Abstract
In yeast cells, transcriptional activation occurs when the RNA polymerase II (Pol II) machinery is artificially recruited to a promoter by fusing individual components of this machinery to a DNA-binding domain. Here, we show that artificial recruitment of components of the TFIID complex can activate transcription in mammalian cells. Surprisingly, artificial recruitment of TATA-binding protein (TBP) activates transiently transfected and chromosomally integrated promoters with equal efficiency, whereas artificial recruitment of TBP-associated factors activates only chromosomal reporters. In contrast, artificial recruitment of various components of the mammalian Pol II holoenzyme does not confer transcriptional activation, nor does it result in synergistic activation in combination with natural activation domains. In the one case examined in more detail, the Srb7 fusion failed to activate despite being associated with the Pol II holoenzyme and being directly recruited to the promoter. Interestingly, some acidic activation domains are less effective when the promoter is chromosomally integrated rather than transiently transfected, whereas the Sp1 glutamine-rich activation domain is more effective on integrated reporters. Thus, yeast and mammalian cells differ with respect to transcriptional activation by artificial recruitment of the Pol II holoenzyme.
The RNA polymerase II (Pol II) transcription machinery is composed of two basic components, TFIID and large Pol II complexes which are often termed Pol II holoenzymes. TFIID, a complex of TATA-binding protein (TBP) and TBP-associated factors (TAFs), binds specifically to TATA elements, a step that initiates the assembly of the active transcription complex (8). The Pol II holoenzyme, loosely defined, consists of the core Pol II, general transcription factors (e.g., TFIIB, TFIIF, and TFIIH), and other associated components including Srb, Med, and (in mammalian cells) Trap proteins (22, 32, 40). The Pol II machinery is sufficient for efficient and accurate initiation in vitro on core promoters containing TATA and initiator elements, whereas these core promoters are virtually inactive in yeast and mammalian cells (50). The failure of core promoters to function in vivo is almost certainly due to the repressive effects of chromatin structure (50), particularly the inability of TBP to bind TATA elements in the context of nucleosomal substrates (25).
The virtual inactivity of core promoters in vivo means that transcription of essentially all eukaryotic genes requires activator proteins binding to enhancer elements. Eukaryotic activators bind enhancer elements through a DNA-binding domain, whereas they stimulate transcription through a functionally distinct, and typically physically separate, activation domain (43, 48). Activation domains are functionally autonomous in that they stimulate transcription when fused at various positions to heterologous DNA-binding domains. However, in yeast cells, activation domains do not stimulate transcription when fused to a variety of components of the Pol II machinery, indicating that activator-dependent recruitment of the Pol II machinery is the predominant mechanism for transcriptional activation (31). Consistent with this view, TBP (and hence the entire Pol II machinery) is not associated with the vast majority of yeast promoters in vivo in the absence of a functional activator (34, 37).
In principle, activation domains could recruit the Pol II machinery directly by contacting components of TFIID or Pol II holoenzyme and/or indirectly by associating with chromatin modifying activities or other coactivators. Evidence that both of these mechanisms occur in vivo is provided by artificial recruitment (also known as activator bypass, or nonclassical activator) experiments in yeast cells (44, 49). In such experiments, the requirement for an activation domain is bypassed by directly connecting a DNA-binding domain to a component of the Pol II machinery or to a subunit of a chromatin-modifying activity. Specifically, transcription is activated upon artificial recruitment of TBP (12, 33, 53), various TAFs (2, 21, 31), TFIIA (47), TFIIB (21, 36), Pol II holoenzyme subunits such as Sin4, Gal11, Srb2, Srb6, and Srb7 (5, 17, 20, 28), and a kinase-defective version of Srb10, a Pol II-associated kinase that normally represses transcription by prematurely phosphorylating the C-terminal tail of Pol II (24). In addition, transcription is activated upon artificial recruitment of Snf2 (Swi2) (35) or Gcn5 (9), which are the catalytic subunits of the Swi/Snf nucleosome remodeling or the SAGA histone acetylase complexes, respectively; in both cases, transcriptional activation is eliminated by mutations that abolish catalytic activity. Taken together, these results indicate that a covalent interaction between an activator and a single component of the Pol II machinery or a chromatin-modifying activity is sufficient to activate transcription. However, unlike natural activation domains, transcriptional activation by artificial recruitment is strongly influenced by promoter architecture, thereby suggesting that natural activators interact with multiple targets in vivo (20).
Artificial recruitment experiments have been performed to a much more limited extent in mammalian cells. In accord with the results in yeast, artificial recruitment of human TBP activates transcription in transiently transfected mammalian cells (23, 39, 41). Artificially recruited TBP synergistically activates transcription in combination with the VP16 and E1A activation domains but not with the Sp1 activation domain, suggesting that these activators function at distinct steps with respect to TBP recruitment (23, 39). The only mammalian holoenzyme components tested in an artificial recruitment experiment are human Srb7 (hSrb7) and hTFIIB, which were reported to weakly activate transcription and to synergize with classical activation domains (41). However, these fusions behaved indistinguishably from comparable fusions to a variety of yeast components (Srb2, Srb6, Srb7, and Srb11), which are unlikely to function in combination with mammalian components or to assemble into the mammalian Pol II holoenzyme. Specifically, hSrb7 does not complement a yeast srb7 mutant strain (11), and the other yeast Srbs have limited or no sequence similarity with components of the mammalian holoenzyme. Thus, the observed transcriptional effects mediated by the yeast proteins are unlikely to be due to artificial recruitment, and the comparable effects mediated by hSrb7 and hTFIIB are difficult to interpret.
These previous artificial recruitment experiments, as well as nearly all analyses of transcriptional regulatory mechanisms in mammalian cells, involved assays of transiently transfected reporter genes. However, transiently transfected promoters often behave differently from the same promoter that is integrated in a single site in the mammalian genome (1, 3, 16). Numerous variables, such as the chromatin state (27) or the extraordinarily high copy number of the transfected reporter plasmid, could explain these differences. To date, systematic studies comparing the functions of different transcriptional activation domains on integrated versus transiently transfected reporter genes have not been reported.
