Abstract
Thrombi form a micro-scale fibrin network consisting of an interlinked structure of nanoscale protofibrils, resulting in haemostasis. It is theorized that the mechanical effect of the fibrin clot is caused by the polymeric protofibrils between crosslinks, or to their dynamics on a nanoscale order. Despite a number of studies, however, it is still unknown, how the nanoscale protofibril dynamics affect the formation of the macro-scale fibrin clot and thus its mechanical properties. A mesoscopic framework would be useful to tackle this multi-scale problem, but it has not yet been established. We thus propose a minimal mesoscopic model for protofibrils based on Brownian dynamics, and performed numerical simulations of protofibril aggregation. We also performed stretch tests of polymeric protofibrils to quantify the elasticity of fibrin clots. Our model results successfully captured the conformational properties of aggregated protofibrils, e.g., strain-hardening response. Furthermore, the results suggest that the bending stiffness of individual protofibrils increases to resist extension.
Keywords: fibrin clot, protofibril, mesoscopic dynamics, computational modelling
1. Introduction
Fibrin clots are one of dominant components of thrombi (others are, e.g. red cells or platelets), which play a crucial role in events such as haemostasis [1] and pulmonary thromboembolism [2]. Studies of fibrin clots have been conducted to clarify both physiological and pathological significance [1,3]. Particular attention has been paid to multi-scale spatiotemporal processes connecting the small components of fibrin clots (e.g. fibrinogen) to macro-scale thrombi [4–7]. Fibrin clots themselves consist of components on multiple scales. Each clot, over several microns in size, is the product of protofibril aggregation mediated by fibrinogen, which is a nano-scale 45-nm-long plasma protein consisting of six paired polypeptide chains (two pairs each of Aα-chains, Bβ-chains and γ-chains) [8–10]. Fibrinogen is activated by thrombin and polymerizes into a double-stranded fibre, the so-called protofibril [1,10]. Architecturally, the protofibril is a regularly repeating 22.5-nm unit corresponding to one-half the length of the fibrinogen protein [10–12].
Since a fibrin clot is an interlinked, branching structure consisting of protofibrils [13], it is expected that clot elasticity can result from the reaction of the polymeric filaments between cross-links, from alterations in the network structure, or both [7,14]. Researchers have shown that the mechanical properties of the fibrin network structure are altered by chemical and mechanical conditions (e.g. fibrinogen concentrations and the solvent flow field) [6,13,15,16]. For instance, it is well known that factor XIIIa increases the elasticity of individual fibrin fibres [17], thereby enhancing the stability of clots by increasing their stiffness and resistance against deformation [13,18,19]. It is also known that some medications commonly used to prevent and treat cardiovascular diseases (e.g. aspirin and heparin) affect fibrin polymerization and clot structure, making fibrin more porous, permeable, and susceptible to lysis [20,21]. Piechocka et al. [22] recently showed that γ-chain cross-linking contributes to clot elasticity by changing the force-extension behaviour of protofibrils, whereas α-chain cross-linking stiffens the clot, as a consequence of tighter coupling between the constituent protofibrils [22]. Despite a number of studies of fibrin clot formation, much is still unknown, in particular about how nanoscale protofibril dynamics affect macro-scale fibrin clot formation and then clot mechanical properties. Considering that the clotting time (or gel point) has been used in clinical assays as an indication of altered coagulation, understanding the mechanisms of fibrin polymerization may provide a basis for informative diagnostic tools, such as molecular markers of thrombin generation and intravascular fibrin deposition.
To comprehensively investigate the fibrin clot architecture and its mechanical properties, Onck et al. [23] numerically investigated the dynamics of individual (actin) fibrous components under shear flow using a two-dimensional model, and showed that stiffening of non-affine, cross-linked semiflexible networks is caused by the transition of a bending-dominated response at small strains to a stretching-dominated response at large strain [23]. Moiseyev et al. [24] used a theoretical and statistical model of fibrin clot growth to quantify the relationship between the density of fibre cross-linking and clot elasticity [24]. These models, however, cannot fully deal with the dynamics of fibrin clot components. Yesudasan et al. proposed the coarse-grained molecular dynamics model to investigate the complex network of fibrin clots [25,26]. However, the model focuses on the dynamics of fibrinogen molecules on a scale of several hundreds nanometres, whereas the network structures of fibrin clots on the scale of several micrometres, corresponding to the cellular scale level, have not yet been fully described.
A mesoscopic framework is needed to clarify the spatio-temporal relationship between the nanoscale protofibril behaviours and the mechanical properties of micrometre-scale fibrin clots, but no such framework has been established yet. Therefore, the objective in this study was to develop a framework to investigate fibrin clot formation on multiple scales, with a focus on nanoscale protofibrils behaviour. We proposed a minimal mesoscopic model for protofibrils based on Brownian dynamics. Using this model, we performed simulations of protofibril aggregation. For these polymeric protofibrils, we also performed numerical stretch tests to quantify the elasticity of fibrin clots. Based on these numerical approaches, we discuss the feasibility of a proposed model to study the multi-scale relationship between protofibril molecular dynamics and the mechanical response of fibrin clots.
2. Methods
2.1. Coarse-grain protofibril model
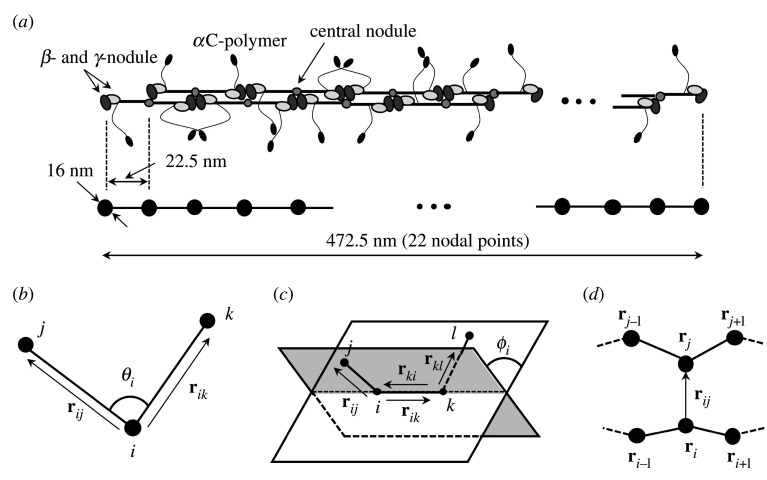
We consider the dynamics of protofibrils with a length of 472.5 nm in a static medium. Based on electron microscope observations [19], each protofibril is modelled as 22 nodal points in series, where the length of the segment consisting of two nodal points is set to 22.5 nm [11], which is the half the length of the 45-nm-long fibrinogen molecule [27]. The model schematic is shown in figure 1(a). Each nodal point is governed by the following overdamped Langevin equation:
| 2.1 |
where ri is the position of the ith node, W is the total potential energy in the computational domain, is the random force of the ith node, characterized as thermal fluctuations of solvent molecules and c is the frictional coefficient estimated by Stokes’ Law, i.e. c = 6πμa. Here, μ (= 1.2 mPa · s) is the solvent viscosity of human plasma. Based on transmission electron microscopy, atomic force microscopy (AFM) and X-ray crystallographic data, the fibrinogen molecule is approximately 2–7 nm in diameter [28–31]. Recent experimental observations using AFM reported that the length of γ-nodule is 5 nm in the naturally folded state (i.e. force-free condition). Considering a double-stranded fibre of protofibril [1,10], we define the characteristic radius of fibrinogen as 8 nm.
Figure 1.
(a) Schematic of a protofibril model with length 472.5 nm, consisting of 22 nodal points in each 22.5-nm-long segment (node to node). Schematic images of (b) bending, (c) torsion and (d) aggregation energy, where superscript (·) represents two adjacent protofibrils.
In this study, the total potential energy W is expressed as five different components, specifically stretch WS, bend WB, torsion WT, aggregation WA and repulsive energy WR
| 2.2 |
The stretch energy WS and stretch force of the ith node can be expressed as
| 2.3a |
| 2.3b |
| 2.3c |
where kS is the stretch energy constant, (= 22.5 nm) is the reference length, which is half the length of the fibrinogen molecule [11], and rj is the jth node adjacent to the ith node in the same protofibril. From both AFM experiments and molecular dynamics simulations, the stretch energy constant can be estimated from the extension dynamics of a single fibrinogen molecule [10,32]. For instance, Lim et al. [32] estimated the longitudinal spring constant of protofibrils as O(kS) ∼ 10−3 N m−1 [32]. While in more recent coarse-grained molecular simulations by Tan et al. [33], the force constant for neighbouring bonds corresponding to kS in this study, was set to be 250 kJ mol−1 Å−2, which can be rewritten as 0.42 N m−1 [33]. Hence, in this study, we set as kS = 0.01 N m−1.
The bending energy WB and bending force of the ith node between the jth and kth nodes can be expressed as
| 2.4a |
| 2.4b |
| 2.4c |
where kB is the bending energy constant, θijk is the angle at the ith node between the jth and kth nodes (figure 1b), and θ0 (=π) is the reference angle [10]. In previous coarse-grained molecular dynamics simulations by [25,33], the bending energy constant was used in the range between 10−19 and 10−17 J rad−2. Hence, in this study, the bending energy constant is set as kB = 0.1–10 × 10−18 J rad−2. The final form of equation (2.4c) is described in appendix A.
