Abstract
Many proteins that contain a carboxyl-terminal CaaX sequence motif, including Ras and yeast a-factor, undergo a series of sequential posttranslational processing steps. Following the initial prenylation of the cysteine, the three C-terminal amino acids are proteolytically removed, and the newly formed prenylcysteine is carboxymethylated. The specific amino acids that comprise the CaaX sequence influence whether the protein can be prenylated and proteolyzed. In this study, we evaluated processing of a-factor variants with all possible single amino acid substitutions at either the a1, the a2, or the X position of the a-factor Ca1a2X sequence, CVIA. The substrate specificity of the two known yeast CaaX proteases, Afc1p and Rce1p, was investigated in vivo. Both Afc1p and Rce1p were able to proteolyze a-factor with A, V, L, I, C, or M at the a1 position, V, L, I, C, or M at the a2 position, or any amino acid at the X position that was acceptable for prenylation of the cysteine. Eight additional a-factor variants with a1 substitutions were proteolyzed by Rce1p but not by Afc1p. In contrast, Afc1p was able to proteolyze additional a-factor variants that Rce1p may not be able to proteolyze. In vitro assays indicated that farnesylation was compromised or undetectable for 11 a-factor variants that produced no detectable halo in the wild-type AFC1 RCE1 strain. The isolation of mutations in RCE1 that improved proteolysis of a-factor-CAMQ, indicated that amino acid substitutions E139K, F189L, and Q201R in Rce1p affected its substrate specificity.
Traditionally, the CaaX sequence motif has been defined as a cysteine (C) four amino acids from the C terminus, followed by two amino acids that are often aliphatic (aa), and a C-terminal amino acid (X). Proteins containing a CaaX sequence motif undergo a series of sequential posttranslational modifications that are important for their localization and function. Processing of most CaaX proteins, including Ras, Rho, G protein gamma subunit, and yeast a-factor, involves prenylation of the cysteine four amino acids from the C terminus, endoproteolytic removal of the three C-terminal amino acids, and carboxymethylation of the newly formed prenyl cysteine. The specific amino acids that comprise the CaaX sequence influence whether the 15-carbon farnesyl group or the 20-carbon geranylgeranyl group is attached to the cysteine and whether the aaX sequence is proteolytically removed. Considerable effort has been made to define the sequence features that influence prenylation by the farnesyltransferase and the geranylgeranyltransferase I (7, 8, 30, 35). More recently, two CaaX proteases, Afc1p and Rce1p, have been identified in yeast (4), and homologs of these enzymes have been found in mammals (12, 23, 32, 40; D. H. Wong, C. E. Trueblood, D. Dimster-Denk, J. W. Phillips, P. M. Lagaay, J. Rine, and M. N. Ashby, unpublished data). Since activated Ras function in yeast is attenuated when the aaX sequence is not removed (4), the CaaX proteases that process Ras may be targets for anticancer drug discovery. A number of CaaX protease inhibitors are currently being developed (10, 27).
There is relatively little information about substrate sequence requirements for the CaaX proteases. Even though the aaX sequence is removed from most CaaX proteins, including Ras and Rho proteins (11, 15, 16), some CaaX proteins, such as the alpha and beta subunits of phosphorylase kinase from rabbit muscle (17), retain the aaX sequence after farnesylation. For most CaaX proteins, the identity of the CaaX proteases responsible for cleavage has not been determined. Despite great interest in biochemical characterization of the CaaX proteolytic activity, only partial purification has been achieved (2, 10, 20, 31), presumably because the CaaX proteases are integral membrane proteins. A recent in vitro study examined the ability of a mammalian membrane fraction to proteolyze a large set of CaaX peptide substrates (20). Yet, it is not clear which CaaX protease was being assayed or whether more than one CaaX protease was present in the membrane fraction.
The isolation of the Saccharomyces cerevisiae AFC1 and RCE1 genes (4) clarified the identity of the various yeast CaaX protease activities that had been reported (3, 11, 18) and demonstrated that Afc1p and Rce1p have distinct but overlapping CaaX sequence specificities. Despite a lack of sequence similarity, both Afc1p and Rce1p process the a-factor CaaX sequence, CVIA. However, the CaaX sequences CAMQ and CTLM, when substituted into a-factor, can be processed only by Afc1p and Rce1p, respectively (4). Rce1p also proteolyzes the CaaX sequence of yeast Ras2 protein, CIIS, but apparently Afc1p does not (4).
In our study, a-factor variants with all possible single amino acids substitutions at either the a1, a2, or X position of the Ca1a2X sequence were expressed in MATa yeast strains that lacked one, both, or neither of the CaaX protease genes, AFC1 and RCE1. The processing of a-factor was measured by halo assay, a biological readout in which secretion of fully processed a-factor leads to growth arrest of a lawn of MATα cells. Halo data, together with in vitro farnesylation information, were used to determine which CaaX sequences were farnesylated and which were proteolyzed by Acf1p and Rce1p. In addition, a region of the Rce1p CaaX protease that affects substrate recognition was identified.
MATERIALS AND METHODS
Plasmids.
A series of plasmids encoding a-factor variants with single amino acid substitutions at either the a1, a2, or X position of the CaaX sequence were generated by insertion of PCR products (encoding the last seven amino acids of the a-factor precursor and the 3′ flanking sequence) between the BamHI and EcoRI sites of YCpL-MFA1′, a CEN LEU2 plasmid that carries the MFA1 promoter and coding sequence for the first 29 amino acids of the a-factor precursor (43). The BamHI site in the MFA1 gene of YCpL-MFA1′ was created by site-directed mutagenesis of nucleotide 90 from C to T. The 5′ oligonucleotides used in PCR with Perkin-Elmer Taq polymerase were derivatives of C-TGG-GAT-CCA-GCA-TGT-GTT-ATT-GCT-TAG-TTT-C that differ in the nucleotides present in the a1, a2, or X codon (in boldface). For the a1, a2, and X substitution series, the oligonucleotide synthesis included a mixture of all four nucleotides at the a1, a2, and X codons, respectively. The BamHI site that was used to clone the PCR products is underlined. The 3′ oligonucleotide used in the PCR (TCA-CTG-TAT-ACG-GAA-TTC-TCA-TCA-GC) contained an EcoRI site (underlined) and was complementary to the 3′ flanking sequence of the MFA1 gene, except for the C in boldface. After ligation into the YCpL-MFA1′ BamHI-EcoRI vector fragment and transformation of Escherichia coli, individual plasmids were sequenced. Approximately 75% of the a1, a2, and X substitutions were obtained by sequencing about 60 plasmids from each of the three transformant pools. The remaining a-factor variants were constructed by a similar strategy using specific oligonucleotides. a-factor variants with G, A, V, L, I, S, T, D, N, E, Q, K, R, H, W, F, Y, P, C, and M at the a1 position are encoded by plasmids pJR2047 to pJR2066, respectively. a-factor variants with G, A, V, L, I, S, T, D, N, E, Q, K, R, H, W, F, Y, P, C, and M at the a2 position are encoded by plasmids pJR2067 to pJR2086, respectively. a-factor variants with G, A, V, L, I, S, T, D, N, E, Q, K, R, H, W, F, Y, P, C, and M at the X position are encoded by plasmids pJR2087 to pJR2106, respectively.
a-factor variants with the CaaX sequences CASQ (pJR2111), CTVM (pJR2112), and CSVM (pJR2113) were made using the same strategy with 5′ oligonucleotides encoding these specific CaaX sequences. A gene encoding an a-factor variant with the CaaX sequence CAMQ was constructed using oligonucleotide-directed mutagenesis to change the CaaX sequence of the MFA1 gene in pJR1457, a plasmid with the 1.57-kb EcoRI-XbaI MFA1 fragment inserted at the BamHI site of pRS426. The resulting plasmid, pJR1561, was used as a source of the gene encoding a-factor-CAMQ, which was cloned into the polylinker regions of pRS416 and pRS415 to yield pJR1556 and pJR2114, respectively.
MFA1 plasmids that encoded variants of a-factor that lack amino acids 2 through 5 were created by PCR using two oligonucleotides (5-M-5E [GAAATGCAGAATTCTATGGCTACCGCCGCTCCAAAAG] and 3-MFA1S [GAATGGACAGTCGACAATTAACTGG]) annealed to the 5′ coding region and 3′ flanking sequence of the MFA1 genes, respectively. To make the desired constructs, plasmids with different CaaX sequences were used as templates for PCR. The PCR fragments were inserted between the EcoRI and SalI sites of pJR1133, a 2μm URA3 expression vector that has the TDH3 promoter and PGK1 terminator. pJR1133 was constructed by deleting the BamHI-EcoRI region of YEplac195 (14) and inserting, between the HindIII and XbaI sites, a ∼3.4-kb HindIII-XbaI fragment from pG-3 (37) that has the TDH3 promoter and PGK terminator, separated by multiple cloning sites that flank a 1.7-kb HindII fragment from the lac operon. MFA1 plasmids that encode a-factor variants lacking amino acids 2 through 5 were constructed with the following CaaX sequences: CVIA (pJR1995), CVAA (pJR1980), CVLA (pJR1981), CVSA (pJR1982), CVTA (pJR1983), CVQA (pJR1984), CVHA (pJR1985), CVWA (pJR1986), CVFA (pJR1987), CVYA (pJR1988), CVCA (pJR1989), CVIG (pJR1990), CVID (pJR1991), CVIE (pJR1992), and CVIK (pJR1993).
Strains.