Here, we investigate the ability of components of the mammalian Pol II machinery to activate transcription when artificially recruited to promoters in mammalian cells. In addition, we examine these artificial recruitment constructs and a number of natural activation domains for their ability to function on transiently transfected or chromosomally integrated reporter genes. Our results indicate that artificial recruitment of TFIID components activates transcription in mammalian cells, but unlike the results in yeast cells, artificial recruitment of Pol II holoenzyme components fails to activate transcription. Further, we demonstrate that some, but not all, activators have different activities on transiently transfected or integrated reporters.
MATERIALS AND METHODS
Plasmids.
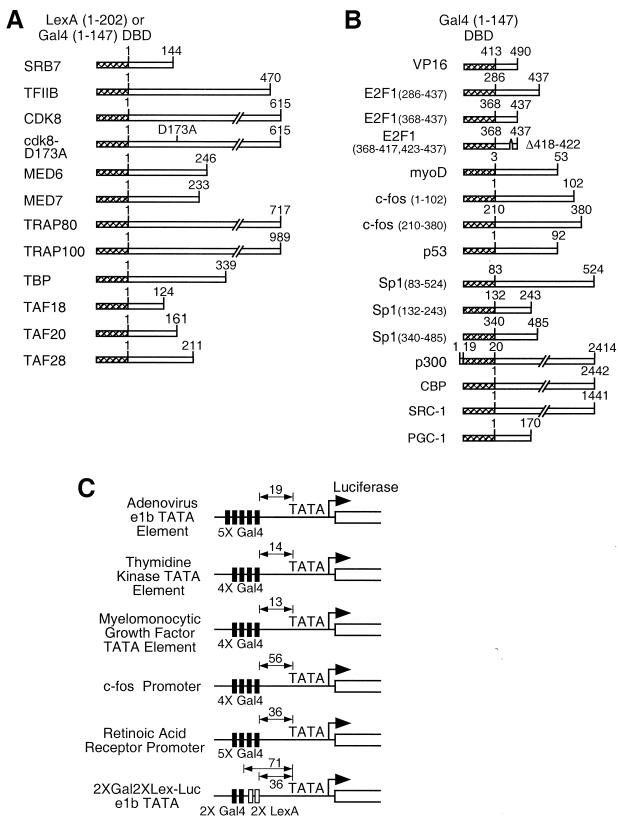
Genes encoding components of the Pol II machinery were generated by PCR of the entire open reading frame from either plasmids harboring the gene or from cDNA libraries and were then inserted into pSG424, which contains a Gal4(1-147) expression cassette under the control of the simian virus 40 promoter (46), or a pCS2+ expression vector containing LexA(1-202) and the simian virus 40 T-antigen nuclear localization signal (29) inserted at codon 3 of LexA. The 5XGal-e1bTATA-luciferase reporter, which contains the luciferase gene in place of the chloramphenicol acetyltransferase gene of 5XGal-e1bTATA-CAT (38) and is harbored on plasmid pBS226 (BRL/Life Technologies), the 4XGal4-thymidine kinase-luciferase reporter (14), the 5XGal-myelomonocytic growth factor-luciferase reporter (4), and the 4XGal-c-fos promoter-luciferase and 5XGal-retinoic acid receptor (RAR) promoter-luciferase reporters (7) have been previously described. The 2XGal2XLex-e1bTATA-luciferase reporter was constructed by removing the five Gal4 binding sites from the 5XGal-e1bTATA-luciferase reporter and then adding two Gal4 DNA-binding sites (CGGAGTACTGTCCTCCG) and two LexA DNA-binding sites (CTGTATATATATACAG). The Gal4 fusions to E2F1 derivatives (18), VP16 (38), MyoD (52), CREB-binding protein (CBP) (10), PGC-1 (45), and p300 (55) have been described. Other Gal4-based activators were constructed by cloning PCR products into pSG424. Plasmid pSG5 expressing wild-type hTBP has been described previously (30).
Cell lines.
CHO14-1-2, CHO14-1-18, and CHO14-1-19, which were obtained from Brian Sauer, are neomycin-sensitive cell lines that each harbor a single Lox site for integration of plasmids at a defined position within the mammalian genome (19). These cells were maintained in minimal essential medium alpha with nucleotides, 10% fetal bovine serum, penicillin, and streptomycin. To construct cell lines harboring the different reporter plasmids, cells were electroporated with 10 μg of Cre recombinase expressing plasmid and 10 μg of the reporter plasmid at 450 V at 500 μF as described elsewhere (19). Cells were selected 2 days after plating in the above medium plus Geneticin (400 μg/ml; Life Technologies). Independent clones were expanded and checked for a proper single-copy integration event by Southern blotting. The CHO2-219, CHO18-219, and CHO19-219 cell lines were constructed in this manner and contain the 2XGal2XLex-luciferase reporter gene integrated into the CHO14-1-2, CHO14-1-18, and CHO14-1-19 parental cell lines, respectively.
Transcriptional analysis.
The CHO cell lines were seeded at a density of 105 cells/well in 12-well plates (1.75-cm-diameter wells) 18 to 24 h before the transfection. Cells (40 to 50% confluent) were transfected with 0.5 or 0.6 μg of DNA/well in the presence of 1.5 μl of FuGENE6 (Boehringer Mannheim) per well according to the manufacturer's instructions. The transfected cells were harvested 48 h after the addition of DNA by washing the cells twice with 1× phosphate-buffered saline and then adding 100 μl of 1× reporter lysis buffer (Promega); 100 μl of luciferase reagent (Promega) was added to 20 μl of extract, and then light units were immediately measured in a Turner luminometer using a 2-s delay and 15-s integration. Each data point is the average of duplicate transfections. Individual experiments are shown, but similar results were obtained at least twice. The error for each determination in the figures is approximately ±25%.
Cell sorting.
