Abstract
Assembly of enhanceosomes requires architectural proteins to facilitate the DNA conformational changes accompanying cooperative binding of activators to a regulatory sequence. The architectural protein HMG-1 has been proposed to bind DNA in a sequence-independent manner, yet, paradoxically, it facilitates specific DNA binding reactions in vitro. To investigate the mechanism of specificity we explored the effect of HMG-1 on binding of the Epstein-Barr virus activator ZEBRA to a natural responsive promoter in vitro. DNase I footprinting, mutagenesis, and electrophoretic mobility shift assay reveal that HMG-1 binds cooperatively with ZEBRA to a specific DNA sequence between two adjacent ZEBRA recognition sites. This binding requires a strict alignment between two adjacent ZEBRA sites and both HMG boxes of HMG-1. Our study provides the first demonstration of sequence-dependent binding by a nonspecific HMG-box protein. We hypothesize how a ubiquitous, nonspecific architectural protein can function in a specific context through the use of rudimentary sequence recognition coupled with cooperativity. The observation that an abundant architectural protein can bind DNA cooperatively and specifically has implications towards understanding HMG-1's role in mediating DNA transactions in a variety of enzymological systems.
An emerging theme in eukaryotic gene expression is that promoter- and cell-specific transcription is achieved through regulated assembly of activators into nucleoprotein structures termed enhanceosomes (6, 22, 37). Enhanceosome assembly is mediated by cooperative protein-protein interactions dictated by the positioning of activator binding sites on a regulatory sequence and the concentration of relevant activators in a cell (6, 43). Because interactions between activators generate energetically unfavorable DNA bends, architectural proteins that bend and twist the DNA are necessary to facilitate cooperative binding. An important issue in the field is how such flexure can be provided on a global level for the thousands of combinatorial activator arrays bound to genes in a eukaryotic nucleus (6, 22, 32, 37, 38, 42).
Both sequence-specific and nonspecific DNA architectural proteins have been identified. The largest family of eukaryotic architectural proteins contains the conserved HMG box, a 75-amino-acid sequence of known structure. Numerous examples exist of HMG-box proteins that bind DNA either specifically (e.g., LEF-1) or nonspecifically (e.g., HMG-1 and -2) (4). The function and mechanism of some sequence-specific architectural proteins have been established, while the nonspecific proteins have remained enigmatic. In this paper we examine how the abundant and relatively nonspecific HMG-1 and -2 proteins can function in a specific context.
To provide a framework for the problem, consider the action of LEF-1 on the T-cell receptor alpha (TCR-α) enhanceosome. LEF-1 or TCF-1 binds to an 8-bp sequence within the 75-bp TCR-α enhancer, bends the DNA, and stimulates cooperative binding of the flanking activators, PEBP2α–Ets-1 and ATF-CREB (18). LEF-1 binds the DNA using three closely packed alpha-helices, which constitute the L-shaped HMG domain. The HMG domain intercalates a methionine between 2 bp in the minor groove of the recognition site, rolling the base pairs and widening the minor groove. This effect in turn generates a 90° bend towards the major groove (36). A central feature of LEF-1 is its ability to bind DNA specifically and independently to generate complexes sensitive to mutagenesis and identifiable by DNase I footprinting or electrophoretic mobility shift assay (EMSA).
By the same criterion HMG-1 and -2 are considered to bind DNA nonspecifically, yet, paradoxically, they facilitate specific DNA interactions by other proteins. Examples include the binding of several sequence-specific transcription factors (steroid receptors, Hox proteins, and p53), recombination by RAG-1 and -2 of the VDJ junctions, and integration by human immunodeficiency virus type 1 integrase (3, 12, 29, 32, 38, 39, 41, 51, 60, 62). HMG-1 and -2 contain two HMG DNA binding motifs, termed box A and box B. The individual boxes have been generated in recombinant form and studied. Both boxes fold (44), bind, and bend DNA (42, 46, 47), although they possess different DNA affinities and bending potentials (47). Intact HMG-1 is believed to bind a 15- to 18-bp region of DNA (46).
The lack of sequence-specific binding by HMG-1 is perplexing. The solution structures of HMG boxes A and B reveal a domain comprising three alpha-helices folded in the shape of an L, remarkably similar to the domain of LEF-1 (24, 45, 57). A 2.5-Å crystal structure of the HMG-1 box A complexed to cisplatin-modified DNA reveals that DNA binding by HMG-1 shares many features with that of LEF-1 (40). Cisplatin is an antitumor drug that binds in the major groove and induces an intrastrand cross-link, resulting in a bend towards the major groove. Box A recognizes the widened minor groove of cisplatinated DNA and induces an additional kink at the cross-link. A phenylalanine residue at position 37 intercalates into the hydrophobic notch created by the drug and the remainder of the protein engages in a series of contacts along the DNA 3′ to the adduct.
Structural studies indicate that nonspecific HMG-1 and -2 family members possess rudiments of sequence recognition. Nuclear magnetic resonance (NMR) analysis revealed that NHP6A, a Saccharomyces cerevisiae homologue of HMG-1, interacted with a DNA fragment in a specific fashion via partial intercalations of methionine and phenylalanine residues (2, 58). A recent crystal structure of HMG-D showed that it also bound to a specific AT-rich sequence in a manner involving partial intercalations of methionine and valine residues (31). In both cases the methionine intercalated into a centrally located pyrimidine-purine step, consistent with a previous binding site selection study with HMG-D (9). Moreover, in both cases the HMG domain structure closely resembled that of LEF-1.
How might such minimal specificity be employed to influence a specific binding reaction? We propose that HMG-1 exhibits a rudimentary sequence recognition capability and that cooperative binding with nearby activators stabilizes the binding of HMG-1. To address this hypothesis we have instituted an analysis of HMG-1 action in assembly of enhanceosomes over Epstein-Barr virus (EBV) lytic genes (14–16, 25, 34). Our previous study revealed that HMG-1 and -2 facilitated cooperative binding of ZEBRA to two pairs of sites in the viral BHLF-1 gene promoter, from positions −50 to −74 (Z-1 and Z-2) and from positions −106 to −146 (Z-3 and Z-4). BHLF-1 controls transcription of abundant early mRNAs, and we focused on it because of its potent promoter. We chose to study the distal set of sites, Z-3 and Z-4, because we previously observed a specific DNase I footprint between them that might be a binding site for HMG-1. This would represent the first example of sequence-dependent HMG-1 binding and would form a model for understanding other reactions where HMG-1 has been shown to influence DNA binding or catalytic activity by transcription factors and recombinases, respectively. We show that HMG-1 does indeed bind this sequence cooperatively and specifically. Efficient binding of HMG-1 requires a specific DNA sequence between Z-3 and Z-4, both boxes (A and B) of HMG-1, the ZEBRA DNA binding domain, both ZEBRA binding sites, and a precise alignment of the two sites.
MATERIALS AND METHODS
Generation of the 100-bp minimal BHLF-1 promoter and mutants.
PCR was employed to amplify a fragment from −990 to +90 of the EBV BHLF-1 promoter using pBamW2 YFSal G, which contains a segment of the B95-8 EBV genome spanning 40 to 61 kb (48). This fragment was subcloned into a mammalian CAT expression vector, generating HLCAT as previously described (13). A DNase I footprint between Z-3 and Z-4, from positions −116 to −128, was observed on this promoter when it was incubated with both ZEBRA and HMG-1. HLCAT was then used as a template to PCR-amplify a 97-bp fragment containing the third (Z-3) and fourth (Z-4) ZEBRA binding sites along with the region encompassing the footprint. The primers, HLZ3,4 up and HLZ3,4 down, contain SacI and PstI restriction sites, respectively. This fragment was subcloned, via SacI and PstI, into a reporter construct bearing the adenovirus E4 core promoter upstream of luciferase (30).
The 97-bp region was mutated by two-step PCR to generate a series of derivatives termed HMGΔ1-7. These mutants contain base pair substitutions in the region encompassing the HMG-1 footprint (HMGΔ1-5) or contain 5- or 10-bp insertions (HMGΔ6-7). The substituted or inserted sequences are underlined in each mutant primer set listed below. The first round of PCR was performed in two reactions, one using the original HLZ3,4 up (SacI) primer along with one of the mutant down primers, and the other using the original HLZ3,4 down (PstI) primer with one of the mutant up primers. The two PCR products were purified and employed in a second round of PCR with the original HLZ3,4 up (SacI) and HLZ3,4 down (PstI) primers to generate the final mutant fragment. The wild-type promoter and the mutants were then subcloned into the SacI and PstI sites of pGL3-E4 Lux, which contains the adenovirus E4T core promoter (30). The primer sets were as follows: HLZ3,4 up (SacI) (5′ GGGGAGCTCGAATAACCTCCAGGTACCACCC 3′) and HLZ3,4 down (PstI) (5′ GGGCTGCAGGTGGGGGCTTCTTATTGGTTAATTC 3′), HL HMG1 down (5′ CATTTTAGCCCCCCCCCTTTCATTAAGGTGTGTCACCAGGTGGG 3′) and HL HMG1 up (5′ CCTTAATGAAAGGGGGGGGGCTAAAATGACACACCTGAATTAACC 3′), HL HMG2 down (5′ GCCCGTTGGGCCCCCCCAAGGTGTGTCACCAGGTGGG 3′) and HL HMG2 up (5′ GGTGACACACCTTGGGGGGGCCCAACGGGCTAAAATGACACACCTG 3′), HL HMG3 down (5′ GCCCGTTCCCTTTCATTAAGGTGTGTCACCAGGTGGG 3′) and HL HMG3 up (5′ CCTTAATGAAAGGGAACGGGCTAAAATGACACACCTG 3′), HL HMG4 down (5′ GCCCGTTGGGCCCCATTAAGGTGTGTCACCAGGTGGG 3′) and HL HMG4 up (5′ CCTTAATGGGGCCCAACGGGCTAAAATGACACACCTG 3′), HL HMG5 down (5′ GCCCGTTGGGTTTCCCTAAGGTGTGTCACCAGGTGGGTGG 3′) and HL HMG5 up (5′ CACACCTTAGGGAAACCCAACGGGCTAAAATGACAC 3′), HL HMG6 down (5′ CATTTTAGCCCCCCCCGTTGGGTTTCATTAAGGTGTGTCACC 3′) and HL HMG6 up (5′ GAAACCCAACGGGGGGGGCTAAAATGACACACCTGAATTAACC 3′), and HL HMG7 down (5′ CATTTTAGCCCCCCCCCCCCCGTTGGGTTTCATTAAGGTGTGTCACCA 3′) and HL HMG7 up (5′ GAAACCCAACGGGGGGGGGGGGGCTAAAATGACACACCTGAATTAACC 3′).