Strains used in these studies were constructed by standard genetic manipulations and grown on standard media (1). The AFC1 RCE1 (JRY5460), afc1Δ RCE1 (JRY5461), AFC1 rce1Δ (JRY5462), and afc1Δ rce1Δ (JRY5463) strains used for these studies are closely related to W303-1a (41) and have the following alleles: MATa his3 leu2 trp1 ura3 ram1H83Y mfa1::hisG mfa2Δ::hisG. The construction of these strains is described below. The mfa2Δ::hisG::URA3::hisG and mfa1::hisG::URA3::hisG alleles were constructed and transformed sequentially into JRY2640 to create a mata1 mfa1::hisG mfa2Δ::hisG ade2 leu2 lys2 ura3 can1 strain, JRY4276 (4). JRY4276 was transformed with MATα plasmid pJR157 to create strain JRY5390, which was then crossed to a MATa his3 leu2 trp1 ura3 afc1Δ::HIS3 strain (JRY5315) (4). The resulting diploid was sporulated to obtain a MATa his3 leu2 trp1 ura3 afc1Δ::HIS3 mfa1::hisG mfa2Δ::hisG segregant that was then crossed to a W303 strain that had the MATa promoter deleted (mataΔp) and had a MATα plasmid. This diploid yielded a mataΔp his3 leu2 trp1 ura3 afc1Δ::HIS3 mfa1::hisG mfa2Δ::hisG segregant, JRY5459, which was transformed with a MATα plasmid and then crossed to JRY5316 (MATa his3 leu2 trp1 ura3 rce1Δ::TRP1) (4). The AFC1 RCE1 (JRY5460), afc1Δ RCE1 (JRY5461), AFC1 rce1Δ (JRY5462), and afc1Δ rce1Δ (JRY5463) strains were segregants from the resulting diploid strain. These four strains were transformed with MFA1 plasmids (pJR2047 to pJR2106) that encode the a-factor variants with all possible single amino acids substitutions at either the a1, a2, or X position.
During the course of these studies, we discovered that the RAM1 gene in strain W303 had a mutation that changes codon 83 from a histidine codon to a tyrosine codon. This mutant form of Ram1p (the beta subunit of farnesyltransferase) reduced farnesyltransferase activity in crude extracts approximately 10-fold (W. Schafer and J. Rine, unpublished results). Since farnesyltransferase plays a central role in a-factor processing, it was important to determine whether the AFC1 RCE1 (JRY5460), afc1Δ RCE1 (JRY5461), AFC1 rce1Δ (JRY5462), and afc1Δ rce1Δ (JRY5463) strains, which are closely related to W303, carried the same allele of RAM1. Strains JRY5460, JRY5461, JRY5462, and JRY5463 were all found to have the allele of RAM1 from W303, which we designated ram1H83Y. Thus, differences in halo sizes seen between these four strains were not due to differences in farnesyltransferase activity.
To generate strains that overproduced wild-type Ram1p and thereby minimize the limiting effect of farnesyltransferase on a-factor production, the series of AFC1 RCE1 (JRY5460) strains carrying the CaaX variants of MFA1 (pJR2047 to pJR2106) was transformed with a high-copy-number RAM1 plasmid, pJR856. An afc1Δ::HIS3 derivative of JRY5460 was created by transformation of JRY5460 with a StuI-EcoRI fragment of pVB38 that carries the afc1Δ::HIS3 allele. The resulting strain, JRY6095, was transformed with a high-copy-number RAM1 plasmid, pJR856, to generate strain JRY6529. JRY6095 and JRY6529 were transformed with plasmids carrying the CaaX variants of MFA1 (pJR2047 to pJR2106).
AFC1 RCE1 (JRY5460), afc1Δ RCE1 (JRY6095), AFC1 rce1Δ (JRY5462), and afc1Δ rce1Δ (JRY5463) strains were transformed with the series of MFA1 plasmids, pJR1980 to pJR1993 and pJR1995, which encode variants of a-factor that lack amino acids 2 through 5.
The afc1Δ rce1Δ strain JRY5463 was transformed with plasmids pJR1968, pJR1969, pJR1970, and pJR1971, which encode wild-type Rce1p and three mutant forms of Rce1p that alter substrate recognition: F189L, E139K F189L, and Q201R. The resulting strains (JRY6811, JRY6812, JRY6813, and JRY6814) were transformed with MFA1 plasmids (pJR2047 to pJR2106) that encode the a-factor variants with all possible single amino acids substitutions either at the a1, a2, or X position.
Halo assays.
The relative levels of a-factor produced by various MATa strains were evaluated by pheromone diffusion (halo) assay. One microliter of a yeast cell pellet (approximately 106 cells) was spotted onto a solid rich medium (YPD) plate containing 0.04% Triton X-100 that had been spread with a lawn (approximately 2 × 106 cells) of the MATα sst2 strain, JRY3443. After 2 to 3 days of growth at 30°C, the relative amounts of a-factor produced by each MATa strain were evident from the size of the zone of growth inhibition (or halo) surrounding the MATa cells. The sst2 mutation facilitates measurement of a-factor production by making MATα cells more sensitive to a-factor-induced arrest (9). For the hydrophilic α-factor peptide, halo diameter is directly proportional to the log of pheromone concentration (35). This simple relationship between halo size and pheromone concentration is not observed for a-factor because it is hydrophobic and does not diffuse freely through the medium. The addition of Triton X-100 increases the rate of diffusion of a-factor, thereby increasing the sensitivity range of the halo assay. The relative halo sizes are reflective of the amount of a-factor exported from the MATa cells.
Farnesyltransferase enzyme assays.
The assay used for farnesylation of peptides was described previously (36). [3H] farnesyl diphosphate (9 μM, 80 μCi/μmol), various peptides of the sequence RTRCxxx (0 to 2.0 mM), and protein farnesyltransferase (116 nM) in buffer containing 50 mM Tris-HCl (pH 7.0), 5 mM MgCl2, 5 mM dithiothreitol, and 0.04% (wt/vol) dodecyl-β-d-maltoside with a total volume of 50 μl were incubated for 4 to 20 min at 25°C. Background levels were determined by reactions containing no peptide. The reaction mixtures were then spotted on a 1- by 3-cm strip of phosphocellulose P81 filter paper (Whatman). The filter papers were immersed in a solution of 1:1 75 mM H3PO4–95% ethanol (10 ml/strip) and gently swirled on a rotary platform for 10 min, followed by two additional 10-min wash cycles with fresh wash solution. The individual wet strips were transferred to scintillation vials containing 10 ml of Cytoscint (ICN) and 0.5 ml of 6 M HCl. The concentration of farnesylated peptide for each RTRCxxx peptide was then calculated by subtraction of background radioactivity from the radioactivity for each peptide.
In vitro PCR mutagenesis.
A DNA fragment containing the RCE1 open reading frame cloned into pRS315 LEU2 CEN vector was used as a template for PCR-based random mutagenesis of RCE1. The mutagenesis was performed in the PCR by increasing the final concentration of three of the four nucleotides to 1 mM, while maintaining the final concentration of the fourth dropout nucleotide at 0.1 mM. The dropout reaction mixtures were set up separately for all four nucleotide combinations. The final concentration of magnesium used in mutagenic PCR reactions was 7 mM. Commercially available M13 forward and reverse sequencing primers (New England Biolabs) were used at the final concentration of 1 μM in a 100 μl of RCE1 amplification reaction mixture. PCR products obtained with each dropout nucleotide mix were purified and pooled.
Identification of Rce1p variants with altered substrate specificity.
The pRS315 LEU2 CEN vector (5 μg) was digested with restriction enzymes PstI and HindIII to generate a linear backbone. The pooled PCR product (10 μl) obtained in the mutagenic PCR was cotransformed with 0.5 μg of the linearized backbone into the afc1Δ rce1Δ mfa1 mfa2Δ yeast strain, JRY5463, which had been transformed with the CAMQ version of MFA1 on a CEN URA3 pRS316 plasmid (pJR1556). Each of six independent transformation reactions was subdivided and plated on five plates. After incubation at 30°C for 3 days, transformants (approximately 250/plate) were replica plated directly onto a lawn of α sst2 cells to identify strains containing the versions of RCE1 displaying improved processing of the a-factor-CAMQ substrate. Halos were visible after 36 h of incubation at 30°C. The second round of selection was performed in a similar fashion using the F189L RCE1 mutant as a template for mutagenesis.
RESULTS
Substrate specificities of Afc1p and Rce1p CaaX proteases.
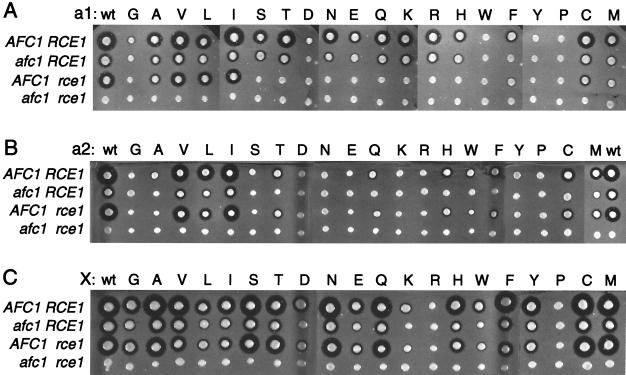
To investigate the protein substrate specificities of the Afc1p and Rce1p CaaX proteases, we examined the posttranslational processing of a-factor variants that had all possible single amino acid substitutions at either the a1, a2, or X position of the a-factor CaaX sequence, CVIA. Plasmids encoding all 57 single amino acid CaaX variants and wild-type a-factor were transformed into four MATa yeast strains that differ in their CaaX protease genes: AFC1 RCE1 (JRY5460), afc1 RCE1 (JRY5461), AFC1 rce1 (JRY5462), and afc1 rce1 (JRY5463). The relative levels of a-factor produced by these strains were evaluated by a-factor pheromone diffusion (halo) assay (Fig. 1). In this assay, secretion of fully processed a-factor leads to growth arrest of a lawn of MATα cells. The size of the a-factor halo, a zone of growth inhibition of MATα cells, reflects the amount of functional a-factor exported from the MATa cells. To be exported and functional, the 36-amino-acid a-factor precursor undergoes farnesylation of the cysteine four amino acids from the C terminus, followed by proteolytic removal of the aaX sequence, carboxylmethylation of the newly formed C terminus, and two N-terminal proteolytic cleavage events. Mature a-factor is a 12-amino-acid protein that is farnesylated and carboxylmethylated. As discussed below, the in vivo halo results provided valuable information about the specificities of the Afc1p and Rce1p CaaX proteases. Note that for a-factor CaaX sequence variants that produced no halos, interpretation of the a-factor halo results requires knowledge about the extent of farnesylation, since production of mature a-factor requires multiple processing steps and farnesylation must occur prior to the other steps. In vitro farnesylation data, presented below, aided in the interpretation of a-factor halo data for the CaaX sequence that produced no halos.
