One-way translocation and processive cleavage of substrate polypeptide occur in each of the Lon proteolytic active sites.
Abstract
The Lon protease is the prototype of a family of proteolytic machines with adenosine triphosphatase modules built into a substrate degradation chamber. Lon is known to degrade protein substrates in a processive fashion, cutting a protein chain processively into small peptides before commencing cleavages of another protein chain. Here, we present structural and biochemical evidence demonstrating that processive substrate degradation occurs at each of the six proteolytic active sites of Lon, which forms a deep groove that partially encloses the substrate polypeptide chain by accommodating only the unprimed residues and permits processive cleavage in the C-to-N direction. We identify a universally conserved acidic residue at the exit side of the binding groove indispensable for the proteolytic activity. This noncatalytic residue likely promotes processive proteolysis by carboxyl-carboxylate interactions with cleaved intermediates. Together, these results uncover a previously unrecognized mechanism for processive substrate degradation by the Lon protease.
INTRODUCTION
The Lon protease is a highly conserved protease present in bacteria, archaea, and eukaryotic organelles. Bacterial Lon plays critical roles in general quality control of proteostasis by degrading abnormal proteins and in the specific cellular physiological control by degrading specific regulatory proteins involved in stress response, virulence, and group behavior (1).
Lon assembles into a homo-hexameric complex with covalently linked N-terminal region, the AAA+ [adenosine triphosphatases (ATPases) associated with diverse cellular activities] domain, and the protease domain in each protomer (2). Crystal structures have shown that the AAA+ and the protease domains form a closed chamber with six ATPase sites facing out and six proteolytic active sites facing in (3–5). The proteolytic active site can adopt a blocked inactive conformation (6–9); in its active conformation, it forms a deep groove open on two ends, with the catalytic Lys-Ser dyad located at one open end of the groove (3, 4, 10, 11). Previous structures are available of the proteolytic active site bound to covalent inhibitors (4, 11). However, structures of the substrate bound to the protease active site have not been determined yet.
So far, much effort has been devoted to understanding how protein substrates are recognized, unfolded, and translocated by the AAA+ ring of AAA+ proteases (12, 13). However, little is known about how the substrates are hydrolyzed at the proteolytic active sites after they reach inside the proteolytic chamber. Previously, protein substrate degraded by several AAA+ proteases including Lon is known to undergo processive proteolysis, by which the protein substrate is cut into small peptides without the release of partially degraded intermediates (14–17). The enclosed chambers formed by these oligomeric enzymes were previously proposed to sequester protein substrates, thereby providing an immediate explanation for the observed processivity (18).
Here, we present multiple lines of evidence to demonstrate that forming a secluded chamber is not the mechanistic reason for the observed processive substrate degradation by Lon. Three high-resolution cryo–electron microscopy (cryo-EM) maps reveal clear density for translocating substrate polypeptide in each individual proteolytic active site. Crystal structures of various substrate polypeptides bound to Lon, invariably via their C-terminal ends, show that each of the six active sites forms a narrow binding groove accommodating only the unprime residues of a substrate, and provide further clues to a C-to-N processive cleavage mechanism. Assays of protein degradation confirm that processivity occurs at a single proteolytic active-site level. In support of these observations, a previously unrecognized acidic residue is identified at the exit side of the substrate-binding groove; this conserved noncatalytic residue is essential for processive cleavage activity, and its potential role is explained on the basis of the crystal structural results.
RESULTS
Cryo-EM structures of Lon bound to endogenous substrate
Cryo-EM data collection was conducted on the wild-type adenosine triphosphate (ATP)–dependent Lon protease from Meiothermus taiwanensis (MtaLonA), purified in the presence of ATPγS (see Materials and Methods). We obtained three cryo-EM maps, at the resolutions of 2.4, 2.4, and 3.3 Å, by single-particle cryo-EM analysis (figs. S1 and S2 and table S1). The atomic models of Con1 and Con2 were built on the basis of the two 2.4-Å maps (Fig. 1 and fig. S3), which show the N-terminal domains (NTDs) and the hexameric core containing the ATPase and the protease domains (Fig. 1 and fig. S4). Con3 was built on the basis of the 3.3-Å map and is overall comparable to Con2, but the map quality is lower. Therefore, we present and discuss mainly the structures of Con1 and Con2.
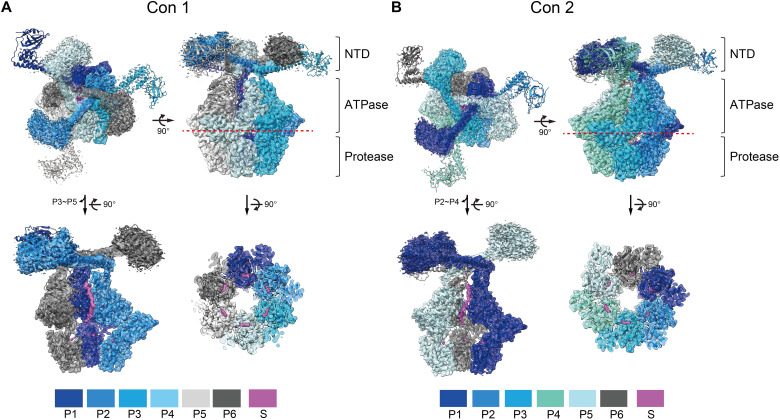
Fig. 1. Overall structures of MtaLonA bound to endogenous substrate.
The fitting of Con1 (A) and Con2 (B) in ribbon models to the 2.4-Å cryo-EM maps (semitransparent) and their cross sections are presented in the top and side views. The six protomers (P1 to P6) of each conformation are arranged in a clockwise direction and displayed in specified colors. In the two lower–left side–view figures, three protomers (P3 to P5 in Con1 and P2 to P4 in Con2) were removed to show the substrate densities inside. The red dotted lines indicate the position of the cross sections to reveal the densities of the substrate (S) in the protease domains (lower right). The contour levels for the NTD regions and the hexameric ATPase-protease core were adjusted to 0.1 and 0.4 in ChimeraX, respectively.
