Abstract
Multiple lines of evidence currently indicate that the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) gains entry into human host cells via a high-affinity interaction with the angiotensin-converting enzyme 2 (ACE2) transmembrane receptor. Research has further shown the widespread expression of the ACE2 receptor on the surface of many different immune, non-immune and neural host cell types, and that SARS-CoV-2 has the remarkable capability to attack many different types of human-host cells simultaneously. One principal neuroanatomical region for high ACE2 expression patterns occurs in the brainstem, an area of the brain containing regulatory centers for respiration, and this may in part explain the predisposition of many COVID-19 patients to respiratory distress. Early studies also indicated extensive ACE2 expression in the whole eye and the brain’s visual circuitry in aged humans. In this study we analyzed ACE2 receptor expression at the mRNA and protein level in multiple cell types involved in human vision, including cell types of the external eye and several deep brain regions known to be involved in the processing of visual signals. Here we provide evidence: (i) that many different optical and neural cell types of the human visual system provide receptors essential for SARS-CoV-2 invasion; (ii) of the remarkable ubiquity of ACE2 presence in cells of the eye and anatomical regions of the brain involved in visual signal processing; (iii) that ACE2 receptor expression in different ocular cell types and visual processing centers of the brain provide multiple compartments for SARS-CoV-2 infiltration; and (iv) of a gradient of increasing ACE2 expression from the anterior surface of the eye to the visual signal processing areas of the occipital lobe and the primary visual neocortex. A gradient of ACE2 expression from the eye surface to the occipital lobe may provide the SARS-CoV-2 virus a novel pathway from the outer eye into deeper anatomical regions of the brain involved in vision. These findings may explain, in part, the many recently reported neuro-ophthalmic manifestations of SARS-CoV-2 infection in COVID-19 affected patients.
Keywords: Alzheimer’s disease; angiotensin-converting enzyme 2 (ACE2); Occipital lobe; Ocular cells; Optic nerve, retinal pigment epithelial (RPE) cells, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic originated late in the year 2019 in an open air market in the city of Wuhan in China’s Hubei province. Early reports suggested that the emergence of SARS-CoV-2 may have occurred via ‘zoonotic spillover’ from the fruit bat (Pteropus scapulatus) and/or other mammalian and/or avian vectors and an interspecies viral transmission [1–6]. Three key findings on the mechanism of SARS-CoV-2 infection were that: (i) direct contact of the novel enveloped, positive single-stranded RNA (ssRNA) SARS-CoV-2 beta coronavirus that causes COVID-19 with host cell surface receptors is the primary suspected route of transmission; (ii) the SARS-CoV-2 virus, via its surface spike (S1) protein requires the single-pass angiotensin-converting enzyme 2 (ACE2) transmembrane receptor to gain entry into human host cells; and (iii) that ACE2, a zinc containing surface receptor protein and metallo-carboxypeptidase (EC 3.4.17.23) normally involved as a key component of the renin-angiotension system (RAS), is ubiquitously expressed in many different human cell types. Immuno-histochemical analysis has recently confirmed expression of ACE2 at the protein level in alveolar and epithelial cells of the respiratory system, enterocytes and intestinal epithelial and endothelial cells, kidney cells of the renal tubule and immune cells, such as alveolar monocytes and macrophages, and many different cell types of the CNS including those of the cerebral cortex, brainstem, the conjunctival epithelium of the eye, the optic nerve, cornea, the corneal epithelial surface and multiple cell types of the brain involved in vision and visual signal processing [5–12]. This may in part explain SARS-CoV-2-mediated invasion of a very wide spectrum of different host cell types including sensory and support cells of the eye and specific anatomical regions of the brain. This viral-mediated elicitation of a multipronged attack underscores the ubiquity, severity and extensive variety of signs and symptoms observed in COVID-19 patients.
Most recently emerging observations indicate widespread neuro-ophthalmic disruptions in COVID-19 infected patients [13–26]. Coronaviruses are known for their neurotropism towards brain cells and cells of the visual system, and COVID-19 infection is currently positively associated with ocular abnormalities including blurred vision, kerato-conjunctivitis (an inflammation of the conjunctiva), conjunctival hyperemia, chemosis (a swelling of the conjunctiva) [13–16], epiphora (increased tear secretions), anosmia, diplopia (monocular or binocular), acute-onset vision loss, acquired cortical blindness caused by damage to the brain’s occipital cortex in both primary and secondary brain visual processing centers [15–18], progressive monocular and binocular/bilateral blindness, eye pain with photophobia, eye pain with disturbances in extraocular movements and cranial nerve involvement [17–20], decreased visual acuity, optic neuritis, visual-associated disturbances in balance and gait issues, total or partial loss of vision in an otherwise normal-appearing eye [19–22], acute uveitis and other neurologic-ophthalmic symptoms including cranial neuropathies in the visual circuitry and a virally-induced Miller-Fisher syndrome [13–15,19–25]. COVID-19 infection is further associated: (i) with alterations in the ganglion cell and plexiform cell layers of the retina and reduced vessel density of the retinal capillary plexus as evidenced by using non-invasive imaging techniques including optical coherence tomography (OCT) angiography [15,26]; and (ii) the observation of increased secretion of tear fluids from the eye (epiphora) during COVID infection; ocular biofluids are known to contain viral particles and have been implicated in the passive transmission of COVID-19 via the nasolacrimal duct through the nasal cavity and into susceptible ACE2 receptor-enriched cells of the upper respiratory tract (see below) [6,20,23,24].