FIG. 1.
a-factor CaaX variants expressed in strains with and without AFC1 and RCE1 produce different amounts of mature a-factor. Plasmids encoding a-factor variants with all possible single amino acid substitutions at either the a1 (A), a2 (B), or X (C) position of the wild-type a-factor CaaX sequence, CVIA, were transformed into four MATa yeast strains that differ in their CaaX protease genes: AFC1 RCE1 (JRY5460), afc1 RCE1 (JRY5461), AFC1 rce1 (JRY5462), and afc1 rce1 (JRY5463). The relative levels of a-factor produced by these strains were evaluated by a-factor pheromone diffusion (halo) assay, a biological assay in which secretion of fully processed a-factor leads to growth arrest of MATα cells (see Materials and Methods). Biologically active a-factor exported from the MATa strains arrested growth of the MATα sst2 cells, forming a zone of growth inhibition (halo) that reflects the amount of a-factor produced.
Amino acid substitutions at the a1 position of the a-factor CaaX sequence (Fig. 1A) revealed that both CaaX proteases were able to accept a common set of amino acids at this position. Rce1p was also able to accept additional amino acids at the a1 position. Specifically, a-factor variants with A, V, L, I, C, or M at the a1 position were substrates of both Afc1p and Rce1p CaaX proteases, as shown by the presence of halos in strains that had either AFC1 or RCE1 and the absence of halos in strains lacking both AFC1 and RCE1. The a-factor variant with F at the a1 position appeared to be a substrate of Afc1p but not Rce1p, since no halo was observed in the afc1Δ strain. In contrast, substitutions of S, T, N, E, Q, K, R, and H at the a1 position created substrates that could be processed by Rce1p but not by Afc1p. It should be noted that the Rce1p-dependent halos in the afc1Δ strain were smaller than the halos produced by the same substrates in wild-type cells (Fig. 1A). At first glance, one might wonder why these halo sizes were smaller in the absence of Afc1p, if these CaaX sequences are not processed by Afc1p. The explanation for this apparent discrepancy is that Afc1p has two roles in a-factor processing: it cleaves the CaaX sequence at the carboxyl terminus and also contributes to N-terminal processing (5, 40). Thus, in afc1Δ strains, the size of all Rce1p-dependent halos was reduced due to loss of the Afc1p N-terminal contribution.
Substitutions of G, D, W, Y, and P at the a1 position created substrates that produced either very small halos (G, D, Y) or no halos (W, P). These deficiencies in a-factor production could result from poor proteolytic processing and/or poor prenylation. The halo data alone did not allow us to distinguish between these possibilities, but in vivo and in vitro results, described below, indicate that poor prenylation is at least partially responsible for the decreased a-factor production of these variants. Introduction of W, Y, and P at the a1 position caused a decrease in the efficiency of farnesylation of synthetic peptides in vitro, suggesting that inefficient prenylation was partially responsible for poor a-factor production in vivo. However, a comparison of halo sizes and in vitro farnesylation data with findings for other peptides indicates that W, Y, and P at the a1 position must also decrease the efficiency of CaaX proteolysis. No in vitro farnesylation assays have been performed on yeast farnesyltransferase with peptides with G or D at the a1 position, and there are conflicting data about whether mammalian farnesyltransferase can effectively farnesylate CaaX substrates with G or D at the a1 position (22, 29, 30, 34). If a-factor with G or D at the a1 position was not farnesylated efficiently, it would not be proteolyzed by Afc1p or Rce1p, due to the absence of a farnesylated cysteine. Alternatively, if we assume that the yeast farnesyltransferase was able to farnesylate a-factor-CDIA or a-factor-CGIA, then neither Afc1p nor Rce1p could proteolyze these proteins efficiently.
Substitutions at the a2 position (Fig. 1B) revealed that there was a restricted set of amino acids that allowed efficient prenylation and subsequent proteolysis by both CaaX proteases. Moreover, Afc1p tolerated a wider range of amino acids at the a2 position than at the a1 position. a-factor variants with V, L, I, C, or M at the a2 position produced medium-sized to large halos in the AFC1 RCE1 strain and were substrates for both Afc1p and Rce1p, as shown by the presence of halos in strains that had either AFC1 or RCE1, but not in a strain that lacked both AFC1 and RCE1. The remaining a2 variants of a-factor produced small halos (T, Q, H, W, F), very small halos (A, S, Y), or no halos (G, D, N, E, K, R, P) in the AFC1 RCE1 strain. Note that the halos produced by a-factor variants with A, S, and Y at the a2 position were marginally detectable and were not always visible in the photographs. Overproduction of the farnesyltransferase beta subunit, Ram1p, increased the size of these halos such that they were consistently detectable, though still quite small (see below). Clearly, inefficient farnesylation of these a-factor variants, and for a-factor variants with T, Q, H, W, and F at the a2 position, contributed to the low production of mature a-factor. As discussed below, the a2 variants that produced no detectable halo, and at least some of the a2 variants that produced very small halos, were defective for farnesylation in vitro. a-factor variants with A, S, T, Q, H, W, F, or Y at the a2 position produced detectable halos in strains that had Afc1p (AFC1 RCE1 and AFC1 rce1Δ) but produced no detectable halos in strains that lacked Afc1p (afc1Δ and afc1Δ rce1Δ), indicating that these a-factor variants were substrates of Afc1p. The absence of a halo in the afc1Δ strains expressing a-factor variants with A, S, T, Q, H, W, F, or Y at a2 may be due to a lack of Rce1p-mediated CaaX proteolysis but could be due to the combined effect of a reduction in prenylation and a lack of Afc1p-mediated N-terminal a-factor processing, resulting from the deletion of AFC1 (40; see below).
Amino acid substitutions at the X position revealed that the farnesyltransferase and both Afc1p and Rce1p CaaX proteases were able to accept a wide range of amino acids at this position. a-factor variants with G, A, V, L, I, S, T, N, E, Q, H, W, F, Y, C, or M at the X position were substrates of both Afc1p and Rce1p, as shown by the presence of halos in strains that have either AFC1 or RCE1 but not in a strain that lacks both AFC1 and RCE1. a-factor variants with R or P at the X position produced no detectable halo in the wild-type strain, indicating that these substrates were not adequately prenylated and/or proteolyzed. The a-factor variants with D or K at the X position produced small halos in strains that have AFC1 (wild type and rce1Δ) and no detectable halo in strains that lack AFC1 (afc1Δ and afc1Δ rce1Δ), indicating that a-factor-CVID and a-factor-CVIK were substrates of the Afc1p CaaX protease. It was not clear whether or not Rce1p could cleave a-factor-CVID or a-factor-CVIK, since the absence of a halo in the afc1Δ strain could be due to a combined effect of a partial loss of CaaX proteolysis, a loss of Afc1p-mediated N-terminal a-factor processing (40; see below), and a decrease in farnesylation efficiency.
N-terminal role of Afc1p in a-factor processing accounted for the absence a halo in afc1Δ strains carrying certain a-factor variants.
In addition to its role in C-terminal proteolysis, Afc1p also plays an important role in maturation of a-factor (40). Afc1p appears to be directly involved in the N-terminal cleavage which removes the first seven amino acids of the a-factor precursor (40), although the precise nature of its N-terminal role is unknown. Both Afc1p and the N-terminal seven amino acids of the a-factor precursor are required for efficient processing of a-factor, even for a-factor variants with CaaX sequences, such as CTLM, that cannot be proteolyzed by Afc1p (5). Deletion of the N-terminal region of the a-factor-CTLM precursor reduces the halo observed in a wild-type strain to a small size, similar to the halo observed in an afc1Δ strain (5).
To estimate the impact of the loss of the N-terminal function of Afc1p on the efficiency of a-factor production from particular a-factor variants, we deleted amino acids 2 to 5 of the a-factor precursors that had A, L, S, T, Q, H, W, F, Y, and C at the a2 position or G, D, E, and K at the X position (see Materials and Methods). The halos produced in the wild-type strain expressing truncated variants of a-factor-CVIA, a-factor-CVLA, a-factor-CVCA, a-factor-CVIG, and a-factor-CVIE were reduced to a size similar to that of halos produced in the afc1Δ strain expressing the same a-factor variants (data not shown). No halos were detected in either the wild-type or the afc1Δ strains expressing truncated a-factor with A, S, T, Q, H, W, F, or Y at the a2 position or with D or K at the X position (data not shown). These observations indicated that loss of the N-terminal Afc1p function alone reduced a-factor production from these variants below a detectable limit. Therefore, the absence of a detectable halo in afc1Δ strains expressing these a-factor variants could not be attributed unambiguously to lack of Rce1p-mediated proteolysis. Whether or not Rce1p can cleave these substrates remains unresolved.
Farnesylation was limiting for some a-factor variants.
A number of a-factor CaaX sequence variants produced no halo or a very small halo in the wild-type strain. Since farnesylation is required for production of mature a-factor and farnesylation is a prerequisite for CaaX proteolysis, two approaches were taken to determine whether farnesylation was limiting for a-factor production for these CaaX sequence variants: (i) the farnesyltransferase beta subunit was overproduced in strains carrying the a-factor variants and (ii) in vitro farnesylation assays were performed on a selected set of peptides.
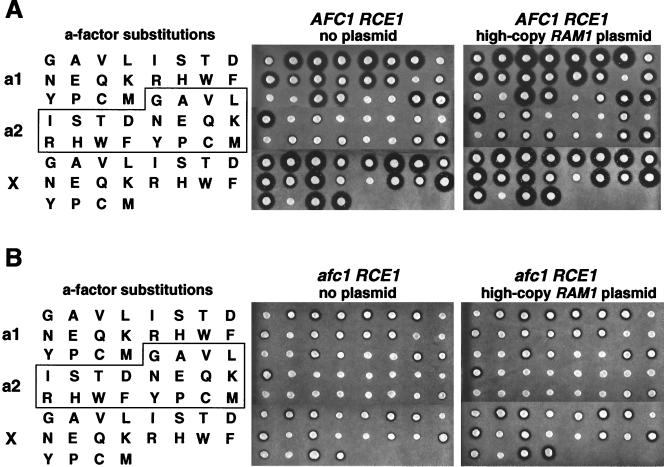
The levels of farnesyltransferase in each of the 58 AFC1 RCE1 strains carrying a-factor variants were increased by introduction of a high-copy-number RAM1 plasmid (encoding the farnesyltransferase beta subunit). Overproduction of the farnesyltransferase beta subunit, Ram1p, increased the halo size in nearly all of the strains that started with a small or medium-sized halo (Fig. 2A). Several a-factor variants that produced marginally detectable halos in the AFC1 RCE1 strain (D and Y at a1; A, S, and Y at a2) were able to produce small but easily detected halos when the wild-type Ram1p was overexpressed. In two cases, CVNA and CVIR, small halos were detected in the strains carrying the high-copy-number RAM1 plasmid, whereas no halo was detected before the introduction of the RAM1 plasmid. Thus, inefficient prenylation contributed to the decreased a-factor production observed for many of the a-factor CaaX variants. Note that for these a-factor variants, inefficient prenylation was presumably due in part to the presence of a hypomorphic RAM1 allele (ram1H83Y) in these yeast strains, which were derived from W303 (see Materials and Methods).