The torsion energy WT and torsion force of the ith node can be expressed as
| 2.5a |
| 2.5b |
| 2.5c |
where kT is the torsion energy constant, ϕi is the torsion angle (or dihedral angle) of the ith node among the jth, kth and lth nodes (figure 1c), ϕ0 (=0) is the reference torsion angle, and m and n are the normal vectors, which are defined by four adjacent nodes on two planes (figure 1c)
| 2.6a |
and
| 2.6b |
The final forms of equation (2.5c) at each of the four nodes are described in the appendix A. In previous coarse-grained modelling for DNA [34] and biomoleculars [33], the energy constant in equation (2.5) was considered in the range between 10−23–10−21 J rad−2. Although tightly coupled studies between experiment and simulation are needed to identify the value of the constant, the torsion energy constant kT was set as kT = 1.0 × 10−23 J rad−2 in this study.
Two protofibrils are bound to each other at the αC regions [1], and hence we define the aggregation force applied to two nodal points of different protofibrils, within a threshold ath (), corresponding to the length of the fibrinogen molecule [11]. Considering two adjacent nodal points of different protofibrils (see figure 1d), the aggregation energy WA and aggregation force of the ith node can be written as
| 2.7a |
| 2.7b |
| 2.7c |
where kA is the aggregation energy constant and (=2a) is the reference length between two adjacent nodes that are not present on the same protofibril. For simplicity, WA is modelled as a simple harmonic potential form as shown in equation (2.7a). The formation could be written in light of the fact that lateral aggregation of protofibrils is responsible for local (several nanometre-scale) conformational changes [35], but this is beyond the scope of the present work. Instead of aggregation force constant between two fibrinogen, molecular interaction between platelet glycoprotein receptor GPIIb/IIIa (integrin αIIbβ3) and fibrinogen have been well investigated. For instance, Litvinov et al. (2011) experimentally investigated thermodynamics and kinetics of bonds between GPIIb/IIIa and fibrinogen, where the molecular spring (or aggregation) constant was used as 12 pN nm−1 (=0.12 N m−1) to estimate the energy needed to dissociate αIIbβ3 from fibrinogen in the long-duration state [36]. While in simulation work by [37], platelet adhesion and aggregation were modelled focusing on the interaction between the GPIIb/IIIa and its ligand, where the aggregation force constant for binding between GPIIb/IIIa and fibrinogen was set to as kA = 1.0 × 10−4 N m−1. According to those studies, the aggregation energy constant was set as kA = 0.2–20 × 10−3 N m−1.
Short distance between two charged particles leads to ‘bouncing off’, resulting in repulsive forces between the two, which corresponds to charged particles interacting through colloidal forces at constant surface charge. Several potential models have been proposed to represent such near-field dynamics, and the Lennard–Jones potential is one of the most classical and well-known models. This was also applied to previous coarse-grained molecular dynamics simulations of protofibrils, e.g. by [25,26]. In this study, however, we focus on developing a minimal mesoscopic model for protofibrils, and hence a non-hydrodynamic inter-node repulsive force is modelled as a simple linear function of two-nodes distance, which is defined only when the two node points on different protofibrils are within the diameter of the fibrinogen (=2a). This force practically allows us to avoid the prohibitively small time step needed to overcome the problem of overlapping nodes on different protofibrils. The repulsive energy WR and repulsive force of the ith node can be expressed as
| 2.8a |
| 2.8b |
| 2.8c |
where kR (=2 × 10−4 N m−1) is the repulsive resistance, and (=2a) is the reference length. The effect of the repulsive force on the trajectories of fibres is very small, because it changes the distance between discrete nodes only when the nodes approach within the distance corresponding to the diameter of fibrinogen, which two order of magnitude smaller than system length. Indeed, the order of the magnitude of repulsive energy was two or three orders magnitude smaller than in the total energy (figures 3a and 6a). The linear repulsive model (equation 2.8) has been also applied to cellular interaction problem, and successfully represented a cellular flow in microchannels [38].
Figure 3.
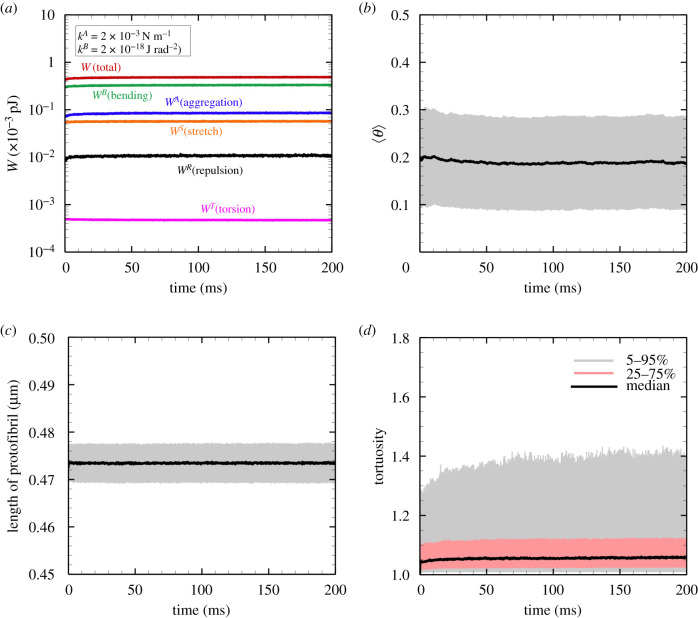
(a) One of the examples of the time history of each energy type; total energy W = WB + WA + WS + WR + WT (red), bending energy WB (green), aggregation energy WA (blue), stretch energy WS (orange), repulsive energy WR (black) and torsion energy WT (pink). The time histories of the mean (b) protofibril orientation angle and (c) protofibril length, where the errors represent the standard deviation of individual protofibrils (=1215). (d) The time history of the median of the tortuosity, where the errors represent 5–95% percentile (light grey) and 25–75% percentile (red), respectively. The results were obtained with kB = 1 × 10−18 J rad−2 and kA = 2 × 10−3 N m−1.
Figure 6.
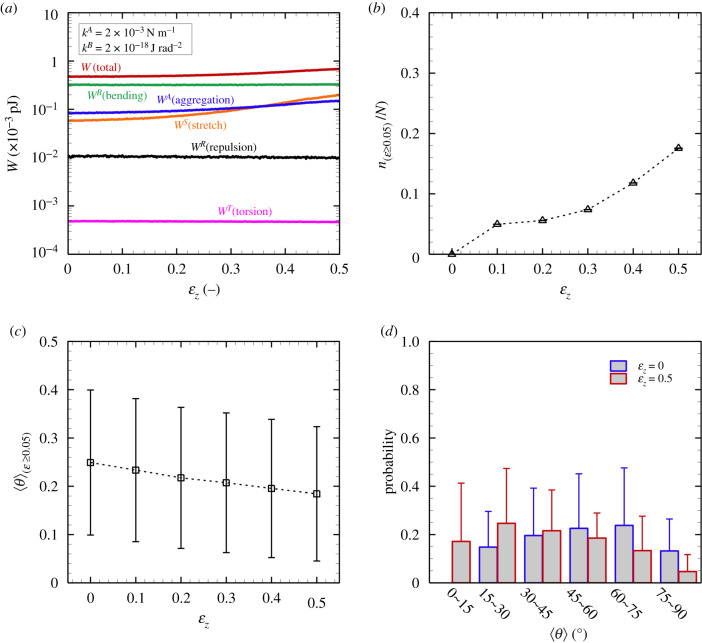
(a) Each averaged energy state (without the standard deviation of run cases), (b) the ratio of the stretched fibres with ɛ ≥ 0.05 n(ɛ≥0.05)/N, and (c) the ensemble average of the orientation angle of protofibrils with ɛ ≥ 0.05 〈θ〉(ɛ≥0.05), as a function of the engineering strain ɛz. (d) Probability of the orientation angle of protofibrils at ɛz = 0 (blue) and ɛz = 0.5 (red). The results were obtained with kB = 1 × 10−18 J rad−2 and kA = 2 × 10−3 N m−1 (M ± SD, ).
The protofibrils experience thermal fluctuations derived both from themselves and from solvent molecules. The random force at the ith node is described as
| 2.9a |
and
| 2.9b |
where σ is the magnitude of the random force, j is the random vector, kb is the Boltzmann constant (=1.38 × 10−23 J K−1) and T (=300 K) is the absolute temperature in the whole system. The temporal direction satisfies the following dynamic statistics:
| 2.10a |
and
| 2.10b |
where 〈 · 〉 represents random average procedures.
Ideally, model parameters characterizing each of the energy components in (2.2) should be determined from molecular dynamic simulations. However, this type of bottom-up approach based on energy trajectories (e.g. [39,40]) is still challenging due to the heavy computational load. In this study, we therefore proposed mesoscopic model properties to qualitatively represent the experimental observations [7,13,41], and in particular focused on the effect of the bending energy constant kB and the aggregation energy constant kA on fibrin clot formation. The parameter values are summarized in table 1.