Despite being purified in the absence of exogenous protein substrate, all the reconstructed cryo-maps of MtaLonA showed blobs of density identified as derived from a copurified endogenous substrate, in multiple locations inside the homo-hexameric Lon complex. A substantial piece of density, corresponding to a 22-residue-long substrate polypeptide, is threaded through the helix triangle pore formed by NTDs and bound to the ATPase domains (Fig. 2A). In Con1, the endogenous substrate is engaged by four protomers in conformational states P1 to P4, arranged clockwisely in descending order; the ascending P5 and P6 protomers do not engage the substrate. Protomers in P1 to P3 states bind ATPγS, whereas the P4 to P6 protomers are adenosine diphosphate (ADP) bound (Fig. 2A and fig. S4B). In Con2, the substrate is gripped by the pore-loop-1 residue Y397 from five protomers P1 to P5, arranged in spirally descending order (Fig. 2A). The P6 protomer breaks the helical arrangement, and its pore-loop-1 detaches from the substrate polypeptide. However, the pore-loop-2 residue W431 from all six protomers engages the bound polypeptide (Fig. 2A). In this structure, protomers P1 to P4 are bound to ATPγS, and P5 and P6 are ADP bound (Fig. 2A and fig. S4C). This substrate binding mode has not been observed previously for bacterial Lon (19, 20). In the recent structure of human Lonp1, the bound substrate is similarly gripped by pore-loop-1 from five protomers; however, the bound polypeptide is engaged only by pore-loop-2 from two protomers (21).
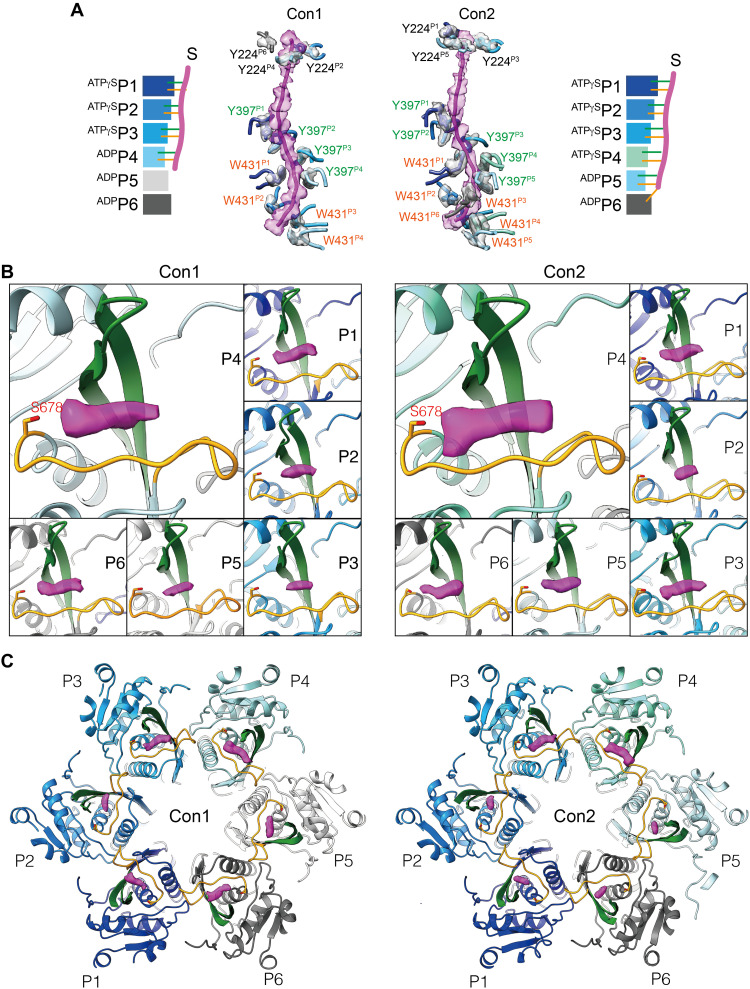
Fig. 2. Translocating substrate in the MtaLonA complex.
(A) The density of the substrate engaging with gate residue Y224 in the helix triangle structure and the pore-loop-1 residue Y397 and -loop-2 residue W431 in the hexameric complex. (B and C) The substrate densities (magenta) occupy the groove formed by the β-hairpin (green) and the extension substrate-binding loop (gold) in the protease domain of each protomer. The locations of each of the substrate densities in the protease domains are shown in the close-up (B) and top views (C). S678 is the catalytic residue.
Unexpectedly, in these maps, clear electron density blobs were also found in each of the six proteolytic active sites. These densities show high occupancies (> 0.75; fig. S5), according to the occupancy maps obtained with LocOccupancy (22). The proteolytic active site forms a deep substrate-binding groove consisting of residues from a β-hairpin and an extended loop (Fig. 2, B and C). Assuming that the density blobs shown in the cryo-EM maps of wild-type Lon with the slowly hydrolyzable ATPγS represent the translocating polypeptide derived from an endogenous substrate, these results reveal two distinct locations for substrate translocation processes occurring inside the Lon complex, one is engaged by the AAA+ ring at the hexamer level and the other by the substrate-binding groove in each protease domain at the monomer level.
Analysis of processive protein degradation
The presence of substantial density blobs in the substrate-binding groove of the protease domain has prompted us to ask the question whether a single Lon proteolytic active site cleaves the substrate in a processive manner, which does not require forming an enclosed oligomeric chamber with ATP-powered substrate translocation activity. To this end, we chose MtaLonC as the study target to investigate substrate degradation. MtaLonC is an ATP-independent protease in M. taiwanensis, which contains an inactive AAA-like domain and a homologous Lon protease domain. MtaLonC forms a symmetrical open barrel assembled by six Lon-like protomers; its AAA-like domains lack the substrate-gripping pore-loops and forms an open pore with a diameter of 13 Å (fig. S6A). The open pore of MtaLonC allows the access of unstructured substrates by passive diffusion to six Lon-like proteolytic active sites inside the barrel (4). Therefore, the open-chambered ATP-independent MtaLonC may offer an opportunity to clarify the substrate-cleavage mechanism by Lon.