FIG. 2.
Overexpression of the beta subunit of farnesyltransferase increased production of many a-factor CaaX variants expressed in AFC1 RCE1 and afc1Δ RCE1 strains. The AFC1 RCE1 strain (JRY5460) and an isogenic afc1Δ RCE1 strain (JRY6095) carrying plasmids encoding a-factor variants with all possible single amino acid substitutions at either the a1, a2, or X position of the wild-type a-factor CaaX sequence, CVIA, were assayed for a-factor production in the absence (no plasmid) and the presence (high-copy-number RAM1 plasmid) of overexpression of the wild-type farnesyltransferase beta subunit. The relative levels of a-factor produced by these strains were evaluated by a-factor pheromone diffusion (halo) assay.
Inefficient prenylation, caused either by limiting amounts of Ram1p or by the ram1H83Y mutation, did not alter any conclusions about substrate specificities of the CaaX proteases. The overexpression of Ram1p in the afc1Δ RCE1 strain resulted in modest increases in halo size for many of the a-factor CaaX variants, particularly those with substitutions at the X position (Fig. 2B). However, there were no cases in which Ram1p overexpression in the afc1Δ RCE1 strain changed a-factor production qualitatively, from undetectable to detectable. Note that the increases in halo size resulting from Ram1p overexpression were much more dramatic in the AFC1 RCE1 strain (Fig. 2A) than in the isogenic afc1Δ RCE1 strain (Fig. 2B). Thus, farnesylation was not rate limiting for a-factor production in the afc1Δ RCE1 strain, presumably because lack of the Afc1p N-terminal function limited a-factor production.
a-factor CaaX variants with severe halo defects were poor substrates for in vitro farnesylation.
To determine directly whether the primary defect in processing of certain CaaX variants was in farnesylation, in vitro farnesylation assays were performed. A selected set of peptides was synthesized that had CaaX sequences identical to each of the a-factor variants that produced no halo and to some of the a-factor variants that produced very small halos. Relative to the natural a-factor CaaX sequence, CVIA, each of the peptides tested was poorly farnesylated in vitro (Table 1); the km values were 8- to 800-fold higher, and the kcat values were 5- to 300-fold lower, than for the wild-type CVIA. Thus, poor farnesylation of these a-factor CaaX variants was at least partially responsible for the low levels of mature a-factor in the in vivo halo assay. Peptides with D, K, or R at the a2 position and the peptide with P at the X position were unreactive, even at peptide concentrations of 50 mM. A 10-fold increase in the concentration of the enzyme did not result in detectable farnesylation of these peptides. Clearly, for a-factor with D, K, and R at the a2 position or P at the X position, the farnesylation defects were sufficient to account for the lack of a halo in vivo. Whether or not these substrates could be recognized by either CaaX protease is irrelevant since CaaX proteolysis is dependent on prenylation. The peptides with substitutions G, N, or E at the a2 position and R at the X position were very poor farnesylation substrates, exhibiting 100- to 700-fold increases in Km, 90- to 300-fold decreases in kcat, and catalytic efficiencies (kcat/Km) between 10−5 and 4 × 10−5, compared to 0.8 for CVIA. Peptides with substitution of Y or P at the a2 position exhibited 80- and 30-fold increases in Km, 20- and 40-fold decreases in kcat, and catalytic efficiencies of 10−3 and 3 × 10−3, respectively. Peptides with W, Y, or P at the a1 position exhibited 8- to 10-fold increases in Km, 5- to 10-fold decreases in kcat, and catalytic efficiencies that ranged from 8 × 10−3 to 2 × 10−2.
TABLE 1.
In vitro farnesylation of a-factor peptide variantsa
| Peptide | Km (μM) | Kcat (s−1) | kcat/Km (s−1 μM−1) |
|---|---|---|---|
| RTRCVIA | 3.1 ± 0.2 | 2.6 ± 0.1 | 0.8 |
| Substitutions at a1 | |||
| RTRCWIA | 28 ± 9 | 0.23 ± 0.02 | 8 × 10−3 |
| RTRCYIA | 23 ± 10 | 0.17 ± 0.04 | 7 × 10−3 |
| RTRCPIA | 32 ± 10 | 0.53 ± 0.09 | 0.02 |
| Substitutions at a2 | |||
| RTRCVGA | 2,223 ± 829 | 0.030 ± 0.007 | 10−5 |
| RTRCVDA | NDb | ND | ND |
| RTRCVNA | 334 ± 141 | 0.0078 ± 0.009 | 2 × 10−5 |
| RTRCVEA | 1,684 ± 316 | 0.018 ± 0.002 | 10−5 |
| RTRCVKA | ND | ND | ND |
| RTRCVRA | ND | ND | ND |
| RTRCVYA | 103 ± 17 | 0.15 ± 0.01 | 10−3 |
| RTRCVPA | 245 ± 102 | 0.069 ± 0.009 | 3 × 10−3 |
| Substitutions at X | |||
| RTRCVIR | 715 ± 250 | 0.028 ± 0.004 | 4 × 10−5 |
| RTRCVIP | ND | ND | ND |
Protein farnesyltransferase (116 nM) and [3H]FPP (9 μM, 80 μCi/μmol) were incubated with the indicated peptides (0 to 2.0 mM) for 4 to 20 min at 25°C in buffer containing 50 mM Tris-HCl (pH 7.0), 5 mM MgCl2, 5 mM DTT, and 0.04% (wt/vol) dodecyl-β-d-maltoside. Transfer of the 3H-labeled farnesyl group to the peptides was measured as described in Materials and Methods.
ND, farnesyltransferase not detectable, even at a peptide concentration of 50 mM.
Some a-factor variants are poorly farnesylated and poorly proteolyzed.
A more detailed comparison of the in vitro farnesylation data and the in vivo halo data suggested that certain a-factor CaaX variants that were inefficiently farnesylated were also poorly proteolyzed in vivo. The ability of CVNA and CVIR to produce detectable halos in the AFC1 RCE1 strain carrying high-copy-number RAM1 (Fig. 2) was remarkable given the very low catalytic efficiencies for farnesylation of CVNA and CVIR in vitro: 100- to 200-fold-higher Km values and 100- to 300-fold-lower kcat values, resulting in catalytic efficiencies more than 10,000-fold lower than for CVIA. Presumably, any substrates that were farnesylated more effectively than CVNA and CVIR in vitro and had comparable or smaller halos in vivo were poor substrates for CaaX proteolysis in vivo. Three a-factor CaaX variants (CWIA, CPIA, and CVPA) had lower Km values and higher kcat values than CVNA and CVIR, yet failed to produce a detectable halo. By deduction, these variants had a CaaX proteolysis defect in addition to their farnesylation defect. The catalytic efficiency for farnesylation of CVPA was about 10-fold higher than for CVNA and CVIR, suggesting that CVPA was proteolyzed somewhat less efficiently than CVNA and CVIR. The catalytic efficiencies for CWIA and CPIA farnesylation were 400- to 1,000-fold higher than for CVNA, strongly suggesting that the latter two sequences were at least partially farnesylated in vivo but were not proteolyzed by either CaaX protease. Two a-factor CaaX variants (CYIA and CVYA) produced halos that were comparable in size to CVNA and CVIR (Fig. 2), yet they had catalytic efficiencies that were 30- to 300-fold higher than for CVNA and CVIR. From this analysis, we deduce that CaaX proteolysis of CYIA and CVYA was partially defective. It is quite possible that, in addition to the established defects in farnesylation, CVGA and CVEA have a defect in CaaX proteolysis, since these CaaX sequences produced no halo and exhibited catalytic efficiencies that are only twofold lower than for CVNA.
Other a-factor variants (G, D, or E at the a1 position; A, S, T, Q, H, W, or F at the a2 position; D, E, K, or W at the X position) also have farnesylation defects, based on their small halo size and the increase in halo size that results from overexpression of the farnesyltransferase beta subunit (Fig. 2). However, because we did not assay in vitro farnesylation of these CaaX sequences, we could not determine whether these substitutions also decrease the efficiency of CaaX proteolysis.
a-factor variants were not substrates for geranylgeranyltransferase I.
We considered the possibility that changes in the CaaX sequence would make a-factor variants substrates of the geranylgeranyltransferase I. In fact, previously published studies show that a-factor-CVIL is both farnesylated and geranylgeranylated in vivo, with both forms being exported and functional (6). However, a-factor-CVIL did not produce a halo in a ram1Δ strain, which lacks the β subunit of the farnesyltransferase (C. Trueblood, unpublished results). Taken together, these data suggest that farnesyltransferase is able to farnesylate and geranylgeranylate a-factor-CVIL to a physiologically significant extent and that a-factor is not accessible to geranylgeranyltransferase I in vivo. It is notable that CVIL and CVII halo sizes increased significantly when Ram1p was overproduced (Fig. 2A). Ram1p overproduction would be expected to increase farnesyltransferase activity and decrease geranylgeranyltransferase I activity due to competition for the α subunit, which is shared by the two prenyltransferases. These data were consistent with the supposition that the farnesyltransferase, rather than the geranylgeranyltransferase I, was responsible for functional prenylation of a-factor-CVIL and a-factor-CVII.
Predictive value of specificity data.
