Summary
Sexual selection results in sex-specific characters like the conspicuously pigmented extension of the ventral tip of the caudal fin - the “sword” - in males of several species of Xiphophorus fishes. To uncover the genetic architecture underlying sword formation and to identify genes that are associated with its development, we characterized the sword transcriptional profile and combined it with genetic mapping approaches. Results showed that the male ornament of swordtails develops from a sexually non-dimorphic prepattern of transcription factors in the caudal fin. Among genes that constitute the exclusive sword transcriptome and are located in the genomic region associated with this trait we identify the potassium channel, Kcnh8, as a sword development gene. In addition to its neural function kcnh8 performs a known role in fin growth. These findings indicate that during evolution of swordtails a brain gene has been co-opted for an additional novel function in establishing a male ornament.
Keywords: Sexual selection, ornamental trait, bioelectric signaling, fin development, transcription factor pre-pattern, potassium channel, QTL-mapping, swordtail
eTOC Blurb Schartl et al.
Schartl et al. study the sexually selected ornament of male Xiphophorus fish. Combining expression profiling and genetic mapping they find the sword-like extension of the tail fin develops on a non-sex biased pre-pattern of transcription factors. A potassium channel specifically expressed in males is identified as main regulator of sword outgrowth.
Introduction
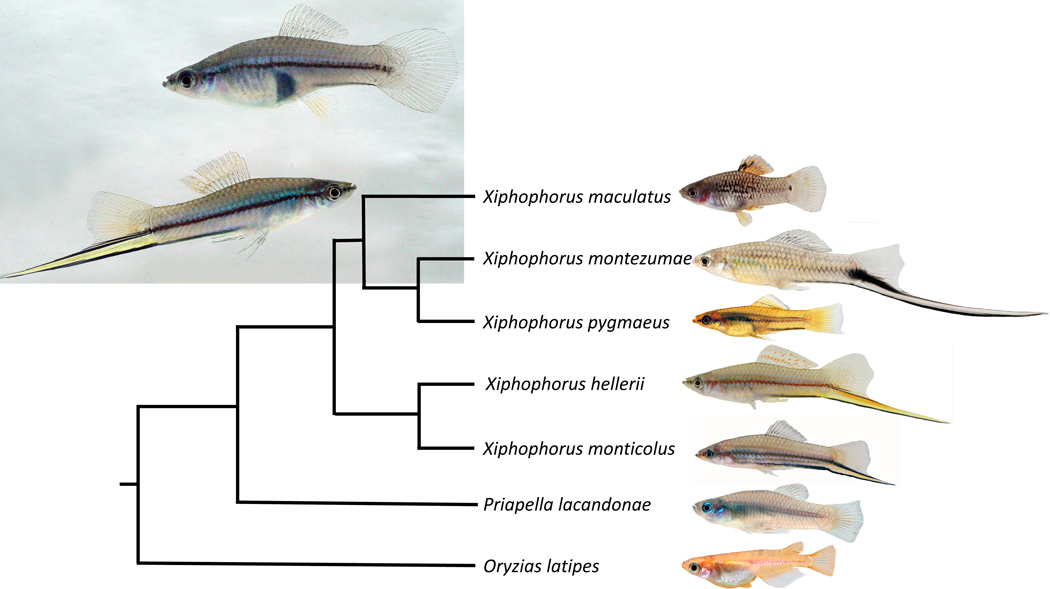
The evolution of male ornaments has intrigued biologists ever since Charles Darwin struggled to explain how exaggerated, expensive and likely deleterious structures such as the peacock’s tail or the horn of male unicorn beetles might have arisen by natural selection. Twelve years after the publication of his book “On the origin of species”, Darwin wrote his second most influential book not on the role of natural selection, but on sexual selection in evolution [1]. He described the “sword” of the green swordtail, Xiphophorus hellerii, as an example for his theory on sexual selection and postulated that selection by female choice can be a strong mechanism that could explain the evolution of traits that are clearly otherwise detrimental in terms of natural selection [1]. In several species of the genus Xiphophorus (Greek for dagger bearer) males carry a sword, a conspicuous extension of the ventral fin rays of the caudal fin that is brightly colored yellow, orange or red and is surrounded by a dark black margin (Figure 1). The sword develops at puberty and can be as long as the fish itself in some species. Its morphogenesis is instructed by the “sword organizer”, the proximal ventral region of caudal fin, where the fin rays connect to body musculature of the peduncle [2]. The sword is a male restricted trait, but female swordtails develop swords like males when treated with testosterone [3, 4]. This finding suggests that a potential sexual conflict has been solved by a strict androgen dependency for expression of the phenotype. Females of Xiphophorus hellerii and several other species preferentially associate with males carrying a longer sword over males with shorter swords, which is thought to result in a higher mating success of long-sworded males [5, 6]. This process exemplifies run-away Fisherian evolution for exaggerated male traits [7]. However, there are also trade-offs [8, 9], because swords attract not only females, but also predators [10], and escape from predators is more difficult because the sword reduces swimming performance [11]. Several species of the genus Xiphophorus, including the so-called platyfishes, do not have this sexually dimorphic character (Figure 1), even though, surprisingly, platyfish females nevertheless prefer heterospecific sworded males over their own swordless conspecifics [5]. This observation supported a major hypothesis in evolutionary ecology, namely that female preference may drive sexual selection by sensory exploitation since the bias in females was thought to be older than the sword itself [12, 13]. In contrast, however, molecular phylogenies showed that the sword is the ancestral state for the genus Xiphophorus [8, 14–16] and implied that derived swordless species had lost the male ornament secondarily, but retained the female preference for them. This phylogenetic inference fueled the discussion about which evolutionary forces drove the evolution and loss of this conspicuous trait [17–19].
Figure 1. Phylogenetic relationships of sworded and non-sworded Xiphophorus species.
The swordless Priapella lacandonae is the nearest (sister genus) and medaka, Oryzias latipes, a distant outgroup. Insert shows female (upper) and male (lower) of the green swordtail, Xiphophorus hellerii.
Females of different Xiphophorus species show differences in their preference for sword [5, 20]. Female preferences such as this are considered to potentially not only drive the evolution of male ornaments, but also to result in speciation [21–23]. In the genus Xiphophorus, the widespread propensity to prefer sworded males led to the formation of two hybrid species X. clemenciae [8, 19] and X. monticolus [16] where, due to the preference for swords females of non-sworded species hybridized with males of sworded species to bring about new, sworded hybrid species.
A huge body of literature on how both sexual and natural selection can lead to speciation exists [24, 25] but almost nothing is known about the genetic basis of these male ornaments [26, 27]. To identify the genes on which female preferences act on is an important task that is necessary to permit the testing of hypotheses regarding the roles of sexual selection at the molecular genetic level.
The swords of swordtails became a textbook example of a sexually selected trait, yet despite research efforts for more than two decades the molecular genetic basis of sword development remained unknown. So far, candidate gene approaches involving known genes of fish fin growth and development [28] [29] and suppression subtractive hybridization cloning [30] have not revealed the secret of the sword.
To identify the genetic basis for sword formation, we combined genome-wide expression analysis during sword development and regeneration with a genetic association study for sword length in a cross of a non-sworded species to a sworded species.
Results
To obtain a comprehensive list of protein coding genes that are involved in the formation of the sword, we compared expression levels using several RNA-seq datasets from the green swordtail, Xiphophorus hellerii (Figure 1). We reasoned that sword genes should be differentially expressed (i) during growth of the developing sword at puberty (Figure S1A) and (ii) during the course of sword regeneration (Figure S1B). Because immature fish and adult females also develop a sword indistinguishable from the male structure following treatment with androgens [3, 4] we generated (iii) an RNA-seq dataset from the sword of testosterone-treated adult females; and added (iv) our previous dataset from testosterone-induced swords in pre-pubertal juveniles [3]. Small biopsies from the dorsal and ventral fin margin during a timed series of growth and of regeneration and from the hormone-induced and naturally developed swords from 15–20 individuals were pooled and used for transcriptome sequencing. To exclude genes that are not involved in sword development but have a more general function during natural and hormone induced caudal fin growth or in regeneration, differential expression was deduced from comparison of the ventral compartment to the corresponding dorsal part of the caudal fin. The four datasets were overlapped to identify genes that are commonly regulated in all four processes of sword development (Figure S2). This process yielded a set of 329 differentially expressed genes (log2FC >=1) in all sword transcriptomes (51 down- and 278 upregulated, table S1).
We expected differentially expressed genes to be of two main categories: those primarily responsible for inducing the sword and those that execute the instruction process by actually building the components of the sword. The sword, like other parts of the caudal fin, consists of bony fin rays, skin, pigment cells, sensory neurons, blood vessels and mesenchyme. Amongst genes upregulated in sword vs control fin regions, four genes (xdh, tyr, myrip, asip) are obviously connected to sword pigmentation; several other upregulated genes are related to increased vascularization (agtr1, angptl5, neurexins) and fin-ray rigidity (collagens and collagen metabolizing enzymes, extracellular matrix, bone and cartilage proteins) that support the sword structure as an extremely long outgrowth of ventral fin rays. It is unclear whether these genes are also critical for the primary process of induction and development of the sword, but all are reasonably predicted to be involved in later differentiation processes. The sword transcriptome was also enriched for genes often expressed in neurons (pdyn, draxin, kcnh8, kcng2, kcns1, kcns22, chrna7, ncan, lypd6, gfra1) or in Ca2+ signaling (stc2, efcc1, fkbp9, −10, −11, −14, trpc1, anxa6) (table S1, Figure S2C).