Because of the extraordinary and singular importance of the ACE2 transmembrane receptor in the recognition, attachment and entry of SARS-CoV-2 into host cells and requisite for SARS-CoV-2 infection, in the current report we investigated the expression of ACE2 at the messenger RNA (mRNA) and protein level in 20 different types of cells involved in human vision, including 11 cell types of the anterior eye and 9 brain regions involved in the processing of visual signals employing a novel highly sensitive radiolabel-hybridization-based detection system using gamma 32P-adenosine tri-phosphate ([γ-32P]dATP) radiolabeled ACE2 probes; see below [6,27,28]. In our 52 years of experience of researching gene expression abundance, speciation and complexity in the human brain and eye in our hands this technique has been found to be the most accurate and quantitative abundance-based analysis [6,27,28]. ACE2 protein levels were also assayed in 14 different eye cell types and anatomical regions of the brain involved in visual signal processing. As discussed more fully below, major findings include: (i) a very strong correlation between ACE2 mRNA abundance with ACE2 protein abundance in the eye and brain cells studied; (ii) the observation of detectable levels of ACE2 receptor expression found throughout eye cells and anatomical regions of the human brain involved in visual signal processing; (iii) that many different cell and tissue types of the visual system and brain provide multiple points of potential SARS-CoV-2 viral entry and host cell invasion and infectivity; and (iv) that a downhill gradient of ACE2 receptor abundance on ocular and brain cell membrane surfaces may, in part, favor translocation of the SARS-CoV-2 coronavirus into deeper anatomical regions of the visual pathways. The ubiquity, abundance and positioning of the ACE2 receptor throughout the visual system of the eye and brain may have implications relevant to the increasing number of reports linking SARS-CoV-2-association with visual disturbances and dysfunction and neuro-ophthalmic manifestations in COVID-19 patients.
MATERIALS AND METHODS
ACE2 mRNA analysis
The entire analytical protocol has been recently reported and updated in some detail by our laboratories [6,27,28]. Briefly, human multiple tissue expression (MTE) array panels (Invitrogen-Clontech, Palo Alto CA; cat no. 7775-1) or eye and brain cells or tissues were utilized to analyze brain- or visual-relevant expression of ACE2 receptor mRNA. The MTE arrays utilized contain polyA+ mRNA samples (each dot-shaped sector containing ~2.0 μg of polyA+ RNA) enriched from 9 human brain regions with 8 independent DNA and RNA controls where the ACE2 receptor is not expressed; polyA+ RNA samples were further extracted from ocular cells (including epithelial, conjunctival, keratocytes (corneal fibroblasts), iris fibroblasts, retinal pigment epithelial (RPE) cells and trabecular meshwork cells obtained from ScienCell (https://www.sciencellonline.com/productsservices/primary-cells/human/cell-systems/ocular-cell-system.html) or were kind gifts from Dr. PN Alexandrov (Figure 1).
Figure 1:
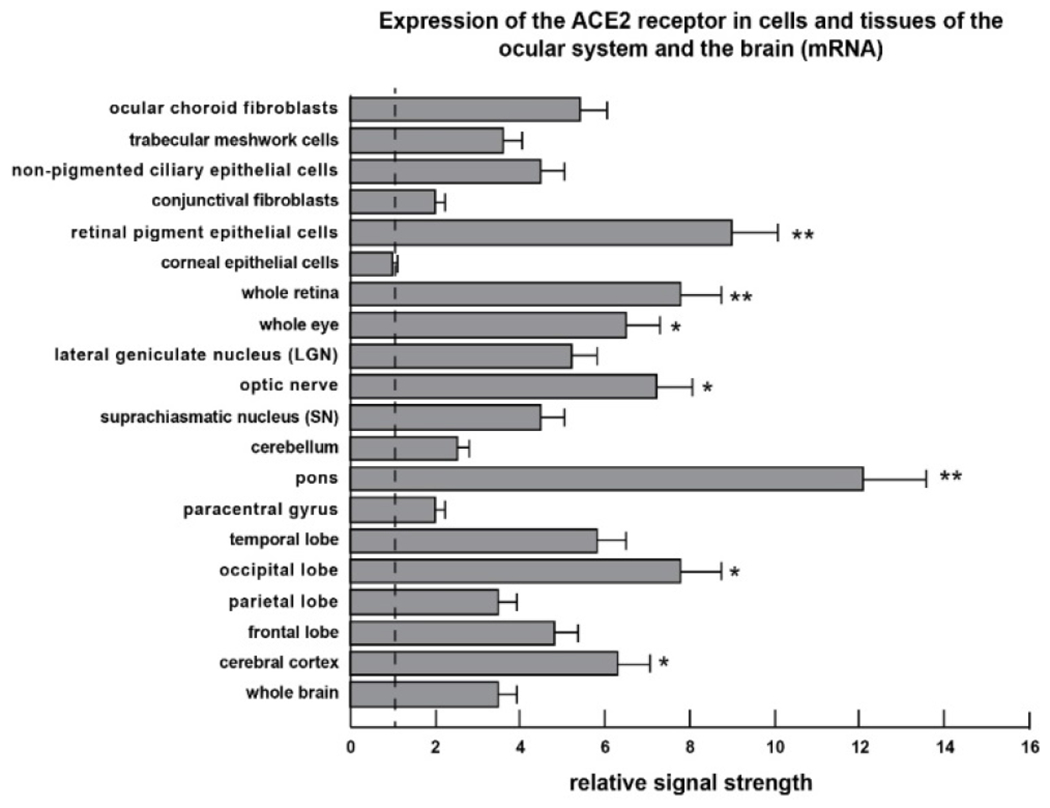
Cell- and tissue-specific patterns of ACE2 expression (mRNA) in human ocular cells and in selected areas of the brain involved in visual processing. The extent of ACE2 transmembrane receptor expression in the plasma membrane is an important indicator of ACE2 gene function, the susceptibility to SARS-CoV-2 invasion and the development of COVID-19. Significant ACE2 receptor expression was initially observed in the whole brain, whole eye and whole retina and subsequently cells and tissues involved in visual signal acquisition and processing were analyzed for ACE2 abundance. ACE2 acts as a receptor for the spike (S1) glycoprotein of the human coronavirus HCoV-NL63 and the human SARS-CoV and SARS-CoV-2 virus that causes COVID-19 (see: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=59272; last accessed 9 September 2021); in the mRNA experiments both G3PDH and β-actin DNA probes were used as abundance controls; in this experiment relative signal strength refers to control β-actin mRNA levels in the same tissues [6]; in this and one previous study [6] the highest expression of the SARS-CoV-2 ACE-2 receptor was found in the cerebral cortex and the occipital lobe, the pons and medulla oblongata of the brain and in the whole retina, the optic nerve and the ocular choroid and RPE cells of the eye. High expression of the ACE2 receptor in the eye and neural pathways involved in vision and visual processing may predispose this circuitry of the visual system to attracting SARSCoV-2 and viral invasion. A vertical dashed line at 1.0 is included for ease of comparison; a minimum of N=3 HyBond N+ or MTE filters were used for each tissue determination; *p<0.05; **p<0.01 (ANOVA); error bars represent one standard deviation of the mean.