Table 1.
Nomenclature for the parameters and variables.
| symbol | physical meaning | values (dimension) | reference |
|---|---|---|---|
| a | fibrinogen radius | 8 nm | [42,43] |
| reference stretch length | 22.5 nm | [11] | |
| ath | threshold of aggregation length | 2 | — |
| reference aggregation length | 2a | — | |
| reference repulsive length | 2a | — | |
| θ0 | reference bending angle | π | [10] |
| ϕ0 | reference torsion angle | 0 | — |
| kb | Boltzmann constant | 1.38 × 10−23 J K−1 | — |
| T | temperature | 300 K | — |
| kA | aggregation energy constant | 0.2–20 × 10−3 N m−1 | [36,37] |
| kB | bending energy constant | 0.1–10 × 10−18 J rad−2 | [25,33] |
| kR | repulsive energy constant | 1.0 × 10−3 N m−1 | — |
| kS | stretch energy constant | 0.01 N m−1 | [10,32,33] |
| kT | torsion energy constant | 1.0 × 10−23 J rad−2 | [33,34] |
2.2. Discretization
Following the study by Ermak & McCammon [44], the discrete form of equation (2.2) is rewritten as
| 2.11 |
where ( ∈ [0, 1]) is the probability density function following equation (2.10). Based on a uniform random number between 0 and 1 that is given by the XORWOW method [45], we use the Box–Muller method to obtain the value of that satisfies the Gauss distribution [46]. The ith nodal point of the protofibril at the n time step is updated by Lagrangian tracking, i.e.
| 2.12 |
The Euler method is used for time integration except for the stretch term in equation (2.2), which is solved by the explicit fourth-order Runge–Kutta method.
2.3. Numerical conditions and analysis
As an initial state, 1215 straight protofibrils (22 nodes/protofibril) are randomly placed in a cubic domain of size 3 μm × 3 μm × 3 μm. Periodic boundary conditions are imposed for all directions. The protofibrils aggregate with each other as time passes, and eventually attain a steady state where they fluctuate only as a result of thermal energy. All simulation cases reached this steady state condition by 50 ms or later (see also figure 3), and hence simulations were performed during a period of 200 ms. In addition, for each run we performed 10 replica simulations from a different initial fibre configuration to ensure that our simulation of the fibre network did not occur in a local energy minimum far from the global minimum. Although it is known that the physiological condition of human fibrinogen is 3–4.5 mg ml [47], some of experimental observations of fibrin clots using scanning electron microscopy (SEM) were conducted in dilute fibrinogen concentrations such as 0.5 mg ml−1 [7,13]. For comparisons between calculated steady or extended fibrin clots with those in experimental observations in, e.g. [7,13,41], a fibrinogen concentration was set to 0.5 mg ml−1.
For steady-state fibrin clots, we also performed stretch testing. Stretching is expressed simply by the proportional scaling of coordinate z and the current computational box length Lz for the z-direction from current z to ξz and Lz to ξLz [39,40] with:
| 2.13 |
where cs (=2.5 mm s−1) is the stretching speed in the z-direction, Δτ (=2 μs) is the relaxation time period. We tested 10 times slower stretching speed (i.e. cs = 0.25 mm s−1), and confirmed that the trajectory of the total energy state against the strain ɛz shown in figure 6a did not change. The nodal position for lateral directions (i.e. x- and y-directions) is not scaled during the extension. The extension is applied until the z-directional engineering strain reaches 1.5, where Lz is the present length of the computational domain and is the initial computational length for the z-direction (=3 μm). Since we assume inertia-less protofibril dynamics represented as equation (2.1), and also ensure that fibrin clot dynamics have reached the equilibrium state during Δτ, we can assume that the effect of stretch extension speed on the fibrin clot conformation is negligible. The engineering stress σz on the stretch direction (z-direction) is calculated from the energy state during fibre extension:
| 2.14 |
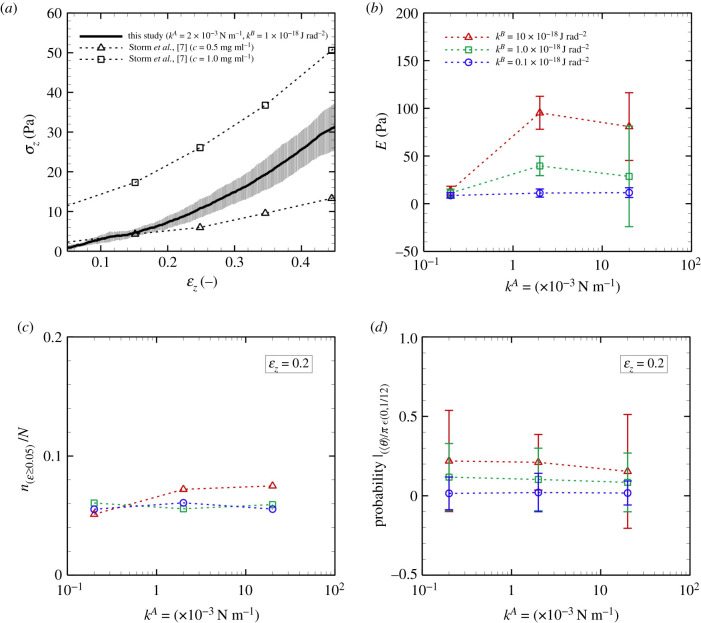
where (=3 × 3 μm2) is the cross-sectional area (x–y plane) and Fz is the force. A representative snapshot of aggregated protofibrils in the steady state (t = 200 ms) and its stretched fibrin clot formation are shown in figure 2a and 2b, respectively. These results are obtained with kA = 2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2.
Figure 2.
Computational domain consisting of a 3 μm × 3 μm × 3 μm cube. A total of 1215 protofibrils (red rods) are randomly placed and orientated in the initial state. Periodic boundary conditions are imposed for all directions. (a) Snapshots of fibrin clot in the steady-state (200 ms after the initial state) and (b) fibrin clot extension in the z-direction. The results are obtained with kA = 2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2.
The orientations of individual protofibrils are defined as
| 2.15 |
| 2.16 |
| 2.17 |
where and are the orientation angle and protofibril length of the jth segment (j ∈ [1, 21]) in the ith protofibril (i ∈ [1, 1215]), respectively, ez is a unit vector along the z-direction in Euclidean space, 〈 · 〉 denotes the ensemble average and N is the total number of protofibrils. Therefore, indicates that the protofibril is oriented in the z-direction. The probability of the orientation of the ith protofibril 〈θi〉 should be normalized by the small band area ΔS on the unit sphere, and can be defined as
| 2.18 |
For instance, the probability of aligned protofibrils with orientations of ∈[0, π/12] is defined as
| 2.19 |
3. Results
3.1. Aggregation of protofibrils
First, we investigated the spontaneously aggregated form of protofibrils (i.e. no external force) for different kA and kB. An example of the temporal history of each energy state is shown in figure 3a, which was obtained with kA = 2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2. The energy state reached a plateau at a relatively early time period of approximately 50 ms. The results regarding fibre conformation, as quantified by the orientation angle, length and tortuosity of individual protofibrils, are shown in figure 3b–d, respectively. Here, the tortuosity (=L/L0 ≥ 1) is defined as the ratio between the length of protofibril L and the Euclidean distance from the start to the end points of protofibril L0. If the fibre is oriented in the length direction, the tortuosity is L/L0 = 1. These results showed that steady conformation has been reached within 50 ms, and the results are consistent with other cases. Hence, the time average is hereafter uniformly calculated over 100 ms after t = 50 ms for all simulations to reduce the influence of the initial conditions.
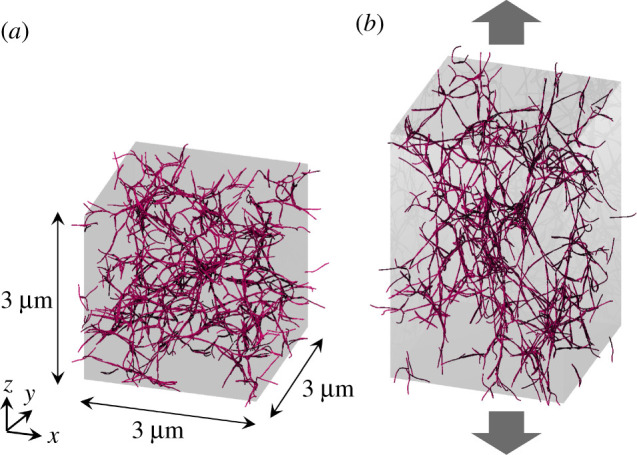
For comparison with a SEM image [13] and calculated three-dimensional fibrin clots, we projected steady-state fibrin clots onto a two-dimensional plane (x–y plane at z = 0) with 10 nm pixel−1. Here, we assigned 10 different luminosity values to the nodes within every 0.3-μm segment in the height z-direction, with higher luminosity associated with increased z-position. For these projected networks, we applied the Sobel filter to obtain the configuration of protofibrils from calculated nodal points, and also apply the Gaussian filter to obtain smoothed fibre structures. The stack images of steady fibrin clots for different kA and kB were obtained using the aforementioned process and are shown in figure 4. Although there was no significant difference in fibre conformation for different kA, the fibres tended to be straighter as kB increased, a finding that is quantitatively shown in figure 5.