DNase I footprinting.
DNase I footprinting with ZEBRA and HMG-1 was performed on the BHLF-1 promoter as previously described (8, 13). The HLZ3,4 down primer was 32P-labeled with T4 polynucleotide kinase and [γ-32P]ATP. The 97-bp template was generated by PCR using 32P-labeled HLZ3,4 down and unlabeled HLZ3,4 up primers. The radiolabeled promoter fragment was fractionated on a 12% native polyacrylamide gel and purified. Full-length ZEBRA and its DNA binding domain (Δ161) were purified as previously described (7). The 13-μl reaction mixtures contained 5 fmol of the 32P-end-labeled probe, a range of wild-type ZEBRA and Δ161 (from 0.6 to 200 ng) and 250 to 450 ng of HMG-1 in binding buffer containing 12.5 mM HEPES (pH 7.9), 60 mM KCl, 12.5% glycerol, 5 mM MgCl2, 0.2 mM EDTA, 60 mM β-mercaptoethanol, 0.5 mg of bovine serum albumin per ml, and 30 μg of poly(dGdC) per ml.
Cloning and purification of HMG-1: wild type, deletion derivatives, and the FLAG-tagged version.
Primers to the 215-amino-acid rat HMG-1 gene were generated. The amino-terminal primer contained an NcoI restriction site, and the carboxyl-terminal primer contained a BamHI site for subcloning. The primer sequences were as follows: for HMG-1(N-term), 5′ CCCCCATGGGCAAAGGAGATCCTAAGAAGCC 3′, and for HMG-1(C-term), 5′ CCCGGATCCTTATTCATCATCATCATCTTCT 3′. PCR was used to amplify the gene. The 650-bp PCR product was fractionated on and purified from a low-melting-point agarose gel and subcloned into the pET11d bacterial expression vector by using NcoI and BamHI. The HMG-1 gene was also inserted between the NdeI and BamHI sites of pET11a by cloning the product of a two-step PCR of the pET11d-HMG-1. This latter step was done to remove an internal NdeI site, creating pRJ1576. HMG-1 mutations were created by PCR of pRJ1576 using the primers listed below and then subcloned into pET11a. The following primers were used: HMG-1 Top (5′ GCGCGCGCATATGGGCAAAGGAGATCCTAAG 3′) (Met 1 at N terminus), HMG-1 Bot (5′ CGCGGATCCAGGAGTGAGTTGTGTACAGGGGGGTTA 3′) (Glu 215 at C terminus), HMG-1 Nde deletion (5′ CACAAAGAATGCGTATGAGGACATTTT 3′) (Ser 17-Tyr 18-Ala 19 silent mutation), HMG-1 Box A Bottom (5′ CGCGGATCCTTAGGGGGGGATGTAGGTTTT 3′) (Pro 81 at C terminus), HMG-1 Box A* Bottom (5′ CGCGGATCCTTACTTCTTTTTGGTCTCCCC 3′) (Lys 88 at C terminus), HMG-1 Box B Top (5′ GCGCGCGCATATGTTCAAGGACCCCAATGCCCCCAAG 3′) (Phe 89 at N terminus), HMG-1 Box B Bottom (5′ CGCGGATCCTTATTTAGCTCTGTAGGCAGCAAT 3′) (Lys 161 at C terminus), and HMG-1 Box B′ Bottom (5′ CGCGGATCCTTACTTCTTTTTCTTGCTCTTCTC 3′) (Lys 185 at C terminus).
A FLAG-tagged version of HMG-1 was also constructed to enable immunodetection of HMG-1 by EMSA. The FLAG tag was introduced by ligation of a double-stranded oligonucleotide encoding the FLAG peptide sequence (MDYKDDDDKV) flanked by the NcoI and BspHI restriction sites. NcoI and BspHI have compatible ends, and ligation of the C-terminal BspHI sequence of the FLAG oligonucleotide to the N terminus of the NcoI-digested HMG-1 PCR product creates a NcoI-FLAG-HMG-1 insert. Ligation of FLAG-BspI to the NcoI HMG-1 PCR product destroys the NcoI site between the FLAG and HMG-1 gene and leaves only the N-terminal NcoI located 5′ to the FLAG tag sequence. The NcoI-FLAG-HMG-1 gene was subcloned both into the pBXG0 mammalian expression vector (13), which contains the simian virus 40 (SV40) promoter and enhancer, and into pET11d using NcoI and BamHI. The primers used were as follows: FLAG Top (5′ CATGGACTACAAGGACGACGACGACAAGGCCTCCGT 3′) and FLAG Bot (5′ CATGACGGAGGCCTTGTCGTCGTCGTCCTTGTAGTC 3′).
Recombinant HMG-1, HMG-1 derivatives, and FLAG–HMG-1 were expressed in RJ1878 (BL21 DE3 hupA::cm hupB::kn) (41). HMG-1 synthesis was induced for 3 h at 37°C in Luria broth when the cells reached an optical density at 595 nm of 0.5 by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Two liters of cells were disrupted by sonication in a 1/10 volume of 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM dithiothreitol (DTT), 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1 mM benzamidine. The extract was clarified by centrifugation at 30,000 × g for 20 min, and the NaCl concentration was increased to 1 M. Polyethyleimine (Sigma) was added to a concentration of 0.3%, and the nucleic acids were removed by centrifugation at 30,000 × g for 20 min. Residual polyethyleimine was removed by the addition of 20% (vol/vol) cellulose phosphate P-11 (Whatman) and cleared by centrifugation at 20,000 × g for 20 min. The supernatant was dialyzed overnight against 0.1 M buffer A (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 1 mM EDTA, 10% glycerol plus 0.1 M NaCl). The dialysate was passed through a 4-ml S-Sepharose (Pharmacia) column equilibrated with the same buffer. HMG-1 proteins were eluted in a 30-ml linear gradient from 0.1 M to 1.0 M NaCl in buffer A. Fractions were analyzed on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel by staining with Coomassie blue.
Fractions that contained HMG-1 box A, A*, B, B′, AB, and AB′ were pooled and subjected to 2% trichloroacetic acid precipitation at 0°C for 30 min to remove contaminating proteins. After centrifugation for 30 min at 30,000 × g the supernatant was adjusted to 10% trichloroacetic acid and the homogenous HMG-1 proteins were recovered by centrifugation as before. The precipitate was washed with acetone, dried briefly, and resuspended in buffer B (20 mM HEPES [pH 7.5], 0.1 M NaCl, 1 mM DTT, 1 mM EDTA, and 50% glycerol) and dialyzed overnight into the same buffer.
Fractions containing HMG-1, HMG-1 box B", AB", and FLAG-HMG-1 were pooled and dialyzed overnight against 0.1 M buffer A. The dialysate was passed through a 4-ml DEAE-Sepharose (Pharmacia) column equilibrated with the same buffer. HMG-1 proteins were eluted in a 30-ml linear gradient from 0.1 M to 1.0 M NaCl in buffer A. Fractions were analyzed on an SDS–12% polyacrylamide gel by staining with Coomassie blue. Fractions containing the HMG-1 proteins were subjected to trichloroacetic acid precipitation as described above. Proteins were quantitated by laser densitometry of SDS-polyacrylamide gels stained with Coomassie blue using a titration of native bovine HMG-1 as a standard (42). The presence of FLAG-HMG-1 was confirmed by Western blotting with anti-FLAG antibodies.
Ligase-mediated circularization assays.
Ligation assays were conducted to determine the functional activity of HMG-1 and HMG-1 derivatives as described by Yen et al. (58). Briefly, 98-bp fragments were created by PCR with reaction mixtures containing [α-32P]dATP of pRJ551-76 as described previously (26). After digestion with EcoRI, the 98-bp fragments were purified in a 10% polyacrylamide gel. A total of 0.2 ng of the 98-bp DNA was incubated with HMG-1 derivatives in 50 mM HEPES (pH 7.5), 50 mM potassium glutamate, 10 mM magnesium acetate, and 1 mM ATP. Four units of T4-DNA ligase (New England Biolabs) was added for 10 min, and the reactions were terminated by incubation at 65°C for an additional 10 min. Exonuclease III (10 U; New England Biolabs) was added to the reaction mixtures to confirm the circularization of the DNA. Products were electrophoresed on an 8% acrylamide gel, dried, and subjected to quantitation by ImageQuant software.
Transient transfections.