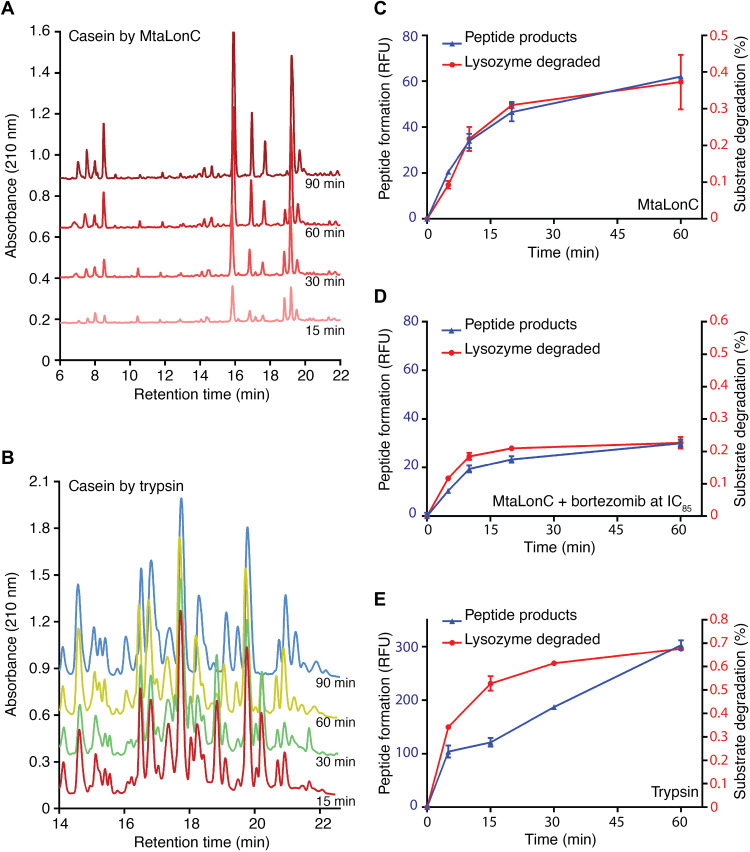
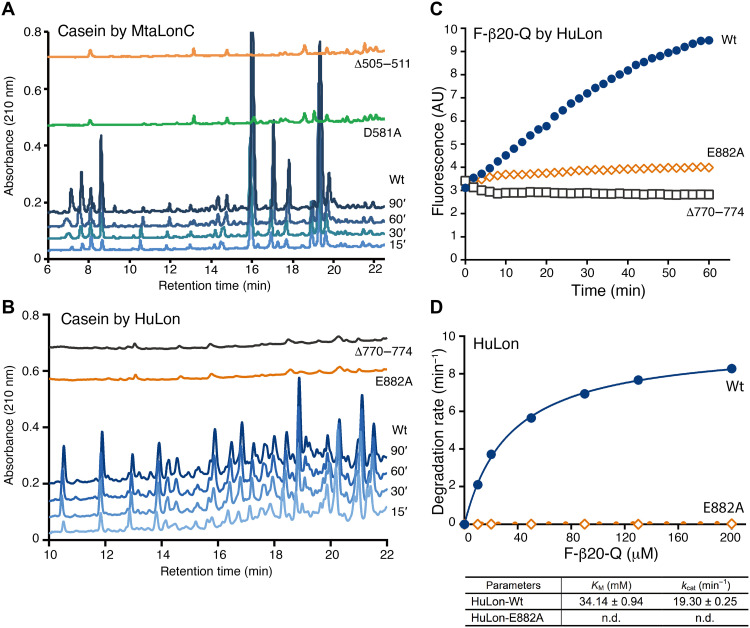
The degraded products of the model substrate casein fluorescein isothiocyanate (FITC-casein), generated at different reaction times, were analyzed by reverse-phase high-performance liquid chromatography (HPLC). Similar experiments have previously been performed to reveal the processive substrate degradation by Lon (14, 16). The substrate degradation profile by MtaLonC shows a time-dependent increase in most peptide-product peaks, which persist throughout the incubation period (Fig. 3A). Notably, the relative ratios of each peak were similar in different reaction times. These results are indicative of processive substrate proteolysis, in which no release of partially degraded intermediates by the enzyme occurs during the degradation process. By contrast, the degraded products generated by trypsin, a nonprocessive peptidase, varied markedly at different reaction times, as shown by the varied product profiles during the reaction (Fig. 3B).
Fig. 3. Processive cleavage of protein substrates by MtaLonC.
(A and B) HPLC profiles of FITC-casein degraded by MtaLonC (A) or the nonprocessive trypsin (B) for 15, 30, 60, and 90 min. (C and D) Substrate degradation assay of reduced lysozyme by MtaLonC in the absence (C) or presence (D) of inhibitor bortezomib at the concentration of IC85 (see Results). (E) Substrate degradation assay of reduced lysozyme by trypsin. The assays were performed by measuring the reaction of the cleaved products with fluorescamine and the consumption of the substrate by integration of the full-length bands on the SDS–polyacrylamide gel electrophoresis gel.
The processivity was also revealed by a different assay, where fluorescamine was used to detect nascent primary amine groups that are generated as a result of peptide-bond cleavage in a substrate, allowing comparison of the rates of the disappearance of the full-length substrate and appearance of the cleaved products (15). If MtaLonC is nonprocessive, then the released polypeptide intermediates after each cleavage reaction may be degraded further in subsequent proteolytic rounds, which would release free nascent amino groups without consumption of any full-length substrate. Consequently, the two rates would not be comparable. Using denatured lysozyme as a model substrate, the appearance of new peptide products reactive to fluorescamine was found to coincide exactly with the disappearance of the full-length substrate (Fig. 3C). The results also confirm the processive substrate cleavage by MtaLonC.
To test whether MtaLonC may exhibit processivity even when most of the six proteolytic sites in the chamber are blocked, we incubated 5/6 maximal inhibitory concentration (i.e., IC85) of the covalent inhibitor bortezomib with MtaLonC to allow only one proteolytic active site per MtaLonC hexamer on average (4). In this inhibitory state, the bortezomib-inhibited MtaLonC still maintained similar processive activity, although the rate of substrate degradation was reduced as expected (Fig. 3D). By contrast, the rates of disappearance of full-length substrate and appearance of product peptides generated by trypsin were not proportional to each other at the reaction times studied (Fig. 3E). Together, these results show that the open-chambered MtaLonC is processive; importantly, the processivity of Lon may occur at the individual proteolytic active site.