Eye and brain tissue-sourced mRNA were obtained from pools of aged control eye and brain tissue sources [6,27,28]. All postmortem tissues utilized in this study had a mean age and one standard deviation of 68.5 +/− 12.2 yrs. RNA concentrations were quantified using a Agilent 2100 Bioanalyzer and Agilent test chips (ThermoFisher Scientific, Waltham MA, USA) and ~2.0 to ~5.0 μg polyA+ RNA were spotted onto HyBond N+ nylon membrane filters using the manufacturer’s protocols as previously described (ThermoFisher Scientific) [6,27,28]; Alexandrov et al., 2012; Tukiw et al., 2020). A unique human-specific 26 nucleotide (nt) ACE2 receptor-specific DNA probe 5′-CTTGCAGCTACACCAGTTCCCAGGCA-3′ (US NIH/NLM Sequence ID: AY217547.1; https://blast.ncbi.nlm.nih.gov/Blast.cgi#27978647; see also accession AB046569.1; https://www.ncbi.nlm.nih.gov/nuccore/AB046569.1); last accessed 9 September 2021) exhibited no homology to ACE1 [6,28–30]; while other ACE2-specific probes have been used previously that gave almost identical hybridization signals, data for the 26 nt ACE2 DNA probe is presented in this report. The 26 nt ACE2 receptor DNA probe was designed so that the sequence crosses two exons, thereby detecting only mature ACE2 mRNA and not ACE2 genomic DNA or heterogeneous RNA (hnRNA) containing the ACE2 DNA or RNA sequence. DNA probes were used due to their stability (as compared to unprotected RNA probes) and RNA-DNA hybrids are known to elicit a more accurate and stable hybridization with a higher energy of association (EA) between the ACE2 DNA probe and the membrane-bound polyA+ mRNA target [6,31,32]; https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=59272; last accessed 9 September 2021). In these experiments ACE2 DNA probes were end-labeled by using [γ-32P]dATP (3,000 Ci/mmol, Amersham Redivue™); either glyceraldehyde 3-phosphate dehydrogenase (G3PDH) or beta-actin (β-actin) were used for control mRNA normalization normalization (see http://www.takara.co.kr/file/manual/pdf/PT3307-1.pdf [6,26–29,32]. To obtain maximal signal quantitation accuracy hybridized and washed membranes were overlaid on the previously marked MTE or polyA+ RNA spotted HyBond N+ nylon templates, excised and each sector counted separately in 10 ml liquid scintillation fluid (Ultima Gold, PerkinElmer, Waltham MA) using a LS600016 LSC Liquid Scintillation System, Beckman, Fullerton CA); relative signal intensities were expressed by comparing the ACE2 hybridization signal to the G3PDH and/or β-actin signal for each polyA+ mRNA on the MTE or polyA+ array [6,28–35].
ACE2 protein analysis
Protein extracts from cells and/or tissues involved in human vision, including ocular choroidal fibroblasts, trabecular meshwork cells, non-pigmented ciliary epithelial cells, retinal pigment epithelial cells, corneal epithelial cells, whole retina, whole eye and tissues from brain visual processing pathways including the optic nerve, cerebellum, pons, temporal lobe, occipital lobe (Brodmann Area 17, containing the primary visual cortex), cerebral cortex and whole brain were generated using a ProteoExtract Complete Mammalian Proteome Extraction Kit (cat no. 539779, Calbiochem/Millipore-Sigma Burlington MA) and were assayed for protein concentration using a Non-Interfering Protein Assay kit (cat no. 488250, Calbiochem/Millipore-Sigma) at 480 nm; protein samples were stored in at −81°C according to the manufacturers protocol (Millipore-Sigma); ACE2 protein [UniProtKB - Q9BYF1 (ACE2_HUMAN)] abundance these selective cell extracts of the visual system and brain were analyzed using a quantitative colorimetric (450 nm) sandwich ELISA specific for human ACE2 using a Fluoroskan Ascent FT Microplate Fluorometer and Luminometer (Cat no. 5200220, ThermoFisher Scientific, Waltham MA; sensitivity 1052 pg/ml; detection range 1.5 ng/ml - 255 ng/ml (Human ACE2 ELISA Kit ab235649; Abcam Cambridge MA, USA); human recombinant ACE2 protein (Abcam ab151852) and human beta-actin (β-actin; anti-beta actin antibody (Abcam ab8227) were used as internal controls to quantify relative ACE2 and β-actin protein abundance in each sample according to the manufacturer’s instructions (Figure 2); concentrations of ACE2 were measured in triplicate and interpolated from the ACE2 standard curve and corrected for sample dilution as according to manufacturer’s protocol; human ACE2 was expressed as ng/ml (Abcam; https://www.abcam.com/human-ace2-elisa-kit-ab235649.html; last accessed 9 September 2021; see Supplementary file 1).