The mammalian SV40-based expression plasmids pBXG0-ZEBRA and pBXG0-HMG-1 (13) were employed in the transient transfection assays. The wild-type HLZ3,4 enhancer region of BHLF-1, from positions −77 to −174, and promoter mutants generated within this region were subcloned into the previously described E4T-Lux construct (30) by using SacI and PstI restriction sites. A total of 50 ng of each reporter template was cotransfected into the baby hamster kidney cell line (BHK21) by using TRX-10 (Promega) with 500 ng of pBXG0-ZEBRA or 1 μg of pBXG0-HMG-1 or both. All DNA concentrations were normalized on an agarose gel before transfection, and the total effector DNA in each experiment was normalized with pBXG0. Cells were harvested 48 h after transfection, lysed using a Luciferase Assay System kit (Promega), and assayed for luciferase activity according to instructions from the manufacturer. The level of transcription generated by the reporter template alone was considered basal transcription. The relative activation was calculated by subtracting the level of transcription achieved in the presence of effector plasmids from the basal level of transcription. All experiments were done in triplicate, and the results are averages from three sets of transfections.
EMSAs.
EMSAs were performed on the wild-type HLZ3,4 and the HMGΔ2 promoters with the conditions described for the DNase I footprinting reactions (13). A saturating amount of Δ161 (7) was 25 ng, while a subsaturating amount was 0.4 ng. The amount of HMG-1 or FLAG-HMG-1 used was 12 ng. The 32P-end-labeled promoters were generated by PCR and incubated with the indicated amounts of protein for 30 min at 30°C. FLAG antibody (Sigma) was added to the reaction mixtures as indicated in the figure legends and incubated for an additional 30 min at 30°C. The samples were fractionated on a 6% native polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE) containing 1% glycerol, dried, and exposed to XAR-5 film with an intensifying screen.
Hydroxyl radical footprinting.
Hydroxyl radical footprinting was performed as described previously (28), and the cleavage products were resolved on a 10% polyacrylamide–7 M urea sequencing gel electrophoresed in 1× TBE. A 5-fmol quantity of either the 32P-end-labeled wild-type HLZ3,4 promoter or the HMGΔ2 promoter mutant was incubated for 1 h at 30°C in binding buffer containing 12.5 mM HEPES (pH 7.9), 60 mM KCl, 0.3 mM MgCl2, 0.2 mM EDTA, 0.01 mM β-mercaptoethanol, 0.5 mg of bovine serum albumin per ml, and 30 μg of poly(dGdC) per ml, with saturating amounts of Δ161 (800 ng) or a titration of subsaturating amounts from 50 ng to 200 ng. Glycerol was removed from the HMG-1 preparation by passing it through a Bio-Spin 6 (Bio-Rad) chromatography column. Note that to observe protein protections by using hydroxyl radical the concentrations of all the proteins were increased. The amount of Δ161 ranged from 50 to 800 ng and the amount of HMG-1 equaled 450 ng. DNase I footprinting was performed in parallel under the hydroxyl radical footprinting conditions to ensure that HMG-1 was binding and maintained its cooperative effect on Δ161.
To quantitate the hydroxyl radical protections, individual bands generated by the cleavage reaction were quantitated using ImageQuant software. The values obtained represented the intensities of each band and were normalized in each experiment to the lane which contained the darkest bands. These values were then compared to the intensity of the bands generated in the absence of protein and the percent saturation or protection was calculated. The results shown are averages of three experiments.
RESULTS
HMG-1 generates a specific DNase I footprint on the BHLF-1 promoter.
Our rationale for studying the EBV lytic cycle as a model for differential transcription by polymerase (pol II) is based on three facets of the virus, as follows: a well-established genetic profile; compact enhancers and promoters; and the observation that lytic genes are controlled largely by two activators called ZEBRA and Rta, although in some instances, cellular activators like Sp-1 contribute to the regulation (13). In the course of analyzing HMG-1-mediated binding of recombinant ZEBRA to the viral BHLF-1 promoter, we observed a novel DNase I footprint between two adjacent ZEBRA binding sites. We tentatively attributed the protection to HMG-1 because it was not observed with saturating concentrations of ZEBRA alone (13). The possibility that a purportedly nonspecific DNA binding protein was binding in a sequence-specific fashion was intriguing. Previous studies had suggested that HMG-1 binds DNA with little sequence dependence (4, 5). An understanding of how HMG-1 influences ZEBRA binding could provide a framework for determining how HMG-1 affects other specific DNA binding reactions (3, 13, 20, 29, 38, 39, 51).
BHLF-1 is expressed early in the EBV lytic cycle and encodes an abundant mRNA. The regulatory region bears, in addition to a pol II promoter, the EBV origin of replication. BHLF-1 is expressed at high levels and contains a potent pol II promoter when studied in vitro and in vivo (27). The BHLF-1 proximal promoter region contains two pairs of ZEBRA sites, Z-1 and Z-2, between position −50 and −74, and Z-3 and Z-4, between positions −106 and −146 (Fig. 1A). We have previously shown that incubation of recombinant ZEBRA and HMG-1 led to stimulation of ZEBRA binding to Z-1 through Z-4. As we noted previously (13), a putative HMG-1 DNase I footprint was observed between Z-3 and Z-4.
FIG. 1.
HMG-1 binds to a specific site in the BHLF-1 proximal promoter. (A) Schematic representation of the BHLF-1 promoter. The ZEBRA sites are numbered Z-1 through Z-4, where Z-1 represents the site most proximal to the start of transcription. A bracket represents the 97-bp region (HLZ3,4) subcloned for use in Fig. 2 to 6. (B) HMG-1 induces cooperative binding of ZEBRA and generates a novel DNase I footprint from positions −122 to −134. A 32P-labeled promoter fragment from positions +38 to −174, bearing Z-1 through Z-4, was incubated with the proteins indicated and subjected to DNase I footprinting analysis. An autoradiograph of the polyacrylamide-urea sequencing gel surrounding the Z-3 and Z-4 footprints is shown. Lane 1, cleavage pattern of naked DNA; lanes 2 to 6, twofold decreasing titration of ZEBRA from 10 to 0.6 ng of protein; lanes 7 to 10, the last four points of the same titration of ZEBRA in the presence of 250 ng of HMG-1; lane 11, 250 ng of HMG-1 alone. (C) ZEBRA induces cooperative binding of HMG-1. Lane 1, DNase I cleavage ladder of naked DNA; lane 2, cleavage pattern generated in the presence of a subsaturating concentration of ZEBRA (2.5 ng) alone; lanes 3 to 7, footprints elicited by twofold increasing concentrations of HMG-1 (31 to 500 ng); lanes 8 to 12, the same titration of HMG-1 in the presence of 2.5 ng of ZEBRA. In the volumes used, 2.5 ng of ZEBRA corresponds to a dimer concentration of 3.5 nM.
The HMG-1 footprint is best illustrated by comparing the binding of ZEBRA to Z-3 and Z-4 of BHLF-1 in the presence and absence of a fixed concentration of HMG-1 (Fig. 1B). Although the footprinting was performed in the context of Z-1 and Z-2, Fig. 1B and C show only the region bearing Z-3 and Z-4. Figure 1B, lane 2, shows the two distinct ZEBRA footprints observed with saturating concentrations of ZEBRA in the absence of HMG-1. As the ZEBRA concentration was lowered HMG-1 elicited two effects. First, it permitted ZEBRA to fill Z-3 and Z-4 at significantly lower concentrations (16-fold), illustrating the cooperative effect of HMG-1 on ZEBRA binding (Fig. 1B, compare lanes 3 to 6 with lanes 7 to 10). Second, HMG-1 led to a new footprint between Z-3 and Z-4 (Fig. 1B, compare lanes 2 and 10). Even at extremely high concentrations ZEBRA did not occupy the region between Z-3 and Z-4, suggesting that the new footprint was due to binding by HMG-1. A reciprocal titration with HMG-1 is presented in Fig. 1C. Here, ZEBRA was present at subsaturating concentrations and HMG-1 concentration was varied (Fig. 1C, compare lanes 3 to 7 with lanes 8 to 12). As the concentration of HMG-1 was increased it promoted both ZEBRA and its own binding. Although ZEBRA could bind on its own at high concentrations, we did not observe a strong HMG-1 footprint in the absence of ZEBRA (Fig. 1B, compare lanes 10 and 11, and Fig. 1C, compare lanes 7 and 12). The cooperative effect was specific for HMG-1 (or HMG-2; data not shown) because ZEBRA binding was not stimulated by titration over a wide range of three other HMG box-containing proteins, including LEF-1 and the yeast HMG-box proteins NHP6A and HMO-1 (data not shown). Based on these observations, we conclude that HMG-1 and ZEBRA are binding cooperatively and specifically.
HMG-1 mediates pairwise cooperativity by ZEBRA.
