FGF and Wnt signaling controls the phase transition of the ocular fate.
Abstract
The developing vertebrate eye cup is partitioned into the neural retina (NR), the retinal pigmented epithelium (RPE), and the ciliary margin (CM). By single-cell analysis, we showed that fibroblast growth factor (FGF) signaling regulates the CM in its stem cell–like property of self-renewal, differentiation, and survival, which is balanced by an evolutionarily conserved Wnt signaling gradient. FGF promotes Wnt signaling in the CM by stabilizing β-catenin in a GSK3β-independent manner. While Wnt signaling converts the NR to either the CM or the RPE depending on FGF signaling, FGF transforms the RPE to the NR or CM dependent on Wnt activity. The default fate of the eye cup is the NR, but synergistic FGF and Wnt signaling promotes CM formation both in vivo and in human retinal organoid. Our study reveals that the vertebrate eye develops through phase transition determined by a combinatorial code of FGF and Wnt signaling.
INTRODUCTION
The vertebrate neural retina (NR) is insulated from extraocular tissue by the retinal pigmented epithelium (RPE) and circumscribed by the ciliary body (CB) and the iris in the periphery; the latter two structures control the intraocular pressure and the light intake, respectively (1, 2). These ocular structures arise in the embryonic eye cup from corresponding zones of the NR, RPE, and ciliary margin (CM) (Fig. 1A). Similarly, the Drosophila eye is insulated optically by the subretinal pigment layer (SRP) at its base and the pigment rim (PR) on the side, which is derived from the pupal eye disc similar to the NR (3). The molecular mechanism specifying the peripheral eye structure also appears to be conserved. In Drosophila, Wingless secreted by the eye-encasing head capsule generates a gradient of Wnt signaling required for PR formation (3). In mice, Wnt signaling mediated by β-catenin is necessary for development of both the RPE and the CM (4–7). Constitutively active β-catenin transforms the NR to the PR in Drosophila, but only to the CM in mice (3, 7, 8). Why Wnt signaling in mice fails to produce ectopic RPE in the retina is not known.
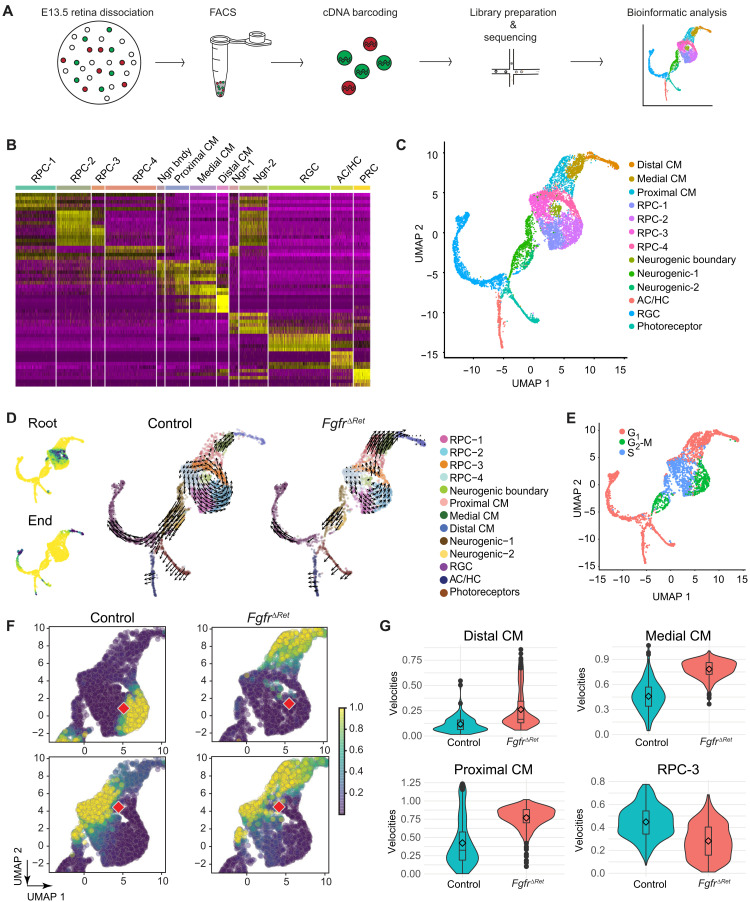
Fig. 1. Genetic ablation of FGFRs disrupted CM development.
(A) Evolutionary conservation of peripheral ocular structures. The vertebrate eye cup is partitioned into the NR, the RPE, and the CM, the latter of which gives rise to the CB and the iris. This resembles the Drosophila eye where the NR is shielded by the pigment rim (PR), the head capsule (HC), and the subretinal pigment (SRP). (B) Deletion of FGFRs in the peripheral retina abolished FGF signaling as indicated by the loss of pERK, Etv5, and Spry2 expression. (C) NR domain (dotted lines) marked by Vsx2, Sox2, Pax6, NICD, Gli1, Sfrp2, and Atoh7 expression was reduced in FgfrΔRet mutants. (D) FgfrΔRet mutant retina displayed ectopic expression of Mitf, Pcad, and Cx43 (dotted lines) but down-regulated Wfdc1, indicating loss of the CM domain. (E) FgfrΔRet mutant animals exhibited dysmorphic CB (arrows) and iris hypoplasia (arrowheads) at P7 and aniridia (white arrowheads) in adulthood. Scale bars, 100 μm.
In vertebrate eye development, fibroblast growth factor (FGF) is another transforming signal like Wnt. We and others have shown that genetic inactivation of FGF signaling disrupts formation of the NR and the optic disc (9–11). In contrast, ectopic expression of FGF not only transforms the RPE to the NR (12–15) but also induces markers of the CM in the junction between the ectopic NR and the remaining RPE (16). It is unclear, however, whether the ectopic CM is induced directly by FGF or indirectly by the ectopic NR. On the basis of experiments performed in retinal organoids, Sasai and colleagues (17) proposed that mutual inhibition of FGF and Wnt pivots the retinal progenitor cells to either the NR or RPE fate, leaving the NR-RPE boundary tissue to self-organize into the CM. In the current study, we show that, unlike in Drosophila, murine embryos have opposing morphogen gradients of FGF and Wnt in the peripheral eye cup induced by their distinctive sources of ligands in the NR and the lens ectoderm, respectively. Single-cell analysis further revealed that FGF signaling promotes subdivision of the CM zone in a dose-dependent manner. Unexpectedly, FGF signaling is required to maintain the high level of Wnt activity necessary for CM fate. By genetic manipulation in mouse models and chemical induction in retinal organoids, we demonstrate that the vertebrate eye cup is specified in a phase transition mode that can be programed reversibly by a binary code of FGF and Wnt signaling.
RESULTS
FGF signaling is required for CM development
During embryonic development, we observed a proximalhigh-distallow pattern of phosphorylated extracellular signal–regulated kinase (pERK) in embryonic day 13.5 (E13.5) retinae (Fig. 1B), suggesting a gradient of FGF signaling (18). To examine its functional significance, we ablated FGF receptors (FGFRs) 1 and 2 using Pax6 α-Cre, which is active in the peripheral retina beginning at E10.5 (19). In concordance with the role of FGF in inducing ERK signaling, both phosphorylation of ERK and expression of FGF response genes Etv5 and Spry2 were lost in distal retinae of α-Cre;Fgfr1flox/flox;Fgfr2flox/flox (FgfrΔRet) mutants (Fig. 1B and fig. S1), leading to a significant reduction in the retinal progenitor cell domain marked by Vsx2, Sox2, Pax6, Notch1 intracellular domain (NICD), Gli1, and Sfrp2 (Fig. 1C and fig. S1). As a result, the zone of the Atoh7+ NR was also contracted, whereas the RPE markers Mitf, Pcad, and Cx43 invaded deeply into FgfrΔRet mutant retinae, followed by a notable reduction in the pan-CM marker Wfdc1 (Fig. 1D and fig. S1). At post natal day 7 (P7), control pups displayed well-organized CBs and elongated iris, but only clumps of pigmented cells were present at the tip of FgfrΔRet mutant retinae (Fig. 1E). In accordance with this, adult FgfrΔRet mice displayed almost complete aniridia (lacking the iris), demonstrating the critical role of FGF signaling in CM development.