The CHO19-219 cell line was seeded at a density of 3 × 106 cells/10-cm-diameter dish 18 h prior to transfection. Each plate of cells (40 to 50% confluent) was transfected with 7.5 μg of the LexA fusion and 7.5 μg of the green fluorescent protein expression plasmid pGREENLANTERN (BRL/Life Technologies) in the presence of 60 μl of LipofectAMINE (BRL/Life Technologies) under serum-free conditions in OptiMEM (BRL/Life Technologies). This transfection solution was removed after 4 h and replaced with growth medium without antibiotics. Cells were harvested 48 h after transfection by trypsinization and separated by fluorescence-activated cell sorting on a MoFlo instrument (Cytomation) at a cutoff where less than 0.5% of nontransfected cells displays fluorescence. A total of 5 × 105 cells were isolated for each LexA fusion, and luciferase values were determined as above.
Western blotting.
CHO14-1-2 cells were transiently transfected as above with plasmids containing the fusion gene only. Protein extracts were made from cells 24 h after transfection by washing cells once in 1× phosphate-buffered saline, adding 150 μl of sodium dodecyl sulfate sample buffer, and then boiling the extract. Twenty microliters of each sample was resolved by sodium dodecyl sulfate-gel electrophoresis on 10% gels and blotted to nitrocellulose at 30 V for 15 h. The blots were probed with a polyclonal anti-LexA antiserum (Upstate Biotechnology) used at a 1:1,000 dilution in blocking solution (5% Carnation nonfat milk, 0.2% Triton X-100, 25 mM Tris-Cl [pH 7.4], 137 mM NaCl, 2.7 mM KCl), and then treated with a peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibody diluted to 1:10,000 in blocking solution. The antisera were detected using Luminol reagent (Santa Cruz Biotechnology).
Coimmunoprecipitation experiments.
CHO14-1-2 cells were seeded at a density of 3 × 106 cells/10-cm-diameter dish 18 h before transfection. Cells were transfected with 10 μg of plasmid DNA expressing LexA-Srb7 or LexA and with 0.06 ml of LipofectAMINE per plate (Gibco/BRL). At 24 h after transfection, cells were harvested in immunoprecipitation buffer (50 mM Tris-acetate [pH 7.9], 2 mM EDTA, 150 mM KCl, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2.5 μg each of pepstatin A, leupeptin, and aprotinin per ml) and then sonicated. The resulting extract was incubated with 3 μg of anti-LexA antiserum (Upstate Biotechnology) overnight at 4°C with rotation. Antibody complexes were collected after a 2-h incubation with 35 μl of a 50% slurry of Ultralink Protein A/G (Pierce) at 4°C, followed by centrifugation and four washes in immunoprecipitation buffer. Immunoprecipitates were analyzed by Western blotting as described above except that anti-Med7 antiserum (a gift from Jeffrey Parvin) was used at a 1:500 dilution. Under these conditions, approximately 30% of the LexA derivatives are immunoprecipitated and approximately 10% of the cellular Med7 is coimmunoprecipitated with LexA-Srb7.
Chromatin immunoprecipitation.
CHO19-219 cells were seeded 18 h before transfection at a density of 6.75 × 106 cells/15-cm-diameter plate. Two plates of cells for each fusion were transfected with 22.5 μg of LexA-TBP or LexA-Srb7 expression plasmid and 0.135 ml of LipofectAMINE per plate. Chromatin immunoprecipitation was performed by standard methods (6, 13, 42, 51), with the following modifications. Briefly, cells were removed from the incubator 24 h after transfection and fixed for 10 min by adding formaldehyde directly to the growth medium to a final concentration of 1%. Cell extracts were sonicated until the DNA fragment length ranged from 500 to 1,000 bp. One-third of the cell sonicate was diluted 10-fold in lysis buffer (13), and 2 μg of anti-LexA antiserum was added. After incubation with rotation at room temperature for 3 h, 50 μl of a 50% slurry of Ultralink Protein A/G beads (Pierce) was added, and the mixture was incubated for another 2 h at room temperature with rotation. Beads were harvested by a brief centrifugation and washed, and the immunoprecipitated material was eluted as described previously (34). The formaldehyde cross-links were reversed by heating at 65°C for 5 h, and DNA was detected by PCR using primers that amplify fragments that contain the LexA operator or sequences within the neomycin resistance gene. PCR was performed in the presence of [32P]dATP-dATP for 30 cycles of 1 min each at 94, 50, and 72°C, using Taq polymerase (Boehringer) and TaqStart anti-Taq antibody (Clontech) for hot-start PCR according to the manufacturer's protocol. PCR products were separated by 6% polyacrylamide gel electrophoresis and quantitated by PhosphorImager analysis. Assays of serial dilutions of immunoprecipitated and control DNA samples indicate that the intensity of the PCR products is directly related to the amount of DNA and hence that the assays represent quantitative measurements of promoter association in vivo.
RESULTS
Artificial recruitment on transiently transfected reporters.
We fused various components of the human Pol II machinery to the Gal4 DNA-binding domain (residues 1 to 147) or intact LexA (Fig. 1A). These components include TBP, TFIIB, Srb7, Med6, Med7, Trap80, Trap100, and several TAFs (TAF18, -20, and -28, which are homologous to yeast TAF19, -61, and -40, respectively). In addition, we examined the wild-type and kinase-inactive (D173A) version of human CDK8, the homolog of yeast Srb10. Although artificial recruitment of Srb10 represses transcription due to premature phosphorylation of the C-terminal tail of Pol II, the kinase-deficient mutant activates transcription in yeast by analogy with other components of the Pol II holoenzyme (24).
FIG. 1.
Gene fusions and reporter genes used in this study. (A) Fusions of the Pol II machinery components to the heterologous DNA-binding domains of LexA or Gal4 (hatched box). (B) Activation domain fusions. (C) Reporter plasmids. The distance between the distal end of the Gal4 or LexA DNA-binding sequence and the TATA sequence is shown.
