro. J Mol Biol. 1998;276:887–902. doi: 10.1006/jmbi.1997.1565. [DOI] [PubMed] [Google Scholar]
- 57.Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takemaru K, Harashima S, Ueda H, Hirose S. Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol Cell Biol. 1998;18:4971–4976. doi: 10.1128/mcb.18.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toda T, Cameron S, Sass P, Zoller M, Scott J D, McMullen B, Hurwitz M, Krebs E G, Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 62.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 co-repressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 63.van Dam H, Wilhelm D, Herr I, Steffan A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to meiate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Heeckeren W J, Dorris D R, Struhl K. The mating-type proteins of fission yeast induce meiosis by directly activating mei3 transcription. Mol Cell Biol. 1998;18:7317–7326. doi: 10.1128/mcb.18.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincent A C, Struhl K. ACR1, a yeast ATF/CREB repressor. Mol Cell Biol. 1992;12:5394–5405. doi: 10.1128/mcb.12.12.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe Y, Yamamoto M. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol. 1996;16:704–711. doi: 10.1128/mcb.16.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilkinson M G, Samuels M, Takeda T, Toone W M, Shieh J-C, Toda T, Millar J B A, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]