Figure 2:
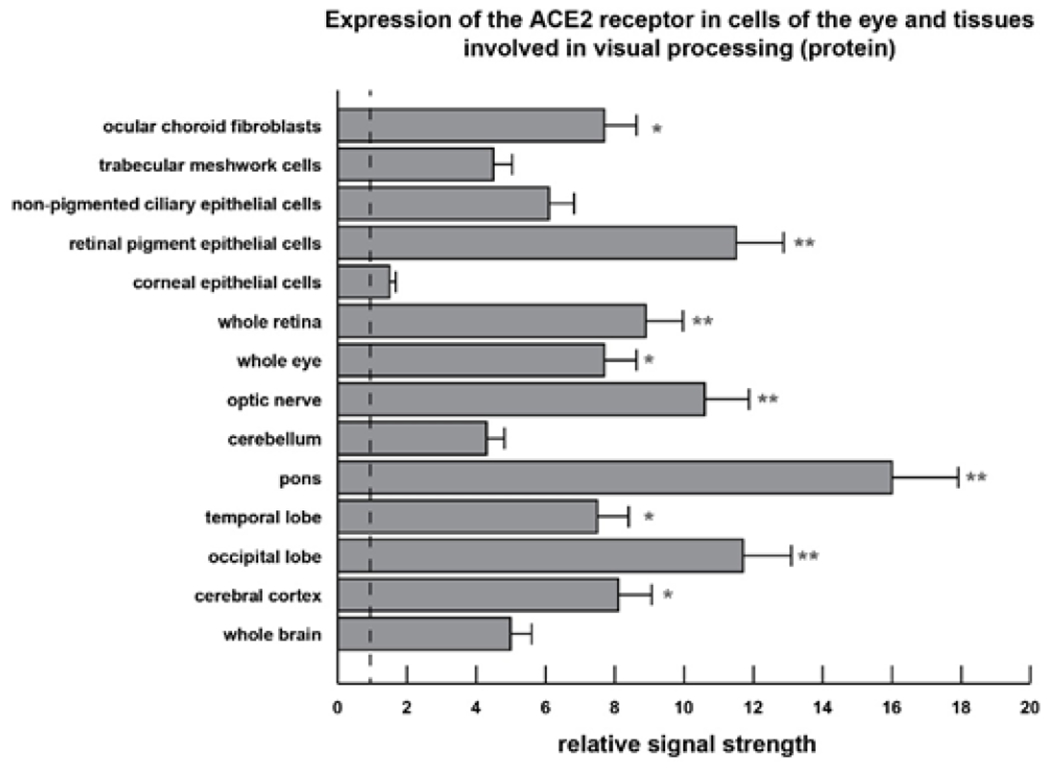
Cell- and tissue-specific patterns of ACE2 expression at the protein level in selected human ocular cells and in anatomical areas of the brain involved in visual processing; bar graph of ELISA analysis (see Supplementary File 1); we observed a very strong correlation between ACE2 mRNA abundance (Figure 1) with ACE2 protein abundance (Figure 2); the highest expression of the SARS-CoV-2 receptor ACE-2 protein was found in RPE cells, the whole retina, optic nerve, the pons and the occipital lobe that contains the primary visual cortex and main visual processing area (Brodmann Area17); relative signal strength refers to control β-actin protein levels in the same tissues; see text for further details; a vertical dashed line at 1.0 is included for ease of comparison; a minimum of N=3 ELISA analyses were performed for each protein determination in cells or tissues; *p<0.05; **p<0.01 (ANOVA); error bars represent one standard deviation of the mean.
Statistical significance
The analysis of statistical significance was evaluated using a two-way factorial analysis of variance (p, ANOVA; SAS Institute, Cary NC, USA). A p<0.05 (ANOVA) was deemed as statistically significant; a p<0.01 (ANOVA) was deemed as very highly significant; experimental values were expressed as the means ± one standard deviation (SD) of that mean (Figures 1 and 2).
RESULTS
In the current study all ocular and brain cell types and tissues exhibited easily detectable ACE2 receptor signals, and in agreement with previous reports underscore the ubiquitous nature of ACE2 receptor expression throughout the human visual system and CNS (Figures 1 and 2) [5,6,24]. Importantly, and as indicated in previous studies, negative controls, yeast and microbial RNA controls, synthetic homo-ribonucleotide polymers and random oligonucleotides showed no expression of the ACE2 receptor mRNA [6]. The highest expression of ACE2 mRNA in the brain was found in the brainstem region known as the pons, situated anterior to the cerebellum between the midbrain and medulla oblongata, and an important relay center known to conduct signals from the cerebrum, through to the cerebellum and medulla oblongata, including fiber tracts that involve the transmission of visual sensory signals into the thalamus [36,37]. The pons is known to contain neural circuits that deal primarily with the regulation of respiration, taste (aguesia), audition, saccadic eye movement, facial sensation and expression and equilibrium, all of which are known to be affected or disrupted in COVID-19 disease [7,17,24,25,36–38]. The respiratory tract connects to the brain without the protection of a blood–brain barrier, and that SARS-CoV-2 might in the early invasive phase attack the cardiorespiratory regulatory nodes located in the pons and medulla oblongata, giving rise to both respiratory and cardiac disruption, as is also commonly observed in COVID-19 patients [25,38–39]. The highest expression of the ACE2 receptor protein in the ocular cells and brain tissues involved in visual signaling and examined in this study were found in ocular choroid fibroblasts and retinal pigment epithelial (RPE) cells, a monolayer of pigmented cells of neuro-ectodermal origin situated between the neurosensory retina and the choroid that nourishes the visual photoreceptor cells.