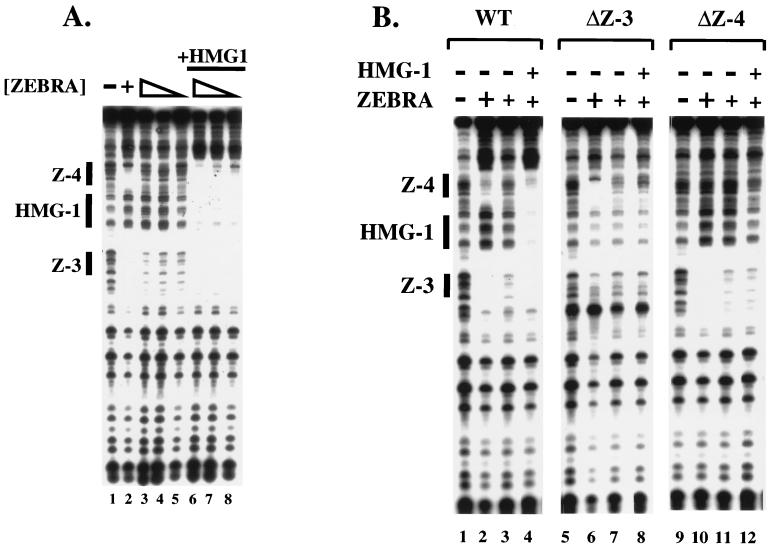
To demonstrate that the HMG-1 DNase I footprint was indeed dependent on the flanking ZEBRA sites we performed two experiments. Figure 2A shows that a 97-bp DNA fragment bearing Z-3 and Z-4, in the absence of Z-1 and Z-2, was sufficient for the HMG-1-mediated cooperative binding of ZEBRA (Fig. 2A). High concentrations of pure recombinant ZEBRA (amino acids 2 to 245) fully protected Z-3 and Z-4 from DNase I digestion (Fig. 2A, lane 2), while lower concentrations (Fig. 2A, lanes 3 to 5) revealed significantly weaker protection. At these lower levels, HMG-1 promoted cooperative binding of ZEBRA to Z-3 and Z-4 and generated the putative HMG-1 footprint between the sites (Fig. 2A, lanes 6 to 8). We conclude that Z-1 and Z-2 are not necessary for the HMG-1 footprint or its effect on Z-3 and Z-4.
FIG. 2.
Both ZEBRA binding sites are required for HMG-1-induced cooperative binding of ZEBRA. (A) HMG-1 and ZEBRA bind cooperatively to Z-3 and Z-4. The 32P-labeled 97-bp (Fig. 1A) DNA fragment, bearing Z-3 and Z-4, was incubated with the proteins indicated and subjected to DNase I footprinting analysis. Lane 1, DNase I cleavage pattern of naked DNA; lane 2, protection of Z-3 and Z-4 at saturating concentrations of ZEBRA (100 ng); lanes 3 to 5, footprints generated by twofold decreasing steps of ZEBRA (2.5 to 0.6 ng); lanes 6 to 8, footprints generated by ZEBRA (2.5 to 0.6 ng) in the presence of 250 ng of recombinant HMG-1. Note the HMG-1 footprint between the ZEBRA sites in lanes 6 to 8. (B) Mutation of individual ZEBRA binding sites abolishes cooperative binding of ZEBRA and HMG-1. The cooperative binding effect of HMG-1 on ZEBRA to the wild-type (WT) 97-bp promoter fragment is shown in lanes 1 to 4. In the context of the ΔZ-3 and ΔZ-4 promoter mutants, HMG-1 was no longer able to facilitate cooperative binding of ZEBRA (2.5 ng), as shown in lanes 5 to 8, where Z-3 has been mutated (ΔZ-3), or in lanes 9 to 12, where Z-4 has been mutated (ΔZ-4). Note in lanes 6 and 10 that ZEBRA binds to the remaining site in the mutant promoters when saturating concentrations of protein (100 ng) were used. (C) The Z-DBD is sufficient to mediate cooperative binding. The Z-DBD (Δ161) was not added (lane 1) or was added in twofold decreasing steps ranging from 24 to 3 ng either alone (lanes 2 to 5) or in the presence of 250 ng of HMG-1 (lanes 6 to 9). A 3-ng quantity of the Z-DBD corresponds to a dimer concentration of approximately 12 nM.
In the second experiment, we demonstrated that both ZEBRA sites were necessary for the HMG-1 effect (Fig. 2B). Promoter mutants were generated bearing base substitutions in one or the other site, creating ΔZ-3 and ΔZ-4, respectively. Neither of the mutant sites prevented high concentrations of ZEBRA from binding to the remaining site in the absence of HMG-1 (Fig. 2B, lanes 6 and 10). However, in contrast to the wild-type promoter (Fig. 2B, lane 4), both the HMG-1 DNase I footprint and HMG-1's cooperative effect on ZEBRA binding were abolished on the mutants when ZEBRA concentrations were limiting (Fig. 2B, lanes 8 and 12). We conclude that a pair of ZEBRA sites is necessary for the cooperative effect of HMG-1 on ZEBRA binding to Z-3 and Z-4 and for the reciprocal stimulation by ZEBRA of HMG-1 binding to the intervening DNA segment.
Z-DBD is sufficient for cooperative binding.
ZEBRA belongs to the bZIP family of transcriptional activators and contains an N-terminal activation domain from amino acids 1 to 167. To determine if the activation domain of ZEBRA was required for cooperative binding, we employed a ZEBRA deletion mutant, which lacks the first 161 amino acids, called Δ161 (7). Increasing concentrations of Δ161 (ZEBRA DNA binding domain [Z-DBD]), bearing amino acids 161 to 245 of ZEBRA, were incubated alone or with recombinant HMG-1 in a standard DNase I footprinting assay. Fig. 2C shows that HMG-1 can facilitate cooperative binding of Δ161 up to 16-fold, a response analogous to the effect observed with intact ZEBRA. This observation suggests that HMG-1 acts as a loading factor for ZEBRA and that its role in enhanceosome assembly is to facilitate cooperative binding to the promoter when ZEBRA is limiting (for elaboration on this point, see Discussion).
Domain requirements of HMG-1.
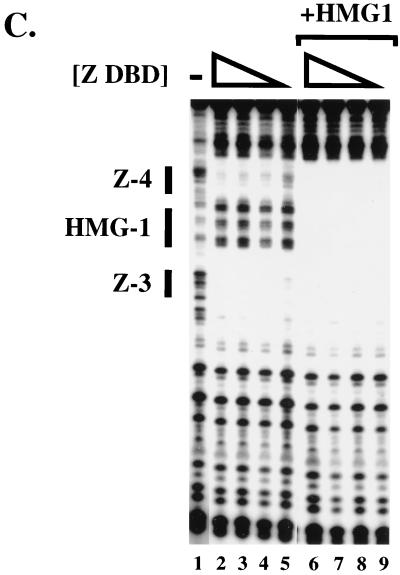
Both HMG boxes (A and B) are required for cooperative binding. Previous studies have suggested that the A and B domains function independently in DNA binding and bending assays (21, 47, 56, 59). To identify the domains required for the cooperative effect, we constructed a series of recombinant HMG-1 deletion derivatives. Each of the derivatives was purified to near homogeneity and assayed for DNA bending activity by ligase-mediated circularization assays on a 98-bp DNA fragment. Intact HMG-1 promoted bending of more than 80% of input DNA at a molar ratio of 160:1. HMG-1, AB, and AB′ bent DNA identically to wild-type, while single HMG boxes required higher concentrations of protein to facilitate bending (Fig. 3A). Other groups have also demonstrated that intact HMG-1 can form circles more effectively than single HMG-1 domains and that slight variations of bending may be attributed to exact boundaries of each HMG-1 domain and to the preparation of protein (21, 50, 54).
FIG. 3.
Both HMG boxes of HMG-1 are required to mediate cooperative binding of ZEBRA. (A) The relative DNA-bending activities of the deletion derivatives in ligase-mediated circularization assays are given. +++, wild-type levels of bending; ++, a threefold increase in protein concentration is required to reach wild-type bending efficiency; +, an approximately 10-fold increase in protein concentration is required to reach wild-type bending efficiency. (B) DNase I footprinting reveals that both HMG boxes (A and B) are required for cooperative binding of ZEBRA. Lane 1, cleavage ladder generated by DNase I on naked DNA; lane 2, protections of Z-3 and Z-4 in the presence of saturating concentrations of ZEBRA (100 ng); lane 3, protections observed with subsaturating concentrations of ZEBRA (2.5 ng); lanes 4 to 11, protections induced upon incubation of 250 ng of each HMG-1 deletion derivatives in the presence of 2.5 ng of ZEBRA.
Only HMG-1 derivatives that contained both boxes A and B were able to facilitate cooperative binding and generate specific protections between Z-3 and Z-4 in DNase I footprinting experiments (Fig. 3B, lanes 4 to 11). Addition of up to 10 times more of each of the remaining single-box derivatives also failed to stimulate ZEBRA binding, suggesting that their lack of activity in this assay is not due to reduced binding. Moreover, each of these derivatives was able to promote cooperative binding of the Rta activator to the distal enhancer region of BHLF-1 (K. Mitsouras and M. Carey, unpublished data). The yeast HMG homologue NHP6A, which has only a single HMG box but a greater specific activity in circularization assays than HMG-1, was also unable to stimulate ZEBRA binding (data not shown). We conclude that both domains of HMG-1 are required to promote cooperative binding of ZEBRA at Z-3 and Z-4.
Context-dependent, sequence-specific binding of HMG-1.