Single-cell analysis reveals that FGF signaling controls self-renewal, differentiation, and survival of CM progenitors
To investigate the molecular basis of FGF signaling in regulating CM development, we performed single-cell RNA sequencing (scRNAseq) of E13.5 eye cups. Taking advantage of an internal ribosome entry site (IRES)–green fluorescent protein (GFP) cassette embedded in the Pax6 α-Cre driver, we used flow cytometry to enrich for Cre/GFP-expressing cells from peripheral retinae (Fig. 2A) and sequenced 6628 control and 4607 FgfrΔRet mutant cells at the mean depth of 2811 genes per cell. As expected, unsupervised clustering analysis identified distinctive groups of retinal progenitor cells (RPC-1 to RPC-4), neurogenic cells (Ngn-1, Ngn-2, and boundary), and differentiated retinal neurons [retinal ganglion cells (RGCs), amacrine/horizontal cells (ACs/HCs), and photoreceptor cells (PRCs)], which were confirmed by expression of known molecular markers (Fig. 2B and fig. S2A). Unlike previous scRNAseq analyses (20, 21), however, we were also able to discern three clusters that express CM-specific markers Mitf, Wls, Msx1, and Wfdc1 (Fig. 2B and fig. S2B). Projected onto the two-dimensional uniform manifold approximation and projection (UMAP) plot, these CM cells formed a separate branch connected to the RPC clusters, diametrically opposite to the branch of retinal neurons (Fig. 2C). We next examined the trajectory of cell differentiation by RNA velocity analysis, which exploits dynamics of mRNA splicing to uncover the rate and direction of transcriptomic changes (22). It correctly identified the differentiation path from the RPC to either the NR or the CM fates (Fig. 2D), which correlated with the transitioning of the cell cycle from the S phase via the G2-M to G1 phase (Fig. 2E). Our analysis showed that once progenitor cells on the NR trajectory entered the neurogenic state in the G2-M phase, they irreversibly progressed toward terminal differentiation. On the CM trajectory, however, progenitor cells (RPC-3) residing in the G2-M phase could either move toward the terminal CM state or re-enter the cell cycle to replenish the RPC pool. Thus, CM progenitor cells displayed the hallmark of stem cells in their capacity to either self-renew or differentiate. In FgfrΔRet mutants, however, RPCs were biased against self-renewal in favor of the terminal CM fate. This was confirmed by comparing transition probabilities between control and mutant progenitor cells at similar locations within RPC clusters (Fig. 2F). Consistent with this, cell proliferation was significantly reduced in FgfrΔRet mutants (fig. S3A) and the overall RNA velocity slowed down considerably in the circling RPC pool, but it accelerated in proximal and medial CM clusters before ramping down in the distal CM (Fig. 2G). This accounted for the increase in the percentage of CM cells at the expense of RPC cells (fig. S3B). Therefore, FGF signaling controls the decision of self-renewal versus differentiation of CM progenitors.
Fig. 2. Single-cell analysis showed that FGF signaling regulates self-renewal and differentiation of CM progenitors.
(A) Schematic diagram of scRNAseq. (B) Heatmap of differential gene expression in single-cell clusters. RPC, retinal progenitor cell; RGC, retinal ganglion cell; AC, amacrine cell; HC, horizontal cell; PR, photoreceptor. (C) UMAP representation of single-cell clusters. (D) Left: Diffusion analysis of RNA velocities identified the root and the end of cell differentiation as represented by high-density regions (dark green) after forward and reverse Markov processes. Right: Cell differentiation trajectories were revealed by the velocity field projected on the UMAP plot. Arrows indicate local RNA velocities on a regular grid. (E) State of the cell cycle represented on the UMAP plot. (F) Single-step transition probabilities from the starting cells (red square) to neighboring cells showed the bias of CM progenitors in FgfrΔRet mutants toward differentiation against self-renewal. (G) Quantification of single-cell velocities in each cluster showed the accelerated differentiation and reduced proliferation of CM progenitor cells.
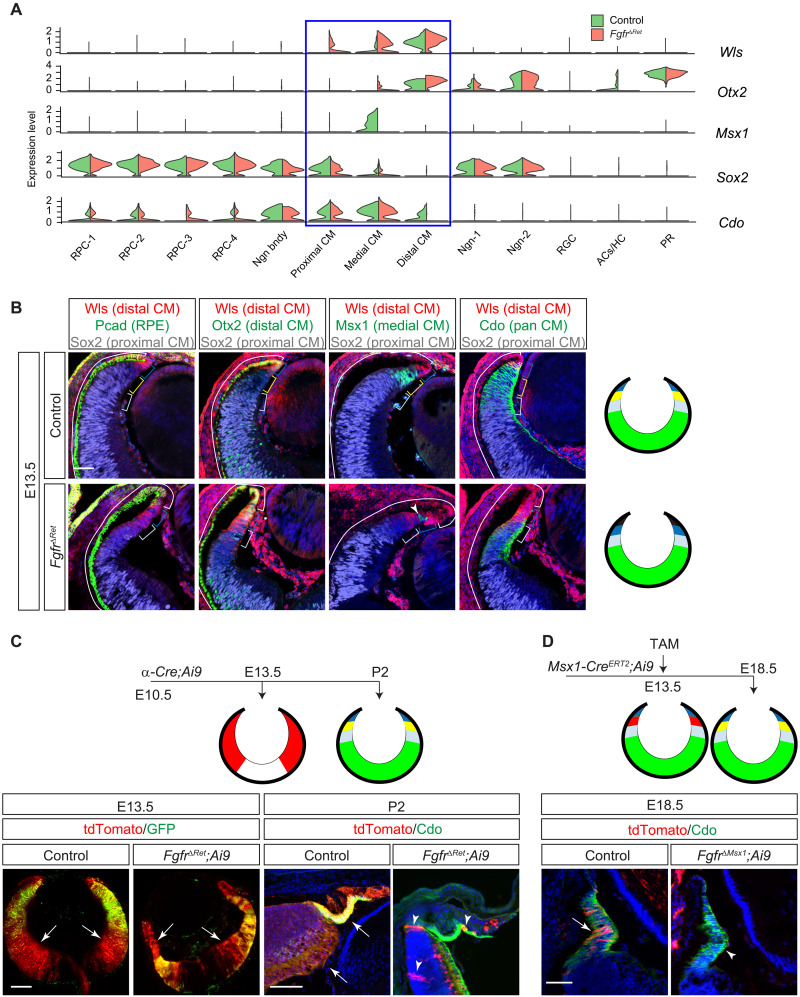
We next asked whether FGF signaling is also required for terminal differentiation of the CM. Our scRNAseq analysis classified CM cells into three clusters: the distal CM defined by expression of Wls and Otx2, the medial CM by Msx1, and the proximal CM by overlapping expression of Sox2 and Cdo (Fig. 3A). We showed that this in silico segregation of CM cells corresponds to their spatial separation in situ (23), as expression of Wls and Otx2 extended beyond the Pcad+ RPE into the distal tip of the retina, bordering the domain of Msx1 expression (Fig. 3B). Msx1, in turn, overlapped with Sox2 and Cdo, the latter two reaching further into the center of the retina. In contrast, FgfrΔRet mutant retinae displayed encroachment of Pcad, expansion of Wls and Otx2, loss of Msx1, and retreat of Sox2 and Cdo. This was in agreement with scRNAseq analysis of FgfrΔRet mutants that Wls and Otx2 expanded from the distal CM into the medial or even proximal CM domains at the expense of Msx1, Sox2, and Cdo. These results demonstrated that FGF signaling is required for spatial demarcation and molecular distinction of the CM subdivisions.
Fig. 3. FGF signaling is required for CM subdivision and survival.
(A) Violin plots showed that the CM can be subdivided by the overlapping expression of Wls, Otx2, Msx1, Sox2, and Cdo, all of which were dysregulated in FgfrΔRet mutants. (B) Immunostaining confirmed that the in silico clustering of CM cells matches the spatial separation of CM subdomains. Aberrant invasion of Pcad, expansion of Wls and Otx2, loss of Msx1, and reduction in Sox2 and Cdo in FgfrΔRet mutants demonstrated CM differentiation defects. Brackets indicate the domain of the RPE and the distal, medial, and proximal CM, corresponding to black, dark blue, yellow, and light blue regions, respectively, in diagrams on the right. The NR is indicated in green. Arrowhead marks residual wild-type cells still expressing Msx1. Scale bar, 50 μm. (C) At E13.5, although Pax6 α-Cre was expressed only in the peripheral retina indicated by its GFP reporter, it has already activated tdTomato expression (arrows) from the Ai9 Cre reporter throughout the retina. At P2, these tdTomato+ progenies remained in control retinae, but only a few (arrowheads) were left in FgfrΔRet mutants. The tdTomato expression is indicated in red in diagrams. Scale bars, 100 μm. (D) CM cells were pulse-labeled by tamoxifen induction of Msx1-CreERT2 at E13.5 and detected at E18.5 by tdTomato expression from the Ai9 Cre reporter. Although these cells remained in the control CM identified by Cdo expression (arrow), they had largely disappeared in the FgfrΔRet mutant (arrowhead), suggesting cell survival defects. Scale bar, 50 μm.