Figure 4.
Representative snapshots of fibrin clots at the steady state for different kA (=0.2, 2 and 20 × 10−3 N m−1) and kB (=0.1, 1 and 10 × 10−18 J rad−2). The scale bar length is 1 μm.
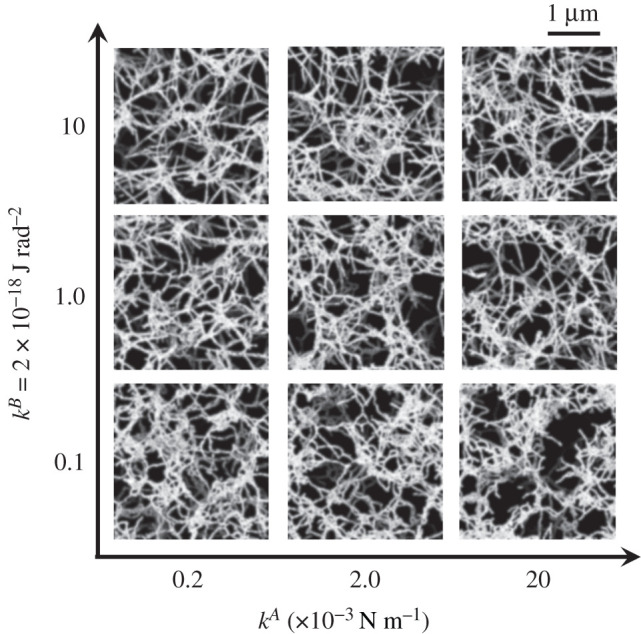
Figure 5.
Time average of the tortuosity of individual protofibrils as a function of kA for different kB (M ± s.d., run cases ). Experimental data [41] of tortuosity of segments and aggregated fibres in collagen-fibrin co-gels with PBS are also displayed.
Protofibril conformation was quantified by the tortuosity of individual protofirbrils. The time average of the tortuosity is shown in figure 5, where error bars represent standard deviation of the run cases (M ± s.d., ). The results show that at each specific kA, the tortuosity decreases (i.e. fibre linearity is enhanced) as kB increases, which qualitatively agrees with previous experimental findings that fibre linearity was positively associated with individual protofibrils’ bending resistance [13,17,22]. Experimental data of collagen-fibrin co-gels in phosphate-buffered saline (PBS) are also displayed [41], as quantified from SEM images. Here, in [41], ‘fibres’ are defined as multiple adjacent segments of protofibrils, and a ‘segment’ is the part of a fibre between cross-links. Especially at kA = 2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2, calculated tortuosity is highly consistent with the experimental result of a ‘segment’ in [41]. Since SEM requires dehydration, a stricter comparison may be needed to consider the effects of such an imaging process, but this is beyond the scope of the present work.
3.2. Stretch tests of fibrin clots
Next, we performed stretch tests for the aforementioned fibrin clots under the same parameter ranges: kA = 0.2–20 × 10−3 N m−1 and kB = 0.1–10 × 10−18 J rad−2. An example of the energy state for each engineering strain is shown in figure 6a. The result shown in figure 6a was obtained with kA = 2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2. As the z-directional engineering strain ɛz increased, the stretch energy WS gradually increased, and then finally overcame the aggregation energy WA (figure 6a). This tendency was more obvious when kA increased (data not shown). The decrease of the contribution of WA to the total energy could be explained by the simple approximation of WA/(WA + WS) = 1/(1 + kA/kS) if the aggregated protofibrils were oriented to the z-direction.
Stretched fibres for different kA and kB are shown in figure 7, where protofibrils coloured red are extended beyond the fibre strain ɛ (=l/l0 − 1) ≥ 0.05 (l and l0 are the current length and the steady-state length of protofibril, respectively). The number of these extended fibres clearly increased when kA increased from 0.2 × 10−3 to 2 × 10−3 N m−1 (figure 7). The structures of stretched protofibrils are quantified by the number of such protofibrils, the fibre orientation θ, and the probability (Prob.) of the orientation. The ratio of the stretched fibres with ɛ ≥ 0.05 to the total number of protofibrils n(ɛ≥0.05)/N is shown in figure 6b. The percentage of fibres with ɛ ≥ 0.05 was only 5% at ɛz = 0.2, but it increased to 20% at ɛz = 0.5. These fibres’ orientations gradually decreased, i.e. the fibres became oriented to the stretch direction, as shown in figure 6c. Since the range of SD almost remains for ɛz, the result indicates that a distribution of fibre orientation angles exists even under extension. The probability of each orientation was therefore examined, and the results at ɛz = 0 and 0.5 are shown in figure 6d. At the steady state (ɛz = 0), the percentage of fibres oriented towards was less than 50%, although it surpassed 60% at ɛz = 0.5 (figure 6d).
Figure 7.
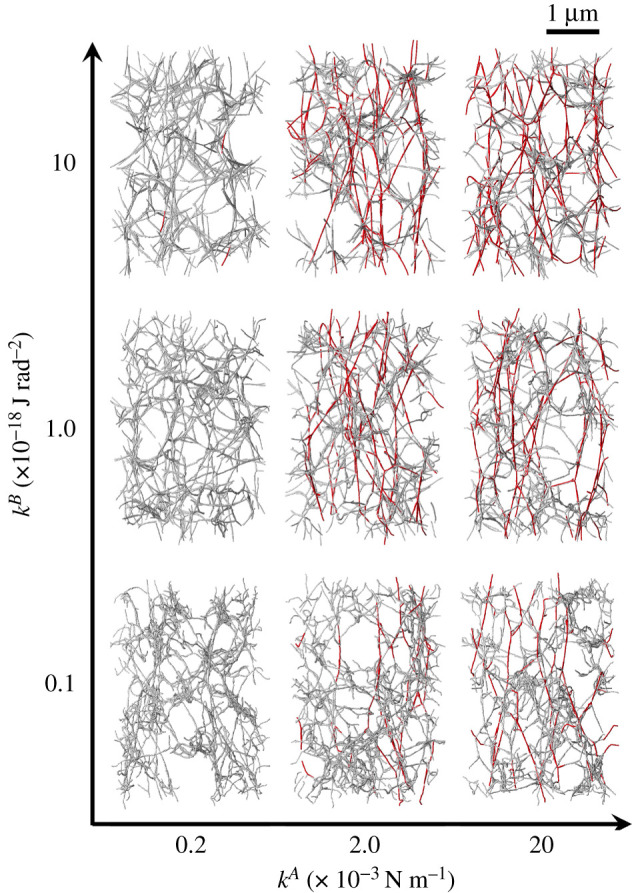
Snapshots of extended fibrin clots at ɛz = 0.5 for different kA and kB. The scale bar length is 1 μm. Protofibrils coloured red are extended beyond the fibre strain ɛ (=l/l0 − 1) ≥ 0.05: l and l0 are the current length and at the steady-state length, respectively.
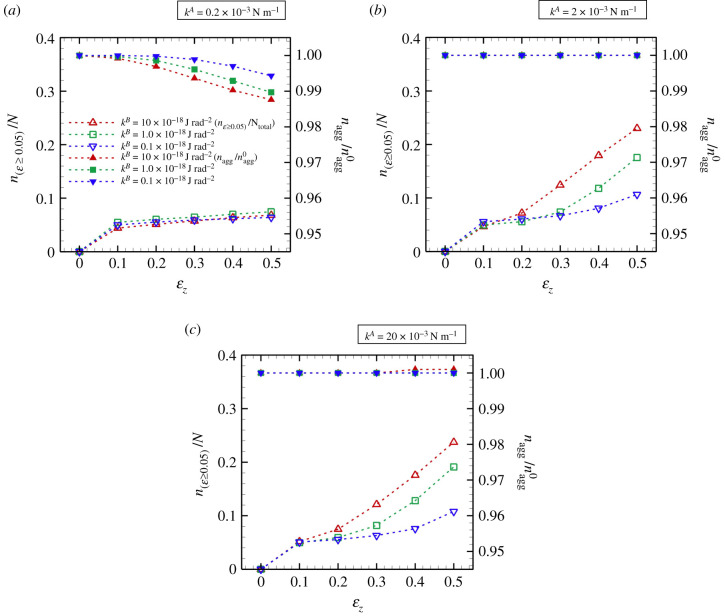
The population of effectively stretched and aggregated fibres was investigated, and the results are shown in figure 8. At the lowest kA (=0.2 × 10−3 N m−1), the ratio n(ɛ≥0.05)/N remained almost constant independent of kB during extension (figure 8a). The aggregated fibres started to dissociate for ɛz ≥ 0.1 (figure 8a), and the dissociation rate was approximately 1%, i.e. at ɛz ≥ 0.5, where nagg is the number of aggregated points, and is the number of initial aggregated points. For relatively large kA ( ≥2 × 10−3 N m−1), the ratio n(ɛ≥0.05)/N increased with kB, and the fibres remained aggregated even at ɛz = 0.5, i.e. (figure 8b,c). Furthermore, there was no marked difference in n(ɛ≥0.05)/N until ɛz ≤ 0.2, independent of kA (figure 8a–c). Note that, at the largest kA (=20 × 10−3 N m−1), only less than 1% fibres still aggregated even for ɛz = 0.5 (figure 8c).