We further note the relatively high expression of the ACE2 receptor mRNA and protein throughout the association neocortex and especially the occipital lobe of the brain that contains the primary visual processing areas. Interestingly ACE2 receptor expression at the protein level was in the order of the corneal epithelial cells < trabecular meshwork cells << non-pigmented ciliary epithelial cells < ocular choroid fibroblasts < whole retina << optic nerve << occipital lobe <<< pons, indicating an increasing ACE2 expression gradient along a pathway from the exterior of the eye (i.e. the anterior sensory aspect of the human visual system) that is exposed to the environment into to deeper vision-relevant regions of the association neocortex located in the occipital lobe (Brodmann area 17; the primary visual cortex).
DISCUSSION
At 29,811 nucleotides the unusually large SARS-CoV-2 virus possesses an enveloped, positive-stranded ssRNA genome that shares significant homology to previously described SARS-CoV and the middle east respiratory syndrome coronavirus (MERS-CoV) [40–42]. SARS-CoV-2 has an extremely high binding affinity for a ubiquitous 92.5 kDa, zinc-containing, membrane-integral ACE2 cell surface receptor (EC 3.4.17.23). SARS-CoV-2-ACE2 binding results in the endocytosis and translocation of both SARS-CoV-2 and the ACE2 receptor into the endosomes of infected cells [5,7,9,22,43–47]. Other ACE2-associated proteins such as the serine protease 2 TMPRSS2 (epitheliasin; EC 3.4.21.109) may facilitate SARS-CoV-2 entry [11,43,48]. Importantly, ACE2 receptor gene expression, both at the level of mRNA and protein is detectable in all eye and brain cells and tissues so far examined and underscores the potential for SARS-CoV-2 entry and infectivity into a remarkably wide variety of host cells populating the human visual and central nervous systems.
Many neurotropic viruses that exhibit ocular tropism, such as the double-stranded DNA (dsDNA) virus herpes simplex 1 (HSV-1) and the ssRNA virus SARS-CoV-2, are also abundant in shed tears and ocular secretions from infected persons [23,24,49–55]. Vital for the health of the outer eye, tear production and drainage via the nasolacrimal duct may provide another novel conduit for SARS-CoV-2 translocation from the eye to the nasal cavity and into the upper respiratory tract that is enriched in ACE2 receptors [23,24,55]. As suggested by the current data another available route of SARS-CoV-2 entry and proliferation in the eye and brain may be via structural support and sensory cells of the eye, through the optic nerve across the optic chiasm to the lateral geniculate nucleus (LGN) and via optic radiations on into the primary visual cortex located in the occipital lobe (Brodmann area 17) of the brain (Figures 1 and 2).
Currently the pathophysiology and ocular pathomechanism for the transmission of SARS-CoV-2 remains an understudied area of viral and retinal neurobiology, and even basic questions, like the initial incapacitating effects of the potent glycoside hydrolase lysozyme (EC 3.2.1.17), a major component of the tear biofluid, on SARS-CoV-2 viability and infectivity are not well understood. Interestingly, ACE2 receptor abundance has been shown to be altered in progressive neurological disorders that include Alzheimer’s disease (AD), but whether or not AD patients have altered susceptibility to SARS-CoV-2 infectivity is another understudied research area [54,55]. Fortunately, the human eye has developed a wide array of defense mechanisms against microbial infection and has evolved a number of anti-microbial (both anti-bacterial and anti-viral) strategies to protect and preserve ocular surfaces and visual system structure and function. Firstly, the surface of the eye is directly exposed to the external environment and hence is susceptible to airborne microbial contamination from potentially pathogenic microbes including viruses, bacteria, fungi and parasites and their pathogen-associated molecules. Airborne contamination includes SARS-CoV-2, often as an aerosolized spray of microdroplets, and is the major known form of a highly infective SARS-CoV-2 viral transmission [24,51,53,56]. Besides different protective anti-microbial substances in the tear fluid such as mucins and globular glycoproteins such as lactoferrin, the tear mucosal film which coats the cornea and conjunctiva contains multiple anti-microbial components including lysozyme (also known as muramidase or N-acetylmuramide glycan-hydrolase), cationic anti-microbial peptides, surfactant protein-D, several RNAse enzymes, S100A peptides such as psoriasin (S100A7) and others, and these are important components of the innate-immune defense system of the eye providing protection against a wide range of potential airborne ocular pathogens [57,58; unpublished observations]. Interestingly, there is recent evidence that the ocular surface microbiota, the resident non-pathogenic symbiotic microorganisms that colonize the conjunctiva and cornea, and the microbiota of other areas of the body, such as the gastrointestinal (GI) tract and oral microbiome are involved in the development and pathophysiology of several ophthalmic diseases including the susceptibility to microbial, and in particular, viral transmission and infection [49–53,59]. Of further emerging interest are the effects of SARS-CoV-2 invasion on the biochemistry, molecular genetics and innate-immunity of neural and ocular host cells and the viral-mediated induction of post-transcriptional and pro-inflammatory signaling factors including certain microRNAs (miRNAs) that modulate the expression of genes involved in viral replication, neuro-inflammation, innate-immune signaling and progressive and age-related aspects of inflammatory neurodegeneration [50–53,59].