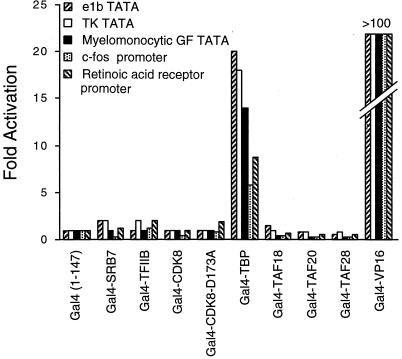
Initially, plasmids expressing Gal4 fusion proteins and reporter plasmids containing four or five Gal4 DNA-binding sites upstream of a TATA element and luciferase structural gene were transiently cotransfected into CHO cells. To address the possibility of promoter specificity, we examined reporters that contain Gal4 binding sites immediately (13 to 19 bp) upstream of TATA elements from the adenovirus e1b, herpesvirus thymidine kinase, and human myelomonocytic growth factor genes. Additionally, we tested regions of natural promoters originating from the c-fos or RAR gene in which Gal4 DNA-binding sites were introduced 56 or 36 bp upstream of the TATA elements (Fig. 1C). Consistent with earlier reports (23, 39, 41), the Gal4-TBP fusion activates transcription from all promoters tested (Fig. 2). Activation of the c-fos and RAR reporters (6- to 9-fold) was slightly less efficient than activation of the other reporters (15- to 20-fold), perhaps due to increased spacing between the Gal4 binding sites and TATA elements. In all cases, activation by Gal4-TBP was considerably less robust than the extremely strong Gal4-VP16 activator.
FIG. 2.
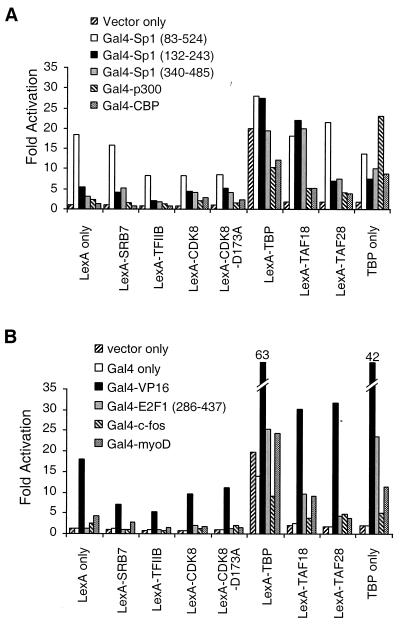
Artificial recruitment assay involving Gal4 fusions to components of the Pol II machinery. CHO14-1-19 cells were transiently transfected with the indicated Gal4 fusion and reporter plasmids and assayed for luciferase activity. Fold activation represents the increase of transcription of the Gal4 fusion compared to the Gal4 DNA-binding domain alone.
Surprisingly, no other Gal4 fusion to a Pol II machinery component activates transcription from any of these reporters (Fig. 2). Addition of trichostatin A, an inhibitor of histone deacetylases (54), does not permit any of these Gal4 fusions to activate transcription from the reporter with the e1b TATA element (data not shown). Thus, deacetylated histones are not solely responsible for the failure of the Pol II machinery components other than TBP to activate transcription when artificially recruited to promoters.
Artificial recruitment on chromosomally integrated reporters.
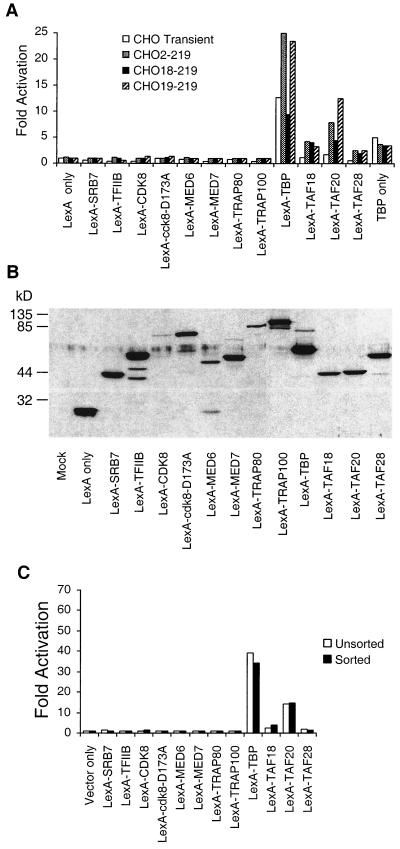
Transiently transfected reporter genes are present in many copies per cell and have chromatin structures that are distinct from those of normal chromosomal genes. To analyze transcriptional activation under more physiological conditions, we used the Cre/Lox recombination system to construct CHO cell lines that harbor a reporter gene at a single chromosomal locus per cell (19). Specifically, we cotransfected a plasmid expressing Cre recombinase along with luciferase reporter plasmids containing a Lox site into three CHO cell lines that have a single chromosomal Lox site. By selecting for neomycin-resistant clones and using Southern blot analysis to confirm the desired recombination event, we obtained CHO cell lines with two Gal4 and two LexA sites upstream of the e1b TATA element and luciferase reporter gene (2XGal2XLex-Luc [Fig. 1]) at the three chromosomal positions defined by the location of the Lox sites in the parental cell lines.
Interestingly, although LexA fusions to Pol II holoenzyme components fail to activate transcription, LexA fusions to TAFs activate transcription from the chromosomal 2XGal2XLex reporter (Fig. 3A). The level of activation is approximately three- to eightfold depending on the TAF tested. Overexpression of equivalent amounts of a plasmid expressing TBP alone activates transcription three- to fivefold, although TBP levels are about fivefold higher than the LexA-TBP protein levels, as judged by immunoblot analysis using antibodies to human TBP (data not shown). Experiments where TBP levels are comparable to those of LexA-TBP result in no activation, demonstrating that activation by LexA-TBP occurs by artificial recruitment. Activation is not significantly affected by the chromosomal location of the reporter, which differs in each cell line, because comparable results were obtained in the three cell lines using conventional activators (data not shown).
FIG. 3.