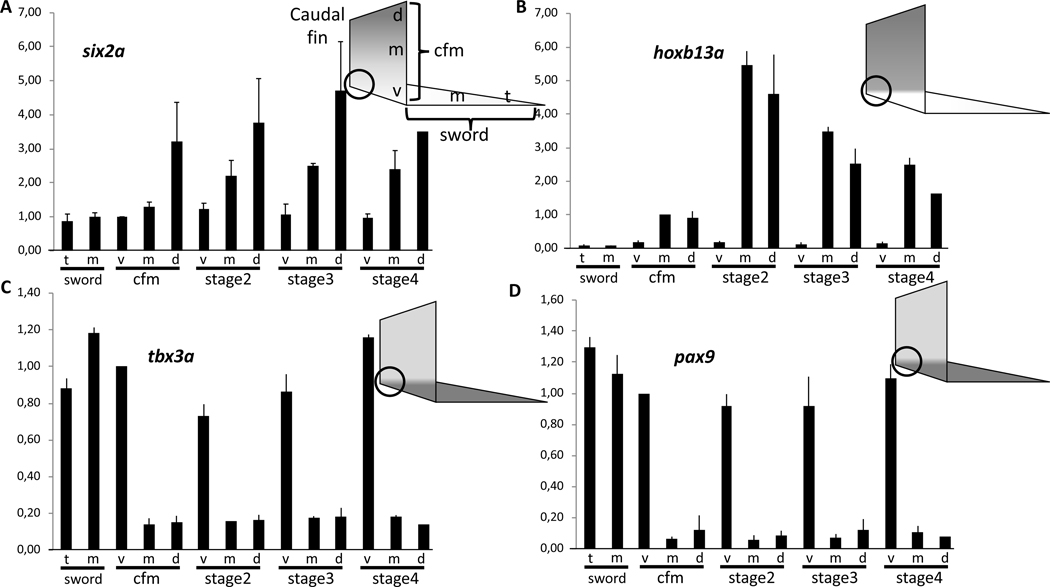
Intriguingly, several transcription factors were included in the differentially expressed genes list and provide strong candidates for having a critical function in regulating caudal fin development and consequently also sword formation. Homeobox protein six2a, which plays a role in chicken hindlimb development [31], forms a continuous dorsoventral expression gradient in the swordtail tail fin (Figure 2A, table S2), similar to several developmental transcriptional regulators in the establishment of the zebrafish pectoral fin anterior-posterior axis [32]. The dorsalizing factor zinc finger protein zic1, which is critical for the development of the homocercal fin shape in fish [33] is highly expressed in the dorsal compartment of the caudal fin, but expression is absent from the medial region and all sword transcriptomes (table S2). More strikingly, homeobox protein hoxb13a, which is the most caudally expressed hox gene in fish [34], has high expression in the non-sword regions of the X. hellerii caudal fin, but is not expressed in the sword or the sword-organizer (table S2). During tail fin regeneration, hoxb13a is upregulated in the median and dorsal region but not expressed in the outgrowth leading to the sword (Figure 2B). The t-box transcription factor tbx3a gene, which promotes formation of the mesoderm cell lineage [35] and is involved in vertebrate limb pattern formation [36], is weakly expressed in the non-sword regions of the tail fin, but abundant in the sword organizer region at the base of the fin, and in the sword during regeneration, natural sword development and hormone-induced sword (Figure 2C, table S2). The same expression pattern is displayed by paired box protein pax9, which in fish is a critical factor for development of the hypural plate supporting the peduncle [37], where the caudal fin is inserted (Figure 2D, table S2). Interestingly, leukocyte tyrosine kinase receptor (ltk), which normally is only lowly expressed without a pronounced spatial expression pattern in the caudal fin of X. hellerii males, builds up a local expression pattern in the sword producing blastema similar to that of hoxb13a during caudal fin regeneration and natural and hormone induced sword development (table S2).
Figure 2. Spatial expression pattern of transcription factor genes in the caudal fin and sword of male Xiphophorus hellerii.
Expression of six2a (A), hoxb13a (B), tbx3a (C) and pax9 (D) in the caudal fin margin (cfm) of the tail fin of adult Xiphophorus hellerii males, the median sector (m) and tip (t) of the sword and during sword regeneration (v, ventral, m, median, d, dorsal compartment). The vertical axis indicates fold change of expression normalized to the ventral cfm (six2a, tbx3a, pax9) or median cfm (hoxb13a). The circle indicates the position of the sword organizer. Data are represented as mean ± SEM. See also Figures S1, S2, S3, S4.
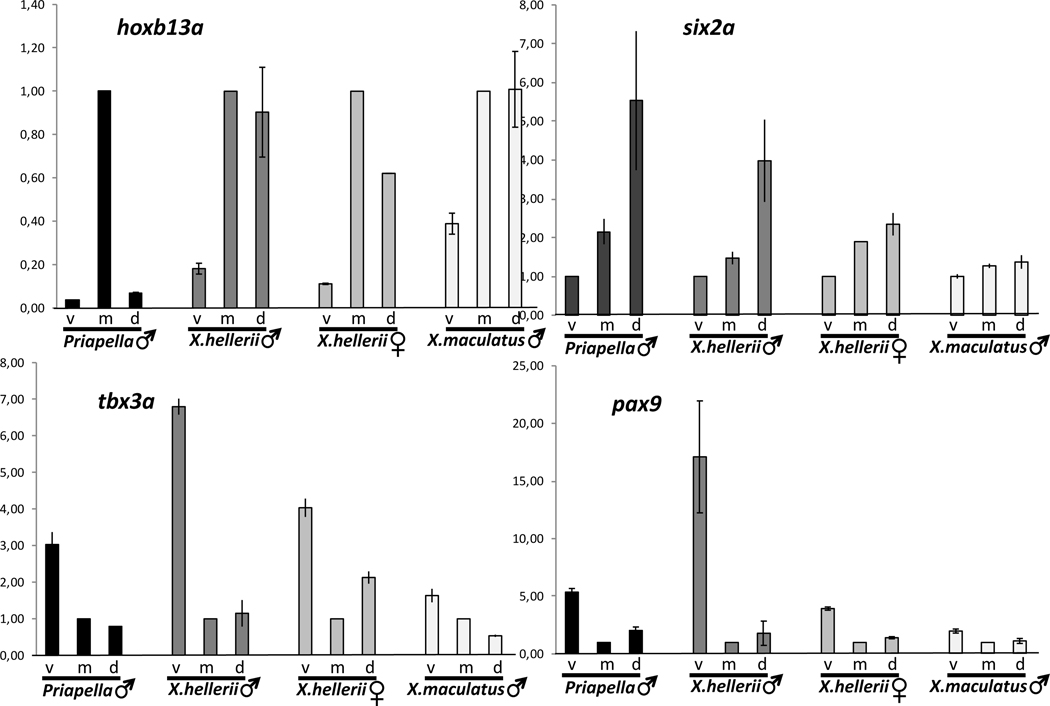
Males of two other swordtail species, X. montezumae and X. monticolus (Figure S3) showed the same expression gradients and temporal pattern during sword regeneration. Of note, analysis in X. montezumae, the species with the longest sword (sword index (sword length/standard body length) up to 1.6), revealed that the transcription factor expression pattern is immediately initiated in the blastema of the regenerating caudal fin and builds up to the levels of the caudal fin margin and sword during the first days of growth. The platyfish X. maculatus, a species that does not develop a sword, and the pygmy swordtail, X. pygmaeus, where males have only a tiny unpigmented ventral protrusion of the tail fin but no sword, display the transcription factor gene expression gradients in the caudal fin, but these gradients are much less pronounced and at lower detectable transcript levels (Figure 3, S4). Phylogenetic evidence suggested that these species have lost the sword secondarily [8, 14]. Apparently, the loss of the male ornamental trait is associated with a decay of this gene expression pattern.
Figure 3: Comparison of transcription factor expression patterns in different species.
Expression of transcription factor genes hoxb13a (A), six2a (B), tbx3a (C) and pax9 (D) in the caudal fin margin of the tail fin of adult males of Priapella lacandonae, Xiphophorus hellerii and X. maculatus, and X. hellerii females. (v, ventral, m, median, d, dorsal compartment). Vertical axis indicates fold change of expression normalized to cfm, m (A, C, D) or cfm, v(B). Data are represented as mean ± SEM. See also Figures S1, S2, S4.
The sword arose at the basis of the genus Xiphophorus [8, 14]. In the sword-less Priapella, the closest sister genus, the tail fin pattern on which the sword is built is already present to a large extent. The expression patterns of pax9, tbx3 and six2a are conserved, except hoxb13a is expressed only in the median compartment (Figure 3, S4). In the very distantly related medaka, Oryzias latipes, the tail fin spatial expression patterns of hoxb13 and pax9 are like in Xiphophorus, however, at much lower transcript levels. Expression of the medaka orthologs of tbx3 and six2a was not detected in the caudal fin (Figure S4).