Structural basis for processive cleavage of Lon substrate
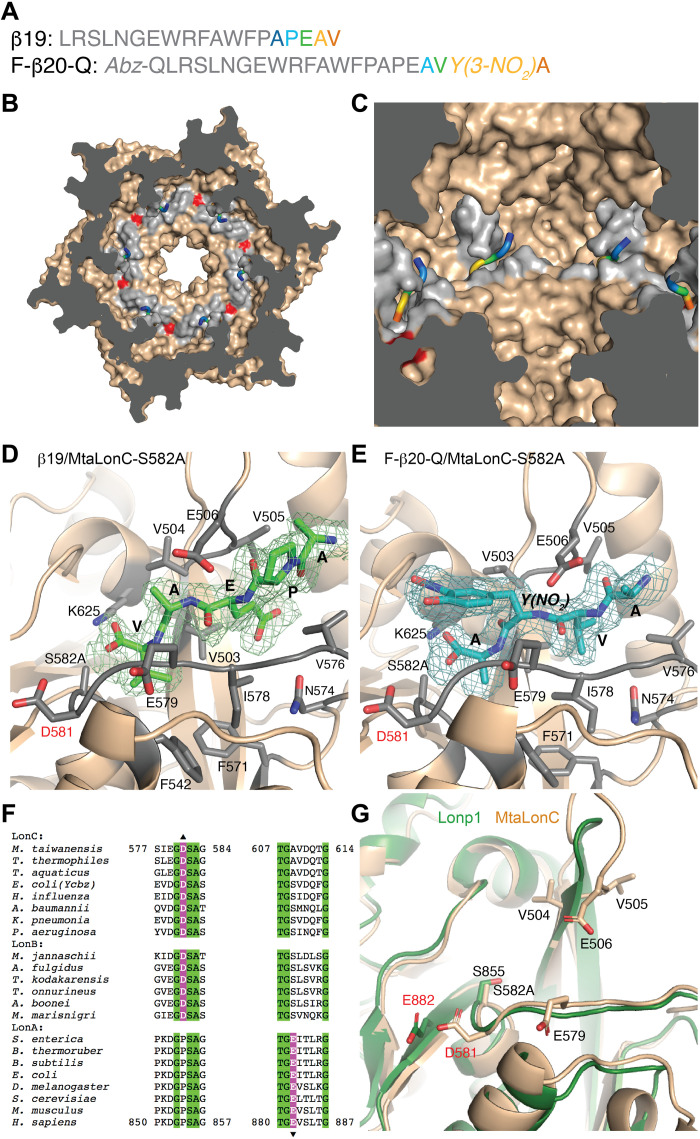
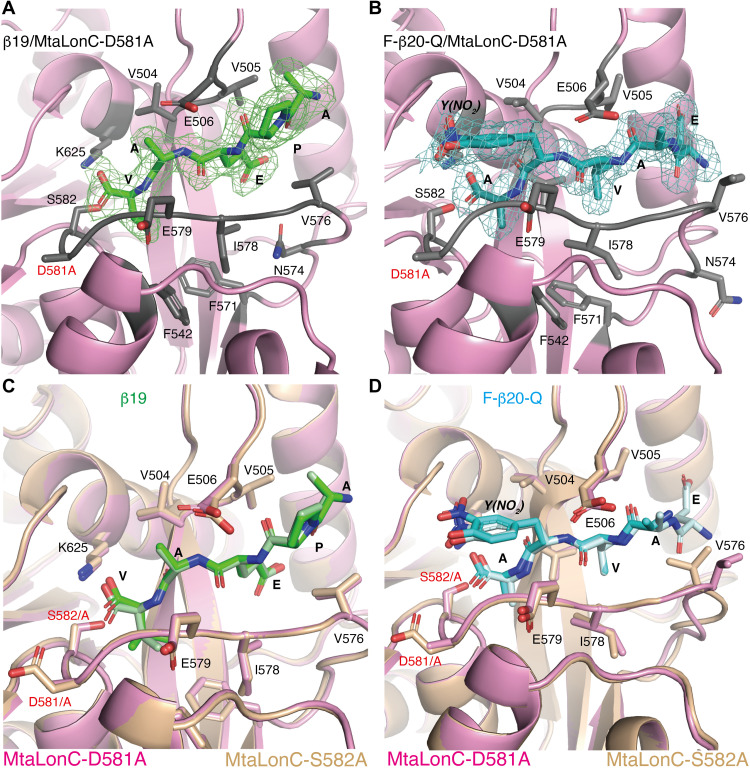
To understand how the substrate polypeptide interacts with the substrate-binding groove of the protease domain, we determined the crystal structures of MtaLonC-S582A, with the catalytic Ser582 mutated to Ala, bound to β19, a known peptide substrate of Lon derived from an internal segment of β-galactosidase, and F-β20-Q, a fluorogenic derivative (Fig. 4A and table S2) (23). MtaLonC effectively cleaves F-β20-Q without ATP and/or Mg2+ (fig. S6B) (24). Both structures show only the C-terminal portions of the polypeptides bound to the groove (Fig. 4, B and C), with well-defined electron density maps for five terminal residues (Ala-Pro-Glu-Ala-Val-COOH) of β19 and four terminal residues (Ala-Val-3-nitro-Tyr-Ala-COOH) of F-β20-Q (Fig. 4, D and E). The substrate peptide engages only backbone hydrogen-bonding interaction with a β-hairpin above and adapts a parallel β sheet conformation with the extended loop below, which forms the substrate-binding groove (Fig. 4, D and E). The C terminus of the peptides is positioned with the two carboxyl oxygens interacting with the catalytic Lys625Nz and Ala(Ser)582N. The homologous binding modes of the two peptides thus suggest that the substrate-binding groove of Lon bound to at least four unprimed substrate residues, which are defined by their location on the N-terminal side of the scissile peptide bond undergoing nucleophilic attacked by the catalytic Ser582. Backbone hydrogen-bonding interactions without involving specific side-chain interactions are also featured in previous crystal structures of other processive enzymes bound to the terminal cleavage recognition units of the substrates (25–27). These results are consistent with a model that the substrate polypeptide in a partially enclosed binding groove of the Lon protease domain is translocated and cleaved processively at the active site in a C-to-N direction, without the intermediates released until multiple rounds of cleavage.
Fig. 4. Crystal structures of MtaLonC-S582A with peptide substrates.
(A) Amino acid sequences of the substrate peptides β19 and F-β20-Q. The C-terminal residues of the peptides, which bind to MtaLonC-S582A, are highlighted by rainbow coloring. (B) Cutaway view showing the location of the six substrate-binding grooves inside the MtaLonC hexamer. Each of the grooves bound to the C-terminal portion of the peptide β19. D581 is colored in red. (C) Close-up view showing four substrate-binding grooves bound to β19 shown as the rainbow-colored coils. (D and E) Structure of the C-terminal residues of β19 (D) and F-β20-Q (E), shown in sticks, bound to MtaLonC-S582A in ribbon models. The simulated-annealing omit maps were contoured at 3.0σ and 2.6σ levels for β19 and F-β20-Q, respectively. D581 and other residues forming the binding groove are also shown in sticks. (F) Sequence alignment showing the conserved acidic residues, Asp or Glu, in the three classes of the Lon proteins. (G) Superimposition of the substrate-binding groove in the protease domains of human Lonp1 and MtaLonC.
By comparing the structures of MtaLonC and those of the canonical ATP-dependent Lon proteases including MtaLonA and Lonp1 (11, 21), we identified Asp581, which is located at the C-end of the substrate-binding loop (Fig. 4, D and E). This acidic residue resides in a hydrophobic patch and is likely protonated. Although the corresponding residue of Asp581 is a proline conserved in MtaLonA and Lonp1 (Fig. 4F), these canonical Lon proteases all have an equivalent acidic Glu residue adjoining the proline residue (Fig. 4G). This suggests that the acidic Asp/Glu abutting upon the substrate-binding groove may be functionally important.