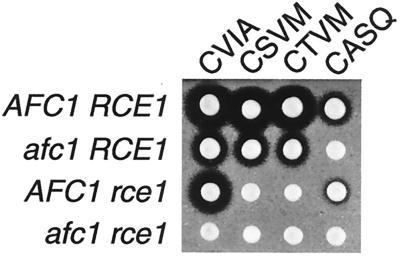
There are 98 proteins encoded in the S. cerevisiae genome that have a cysteine as the fourth amino acid from the C terminus (http://genome-www.stanford.edu/cgi-bin/SGD/search). To test whether the single amino acid substitution series in a-factor had predictive value for determining which of the 98 potential yeast CaaX proteins can serve as substrates of Afc1p and Rce1p, several additional a-factor variants were constructed. Based on our results, substrates with serine or threonine at the a1 position would be predicted to be proteolyzed by Rce1p but not by Afc1p, whereas substrates with serine or threonine at the a2 position would be predicted to be proteolyzed by Afc1p. The yeast genome encodes 18 proteins with serine or threonine at the a1 position. Nine of these proteins have an amino acid at the a2 position (G, D, N, E, K, or R) that resulted in no halo in our substitution series (Fig. 1) and in very poor or no farnesylation in vitro (Table 1). These proteins are unlikely to be adequately prenylated in vivo and therefore were not considered candidates for further study. The nine remaining proteins were considered likely to be Rce1p-specific substrates. One of these CaaX sequences, CTLM from Ste18p (the gamma subunit of the heterotrimeric G protein), was previously placed on a-factor and found to be proteolyzed by Rce1p but not Afc1p (5), in agreement with our predictions. Similarly, a-factor variants with CSVM and CTVM were tested and found to be proteolyzed by Rce1p but not Afc1p, as predicted (Fig. 3). The yeast genome encodes three proteins with serine or threonine at the a2 position that would be predicted to be proteolyzed by Afc1p. One of these three C-terminal sequences, CASQ, which is derived from Ydj1p (YNL064C), was tested in the context of a-factor and found to be proteolyzed by Afc1p but not Rce1p (Fig. 3), as predicted. Thus, to a first approximation, the information from the a-factor single amino acid substitution series appeared to be valuable in predicting which CaaX protease could proteolyze a-factor variants with CaaX sequences derived from bona fide proteins.
FIG. 3.
Afc1p and Rce1p CaaX proteases differ in the ability to proteolyze CaaX sequences with serine and threonine at the a1 and a2 positions. Plasmids encoding wild-type a-factor (CVIA) and a-factor variants with the CaaX sequences CSVM, CTVM, and CASQ were transformed into four MATa yeast strains that differ in their CaaX protease genes: AFC1 RCE1 (JRY5460), afc1 RCE1 (JRY6095), AFC1 rce1 (JRY5462), and afc1 rce1 (JRY5463). The relative levels of a-factor produced by these strains were evaluated by a-factor pheromone diffusion (halo) assay.
Mutations that alter Rce1p substrate specificity.
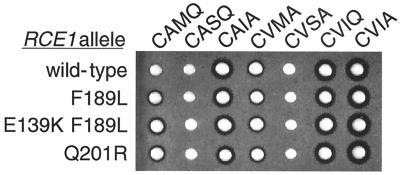
The lack of obvious sequence similarity of Rce1p to known proteases has precluded predictions of the position of the Rce1p active site or substrate binding site. A genetic screen was performed to identify mutations in RCE1 that would alter Rce1p substrate specificity and allow proteolysis of a-factor-CAMQ, a substrate that was very poorly recognized by the wild-type Rce1p (4, 5). The logic behind the screen was that mutations altering the substrate recognition properties of the enzyme would identify key regions involved in substrate recognition and/or binding. RCE1 PCR products, generated by PCR-based random mutagenesis, and a linearized CEN LEU2 plasmid were cotransformed into a afc1Δ rce1Δ yeast strain that expressed a-factor-CAMQ as the only form of a-factor precursor (JRY5463 transformed with pJR1556). Transformants were obtained as a result of recombination between the ends of the RCE1 PCR product and the homologous ends of the linear plasmid. Mutant forms of Rce1p capable of cleaving a-factor-CAMQ were identified by their ability to produce mature a-factor and thereby form halos on a lawn of α sst2 cells. Two different single amino acid changes in Rce1p led to small but reproducible increases in the halo size (Fig. 4): a phenylalanine-to-leucine substitution at position 189 (F189L) and a glutamine-to-arginine substitution at position 201 (Q201R). The mutated RCE1 gene encoding F189L-Rce1p was subjected to a second round of PCR mutagenesis to search for an additional mutation that could further increase the a-factor-CAMQ processing. A change of the glutamic acid codon at position 139 to a lysine codon (E139K) led to a notable increase in halo size (Fig. 4).
FIG. 4.
Amino acid substitutions in Rce1p that increased proteolysis of a-factor-CAMQ but did not alter proteolysis of other a-factor CaaX variants. Plasmids encoding a-factor variants with the indicated CaaX sequences (CAMQ, CASQ, CAIA, CVMA, CVSA, and CVIQ) or wild-type a-factor (CVIA) were transformed into MATa afc1 rce1 yeast strains that differ in the RCE1 allele carried on a second plasmid: wild-type RCE1 (wild type) or a mutant allele with the indicated amino acid substitution (F189L, E139K F189L, or Q210R). The relative levels of a-factor produced by these strains were evaluated by a-factor pheromone diffusion (halo) assay.
To assess whether the mutant forms of Rce1p had altered specificity for other a-factor variants, afc1Δ rce1Δ strains expressing either the E139K F189L mutant form of Rce1p or wild-type Rce1p were transformed with plasmids encoding wild-type a-factor and the 57 a-factor variants that have all possible substitutions at either the a1, a2, or X position. Halo assays on the resulting strains revealed no differences in the ability of the wild-type and mutant Rce1p enzymes to process these a-factor variants (data not shown). The failure of the mutant Rce1p enzymes to increase halo sizes for these a-factor variants ruled out the possibility that a general increase in Rce1p activity was responsible for the increased halo observed for a-factor-CAMQ. These data also indicated that the Rce1p mutations broadened the substrate specificity of the enzyme but did not result in any loss in the range of substrates it could process. In addition, afc1Δ rce1Δ strains expressing either wild-type Rce1p, the F189L mutant form of Rce1p, the E139K F189L mutant form of Rce1p, or the Q210R mutant form of Rce1p were transformed with plasmids encoding a-factor variants with the following CaaX sequences: CAMQ, CASQ, CAIA, CVMA, CVSA, CVIQ, and CVIA. Among these a-factor substrates, only a-factor-CAMQ was processed more effectively by the mutant forms of Rce1p (Fig. 4). Clearly the mutant forms of Rce1p did not exhibit an enhanced ability to proteolyze an a-factor variant sequence closely related to CAMQ (CASQ), nor a-factor variants with individual substitutions of A, M, or Q at the a1, a2, or X position, respectively. These observations indicate that the combination of amino acids in the CaaX sequence can sometimes influence Rce1p recognition in a way that cannot be predicted from the individual amino acid substitutions.
DISCUSSION
The goal of this work was to provide a comprehensive evaluation of the substrate specificities for the S. cerevisiae CaaX proteases, Afc1p and Rce1p, and to identify amino acids in Rce1p that affect substrate specificity. Part of the interest in CaaX proteases stems from their potential utility in modulating the activity of Ras proteins and other prenylated proteins that contribute to the growth and division of cancer cells. There is considerable interest in developing inhibitors against the human CaaX protease(s) that proteolyze the human N-, Ki-, and Ha-Ras proteins. Like farnesyltransferase inhibitors, which are currently being developed (13, 45), CaaX protease inhibitors could modulate Ras activity and thereby potentially function as anticancer agents. Even in cancer cells that do not carry activated Ras, Ras and other prenylated proteins, including RhoB, contribute to cell propagation and are potential targets for therapeutic intervention (24, 26, 39). Previously, there has been little information about the substrates and substrate specificities of the yeast and mammalian CaaX proteases.
Single amino acid substitution data serve as a predictive guide for which yeast CaaX sequences are prenylated and which are substrates of Afc1p and/or Rce1p.
The in vivo halo and in vitro farnesylation data from single amino acid substitutions in the a-factor CaaX sequence are summarized in Table 2. Afc1p and Rce1p exhibited substantial overlap in specificity at the a1, a2, and X positions (Table 2, row 1). However, Rce1p was able to accept additional amino acids at the a1 position that Afc1p was unable to accept (Table 2, row 2), and Afc1p was able to accept additional amino acids that Rce1p may not accept (Table 2, row 3). It was not possible to determine whether or not Rce1p was able to act on these latter a-factor variants, since the loss of the N-terminal role of Afc1p in a-factor processing, in combination with a partial defect in prenylation, reduced a-factor production below a detectable level. Both Afc1p and Rce1p accepted all amino acids at the X position that allowed efficient farnesylation. These results are in agreement with the observation that partially purified CaaX proteases can accept d-amino acids at the X position, but not at the a1 or a2 position (27), and can proteolyze farnesylated tripeptides nearly as well as farnesylated tetrapeptides (20, 27).
TABLE 2.
Summary of prenylation and proteolysis results on a-factor variants
| Results | Single amino acid substitution(s) in a-factor at the indicated position
|
||
|---|---|---|---|
| a1 | a2 | X | |
| Proteolyzed by Afc1p and Rce1p | A V L I C M | V L I C M | G A V L I S T N E Q H W F Y C M |
| Proteolyzed by Rce1p but not Afc1p | S T N E Q K R H | ||
| Proteolyzed by Afc1pa | F | A S T Q H W F Y | D K |
| Weak prenylation and/or proteolysisb | G D F | A S T Q H W F | D K W |
| Weak prenylation and proteolysisc | W Y P | Y P | |
| In vitro farnesylation defectd | W Y P | G D N E K R Y P | P R |
It was not clear whether Rce1p was able to act on these substrates. No halo was detected for these a-factor variants in strains lacking Afc1p, but the loss of Afc1p-dependent N-terminal processing of a-factor, in combination with a partial defect in prenylation, was sufficient to explain the lack of a halo.
The halo sizes produced by these a-factor variants indicated a defect in a-factor production. The lack of prenylation data precluded any conclusions about whether the defect was in prenylation and/or proteolysis. For the variants with X = D K W, in vivo data indicate a deficit in farnesylation (43), but there could also be a proteolysis defect.
The magnitude of in vitro farnesylation defect did not fully explain the observed reduction in a-factor production, indicating that there was also a proteolysis defect.
In vitro farnesylation assays were performed on these 13 a-factor CaaX variants. Each peptide exhibited at least a 50-fold decrease in farnesylation (50- to 100-fold decrease for a1 = W Y P; 103-fold decrease for a2 = Y P; 105-fold decrease for a2 = G N E and X = R; no reactivity for a2 = D K R and X = P).