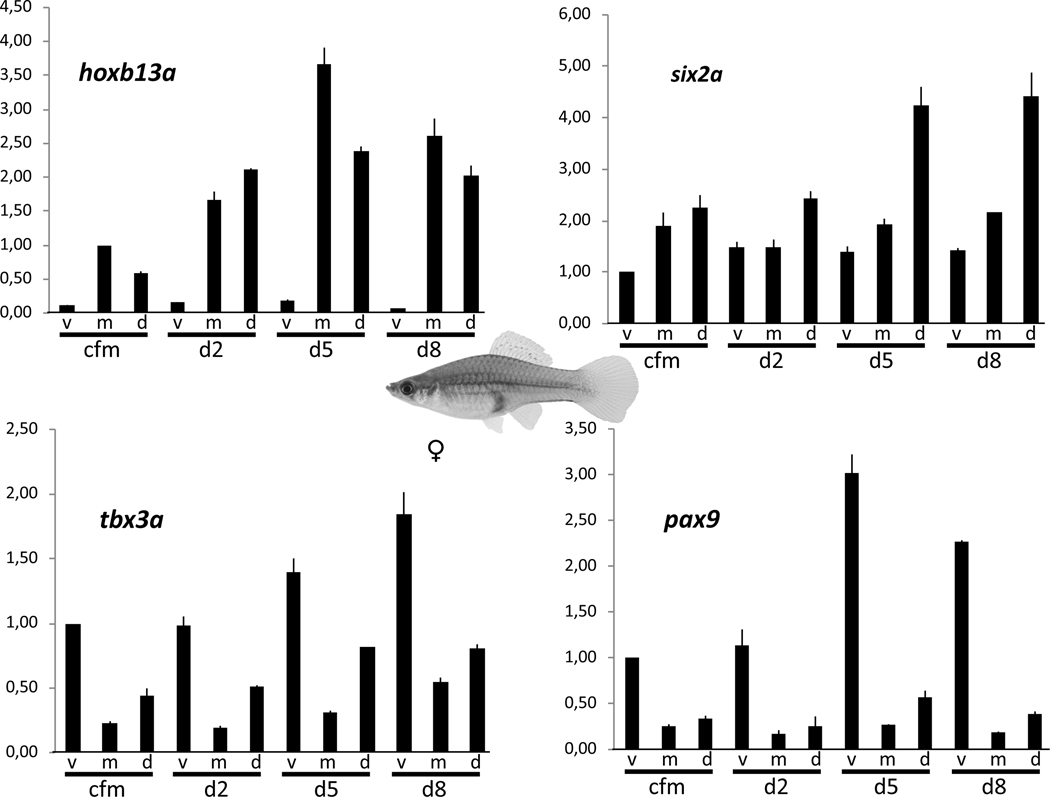
Importantly, the same expression profile of the five transcription factors described above for males of Xiphophorus was also observed in female swordtail caudal fins (Figure 4, table S1, S2), although at lower expression levels for six2a, tbx3a and pax9 (Figure 4, table S2). However, this finding indicates that a pre-pattern of transcription factors exists in the caudal fin of both sexes that provides in males the positional information for sword development, but it rules out these genes as candidates for sword induction.
Figure 4: Spatial expression pattern of transcription factor genes in the caudal fin of Xiphophorus hellerii females.
Expression of hoxb13a (A), six2a (B), tbx3a (C) and pax9 (D) in the caudal fin margin of the tail fin (cfm) and during tail fin regeneration (v, ventral, m, median, d, dorsal compartment). Vertical axis indicates fold change of expression normalized to cfm, v (six2a, tbx3a, pax9) or cfm, m (hoxb13a). Data are represented as mean ± SEM. See also Figure S1.
Reasoning that genes that are responsible for the sword would be expressed only in males, we thus generated transcriptomes from upper and lower terminal caudal fin compartments of females and used these to eliminate genes from candidate status in the sword transcriptome if they showed the same regulation in male and female caudal fin regeneration. Overlap with the DEGs of caudal fin regeneration of swordless platyfish males removed another 3 genes. This process still left us with 255 of the original 329 candidate genes (table S1). To further reduce the number of candidate sword genes we performed a genetic mapping approach.
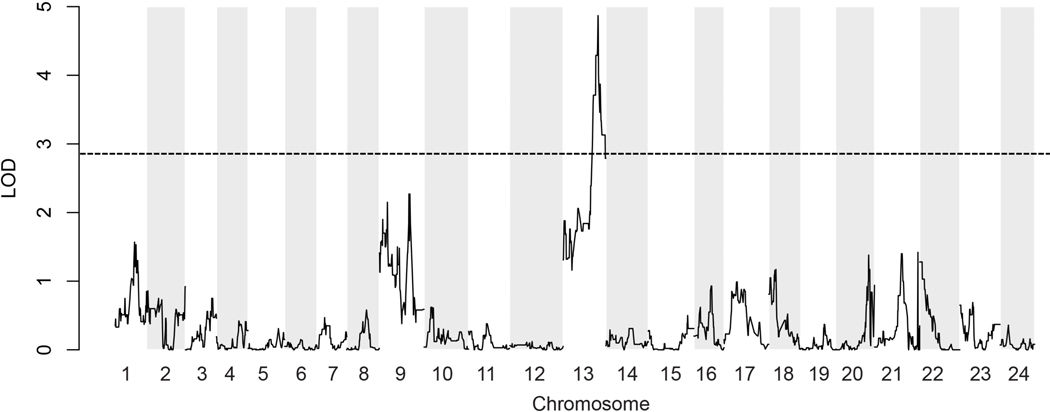
QTL mapping was performed using RAD-tags. Because crossing of a swordtail to a nearest outgroup species prior to evolution of this character (e.g. Priapella sp.) does not produce offspring, we used a congeneric species that has lost the sword. We choose the swordless platyfish, X. maculatus, as a most distantly related species belonging to the clade of platyfishes, while X. hellerii is a member of the clade of Southern swordtails. Both clades are estimated to have diverged 2–3 million years ago [14]. A backcross was generated between X. maculatus and the green swordtail X.hellerii using X.hellerii as the recurrent parent [38]. Mapping the sword-index of 85 backcross males against genetic polymorphisms in the reference swordtail genome (data S1) revealed significant association with a region on linkage group (LG) 13 (LOD score standard interval mapping em = 3.86, non-parametric np = 4.87, 2 LOD interval 3.6 Mb, 179 genes) (Figure 5). A region on LG 1 (LOD score em = 3.17, np = 1.57, 8.1 Mb, 306 genes) and LG 9 (LOD score em = 2.54, np =2.15, 2.7 Mb, 147 genes) failed to reach the significance level. Several minor peaks also appeared on LG’s 20 – 24. This result defines the sword as a highly polygenic trait, which is in accordance with the distribution of sword lengths in platyfish/swordtail hybrids [39].
Figure 5. Results of quantitative trait loci (QTL) mapping for sword length.
The highest QTL peak is located on chromosome 13 and two minor peaks on chromosomes 1 and 9. The plot depicts aligned RAD-tag marker positions on the Xiphophorus hellerii genome version 4.1 with non-parametric statistics. The broken horizontal line indicates the 5% genome-wide threshold for significance (LOD 2.86). Significance levels were determined by permutation testing. See also Data S1.
When the positions of sword-specific differentially expressed genes (table S1) were examined with respect to the QTL peaks in the 2.0 LOD interval, none of the genes involved in establishing the prepattern and none of the pigmentation, angiogenesis, or ECM genes that were differentially regulated during sword development were found to be encoded in any of the regions identified in the QTL analysis. Only three differentially expressed genes with log2FC >=1 mapped to the main QTL peak on chromosome 13. These are nkain1, fkbp9 and kcnh8.
The nkain1 gene codes for an uncharacterized sodium/potassium transporting ATPase-interacting protein. It is weakly upregulated in the sword transcriptomes but not in the sword organizer, where a gene expected to initiate sword development should be expressed.
The gene encoding the chaperone peptidyl-prolyl cis-trans isomerase Fkbp9 is 2- to 3-fold higher expressed in the developing sword than in control tissue and becomes upregulated in sword regeneration at stages 3–4 and testosterone-induced swords (table S2). Expression is not elevated in the sword organizer, which weakens its candidacy as a gene responsible for induction of sword development.
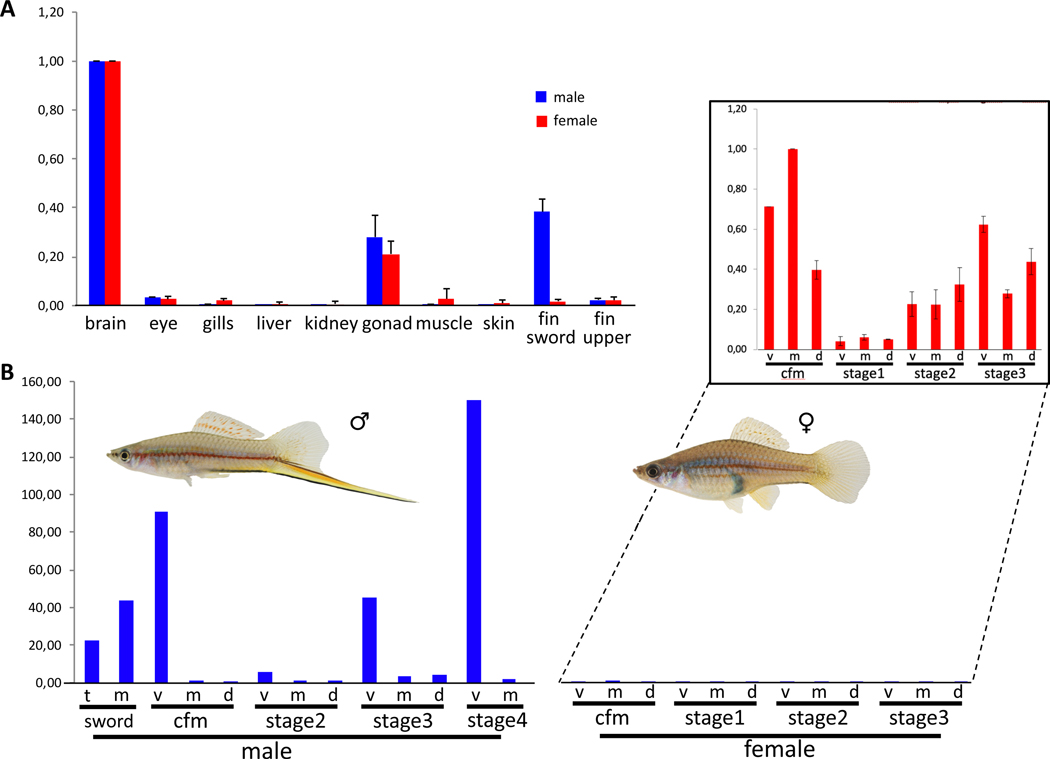
The other gene with overlapping candidate status from both gene expression and mapping studies is kcnh8. Kcnh8 is a potassium channel of the ether-à-go-go (EAG) type that is expressed abundantly in brain and at intermediate levels in ovary and testis (Figure 6A). Importantly, kcnh8 is strongly upregulated in the sword during normal development and following androgen treatments, in the sword organizer region, and in the fully developed sword, and becomes strongly upregulated during sword regeneration (Figure 6B, table S2). It is amongst the 0.3% most differentially expressed genes (>21,000 total). Transcripts of kcnh8 are almost absent from all other fin areas of males and kcnh8 is only expressed at background levels in female caudal fins (Figure 6B, table S2).