Last, we examined the potential role of FGF signaling in retinal cell survival. To this end, we permanently labeled α-Cre–expressing cells at E10.5 using the Ai9 reporter, which expressed tdTomato protein after Cre-mediated recombination (Fig. 3C). At E13.5, we observed that tdTomato+ cells spread to much of the retina in both control and FgfrΔRet mutants, even beyond the α-Cre–expressing domain marked by GFP expression. Although control retinae at P2 still displayed ubiquitous expression of tdTomato, only a handful of tdTomato+ cells persisted in FgfrΔRet mutants, likely a result of incomplete recombination due to Cre mosaicism. To determine whether loss of tdTomato+ cells in FgfrΔRet mutants was solely due to depletion of the RPCs, we selectively labeled medial CM cells by inducing Msx1-CreERT2 with tamoxifen at E13.5 and examined their fates at E18.5 (Fig. 3D). In contrast to abundant expression of tdTomato reporter in control CBs marked by Cdo, Msx1-CreERT2;Fgfr1flox/flox;Fgfr2flox/flox;Ai9 (FgfrΔMsx1;Ai9) mutants were largely devoid of tdTomato+ cells. Therefore, FGF signaling is required for survival of CM cells even after their onset of differentiation.
The CM is patterned by graded FGF signaling
We next focused on how FGF signaling regulates patterning of the CM. Our scRNAseq analysis revealed that the retina expresses three FGF ligands—Fgf3 restricted to the RPCs, Fgf9 extending into the proximal CM, and Fgf15 further encompassing the medial CM (Fig. 4A). This nested pattern of Fgf expression coincides with the proximal-distal gradient of FGF signaling (Fig. 1B) and subdivision of the CM in the distal retina. We hypothesized that sequential removal of Fgfs may flatten the gradient of FGF signaling and disrupt the patterning of the CM (Fig. 4B). To test this model, we first deleted Fgf9, which was previously shown to control the anterior boundary of the RPE (24). In α-Cre;Fgf9flox/flox (Fgf9ΔRet) mutants, Mitf expression advanced from the distal CM toward the central retina, accompanied by the down-regulation of Vsx2, Wfdc1, and Msx1 expression in the proximal CM domains (Fig. 4, B and D). We next ablated both Fgf3 and Fgf9 in α-Cre;Fgf3flox/flox;Fgf9flox/flox (Fgf3/9ΔRet) mutants, which led to further encroachment of Mitf into the retina accompanied by a reduction in Vsx2 and Wfdc1 and loss of the medial CM maker Msx1. These results suggested that the graded expression of Fgfs in the distal retina is necessary for the subdivision of the CM.
Fig. 4. CM development is dependent on the dosage of FGF signaling.
(A) scRNAseq analysis revealed a nested pattern of Fgf3, Fgf9, and Fgf15 expression in the retina. (B) By reducing the dosage of Fgf ligands, Fgf9ΔRet and Fgf3/9ΔRet exhibited progressive expansion of the distal CM/RPE marker Mitf, reduction in the proximal CM/NR marker Vsx2, and loss of pan-CM marker Wfdc1 and medial CM marker Msx1. (C) Overexpression of Fgf8 in Fgf8OE mutants stimulated pERK and FGF-responsive gene Spry2 in the presumptive RPE territory, which was transformed to NR as indicated by Atoh7 expression. Reducing the strength of FGF signaling by deleting Fgfr1 and Fgfr2 in FgfrΔRet;Fgf8OE mutants down-regulated pERK. This prevented expression of Atoh7 but induced the CM markers Otx1 and Msx1, indicating the transformation to the CM fate. The Cre expression in the RPE territory was indicated by GFP expression from the α-Cre driver. Scale bars, 100 μm. (D) Relative area expressing each marker gene was normalized against the entire inner (left) or outer (right) eye cup, and the statistical significance was evaluated using one-way analysis of variance (ANOVA) test. *P < 0.01, **P < 0.001, and ***P < 0.0001; n = 3 for all markers.
Previous studies have shown that overactivation of FGF signaling can transform the RPE into the NR. Because our above result indicated that the dosage of Fgfs is important for the specification of the CM in the retina, we wondered if titration of FGF signaling may redirect the RPE to the CM fate. To this end, we first crossed α-Cre with R26LSL-Fgf8 to induce ectopic expression of Fgf8 in the retina, which is expected to diffuse to the RPE to activate FGFRs (Fgfr1 to Fgfr4). This was confirmed by the ectopic induction of pERK and FGF response gene Spry2 throughout the GFP-expressing RPE/NR double-layered eye cup in α-Cre;R26LSL-Fgf8 (Fgf8OE) embryos (Fig. 4C). Furthermore, the original RPE layer now resembled the NR in both the thickness and the Vsx2+Atoh7+Mitf −Gja1− expression pattern, confirming its conversion into the NR. We next attenuated the ectopic FGF signaling in the former RPE territory by ablating Fgfr1 and Fgfr2, leaving only Fgfr3 and Fgfr4 available to transmit the Fgf8 signal. This genetic manipulation reduced but did not eliminate pERK and Spry2 staining in the presumptive RPE in the α-Cre;Fgfr1flox/flox;Fgfr2flox/flox;R26LSL-Fgf8 (FgfrΔRet;Fgf8OE) embryo. Although this region remained thickened compared to the control RPE, it no longer expressed the NR marker Atoh7 but instead acquired medial/distal CM markers Msx1 and Otx1 (Fig. 4, C and D). While the Cre activity was extended throughout the RPE domain as shown by both the GFP expression from the α-Cre driver and the tdTomato expression from the Ai9 Cre reporter, the proximal CM marker Cdo was confined to the distal eye cup, indicating preservation of the spatial relationship to Msx1 and Otx1 (fig. S4). Together, these results showed that graded FGF signaling is important for CM differentiation genes in both the RPE and the retinal domains.
FGF signaling determines the transformative activity of Wnt signaling
Because the above results demonstrate an essential role of FGF signaling in CM development, we wondered whether FGF interacts with Wnt signaling, which is also known to promote the CM fate (7). Analyzing our scRNAseq data, we noticed that Lef1 and Axin2, two response genes for canonical Wnt signaling, were down-regulated in FgfrΔRet mutants, suggesting that FGF signaling may be required for Wnt activity in the peripheral retina (Fig. 5A). This was unexpected because previous studies have suggested that FGF and Wnt play opposing roles in early eye cup patterning (6, 17). We thus sought to confirm our findings by mosaic analysis. As described above, inactivation of FGF signaling resulted in expansion of Mitf and Otx2 expression into the FgfrΔRet peripheral retina (Fig. 5B). Because of the mosaic activity of α-Cre, however, we occasionally observed patches of cells still negative for Mitf and Otx2 in distal retinae, suggesting that they were residual wild-type cells. Lef1 expression was retained in these Mitf−/Otx2− cells but lost in their neighbors (Fig. 5B, arrowheads and dotted lines), demonstrating that FGF signaling cell-autonomously regulates Wnt signaling.
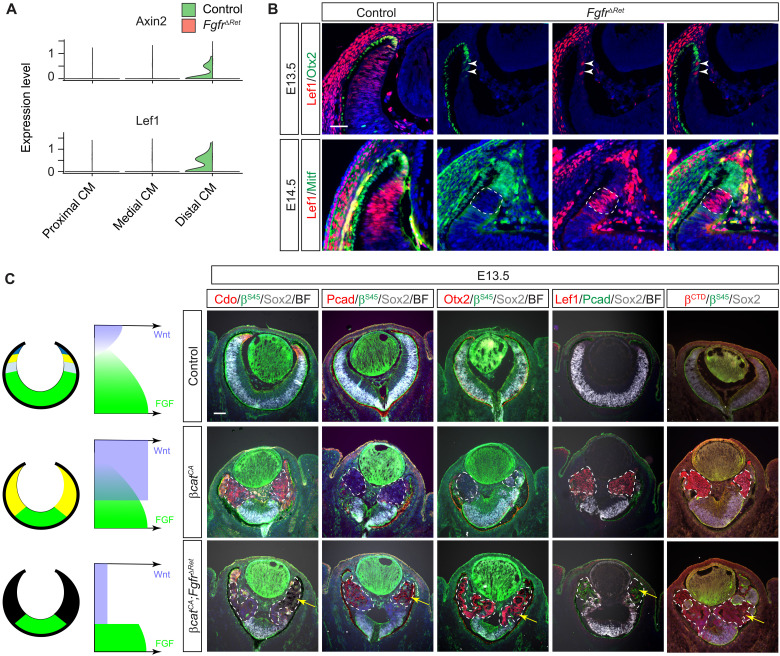
Fig. 5. FGF signaling determines the CM fate by promoting Wnt signaling.