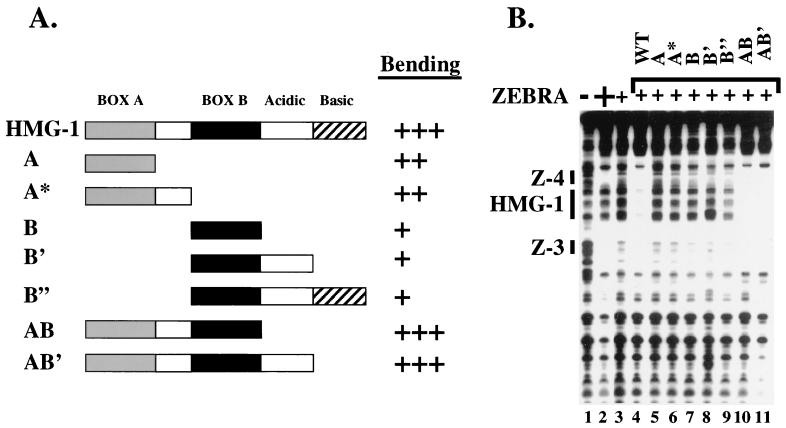
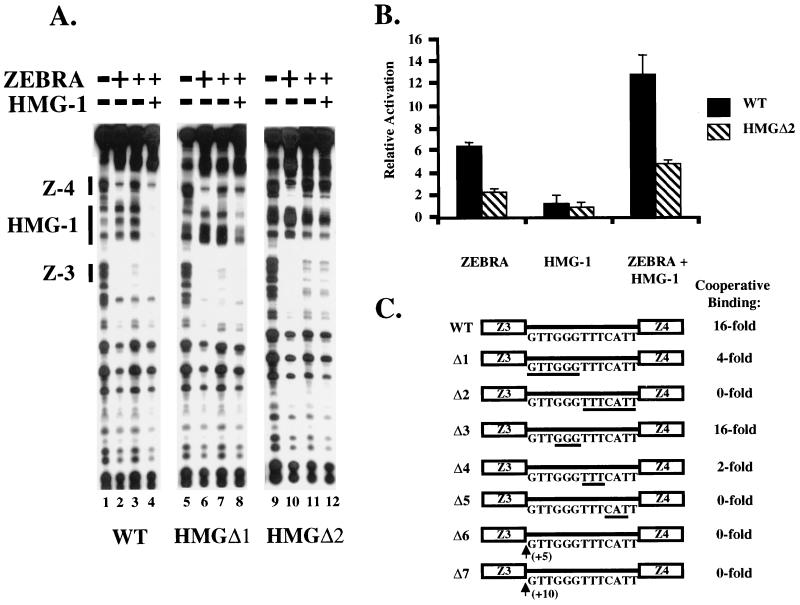
Mutagenesis revealed that HMG-1 bound DNA specifically between Z-3 and Z-4. We constructed two promoter mutants, HMGΔ1 and HMGΔ2, that spanned the 13-bp HMG-1 DNase I footprint. The footprint is a minimum estimate because it overlaps the ZEBRA protection of Z-3 and Z-4. Nevertheless, the size is consistent with previous data suggesting that HMG-1 can bind to a 15- to 18-bp region (46). The mutations were constructed by substituting cytosine residues for the natural DNA sequence. The rationale was twofold. First, inserting AT-rich sequence is believed to generate intrinsic flexibility in the DNA (10), an effect that might influence HMG-1 binding. Second, we did not wish to generate pyrimidine/purine (Y/R) steps, which were hypothesized to serve as weak recognition sites for the HMG-1 or -2 class of protein (2, 9). In HMGΔ1 the first 6 bp of the HMG-1 binding site were substituted, and in HMGΔ2, the last 7 bp were substituted (see Fig. 4C). The substitution mutations in HMGΔ1 and HMGΔ2 did not disrupt binding of ZEBRA when it was present at saturating concentrations (Fig. 4A, lanes 2, 6, and 10). However, when subsaturating concentrations of ZEBRA were used, HMG-1 failed to stimulate strong cooperative binding (Fig. 4A, compare lanes 3 and 4, 7 and 8, and 11 and 12). HMGΔ1 elicited a weak effect on ZEBRA binding, while HMGΔ2 eliminated cooperative binding altogether.
FIG. 4.
HMG-1 binding to DNA requires a specific sequence. (A) Mutations to sequences between Z-3 and Z-4 are deleterious to HMG-1 binding and reduce its ability to stimulate cooperative binding of ZEBRA. The effect of HMG-1 was measured on the unaltered promoter (WT) and two mutants, termed HMGΔ1 and HMGΔ2, encompassing the HMG-1 DNase I footprint. No protein (lanes 1, 5, and 9) or saturating amount of ZEBRA (100 ng) (lanes 2, 6, and 10), a subsaturating amount of ZEBRA (2.5 ng) (lanes 3, 7, and 11), or 2.5 ng of ZEBRA and 250 ng of HMG-1 (lanes 4, 8, 12) were incubated with the DNA fragments indicated (see text for details about the promoter mutants) and subjected to cleavage by DNase I. HMGΔ1 and HMGΔ2 failed to support either HMG-1 binding or cooperative binding of ZEBRA (lanes 8 and 12). (B) HMGΔ2 failed to support HMG-1-stimulated transcription. Quantities (50 ng each) of reporter templates bearing the wild-type HLZ3,4-promoter fragment upstream of E4-Lux or the HMGΔ2 mutant were cotransfected via lipofection into BHK21 cells either in the presence or absence of 500 ng of pBXG0-ZEBRA or 1,000 ng of pBXG0-HMG-1 or both together. Luciferase activity was determined (relative luciferase units), and the relative activation of each reporter in the presence or absence of a specific effector was calculated by subtracting out the basal level of activity. The results shown are the averages of three experiments. (C) Refining the HMG-1 target sequence. DNase I footprinting was performed on the listed promoter mutants (see text for details) and the fold cooperativity was calculated based on titrations of ZEBRA (10 to 0.6 ng) in the presence and absence of 250 ng of HMG-1. The underlined regions represent the positions of the mutations in Δ3, Δ4, and Δ5. Δ6 and Δ7 represent insertion mutants of +5 and +10 bp. (D) Increased DNA flexibility does not bypass the requirement for HMG-1 in promoting cooperative binding of ZEBRA. Heteroduplex DNA was generated by mixing equal volumes of the wild-type 32P-end-labeled HLZ3,4 promoter with either the HMGΔ1 or HMGΔ2 mutant promoters. The mixtures were then heated at 93°C for 3 min and allowed to cool slowly to room temperature. Samples were then fractionated on a mutation detection enhancement gel, which separates the parental homoduplexes from the heteroduplexes (49). The heteroduplex DNAs were purified and validated by digestion with 1 U of mung bean nuclease for 10 min, followed by polyacrylamide-urea gel analysis. Arrows point to the positions of cleavage at the heteroduplex region. These probes were employed in DNase I footprinting assays. The heteroduplex joints did not stimulate ZEBRA binding versus the parental probes and supported only a modest effect when HMG-1 was added. The fold cooperativity in the presence of HMG-1 was calculated and is summarized.
The binding defect of the HMGΔ2 mutant was paralleled by reduced transcription in transient transfection assays. On a luciferase reporter template bearing Z-3 and Z-4 upstream of the adenovirus E4 core promoter (HLZ3,4-E4-Lux), we observed a 12- to 15-fold increase in the level of transcription when the effector plasmids expressing ZEBRA and HMG-1 were cotransfected (Fig. 4B). The activation was not due to augmented levels of HMG-1 or ZEBRA. Both proteins were expressed from the SV40 enhancer, and neither influenced transcription from an SV40–β-galactosidase reporter (data not shown). In contrast, when the HMGΔ2 mutant promoter was cotransfected, the reporter response was five- to sixfold. The mutation also decreased activation by ZEBRA alone, suggesting that endogenous HMG-1 was influencing activity. Taken together, these data demonstrate a correlation between the HMG-1 binding in vitro and activation in transfection assays.
To further delineate the DNA sequences required for HMG-1 binding, we constructed a series of 3-bp, cytosine substitution mutations, which spanned the original HMGΔ1 and HMGΔ2 promoter mutants. These additional mutants are called HMGΔ3, -4, and -5 (Fig. 4C). The mutants were subjected to DNase I footprinting analysis. The HMG-1-induced cooperative binding of ZEBRA (specifically to Z-4) was quantitated by densitometry. On the intact wild-type promoter, 16-fold-higher concentrations of ZEBRA were required to fill Z-3 and Z-4 in the absence of HMG-1. The HMGΔ3 mutant elicited approximately the same level of cooperativity as the intact promoter. However, the HMGΔ4 and HMGΔ5 promoter mutants, which span the original HMGΔ2 mutant, were severely compromised (twofold and no cooperativity, respectively).
The precise spacing of the ZEBRA sites is necessary for the cooperativity. When the helical phase of the DNA was altered by inserting 5 bp between Z-3 and Z-4 (HMGΔ6) (Fig. 4C), the cooperative effect of HMG-1 was abolished. Paradoxically, the cooperative effect was not restored by addition of 10 bp (HMGΔ7) (Fig. 4C), which would restore the helical phase but further increase the spacing between the ZEBRA binding sites. This result contrasts with that of a similar experiment on the TCR-α enhancer with LEF-1 (18). The cooperative effect of LEF-1 on PEBP2α-Ets-1 and ATF-CREB binding could be abolished by insertion of a helical half increment but restored upon insertion of a full increment.
Figure 4D summarizes the effect of creating heteroduplex joints in the HMG-1 binding site. Studies by Kahn and Crothers have demonstrated that heteroduplex joints can enhance flexibility of DNA, and we predicted that such enhanced flexibility might contribute to cooperative ZEBRA binding in the absence of HMG-1 or an increased cooperative effect of HMG-1 (33). Figure 4D shows the results of an experiment confirming that the purified heteroduplexes contained melted regions at the appropriate positions. In this experiment, 32P-labeled probes bearing the wild-type and either Δ1 or Δ2 mutants were mixed, heated, and reannealed. The heteroduplexes were separated from the parental molecules with mutation detection enhancement gels (49). The purified heteroduplexes were subjected to mung bean nuclease cleavage to confirm the melting at the predicted locations. We found that the heteroduplex did not enhance binding of ZEBRA in the absence of HMG-1 (data not shown). Notably, however, a small effect of HMG-1 on ZEBRA binding was observed on both heteroduplex molecules. This may have been due to HMG-1's ability to bind residually to the specific sequence or to HMG-1's ability to bind single-stranded DNA within the heteroduplex. Nevertheless, we conclude that enhanced flexibility does not substitute for or enhance the HMG-1 effect on ZEBRA.