Figure 6. Expression of kcnh8 in adult males and females of Xiphophorus hellerii.
(A) Organ-specific expression profile in adult females and males. (B) Expression of kcnh8 in the caudal fin margin (cfm) of the tail fin of adult Xiphophorus hellerii males and females, the median sector (m) and tip (t) of the sword and during caudal fin regeneration (v, ventral, m, median, d, dorsal compartment). Insert: expression in females upscaled, note the difference scale for the Y-axis. Vertical axis indicates fold change of expression normalized to brain (A), cfm, m (B). Data are represented as mean ± SEM. See also Figures S1, S6, S7.
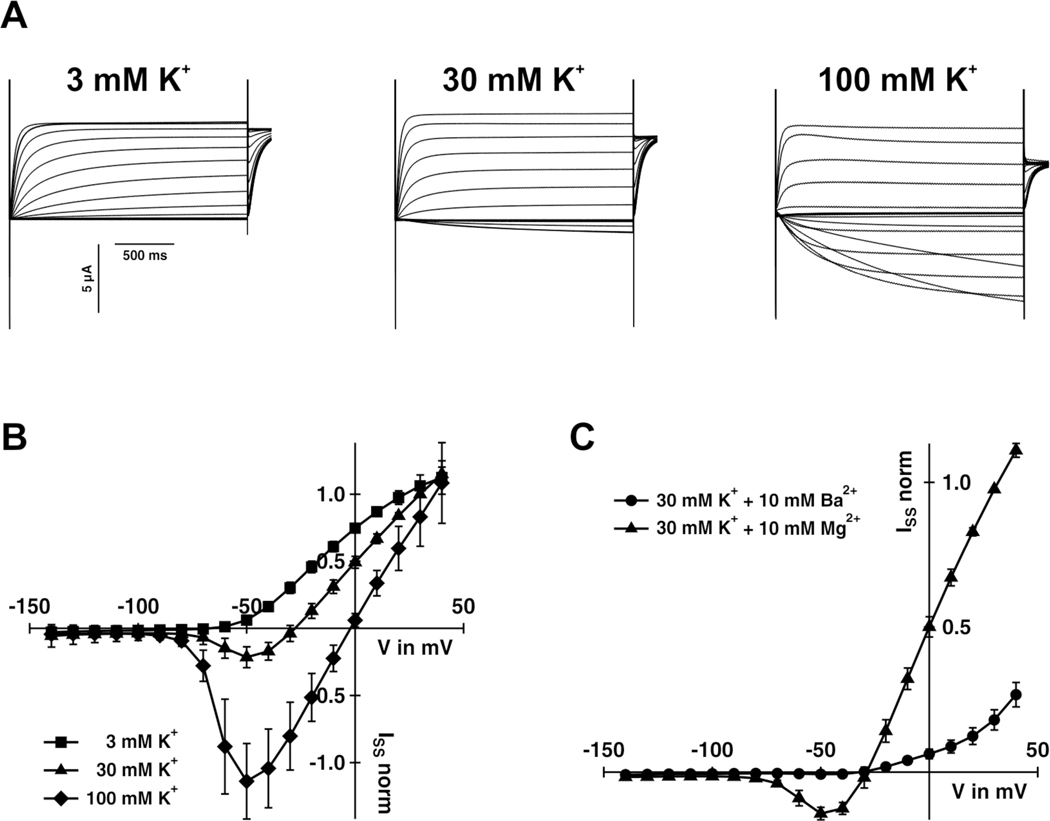
To test the function of the swordtail Kcnh8 protein, we expressed kcnh8 in the Xenopus oocyte system and performed two-electrode voltage clamp analyses. Results revealed that the protein has the hallmark characteristics of a fully functional voltage gated potassium channel member of the Kv12.1 family [40] in terms of voltage activation characteristics, time-dependent activation kinetics, potassium selectivity and inhibition by Ba2+ ions (Figure 7). Exposure to a potassium channel inhibitor during caudal fin regeneration resulted in a shorter sword (Figure S5).
Figure 7. Electrical features of Xiphophorus hellerii Kcnh8.
(A) Representative TEVC recordings of Kcnh8-expressing Xenopus oocytes at the indicated potassium concentrations. Test voltages ranged between +40 to −140 mV in 10 mV decrements. (B) Steady-state currents (ISS) extracted from recordings as shown in (A) of Kcnh8-expressing oocytes were plotted as a function of the applied membrane potential (mean of n ≥ 7 oocytes ± SD of ≥ 3 independent experiments). (C) Application of 10 mM BaCl2 in the presence of 30 mM KCl inhibited the KCNH8-mediated ISS (mean of n = 6 oocytes ± SD of ≥ 2 independent experiments). (B) and (C) ISS were normalized to the currents at +30 mV in standard bath medium (30 mM KCl). See also Figure S5.
We also found that X. montezumae, which has an even longer sword than X. hellerii, shows also the high expression of kcnh8 in the sword and during sword regeneration (Figure S6A). Interestingly, in X. monticolus, a species that develops a much shorter sword than X.hellerii, kcnh8 expression during sword regeneration is only weakly upregulated (Figure S6B). In X. pygmaeus, where males have only a tiny protrusion of the ventral fin rays, but no sword, kcnh8 expression is upregulated only to low extent (Figure S6C). In the swordless platyfish X. maculatus, no expression above background levels of kcnh8 was noted in the lower and upper compartment and during regeneration of the caudal fin (table S2).
Discussion
Sexually selected traits are present in many animals are a hallmark of sexual dimorphism. The evolutionary mechanism driving their origin, maintenance and role in speciation have been intensively studied, but still today little is known about the proximate causes, i.e. the genes encoding sexually selected traits and their function in development of the structure, aside from a few examples from Drosophila [41, 42]. The sword is a male-specific outgrowth of the lower margin of the caudal fin and we wanted to know what genes provoke its sex-specific elongation. The fins of fish are intricate three-dimensional structures composed of numerous cell types. Size, shape, pigmentation and other features of fins are generally highly fixed and specific for different species and certain ontogenetic stages. In many lineages of fish fins are sexually dimorphic traits [43]. In zebrafish it has been shown that pectoral fins have a regionalized gene expression pattern that creates gradients of transcription factors [32]. We conclude that also in the caudal fin of male swordtails a similar specific regionalized gene activity pattern provides positional information for development of the sword. The regional expression of the transcription factors Hoxb13a, Six2a, Tbx3a and Pax9 produces a prepattern in the tail fin that is connected to sword development since their expression s vanishes in species that have secondarily lost the sword. This pattern is established before the sword develops during puberty and its presence (although for some factors at lower levels) in adult females may allow the development of a sword after experimental androgen treatment or as a natural phenomenon in old post-reproductive females [44, 45]. In X. maculatus the decay of the prepattern appears to be a consequence of the loss of the sword in this species, while absence of kcn8 expression can be interpreted to be causal for the failure to develop the trait. It is interesting to note, that the pre-pattern is almost completely conserved in Priapella, a genus basal to Xiphophorus where females also show a preference for swords although not displayed by their conspecific males [46]. Obviously, establishment of the prepattern preceded the evolution of the sword as a preadaptation. It will be interesting to search for the conditions that maintain the prepattern in the primarily swordless genus, while it degenerates in Xiphophorus species that have secondarily lost the sword.
To identify those genes whose expression results in the development of the sword in males we reasoned that such genes should be differentially expressed in sword development spatially and between males and females and should be encoded in genomic regions that are linked to this trait. Our QTL analysis, consistent with earlier genetic findings [39], uncovered that several chromosomal regions contribute to the polygenic basis of the male structure. Consistently, our major locus on chromosome 13 fully overlaps a similar broader QTL that was obtained in an independent study for the character sword length in natural hybrids between two sister species from the Northern swordtail clade, the swordless species X. birchmanni and X. malinche, which develops a sword. [47]. That study forwarded three candidate “sword” genes: the transcription factor sp8a, the cytoskeletal motor protein dynein subunit dync1i1 and the signal protein precursor progranulin grn2. In our RNA-seq datasets of X. hellerii dync1i1 is not expressed in the caudal fin, including the sword, but is upregulated during fin regeneration in females and males. Although grn2 is upregulated during sword regeneration in the Northern swordtail, both grn2 and sp8 are downregulated in the X. hellerii sword transcriptomes (table S2). These divergent findings might be explained by the phylogenetic distance between X. hellerii and X. malinche, which diverged at least one to two million years ago[14].
We identified in the QTL region genes that appear to be involved in the development of the sword. Rather than being typical regulators of development and differentiation such as transcription factors or extracellular diffusible growth factors, the conjunction of genetic mapping and gene expression analyses converged on a channel protein gene, kcnh8, and a chaperone gene, fkpb9.
In zebrafish long fin mutants, mutations in several potassium channel genes, including kcnh2a, kcnk5b, kcnj13 and kcc4a cause various types of fin overgrowth [48–51]. In fighting fish, Betta splendens, kcnh8 mis-expression is associated with pectoral fin overgrowth (Wang et al. submitted). A hyperpolarizing mutation in kcnk5b causes the long fin phenotype in ornamental goldfish [52]. Mutations disrupting ion channels and ion-dependent signaling are extensively related to abnormal organ development and regeneration via bioelectrical regulation [53]. Bioelectricity has been proposed as a mechanism for fin patterning from the dermomytome [34]. Potassium channels of the Kcnh family have been implicated in cell proliferation by influencing membrane polarization and thus calcium signaling [54, 55]. Increased intracellular calcium levels activate osteoblasts and their precursors [56, 57], which build the fin rays of the overgrowing structures of the long-fin mutants and the Xiphophorus sword. Potassium channels can also play a role in cell cycle and proliferation control by mechanisms unrelated to ion channel permeability [55]. Despite this wide spectrum of biological functions of potassium channels besides the classical channel properties, their transcriptional regulation and biochemical interactions are not well understood.