Our single amino acid substitution data tested 57 a-factor CaaX sequence variants, which are a small fraction of the 203 possible CaaX sequences. Nevertheless, rules emerged that had predictive power. For example, the data suggested that CaaX sequences with serine or threonine at the a1 position would be substrates of Rce1p, but not Afc1p, and that CaaX sequences with serine or threonine at the a2 position would be substrates of Afc1p. In agreement with these predictions, a-factor variants with the sequences CTVM, CSVM, and CTLM were found to be proteolyzed by Rce1p, but not Afc1p, whereas a-factor with CASQ was proteolyzed by Afc1p (Fig. 3) (4).
Although caveats concerning the accessibility of substrates and the potential influence of sequences outside the CaaX motif must be kept in mind, the a-factor single amino acid substitution data, together with in vitro farnesylation data, can help guide predictions of which yeast proteins are substrates of Afc1p and/or Rce1p. There are 98 yeast proteins that have a cysteine as the fourth amino acid from the C terminus. Predictions concerning prenylation and proteolysis of these proteins are presented in Table 3. According to our data, 24 of the 98 proteins would be predicted to be adequately prenylated and proteolyzed by both Afc1p and Rce1p, and 14 proteins would be predicted to be adequately prenylated and proteolyzed by Rce1p but not Afc1p. Nine proteins would be predicted to have a partial defect in prenylation, as well as a defect in proteolysis by both Afc1p and Rce1p. Twenty-two proteins would be predicted to have deficiencies in prenylation and/or proteolysis. Each of these 22 proteins has at least one amino acid at the a1, a2, or X position that caused inefficient a-factor production in the a-factor single amino acid substitution series. The available data do not allow a clear determination of the relative deficits in prenylation versus proteolysis for these amino acids. These proteins each have an amino acid that did not allow detectable Rce1p cleavage in the a-factor substitution series (Table 2, row 4). Our data cannot predict whether or not Rce1p could process these proteins, since loss of the N-terminal role of Afc1p in a-factor processing, in combination with decreased prenylation, reduced a-factor production below a detectable threshold in afc1Δ strains. Half of these 22 proteins have an amino acid at the a1 position that was accepted by Afc1p, whereas the other half have an amino acid at the a1 position that precludes Afc1p proteolysis in the a-factor series.
TABLE 3.
Predictions of prenylation and proteolysis of potential yeast CaaX proteinsa
| Prenylated, cleaved by Afc1p and Rce1pb | Prenylated, cleaved by Rce1p but not Afc1pc | Weakly prenylated, not cleaved by Afc1p and Rce1pd | Compromised for prenylation and/or proteolysise | Prenylation unlikelyf |
|---|---|---|---|---|
| YBL061C CVIM | YBL018C CKCI | YAL014C CWLI | Cleaved by Afc1p | YBR096W CSEI |
| YDL009C CAVS | YBL049W CKCT | YDR455C CPLS | YBR033W CFFN | YBR134W CFRV |
| YDR461W CVIA | YCR004C CTVM | YDL151C CYPA | YBR042C CFIF | YBR150C CVKM |
| YFL066C CCVC | YCR027C CSIM | YFL065C CCPS | YBR087W CCLD | YBR209W CSKP |
| YGL069C CVCC | YGR152C CTIL | YJR066W CPFW | YDR261C CASL | YCR020C CYNA |
| YGL082W CVIM | YIL118W CTIM | YKL203C CPFW | YDR307W CLAK | YDR257C CVKK |
| YIR007W CVIS | YJL059W CRME | YHL049C CCPS | YGL169W CIQF | YDR301W CQGK |
| YIR032C CIII | YJR086W CTLM | YNL106C CDPN | YJL062W CALD | YDR528W CTRK |
| YJL118W CCCS | YLL044W CSLY | YPR203W CCPS | YKL069W CVFK | YGL045W CDDY |
| YJL204C CCIM | YLR322W CRIF | YMR060C CKYI | YGL263W CNDV | |
| YJR128W CMMI | YLR444C CRCG | YNL064C CASQ | YGR068C CDDD | |
| YKL196C CIIM | YML116W CTVA | YPR099C CVST | YHL049C CCPS | |
| YKR055W CIIM | YNL234W CSIM | Not cleaved by Afc1p | YHR053C CSGK | |
| YLR090W CCIQ | YPR092W CKIS | YDL065C CKQQ | YHR055C CSGK | |
| YLR229C CAIL | YDL186W CHHD | YJR107W CSGL | ||
| YML006C CAIM | YDR034W-B CDVF | YML041C CRNR | ||
| YNL090W CIIL | YDR094W CDMI | YML075C CIKS | ||
| YNL098C CIIS | YGR236C CDFT | YMR023C CIGK | ||
| YNL145W CVIA | YGR282C CDFS | YMR187C CKGE | ||
| YNL180C CVIL | YKL176C CNAG | YMR265C CSNA | ||
| YOL014W CIIL | YKR048C CKQS | YMR300C CADY | ||
| YOR101W CIIC | YMR158W CRVK | YNL255C CPKA | ||
| YPL191C CVVM | YMR193C-A CSTS | YOR031W CEKC | ||
| YPR165W CVLL | YPR087W CQAS | YOR231W CWKD | ||
| YOR239W CLRH | ||||
| YOR242C CIDL | ||||
| YOR257W CTDS | ||||
| YOR299W CYDA | ||||
| YOR392W CSDL | ||||
| YPR093C CHDE |
There are 98 S. cerevisiae proteins in the Saccharomyces Genome Database (http://genome www.stanford.edu/Saccharomyces/) that have a cysteine four amino acids from the C terminus. The systematic gene names and the last 4 amino acids of the 98 predicted protein sequences are listed. Predictions concerning farnesylation and proteolysis are based on in vitro farnesylation data and in vivo halo data from a-factor variants with single amino acid substitutions. The following caveats must be kept in mind when considering these predictions: (i) single amino acid substitution data may not accurately predict whether or not a particular substrate with multiple amino acid differences relative to a-factor will be a farnesyltransferase or CaaX protease substrate; (ii) sequences outside the CaaX sequence could influence substrate-protease interactions; and (iii) subcellular localization could make some potential CaaX protease substrates inaccessible to the farnesyltransferase, the Afc1p CaaX protease, or the Rce1p CaaX protease.
These 24 proteins meet the criteria derived from the a-factor variants that were proteolyzed by Afc1p and Rce1p (Table 2, row 1). Note that at least one of these proteins, Ras2p (CIIS [YNL098C]), appears to be cleaved by Rce1p but not Afc1p (4), despite having a CaaX sequence that was predicted to be cleavable by Rce1p and Afc1p. Therefore, Ras does not appear to be accessible to Afc1p (see footnote a).
These 14 proteins meet the criteria derived from the a-factor variants that were permissive for Rce1p proteolysis but not for Afc1p proteolysis (Table 2, row 2).
These nine yeast proteins, which have W or P at the a1 position or P at the a2 position, are predicted to be weakly prenylated and to be uncleavable by either Afc1p or Rce1p. Note that the CPFW CaaX sequence may not be prenylated at all because of the combined effect of P, F, and W at the a1, a2, and X positions, respectively. Note also that the D at the a1 position of the CDPN CaaX sequence is predicted to negatively influence prenylation and/or proteolysis.
All of these proteins meet the criteria listed in Table 2, row 4. Since those a-factor variants produced a small in vivo halo but were not assayed for in vitro farnesylation, the defect in a-factor production could be in farnesylation and/or proteolysis. Assuming substrate accessibility and adequate prenylation, the CaaX sequences of the first 11 proteins are predicted to be cleaved by Afc1p. No prediction can be made about whether or not Rce1p would also cleave these CaaX sequences. However, note that when CASQ was substituted into a-factor, it was cleaved by Afc1p but not Rce1p. Therefore, it is unlikely that Rce1p can cleave the Ydj1p CaaX sequence (CASQ [YNL064C]). The second set of 11 proteins, which are predicted not to be cleaved by Afc1p, have an amino acid at the a1 position that precluded cleavage by Afc1p (Table 2, row 2) and an amino acid that decreased prenylation and/or precluded Rce1p proteolysis (Table 2, row 3). If these CaaX sequences are adequately prenylated, they are predicted not to be cleaved efficiently by either Afc1p or Rce1p.
These predictions were based on in vitro farnesylation data for a subset of a-factor variants that exhibited greater than a 104-fold decrease in catalytic efficiency (Table 1). The 29 yeast proteins listed here have G, D, N, E, K, or R at the a2 position of the CaaX sequence. Nearly half of these CaaX sequences also have an amino acid at the a1 or X position that adversely affected prenylation.
Deductions about which potential CaaX proteins in yeast are unlikely to be farnesylated.
In vitro farnesylation data, together with the in vivo a-factor production (halo assay) data, suggest that at least 29 out of 98 yeast proteins with a cysteine four amino acids from the C terminus are unlikely to be farnesylated (Table 3). These 29 proteins have an amino acid at the a2 position that resulted in very poor farnesylation (G, N, or E) or no farnesylation (D, K, or R) in vitro. Note that even among the remaining 69 proteins, some may be poorly prenylated due to the presence of W, Y, or P at the a1 position and Y or P at the a2 position, which exhibited a partial in vitro farnesylation defect. In addition, G or D at the a1 position, A, S, T, Q, H, W, or F at the a2 position, or D, E, K, or W at the X position may decrease farnesylation efficiency. In the a-factor substitution series, a-factor variants with these substitutions produced small halos that increased in size when the beta subunit of farnesyltransferase was overexpressed, supporting the idea that inefficient farnesylation is at least partially responsible for the low level of a-factor production from these variants (Fig. 2).
Note that 17 of the 98 potential CaaX proteins have leucine or isoleucine at the X position and therefore are likely to be preferred substrates of geranylgeranyltransferase I, although farnesyltransferase can also act on substrates with a C-terminal leucine (7, 44). However, this fact does not change our predictive analysis with respect to CaaX proteolysis, since both yeast (10a) and human (32) Rce1p CaaX proteases are able to cleave both farnesylated and geranylgeranylated peptides in vitro. Since the CaaX sequence requirements for geranylgeranyltransferse I are not as well defined as those of the farnesyltransferase, it is more difficult to predict which of these sequences would be prenylated. From the studies that have been done, this enzyme appears to accept fewer amino acids at the a1 and a2 positions than farnesyltransferase (29). Based on known geranylgeranyltransferase I substrates (28, 29), 7 of these 17 C termini (CTIL, CAIL, CIIL, CVIL, CIIL, CVLL, and CIII) are expected to be substrates of geranylgeranyltransferase I. Five of the C termini (CASL, CKCI, CDMI, CMMI, and CKYI) have amino acids at the a1 and a2 positions that allow farnesylation but may not allow geranylgeranylation. Five of the C termini were included in the set of 29 proteins that are unlikely to be substrates of the farnesyltransferase. Due to suboptimal amino acids at the a1 and/or a2 position, it is unlikely that these sequences (CSGL, CIDL, CSDL, CWLI, and CSEI) are efficiently prenylated by either farnesyltransferase or geranylgeranyltransferase I.