EMSA of HMG-1-ZEBRA complexes.
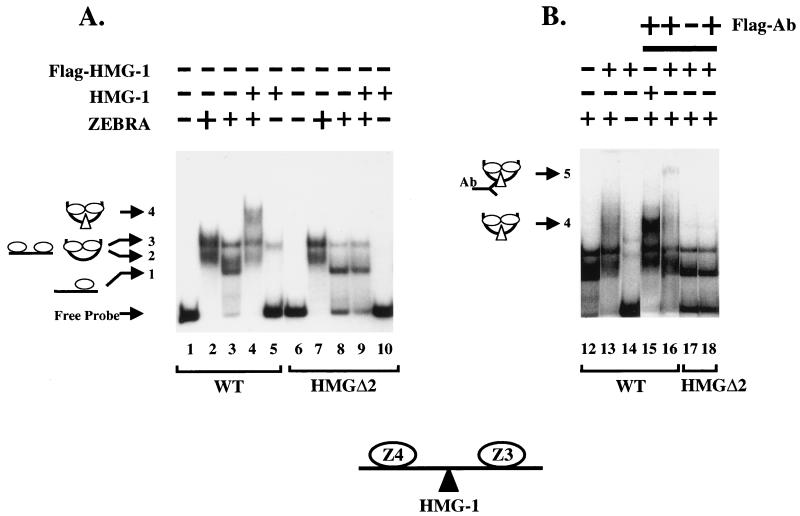
Figure 5 shows the result of an EMSA employed to further demonstrate that HMG-1 binds cooperatively with ZEBRA. HMG-1 was subcloned and tagged with the FLAG antigen at its amino terminus to enable detection by antibody supershifts. The ZEBRA DNA binding domain Δ161 was used in place of ZEBRA because we reasoned that its small size would facilitate detection of ternary complexes by EMSA.
FIG. 5.
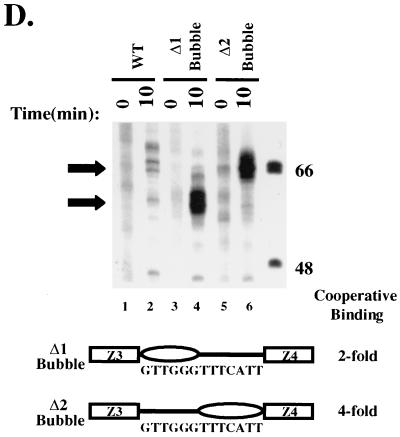
HMG-1 forms a ternary complex with ZEBRA and the HLZ3,4 promoter fragment. EMSAs on the wild-type (WT) HLZ3,4 and mutant HMGΔ2 promoters were performed as described in Materials and Methods. Panels A and B represent lanes from the same gel except panel B is a darker exposure of selected lanes. Lane 1, the migration of free probe through a native 6% polyacrylamide gel; lanes 2 and 3, complexes formed on the wild-type HLZ3,4 promoter with saturating (25 ng) and subsaturating amounts (0.4 ng) of Δ161, respectively; lanes 4, 9, and 13, 12.5 ng of HMG-1 or FLAG-HMG-1 incubated with subsaturating Δ161 (0.4 ng); lanes 5 and 14, binding of HMG-1 and FLAG-HMG-1 alone (12.5 ng) to the wild-type HLZ3,4 probe; lane 10, HMG-1 (12.5 ng) binding to the mutant HMGΔ2 probe; lanes 16 and 18, FLAG antibody added to complexes incorporating FLAG-HMG-1 with either the WT (lane 16) or the HMGΔ2 (lane 18) promoter. Note that the HMGΔ2 DNA template failed to induce cooperative binding of ZEBRA and the HMG-1-supershifted complexes seen with the wild-type template (compare lanes 3 and 4 and 8 and 9). A schematic diagram of the various complexes is shown to the left and is explained in Results.
Lanes 2 and 3 of Fig. 5A show the complexes resulting from addition of saturating and subsaturating concentrations of Δ161 on the wild-type DNA fragment. At subsaturating concentrations, complex 1 was the predominant species, while complexes 2 and 3 were present at lower levels. Complex 1 appears to represent binding of a single ZEBRA molecule since it was the only complex observed when mutants in one or the other ZEBRA site were used (ΔZ-3 and ΔZ-4 mutants from Fig. 2B, data not shown). On the wild-type fragment, saturating concentrations of ZEBRA led to increased amounts of complexes 2 and 3. We believe, based on binding studies of the lac repressor and Escherichia coli CAP proteins (17, 35), that complexes 2 and 3 represent DNAs bound to two molecules of ZEBRA, which are either interacting or not. Although we cannot unambiguously distinguish between the complexes, to do so is not critical to our argument.
Three observations suggest that HMG-1 is a component of the complex with ZEBRA and DNA, as follows: (i) when HMG-1 was added to the reactions it enhanced Δ161 binding and generated a supershift to complex 4 (Fig. 5A, compare lanes 3 and 4). The supershift coincided with disappearance of complex 1 and a diminution in the amounts of complexes 2 and 3. Complex 4 was not evident at high concentrations of ZEBRA alone, supporting the idea that complex 4 reflected binding of HMG-1 (Fig. 5A, compare lanes 2 and 4). HMG-1 is a relatively nonspecific DNA binding protein on its own, and we were concerned that it might be supershifting the complexes in a nonspecific manner. However, we believe that the small amount of HMG-1 binding nonspecifically in lane 5 does not account for the larger amount of complex 4 formed in the presence of ZEBRA. (ii) Complex 4 did not form when the HMGΔ2 mutant was employed in the binding assays (Fig. 5, compare lanes 8 and 9), demonstrating that HMG-1 did not influence Δ161 binding nonspecifically. (iii) Finally, when FLAG-HMG-1 was used in place of HMG-1 it also promoted cooperative binding of Δ161 to Z-3 and Z-4 and induced a supershift to complex 4 (Fig. 5B, lanes 12 to 16). Addition of FLAG antibody led to a further supershift to complex 5 on the wild-type DNA fragment but not on the HMGΔ2 promoter mutant (Fig. 5B, compare lanes 16 and 18). For reasons that we do not understand, despite the addition of large amounts of antibody, complex 4 was not quantitatively shifted to complex 5, although complex 4 was diminished in intensity on the autoradiograph. We conclude that HMG-1, ZEBRA, and the DNA fragment form a ternary complex.
Hydroxyl radical footprint of HMG-1.
Hydroxyl radical footprinting confirmed that HMG-1 was binding to specific sequences in the region between Z-3 and Z-4. Hydroxyl radical footprinting is used to probe DNA-protein interactions along the sugar-phosphate backbone. Hydroxyl radical is a particularly appropriate footprinting reagent because HMG-1 binds predominantly in the minor groove. Furthermore, a previous structural study had shown that HMG-1 generates a strong hydroxyl radical footprint on cisplatinated DNA (40). Conversely, ZEBRA, a bZIP family protein, binds primarily in the major groove (11, 19), where hydroxyl radical does not typically generate strong footprints (53).
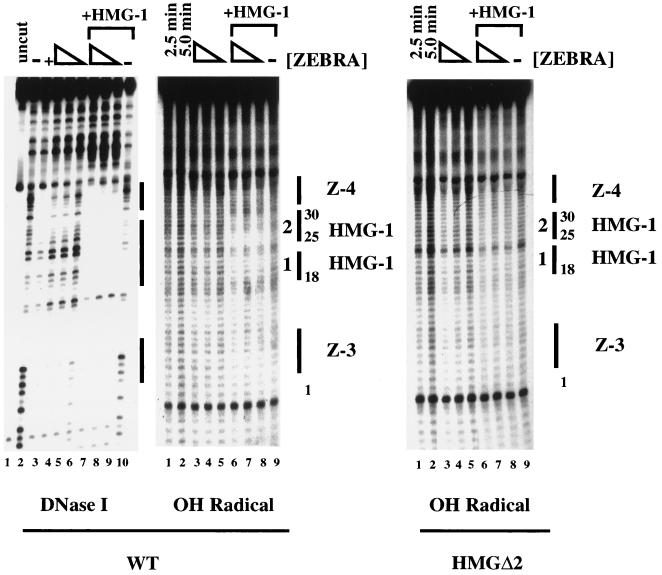
A DNase I footprint was performed alongside the hydroxyl radical as a control. Saturating concentrations of Δ161 fully protected Z-3 and Z-4 (Fig. 6, left panel, lane 3), while lower concentrations generated weaker protection (Fig. 6, lanes 4 to 6). Moreover, HMG-1 promoted cooperative binding of Δ161 binding (Fig. 6, compare lanes 4 to 6 with lanes 7 to 9). Although these results were consistent with those of previous experiments, interestingly, at the high concentrations of HMG-1 used here, several weak protections were observed along the length of the fragment (Fig. 6, lane 10) even in the absence of ZEBRA. One of these protections was over the HMG-1 site, a point we return to (see Discussion) because it bears on the issue of rudimentary sequence preferences by HMG-1.
FIG. 6.