Voltage gated channels of the EAG family are inhibited by intracellular calcium [58]. One function of Fkpb9 besides acting as a prolyl cis-trans isomerase is mediated through its calcium binding Ef-H domain [59]. In zebrafish tailfin growth, the calcium activated protein phosphatase calcineurin plays a predominant role and a mechanism was proposed for calcineurin to operate as a molecular switch between allometric and position associated isometric growth [60]. Interestingly, a Kcnk5b potassium channel bioelectric calcineurin signaling module was identified to regulate fin growth in zebrafish [61]. For the pectoral fin overgrowth mutation of the fighting fish a calcium binding peptidyl-prolyl cis-trans isomerase gene, fkbp14, is linked to kcnh8 at the mutant locus and shows co-regulation with the potassium channel in the overgrowing fin (Wang et al., submitted).
The involvement of kcnh8 in the mechanism that leads to formation of the male sword is supported by several reports that levels of channel expression are directly related to the extent of growth of the fins in zebrafish [48, 50, 62]. Mutants involve gain of function or overexpression of wildtype channels, leading to overgrowth. Transgenic overexpression of a set of potassium channels phenocopied long fin mutations, even when the transgene was ectopically expressed in the dermomyotome of the developing embryo, which later constitutes the peduncle [48, 62]. The expression of kcnh8 in the sword organizer close to the peduncle and its upregulation in the sword during normal and hormone treatment -induced growth and during regeneration, as well as the correlation of kcnh8 expression with sword length in several Xiphophorus species is consistent with an important role of bioelectrical signaling in regulation of fin growth. Of note, in our sword transcriptomes other genes involved in this process are co-regulated with kcnh8, e.g. the gap junction component gja1, ion channels (cac1g, scn4b, kcng2, cacna1), transporters (slc26a1, slc12a5, slc8a1), interacting proteins (nkain1, kcnip1) and other regulators of the membrane potential (chrna7, gabr1) (table S1).
Kcnh8 is the pore-forming unit of some voltage-gated potassium channels, which have broad functions mainly in neurotransmitter release and neuronal excitability, but also in epithelial electrolyte transport and cell volume regulation [55, 63]. In zebrafish, due to the presence of duplicate versions of the channel protein coding genes, one paralog obviously can fulfill functions restricted to the fin. Mutations of the “fin” paralog only affect fin growth, while the other channel functions are executed by the second paralog. However, kcnh8 is present only as a single copy and it is abundantly expressed in the brain and to a lesser extent in the gonads of both sexes. Additionally, we found expression in the male sword of Xiphophorus but importantly not in the corresponding part of the female caudal fin. These expression domains imply that a neuronal gene was recruited during the evolution of the male ornament about 3–5 million years ago, early during the diversification of swordtail fish through a rewiring of its regulatory network rather than by selection on its protein function. The Kcnh8 proteins of Xiphophorus species have a few amino acid changes, which, however, do not correlate with the presence or absence of a sword in males (Figure S7). Thus, it is more likely that the novel function for sword development has been added to the kcnh8 gene through changes its gene regulation. Transcription factor binding site prediction did not uncover an androgen receptor responsive element, consistent with the equal expression of kcnh8 expression in male and female brains. But several binding sites were found for the transcription factors that make up the pre-pattern (data S2) and await further experimental evaluation. Although these motifs are conserved between X. hellerii and X. maculatus, the fact that pax9, tbx3a and six2a have lower expression levels in X. maculatus and also in females of X. hellerii may indicate a quantitative rather than qualitative effect that leads to the sword specific expression of kcnh8.
The implication of Kcnh8 activity in natural sword development provides the first case of an evolutionary mutant for a potassium channel being involved in regulation of fin growth, which thus far was only seen in laboratory mutants and domesticated fish. It appears that in fish fins at least five genes - kcnh2a, kcnk5b, kcc4a, kcnj13 and kcnh8 - govern a common pathway of downstream signaling that connects membrane potential, K+ permeability, conductance and calcium homeostasis to the ubiquitous machinery of cell growth and proliferation. A general role of integrated bioelectric signals that act in concert as organizer or coordinating regulator of growth has been proposed based on findings in zebrafish [48, 49, 64]. In this model the bioelectric signal is translated into growth through the action of potassium channels, acting as central rheostat to modulate calcium-dependent proliferation [49]. Calcineurin may provide a molecular toehold [60] by which to probe these connections, because it appears to bind and regulate at least two different potassium channels [61]. These changes in membrane potential can also be transduced within tissue layers by cellular junctions to create tissue-wide bioelectric gradients that effect changes in large-scale patterning[53, 65]. The role of Kcnh8 in the development of the ventral tail fin outgrowth in male swordtails is well in line with these models.
Star Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Manfred Schartl (phch1@biozentrum.uni-wuerzburg.de)
Materials Availability
This study did not generate new unique reagents or animal strains.
Data and Code Availability
Datasets supporting the current study (multi sequence alignment, marker sequences and mapping data) are supplied as supplementary tables. RNA-seq data can be accessed at PRJNA673149.
Experimental Model and Subject Details
Experimental Animals
All fish were reared under a standard conditions [80] with a light/dark cycle of 14/10 h at 26 °C in the fish facility of the Biocenter at the University of Wuerzburg, Germany. All animals were kept and sampled in accordance with the applicable EU and national German legislation governing animal experimentation. In particular, all experimental protocols were approved through an authorization (568/300–1870/13) of the Veterinary Office of the District Government of Lower Franconia, Germany, in accordance with the German Animal Protection Law (TierSchG).
For regeneration experiments, fish were immobilized by dipping into 4°C water, and the caudal margin (1mm) of the tail fin was resected with a razor blade. Tissues were collected at different stages of regeneration (Figure S1). Samples from X. hellerii females and the swordless males of Priapella lacondonae and X.maculatus were taken after caudal fin resection at the same day according to male sword regeneration stages. Tissues from naturally developing swords and the median and upper caudal fin margin of male X. hellerii were sampled at different stages according to figs. S1, 17. Induction of the sword in mature female X. hellerii (4–5 months old) was accomplished by addition of 17-methyl testosterone to the tank water (1μMol, replenished daily). The dorsal, median and ventral caudal fin margins, including the sword were collected after 11 days of treatment at a stage corresponding to naturally developing sword stage 4 (Figure S1). Areas used for RNA-seq and qPCR experiments are depicted in Figure S1. Samples from 10 – 30 individuals were pooled for RNA extraction.
For modulating potassium channels fish (N=3) were kept during sword regeneration for 50 days in 1μM of EAG potassium channel inhibitor 4-aminopyridine [81] with water changes every 48h. Due to the high toxicity of the substance only a mild treatment could be performed (see Figure S5).
Method Details
RNA-seq transcriptomics
Total RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific, Waltham, USA) according to the supplier’s recommendation. Custom sequencing (BGI, Shenzen, China) of TruSeq libraries generated 25–30 million 100bp paired end reads for each sample on the Illumina Hiseq4000 platform.
Differential gene expression analysis
After duplicate and barcode removal reads were aligned to the Xiphophorus_hellerii-4.1 genome (https://www.ncbi.nlm.nih.gov/genome/15325?genome_assembly_id=7477339) using the STAR aligner version 2.5 (--runMode alignReads --quantMode GeneCounts) [66]. Resulting read counts were used by DESeq2 [82] for differential gene analysis. Datasets generated at different time points were analyzed separately.
For further analysis (tables S1, S2), only expressed genes were considered. “Expressed” was defined as normalized read count >= 10 in at least one sample in datasets “female” (regeneration of caudal fin in adult females), “sword development” (normal sword development in young males at puberty), testosterone induced sword in adult females (“testosterone induced sword”), “sword regeneration” (regeneration of tail fin and sword in adult males), “X. maculatus, male” (regeneration of caudal fin in adult male platyfish, “sword organizer males” (proximal ventral caudal fin segment) and “sword organizer females” (corresponding region in female caudal fin to the sword organizer in males). We added a published dataset (“testosterone treated juveniles”) [3] of an independent testosterone treatment for sword induction in 3 months old undifferentiated juvenile X. hellerii. Because of the lower sequencing depth and because dataset “testosterone treated juveniles” has four replicates for each sample a gene was required to have a normalized read count>= 3 in at least two samples. To exclude genes that have general functions during physiological growth and regeneration, genes were further considered only if they were differentially expressed between the ventral compartment of the caudal fin, which is the sword development region, and the dorsal compartment, which does not develop a pigmented outgrowth. Subsequently all datasets were filtered for genes with a log2 fold change >=1 up or down, respectively, in at least one time point. Differentially expressed genes of the four male datasets were represented in a Venn diagram (https://bioinfogp.cnb.csic.es/tools/venny/) (Figure S2) and the overlap of all four datasets generated dataset “common in all male” (table S1). Next, all genes that showed the same differential regulation in “female” were removed from “common in all male” (table S1), and the remaining 54 genes were annotated for their chromosomal location.
qPCR expression analysis
Total RNA was isolated from pooled samples using TRIzol Reagent (Thermo Fisher Scientific, Waltham, USA) according to the supplier’s recommendation. After DNase treatment, total RNA (1–2 μg) was reverse transcribed using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, USA) and random hexamer primers, according to the manufacturer’s instructions. For real-time qRT-PCR, cDNA from 50 ng of total RNA was used. All results reported here are averages of at least two independent reverse transcription (RT) reactions and two PCR experiments from each such reaction. Primer sequences are listed in table S3. Amplification was monitored using a Mastercycler ep realplex2 (Eppendorf, Hamburg, Germany). For quantification, expression of each gene was normalized to the housekeeping gene ef1a1 (elongation factor 1 alpha 1) using the delta Ct method [83].
qPCR expression analysis was performed to confirm differential expression results from the RNA-seq datasets from X. hellerii and to monitor differential expression in other species (X. maculatus, X. montezumae, X. monticolus, X. pygmaeus, P. lacandonae, O. latipes) (Figures 2–4, 6, S3, S4, S6).