Artificial recruitment assay involving LexA fusions using chromosomally integrated or transiently transfected reporters. (A) For transiently transfected reporters (unfilled boxes), CHO14-1-19 cells were transfected with equal amounts of the indicated LexA fusion and the 2XGal2XLex-e1bTATA-luciferase reporter plasmid. Otherwise, cells that harbor a single chromosomal copy of the 2XGal2XLex-e1bTATA-luciferase reporter, CHO2-219, CHO18-219, or CHO19-219 (hatched or filled boxes), were transiently transfected with LexA fusion plasmids. Fold activation is the difference in the luciferase activity compared to cells transfected with the reporter plasmid only (transient reporter) or with an empty vector only (integrated reporter). (B) Immunoblot analysis of CHO14-1-19 cells transiently transfected with the indicated LexA fusion plasmids using LexA-specific antisera. (C) CHO19-219 cells were transfected with plasmids expressing the indicated LexA fusions and green fluorescent protein, and luciferase activity was monitored in cells that were unsorted (filled box) or were sorted (black box) by fluorescence activation. The transfection efficiency was estimated to be at least 80%.
The failure of the LexA hybrid proteins to activate transcription is not due simply to failure to express the fusion protein. Immunoblot analysis using LexA antibodies indicate that LexA fusion proteins are comparably expressed (except for the CDK8 fusion) in the transfected cells (Fig. 3B). To avoid the contribution from the chromosomal reporter in nontransfected cells, we cotransfected cells with plasmids expressing the LexA fusion of interest and the green fluorescent protein and then purified the transfected cells by fluorescence-activated cell sorting. Under the conditions used, at least 80% of the cells were transfected (data not shown), and the sorted and unsorted activation values are not significantly different (Fig. 3C). Taken together, these results indicate that artificial recruitment of TFIID, but not components of the mammalian Pol II holoenzyme, results in transcriptional activation in mammalian cells.
Synergy experiments.
To examine whether the artificial recruitment constructs could synergistically active transcription, most possible combinations of LexA and Gal4 hybrid proteins were examined on the transiently transfected or chromosomally integrated 2XGal2XLex reporter (fusions to Med6, Med7, Trap80, and Trap100 were not tested). Excluding combinations containing either Gal4-TBP or LexA-TBP, no other combination of fusions activates transcription more than twofold (data not shown). Although Gal4-TBP and LexA-TBP can independently activate transcription, the combination of these proteins results in luciferase levels that are at best additive.
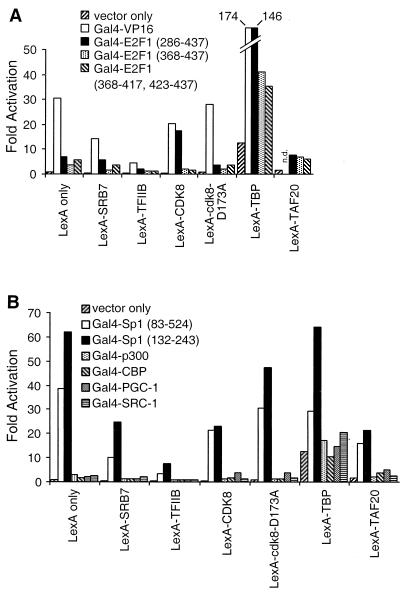
Next, we asked whether Gal4 fusions to natural activation domains would synergistically activate transcription in combination with LexA fusions to components of the Pol II machinery. When assayed on the transiently transfected 2XGal2XLex reporter, LexA fusions to components other than TBP are incapable of synergizing with any activation domain tested, and the TFIIB fusion actually decreases the level of transcription (Fig. 4). However, LexA-TBP synergistically activates transcription in combination with Gal4-VP16 and various derivatives of Gal4-E2F1 (Fig. 4A), both of which contain acidic activation domains. Synergistic activation with these Gal4 activators is not completely dependent on artificial recruitment of TBP; synergy is also observed with unfused TBP, although higher concentrations of TBP than of comparison to LexA-TBP are required. A titration analysis comparing TBP alone to LexA-TBP showed that comparable synergy can be detected for the LexA-TBP fusion at protein levels that are 25-fold lower than the level of TBP alone (data not shown). Synergistic activation between VP16 and directly recruited TBP has been observed previously (23). In contrast to the synergy observed with acidic activators, LexA-TBP does not synergize with various versions of Gal4-Sp1, which contains a glutamine-rich activation domain, or with Gal4 fusions to p300 or CBP, which are coactivators with histone acetylase activity (Fig. 4B).
FIG. 4.
Synergy between artificial and natural activators on a transiently transfected promoter. (A) Analysis of Gal4 fusions to the acidic activation domains from VP16 or E2F1. n.d., not determined. (B) Analysis of Gal4 fusions to the glutamine-rich activation domain of Sp1 or the histone acetylase CBP or p300. CHO14-1-19 cells were transiently transfected with the 2XGal2XLex-e1bTATA-luciferase reporter and the indicated Gal4 and LexA fusion plasmids. Fold activation is the difference in the luciferase activity compared to cells transfected with the reporter plasmid only.
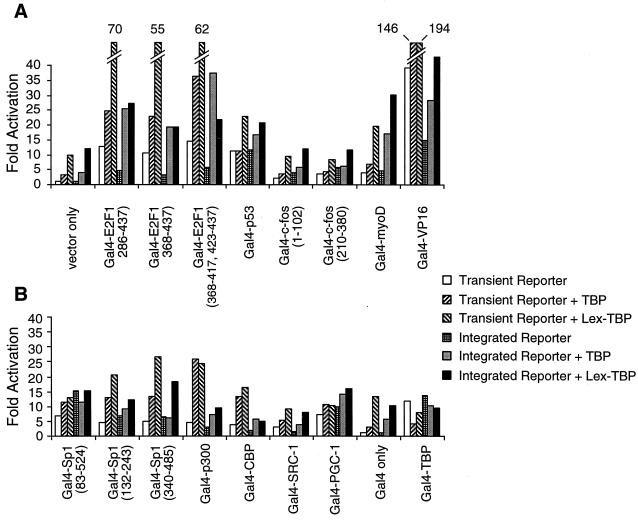
Last, we examined most of the above combinations for synergistic activation in the context of a chromosomal reporter gene (Fig. 5). In accord with results on transiently transfected reporters, LexA-TBP could synergize with Gal4-VP16 activator in CHO19-219 cells (the increases in transcription observed in combination with other activators are additive). Synergy with unfused TBP is observed with VP16, E2F1, and p300. In addition, LexA-TAF18 and LexA-TAF28 appear to synergize with Gal4-VP16, and LexA-TAF18 synergizes with two truncated derivatives of Gal4-Sp1 (132-243 and 340-485). Thus, as is the case with activation per se, synergy that occurs with artificially recruited TAFs is observed only on chromosomal reporters.