SUMMARY
Our scientific understanding of the factors involved in SARS-CoV-2 viral transmission and affinity for multiple human host cell targets continues to evolve. There is now an expanding list of documented impairments of the human visual system and multiple types of visual dysfunction and/or vision deficiency associated with COVID-19 infection [15–25,46,51–53,60–64]. The current study provides new insight into the distribution of the ACE2 receptor in eye and brain cells and tissues involved in visual signal processing. The ACE2 receptor is acknowledged as the major molecular receptor for SARS-CoV-2, and specific anatomical regions of the eye and the visual circuitry of the brain contain abundantly detectable ACE2 receptor mRNA and protein. If the abundance of the ACE2 receptor has any bearing on the ability of attracting and binding SARS-CoV-2, allowing viral entry into human host cells, high ACE2 receptor expression along a gradient from the most exterior surface of the eye via multiple visual processing centers to the primary visual cortex in the occipital lobe provides another novel route for SARS-CoV-2 viral transmission and potential translocation into deeper anatomical regions of the visual brain [64].
In summary the remarkable ubiquity of ACE2 receptor expression in the trabecular meshwork and ocular choroid cells of the outer eye, RPE cells, the optic nerve and optic radiations to the occipital cortex suggests: (i) that multiple cell types of the visual system provide multiple potential entry points for SARS-CoV-2 invasion; (ii) that this broad spectrum of cell types involved in vision and potentially susceptible to SARS-CoV-2 infection reflects on the equally wide range of visual functions that may be impacted by this lethal virus; (iii) that a gradient of increasing abundance of ACE2 receptor expression in several different ocular cell types along the visual processing pathways of the brain provides SARS-CoV-2 access to anatomical regions of the brain involved in visual processing functions; and (iv) that as reflected by ACE2 abundance there are multiple potential compartments that support SARS-CoV-2 accessibility and infiltration in the human visual system. These results further indicate that under certain circumstances that eyeglasses or face-shields may be as important as face-masks in reducing the transmission and spread of the SARS-CoV-2 virus, and that COVID-19 healthcare workers and ophthalmologists may need to take additional care when dealing with both SARS-CoV-2 carriers and COVID-19 patients.
Supplementary Material
ACKNOWLEDGEMENTS
Sincere thanks are extended to Drs. Jian-Guo Cui, Christopher Eicken, Elizabeth Head, Christopher Hebel, Wayne Poon and Piotr N. Alexandrov for short PMI human brain, eye and/or retinal tissues or extracts, ACE2 DNA and cDNA probes, recombinant ACE2 protein control standards, miRNA array work and initial data interpretation, and to Aileen I. Pogue and Darlene Guillot for expert technical assistance and medical artwork. Dr. James M Hill (deceased) had a life-long interest in virology, viral transmission and in ACE2 expression in the aging CNS and in neurological disease and the renin-angiotensin system (RAS) in Alzheimer’s disease (AD). This communication is dedicated to Dr JM Hill’s many years of scientific work in this fascinating research area. Thanks are further extended to the many neuropathologists, physicians and researchers of Canada and the USA who have provided high quality, short postmortem interval (PMI) human CNS, eye and retinal tissues or extracted total brain and eye RNA for scientific study and quantitative analysis.
FUNDING
Research on high quality short PMI human brain, eye and brain and host cell transcriptomics in the Lukiw laboratory involving total human RNA transcription and gene expression, the innate-immune response in AD and in other forms of neurological or retinal disease, amyloidogenesis and neuro-inflammation was supported through an unrestricted grant to the LSU Eye Center from Research to Prevent Blindness (RPB); the Louisiana Biotechnology Research Network (LBRN) and NIH grants NEI EY006311, NIA AG18031 and NIA AG038834 (WJL).
ETHICAL CONSIDERATIONS
The acquisition, handling, experimental, and analytical procedures involving short post-mortem interval (PMI) human eye and brain tissues were carried out in strict accordance with the ethics review board policies at eye, brain and tissue donor institutions and at the Louisiana State University (LSU) Health Sciences Center. Informed consent from next of kin was obtained at eye and brain tissue donor institutions for all tissue samples prior to autopsy and donation; coded short post-mortem interval (PMI) tissue samples (containing no personal identifying information of the donors) were obtained from eye and brain tissue banks used in these studies (see below). The ethical use of short PMI human eye and brain tissues and their analyses were performed in accordance with the Declaration of Helsinki and carried out in strict accordance with the Institutional Biosafety Committee and the Institutional Review Board Committee (IBC/IRBC) ethical guidelines IBC#18059 and IRBC#6774 at the LSU Health Sciences Center, New Orleans LA 70112 USA. Thanks are extended to the Alzheimer’s Disease Research Center (ADRC) at the UC Irvine Institute for Memory Impairments and Neurological Disorders (UCI MIND); a designated Center of Excellence funded by the National Institute on Aging (NIA), a division of the National Institutes of Health (NIH) Bethesda MD USA for high quality short post-mortem human brain tissues. Eye and brain tissues and/or total RNA extracts were also obtained from the University of Toronto, Canada, from the Oregon Health and Sciences University Brain Bank, Portland OR, USA and from eye and brain tissue archives located at the LSU Neuroscience Center, New Orleans LA, USA. Project identification codes: National Institutes of Health grants NEI EY006311; NIA AG18031; NIA AG038834.
Footnotes
DECLARATIONS
Ethics approval and consent to participate – as pointed out in the manuscript text in multiple places, no live humans were used in these studies, only high quality short post-mortem interval (PMI) tissues; please see the ‘Acknowledgements’ and ‘Ethical Considerations’ section (below) for further details;
CONFLICT OF INTEREST AND COMPETING INTERESTS
The authors declare that they have no conflict of interest of any kind with any of the material presented in this research report. The authors declare that they have no competing interests associated with any data in this report.
AVAILABILITY OF DATA AND MATERIALS
All data regarding ‘Methodology’ is available in the ‘Materials and Methods’ Section of the current manuscript. All relevant experimental data in this study are presented in 2 Figures and 2 Supplementary data files appended to this research communication. Any relevant queries on data, materials and methodology should be requested in writing and consultation with the corresponding author. We thank you for your interest in advance and would be happy to provide any ancillary information that you may need.