(A) Violin plots showed down-regulation of Wnt response genes Lef1 and Axin2 in the FgfrΔRet mutant transcriptome. (B) In mosaic analysis, FgfrΔRet mutant cells acquired ectopic expression of Otx2 and Mitf at the expense of Lef1. In contrast, the remaining wild-type cells identified by the lack of Otx2 and Mitf expression still maintained Lef1 expression. Scale bar, 50 μm. (C) Constitutive activation of Wnt signaling by deleting the β-cateninSer45 (βSer45) motif transformed βcatCA retinae to the CM as indicated by Cdo expression. Further deletion of FGFRs in FgfrΔRet;βcatCA mutant retinae converts them to the RPE as indicated by ectopic expression of Pcad and Otx2 as well as the appearance of pigmentation (arrows). Both the level of Lef1 and the amount of β-catenin detected by the β-catenin C-terminal antibody (βCTD) arose in βcatCA retinae but declined in FgfrΔRet;βcatCA mutants, showing that the Wnt–β-catenin signaling is dependent on FGF signaling. The mutant regions are marked by dotted lines. Scale bar, 100 μm.
We next asked whether FGF signaling is also required functionally for Wnt signaling to promote CM formation. To test this hypothesis, we first induced constitutive Wnt signaling in the retina by deleting exon 3 of β-catenin, which encodes the Ser45 phosphorylation site necessary for glycogen synthase kinase 3β (GSK3β)–regulated degradation (25). Identified by an antibody specific to the β-cateninSer45 (βSer45) residue, β-catenin mutant cells in α-Cre;β-cateninfl3/fl3 (βcatCA) retinae up-regulated the expression of the CM marker Cdo at the expense of Sox2 (Fig. 5C). This is consistent with previous reports that activation of Wnt signaling transforms the NR to the CM (7, 8). The Cdo expression, however, was abolished after the inactivation of FGF signaling in α-Cre;Fgfr1flox/flox;Fgfr2flox/flox;β-cateninfl3/fl3 (FgfrΔRet;βcatCA) retinae. Instead, these FgfrΔRet;β-cateninCA cells acquired expression of Otx2 and Pcad as well as RPE-like pigmentation (Fig. 5C, arrows), suggesting that they have taken on an RPE identity. Together, these results demonstrated that the level of FGF signaling dictates whether Wnt signaling transforms the NR to either the CM or the RPE fate.
To understand the mechanism by which FGF signaling regulates Wnt activity, we examined Lef1 expression induced by the stabilized β-catenin. As expected, Lef1 expression was greatly elevated in βcatCA retinae, but it became significantly attenuated after the loss of FGF signaling in FgfrΔRet;βcatCA retinae (Fig. 5C). This is consistent with our above finding that FGF signaling influences Lef1 expression in wild-type retina (Fig. 5, A and B). Moreover, we observed that βcatCA mutants exhibited strong cytoplasmic β-catenin staining as revealed by a β-catenin C-terminal antibody (βCTD), but this staining was markedly reduced in FgfrΔRet;βcatCA mutants. Because βcatCA mutants expressed the truncated β-catenin resistant to GSK3β phosphorylation, this result suggested that FGF signaling promotes the stability of β-catenin in a GSK3β-independent manner. To determine the downstream effector of FGF signaling, we further disrupted the mitogen-activated protein kinase (MAPK) pathway by deleting Mek1 and Mek2 in α-Cre;Mek1flox/flox;Mek2−/− (MekΔRet) mutants, which was confirmed by loss of pERK in the peripheral retina. MekΔRet animals phenocopied FgfrΔRet mutants in their aberrant expression of Vsx2, Atoh7, Wfdc1, Msx1, and Mitf during the embryogenesis and dysgenesis of the CB and iris in adults (fig. S5). Together, these results show that FGF-MAPK signaling stabilizes β-catenin to promote Wnt signaling in CM development.
The lens ectoderm and periocular mesenchyme induce a Wnt signaling gradient to pattern the eye cup
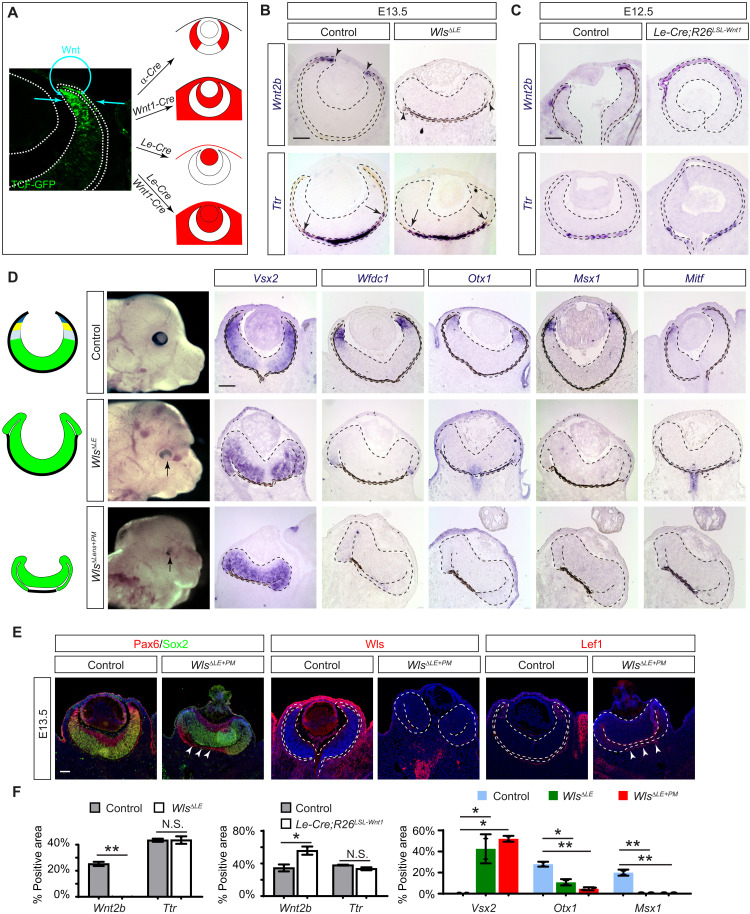
In Drosophila development, the head capsule patterns the peripheral eye structures by secreting Wnt ligand to induce the Wnt signaling gradient (3). We wondered whether this mechanism of Wnt signaling induction is conserved in vertebrates. The vertebrate RPE consists of the proximal and distal domains (2); the latter forms the pigmented epithelium of the CB lining the nonpigmented epithelium derived from the CM. Consistent with previous reports (7, 26), we found that Wnt signaling revealed by a highly sensitive T cell factor (TCF)–GFP reporter is strongest in the distal CM and distal RPE before tapering off toward the center of the eye cup (Fig. 6A) (27). We reasoned that there could be three potential sources of Wnt—the distal retina, the periocular mesenchyme, and the lens ectoderm (the surface ectoderm and the lens). These tissues can be targeted by α-Cre, Wnt1-Cre, and Le-Cre, respectively (Fig. 6A), allowing us to eliminate their Wnt production by genetically ablating Wntless, a cytoplasmic transporter necessary for Wnt secretion (19, 26, 28, 29). In both α-Cre;Wntlessflox/flox (WlsΔRet) and Wnt1-Cre;Wntlessflox/flox (WlsΔPM) embryos, we did not observe any ocular defects (fig. S6). In line with a previous report, however, Le-Cre;Wntlessflox/flox (WlsΔLE) mutants exhibited a posterior shift of the RPE-retina boundary (Fig. 6B) (26). In addition, we observed that WlsΔLE mutants lost the distal RPE marker Wnt2b, but not the proximal marker Ttr, suggesting a role of lens ectoderm–derived Wnt ligands in RPE regionalization (Fig. 6, B and F) (30). We further generated Le-Cre;R26LSL-Wnt1 to overexpress Wnt1 in the lens ectoderm, which abolished lens development because of its inhibitory effect in lens induction (Fig. 6, C and F) (31). In the eye cup, however, this led to expansion of the Wnt2b+ domain without affecting Ttr expression. Moreover, the distal eye cup in WlsΔLE mutants expressed the RPC marker Vsx2, but none of the pan-CM markers Wfdc1 and Otx1, the medial CM marker Msx1, and the distal CM marker Mitf (Fig. 6, D and F). Together, these results showed that the lens ectoderm–derived Wnt is responsible for specification of both the distal RPE and the CM.
Fig. 6. Paracrine Wnt signaling patterns the CM and the distal RPE.