FIG. 5.
Synergy between artificial and natural activators on a chromosomally integrated promoter. CHO19-219 cells were transiently transfected with plasmids expressing the indicated LexA fusions and Gal4 fusions. (A) Analysis of Sp1 activation domains or histone acetylases. (B) Analysis of acidic activation domains. Fold activation is the difference in the luciferase activity compared to cells transfected with an empty vector only.
LexA-Srb7 is associated with Pol II holoenzyme and is recruited to promoters in vivo.
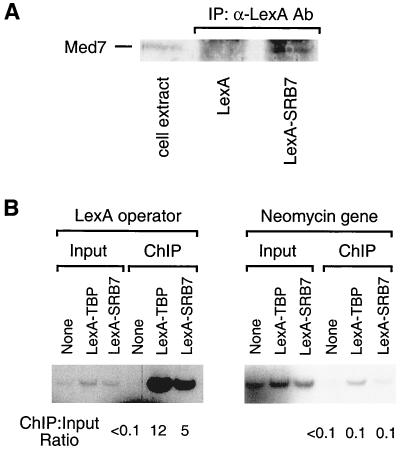
Given the unavailability of genetic complementation assays in mammalian cells, we performed two additional experiments to provide evidence that the LexA-Srb7 fusion is functionally active. First, we used coimmunoprecipitation to assess the ability of LexA-Srb7 to incorporate into the holoenzyme. Extracts from cells transfected with plasmids expressing either LexA alone or LexA-Srb7 were immunoprecipitated with LexA antibodies, and the resulting immunoprecipitates were analyzed for the presence of the holoenzyme component Med7. Med7 is present in immunoprecipitates from cells expressing LexA-Srb7 but not from cells expressing LexA alone (Fig. 6A), indicating that LexA-Srb7 is associated with the holoenzyme in vivo.
FIG. 6.
LexA-Srb7 is associated with Pol II holoenzyme and is recruited to the promoter with LexA operators in vivo. (A) Coimmunoprecipitation. LexA-containing complexes from cells expressing LexA or LexA-Srb7 were immunoprecipitated (IP) with LexA antibodies (Ab) and analyzed for the presence of Med7 by Western blotting. The left lane represents analysis of an extract from untransfected cells. (B) Chromatin immunoprecipitation. Cross-linked chromatin from cells containing a chromosomally integrated reporter and expressing the indicated LexA derivative was (ChIP) or was not (input) immunoprecipitated with LexA antibodies, and the resulting material was analyzed by PCR using primers that amplify the promoter region containing LexA operators or the neomycin resistance gene. The ratios shown were calculated by dividing the counts in the ChIP lanes by counts in the input lane for each sample.
Second, we analyzed occupancy of the LexA-Srb7 fusion at the promoter of the luciferase reporter gene by chromatin immunoprecipitation. Cells containing the integrated reporter, which contains two LexA operators, were transfected with plasmids expressing LexA-TBP or LexA-Srb7 and treated with formaldehyde to cross-link proteins to DNA; the resulting chromatin preparations were immunoprecipitated with LexA antibodies. As shown in Fig. 6B, LexA-Srb7 associates with the promoter at levels roughly comparable to that of LexA-TBP, whereas neither protein shows significant occupancy within the coding region of the neomycin resistance gene, which does not contain LexA binding sites. As defined by the ratio of the immunoprecipitated to input DNA samples, specific binding of the LexA fusion proteins to the reporter promoter is 50- to 100-fold higher than the presumed nonspecific binding to the neomycin resistance gene. Taken together, the coimmunoprecipitation and chromatin immunoprecipitation experiments strongly suggest that the LexA-Srb7 fusion recruits the holoenzyme to the reporter gene promoter.
Integrated versus transiently transfected reporters.
A noteworthy finding from the above experiments is that transcriptional activators can function with different levels of efficiency on integrated and transiently transfected reporters (Fig. 7). Some acidic activation domains (VP16 and E2F1) have lower activation ability from integrated reporter genes. In striking contrast, artificial recruitment of TBP and many other activation domains [e.g., Sp1(83-524) or p53] results in comparable levels of transcription from integrated or transient reporters. Also, artificial recruitment of the TAFs activates only transcription from integrated reporters (Fig. 3A). Addition of trichostatin A did not increase the relative activation by activation domains from integrated reporters, suggesting the histone acetylation status alone of these reporters is unlikely to account for the discrepancies in activation levels. However, activation from integrated promoters was increased upon cotransfection of TBP or LexA-TBP for many activators (Fig. 7), suggesting that TBP is limiting for transcription in these cases. Similar results were obtained using the other two CHO cell lines that harbor chromosomal reporter genes (data not shown), suggesting that this effect is not limited to a single genomic locus.
FIG. 7.
Comparison of a transiently transfected and chromosomally integrated reporter gene. For the transiently transfected reporter, CHO14-1-2 cells were transfected with the indicated Gal4 fusion and the 2XGal2XLex-e1bTATA-luciferase reporter plasmid in the absence (unfilled boxes) or presence of plasmids expressing TBP or LexA-TBP (hatched boxes). For the chromosomally integrated reporter, CHO19-219 cells were transfected with the indicated Gal4 fusion plasmids in the absence (unfilled bars) or presence of a TBP expression plasmid (gray boxes) or LexA-TBP expression plasmid (black boxes). Fold activation is the difference in the luciferase activity compared to cells transfected with the reporter plasmid only (transient reporter) or with an empty vector only (integrated reporter).