REFERENCES
- 1.Hu K, Patel J, Patel BC. Ophthalmic manifestations of coronavirus (COVID-19). 2021. [PubMed] [Google Scholar]
- 2.Rodriguez-Morales AJ, Bonilla-Aldana DK, Balbin-Ramon GJ, Rabaan AA, Sah R, Paniz et al. History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel coronavirus epidemic. Infez Med. 2020;28(1): 3–5. [PubMed] [Google Scholar]
- 3.Tiwari R, Dhama K, Sharun K, Yatoo MI, Malik YS, Singh R, et al. COVID-19: animals, veterinary and zoonotic links. Vet Q. 2020:40(1): 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16(10): 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukiw WJ, Pogue AI, Hill JM. SARS-CoV-2 infectivity and neurological targets in the brain. Cell Mol Neurobiol. 2020; 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan WJ, Ni ZY, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020; 382: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabbani N, Olds JL. Does COVID19 infect the brain? Molecular Pharmacology. 2020;97(5): 351–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clausen TM, Sandovalm DR, Spliid CB, Pihl J, Perrett H, Painter CD, et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183(4): 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zubair AS, McAlpine LS, Gardin T, Farhadian S, Kuruvilla DE, Spudich S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review Neurol. 2019;77(8): 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L, Xu Z, Castiglione GM, Soiberman US, Eberhart CG, Duh EJ. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. bioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senanayake P, Drazba J, Shadrach K, Mlisted A, Rungger-Brandle E, Nishiyama K, et al. Angiotensin II and its receptor subtypes in the human retina. Invest Ophthalmol Vis Sci. 2007;48(7): 3301–3311. [DOI] [PubMed] [Google Scholar]
- 13.Cardona GC, Pájaro LDQ, Marzola IDQ, Villegas YR, Salazar LRM. Neurotropism of SARS-CoV-2: mechanisms and manifestations. J Neurol Sci. 2020;412: 116824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong KCK, Mehta PR, Shukla G, Mehta AR. COVID19, SARS and MERS: A neurological perspective. J Clin Neurosci. 2020;77: 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marinho PM, Marcos AAA, Romano AC, Nascimento H, Belfor R. Retinal findings in patients with COVID-19. Lancet. 2020;395(10237); 1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu P, Duan F, Luo C, Liu Q, Qu X, Liang L, et al. Characteristics of ocular findings of patients with COVID-19 in Hubei Province, China. JAMA Ophthalmol. 2020;138: 575–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortiz-Egea JM, Ruiz-Medrano J, Ruiz-Moreno JM. Retinal imaging study diagnoses in COVID-19: a case report. J Med Case Rep. 2021;15(1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Cai D, Huang X, Shen Y. COVID-2019 associated with acquired monocular blindness. Curr Eye Res. 2021;46(8): 1–4. [DOI] [PubMed] [Google Scholar]
- 19.Tisdale AK, Chwalisz BK. Neuro-ophthalmic manifestations of coronavirus disease 19. Curr Opin Ophthalmol. 2020;31(6): 489–494. [DOI] [PubMed] [Google Scholar]
- 20.Sun S, Su W, Lin CQ, Li X, Yan H, Tian B, et al. COVID-19: the novel coronavirus disease and its manifestations and management in ophthalmology. Discov Med. 2020;29( 158): 145–157. [PubMed] [Google Scholar]
- 21.Selvaraj V, Sacchetti D, Finn A, Dapaah-Afriyie K. Acute vision loss in a patient with COVID-19. R I Med J. 2020; 103(6): 37–38. [PubMed] [Google Scholar]
- 22.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015; 1282: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abobaker A, Alzwi A. The Eye: A possible new route of infection in COVID-19. Disaster Med Public Health Prep. 2020;14(4): e25–e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertoli F, Veritti D, Danese C, Samassa F, Sarao V, Rassu N, et al. Ocular findings in COVID-19 patients: a review of direct manifestations and indirect effects on the eye. J Ophthalmol. 2020:4827304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siedlecki J, Brand V, Schworm B, Mayer WJ, Gerhardt M, Michalakis S, et al. COVID-19: ophthalmological aspects of the SARS-COV 2 global pandemic. Klin Monbl Augenheilkd. 2020:237(5): 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zapata MÁ, Banderas-García S, Sánchez-Moltalvá A, Falcó A, Otero-Romero S, Arcos G, Velazquez-Villoria D, et al. Retinal microvascular abnormalities in patients after COVID-19 depending on disease severity. Br J Ophthalmol. 2020. [DOI] [PubMed] [Google Scholar]
- 27.Alexandrov PN, Dua P, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int J Biochem Mol Biol. 2012; 3(4): 365–373. [PMC free article] [PubMed] [Google Scholar]
- 28.Lukiw WJ, Wong L, McLachlan DRC. Cytoskeletal messenger RNA stability in human neocortex: studies in normal aging and in Alzheimer’s disease. Int J Neurosci. 1990;55(2): 81–88. [DOI] [PubMed] [Google Scholar]
- 29.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2005;275(4): 33238–33243. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann H, Geier M, Marzi A, Krumbiegel M, Peipp M, Fey GH, et al. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem Biophys Res Commun. 2004;319(4): 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goulter AB, Goddard MJ, Allen JC, Clark KL. ACE2 gene expression is up-regulated in the human failing heart. BMC Med. 2004; 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem Res. 1990;17(6): 591–597. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2021;29(3): 326–331. [DOI] [PubMed] [Google Scholar]