Figure 8.
The ratio n(ɛ≥0.05)/N, and the ratio of the number of aggregated points nagg to the number of initial aggregated points () as a function of ɛz for (a) kA = 0.2 × 10−3 N m−1, (b) kA = 2 × 10−3 N m−1 and (c) kA = 20 × 10−3 N m−1, respectively (M ± s.d., ).
Multiple examples of σz as a function of ɛz, obtained with kA = 2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2, are shown in figure 9a. The moving-averaged data were obtained for the data of total energy W with a window size of ɛz = 0.08. We also evaluated different window sizes with a moving-average (ɛz = 0.04 and 0.16), and confirmed that there was almost no difference in results between obtained with ɛz = 0.04 and ɛz = 0.08. The values of σz shown in figure 9a indicate strain hardening, which qualitatively agrees with experimental data derived with fibrinogen concentrations of 0.5 and 1.0 mg ml−1 [7] when an isotropic network model was used. Note that for the lowest kA, the clots showed strain softening for ɛz ≥ 0.2 due to dissociation, as shown in figure 8a.
Figure 9.
(a) Stress σz of the fibrin clot as a function of the strain ɛz. The result is obtained with kA = 2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2. Experimental data of the shear modulus of fibrin clots (fibrinogen concentration of 0.5 and 1.0 mg ml−1) [7] are also displayed. (b) Elastic coefficient E of fibrin clots as a function of kA for different kB. Using the data of ɛz = 0.05 to 0.15, E was calculated by least-squares fitting to the plot of {σz − (Eɛz + σ0)}, where σ0 is the residual stress. (c) Ratio n(ɛ≥0.05)/N. (d) Probability with 〈θ〉 ∈ [0, π/12] at ɛz = 0.2 as a function of kA for different kB (M ± s.d., ).
Especially for ɛz < 0.2, we confirmed an almost proportional relationship between σz and ɛz in all conditions that were investigated (kA = 0.2–2 × 10−3 N m−1 and kB = 1 × 10−18 J rad−2), i.e. the effect of dissociation of aggregated fibres on σz was negligible. Thus, using the data of ɛz = 0.05 to 0.15, the elastic coefficient of fibrin clots E was calculated by least-squares fitting [48] to the plot of {σz − (Eɛz + σ0)}, where σ0 is the residual stress. The calculated E for different kA and kB is summarized in figure 9b. Although there was no marked difference in E at the smallest kA (=0.2 × 10−3 N m−1), E increased as kA rose from 0.2 × 10−3 to 2 × 10−3 N m−1, and the rate of its increase decreased thereafter. the error bars were relatively large, especially at the largest kA (=20 × 10−3 N m−1), representing variations of the energy state depending on the initial (quasi-steady) state. For each specific kA, the value of E became large with kB, which qualitatively agrees with the following conclusion based on experimental studies [13,17,22]: the bending resistance of individual protofibrils enhances not only fibre linearity but also the elasticity of fibrin clots.
Figure 9c shows the ratio n(ɛ≥0.05)/N at ɛz = 0.2 as a function of kA, and indicates that the number of extended fibres with ɛz ≥ 0.05 was insensitive to kA. Figure 9d shows the probability of fibres oriented in the stretch direction, defined as those with 〈θ〉 ∈ [0, π/12] (i.e. Prob.|〈θ〉/π ∈ [0, 1/12]) at ɛz = 0.2. These fibres continued to be oriented in this direction even though kA increased, while the probability of this orientation increased with kB, i.e. the fibrin clot structure was transformed to one that withstood extension as a result of increased kB (figure 9d). Hence, the attenuation of the rate of increase in E with increasing kA (figure 9b) may be correlated with the slow increase of effectively stretched fibres, which are defined with ɛ ≥ 0.05 and 〈θ〉 ∈ [0, π/12].
4. Discussion and conclusion
Despite a number of studies of fibrin clots, much is still unknown about the relationship between the behaviour of individual protofibrils and the macro-scale mechanical response of fibrin clots. Yesudasan et al. proposed the coarse-grained molecular dynamics model, featuring several hundred nanometre-scale structures, to investigate a complex network of fibrin clots in terms of fibrinogen molecules [25,26]. However, there remain limitations to the mesoscopic mode focusing on the dynamics of fibrin clots, whose interactions can be characterized on a cellular scale (i.e. several micrometres). Stylianopoulos & Barocas developed a structural model that uses volume-averaging theory to study the mechanical behaviour of collagen networks [49], wherein the bending rigidity of individual fibres was neglected. Although their model successfully demonstrated the strain-hardening character of type I collagen extracellular matrices observed experimentally [50], other experimental studies using X-rays [51] or mechanical (shear strain) testing [52] suggested that the flexibility of fibrin clots may arise from bending of the fibres, a theory that was supported by later studies using electron microscopy [53] and optical tweezers [17]. Despite the insights about bending properties at the fibre level, the relationship between such mechanical properties on individual fibre conformations and the mechanical responses of clots are still unknown. To tackle these challenges, a mesoscopic framework based on protofibril behaviours is necessary, but has not yet been established. We thus propose a minimal mesoscopic model for protofibrils based on Brownian dynamics. Through various numerical simulations, we evaluated the feasibility of our mesoscopic model analysis.
The approach used in our model successfully demonstrated conformation of a fibrin clot (figure 5) which was consistent with experimental data in [13,41]. Further, our numerical results captured the strain-hardening response, e.g. as reported in [7] based on protofibril dynamics (figure 9a). Although there are a number of experimental works about tensile mechanical properties in living materials [54] and also single-molecule biophysics [55], there are few reports about assessments of the stress acting on fibrin clots during extensions. For instance, Roeder et al. performed tensile mechanical tests for matrices purified from type I collagen for different collagen concentrations ranging between 0.3–3 mg ml−1, and showed the strain-hardening character of the matrices [50]. The experimental estimation of the engineering stress obtained with 2 mg ml−1 collagen concentration was one or two orders of magnitude higher (i.e. O(σz) = 102–103 Pa) in the range between 10–20%-strain than our numerical results of fibrin clots with 0.5 mg ml−1 fibrinogen concentration [50].
As with other numerical models, our mesoscopic model required a number of parameters. Although there are many SEM results regarding human fibrin clot structures (e.g. [16]), quantitative data of fibrin fibre parameters are still limited, and thus we could not determine them in a straightforward manner. Yesudasan et al. [25,26] used mathematical methods [56] to characterize and quantify 3D images of fibrin clot networks, and then compared them with simulation results [25]. Although our model parameters were not determined by this type of bottom-up molecular dynamic approach, our methodology potentially characterized the conformation of fibrin clots, as well as their mechanical responses, according to the model parameters. We thus suggest that our microscopic model can be used to estimate the relationship between the nanometre-scale dynamics of individual protofibrils and the mechanical properties of micrometre-scale fibrin clots. For example, simulations of aggregated protofibrils whose degree of extension depends on the aggregation energy constant kA will provide insight into the dependency of factor XIIIa on fibrin clot elasticity [13,18,19]. Since our numerical results showed that the elasticity of the fibrin clots increased with kA, the dependence of factor XIIIa on the elasticity resulting from the fibrous architecture of the clots may be characterized by kA. Previous experimental results showed that stiffening of fibrin clots can result from the response of the polymeric protofibrils between cross-links, from alterations in the network structure, or both [6,7,14,15]. Although we did not fully quantify the architecture of aggregated protofibrils, the simulated protofibrils in the networks also tended to align in the stretch direction as kB increased (figure 9d), resulting in sparse networks (figure 7). Such interlinked protofibril structures may contribute to decreasing the elasticity of fibrin clots, especially at the highest kA (=2.0 × 10−3 N m−1) and kB (=1.0 × 10−18 J rad−2) (figure 9b). Future quantitative analysis of the network structure will provide insight into the relationship between the fibrous architecture and mechanical properties of the clot. Experimental observations using SEM have clarified the precise structure of fibrin clots, while some limitations should be carefully considered. SEM analysis allows us to visualize single fibres even under load [57], but originally hydrated fibrin networks are most often dehydrated and fixed, and hence it remains difficult to capture 3D networks of fibrin clots in dynamics. Spectroscopic tools such as Fourier transform infrared (FTIR) and Raman scattering offer alternative methods to probe structural changes of proteins using molecular vibrations. These methods can be performed on 3D fibrin hydrogels under mechanical deformation in situ. For instance, Litvinov et al. used FTIR spectroscopy to reveal force-induced changes in the secondary structure of hydrated fibrin clots made of human blood plasma in vitro [58]. In more recently, Wang et al. used broadband coherent anti-Stokes Raman scattering (BCARS) microscopy and custom-built loading devices to capture the molecular vibrational signature of fibrin under shear and tensile deformations in situ [59]. Along with those microscopic observation techniques, image analyses of a 3D network of fibrin clots obtained with confocal microscopy have been reported as well as its observation techniques [60,61]. The analyses have also gained insights into the mechanics of a fibrin network in terms of its microscopic structure [60,61].