Note that these predictions of whether the farnesyltransferase and/or the geranylgeranyltransferase I can prenylate a given CaaX sequence on a particular protein are tentative. Although both of these enzymes are able to prenylate four amino acid CaaX peptides and therefore do not require additional sequences, the efficiency of prenylation can be influenced by sequences outside the CaaX sequence (19, 21, 22).
Yeast CaaX protease specificity information aids in the interpretation of mammalian CaaX protease observations.
The yeast and mammalian CaaX proteases have proven extremely difficult to purify, and consequently in vitro studies of substrate specificity on purified proteases have not been done. A partially purified prenyl protein-specific endoprotease (PPEP) activity, which may include one or more CaaX proteases of unknown identity, was used in a recent specificity study (20). By comparing the in vitro specificity of the partially purified activity to the in vivo specificities of yeast Afc1p (Fig. 1), yeast Rce1p (Fig. 1), and human Rce1p expressed in yeast (Wong et al., unpublished), we deduce that the activity is likely to be that of a mammalian Afc1p homolog. One of the key results that led to this deduction was that yeast Rce1p (Fig. 1) and human Rce1p (Wong et al., unpublished) were able to cleave the a-factor variant with R at the a1 position, whereas neither yeast Afc1p (Fig. 1) nor the PPEP activity (20) could cleave substrates with R at the a1 position. Specificity studies have not been performed with the other partially purified enzymes (2, 10, 31), but one enzyme activity (31) is inhibited by o-phenanthroline, as has been observed for yeast Afc1p (4, 10a), whereas the other enzyme activity (10) is not sensitive to o-phenanthroline and has properties expected for Rce1p. Further studies will be necessary to clarify which of these activities are Afc1p, Rce1p, or other CaaX proteases.
In vivo cleavage of Ras proteins by Rce1p, but not Afc1p, is not explained by Afc1p CaaX sequence specificity constraints.
Studies of human Rce1p protein in insect cells establish its ability to cleave farnesyl-Ki-Ras, geranylgeranyl-Ki-Ras, farnesyl-Ha-Ras, farnesyl-N-Ras, farnesyl-Gγ1, and geranylgeranyl-Rap1 (32), which have the CaaX sequences CVIM, CVLS, CVLM, CVIS, and CQLL, respectively. In a complementary study, mouse fibroblasts that are homozygous for an rce1 deletion are unable to process farnesyl-Ki-Ras, geranylgeranyl-Ki-Ras, farnesyl-Ha-Ras, farnesyl-N-Ras, farnesyl-Gγ1, or geranylgeranyl-Rap1 (23). By deduction, the mouse Afc1p homolog appears not to proteolyze these substrates.
The substrate specificity data for yeast Rce1p (Fig. 1) and for human Rce1p (Wong et al., unpublished results) are consistent with the ability of Rce1p to cleave these substrates. Human Rce1p expressed in yeast is able to process a-factor (CVIA) and Ras2 (CIIS) and exhibits a substrate specificity profile remarkably similar to that shown here for yeast Rce1p (Wong et al., unpublished). However, the failure of Afc1p to cleave Ki-Ras, Ha-Ras, N-Ras, or Gγ1 cannot be explained by the available substrate specificity data on yeast Afc1p (Fig. 1), the partially purified PPEP activity (20) (which we deduced is likely to be rat Afc1p), or human Afc1p expressed in yeast, which is able to process a-factor (40) but has not been assayed with any other substrates. A trivial explanation would be that mammalian Afc1p is not expressed in fibroblasts. However, a similar discrepancy in yeast suggests a different interpretation. Specifically, the single amino acid substitution data suggest that the Ras2 CaaX sequence, CIIS, would be cleaved by both Afc1p and Rce1p. However, the genetic data indicate that Ras2p (CIIS) is processed primarily by Rce1p, not by Afc1p (4). Moreover, there are other farnesylated mammalian CaaX proteins that, despite having CaaX sequences that appear acceptable for Afc1p, are not proteolyzed in vivo. The alpha and beta subunits of phosphorylase kinase in rabbit reticulocytes are farnesylated, but the aaX sequences, AMQ and LVS, respectively, are not removed (17). Together, these observations point out that the CaaX sequence alone may not be the only factor that influences the ability of a CaaX protease to cleave a potential substrate. For example, the substrate and the enzyme may reside in different cellular compartments. Alternatively, the CaaX sequence may be masked, either by being folded into the interior of the protein or by being bound to another protein.
In addition to the phosphorylase kinase subunits described above, two other proteins have been identified as being farnesylated but not proteolyzed. CaaX variants of Ki-Ras4B that have CVGM and CVYM are farnesylated less efficiently than Ki-Ras4B (CVIM), and the small amount of Ras protein that is farnesylated remains unproteolyzed (22). Similarly, our combined in vivo halo and in vitro farnesylation results with a-factor-CVGA and a-factor-CVYA demonstrated a significant decrease in farnesylation, as well as a deduced deficit in CaaX proteolysis (Fig. 1; Table 1).
A substrate recognition domain in Rce1p.
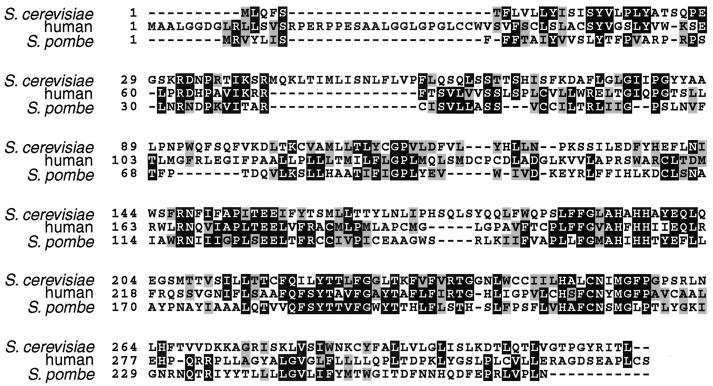
Amino acid substitutions in Rce1p that improved its ability to process a-factor-CAMQ, without affecting processing of other a-factor variants, provided information about the region of Rce1p likely to be involved in substrate recognition. The F189L and Q201R amino acid substitutions in Rce1p each independently increased Rce1p processing of a-factor-CAMQ. F189 is conserved in both the human and Schizosaccharomyces pombe homologs of Rce1p, which share amino acid identity of only 32% with each other (Fig. 5). A conserved histidine that residues between F189 and Q201 (H194 in yeast Rce1p and H208 in human Rce1p) is required for proteolytic activity of yeast (10a) and human (Wong et al., unpublished) Rce1p. Together, these data strongly suggest that this region of Rce1p is involved in substrate binding. This same region contains a conserved HxxE motif which has been proposed to be a Zn binding site (12), by analogy to a previously described class of metalloproteases (33). However, the HxxE sequence is not critical to Rce1p function because human Rce1p remains functional when either histidine 211 or glutamate 214 is replaced with alanine (Wong et al., unpublished). Substitution of the analogous histidine of yeast Rce1p reduced the specific activity about 10-fold (10a). Note also that zinc chelators, such as o-phenanthroline, do not appear to inhibit Rce1p, raising doubt about the role of zinc in Rce1p function (5, 10a).
FIG. 5.
Alignment of Rce1p homologs from S. cerevisiae (4), human (32), and S. pombe (accession numbers: Swiss-Prot Q10071 and GenBank AL035064.1) sequences were aligned with the ClustalW 1.7 program (42), accessed through the BCM sequence launcher (http://www.hgsc.bcm.tmc.edu/SearchLauncher/). Identities between homologs are shaded in black, and similarities are shaded in gray. The region spanning amino acids 147 to 257 of S. cerevisiae is most highly conserved region among the three homologs, with pairwise percent identities of 41% (S. cerevisiae-human), 44% (human-S. pombe), and 35% (S. cerevisiae-S. pombe) and percent similarities of 50 to 58%. The figure was prepared using BOXSHADE (http://www.isrec.isb-sib.ch:8080/software/BOX_form.html).
Processing of a-factor-CAMQ by the F189L mutant form of Rce1p was improved further by an E139K substitution. Although E139 is not conserved in human or S. pombe Rce1p, it is near a conserved region: RNxxxAPxTEE (amino acids 147 to 157 in yeast Rce1p). The penultimate glutamate in this conserved sequence is required for yeast (10a) and human (Wong et al., unpublished) Rce1p function.
Both Afc1p and Rce1p are polytopic membrane proteins of the endoplasmic reticulum (5, 38). Unfortunately, there is no direct information regarding the topology of any portion of the proteins, and so the experimental findings must be interpreted in light of predicted structures rather than established ones. A comparison of the transmembrane domain predictions of Rce1p with homologs from human and S. pombe reveals that although all three proteins are predicted to span a membrane multiple times, there are substantial differences in the structures predicted for the three proteins. Even in the region of strongest homology (corresponding to amino acids 147 to 257 of yeast Rce1p), where one would expect that the structure and topology of the three Rce1p homologs would be best conserved, there was a high degree of variability in both the number and the position of the transmembrane helices predicted for each of the three proteins with different programs.
Perhaps the conflicting models reflect the ability of some of the hydrophobic regions to penetrate into, but not span, the membrane. If so, the active site of the Rce1p proteases may be partially buried in the membrane or at the membrane-cytoplasm interface. Considering that the substrates of the CaaX proteases are prenylated at the cysteine adjacent to the cleavage site and that farnesylation or geranylgeranylation of these CaaX protein substrates is required for proteolysis, perhaps the prenyl lipid helps present the critical peptide bond to the active site close to, or within, the membrane.
It is interesting that S2P, a membrane-associated metalloprotease, has been proposed to have two classical transmembrane helices and three longer hydrophobic regions (30, 37, and 118 amino acids) that reside within the membrane, but not as transmembrane helices (46). This model, which is supported by protease protection and glycosylation site mapping data, contrasts dramatically with the predicted structure (25). The new model, which posits that the active site is within the membrane, is attractive in that the S2P metalloprotease is thought to directly cleave the sterol response element binding protein at a site that is buried within a transmembrane domain (25). Clearly, enzymes that catalyze reactions in a hydrophobic environment offer considerable challenges to structural predictions, as well as novel opportunities for the design of inhibitors.