To understand the molecular basis for the processivity in the substrate-binding groove, we carried out structure-based mutational analyses on MtaLonC and human mitochondrial Lon (Lonp1). Deletion of the β-hairpin in MtaLonC (residues 505 to 511; MtaLonCΔ) and in Lonp1 (residues 770 to 774; Lonp1Δ) almost completely abolished cleavage of protein substrates (Fig. 5, A and B). Therefore, substrate interaction by the β-hairpin is critical for productive proteolysis by Lon. Mutating the conserved acidic residue, in MtaLonC-D581A and Lonp1-E882A, inhibited processive substrate cleavage of casein or F-β20-Q (Fig. 5, C and D, and S6C). D581A in MtaLonC is found to affect kcat but not Km of the protease, which is unlike the control mutations expected to alter the substrate-binding groove (fig. S6, B and C). This essential Asp/Glu residue may thus serve a role to attract the free carboxylic end of the cleaved substrate intermediates without affecting binding. It may thereby facilitate one-way translocation of substrate polypeptide in the proteolytic groove upon each cleavage reaction through carboxyl-carboxylate interaction (28, 29). For example, this noncatalytic residue may contribute to rectified Brownian motions driven by proteolysis to achieve processivity (30, 31). If so, a mutant without this key noncatalytic residue may be able to trap the substrate polypeptide in the “pre-Michaelis” state, similar to the peptide-bound MtaLonC-S582A structures. The crystal structures of MtaLonC-D581A bound to β19 and F-β20-Q show the same binding of the C-terminal residues of the substrates (Fig. 6, A and B); the bound peptides align well with their respective structures complexed to the S582A mutant (Fig. 6, C and D). Overall, these results support a processive-cleavage mechanism operated at the proteolytic groove of Lon.
Fig. 5. A conserved acidic residue essential for processive cleavage of substrates.
(A and B) HPLC profiles of the degradation products after 15-, 30-, 60-, and 90-min incubations of FITC-casein with either wild-type (Wt) or mutants of MtaLonC (A), and with either Wt or mutants of Lonp1 (B). AU, arbitrary units. (C) Peptidase activity of Wt and mutants of Lonp1. Cleavage of the intramolecularly quenched fluorogenic peptide F-β20-Q was monitored by real-time fluorescence detection. (D) Enzyme kinetic analysis of Lonp1 and the E882A mutant. The reactions were carried out by incubation at 37°C for 30 min and then stopped before fluorescence measurement. n.d., nondetected values.
Fig. 6. Crystal structures of MtaLonC-D581A with peptide substrates.
(A and B) Structure of the C-terminal residues of β19 (A) and F-β20-Q (B) in sticks, bound to MtaLonC-D581A in ribbons. The omit maps were contoured at 2.9σ and 2.5σ for β19 and F-β20-Q, respectively. (C and D) Superimposition of the structures of β19 (C) and F-β20-Q (D) bound to MtaLonC-D581A and -S582A, where each peptide is in dark and light coloring, respectively.
DISCUSSION
Processive enzymes have evolved to retain their polymeric substrates and perform many rounds of catalysis without dissociation. The structures have been determined of many processive enzymes, which act on either nucleic acids or polysaccharides. Some have a shape like a ring or toroid to completely enclose their substrates; others form a saddle- or hand-shaped groove to partially enclose their substrates (18). In the present work, we show cryo-EM structural evidence that the hand-shaped proteolytic groove of Lon, consisting of a β-hairpin and an extended loop, appears to remain attached to a translocating polypeptide. Our high-resolution cryo-EM maps show multiple blobs of substrate density not only in the center of the AAA+ ring but also in all of the six proteolytic grooves formed in the hexameric core. Presumably, they represent the average density from multiple translocating substrate polypeptides captured in the ~350,000 Lon specimen particles in the grids. In each specimen particle, a substrate polypeptide was translocated through the only pathway formed by the AAA+ ring; however, the C-terminal tail of the translocated substrate was subsequently captured at random by one of the six different proteolytic active sites inside the chamber to undergo processive degradation.
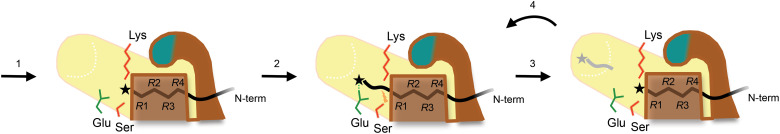
We followed this observation by showing time-independent product profiles of model substrates by a Lon-like protease that does not form a substrate-sequestering chamber, which supports that processivity may occur at the binding groove in the Lon protease domain. The cocrystal structures with four substrate polypeptides collectively show a binding preference of the Lon proteolytic groove for the C-terminal tail of the substrate. Last, structural analysis reveals a strategically arranged acidic residue universally present in all Lon proteins, which is essential for the cleavage activity. Together with proteolysis, this noncatalytic residue may form an “electrostatic ratchet” mechanism for achieving unidirectional processivity (Fig. 7). The size range of peptide products generated by Lon spans from mono- or dipeptides up to those with ~30 residues (16, 32, 33). While the large peptide fragments may result from endoproteolytic cuts of a completely translocated substrate polypeptide by the six spatially arranged active sites in the Lon complex, the shorter peptide products in majority may be generated by the processive feature of each active site, which effectively reduces a three-dimensional (3D) search for a cleavage target site into a 1D process.
Fig. 7. Electrostatic ratchet mechanism proposed for processive cleavage of substrate by the Lon protease.
In step 1, the C terminus of the substrate polypeptide chain (black coil) binds to the proteolytic groove, which accommodates four residues (denotes R1 to R4), resulting in the formation of the complex seen in the crystal structures. In step 2, the substrate polypeptide chain translocates to form a productive complex allowing proteolytic hydrolysis (indicated by a lightning bolt), driven by the electrostatic interaction between the C-terminal carboxylate group of the substrate (represented by a star) and the protonated carboxyl group of Glu residing in a hydrophobic area. In step 3, following the release of the cleaved product, the nascent C-terminal carboxylate group of the bound substrate intermediate is once more attracted by Glu, undergoing another round of rectified Brownian movement to form the next productive complex (step 4). Cycles of steps 3 and 4 yield the substrate density blobs seen in the reconstructed cryo-EM maps. The catalytic Lys-Ser dyad and the conserved Glu residue essential for the cleavage activity are shown as red and green sticks, respectively. The β-hairpin of the substrate-binding groove is highlighted in teal.