Venous thromboembolism, including deep-vein thrombosis and pulmonary embolism, is the leading cause of lost disability-adjusted life years and the third leading cause of cardiovascular death in the world [62]. Potential risk analysis for thromboembolism is of fundamental importance in diagnosis or therapeutic decisions. Recent experimental observations showed that there are differences in structure and composition between arterial and venous thrombi and pulmonary emboli [63]. Therefore, the tensile mechanical properties of those thrombi characterized as volume fraction of fibrin fibres or fibrin bundles would be one of risks in thromboembolism. Since our numerical model enables the investigation of tensile stress as well as elastic coefficient for different fibrinogen concentrations, calculated elastic coefficients of fibrin clots with specific fibrinogen concentrations based on blood samples would provide additional bioengineering insights in the diagnosis of thromboembolism. While nonlinear mechanical properties of fibrin clots can be useful in bioengineering applications not only in wound repair but also drug delivery, cell delivery, cell differentiation, tissue engineering and patterning [59,64,65]. Our numerical model will gain insights into their designs, in terms of mechanical properties.
In in vitro experiments, clots are often obtained under conditions of purified fibrinogen or plasma with low platelet counts. However, in the actual human circulation, haemostatic clots or obstructive thrombi are formed in the presence of blood flow that profoundly affects the formation of the fibrin network and its structure and properties. For instance, it is known that the mechano-sensitive plasma protein von Willebrand factor (vWF) and its binding with the platelet receptor glycoprotein Ibα (GPIbα) play a crucial role in haemostasis, yet is also key to pathological thrombus initiation and propagation, which are well reviewed in, e.g., [62,66]. Furthermore, the circulation continually brings more fibrinogen to the developing clot, and as a result it becomes denser, with thicker, bundled fibres [67]. Fluid flow can also cause the fibrin fibres to orient along the direction of flow both in vitro [68,69] and in vivo [70], which has important consequences for the mechanical properties of the clots [69,71]. Flow fields would also affect not only the structure of thrombi but also the composition. Recent clinical observation using high-resolution SEM clearly showed that venous thrombi are fibrin-rich, and arterial thrombi are fibrin- and platelet-rich [63]. The evidence that arterial thrombi contain a large amount of fibrin [63] is in contrast to previous animal experiments by [72,73], where high wall shear rates over the range 50–2500 s−1 impede fibrin deposition in subendothelium [72] and extracellular matrix [73]. Hence, further investigations about mechanical factors regulating thrombus growth are needed while paying attention to haemodynamic flow as well as mechanical reactions of plasma proteins. In our simulation, we did not consider the flow of background medium, and also neglected the presence of red cells/platelets and solvent proteins. Since there are only a few reports on the modelling of protein activation under cellular blood flow [74–76], it would be interesting to study how fibrous interactions affect the suspension rheology of red blood cells (RBCs) [77] or the flow behaviours of RBCs and platelets in microvessels depending on shear rates [38,78].
In summary, we proposed a minimal mesoscopic model of protofibrils based on Brownian dynamics. Since our results successfully captured the conformation of aggregated protofibrils (e.g. tortuosity [41]) as well as their mechanical response (e.g. strain-hardening response [7]) depending on model parameters, we suggest that our microscopic model approach can estimate the relationship between nanometre-scale dynamics of individual protofibrils and the mechanical properties of micrometre-scale fibrin clots. For example, the dependence of specific mediators such as factor XIIIa on the network architecture, and thus on the elasticity of fibrin clots, can be estimated by modelling the aggregation or bending stiffness on the level of protofibril fibres. Our numerical results suggest that the aggregated protofibril structure was altered to withstand extension by increasing the bending stiffness of individual protofibrils, which is consistent with experimental results [13,17,22]. In the future, we will report the relationship between individual protofibril behaviours and the macro-scale mechanical response of fibrin clots, including the fibrous network structure.
Acknowledgements
The authors thanks Dr Hirohiko Niioka, Dr Hideo Hirakata, Dr Naoko Sugita, Dr Kazuya Tatsumi and Dr Ryo Torii for their helpful discussions.
Appendix A
A.1. Bending energy
For equations (2.4c), we redefine the position vectors as
| A 1a |
| A 1b |
| A 1c |
and also
| A 2a |
and
| A 2b |
Then, the bending energy can be expressed as
| A 3a |
| A 3b |
| A 3c |
where is the total set of indices for bending elements. The nodal force (i ∈ [1, N]; N is the number of nodal points) is calculated by
| A 4a |
and
| A 4b |
where is the subset of involving the nodal point ri. Focusing on the formulation of a bending element e, we calculate as
| A 5 |
and
| A 6a |
| A 6b |
| A 6c |
Introducing , we obtain
| A 7 |
and analogously
| A 8 |
Consequently, the nodal forces in a bending element e are given by
| A 9 |
| A 10 |
| A 11 |
A.2. Torsion energy
For equation (2.5c), we redefine the position vectors as
| A 12a |
| A 12b |
| A 12c |
| A 12d |
and
| A 13a |
| A 13b |
| A 13c |
| A 13d |
Then, we rewrite equation (2.6) as
| A 14a |
and
| A 14b |
The torsion energy WT is given by
| A 15a |
| A 15b |
| A 15c |
where is the total set of indices for torsional elements. The nodal force (i ∈ [1, N]) is calculated by
| A 16a |
and
| A 16b |
where is the subset of involving the nodal point ri. Focusing on the formulation of a torsional element e, we calculate as
| A 17 |
and
| A 18 |
Introducing , and referring to equations (A7) and (A8), we obtain
| A 19a |
and
| A 19b |
We derive the index notation with the summation convention
| A 20a |
and
| A 20b |
where
| A 21a |
| A 21b |
| A 21c |
and
| A 21d |
Thus,
| A 22a |
and
| A 22b |
for the arbitrary vector β. Consequently, the nodal forces in a torsional element e are given by
| A 23a |
| A 23b |
| A 23c |
and
| A 23d |
Note that in practical application, we take
| A 24 |
Data accessibility
The data used in the figures are provided in the electronic supplementary material.
Authors' contributions
S.Ii. and S.W. conceptualized the topic. S.Ii., R.E., N.T., T.S. and S.Is. developed the methodology. N.T., R.E., T.S., S.Is. and S.W. designed the analysis conditions. N.T. and R.E. performed numerical simulation. N.T., R.E. and T.S. analysed data. S.Is. developed the software. S.W. and S.Ii. supervised the project. N.T. and R.E. drafted the manuscript; and all authors edited the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by JSPS KAKENHI grant nos. JP15K12510, JP19H01175, JP20H04504, JP20K20169, and by MEXTas a ‘Program for Promoting Researches on the Supercomputer Fugaku’ (hp200136, hp210181). N.T. and S.W. are grateful for the financial support of UCL-Osaka Partner Funds.