(A) Potential source of the Wnt signaling gradient revealed by the TCF-GFP reporter may be targeted by α-Cre for retina, Wnt1-Cre for periocular mesenchyme, and Le-Cre for the lens ectoderm (the surface epithelium and the lens). (B) Ablation of Wnt transporter Wls in the lens ectoderm specifically disrupted the proximal RPE differentiation in WlsΔLE mutants as indicated by loss of Wnt2b but not the proximal RPE marker Ttr. (C) Overexpression of Wnt1 in the lens ectoderm also affected the proximal RPE by expanding the domain of Wnt2b expression in Le-Cre;R26LSL-Wnt1 eye cup without affecting Ttr expression. (D) CM was lost in WlsΔLE mutants as shown by the expansion of Vsx2 and the absence of Wfdc1, Otx1, Msx1, and Mitf. The band of the RPE was further diminished in WlsΔLE+ΔPM mutants. (E) WlsΔLE+ΔPM mutant eyes lost Wls and Lef1 in both the lens ectoderm and the periocular mesenchyme, leaving only a small band of Lef1 expression next to the Pax6-expressing RPE (arrowheads). Scale bars, 100 μm. (F) Relative area expressing Wnt2b, Ttr, or Vsx2 was normalized against the entire RPE region, while the Otx1- or Msx1-expressing area was normalized against the NR region. Student’s t test for Wnt2b and Ttr and one-way ANOVA test for Vsx2, Otx1, and Msx1. *P < 0.05 and **P < 0.01; n = 2 for all markers. N.S., not significant.
We next explored the source of Wnt ligands for specifying the proximal RPE by combining Wnt1-Cre and Le-Cre to abolish Wnt secretion from both the lens ectoderm and the periocular mesenchyme. Compared to WlsΔLE embryos, Le-Cre;Wnt1-Cre;Wntlessflox/flox (WlsΔLE+ΔPM) mutants exhibited further shortening of the RPE that led to a severe hypopigmentation of the eye (Fig. 6D). This was confirmed by Pax6 and Sox2 staining, which showed extensive invasion of the NR into the backside of the WlsΔLE+ΔPM mutant eye cup, leaving only a vestigial segment of the RPE in the center (Fig. 6E, arrowheads). As Wls was ablated in the lens, the surface ectoderm, and the periocular mesenchyme, Lef1 expression was also largely eliminated in these regions, except in a small band of mesenchymal cells next to the remaining RPE (Fig. 6E, arrowheads). These results demonstrated that the lens ectoderm, in partial redundancy with the periocular mesenchyme, induces a Wnt signaling gradient in the eye cup to specify the CM and the distal RPE.
Balanced FGF and Wnt signaling drives CM fate
Our results thus far demonstrated that the eye cup is biased toward the fate of the NR, RPE, or CM depending on the availability of FGF and Wnt signaling. What then is the default fate of the eye cup when both FGF and Wnt signaling are absent? To answer this question, we first abolished Wnt signaling by deleting β-catenin in α-Cre;β-cateninflox/flox (βcatΔRet) mutants. Consistent with previous studies (7, 8), this transformed the CM into the NR-like tissue as evident by the expansion of Sox2 and loss of Cdo and Msx1 without incurring invasion of Otx2 and Pcad (Fig.7A). As a result, the pERK level was also elevated in the distal retina. We next ablated both FGFRs and β-catenin in α-Cre;Fgfr1flox/flox;Fgfr2flox/flox;β-cateninflox/flox3 (FgfrΔRet;βcatΔRet) embryos. Despite the loss of pERK in the β-catenin–negative domain, these FgfrΔRet;βcatΔRet mutant cells retained the same NR-like expression patterns as βcatΔRet mutants (Fig. 7A), indicating that the eye cup developed into the NR in the absence of FGF and Wnt signaling.
Fig. 7. FGF and Wnt induce phase transition to generate the CM.
(A) Deletion of either β-catenin alone (βcatΔRet) or β-catenin and Fgfrs together (FgfrΔRet;βcatΔRet) transformed the distal retina into the NR as indicated by loss of Cdo and Msx1, lack of Otx2 and Pcad, and expansion of Sox2 (all at 100% in the β-catenin or β-catenin/pERK–deficient regions marked by dotted lines; n = 3). (B) Although overexpression of Wnt1 in Wnt1OE embryos did not produce any phenotype in the retina, when combined with overexpression of Fgf8, it transformed the RPE domain of the Wnt1OE;Fgf8OE eye cup into the CM as indicated by the up-regulation of the CM-specific genes Otx1 and Cdo and the lack of the RPE marker Mitf and the NR marker Atoh7. (C) Untreated hiPSC organoid culture contained few MSX1+ or CDO+ CM-like cells. Addition of the 3 μM WNT agonist CHIR99021 significantly expanded the number of MSX1+ and CDO+ cells. (D) Quantification of the MSX1+ and CDO+ area as the percentage of the total organoid culture. (E) FGF and Wnt signaling promotes the abrupt and reversible transition of eye cup progenitors into three phases: the NR, the CM, and the RPE. Our study showed that the NR can be transformed by constitutive activation of Wnt signaling (βcatCA) into the CM, which is converted to the RPE after the loss of FGF signaling (FgfrΔ). The RPE reverts back to the CM by titrating FGF signaling (Fgf8OE;FgfrΔ). Otherwise, ablation of Wnt signaling (βcatΔ) or overexpression of Fgf8 (Fgf8OE) can turn the RPE into the NR, which may transition further into the CM after overexpression of Wnt1 (Wnt1OE). Scale bars, 100 μm.
We next explored the fate of the eye cup when FGF and Wnt signaling is elevated simultaneously. We first bred α-Cre;R26LSL-Wnt1 (Wnt1OE) animals to overexpress Wnt1 in the retina. Likely because the Wnt transporter Wls is restricted to the distal CM, Wnt1OE embryos did not exhibit any ocular phenotypes (Fig. 7B), consistent with the above result that the retinal Wnt is dispensable for eye cup patterning (fig. S4). In contrast, coexpression of both Wnt1 and Fgf8 in α-Cre;R26LSL-Wnt1;R26LSL-Fgf8 (Wnt1OE;Fgf8OE) mutants led to thickening of the presumptive RPE, which exhibited ectopic FGF signaling activity as indicated by the expression of Spry2 (Fig. 7B). Unlike Fgf8OE mutants (Fig. 4C), however, the transformed RPE region in Wnt1OE;Fgf8OE mutants failed to express the NR marker Atoh7 but up-regulated the pan-CM markers Otx1 (control: 0%, mutant: 78%, P < 0.0001, n = 3) and Cdo at the expense of the RPE marker Mitf (control: 100%, mutant: 40%, P < 0.0001, n = 3). As reducing the dosage of ectopic Fgf8 signaling can also transform the RPE into the CM-like tissue, these results demonstrated that the CM fate is specified by the relative, not the absolute, levels of FGF and Wnt signaling.
With this understanding of FGF and Wnt function in CM development, we sought to improve Sasai and colleagues’ (17) induction-reversal method to generate CM-like stem cells from human embryonic stem cells. Instead of sequential induction of RPE and NR tissues by adding and then removing GSK and FGFR inhibitors, we hypothesized that, in the absence of an FGFR inhibitor, an exogenous WNT agonist may cooperate with endogenous FGF produced by the retinal organoid to directly promote the CM fate (32). Using human induced pluripotent stem cells (hiPSCs), we confirmed that the standard retinoid differentiation protocol without GSK and FGFR inhibitors produced only a small amount of CM-like tissue as indicated by the limited MSX1 and CDO expression (Fig. 7C). Addition of WNT agonist CHIR99021 (3 μM) drastically increased the number of MSX1+ and CDO+ cells in the organoid culture (Fig. 7, C and D). Therefore, WNT signaling in the presence of FGF promotes expression of CM differentiation genes in human retinal organoids.
DISCUSSION
In this study, we have shown that both FGF and Wnt are transformative signals in eyecup patterning, forming the basis of a combinatorial code that determines the ocular cell fate. This can be depicted in a two-dimensional map on the strength of FGF and Wnt signaling (Fig. 7E). As demonstrated by our genetic analysis, stimulation of Wnt signaling by constitutively active β-catenin transforms the NR into the CM, which can be further converted to the RPE after removal of FGFRs. The RPE can revert back to the NR by deleting β-catenin or overexpressing FGF. Otherwise, FGF overexpression may be co-opted to transform the RPE to the CM by either the overexpression of Wnt1 or the ablation of FGFRs, which either balances or reduces the strength of FGF signaling, respectively. Thus, cell fate determination at the eye cup stage is a reversible process dictated by the balance between FGF and Wnt signaling, but it is also highly tolerant of extrinsic noise as long as these signals stay within certain confines. Such a switch-like behavior resembles the principle of phase transition in physics, in which matter is maintained in discrete states of solid, liquid, or gas within broad ranges of temperature and pressure. Once the boundary of these domains is crossed, however, these physical matters abruptly transition into another phase in an all-or-none manner. We propose that eye cup patterning also acts in a mode of phase transition, a mechanism that may be broadly applicable to other biological systems to achieve versatile yet stable outcomes. As demonstrated by our success in inducing hiPSCs into the CM-like fate, systematic exploration of biological phase maps may provide insightful guidance for tissue engineering.