MATERIALS AND METHODS
Cloning, protein expression, and purification
The full-length MtaLonA and MtaLonC were cloned into the bacterial expression vector pET21a(+) with a C-terminal 6xHis tag, and the recombinant proteins were expressed and purified by following previously described procedures (4, 11). All plasmids were transformed into BL21(DE3) cells, which were grown in LB medium; protein expression was induced by adding 1 mM isopropyl-thio-β-d-galactoside overnight at 20°C. The cell pellets were resuspended in the lysis buffer [50 mM tris-HCl (pH 8.0) and 500 mM NaCl] and ruptured by a high-pressure homogenizer. The cleared cell lysate was mixed with nickel chelate affinity resins (Ni–nitrilotriacetic acid, Qiagen) for 1 to 2 hours at 4°C. The resins were washed with a binding buffer containing 20 mM imidazole, and the protein was eluted with a binding buffer containing 250 mM imidazole. For the purification of MtaLonA/C and mutants, the eluted proteins were concentrated and further purified by a Superose 6 column (GE Healthcare) equilibrated in 20 mM Hepes (pH 7.5) with 1 mM dithiothreitol. Cloning, expression, and purification of human Lonp1 protease were carried out with the same procedures.
Site-directed mutagenesis
All point and deletion mutations of MtaLonC-Δ505-511, MtaLonC-E579A, MtaLonC-Q573L, MtaLonC-S582A, MtaLonC-D581A, Lonp1-Δ770-774, and Lonp1-E882A were generated by a polymerase chain reaction–based strategy using the QuikChange kit (Stratagene), and the identities of the mutagenized products were verified by sequencing.
Peptidase assays
The peptidase assays were performed using the intramolecularly quenched fluorogenic peptide F-β20-Q {ortho-aminobenzoicacid (Abz)-QLRSLNGEWRF AWFPAPEAV[Tyr(3-NO2)]A}, synthesized by the peptide core of IBC, Academia Sinica. The reaction mixture contains 50 mM tris-HCl (pH 8.0), 10 mM NaH2PO4, 3 to 100 mM F-β20-Q, and 100 nM wild-type or mutant MtaLonC (hexamer). For real-time fluorescence detection of substrate cleavage, the reaction was monitored at 55°C using a FluoroMax-4 spectrofluorometer (Horiba, CA). For enzyme kinetic analysis, the reactions were carried out by incubation at 55°C (for MtaLonC) or 37°C (for Lonp1) for 30 min and were then stopped by adding an equal volume of 7.4 M guanidine hydrochloride before fluorescence measurement (excitation, 320 nm; emission, 420 nm) using a FlexStation 3 microplate reader (Molecular Devices, Sunnyvale, CA). The fluorescent cleavage product of F-β20-Q was estimated on the basis of a standard curve created using known concentrations of F-β20-Q fully cleaved by trypsin. The protocol of huLon protease assay is the same as described above. The kinetic parameters Km and kcat were calculated using the program Prism 6.0 (GraphPad Software Inc., San Diego). Each assay was performed in triplicate.
Substrate degradation assays
For analysis by fluorescamine to detect an increase in primary amine groups caused by substrate degradation, 6 mM reduced lysozyme [by treating with 1 mM tris-(2-carboxyethyl)phosphine (TCEP)] was incubated with 50 nM MtaLonC (hexamer), in the absence or presence of 15 mM of the covalent inhibitor bortezomib, in a buffer containing 20 mM Hepes (pH 7.5), 100 mM NaCl, and 10 mM NaH2PO4, for 0, 5, 10, 20, and 60 min at 55°C. The 5/6 maximal inhibitory concentration (i.e., the IC85) of bortezomib against 50 nM MtaLonC (hexamer) was determined using the F-β20-Q as the substrate. Degradation by trypsin was carried out by mixing reduced lysozyme (5 mg/ml) with trypsin (5 mg/ml) in 50 mM Hepes buffer (pH 7.5) for 0, 5, 15, 30, and 60 min at 37°C. Twenty microliters of each reaction sample was mixed with 80 ml of 100 mM Hepes (pH 7.5) and 50 ml of fluorescamine (0.3 mg/ml in dimethyl sulfoxide) by vortexing. Fluorescence was then measured with an excitation wavelength of 390 nm and emission wavelength of 475 nm using a FlexStation 3 microplate reader (Molecular Devices, Sunnyvale, CA). For product analysis by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), 16 μl of each reaction sample obtained as described above from the fluorescamine assays was loaded onto a NuPAGE gel (4 to 12% bis-tris; Invitrogen). Substrate protein bands stained by Coomassie blue were quantified using a Gel Doc XR+ imaging system (Bio-Rad).
Analysis of protein degradation by reverse-phase chromatography
For the degraded product analysis by reverse-phase HPLC, degradation of S2-casein (Sigma-Aldrich) and reduced lysozyme (Sigma-Aldrich), which were prepared as described (24), by wild type or mutants of MtaLonC or huLon (with 10 mM MgCl2 and 15 mM ATP) was carried out by incubating the substrate (300 μg) with the enzyme (30 μg) in a buffer containing 50 mM tris-HCl (pH 8.0) and 10 mM NaH2PO4 for 15, 30, 60, and 90 min at 55°C (for MtaLonC) or 37°C (for huLon). The reaction was stopped by adding an equal volume of 7.4 M guanidine hydrochloride before injecting onto a Hypersil BDS C18 3μ column (150 × 2.1 mm; Thermo Fisher Scientific), and elution was performed at 0.2 ml/min with a gradient of acetonitrile (from 0 to 100% in 60 min) in 0.1% trifluoroacetic acid using an UltiMate 3000 LC system (Dionex, CA). Peaks were detected at 210 nm.
Protein crystallization and soaking experiments
Crystallization of MtaLonC-S582A and MtaLonC-D581A was carried out at 22°C by the sitting-drop vapor diffusion technique. MtaLonC-S582A and MtaLonC-D581A formed plate-like crystals over a reservoir containing 10% isopropanol, 100 mM NaH2PO4, and 100 mM sodium citrate at pH 4.6. Crystals of the enzyme-substrate complex were obtained by soaking the apo crystals in mother-liquid drops containing 0.5 mM of either β19 or F-β20-Q, which contains the fluorophore (F; Abz) and the quencher (Q; nitrotyrosine) residues, synthesized by the in-house peptide core laboratory. Crystals were harvested and immersed briefly in the mother liquid containing 30% glycerol before data collection.