Mapping the HMG-1 binding site by hydroxyl radical footprinting. The figure compares HMG-1-mediated cooperative binding of ZEBRA on the wild-type (WT) HLZ3,4 probe by DNase I and hydroxyl (OH) radical footprinting or on the HMGΔ2 mutant by hydroxyl radical. Lane 3 of the DNase I footprint shows the protections to Z-3 and Z-4 generated by Δ161 using saturating concentrations of protein (800 ng). The protections generated by twofold-decreasing steps of Δ161 ranging from 200 to 50 ng are shown in lanes 4 to 6, while lanes 7 to 9 illustrate the cooperative effect of 450 ng of HMG-1. Lane 10 represents the DNase I footprint of 450 ng of HMG-1 alone. The hydroxyl radical footprints on HLZ3,4 are also shown. Lanes 1 and 2 show the cleavage ladder of naked DNA generated in 2.5- and 5-min reactions. Lanes 3 to 9 show 5-min cleavage reactions performed on twofold-decreasing concentrations (200 to 50 ng) of Δ161 in the absence (lanes 3 to 5) or presence (lanes 6 to 8) of 450 ng of HMG-1. Lane 9 represents the hydroxyl radical protections generated by 450 ng of HMG-1 alone. The HMG-1 footprints between Z-3 and Z-4 are numbered 1 and 2. Identical reactions performed on the HMGΔ2 promoter mutant are shown. The individual bands generated by hydroxyl radical cleavage were boxed and quantitated using ImageQuant software. The values obtained from three different footprints were normalized to the most intense lane in each experiment and the percent saturation was calculated; strongest protections were mapped in Fig. 7. Note, higher concentrations of both HMG-1 and Δ161 were used in the hydroxyl radical footprinting experiments (see text).
Hydroxyl radical footprinting reactions performed in parallel are shown in the middle panel of Fig. 6, where lanes 1 and 2 represent naked DNA cleaved for 2.5 or 5 min. When Δ161 and HMG-1 were incubated together as shown in Fig. 6, middle panel, lanes 6 to 8, two ∼5-bp protected regions were observed within the HMG-1 binding site (i.e., defined in Fig. 2 to 4 by DNase I footprinting and mutagenesis). Previous studies showed that hydroxyl radical footprints of Box A on cisplatinated DNA were 4 to 5 bp in size (40). This observation suggested that the ∼5-bp protections spanning the HMG-1 footprint in BHLF-1 might represent binding of each HMG box.
Hydroxyl radical footprinting on the HMGΔ2 mutant promoter confirmed the specificity of the footprints. As shown in Fig. 6, right panel, lanes 1 to 9, the protections that map to the HMG-1 site on the wild-type probe were not observed on HMGΔ2. The intensity of the bands generated by hydroxyl radical footprinting on both the wild-type and HMGΔ2 promoters were quantitated and normalized, and the percent saturation of each band was determined. The wild-type promoter displayed protections dependent upon HMG-1 from positions 18 to 30, where base pairs at positions 20 and 25 to 27 were nearly 100% protected as measured by densitometry. Taken together, the data suggest that HMG-1 binds at specific locations along the DNA backbone between Z-3 and Z-4.
DISCUSSION
The ability of HMG-1 to bind cooperatively and specifically in certain contexts was predicted, but direct evidence for specific binding by HMG-1 has not been reported in the literature (22, 23). Previously, HMG-1 and -2 have been shown to stimulate DNA binding by several sequence-specific transcription factors, including p53, the steroid receptors, and Hox proteins (3, 32, 52, 60, 61). In those cases, however, HMG-1 and -2 bound the transcription factor in solution and assisted the factor in targeting its site—sequence-specific HMG-1 binding was not demonstrated. Here, we show that HMG-1 is binding DNA at a specific site and stimulating cooperative interaction of the two flanking ZEBRA dimers. This result provides a system for elucidating the mechanism of the effect of HMG-1 and -2.
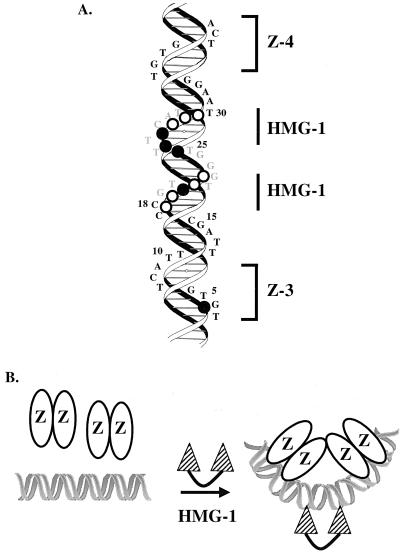
To model the binding of HMG-1 we superimposed the ZEBRA and HMG-1 sites onto a schematic of a typical B-DNA helix (Fig. 7A). The sequences altered in the HMGΔ1 and HMGΔ2 promoter mutants (from Fig. 4) are shown in gray. The numbers on the side of the helix correspond to the sequence positions quantitated in the hydroxyl radical footprinting experiment of Fig. 6. The minor groove hydroxyl radical protections are projected onto the helix as open (weak) and closed (strong) circles. The protections position HMG-1 on the opposite side of the helix as ZEBRA, when measuring from the centers of Z-3 and Z-4. By assuming that HMG-1 induces a bend towards the major groove, as shown schematically in Fig. 7B, the binding of HMG-1 would be predicted to bring the ZEBRA sites into direct apposition, facilitating an interaction that would lead to cooperative binding. A similar model has been proposed for HMG-1 in VDJ recombination by RAG-1 and -2 at the 23-bp recombination signal sequence (1, 51).
FIG. 7.
Modeling the interaction of HMG-1 with its site. (A) A helical projection of the promoter sequences from Z-3 to Z-4 of BHLF-1. The 7-bp ZEBRA binding sites are indicated by brackets. Hydroxyl radical protections are projected onto the helical backbone. Open circles, base pairs that were partially (50 to 70%) saturated; closed circles, complete (80 to 100%) protections in the presence of ZEBRA and HMG-1. (B) Model depicting how HMG-1 induces cooperative binding of ZEBRA to the Z-3 and Z-4 pair of binding sites. In the presence of both proteins, ZEBRA stabilizes HMG-1 interaction with DNA between Z-3 and Z-4 and HMG-1 induces cooperative binding of ZEBRA.
A strict level of control must be superimposed if genes are to rely on the non-sequence-specific architectural factors for regulation. We imagine that such regulation is achieved through a combination of a low intrinsic affinity coupled with cooperativity to augment binding. Such a mechanism would be sufficient to generate specific responses in the appropriate contexts. There are two models for how this might work in the case of ZEBRA and HMG-1. In the first model, two ZEBRA molecules bind first to the DNA and weakly interact. This interaction results in a transient bend, which in turn recruits HMG-1. In the second model, HMG-1 binds first and actively induces the bend by binding to a rudimentary recognition sequence. The bend in turn facilitates cooperative binding by ZEBRA, analogous to the effect of LEF-1 on PEBP2α-Ets-1 and ATF-CREB at the TCR-α enhancer (18).
The model postulating a ZEBRA-induced bend is supported by EMSA data revealing weak cooperative binding by ZEBRA even in the absence of HMG-1. When either Z-3 or Z-4 is deleted, a reproducible decrease in affinity of ZEBRA for the remaining site is observed (data not shown). Several observations are consistent with HMG-1-box proteins displaying a modest sequence preference for binding. Yeast NHP6A and Drosophila HMG-D formed specific complexes on duplex oligonucleotides as revealed by NMR and X-ray crystallography (2, 31). Targeting may be influenced by the presence of Y/R steps that have intrinsic flexibility and are thus receptive to inserting hydrophobic amino acids present on the HMG DNA binding domain. There are two Y/R steps within the HMG-1 footprint on BHLF-1, and mutagenesis together with hydroxyl radical footprinting supports the idea that these steps may represent critical interaction points. Indeed, high concentrations of HMG-1 alone weakly protect this region although we have never observed complete protection in the absence of ZEBRA (Fig. 6). Moreover, in further support of a rudimentary recognition mode, HMG-1 generates a weak but discrete complex on the wild-type DNA fragment by EMSA, which is diminished on the mutant (Fig. 5, compare lanes 5 and 10).
Although our data imply that a simple bend would suffice to enhance ZEBRA cooperativity, three results support the view that there is a requirement for a precise spatial alignment between the two ZEBRA sites. First, deletion derivatives of HMG-1 that contained only one of the DNA recognition motifs were unable to facilitate cooperative binding. Because both individual boxes bend DNA in cyclization assays (Fig. 3), albeit less effectively than intact HMG-1, the requirement for both boxes probably reflects an aspect of stereospecificity. Second, cooperative binding was abolished by both 5- and 10-bp alterations in the spacing (Fig. 4). This result implies that the ZEBRA sites do not exhibit a simple requirement for helical periodicity. Finally, enhanced flexibility of the HMG-1 binding site does not circumvent the HMG-1 requirement (Fig. 4). Generation of heteroduplex DNA would theoretically introduce a flexible linker between the pair of ZEBRA sites that could bypass the unfavorable energy requirements caused by bending DNA fragments below the persistence length (55). Promoter mutants containing mismatched sequences in the region of the HMG-1 footprint (−122 to −134) were not, however, able to circumvent the HMG-1 requirement for cooperative ZEBRA binding (Fig. 4). Taken together, the phasing and flexibility experiments suggest that HMG-1 is not merely introducing a bend in the DNA but may position the ZEBRA dimers in a fashion requiring a specific geometry.