In vertebrates from teleost fish to avian, the CM not only contributes to the NR and the peripheral ocular structures during embryogenesis but also maintains stem cells in adulthood that could replenish retinal neurons after injury (33). The regenerative capacity of the CM appears to be lost in adult mammals, but recent studies suggested that, at least in embryonic mice, the CM could give rise to both neuronal and non-neuronal cells (34, 35). Previous scRNAseq analyses of mouse retina have readily identified RPCs and neurogenic progenitors that yield retinal neurons, but the CM progenitor pool remains elusive (20, 21). By enriching for the peripheral retina, our scRNAseq study has mapped the CM progenitor population, showing that they reside in the same G2-M phase of the cell cycle as neurogenic progenitors. Our velocity analysis revealed that neurogenic progenitors inevitably exit the cell cycle to differentiate. In contrast, CM progenitors have the bivalent potential to either differentiate or to resume the cell cycle; the latter decision produces daughter cells that may then branch into the neurogenic route. This explains why the CM progenitors can generate both CM cells and retinal neurons, suggesting that these progenitors resemble stem cells in their ability to either self-renew or differentiate. This stem cell–like capacity apparently requires FGF signaling, because loss of FGF signaling will bias CM progenitors toward terminal differentiation. As the regenerative capacity wanes from lower vertebrates to mammals, it will be interesting to explore whether FGF signaling is important for the adult CM to preserve the stem cell–like capability.
Despite their stark differences in anatomical structures, both vertebrate and Drosophila eyes acquired dark pigments to shield them from unwanted light exposure. In Drosophila eyes, a Wnt signaling gradient induced by the adjacent head capsule is necessary for emergence of the PR (3). In the mouse eye cup, we show that both the lens ectoderm and periocular mesenchyme are sources of Wnt ligands. An earlier study concluded that Wnt ligands from the lens ectoderm regulate proliferation but not differentiation of the optic cup rim (26). Our analysis instead reveals that both CM and distal RPE genes were down-regulated if secretion of Wnt ligands from the lens ectoderm was inhibited. The defect in eye cup patterning was likely aggravated by the aberrant shape of the eye cup, but because we were unable to detect any expression of the CM marker Msx1 and the distal RPE marker Wnt2b, it suggests that the lens ectoderm–derived Wnt is responsible for specification of the distal RPE and the CM. This is consistent with classic embryological studies that the lens epithelium has the capacity to induce the iris and CB in the optic cup (36, 37). Our study thus demonstrates an evolutionarily conserved requirement for paracrine Wnt signaling to induce differentiation of peripheral ocular structures.
In contrast to the conserved role of Wnt signaling, FGF signaling appears to be uniquely required for peripheral eye development in vertebrates. Our study showed that graded FGF signaling is necessary for subdivision of the CM into the proximal, medial, and distal zones, which may correspond to pars plana and pars plicata of the CB and the iris in adult eyes. We found that FGF signaling maintains the strength of Wnt signaling, which may serve to extend the range of Wnt signaling gradient. FGF signaling stabilizes β-catenin independently of its GSK3β degradation motif, revealing a previously unrecognized mechanism of Wnt signaling regulation (38). On the other hand, our study also showed that genetic ablation of both FGF and Wnt signaling transforms the entire retina into the NR. This is reminiscent of NR transformation by deleting β-catenin in the RPE, which lacks FGF signaling (4, 5). Thus, we propose that the ground state of the eye cup is the NR, which explains why the three-dimensional embryonic stem cell culture under the low growth factor (serum-free) condition naturally develops into the NR (39). Only a combination of FGF and Wnt signaling can induce CM formation in the eye cup. These insights are instrumental in our rational design of the CM differentiation protocol for hiPSC culture. Our study shows that FGF and Wnt signaling cooperates to determine the demarcation and subdivision of the eye cup into the NR, CM, and RPE. It demonstrates how cross-talk between two signaling pathways is able to generate remarkable complexity in organ development.
MATERIALS AND METHODS
Mice
All animal procedures were performed according to the protocols approved by Columbia University’s Institutional Animal Care and Use Committee. β-Cateninfl3 was obtained from S. Kousteni (Columbia University), Fgf3 flox was from S. L. Mansour (University of Utah), Fgfr2flox was from D. Ornitz (Washington University Medical School), Mek1flox/flox and Mek2−/− were from J. Charron (Université Laval), Pax6 α-Cre (α-Cre) was from N. Brown (Children’s Hospital Research Foundation), Pax6 Le-Cre (Le-Cre) was from R. Lang (Children’s Hospital Research Foundation), R26LSL-Fgf8 was from Y. Chen (Tulane University), R26LSL-Wnt1 was from T. Carroll (UT Southwestern), and Ai9, β-catenin flox, Fgfr1flox, Msx1-CreERT2, Wls flox, and Wnt1-Cre were from The Jackson Laboratory as listed in table S1 (28, 29, 40–49). Mice were maintained on a mixed genetic background. At least three animals were analyzed for each of the crosses described. We did not observe phenotypic variations between Cre heterozygous controls and no-Cre homozygous controls. These two genotypes are hence described together as controls.
Histology and immunohistochemistry
Histology and immunohistochemistry were performed on the paraffin and cryosections as previously described (50, 51). Briefly, slides were washed in phosphate-buffered saline (PBS) three times for 5 min each, followed by PBST (0.3% Triton) washes three times for 5 min each before blocking in 10% horse serum in PBST (blocking buffer) for 1 hour. Primary antibodies listed in table S2 were diluted in the blocking buffer according to the dilutions and added to the slides at 4°C overnight. Antigen retrieval was performed in 10 mM sodium citrate–0.05% Tween (pH 6.0) in an antigen retriever (Aptum Biologics) for three cycles of heating. Secondary antibodies were added the next day for 2 hours at room temperature, followed by 5-min incubation with 4′,6-diamidino-2-phenylindole (DAPI). Slides were coverslipped using n-propyl gallate–glycerol mounting medium for imaging. The antibodies used are listed in table S2.
In situ hybridization
Cryosections were processed for in situ hybridization using previously established protocols (52). Briefly, Digoxigenin-labeled mRNA in situ probes diluted in hybridization buffer were added to the slides and incubated at 65°C overnight, followed by washes of high Stringency buffers and maleic acid buffer with tween (MABT) before blocking with 10% goat serum in MABT. Anti-Digoxigenin antibody diluted in MABT (with 1% goat serum) was added to the slides at 4°C overnight. Slides were then washed in MABT, followed by alkaline phosphatase buffer (NTMT). BM Purple (Millipore Sigma) was added, and the color reaction was allowed to develop overnight. Slides were then briefly washed in PBS and coverslipped with mounting medium for imaging. The probes used are listed in table S3.
Retinal dissociation and flow cytometry
E13.5 retinae with attached RPE from three control and mutant embryos each were harvested in ice-cold Hanks’ balanced salt solution after removal of the lens. Dissociation was performed using a papain dissociation kit (Worthington Biochemical) for 20 min at 37°C and stopped using Dulbecco’s modified Eagle’s medium (DMEM) + 10% fetal bovine serum (FBS). Deoxyribonuclease I was added to the samples, followed by incubation at 37°C for 5 min. Cells were gently triturated using a P1000 pipette, centrifuged at 300g, and washed with DMEM + 10% FBS. After passing through a 40-μm filter and washing again in DMEM + 10% FBS, the cell suspension was stained with SYTOX to mark dead cells before flow cytometry was performed at the Columbia University stem cell core facility. Single cells were collected with a Bio-Rad S3e cell sorter and gated for live cells, GFP+ cells, and TdTomato+ cells using FlowJo software. Approximately 100,000 cells were collected into 1.5-ml tubes precoated with FBS and DMEM + 10% FBS for submission for single-cell sequencing.