Cryo-EM data acquisition
The MtaLonA sample was diluted at a final concentration of around 0.5 mg/ml. Three microliters of the sample were applied onto glow-discharged 200-mesh R1.2/1.3 Quantifoil copper grids. The grids were blotted for 4 s and rapidly cryo-cooled in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4°C and 100% humidity. The sample was screened using a Talos Arctica cryo-EM (Thermo Fisher Scientific) operated at 200 kV. It was then imaged in a Titan Krios cryo-EM (Thermo Fisher Scientific) at a magnification of ×96,000 (corresponding to a calibrated sampling of 0.82 Å per pixel). Micrographs were recorded by EPU software (Thermo Fisher Scientific) with a Falcon 4 detector, where each image was composed of 40 individual frames in gain-normalized mrc format with an exposure time of 5.8 s and an exposure rate of 8.28 electrons second per Å2. A total of 11,071 movie stacks were collected.
Single-particle image processing and 3D reconstruction
All micrographs were first imported into Relion (34) for image processing. The motion correction was performed using MotionCor2 (35), and the contrast transfer function (CTF) was determined using CTFFIND4 (36). Then, the micrographs with “rlnMotionEarly < 10” and “rlnCtfMaxResolution < 5” were selected using the “subset selection” option in Relion. All particles were autopicked using the NeuralNet option (threshold 1 = 0; threshold 2 = −5) in EMAN2 (37). Then, particle coordinates were imported to Relion, where the poor 2D class averages were removed by several rounds of 2D classification. A total of 1,019,665 particles were transferred to cryoSPARC (38) for ab initio map generation. Then, two good classes with apparent N-termini and better-resolved AAA+ and protease domains, containing 781,063 particles, were subjected to nonuniform refinement in cryoSPARC. One round of heterogeneous refinement was performed to further classify the particles using the map from the nonuniform refinement as starting references, and three good classes (Con1, Con2, and Con3) were generated. Homogeneous refinement and local refinement were then conducted to achieve the final maps. A 2.44-Å map from 324,693 particles for Con1, a 2.36-Å map from 361,692 particles for Con2, and a 3.28-Å resolution map from 77,159 particles for Con3 were lastly obtained. Resolutions for the final maps were estimated with the 0.143 criterion of the Fourier shell correlation curve. Resolution maps were calculated in cryoSPARC using the “Local Resolution Estimation” option.
Structure determination and model building
For model building of the 2.4-Å resolution cryo-EM map of MtaLonA Con1, the atomic structure of a full-length Lon protease bound to casein from a companion work (39) was rigidly fitted into the cryo-EM map. Molecular dynamics flexible fitting (40) was then used to adjust the conformational differences. The resultant model was optimized with Coot (41) and phenix.real_space_refine (42). The type of the bound nucleotides, ADP or ATPγS, was determined by LigandFit in Phenix, with an overall correlation coefficient of the ligand to the map of over 0.7. The complete model consisting of full-length MtaLonA, bound nucleotide, and substrate was optimized using phenix.real_space_refine. The same modeling procedures were also conducted on the 2.4-Å resolution cryo-EM map of MtaLonA Con2 and 3.3-Å resolution cryo-EM map of MtaLonA Con3. The final models were evaluated by MolProbity (43). Statistics of the model building are summarized in table S1.
The crystal structures of polypeptide substrates bound to MtaLonC-S582A and MtaLonC-D581A were determined from datasets collected at the beamlines BL44XU (with F-β20-Q) of SPring-8 (Japan) and at 13B1 (with β19) of NSRRC (Taiwan). Data were processed using HKL2000 (44). The structures were determined by molecular replacement with the program Phaser (45) using the structure of apo-MtaLonC as the search model (4). The models of the substrate peptides were built and fit into the electron density map using the program Coot (46). The structures were refined with iterative cycles of manual refitting in Coot and refinement using the program Refmac5 (47). Crystallographic and refinement statistics are listed in table S2. The figures were prepared using Chimera (48), ChimeraX (49), and PyMol (50).
Acknowledgments
We thank the High-End Cryo-EM Platform, Core Facility Center for Life Sciences, University of Science and Technology of China for the EM facility support and W. Chiu at Stanford University for the support of cryo-EM data collection. We also thank W.-L. Yen for technical assistance on the HPLC and enzymatic assays. We appreciate the assistance from P. P. Lin and D.-M. Chang for scientific computing and Academia Sinica Cryo-EM Facility for support of cryo-grid preparation.
Funding: Academia Sinica (C.-I.C.), Ministry of Science and Technology grant 108-2320-B-001-011-MY3 (C.-I.C.), and Start-up funding by the University of Science and Technology of China KY9100000032 and KJ2070000080 (K.Z.)
Author contributions: Conceptualization: K.Z. and C.-I.C. Methodology: K.Z. and C.-I.C. Investigation: S.L., K.-Y.H., C.-I.K., S.-C.S., K.-F.H., and K.Z. Visualization: S.L., K.-Y.H., and C.-I.C. Funding acquisition: K.Z. and C.-I.C. Project administration: K.Z. and C.-I.C. Supervision: K.Z. and C.-I.C. Writing, original draft: S.L., K.-Y.H., K.Z., and C.-I.C. Writing, review and editing: S.L., K.-Y.H., K.Z., and C.-I.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Cryo-EM maps of MtaLonA have been deposited in the Electron Microscopy Data Bank (EMD-31589, EMD-31590, and EMD-31607); the coordinates of the atomic models of Con1, Con2, and Con3 have been deposited in the Protein Data Bank with the accession codes 7FID, 7FIE, and 7FIZ, respectively. The coordinates and structural factors of the crystals of MtaLonC bound to substrate peptides have been deposited in the Protein Data Bank with PDB codes 7EV4 (S582A/F-β20-Q), 7EUY (S582A/β19), 7EV6 (D581A/F-β20-Q), and 7EUX (D581A/β19). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S2
REFERENCES AND NOTES
- 1.Van Melderen L., Aertsen A., Regulation and quality control by Lon-dependent proteolysis. Res. Microbiol. 160, 645–651 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Rotanova T. V., Melnikov E. E., Khalatova A. G., Makhovskaya O. V., Botos I., Wlodawer A., Gustchina A., Classification of ATP-dependent proteases Lon and comparison of the active sites of their proteolytic domains. Eur. J. Biochem. 271, 4865–4871 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Cha S.-S., An Y. J., Lee C. R., Lee H. S., Kim Y.-G., Kim S. J., Kwon K. K., De Donatis G. M., Lee J.-H., Maurizi M. R., Kang S. G., Crystal structure of Lon protease: Molecular architecture of gated entry to a sequestered degradation chamber. EMBO J. 29, 3520–3530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao J.-H., Ihara K., Kuo C.-I., Huang K.-F., Wakatsuki S., Wu S.-H., Chang C.-I., Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors. Acta Crystallogr. D Biol. Crystallogr. 69, 1395–1402 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Lin C.-C., Su S.-C., Su M.-Y., Liang P.-H., Feng C.-C., Wu S.-H., Chang C.-I., Structural insights into the allosteric operation of the Lon AAA+ protease. Structure 24, 667–675 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Botos I., Melnikov E. E., Cherry S., Kozlov S., Makhovskaya O. V., Tropea J. E., Gustchina A., Rotanova T. V., Wlodawer A., Atomic-resolution crystal structure of the proteolytic domain of Archaeoglobus fulgidus lon reveals the conformational variability in the active sites of lon proteases. J. Mol. Biol. 351, 144–157 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Duman R. E., Löwe J., Crystal structures of Bacillus subtilis Lon protease. J. Mol. Biol. 401, 653–670 (2010). [DOI] [PubMed] [Google Scholar]