What role does HMG-1 play in EBV regulation? By allowing ZEBRA to bind cooperatively and with high affinity, HMG-1 could permit enhanceosome assembly and gene activation at low concentrations of ZEBRA. Cooperative binding also promotes occupancy of a promoter over a significantly narrower range of protein. We speculate, however, that the cooperativity is primarily a means of establishing specificity because only pairwise combinations of ZEBRA in the appropriate spatial alignment on a promoter will cooperate with HMG-1 to facilitate binding. This ensures that ZEBRA does not bind and fortuitously activate nontarget genes.
In contrast to other transcription factors where HMG-1 has been shown to act, we have not been able to identify a specific protein-protein interaction between ZEBRA and HMG-1 using glutathione S-transferase pulldown analysis, although the effect may be weak and not readily detectable in the absence of DNA. The ability of HMG-1 to function in such a context implies it could play a broad role in gene regulation, using minimal sequence requirements and cooperativity to assemble specific nucleoprotein complexes. Additionally, HMG-1 may also participate in global DNA conformational changes related to promoter and enhancer function. Structural studies of the HMG-D complex reveal that a single domain can bend DNA by as much as 111°. Two domains might therefore be able to completely reverse the directionality of a DNA segment. Such dramatic alterations in the path of DNA could serve to align distal upstream enhancers and the core promoter or might be involved in EBV lytic replication which requires the ZEBRA sites in BHLF-1.
ACKNOWLEDGMENTS
This study was supported by grants GM057283 and GM38509 from the National Institutes of Health.
We thank Kristy Johnson and Katherine Mitsouras for commenting on the manuscript.
REFERENCES
- 1.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi M E, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol Cell Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain F H, Yen Y M, Masse J E, Schultze P, Dieckmann T, Johnson R C, Feigon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi M E, Taraseviciene L, Nordeen S K, Allegretto E A, Edwards D P. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 6.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 7.Chi T, Carey M. The ZEBRA activation domain: modular organization and mechanism of action. Mol Cell Biol. 1993;13:7045–7055. doi: 10.1128/mcb.13.11.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 9.Churchill M E, Jones D N, Glaser T, Hefner H, Searles M A, Travers A A. HMG-D is an architecture-specific protein that preferentially binds to DNA containing the dinucleotide TG. EMBO J. 1995;14:1264–1275. doi: 10.1002/j.1460-2075.1995.tb07110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crothers D M. DNA curvature and deformation in protein-DNA complexes: a step in the right direction. Proc Natl Acad Sci. 1998;95:15163–15165. doi: 10.1073/pnas.95.26.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 12.Ellwood K, Chi T, Huang W, Mitsouras K, Carey M. Cooperative assembly of RNA polymerase II transcription complexes. Cold Spring Harb Symp Quant Biol. 1998;63:253–261. doi: 10.1101/sqb.1998.63.253. [DOI] [PubMed] [Google Scholar]
- 13.Ellwood K, Huang W, Johnson R, Carey M. Multiple layers of cooperativity regulate enhanceosome-responsive RNA polymerase II transcription complex assembly. Mol Cell Biol. 1999;19:2613–2623. doi: 10.1128/mcb.19.4.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flemington E K, Goldfeld A E, Speck S H. Efficient transcription of the Epstein-Barr virus immediate-early BZLF1 and BRLF1 genes requires protein synthesis. J Virol. 1991;65:7073–7077. doi: 10.1128/jvi.65.12.7073-7077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartenberg M R, Crothers D M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- 18.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 19.Glover J N, Harrison S C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 20.Golding A, Chandler S, Ballestar E, Wolffe A P, Schlissel M S. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasser K D, Teo S H, Lee K B, Broadhurst R W, Rees C, Hardman C H, Thomas J O. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1) Eur J Biochem. 1998;253:787–795. doi: 10.1046/j.1432-1327.1998.2530787.x. [DOI] [PubMed] [Google Scholar]
- 22.Grosschedl R. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 23.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 24.Hardman C H, Broadhurst R W, Raine A R, Grasser K D, Thomas J O, Laue E D. Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1995;34:16596–16607. doi: 10.1021/bi00051a007. [DOI] [PubMed] [Google Scholar]
- 25.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haykinson M J, Johnson R C. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993;12:2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayward S D, Hardwick J M. Herpes virus transcription and its regulation. Boca Raton, Fla: CRC Press; 1991. [Google Scholar]
- 28.Heumann P S A H. DNA-protein interactions principles and protocols. Vol. 30. Totowa, N.J: Humana Press; 1994. [Google Scholar]
- 29.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Shostak Y, Tarr P, Sawyers C, Carey M. Cooperative assembly of androgen receptor into a nucleoprotein complex that regulates the prostate-specific antigen enhancer. J Biol Chem. 1999;274:25756–25768. doi: 10.1074/jbc.274.36.25756. [DOI] [PubMed] [Google Scholar]
- 31.Iv F V, Sweet R M, Churchill M E. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaraman L, Moorthy N C, Murthy K G, Manley J L, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahn J D, Yun E, Crothers D M. Detection of localized DNA flexibility. Nature. 1994;368:163–166. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- 34.Kieff A B R A E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. p. 2343. [Google Scholar]
- 35.Kramer H, Niemoller M, Amouyal M, Revet B, von Wilcken-Bergmann B, Muller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 38.Melvin V S, Edwards D P. Coregulatory proteins in steroid hormone receptor action: the role of chromatin high mobility group proteins HMG-1 and -2. Steroids. 1999;64:576–586. doi: 10.1016/s0039-128x(99)00036-7. [DOI] [PubMed] [Google Scholar]
- 39.Oettinger M A. V(D)J recombination: on the cutting edge. Curr Opin Cell Biol. 1999;11:325–329. doi: 10.1016/S0955-0674(99)80044-1. [DOI] [PubMed] [Google Scholar]
- 40.Ohndorf U M, Rould M A, He Q, Pabo C O, Lippard S J. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 41.Paull T T, Carey M, Johnson R C. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 42.Paull T T, Haykinson M J, Johnson R C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 43.Ptashne M, Gann A. Imposing specificity by localization: mechanism and evolvability. Curr Biol. 1998;8:R897. doi: 10.1016/s0960-9822(07)00557-x. [DOI] [PubMed] [Google Scholar]
- 44.Ramstein J, Locker D, Bianchi M E, Leng M. Domain-domain interactions in high mobility group 1 protein (HMG1) Eur J Biochem. 1999;260:692–700. doi: 10.1046/j.1432-1327.1999.00185.x. [DOI] [PubMed] [Google Scholar]
- 45.Read C M, Cary P D, Crane-Robinson C, Driscoll P C, Norman D G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993;21:3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito K, Kikuchi T, Shirakawa H, Yoshida M. The stabilized structural array of two HMG1/2-boxes endowed by a linker sequence between them is requisite for the effective binding of HMG1 with DNA. J Biochem (Tokyo) 1999;125:399–405. doi: 10.1093/oxfordjournals.jbchem.a022300. [DOI] [PubMed] [Google Scholar]
- 47.Stros M. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J Biol Chem. 1998;273:10355–10361. [PubMed] [Google Scholar]
- 48.Sugden B, Yates J, Mark W. Transforming functions associated with Epstein-Barr virus. J Investig Dermatol. 1984;83:82s–87s. doi: 10.1111/1523-1747.ep12281491. [DOI] [PubMed] [Google Scholar]
- 49.Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 50.Teo S H, Grasser K D, Thomas J O. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur J Biochem. 1995;230:943–950. doi: 10.1111/j.1432-1033.1995.tb20640.x. [DOI] [PubMed] [Google Scholar]
- 51.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verrier C S, Roodi N, Yee C J, Bailey L R, Jensen R A, Bustin M, Parl F F. High-mobility group (HMG) protein HMG-1 and TATA-binding protein-associated factor TAF(II)30 affect estrogen receptor-mediated transcriptional activation. Mol Endocrinol. 1997;11:1009–1019. doi: 10.1210/mend.11.8.9962. [DOI] [PubMed] [Google Scholar]
- 53.Vinson C R, Sigler P B, McKnight S L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 54.Wagner J P, Quill D M, Pettijohn D E. Increased DNA-bending activity and higher affinity DNA binding of high mobility group protein HMG-1 prepared without acids. J Biol Chem. 1995;270:7394–7398. doi: 10.1074/jbc.270.13.7394. [DOI] [PubMed] [Google Scholar]
- 55.Wang J C, Giaever G N. Action at a distance along a DNA. Science. 1988;240:300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- 56.Webb M, Thomas J O. Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain. J Mol Biol. 1999;294:373–387. doi: 10.1006/jmbi.1999.3150. [DOI] [PubMed] [Google Scholar]
- 57.Weir H M, Kraulis P J, Hill C S, Raine A R, Laue E D, Thomas J O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yen Y M, Wong B, Johnson R C. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J Biol Chem. 1998;273:4424–4435. doi: 10.1074/jbc.273.8.4424. [DOI] [PubMed] [Google Scholar]
- 59.Yoshioka K, Saito K, Tanabe T, Yamamoto A, Ando Y, Nakamura Y, Shirakawa H, Yoshida M. Differences in DNA recognition and conformational change activity between boxes A and B in HMG2 protein. Biochemistry. 1999;38:589–595. doi: 10.1021/bi981834l. [DOI] [PubMed] [Google Scholar]
- 60.Zappavigna V, Falciola L, Citterich M H, Mavilio F, Bianchi M E. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang C C, Krieg S, Shapiro D J. HMG-1 stimulates estrogen response element binding by estrogen receptor from stably transfected HeLa cells. Mol Endocrinol. 1999;13:632–643. doi: 10.1210/mend.13.4.0264. [DOI] [PubMed] [Google Scholar]
- 62.Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]