- 34.Jaber V, Zhao Y, Lukiw WJ. Alterations in micro RNA-messenger RNA coupled signaling networks in sporadic Alzheimer’s disease hippocampal CA1. J Alzheimer Dis Parkinson. 2017;7(2): 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piras IS, Bleul C, Schrauwen I, Talboom J, Llaci L, Both MDD, et al. Transcriptional profiling of multiple system atrophy cerebellar tissue highlights differences between the Parkinsonian and cerebellar subtypes of the disease. Acta Neuropathol Commun. 2020;8: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda K, Kawakami K, Onimaru H, Okada Y, Yokota S, Koshiya N, et al. The respiratory control mechanisms in the brainstem and spinal cord: integrative views of the neuroanatomy and neurophysiology. J Physiol Sci. 2017;67(1): 45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghali MGZ. Respiratory rhythm generation and pattern formation: oscillators and network mechanisms. J Integr Neurosci. 2019; 18(4): 481–517. [DOI] [PubMed] [Google Scholar]
- 38.Riederer P, Ter Meulen V. Coronaviruses: a challenge of today and a call for extended human postmortem brain analyses. J Neural Transm (Vienna). 2020;127(9): 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akhmerov A, Marbán E. COVID-19 and the heart. Circ Res. 2020; 126(10): 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Research. 2008;18(2): 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millet JK, Whittaker GR. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis R COVID-19 vaccine will close in on the spikes. DNA Science Blog. 2020; Public Library of Science. 2020. [Google Scholar]
- 43.Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21(2): 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sah R, Rodriguez-Morales AJ, Jha R, Chu DKW, Gu H, Peiris M, et al. Complete genome sequence of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol Resour Announc. 2020;9(11): e00169–e00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang J, Ye G, Shi K, Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020;581(7807): 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen S, Kannan NB, Kumar J, Rajan RP, Kumar K, Baliga G, et al. Retinal manifestations in patients with SARS-CoV-2 infection and pathogenetic implications: a systematic review. Int Ophthalmol. 2021; 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Wang Y, Luo W, Huang L, Xiao J, Li F, et al. A comprehensive investigation of the mRNA and protein level of ACE2, the putative receptor of SARS-CoV-2, in human tissues and blood cells. Int J Med Sci. 2020;17(11): 1522–1531.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. 2004; 101(12): 4240–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hill JM, Nolan NM, McFerrin HE, Clement C, Foster TP, Halford WP, et al. HSV-1 latent rabbits shed viral DNA into their saliva. Virol J. 2012; 9: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hill JM, Clement C, Zhao Y, Lukiw WJ. Induction of the pro-inflammatory NF-kB-sensitive miRNA-146a by human neurotrophic viruses. Front Microbiol. 2015;6: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho D, Low R, Tong L, Gupta V, Veeraraghavan A, Agrawal R. COVID-19 and the ocular surface: a review of transmission and manifestations. Ocul Immunol Inflamm. 2020; 28: 726–734. [DOI] [PubMed] [Google Scholar]
- 52.Petrillo F, Pignataro D, Lavano MA, Santella B, Folliero V, Zannella C, et al. Current evidence on the ocular surface microbiota and related diseases. Microorganisms. 2020;8(7): 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Yu H, Mei T, Chen B, Chen L, Li S, et al. SARS-CoV-2 on the ocular surface: is it truly a novel transmission route? Br J Ophthalmol. 2020;105(9): 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miners S, Ashby E, Baig S, Harrison R, Tayler H, Speedy E, et al. Angiotensin-converting enzyme levels and activity in Alzheimer’s disease: differences in brain and CSF ACE and association with ACE1 genotypes. Am J Transl Res. 2009; 1(2): 163–177. [PMC free article] [PubMed] [Google Scholar]
- 55.Kehoe PG, Wong S, Al Mulhim N, Palmer LE, Miners JS. Angiotensin-converting enzyme 2 is reduced in Alzheimer’s disease in association with increasing amyloid-β and tau pathology. Alzheimers Res Ther. 2016;8(1): 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khan MM, Parab SR. Simple face shield for public as a crucial factor to slow aerosol transmission during unlock phase of COVID pandemic. Indian J Otolaryngol Head Neck Surg. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garreis F, Gottschalt M, Paulsen FP. Antimicrobial peptides as a major part of the innate immune defense at the ocular surface. Dev Ophthalmol. 2010;45: 16–22. [DOI] [PubMed] [Google Scholar]
- 58.Mohammed I, Said DG, Dua HS. Human antimicrobial peptides in ocular surface defense. Prog Retin Eye Res. 2017;61: 1–22. [DOI] [PubMed] [Google Scholar]
- 59.Lukiw WJ. The microbiome, microbial-generated pro-inflammatory neurotoxins, and Alzheimer’s disease. J. Sport Health Sci. 2016;5(4): 393–396>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16(7): e9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baig AM, Sanders EC. Potential neuroinvasive pathways of SARS-CoV-2: Deciphering the spectrum of neurological deficit seen in COVID-19. J Med Virol. 2020;92(10): 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazzotta C, Giancipoli E. Anterior acute uveitis report in a SARS-CoV-2 patient managed with adjunctive topical antiseptic prophylaxis preventing 2019-ncov spread through the ocular surface route. Int Med Case Rep J. 2020;13: 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gascon P, Briantais A, Bertrand E, Ramtohul P, Comet A, Beylerian M, et al. Covid-19-associated retinopathy: a case report. Ocul Immunol Inflamm. 2020;28(8): 1293–1297. [DOI] [PubMed] [Google Scholar]
- 64.Lukiw WJ. David Hunter Hubel, the ‘Circe effect’, and SARS-CoV-2 infection of the human visual system. J Int Neurosci. 2021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data regarding ‘Methodology’ is available in the ‘Materials and Methods’ Section of the current manuscript. All relevant experimental data in this study are presented in 2 Figures and 2 Supplementary data files appended to this research communication. Any relevant queries on data, materials and methodology should be requested in writing and consultation with the corresponding author. We thank you for your interest in advance and would be happy to provide any ancillary information that you may need.