References
- 1.Weisel JW, Litvinov RI. 2013. Mechanisms of fibrin polymerization and clinical implications. Blood 121, 1712-1719. ( 10.1182/blood-2012-09-306639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heit JA. 2015. Epidemiology of venous thromboembolism. Nat. Rev. Cardiol. 12, 464-474. ( 10.1038/nrcardio.2015.83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kattula S, Byrnes JR, Wolberg AS. 2017. Fibrinogen and fibrin in hemostasis and thrombosis. Arterioscler. Thromb. Vasc. Biol. 37, e13-e21. ( 10.1161/ATVBAHA.117.308564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown AEX, Litvinov RI, Discher DE, Purohit PK, Weisel JW. 2009. Multiscale mechanics of fibrin polymer: gel stretching with protein unfolding and loss of water. Science 325, 741-744. ( 10.1126/science.1172484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen CE, Qiu Y, McCarty OJT, Lam WA. 2018. Platelet mechanotransduction. Annu. Rev. Biomed. Eng. 20, 253-275. ( 10.1146/annurev-bioeng-062117-121215) [DOI] [PubMed] [Google Scholar]
- 6.Lam WA, Chaudhuri O, Crow A, Webster KD, Li T-D, Kita A, Huang J, Fletcher DA. 2011. Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nat. Mater. 10, 61-66. ( 10.1038/NMAT2903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. 2005. Nonlinear elasticity in biological gels. Nature 435, 191-194. ( 10.1038/nature03521) [DOI] [PubMed] [Google Scholar]
- 8.Chernysh IN, Weise JW. 2008. Dynamic imaging of fibrin network formation correlated with other measures of polymerization. Blood 111, 4854-4861. ( 10.1182/blood-2007-08-105247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernysh IN, Nagaswami C, Weisel JW. 2011. Visualization and identification of the structures formed during early stages of fibrin polymerization. Blood 117, 4609-4614. ( 10.1182/blood-2010-07-297671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhmurov A, Protopopova AD, Litvinov RI, Zhukov P, Weisel JW, Barsegov V. 2018. Atomic structural models of fibrin oligomers. Structure 26, 857-868. ( 10.1016/j.str.2018.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisel JW, Medved L. 2001. The structure and function of the alpha C domains of fibrinogen. Ann. NY Acad. Sci. 936, 312-327. ( 10.1111/j.1749-6632.2001.tb03517.x) [DOI] [PubMed] [Google Scholar]
- 12.Yermolenko IS, Lishko VK, Ugarova TP, Magonov SN. 2011. High-resolution visualization of fibrinogen molecules and fibrin fibers with atomic force microscopy. Biomacromolecules 12, 370-379. ( 10.1021/bm101122g) [DOI] [PubMed] [Google Scholar]
- 13.Ryan EA, Mockros LF, Weisel JW, Lorand L. 1999. Structural origins of fibrin clot rheology. Biophys. J. 77, 2813-2826. ( 10.1016/S0006-3495(99)77113-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisel JW, Nagaswami C. 1992. Computer modeling of fibrin polymerization kinetics correlated with electron microscope and turbidity observations: clot structure and assembly are kinetically controlled. Biophys. J. 63, 111-128. ( 10.1016/S0006-3495(92)81594-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z et al. 2019. Microclot array elastometry for integrated measurement of thrombus formation and clot biomechanics under fluid shear. Nat. Commun. 10, 2051. ( 10.1038/s41467-019-10067-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badiei N et al. 2015. Effects of unidirectional flow shear stresses on the formation, fractal microstructure and rigidity of incipient whole blood clots and fibrin gels. Clin. Hemorheol. Microcirc. 60, 451-464. ( 10.3233/CH-151924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collet JP, Shuman H, Ledger RE, Lee S, Weisel JW. 2005. The elasticity of an individual fibrin fiber in a clot. Proc. Natl Acad. Sci. USA 102, 9133-9137. ( 10.1073/pnas.0504120102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg CS, Dobson JV, Miraglia CC. 1985. Regulation of plasma factor XIII binding to fibrin in vitro. Blood 66, 1028-1034. ( 10.1182/blood.V66.5.1028.1028) [DOI] [PubMed] [Google Scholar]
- 19.Weisel JW. 2004. The mechanical properties of fibrin for basic scientists and clinicians. Biophys. Chem. 112, 267-276. ( 10.1016/j.bpc.2004.07.029) [DOI] [PubMed] [Google Scholar]
- 20.He S, Bark N, Wang H, Svensson J, Blombäck M. 2009. Effects of acetylsalicylic acid on increase of fibrin network porosity and the consequent upregulation of fibrinolysis. J. Cardiovasc. Pharmacol. 53, 24-29. ( 10.1097/FJC.0b013e3181953e0f) [DOI] [PubMed] [Google Scholar]
- 21.Yeromonahos C, Marlu R, Polack B, Francois C. 2012. Antithrombin-independent effects of heparins on fibrin clot nanostructure. Arterioscler. Thromb. Vasc. Biol. 32, 1320-1324. ( 10.1161/ATVBAHA.112.245308) [DOI] [PubMed] [Google Scholar]
- 22.Piechocka IK, Kurniawan NA, Grimbergen J, Koopman J, Koenderink GH. 2017. Recombinant fibrinogen reveals the differential roles of - and -chain cross-linking and molecular heterogeneity in fibrin clot strain-stiffening. J. Thromb. Haemost. 15, 938-949. ( 10.1111/jth.13650) [DOI] [PubMed] [Google Scholar]
- 23.Onck PR, Koeman T, van Dillen T, van der Giessen E. 2005. Alternative explanation of stiffening in cross-linked semiflexible networks. Phys. Rev. Lett. 95, 178102. ( 10.1103/PhysRevLett.95.178102) [DOI] [PubMed] [Google Scholar]
- 24.Moiseyev G, Givli S, Bar-Yoseph PZ. 2013. Fibrin polymerization in blood coagulation: a statistical model. J. Biomech. 46, 26-30. ( 10.1016/j.jbiomech.2012.09.019) [DOI] [PubMed] [Google Scholar]
- 25.Yesudasan S, Wang X, Averett RD. 2018. Coarse-grained molecular dynamics simulations of fibrin polymerization: effects of thrombin concentration on fibrin clot structure. J. Mol. Model. 24, 109. ( 10.1007/s00894-018-3642-7) [DOI] [PubMed] [Google Scholar]
- 26.Yesudasan S, Wang X, Averett RD. 2018. Fibrin polymerization simulation using a reactive dissipative particle dynamics method. Biomech. Model. Mechanobiol. 17, 1389-1403. ( 10.1007/s10237-018-1033-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosesson MW, Siebenlist KR, Hainfeld Jf, Wall JS, Soria J, Soria C, Caen JP. 1996. The relationship between the fibrinogen D domain self-association/cross-linking site (gammaXL) and the fibrinogen Dusart abnormality (Aalpha R554C-albumin): clues to thrombophilia in the ‘Dusart syndrome’. J. Clin. Invest. 97, 2342-2350. ( 10.1172/JCI118677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall CE, Slayter HS. 1959. The fibrinogen molecule: its size, shape, and mode of polymerization. J. Biophys. Biochem. Cytol. 5, 11-27. ( 10.1083/jcb.5.1.11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fowler WE, Erickson HP. 1979. Trinodular structure of fibrinogen: confirmation by both shadowing and negative stain electron microscopy. J. Mol. Biol. 134, 241-249. ( 10.1016/0022-2836(79)90034-2) [DOI] [PubMed] [Google Scholar]
- 30.Williams RC. 1981. Morphology of bovine fibrinogen monomers and fibrin oligomers. J. Mol. Biol. 150, 399-408. ( 10.1016/0022-2836(81)90555-6) [DOI] [PubMed] [Google Scholar]
- 31.Weisel JW, Stauffacher CV, Bullitt E, Cohen C. 1985. A model for fibrinogen: domains and sequence. Science 230, 1388-1391. ( 10.1126/science.4071058) [DOI] [PubMed] [Google Scholar]
- 32.Lim BBC, Lee EH, Sotomayor M, Schulten K. 2008. Molecular basis of fibrin clot elasticity. Structure 16, 449-459. ( 10.1016/j.str.2007.12.019) [DOI] [PubMed] [Google Scholar]
- 33.Tan C, Jung J, Kobayashi C, Sugita Y. 2020. A singularity-free torsion angle potential for coarse-grained molecular dynamics simulations. J. Chem. Phys. 153, 044110. ( 10.1063/5.0013089) [DOI] [PubMed] [Google Scholar]
- 34.Freeman GS, Hinckley DM, Lequieu JP, Whitmer JK, de Pablo JJ. 2014. Coarse-grained modeling of DNA curvature. J. Chem. Phys. 141, 165103. ( 10.1063/1.4897649) [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Mochalkin I, Doolittle RF. 2000. A model of fibrin formation based on crystal structures of fibrinogen and fibrin fragments complexed with synthetic peptides. Proc. Natl Acad. Sci. USA 97, 14 156-14 161. ( 10.1073/pnas.97.26.14156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litvinov RI, Barsegov V, Schissler AJ, Fisher AR, Bennett JS, Weisel JW, Shuman H. 2011. Dissociation of bimolecular αIIbβ3-fibrinogen complex under a constant tensile force. Biophys. J. 100, 165-173. ( 10.1016/j.bpj.2010.11.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamada H, Imai Y, Nakamura M, Ishikawa T, Yamaguchi T. 2013. Computational study on thrombus formation regulated by platelet glycoprotein and blood flow shear. Microvasc. Res. 89, 95-106. ( 10.1016/j.mvr.2013.05.006) [DOI] [PubMed] [Google Scholar]
- 38.Ii S, Shimizu K, Sugiyama K, Takagi S. 2018. Continuum and stochastic approach for cell adhesion process based on Eulerian fluid-capsule coupling with lagrangian markers. J. Comput. Phys. 374, 769-786. ( 10.1016/j.jcp.2018.08.002) [DOI] [Google Scholar]
- 39.Koshiyama K, Wada S. 2011. Molecular dynamics simulations of pore formation dynamics during the rupture process of a phospholipid bilayer caused by high-speed equibiaxial stretching. J. Biomech. 44, 2053-2058. ( 10.1016/j.jbiomech.2011.05.014) [DOI] [PubMed] [Google Scholar]