ACKNOWLEDGMENTS
We thank Ashild Vik, Qun Shan, Sara Okamura, and other members of the Rine lab for valuable discussions. We thank Matt Ashby for pJR1561 and JRY4276.
This work was supported by an NSF postdoctoral fellowship (to D.R.) and by NIH grants GM35827 (to J.R.) and GM 21328 (to C.D.P.).
REFERENCES
- 1.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics: a Cold Spring Harbor laboratory course manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 2.Akopyan T N, Couedel Y, Orlowski M, Fournie-Zaluski M C, Roques B P. Proteolytic processing of farnesylated peptides: assay and partial purification from pig brain membranes of an endopeptidase which has the characteristics of E.C. 3.4.24.15. Biochem Biophys Res Commun. 1994;198:787–794. doi: 10.1006/bbrc.1994.1113. [DOI] [PubMed] [Google Scholar]
- 3.Ashby M N, King D S, Rine J. Endoproteolytic processing of a farnesylated peptide in vitro. Proc Natl Acad Sci USA. 1992;89:4613–4617. doi: 10.1073/pnas.89.10.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyartchuk V L, Ashby M N, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 5.Boyartchuk V L, Rine J. Roles of prenyl protein proteases in maturation of Saccharomyces cerevisiae a-factor. Genetics. 1998;150:95–101. doi: 10.1093/genetics/150.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell G A, Wang S H, Naider F, Becker J M. Consequences of altered isoprenylation targets on a-factor export and bioactivity. Proc Natl Acad Sci USA. 1994;91:1275–1279. doi: 10.1073/pnas.91.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caplin B E, Hettich L A, Marshall M S. Substrate characterization of the Saccharomyces cerevisiae protein farnesyltransferase and type-I protein geranylgeranyltransferase. Biochim Biophys Acta. 1994;1205:39–48. doi: 10.1016/0167-4838(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 8.Caplin B E, Ohya Y, Marshall M S. Amino acid residues that define both the isoprenoid and CAAX preferences of the Saccharomyces cerevisiae protein farnesyltransferase. Creating the perfect farnesyltransferase. J Biol Chem. 1998;273:9472–9479. doi: 10.1074/jbc.273.16.9472. [DOI] [PubMed] [Google Scholar]
- 9.Chan R K, Otte C A. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α-factor pheromones. Mol Cell Biol. 1982;2:11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Ma Y T, Rando R R. Solubilization, partial purification, and affinity labeling of the membrane-bound isoprenylated protein endoprotease. Biochemistry. 1996;35:3227–3237. doi: 10.1021/bi952529s. [DOI] [PubMed] [Google Scholar]
- 10a.Dolence J M, Steward L E, Dolence E K, Wong D H, Poulter C D. Overproduction of the yeast CaaX prenyl protease Rce1p in Saccharomyces cerevisiae and its initial biochemical characterization. Biochemistry. 2000;39:4096–4104. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- 11.Farh L, Mitchell D A, Deschenes R J. Farnesylation and proteolysis are sequential, but distinct steps in the CaaX box modification pathway. Arch Biochem Biophys. 1995;318:113–121. doi: 10.1006/abbi.1995.1211. [DOI] [PubMed] [Google Scholar]
- 12.Freije J M, Blay P, Pendas A M, Cadinanos J, Crespo P, Lopez-Otin C. Identification and chromosomal location of two human genes encoding enzymes potentially involved in proteolytic maturation of farnesylated proteins. Genomics. 1999;58:270–280. doi: 10.1006/geno.1999.5834. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs J B, Oliff A. The potential of farnesyltransferase inhibitors as cancer chemotherapeutics. Annu Rev Pharmacol Toxicol. 1997;37:143–166. doi: 10.1146/annurev.pharmtox.37.1.143. [DOI] [PubMed] [Google Scholar]
- 14.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Hancock J F, Cadwallader K, Marshall C J. Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B) EMBO J. 1991;10:641–646. doi: 10.1002/j.1460-2075.1991.tb07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock J F, Hall A. A novel role for RhoGDI as an inhibitor of GAP proteins. EMBO J. 1993;12:1915–1921. doi: 10.1002/j.1460-2075.1993.tb05840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmeyer L M, Jr, Serwe M, Weber C, Metzger J, Hoffmann-Posorske E, Meyer H E. Farnesylcysteine, a constituent of the alpha and beta subunits of rabbit skeletal muscle phosphorylase kinase: localization by conversion to S-ethylcysteine and by tandem mass spectrometry. Proc Natl Acad Sci USA. 1992;89:9554–9558. doi: 10.1073/pnas.89.20.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrycyna C A, Clarke S. Purification and characterization of a novel metalloendopeptidase from Saccharomyces cerevisiae. Biochemistry. 1993;32:11293–11301. doi: 10.1021/bi00093a005. [DOI] [PubMed] [Google Scholar]
- 19.James G, Goldstein J L, Brown M S. Resistance of K-RasBV12 proteins to farnesyltransferase inhibitors in Rat1 cells. Proc Natl Acad Sci USA. 1996;93:4454–4458. doi: 10.1073/pnas.93.9.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang G F, Gelb M H. Substrate specificity of mammalian prenyl protein-specific endoprotease activity. Biochemistry. 1998;37:4473–4481. doi: 10.1021/bi972289b. . (Erratum, 37:5336.) [DOI] [PubMed] [Google Scholar]
- 21.Kalman V K, Erdman R A, Maltese W A, Robishaw J D. Regions outside of the CAAX motif influence the specificity of prenylation of G protein gamma subunits. J Biol Chem. 1995;270:14835–14841. doi: 10.1074/jbc.270.24.14835. [DOI] [PubMed] [Google Scholar]
- 22.Kato K, Cox A D, Hisaka M M, Graham S M, Buss J E, Der C J. Isoprenoid addition to Ras protein is the critical modification for its membrane association and transforming activity. Proc Natl Acad Sci USA. 1992;89:6403–6407. doi: 10.1073/pnas.89.14.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E, Ambroziak P, Otto J C, Taylor B, Ashby M, Shannon K, Casey P J, Young S G. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J Biol Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- 24.Lebowitz P F, Sakamuro D, Prendergast G C. Farnesyl transferase inhibitors induce apoptosis of Ras-transformed cells denied substratum attachment. Cancer Res. 1997;57:708–713. [PubMed] [Google Scholar]
- 25.Lewis A P, Thomas P J. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobell R B, Kohl N E. Pre-clinical development of farnesyltransferase inhibitors. Cancer Metastasis Rev. 1998;17:203–210. doi: 10.1023/a:1006018922878. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y T, Gilbert B A, Rando R R. Inhibitors of the isoprenylated protein endoprotease. Biochemistry. 1993;32:2386–2393. doi: 10.1021/bi00060a033. . (Erratum, 32:5924.) [DOI] [PubMed] [Google Scholar]
- 28.Mayer M L, Caplin B E, Marshall M S. CDC43 and RAM2 encode the polypeptide subunits of a yeast type I protein geranylgeranyltransferase. J Biol Chem. 1992;267:20589–20593. . (Erratum, 268:4568, 1993.) [PubMed] [Google Scholar]
- 29.Moores S L, Schaber M D, Mosser S D, Rands E, O'Hara M B, Garsky V M, Marshall M S, Pompliano D L, Gibbs J B. Sequence dependence of protein isoprenylation. J Biol Chem. 1991;266:14603–14610. [PubMed] [Google Scholar]
- 30.Newman P, Kube E, Gerke V, Weber K. Polyisoprenylation of the CAAX motif—an in vitro protein synthesis study. Biochim Biophys Acta. 1991;1080:227–230. doi: 10.1016/0167-4838(91)90006-l. [DOI] [PubMed] [Google Scholar]
- 31.Nishii W, Muramatsu T, Kuchino Y, Yokoyama S, Takahashi K. Partial purification and characterization of a CAAX-motif-specific protease from bovine brain using a novel fluorometric assay. J Biochem (Tokyo) 1997;122:402–408. doi: 10.1093/oxfordjournals.jbchem.a021767. [DOI] [PubMed] [Google Scholar]
- 32.Otto J C, Kim E, Young S G, Casey P J. Cloning and characterization of a mammalian prenyl protein-specific protease. J Biol Chem. 1999;274:8379–8382. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 33.Rawlings N D, Barrett A J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 34.Reiss Y, Stradley S J, Gierasch L M, Brown M S, Goldstein J L. Sequence requirement for peptide recognition by rat brain p21ras protein farnesyltransferase. Proc Natl Acad Sci USA. 1991;88:732–736. doi: 10.1073/pnas.88.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reneke J E, Blumer K J, Courchesne W E, Thorner J. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- 36.Roskoski R, Jr, Ritchie P, Gahn L G. Farnesyl-protein transferase and geranylgeranyl-protein transferase assays using phosphocellulose paper absorption. Anal Biochem. 1994;222:275–280. doi: 10.1006/abio.1994.1485. [DOI] [PubMed] [Google Scholar]
- 37.Schena M, Picard D, Yamamoto K R. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt W K, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sebti S, Hamilton A D. Inhibitors of prenyl transferases. Curr Opin Oncol. 1997;9:557–561. doi: 10.1097/00001622-199711000-00011. [DOI] [PubMed] [Google Scholar]
- 40.Tam A, Nouvet F J, Fujimura-Kamada K, Slunt H, Sisodia S S, Michaelis S. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 42.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trueblood C E, Boyartchuk V L, Rine J. Substrate specificity determinants in the farnesyltransferase beta-subunit. Proc Natl Acad Sci USA. 1997;94:10774–10779. doi: 10.1073/pnas.94.20.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trueblood C E, Ohya Y, Rine J. Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4260–4275. doi: 10.1128/mcb.13.7.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W, Del Villar K, Urano J, Mitsuzawa H, Tamanoi F. Advances in the development of farnesyltransferase inhibitors: substrate recognition by protein farnesyltransferase. J Cell Biochem Suppl. 1997;27:12–19. [PubMed] [Google Scholar]
- 46.Zelenski N G, Rawson R B, Brown M S, Goldstein J L. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274:21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]