- 8.García-Nafría J., Ondrovicová G., Blagova E., Levdikov V. M., Bauer J. A., Suzuki C. K., Kutejová E., Wilkinson A. J., Wilson K. S., Structure of the catalytic domain of the human mitochondrial Lon protease: Proposed relation of oligomer formation and activity. Protein Sci. 19, 987–999 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Im Y. J., Na Y., Kang G. B., Rho S.-H., Kim M.-K., Lee J. H., Chung C. H., Eom S. H., The active site of a lon protease from Methanococcus jannaschii distinctly differs from the canonical catalytic Dyad of Lon proteases. J. Biol. Chem. 279, 53451–53457 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Botos I., Melnikov E. E., Cherry S., Tropea J. E., Khalatova A. G., Rasulova F., Dauter Z., Maurizi M. R., Rotanova T. V., Wlodawer A., Gustchina A., The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site. J. Biol. Chem. 279, 8140–8148 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Su S.-C., Lin C.-C., Tai H.-C., Chang M.-Y., Ho M.-R., Babu C. S., Liao J.-H., Wu S.-H., Chang Y.-C., Lim C., Chang C.-I., Structural basis for the magnesium-dependent activation and hexamerization of the Lon AAA+ protease. Structure 24, 676–686 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Sauer R. T., Baker T. A., AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Puchades C., Sandate C. R., Lander G. C., The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol. Cell Biol. 21, 43–58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurizi M. R., Degradation in vitro of bacteriophage lambda N protein by Lon protease from Escherichia coli. J. Biol. Chem. 262, 2696–2703 (1987). [PubMed] [Google Scholar]
- 15.Akopian T. N., Kisselev A. F., Goldberg A. L., Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J. Biol. Chem. 272, 1791–1798 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Nishii W., Suzuki T., Nakada M., Kim Y.-T., Muramatsu T., Takahashi K., Cleavage mechanism of ATP-dependent Lon protease toward ribosomal S2 protein. FEBS Lett. 579, 6846–6850 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Thompson M. W., Singh S. K., Maurizi M. R., Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 269, 18209–18215 (1994). [PubMed] [Google Scholar]
- 18.Breyer W. A., Matthews B. W., A structural basis for processivity. Protein Sci. 10, 1699–1711 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S., Hsieh K.-Y., Su S.-C., Pintilie G. D., Zhang K., Chang C.-I., Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease. J. Biol. Chem. 297, 101239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin M., Puchades C., Asmita A., Puri N., Adjei E., Wiseman R. L., Karzai A. W., Lander G. C., Structural basis for distinct operational modes and protease activation in AAA+ protease Lon. Sci. Adv. 6, eaba8404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin M., Watson E. R., Song A. S., Mindrebo J. T., Novick S. J., Griffin P. R., Wiseman R. L., Lander G. C., Structures of the human LONP1 protease reveal regulatory steps involved in protease activation. Nat. Commun. 12, 3239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur S., Gomez-Blanco J., Khalifa A. A. Z., Adinarayanan S., Sanchez-Garcia R., Wrapp D., McLellan J. S., Bui K. H., Vargas J., Local computational methods to improve the interpretability and analysis of cryo-EM maps. Nat. Commun. 12, 1240 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gur E., Sauer R. T., Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 22, 2267–2277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao J.-H., Kuo C.-I., Huang Y.-Y., Lin Y.-C., Lin Y.-C., Yang C.-Y., Wu W.-L., Chang W.-H., Liaw Y.-C., Lin L.-H., Chang C.-I., Wu S.-H., A Lon-like protease with no ATP-powered unfolding activity. PLOS ONE 7, e40226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jedrzejas M. J., Mello L. V., de Groot B. L., Li S., Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate. J. Biol. Chem. 277, 28287–28297 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., McCabe K. A., Bell C. E., Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. U.S.A. 108, 11872–11877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorentzen E., Conti E., Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell 20, 473–481 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Sawyer L., James M. N. G., Carboxyl–carboxylate interactions in proteins. Nature 295, 79–80 (1982). [DOI] [PubMed] [Google Scholar]
- 29.Torshin I. Y., Harrison R. W., Weber I. T., Close pairs of carboxylates: A possibility of multicenter hydrogen bonds in proteins. Protein Eng. 16, 201–207 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Saffarian S., Collier I. E., Marmer B. L., Elson E. L., Goldberg G., Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science 306, 108–111 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Xie P., Molecular motors that digest their track to rectify Brownian motion: Processive movement of exonuclease enzymes. J. Phys. Condens. Matter 21, 375108 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Maehara T., Hoshino T., Nakamura A., Characterization of three putative Lon proteases of Thermus thermophilus HB27 and use of their defective mutants as hosts for production of heterologous proteins. Extremophiles 12, 285–296 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Nishii W., Maruyama T., Matsuoka R., Muramatsu T., Takahashi K., The unique sites in SulA protein preferentially cleaved by ATP-dependent Lon protease from Escherichia coli. Eur. J. Biochem. 269, 451–457 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Li S., Hsieh K.-Y., Kuo C.-I., Lee S.-H., Pintilie G. D., Zhang K., Chang C.-I., Complete three-dimensional structures of the Lon protease translocating a protein substrate. Sci. Adv. 7, eabj7835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trabuco L. G., Villa E., Mitra K., Frank J., Schulten K., Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.P. D. Adams, P. V. Afonine, G. Bunkóczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. -W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, P. H. Zwart, in International Tables for Crystallography (2012), pp. 539–547. [Google Scholar]
- 43.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z., Minor W., [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 45.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phasercrystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rigsby R. E., Parker A. B., Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 44, 433–437 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S2