- 40.Shigematsu T, Koshiyama K, Wada S. 2015. Effects of stretching speed on mechanical rupture of phospholipid/cholesterol bilayers: molecular dynamics simulation. Sci. Rep. 5, 15369. ( 10.1038/srep15369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai VK, Frey CR, Kerandi AM, Lake SP, Tranquillo RT, Barocas VH. 2012. Microstructural and mechanical differences between digested collagen-fibrin co-gels and pure collagen and fibrin gels. Acta Biomater. 8, 4031-4042. ( 10.1016/j.actbio.2012.07.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fowler WE, Hantgan RR, Hermans J, Erickson HP. 1981. Structure of the fibrin protofibril. Proc. Natl Acad. Sci. USA 78, 4872-4876. ( 10.1073/pnas.78.8.4872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhmurov A, Brown AEX, Litvinov RI, Dima RI, Weisel JW, Barsegov V. 2011. Mechanism of fibrin(ogen) forced unfolding. Structure 19, 1615-1624. ( 10.1016/j.str.2011.08.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ermak DL, McCammon JA. 1978. Brownian dynamics with hydrodynamic interactions. J. Chem. Phys. 69, 1352-1360. ( 10.1063/1.436761) [DOI] [Google Scholar]
- 45.Marsaglia G. 2003. Xorshift RNGs. J. Stat. Softw. 8, 1-6. ( 10.18637/jss.v008.i14) [DOI] [Google Scholar]
- 46.Box GEP, Muller ME. 1958. A note on the generation of random normal deviates. Ann. Math. Stat. 29, 610-611. ( 10.1214/aoms/1177706645) [DOI] [Google Scholar]
- 47.Ogston CM, Ogston D. 1966. Plasma fibrinogen and plasminogen levels in health and in ischaemic heart disease. J. Clin. Pathol. 19, 352-356. ( 10.1136/jcp.19.4.352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savitzky AS, Golay MJE. 1964. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627-1639. ( 10.1021/ac60214a047) [DOI] [Google Scholar]
- 49.Stylianopoulos T, Barocas VH. 2007. Volume-averaging theory for the study of the mechanics of collagen networks. Comput. Methods Appl. Mech. Eng. 196, 2981-2990. ( 10.1016/j.cma.2006.06.019) [DOI] [Google Scholar]
- 50.Roeder BA, Kokini K, Sturgis JE, Robinson JP, Voytik-Harbin SL. 2002. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J. Biomech. Eng. 124, 214-222. ( 10.1115/1.1449904) [DOI] [PubMed] [Google Scholar]
- 51.Roska FJ, Ferry JD. 1982. Studies of fibrin film. I. Stress relaxation and birefringence. Biopolymers 21, 1811-1832. ( 10.1002/bip.360210910) [DOI] [PubMed] [Google Scholar]
- 52.Nelb GW, Kamykowski GW, Ferry JD. 1981. Rheology of fibrin clots. V. Shear modulus, creep, and creep recovery of fine unligated clots. Biophys. Chem. 13, 15-23. ( 10.1016/0301-4622(81)80020-8) [DOI] [PubMed] [Google Scholar]
- 53.Muller MF, Ris H, Ferry JD. 1984. Electron microscopy of fine fibrin clots and fine and coarse fibrin films. J. Mol. Biol. 174, 369-384. ( 10.1016/0022-2836(84)90343-7) [DOI] [PubMed] [Google Scholar]
- 54.Guimarães CF, Gasperini L, Marques AP, Reis RL. 2020. The stiffness of living tissues and its implications for tissue engineering. Nat. Rev. Mater. 5, 351-370. ( 10.1038/s41578-019-0169-1) [DOI] [Google Scholar]
- 55.Bustamante CJ, Chemla YR, Liu S, Wnag MD. 2021. Optical tweezers in single-molecule biophysics. Nat. Rev. Methods Primers 1, 25. ( 10.1038/s43586-021-00021-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Norton DG, Fan NK, Goudie MJ, Handa H, Platt MO, Averett RD. 2017. Computational imaging analysis of glycated fibrin gels reveals aggregated and anisotropic structures. J. Biomed. Mater. Res. A 105, 2091-2198. ( 10.1002/jbm.a.36074) [DOI] [PubMed] [Google Scholar]
- 57.Veklich Y, Francis CW, White J, Weisel JW. 1998. Structural studies of fibrinolysis by electron microscopy. Blood 92, 4721-4729. ( 10.1182/blood.V92.12.4721) [DOI] [PubMed] [Google Scholar]
- 58.Litvinov RI, Faizullin DA, Zuev YF, Weisel JW. 2012. The α-helix to β-sheet transition in stretched and compressed hydrated fibrin clots. Biophys. J. 103, 1020-1027. ( 10.1016/j.bpj.2012.07.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Kumar S, Nisar A, Bonn M, Rausch MK, Parekh SH. 2021. Probing fibrin’s molecular response to shear and tensile deformation with coherent Raman microscopy. Acta Biomater. 121, 383-392. ( 10.1016/j.actbio.2020.12.020) [DOI] [PubMed] [Google Scholar]
- 60.Mickel W, Münster S, Jawerth LM, Vader DA, Weitz DA, Sheppard AP, Mecke K, Fabry B, Schröder-Turk GE. 2008. Robust pore size analysis of filamentous networks from three-dimensional confocal microscopy. Biophys. J. 95, 6072-6080. ( 10.1529/biophysj.108.135939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia J, Cai LH, Wu H, MacKintosh FC, Weitz DA. 2021. Anomalous mechanics of Zn2+-modified fibrin networks. Proc. Natl Acad. Sci. USA 118, e2020541118. ( 10.1073/pnas.2020541118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolberg AS, Rosendaal FR, Weitz JI, Jaffer IH, Agnelli G, Baglin T, Mackman N. 2015. Venous thrombosis. Nat. Rev. Disease Primers 1, 15006. ( 10.1038/nrdp.2015.6) [DOI] [PubMed] [Google Scholar]
- 63.Chernysh IN, Nagaswami C, Kosolapova S, Peshkova AD, Cuker A, Cines DB, Cambor CL, Litvinov RI, Weisel JW. 2020. The distinctive structure and composition of arterial and venous thrombi and pulmonary emboli. Sci. Rep. 10, 5112. ( 10.1038/s41598-020-59526-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janmey PA, Winer JP, Weisel JW. 2009. Fibrin gels and their clinical and bioengineering applications. J. R. Soc. Interface 6, 1-10. ( 10.1098/rsif.2008.0327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sivaraj D. 2021. Hydrogel scaffolds to deliver cell therapies for wound healing. Front. Bioeng. Biotechnol. 9, 660145. ( 10.3389/fbioe.2021.660145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rana A, Westein E, Niego B, Hagemeye CE. 2019. Shear-dependent platelet aggregation: mechanisms and therapeutic opportunities. Font. Cardiovasc. Med. 6, 141. ( 10.3389/fcvm.2019.00141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neeves KB, Illing DA, Diamond SL. 2010. Thrombin flux and wall shear rate regulate fibrin fiber deposition state during polymerization under flow. Biophys. J. 98, 1344-1352. ( 10.1016/j.bpj.2009.12.4275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gersh KC, Edmondson KE, Weisel JW. 2010. Flow rate and fibrin fiber alignment. J. Thromb. Haemost. 8, 2826-2828. ( 10.1111/j.1538-7836.2010.04118.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campbell RA, Aleman MM, Gray LD, Falvo MR, Wolberg AS. 2010. Flow profoundly influences fibrin network structure: implications for fibrin formation and clot stability in haemostasis. Thromb. Haemost. 104, 1281-1284. ( 10.1160/TH10-07-0442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whittaker P, Przyklenk K. 2009. Fibrin architecture in clots: a quantitative polarized light microscopy analysis. Blood Cells Mol. Dis. 42, 51-56. ( 10.1016/j.bcmd.2008.10.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varjú I, Sótonyi P, Machovich R, Szabó L, Tenekedjiev K, Silva MMCG, Longstaff, C, Kolev K. 2011. Hindered dissolution of fibrin formed under mechanical stress. J. Thromb. Haemost. 9, 979-986. ( 10.1111/j.1538-7836.2011.04203.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiss HJ, Turitto VT, Baumgartner HR. 1986. Role of shear rate and platelets in promoting fibrin formation on rabbit subendothelium. Studies utilizing patients with quantitative and qualitative platelet defects. J. Clin. Invest. 78, 1072-1082. ( 10.1172/JCI112663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tijburg PN, Ijsseldijk MJ, Sixma JJ, de Groot PG. 1991. Quantification of fibrin deposition in flowing blood with peroxidase-labeled fibrinogen. High shear rates induce decreased fibrin deposition and appearance of fibrin monomers. Arterioscler. Thromb. 11, 211-220. ( 10.1161/01.ATV.11.2.211) [DOI] [PubMed] [Google Scholar]
- 74.Kamada H, Imai Y, Nakamura M, Ishikawa T, Yamaguchi T. 2012. Computational analysis on the mechanical interaction between a thrombus and red blood cells: possible causes of membrane damage of red blood cells at microvessels. Med. Eng. Phys. 34, 1411-1420. ( 10.1016/j.medengphy.2012.01.003) [DOI] [PubMed] [Google Scholar]
- 75.Rack K, Huck V, Hoore M, Fedosov DA, Schneider SW, Gompper G. 2017. Margination and stretching of von Willebrand factor in the blood stream enable adhesion. Sci. Rep. 7, 14278. ( 10.1038/s41598-017-14346-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye H, Shen Z, Li Y. 2019. Interplay of deformability and adhesion on localization of elastic micro-particles in blood flow. J. Fluid Mech. 861, 55-87. ( 10.1017/jfm.2018.89) [DOI] [Google Scholar]
- 77.Takeishi N, Rosti ME, Imai Y, Wada S, Brandt L. 2019. Haemorheology in dilute, semi-dilute and dense suspensions of red blood cells. J. Fluid Mech. 872, 818-848. ( 10.1017/jfm.2019.393) [DOI] [Google Scholar]
- 78.Takeishi N, Imai Y, Wada S. 2019. Capture event of platelets by bolus flow of red blood cells in capillaries. J. Biomech. Sci. Eng. 14, 1-10. ( 10.1299/jbse.18-00535) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in the figures are provided in the electronic supplementary material.