Single-cell sequencing
Single-cell sequencing was performed at the single-cell sequencing core in Columbia Genome Center. Flow-sorted single cells were loaded into chromium microfluidic chips with v3 chemistry and barcoded with a 10× chromium controller (10× Genomics). RNA from the barcoded cells was reverse-transcribed, and sequencing libraries were constructed with a Chromium Single Cell v3 reagent kit (10× Genomics). Sequencing was performed on NovaSeq 6000 (Illumina). The RNAseq data were deposited at the Gene Expression Omnibus (GEO) database (GSE139904) and accessible at the Single-Cell Portal (SCP1618).
Raw reads mapped to the mm10 reference genome by 10× Genomics Cell Ranger pipeline (v2.1.1) using default parameters were used for all downstream analyses using Seurat v3 and Velocyto. Briefly, the dataset was filtered to contain cells with at least 200 expressed genes and genes with expression in more than three cells. Cells were also filtered for mitochondrial gene expression (<20%). The dataset was log-normalized and scaled. Cells with TdTomato reporter expression were extracted, and a separate expression matrix was constructed. This provided the “control” and “mutant” gene expression matrices. Unsupervised clustering was performed initially, followed by manual annotation of Seurat clusters. Biological incompatibility based on gene expression was used to identify doublets. Previously known unique gene expression in extraretinal cells such as the cornea, lens, and mesenchyme was used to identify extraretinal clusters. An integration analysis was performed to compare and analyze the control and mutant gene expression matrices, which included normalization for cell numbers. The unsupervised clustering with a resolution parameter of 0.7 for both control and mutant cells was represented on a common UMAP space, and cluster identity was assigned. Expression of various known genes was used to determine cluster identities. The cell cycle was assessed using the vignette from Seurat.
For RNA velocity analysis, Velocyto.py was run following an example notebook (https://github.com/velocyto-team/velocyto-notebooks/blob/master/python/DentateGyrus.ipynb) using the clustering from Seurat. Briefly, loom files were constructed by extracting the reads from the 10× single-cell dataset. After gene filtering, spliced and unspliced counts were normalized based on total counts per cell. Velocity plots are represented as the transcriptomic integration of control and mutant datasets to represent similarities and differences in their velocities, respectively. Velocity vector fields of control and mutant datasets were calculated by pooling the unspliced and spliced counts from similar cells using the k-nearest neighbors (kNN) imputation method with 120 neighbors.
Retinal organoid culture
The hiPSC line PLA-1-3 was maintained on Matrigel (BD)–coated plates in mTeSR medium (STEMCELL Technologies) and passaged with ReleSR (STEMCELL Technologies) (53). Retinal organoid differentiation was carried out as previously reported with minor modifications (54, 55). Briefly, iPSCs at 90% confluence were checkerboard-scraped, approximately 1 to 1.5 mm2, using a 200-μl pipette tip and lifted using a cell scraper. Colony fragments were collected and incubated with (±)blebbistatin in mTeSR medium overnight before moving to neural induction medium–1 (NIM-1) to form embryoid bodies (EBs) in floating culture over 3 days. All floating culture steps were performed in poly(2-hydroxyethyl methacrylate) (poly-HEMA) (Sigma-Aldrich)–coated wells. On differentiation day (DD) 7, floating EBs were transferred to Matrigel-coated wells until DD28, with transition from NIM-1 to NIM-2 medium at DD16. Neuroepithelia were lifted using the checkerboard-scraping method (56). From DD28 to DD32, retinal organoids were sorted into poly-HEMA–coated wells and maintained in NIM-2 until the end of the experiment. For CM induction, DD32 retinal organoids were treated with the GSK inhibitor CHIR99021 (3 μM) for 10 days of treatment, followed by 2 days of no treatment before collection. Organoids were washed twice with PBS, fixed for 30 min in 4% paraformaldehyde in PBS, and cryoprotected by subsequent incubations of 30 min in 15 and 30% sucrose in PBS. The organoids were embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek), frozen, and stored at −80°C. Sections of 10 μm were generated with a Leica CM1850 cryostat for immunohistochemistry. The CM area was identified by the MSX1+ and CDO+ expression pattern and measured as the percentage of the total area of organoids in ImageJ. The statistical significance was calculated using Student’s t test, and results are expressed as means ± SD. NIM-1: 48.95 ml of DMEM/F12 supplemented with 0.5 ml of 100× N2 supplement, 0.5 ml of 100× minimum essential medium (MEM) nonessential amino acids (NEAAs), and 10 μl of heparin (10 mg/ml; Sigma-Aldrich). NIM-2: 96 ml of DMEM/F12 (3:1) supplemented with 2 ml of 50× B27 supplement, 1 ml of 100× NEAA, and 1 ml of 100× penicillin (10,000 U/ml)–streptomycin (10,000 μg/ml).
Acknowledgments
We thank N. Brown, T. Carroll, J. Charron, Y. Chen, S. Kousteni, D. Ornitz, and S. L. Mansour for mice and H. Arnheiter, A. Joyner, J. Hebert, B. Hogan, T. Jessell, T. Glaser, G. Martin, C. Mason, R. McInnes, and V. Wallace for in situ probes. We also thank C. Mason and A. Tomlinson for reading the manuscript.
Funding: This work was supported by a grant from the NIH (EY025933 to X.Z.). The Columbia Ophthalmology Core Facility is supported by NIH Core grant 5P30EY019007 and unrestricted funds from Research to Prevent Blindness (RPB). R.B. is a recipient of Knights Templar Eye Foundation Career Starter grant and Bright Focus Foundation National Glaucoma Research. P.M.J.Q. is a recipient of Curing Retinal Blindness Foundation grant and Knights Templar Eye Foundation Career Starter grant. B.L.d.C. is a recipient of a Capes PhD scholarship. P.J.F. and Q.L.G. were supported by funds from the Swiss National Fund (Ambizione grant PZ00P3_174032 to P.J.F.). Q.W. is a recipient of Postdoctoral Fellowship from Natural Sciences and Engineering Research Council of Canada. C.T. is a recipient of a Jonas Scholar award. S.H.T. is supported by Jonas Children’s Vision Care. X.Z. is supported by Jules and Doris Stein Research to Prevent Blindness Professorship. Jonas Children’s Vision Care receives the support of U01 EY030580, U54OD020351, R24EY028758, R24EY027285, 5P30EY019007, R01EY018213, R01EY024698, R01EY026682, R01EY031354, and R21AG050437.
Author contributions: C.T. initiated the project. R.B. X.M., C.T., and K.P. generated mouse mutants, performed experiments, and analyzed the data. R.B. performed scRNAseq experiment, and R.B., Q.L.G., and D.B. analyzed the data. P.M.J.Q. and B.L.d.C. performed retinal organoid experiment. N.M., J.P., M.B., Y.M., and Q.W. performed immunohistochemistry. F.W. and L.M. generated Fgf9flox and R26LSL-Fgf8 mice. S.H.T., P.J.F., and X.Z. supervised the work and secured funding. X.Z. conceived the study. R.B. and X.Z. wrote the manuscript with input from the rest of the authors.