Sequence analysis of kcnh8
Protein sequences of Kcnh8 were retrieved for different species: X.hellerii and X.couchianus from NCBI (XP_032437747.1, XP_027893054.1); X.maculatus from Ensembl (ENSXMAP00000000856); X.birchmanni and X.malinche, from a previous study [84]; X.signum, X.mixei, X.montezumae, X.clemenciae, X.monticolus, X.kallmani, X.mayae, X.andersi, X.pygmaeus, X.continens, X.multilineatus, X.nigrensis, X.milleri, X.gordoni, X.meyeri, X.evelynae, X.xiphidium and X.variatus, from raw NGS reads.
To retrieve kcnh8 sequences from raw NGS reads, first, we collected all related reads by aligning them to the existing protein sequences from reference genomes using DIAMOND [77]. The kept reads were then assembled into exon-fragments using CAP3 [78]. For each fragment we determined its best translation frame by mapping it onto the reference protein sequences using GeneWise [85]. Finally, the resulting protein fragments were ordered and merged into a complete sequence according to the alignment.
Putative transcription factor binding sites were detected using the Bioconductor/R package TFBSTools (http://bioconductor.org/packages/release/bioc/html/TFBSTools.html) based on the JASPAR database 2018 release (http://jaspar.genereg.net/blog/2017/10/16/jaspar-database-seventh-release-2018). A threshold of score >10 was used. Intron 1 and the upstream region from the transcription start site to the next gene were searched (Data S2).
For functional classification the DAVID (https://david.ncifcrf.gov/) functional enrichment analysis web tool was used with default settings.
QTL mapping
To identify regions of the genome associated with the sword trait, the Sword Index (SI), which is the sword length divided by standard length, was determined. F1 individuals were obtained from a cross of a female Xiphophorus hellerii (Rio Lancetilla strain) with a male X. maculatus (Jp163A strain) aided by artificial insemination. The low fertility of F1 intercrosses [86] precluded the production of F2 families, so we performed two backcrosses of X. maculatus /X. hellerii F1 males with X. hellerii (Rio Lancetilla strain) females as the recurrent parent. Quantitative trait locus (QTL) analysis was performed in R/qtl v.1.39–5 [87] with phenotype (herein) and genotype data for 85 males and 16,250 RAD-tag loci and the genetic map from Amores et al [38]. 8,487 markers were retained after markers with identical genotypes were removed from the analyses Backcross generation males for mapping were produced by two sires; 60 offspring from male #2059 crossed with four full-sib females (44, 2, 12, and 2 offspring per female) and 25 from male #2074, all from one female. The dataset was coded as homozygous for the genotype of the backcross parent X. hellerii (data code b), or heterozygous (h) with alleles from X. hellerii and X. maculatus. Interval mapping was performed using the non-parametric model (method= “np”) due to the non-normal distribution of the SI phenotype and standard interval mapping as implemented in R/qtl (method = “em”). Genotype probabilities were calculated at a maximum distance of 1 centiMorgan and markers with identical genotypes were removed from the analysis. The genome-wide significance thresholds were determined using a permutation test with 1000 replicates. For the non-parametric analysis, the 5% genome-wide threshold for significance was LOD 2.85, for the standard interval mapping analysis the 5% threshold was LOD 3.07. The marker sequences (table S4) used for QTL mapping (table S5) were later aligned to the X. maculatus genome (NCBI GCF_002775205.1) and the X. hellerii genome (GCA_003331165.2) with GSNAP version 2018–03-25 [67] (data S1). To identify candidate genes the 2 LOD drop was used to define the genomic region of interest.
Electrophysiology
To generate cRNA for functional characterization of Xiphophorus hellerii Kcnh8 in Xenopus oocytes, the coding sequence of Xiphophorus hellerii kcnh8 was cloned into oocyte expression vector pNBI16/pNB1u (pGEM-based vector) using the USER-technique [88]. The construct was verified by sequencing. cRNA of kcnh8 was prepared using the AmpliCap-Max™ T7 High Yield Message Maker Kit (Cellscript, Biozym Scientific GmbH, Hessisch Oldendorf, Germany). Oocyte preparation and cRNA injection have been described elsewhere [89]. Following the injection of 20 ng cRNA per oocyte, oocytes were incubated at 16°C for 24 to 36 hours in ND96 solution (96 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM Hepes pH7,4) supplemented with 50 mg/l gentamycin.
In two-electrode voltage-clamp studies, oocytes were perfused with KCl-containing solutions, based on Tris/Mes buffers. The standard solution contained 10 mM Tris/Mes, pH 7.5, 1 mM CaCl2, 1 mM MgCl2, 30 mM KCl and 70 mM LiCl. If appropriate, osmolarity was adjusted to 220 mOsmol/kg using D-sorbitol. For measurements at varying K+ concentrations, the ionic strength was kept constant by replacing KCl with LiCl and vice versa. Voltage-dependent activation of Kcnh8-expressing oocytes was recorded with voltage-pulse-protocols designed and applied with the acquisition software Patchmaster (HEKA Elektronik GmbH, Lambrecht/Pfalz, Germany). Proceeding from a holding potential (VH) of −20 mV, a series of 4s test voltage pulses ranging from +40 to −140 mV in 10 mV decrements were applied. Steady state currents (ISS) were extracted at the end of the test voltage pulses.
Quantification and Statistical Analysis
For the QTL mapping analyses were performed using standard interval mapping with methods “em” and “np” implemented as scanone in R/qtl analysis (see Key Resources Table). Resulting LOD scores for peaks surpassing analysis-specific 5% genome-wide thresholds for significance (em = 3.07 and np = 2.86) are specified in the results and the results of the np analysis are plotted in Figure 5. Significance levels were determined using a permutation test with 1000 replicates. For RNaseq data statistical analysis was performed using DESeq2[68]. For replicates all values including log fold change and p-values for differential gene expression were calculated using DESeq2. Log fold change for time course data were calculated using an inhouse R script. Only “expressed” genes (normalized read count >= 10 in at least one sample of the dataset) were considered in both cases. Real time qPCR analyses were done with the delta Ct method [83] from at least two independent cDNA samples. All data in Figures 2, 3, 4, 6, S3, S4, and S6 are represented as mean +/− standard deviation. Figures were prepared with Adobe Photoshop 2020 and Microsoft Powerpoint.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRIzol Reagent | Invitrogen | Cat#15596026 |
| dNTPs (Set, 100mM) | Sigma-Aldrich | Cat#DNTP100 |
| DNase, RNase-free | Thermo Scientific | Cat#EN0521 |
| SYBR Green I (10 000x in DMSO) | Invitrogen | Cat#S7563 |
| Q5 High Fidelity DNA Polymerase | New England Biolabs | Cat#M04915 |
| Critical Commercial Assays | ||
| AmpliCap-Max™ T7 High Yield Message Maker Kit | Biozym | Cat#150472 |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific | Cat#K1621 |
| Deposited Data | ||
| kcnh8 sequences | this paper | https://github.com/dukecomeback/swordtail/blob/main/kcnh8_Xiphophorus.fa |
| RNA-seq short read sequences | [16] and this paper | ENA accession code PRJEB8012 and PRJNA673149 |
| Experimental Models: Organisms/Strains | ||
| Xiphophorus hellerii, Rio Lancetilla strain | Biocenter Würzburg | WLC1337 |
| Xiphophorus maculatus, Rio Jamapa strain Jp163A | Biocenter Würzburg | WLC6628 |
| Xiphophorus montezumae, Tamasops strain | Biocenter Würzburg | WLC1052 |
| Xiphophorus monticolus, El Tejon strain | Biocenter Würzburg | WLC3344 |
| Xiphophorus pygmaeus, strain Rio Axtla | Biocenter Würzburg | WLC3015 |
| Priapella lacandonae | Biocenter Würzburg | WLC5281 |
| Oryzias latipes, Carbio strain | Biocenter Würzburg | WLC2674 |
| Oligonucleotides | ||
| Oligonucleotide Primers for PCR | see Table S3 for primers | |
| Recombinant DNA | ||
| pcDNA3 | Thermo Fisher | V79020 |
| pNBI16/pNB1u | [66] | n.a. |
| Software and Algorithms | ||
| GSNAP version 2018-03-25 | [67] | http://research-pub.gene.com/gmap/ |
| STAR | [66] | https://github.com/alexdobin/STAR/releases |
| DESeq2 | [68] | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Ensembl Biomart | [69] | http://www.ensembl.org/biomart/martview/a61c56ebb44f17c0e39ce71a4d79f44a |
| DAVID | [70] | https://david.ncifcrf.gov/ |
| Inparanoid | [71] | http://inparanoid.sbc.su.se/cgi-bin/index.cgi |
| Pal2Nal | [72] | http://www.bork.embl.de/pal2nal/ |
| MUSCLE | [73] | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| Gblocks | [74] | http://molevol.cmima.csic.es/castresana/Gblocks_server.html |
| Phylip | [75] | http://evolution.genetics.washington.edu/phylip.html, |
| FigTree | see website | http://tree.bio.ed.ac.uk/software/figtree/ |
| ETE3 | [76] | http://etetoolkit.org/ |
| Primer3 (version 4.4.0) | see website | https://bioinfo.ut.ee/primer3/ |
| R/qtl v.1.39-5 | see website | https://rqtl.org/ |
| diamond v0.9.24.125 | [77] | https://github.com/bbuchfink/diamond |
| CAP3 | [78] | http://doua.prabi.fr/software/cap3 |
| GeneWise wise2.2.3-rc7 | [79] | http://www.ebi.ac.uk/Wise2/ |
Supplementary Material
Sword QTL mapping data, related to Figure 5 and STAR Methods section “QTL mapping”.