DISCUSSION
Components of TFIID, but not Pol II holoenzyme, activate transcription by artificial recruitment in mammalian cells.
In accord with experiments in yeast, we show that in all four cases tested, artificial recruitment of TFIID components activates transcription in mammalian cells. In striking contrast, seven different components of the Pol II machinery other than TFIID fail to activate transcription when artificially recruited, even though these holoenzyme fusions are expressed to comparable levels as the activation-competent TFIID fusions.
The clear dichotomy between TFIID and holoenzyme components almost certainly reflects an inherent property of transcriptional activation in mammalian cells and is extremely unlikely to arise for trivial reasons. While any individual hybrid protein might be functionally inactive due to the fusion of the DNA-binding domain, it is extremely unlikely that this is the case for all seven holoenzyme components but not for all four TFIID components tested. Although genetic complementation assays are unavailable to determine whether the hybrid proteins are functional in mammalian cells, there is no basis or plausible explanation for why TFIID or holoenzyme components should differ with respect to the probability of being inactivated by fusion of a DNA-binding domain. In this regard, most LexA and Gal4 DNA-binding domain fusions to yeast proteins are functional in genetic complementation assays, and there is no pattern for which kinds of proteins are nonfunctional. Of specific relevance, comparable LexA and Gal4 fusions to yeast TFIID and holoenzyme components are functionally comparable to the wild-type protein, and they activate transcription in artificial recruitment assays (2, 5, 12, 17, 20, 21, 31, 33, 36, 47, 53). It is inconceivable that trivial reasons could account for why seven different fusions to mammalian holoenzyme components could be nonfunctional, whereas numerous comparable fusions to yeast holoenzyme components are functional. Finally, in the one case tested, LexA-Srb7 associates with the Pol II holoenzyme and in vivo, and occupancy of the chromosomally integrated reporter in vivo is comparable to that of the transcriptionally competent LexA-TBP. Thus, the failure of mammalian holoenzyme components to activate transcription when artificially recruited is not a negative result but rather indicates a difference between transcription in yeast and mammalian cells.
Our results appear to conflict with a report concluding that in transiently transfected mammalian cells, artificial recruitment of components other than TBP can weakly activate transcription and can function synergistically with classical activation domains (41). However, the two human components tested (hSrb7 and hTFIIB) behaved qualitatively and quantitatively similarly to four different yeast components (Srb2, Srb6, Srb7, and Srb11). As yeast and human Srb7 are not functionally interchangeable in yeast cells (11), and as the other yeast components have limited sequence similarity with mammalian proteins, it is unlikely that these yeast components function in combination with mammalian components or assemble into the mammalian Pol II holoenzyme. Thus, the observed transcriptional effects of the yeast components in mammalian cells are almost certainly not due to artificial recruitment of the mammalian transcription machinery, thereby making it difficult to interpret the similar transcriptional effects of hSrb7 and hTFIIB. It should also be noted that the unfused DNA-binding domain detectably activated transcription, and that the fusions were only slightly (two- to threefold) more active (41).
Activation domains can differ in the ability to stimulate transcription of transiently transfected or chromosomally integrated reporter genes.
Although analyses of enhancers from natural genes has revealed differences between transiently transfected and chromosomally integrated reporters (1, 3), our study represents the first systematic comparison of transcriptional activation domain function on the same promoter. Unexpectedly, the VP16 and E2F1 activation domains are less effective on integrated reporters compared to transiently transfected reporters. In contrast, the full-length Sp1 activation domain and artificially recruited TFIID components function comparably or better on integrated reporters.
Synergistic activation in combination with excess TBP or with artificially recruited TBP has been observed with VP16 and E1A but not with Sp1, suggesting that Sp1 activates transcription by recruiting TBP (TFIID), whereas VP16 and E1A function at a step(s) subsequent to TFIID recruitment (23, 39). Our observation that Sp1 and artificially recruited TBP both have equivalent activities on integrated and transiently transfected reporters provides additional evidence that Sp1 functions by recruiting TFIID to the promoter. Conversely, the E2F1 and VP16 activation domains show the most pronounced difference between integrated and transfected reporters (i.e., behave differently than artificially recruited TBP), providing further support for the idea that they function primarily by recruiting components other than TFIID to promoters.
Possible explanations for the difference between yeast and mammalian cells.
As defined by artificial recruitment experiments, our results indicate that yeast and mammalian cells differ with respect to transcriptional activation. Specifically, on standard TATA-containing promoters, artificial recruitment of Pol II holoenzyme components activates transcription in yeast cells but not in mammalian cells. Moreover, artificial recruitment of yeast Pol II holoenzyme components often leads to higher levels of transcriptional activation than artificial recruitment of TBP (20).
There are several explanations, not mutually exclusive, for why artificial recruitment of components of the Pol II holoenzyme fails to activate transcription in mammalian cells. First, if recruitment of TFIID were the sole limiting step for transcription, artificial recruitment of other components would not overcome this step and hence would not activate transcription. Second, recruitment of the mammalian Pol II holoenzyme might require the activator to contact multiple targets; in this view, artificial recruitment would fail because only one target was contacted by the enhancer-binding protein. In this regard, activation by artificial recruitment in yeast is strongly influenced by promoter architecture, whereas natural activators have a much broader spectrum of activity (20). Third, the physical connection between the DNA-binding domain and the component of the Pol II machinery might cause structural constraints that preclude both moieties of the fusion protein from functioning at the same time. It is unclear, however, why such structural constraints would occur in mammalian cells but not in yeast cells and would be limited to holoenzyme components but not TFIID components. Fourth, transcriptional activation in mammalian cells might involve a step that occurs after recruitment of Pol II holoenzyme, such that artificial recruitment of Pol II holoenzyme is insufficient for activation.