Competing interests: S.H.T. has received support from Abeona Therapeutics Inc. and Emendo. S.H.T. is the founder of Rejuvitas. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S3
REFERENCES AND NOTES
- 1.Chow R. L., Lang R. A., Early eye development in vertebrates. Annu. Rev. Cell Dev. Biol. 17, 255–296 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Fuhrmann S., Zou C., Levine E. M., Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res. 123, 141–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomlinson A., Patterning the peripheral retina of the fly: Decoding a gradient. Dev. Cell 5, 799–809 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Fujimura N., Taketo M. M., Mori M., Korinek V., Kozmik Z., Spatial and temporal regulation of Wnt/β-catenin signaling is essential for development of the retinal pigment epithelium. Dev. Biol. 334, 31–45 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Westenskow P., Piccolo S., Fuhrmann S., β-Catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development 136, 2505–2510 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bharti K., Gasper M., Ou J., Brucato M., Clore-Gronenborn K., Pickel J., Arnheiter H., A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLOS Genet. 8, e1002757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H., Xu S., Wang Y., Mazerolle C., Thurig S., Coles B. L. K., Ren J. C., Taketo M. M., van der Kooy D., Wallace V. A., Ciliary margin transdifferentiation from neural retina is controlled by canonical Wnt signaling. Dev. Biol. 308, 54–67 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Heavner W. E., Andoniadou C. L., Pevny L. H., Establishment of the neurogenic boundary of the mouse retina requires cooperation of SOX2 and WNT signaling. Neural Dev. 9, 27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Z., Feng G. S., Zhang X., Temporal requirement of the protein tyrosine phosphatase Shp2 in establishing the neuronal fate in early retinal development. J. Neurosci. 30, 4110–4119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Z., Tao C., Li H., Ladher R., Gotoh N., Feng G. S., Wang F., Zhang X., Deficient FGF signaling causes optic nerve dysgenesis and ocular coloboma. Development 140, 2711–2723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S., Li H., Gaudenz K., Paulson A., Guo F., Trimble R., Peak A., Seidel C., Deng C., Furuta Y., Xie T., Defective FGF signaling causes coloboma formation and disrupts retinal neurogenesis. Cell Res. 23, 254–273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakaguchi D. S., Janick L. M., Reh T. A., Basic fibroblast growth factor (FGF-2) induced transdifferentiation of retinal pigment epithelium: Generation of retinal neurons and glia. Dev. Dyn. 209, 387–398 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Pittack C., Grunwald G. B., Reh T. A., Fibroblast growth factors are necessary for neural retina but not pigmented epithelium differentiation in chick embryos. Development 124, 805–816 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Guillemot F., Cepko C. L., Retinal fate and ganglion cell differentiation are potentiated by acidic FGF in an in vitro assay of early retinal development. Development 114, 743–754 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Nguyen M., Arnheiter H., Signaling and transcriptional regulation in early mammalian eye development: A link between FGF and MITF. Development 127, 3581–3591 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Dias da Silva M. R., Tiffin N., Mima T., Mikawa T., Hyer J., FGF-mediated induction of ciliary body tissue in the chick eye. Dev. Biol. 304, 272–285 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwahara A., Ozone C., Nakano T., Saito K., Eiraku M., Sasai Y., Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 6, 6286 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Brewer J. R., Mazot P., Soriano P., Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 30, 751–771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquardt T., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F., Gruss P., Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43–55 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Clark B. S., Stein-O’Brien G. L., Shiau F., Cannon G. H., Davis-Marcisak E., Sherman T., Santiago C. P., Hoang T. V., Rajaii F., James-Esposito R. E., Gronostajski R. M., Fertig E. J., Goff L. A., Blackshaw S., Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Giudice Q., Leleu M., La Manno G., Fabre P. J., Single-cell transcriptional logic of cell-fate specification and axon guidance in early born retinal neurons. Development 146, dev178103 (2019). [DOI] [PubMed] [Google Scholar]
- 22.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M. E., Lönnerberg P., Furlan A., Fan J., Borm L. E., Liu Z., van Bruggen D., Guo J., He X., Barker R., Sundström E., Castelo-Branco G., Cramer P., Adameyko I., Linnarsson S., Kharchenko P. V., RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowan S., Chen C. M., Young T. L., Fisher D. E., Cepko C. L., Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development 131, 5139–5152 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Zhao S., Hung F. C., Colvin J. S., White A., Dai W., Lovicu F. J., Ornitz D. M., Overbeek P. A., Patterning the optic neuroepithelium by FGF signaling and Ras activation. Development 128, 5051–5060 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M., Taketo M. M., Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 18, 5931–5942 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carpenter A. C., Smith A. N., Wagner H., Cohen-Tayar Y., Rao S., Wallace V., Ashery-Padan R., Lang R. A., Wnt ligands from the embryonic surface ectoderm regulate ‘bimetallic strip’ optic cup morphogenesis in mouse. Development 142, 972–982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrer-Vaquer A., Piliszek A., Tian G., Aho R. J., Dufort D., Hadjantonakis A. K., A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC Dev. Biol. 10, 121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashery-Padan R., Marquardt T., Zhou X., Gruss P., Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 14, 2701–2711 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P., Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Iwai-Takekoshi L., Balasubramanian R., Sitko A., Khan R., Weinreb S., Robinson K., Mason C., Activation of Wnt signaling reduces ipsilaterally projecting retinal ganglion cells in pigmented retina. Development 145, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith A. N., Miller L.-A., Song N., Taketo M. M., Lang R. A., The duality of β-catenin function: A requirement in lens morphogenesis and signaling suppression of lens fate in periocular ectoderm. Dev. Biol. 285, 477–489 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Kinoshita H., Suzuma K., Kaneko J., Mandai M., Kitaoka T., Takahashi M., Induction of functional 3D ciliary epithelium–like structure from mouse induced pluripotent stem cells. Invest. Ophthalmol. Vis. Sci. 57, 153–161 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Fischer A. J., Bosse J. L., El-Hodiri H. M., The ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Exp. Eye Res. 116, 199–204 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Belanger M. C., Robert B., Cayouette M., Msx1-positive progenitors in the retinal ciliary margin give rise to both neural and non-neural progenies in mammals. Dev. Cell 40, 137–150 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Marcucci F., Murcia-Belmonte V., Wang Q., Coca Y., Ferreiro-Galve S., Kuwajima T., Khalid S., Ross M. E., Mason C., Herrera E., The ciliary margin zone of the mammalian retina generates retinal ganglion cells. Cell Rep. 17, 3153–3164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beebe D. C., Development of the ciliary body: A brief review. Trans. Ophthalmol. Soc. U.K. 105( Pt. 2), 123–130 (1986). [PubMed] [Google Scholar]
- 37.Giroud A., Phenomena of induction & their perturbation in mammals. Acta Anat. 30, 297–306 (1957). [PubMed] [Google Scholar]
- 38.Clevers H., Nusse R., Wnt/β-Catenin signaling and disease. Cell 149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., Sasai Y., Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Hoch R. V., Soriano P., Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development 133, 663–673 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A., Ornitz D. M., Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–3074 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Bissonauth V., Roy S., Gravel M., Guillemette S., Charron J., Requirement for Map2k1 (Mek1) in extra-embryonic ectoderm during placentogenesis. Development 133, 3429–3440 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Belanger L. F., Roy S., Tremblay M., Brott B., Steff A.-M., Mourad W., Hugo P., Erikson R., Charron J., Mek2 is dispensable for mouse growth and development. Mol. Cell. Biol. 23, 4778–4787 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y., Liu G., Wang F., Generation of an Fgf9 conditional null allele. Genesis 44, 150–154 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Urness L. D., Paxton C. N., Wang X., Schoenwolf G. C., Mansour S. L., FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev. Biol. 340, 595–604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D. H., McMahon A. P., Sommer L., Boussadia O., Kemler R., Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Lin C., Yin Y., Bell S. M., Veith G. M., Chen H., Huh S. H., Ornitz D. M., Ma L., Delineating a conserved genetic cassette promoting outgrowth of body appendages. PLOS Genet. 9, e1003231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll T. J., Park J. S., Hayashi S., Majumdar A., McMahon A. P., Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell 9, 283–292 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Carpenter A. C., Rao S., Wells J. M., Campbell K., Lang R. A., Generation of mice with a conditional null allele for Wntless. Genesis 48, 554–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carbe C., Hertzler-Schaefer K., Zhang X., The functional role of the Meis/Prep-binding elements in Pax6 locus during pancreas and eye development. Dev. Biol. 363, 320–329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbe C., Zhang X., Lens induction requires attenuation of ERK signaling by Nf1. Hum. Mol. Genet. 20, 1315–1323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carbe C., Garg A., Cai Z., Li H., Powers A., Zhang X., An allelic series at the paired box gene 6 (Pax6) locus reveals the functional specificity of Pax genes. J. Biol. Chem. 288, 12130–12141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., Zhang Y., Xu Y., Kittredge A., Ward N., Chen S., Tsang S. H., Yang T., Patient-specific mutations impair BESTROPHIN1’s essential role in mediating Ca2+-dependent Cl- currents in human RPE. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinn P. M., Buck T. M., Mulder A. A., Ohonin C., Alves C. H., Vos R. M., Bialecka M., van Herwaarden T., van Dijk E. H. C., Talib M., Freund C., Mikkers H. M. M., Hoeben R. C., Goumans M. J., Boon C. J. F., Koster A. J., Chuva de Sousa Lopes S. M., Jost C. R., Wijnholds J., Human iPSC-derived retinas recapitulate the fetal CRB1 CRB2 complex formation and demonstrate that photoreceptors and Müller glia are targets of AAV5. Stem Cell Rep. 12, 906–919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M. N., Cao L. H., Peters A., Park T. S., Zambidis E. T., Meyer J. S., Gamm D. M., Yau K. W., Canto-Soler M. V., Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5, 4047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowan C. S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T. M., Krol J., Szikra T., Cuttat R., Waldt A., Papasaikas P., Diggelmann R., Patino-Alvarez C. P., Galliker P., Spirig S. E., Pavlinic D., Gerber-Hollbach N., Schuierer S., Srdanovic A., Balogh M., Panero R., Kusnyerik A., Szabo A., Stadler M. B., Orgül S., Picelli S., Hasler P. W., Hierlemann A., Scholl H. P. N., Roma G., Nigsch F., Roska B., Cell types of the human retina and its organoids at single-cell resolution. Cell 182, 1623–1640.e34 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S3