Transcription factor binding sites predicted for kcnh8, related to STAR Methods section “Sequence analysis of kcnh8”.
Differentially expressed genes, related to STAR Methods section “Differential gene expression analysis”.
Normalized read counts and log2 fold changes of selected genes, related to STAR Methods section “Differential gene expression analysis”.
Primers used for real time qPCR, related to STAR Methods section “qPCR expression analysis”.
Sequences of RAD-tags used for mapping, related to STAR Methods section “QTL mapping”.
Swordtail mapping data, related to STAR Methods section “QTL mapping”.
Highlights:
RNA-seq coupled with QTL mapping identified a genetic network for the sword
A non-sex biased transcription factor pre-pattern underlies sword formation
A main factor for the development of the polygenic sword is a potassium channel
A brain gene was recruited for evolving a sexually selected male ornamental trait
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (grants 5263398, 163418330 and 5446040 to AM and his laboratory) and by NIH grant 5R01OD011116 (JHP), R24OD018555 (JHP, MS, RW, WW).
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Darwin C. (1871). The Descent of Man and Selection in Relation to Sex, (London: John Murray; ). [Google Scholar]
- 2.Eibner C, Pittlik S, Meyer A, and Begemann G. (2008). An organizer controls the development of the “sword,” a sexually selected trait in swordtail fish. Evol Dev 10, 403–412. [DOI] [PubMed] [Google Scholar]
- 3.Kang JH, Manousaki T, Franchini P, Kneitz S, Schartl M, and Meyer A. (2015). Transcriptomics of two evolutionary novelties: how to make a sperm-transfer organ out of an anal fin and a sexually selected “sword” out of a caudal fin. Ecol Evol 5, 848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dzwillo M. (1964). Sekundäre Geschlechtsmerkmale an der Caudalflosse einiger Xiphophorini unter dem Einfluß von Methyltestosteron. Mitt. Hamburg. Zool. Mus. Inst, 15–22. [Google Scholar]
- 5.Basolo AL (1990). Female preference predates the evolution of the sword in swordtail fish. Science 250, 808–810. [DOI] [PubMed] [Google Scholar]
- 6.Basolo AL (1998). Evolutionary change in a receiver bias: a comparison of female preference functions. Proc Biol Sci 265, 2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher RA (1930). The Genetical Theory of Natural Selection, (Oxford: Oxford University Press; ). [Google Scholar]
- 8.Meyer A, Morrissey JM, and Schartl M. (1994). Recurrent origin of a sexually selected trait in Xiphophorus fishes inferred from a molecular phylogeny. Nature 368, 539–542. [DOI] [PubMed] [Google Scholar]
- 9.Wagner WE Jr., Beckers OM, Tolle AE, and Basolo AL (2012). Tradeoffs limit the evolution of male traits that are attractive to females. Proc Biol Sci 279, 2899–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenthal GG, Flores Martinez TY, Garcia de Leon FJ, and Ryan MJ (2001). Shared preferences by predators and females for male ornaments in swordtails. Am Nat 158, 146–154. [DOI] [PubMed] [Google Scholar]
- 11.Basolo AL, and Alcaraz G. (2003). The turn of the sword: length increases male swimming costs in swordtails. Proc Biol Sci 270, 1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endler JA (1992). Signals, signal conditions, and the direction of evolution. American Naturalist 139, 125–153. [Google Scholar]
- 13.Ryan MJ (1990). Sexual selection, sensory systems and sensory exploitation. Oxford Surveys in Evolutionary Biology 7, 157–195. [Google Scholar]
- 14.Jones JC, Fan S, Franchini P, Schartl M, and Meyer A. (2013). The evolutionary history of Xiphophorus fish and their sexually selected sword: a genome-wide approach using restriction site-associated DNA sequencing. Mol Ecol 22, 2986–3001. [DOI] [PubMed] [Google Scholar]
- 15.Cui R, Schumer M, Kruesi K, Walter R, Andolfatto P, and Rosenthal GG (2013). Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution 67, 2166–2179. [DOI] [PubMed] [Google Scholar]
- 16.Kang JH, Schartl M, Walter RB, and Meyer A. (2013). Comprehensive phylogenetic analysis of all species of swordtails and platies (Pisces: Genus Xiphophorus) uncovers a hybrid origin of a swordtail fish, Xiphophorus monticolus, and demonstrates that the sexually selected sword originated in the ancestral lineage of the genus, but was lost again secondarily. BMC Evol Biol 13, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winquist ST, Weary DM, Inman AJ, Mountjoy DJ, and Krebs EA (1991). Male swords and female preferences. Science 253, 1426. [DOI] [PubMed] [Google Scholar]
- 18.Endler JA, and Basolo AL (1998). Sensory ecology, receiver biases and sexual selection. Trends Ecol Evol 13, 415–420. [DOI] [PubMed] [Google Scholar]
- 19.Meyer A, Salzburger W, and Schartl M. (2006). Hybrid origin of a swordtail species (Teleostei: Xiphophorus clemenciae) driven by sexual selection. Mol Ecol 15, 721–730. [DOI] [PubMed] [Google Scholar]
- 20.Wong BB, and Rosenthal GG (2006). Female disdain for swords in a swordtail fish. Am Nat 167, 136–140. [DOI] [PubMed] [Google Scholar]
- 21.Coyne JA, and Orr HA (2004). Speciation, (Sunderland, MA: Sinauer; ). [Google Scholar]
- 22.Mayr E. (1963). Animal species and evolution, (Cambridge, MA: Belknap Press of Harvard University Press; ). [Google Scholar]
- 23.Ritchie MG (2007). Sexual selection and speciation. Annual Review of Ecology, Evolution, and Systematics 38, 79–102. [Google Scholar]
- 24.van Doorn GS, Edelaar P, and Weissing FJ (2009). On the origin of species by natural and sexual selection. Science 326, 1704–1707. [DOI] [PubMed] [Google Scholar]
- 25.Tinghitella RM, Lackey ACR, Martin M, Dijkstra PD, Drury JP, Heathcote R, Keagy J, Scordato ESC, and Tyers AM (2017). On the role of male competition in speciation: a review and research agenda. Behavioral Ecology 29, 783–797. [Google Scholar]
- 26.Gotoh H, Zinna RA, Ishikawa Y, Miyakawa H, Ishikawa A, Sugime Y, Emlen DJ, Lavine LC, and Miura T. (2017). The function of appendage patterning genes in mandible development of the sexually dimorphic stag beetle. Dev Biol 422, 24–32. [DOI] [PubMed] [Google Scholar]
- 27.Kraaijeveld K. (2019). Genetic architecture of novel ornamental traits and the establishment of sexual dimorphism: insights from domestic birds. Journal of Ornithology 160, 861–868. [Google Scholar]
- 28.Zauner H, Begemann G, Mari-Beffa M, and Meyer A. (2003). Differential regulation of msx genes in the development of the gonopodium, an intromittent organ, and of the “sword,” a sexually selected trait of swordtail fishes (Xiphophorus). Evol Dev 5, 466–477. [DOI] [PubMed] [Google Scholar]
- 29.Offen N, Blum N, Meyer A, and Begemann G. (2008). Fgfr1 signalling in the development of a sexually selected trait in vertebrates, the sword of swordtail fish. BMC Dev Biol 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Offen N, Meyer A, and Begemann G. (2009). Identification of novel genes involved in the development of the sword and gonopodium in swordtail fish. Dev Dyn 238, 1674–1687. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto-Shiraishi Y, and Kuroiwa A. (2013). Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Dev Biol 377, 363–374. [DOI] [PubMed] [Google Scholar]
- 32.Nachtrab G, Kikuchi K, Tornini VA, and Poss KD (2013). Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development 140, 3754–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriyama Y, Kawanishi T, Nakamura R, Tsukahara T, Sumiyama K, Suster ML, Kawakami K, Toyoda A, Fujiyama A, Yasuoka Y, et al. (2012). The medaka zic1/zic4 mutant provides molecular insights into teleost caudal fin evolution. Curr Biol 22, 601–607. [DOI] [PubMed] [Google Scholar]
- 34.Corredor-Adamez M, Welten MC, Spaink HP, Jeffery JE, Schoon RT, de Bakker MA, Bagowski CP, Meijer AH, Verbeek FJ, and Richardson MK (2005). Genomic annotation and transcriptome analysis of the zebrafish (Danio rerio) hox complex with description of a novel member, hox b 13a. Evol Dev 7, 362–375. [DOI] [PubMed] [Google Scholar]
- 35.Waghray A, Saiz N, Jayaprakash AD, Freire AG, Papatsenko D, Pereira CF, Lee DF, Brosh R, Chang B, Darr H, et al. (2015). Tbx3 Controls Dppa3 Levels and Exit from Pluripotency toward Mesoderm. Stem Cell Reports 5, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rallis C, Del Buono J, and Logan MP (2005). Tbx3 can alter limb position along the rostrocaudal axis of the developing embryo. Development 132, 1961–1970. [DOI] [PubMed] [Google Scholar]
- 37.Mise T, Iijima M, Inohaya K, Kudo A, and Wada H. (2008). Function of Pax1 and Pax9 in the sclerotome of medaka fish. Genesis 46, 185–192. [DOI] [PubMed] [Google Scholar]
- 38.Amores A, Catchen J, Nanda I, Warren W, Walter R, Schartl M, and Postlethwait JH (2014). A RAD-Tag Genetic Map for the Platyfish (Xiphophorus maculatus) Reveals Mechanisms of Karyotype Evolution Among Teleost Fish. Genetics 197, 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zander CDD, M. (1969). Untersuchungen zur Entwicklung und Vererbung des Caudalfortsatzes der Xiphophorus-Arten(Pisces). Z Wissenschaftliche Zool 178, 267–315. [Google Scholar]
- 40.Zou A, Lin Z, Humble M, Creech CD, Wagoner PK, Krafte D, Jegla TJ, and Wickenden AD (2003). Distribution and functional properties of human KCNH8 (Elk1) potassium channels. Am J Physiol Cell Physiol 285, C1356–1366. [DOI] [PubMed] [Google Scholar]