It is important to note that the artificial recruitment experiments performed here do not address whether Pol II holoenzyme components are targets of natural activators. The multiple-contact and postrecruitment explanations above are based on the idea that Pol II holoenzyme components are physiologically relevant targets. More generally, artificial recruitment constructs are subject to more functional restrictions than natural activation domains (20). Thus, it is difficult to simply extrapolate from the results presented here to activation mechanisms employed by natural activation domains.
The observations presented here are analogous to results in yeast cells involving promoters with defective TATA elements, in which activation occurs upon artificial recruitment of TBP or TAFs but not TFIIB (12, 21, 33). As the critical limitation for transcription from yeast TATA-defective promoters is undoubtedly the association of TFIID, it is not surprising that this limitation can be bypassed by artificial recruitment of TFIID but not Pol II holoenzyme. The unexpected similarity between standard mammalian promoters and yeast TATA-defective promoters prompts the speculation that recruitment of TFIID might be more limiting in mammalian cells than in yeast cells. Aside from explaining the difference between yeast and mammals in artificial recruitment assays, this hypothesis is consistent with results of TBP overexpression experiments. In mammalian cells, the level of transcriptional activation and/or synergy can be increased by overexpression of TBP (15, 23, 39), an observation we have confirmed here. In contrast, there is no evidence that overexpression of TBP increases transcription in yeast cells, and strong acidic activators can stimulate transcription to the maximal level at physiological levels of TBP (26, 34). Although genetic and direct TBP occupancy experiments indicate that association of TBP is limiting at the vast majority of yeast promoters in vivo, TBP association in vivo also depends on Pol II holoenzyme (34, 37). Thus, it is difficult to assess whether the primary limiting factor in yeast is TFIID recruitment per se or recruitment of Pol II holoenzyme which then stabilizes TFIID at the promoter. The above speculation does not imply that yeast and mammalian cells have fundamentally different mechanisms of transcriptional activation but rather suggests the possibility that the relative importance of individual steps in the process might differ.
ACKNOWLEDGMENTS
We thank Brian Sauer for the CHO cell lines crucial for this work, Jeffrey Parvin for antibodies to Med7, and Don Ayer, Irvin Davidson, Robert Eisenman, Ron Evans, William Kaelin, Jr., Marie Keaveney, Andrew Lassar, Mark Ptashne, Doug Spicer, and Bruce Spiegelman for plasmids.
This work was supported by a postdoctoral fellowship to D.R.D. and research grants GM30186 and CA65965 to K.S. from the National Institutes of Health.
REFERENCES
- 1.Alberts A, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 3.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 4.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mad proteins contain a dominant transcriptional repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 6.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 8.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 9.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 11.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee S, Struhl K. Connecting a promoter-bound protein to the TATA-binding protein overrides the need for a transcriptional activation region. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 14.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 15.Colgan J, Manley J L. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 1992;6:304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- 16.Donoviel D B, Shield M A, Bushkin J N, Haugen H S, Clegg C H, Hauschka S D. Analysis of muscle creatine kinase gene regulatory elements in skeletal and cardiac muscles of transgenic mice. Mol Cell Biol. 1996;16:1649–1658. doi: 10.1128/mcb.16.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrell S, Simkovich N, Wu Y B, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 18.Flemington E K, Speck S H, Kaelin W G. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushige S, Sauer B. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci USA. 1992;89:7905–7909. doi: 10.1073/pnas.89.17.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant G O, Struhl K, Ptashne M. Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Couto E, Klages N, Strubin M. Synergistic and promoter-selective activation of transcription by recruitment of TFIID and TFIIB. Proc Natl Acad Sci USA. 1997;94:8036–8041. doi: 10.1073/pnas.94.15.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 23.He S, Weintraub S J. Stepwise recruitment of components of the preinitiation complex by upstream activators in vivo. Mol Cell Biol. 1998;18:2876–2883. doi: 10.1128/mcb.18.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengartner C J, Myer V E, Liao S-M, Wilson C J, Koh S S, Young R A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 25.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 26.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong S, Stein A. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y W, Stillman D J. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalderon D, Richardson W D, Markham A F, Smith A E. Sequence requirements for nuclear location of simian virus 40 large T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 30.Keaveney M, Berkenstam A, Feigenbutz M, Vriend G, Stunnenberg H G. Residues in the TATA-binding protein required to mediate a transcriptional response to retinoic acid in EC cells. Nature. 1993;365:562–566. doi: 10.1038/365562a0. [DOI] [PubMed] [Google Scholar]
- 31.Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 32.Kingston R E. A shared but complex bridge. Nature. 1999;399:199–200. doi: 10.1038/20302. [DOI] [PubMed] [Google Scholar]
- 33.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 34.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;389:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 35.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee M, Struhl K. A severely defective TATA-binding protein-TFIIB interaction does not preclude transcriptional activation in vivo. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X-L, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;389:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 38.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 39.Majello B, Napolitano G, DeLuca P, Lania L. Recruitment of human TBP selectively activates RNA polymerase TATA-dependent promoters. J Biol Chem. 1998;273:16509–16515. doi: 10.1074/jbc.273.26.16509. [DOI] [PubMed] [Google Scholar]
- 40.Myer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 41.Nevado J, Gaudreau L, Adam M, Ptashne M. Transcriptional activation by artificial recruitment in mammalian cells. Proc Natl Acad Sci USA. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 43.Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 44.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 45.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman B M. Activation of PPARg coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 46.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stargell L A, Moqtaderi Z, Dorris D R, Ogg R C, Struhl K. TFIIA has activator-dependent and core promoter functions in vivo. J Biol Chem. 2000;275:12374–12380. doi: 10.1074/jbc.275.17.12374. [DOI] [PubMed] [Google Scholar]
- 48.Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987;49:295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- 49.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 50.Struhl K. Fundamentally different logic of gene expression in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 51.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-b enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 52.Weintraub H, Dwarki V J, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott S J. Muscle-specific transcriptional activation by myoD. Genes Dev. 1991;5:1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- 53.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida M, Kijima M, Akits M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 55.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor myoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]