- 41.Cloud-Richardson KM, Smith BR, and Macdonald SJ (2016). Genetic dissection of intraspecific variation in a male-specific sexual trait in Drosophila melanogaster. Heredity (Edinb) 117, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ichimura K, Kawashima Y, Nakamura T, Powell R, Hidoh Y, Terai S, Sakaida I, Kodera Y, Tsuji T, Ma JX, et al. (2013). Medaka fish, Oryzias latipes, as a model for human obesity-related glomerulopathy. Biochem Biophys Res Commun 431, 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fairbairn D. (2013). Odd Couples: Extraordinary Differences between the Sexes in the Animal Kingdom., (Princeton; ). [Google Scholar]
- 44.Kallman KD (1984). A new look at sex determination in Poeciliid Fishes. In Evolutionary Genetics of Fishes, Turner BJ, ed. (Plenum Publishing Corporation; ), pp. 95–171. [Google Scholar]
- 45.Schartl M, Galiana-Arnoux D, Schultheis C, Böhne A, and Volff J-N (2010). A primer of sex determination in poeciliids. In Ecology and Evolution of Poeciliid Fishes, Pilastro A, Evans J. and Schlupp I, eds., pp. 264–275. [Google Scholar]
- 46.Basolo AL (1995). Phylogenetic evidence for the role of a pre-existing bias in sexual selection. Proc Biol Sci 259, 307–311. [DOI] [PubMed] [Google Scholar]
- 47.Powell DP,C; Keegan M; Banerjee SM; Cui R; Andolfatto P, Schumer M; Rosenthal GG (2020). The genetic architecture of the sexually selected sword ornament. BioRxiv doi: 10.1101/2020.07.23.218164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perathoner S, Daane JM, Henrion U, Seebohm G, Higdon CW, Johnson SL, Nusslein-Volhard C, and Harris MP (2014). Bioelectric signaling regulates size in zebrafish fins. PLoS Genet 10, e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanni JS, Peal D, Ekstrom L, Chen H, Stanclift C, Bowen ME, Mercado A, Gamba G, Kahle KT, and Harris MP (2019). Integrated K+ channel and K+Cl-cotransporter functions are required for the coordination of size and proportion during development. Dev Biol 456, 164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart SLB,HK; Yette GA; Henner AL; Braunstein JA, Stankunas K. (2019). longfin causes cis-ectopic expression of the kcnh2a ether-a-go-go K+ channel to autonomously prolong fin outgrowth. . BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bougourd SM, and Jones RN (1997). B chromosomes: a physiological enigma. New Phytologist 137, 43–54. [Google Scholar]
- 52.Kon T, Omori Y, Fukuta K, Wada H, Watanabe M, Chen Z, Iwasaki M, Mishina T, Matsuzaki SS, Yoshihara D, et al. (2020). The Genetic Basis of Morphological Diversity in Domesticated Goldfish. Curr Biol 30, 2260–2274 e2266. [DOI] [PubMed] [Google Scholar]
- 53.McLaughlin KA, and Levin M. (2018). Bioelectric signaling in regeneration: Mechanisms of ionic controls of growth and form. Dev Biol 433, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sundelacruz S, Levin M, and Kaplan DL (2009). Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev Rep 5, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urrego D, Tomczak AP, Zahed F, Stuhmer W, and Pardo LA (2014). Potassium channels in cell cycle and cell proliferation. Philos Trans R Soc Lond B Biol Sci 369, 20130094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung H, and Akkus O. (2016). Activation of intracellular calcium signaling in osteoblasts colocalizes with the formation of post-yield diffuse microdamage in bone matrix. Bonekey Rep 5, 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goltzman D, and Hendy GN (2015). The calcium-sensing receptor in bone--mechanistic and therapeutic insights. Nat Rev Endocrinol 11, 298–307. [DOI] [PubMed] [Google Scholar]
- 58.Han B, Tokay T, Zhang G, Sun P, and Hou S. (2017). Eag1 K(+) Channel: Endogenous Regulation and Functions in Nervous System. Oxid Med Cell Longev 2017, 7371010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somarelli JA, Lee SY, Skolnick J, and Herrera RJ (2008). Structure-based classification of 45 FK506-binding proteins. Proteins 72, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kujawski S, Lin W, Kitte F, Bormel M, Fuchs S, Arulmozhivarman G, Vogt S, Theil D, Zhang Y, and Antos CL (2014). Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev Cell 28, 573–587. [DOI] [PubMed] [Google Scholar]
- 61.Daane JM, Lanni J, Rothenberg I, Seebohm G, Higdon CW, Johnson SL, and Harris MP (2018). Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Sci Rep 8, 10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silic MR, Wu Q, Kim BH, Golling G, Chen KH, Freitas R, Chubykin AA, Mittal SK, and Zhang G. (2020). Potassium Channel-Associated Bioelectricity of the Dermomyotome Determines Fin Patterning in Zebrafish. Genetics 215, 1067–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jan LY, and Jan YN (2012). Voltage-gated potassium channels and the diversity of electrical signalling. J Physiol 590, 2591–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris MP, Daane JM, and Lanni J. (2020). Through veiled mirrors: Fish fins giving insight into size regulation. Wiley Interdiscip Rev Dev Biol, e381. [DOI] [PubMed] [Google Scholar]
- 65.Levin M, Pezzulo G, and Finkelstein JM (2017). Endogenous Bioelectric Signaling Networks: Exploiting Voltage Gradients for Control of Growth and Form. Annu Rev Biomed Eng 19, 353–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu TD, and Watanabe CK (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875. [DOI] [PubMed] [Google Scholar]
- 68.Anders S, and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. (2011). Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahuja MR, and Anders F. (1976). A Genetic Concept of the Origin of Cancer,. Based in Part upon Studies of Neoplasms. Fish. Prog. Exptl. Tumor Res 20, 380–397. [DOI] [PubMed] [Google Scholar]
- 71.Sonnhammer EL, and Ostlund G. (2015). InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 43, D234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suyama M, Torrents D, and Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar RC (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17, 540–552. [DOI] [PubMed] [Google Scholar]
- 75.Felsenstein J. (1989). Mathematics vs. Evolution: Mathematical Evolutionary Theory. Science 246, 941–942. [DOI] [PubMed] [Google Scholar]
- 76.Huerta-Cepas J, Serra F, and Bork P. (2016). ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol Biol Evol 33, 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buchfink B, Xie C, and Huson DH (2015). Fast and sensitive protein alignment using DIAMOND. Nature methods 12, 59–60. [DOI] [PubMed] [Google Scholar]
- 78.Huang X, and Madan A. (1999). CAP3: A DNA sequence assembly program. Genome research 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff JN, et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46, 253–260. [DOI] [PubMed] [Google Scholar]
- 80.Kallman K. (1975). The platyfish, Xiphophorus maculatus. In Handbook of Genetics, Volume 4, RC K, ed. (New York, N.Y.: Plenum Press; ), pp. 81–132. [Google Scholar]
- 81.Wulff H, and Zhorov BS (2008). K+ channel modulators for the treatment of neurological disorders and autoimmune diseases. Chem Rev 108, 1744–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simpson DA, Feeney S, Boyle C, and Stitt AW (2000). Retinal VEGF mRNA measured by SYBR green I fluorescence: A versatile approach to quantitative PCR. Mol Vis 6, 178–183. [PubMed] [Google Scholar]
- 84.Powell DL, García-Olazábal M, Keegan M, Reilly P, Du K, Díaz-Loyo AP, Banerjee S, Blakkan D, Reich D, and Andolfatto P. (2020). Natural hybridization reveals incompatible alleles that cause melanoma in swordtail fish. Science 368, 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Birney E, Clamp M, and Durbin R. (2004). GeneWise and genomewise. Genome research 14, 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franchini P, Jones JC, Xiong P, Kneitz S, Gompert Z, Warren WC, Walter RB, Meyer A, and Schartl M. (2018). Long-term experimental hybridisation results in the evolution of a new sex chromosome in swordtail fish. Nat Commun 9, 5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Broman KW, Wu H, Sen S, and Churchill GA (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. [DOI] [PubMed] [Google Scholar]
- 88.Nour-Eldin HH, Hansen BG, Norholm MH, Jensen JK, and Halkier BA (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Becker D, Dreyer I, Hoth S, Reid JD, Busch H, Lehnen M, Palme K, and Hedrich R. (1996). Changes in voltage activation, Cs+ sensitivity, and ion permeability in H5 mutants of the plant K+ channel KAT1. Proc Natl Acad Sci U S A 93, 8123–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sword QTL mapping data, related to Figure 5 and STAR Methods section “QTL mapping”.
Transcription factor binding sites predicted for kcnh8, related to STAR Methods section “Sequence analysis of kcnh8”.
Differentially expressed genes, related to STAR Methods section “Differential gene expression analysis”.
Normalized read counts and log2 fold changes of selected genes, related to STAR Methods section “Differential gene expression analysis”.
Primers used for real time qPCR, related to STAR Methods section “qPCR expression analysis”.
Sequences of RAD-tags used for mapping, related to STAR Methods section “QTL mapping”.
Swordtail mapping data, related to STAR Methods section “QTL mapping”.
Data Availability Statement
Datasets supporting the current study (multi sequence alignment, marker sequences and mapping data) are supplied as supplementary tables. RNA-seq data can be accessed at PRJNA673149.